ABSTRACT
It was well known that cancer-associated fibroblasts (CAFs) were an essential factor in tumor progression. However, the actual mechanism of stromal fibroblasts activation and tumor promoting effects remain unclear. Here, we showed that KLF5 expression was more frequently observed in gastric cancer-associated fibroblasts compared with normal mucosal fibroblasts. Moreover, KLF5 expression in tumor stroma was closely associated with clinicopathological features such as tumor size, invasion depth, cell grade and lymph node metastasis, as well as poor prognosis in patients with gastric cancer. In addition, we further demonstrated that KLF5-regulating CAFs affect gastric cancer cells progression by CCL5 secretion and activation of CCR5. Taken together, we concluded that KLF5 expression in gastric cancer-associated fibroblasts contribute to poor survival and promote cancer cells progression by activation of CCL5/CCR5 axis, which suggesting that KLF5 in CAFs might be considered as a promising target for the treatment of gastric cancer.
KEYWORDS: Cancer-associated fibroblasts, Gastric cancer, KLF5, CCL5, CCR5
Introduction
Gastric cancer is a leading cause of cancer-related death in the world.1 It was widely considered that genetic mutations and epigenetic alterations of oncogenes and tumor suppressor genes were closely associated with the initiation and development of human gastric cancer,2 but the actual molecular mechanism of gastric cancer remains to be elucidated.
Recently, it has been increasingly verified in previous studies that tumor microenvironment play a vital role in cancer progression.3 The tumor microenvironment consists of the ECM, growth factors, cytokines, and various types of non-tumor cells, such as fibroblasts, immune cells, as well as endothelial cells.4 Among these cells, the fibroblast is one of the most important stromal cell types in the microenvironment, which forms the basic tissue architecture, contributing to structural integrity and the main components of the extracellular matrix (ECM).5 Cancer associated fibroblasts (CAFs) are very important cells in the tumor stroma, which have crucial effects on cancer development by degrading ECM and secreting a variety of active molecules, such as growth factors and chemokine, which contribute to the promotion of cell proliferation, migration and metastasis, as well as stimulation of angiogenesis.6 Thus, it is necessary to further study the exact mechanism how CAFs promote cancer progression, and targeting CAFs may become a new promising therapy that can improve favorable patient prognosis.
Krüppel-like Zinc-Finger Transcription Factor 5 (KLF5) belongs to the KLF protein family, which contains three peptide modules of the c2h2 zinc finger type.7,8 It is ubiquitously expressed in many human tissues, such as colon9, lung10, breast11 and prostate12, which regulate target gene expression through binding GC boxes. As a basic transcriptional factor, KLF5 also plays an essential role in cell proliferation, differentiation and apoptosis in different human cancer.13
Accumulating data indicate that a variety of factors including genetic variation contribute to the activation of fibroblasts and the induction of tumor progression, whether the KLF5 expression in CAFs affects tumor cell biological behavior especially in gastric cancer is still unknown. In this study, we determined the difference of KLF5 expression between CAFs and normal gastric mucosa fibroblasts, revealed its clinical significance and specific biological function, as well as exact molecular mechanism in human gastric cancer cells.
Materials and methods
Cell lines and tissue samples
Human tumor tissue and adjacent normal gastric mucosa tissue were taken from patients with gastric cancer after surgical resection. All these patients weren't underwent with radiation therapy or chemotherapy before surgery, at the department of general surgery in Shanghai Tenth Peoples’ Hospital. The protocols were approved by the Ethics Committee of Shanghai Tenth Peoples’ Hospital. CAFs and normal fibroblasts (NFs) were obtained as previously described in our previous article.14 All gastric cancer cell lines SGC-7901, BGC-823, MNK-45, including normal gastric mucosa epithelial cell lines GES-1 were obtained from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). All cells were maintained in a 37°C, 5% CO2 humidified incubator with RPMI-1640 medium (Invitrogen, US) containing 10% fetal bovine serum (Invitrogen, US). All cell lines in passage three were used for the experiments.
RNA extraction and quantitative RT-PCR
Total RNA was isolated from cell lines using Trizol Reagent according to the manufacturer's instructions, and then RNA was reversely transcribed into cDNA with Reverse Transcription system (Promega, WI, USA). Real time quantitative RT-PCR was performed to quantify t096he target gene transcripts using Real-time PCR Universal Reagent (GenePharma, Shanghai) following the manufacturer's manual. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was also determined and used as an internal control. Data analyses are performed with the 2-ΔΔCt method.
Western-blot analysis
Proteins were extracted and purified using NE-PER Nuclear and Cytoplasmic Protein Extraction Reagents Kit (Thermo Fisher Scientific, Waltham, US). Briefly, cells were washed with Hanks's solution and lysed with lysis buffer. The protein concentrations of the lysates were determined by Bradford protein assay kit (Bio-Rad, Philadelphia, US), and then resolved for 2.5 hours by 10% SDS PAGE. The protein was transferred to PVDF membranes for 2 hours. Immumoblotted the membranes with primary antibodies solution overnight at 4°C, and incubated with HRP-conjugated secondary antibody for 1 hour at 37°C. The expression level of protein was analyzed using the Lab Work Program (UVP) according to the manufacturer's instructions and normalized to β-actin.
Tissue microarrays (TMAs)
Tumor cores were obtained from 120 patients with gastric cancer who performed operation in the department of surgery, Shanghai Tenth People's Hospital from January 2006 to December 2012. None of the patients had preoperative chemotherapy or radiotherapy. A tissue core section in each FFPE tissue block was removed and arranged into new paraffin blocks for constructing the tissue microarray. Written informed consents were provided for all patients. Clinicopathological features including age, sex, tumor size, histologic grade, Lauren classification, invasion depth, and lymph nodal status were reviewed. TNM staging was designated according to the 7th edition of the American Joint Committee on Cancer (AJCC) cancer staging manual.
Immunohistochemical staining
Sections fixed on microslide were deparaffinized using xylene, hydrated with a series of diluted alcohol, and blocked endogenous peroxidase activity by using methanol. Antigens were retrieved with Tris-EDTA (TE) buffer and treated with bovine serum albumin in phosphate buffer solution (PBS) to reduce nonspecific staining. The sections were incubated in rabbit anti-KLF5 polyclonal antibody (1:200; Abcam, UK) overnight at 4°C in humidified chambers. After three successive rinses with washing buffer, the sections were incubated with anti-rabbit polymer kit (Dako, Carpinteria, CA) for 30 min at 37°C. The sections were cultured with diaminobenzidine, counterstained with hematoxylin after being washed three times with PBS, and then observed using confocal microscope.
cDNA microarray analysis
Total RNAs were isolated from six pairs of CAFs and NFs by primary culture of different gastric cancer and normal mucosal tissues. cDNA was synthesized from total RNA using a reverse transcription kit according to the manufacturer's protocol. To prepare hybridization probes, Cy3-dCTP or Cy5-dCTP was incorporated during reverse transcription. We applied to Human OneArray® Whole Genome Microarrays v 5.1 (Phalanx Biotech, San Diego, CA) to measure gene expression level. Each microarray contain 30225 oligonucleotide probes: 29187 human genome probes, and 1088 experimental control probes. Hybridization was performed at 42°C for 16h. After hybridization, wash were performed with 0.2 × SSC at room temperature for 5 min. And then scanned the fluorescent signals by using Agilent's microarray scanner system (Santa Clara, CA, USA). The images and quantitative data of the gene expression levels were analyzed by Agilent's Feature Extraction software.
Cell transfection and co-culture assay
The assay was performed as we previously described.14 In Brief, fibroblasts were transfected with lentivirus mediated KLF5 gene or empty vector, and KLF5-siRNA or control scrambled siRNA, along with HiPerfect Transfection Reagent, according to the manufacturer's manual. For co-culture assay, gastric cancer cells were seeded in the plates in 1 × 104 cells per well, and then condition media collected from CAFs was added to plates. MTT assay was used to assess he tumor cell growth capacity. In brief, the cells were incubated in MTT at 37°C and then treated with DMSO at room temperature for 15 min. Optical density (OD) value was determined in 580 nm absorbance value at different time points. All experiments were repeated for 3 times.
Clonogenic assay
Clonogenic assay was performed as we previously described.14 In Brief, tumor cells were resuspended with trypsin, and then layered onto 0.6% solidified agar in RPMI-1640 medium containing 10% FBS in 6-well plates and incubated for 14 days at 37°C. Colonies containing at least 50 cells were counted. All experiments were repeated for 3 times.
Cell migration assay
Cell migration assay was also performed as we previously described. Briefly, Cancer cells were incubated in serum-free medium for 24 h, and then added 1 × 104 cells to upper chamber, the lower chamber was added RPMI-1640 medium containing 10% fetal bovine serum (FBS). The membranes were stained with crystal violet after the completion of incubation and put on a glass slide. The cells penetrating across the membrane were counted with microscope. All experiments were repeated for 3 times.
Wound-healing assay
Cancer cells were seeded in six-well plates at 1 × 104 cells per well and then were grown into a monolayer, and make a straight scratch on cell monolayers with sterile pipette tip. Images of the fields were collected with photomicrographs at × 100 magnification and the migration distance was investigated at different time point. All experiments were repeated for 3 times.
Tumor xenograft assay
Male BALB/c nude mice at the age of 4 weeks old were maintained in ventilated caging systems at five mice per group. Fibroblasts mixed with cancer cells at the ratio of 1:4 within 0.1 ml PBS were injected subcutaneously into the flank of the mice. Tumor growth of the mice was observed weekly, and measured the tumor length and width with digital calipers and the tumor volume is calculated by the formula: Volume = 0.5 LW2. After the mice were sacrificed, all tumors were harvested and measured. All animal procedures were consent with the Animal Care and Use Committee of Shanghai Tenth Peoples’ Hospital affiliated Tongji University.
Human cytokine antibody array
We prepared conditioned medium (CM) from fibroblasts by seeding fibroblasts into dishes with 10 mL of RPMI-1640 containing 2% FBS and then incubating the cells for 3 days. To obtain CM, the fibroblasts were washed with PBS and then incubated for an additional 3 days in 4mL of RPMI-1640. The supernatant used as CM was collected from each dish and centrifuged at 1,000 g for 5 minutes at 4°C, and then was used for human cytokine antibody array with Raybio Human Cytokine Antibody Array G series 6 according to the manufacturer's instruction. The experiments were repeated in duplicates.
ELISA assay
CM collected from CAFs were placed at room temperature for 4h before they were centrifuged at 2000g for 20 min, which were stored at −80°C until use for CCL5 measurement by CCL5 ELISA kit (Ocean Science, Beijing, China) according to the manufacturer's instruction.
Statistical analysis
Correlations between the expression levels of KLF5 and different clinicopathological parameters were examined using Pearson χ2. Kaplan-Meier method was used to explore the relationship between the KLF5 expression level and overall survival. P < 0.05 was considered statistically significance. Data analyses were performed with SPSS statistical software, version 17 (SPSS Inc, Chicago, IL).
Results
KLF5 expression is frequently present in CAFs compared with NFs
We have primarily cultured six paired CAFs and NFs from tumor tissues and normal gastric mucosa tissues, respectively, and also determined the gene expression differences between CAFs and NFs. In the result, we identified eight human genes differentially expressed in CAFs compared with corresponding NFs. Of these, CCL18, MMP-9, β-catenin, CCL12, CXCR4, including KLF5 gene, were significantly up-regulated, and other two genes such as PTEN and KCND3 gene were significantly down-regulated (Fig. 1A).
Figure 1.
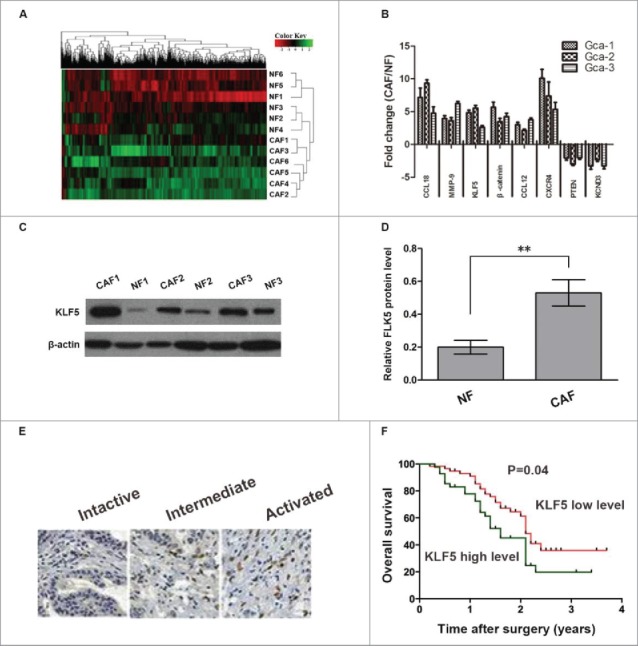
KLF5 expression in gastric cancer associated fibroblasts and gastric cancer stromal tissues. (A) The dysregulated genes were analyzed by cDNA array used 6 paired CAFs and NFs obtained from patients with gastric cancer. (B) Eight selected genes were analyzed by qRT-PCR in CAFs and NFs derived from gastric cancer patients. Data are shown as fold changes between CAFs and NFs. (C, D) The expression of KLF5 protein in 3 paired CAFs and NFs were measured by Western-blot assay. (E, F) Representative images of KLF5 immunostaining assay in gastric cancer stromal tissues. Kaplan-Meier analysis showed that high level of KLF5 expression in stroma is associated with decreased overall survival (P = 0.04). Data are representative of three independent experiments. *P < 0.05 by t test, **P < 0.01 by t test.
We further investigated the mRNA and protein expression of KLF5 in CAFs and NFs using qRT-PCR and Western blotting assay, the results indicated that KLF5 mRNA and protein expression were also all significantly up-regulated in CAFs compared with corresponding NFs (Figure 1B-D).
The overexpression of KLF5 in gastric cancer stroma is related to poor patient prognosis
The tissue microarray contains 120 gastric cancer tissues after radical surgery were stained for KLF5 protein expression via immunohistochemistry. Two independent pathologists were blinded to the specific diagnosis for the stained slides and collection of clinical information. The staining intensity was scored on a scale range from 0 to 3. After dividing the patients into two groups with high or low level of KLF5 expression in gastric cancer stroma according to the median score.
The results showed that the level of KLF5 expression in gastric cancer stroma was significantly related to tumor size (P = 0.004), grade (P = 0.017), invasive depth (P = 0.016), and lymph node metastasis (P = 0.005) (Table 1). Moreover, the higher level of KLF5 expression in the stroma underwent a shorter overall survival (P = 0.04, Fig. 1F) in the analysis of Kaplan-Meier survival, which suggesting that KLF5 in gastric cancer stroma might be a favorable important factor in the development of gastric cancer.
Table 1.
Relationship between KLF5 expression in gastric cancer stroma and clinicopathologic features.
| KLF5 expression in stroma |
|||||
|---|---|---|---|---|---|
| Variable | Total | Low expression (%) | High expression (%) | P value | |
| Age (years) | |||||
| < 60 | 33 | 10 (30.3) | 23 (69.7) | 0.964 | |
| ≥ 60 | 87 | 26 (29.9) | 61 (70.1) | ||
| Sex | |||||
| Female | 41 | 15 (36.6) | 26 (63.4) | 0.174 | |
| Male | 79 | 21 (26.6) | 58 (73.4) | ||
| Tumor size (cm) | |||||
| <5 | 53 | 23 (43.4) | 30 (56.6) | 0.004 | |
| ≥5 | 67 | 13 (19.4) | 54 (80.6) | ||
| Lauren's classification | |||||
| Intestine | 62 | 17 (27.4) | 45 (72.6) | 0.524 | |
| Diffuse | 58 | 19 (32.8) | 39 (67.2) | ||
| Grade | |||||
| G1 | 23 | 14 (60.9) | 9 (39.1) | 0.017 | |
| G2 | 43 | 14 (32.6) | 29 (67.4) | ||
| G3 | 54 | 8 (14.8) | 46 (85.2) | ||
| Invasive depth | |||||
| T1-T2 | 35 | 16 (45.7) | 19 (54.3) | 0.016 | |
| T3-T4 | 85 | 20 (23.5) | 65 (76.5) | ||
| Lymph node metastasis | |||||
| Negative | 47 | 21 (44.7) | 26 (55.3) | 0.005 | |
| Positive | 73 | 15 (20.5) | 58 (79.5) | ||
Down-regulation of KLF5 expression in CAFs inhibit gastric cancer cells growth, migration and invasion
To investigate the function of KLF5 in CAFs, we determined whether abnormal expression of KLF5 in CAFs affect the growth, migration and invasion of tumor cell. We transfected lentivirus-siRNA-KLF5 or Lenti-KLF5 and corresponding control vector into CAFs to regulate the KLF5 expression in vitro. MTT and clonogenic assay showed that the growth ability of gastric cells cultured with CAFs transfected lentivirus-KLF5 was significantly higher than the corresponding control, but the growth ability of gastric cancer cultured with CAFs transfected lentivirus-siRNA-KLF5 was significantly slower than the corresponding control (Fig. 2A and B).
Figure 2.
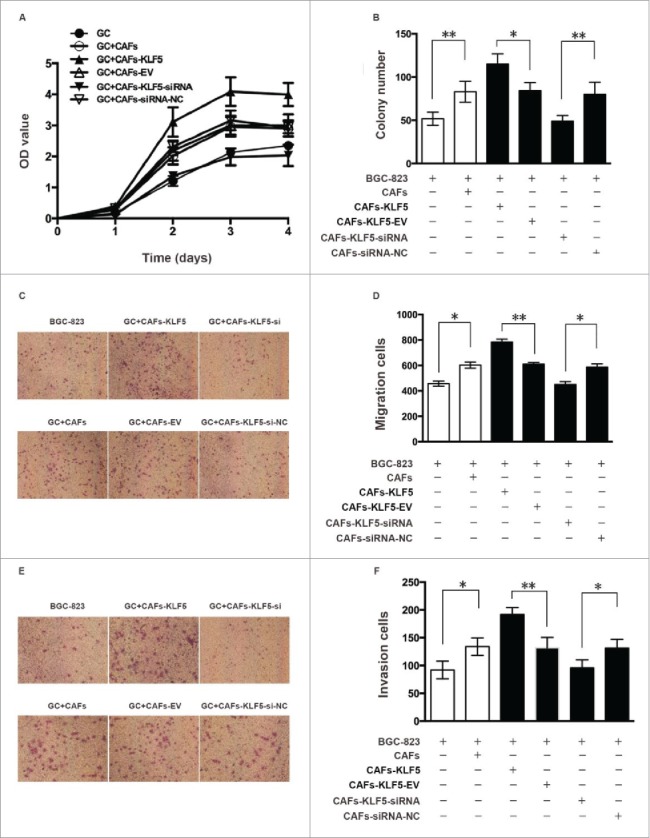
Down-regulation of KLF5 expression in CAFs inhibits the growth, migration and invasion of gastric cancer cells. (A, B) Cell growth ability was measured using MTT and clonogenic assay. The growth ability of gastric cells cultured with CAFs transfected lentivirus-KLF5 was significantly higher than the corresponding control, but the growth ability of gastric cancer cultured with CAFs transfected lentivirus-siRNA-KLF5 was significantly slower than the corresponding control. (C-D) Cell migration ability was measured by Transwell migration assay. The migration ability of gastric cells cultured with CAFs transfected lentivirus-KLF5 and lentivirus-siRNA-KLF5 was significantly greater and weaker than the corresponding control, respectively. (E, F) Cell invasion ability was measured by Transwell invasion assay. The invasion ability of gastric cells cultured with CAFs transfected lentivirus-KLF5 and lentivirus-siRNA-KLF5 was significantly greater and weaker than the corresponding control, respectively. Data represent mean ± SEM from three independent experiments; *P < 0.05 by t test, **P < 0.01 by t test.
Transwell migration assay indicated that the migration ability of gastric cells cultured with CAFs transfected lentivirus-KLF5 and lentivirus-siRNA-KLF5 was significantly greater and weaker than the corresponding control, respectively (Fig. 2C and D). Transwell invasion assay indicated that the invasion ability of gastric cells cultured with CAFs transfected lentivirus-KLF5 and lentivirus-siRNA-KLF5 was significantly greater and weaker than the corresponding control, respectively (Fig. 2E and F).
Wound-healing assay revealed that the wound closure rate of gastric cells cultured with CAFs transfected lentivirus-KLF5 and lentivirus-siRNA-KLF5 was significantly higher and lower than corresponding control, respectively (Fig. 3A and B). All these data suggested that the CM from CAFs with KLF5 low expressions significantly inhibit gastric cancer cells growth, migration and invasion, whereas CAFs with KLF5 high expression could significantly promote the growth, migration and invasion of gastric cancer cells.
Figure 3.
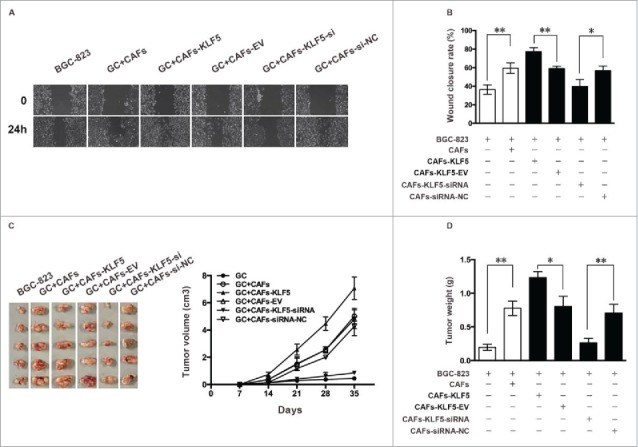
Down-regulation of KLF5 expression in CAFs inhibits the migration of gastric cancer cells and the tumor growth in vivo. (A, B) Cell migration ability was also measured by a wound-healing assay. The wound closure rate of gastric cells cultured with CAFs tranfected lentivirus-KLF5 and lentivirus-siRNA-KLF5 was significantly higher and lower than corresponding control, respectively. (C, D) Tumor growth ability was measured by a tumor xenograft assay. The tumor volume of mice injected gastric cancer cells cultured with CAFs transfected lentivirus-KLF5 and lentivirus-siRNA-KLF5 was significantly bigger and smaller than corresponding control, respectively. The tumor weight of mice injected gastric cancer cells cultured with CAFs transfected lentivirus-KLF5 and lentivirus-siRNA-KLF5 was significantly heavier and lighter than corresponding control, respectively. Data represent mean ± SEM from three independent experiments. *P < 0.05 by t test, **P < 0.01 by t test.
We further tested whether abnormal expression of KLF5 in CAFs affect tumor cells growth in vivo. Tumor xenograft assay showed that the tumor volume of mice injected tumor cells together with CAFs transfected lentivirus-KLF5 and lentivirus-siRNA-KLF5 was significantly bigger and smaller than corresponding control, respectively (Fig. 3C). The tumor weight of mice injected gastric cancer cells cultured with CAFs transfected lentivirus-KLF5 and lentivirus-siRNA-KLF5 was significantly heavier and lighter than corresponding control, respectively (Fig. 3D), which suggesting that CAFs with KLF5 low expression could significant inhibit gastric cancer growth, whereas CAFs with KLF5 high expression promote gastric cancer growth in vivo, consistent with the data obtained from assays in vitro.
KLF5-regulating CAFs affect tumor cell progression by CCL5/CCR5 axis
To further reveal the exact mechanism of the inhibitory effect of KLF5-downregulating CAFs on tumor cell progression, we determined cytokine expression difference between CAFs-CM and CAFs-KLF5-si-CM using human cytokine antibody array. The result showed that three cytokines including CCL5, CCL16 and CXCL12 were significantly down-regulated (Fig. 4A and B) in CAFs-KLF5-si-CM compared with those in CAFs-CM. We also detected the mRNA expression differences of CCL5 between CAFs and NFs using RT-PCR, and secretion differences between CAFs-CM and NFs-CM with ELISA (Fig. 4C and D). The results showed that CCL5 mRNA expression is up-regulated significantly in CAFs compared with NFs, and CCL5 secretion expression is also up-regulated significantly in CAFs-CM compared with NFs-CM.
Figure 4.
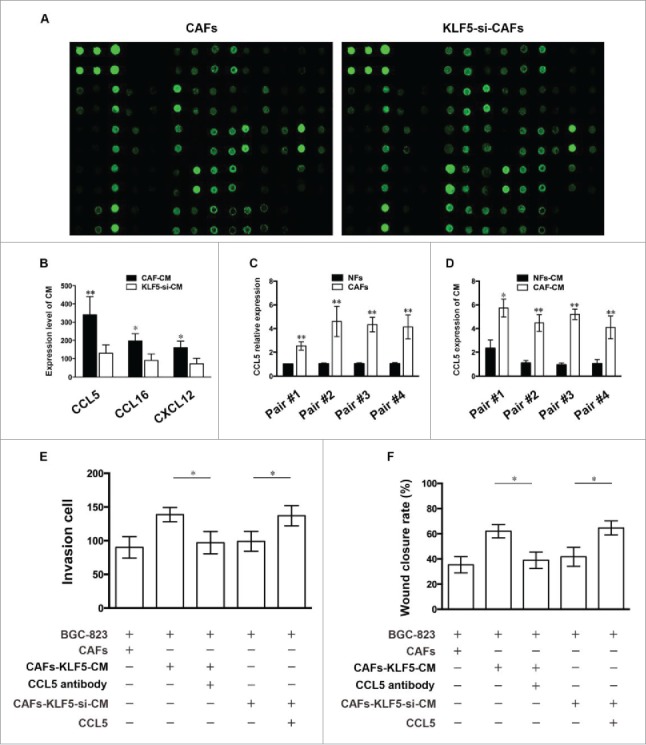
KLF5-downregulating CAFs inhibit tumor cell migration and invasion by reducing CCL5 secretion (A) Representative images of cytokine expression difference between CAFs-CM and CAFs-KLF5-si-CM in human cytokine antibody array. (B) Cytokine screening of conditioned medium collected from CAFs and CAFs-KLF5-si. (C) qRT-PCR detection of CCL5 mRNA levels in four pairs of CAFs and NFs. (D) ELISA detection of CCL5 protein concentration in the medium conditioned by these cells. (E, F) Transwell invasion and wound-healing assay showed the effect of different treated CAFs on tumor cell invasion in the presence of CCL5 neutralizing antibody or exogenous CCL5. Data are representative of three independent experiments. *P < 0.05 by t test, **P < 0.01 by t test.
To determine whether CCL5 directly contributes to the inhibitory effect of KLF5-downregulating CAFs on gastric cancer cells, we detected the effect of CAFs-KLF5-si-CM on gastric cancer cells invasion after the addition of CCL5 antibody and exogenous CCL5, respectively. The results showed that CAFs-KLF5-si-CM added exogenous CCL5 could significantly increase gastric cancer cells invasion, whereas the tumor cells invasion decreased significantly compared with control when we added CCL5 antibody into the co-culture system (Fig. 4E and F), which suggesting that CCL5 is the major factor contributing to the tumor inhibitory properties of KLF5-downregulating CAFs.
To further confirm whether CCL receptor (CCR) is activated by CAF produced CCL5, we detected the expression of CCR1, CCR3 and CCR5 in different group of gastric cancer cells. The results showed that the CCR5 mRNA expression of gastric cancer cells in the CAFs-KLF5-CM and CCL5 group was significantly up-regulated compared with control group, but not CCR1 and CCR3 (Fig. 5A). Western blot results indicated that CCR5 protein expression of cancer cells in these groups were also significantly up-regulated compared with those in control group (Fig. 5B). Moreover, the transwell and wound-healing assay results revealed that the gastric cancer cells invasion decreased significantly compared with control group when we added CCR5 antagonist into the co-culture system (Fig. 5C and D), which suggesting that KLF5-regulating CAFs affected gastric cancer cells progression by CCL5 secretion and activation of CCR5.
Figure 5.
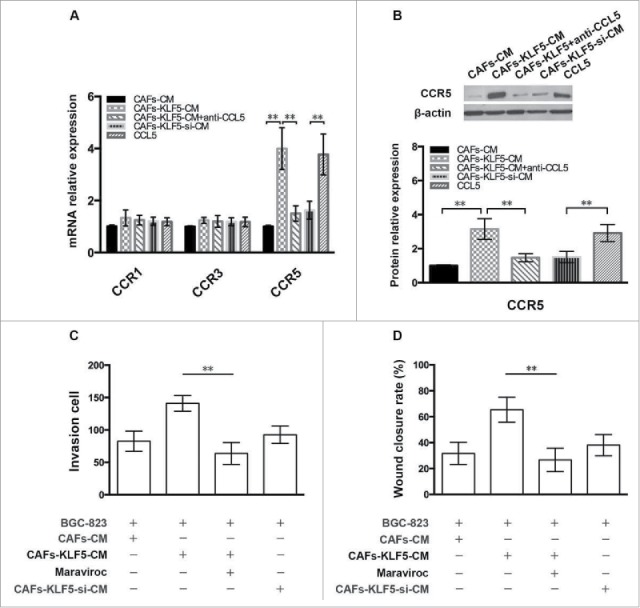
CCR5 is activated by CAFs produced CCL5. (A) The mRNA expression of CCR1, CCR3 and CCR5 in gastric cancer cells treated with different CAFs was examined by qPCR. (B) The protein expression of CCR5 in gastric cancer cells treated with different CAFs was determined by Western blot. (C, D) Transwell invasion and wound-healing assay showed the effect of different treated CAFs on tumor cell invasion in the presence of CCL5 antagonist.
Discussion
A growing number of evidences indicated that the interactions between tumor and stromal cells in the tumor microenvironment play an important promotional role in the initiation and progression of cancer.15-17 Among these stromal cells, cancer associated fibroblasts are the most abundant and important cells, which were confirmed that it was differ from normal fibroblasts in several aspects such as expression profile and behavior, especially different effects on cancer cells.18 However, the exact molecular mechanism underlying aberrant expression remains poorly understood.
There are various factors and steps such as gene variation and microRNA involved in the motility and activity of stromal cells. Several previous studies have indicated that a variety of genes variation play an important role in the tumor microenvironment, suggesting that functional proteins expressed specifically in CAFs might be candidate molecular targets for the treatment of gastric cancer. It has been proven that low Cav-1 protein expression in tumor stroma predicts adverse outcome in breast and prostate cancer. Moreover, it have been also demonstrated that the expression of Cav-1 in CAFs predicts early recurrence and poor survival with gastric cancer patients, suggesting Cav-1 in CAFs may be a novel promising target gene for gastric cancer patients.19,20
Twist1 expression in gastric cancer associated fibroblasts was related to several clinical factors such as tumor size, invasion depth, and lymph node metastasis, which were also related to poor prognosis in gastric cancer patients.21 In addition, up-regulation of twist1 in fibroblasts promoted the migration and invasion of cancer cells. Galectin-1 (Gal-1) is also highly expressed in gastric cancer associated fibroblasts, and increased expression of Gal-1 enhanced the migration and invasion of gastric cancer cells via regulating the expression of integrin β1 in gastric cancer.22,23 Tumor endothelial marker (TEM1) was over-expressed in the stromal tissues of gastric cancer, and the expression intensity of TEM1 in CAFs was associated with overall survival and relapse free survival. Moreover, the expression of TEM1 can be determined in both tumor cells and stromal cells of cancer tissue, suggesting that TEM1 might also be a novel promising target for gastric cancer.24
Here, we investigated whether it had gene expression differences between NFs and CAFs in gastric cancer, which could regulate the biological behavior of fibroblasts and tumor cells. In our present study, we demonstrated that KLF5 expression was more frequent in gastric cancer associated fibroblasts. Moreover, the expression of KLF5 in the gastric cancer stromal tissue is closely associated with clinical pathological factors, such as tumor size, grade, invasive depth, and lymph node metastasis, which are also related to poor survival in patients with gastric cancer. In addition, up-regulation of KLF5 in CAFs also could promote the growth, migration and invasion of gastric cancer cells in vitro and in vivo, which suggesting that KLF5 expression in CAFs may play a key role in gastric cancer cells progression.
KLF5 belongs to the human Sp1/KLF family of transcription factors containing three conserved C2H2-type zinc fingers in the C-terminal domain, which promotes cell proliferation, migration and invasion by regulating a number of downstream target genes or microRNA, such as mPGES125, p2126, Survivin27, VEGFA28, TNFAIP229, miR-200 family30, and so on, which binds to the promoters of these genes through GC-rich DNA sequences using its zinc finger domains. It was reported previously that KLF5 plays a vital role in fibroblasts from different tissue. The expression of KLF5 in cardiac fibroblasts is upregulated under pressure overload, and KLF5 is also an attractive target as it appears to be required in essentially providing a novel strategy for treating heart failure through the regulation of cardiac fibroblasts function.31 Overexpression of KLF5 in transfected NIH3T3 cells significantly promote cellular proliferation and give rise to transformed phenotype.32 Additionally, KLF5 is highly expressed in bronchial fibroblasts from the COPD patients and contributes to the remodeling of COPD.33
Cancer associated fibroblasts are most important non-tumor stromal cells of in various tumor microenvironment by secreting a variety of growth factors and chemokines, such as transforming growth factor-b (TGF-b)34, hepatocyte growth factor (HGF)35, platelet-derived growth factor (PDGF)36, fibroblast growth factor-3 (FGF-3)37 and stromal cell-derived factor-1 (SDF-1)38 into the tumor microenvironment, and stimulate tumor cells proliferation, invasion, metastasis and angiogenesis.
C-C chemokine ligand 5 (CCL5), also known as regulated upon activation, normal T-cell expressed, and secreted (RANTES), belongs to the C-C chemokine family, which is expressed by T lymphocytes, macrophages, platelets, synovial fibroblasts, tubular epithelium, and certain types of tumor cells.39 CCL5 plays a vital role in certain tumor progression including breast cancer39, melanoma40, colon cancer41 and prostate cancer42, as well as gastric cancer.43 The serum elevated CCL5 level in gastric cancer patients were associated with a poor prognosis,44 and the higher expression of CCR5 in tumor tissue was related to a lower survival rate in gastric cancer.45
CCL5 is also an essential factor in the tumor-promoting effect of CAFs on tumor cells. The down-regulation expression of miR-214 in CAFs increases CCL5 secretion, which leads to increased tumor growth. However, CCL5 antibodies block the promoting effect of CAFs on tumor growth and cell migration and CCL5-transfected normal fibroblasts increase the invasion of ovarian cancer cells, suggesting that CCL5 is a candidate molecule in CAFs, contributing to tumor cell recruitment and growth. 46
In our study, we revealed that CCL5 was the major factor contributing to the tumor inhibitory properties of KLF5-downregulating CAFs. The down-regulation expression of KLF5 in CAFs reduces CCL5 production, leading to inhibit gastric cancer cells invasion, whereas CCL5 antibody could block the promoting effect of CAFs on tumor cells invasion. Moreover, the addition of exogenous CCL5 could further increase the enhance effects of CAFs on tumor cell invasion. Thus, KLF5-regulating in CAFs affected tumor cells invasion through the reduction of CCL5 secretion.
It was well known that CCL5 activity was mediated through its binding to CCR1, CCR3, and mainly CCR5.39 Here, we also demonstrated that CCR5 expression of gastric cancer cells in the CAFs with KLF5 overexpression were significantly up-regulated, but not CCR1 and CCR3. Moreover, the addition of CCR5 antagonist to co-culture system could significantly inhibit gastric cancer cells invasion, which suggesting that KLF5-regulating CAFs affected gastric cancer cells progression by CCL5 secretion and activation of CCR5.
In summary, our results revealed that KLF5 was overexpressed in gastric cancer associated fibroblasts and tumor stroma, which is closely associated with clinical pathological features and poor prognosis in gastric cancer patients. Moreover, KLF5-regulating in CAFs could significantly affect gastric cancer cell growth, migration and invasion by activation of CCL5/CCR5 axis. Based on these experimental findings, we suggested that KLF5 in cancer-associated fibroblasts might be considered as a novel promising therapeutic target for the treatment of gastric cancer.
Conflict of Interests
No potential conflicts of interest were disclosed.
Funding support
The project is supported by Natural Science Foundation of Shanghai (No.15ZR1432900) awarded to TS Yang.
References
- 1.Siegel RL, Miller KD, Jemal A: Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7-30. doi: 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 2.Tamura G: Alterations of tumor suppressor and tumor-related genes in the development and progression of gastric cancer. World J Gastroenterol. 2006;12:192-8. doi: 10.3748/wjg.v12.i2.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quail DF, Joyce JA: Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423-37. doi: 10.1038/nm.3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo H, Tu G, Liu Z, Liu M. Cancer-associated fibroblasts: a multifaceted driver of breast cancer progression. Cancer Lett. 2015;361:155-63. doi: 10.1016/j.canlet.2015.02.018 [DOI] [PubMed] [Google Scholar]
- 5.Cirri P, Chiarugi P: Cancer associated fibroblasts: the dark side of the coin. Am J Cancer Res. 2011;1:482-97. [PMC free article] [PubMed] [Google Scholar]
- 6.Pietras K, Ostman A: Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res. 2010;316:1324-31. doi: 10.1016/j.yexcr.2010.02.045 [DOI] [PubMed] [Google Scholar]
- 7.Noto JM, Khizanishvili T, Chaturvedi R, Piazuelo MB, Romero-Gallo J, Delgado AG, Khurana SS, Sierra JC, Krishna US, Suarez G, et al.. Helicobacter pylori promotes the expression of Kruppel-like factor 5, a mediator of carcinogenesis, in vitro and in vivo. PLoS One. 2013;8:e54344. doi: 10.1371/journal.pone.0054344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bieker JJ: Kruppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355-8. doi: 10.1074/jbc.R100043200 [DOI] [PubMed] [Google Scholar]
- 9.McConnell BB, Kim SS, Yu K, Ghaleb AM, Takeda N, Manabe I, Nusrat A, Nagai R, Yang VW. Kruppel-like factor 5 is important for maintenance of crypt architecture and barrier function in mouse intestine. Gastroenterology. 2011;141:1302-13, 1313.e1-6. doi: 10.1053/j.gastro.2011.06.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan H, Luo F, Wert SE, Zhang L, Xu Y, Ikegami M, Maeda Y, Bell SM, Whitsett JA. Kruppel-like factor 5 is required for perinatal lung morphogenesis and function. Development. 2008;135:2563-72. doi: 10.1242/dev.021964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tong D, Czerwenka K, Heinze G, Ryffel M, Schuster E, Witt A, Leodolter S, Zeillinger R. Expression of KLF5 is a prognostic factor for disease-free survival and overall survival in patients with breast cancer. Clin Cancer Res. 2006;12:2442-8. doi: 10.1158/1078-0432.CCR-05-0964 [DOI] [PubMed] [Google Scholar]
- 12.Chen C, Bhalala HV, Vessella RL, Dong JT. KLF5 is frequently deleted and down-regulated but rarely mutated in prostate cancer. Prostate. 2003;55:81-8. doi: 10.1002/pros.10205 [DOI] [PubMed] [Google Scholar]
- 13.Dong JT, Chen C: Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell Mol Life Sci. 2009;66:2691-706. doi: 10.1007/s00018-009-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang TS, Yang XH, Chen X, Wang XD, Hua J, Zhou DL, Zhou B, Song ZS. MicroRNA-106b in cancer-associated fibroblasts from gastric cancer promotes cell migration and invasion by targeting PTEN. FEBS Lett. 2014;588:2162-9. doi: 10.1016/j.febslet.2014.04.050 [DOI] [PubMed] [Google Scholar]
- 15.Herrera M, Islam AB, Herrera A, Martín P, García V, Silva J, Garcia JM, Salas C, Casal I, de Herreros AG, et al: Functional heterogeneity of cancer-associated fibroblasts from human colon tumors shows specific prognostic gene expression signature. Clin Cancer Res. 2013;19:5914-26. doi: 10.1158/1078-0432.CCR-13-0694 [DOI] [PubMed] [Google Scholar]
- 16.Li H, Fan X, Houghton J: Tumor microenvironment: the role of the tumor stroma in cancer. J Cell Biochem. 2007;101:805-15. doi: 10.1002/jcb.21159 [DOI] [PubMed] [Google Scholar]
- 17.Bhowmick NA, Neilson EG, Moses HL: Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332-7. doi: 10.1038/nature03096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orimo A, Weinberg RA: Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle. 2006;5:1597-601. doi: 10.4161/cc.5.15.3112 [DOI] [PubMed] [Google Scholar]
- 19.Shen XJ, Zhang H, Tang GS, Wang XD, Zheng R, Wang Y, Zhu Y, Xue XC, Bi JW. Caveolin-1 is a modulator of fibroblast activation and a potential biomarker for gastric cancer. Int J Biol Sci. 2015;11:370-9. doi: 10.7150/ijbs.10666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao X, He Y, Gao J, Fan L, Li Z, Yang G, Chen H. Caveolin-1 expression level in cancer associated fibroblasts predicts outcome in gastric cancer. PLoS One. 2013;8:e59102. doi: 10.1371/journal.pone.0059102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sung CO, Lee KW, Han S, Kim SH. Twist1 is up-regulated in gastric cancer-associated fibroblasts with poor clinical outcomes. Am J Pathol. 2011;179:1827-38.. doi: 10.1016/j.ajpath.2011.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang D, Gao J, Wang S, Ye N, Chong Y, Huang Y, Wang J, Li B, Yin W, Wang D. Cancer-associated fibroblasts promote angiogenesis in gastric cancer through galectin-1 expression. Tumour Biol. 2016;37:1889-99. doi: 10.1007/s13277-015-3942-9 [DOI] [PubMed] [Google Scholar]
- 23.He XJ, Tao HQ, Hu ZM, et al: Expression of galectin-1 in carcinoma-associated fibroblasts promotes gastric cancer cell invasion through upregulation of integrin beta1. Cancer Sci. 2014;105:1402-10. doi: 10.1111/cas.12539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujii S, Fujihara A, Natori K, Abe A, Kuboki Y, Higuchi Y, Aizawa M, Kuwata T, Kinoshita T, Yasui W. et al.. TEM1 expression in cancer-associated fibroblasts is correlated with a poor prognosis in patients with gastric cancer. Cancer Med. 2015;4:1667-78. doi: 10.1002/cam4.515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia H, Wang C, Chen W, Zhang H, Chaudhury L, Zhou Z, Liu R, Chen C. Kruppel-like factor 5 transcription factor promotes microsomal prostaglandin E2 synthase 1 gene transcription in breast cancer. J Biol Chem. 2013;288:26731-40. doi: 10.1074/jbc.M113.483958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He M, Han M, Zheng B, Shu YN, Wen JK. Angiotensin II stimulates KLF5 phosphorylation and its interaction with c-Jun leading to suppression of p21 expression in vascular smooth muscle cells. J Biochem. 2009;146:683-91. doi: 10.1093/jb/mvp115 [DOI] [PubMed] [Google Scholar]
- 27.Zhu N, Gu L, Findley HW, Chen C, Dong JT, Yang L, Zhou M. KLF5 Interacts with p53 in regulating survivin expression in acute lymphoblastic leukemia. J Biol Chem. 2006;281:14711-8. doi: 10.1074/jbc.M513810200 [DOI] [PubMed] [Google Scholar]
- 28.Gao Y, Wu K, Chen Y, Zhou J, Du C, Shi Q, Xu S, Jia J, Tang X, Li F, et al: Beyond proliferation: KLF5 promotes angiogenesis of bladder cancer through directly regulating VEGFA transcription. Oncotarget. 2015;6:43791-805. doi: 10.18632/oncotarget.6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia L, Zhou Z, Liang H, Wu J, Shi P, Li F, Wang Z, Wang C, Chen W, Zhang H, et al.. KLF5 promotes breast cancer proliferation, migration and invasion in part by upregulating the transcription of TNFAIP2. Oncogene 2016;35:2040-51. doi: 10.1038/onc.2015.263 [DOI] [PubMed] [Google Scholar]
- 30.Zhang B, Zhang Z, Xia S, Xing C, Ci X, Li X, Zhao R, Tian S, Ma G, Zhu Z, et al.. KLF5 activates microRNA 200 transcription to maintain epithelial characteristics and prevent induced epithelial-mesenchymal transition in epithelial cells. Mol Cell Biol. 2013;33:4919-35. doi: 10.1128/MCB.00787-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeda N, Manabe I, Uchino Y, Eguchi K, Matsumoto S, Nishimura S, Shindo T, Sano M, Otsu K, Snider P, et al: Cardiac fibroblasts are essential for the adaptive response of the murine heart to pressure overload. J Clin Invest. 2010;120:254-65. doi: 10.1172/JCI40295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun R, Chen X, Yang VW: Intestinal-enriched Kruppel-like factor (Kruppel-like factor 5) is a positive regulator of cellular proliferation. J Biol Chem. 2001;276:6897-900. doi: 10.1074/jbc.C000870200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abe K, Sugiura H, Hashimoto Y, Ichikawa T, Koarai A, Yamada M, Numakura T, Onodera K, Tanaka R, Sato K, et al.. Possible role of Kruppel-like factor 5 in the remodeling of small airways and pulmonary vessels in chronic obstructive pulmonary disease. Respir Res. 2016;17:7. doi: 10.1186/s12931-016-0322-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Chen S, Wang W, et al: Cancer-associated fibroblasts promote hepatocellular carcinoma metastasis through chemokine-activated hedgehog and TGF-beta pathways. Cancer Lett. 2016;379:49-59. doi: 10.1016/j.canlet.2016.05.022 [DOI] [PubMed] [Google Scholar]
- 35.Tyan SW, Kuo WH, Huang CK, Pan CC, Shew JY, Chang KJ, Lee EY, Lee WH. Breast cancer cells induce cancer-associated fibroblasts to secrete hepatocyte growth factor to enhance breast tumorigenesis. PLoS One. 2011;6:e15313.. doi: 10.1371/journal.pone.0015313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rizvi S, Mertens JC, Bronk SF, Hirsova P, Dai H, Roberts LR, Kaufmann SH, Gores GJ. Platelet-derived growth factor primes cancer-associated fibroblasts for apoptosis. J Biol Chem. 2014;289:22835-49. doi: 10.1074/jbc.M114.563064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai YP, Shang K, Chen H, Ding F, Wang Z, Liang C, Xu Y, Sun MH, Li YY. FGF-1/-3/FGFR4 signaling in cancer-associated fibroblasts promotes tumor progression in colon cancer through Erk and MMP-7. Cancer Sci. 2015;106:1278-87. doi: 10.1111/cas.12745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.David L, Dulong V, Coquerel B, Le Cerf D Cazin L, Lamacz M, Vannier JP. Collagens, stromal cell-derived factor-1alpha and basic fibroblast growth factor increase cancer cell invasiveness in a hyaluronan hydrogel. Cell Prolif. 2008;41:348-64. doi: 10.1111/j.1365-2184.2008.00515.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soria G, Ben-Baruch A: The inflammatory chemokines CCL2 and CCL5 in breast cancer. Cancer Lett. 2008;267:271-85. doi: 10.1016/j.canlet.2008.03.018 [DOI] [PubMed] [Google Scholar]
- 40.Mrowietz U, Schwenk U, Maune S, Bartels J, Küpper M, Fichtner I, Schröder JM, Schadendorf D. The chemokine RANTES is secreted by human melanoma cells and is associated with enhanced tumour formation in nude mice. Br J Cancer. 1999;79:1025-31. doi: 10.1038/sj.bjc.6690164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cambien B, Richard-Fiardo P, Karimdjee BF, et al: CCL5 neutralization restricts cancer growth and potentiates the targeting of PDGFRbeta in colorectal carcinoma. PLoS One. 2011;6:e28842. doi: 10.1371/journal.pone.0028842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaday GG, Peehl DM, Kadam PA, Lawrence DM. Expression of CCL5 (RANTES) and CCR5 in prostate cancer. Prostate. 2006;66:124-34. doi: 10.1002/pros.20306 [DOI] [PubMed] [Google Scholar]
- 43.Fukui R, Nishimori H, Hata F, Yasoshima T, Ohno K, Nomura H, Yanai Y, Tanaka H, Kamiguchi K, Denno R, et al.. Metastases-related genes in the classification of liver and peritoneal metastasis in human gastric cancer. J Surg Res. 2005;129:94-100. doi: 10.1016/j.jss.2005.04.030 [DOI] [PubMed] [Google Scholar]
- 44.Wang T, Wei Y, Tian L, Song H, Ma Y, Yao Q, Feng M, Wang Y, Gao M, Xue Y. C-C motif chemokine ligand 5 (CCL5) levels in gastric cancer patient sera predict occult peritoneal metastasis and a poorer prognosis. Int J Sur. 2016;32:136-42.. doi: 10.1016/j.ijsu.2016.07.008 [DOI] [PubMed] [Google Scholar]
- 45.Sugasawa H, Ichikura T, Tsujimoto H, Kinoshita M, Morita D, Ono S, Chochi K, Tsuda H, Seki S, Mochizuki H. Prognostic significance of expression of CCL5/RANTES receptors in patients with gastric cancer. J Surg Oncol. 2008;97:445-50. doi: 10.1002/jso.20984 [DOI] [PubMed] [Google Scholar]
- 46.Mitra AK, Zillhardt M, Hua Y, Tiwari P, Murmann AE, Peter ME, Lengyel E. MicroRNAs reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov. 2012;2:1100-8. doi: 10.1158/2159-8290.CD-12-0206 [DOI] [PMC free article] [PubMed] [Google Scholar]


