Abstract
Treatment of BRAF mutant melanoma with kinase inhibitors has been associated with rapid tumor regression; however, this clinical benefit is short-lived, and most patients relapse. A number of studies suggest that the extracellular environment promotes BRAF inhibitor resistance and tumor progression. Extracellular vesicles, such as exosomes, are functional mediators in the extracellular environment. They are small vesicles known to carry a concentrated group of functional cargo and serve as intercellular communicators not only locally but also systemically. Increasingly, it is reported that extracellular vesicles facilitate the development of drug resistance in cancer; however, their role in BRAF inhibitor resistance in melanoma is unclear. Here we investigated if extracellular vesicles from BRAF inhibitor–resistant melanoma could influence drug sensitivity in recipient melanoma cells. We demonstrate that the resistance driver, PDGFRβ, can be transferred to recipient melanoma cells via extracellular vesicles, resulting in a dose-dependent activation of PI3K/AKT signaling and escape from MAPK pathway BRAF inhibition. These data suggest that the BRAF inhibitor–sensitive phenotype of metastatic melanoma can be altered by delivery of PDGFRβ by extracellular vesicles derived from neighboring drug-resistant melanoma cells.
Introduction
BRAF inhibitors (BRAFis) have contributed to a significant improvement in survival rate for melanoma patients harboring tumors with V600 activating mutation in the BRAF oncogene [1], [2], [3]. BRAF is a component of the mitogen-activated protein kinase (MAPK) pathway involved in cell differentiation and survival. Around 40% to 60% of cutaneous melanomas express somatic mutations in BRAF, resulting in constitutive activation of the MAPK pathway and cell proliferation [4].
Treatment of BRAF mutant melanoma with BRAF kinase inhibitors, such as vemurafenib and dabrafenib, has been associated with rapid tumor regression in many patients; unfortunately, clinical benefit is short-lived, and most patients relapse within 6 to 9 months [5], [6]. Multiple mechanisms of resistance have been described, including activation of NRAS, KRAS, and MEK; amplification of the BRAF gene; alternative splicing of BRAF; upregulation of CRAF and COT (MAP3K8), an ERK upstream component; or upregulation of receptor tyrosine kinases, such as EGFR and PDGFRβ and PDGFRα, which induce activation of the phosphatidylinositol-3-OH kinase (PI3K)–AKT signaling pathway, bypassing BRAF inhibition of the MAPK pathway (for a review, see [7]).
A number of recent studies on BRAFi resistance suggest a role for the tumor microenvironment in mediating escape from BRAF inhibition [8], [9], [10], with the stromal secretome and recombinant RTK ligands capable of rescuing melanoma cells from BRAFi [8], [9], [11]. Extracellular membrane vesicles, namely, exosomes, have become recognized as important in cellular communication [12] and tumor microenvironment regulation [13]. Unlike soluble secreted factors, extracellular vesicles (EVs) carry a concentrated group of functional cargo, provide protection to the transported molecules, and serve as intercellular communicators not only locally but also systemically.
Here we evaluated whether EVs released from patient-derived melanoma cells could influence drug sensitivity in recipient cells. We found that EVs can mediate resistance in melanoma cells that are susceptible to BRAF inhibition by transfer of the RTK PDGFRβ. Together, our results support the hypothesis that EVs released from BRAFi-resistant cells could spread drug resistance by transferring protein cargo to susceptible cells at distant sites.
Materials and Methods
Reagents and Antibodies
Antibodies for ERK, pERK, AKT, pAKT, EGFR, α/β tubulin, PDGFRβ, ALIX, BAD, and calnexin were purchased from Cell Signaling Technologies (Danvers, MA) and flotillin-1 from BD Biosciences. The receptor tyrosine kinase antibody array and PDGFRβ neutralizing antibody were obtained from R&D systems. The BRAFi, PLX4720, was synthesized by Selleck Chemicals and solubilized in DMSO to a stock concentration of 1 M. Phospho-RTK arrays were performed according to the manufacturer's recommendations (Human Phospho-RTK Array Kit, R&D Systems).
Cell Culture
Melanoma cell lines were established from resected melanoma metastases by mechanical dissociation of tissue with subsequent overnight digestion in media containing collagenase IV at 37°C. The human melanoma cell line LM-MEL-64 expressing the V600E BRAF mutation has been described previously [14]. Melanoma cell lines and A431 (ATCC) were maintained at 37°C in a humidified atmosphere at 5% CO2 grown in RPMI 1640 supplemented with 10% fetal bovine serum, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 2 nmol/l glutamine (Life Technologies, USA). LM-MEL-64R3 was maintained in the above-mentioned medium and 1 μmol/L of PLX4720, except where otherwise indicated. Established cell lines were mycoplasma-tested using the MycoAlert test (Lonza Rockland, Inc., Rockland, ME). All tissue donors provided written informed consent for tissue collection and research, which was covered by protocols approved by the Austin Health Human Research Ethics Committee, Melbourne, Australia.
Generation of PLX4720-Resistant Cell Lines
LM-MEL-64 cells were plated, and after overnight incubation, the medium was removed and fresh medium was added together with PLX4720 at 1 μmol/l. Fresh medium containing drug was added to the cells every 3 days for 10 weeks to generate LM-MEL-64R3.
Proliferation Assays
Melanoma cells were plated out and treated as described. Cellular viability was assessed using the CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega Corporation, Madison, WI) according to the manufacturers' protocol.
Co-Culture Assays
To determine if EVs can rescue cells from BRAF inhibition 24 hours after plating, LM-MEL-64 (4 × 105 cells per well in a 6-well plate or 2.5 × 105 cells in a 12-well plate) was incubated with 50, 100, or 200 μg/ml of EVs derived from LM-MEL-64 or LM-MEL-64R3 for 1 hour followed by addition of 500 nm or 1 μM PLX4720 with and without PDGFR neutralizing antibody (50 μg/ml of EVs was used for the PDGFR neutralizing antibody experiments). After 15 minutes or 24 hours, cell signaling was examined by Western blot. To determine changes in proliferation, cells were incubated with an additional dose of EVs (50 μg/ml) 8 hours after the initial treatment, and cell viability was determined by MTS assay.
Isolation of Crude Exosomes (EVs) from Cell Culture Supernatants
Exosomes of bovine origin were removed from FCS prior to use in cell culture by overnight (18 hours, 4°C) ultracentrifugation at 100,000 ×g (45Ti rotor; Beckman Coulter Inc., USA). Cells (~2 × 107) were cultured for 3 to 4 days in this FCS-depleted media prior to exosome isolation by differential centrifugation. Cellular debris was removed by centrifugation at 2000 ×g for 10 minutes, and the supernatant was centrifuged at 10,000 ×g for 30 minutes at 4°C before crude exosomes were pelleted by ultracentrifugation at 120,000 ×g for 1 hour 30 minutes at 4°C. Exosomes were pooled, washed in PBS, repelleted, and resuspended in PBS.
Western Immunoblotting
Confluent cell cultures or exosomes were lysed in RIPA buffer (Pierce) on ice for 20 minutes, and protein concentration was determined for cell lysates and exosomes (prepared as described above) by BCA assay (Pierce). Equivalent amounts of cell or exosomes were electrophoresed on 4% to 12% Bis-Tris gels (NuPage; Invitrogen) and then transferred onto nitrocellulose membrane (iBlot, Life Technologies). Membranes were blocked in LiCor blocking buffer (LI-COR, Lincoln, NE), probed with primary antibody diluted in PBS-T overnight at 4°C, and then incubated with either IR labeled secondary antibodies (IRDye 800CW or IRDye 680LT, 1:20,000, LI-COR). Immunoblots were analyzed with the Odyssey Infrared imaging system (LI-COR).
Exosome Purification by Sucrose Density Gradient Centrifugation
Crude exosomes (20 μg) were resuspended in 1 ml 0.25 M sucrose and 20 mM HEPES pH 7.2, overlaid on top of 6-ml linear sucrose gradient (2.0-0.25 M sucrose, 20 mM HEPES pH 7.2), and ultracentrifuged (SW41 rotor) at 70,000 ×g for 16 hours at 4°C. The refractive index of each fraction before and after ultracentrifugation was measured using a Palm Abbe Digital Refractometer. Eight 1-ml gradient fractions were collected, diluted in 10 ml PBS, and ultracentrifuged for 1 hour at 200,000 ×g. Pellets were solubilized in sample buffer, electrophoresed, and immunoblotted as described above to determine enriched exosomal-containing fractions.
Electron Microscopy
An aliquot of 1 μg of exosomes in PBS was fixed with 1% glutaraldehyde for 30 minutes or O/N at 4°C, 6 μl was absorbed onto glow-discharged 300-mesh heavy-duty carbon-coated formvar Cu grids (ProSciTech, Kirwan, QLD, Australia) for 5 minutes, and excess was blotted on filter paper (Whatman, Maidstone, UK).Grids were washed twice with MilliQ water and negative stained with 2.5% uranyl acetate. Images were taken on a Tecnai G2F30 (FEI, Eindhoven, the Netherlands) transmission electron microscope operating at 300 kV (Bio21 Molecular Science and Biotechnology Institute, University of Melbourne).
qRT-PCR
RNA for qPCR was extracted using the RNeasy kit (Qiagen, Germany). Reverse transcription was carried out using the High Capacity cDNA RT kit (Applied Biosystems, Life Technologies, USA). Following reverse transcription, qRT-PCR was performed using SYBR Green (Qiagen, Germany). beta-Actin (ActB) was used as internal control. The following primers (Sigma-Aldrich) were used: ActB (forward) 5′-ctg gaa cgg tga agg tga ca-3′ and (reverse) 5′-cgg cca cat tgt gaa ctt tg-3′, and PDGFRβ (forward) 5′-TTCCATGCCGAGTAACAGAC-3′ and (reverse) 5′-CGTTGGTGATCATAGGGGAC-3′.
Statistical Analysis
The percentage difference was calculated relative to DMSO-treated control. *P ≤ .05, **P ≤ .01, and *P ≤ .001; repeated-measures ANOVA followed by Holm-Sidak's multiple-comparisons test was performed.
Results
Generation of a Melanoma Cell Line with Acquired Resistance to PLX4720
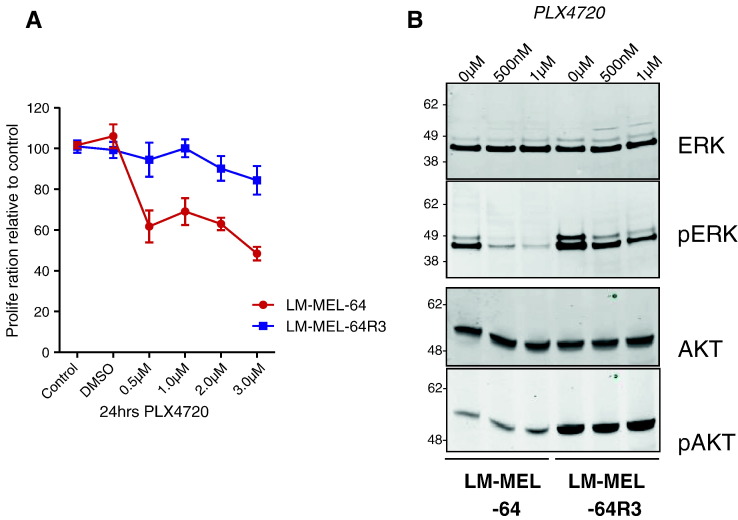
To investigate whether exosomes could mediate acquired resistance to BRAFis in melanoma, we first generated the BRAFi-resistant line LM-MEL-64R3 by culturing the BRAF V600E melanoma cell line LM-MEL-64 in 1 μmol/l of the BRAFi PLX4720 for 10 weeks. The resistant line was less sensitive to the growth-inhibitory effects of PLX4720 than the parental line as shown in a proliferation assay (Figure 1A). Because acquired BRAFi resistance can be mediated by reactivation of the MAPK pathway or by activation of the PI3K/AKT/AKT pathway, we evaluated the phosphorylation of the RAF downstream effector, ERK1/2, and P13K effector, AKT, in LM-MEL-64 versus LM-MEL-64R3 with and without PLX4720. Following 4-hour BRAF inhibition, p-ERK1/2 was reduced in LM-MEL-64 and partially reduced in LM-MEL-64R3 (Figure 1B), suggesting that reactivated MAPK signaling was not the sole survival pathway in these cells. Indeed, p-AKT was highly elevated in LM-MEL-64R3, suggesting that resistance was associated with activation of the PI3K/AKT/AKT pathway.
Figure 1.
Generation of a patient-derived melanoma cell line with acquired resistance to the BRAF kinase inhibitor PLX4720. (A) LM-MEL-64R3 is less sensitive to BRAF inhibition. Proliferation as measured by MTS assay of LM-MEL-64 and LM-MEL-64R3 cells in the absence or presence of increasing concentrations of the BRAFi PLX4720 (0, 500 nM, 1 μM, 2 μM, and 3 μM) for 24 hours. Data show the mean of three experiments performed in triplicate. (B) LM-MEL-64R3 displays differential MAPK reactivation. Western blot analysis for total and phosphorylated ERK and AKT in LM-MEL-64 and LM-MEL-64R3 in the absence or presence of 500 nM and 1 μM PLX4720 for 4 hours. The cell lines were treated with increasing PLX4720 concentrations (0, 500 nM, and 1 μM), and the effects on MAPK and PI3K signaling were determined by immunoblotting for total ERK and p-ERK1/2, and total AKT and p-AKT levels. Data show representative immunoblots of four independent experiments.
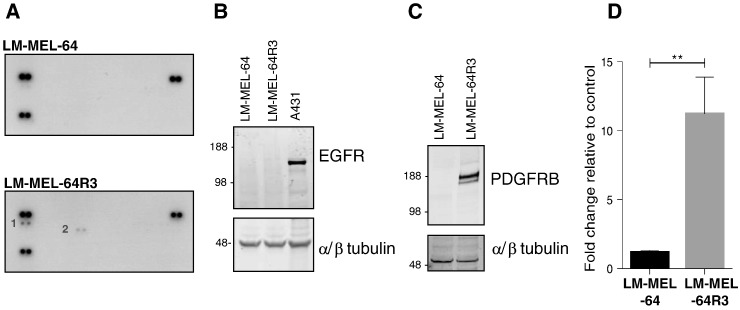
Adaptive resistance, mediated by PI3K/AKT/AKT pathway activation, can result from RTK signaling (for a review, see [7]), so we sought to identify RTKs hyperactivated in LM-MEL-64R3 using RTK phospho-antibody arrays. Consistent with previous studies [15], phosphorylation of two RTKs, EGFR and PDGFRβ, was greater in LM-MEL-64R3 relative to the parental cell line following short-term treatment with BRAFi (Figure 2A).
Figure 2.
Acquired resistance to PLX4720 is mediated by PDGFRβ. Hyperactivated RTK signaling in LM-MEL-64R3. (A) LM-MEL-64 or LM-MEL-64R3 whole-cell extracts (300 ng) were incubated on RTK antibody arrays, and phosphorylation status was determined by subsequent incubation with anti-phosphotyrosine horseradish peroxidase. Each RTK is spotted in duplicate; positive controls in corners. Gene identity is (1) p-Y-EGFR and (2) p-Y-PDGFRβ. (B) EGFR is not elevated in late-passage LM-MEL-64R3 relative to LM-MEL-64. Total protein levels of EGFR were determined by Western blotting LM-MEL-64 and LM-MEL-64R3 cell lysates; alpha/β tubulin was used as a loading control. The A431 cell line, which expresses high levels of EGFR, was used as a positive control. (C) PDGFRβ expression is elevated in LM-MEL-64R3. Total protein levels of PDGFRβ were determined by Western blotting LM-MEL-64 and LM-MEL-64R3 cell lysates; alpha/β tubulin was used as a loading control. (D) Relative RNA levels of PDGFRβ in LM-MEL-64 and LM-MEL-64R3 as determined by real-time, quantitative PCR (four independent experiments performed in triplicate) (normalized to beta-actin). Immunoblots are representative of three independent experiments.
Of these two candidate RTKs, only PDGFRβ RNA and protein levels were consistently overexpressed in long-term–treated LM-MEL-64R3 cells (Figure 2, B and C). Short-term exposure of melanoma cells to BRAFi is known to initially drive hyperphosphorylation of EGFR [16]. However, after prolonged drug exposure, cells undergo adaptive changes which allow long-term stable resistant cells to survive [16].
This suggested PDGFRβ-mediated resistance to BRAF inhibition, and not EGFR (Figure 2B), in LM-MEL-64R3, consistent with previous studies [15], [17], [18]. No changes were detected in NRAS, KRAS, and MEK sequences, and the BRAF (V600E) mutation was present in both parental and resistant cell lines (data not shown).
LM-MEL-64 and LM-MEL-64R3 Release EVs
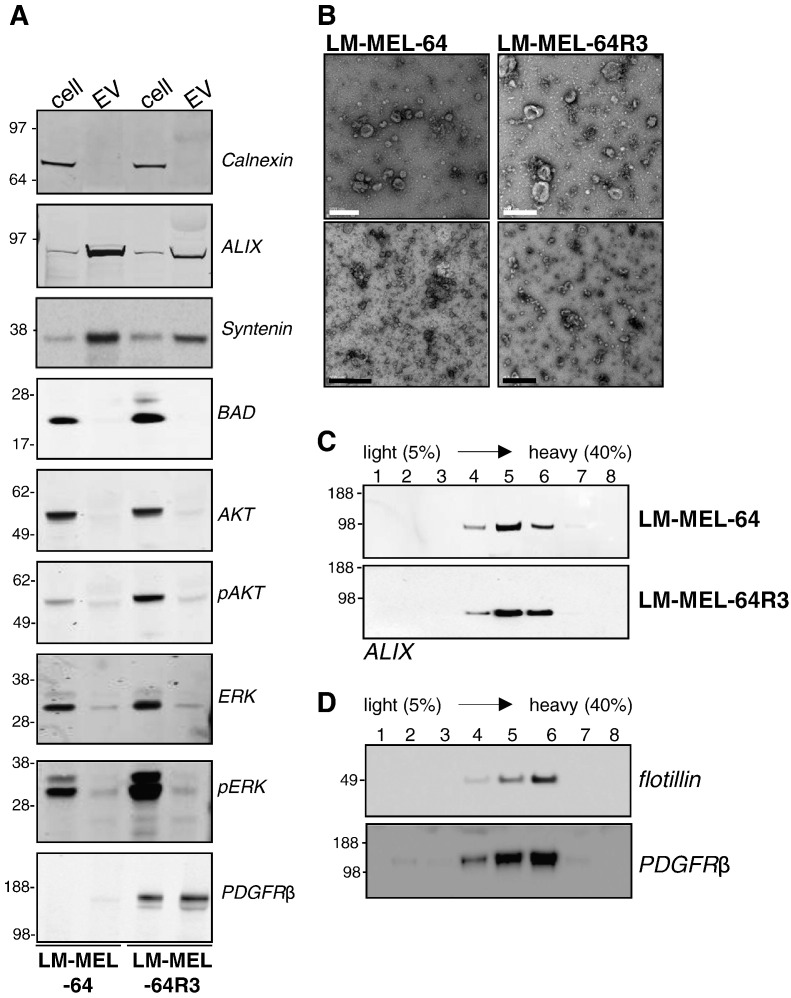
Because soluble mediators in the microenvironment can promote therapy resistance [8], [9], [10], [11], [19], [20], we hypothesized that EVs might aid in promoting escape from BRAF inhibition. To evaluate EV release from LM-MEL-64 cell lines, conditioned cell culture media were subjected to differential ultracentrifugation, and the resulting pellet was examined by Western blotting, electron microscopy, and density gradient as previously described [21] (Figure 3).
Figure 3.
Characterization of exosomes released from LM-MEL-64 cell lines. (A) Western blot analysis of equivalent total cell lysate and exosomal protein. A. whole cell lysates or exosomes were analyzed by immunoblotting using antibodies to; Calnexin is a resident ER protein not in exosomes, ALIX and Syntenin are involved in endosomal-lysosomal sorting and present in exosomes and BAD, a mitochondrial protein, not in exosomes. AKT, pAKT, ERK and pERK are cytosolic protein not enriched in exosomes. Exosomes derived from LM-MEL-64R3 overexpress PDGFRβ. (B) Electron micrograph of exosome preparations. Transmission electron micrographs of LM-MEL-64 exosomes and LM-MEL-64R3 exosomes. EM showed small vesicles with a morphology that is consistent for exosomes. White scale bar = 200 nm; black scale bar = 500 nM. (C) Sucrose density gradient analysis of exosomes. LM-MEL-64 and LM-MEL-64R3 exosome preparations were loaded on top of a 0.25- to 2.5-M sucrose gradient and ultracentrifuged to equilibrium. The exosome marker, ALIX, was recovered in fractions 4, 5, and 6 corresponding to densities of 1.12 to 1.18 g/ml. (D) Sucrose density gradient analysis of exosomes. LM-MEL-64R3 exosome preparations were loaded on top of a 0.25- to 2.5-M sucrose gradient and ultracentrifuged to equilibrium. The exosome marker, flotillin, and PDGFR were recovered in fractions 4, 5, and 6 corresponding to densities of 1.12 to 1.18 g/ml. Immunoblots are representative of at least three independent experiments.
Exosomes from both drug-sensitive and -resistant cell lines were found to contain the established exosomal marker proteins ALIX, Syntenin, and flotillin-1 and were negative for calnexin and BAD. Neither was enriched for the cytosolic signaling proteins ERK, pERK, AKT, and pAKT, confirming that the preparations were not heavily contaminated with cellular debris (Figure 3A). The purified vesicles were examined by negative staining transmission electron microscopy which revealed that both preparations contained vesicles which were membrane bound, were “cup shaped,” and had a similar size (~50-150 nm diameter) to previously described exosomes (Figure 3B) [22]. Exosomes float in sucrose gradients with a density ranging from 1.13 g/ml to 1.19 g/ml depending on the cell type [23]. Immunoblotting of sucrose gradient fractions showed that LM-MEL vesicles migrated to a density of 1.12 to 1.18 g/ml with a concomitant enrichment in ALIX (Figure 3C). Thus, the EVs fulfill the established criteria for exosomes [22].
Exosomes Derived from LM-MEL-64R Are Enriched for PDGFRβ
Exosomes contain a common set of molecules that reflect their endosomal origin, as well as cell type–specific components, such as overexpressed proteins. Because LM-MEL64R demonstrated increased PDGFRβ expression, exosomes derived from both the sensitive and resistant lines were examined for PDGFRβ protein by Western blotting (Figure 3A).
To confirm that exosomes released from LM-MEL-64R3 contained PDGFRβ, exosomes derived from LM-MEL-64R3 were floated into a sucrose gradient and probed for PDGFRβ and the exosome marker flotillin (Figure 3D). PDGFRβ was detected at the same density as exosomes, indicating that PDGFRβ was likely to be exosome associated. These results are analogous to previous findings in the commonly used BRAF V600E mutant M229 melanoma cell line and its PLX4032-resistant counterpart, M229AR [15]. Similar to LM-MEL-64R3, the M229AR cell line acquired BRAFi resistance by upregulating PDGFRβ. Exosomes released from the resistant cell line, M229AR, contained PDGFRβ, whereas expression was not detected in exosomes released from the parental cell line [24]. Together, these studies demonstrate that PDGFRβ-driven BRAFi-resistant cells secrete exosomes carrying the receptor and represent a unique oncogenic PDGFRβ protein delivery system.
Exosomes Derived from BRAFi Resistance Cells Rescue Growth Inhibition
Melanomas are heterogeneous tumors composed of subpopulations of melanoma cells with distinct phenotypes that can arise because of, or independently from, genetic mutations [25]. This has implications for the emergence of resistance during treatment because subpopulations are differentially sensitive to drugs [7].
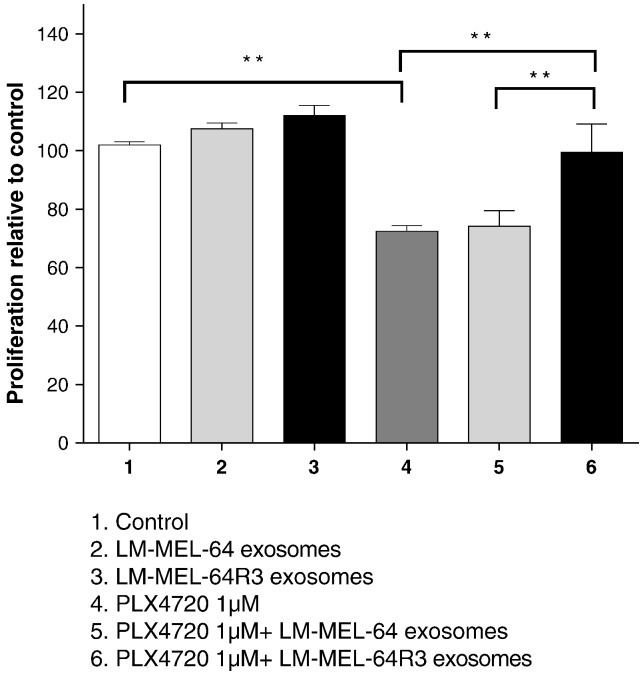
We assessed whether exosomes could transfer PDGFRβ from drug-resistant to -sensitive cell lines, thereby enabling escape from growth inhibition. The LM-MEL-64 BRAFi-sensitive cell was treated with PLX4720 in the presence of exosomes derived from both sensitive and resistant lines. PLX4720 suppressed growth of LM-MEL-64 cell line compared with DMSO-treated control. This effect was completely abrogated by exosomes derived from LM-MEL-64R3 (Figure 4).
Figure 4.
Exosomes derived from BRAFiR cells rescue cell from BRAF inhibition. The LM-MEL-64 cell line was incubated with LM-MEL-64 exosomes or exosomes derived from LM-MEL-64R3 in the presence or absence of PLX4720 (500 nM). Proliferation of LM-MEL-64 was measured after 24 hours by MTS assay. Percentage survival was quantified from absorbance at 490 nm. Bars are mean values ± S.D. values of three independent experiments. **P ≤ .01 and ***P ≤ .001; repeated-measures ANOVA followed by Holm-Sidak's multiple-comparisons test.
Exosomes Derived from Drug-Resistant Cells Rescue Recipient Cells from BRAF Inhibition
Cancer cell proliferation typically results from signaling via the P13K/AKT or MAPK/ERK pathways, and exosomes have been shown to promote proliferation by directly activating these pathways in recipient cells [26].
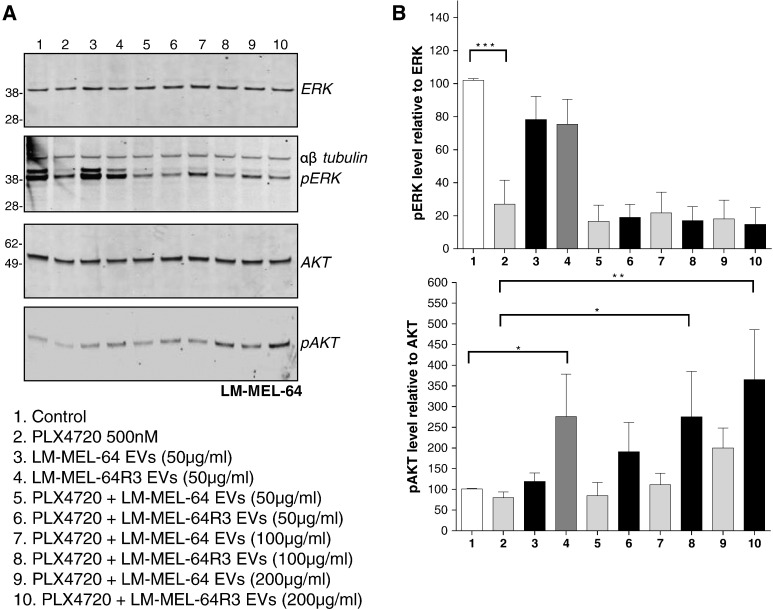
To determine which signaling pathway was activated by LM-MEL-64R3 exosomes in BRAF-inhibited LM-MEL-64 cells, exosomes were incubated with LM-MEL-64 in the presence of PLX4720, and pERK and pAKT levels were determined by Western blotting. Drug-suppressed ERK phosphorylation in LM-MEL-64 and the addition of exosomes did not affect this, whether from LM-MEL64 or from LM-MEL-64R3. In contrast, the addition of LM-MEL-64R3 exosomes to drug-inhibited cells resulted in a dose-dependent increase in pAKT, indicating that exosome activation of PI3K/AKT signaling was the likely mechanism mediating escape from MAPK pathway inhibition (Figure 5).
Figure 5.
Exosomes derived from BRAFiR cells rescue recipient cells from BRAF inhibition by activation of PI3K signaling pathways. (A) Western blot analysis of LM-MEL-64 cell lysate for total and phosphorylated ERK and AKT. LM-MEL-64 was incubated with or without 500 nM PLX4720 (lanes 1 and 2) and increasing doses of exosomes for 15 minutes. Exosomes derived from the BRAFi-resistant cell line, LM-MEL-64R3, enhanced pAKT signaling in BRAF-inhibited LM-MEL-64 cells, indicating that exosomes derived from resistance cell lines are capable of activating alternate cell signaling pathways in BRAFi cells. Images are representative of three independent experiments. (B) Densitometric analysis. Expressed as mean ± S.E.M. values of three independent experiments. The relative level of pERK relative to total ERK or pAKT relative AKT are shown. The percentage difference was calculated relative to DMSO-treated control. *P ≤ .05, **P ≤ .01; repeated-measures ANOVA followed by Holm-Sidak's multiple-comparisons test.
Resistance Drivers Are Transferred via Exosomes and Activate PI3K/AKT Signaling in Recipient Cells
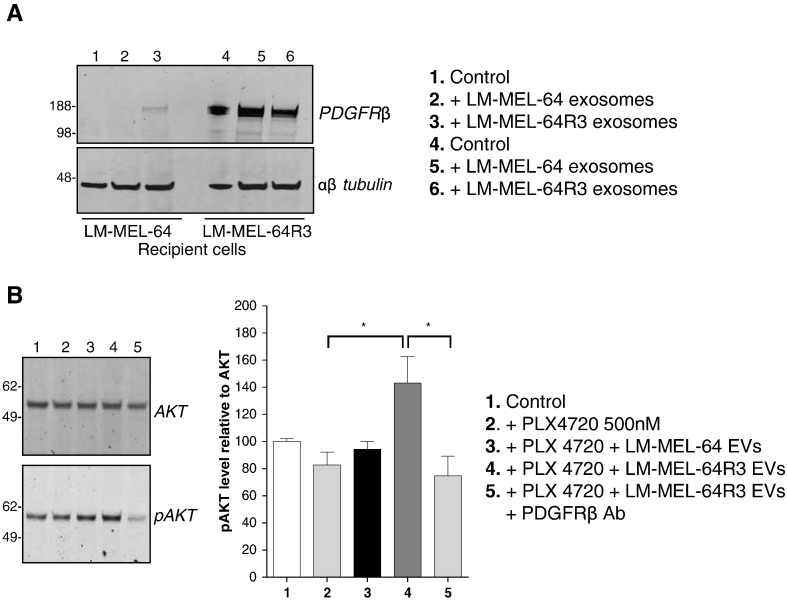
To assess whether exosomal PDGFRβ derived from LM-MEL-64R3 cells could be transferred to recipient cells, LM-MEL-64 was incubated with preparations of exosomes derived from LM-MEL-64 or LM-MEL64R3 for 15 minutes and then extensively washed and then lysed, and PDGFRβ was detected by Western blotting. Figure 6A shows that PDGFRβ was detected in the recipient cell LM-MEL-64 cell lysate. To determine whether incorporation of PDGFRβ resulted in AKT phosphorylation, LM-MEL-64 was treated with PLX4720 and LM-MEL-64R3 exosomes in the presence of a neutralizing PDGFRβ antibody. By blocking PDGFRβ signaling, exosome-mediated activation of the PI3K/AKT pathway in LM-MEL-64R3 was markedly reduced (Figure 6B). These results indicate that exosomes released from BRAFi-resistant cells transfer PDGFRβ and activate PI3K/AKT pathway signaling in recipient cells. This likely accounts for the escape from BRAF inhibition seen in these cells.
Figure 6.
Resistance drivers are transferred via exosomes and activate PI3K signaling in recipient cells. (A) Exosomal PDGFRβ is transferred to recipient LM-MEL-64 cells. LM-MEL-64 and LM-MEL-64R3 were incubated in the presence or absence of exosomes for 15 minutes, washed 5 times, and then analyzed for expression of PDGFRβ by Western blot. Image is representative of three independent experiments. (B) Exosomal PDGFRβ activates PI3K signaling in BRAF-inhibited cells. LM-MEL-64 cells were incubated with PLX4720 (500 nM) in the presence of exosomes derived from LM-MEL64 or LM-MEL-64R3, or LM-MEL-64R3 in addition to a PDGFRβ neutralizing antibody for 15 minutes. Images are representative of three independent experiments. Densitometric analysis is expressed as mean ± S.D. values of three independent experiments. The relative level of pAKT relative to total AKT is shown. The percentage difference was calculated relative to DMSO-treated control. *P ≤ .05; repeated-measures ANOVA followed by Holm-Sidak's multiple-comparisons test.
The transfer of resistance via exosomal PDGFRβ is anticipated to occur in other melanomas and not be restricted to LM-MEL-64. The well-established cell line M229AR, with acquired BRAFi resistance via PDGFRβ upregulation, also releases exosomes enriched in PDGFRβ, suggesting that a common mechanism exists for PDGFRβ-driven resistant melanoma [24]. Thus, the transfer of resistance via exosomal PDGFRβ may occur in other melanomas, and further research is warranted to determine the relevance of this finding in vivo.
Discussion
Clinical benefit from BRAFis in metastatic melanoma is often short-lived due to the development of drug resistance. This resistance can involve mechanisms that are intrinsic to the cancer cell or that involve extracellular factors [11]. Here we report that EVs, namely exosomes, can contribute to the development of BRAFi resistance.
Soluble mediators in the microenvironment, first shown by Straussman et al. and Wilson et al. in 2012 [8], [9], can support cancer growth and resistance to BRAFi therapy. Secreted hepatocyte growth factor (HGF) was shown to activate the HGF receptor MET, reactivating the MAPK and PI3K/AKT-AKT signaling pathways and bypassing BRAF inhibition [8], [9]. These and other studies laid the foundations for the current work by showing that paracrine interactions promote resistance to BRAF blockade [10], [11], [19], [27], [28], [29].
We now demonstrate that, in addition to soluble mediators in the extracellular environment, exosomes can also modulate BRAFi sensitivity. Parental LM-MEL-64 cells pretreated with exosomes derived from the resistant cell line had significantly increased cell viability which was associated with an increase in PI3K/AKT signaling. Further investigation revealed that this phenomenon was a result of exosomal PDGFRβ transfer between cells. Of note, this effect could be prevented with PDGFRβ neutralizing antibodies This suggests that exosomal transfer of PDGFRβ could be a potential mechanism for the transfer of resistance between subpopulations of cells in BRAF mutant melanoma. [25].
All cancer cells release exosomes [30]. Functions that have been attributed to melanoma exosomes include stimulation of endothelial signaling [31], “preparation” of lymph nodes and bone marrow cells to create a niche for metastasis [32], [33], and acceleration of lung metastasis [34]. Exosomes facilitate resistance to therapeutics in many cancers including breast, prostate, glioblastoma, multiple myeloma, lung, and ovarian cancer (for review, see [35]). We now propose that resistance to BRAFi can be added to this list.
Exosomes from different cellular origins contain both common (shared) and cell type–specific components. The former includes proteins that reflect the endosomal origin of exosomes; and the latter, cancer cell–specific molecules such as RTKs, oncoproteins, phosphorylated proteins, and miRNA species [26]. Consistent with this observation, we found high expression of PDGFRβ in LM-MEL-64R3 which was reflected in the exosomes released by this cell line.
Following release from a cell, exosomes can transfer receptors with oncogenic activity to recipient cells, resulting in signal pathway activation. Cancer cell–derived exosomes have been shown to carry growth factor receptors such as EGFR, EGFRVIII [36], HGFR (MET) [33], and cKIT (normal cellular homolog of the viral oncoprotein v-Kit) [37]. EGFRvIII has been shown to be “shared” between glioma cells by intercellular transfer of exosomes and merging with the plasma membranes of cancer cells lacking EGFRvIII [38]. This event leads to the transfer of oncogenic activity, including activation of MAPK signaling pathways [38]. In this study, we now show that another RTK, PDGFRβ, can be trafficked via exosomes.
Exosomes were first implicated in drug resistance in cancer following the discovery that they could mediate chemotherapeutic drug expulsion from the cell (for a review, see [35]). Many anticancer drugs, including cisplatin and doxorubin, can be encapsulated and transported out of cells via exosomes, resulting in a chemoresistant tumor. Other exosome-mediated mechanisms of resistance have recently come to light; these include the exchange of cellular material such as stroma-derived RNA driving chemotherapy and radiation resistance in breast cancer [39] and bone marrow stromal cell–induced drug resistance to bortezomib in multiple myeloma cells [40]. However, the exosome isolation method used in this study and many others (ultracentrifugation) isolates a heterogenous population of exosomes and other small EVs [41], [42], so further dissection of exosome heterogeneity is necessary to determine the role of each vesicle subpopulations. Although PDGFRβ appears to be the predominant driver of exosome-mediated resistance in this model, we cannot rule out a role for other exosomal cargo such as protein, lipid, or small RNA, and further studies will need to be performed to determine the relative contribution of these factors.
Detecting preexisting intrinsic BRAFi resistance and monitoring the development of resistance following treatment are critical clinical challenges. To guide clinical management and to minimize clinical deterioration and distress, it would be extremely valuable to both detect emerging drug resistance and identify underlying resistance mechanisms early, before treatment failure becomes clinically apparent. Because exosomes are actively secreted, extracellular sampling of exosomes could provide insights into preexisting or developing resistance.
The presence of tumor-derived exosomes in the circulation and stability in blood provides a unique opportunity to develop a blood test to interrogate intratumoral biology. Thus, exosomes may be biomarkers for cancer behavior, making it possible to monitor the evolution of the tumor's biological characteristics ahead of clinical failure, thereby facilitating prompt clinical decision making.
In conclusion, we have shown in a cell model that exosomes facilitate the escape of melanoma cells from BRAF inhibition. In light of the complexities of the microenvironment and the difficulties associated with studying exosomal trafficking, it is difficult to unravel the importance of exosome-mediated BRAF resistance in vivo. Nonetheless, the recognition that exosomes can traffic widely throughout the body as stable entities has substantive implications. Indeed, the possibility that drug resistance might be transferred in an infectious manner between sites of metastatic disease warrants investigation. A greater understanding of such mechanisms may prove critical to understanding, countering, and detecting exosome-associated drug resistance.
Disclosures
The authors have declared no conflicts of interest.
Footnotes
Funding: This work was funded in part by Ludwig Cancer Research and by Operational Infrastructure Support Program funding of the Victorian State Government.
References
- 1.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, McArthur GA, Hutson TE, Moschos SJ, Flaherty KT. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parakh S, Murphy C, Lau D, Cebon JS, Andrews MC. Response to MAPK pathway inhibitors in BRAF V600M-mutated metastatic melanoma. J Clin Pharm Ther. 2015;40:121–123. doi: 10.1111/jcpt.12229. [DOI] [PubMed] [Google Scholar]
- 4.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 5.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–599. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flaherty KT, McArthur G. BRAF, a target in melanoma: implications for solid tumor drug development. Cancer. 2010;116:4902–4913. doi: 10.1002/cncr.25261. [DOI] [PubMed] [Google Scholar]
- 7.Welsh SJ, Rizos H, Scolyer RA, Long GV. Resistance to combination BRAF and MEK inhibition in metastatic melanoma: Where to next? Eur J Cancer. 2016;62:76–85. doi: 10.1016/j.ejca.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, Peng J, Lin E, Wang Y, Sosman J. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seifert H, Hirata E, Gore M, Khabra K, Messiou C, Larkin J, Sahai E. Extrinsic factors can mediate resistance to BRAF inhibition in central nervous system melanoma metastases. Pigment Cell Melanoma Res. 2016;29:92–100. doi: 10.1111/pcmr.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fedorenko IV, Wargo JA, Flaherty KT, Messina JL, Smalley KS. BRAF Inhibition Generates a Host-Tumor Niche that Mediates Therapeutic Escape. J Invest Dermatol. 2015;135:3115–3124. doi: 10.1038/jid.2015.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tkach M, Thery C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 13.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behren A, Anaka M, Lo PH, Vella LJ, Davis ID, Catimel J, Cardwell T, Gedye C, Hudson C, Stan R. The Ludwig institute for cancer research Melbourne melanoma cell line panel. Pigment Cell Melanoma Res. 2013;26:597–600. doi: 10.1111/pcmr.12097. [DOI] [PubMed] [Google Scholar]
- 15.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fattore L, Marra E, Pisanu ME, Noto A, de Vitis C, Belleudi F, Aurisicchio L, Mancini R, Torrisi MR, Ascierto PA. Activation of an early feedback survival loop involving phospho-ErbB3 is a general response of melanoma cells to RAF/MEK inhibition and is abrogated by anti-ErbB3 antibodies. J Transl Med. 2013;11:180. doi: 10.1186/1479-5876-11-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rebecca VW, Wood E, Fedorenko IV, Paraiso KH, Haarberg HE, Chen Y, Xiang Y, Sarnaik A, Gibney GT, Sondak VK. Evaluating melanoma drug response and therapeutic escape with quantitative proteomics. Mol Cell Proteomics. 2014;13:1844–1854. doi: 10.1074/mcp.M113.037424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi H, Kong X, Ribas A, Lo RS. Combinatorial treatments that overcome PDGFRbeta-driven resistance of melanoma cells to V600EB-RAF inhibition. Cancer Res. 2011;71:5067–5074. doi: 10.1158/0008-5472.CAN-11-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Filitis DC, Rauh J, Mahalingam M. The HGF-cMET signaling pathway in conferring stromal-induced BRAF-inhibitor resistance in melanoma. Melanoma Res. 2015;25:470–478. doi: 10.1097/CMR.0000000000000194. [DOI] [PubMed] [Google Scholar]
- 20.Kemper K, Krijgsman O, Cornelissen-Steijger P, Shahrabi A, Weeber F, Song JY, Kuilman T, Vis DJ, Wessels LF, Voest EE. Intra- and inter-tumor heterogeneity in a vemurafenib-resistant melanoma patient and derived xenografts. EMBO Mol Med. 2015;7:1104–1118. doi: 10.15252/emmm.201404914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vella LJ, Sharples RA, Lawson VA, Masters CL, Cappai R, Hill AF. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J Pathol. 2007;211:582–590. doi: 10.1002/path.2145. [DOI] [PubMed] [Google Scholar]
- 22.Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 24.Goedert L, Koya R, Hu-Lieskovan S, Ribas A. Exosomes as a predictor tool of acquired resistance to melanoma treatment. BMC Proc. 2014;8:1–2. [Google Scholar]
- 25.Anaka M, Hudson C, Lo PH, Do H, Caballero OL, Davis ID, Dobrovic A, Cebon J, Behren A. Intratumoral genetic heterogeneity in metastatic melanoma is accompanied by variation in malignant behaviors. BMC Med Genet. 2013;6:40. doi: 10.1186/1755-8794-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meehan K, Vella LJ. The contribution of tumour-derived exosomes to the hallmarks of cancer. Crit Rev Clin Lab Sci. 2016;53:121–131. doi: 10.3109/10408363.2015.1092496. [DOI] [PubMed] [Google Scholar]
- 27.Wang T, Xiao M, Ge Y, Krepler C, Belser E, Lopez-Coral A, Xu X, Zhang G, Azuma R, Liu Q. BRAF Inhibition Stimulates Melanoma-Associated Macrophages to Drive Tumor Growth. Clin Cancer Res. 2015;21:1652–1664. doi: 10.1158/1078-0432.CCR-14-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirata E, Girotti MR, Viros A, Hooper S, Spencer-Dene B, Matsuda M, Larkin J, Marais R, Sahai E. Intravital Imaging Reveals How BRAF Inhibition Generates Drug-Tolerant Microenvironments with High Integrin beta1/FAK Signaling. Cancer Cell. 2015;27:574–588. doi: 10.1016/j.ccell.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menon DR, Schaider H. Microenvironment-Driven Resistance to BRAF Inhibition Comes of Age. J Invest Dermatol. 2015;135:2923–2925. doi: 10.1038/jid.2015.373. [DOI] [PubMed] [Google Scholar]
- 30.Zhang M, Schekman R. Cell biology. Unconventional secretion, unconventional solutions. Science. 2013;340:559–561. doi: 10.1126/science.1234740. [DOI] [PubMed] [Google Scholar]
- 31.Hood JL, Pan H, Lanza GM, Wickline SA. Paracrine induction of endothelium by tumor exosomes. Lab Investig. 2009;89:1317–1328. doi: 10.1038/labinvest.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011;71:3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- 33.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar C. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hao S, Ye Z, Li F, Meng Q, Qureshi M, Yang J, Xiang J. Epigenetic transfer of metastatic activity by uptake of highly metastatic B16 melanoma cell-released exosomes. Exp Oncol. 2006;28:126–131. [PubMed] [Google Scholar]
- 35.Zhao L, Liu W, Xiao J, Cao B. The role of exosomes and "exosomal shuttle microRNA" in tumorigenesis and drug resistance. Cancer Lett. 2015;356:339–346. doi: 10.1016/j.canlet.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 36.Al-Nedawi K, Meehan B, Kerbel RS, Allison AC, Rak J. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc Natl Acad Sci U S A. 2009;106:3794–3799. doi: 10.1073/pnas.0804543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atay S, Banskota S, Crow J, Sethi G, Rink L, Godwin AK. Oncogenic KIT-containing exosomes increase gastrointestinal stromal tumor cell invasion. Proc Natl Acad Sci U S A. 2014;111:711–716. doi: 10.1073/pnas.1310501111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 39.Boelens MC, Wu TJ, Nabet BY, Xu B, Qiu Y, Yoon T, Azzam DJ, Twyman-Saint Victor C, Wiemann BZ, Ishwaran H. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell. 2014;159:499–513. doi: 10.1016/j.cell.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang J, Hendrix A, Hernot S, Lemaire M, De Bruyne E, Van Valckenborgh E, Lahoutte T, De Wever O, Vanderkerken K, Menu E. Bone marrow stromal cell-derived exosomes as communicators in drug resistance in multiple myeloma cells. Blood. 2014;124:555–566. doi: 10.1182/blood-2014-03-562439. [DOI] [PubMed] [Google Scholar]
- 41.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Thery C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113:968–977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willms E, Johansson HJ, Mager I, Lee Y, Blomberg KE, Sadik M, Alaarg A, Smith CI, Lehtio J, El Andaloussi S. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci Rep. 2016;6 doi: 10.1038/srep22519. [2-12] [DOI] [PMC free article] [PubMed] [Google Scholar]