Abstract
MicroRNAs (miRNAs) are 22 nucleotide long RNA transcripts, their synthesis starts in nucleus and continues in cytoplasm. As being critical for post-transcriptional regulators of gene expression they have been investigated in psychiatric disorders. There are numerous studies performed in peripheral tissues for psychiatric disorders. Here in this article, we aimed to review some common miRNAs denoted significant in at least two studies and their relevance to psychiatric research. We focused on miR-320, miR-106, miR-34, miR-223, miR-107, and miR-134.
Keywords: MicroRNA, Psychiatry, Blood, Plasma, Serum, Biomarker
INTRODUCTION
Diagnosis of psychiatric disorders depends on clinical observations and the subjective experiences of patients. Diagnostic and Statistical Manual of Mental Disorders (DSM) and International Classification of Diseases (ICD) are the most common classification systems for diagnosis. On the other hand, same clinical manifestations may be defined as a different disorder in different classification systems. Also, classification systems may give different names to same clinical manifestations over the years. In addition, the neurobiology of any psychiatric disorders has not yet been determined exactly. In this context, many biomarker studies have been made in recent years to determine both diagnosis and prognosis of psychiatric disorders. Aims of these studies are making diagnosis, determining prognosis, predicting treatment response, evaluating mental and emotional capacity.1)
Many studies have been conducted in peripheral tissues so far. In these studies, oxidative stress parameters, anti-oxidant enzyme levels, interleukins and brain derived neurotrophic factor (BDNF) have been studied.2–5) However, a certain biomarker has not been determined yet.
MAIN SUBJECTS
MicroRNA (miRNA) Systhesis
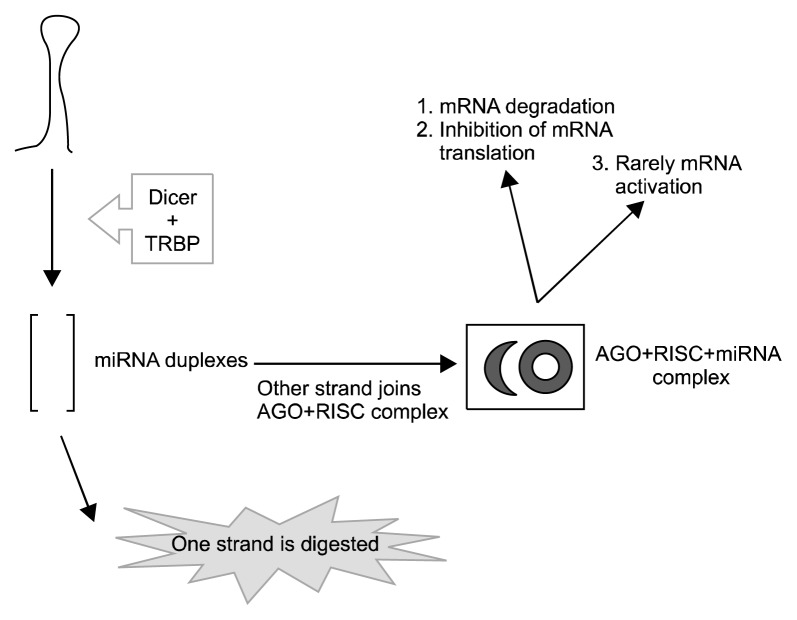
The synthesis of miRNAs starts in the nucleus. RNA polymerase II synthesizes 100–1,000 nucleotide long primiRNAs in the nucleus, these primiRNAs contain 5′ cap and 3′ polyA tail.6) Drosha (which is a nuclear RNase III), its co-factor DiGeorge syndrome critical region protein 8 and several enzymes remove 5′ cap and 3′ polyA tail from primiRNAs, and form premiRNAs in the nucleus.7) The transport of premiRNAs is performed by exportin 5/RanGTP.8) The maturation of miRNAs continues in the cytoplasm. Dicer and its co-factor trans-activator RNA binding protein (TRBP) synthesize mature miRNAs (miRNA duplex) consisted of 20 bp (Dicer clevage).9) Mature miRNAs incorporate to RNA-induced silencing complex (RISC) for gene regulation.10) miRNAs usually point mRNAs to degrade or prevents them from translation.11) With this mechanism increase of miRNAs usually results with decreased expression of target gene. In rare cases miRNAs may increase gene expression. In this way, miRNAs are thought to be capable of affecting the expressions of over 10,000 genes (Figs. 1, 2).12)
Fig. 1.
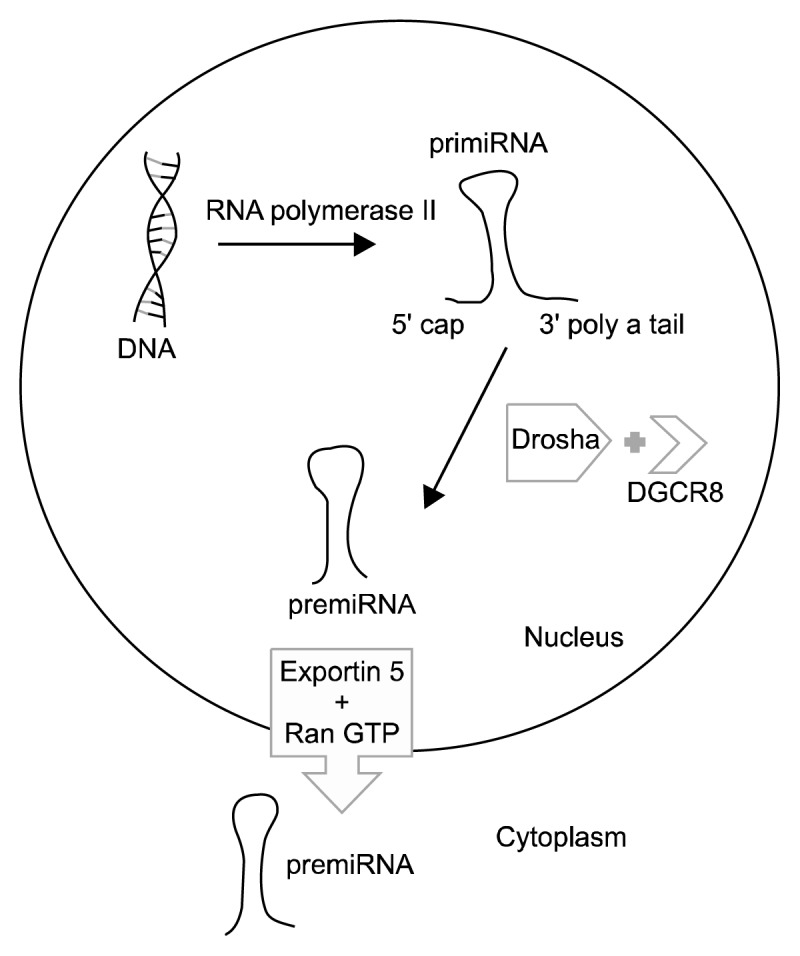
The synthesis of microRNAs (miRNAs) starts in the nucleus. RNA polymerase II synthesizes primiRNAs with 5′ cap and 3′ polyA tail. Drosha and its co-factor DiGeorge syndrome critical region protein 8 (DGCR8) remove 5′ cap and 3′ polyA tail and form premiRNAs. PremiRNAs are transported by exportin 5/Ran GTP system.
Fig. 2.
The maturation of microRNAs (miRNAs) in the cytoplasm. Dicer and its co-factor trans-activator RNA binding protein (TRBP) perform cleavage. Mature miRNAs incorporate to RNA-induced silencing complex (RISC). miRNAs usually point mRNAs 1) to degrade or 2) prevents them from translation. In rare cases miRNAs may 3) increase gene expression.
Role of miRNAs in Neural Development and Maintenance
miRNAs take part in almost every biological function in our body, especialy in brain. A zebrafish study pointed out mature miRNA injection is beneficial for correcting improper brain development.13) Furthermore, miR-133b is essential for proper development of dopaminergic neurons.14) Transform of stem cells into mature neurons require repression of Sox9 by miR-124 and miR-137 found to be important in development of neural stem cells into mature neurons.15,16) miRNAs are important for not only neurodevelopment but also for maintenance of functioning of neurons. Missing of Dicer was associated with death of neurons.17) The RISC pathway is crucial for synaptic plasticity.18) Increased expression miR-132—which is related to BDNF—has facilitating effects on migration of neurons and dendritic maturation.19,20) For detailed information about the synthesis and functions of miRNAs, the importance of miRNAs for synaptic plasticity and neurogenesis please check reviews by Dwivedi21) and O’Connor et al.22)
miRNAs and Treatment
Limited data exist about therapeutic potential of miRNAs in various diseases. Current focus depends on antagomir-based knock down.23) Although miRNA based treatment approach found beneficial for viral infections there are several challenges in transportation of miRNAs (or nucleic acids) to central nervous system. An oral taken drug should pass through gastrointestinal system without deterioration, cross the blood-brain barrier, reach to target region of the brain without causing a toxic effect. The blood brain barrier is consisted of tight junctions and it is somehow impossible for nucleic acids to cross.24,25) On the other hand, brain itself contains many kind of cells and it has a heterogeneous nature.26) To overcome this issue viral or non-viral vectors may be beneficial with specific advantages and disadvantages. Viral vectors integrates host chromosome well but safety is a major issue.27) In spite of being more safe, non-viral vectors don’t have the ability to cross many barriers to reach out the brain.28)
miRNAs are more stable than mRNAs and their stability makes them as potential diagnostic markers for diseases. Although source of circulating miRNAs isn’t clear, they are considered to be functioning in the communication between cells. This feature of miRNAs increase their likelihood as a biomarker.29) In this review, we are going to discuss some miRNAs and their importance that have been found significant in psychiatric disorders and in peripheral tissue studies (Table 1). We used Google Scholar and PubMed databases to find relevant articles for our review. Main criterion for selection of miRNAs is being significantly dysregulated (up or down) in at least two studies performed in peripheral tissues.
Table 1.
Importance of microRNAs subject to this review
| MicroRNA | Psychiatric disorder | Importance |
|---|---|---|
| miR-320 | Major depression | miR-320 family may be important as a peripheral biomarker |
| Autism | miR-320 was dysregulated in lymphoblastoid cell lines of autism patients | |
| miR-106 | Major depression | miR-106a-5p levels was higher in depressed patients |
| Schizophrenia | miR-106b-5p levels was higher in schizophrenia patients | |
| Attention deficit hyperactivity disorder | Lower miR-106b-5p levels were determined in the control group | |
| Autism | miR-106-5p showed higher expression in autism | |
| miR-34 | Major depression | miR-34b-5p and miR-34c-5p levels in peripheral blood leukocytes were closely related to suicide idea and cognitive function |
| Schizophrenia | miR-34 were presented as peripheral biomarkers | |
| Bipolar disorder | miR34-a may be critical for bipolar disorder in terms of neurodevelopment | |
| Alzheimer’s disease | miR-34a was observed to be decreased in plasma and cerebrospinal fluid according to the control group | |
| miR-223 | Major depression | miR-223-3p levels were reported to be significantly higher in patients than in controls |
| Posttraumatic stress disorder | miR-223 could be a potential biomarker | |
| miR-107 | Amnestic mild cognitive impairment | miR107 expressions in plasma had a high capability to distinguish individuals with amnestic mild cognitive impairment from healthy controls |
| Major depression | Positive correlation between N1, P2latency of P300 and miR-107 in depressed patients | |
| miR-134 | Mild cognitive impairment | miRNA biomarker pairs successfully detected MCI in most of the patients at asymptomatic stage |
| Bipolar disorder | Decreased miR-134 level was directly associated with bipolar disorder |
miR-320
Four studies have been showed differences in the expressions of miR-320 family in psychiatric disorders so far. One of these studies was made by Mundalil Vasu et al.,30) which compared serum miRNA expressions of 55 autistic children with 55 healthy controls. In this study 8 miRNA levels, including miR-320, were found to be decreased compared to the control group, and 5 miRNAs were found to be increased. Moreover, Talebizadeh et al.31) investigated 470 miRNAs including miR-320 in lymphoblastoid cell lines (LCL) of autism patients. They found miR-320 was dysregulated in LCL of autism patients and they propound MECP2, NLGN3, PTEN, AUTS2, TSC1, SLITRK3, SLITRK5, SNRPN as related pathways to miR-320.
In another study comparing the differences of plasma miRNA levels of 16 depressed patients with 14 healthy controls, miR-320d was higher in depression group.32) Study of Camkurt et al.33) including 50 patients with depression and 41 healthy individuals reported that miR-320a levels were significantly lower in patient group. All these results suggest that miR-320 family may be important as a peripheral biomarker in psychiatric disorders. Another important feature of miR-320a is affecting GRIN2A and DISC1 genes that are known to be significant in the etiology of major depression.33)
miR-106
miR-106 family has been studied quite extensively in psychiatric disorders. In an autism study we mentioned above, miR-320a was found to be decreased, but miR-106-5p showed higher expression in patients with autism.30) Liu et al.32) reported that miR-106a-5p was higher in patients with major depression than in control group. In another study, children with attention deficit hyperactivity disorder showed lower miR-106b-5p levels than healthy controls.34) In the study of Camkurt et al.35) comparing 16 schizophrenia patients with 16 healthy individuals, miR-106b-5p was higher in the schizophrenia group. At this point, giving some information about miR-106 may be useful. As known, interleukin (IL) levels have been studied in psychiatric disorders so far. IL-10 has anti-inflammatory properties, and it has been studied intensively in psychiatric disorders. According to a meta-analysis study, IL-10 has been found to be increased in bipolar disorder. Consistently, it has been showed that miR-106 has post-transcriptional regulatory effects for IL-10.36)
ABCA1 (member 1 of human transporter sub-family ABCA) is a protein that plays a significant role in taking out cholesterol from cells, and it is inhibited by miR-106b. Levels of pregnenolone and dehydroepiandrosterone, which are important in cholesterol metabolism, have been showed to be higher in some psychiatric disorders.37) All of these findings show that miR-106 family is very important for psychiatric disorders, and it is necessary to investigate miR-106 family in detail in future studies.
miR-34
miR-34 family is probably the most intensely researched group of miRNAs in psychiatric disorders. In a study made by Bocchio-Chiavetto et al.,38) beginning of treatment and 12 weeks of escitalopram treatment were compared for miRNA levels in peripheral blood, and miR-34c-5p was shown to be decreased. In another study, lymphoblastoid cells were exposed to lithium for 16 days, and as a result, miR-34a expression was altered.39)
In a study comparing schizophrenia patients’ miRNA levels with healthy individuals, plasma miR-34 levels were higher than the control group.40) Moreover, miRNA expression levels in mononuclear leukocytes in patients with schizophrenia were evaluated, and miR-34 and other miRNAs were presented as peripheral biomarkers. Further important feature of this study is application of neuro-cognitive tests for phenotyping patients.41) A recent study investigated miR-34b-5p and miR-34c-5p in peripheral blood leukocytes of 32 major depressive disorder (MDD) patients and 32 healthy controls. They found the miR-34b-5p and miR-34c-5p were significantly higher in depressed patients, and Notch1 gene were significantly lower in MDD patients. They draw a conclusion as following; miR-34b-5p and miR-34c-5p levels in peripheral blood leukocytes were closely related to suicide idea and cognitive function and this may serve as biomarkers for the diagnosis of MDD.42)
A study performed for the detection of clinical bio-markers in patients with schizophrenia by Sun et al.43) investigated 10 miRNAs, including miR-34a. Combination of 5 microRNAs including miR-34a revealed a diagnostic specificity of 90.2%, and these 5 miRNAs were suggested to be useful in terms of diagnosis. Likewise miR-34a was determined to be increased in Alzheimer’s patients’ mononuclear cells.44) Besides, miR-34a was observed to be decreased in plasma and cerebrospinal fluid of Alzheimer’s patients.45)
In their study investigating miR-34c as a peripheral bio-marker in plasma for Alzheimer’s disease, Bhatnagar et al.46) showed that the area under the curve was 0.99 in receiver operating characteristic (ROC) analysis, and the r value was 0.7 in correlation analysis. In conclusion, they revealed that miR-34c levels and mini-mental examination showed inverse correlation, and miR-34c might be a good biomarker.
miR-34 family is one of the best examples for understanding the etiology of psychiatric disorders. Peripheral and other biomarker studies suggest that miR-34 family frequently shows expression changes in peripheral tissues, and they are the targets of mood stabilizing drugs.
This intensive knowledge about the miR-34 family probably lead the study of Bavamian et al.47) They showed higher miR-34a expressions in brains of patients with bipolar disorder. Along with the investigation of post-mortem brain tissues, on the other hand they used stem cell modeling to identify the role of miR-34a. They suggest that miR34-a is critical for bipolar disorder in terms of neurodevelopment. Evaluation of their results with peripheral data, it is a clear demonstration of importance of miRNAs for psychiatric disorders and probable correlation between brain and peripheral tissue.
miR-223
Two studies showed the significance of miR-223 family in peripheral tissues. The first is the previously mentioned study including 50 depressed patients and 41 healthy controls. In this study, miR-223-3p levels were reported to be significantly higher in patients than in controls.33) In another study examining the value of miRNAs as a peripheral biomarker in posttraumatic stress disorder, Balakathiresan et al.48) investigated miRNA changes in amigdala and peripheral blood of animal experiments with fear and stress. Results of this study suggested that 9 miRNAs, including miR-223, could be a potential biomarker.
miR-107
Wang et al.49) investigated the relationship between miR-107 and BACE1 mRNA gene expressions in plasma for clinical diagnosis of amnestic mild cognitive impairment. They reported that miR-107 expressions in plasma had a high capability to distinguish individuals with amnestic mild cognitive impairment from healthy controls. Furthermore, a study performed in peripheral leukocytes of depressed patients by Sun et al.42) found a positive correlation between N1, P2 latency of P300 and miR-107.
miR-134
Sheinerman et al.50) evaluated the feasibility of using pairs of brain-enriched plasma miRNA, at least one of which is enriched in synapses and neurites, as biomarkers that could discriminate between patients with mild cognitive impairment and age-matched controls. The identified biomarker pairs fall into two sets: the miR-132 family and the miR-134 family. The area under the curve was 0.91–0.95 in ROC analysis, with sensitivity and specificity at 79–100% (miR-132 family) and 79–95% (miR-134 family), and p<0.001. In a separate study, these miRNA bio-marker pairs successfully detected mild cognitive impairment in most of the patients at asymptomatic stage.50) Rong et al.51) evaluated miR-134 levels before and after treatment for bipolar manic episode, their conclusion was decreased miR-134 level was directly associated with bipolar disorder and miR-134 level was significantly increased after treatment.
CONCLUSION
The field of miRNA research is constantly getting stronger and expanded. As being epigenetic regulators of gene expression, deeper investigation of miRNAs will probably be promising. Our current knowledge points out miR-34 family as a promising peripheral marker. However there are some points, limitations and obstacles, which need to be overcome in this research field. First, miRNA levels are very sensitive to external factors like diet, exercise, drug use, acute-chronic medical conditions. While including patients, providing homogenous groups is crucial. Moreover, for psychiatric investigations phenotyping of patients according to rating scales, imaging methods or symptomatology may help researchers to acquire better results. Especially, studies focusing on neuro-imaging techniques and association of peripheral miRNA levels in peripheral tissue could be a novel research topic in this regard. Further issue seems to be tissue type. There are diverse peripheral tissue types like mononuclear leukocytes, lymphocytes, plasma and serum. As is known, same miRNA tends to express diversely between tissues or different parts of the same tissue. Future studies should investigate same miRNAs in different peripheral tissue types simultaneously to clarify inter-tissue expression differences. Another point is the source of peripheral circulating miRNAs in plasma or sera of patients. The source of circulating miRNAs is known to be secretion of different tissues.29,52) Although obtaining serum or plasma is feasible, variety of secreting tissues decreases reliability of the results. On the other hand, exosomal circulating miRNA investigations may be helpful to outface this problem.53)
Currently, there are two techniques in miRNA research. One of them is identifying few miRNAs and investigating them in study samples. Limited number of investigated miRNAs is the major limitation for this method.54) Other technique is array method. In this method researcher can study tens or hundreds of miRNAs in one panel. However, some arrays prevent researchers from selecting specific miRNAs to study and that is a limitation. On the other arm, there are some arrays focusing on disease pathways, which we think more reliable to use in research.55)
Before starting a research, miRNA databases should be deeply investigated to detect the most important miRNAs and target genes. There are “validated targets” and “predicted targets” for each miRNA. Although the number of validated targets is very low, researchers may also draw conclusions according to predicted targets, see Table 2 for databases. One more issue is epistatic interactions, which is associated with complexity.56) In general the regulation of miRNAs-genes is not like one miRNA-one gene interaction. In fact, one single miRNA can regulate several genes, and one gene may be target for several miRNAs, check databases mentioned in Table 2.
Table 2.
Target detection databases for miRNAs
| Database name | Website |
|---|---|
| miRBase* | http://www.mirbase.org |
| DIANA-MICROT† | http://diana.imis.athena-innovation.gr/DianaTools/index.php |
| MICRORNA.ORG† | http://www.microrna.org |
| MIRDB† | http://mirdb.org/miRDB/ |
| RNA22-HAS† | https://cm.jefferson.edu/ |
| TARGETSCAN-VERT† | http://www.targetscan.org |
| PICTAR-VERT† | http://pictar.mdc-berlin.de |
| TARGETMINER† | http://www.isical.ac.in |
| MIRECORDS‡ | http://mirecords.biolead.org/ |
| MIRTARBASE‡ | http://mirtarbase.mbc.nctu.edu.tw |
| TARBASE‡ | http://diana.imis.athena-innovation.gr |
Guideline website for detection;
predicted target detection databases;
validated target detection databases.
The types or miRNAs still keep increasing and subtypes are evolving continuously. The more miRNA types increase the more research we will need in this field.
REFERENCES
- 1.Singh I, Rose N. Biomarkers in psychiatry. Nature. 2009;460:202–207. doi: 10.1038/460202a. [DOI] [PubMed] [Google Scholar]
- 2.Nurjono M, Lee J, Chong SA. A review of brain-derived neurotrophic factor as a candidate biomarker in schizophrenia. Clin Psychopharmacol Neurosci. 2012;10:61–70. doi: 10.9758/cpn.2012.10.2.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neelamekam S, Nurjono M, Lee J. Regulation of inter-leukin-6 and leptin in schizophrenia patients: a preliminary analysis. Clin Psychopharmacol Neurosci. 2014;12:209–214. doi: 10.9758/cpn.2014.12.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma SL, Lam LC. Panel of genetic variations as a potential non-invasive biomarker for early diagnosis of alzheimer’s disease. Clin Psychopharmacol Neurosci. 2011;9:54–66. doi: 10.9758/cpn.2011.9.2.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camkurt MA, Fındıklı E, İzci F, Kurutaş EB, Tuman TC. Evaluation of malondialdehyde, superoxide dismutase and catalase activity and their diagnostic value in drug naïve, first episode, non-smoker major depression patients and healthy controls. Psychiatry Res. 2016;238:81–85. doi: 10.1016/j.psychres.2016.01.075. [DOI] [PubMed] [Google Scholar]
- 6.Davis BN, Hata A. Regulation of MicroRNA biogenesis: A miRiad of mechanisms. Cell Commun Signal. 2009;7:18. doi: 10.1186/1478-811X-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 8.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 10.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 11.Camkurt MA. MicroRNAs as a new hope for depression. J Mood Disord. 2015;5:23–30. doi: 10.5455/jmood.20141013124845. [DOI] [Google Scholar]
- 12.Zhang J, Liu Q, Zhang W, Li J, Li Z, Tang Z, et al. Comparative profiling of genes and miRNAs expressed in the newborn, young adult, and aged human epididymides. Acta Biochim Biophys Sin (Shanghai) 2010;42:145–153. doi: 10.1093/abbs/gmp116. [DOI] [PubMed] [Google Scholar]
- 13.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12:399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaefer A, O’Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, et al. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med. 2007;204:1553–1558. doi: 10.1084/jem.20070823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 19.Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HY, et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci U S A. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, et al. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dwivedi Y. Evidence demonstrating role of microRNAs in the etiopathology of major depression. J Chem Neuroanat. 2011;42:142–156. doi: 10.1016/j.jchemneu.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Connor RM, Dinan TG, Cryan JF. Little things on which happiness depends: microRNAs as novel therapeutic targets for the treatment of anxiety and depression. Mol Psychiatry. 2012;17:359–376. doi: 10.1038/mp.2011.162. [DOI] [PubMed] [Google Scholar]
- 23.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braasch DA, Paroo Z, Constantinescu A, Ren G, Oz OK, Mason RP, et al. Biodistribution of phosphodiester and phosphorothioate siRNA. Bioorg Med Chem Lett. 2004;14:1139–1143. doi: 10.1016/j.bmcl.2003.12.074. [DOI] [PubMed] [Google Scholar]
- 25.O’Brien FE, Dinan TG, Griffin BT, Cryan JF. Interactions between antidepressants and P-glycoprotein at the blood-brain barrier: clinical significance of in vitro and in vivo findings. Br J Pharmacol. 2012;165:289–312. doi: 10.1111/j.1476-5381.2011.01557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thakker DR, Hoyer D, Cryan JF. Interfering with the brain: use of RNA interference for understanding the pathophysiology of psychiatric and neurological disorders. Pharmacol Ther. 2006;109:413–438. doi: 10.1016/j.pharmthera.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 28.Gao K, Huang L. Nonviral methods for siRNA delivery. Mol Pharm. 2009;6:651–658. doi: 10.1021/mp800134q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Etheridge A, Lee I, Hood L, Galas D, Wang K. Extracellular microRNA: a new source of biomarkers. Mutat Res. 2011;717:85–90. doi: 10.1016/j.mrfmmm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mundalil Vasu M, Anitha A, Thanseem I, Suzuki K, Yamada K, Takahashi T, et al. Serum microRNA profiles in children with autism. Mol Autism. 2014;5:40. doi: 10.1186/2040-2392-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Talebizadeh Z, Butler MG, Theodoro MF. Feasibility and relevance of examining lymphoblastoid cell lines to study role of microRNAs in autism. Autism Res. 2008;1:240–250. doi: 10.1002/aur.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Zhang L, Cheng K, Wang X, Ren G, Xie P. Identification of suitable plasma-based reference genes for miRNAome analysis of major depressive disorder. J Affect Disord. 2014;163:133–139. doi: 10.1016/j.jad.2013.12.035. [DOI] [PubMed] [Google Scholar]
- 33.Camkurt MA, Acar Ş, Coşkun S, Güneş M, Güneş S, Yılmaz MF, et al. Comparison of plasma MicroRNA levels in drug naive, first episode depressed patients and healthy controls. J Psychiatr Res. 2015;69:67–71. doi: 10.1016/j.jpsychires.2015.07.023. [DOI] [PubMed] [Google Scholar]
- 34.Kandemir H, Erdal ME, Selek S, Ay Öİ, Karababa IF, Kandemir SB, et al. Evaluation of several micro RNA (miRNA) levels in children and adolescents with attention deficit hyperactivity disorder. Neurosci Lett. 2014;580:158–162. doi: 10.1016/j.neulet.2014.07.060. [DOI] [PubMed] [Google Scholar]
- 35.Camkurt MA, Karababa İF, Erdal ME, Bayazıt H, Kandemir S, Kandemir H, et al. Investigation of dysregulation of several microRNAs in peripheral blood of schizophrenia patients. Clin Psychopharmacol Neurosci. 2016;14:256–260. doi: 10.9758/cpn.2016.14.3.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma A, Kumar M, Aich J, Hariharan M, Brahmachari SK, Agrawal A, et al. Posttranscriptional regulation of interleukin-10 expression by hsa-miR-106a. Proc Natl Acad Sci U S A. 2009;106:5761–5766. doi: 10.1073/pnas.0808743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marx CE, Trost WT, Shampine LJ, Stevens RD, Hulette CM, Steffens DC, et al. The neurosteroid allopregnanolone is reduced in prefrontal cortex in Alzheimer’s disease. Biol Psychiatry. 2006;60:1287–1294. doi: 10.1016/j.biopsych.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 38.Bocchio-Chiavetto L, Maffioletti E, Bettinsoli P, Giovannini C, Bignotti S, Tardito D, et al. Blood microRNA changes in depressed patients during antidepressant treatment. Eur Neuropsychopharmacol. 2013;23:602–611. doi: 10.1016/j.euroneuro.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 39.Chen H, Wang N, Burmeister M, McInnis MG. MicroRNA expression changes in lymphoblastoid cell lines in response to lithium treatment. Int J Neuropsychopharmacol. 2009;12:975–981. doi: 10.1017/S1461145709000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song HT, Sun XY, Zhang L, Zhao L, Guo ZM, Fan HM, et al. A preliminary analysis of association between the down-regulation of microRNA-181b expression and symptomatology improvement in schizophrenia patients before and after antipsychotic treatment. J Psychiatr Res. 2014;54:134–140. doi: 10.1016/j.jpsychires.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 41.Lai CY, Yu SL, Hsieh MH, Chen CH, Chen HY, Wen CC, et al. MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS One. 2011;6:e21635. doi: 10.1371/journal.pone.0021635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun N, Lei L, Wang Y, Yang C, Liu Z, Li X, et al. Preliminary comparison of plasma notch-associated microRNA-34b and -34c levels in drug naive, first episode depressed patients and healthy controls. J Affect Disord. 2016;194:109–114. doi: 10.1016/j.jad.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 43.Sun XY, Zhang J, Niu W, Guo W, Song HT, Li HY, et al. A preliminary analysis of microRNA as potential clinical biomarker for schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2015;168B:170–178. doi: 10.1002/ajmg.b.32292. [DOI] [PubMed] [Google Scholar]
- 44.Schipper HM, Maes OC, Chertkow HM, Wang E. MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul Syst Bio. 2007;1:263–274. doi: 10.4137/grsb.s361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiko T, Nakagawa K, Tsuduki T, Furukawa K, Arai H, Miyazawa T. MicroRNAs in plasma and cerebrospinal fluid as potential markers for Alzheimer’s disease. J Alzheimers Dis. 2014;39:253–259. doi: 10.3233/JAD-130932. [DOI] [PubMed] [Google Scholar]
- 46.Bhatnagar S, Chertkow H, Schipper HM, Yuan Z, Shetty V, Jenkins S, et al. Increased microRNA-34c abundance in Alzheimer’s disease circulating blood plasma. Front Mol Neurosci. 2014;7:2. doi: 10.3389/fnmol.2014.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bavamian S, Mellios N, Lalonde J, Fass DM, Wang J, Sheridan SD, et al. Dysregulation of miR-34a links neuronal development to genetic risk factors for bipolar disorder. Mol Psychiatry. 2015;20:573–584. doi: 10.1038/mp.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balakathiresan NS, Chandran R, Bhomia M, Jia M, Li H, Maheshwari RK. Serum and amygdala microRNA signatures of posttraumatic stress: fear correlation and biomarker potential. J Psychiatr Res. 2014;57:65–73. doi: 10.1016/j.jpsychires.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 49.Wang T, Chen K, Li H, Dong S, Su N, Liu Y, et al. The feasibility of utilizing plasma miRNA107 and BACE1 messenger RNA gene expression for clinical diagnosis of amnestic mild cognitive impairment. J Clin Psychiatry. 2015;76:135–141. doi: 10.4088/JCP.13m08812. [DOI] [PubMed] [Google Scholar]
- 50.Sheinerman KS, Tsivinsky VG, Crawford F, Mullan MJ, Abdullah L, Umansky SR. Plasma microRNA biomarkers for detection of mild cognitive impairment. Aging (Albany NY) 2012;4:590–605. doi: 10.18632/aging.100486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rong H, Liu TB, Yang KJ, Yang HC, Wu DH, Liao CP, et al. MicroRNA-134 plasma levels before and after treatment for bipolar mania. J Psychiatr Res. 2011;45:92–95. doi: 10.1016/j.jpsychires.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 52.De Rosa S, Fichtlscherer S, Lehmann R, Assmus B, Dimmeler S, Zeiher AM. Transcoronary concentration gradients of circulating microRNAs. Circulation. 2011;124:1936–1944. doi: 10.1161/CIRCULATIONAHA.111.037572. [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, Li S, Li L, Li M, Guo C, Yao J, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang SW, Vosch T. Rapid detection of microRNA by a silver nanocluster DNA probe. Anal Chem. 2011;83:6935–6939. doi: 10.1021/ac201903n. [DOI] [PubMed] [Google Scholar]
- 55.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phillips PC. Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]