ABSTRACT
Objective: Hormonal therapy is an important component of first line of treatment for breast cancer. Response to hormonal therapy is influenced by the progesterone receptor (PR)-status of breast cancer patients. However as an early effect, exposure to progesterone decreases expression of PR in breast cancer cells. An understanding of the mechanism underlying down-regulation of PR could help improve response to hormonal therapy. Methods: We performed small RNA sequencing of breast cancer cells for identification of microRNAs targeting PR in response to progesterone treatment. Biochemical approaches were used to validate the findings in breast cancer cells. Results: Analysis of small RNA sequencing of four breast cancer cell lines treated with progesterone revealed an up-regulation of miR-129-2 independent of the PR status of the cells. We show that miR-129-2 targets 3′UTR of PR to down-regulate its expression. Furthermore, inhibition of miR-129-2 expression rescues the down-regulation of PR in breast cancer cells. Also, the expression levels of miR-129-2 was observed to be elevated in patients with low expression of PR in the TCGA cohort (n = 359). Conclusion: miR-129-2 mediates down-regulation of PR in breast cancer cells in response to progesterone, while anti-miR-129-2 could potentiate PR expression levels among patients with inadequate PR levels. Thus, modulation of activity of miR-129-2 could stabilize PR expression and potentially improve response to hormonal therapy under adjuvant or neo-adjuvant settings.
KEYWORDS: Breast cancer, hormonal therapy, progesterone, progesterone receptor, microRNA, cancer genomics, small RNA sequencing
Introduction
Breast cancer is the most prevalent cancer among women worldwide. Despite all advances in early diagnosis and treatment, nearly 30% node-negative and 70% node-positive patients relapse with metastatic disease.1 Treatment of breast cancer patients is influenced by the presence of estrogen receptor (ER) and progesterone receptor (PR). ER/PR positive patients tend to respond better to hormonal therapy and have a lower risk of relapse compared to ER-positive, PR-negative patients.2 The down-regulation of PR expression in breast cancer cells is caused either by methylation at PR promoter3, or in response to progesterone by post translational modification of the PR protein by CUEDC2 and MAPK.4,5 Growing evidence also suggest microRNAs to respond to steroid hormones and suppress the activity of respective hormone receptor.6 For instance, miR-18a, miR-19b and miR-20b (paralogous pri-microRNAs) down-regulate the expression of ER in response to estrogen in breast cancer.7 Comparatively, similar regulation of microRNA expression in response to progesterone has been less explored.8 In order to study the progesterone-regulated microRNAs targeting PR, we performed small RNA sequencing of breast cancer cell lines treated with progesterone. The differentially expressed microRNAs were used to identify microRNAs that target 3′UTR of PR. Our analysis reveals miR-129-2 targets PR and is up-regulated in response to progesterone. The association of miR-129-2 and PR was functionally validated by luciferase assay. Also western blot analysis suggests that inhibition of miR-129-2 stabilizes PR in breast cancer cells even in presence of progesterone. Moreover, patients with high miR-129-2 levels had significantly lower expression of PR as compared to patients with no miR-129-2 expression in The Cancer Genome Atlas cohort.
Results
Identification of progesterone responsive microRNAs targeting PR expression in breast cancer cells
Consistent with earlier reports, we observed down-regulation of PR transcripts when T47D and BT474 cells were treated with 10 nM progesterone for 6 h (Fig. 1a). Similarly, progesterone reduced the expression level of PR protein in T47D cells (Fig. 1c). To understand the role of microRNA's involved in regulation of PR expression, we performed small RNA sequencing of three PR-positive T47D, BT474, MCF7 and one PR-negative MDA-MB-231 cell line for identifying microRNAs which could down-regulate PR expression in response to progesterone treatment for 6 h. On an average we obtained 22 million reads per sample per cell line. The sequence reads were mapped to human microRNA sequences obtained from miRBase (version 21) to identify median 800 mature microRNA sequences. These microRNA reads were used for identification of differentially expressed microRNAs. We used a fold-change cut-off of 3-fold difference and observed that progesterone had an effect in both directions by up-regulating and down-regulating the microRNAs and all the four cells had different number of de-regulated microRNAs (Supplementary Table 1). Of these, 98 microRNAs were up-regulated in T47D, 96 in BT474, 189 in MCF7 and 106 in MDA-MB-231 cells in response to progesterone. Intriguingly, expression of miR-513a-5p shown to be differentially up-regulated in response to synthetic progestin (medroxy progesterone acetate, MPA) by microarray-based analysis in T47D cells was not observed in any of the four breast cancer cells in this study8, possibly due to variable downstream effects elicited by synthetic progestin (MPA) and progesterone9 or distinct platform specific threshold involved in these studies. The up-regulated microRNAs found across the four breast cancer cells were further used to search microRNAs targeting 3′UTR of PR gene. Of the 6 different algorithms used, we found three microRNAs (miR-3908, miR-129-2-3p and miR-3140-3p) that were predicted to target 3′ UTR of PR and showed an increased expression relative to levels in control. When expression of these microRNAs was checked in the TCGA breast cancer cohort (n = 359), only miR-129-2 was found to be expressed. Next, the up-regulation of miR-129-2 in response to progesterone could be validated by real-time PCR in our panel of cells (Fig. 1b). While the progestin-regulated miR-513a-5p could be validated only at 100 nM progestin (MPA) as reported by Cochrane et al.8, we could validate consistent up-regulation of miR-129-2 in response to 10 nM progesterone that was used for small RNA sequencing analysis, inclusive of PR-negative breast cancer cells (Fig. 1b). Thus we observed that progesterone mediated up-regulation of miR-129-2 was independent of the PR expression of cells.
Figure 1.
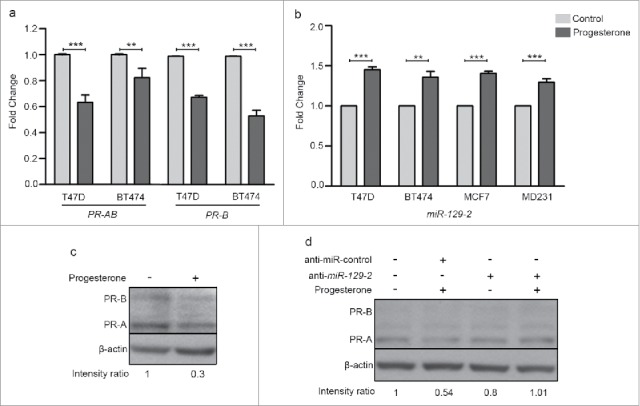
Progesterone receptor is down-regulated in breast cancer cell lines in response to progesterone. (a) Transcript levels of PR were measured using real-time PCR in T47D and BT474 cells treated with 10 nM progesterone for 6 h. Graph has been plotted as fold change expression of PR normalized to GAPDH in progesterone-treated versus control. Analysis is representative of three independent experiments and P-value was calculated using student's unpaired t-test. (b) Transcript levels of miR-129-2 under similar progesterone treatment conditions were measured by real-time PCR and plotted as fold change in progesterone versus control of T47D, BT474, MCF7 and MDA-MB-231 cells obtained after normalization to expression of U6 small RNA. Transcript levels in both control and progesterone-treated cells have been shown. Analysis is representative of three independent experiments and P-value was calculated using student's unpaired t-test. ** indicates P-value <0.001; *** indicates P-value <0.0001. (c) Western blot analysis of PR (PR-A and PR-B) in response to progesterone treatment in T47D cells. β-actin was used internal protein loading control. Numbers on blot indicate ratio of intensity of PR with respect to β-actin for each lane. Western blot analysis is representative of two independent experiments. (d) Western blot analysis of PR (PR-A and PR-B) in T47D cells treated with either anti-miR-control or anti-miR-129-2. As indicated in the panel, cells were either treated with progesterone or untreated. β-actin was used as internal protein loading control. Numbers on blot indicate ratio of intensity of PR with respect to β-actin for each lane. Western blot analysis is representative of two independent experiments.
Functional validation of miR-129-2 based regulation of progesterone receptor
In our attempt to functionally characterize the association of miR-129-2 with PR, a ∼1000 bp 3′UTR of PR (containing seed sequence for miR-129-2) was cloned downstream to luciferase gene in a pGL3-promoter vector. The sequence for premature miR-129-2 (∼400bp) was cloned in pcDNA3.1 vector. Three set of transfections were performed— pGL3-PR 3′UTR; pGL3-PR 3′UTR with pcDNA-3.1-miR-129-2; and, combination of pGL3-PR 3′UTR, pcDNA3.1-miR-129-2 and anti-miR-129-2 (Fig. 2a). The expression of firefly luciferase gene was analyzed using firefly luciferase reporter assay system and normalized to renilla expression, which was used as internal control, in each of these sets. Our analysis suggests that upon over-expression of miR-129-2, the luciferase signal was significantly reduced as compared to signal in vector only cells. Addition of microRNA inhibitor against miR-129-2 (anti-miR-129-2), a double stranded RNA sequence which is complimentary to and specifically targets miR-129-2, reversed the repression and showed an increase in luciferase signal (Fig. 2b). Next, we inhibited miR-129-2 in T47D cells and compared the PR expression in these cells with the expression in cells transfected with negative control (targeting miR-29a) inhibitor. Our western blot analysis suggests that upon exposure to progesterone, T47D cells transfected with negative control showed decrease in PR expression, while PR showed stable expression in cells transfected with miR-129-2 inhibitor even in the presence of progesterone treatment (Fig. 1d). Thus our results provide basis for direct interaction of miR-129-2 with PR, where in addition to previous findings, we demonstrate that over-expression of miR-129-2 mimics the effect of progesterone treatment to down-regulate PR and that inhibition of miR-129-2 abrogates its interaction with PR in breast cancer cells. Taken together, these studies emphasize the plurality in microRNA-mediated feedback regulation of PR.
Figure 2.
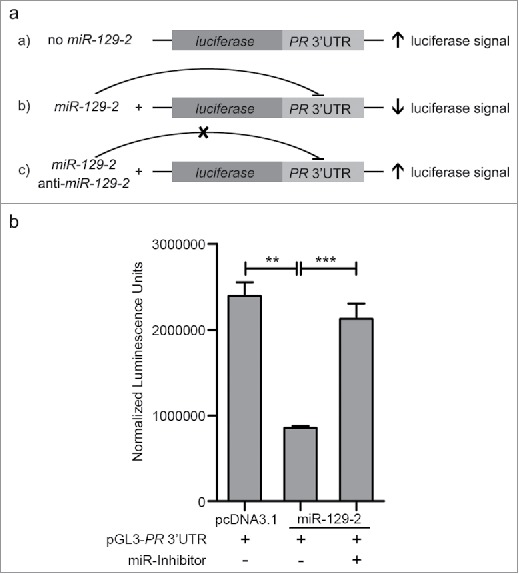
Validation of miR-129-2-based regulation of PR. (a) pCDNA3.1-miR-129-2 and pGL3- PR 3′UTR in different combinations with anti-miR-129-2 were co-transfected in 293FT cells and luciferase signal in each condition was measured, as shown in the figure. (b) Quantified luminescence units normalized to renilla expression was plotted for each of the sets mentioned above. Analysis is representative of three independent experiments and the P-value was calculated using student's t-test. ** indicates P-value <0.001; *** indicates P-value <0.0001.
Next, we analyzed the TCGA breast cancer cohort (n = 359) for studying expression of PR in breast cancer patients with high miR-129-2 expression and in absence of miR-129-2 expression. When expression of PR was checked in patients with high miR-129-2 expression (n = 134) versus patients with absence of miR-129-2 expression (n = 225) we observed a significantly higher expression of PR in patients with absence of miR-129-2 expression as compared to patients with high expression of miR-129-2 (P = 0.0002) as shown in Fig. 3. Thus a further in-depth analysis needs to be carried out to ascertain the exact role of miR-129-2 in survival of breast cancer patients.
Figure 3.
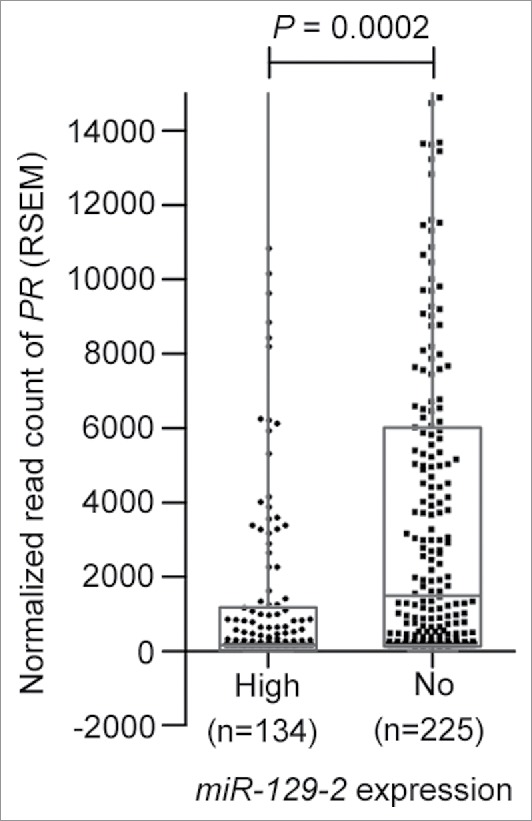
Expression of miR-129-2 in breast cancer patients in TCGA dataset. Expression plot for PR in breast cancer patients with high miR-129-2 expression (n = 134) and with absence of miR-129-2 expression (n = 225) in the TCGA cohort. The box-plot is overlaid with dot-plot wherein each point represents patient sample. Y-axis indicates normalized read count (RSEM) values for PR in a total of 359 breast cancer patients where expression of PR and miR-129-2 was available. P-value (P = 0.0002) was calculated using student's unpaired t-test with Welch's correction.
Discussion
Small RNA sequencing analysis of progesterone treated breast cancer cell lines led to the identification of a novel PR-targeting microRNA miR-129-2. Since the increased expression of miR-129-2 was independent of the PR-status of breast cancer cells, a possible role of other steroid hormone receptors like membrane progesterone receptor or glucocorticoid receptor as suggested in literature10,11 to mediate the role of progesterone in these cells remains to be systematically analyzed. Consistent with our finding, analysis of TCGA breast cancer dataset suggests a significantly decreased expression of PR in patients with elevated expression of miR-129-2 as compared to patients with no miR-129-2 expression, indicating a possibility for the decreased expression of PR in patients with low PR expression. It has been observed that factors like loss of PR or menopausal status of women can alter the response to hormonal therapy.12 Some studies have indicated that the absence of PR could underlie tumors resistance to hormonal therapy2, or could potentially increase the risk of relapse13. Hence we propose that stabilization of PR expression in patients with tumors expressing low PR levels by blocking activity of such microRNAs using specific microRNA inhibitors, along with other treatment modalities, could be potentially helpful in enhancing the response of patients to hormonal therapies. In support of this notion, our in vitro luciferase assay and western blot results using miR-129-2 inhibitor suggest that inhibition of miR-129-2 can increase the expression of PR. Thus, we validate under in vitro settings that addition of progesterone leads to up-regulation of miR-129-2, which suppresses the expression of PR in breast cancer cells; and, the inhibition of miR-129-2 reinstates the PR expression in these breast cancer cells even in presence of progesterone. Also since microRNAs are being assessed for their use in clinics14, strategies like microRNA sponges and chemically modified antisense oligonucleotides (inhibitors) hold promise as a promising line of treatment of breast cancer that need to be exhaustively explored with larger datasets.15 Thus, our study suggests an underlying mechanism to a possible clinical consequence in response to progesterone treatment among patients with varying PR expression levels. Also, it is suggestive of treatment with anti-miR-129-2 among those patients expressing inadequate PR levels, under adjuvant and neo-adjuvant settings, before considering for hormonal therapy. Whether modulation of activity of miR-129-2 could stabilize PR expression and potentially improve response to hormonal therapy remains to be validated as an immediate follow up to this pilot study.
Materials and methods
Breast cell lines
T47D, BT474, MDA-MB-231 and MCF7 breast cancer cell lines were obtained from Dr. Slamon's laboratory (Department of Medicine, UCLA, USA). Human embryonic kidney 293FT cells were obtained from Invitrogen. The cell lines were authenticated by DNA Short Tandem Repeat (STR) profiling using the Promega GenePrint 10 system and the analysis was performed using the GeneMarker HID software and the ATCC database. Cells in culture were tested for mycoplasma and were made mycoplasma-free using EZKill Mycoplasma Removal reagent (HiMedia). All the cell lines were grown in DMEM medium (Gibco) supplemented with 10% (v/v) FBS (Gibco), 2.5 mg/ml Amphotericin-B (Abbott) and 1.25 µl/ml Gentamycin (Abbott) and were cultured at 37°C in a 5% CO2 incubator. The ER/PR/Her2 receptor status of breast cancer cells was validated by RT-PCR.
Progesterone treatment, RNA isolation and protein sample preparation
Cells were grown to 70–80% confluence and then serum starved in a phenol-red free DMEM low glucose medium (HiMedia) for a period of 24h. In the same medium, cells were treated with 10 nM concentration of 17-α hydroxy-progesterone caproate (progesterone) (MP Biomedicals) for 6 h. Equal amount of alcohol was used as vehicle control. After 6 h of progesterone treatment, TRIzol reagent (Invitrogen) was used to lyze the cells. RNA was isolated according to the manufacturer's protocol. The RNA concentration was measured using NanoDrop. For western blotting, proteins were isolated from cells using RIPA buffer and separated on a 10% SDS-PAGE and transferred onto PVDF membrane for probing with primary antibody. Primary antibodies used were PR-AB (sc-810, 1:300 dilution) and β-actin (sc-1616-R, 1:4000 dilution). Secondary antibody used was goat anti-mouse (sc-2005, 1:3000 dilution) and goat anti-rabbit (sc-2004, 1:3000 dilution).
Small RNA sequencing analysis
Small RNA sequencing was performed on single lane of Illumina HiSeq 1000 with eight multiplex libraries from the four breast cancer cell lines. The reads obtained from deep sequencing of small RNAs were subjected to Illumina adaptor trimming using FastX tool kit and were size filtered to select for candidate miRNA's (14 to 24 bases) from a pool of small RNA sequences using in-house perl script. The size separated reads were then mapped onto human miRNA reads obtained from miRBase (version 21) using Bowtie2 (version 2.1.0)16 with 0 mismatches in the first 8 bases. MicroRNAs were quantified followed by normalisation by read per million using in-house script. Deregulated miRNAs with > = 3 fold change were retained for further analysis. For searching microRNAs targeting PR 3′UTR, differentially expressed microRNAs in response to progesterone were compared to microRNAs predicted to target PR using 6 algorithms (TargetScan, miRanda, miRWalk, miRMap, RNA22 and RNAhybrid).
Quantitative real-time PCR
Transcript levels of candidate microRNA's were analyzed by quantitative real time PCR. 1 µg total RNA was used for cDNA synthesis using Mir-X miRNA First-Strand Synthesis Kit (Clontech Takara). For analyzing transcript levels of de-regulated genes, cDNA was synthesized using High capacity cDNA reverse transcription kit (Applied Biosystems). cDNA from each cell line with the two treatment conditions were then subjected to quantitative real-time PCR analysis using Roche Light-Cycler-II 480 instrument using the Mir-X miRNA qRT-PCR SYBR Kit (2X) Master Mix (Clontech Takara) for microRNAs and Roche real-time master mix (Roche) for genes. Expression change of candidate miRNAs and genes de-regulated by progesterone was calculated by the 2−ΔΔCT method. U6 small RNA (primers provided by Clontech Takara) was used as an internal control for microRNAs and GAPDH was used for genes. Primer sequences for each microRNA and gene used for validation purpose are given in Supplementary table 2.
Cloning of microRNA/PR 3′UTR and luciferase assay
A 400bp sequence of miR-129-2 containing the seed sequence was PCR amplified using genomic DNA isolated from T47D. Amplicons were cloned in a T/A cloning vector (Fermentas, USA) followed by sub-cloning in BamHI and HindIII sites of pCDNA 3.1 (−) expression vector (In vitrogen). PR-3′UTR of 1000bp was PCR amplified using T47D cDNA. Amplicons were cloned in a T/A cloning vector followed by sub-cloning between XbaI sites in pGL3-promoter vector (Luciferase Expressing vector, Promega). For the Luciferase assay, 293FT cells (50,000 cells/well) were transfected using lipofectamine 2000 reagent (Life Technologies) with the combination of these constructs along with Renilla luciferase vector (for normalizing transfection efficiency) and 5 nM miR-inhibitors (SIGMA, HSTUD0162) in separate wells. 48 hours post-transfection cells were lyzed and luciferase assay was performed to measure luminescence (Berthold Luminometer, Germany). Experiment was performed in triplicates and differences between group showing p-values <0.05 (calculated using an unpaired student's t-test) were considered significant.
Transfection of microRNA inhibitor in breast cancer cells
T47D cells were grown up to 60% confluence and transfected with 25 nM negative control miRNA inhibitor (against miR-29a) and miR-129-2 inhibitor (SIGMA, HSTUD0162). Post-transfection, cells were incubated for 48 h and then treated with progesterone for 6 h. Cell lysate was prepared and western blot analysis was performed.
Supplementary Material
Disclosure of potential conflict of interest
No potential conflict of interest was reported by the authors.
Acknowledgments
All members of the Dutt laboratory for critically reviewing the manuscript and Pawan Upadhyay for STR profiling of breast cancer cells. Small RNA sequencing were performed at Genotypic Pvt Ltd, Bangalore, India.
Authors' contributions
M.G. and A.D designed the research; M.G., P.C., N.G., H.D., K.P. and N.Y. performed the research; M.G., P.C. and A.D. analyzed the data; M.G., S.G., R.B. and A.D. wrote the paper. All the authors have read and approved the manuscript.
Funding
A.D. is supported by an Intermediate Fellowship from the Wellcome Trust/DBT India Alliance (IA/I/11/2500278), intramural grants [IRB project 2712], and by a grant from DBT (BT/MED/30/VNCI-Hr-RCA/2015). M.G., P.C., N.G. and N.Y. are supported by research fellowship from Homi Bhabha National Institute (HBNI), ACTREC-TMC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cardoso F, Senkus-Konefka E, Fallowfield L, Costa A, Castiglione M. Locally recurrent or metastatic breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21(Suppl 5):v15-9. doi: 10.1093/annonc/mdq160 [DOI] [PubMed] [Google Scholar]
- 2.Cui X, Schiff R, Arpino G, Osborne CK, Lee AV. Biology of progesterone receptor loss in breast cancer and its implications for endocrine therapy. J Clin Oncol. 2005;23:7721-35. doi: 10.1200/JCO.2005.09.004 [DOI] [PubMed] [Google Scholar]
- 3.Ren Y, Liu X, Ma D, Feng Y, Zhong N. Down-regulation of the progesterone receptor by the methylation of progesterone receptor gene in endometrial cancer cells. Cancer Genet Cytogenet. 2007;175:107-16. doi: 10.1016/j.cancergencyto.2007.02.002 [DOI] [PubMed] [Google Scholar]
- 4.Zhang PJ, Zhao J, Li HY, Man JH, He K, Zhou T, Pan X, Li AL, Gong WL, Jin BF, et al.. CUE domain containing 2 regulates degradation of progesterone receptor by ubiquitin-proteasome. EMBO J. 2007;26:1831-42. doi: 10.1038/sj.emboj.7601602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lange CA, Shen T, Horwitz KB. Phosphorylation of human progesterone receptors at serine-294 by mitogen-activated protein kinase signals their degradation by the 26S proteasome. Proc Natl Acad Sci U S A. 2000;97:1032-7. doi: 10.1073/pnas.97.3.1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cochrane DR, Cittelly DM, Richer JK. Steroid receptors and microRNAs: relationships revealed. Steroids. 2011;76:1-10. doi: 10.1016/j.steroids.2010.11.003 [DOI] [PubMed] [Google Scholar]
- 7.Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P, Barton G, Jiao LR, Wait R, Waxman J, et al.. The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci U S A. 2009;106:15732-7. doi: 10.1073/pnas.0906947106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cochrane DR, Jacobsen BM, Connaghan KD, Howe EN, Bain DL, Richer JK. Progestin regulated miRNAs that mediate progesterone receptor action in breast cancer. Mol Cell Endocrinol. 2012;355:15-24. doi: 10.1016/j.mce.2011.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seeger H, Wallwiener D, Mueck AO. The effect of progesterone and synthetic progestins on serum- and estradiol-stimulated proliferation of human breast cancer cells. Horm Metab Res. 2003;35:76-80. doi: 10.1055/s-2003-39061 [DOI] [PubMed] [Google Scholar]
- 10.Xie M, You S, Chen Q, Chen X, Hu C. Progesterone inhibits the migration and invasion of A549 lung cancer cells through membrane progesterone receptor alpha-mediated mechanisms. Oncol Rep. 2013;29:1873-80. doi: 10.3892/or.2013.2336 [DOI] [PubMed] [Google Scholar]
- 11.Lei K, Chen L, Georgiou EX, Sooranna SR, Khanjani S, Brosens JJ, Bennett PR, Johnson MR. Progesterone acts via the nuclear glucocorticoid receptor to suppress IL-1beta-induced COX-2 expression in human term myometrial cells. PLoS One. 2012;7:e50167. doi: 10.1371/journal.pone.0050167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chlebowski RT, Rohan TE, Manson JE, Aragaki AK, Kaunitz A, Stefanick ML, Simon MS, Johnson KC, Wactawski-Wende J, O'Sullivan MJ, et al.. Breast Cancer After Use of Estrogen Plus Progestin and Estrogen Alone: Analyses of Data From 2 Women's Health Initiative Randomized Clinical Trials. JAMA oncology. 2015;1:296-305. doi: 10.1001/jamaoncol.2015.0494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartlett JM, Rea D, Rimm DL. Quantification of hormone receptors to guide adjuvant therapy choice in early breast cancer: better methods required for improved utility. J Clin Oncol. 2011;29:3715-6. doi: 10.1200/JCO.2011.37.3704 [DOI] [PubMed] [Google Scholar]
- 14.Bertoli G, Cava C, Castiglioni I. MicroRNAs: New biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5:1122-43. doi: 10.7150/thno.11543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3:1. doi: 10.1186/1758-907X-3-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357-9. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


