ABSTRACT
Background: Probiotic formulations can be single- or multi-strain. Commercially, multi-strain preparations have been suggested to have improved functionality over single-strain cultures. Probiotics are often tested as single-strain preparations but may subsequently be commercially formulated as multi-strain products.
Objective: The aim of this study was to determine what happens at the site of action, the intestine, with probiotics as single- compared to multi-strain preparations. The human gastrointestinal tract contains a broad mixture of different microbes which may affect the survival of different probiotics in different ways.
Design: The current study was performed to evaluate, in an in vitro colon simulation, whether probiotics influence each other’s survival when they are taken as a combination of several strains (HOWARU Restore; Lactobacillus acidophilus NCFM, Lactobacillus paracasei Lpc-37, Bifidobacterium lactis Bl-04 and B. lactis Bi-07) compared to the strains as single preparations.
Results: All strains could be detected after the colon simulations and there were no substantial differences in levels of the same strain when comparing single- and multi-strain products.
Conclusions: It can be concluded that probiotics do not have an antagonistic effect on each other’s survival when used in a multi-strain product compared to a single-strain product, at least within a microbiota in a simulated colonic environment.
KEYWORDS: Lactobacillus, Bifidobacterium, multi-strain probiotic, single-strain probiotic, survival, interaction, in vitro, colonic simulator
Introduction
Probiotics are defined as ‘live microorganisms that, when administered in adequate amounts, confer a health benefit on the host’ [1,2]. The definition assumes the viability of the microbial content of each probiotic component in a product until consumption. The viability and survival of microbes in the different environments that are presented by the probiotic products and the environments they will encounter during manufacture, transportation and storage are critical in ensuring their effectiveness. The definition of a probiotic does not stipulate that probiotics should survive transit through the gastrointestinal tract, although this is often assumed. Although probiotics are commonly selected to withstand low pH and bile acid, transit through the gastrointestinal tract may result in a substantial loss of viable organisms. Physical and chemical barriers, such as low pH, bile, digestive enzymes and the potential presence of antimicrobial components from foods, endogenous microbes and the host may lead to a loss of viability of the consumed probiotic. The ability of different probiotics to survive passage through the gastrointestinal tract has been demonstrated in complex microbial communities [3–5]. However, the extent to which the survival of a probiotic strain is influenced by whether it is formulated as a single-strain or a multi-strain product has hitherto not been investigated. In other words, are probiotic strains antagonistic or even synergistic towards each other, when combined and in the gastrointestinal tract? In the marketplace, especially when considering dietary supplements, probiotics are often formulated as multi-strain products, whereas their survival and efficacy have often been investigated as single-strain products.
The aim of the current study was to investigate in vitro, using a simulated colon model, whether probiotic strains have an agonistic effect upon each other and whether survival of a given strain differs when it is used alone or in combination with other probiotics. An in vitro model of the human colon enables us to test the survival and growth dynamics of single- and multi-strain probiotic formulations in a controlled and replicable manner, allowing the detection of even subtle differences.
Materials and methods
Probiotic bacteria
For the in vitro colon simulations, the following probiotic strains were included: Lactobacillus acidophilus NCFM (ATCC 700396), Lactobacillus paracasei Lpc-37 (ATCC SD5275), Bifidobacterium animalis subsp. lactis Bl-04 (ATCC SD5219) and B. lactis Bi-07 (ATCC SD5220). The strains were included either as single freeze-dried cultures at a dose of 2.5 × 109 colony-forming units (cfu), or in a combination of two strains (L. acidophilus NCFM + B. lactis Bl-04, 5 × 109 cfu [6]) or as all four strains combined (HOWARU Restore, 1010 cfu [7]). The dose of the strains used in the simulations was similar to the daily doses used in two human intervention studies, of 5 × 109 and 1.7 × 1010 cfu, respectively [6,7]. Counts of the single strains were controlled by flow cytometry of the bulk material [8], and the multi-strain products were composed accordingly.
Colon simulations
In vitro colon simulations were performed as described by Mäkivuokko et al. [9], and upper gastrointestinal tract digestion has been described by Mäkeläinen et al. [5]. In short, each simulator unit consists of four connected glass vessels (V1–V4) which mimic different compartments of the human colon from the proximal to the distal part, each having a different pH and flow rate. At the start of a simulation, each unit was inoculated with faecal microbes which form the microbiota of the colonic model. The faecal microbes, from an apparently healthy donor, had been suspended with three parts (w/w) anaerobic synthetic ileal medium which contains cysteine and resazurin to indicate sustained anaerobic conditions [9]. The probiotics (combinations) were added to the synthetic ileal medium and fed semi-continuously in 3 h cycles to the colon model for 48 h, during which transition of fermented fluids and microbes and feeding of fresh medium occurred. Anaerobiosis was maintained by flushing the simulator with nitrogen. After simulation, the simulated digesta were collected and bacterial DNA was extracted and analysed by quantitative polymerase chain reaction (qPCR) for quantification of the probiotic bacteria. As a control, the synthetic ileal medium without freeze-dried bacteria was used.
Extraction and quantification of bacterial DNA
The DNA from the simulation samples was extracted and purified with an automated MagMAX™ Sample Preparation System (Life Technologies, Halle, Belgium), using the MagMAX Nucleic Acid Isolation Kit. The amount of extracted DNA was determined with a Qubit® dsDNA HS Assay Kit (Thermo Fisher Scientific, Vantaa, Finland). To produce qPCR standards of B. lactis Bi-07, B. lactis Bl-04, L. acidophilus NCFM and L. paracasei Lpc-37, the strains were grown in liquid culture overnight, and subsequently genomic DNA was extracted using the MagMAX Sample Preparation System. Ten-fold dilution series of genomic DNA from standard strains were used to generate standard curves for the qPCR in quantities ranging from 100 fg to 10 ng (corresponding to 1 × 101 to 7 × 106 copies).
For absolute quantification, qPCR reactions were performed in a total volume of 25 µl with an ABI-PRISM 7500 sequencing detection system (Applied Biosystems, Bridgewater, NJ, USA). The SYBR Green methodology (Applied Biosystems) was used for B. lactis Bi-07, B. lactis Bl-04, Lactobacillus spp. [10] and L. acidophilus, while the TaqMan methodology was used for Bifidobacterium spp. [9], L. acidophilus and L. paracasei [11]. Ten-fold dilution series (10 pg and 100 fg) of DNA from standard strains were used for the standard curves. For the determination of the quantity of genome copies DNA, triplicate samples were used, and the mean quantity (log10) per gram of faecal wet weight was calculated.
Statistical analysis
Simulations were performed in triplicate, independent from each other, with faecal inocula from different donors. Simulations were compared to each other using Student’s t test. A p-value of 0.05 or less was considered significant.
Results
All of the probiotic strains were detected at elevated levels, 1–2 log10 above baseline, in the simulation samples to which they had been fed, while none of them could be detected above baseline in the simulations to which they had not been added (Figure 1).
Figure 1.
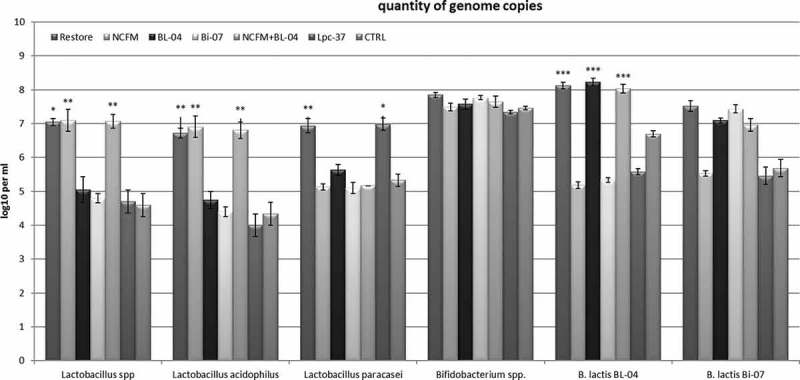
Microbial quantities presented as mean ± SEM for the pooled vessels (V1–V4) from three separate colon simulations. The x-axis represents the analysed species or strains, while the quantity is expressed as log10 copies per millilitre. Restore, HOWARU Restore; NCFM, Lactobacillus acidophilus; Bl-04, Bifidobacterium lactis; Bi-07, B. lactis; Lpc-37, Lactobacillus paracasei. The control (CTRL) represents the same synthetic ileal medium used for feeding the simulator but without freeze-dried bacteria. Statistical differences between treatment and control: *p < 0.01, **p = 0.02, ***p < 0.05.
As seen in Figure 1, when Lactobacillus was quantified at genus level, an increase was detected for simulations with the HOWARU Restore product (p < 0.01), L. acidophilus NCFM (p = 0.02) and the combination of L. acidophilus NCFM + B. lactis Bl-04 (p = 0.02). Lactobacillus paracasei Lpc-37 inclusion was not observed to lead to an increase in lactobacilli (p > 0.05); this is due to the fact that L. paracasei is not a target species for the primers used with the Lactobacillus spp. detection. When L. acidophilus was quantified, substantial increases were detected in the simulations with HOWARU Restore (p = 0.02), L. acidophilus NCFM (p = 0.02) and L. acidophilus NCFM + B. lactis Bl-04 (p = 0.02) compared to the control (Figure 1). Similarly, quantification of L. paracasei was observed to lead to substantial increases in simulations with L. paracasei Lpc-37 (p = 0.03) and HOWARU Restore (p = 0.0002) compared to the control (Figure 1). However, quantification of Bifidobacterium spp. was not associated with any difference in Bifidobacterium levels in those simulations that contained bifidobacteria and the control (p > 0.05) (Figure 1). On the other hand, quantification of B. lactis Bl-04 was associated with increased levels in simulations with HOWARU Restore, B. lactis Bl-04 and L. acidophilus NCFM + B. lactis Bl-04 compared to the control (p < 0.05) (Figure 1). Similarly, quantification of B. lactis Bi-07 was associated with increased levels in simulations with HOWARU Restore and B. lactis Bi-07 compared to the control (p < 0.05) (Figure 1). However, the assay also indicated an increase in simulations with products containing B. lactis Bl-04 (p < 0.05) (Figure 1). This indicates that the assay is not sufficiently strain specific. For all assays, the detection limit is approximately 5 log10/ml for all analysed targets (strain, species and genus).
When comparing simulations with single strains (L. acidophilus NCFM, L. paracasei Lpc-37, B. lactis Bl-04 and B. lactis Bi-07) with simulations of strain combination products (HOWARU Restore and L. acidophilus NCFM + B. lactis Bl-04), it is clear that the levels of single and combination products were not different. Lactobacillus acidophilus levels were not different for L. acidophilus NCFM, HOWARU Restore and L. acidophilus NCFM + B. lactis Bl-04 simulations (p > 0.05). Similarly, L. paracasei levels were not different between L. paracasei Lpc-37 and HOWARU Restore simulations (p > 0.05). Likewise, B. lactis Bl-04 levels were not different between B. lactis Bl-04, HOWARU Restore and L. acidophilus NCFM + B. lactis Bl-04 simulations (p > 0.05), nor were there differences in B. lactis Bi-07 levels for B. lactis Bi-07 and HOWARU Restore simulations (p > 0.05).
In the supplementary material (Figure S1), levels of the tested genus, species and strain can be seen in the separate vessels of the respective probiotic (combination) simulations and the control. It can also be seen that strain levels were similar in the respective vessels, regardless of whether they were included as single or multiple strains.
Discussion
The efficacy of probiotics has been assessed in human intervention trials. The products in such studies may consist of single or multiple probiotic strains. To assess the potential interaction between probiotic strains in a multi-strain product, studies would have to be performed with additional arms to investigate single strains and strain combinations. This would be a highly resource-demanding effort. Here, we determined whether and what interactions between probiotic strains can be observed, affecting their survival, in a simulated colonic environment. This does not answer the question of the efficacy of single- versus multi-strain probiotics. Nor does it allow the effect of a particular strain in a probiotic combination to be isolated. However, it may give an indication of whether survival is influenced, and if reduced survival is observed, one could hypothesise that it may also influence efficacy.
The colon simulator model enables the simulation of the colonic conditions and especially the microbiota. In addition to environmental parameters, e.g. variations in acidity and substrate limitation, the strain balance and activities of microorganisms are determined by interplay between members of the microbiota, including antagonism, competition for substrates and symbiosis by cross-feeding [12].
The present study showed that all the included probiotic strains survived colon simulation. Levels of the particular strains were significantly higher than background levels, with the exception of Bifidobacterium spp., where no change in levels was observed. This can be explained by the overall prevalence of relatively high levels of bifidobacteria in humans and therefore also in the simulations. The addition of the probiotic bifidobacteria could not be detected against the already high background. Whether strains were administered to the simulators alone or as part of a multi-strain product was found not to influence the detected levels.
Co-feeding of several strains showed that all strains were detected after the colon simulations and there were no substantial differences in levels when comparing single- and multi-strain products. Thus, it can be concluded that the tested strains have neither an antagonistic nor a synergistic effect upon quantity, even though they are present in mixed populations, at least under the in vitro simulation conditions tested here.
Supplementary Material
Acknowledgements
Kirsi Stenström and Minna Eskola are thanked for their excellent technical assistance with the simulations.
Biography
Both authors designed the study. SF managed the execution of the study. Both authors interpreted the results and wrote the manuscript.
Disclosure statement
Both authors are employees of DuPont. DuPont manufactures and markets the investigated probiotic strains.
Supplemental data
Supplemental data for this article can be accessed here.
References
- [1]. FAO/WHO , editor. Guidelines for the evaluation of probiotics in food. 2002. http://www.who.int/foodsafety/publications/fs_management/probiotics2/en/
- [2]. Hill C, Guarner F, Reid G, et al. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506–4. [DOI] [PubMed] [Google Scholar]
- [3]. Goldin BR, Gorbach SL, Saxelin M, et al. Survival of Lactobacillus species (strain GG) in human gastrointestinal tract. Dig Dis Sci. 1992;37(1):121–128. [DOI] [PubMed] [Google Scholar]
- [4]. Godward G, Kailasapathy K.. Viability and survival of free and encapsulated probiotic bacteria in Cheddar cheese. Milchwissenschaft-Milk Sci Int. 2003;58(11–12):624–627. [Google Scholar]
- [5]. Mäkeläinen H, Olli K, Forssten S, et al. Probiotic lactobacilli in a semi-soft cheese survive in the simulated human gastrointestinal tract. Int Dairy J. 2009;19(11):675–683. [Google Scholar]
- [6]. Ouwehand AC, Nermes M, Collado MC, et al. Specific probiotics alleviate allergic rhinitis during the birch pollen season. World J Gastroenterol. 2009;15(26):3261–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Ouwehand AC, DongLian C, Weijian X, et al. Probiotics reduce symptoms of antibiotic use in a hospital setting: a randomized dose response study. Vaccine. 2014;32(4):458–463. [DOI] [PubMed] [Google Scholar]
- [8]. ISO-IDF Milk and milk products – Starter cultures, probiotics and fermented products – Quantification of lactic acid bacteria by flow cytometry. Geneva: International Organization for Standardization; 2015. [Google Scholar]
- [9]. Mäkivuokko H, Nurmi J, Nurminen P, et al. In vitro effects on polydextrose by colonic bacteria and Caco-2 cell cyclooxygenase gene expression. Nutr Cancer. 2005;52(1):94–104. [DOI] [PubMed] [Google Scholar]
- [10]. Delroisse JM, Boulvin AL, Parmentier I, et al. Quantification of Bifidobacterium spp. and Lactobacillus spp. in rat fecal samples by real-time PCR. Microbiol Res. 2008;163:663–670. [DOI] [PubMed] [Google Scholar]
- [11]. Haarman M, Knol J. Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. Appl Environ Microbiol. 2006;72(4):2359–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. De Vuyst L, Leroy F. Cross-feeding between bifidobacteria and butyrate-producing colon bacteria explains bifdobacterial competitiveness, butyrate production, and gas production. Int J Food Microbiol. 2011;149(1):73–80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


