Abstract
Aims: The aim of this study was to characterize redox changes in the nuclei and cytosol occurring during the mitotic cell cycle in the embryonic roots of germinating Arabidopsis seedlings, and to determine how redox cycling was modified in mutants with a decreased capacity for ascorbate synthesis.
Results: Using an in vivo reduction-oxidation (redox) reporter (roGFP2), we show that transient oxidation of the cytosol and the nuclei occurred at G1 in the synchronized dividing cells of the Arabidopsis root apical meristem, with reduction at G2 and mitosis. This redox cycle was absent from low ascorbate mutants in which nuclei were significantly more oxidized than controls. The cell cycle-dependent increase in nuclear size was impaired in the ascorbate-deficient mutants, which had fewer cells per unit area in the root proliferation zone. The transcript profile of the dry seeds and size of the imbibed seeds was strongly influenced by low ascorbate but germination, dormancy release and seed aging characteristics were unaffected.
Innovation: These data demonstrate the presence of a redox cycle within the plant cell cycle and that the redox state of the nuclei is an important factor in cell cycle progression.
Conclusions: Controlled oxidation is a key feature of the early stages of the plant cell cycle. However, sustained mild oxidation restricts nuclear functions and impairs progression through the cell cycle leading to fewer cells in the root apical meristem. Antioxid. Redox Signal. 27, 1505–1519.
Keywords: : antioxidant, ascorbate, cell cycle, germination, redox regulation, root apical meristem, seed dormancy, transcript profiling
Introduction
Concepts of the roles of reactive oxygen species (ROS) in plants and animals have shifted in recent years from focusing on oxidative damage effects to the current view of ROS as universal signaling metabolites (26). Reduction/oxidation (redox) signaling is an essential component of cellular energy homeostasis and responses to the environment in animals and plants and redox-sensitive proteins functioning as sensors that trigger acclimation, growth, and defense processes. Cells rely on proliferative signaling pathways and stress surveillance systems to regulate entry and progress through the mitotic cell cycle (7, 9, 42).
Innovation.
Cellular redox status exerts control over many aspects of plant biology, but the effects of the redox status experienced by the mother plant on seed vigor remain poorly characterized. In this study, we show that low redox buffering capacity in mutants with low ascorbate has a strong influence on the seed transcriptome without marked effects on germination rates. We provide evidence that the cytosol and nuclei become more oxidized during the G1 phase of the cell cycle in synchronized dividing cells of the Arabidopsis embryonic root meristem. Mild oxidation impaired these redox fluctuations and delayed the required increase in nucleus size.
ROS are important regulators of both the proliferative signaling and stress surveillance systems in plants, as they are in animals (10, 24, 59, 63, 66). In mammals, interactions between growth factors and receptors generate ROS, which activate proliferative signaling for cell division by modulation of the redox state of certain protein cysteine residues to elicit oxidative signaling pathways in cells (7, 9, 42). While proliferative oxidative signals flow through intracellular signaling pathways to activate the cell cycle, stress-related oxidative signals from within or outside the cell oppose proliferation. The mechanisms by which biotic and abiotic stresses lead to an arrest of the mitotic cell cycle are largely uncharacterized, although the notion of an oxidative stress checkpoint has long been postulated in plants and animals (16, 53).
The complex redox control of the cell cycle observed in animals is often explained very simply in terms of a given threshold ROS level required to generate cell proliferation or cell cycle arrest (9). However, the outcomes of cellular oxidative signaling pathways depend on a number of parameters, principally the chemical nature of ROS form produced (i.e., superoxide, hydrogen peroxide, or singlet oxygen) and the nature of the interacting partner (protein thiol, metabolite, lipid, or DNA molecule), as well as cell identity. The complex interplay between proliferative oxidative signals and stress-related oxidative signals may be particularly important in kick-starting new cell proliferation after a period of quiescence, as occurs upon seed germination.
Seeds are a vital part of the life cycle of flowering plants, which rely on them for reproduction. Seed vigor (4, 44, 52, 56) is important because the capacity for germination and seedling establishment have a profound impact on subsequent crop productivity and yield. Mature dry seeds are desiccation tolerant and the internal environment is essentially hypoxic (4, 38). However, ROS generation and redox regulation of seed proteins are essential to the success of the germination process (2, 5, 22, 23). While ROS accumulation is considered to be a positive signal for dormancy release (2, 22, 24), relatively little is known about the regulation of cellular redox state at the early stages of seedling development.
Mature Arabidopsis thaliana seeds house the embryo in a desiccated state, which is entrained at the final stages of seed development, when the accumulation of abscisic acid (ABA) and late embryogenesis abundant proteins accompany decreases in water content to about 10% of seed weight (1). In the dry state, seeds have little metabolic activity. Metabolism is reactivated upon seed imbibition and germination. From the initial rapid period of water uptake, imbibition progresses to the emergence of the embryo root or radical (4).
Seed germination is considered to be one of the most critical stages in plant development because of the innate vulnerability of young seedlings to abiotic stress from the environment as well as from disease (52, 60). Germination is regulated by many factors, including the composition of messenger RNAs (mRNAs) stored in the embryo during maturation in the mother plant (52), and the levels of metabolites and hormones (44). Imbibition provides rapid relief from desiccation (28, 34). In dry seeds, ROS are produced mainly through chemical processes such as Amadori–Maillard reactions; hydration results in rapid ROS synthesis by apoplastic peroxidases and NADPH oxidases, followed by local oxidation within the embryonic axis and peripheral tissues (62). Regulated oxidation is required for extension growth of the cell walls in the developing radicle as well as dormancy release (2, 22). Once root cells have moved from the region of cell division to the differentiation zone, proteins such as ALF4 are important in maintaining the pericycle cells next to the xylem in the mitotically-active state that is required for lateral root development, in an environment-responsive but auxin-independent manner (18).
In the dry state, orthodox seeds have little antioxidant capacity, relying on reduced glutathione and tocochromanols for antioxidant defenses rather than ascorbate or ascorbate peroxidase (APX). Ascorbate is synthesized de novo within a few hours of imbibition, resulting in rapid increases in the ascorbate pool in the developing embryo (15, 57). Ascorbate has been used to prime seeds for improved stress tolerance (25) and to prevent preharvest sprouting (24). Moreover, mutants lacking cytosolic APX6 have poor germination and altered responses to ABA, suggesting that this enzyme plays a role in redox–hormone crosstalk during seed germination (8). Although a small amount of dehydroascorbate (DHA) is present in dry seeds, its functions are uncertain (15, 57).
Arabidopsis vitamin C defective 2 (vtc2) vtc5 double mutants have no ascorbate and show a seedling lethal phenotype, which can be rescued by the addition of ascorbate or its precursor, l-galactose (21, 39). While the effects of low antioxidant buffering capacity on shoot growth and leaf parameters have been extensively studied (12, 21, 32, 36, 45–49, 61, 65), the knockon effects of enhanced oxidation on seeds before and after imbibition remain poorly characterized.
This study characterizes the effect of the low redox buffering capacity in the ascorbate-deficient vtc2-1 and vtc2-4 mutants on seed germination and storage life. The findings demonstrate the presence of a redox cycle within the cell cycle, which is adversely affected by the redox state of the cell in the expanding embryonic root. The paralogous genes VTC2 and VTC5 encode GDP-l-galactose phosphorylase, which is considered to be an important control point in the ascorbate synthesis pathway. The vtc2-1 mutant has less than 2% of the leaf GDP-l-galactose phosphorylase activity than is observed in the wild type. This arises from a single base substitution (G to A) in the predicted 3′ splice site of the fifth intron of the VTC2 gene, resulting in a 90% reduction in transcript levels compared with the wild type (21).
Unlike vtc2-1, which has detectable levels of full-length VTC2 transcript (21), vtc2-4 is a T-DNA insertion mutant with complete loss of function. In the vtc2-4 mutants, the VTC5 gene still provides sufficient residual GDP-l-galactose phosphorylase activity to allow a reduced level of ascorbate synthesis and accumulation (21). The vtc2-1 and vtc2-4 mutants have only about 20% of the leaf ascorbate level of the wild type when plants were grown under short-day (10-h photoperiod) growth conditions (17).
The rosette phenotypes were similar in the wild type and vtc2-1 and vtc2-4 mutants at 14 days under all growth conditions. However, under short-day growth conditions, rosette biomass was significantly lower in the vtc2-1 and vtc2-4 mutants than the wild type at later stages of development (17). When plants were grown under continuous light conditions, leaf ascorbate contents were significantly increased in all lines compared with short-day conditions (17). The vtc2-1 and vtc2-4 mutants had 50% less leaf ascorbate than the wild type (17). The plants flowered at 21 days when grown under continuous light. At this point, the wild type and vtc2-4 mutants had a similar rosette size, but the vtc2-1 mutants were significantly smaller (17).
In this study, we present data demonstrating the presence of a redox cycle within the cell cycle. We show that the nuclei and cytosol become more oxidized when redox buffering capacity is low and this mild oxidation eliminates the natural redox rhythm observed during cell cycle progression. In addition, the data presented here show that oxidation in the mother plant, resulting from low redox buffering capacity, alters the seed transcriptome and results in enhanced embryo growth, without consistently affecting dormancy and longevity. However, once seeds are imbibed and when ascorbate synthesis is initiated, low redox buffering capacity results in oxidation of the nucleus, slowing the cell cycle and limiting cell numbers in the proliferation zone.
Results
Effects of low redox buffering capacity on seed size, dormancy, and sensitivity to aging
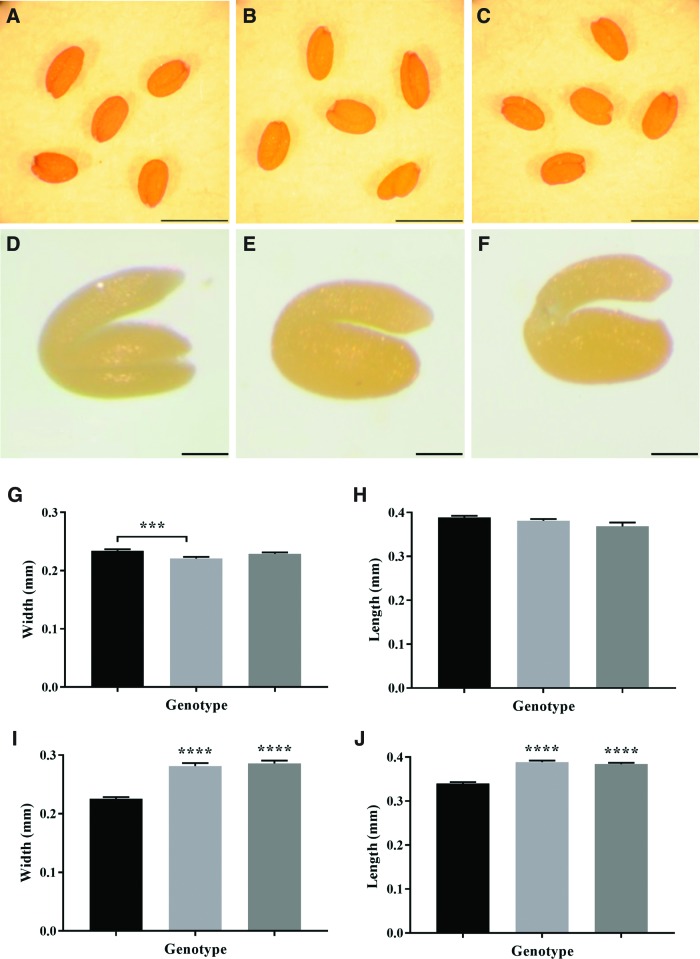
The dry vtc2 mutant seeds showed no phenotypic differences to the wild type, the low ascorbate mutants having similar dimensions to the wild type (Fig. 1). There were no consistent differences in the 1,000-seed mass between the low ascorbate mutants and the wild type. While the embryos of the dry vtc2 mutant seeds had similar phenotypes to the wild type, they were significantly larger than the wild type when imbibed (Fig. 1).
FIG. 1.
A comparison of seed parameters in wild-type (Col-0), vtc2-1, and vtc2-4 mutants. Images of dry (A–C) and imbibed (D–F) wild-type (Col-0) (A, D), vtc2-1 (B, E), and vtc2-4 (C, F) mutants. Width of dry and imbibed seeds (G, I); length of dry and imbibed seeds (H, L): Col-0 (black bars), vtc2-1 (light gray bars), and vtc2-4 (dark gray bars) dry seeds. (A–C) Scale bar = 500 μm. (D–F) Scale bar = 100 μm. ***p < 0.001 and ****p < 0.0001. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
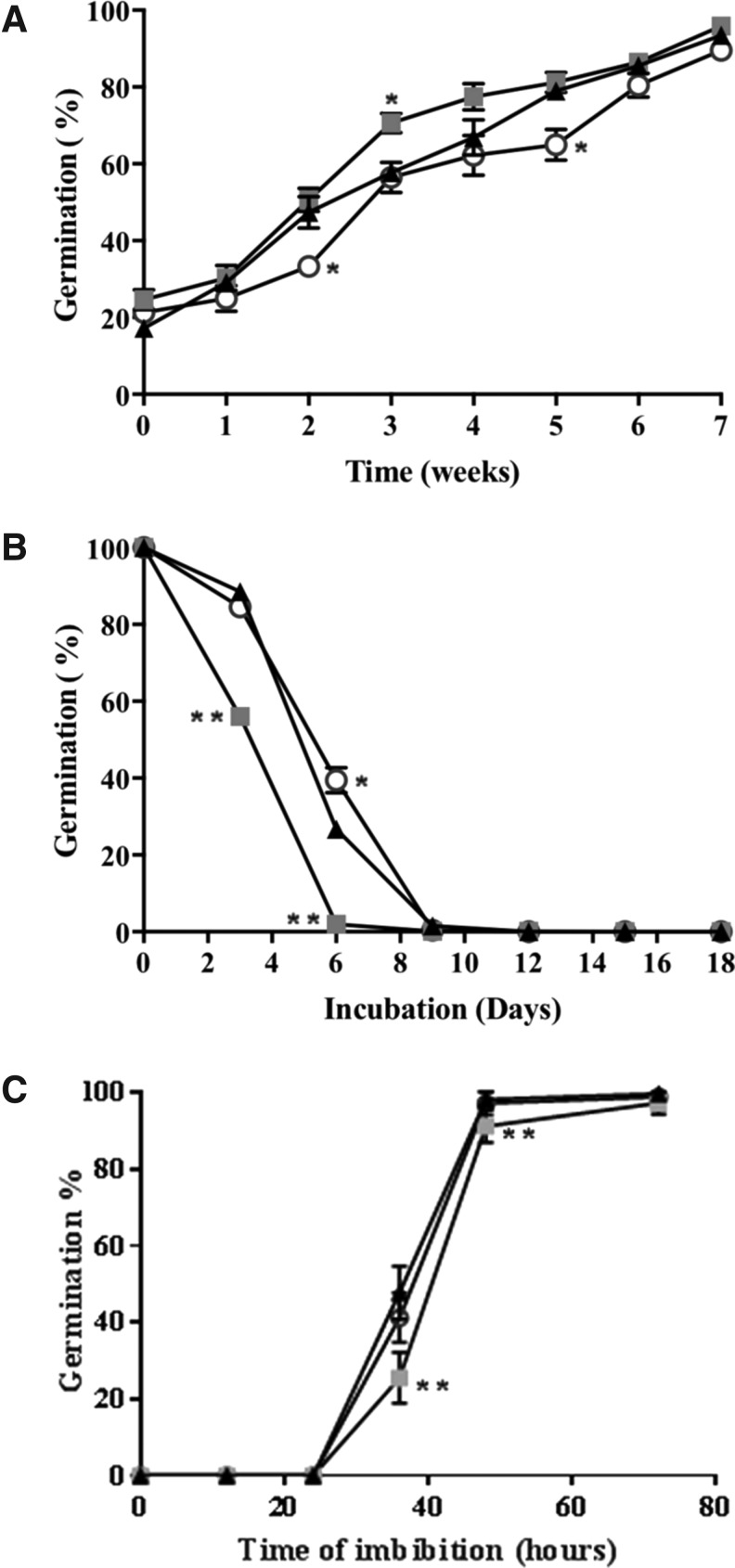
Seed dormancy, measured as percentage of seed germination during seed storage, showed no significant differences between the mutants with low antioxidant capacity and the wild type (Fig. 2A). Moreover, the controlled deterioration test (CDT) showed consistent effect of low antioxidant buffering capacity on sensitivity to seed aging compared with the wild type, specifically vtc2-1 seeds were more susceptible to aging than the wild type or vtc2-4 (Fig. 2B). Analysis of germination speed of fully after-ripened seeds showed that vtc2-1 seeds germinated significantly slower than wild-type and vtc2-4 seeds (Fig. 2C).
FIG. 2.
Germination characteristics of vtc2 mutants. Seed germination after increased storage time after harvest (A) or aging (B). (A) Dormancy and (B) seed longevity. (C) Percentage of germinating seeds during imbibition. Col-0 wild type (triangle), vtc2-1 (square), and vtc2-4 (round). Significance levels: *p < 0.01; **p < 0.001 compared with Col wild type by Student's t-test. Error bars represent standard error (SE) of 12 (A), 9 (B), or 10 (C) biological replicates. vtc2, vitamin C defective 2.
Low redox buffering capacity affects the transcriptome profiles of dry seeds
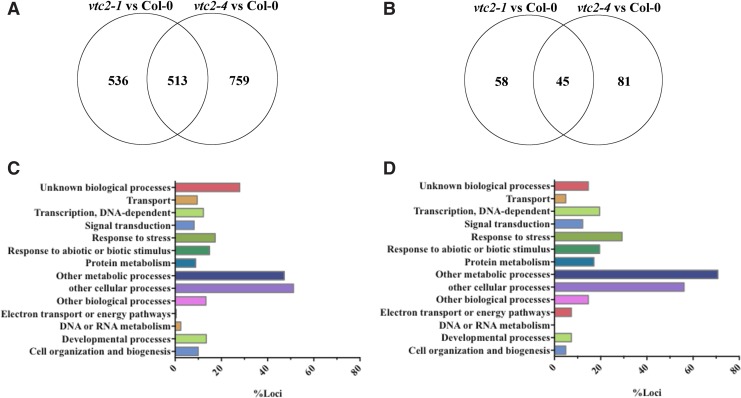
The decrease in redox buffering capacity in low ascorbate mutants exerted a strong effect on the transcriptome profiles of dry seeds (Supplementary Tables S1 and S2; Supplementary Data are available online at www.liebertpub.com/ars). In total, 1,049 transcripts were differentially expressed in the dry seeds of the vtc2-1 mutant and 1,272 transcripts were differentially expressed in the dry seeds of the vtc2-4 mutant relative to the wild type. Of these, 513 mRNAs were differentially expressed in the same manner in both ascorbate-deficient mutants relative to the wild type (Fig. 3A).
FIG. 3.
A comparison of the transcriptome profiles of dry and imbided ascorbate-deficient mutant seeds with the Col-0 wild type. Differentially expressed transcripts in dry (A) and imbibed (B) seeds vtc2-1 and vtc2-4 relative to Col-0. Functional categorization of transcripts that were differentially expressed in dry (C) and imbibed (D) seeds. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Many of the transcripts that were highly expressed in the dry seeds of the low ascorbate mutants are important in plant defense (Supplementary Tables S1 and S2 and Table 1). For example, the levels of transcripts encoding flavin-containing monooxygenase 1, which is an important component in systemic acquired resistance (SAR) controlling immunity and cell death, were much higher in the dry seeds of ascorbate-deficient mutants than the wild type. Similarly, the levels of transcripts encoding the cytochrome P450 monooxygenase CYP82C2 were significantly higher in the dry seeds of low ascorbate mutants than the wild type. The CYP82C2 protein is required for accumulation of jasmonate (JA)-induced indole glucosinolates and for JA-induced defense gene expression.
Table 1.
Transcripts Whose Abundance Was Most Influenced by Low Redox Buffering Capacity in Dry Seeds of the vtc2-1 and vtc2-4 Mutants Relative to the Wild Type
| Log fold change | |||
|---|---|---|---|
| Accession | Description | vtc2-1 | vtc2-4 |
| At1g19250 | FMO1 | 5.7 | 6.3 |
| At5g17800 | MYB56 | 4.7 | 4.4 |
| At1g69920 | GSTU12 | 4.4 | 2.4 |
| At1g26400 | FOX3 | 4.3 | 3.8 |
| At2g38620 | CDKB1;2 | 4.0 | 3.2 |
| At4g37010 | CML19 | 4.0 | 4.4 |
| At3g09270 | GSTU8 | 3.5 | 4.4 |
| At4g16970 | CDC7 | 3.5 | 3.7 |
| At1g14750 | SDS | 3.5 | 5.1 |
| At5g04140 | GLU1 | 3.0 | 3.9 |
| At4g31970 | CYP82C2 | 2.6 | 5.3 |
| At1g19050 | ARR7 | 2.6 | 4.1 |
| At5g14070 | GRX-C8 | 2.6 | 3.2 |
| At4g38420 | SKU5 similar 9 | 2.6 | 2.4 |
| At2g28760 | UXS6 | 2.6 | 3.0 |
| At5g61420 | MYB28 | −2.3 | −2.3 |
| At3g49690 | MYB84/RAX3 | −2.3 | −2.3 |
| At5g01420 | GRX-Like | −2.3 | −2.3 |
| At2g30540 | GRX-S9 | −2.3 | −2.3 |
| At1g66390 | MYB90 | −2.5 | −5.1 |
In contrast, transcripts encoding components involved in secondary metabolism were decreased in abundance in dry seeds with low redox buffering capacity compared with the wild type (Supplementary Tables S1 and S2). For example, the levels of transcripts that encode 4-coumarate-CoA5 ligase and anthocyanin pigment 2 protein (PAP2), which are involved in phenylpropanoid metabolism and anthocyanin synthesis, were much lower in the dry seeds of ascorbate-deficient mutants than the wild type. In addition, transcripts encoding two stress-inducible glycosyltransferases (UGT79B5 and UGT72B2) were significantly lower in the dry seeds of both ascorbate-deficient mutants than the wild type. UGTs catalyze the glycosylation of small molecules in the biosynthesis of secondary metabolites (Supplementary Tables S1 and S2).
The abundance of a number of transcripts involved in cell division and DNA repair was significantly increased in dry seeds of the low ascorbate mutants relative to the wild type (Table 1). For example, CENTRIN 2, also called calcium-binding 19 protein (CML19), was highly expressed in dry seeds of low ascorbate mutants. This protein is important in nucleotide excision repair during homologous recombination. The levels of Arabidopsis response regulator (ARR7) transcripts were also significantly higher in the dry seeds of the low ascorbate mutants. ARR7 has been shown to inhibit the ABA response and so promote germination (28).
In addition, transcripts encoding the R2R3-MYB transcription factor, MYB 56, were significantly higher in dry seeds of low ascorbate mutants compared with the wild type. MYB56, also called brassinosteroids at vascular and organizing center (BRAVO), is involved in brassinosteroid (BR) perception linked to cell growth, acting as a cell-specific repressor of BR-mediated cell division in the Arabidopsis root (60). This transcription factor is predominantly expressed in developing seeds, regulating the expression of genes involved in cell wall metabolism as well as cell division and expansion in a manner that controls seed size. Taken together, the high levels of ARR7 and MYB 56 transcripts might explain, at least in part, the observed larger size of the imbibed seeds/embryos observed in the mutants with low antioxidant buffering capacity.
Cell cycle progression is driven and regulated by an ancient molecular module of cyclin-dependent kinases (CDKs) and cyclins. A number of transcripts involved in this cell cycle regulation such as solo dancers (SDS), a plant cyclin related to the A- and B-type cyclins, which is required for homologous chromosome pairing (6), were found at high levels in seeds with low antioxidant capacity (Table 1). The levels of CDC7 and CDKB1;2 transcripts were significantly higher in the low ascorbate mutants than the wild type. CDC7, which is predominantly expressed at the G1 to S transition, recruits the enzymes required for replication to the DNA replication site (41). While CDC7 expression begins at the G1 to S transition and it is constant thereafter, CDKB1;2 regulates the G2 to M transition, interacting with A2-type cyclins and preventing endoreduplication (41).
The levels of transcripts encoding several glutathione S-transferases (GSTs) and glutaredoxins (GRX) were higher in the dry seeds of the ascorbate-deficient mutants than the wild type. For example, transcripts encoding 2 tau class GSTs (GSTU8 and GSTU12) were abundant in the ascorbate-deficient mutants. GSTU8 has high peroxidase activity and can form glutathione-oxylipin conjugates, linking this GST to JA signaling. Like GSTU8, GSTU12 is a hydrogen peroxide-responsive gene that localizes to the nucleus. The GSTU12 protein has been suggested to function as a transcriptional regulator in the nucleus. In contrast, the levels of transcripts encoding GRXS9 and a GRX-like protein (At2g30540 and At5g01420, respectively) were significantly lower in the ascorbate-deficient mutants than the wild type. The expression of GRX9 suppresses defense responses mediated by jasmonic acid.
Transcriptome profiles of imbibed seeds with low redox buffering capacity
Only about 100 transcripts were differentially expressed in the imbibed seeds of the ascorbate-deficient mutants relative to the wild type (Supplementary Tables S3 and S4 and Fig. 3). Only two transcripts decreased in abundance in imbibed seeds with low redox buffering capacity relative to the wild type (Table 2). The levels of a greater number of transcripts such as CEP1 were significantly increased in the imbibed seeds with low redox buffering capacity compared with the wild type. The levels of transcripts related to secondary metabolism were also significantly increased in the imbibed seeds with low redox buffering capacity relative to the wild type (Table 2). Moreover, transcripts encoding mitochondrial complex I subunit, NADH-ubiquinone oxidoreductase chain 1 (ND1) and chain 2 (ND2), were significantly increased in both ascorbate-deficient mutants relative to the wild type.
Table 2.
Transcripts Whose Abundance Was Most Influenced by Low Redox Buffering Capacity in Imbibed Seeds of the vtc2-1 and vtc2-4 Mutants Relative to the Wild Type
| Log fold change | |||
|---|---|---|---|
| Accession | Description | vtc2-1 | vtc2-4 |
| At5g50260 | CEP1 | 4.9 | 3 |
| At3g26200 | CYP450 71B22 | 4.4 | 2.2 |
| At1g26400 | FOX3 | 4.3 | 3.8 |
| AtMg00516 | ND1 | 4 | 4.1 |
| At3g44860 | FAMT | 3.9 | 4.3 |
| At1g30135 | JAZ8 | 3.7 | 3 |
| At3g55210 | NAC63 | 3 | 3.3 |
| At4g34410 | RRTF1 | 2.8 | 3.4 |
| At4g34410 | ERF109 | 2.8 | 3.4 |
| At4g23600 | CORI3 | 2.2 | 3.8 |
| AtMg00285 | ND2 | 2.2 | 2.7 |
| At1g17380 | JAZ5 | 2.2 | 3 |
| At1g80840 | WRKY40 | 2.1 | 2.3 |
| AtCg00730 | PETD | −2.1 | −2.4 |
| AtCg00730 | PSBJ | −2.7 | −2.6 |
As was observed in dry seeds, the levels of several JA-associated transcripts, including coronatine induced3 (CORI3), jasmonate ZIM-domain (JAZ)5, and JAZ8, were increased in the imbibed seeds of the ascorbate-deficient mutants relative to the wild type. The levels of ethylene response factor (ERF) 109 (also called redox-responsive transcription factor 1) transcripts were significantly increased in the imbibed seeds of the low ascorbate mutants relative to the wild type. This transcription factor regulates shoot and root development as well as plant defense responses through the propagation of ROS signals and regulation of redox homeostasis under stress.
A comparison of low ascorbate transcriptome profiles of dry and imbibed seeds with those of rosette leaves
We compared the data obtained in the present experiments with the available datasets for vtc2-1 rosette leaves. The degree of overlap between the genes that are significantly differentially expressed between vtc2 and the wild type for each of vtc2 rosette, vtc2-1 dry seeds, vtc2-1 imbibed, vtc2-4 dry seeds, and vtc2-4 imbibed are shown in a five-way Venn diagram (Supplementary Fig. S1). The significantly different genes between the alleles for dry seeds and imbibed, respectively, are shown in Supplementary Tables S5 and S6.
Effects of low redox buffering capacity on cell numbers and nucleus size during the cell cycle
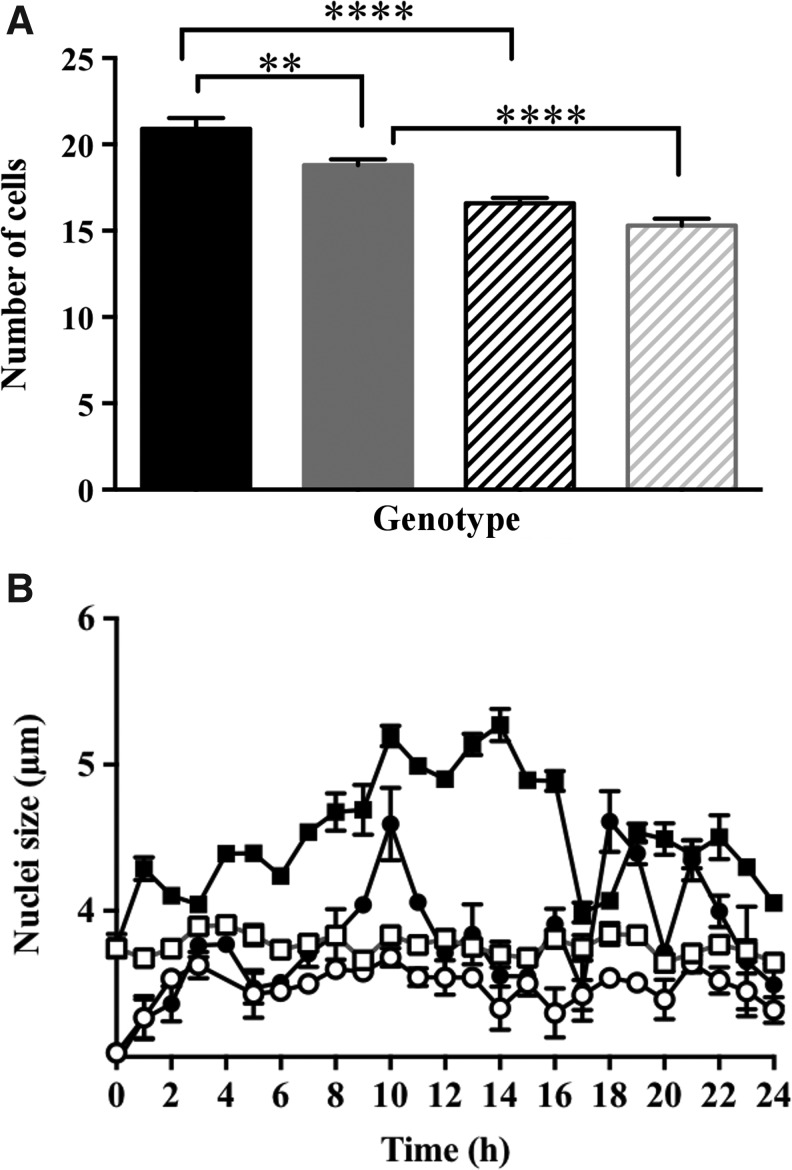
The proliferating cells within the developing embryonic root meristem do not divide synchronously. We therefore used hydroxyurea (HU), which inhibits the small subunit of the ribonucleotide reductase, leading to stalling of the replication fork and cell cycle arrest between G1 and S phases, to synchronize the cell cycle in embryonic roots of germinating seeds (13). The number of cells in the proliferation zone of embryonic roots was significantly lower after 24 h in the presence of HU compared with untreated samples (Fig. 4A). Moreover, the roots with low redox buffering capacity had fewer cells per unit area than the wild type (Fig. 4A). The nuclei were smaller in the proliferation zone of low redox buffering capacity roots than those of the roots of roGFP-expressing wild type in the absence of HU (Fig. 4B).
FIG. 4.
A comparison of cell numbers and the size of the nuclei in the proliferation zones of the embryonic roots of ascorbate-deficient seedlings in comparison to controls in the absence or presence of cell cycle synchronization with hydroxyurea (HU). Effects of HU treatment on cell numbers (A) and the diameter of nuclei (B) within the proliferation zone of embryonic roots of vtc2-1-roGFP2 seedlings. (A) Cell number in roGFP2 (black) and vtc2-roGFP2 (gray) roots in the absence (solid) or presence of HU (stripes). (B) Nuclear diameter in roGFP2 (square) and vtc2-1 (round) roots in the absence (open bars) or presence (striped bars) of HU.
The effect of low antioxidant buffering capacity on size of the nuclei was most marked in cells in which the cell cycle was synchronized using HU. Over the 24-h time course of cell cycle synchronization, the size of the nuclei in the proliferation zone increased progressively from the first hour of HU treatment to maximum values between 10 and 15 h. Thereafter, there was a decrease in the size of the nuclei, but overall the nuclei were significantly larger in the presence of HU compared with untreated controls (Fig. 4B). This was not the case for the nuclei in the roots with low redox buffering capacity, where the nuclei remained small except for a transient increase in size between 8 and 12 h after onset of HU treatment.
Oxidation of the cytosol at G1, followed by oxidation of the nucleus in the S phase of the cell cycle
We used cell-type-specific markers (WOX5:GFP, WOL:GFP, and PLT3:GFP) to identify the cell proliferation zone, in which all the measurements of the redox state of the nuclei and cytosol were made (Supplementary Fig. S2). Cell cycle progression was determined in the absence or presence of HU using a range of different markers (Fig. 5 and Supplementary Figs. S2–S5). For example, cell cycle tracking in plant cells (Cytrap) allows simultaneous monitoring of the S and G2/M phases (Supplementary Fig. S2). Expression of these markers shows that cells in the proliferation zone accumulated in the G1/S phase between 6 and 8 h after the start of the HU treatment, with G2/M phase accumulation occurring between 16 and 24 h after the onset of HU treatment. Incorporation of 5-ethynyl-2′-deoxyuridine (EdU), which was used as a marker for S phase, occurred at 17 h after the onset of HU treatment phases (Supplementary Fig. S3).
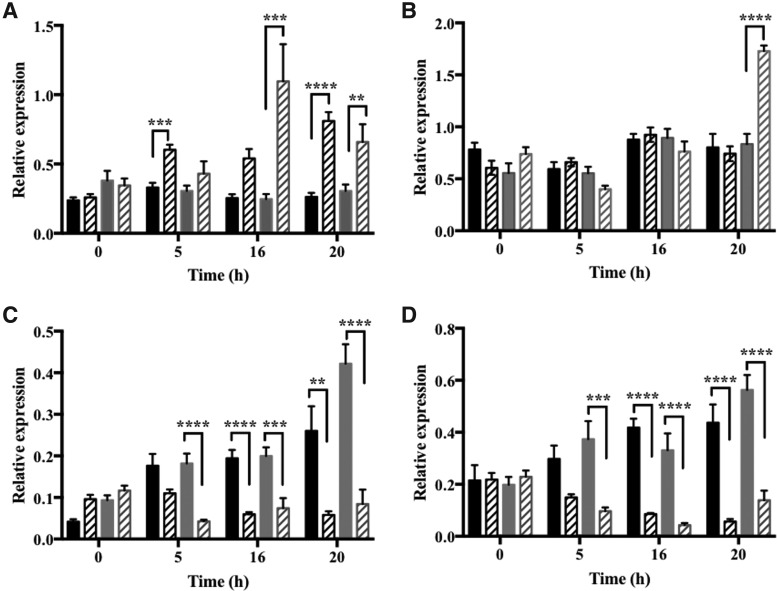
FIG. 5.
The effect of HU on the expression of cell cycle markers in synchronized cells in proliferation zones of embryonic roGFP2 and vtc2-1-roGFP roots. (A) CYCA3.1, (B) CYCD3.1, (C) CYCA.1, (D) CYCB2.1 mRNAs in roGFP2 (black) and vtc2-1-roGFP (gray) in the absence (black stripes) or presence (gray stripes) of HU. **p < 0.01, ***p < 0.001 and ****p < 0.0001. mRNAs, messenger RNAs.
The levels of CYCD3.1 transcripts were maximal between the 8- and 17-h time points after the start of HU treatment, suggesting that there was strong enrichment of S-phase cells at this time point. The expression of the CYCB1.1:DB-GUS marker was maximum 18 h after the start of HU treatment (Supplementary Fig. S4). The abundance of cyclin mRNAs was used as an indicator of progression through the cell cycle (Fig. 5): cyclin CYCA3.1 (Fig. 5A) was used as a marker for the G1/S phase of the cell cycle, CYCD3.1 (Fig. 5B) was used as an S phase marker, and cyclins, CYCA1.1 (Fig. 5C) and 1 CYCB2.1 (Fig. 5D), were used as markers for the G2/M phase of the cell cycle (11, 13, 14, 40, 41). The levels of CYCA3.1 transcripts were much higher in roots 5, 16, and 20 h after the onset of HU treatment than in the roGFP2 roots (Fig. 5A).
Taken together, these data show that the HU treatment led to synchronization of the progression of cell cycle in the proliferation zone such that the majority of cells were in the G1 phase between 6 and 8 h after the start of the HU treatment, in S phase between 8 and 16 h, and in the G2/M phase thereafter.
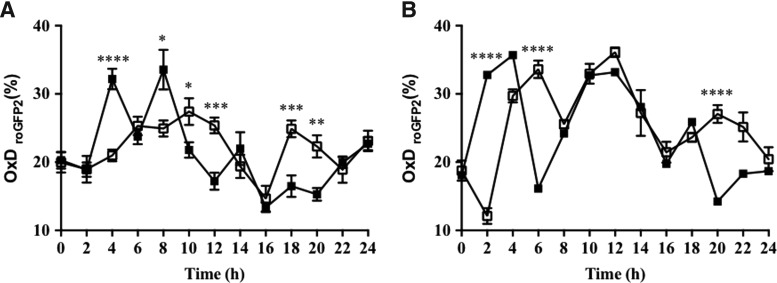
The degree of oxidation of nuclei and cytosol in cells in the proliferation zone of embryonic roots of germinating Arabidopsis seeds was measured using roGFP2 (Fig. 6). In the absence of synchronization, the nuclei and cytosol of dividing cells in the Arabidopsis root tip had similar glutathione redox potentials, with values of −297.5 mV ±0.7 and −292.8 mV ±0.6, respectively. There was a significant increase in the degree of oxidation in the cytosol within the first h after the onset of HU treatment. A similar, but later (between 5 and 8 h), increase in the degree of oxidation was observed in the nuclei. Significant oxidation was therefore observed in both the cytosol and nuclei within the first hour of HU treatment, when most of the cells in the proliferation zone were in the G1 and S phases of the cell cycle. In contrast, the degree of oxidation in the nuclei and cytosol was lower within the final hours of HU treatment where cells accumulate in the G2 and M phases (Fig. 6).
FIG. 6.
Redox cycling within the cell cycle measured as changes in the degree of oxidation occurring in the proliferating cells of the embryonic roots of germinating Arabidopsis seeds. Effects of HU on the degree of oxidation of the nuclei (A) and cytosol (B) of proliferation zone cells in roGFP2-expressing roots. Fluorescence was measured in the absence (open squares) or presence (filled squares) of HU. *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001.
Low redox buffering capacity decreases the oxidation degree of the nucleus and delays cell cycle progression
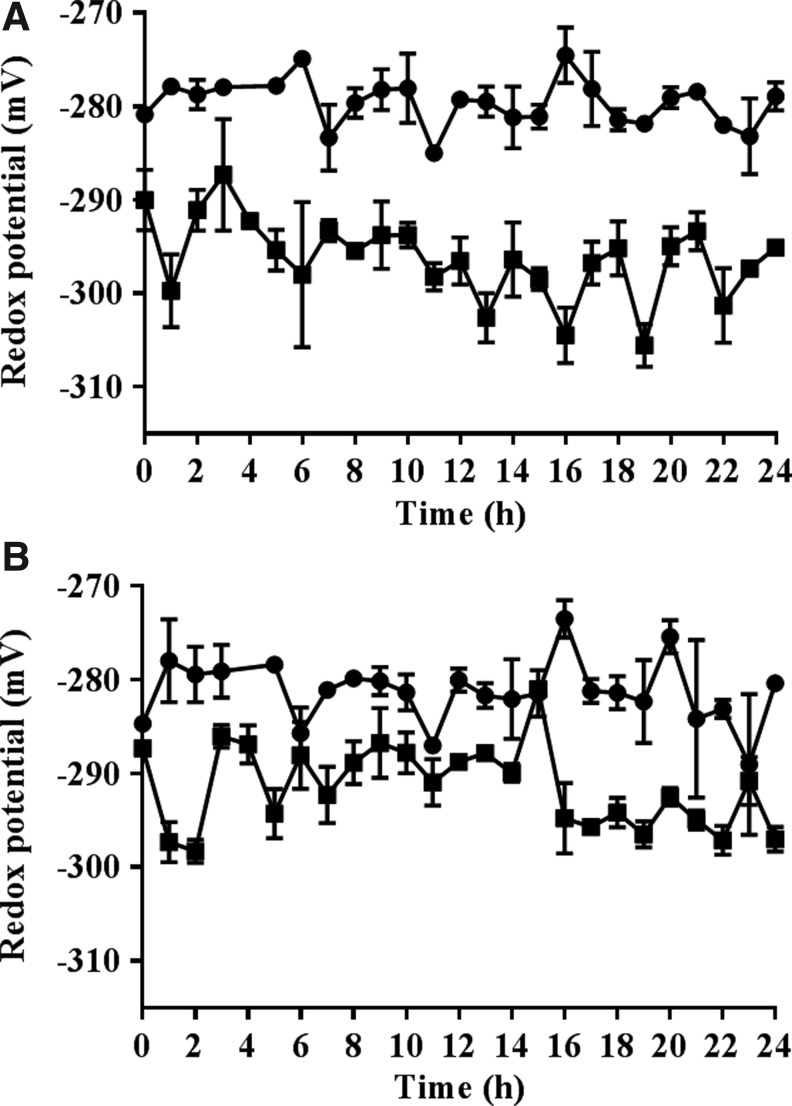
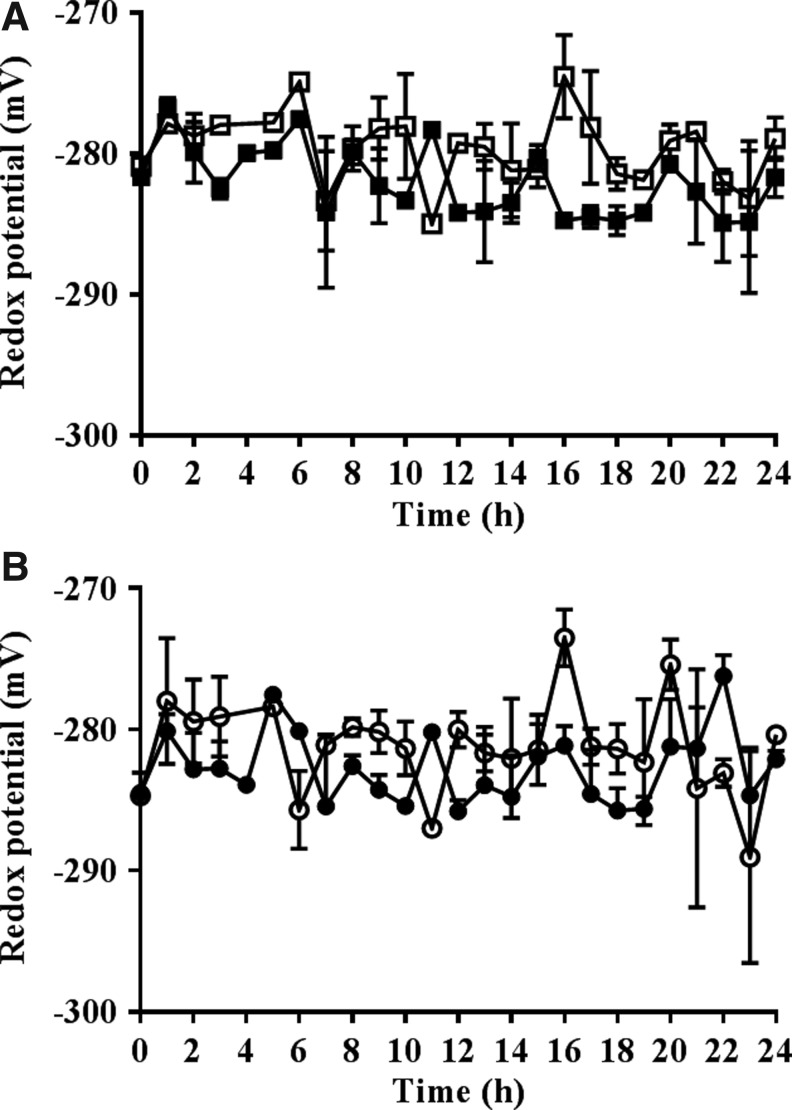
The calculated redox potentials of the nuclei of dividing cells in the embryonic roots with low redox buffering capacity were significantly more oxidized than those of the roGFP2 control roots (Fig. 7). The addition of HU caused a slight reduction in the redox state of the nuclei and the cytosol within the first and final hours of HU treatment where cells accumulate in the G1/S phases and G2/M phases, respectively (Fig. 8).
FIG. 7.
A comparison of the glutathione redox potentials of the nuclei and cytosol in the proliferating cells of the embryonic roots of ascorbate-deficient seedlings and controls. Calculated glutathione redox potentials in the nuclei (A) and cytosol (B) of the dividing cells of roGFP2 (squares) and vtc2-1-roGFP-expressing embryonic roots (circles) over the period of measurement in the absence of synchronization.
FIG. 8.
Transient oxidation is replaced by transient reduction of the nuclei and cytosol in cells in the proliferation zone of the embryonic roots of ascorbate deficient seedlings over the first seven hours after the onset of HU treatment. The effects of HU treatment on the calculated glutathione redox potentials of the nuclei (A) and cytosol (B) in proliferation zones of embryonic roots of vtc2-1-roGFP plants in the absence (open symbols) and presence (closed symbols) of HU.
As discussed above, treatment with HU resulted in an accumulation of cells in G1/S phase of the cell cycle for up to 8 h after addition of the inhibitor in the roGFP2 control roots (Fig. 5 and Supplementary Figs. S2–S4), in the S phase for up to 16 h, and the G2/M phase after 16 h. This pattern was changed in vtc2-1-roGFP2 roots (Fig. 5) such that CYCA3.1 transcript levels were significantly higher only between 16 and 20 h after the onset of HU treatment (Fig. 5A).
Treatment with HU had no effect on the levels of CYCD3.1 transcripts, except that the abundance of CYCD3.1 transcripts significantly increased in the vtc2-1-roGFP2 roots at 20 h after the onset of HU treatment (Fig. 5B). The abundance of CYCA1.1 (Fig. 5C) and CYCB2.1 (Fig. 5D) transcripts, which were used here as markers for the G2/M phase of the cell cycle, was significantly lower in HU-treated vtc2-1-roGFP2 roots than the roGFP2 control roots at the 5-h time point (Fig. 5B). The levels of CYCA1.1 (Fig. 5C) and CYCB2.1 (Fig. 5D) transcripts were also lower in both the roGFP2 and vtc2-1-roGFP2 roots at 16 and 20 h after the onset of HU treatment (Fig. 5C, D).
Discussion
Redox cycling during the cell cycle is well established in animals. A transient accumulation of ROS is required to activate key proteins involved in the initiation and maintenance of cell division (9, 42, 50, 66). To date, this type of regulation has not been described in plants, although we (19, 20) and others (24) have postulated such regulation. The data presented here provide the first evidence that transient oxidation occurs at G1 in the cytosol and nuclei of proliferating cells in the embryonic root, showing that redox cycling occurs within the plant cell cycle as it does in animal cells. Moreover, the evidence presented here clearly demonstrates that transient oxidation of the cytosol occurring early in the G1 phase of the cell cycle precedes transient oxidation of the nuclei.
ROS accumulation has been linked to an increased rate of cell division and an enhancement in root cell differentiation via signaling through the P-loop NTP hydrolase family protein called APP1, which influences the activity of the mitochondrial complex I and related ROS signaling, together with downstream targets such as SCARECROW and SHORT ROOT (63). ROS are considered to be important regulators of stem cell identity in mammals, regulating hematopoietic stem cell function (66), as well as controlling stem cell differentiation (59). The enhanced oxidation of both the cytosol and nuclei observed at the G1 phase may explain previous observations showing that plant cells are more sensitive to mild oxidative stress in G1 than at the S phase of the cell cycle (10).
The increase in oxidation of the cytosol and nuclei in G1 reported here was only transient and was rapidly followed by rereduction at later stages of the cell cycle. Although the mechanisms that link changes in cytoplasmic redox state to cell cycle regulation are unknown, the proliferating cell nuclear antigen (PCNA) protein, which is involved in DNA replication and has cell cycle-dependent properties, the E2F and MYB3R transcription factors, and the retinoblastoma-related (RBR) proteins are likely candidates because they regulate the suites of genes involved in the G1/S and G2/M transitions in plants.
We used solvent accessibility prediction (NetSurfP) (51) to identify cysteine residues, which are potentially accessible in a core set of 60 Arabidopsis cell cycle proteins. Of these, 14 of the proteins either had no cysteines or are predicted to have only buried cysteines. However, of the remaining 46, the majority (34) had between 1 and 3 accessible cysteine residues and the remaining 12 had between 4 and 8 potentially exposed cysteines. Our list of putative redox-regulated cell cycle proteins includes RBR1, WEE1, and several members of the cyclin A, B, and D families. The cyclin D family shows members with several (up to four) exposed cysteines or with no exposed cysteine residues, which may be indicative of a redox-sensitive and redox-insensitive population.
RBR, which is a signal-dependent scaffold protein, is the plant ortholog of the animal retinoblastoma (Rb) protein and is of central importance in modulating cell proliferation and meristem activity to prevailing environmental conditions (27). Like Rb, RBR is regulated by post-translational modifications, particularly by multisite phosphorylation, which determines binding to the E2F transcription factors that regulate the cell cycle (27). During G1, Rb becomes phosphorylated on alternative individual sites, leading to monophosphorylated Rb forms with distinct properties, such as the association with activator E2F1 to regulate checkpoint arrest or dissociation from repressor E2F4 to enable entry into G1/S.
In animal tumors, cell cycle regulation through the Rb pathway is coupled to cellular metabolism through ROS signaling in a manner that links mitochondrial biogenesis and functions to the cell cycle (40), a process that may also occur in plants. In addition, hydrogen peroxide is a potent activator of plant mitogen-activated protein kinases (33). Redox signaling pathways in plants interact directly with cell division and organ growth regulation by phytohormones such as auxin (3). The outstanding question remains the identification of specific factor(s) required for redox-dependent regulation of plant cell cycle progression.
Changes in the abiotic environment that are perceived by the mother plant have long been considered to influence the seed, but the mechanisms involved and their impact on seed properties remain poorly characterized. One way in which signals from the environment may influence seed characteristics is through effects on cellular redox status and redox signaling. The transcriptome profiling data presented here show that low redox buffering capacity in the mother plant has a strong influence on the levels of specific suites of transcripts in the dry seed. In particular, unlike a number of defense-related transcripts, which were increased by low redox buffering capacity, transcripts encoding some components involved in secondary metabolism were decreased in the dry seeds with low ascorbate.
These findings are consistent with previous observations that ascorbate deficiency constitutively increases pathogen defenses through salicylic acid, ethylene, and JA-associated pathways (32, 40, 42, 48). However, the pathways of synthesis of flavonoids and related metabolites have redox-sensitive steps (47). The ascorbate-deficient mutants have previously been shown to accumulate less anthocyanin and show impaired light-mediated induction of transcripts encoding anthocyanin biosynthesis enzymes such as transcription factors that activate the pathway. Hence, the finding that transcripts involved in phenylpropanoid metabolism and anthocyanin synthesis were much lower in the dry seeds of ascorbate-deficient mutants than the wild type is consistent with previous observations (47).
While the effects of low ascorbate resulted in marked changes in the transcriptome of dry seeds, taken together, the vtc2-1 and vtc2-4 data show that low ascorbate had no effect on the germination rate and the imbibed seeds were significantly larger than the wild type even though the embryonic root had fewer cells per unit area, suggesting that enhanced oxidation experienced by the developing seed within the mother plant leads to a smaller number of larger cells in the embryo. Seed maturation involves a certain degree of redox stress during the desiccation phase, but this may be enhanced when the antioxidant buffering capacity of the parental tissues is low. The stimulatory effect of germination is relatively short-lived, however, because the transcript profiles of imbibed seed from plants with low antioxidant capacity have a very similar transcript profile to the wild type and the level of germination was similar in all lines.
The level of oxidation experienced by seeds influences traits such as dormancy and longevity. Low levels of oxidation are required to release seed dormancy, while higher levels can be detrimental and cause a reduction in seed longevity (22, 52). The decreased ascorbate levels in the vtc2-1 and vtc2-4 mutants are not associated with altered seed dormancy levels (Fig. 2A). This suggests that ascorbate does not play an important role in seed dormancy. This might be expected given that dry wild-type seeds are largely devoid of ascorbate, which is synthesized de novo upon germination. The absence of ascorbate from wild-type dry seeds therefore precludes alternative explanations regarding the role of ascorbate, for example, as a cofactor in many reactions. It is therefore possible to distinguish the effects of low redox buffering capacity caused by low ascorbate in the mother plant from effects that would be caused directly in the seed, for example, by impaired ascorbate-dependent biochemical reactions (e.g., proline hydroxylase, other ascorbate-dependent enzymes).
A quantitative analysis of the relative distribution of ascorbate between the organelles of leaf cells using immunogold labeling revealed that the highest ascorbate-specific labeling was detected in the nuclei and the cytosol, leading to a calculated ascorbate concentration of ascorbate in the nuclei of about 16 mM (64). The decrease in ascorbate in the nuclei in the leaves of the vtc2-1 mutant was between 54% and 65% (64). While dry seeds are largely devoid of ascorbate, ascorbate is synthesized upon imbibition and germination. The data presented here show that an inability to synthesize ascorbate following imbibition has a profound impact on the redox state of nuclei in the proliferation zone of the embryonic root.
Very few measurements of the redox state of root cells are available in the literature. A recent study showed that the cells in the root tip reside in a highly reduced state and that the redox state of the cytosol changes little (only 5–10 mV) as the cells expand away from the proximal meristem, the redox state of the cells remaining more or less unchanged throughout the transition zone (31). The data presented here show that the redox state of nuclei in the proliferation zone decreased to −279.7 ± 0.5 in the vtc2-1 roots with low redox buffering capacity compared with 297.5 ± 0.7 in the wild type. Similarly, the redox state of the cytosol of cells in the proliferation zone decreased from −292.8 ± 0.6 in the wild type to −281.2 ± 0.8 in the vtc2-1 roots. Hence, the redox state of nuclei was altered significantly more by low redox buffering capacity than the cytosol, resulting in a persistent state of oxidation in the nuclei that was more severe than the low redox buffering capacity-induced oxidation of the cytosol. A similar oxidation of the cytoplasm, which was accompanied by changes in the distribution of auxin transporters such as AUX1 and PIN1/2, was observed upon the imposition of salt stress (31).
As discussed above, the data presented here demonstrate that low antioxidant capacity in the imbibed seed not only eliminated the redox cycle of oxidation, followed by rereduction observed during the cell cycle in the proliferation zone of the embryonic root, but it also resulted in sustained oxidation of nuclei in the proliferation zone of the embryonic root, and the redox state was changed by a substantial amount (20 mV). The enhanced oxidation of nuclei determined in cells with low redox buffering capacity was associated with a failure of the nuclei to increase in size during G1, except in a very transient manner, during the cell cycle. This finding confirms previous observations showing oxidation-dependent inhibition of the induction of DNA synthesis in plant cells experiencing mild oxidative stress (10).
Oxidation-mediated inhibition of DNA replication resulting in decreased nuclear DNA content has also been observed in meristematic zones (10, 55) in a manner that is consistent with the smaller nuclear size observed in the cells of roots with decreased antioxidant capacity. The high level of oxidation of nuclei in roots with low antioxidant buffering capacity shows that a change in the nuclear redox state of about 20 mV is sufficient to delay G1 and progression through the cell cycle. This finding is consistent with previous observations showing that mild oxidative stress experienced during the S phase delayed entry into mitosis because the cells had a longer G2 phase (10).
It has recently been suggested than an independent cryptic mutation may be responsible for the slow shoot growth phenotype reported in vtc2-1 (37). However, no quantitative evidence for this conclusion was provided in support of this conclusion under short-day conditions (37). While no phenotype has been reported for germinating seeds that have low ascorbate, it has recently been shown that when the interaction between the photomorphogenic factor COP9 signalosome subunit 5 and VTC1 is impaired, ascorbate accumulation is enhanced in Arabidopsis seedlings, stimulating seedling growth (36). Therefore, any influence of secondary mutations in vtc2-1 on plant growth remains to be substantiated. Moreover, we have focused only on the common features of the vtc2-1 and vtc2-4 transcriptome signatures to link antioxidant capacity to cell cycle regulation.
Taken together, the data presented not only provide unequivocal evidence for the existence of a redox cycle within the cell cycle in plants but they also show that failure to restore the redox poise of dividing cells leads to slowdown of the cell cycle. However, the data also show that the cell cycle can continue even when the redox state of nuclei increases by as much as 20 mV. This level of oxidation is not enough to cause complete arrest of cell division. Oxidation-dependent cell cycle arrest is generally associated with an inhibition of the activity of CDKs, cell cycle gene expression, and concomitant activation of stress genes. However, redox regulation of plant CDKs has not yet been described.
Materials and Methods
Seeds of A. thaliana ([L.] Heynh.) wild type (Col-0) and vtc2-4 (SAIL_769_H05) (37) were obtained from the Nottingham Arabidopsis Stock Centre. Seeds of the vtc2-1 mutant (11, 26, 30) were obtained from Dr. Robert Last. A redox-sensitive roGFP2 reporter line (43) was used to estimate the redox state of the nuclei and cytosol in the proliferation zone of the embryonic root during the cell cycle. The vtc2-1 mutant lines expressing roGFP2 (vtc2-1-roGFP2) ecotype Columbia (Col-0) were produced by Till K. Pellny (Rothamsted Research). Seeds of WUSCHEL (WOX5::GFP), WOODEN LEG 1 (WOL::GFP), and PLETHORA 3 (PLT3::GFP) were provided by Dr. Yoselin Beñitez Alfonso (University of Leeds). Seeds of CYCB1;1::GUS reporter line (15) were provided by Dr. Chris West (University of Leeds). Seeds of the dual-core marker system (Cytrap) expressing pHTR2::CDT1a (C3)-RFP and pCYCB1::CYCB1-GFP were provided by Dr Masaaki Umeda (Nara Institute of Science and Technology).
Seed properties
The 1,000-seed weights were calculated by counting out samples of 1,000 dry seeds from each line and measuring their weight (g). Dry seeds were viewed under a dissecting microscope. Seed dimensions (width and length) were then measured using the ImageJ/Fiji software (Version: 2.0.0-rc-49/1.51a). Approximately 30–50 seeds of each line were then placed in Petri dishes on absorbent filter paper soaked with 5 ml distilled water and left for 2 days at 4°C. The imbibed seeds were then imaged using a dissection microscope. The seed coat was removed with forceps and the embryos photographed as per the dry seeds. Fiji (ImageJ) software was used to measure the size of the embryos.
Seed germination and aging analysis
Wild-type and mutant plants were grown in an Elbanton controlled environment chamber with a 16-h photoperiod at 22°C and 8-h darkness at 16°C (50% relative humidity [RH]) until the start of flowering. For germination, speed assay plants stayed in the Elbanton until seed harvest, and for dormancy and CDT, plants were transferred after the start of flowering to a Percival controlled environment chamber with 16-h light at 16°C and 8-h darkness at 14°C (50% RH). Seeds of 10–16 plants were individually harvested per line and assayed as below.
Germination measurement for dormancy
Fifty to 100 seeds were sown in small Petri dishes on filter paper with 600 μl water, germinated in an incubator (12-h light at 25°C–12-h darkness at 20°C). Germinated seeds were counted after 7 days.
Controlled deterioration test
Seeds (three biological replicates per line [consisting of seeds from five to six plants] and three technical replicates for every biological replicate) were incubated for 0/3/6/9/12/15/18 days in open 0.5-ml reaction tubes in a box with 83% RH (saturated KCl solution) at 37°C. After the treatment, seeds were dried for 3–5 days in a box with drying pearls.
Germination measurement for CDT
Fifty to 100 seeds were sown in small Petri dishes on filter paper with 600 μl H2O, stratified for 2 days, germinated in an incubator (12-h light at 25°C–12-h darkness at 20°C), and counted after 5 days.
Germination speed assay
Seeds were stored after harvest in a controlled cabinet (MMM Medcenter, Brno, Czech Republic) (21°C; 50% RH; in darkness) until they were fully after-ripened. Germination tests were carried out as in the dormancy test except that germinated seeds (with a visible radicle) were counted every 12 h.
Cell cycle synchronization and roGFP2 measurements
Seeds were surface-sterilized and transferred to plates containing half-strength Murashige and Skoog 1% agar medium (½MS). Seeds were stratified for 48 h at 4°C and allowed to germinate at 21°C for further 48 h in the dark. The germinated seeds were transferred to fresh ½MS medium in the absence (control) or presence of 3 mM HU to allow cell cycle synchronization. Plates were then incubated at 21°C for 24 h. roGFP2 fluorescence measurements were then performed over a 24-h period. Germinated seeds were placed on a slide in a drop of sterile water and fluorescence imaged using a Carl Zeiss confocal microscope LSM700 equipped with 405 and 488 nm lasers for excitation. Images were taken with 40 × /1.3 Oil DIC M27 lens in a multitrack mode. Ratiometric analyses were performed using ImageJ software (http://rsbweb.nih.gov/ij/). The fluorescence at 405 nm was corrected by subtraction of the background and the corrected image was divided by the 488 nm fluorescence image to give a threshold of 3. The roGFP2 signal was calibrated at the end of each experiment using 2.5 mM dithiothreitol (reduced) and 2 mM hydrogen peroxide (oxidized). The oxidation degree and glutathione redox potential values were calculated as described by Meyer et al. (43).
Cell number and nuclear size measurements
The number of cells and the size of the nuclei were measured in the proliferation zone (2.5 mm area per sample) of the embryonic roots. The number of cells was measured at the 24-h time point after the start of HU treatment. Nuclear size measurements were performed every hour over the 24-h period of the experiment in the absence or presence of HU.
Cell cycle markers
The pHTR2::CDT1a (C3)-RFP and pCYCB1::CYCB1-GFP fluorescence was visualized by confocal laser scanning microscopy (CLSM), exciting at 488 nm and at 559 nm, respectively.
Selection of vtc2-1-roGFP2 lines
The vtc2-1-roGFP2 line used in these studies was prepared by crossing the roGFP2-expressing Arabidopsis reporter line with vtc2-1 mutants. All plants were grown on compost in controlled environment cabinets under a light intensity of 250 μmol m−2 s−1, with a 16-h photoperiod at a constant temperature of 20°C and an RH of 60%. F2 generation seeds from the vtc2-1 x roGFP2 crosses were surface sterilized and germinated on plates supplemented with 50 μg/ml kanamycin. Seedlings were transferred to moist compost and grown as above. Lines were selected on the basis of low ascorbate content. Homozygous vtc2-1-roGFP2 plants were verified by sequencing and selected for further measurements.
Polymerase chain reaction (PCR) was performed using ReadyMix™ Taq PCR Reaction Mix (Sigma) to amplify regions where mutations were localized. Primers used for amplification of the region containing the vtc2 mutation were 5′-CCTTTTGCTTGCAGTTCACA-3′ and 5′-TGAAGGCAAACACAGCAGTC-3′, while those used for amplification for GFP were 5′-GCAAGCTGACCCTGAAGTTC-3′ and 5′-CTTCTCGTTGGGGTCTTTGC-3′. The validated homozygous vtc2-1-roGFP2 plants were grown under the conditions described below and seeds collected from individual plants.
RNA seq analysis
Three biological replicates were taken from dry and imbibed seeds of the genotypes Col0, vtc2-1 (EMS), and vtc2-4 (SAIL_769_H05). RNA was extracted using RNAqueous small-scale phenol-free total RNA isolation kit (catalog No. AM1912; Ambion) according to the manufacturer's protocol. Sequencing was performed on the Illumina GAII platform. Unpaired 100 bp Illumina reads were aligned using TopHat (v2.0.10) against a Bowtie2 (v2.2.8) index built from the TAIR10 reference sequence to create SAM alignment files. SAM files were then sorted and converted into BAM alignment files using Samtools (v1.3) (35, 58). The aligned read replicates were counted using featureCounts to create gene count matrix and tested for differential gene expression using EdgeR in R/Bioconductor. Differentially expressed genes were those showing fold changes of >2 and an FDR-corrected p-value of 0.05 or less (54).
Real-time PCR
Real-time quantitative PCR (qPCR) was performed as described previously (46). Total RNA was extracted as described above. RNA reverse transcription and qPCR were performed on an Eppendorf Realplex2 real-time PCR system by one-step real time-polymerase chain reaction (RT-PCR) using Quantifast SYBR Green RT-PCR Kit (Qiagen), following the manufacturer's instructions. The expression of the genes of interest was normalized using A. thaliana TIP41 as endogenous control. Accessions and primer sequences used are given in Supplementary Tables S7 and S8: (At4g34270) sense 5′-GGCGAAGATGAGGCACCAA-3′ and antisense 5′-GCCTCTGACTGATGGAGCT-3′.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: VTC2 (At4g26850), VTC5 (At5g55120), CYP82C2 (At4g31970), CENTRIN 2 (At4g37010), GSTU12 (At1g69920), GSTU8 (At3g09270), GLU1 (At3g09270), MYB56 (At5g17800), SDS (At1g14750), CDC7 (At4g16970), CDKB1;2 (At2g38620), GRX-C8 (At5g14070), GRXS9 (At5g01420), SKU5 (At4g38420), UXS6 (At2g28760), PAP2 (At1g66390), UGT72B2 (At1g01390), UGT79B5 (At1g50580), CEP1 (At5g50260), AAE12 (At1g65890), CYP71B22 (At3g26200), FOX3 (At1g26400), ND1 (AtMg00516), CORI3 (At4g23600), JAZ5 (At1g17380), JAZ8 (At1g30135), ARR7 (At1g19050), ACOS5 (At1g62940), ERF109 (At4g34410), NAC063 (At3g55210), and FAMT (At3g44860). Sequences and accession numbers for primers for selected cell cycle markers are given in Supplementary Table S7 and for housekeeping genes in Supplementary Table S8.
All RNAseq data from this article are available at ArrayExpress (www.ebi.ac.uk/arrayexpress/) under the accession number E-MTAB-5103.
Supplementary Material
Abbreviations Used
- ABA
abscisic acid
- APX
ascorbate peroxidase
- ARR
Arabidopsis response regulator
- BR
brassinosteroid
- CDK
cyclin-dependent kinase
- CDT
controlled deterioration test
- CORI3
coronatine induced3
- Cytrap
cell cycle tracking in plant cell
- ERF
ethylene response factor
- GRX
glutaredoxin
- GST
glutathione S-transferase
- HU
hydroxyurea
- JA
jasmonate
- JAZ
jasmonate ZIM-domain
- mRNA
messenger RNA
- PAP2
anthocyanin pigment 2 protein
- PCR
polymerase chain reaction
- qPCR
quantitative PCR
- Rb
retinoblastoma
- RBR
retinoblastoma-related
- RH
relative humidity
- ROS
reactive oxygen species
- SDS
solo dancers
- vtc2
vitamin C defective 2
Acknowledgments
The authors thank the European Union (KBBE-2012–6-311840; ECOSEED) for financial support and acknowledge additional funding by the Australian Research Council (DP150103211). They thank Robbie Gillet (University of Leeds) for isolating homozygous seed of the vtc2 (T-DNA) line SAIL_769_H05 for At4g26850, Wanda Waterworth for help with the seed dimension calculations, Pedro Diaz Vivancos for help with the measurements of nuclear size, Dorothee Wozny for help with the RNAseq data analysis, and Christina Philipp for performing the seed counting experiments. The authors also thank the Genome Centre at Max Planck Institute for Plant Breeding Research (Cologne, Germany) for the RNAseq analysis. The roGFP2 seeds used in these studies were originally obtained from Dr. Andreas Meyer (University of Bonn, Bonn, Germany). Y.V. and M.C. acknowledge the University of Western Australia for a postgraduate research scholarship and stipend, the Australian Grape and Wine Authority postgraduate top-up scholarship and the Organisation of Vine and Wine (OIV) for a research travel grant.
Authors' Contributions
A.D.S. performed the experimental work, except where attributed to other authors, and produced the final figures. R.H. produced the data in Figure 1, N.V.D.L.T. produced the data in Figure 3, Y.V. produced the data in Figure 6, M.W. undertook all bioinformatic analyses of the data, M.J.C. checked the final version of the manuscript, W.J.J.S. produced the data in Figure 2, and C.H.F. directed the work and wrote the manuscript.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Amara I, Zaidi I, Masmoudi K, Ludevid MD, Pagés M, Goday A, and Brini F. Insights into late embryogenesis abundant (LEA) proteins in plants: From structure to the functions. Am J Plant Sci 5: 3440–3455, 2014 [Google Scholar]
- 2.Bailly C. Oxidative signaling in seed germination and dormancy. Plant Signal Behav 3: 175–182, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berkowitz O, De Clercq I, Van Breusegem F, and Whelan J. Interaction between hormonal and mitochondrial signalling during growth, development and in plant defence responses. Plant Cell Environ 39: 1127–1139, 2016 [DOI] [PubMed] [Google Scholar]
- 4.Bewley JD. Seed germination and dormancy. Plant Cell 9: 1055–1066, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borisjuk L. and Rolletschek H. The oxygen status of the developing seed. New Phytol 182: 17–30, 2009 [DOI] [PubMed] [Google Scholar]
- 6.Bulaankova P, Akimcheva S, Fellner N, and Riha K. Identification of Arabidopsos meiotic cyclins reveals a functional diversification among plant cyclin genes. PLoS Genet 9: e1003508, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burhans WC. and Heintz NH. The cell cycle is a redox cycle: Linking phase-specific targets to cell fate. Free Radic Biol Med 47: 1282–1293, 2009 [DOI] [PubMed] [Google Scholar]
- 8.Chen CM, Letnik I, Hacham Y, Dobrev P, Ben-Daniel BH, Vankova R, Amir R, and Miller G. ASCORBATE PEROXIDASE6 protects Arabidopsis desiccating and germinating seeds from stress and mediates cross talk between reactive oxygen species, abscisic acid, and auxin. Plant Physiol 166: 370–383, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu J. and Dawes IW. Redox control of cell proliferation Trends Cell Biol 22: 592–601, 2012 [DOI] [PubMed] [Google Scholar]
- 10.Clopton DA. and Saltman P. Low-level oxidative stress causes cell-cycle specific arrest in cultured cells. Biochem Biophys Res Commun 210: 189–196, 1995 [DOI] [PubMed] [Google Scholar]
- 11.Combettes B, Reichheld JP, Chaboute ME, Philipps G, Shen WH, and Chaubet-Gigot N. Study of phase-specific gene expression in synchronized tobacco cells. Methods Cell Sci 21: 109–121, 1999 [DOI] [PubMed] [Google Scholar]
- 12.Conklin PL, Saracco SA, Norris SR, and Last RL. Identification of ascorbic acid-deficient Arabidopsis thaliana mutants. Genetics 154: 847–856, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cools T, Iantcheva A, Maes S, Van den Daele H, and De Veylder L. A replication stress-induced synchronization method for Arabidopsis thaliana root meristems. Plant J 64: 705–714, 2010 [DOI] [PubMed] [Google Scholar]
- 14.Culligan K, Tissier A, and Britt A. ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16: 1091–1104, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeGara L, dePinto MC, and Arrigoni O. Ascorbate synthesis and ascorbate peroxidase activity during the early stage of wheat germination. Physiol Plant 100: 894–900, 1997 [Google Scholar]
- 16.De Schutter K, Joubles J. Cools T, Verkest A, Corellou F, Babiychuk E, Van Der Schueren E, Beckman T, Kushnir S, Inze D, and De Veylder L. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19: 211–225, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Simone A, Pellny TK, and Foyer CH. Accumulation of ascorbate in the leaves of Arabidopsis thaliana wildtype and vtc2 mutants under continuous light and short photoperiod conditions. Asp Appl Biol 124: 123–130, 2015 [Google Scholar]
- 18.Di Donato RJ, Arbuckle E, Buker S, Sheets J, Tobar J, Totong R, Grisafi P, Fink GR, and Celenza JL. Arabidopsis ALF4 encodes a nuclear-localized protein required for lateral root formation. Plant J 37: 340–353, 2004 [DOI] [PubMed] [Google Scholar]
- 19.Diaz-Vivancos P, de Simone A, Kiddle G, and Foyer CH. Glutathione—Linking cell proliferation to oxidative stress. Free Radic Biol Med 89: 1154–1164, 2015 [DOI] [PubMed] [Google Scholar]
- 20.Diaz Vivancos P, Wolff T, Markovic J, Pallardo FV, and Foyer CH. A nuclear glutathione cycle within the cell cycle. Biochem J 431: 169–178, 2010 [DOI] [PubMed] [Google Scholar]
- 21.Dowdle J, Ishikawa T, Gatzek S, Rolinski S, and Smirnoff N. Two genes in Arabidopsis thaliana encoding GDP-L-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J 52: 673–689, 2007 [DOI] [PubMed] [Google Scholar]
- 22.El-Maarouf-Bouteau H. and Bailly C. Oxidative signaling in seed germination and dormancy. Plant Signal Behav 3: 175–182, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Maarouf-Bouteau H, Sajjad Y, Bazin J, Langlade N, Cristescu S M, Balzergue S, Baudouin E, and Bailly C. Reactive oxygen species, abscisic acid and ethylene interact to regulate sunflower seed germination. Plant Cell Environ 38: 364–74, 2015 [DOI] [PubMed] [Google Scholar]
- 24.Feher A, Otvos K, Pasternak TP, and Szandtner AP. The involvement of reactive oxygen species (ROS) in the cell cycle activation (G(0)-to-G(1) transition) of plant cells. Plant Signal Behav 3: 823–826, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fercha A, Capriotti AL, Caruso G, Caualiere C, Samperi R, Stampachiacchiere S, and Lagana A. Comparative analysis of metabolic proteome variation in ascorbate-primed and unprimed wheat seeds during germination under salt stress. J Proteomics 108: 238–257, 2014 [DOI] [PubMed] [Google Scholar]
- 26.Foyer CH, Ruban AV, and Noctor G. Viewing oxidative stress through the lens of oxidative signalling rather than damage. Biochem J 474: 877–883, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harashima H. and Sugimoto K. Integration of developmental and environmental signals into cell proliferation and differentiation through RETINOBLASTOMA-RELATED 1. Curr Opin Plant Biol 29: 95–103, 2016 [DOI] [PubMed] [Google Scholar]
- 28.Huang X, Zhang X, Gong Z, Yang S, and Shi Y. ABI4 represses the expression of type-A ARRS to inhibit seed germination in Arabidopsis. Plant J 89: 354–365, 2017 [DOI] [PubMed] [Google Scholar]
- 29.Ishibashi Y. and Iwaya-Inoue M. Ascorbic acid suppresses germination and dynamic states of water in wheat seeds. Plant Prod Sci 9: 172–175, 2006 [Google Scholar]
- 30.Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, and Last RL. Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129: 440–450, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang K, Moe-Lange J, Hennet L, and Feldman LJ. Salt stress affects the redox status of Arabidopsis root meristems. Front Plant Sci 7: 81, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerchev PI, Pellny TK, Vivancos PD, Kiddle G, Hedden P, Driscoll S, Vanacker H, Verrier P, Hancock RD, and Foyer CH. The transcription factor ABI4 is required for the ascorbic acid-dependent regulation of growth and regulation of jasmonate-dependent defense signaling pathways in Arabidopsis. Plant Cell 23: 3319–3334, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovtun Y, Chiu WL, Tena G, and Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci U S A 97: 2940–2945, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kranner I, Roach T, Beckett RP, Whitaker C, and Minibayeva FV. Extracellular production of reactive oxygen species during seed germination and early seedling growth in Pisum sativum. J Plant Physiol 167: 805–811, 2010 [DOI] [PubMed] [Google Scholar]
- 35.Langmead B. and Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S, Wang J, Yu Y, Wang F, Dong J, and Huang R. D27E mutation of VTC1 impairs the interaction with CSN5B and enhances ascorbic acid biosynthesis and seedling growth in Arabidopsis. Plant Mol Biol 92: 473–482, 2016 [DOI] [PubMed] [Google Scholar]
- 37.Lim B, Smirnoff N, Cobbett CS, and Golz JF. Ascorbate-deficient vtc2 mutants in Arabidopsis do not exhibit decreased growth. Front Plant Sci 7: 1025, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C, Ding F, Hao F, Yu M, Lei H, Wu X, Zhao Z, Guo H, Yin J, Wang Y, and Tang H. Reprogramming of seed metabolism facilitates pre-harvest sprouting resistance of wheat. Sci Rep 6: 20593, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lukowitz W, Nickle TC, Meinke DW, Last RL, Conklin PL, and Somerville CR. Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc Natl Acad Sci U S A 98: 2262–2267, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacLeod KF. The role of the RB tumour suppressor pathway in oxidative stress responses in the haematopoietic system. Nat Rev Cancer 8: 769–781, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menges M. and Murray JA. Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J 30: 203–212, 2002 [DOI] [PubMed] [Google Scholar]
- 42.Menon SG. and Goswami PC. A redox cycle within the cell cycle: Ring in the old with the new. Oncogene 26: 1101–1109, 2007 [DOI] [PubMed] [Google Scholar]
- 43.Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, and Hell R. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J 52: 973–986, 2007 [DOI] [PubMed] [Google Scholar]
- 44.Miransari M. and Smith DL. Plant hormones and seed germination. Environ Exper Bot 99: 110–121, 2014 [Google Scholar]
- 45.Mukherjee M, Larrimore KE, Ahmed NJ, Bedick TS, Barghouthi NT, Traw MB, and Barth C. Ascorbic acid deficiency in Arabidopsis induces constitutive priming that is dependent on hydrogen peroxide, salicylic acid, and the NPR1 gene. Mol Plant Microbe Interact 23: 340–351, 2010 [DOI] [PubMed] [Google Scholar]
- 46.Muller-Moule P, Golan T, and Niyogi KK. Ascorbate-deficient mutants of Arabidopsis grow in high light despite chronic photooxidative stress. Plant Physiol 134: 1163–1172, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Page M, Sultana N, Paszkiewicz K, Florance H, and Smirnoff N. The influence of ascorbate on anthocyanin accumulation during high light acclimation in Arabidopsis thaliana: Further evidence for redox control of anthocyanin synthesis. Plant Cell Environ 35: 388–404, 2012 [DOI] [PubMed] [Google Scholar]
- 48.Pastori GM, Kiddle G, Antoniw J, Bernard S, Veljovic-Jovanovic S, Verrier PJ, Noctor G, and Foyer CH. Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling. Plant Cell 15: 939–951, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pavet V, Olmos E, Kiddle G, Mowla S, Kumar S, Antoniw J, Alvarez ME, and Foyer CH. Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiol 139: 1291–1303, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pellny TK, Locato V, Vivancos PD, Markovic J, De Gara L, Pallardo FV, and Foyer CH. Pyridine nucleotide cycling and control of intracellular redox state in relation to poly (ADP-ribose) polymerase activity and nuclear localization of glutathione during exponential growth of Arabidopsis cells in culture. Mol Plant 2: 442–456, 2009 [DOI] [PubMed] [Google Scholar]
- 51.Petersen B, Nordahl Petersen T, Andersen P, Nielsen M, and Lundegaard C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol 9: 51, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rajjou L, Duval M, Gallardo K, Catusse J, Bally J, Job C, and Job D. Seed germination and vigor. Annu Rev Plant Biol 63: 507–533, 2012 [DOI] [PubMed] [Google Scholar]
- 53.Reichheld J-P, Vernoux T, Lardon F, Van Montagu M, and Inze D. Specific checkpoints regulate plant cell cycle progression in response to oxidative stress. Plant J 17: 647–656, 1999 [Google Scholar]
- 54.Robinson MD, McCarthy DJ, and Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russo T, Zambrano N, Esposito F, Ammendola R, Cimino F, Fiscella M, Jackman J, O'Connor PM, Anderson CW, and Appella E. A p53-independent pathway for activation of WAF1/CIP1 expression following oxidative stress. J Biol Chem 270: 29386–29391, 1995 [DOI] [PubMed] [Google Scholar]
- 56.Sano N, Rajjou L, North HM, Debeaujon I, Marion-Poll A, and Seo M. Staying alive: Molecular aspects of seed longevity. Plant Cell Physiol 57: 660–674, 2016 [DOI] [PubMed] [Google Scholar]
- 57.Tommasi F, Paciolla C, De Pinto MC, and De Gara L. A comparative study of glutathione and ascorbate metabolism during germination of Pinus pinea L. seeds. J Exp Bot 52: 1647–1654, 2001 [PubMed] [Google Scholar]
- 58.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Varum S, Momcilovic O, Castro C, Ben-Yehudah A, Ramalho-Santos J, and Navara CS. Enhancement of human embryonic stem cell pluripotency through inhibition of the mitochondrial respiratory chain. Stem Cell Res 3: 142–156, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vilarrasa-Blasi J, Gonzalez-Garcia M-P, Figola D, Fabregas N, Alexiou KG, |Lopez-Bigas N, Rvas S, Jauneau A, Lohmann JU, Benfey PN, Inanes M, and Cano Delgado AI. Regulation of stem cell quiescence by a brassinosteroid signalling module. Dev Cell 30: 36–47, 2014 [DOI] [PubMed] [Google Scholar]
- 61.Wang J, Yu YW, Zhang ZJ, Quan RD, Zhang HW, Ma LG, Deng XW, and Huang RF. Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis. Plant Cell 25: 625–636, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wojtyla Ł, Garnczarska M, Zalewski T, Bednarski W, Ratajczak L, and Jurga S. A comparative study of water distribution, free radical production and activation of antioxidative metabolism in germinating pea seeds. J Plant Physiol 163: 1207–1220, 2006 [DOI] [PubMed] [Google Scholar]
- 63.Yu Q, Tian H, Yue K, Liu J, Zhang B, Li X, and Ding Z. A P-loop NTPase regulates quiescent center cell division and distal stem cell identity through the regulation of ROS homeostasis in Arabidopsis root. PLoS Genet 12: e1006175, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zechmann B, Stumpe M, and Mauch FImmunocytochemical determination of the subcellular distribution of ascorbate in plants Planta 233: 1–12, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z, Wang J, Zhang R, and Huang R. The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J 71: 273–287, 2012 [DOI] [PubMed] [Google Scholar]
- 66.Zhou D, Shao L, and Spitz DR. Reactive oxygen species in normal and tumor stem cells. Adv Cancer Res 122: 1–67, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.