Abstract
Background
Epithelial-mesenchymal transition (EMT) is an early event in tumour invasion and metastasis, and widespread and distant metastasis at early stages is the typical biological behaviour in small cell lung cancer (SCLC). Our previous reports showed that high expression of the transcription factor E2F1 was involved in the invasion and metastasis of SCLC, but the role of E2F1 in the process of EMT in SCLC is unknown.
Methods
Immunohistochemistry was performed to evaluate the expressions of EMT related markers. Immunofluorescence was used to detect the expressions of cytoskeletal proteins and EMT related markers when E2F1 was silenced in SCLC cell lines. Adenovirus containing shRNA against E2F1 was used to knock down the E2F1 expression, and the dual luciferase reporter system was employed to clarify the regulatory relationship between E2F1 and ZEB2.
Results
In this study, we observed the remodelling of cytoskeletal proteins when E2F1 was silenced in SCLC cell lines, indicating that E2F1 was involved in the EMT in SCLC. Depletion of E2F1 promoted the expression of epithelial markers (CDH1 and CTNNB1) and inhibited the expression of mesenchymal markers (VIM and CDH2) in SCLC cell lines, verifying that E2F1 promotes EMT occurrence. Next, the mechanism by which E2F1 promoted EMT was explored. Among the CDH1 related inhibitory transcriptional regulators ZEB1, ZEB2, SNAI1 and SNAI2, the expression of ZEB2 was the highest in SCLC tissue samples and was highly consistent with E2F1 expression. ChIP-seq data and dual luciferase reporter system analysis confirmed that E2F1 could regulate ZEB2 gene expression.
Conclusion
Our data supports that E2F1 promotes EMT by regulating ZEB2 gene expression in SCLC.
Electronic supplementary material
The online version of this article (10.1186/s12885-017-3701-y) contains supplementary material, which is available to authorized users.
Keywords: Epithelial-mesenchymal transition, E2F1, Small cell lung cancer, ZEB2
Background
The cancer statistics for China in 2015 showed that there were 733,330 new cases of lung cancer (509,300 cases in males and 224,000 in females), and 610,200 deaths (432,400 males and 177,800 females) due to lung cancer. The incidence and mortality rate for lung cancer ranked first out of all tumours [1]. Small cell lung cancer (SCLC) is one of the most malignant tumours and accounts for 20–25% of all lung cancers. With the aggravation of environmental pollution, the incidence of SCLC is increasing year by year, and the five-year survival rate remains at approximately 10% [2]. One of the typical biological behaviours of SCLC is widespread, distant metastases at early stages [3]. In addition, RB1 (RB transcriptional corepressor 1) gene loss, which is a typical genetic characteristic of SCLC, leads to the deregulation of E2F1 [4]. Our previous research showed that E2F1 is highly expressed in SCLC [2], indicating that E2F1 plays a role in SCLC.
E2F1 is a transcription factor that is involved in the cell cycle, proliferation, apoptosis and differentiation [5]. Recent reports showed that E2F1 took part in tumour invasion and metastasis by regulating thrombospondin 1 [6], PDGFR [7] and VEGFR [8]. Our previous research showed that E2F1 was also involved in invasion and metastasis by controlling MMP-9, MMP-16 and ADAM-12 [2, 3, 9]. We also observed changes in cell morphology when E2F1 was silenced, indicating that E2F1 may regulate the epithelial-mesenchymal transition (EMT).
EMT is an early event in the process of tumour invasion and metastasis [10]. E2F1 can suppress the Wnt/β-catenin signalling pathway by regulating ICAT [11] and GSK-3 [12] in colorectal cancer and can also drive EMT by inducing miR-224/452 in malignant melanoma [13]. These results showed that E2F1 is closely associated with EMT, but the detailed mechanism of EMT regulation by E2F1 in SCLC is unknown.
In this study, we analysed the expression pattern of EMT-related proteins in SCLC tissue samples. We then examined the changes in cell morphology and cytoskeleton remodelling processes when E2F1 was knocked down by shRNA in SCLC cells. In these cells, epithelial markers were significantly increased and mesenchymal markers were significantly decreased. ChIP-seq and dual-luciferase reporter experiments indicated that E2F1 directly regulated the expression of ZEB2 by binding to its promoter. Our results suggest that E2F1 promotes EMT by regulating ZEB2 in SCLC.
Methods
Patients and cell lines
Sixty SCLC biopsy tissue samples before treatment were obtained from the affiliated hospital of Binzhou Medical University from January 2014 to January 2015. All patients signed informed consent forms before providing tissue samples. This research was approved by the Medical Ethics Committee of Binzhou Medical University (No. 2013027). This study was performed according to the Declaration of Helsinki and to the relevant ethical guidelines for research on humans. The basic patient information is listed in Table 1. Human SCLC cell lines H446 (TCHu196, Chinese Academy of Sciences cell bank) and H1688 (TCHu154, Chinese Academy of Sciences cell bank) were stored in our lab. All cells were cultured in RPMI 1640 media (Gibco, Cat:89,984) with 10% FBS (Gibco, Cat: 26,140,079) and 100 U/ml penicillin and 100 μg/ml streptomycin.
Table 1.
Clinicopathologic features for patients suffering SCLC (n = 60)
| Variables | N (%) | Statistical analysis | |||
|---|---|---|---|---|---|
| HR | 95%CI | P | |||
| Age | <60 | 16(26.67) | 1 | 0.297–1.558 | 0.362 |
| ≥ 60 | 44(73.33) | 0.681 | |||
| Gender | Male | 55(91.67) | 1 | 0.900–8.409 | 0.076 |
| Female | 5(8.33) | 2.752 | |||
| Smoking | Non-smoker | 4(6.67) | 1 | 0.603–4.697 | 0.321 |
| Smoker | 56(93.33) | 1.682 | |||
| Tumor size | < 4 cm | 24(40.00) | 1 | 0.344–1.397 | 0.306 |
| ≥ 4 cm | 36(60.00) | 0.693 | |||
| Clinical stage | Limited disease | 8(13.33) | 1 | 0.072–0.831 | 0.024 |
| Extensive disease | 52(86.67) | 0.224 | |||
*represents p<0.05
Immunohistochemistry
All tissue sections were dewaxed in dimethylbenzene and rehydrated in an alcohol gradient ranging from 100% to 75%. Antigen retrieval was performed by placing samples in an EDTA antigen repair solution (ph = 9.0). Sections were then washed three times with PBS buffer (ph = 7.0). Next, the sections were incubated with a primary antibody at 4 °C overnight. The antibodies were used at the following dilutions: 1:100 for VIM (Cell signalling technology, Cat:5741), 1:150 for CDH2 (Cell signalling technology,Cat:13,116), 1:250 for CLDN1 (Cell signalling technology, Cat:13,255), 1:100 for CTNNB1 (Cell signalling technology, Cat:8480), 1:400 for CDH1 (Cell signalling technology, Cat:3195), 1:50 for E2F1 (Santa Cruz, Cat:sc-251), 1:200 for ZEB2 (Santa Cruz, Cat:271,984), ZEB1 (Cell signalling technology, Cat:3396), SNAI1 (Cell signalling technology, Cat:3879), and SNAI2 (Cell signalling technology, Cat:9585). The sections were washed three times with PBS and incubated with HRP-conjugated secondary antibodies for 40 min at 37 °C. Then, the DAB (diaminobenzidine) reaction, hematoxylin staining, differentiation with hydrochloric acid alcohol, dehydration and transparency steps were conducted in turn. All images were captured by Leica Microsystems CMS (DFC365 FX). All staining was scored according to our previous reports [2, 3]. In brief, the staining was quantified using a 4-value intensity score: 0 as negative; 1+ as weak; 2+ as moderate, 3+ as strong, and the percentage (0–100) of the extent of reactivity. A final score was obtained by multiplying the intensity and reactivity extension values (range, 0–300) [14].
H446 cells in which E2F1 was stably knocked down by adenovirus containing E2F1 specific shRNA were constructed
Adenovirus (1 × 109 titers) containing shRNA against E2F1 was provided by Gene Pharma Company. H446 cells were conventionally cultured in six-well plates. At a confluence of approximately 70%–80%, 1 × 106 virus titers were added and mixed gently. After 12 h, the completed medium was added to cells and the previous medium was removed. After 72 h, puromycin (5 μg/ml) was added to the media. After 5 days, all cells were digested, diluted, and cultured in a 96-well plate to form monoclonal colonies. The medium containing puromycin (1 μg/ml) was changed every 3 days until the confluence was approximately 90%–100%. Cells were then digested and inoculated into 48-well, 24-well and 6-well plates. These cells were named H446-E2F1sh.
siRNA and transfection
siRNAs targeting E2F1 was transfected into H1688 according to the methods stated in our previous report [2]. H1688 cells in which E2F1 was transiently silenced were named H1688-E2F1si.
Real-time PCR.
Total RNA was extracted with RNAiso Plus (Takara, Cat: 9108), and cDNA was synthesized with the Prime Script RT reagent kit with gDNA Eraser (Takara, Cat: RR047A). Real-time PCR was performed according to the instructions provided for the SYBR Fast qPCR Mix (Takara, Cat: RR430A). The primers used are listed in additional file 1.
Western blotting
All cells were lysed with RIPA lysis buffer containing a protease inhibitor cocktail (Sigma, Cat: S8820). The protein concentrations were measured, and 50 μg of protein was run on an SDS-PAGE gel and electrophoretically transferred onto NC membranes. The membranes were blocked with 5% fat-free milk and incubated overnight at 4 °C with primary antibodies, including VIM (1:1000), CDH2 (1:1000), CTNNB1 (1:1000), CDH1 (1:1000), E2F1 (1:500), ZEB2 (1:500), α-tubulin (1:1000), β-actin (1:1000) and GAPDH (1:2000). The membranes were then washed three times and incubated for 40 min with HRP-conjugated secondary antibodies. Protein bands were detected by the ECL system [2, 3].
Immunofluorescence
Cells were placed on slides, washed three times with PBS buffer, and fixed with ice-cold methanol and acetone (1:1). Next, the slides were blocked for 30 min with goat serum, washed three times with PBS-TX (PBS containing 1% Tritonx-100), and incubated overnight at 4 °C with primary antibodies. The slides were then incubated with Rhodamine or FITC-labelled fluorescent secondary antibodies and DAPI. Laser scanning confocal microscope (leica-LM7000) was used to observe cell morphology and capture pictures.
Construction of ZEB2 luciferase reporter vector and activity analysis
Genomic DNA was extracted from H446 cells, and the ZEB2 promoter was amplified by PCR and purified. The PCR fragment and pGL3-basic vector were digested with Nhe I and Bgl II enzymes, and T4 DNA ligase was used to ligate these two fragments together to construct the ZEB2 promoter reporter vector. Luciferase activity analysis was performed according to methods described in our previous report [2].
Statistical analysis
SPSS.17.0 statistical software and GraphPad Prism 7 was used. The results from the IHC experiments were analysed by a Chi Square test. The expression differences among target genes were analysed by a t-test. Multivariate survival analysis was performed by Cox’s regression.
Results
The expression of EMT markers in SCLC tissue samples
Distant metastasis in early stages is a typical feature of SCLC, and the occurrence of EMT is considered to be an early event in the process of tumour invasion and metastasis. Therefore, it is important to determine the expression pattern of EMT markers in SCLC tissue. We examined CDH1 expression and found that it was highly related with CTNNB1 expression (r = 0.9985, (p < 0.001) and that both proteins were located on the cellular membrane and absent from the cytoplasm and nucleus. CTNNB1 and CDH1 were expressed in 90% (54/60) of small cell lung cancer cells (Fig. 1a), in which weak expression (score between 10 and 20) was 11.11%, moderate (score between 20 and 100) was 70.37% and strong (score between 100 and 300) was 18.52% (Additional file 2: Figure S1). This result was consistent with other reports [15–17]. CDH1 and CTNNB1 in bronchial epithelial cells as positive control were presented in Additional file 3: Figure S2. The Spearman analysis showed that CDH1, CTNNB1 were significantly relevant with tumour sizes (p = 0.03) and clinical stage (p < 0.001) not age (p = 0.461), gender (p = 0.335), smoking (p = 0.224). CTNNB1 has been previously observed in the nucleus when CDH1 was deregulated [18, 19]; however, we did not detect CTNNB1 in the nucleus (Fig. 1a, Additional file 2: Figure S1 and Additional file 3: Fig. S2), which was consistent with other reports [20]. This result supported that CTNNB1 did not transfer into the nucleus in SCLC samples. In additional, we analysed CDH1 and CTNNB1 expressions from the gene expression database of SCLC cell lines (https://sclccelllines.cancer.gov/) [21] (Additional file 4: Figure S3), and found that their expressions were not related in SCLC cell lines (r = 0.01671, p = 0.443). This told us that the expression pattern of CDH1 and CTNNB1 was different between SCLC tissues and SCLC cell lines.
Fig. 1.
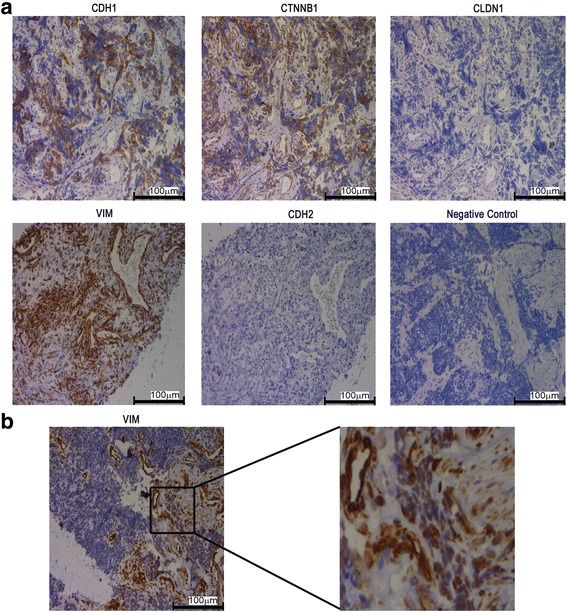
The immunohistochemical (IHC) staining of EMT related markers. a. EMT related markers, including CDH1, CTNNB1, CLDN1, VIM and CDH2, were detected by IHC in 60 SCLC tissue samples. b VIM expression was inconsistent in the same tissue sample. Representative image was presented, and the magnification is 200 X
CLDN1 and CDH2 were not detected in any SCLC samples (0/60) (Fig. 1a). VIM was observed in all stromal cells (Fig. 1a) and 25% (15/60) tumour cells (Fig. 1b), and its expression was significantly related with tumour differentiation (p < 0.001) [22].
Our previous paper reported that E2F1 was highly expressed and was an independent and adverse prognostic factor for SCLC. We also found that E2F1 could directly regulate the expressions of RELA, MMPs and ADAM12 [2, 3, 9]. NF-κB, MMPs and ADAMs were found to be closely associated with EMT [23], indicating that E2F1 might promote EMT in SCLC.
Depletion of E2F1-induced cell morphology changes in H446 and H1688 cells
In our previous research, we found that the cellular morphology was changed when E2F1 gene was silenced by specific siRNAs in SCLC. To further study cellular morphology, stable E2F1 gene knockdown cells were generated using an adenovirus containing shRNA against E2F1 in H446 cells (H446-E2F1sh cells). Eight clones were randomly tested for E2F1 expression. E2F1 was efficiently knocked down in clone-4 (Fig. 2a and Additional file 5: Figure S4), and clone-4 was then used to explore the relationship between E2F1 and EMT. The cellular morphology was significantly changed in clone-4 compared to negative control cells. When E2F1 was knocked down, cells changed from being grain-shaped to appearing slender and fibrous (Fig. 2b). The same change was observed when E2F1 was silenced using siRNA against E2F1 in H1688 cells (Fig. 2a, c and Additional file 5: Figure S4). Combined with our previous results [2, 3], we considered that E2F1 might regulate EMT to promote invasion and metastasis in SCLC.
Fig. 2.
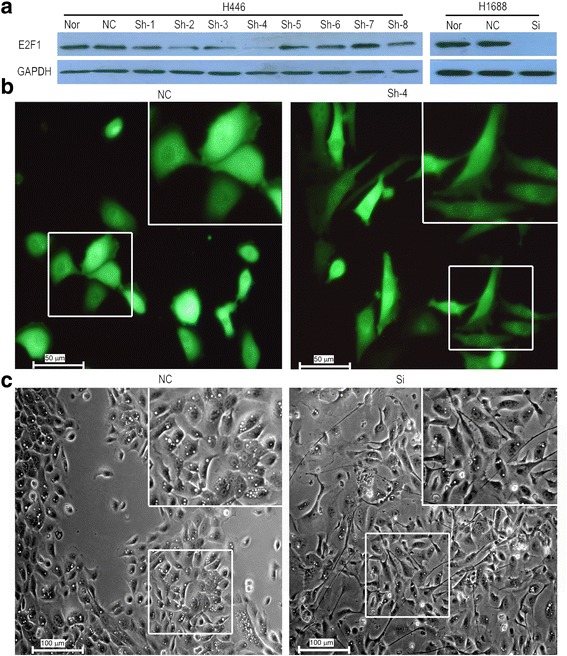
The cellular morphology was changed when E2F1 was depleted in SCLC cells. a E2F1 protein was examined in 8 clones where E2F1 was knocked down by shRNA in H446 cells (H446-E2F1sh) and transiently silenced by E2F1 specific siRNA in H1688 cells (H1688-E2F1si). b Cellular morphology was changed from grain-shaped to slender and fibrous in clone-4 of H446-E2F1sh and H1688-E2F1si cells. Because shRNA vector contained eGFP, H446-E2F1sh cells were green. The magnification of H446 (panel b) was 400 X, and the magnification of H1688 (panel c) was 200 X
The expression of cytoskeletal proteins was changed when E2F1 was knocked down.
Because a depletion of E2F1 changed the cell morphology, the expression of cytoskeletal proteins was tested. The expression levels of α-tubulin and β-actin were examined in H446 and H1688 cells by western blot and immunofluorescence. When E2F1 was knocked down in H446 cells, α-tubulin and β-actin were decreased (Fig. 3a and b). The same results were observed in H1688 cells when E2F1 was silenced by E2F1 specific siRNA (Fig. 3a and b). These results coincide with those shown in Fig. 2, indicating that E2F1 affects cytoskeletal protein expression in SCLC, further implying that the role of E2F1 is important in the process of EMT.
Fig. 3.
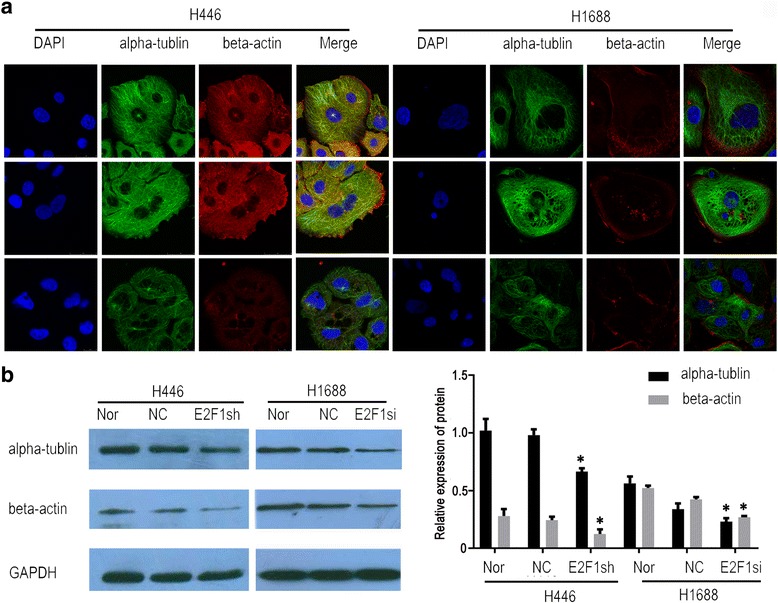
Depletion of E2F1 influenced the expression of cellular cytoskeletal proteins. The expression levels of cellular cytoskeletal proteins including α-tublin and β-actin were decreased by immunofluorescence (a) and western blotting (b) in H446-E2F1sh and H1688-E2F1si cells. The immunofluorescence magnification was 630 X, and the levels of alpha-tubulin and beta-actin were calculated by mean ± standard deviation in 3 independent experiments. * represents p < 0.05
Expression of E2F family members when E2F1 was knocked down in H446 cells and silenced in H1688 cells
The E2F family has 8 members, from E2F1 to E2F8. Although they have similar DNA binding domains, the target genes controlled by E2F family members are different [24]. When E2F1 was stably knocked down in H446 cells, other E2F family members could have compensated for E2F1 function. We therefore tested the expression of other E2F family members with qPCR in H446 and H1688 cells. The results showed that the E2F1 expression was significantly knocked down (p = 0.0007), and the expressions of E2F2, E2F3, E2F5 and E2F8 mRNA was up-regulated, but not to statistically significant levels in H446-E2F1sh cells (Fig. 4). In H1688-E2F1si cells, E2F1 expression was significantly silenced (p = 0.00026) and the mRNA levels of other E2F family members were almost no changed (Fig. 4). These results showed that E2F family members did not compensate for E2F1 when E2F1 gene was stably knocked out in SCLC cell line.
Fig. 4.
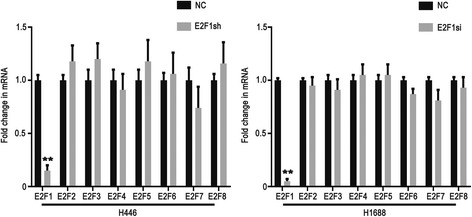
Other E2F family members were detected by real-time PCR when E2F1 was depleted. The expression levels of other E2F family members were examined by real-time PCR in H446-E2F1sh and H1688-E2F1si cells. The p value and error bar was calculated in 3 independent experiments. ** represents p < 0.001
E2F1 promoted EMT occurrence in SCLC
A depletion of E2F1 changed the expression of cytoskeletal proteins, and other members of E2F1 family did not compensate E2F1 function. Next, epithelial and mesenchymal markers (CDH1, CTNNB1 and VIM, CDH2) were examined in SCLC cell lines. Real-time PCR showed that CTNNB1 was significantly increased, while VIM and CDH2 were significantly decreased in H446-E2F1sh cells. Additionally, CDH1 was decreased, but this change was not statistically significant (Fig. 5a). The protein levels of these EMT markers were consistent with the mRNA levels, with the exception of CDH1 (Fig. 5b, c and Additional file 6: Figure S5A). In H446-E2F1sh cells, we used two different CDH1 antibodies, but CDH1 was not detected by western blot and immunofluorescence. Although CDH1 was highly expressed in SCLC tissue samples (Fig. 1a), it was not present in H446 cells. Therefore, we selected the other SCLC cell line, H1688, to test for CDH1 expression. When E2F1 was transiently silenced by siRNAs in H1688 cells, CDH1 expression was significantly increased at both the mRNA and protein levels (Fig. 5d, Additional file 6: Figure S5B). To further certify that E2F1 could affect the expressions of EMT markers, we selected the lower E2F1 cell line A549 to verify this result. Compared with H1688 and H446, E2F1 expression was lower in A549 cells [2]. Next, E2F1 was transfected into A549 cells (named A549-E2F1). In A549-E2F1 cells, we found that CDH1 and CTNNB1 were decreased, and VIM and CDH2 were increased in mRNA and protein levels (Additional file 7: Figure S6). These results further indicated that E2F1 could affect the expressions of EMT related markers, and promote EMT occurrence.
Fig. 5.
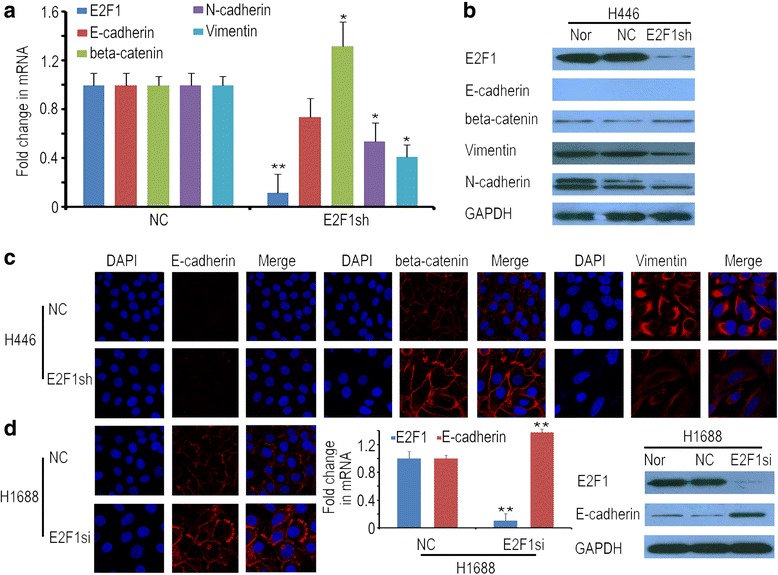
E2F1 influenced the expression of EMT-related proteins. The expression levels of CDH1, CTNNB1, CDH2 and VIM were tested by real-time PCR (a), western blotting (b) and immunofluorescence (c). Because CDH1 protein was not been detected in H446 cells, we selected H1688 to detect the CDH1 expression (d). * represents p < 0.05, ** represents p < 0.001
The expression of CDH1 related inhibitory transcription factors in SCLC tissue samples
The above results showed that E2F1 promotes EMT, but the mechanism by which it does so is unknown. Previous ChIP-seq studies that examined the target genes of E2F1 in SCLC found that CDH1, CTNNB1, VIM and CDH2 were not directly regulated by E2F1 [2, 25], indicating that E2F1 does not directly control EMT. In addition, ChIP-seq data showed that only ZEB2 not other known EMT transcription factors was a target of E2F1. We then investigated the expression of CDH1 inhibitory transcription factors ZEB1, ZEB2, SNAI1 and SNAI2 by immunohistochemistry. The results showed that ZEB2 was the highest among transcriptional repressors of CDH1 (Fig. 6). ZEB2 was found to be strong positive (score between 100 and 300) in 76.67% (46/60) SCLC tissue samples, moderate positive (score between 20 and 100) in 5% (3/60), weak expression (score between 10 and 20) in 6.67% (4/60), and negative expression (score 0) in 11.67% (7/60, Additional file 8: Figure S7). ZEB1 was mainly localized to mesenchymal cells (60/60), not tumour cells (0/60). SNAI1 was mainly localized to the vascular endothelium (55/60). SNAI2 was very weak and only found in a small number of tumour cells (2/60) (Fig. 6). These results were consistent with other reports [15, 18], and indicated that ZEB2 might play an important role in promoting EMT in SCLC.
Fig. 6.
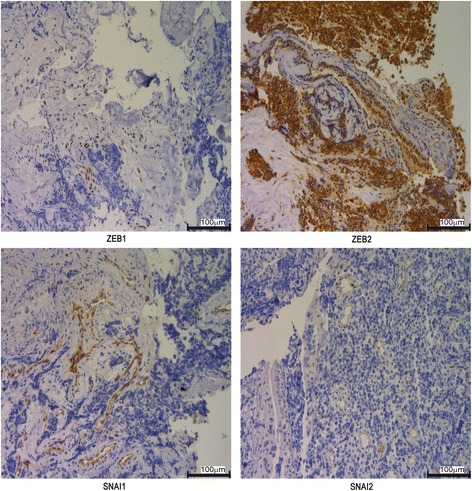
Expressions of ZEB1, ZEB2, SNAI1 and SNAI2 were analysed by IHC in SCLC tissue samples. ZEB1, ZEB2, SNAI1 and SNAI2 were detected in 60 SCLC tissue samples by IHC. Representative image was presented, and the magnification is 200 X
E2F1 promoted EMT occurrence in SCLC by regulating ZEB2 expression
Although ZEB1 and SNAI1 have been found to promote invasion and metastasis in SCLC [26, 27], ZEB1 and SNAI1 was very low in SCLC tissue. We analysed the expression of ZEB1, ZEB2, SNAI1 and SNAI2 in H446-E2F1sh and H1688-E2F1si cells by real-time PCR. ZEB2 mRNA was reduced by 60% and 79%, respectively in H446-E2F1sh and H1688-E2F1si cells, and ZEB2 protein was significantly decreased (Fig. 7a). These results suggest that E2F1 might promote EMT in SCLC by regulating ZEB2 expression. To test this hypothesis, we found that E2F1 was higher in these SCLC tissues where ZEB2 was strong positive (Fig. 7 b). Our ChIP-seq data (Additional file 9) [2] showed that ZEB2 was the only target EMT known transcription repressor factor regulated by E2F1. To further certify that E2F1 could regulate ZEB2 expression in SCLC, we constructed the dual luciferase reporter vectors containing ZEB2 promoter. After transfection, luciferase activity analysis showed that E2F1 could regulate ZEB2 expression in H446 and H1688 cells (Fig. 7c). These results further certified our hypothesis that E2F1 promotes EMT by regulating ZEB2 in SCLC.
Fig. 7.
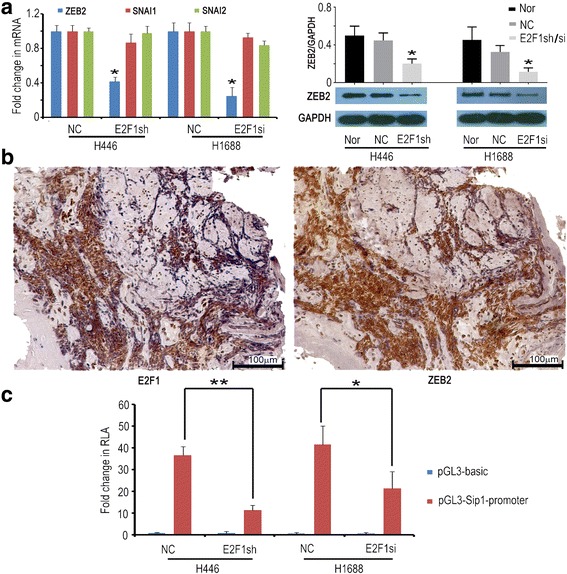
E2F1 could regulate ZEB2 expression in SCLC cell lines. a The expression levels of ZEB2, SNAI1 and SNAI2 mRNA were detected by real-time PCR in H446 and H1688 cells. ZEB2 protein was detected in H446-E2F1sh and H1688-E2F1si cells. mRNA levels were calculated in 3 independent experiments. * represents p < 0.05. b ZEB2 was highly expressed in tissue samples in which E2F1 was also high. Representative image was presented, and the magnification is 200 X. c Dual luciferase activity analysis from 3 independent experiments showed that E2F1 could directly regulate ZEB2 expression in H446 and H1688 cells, * represents p < 0.05, ** represents p < 0.001. RLA: Relative luciferase activity
Discussion
E2F1 is a transcription factor that takes part in regulating various biological activities, including the cell cycle [28], apoptosis [29], proliferation [30], angiogenesis [31], tumour drug resistance [32, 33], invasion and metastasis [34, 35] and so on. In our previous studies, we found that E2F1 could regulate MMPs and ADAM-12 to promote invasion and metastasis in SCLC [2, 3]. Recently, several papers have reported that E2F1 can induce EMT in different tumour cells, but the mechanisms described in these studies are inconsistent [13, 36]. In addition, E2F1 was shown to be deregulated upon the loss of Rb in SCLC [37]. Therefore, a role for E2F1 in SCLC has been indicated. Distant metastasis in early stages is a classic biological feature of SCLC, and EMT is considered to be an early, essential step for invasion and metastasis. It is unknown how E2F1 regulates EMT in SCLC. In this study, we detected and analysed the expression of EMT markers in SCLC tissue samples and found that CDH1, CTNNB1 and VIM were closely related with clinical stage and differentiation [38]. When E2F1 was depleted, the cellular morphology changed from grain-shaped to slender and fibrous. Additionally, cytoskeletal proteins underwent remodelling and EMT markers were significantly changed, indicating that E2F1 plays a role in EMT in SCLC. Next, we found that ZEB2 was highly expressed in the same tissues in which E2F1 was highly expressed. ChIP-seq data and luciferase analysis indicated that E2F1 could control ZEB2 expression in SCLC.
EMT is a process whereby epithelial cell polarity is lost, cellular adhesions are weakened, and the cytoskeleton is remodelled [10]. Tumour cells are surrounded by mesenchymal cells. For tumour cells to move a distance, they must break through the surrounding mesenchymal cells. To do this, they have to disguise themselves as mesenchymal cells. EMT is considered to be the first step in breaking through the mesenchymal cell defence. A loss of the CDH1/CTNNB1 complex is a marker of EMT occurrence. This complex plays an important role in mediating intercellular adherence junction and maintaining epithelial integrity [39]. A “Cadherin Switch” refers to the change from CDH1 to CDH2 and is considered to be the key factor in tumour invasion and metastasis [40]. The expressions of CDH1 and CTNNB1 were significantly weaker in tumour tissue than normal bronchial epithelial cells [41]. In our study, CDH2 was not detected in SCLC tissue samples, but was decreased in H446-E2F1sh cells. In H1688 cells, when E2F1 was silenced, CDH1 was increased and CDH2 was decreased. In addition, CDH1 expression was decreased and CDH2 expression was increased in A549-E2F1 cells. A previous study showed that ZEB2 played an important role in the “Cadherin Switch” during cranial neural crest EMT [42]. In our study, we found that ZEB2 was regulated by E2F1 in SCLC, and we speculate that E2F1 might drive the “Cadherin Switch” to further promote EMT in SCLC.
Interestingly, some papers reported that EMT rarely occurs homogenously across the whole tumour, and it is hypothesized that EMT is transient and occurs at the tumour margin tissues [23, 43, 44]. In our study, we found that VIM is higher in adjacent mesenchymal tumour cells, and is very lower in the tumour central sites. This result further confirmed this conclusion that EMT is transient and occurs at the tumour margin tissues. The same results were also observed in non-small cell lung cancer (NSCLC). Mahmood MQ et al. found that VIM and CDH2 were higher in tumour cells located at the peripheral leading edge of NSCLC when compared with centrally located tumour cells of same subjects [45].
ZEB2 is expressed in various human tumours, including liver cancer [46], colorectal cancer [47] and breast cancer [48]. Our results showed that ZEB2 was higher than ZEB1, SNAI1 and SNAI2 in SCLC tissue samples, which is inconstant with other reports [49, 50]. Additionally, research on ZEB2 as an EMT facilitator has been focused on because of its role as a transcriptional repressor of CDH1 [51], with little research being done on the regulation of ZEB2 expression. SNAI11 could increase ZEB2 expression at the translational level, as opposed to the transcriptional level, by inhibiting the splicing of the 5′-UTR in the ZEB2 intron [52]. Integrative genomic analyses showed that SMAD, ETS1, HIF1α, POU/OCT and NF-κB could affect ZEB2 transcription [53]. In our study, we found that E2F1 expression was highly consistent with ZEB2 expression. ChIP-seq data and luciferase activity also showed that E2F1 regulated ZEB2 expression through E2F1 binding sites on the promoter of ZEB2 in SCLC.
In summary, our data supports the idea that E2F1 promotes EMT by regulating ZEB2 expression in SCLC.
Conclusion
Transcript factor E2F1 promotes EMT by regulating ZEB2 gene expression, and then participates in invasion and metastasis of SCLC.
Additional files
The specific primers for target genes. (DOCX 19 kb)
The differential expression intensity of CDH1 and CTNNB1 in SCLC tissue samples. (TIFF 10260 kb)
The staining of CDH1 and CTNNB1 in bronchial epithelial cells. (TIFF 6069 kb)
The expression levels of CDH1 and CTNNB1 in differential SCLC cell lines. (TIFF 13906 kb)
E2F1 expression was quantified in H446-E2F1sh and H1688-E2F1si cells. * represents p < 0.05, ** represents p < 0.001. (TIFF 3949 kb)
The relative protein levels of E2F1, CTNNB1, VIM and CDH2 in H446-E2F1sh cells, and E2F1, CTNNB1 in H1688-E2F1si cells. (TIFF 533 kb)
E2F1 overexpression in A549 cells could inhibit the expression of CDH1 and CTNNB1, and promote the expression of CDH2. (TIFF 6963 kb)
The differential expression intensity of ZEB2 in SCLC tissue samples. (TIFF 7166 kb)
The target genes of E2F1 by ChIP-seq in H1688 cell line. (XLSX 431 kb)
Acknowledgements
None.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81302017) and Natural Science Foundation of Shandong (grant no. ZR2013HL004). The funder was not involved in designing the study, collecting or analysing the data, or preparing the manuscript.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Author’s contributions
TW performed the experimental operations including cell culture, immunohistochemistry, real-time PCR, western blotting and immunofluorescence. XC participated in the design and was involved in drafting the manuscript. WQ and LK constructed the vectors, performed the transfection, siRNA and shRNA experiments and the luciferase analysis. DS and ZL conceived and designed this study, and ZL wrote this manuscript. The revision of the manuscript was performed by all of the authors. The approval of publication was obtained from all of the authors.
Abbreviations
- ADAM
A disintegrin and metalloprotease
- ChIP-seq
Chromosome immunoprecipitation to sequencing
- EMT
Epithelial-mesenchymal transition
- MMPs
matrix metalloproteinases
- PDGFR
platelet-derived growth factor receptor
- SCLC
small cell lung cancer
- VEGFR
vascular endothelial growth factor receptor
Ethics approval and consent to participate
Sixty SCLC biopsy tissue samples were obtained from the affiliated hospital of Binzhou Medical University from January 2014 to January 2015. All patients signed informed consent forms before providing tissue samples. This research was approved by the Medical Ethics Committee of Binzhou Medical University (No. 2013027). This study was performed according to the Declaration of Helsinki and to the relevant ethical guidelines for research on humans.
Consent for publication
Not applicable.
Competing interests
All authors declare that there are no conflicts of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12885-017-3701-y) contains supplementary material, which is available to authorized users.
Contributor Information
Tingting Wang, Email: christinawang0206@163.com.
Xufang Chen, Email: ytchenxf@163.com.
Weiwei Qiao, Email: qwwi5@126.com.
Lijun Kong, Email: kong_lijun@163.com.
Daqing Sun, Email: sundaqingchris@126.com.
Zunling Li, Email: lizunling@bzmc.edu.cn.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, XQ Y, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Li Z, Guo Y, Jiang H, Zhang T, Jin C, Young CY, Yuan H. Differential regulation of MMPs by E2F1, Sp1 and NF-kappa B controls the small cell lung cancer invasive phenotype. BMC Cancer. 2014;14:276. doi: 10.1186/1471-2407-14-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Z, Wang Y, Kong L, Yue Z, Ma Y, Chen X. Expression of ADAM12 is regulated by E2F1 in small cell lung cancer. Oncol Rep. 2015;34(6):3231–3237. doi: 10.3892/or.2015.4317. [DOI] [PubMed] [Google Scholar]
- 4.Schaffer BE, Park KS, Yiu G, Conklin JF, Lin C, Burkhart DL, Karnezis AN, Sweet-Cordero EA, Sage J. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res. 2010;70(10):3877–3883. doi: 10.1158/0008-5472.CAN-09-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chong JL, Wenzel PL, Saenz-Robles MT, Nair V, Ferrey A, Hagan JP, Gomez YM, Sharma N, Chen HZ, Ouseph M, et al. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature. 2009;462(7275):930–934. doi: 10.1038/nature08677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji W, Zhang W, Xiao W. E2F-1 directly regulates thrombospondin 1 expression. PLoS One. 2010;5(10):e13442. doi: 10.1371/journal.pone.0013442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minato Y, Tashiro E, Kanai M, Nihei Y, Kodama Y, Imoto M. Transcriptional regulation of a new variant of human platelet-derived growth factor receptor alpha transcript by E2F-1. Gene. 2007;403(1–2):89–97. doi: 10.1016/j.gene.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Pillai S, Kovacs M, Chellappan S. Regulation of vascular endothelial growth factor receptors by Rb and E2F1: role of acetylation. Cancer Res. 2010;70(12):4931–4940. doi: 10.1158/0008-5472.CAN-10-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shao S, Li Z, Gao W, Yu G, Liu D, Pan F. ADAM-12 as a diagnostic marker for the proliferation, migration and invasion in patients with small cell lung cancer. PLoS One. 2014;9(1):e85936. doi: 10.1371/journal.pone.0085936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang Y, Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118(3):277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 11.Wu Z, Zheng S, Li Z, Tan J, Yu Q. E2F1 suppresses Wnt/beta-catenin activity through transactivation of beta-catenin interacting protein ICAT. Oncogene. 2011;30(37):3979–3984. doi: 10.1038/onc.2011.129. [DOI] [PubMed] [Google Scholar]
- 12.Morris EJ, Ji JY, Yang F, Di Stefano L, Herr A, Moon NS, Kwon EJ, Haigis KM, Naar AM, Dyson NJ. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455(7212):552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knoll S, Furst K, Kowtharapu B, Schmitz U, Marquardt S, Wolkenhauer O, Martin H, Putzer BM. E2F1 induces miR-224/452 expression to drive EMT through TXNIP downregulation. EMBO Rep. 2014;15(12):1315–1329. doi: 10.15252/embr.201439392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang X, Kadara H, Behrens C, Liu DD, Xiao Y, Rice D, Gazdar AF, Fujimoto J, Moran C, Varella-Garcia M, et al. Abnormalities of the TITF-1 lineage-specific oncogene in NSCLC: implications in lung cancer pathogenesis and prognosis. Clin Cancer Res. 2011;17(8):2434–2443. doi: 10.1158/1078-0432.CCR-10-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassan WA, Yoshida R, Kudoh S, Motooka Y, Ito T. Evaluation of role of Notch3 signaling pathway in human lung cancer cells. J Cancer Res Clin Oncol. 2016;142(5):981–993. doi: 10.1007/s00432-016-2117-4. [DOI] [PubMed] [Google Scholar]
- 16.Pore M, Meijer C, de Bock GH, Boersma-van Ek W, Terstappen LW, Groen HJ, Timens W, Kruyt FA, Hiltermann TJ. Cancer stem cells, epithelial to mesenchymal markers, and circulating tumor cells in small cell lung cancer. Clinical lung cancer. 2016;17(6):535–542. doi: 10.1016/j.cllc.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton G, Hochmair M, Rath B, Klameth L, Zeillinger R. Small cell lung cancer: circulating tumor cells of extended stage patients express a mesenchymal-epithelial transition phenotype. Cell Adhes Migr. 2016;10(4):360–367. doi: 10.1080/19336918.2016.1155019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galvan JA, Astudillo A, Vallina A, Crespo G, Folgueras MV, Gonzalez MV. Prognostic and diagnostic value of epithelial to mesenchymal transition markers in pulmonary neuroendocrine tumors. BMC Cancer. 2014;14:855. doi: 10.1186/1471-2407-14-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klymkowsky MW. Beta-catenin and its regulatory network. Hum Pathol. 2005;36(3):225–227. doi: 10.1016/j.humpath.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez-Salas N, Palacios J, de Castro J, Moreno G, Gonzalez-Baron M, Gamallo C. Beta-catenin expression pattern in small cell lung cancer: correlation with clinical and evolutive features. Histol Histopathol. 2001;16(2):353–358. doi: 10.14670/HH-16.353. [DOI] [PubMed] [Google Scholar]
- 21.Polley E, Kunkel M, Evans D, Silvers T, Delosh R, Laudeman J, Ogle C, Reinhart R, Selby M, Connelly J, et al. Small cell lung cancer screen of oncology drugs, investigational agents, and gene and microRNA expression. J Natl Cancer Inst. 2016;108(10) [DOI] [PMC free article] [PubMed]
- 22.Pore M, Meijer C, de Bock GH, Boersma-van Ek W, Terstappen LW, Groen HJ, Timens W, Kruyt FA, Hiltermann TJ. Cancer stem cells, epithelial to mesenchymal markers, and circulating tumor cells in small cell lung cancer. Clinical lung cancer. 2016; [DOI] [PubMed]
- 23.Felipe Lima J, Nofech-Mozes S, Bayani J, Bartlett JM. EMT in breast carcinoma-a review. J clin med. 2016;5(7) [DOI] [PMC free article] [PubMed]
- 24.Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, Farnham PJ. A comprehensive ChIP-Chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17(11):1550–1561. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li ZL, Jiao F, Ma Y, Yue Z, Kong LJ. Target genes regulated by transcription factor E2F1 in small cell lung cancer. Sheng li xue bao : [Acta physiologica Sinica] 2016;68(3):276–284. [PubMed] [Google Scholar]
- 26.Liu Y, Zhang N, Wang Y, Xu M, Liu N, Pang X, Cao J, Ma N, Pang H, Liu L, et al. Zinc finger E-box binding homeobox 1 promotes invasion and bone metastasis of small cell lung cancer in vitro and in vivo. Cancer Sci. 2012;103(8):1420–1428. doi: 10.1111/j.1349-7006.2012.02347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merikallio H, Turpeenniemi-Hujanen T, Paakko P, Makitaro R, Riitta K, Salo S, Salo T, Harju T, Soini Y. Snail promotes an invasive phenotype in lung carcinoma. Respir Res. 2012;13:104. doi: 10.1186/1465-9921-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ertosun MG, Hapil FZ, Osman Nidai O. E2F1 transcription factor and its impact on growth factor and cytokine signaling. Cytokine Growth Factor Rev. 2016; [DOI] [PubMed]
- 29.Kumari A, Iwasaki T, Pyndiah S, Cassimere EK, Palani CD, Sakamuro D. Regulation of E2F1-induced apoptosis by poly(ADP-ribosyl)ation. Cell Death Differ. 2015;22(2):311–322. doi: 10.1038/cdd.2014.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang E, Yin D, Han L, He X, Si X, Chen W, Xia R, Xu T, Gu D, De W, et al. E2F1-induced upregulation of long noncoding RNA LINC00668 predicts a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically silencing of CKIs. Oncotarget. 2015; [DOI] [PMC free article] [PubMed]
- 31.Engelmann D, Mayoli-Nussle D, Mayrhofer C, Furst K, Alla V, Stoll A, Spitschak A, Abshagen K, Vollmar B, Ran S, et al. E2F1 promotes angiogenesis through the VEGF-C/VEGFR-3 axis in a feedback loop for cooperative induction of PDGF-B. J Mol Cell Biol. 2013;5(6):391–403. doi: 10.1093/jmcb/mjt035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan LH, Wei WY, Cao WL, Zhang XS, Xie YB, Xiao Q. Overexpression of E2F1 in human gastric carcinoma is involved in anti-cancer drug resistance. BMC Cancer. 2014;14:904. doi: 10.1186/1471-2407-14-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alla V, Kowtharapu BS, Engelmann D, Emmrich S, Schmitz U, Steder M, Putzer BM. E2F1 confers anticancer drug resistance by targeting ABC transporter family members and Bcl-2 via the p73/DNp73-miR-205 circuitry. Cell Cycle. 2012;11(16):3067–3078. doi: 10.4161/cc.21476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hollern DP, Honeysett J, Cardiff RD, Andrechek ER. The E2F transcription factors regulate tumor development and metastasis in a mouse model of metastatic breast cancer. Mol Cell Biol. 2014;34(17):3229–3243. doi: 10.1128/MCB.00737-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang YX, JM L, Mo RJ, He HC, Xie J, Jiang FN, Lin ZY, Chen YR, YD W, Luo HW, et al. E2F1 promotes tumor cell invasion and migration through regulating CD147 in prostate cancer. Int J Oncol. 2016;48(4):1650–1658. doi: 10.3892/ijo.2016.3364. [DOI] [PubMed] [Google Scholar]
- 36.Pillai S, Trevino J, Rawal B, Singh S, Kovacs M, Li X, Schell M, Haura E, Bepler G, Chellappan S. beta-arrestin-1 mediates nicotine-induced metastasis through E2F1 target genes that modulate epithelial-mesenchymal transition. Cancer Res. 2015;75(6):1009–1020. doi: 10.1158/0008-5472.CAN-14-0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niederst MJ, Sequist LV, Poirier JT, Mermel CH, Lockerman EL, Garcia AR, Katayama R, Costa C, Ross KN, Moran T, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun. 2015;6:6377. doi: 10.1038/ncomms7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao QJ, Panetti C, Marconi S, Krebs P, Lorenzana RR, Goulart RA, Bur ME. Vimentin and leukocyte common antigen-negative molding cells in pleural effusions of patients with small cell lung carcinoma. Acta Cytol. 2003;47(6):1033–1037. doi: 10.1159/000326641. [DOI] [PubMed] [Google Scholar]
- 39.Gavilan MP, Arjona M, Zurbano A, Formstecher E, Martinez-Morales JR, Bornens M, Rios RM. Alpha-catenin-dependent recruitment of the centrosomal protein CAP350 to adherens junctions allows epithelial cells to acquire a columnar shape. PLoS Biol. 2015;13(3):e1002087. doi: 10.1371/journal.pbio.1002087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Priya R, Yap AS. Making a choice: how cadherin switching controls cell migration. Dev Cell. 2015;34(4):383–384. doi: 10.1016/j.devcel.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Miao Y, Li AL, Wang L, Fan CF, Zhang XP, HT X, Han Y, Liu Y, Wang E. Expression of p130cas, E-cadherin and beta-catenin and their correlation with clinicopathological parameters in non-small cell lung cancer: p130cas over-expression predicts poor prognosis. Folia Histochem Cytobiol. 2012;50(3):392–397. doi: 10.5603/FHC.2012.0053. [DOI] [PubMed] [Google Scholar]
- 42.Rogers CD, Saxena A, Bronner ME. Sip1 mediates an E-cadherin-to-N-cadherin switch during cranial neural crest EMT. J Cell Biol. 2013;203(5):835–847. doi: 10.1083/jcb.201305050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matejka VM, Finek J, Kralickova M. Epithelial-mesenchymal Transition in Tumor Tissue and Its Role for Metastatic Spread of Cancer. Klinicka onkologie : casopis Ceske a Slovenske onkologicke spolecnosti. 30(1):20–7. [DOI] [PubMed]
- 44.Alkatout I, Wiedermann M, Bauer M, Wenners A, Jonat W, Klapper W. Transcription factors associated with epithelial-mesenchymal transition and cancer stem cells in the tumor centre and margin of invasive breast cancer. Exp Mol Pathol. 2013;94(1):168–173. doi: 10.1016/j.yexmp.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Mahmood MQ, Ward C, Muller HK, Sohal SS, Walters EH. Epithelial mesenchymal transition (EMT) and non-small cell lung cancer (NSCLC): a mutual association with airway disease. Med Oncol. 2017;34(3):45. doi: 10.1007/s12032-017-0900-y. [DOI] [PubMed] [Google Scholar]
- 46.Li YM, Xu SC, Li J, Han KQ, Pi HF, Zheng L, Zuo GH, Huang XB, Li HY, Zhao HZ, et al. Epithelial-mesenchymal transition markers expressed in circulating tumor cells in hepatocellular carcinoma patients with different stages of disease. Cell Death Dis. 2013;4:e831. doi: 10.1038/cddis.2013.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kahlert C, Lahes S, Radhakrishnan P, Dutta S, Mogler C, Herpel E, Brand K, Steinert G, Schneider M, Mollenhauer M, et al. Overexpression of ZEB2 at the invasion front of colorectal cancer is an independent prognostic marker and regulates tumor invasion in vitro. Clin Cancer Res. 2011;17(24):7654–7663. doi: 10.1158/1078-0432.CCR-10-2816. [DOI] [PubMed] [Google Scholar]
- 48.Elloul S, Elstrand MB, Nesland JM, Trope CG, Kvalheim G, Goldberg I, Reich R, Davidson B. Snail, slug, and Smad-interacting protein 1 as novel parameters of disease aggressiveness in metastatic ovarian and breast carcinoma. Cancer. 2005;103(8):1631–1643. doi: 10.1002/cncr.20946. [DOI] [PubMed] [Google Scholar]
- 49.Fang S, Zeng X, Zhu W, Tang R, Chao Y, Guo L. Zinc finger E-box-binding homeobox 2 (ZEB2) regulated by miR-200b contributes to multi-drug resistance of small cell lung cancer. Exp Mol Pathol. 2014;96(3):438–444. doi: 10.1016/j.yexmp.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 50.Krohn A, Ahrens T, Yalcin A, Plones T, Wehrle J, Taromi S, Wollner S, Follo M, Brabletz T, Mani SA, et al. Tumor cell heterogeneity in small cell lung cancer (SCLC): phenotypical and functional differences associated with epithelial-mesenchymal transition (EMT) and DNA methylation changes. PLoS One. 2014;9(6):e100249. doi: 10.1371/journal.pone.0100249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vandewalle C, Comijn J, De Craene B, Vermassen P, Bruyneel E, Andersen H, Tulchinsky E, Van Roy F, Berx G. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005;33(20):6566–6578. doi: 10.1093/nar/gki965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22(6):756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katoh M, Katoh M. Integrative genomic analyses of ZEB2: transcriptional regulation of ZEB2 based on SMADs, ETS1, HIF1alpha, POU/OCT, and NF-kappaB. Int J Oncol. 2009;34(6):1737–1742. doi: 10.3892/ijo_00000304. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The specific primers for target genes. (DOCX 19 kb)
The differential expression intensity of CDH1 and CTNNB1 in SCLC tissue samples. (TIFF 10260 kb)
The staining of CDH1 and CTNNB1 in bronchial epithelial cells. (TIFF 6069 kb)
The expression levels of CDH1 and CTNNB1 in differential SCLC cell lines. (TIFF 13906 kb)
E2F1 expression was quantified in H446-E2F1sh and H1688-E2F1si cells. * represents p < 0.05, ** represents p < 0.001. (TIFF 3949 kb)
The relative protein levels of E2F1, CTNNB1, VIM and CDH2 in H446-E2F1sh cells, and E2F1, CTNNB1 in H1688-E2F1si cells. (TIFF 533 kb)
E2F1 overexpression in A549 cells could inhibit the expression of CDH1 and CTNNB1, and promote the expression of CDH2. (TIFF 6963 kb)
The differential expression intensity of ZEB2 in SCLC tissue samples. (TIFF 7166 kb)
The target genes of E2F1 by ChIP-seq in H1688 cell line. (XLSX 431 kb)
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files.


