Abstract
BACKGROUND: Current risk stratification systems for neuroblastoma patients consider clinical, histopathological, and genetic variables, and additional prognostic markers have been proposed in recent years. We here sought to select highly informative covariates in a multistep strategy based on consecutive Cox regression models, resulting in a risk score that integrates hazard ratios of prognostic variables. METHODS: A cohort of 695 neuroblastoma patients was divided into a discovery set (n = 75) for multigene predictor generation, a training set (n = 411) for risk score development, and a validation set (n = 209). Relevant prognostic variables were identified by stepwise multivariable L1-penalized least absolute shrinkage and selection operator (LASSO) Cox regression, followed by backward selection in multivariable Cox regression, and then integrated into a novel risk score. RESULTS: The variables stage, age, MYCN status, and two multigene predictors, NB-th24 and NB-th44, were selected as independent prognostic markers by LASSO Cox regression analysis. Following backward selection, only the multigene predictors were retained in the final model. Integration of these classifiers in a risk scoring system distinguished three patient subgroups that differed substantially in their outcome. The scoring system discriminated patients with diverging outcome in the validation cohort (5-year event-free survival, 84.9 ± 3.4 vs 63.6 ± 14.5 vs 31.0 ± 5.4; P < .001), and its prognostic value was validated by multivariable analysis. CONCLUSION: We here propose a translational strategy for developing risk assessment systems based on hazard ratios of relevant prognostic variables. Our final neuroblastoma risk score comprised two multigene predictors only, supporting the notion that molecular properties of the tumor cells strongly impact clinical courses of neuroblastoma patients.
Introduction
Neuroblastoma is the most common extracranial solid cancer in childhood [1]. The clinical courses of the disease are remarkably diverse, ranging from spontaneous regression to fatal progression [2], [3], [4]. Accordingly, current treatment stratification systems cover a broad spectrum of therapeutic strategies. These may vary between a “wait-and-see” approach for patients in whom the tumor is expected to regress spontaneously and intensive multimodal treatment for patients who are at high risk to die from the disease. Accurate risk assessment of neuroblastoma patients at diagnosis is thus essential for selecting the most appropriate first-line therapy. In current treatment stratification systems, the clinical prognostic variables “age at diagnosis” and “stage of disease” are placed in the center of neuroblastoma risk estimation [5], [6], [7], although their exact definition is under continuous discussion [6], [8].
A number of molecular markers have been established for neuroblastoma risk assessment, some of which are currently in clinical use. Among others, these markers include amplification status of the proto-oncogene MYCN [9], copy number status of chromosomes 1p and 11q [5], [10], [11], ploidy of the tumor cells [12], [13], and numerical and segmental copy number alterations [14], [15]. Furthermore, several other genetic alterations, such as activating ALK mutations [16], [17], [18], inactivating mutations of the ATRX gene [19], [20], and rearrangements of the TERT locus [21], [22], have been reported to impact clinical outcome. In addition to genomic alterations, gene expression–based classifiers have been supposed to predict patient outcome with high accuracy [23], [24], [25], [26], [27]. In previous studies, the prognostic value of such novel biomarkers has been examined in patient subgroups defined by well-established markers, such as stage, age, and MYCN status, to assess whether the new variables can contribute to already existing risk estimation systems [23], [24], [25], [26], [27]. Due to the plethora of potentially relevant markers, however, it has remained challenging to determine the most appropriate combination of prognostic variables for optimal risk assessment in neuroblastoma.
Here, we developed an alternative approach to integrate prognostic markers for risk estimation of neuroblastoma patients. We implemented a multistep strategy based on consecutive Cox regression models, thereby avoiding subgroup analyses completely. We considered all prognostic variables currently used for treatment stratification in Germany as well as four distinct multigene classifiers developed previously [23]. Relevant variables were first selected in a stepwise procedure and then integrated in a prognostic index that was translated into a new risk score. The final risk score consists of two gene expression–based classifiers only, thus emphasizing the power of gene expression–based classification for outcome prediction in neuroblastoma.
Patients and Methods
Patient Cohort
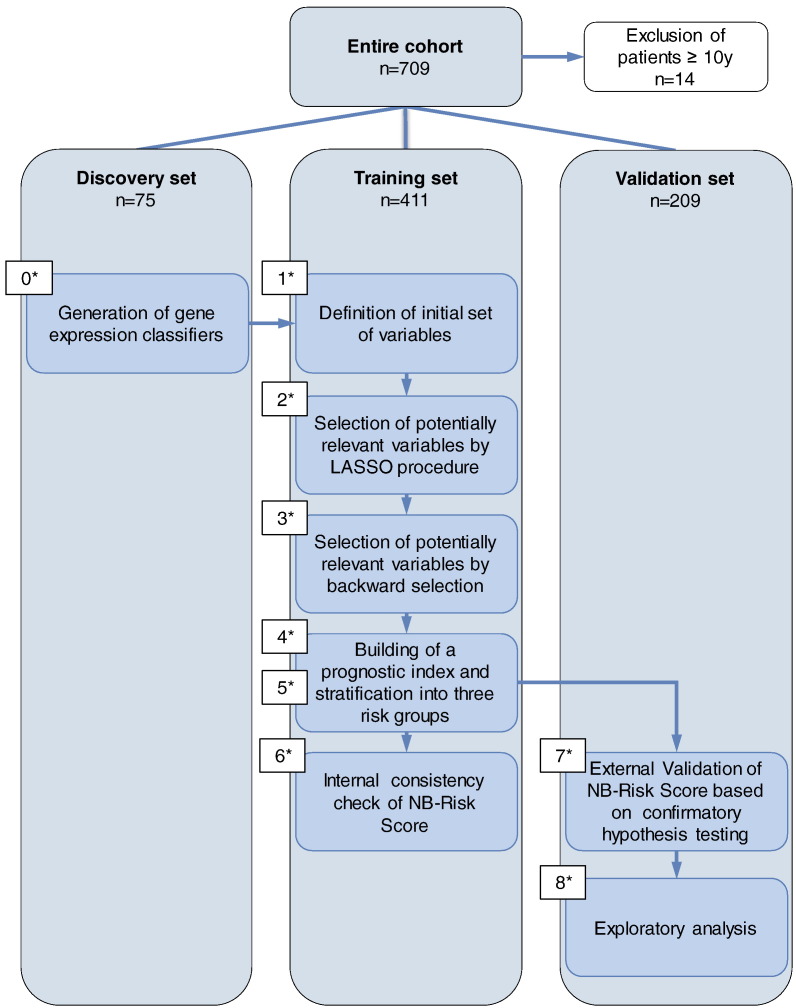
The study was performed on a neuroblastoma patient cohort that has been described previously (n = 709) [23]. All patients were registered in the respective clinical trials with informed consent. We considered only patients below 10 years of age since neuroblastoma in adolescents and adults is rare (<5% of all neuroblastomas [28]), and both patient clinical courses and molecular profiles of the tumors differ from neuroblastoma of younger children [29], [30]. In order to prevent outlier effects of these atypical courses on model development, we therefore excluded 14 patients older than 10 years at diagnosis (2%), leaving a cohort of 695 patients for analysis. The entire cohort was divided into discovery, training, and validation sets (Table 1 and Figure 1). The discovery set (n = 75) was used for generation of gene expression–based classifiers, the training set (n = 411) for score building, and the validation set (n = 209) for external validation of the score. Thus, score building and validation were performed on two independent data sets to address multiple testing issues and potential overfitting of underlying models.
Table 1.
Patient Characteristics of the Entire Cohort, the Discovery Set, the Training Set, and the Validation Set; Absolute and Relative Frequencies Are Indicated
| Entire Cohort |
Discovery Set |
Training Set |
Validation Set |
P† |
|||||
|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | ||
| 695 | 75 | 411 | 209 | ||||||
| Age at diagnosis | |||||||||
| <18 months | 433 | 62 | 51 | 68 | 251 | 61 | 131 | 63 | .528 |
| ≥18 months | 262 | 38 | 24 | 32 | 160 | 39 | 78 | 37 | |
| <30 months | 512 | 74 | 58 | 77 | 300 | 73 | 154 | 74 | .775 |
| ≥30 months | 183 | 26 | 17 | 23 | 111 | 27 | 55 | 26 | |
| Stage | .002 | ||||||||
| Stage 1 | 156 | 22 | 27 | 36 | 90 | 22 | 39 | 19 | |
| Stage 2 | 116 | 17 | 14 | 19 | 68 | 17 | 34 | 16 | |
| Stage 3 | 91 | 13 | 3 | 4 | 61 | 15 | 27 | 13 | |
| Stage 4 | 252 | 36 | 18 | 24 | 156 | 38 | 78 | 37 | |
| Stage 4S | 80 | 12 | 13 | 17 | 36 | 9 | 31 | 15 | |
| MYCN⁎ | .281 | ||||||||
| Nonamplified | 571 | 82 | 66 | 88 | 337 | 82 | 168 | 80 | |
| Amplified | 118 | 17 | 8 | 11 | 71 | 17 | 39 | 19 | |
| N.D. | 6 | 1 | 1 | 1 | 3 | 1 | 2 | 1 | |
| 1p⁎ | .253 | ||||||||
| No deletion | 420 | 60 | 60 | 80 | 238 | 58 | 122 | 58 | |
| Deletion | 146 | 21 | 13 | 17 | 88 | 21 | 45 | 22 | |
| N.D. | 129 | 19 | 2 | 3 | 85 | 21 | 42 | 20 | |
| Gene expression–based classifier NB-th10 | 1.000 | ||||||||
| Favorable | 248 | 60 | 126 | 60 | |||||
| Unfavorable | 163 | 40 | 83 | 40 | |||||
| Gene expression–based classifier NB-th24 | 1.000 | ||||||||
| Favorable | 228 | 56 | 116 | 56 | |||||
| Unfavorable | 183 | 45 | 93 | 45 | |||||
| Gene expression–based classifier NB-th26 | .733 | ||||||||
| Favorable | 228 | 56 | 119 | 57 | |||||
| Unfavorable | 183 | 44 | 90 | 43 | |||||
| Gene expression–based classifier NB-th44 | .726 | ||||||||
| Favorable | 257 | 63 | 127 | 61 | |||||
| Unfavorable | 154 | 38 | 82 | 39 | |||||
Missing information on MYCN in 3/2 (1%/1%) and on chromosome 1p status in 85/42 (21%/20%) patients of the training/validation cohort, respectively.
P value of Fisher's exact test comparing the distribution of the respective variables in the cohorts.
Figure 1.
Flow diagram of the study. The entire patient cohort was divided into three subsets. All patients older than 10 years of age were excluded from the study (n = 14). In a previous step (step 0), gene expression–based classifiers had been generated on a discovery set. A risk score (referred to as NB-Risk Score) was developed and evaluated on a separate training set (steps 1-6). Finally, the performance of the risk score in predicting patient outcome was evaluated in an independent validation set (steps 7 and 8). The different steps of NB-Risk Score building and validation procedure are numbered in the appropriate order.
Generation of Gene Expression–Based Classifiers
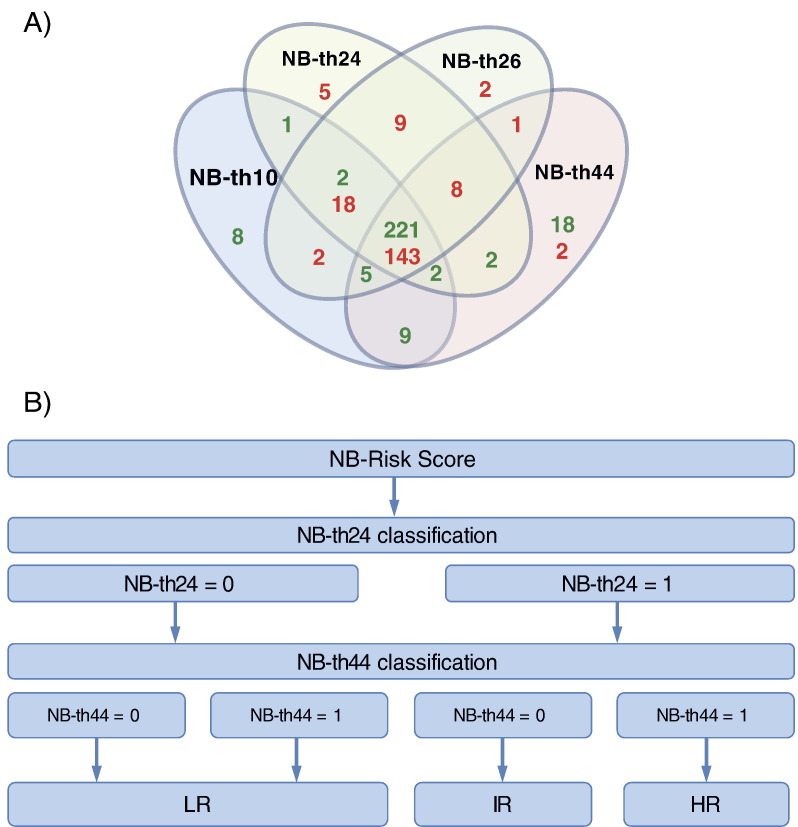
Gene expression profiles of all 695 neuroblastoma samples had been generated previously using customized 4x44K oligonucleotide microarrays (Agilent Technologies) [23]. Gene expression–based classifiers had been developed on a cohort of 75 tumors (discovery set) from patients with maximally divergent clinical courses as described elsewhere [23] (see Supplementary Material). Expression data and basic clinical information are available through ArrayExpress (http://www.ebi.ac.uk/arrayexpress; accession: E-MTAB-1781). As the classifiers led to discrepant results for a number of patients in the training set (Figure 2A), we considered all four predictors for risk score building.
Figure 2.
(A) Venn diagram of concordant and discordant classification results of the four different gene expression classifiers (NB-th10, -th24, -th26, and -th44) within the training set. The numbers of tumors classified as favorable or unfavorable are highlighted in green and red, respectively. (B) Schematic representation of the NB-Risk Score that considers classification results and hazard ratios of the two gene expression–based classifiers, NB-th24 and NB-th44. Favorable classification, 0; unfavorable classification, 1; LR, low risk; IR, intermediate risk; HR, high risk.
NB-Risk Score Building and Validation Procedure
A stepwise modeling strategy was implemented to ensure reliable and robust model development and validation. The endpoint event-free survival (EFS) was used for score building. Score validation was performed on both EFS and overall survival (OS). EFS was calculated from time of diagnosis until event, defined as recurrence, progression ,or death from disease, and OS was calculated from time of diagnosis until death from disease or last contact for patients alive. Cases of death due to other causes occurred in 10 patients of the entire cohort and were considered as censoring events. Selection of prognostic markers for model building consisted of consecutive univariable Cox regression, multivariable L1-penalized least absolute shrinkage and selection operator (LASSO) Cox regression, and backward selection based on Wald test in a multivariable Cox model. We used the LASSO procedure [31], which simultaneously performs variable selection and shrinkage of regression coefficients, to reduce the number of potentially relevant variables to the most relevant ones. This step enabled backward selection on the remaining variables, leading to further reduction to the final set of variables that are relevant and can be estimated with adequate precision. The parameter estimates from the final model were used to build a prognostic index defined as the linear predictor function of the final Cox regression model, and three risk groups were built based on an optimal stratification of the prognostic index, referred to as NB-Risk Score. Detailed information on the consecutive steps as well as the statistical analyses of the NB-Risk Score building and validation procedure is given in the Supplementary Material.
Analysis of Genetic Alterations
Global copy number alterations of the tumors were assessed by array-based comparative genomic hybridization (aCGH) as described previously [32]. Copy number alterations at chromosome 11q and the ALK locus were determined by fluorescence in situ hybridization as described [33]. Whole-genome sequencing data of tumors and matched normal controls have been published elsewhere [21]. Single nucleotide variants of ALK were determined by dideoxy-sequencing or targeted massively parallel sequencing as described previously [17], [21].
Results
Development of the NB-Risk Score
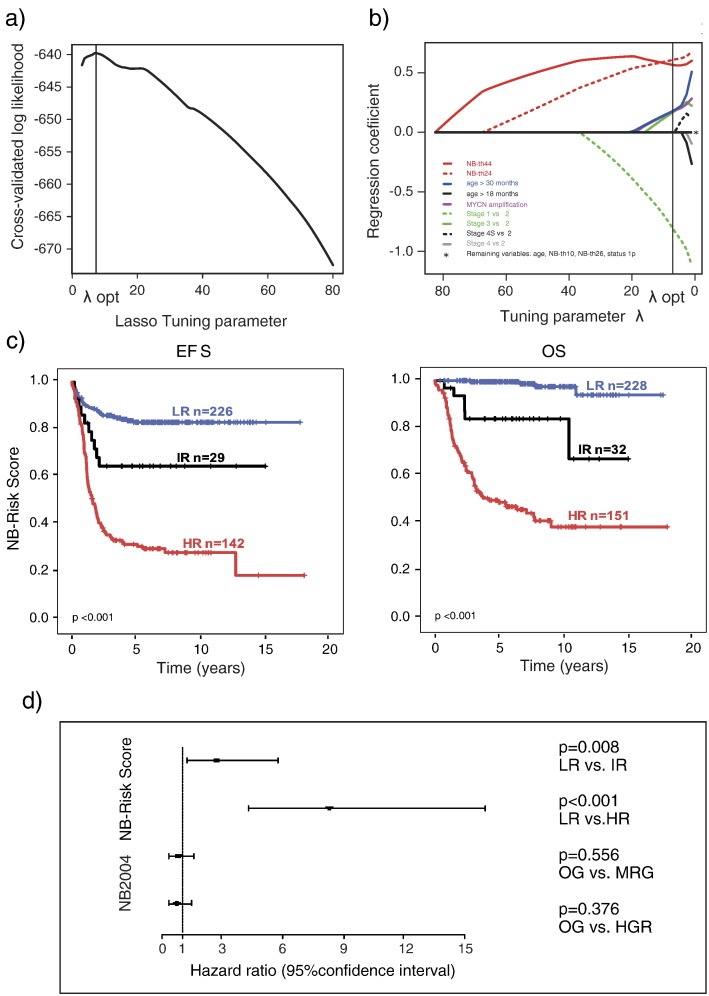
Gene expression–based classifiers had been developed in our previous work on the discovery set [23] (step 0). As a first step in NB-Risk Score development, we determined the prognostic value of each variable separately in the training set by univariable analysis (step 1). As expected, all markers turned out to be highly informative and robustly discriminated patients with favorable and unfavorable EFS (P < .001 each; Supplementary Table 1). At step 2 of the score building procedure, we performed multivariable Cox regression analysis according to the LASSO procedure [31]. A LASSO parameter of λopt = 7.01 turned out to be optimal, maximizing the cross-validated log likelihood (Supplementary Figure 1A). We found that only five variables were retained after this step at the optimal LASSO tuning parameter: stage of disease, age at diagnosis (≤30 vs >30 months), MYCN amplification status, and the gene expression classifiers NB-th24 and NB-th44 (Supplementary Figure 1B).
Supplementary Figure 1.
(A) Optimization of the LASSO tuning parameter regarding the criterion of cross-validated log likelihood in step 2 of the score building procedure. (B) Results of multivariable L1 penalized (LASSO) Cox regression with time-to-event variable EFS (step 2 of the score building procedure). The fitted regression coefficients are shown in dependence of the LASSO tuning parameter. Variables not shrunk to zero at the optimal tuning parameter λopt are highlighted in colors and were included in the next step of score building. (C) EFS and OS of patients of the training set (n = 411) stratified according to the NB-Risk Score. LR, low risk; IR, intermediate risk; HR, high risk. (D) Multivariable Cox regression analysis of risk groups defined by the German NB2004 trial and the NB-Risk Score for EFS in patients of the training set. OG, observation group; MRG, intermediate-risk group; HRG, high-risk group according to NB2004 stratification and LR, low risk; IR, intermediate-risk; HR, high risk according to NB-Risk Score stratification.
To improve model precision, we next performed backward selection based on Wald test in a Cox regression model including the five previously identified variables (step 3). We observed that only the gene expression–based classifiers NB-th24 and NB-th44 contributed independently to the final model (Table 2). As both classifiers discriminate only between favorable (0) and unfavorable (1) outcome, we thus obtained a simple prognostic index (PI, step 4) that is calculated for every patient considering both the individual classification result (0 vs 1) and the hazard ratio of each gene expression–based classifier (Supplementary Material). Several of the genes included by the classifiers NB-th24 and NB-th44 have been related to neuroblastoma previously (e.g., CD47 [34], CNR1 [35], [36], and DST [36]; Supplementary Table 2). To determine which molecular pathways or processes may be represented by the genes of the classifiers, we performed Gene Ontology enrichment analysis; however, no functional category was identified by this approach.
Table 2.
Selection of Prognostic Variables in Step 3 of the Score Building Procedure Using Multivariable Backward Selection in a Cox Regression for EFS
| Variable | Available Cases (n)* | HR | 95% CI | P |
|---|---|---|---|---|
| NB-th24† | ||||
| Unfavorable vs favorable | 144 vs 253 | 2.64 | 1.38-5.08 | .004 |
| NB-th44† | ||||
| Unfavorable vs favorable | 171 vs 226 | 2.40 | 1.31-4.40 | .005 |
| MYCN amplification‡ | ||||
| Yes vs no | 394 | - | - | N/S (.246) |
| Stage | ||||
| 1, 4S, 2, 3, or 4, ref.: 1 | 397 | - | - | N/S (.092) |
| Age at diagnosis | ||||
| ≥30 vs < 30 months | 397 | - | - | N/S (.364) |
N/S, not selected; P, P value of Wald/score test at the final step of backward selection for selected/not selected variables; HR, hazard ratio; CI, confidence interval.
Available cases n = 397 (145 events), missing information on EFS in 14 patients.
No interaction terms were selected, indicating that the simultaneous risk of both NB-th24 and NB-th44 results in a multiplied HR of 6.4.
Missing information on MYCN in 3 patients.
We next used the prognostic index to define three risk groups based on optimal stratification of the training set patients according to EFS (step 5). The final risk stratification, referred to as NB-Risk Score, is thus a two-step dichotomous decision process that considers NB-th24 prediction on the first level and NB-th44 prediction on the second level to allocate neuroblastoma patients to three distinct risk groups (Figure 2B).
Validation of the Prognostic Value of the NB-Risk Score
We first evaluated the prognostic accuracy of the NB-Risk Score internally in the training set that had been used for model development (step 6). In this cohort, clinical outcome of patients in the three risk groups differed substantially in terms of both EFS (5-year EFS, 0.832 ± 0.026 vs 0.648 ± 0.090 vs 0.320 ± 0.040; P < .001) and OS (5-year OS, 0.995 ± 0.005 vs 0.840 ± 0.060 vs 0.490 ± 0.042; P < .001; Supplementary Figure 1C). We also assessed the prognostic value of the NB-Risk Score and NB2004 risk stratification by multivariable Cox regression analysis and observed that only the NB-Risk Score variables “intermediate risk” (P = .008) and “high risk” (P < .001) were retained in the final model based on EFS (Supplementary Figure 1D). Analogous models for OS could not be fitted, indicating substantial convergence of the NB-Risk Score and NB2004 stratification system for OS.
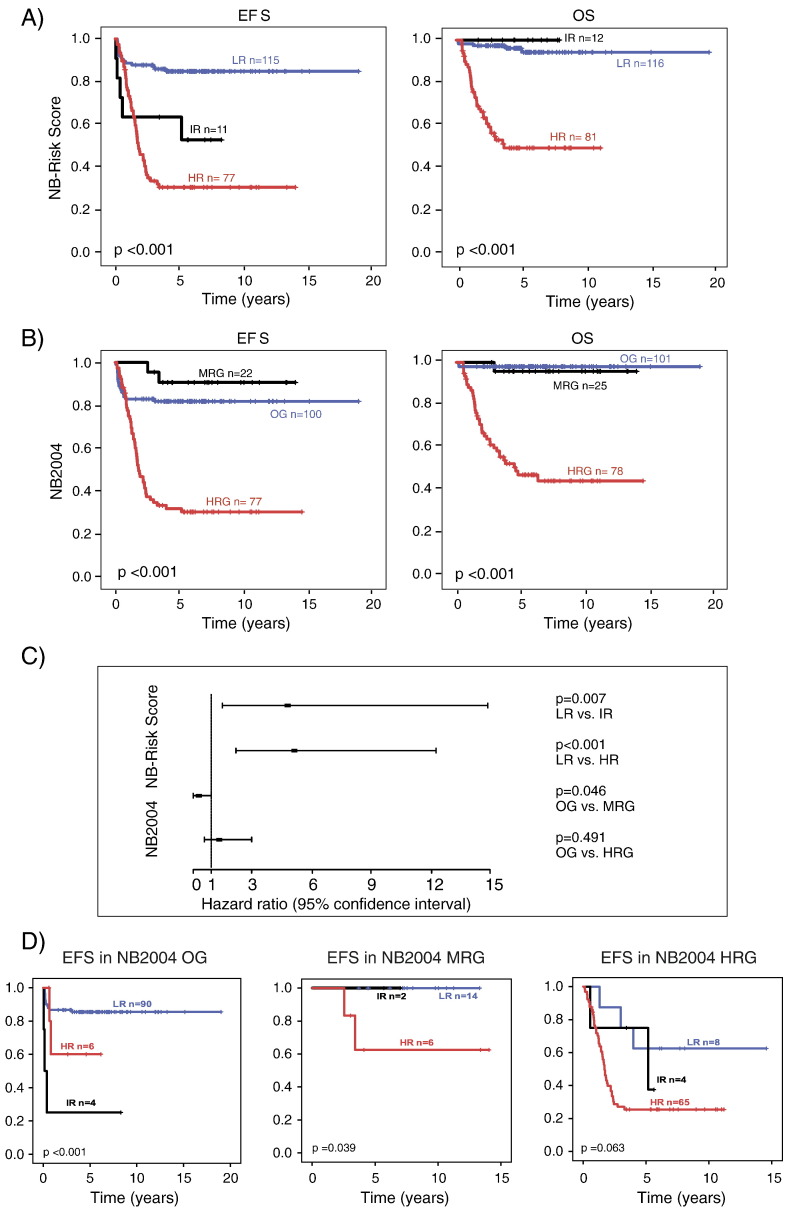
To validate the NB-Risk Score, we examined the performance of the score externally in the validation set (n = 209, step 7) by confirmatory hypothesis testing (Supplementary Material). The validity of the prognostic index in predicting EFS was substantiated by Cox regression analysis (step 8; βPI = 1.000; 95% confidence interval = 0.702-1.230). Allocation of patients to low-, intermediate-, and high-risk groups according to the NB-Risk Score demonstrated that clinical outcome of validation set patients differed substantially between the three risk groups for both EFS (5-year EFS, 0.849 ± 0.034 vs 0.636 ± 0.145 vs 0.310 ± 0.054) and OS (5-year-OS, 0.962 ± 0.019 vs 1.000 ± 0.000 vs 0.498 ± 0.060; Figure 3A). Patients of the low-risk group had significantly better EFS and OS than high-risk patients (P < .001 each; rejection of null hypotheses H2 and H5, see Supplementary Material). In addition, intermediate-risk patients had significantly worse EFS than low-risk patients, while OS of intermediate-risk patients was significantly better than that of high-risk patients (P = .011 each; rejection of hypotheses H1 and H6). We did not observe differences in terms of EFS between the intermediate- and high-risk group and in terms of OS between the low- and intermediate-risk group (P = .283 and P = .773, respectively; no rejection of hypotheses H3 and H4). This finding may suggest that the NB-Risk Score identifies children as intermediate-risk patients who are at increased risk for disease progression or relapse but may have excellent overall survival with appropriate treatment.
Figure 3.
(A) EFS and OS of patients of the validation set (n = 209) stratified according to the NB-Risk Score. (B) EFS and OS of patients of the validation set (n = 209) stratified according to the NB2004 Risk assessment. (C) Multivariable Cox regression analysis of risk groups defined by the German NB2004 trial and the NB-Risk Score for EFS within the validation set. OG, observation group; MRG, intermediate-risk group; HRG, high-risk group according to NB2004 stratification and LR, low risk; IR, intermediate risk; HR, high risk according to NB-Risk Score stratification. (D) EFS of patients of the validation set stratified according to the NB-Risk Score within NB2004 subgroups OG (observation group), MRG (intermediate-risk group) and HRG (high-risk group): LR, low risk; IR, intermediate risk; HR, high risk group.
We next aimed to assess the prognostic value of NB-Risk Score classification in comparison to stratification by the NB2004 system. Kaplan-Meier estimates for EFS and OS were similar between both stratification systems (Figure 3, A and B), except that EFS of patients of the NB2004 intermediate-risk group (MRG) was exceptionally favorable. This observation may be due to a sampling bias of this minor patient subgroup. Alternatively, it has to be considered that the majority of patients in the NB2004 observation group (OG) did not receive chemotherapy, which may have resulted in relatively worse EFS of OG patients in comparison to MRG patients. We finally assessed performances of the NB-Risk Score and the NB2004 system in the validation cohort by multivariable Cox regression analysis built on EFS. We found that only the NB-Risk Score strata “intermediate risk” (P = .007) and “high risk” (P < .001) were independent prognostic variables (Figure 3C). Examination of patients who were discordantly classified by the two risk estimation systems revealed that, within the NB2004 observation group, patients classified as intermediate or high risk by the NB-Risk Score had worse outcome than those classified as low risk (Figure 3D). Conversely, patients classified as low or intermediate risk by the NB-Risk Score within the NB2004 high-risk group had better outcome than those classified as high risk, although statistical significance was marginal in this subgroup (P = .063; Figure 3D). Together, these findings demonstrate that the NB-Risk Score consisting of two multigene predictors only is able to accurately predict outcome of neuroblastoma patients.
Comparison of Prognostic Genomic Alterations with NB-Risk Score Classification
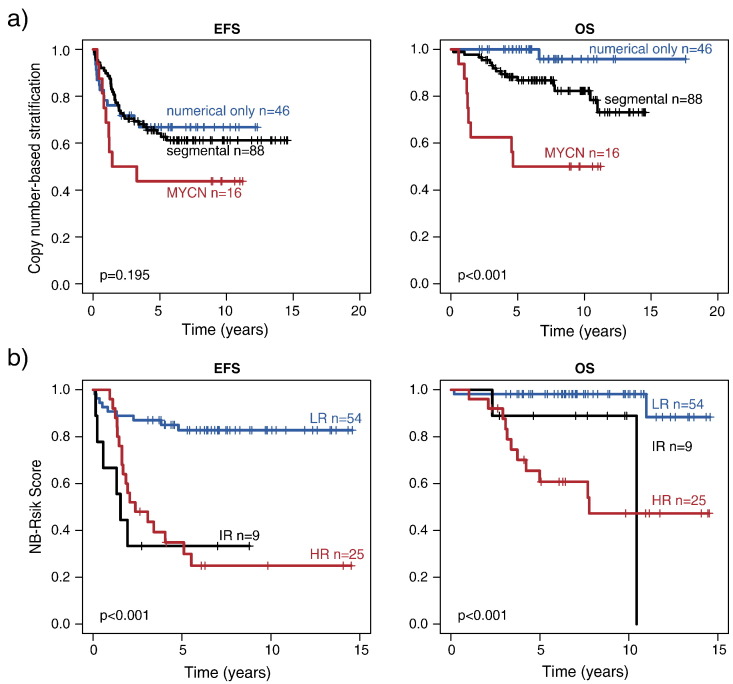
We finally evaluated the association of NB-Risk Score classification with genomic alterations that are supposed to impact clinical courses in neuroblastoma. Genomic loss of 11q occurred in 73/334 cases of the combined training and validation set, and was significantly associated with high-risk classification by the NB-Risk Score (P < .001; Supplementary Table 3). We also assessed the prognostic impact of numerical and segmental copy number alterations according to Janoueix-Lerosey and coworkers [14] in 150 cases that had been analyzed by aCGH. In line with that study, we found that numerical copy number alterations were associated with excellent outcome, while outcome of patients whose tumors harbored segmental alterations or MYCN amplification was significantly worse (Supplementary Figure 2A). Comparison of the copy number–based classification with that of the NB-Risk Score revealed that genomic subgroups bearing numerical alterations only and MYCN amplification were significantly associated with the low-risk and high-risk groups of the NB-Risk Score, respectively (both P < .001; Supplementary Table 4). By contrast, the genomic subgroup bearing segmental alterations was separated by the NB-Risk Score into low-, intermediate-, and high-risk subgroups, corresponding to favorable, intermediate, and poor patient outcome (Supplementary Figure 2B).
Supplementary Figure 2.
(A) EFS and OS of 150 patients stratified according to a genomic copy number–based classification as proposed by Janoueix-Lerosey and coworkers [14]. (B) EFS and OS of patients with segmental copy number alterations (n = 88) stratified according to the NB-Risk Score. LR, low risk; IR, intermediate risk; HR, high risk.
Genomic alterations of several genes have been suggested to impact clinical outcome of neuroblastoma patients. We determined single nucleotide variants and copy numbers of ALK in 244 and 185 cases, respectively. While ALK amplification occurred exclusively in the high-risk group (n = 4, P = .002; Supplementary Table 5), we did not observe a significant association of ALK single nucleotide variants with risk groups defined by the NB-Risk Score (P = .644; Supplementary Table 5). While ALK mutations have been previously associated with unfavorable clinical courses in general, it has been also demonstrated that such alterations occur in neuroblastomas covering the entire spectrum of the disease [16], [17], which may explain the lack of significance in our cohort. In addition, we took whole-genome sequencing data of 32 cases into account and found that rearrangements of the TERT locus (high risk, n = 5; intermediate risk, n = 1), ATRX mutations (high risk, n = 2), PTPRD deletion (high risk, n = 1), and chromothripsis (high risk, n = 2; intermediate risk, n = 1) were predominantly detected in tumors classified as high risk by the NB-Risk Score. Similarly, evidence for alternative lengthening of telomeres was mainly found in patients classified to be at high risk (high risk, n = 4; intermediate risk, n = 1; low risk, n = 1). While these findings may be limited by the small sample size, they point towards association of unfavorable molecular alterations with high-risk classification by the NB-Risk Score.
Discussion
Current risk assessment systems for neuroblastoma patients are taking combinations of clinical, histopathological, and genetic prognostic markers into account [5], [6]. Results from clinical and molecular studies suggest that current risk assessment systems are still imperfect [4], [11], [23], [37], resulting in over- or undertreatment of a fraction of patients. Over recent years, several studies have proposed that molecular markers such as gene expression classifiers [23], [24], [25], [26], [27], copy number alteration alterations [10], [11], [14], [33], [38], and somatic mutation patterns [16], [17], [19], [20], [21], [22] may precisely reflect the biology of the tumor. In a study of Tomioka and coworkers, both gene expression– and copy number–based signatures were accurate and independent prognostic markers, suggesting that combination of both may improve treatment stratification of the patients [15]. It has remained unclear to date, however, how these novel biomarkers can be integrated best into existing risk estimation systems.
In a classical approach, the potential clinical utility of prognostic biomarkers is being examined by determining their predictive power in patient subgroups, defined by established prognostic variables [14], [23], [24], [25], [26], [27]. While this strategy can provide valuable information on how to integrate such biomarkers in existing risk stratification, it may prevent substitution of established markers by novel variables. In contrast to this practice, we here present an unbiased strategy to develop risk assessment systems that renounce completely on subgroup analyses. In this approach, we selected the most informative variables in a stepwise procedure in consecutive Cox regression analyses [39], [40] and integrated them along with their hazard ratios in a prognostic index. We hypothesized that the combination of different statistical approaches in a sequential model building strategy may multiply their benefits and compensate for potential shortcomings of the approaches, thus resulting in increased model stability.
Using our stepwise model building approach, we demonstrate that risk stratification of neuroblastoma patients is highly accurate when considering gene expression information only, even in the absence of established prognostic markers such as stage, age, and MYCN status. In addition, we show that integration of more than one gene expression–based classifier in a risk estimation score may improve outcome prediction of patients. Stratification according to the NB-Risk Score readily discriminated two major subgroups, consisting of patients with excellent outcome and patients who are at high risk to die from the disease. In addition, the NB-Risk Score delineated a small group of intermediate-risk patients, similar to current risk stratification systems. In the validation set, these patients had relatively poor EFS but excellent OS. Events had occurred in 5 of the 12 patients in this subgroup, 3 of which were localized progressions in clinically low-risk patients who had not been treated with chemotherapy upfront but only at the time of progression. Today, all three patients have survived without further events for 4 years or more. One event occurred as localized progression in a 17-month-old patient with stage 4 disease following chemotherapy. The tumor was resected after one additional cycle of chemotherapy. Since histology turned out to be ganglioneuroblastoma, cytotoxic treatment was discontinued, and the patient has survived event-free for more than 9 years to date. Finally, one event occurred in a 4-year-old patient with stage 4 disease, who relapsed after treatment according to the high-risk protocol. After having received second-line chemotherapy, the patient has now survived event-free for more than 3 years. Four of the remaining 7 patients had received chemotherapy upfront, one patient had been resected completely, and one had been observed only; for one patient, no information on treatment was available. Together, these data suggest that the majority of patients classified as intermediate risk by the NB-Risk Score may have favorable outcome with appropriate therapy.
We found that genomic alterations associated with unfavorable disease, such as 11q loss, occurred predominantly in patients classified as high risk by the NB-Risk Score, thus supporting the notion that molecular characteristics of the tumor strongly impact clinical outcome of the patients. As an exception, we observed that patients whose tumors harbored segmental copy number alterations [14] were separated into subgroups with favorable, intermediate, and poor outcome. These data suggest that gene expression–based classification may be able to improve stratification based on copy number alterations; however, this finding needs to be validated in larger and independent patient cohorts.
Various gene expression–based classifiers have been proposed by different research groups [23], [24], [25], [26], [27]. As gene signatures of the distinct classifiers hardly overlap, concerns have been raised on their general applicability. Taking the enormous number of genes differentially expressed between favorable and unfavorable neuroblastoma subtypes into account [41], however, it is comprehensible that different bioinformatics algorithms applied on different gene expression datasets generated on different platforms may not yield exactly the same results. In fact, the reliability and robustness of gene expression–based risk assessment in neuroblastoma have been demonstrated by 1) validation of the prognostic value of an expression signature in a prospective setting [24] and 2) demonstrating that prognostic signatures developed on microarray-based expression profiles can be readily transferred to RNA-seq data and vice versa [42].
It has to be considered that our study has been performed in retrospect on neuroblastoma patients who had been treated in different trials, both from Germany and from other countries [23]. The clinical value of risk estimation by evaluating prognostic variables retrospectively may be limited by the fact that treatment effects on patient outcome cannot be taken fully into account, which applies not only to the NB-Risk Score but also to the NB2004 stratification system in this study. The fact that the NB-Risk Score performed excellently in the independent validation cohort, however, substantiates its robustness. Nevertheless, the clinical relevance of our novel risk stratification system needs to be validated in prospective clinical trials. Based on our previous studies [24], we expect that about 75% of neuroblastoma samples in Germany will currently meet the requirements for microarray-based expression analysis (e.g., sufficient amount of tumor tissue, tumor infiltration grade of at least 50%, high-quality RNA). Implementation of a risk estimation system based on multigene predictors thus appears to be feasible.
Conclusion
Together, we here propose a multistep strategy to establish risk estimation systems by identifying and integrating prognostic variables using consecutive multivariable Cox regression analyses. Using this strategy, we developed a highly accurate neuroblastoma risk assessment system consisting of prognostic information from two gene expression–based classifiers only. Our results support the notion that neuroblastoma is a molecularly defined disease [1], [14], [15], [19], [20], [21], [24], [26], [27] and that clinical variables, such as stage of disease or age at diagnosis, are essentially reflected by the genetic properties of the tumor cells.
The following are the supplementary data related to this article.
Annotations of All Probes of the Two Gene Expression–Based Classifiers NB-th44 and NB-th24
Supplementary material
Acknowledgments
Acknowledgements
This work was supported by the German Ministry of Science and Education as part of the e:Med initiative (grant 01ZX1303A and 01ZX1307D to M.F.), the Fördergesellschaft Kinderkrebs-Neuroblastom-Forschung e.V. (to M.F. and F.B.), and the Center for Molecular Medicine Cologne.
Conflict of Interest
All authors declare that there is no conflict of interests.
Footnotes
Funding: This work was supported by the German Ministry of Science and Education as part of the e:Med initiative (grant 01ZX1303A and 01ZX1307D to M.F.), the Fördergesellschaft Kinderkrebs-Neuroblastom-Forschung e.V. (to M.F. and F.B.), and the Center for Molecular Medicine Cologne.
References
- 1.Bosse KR, Maris JM. Advances in the translational genomics of neuroblastoma: From improving risk stratification and revealing novel biology to identifying actionable genomic alterations. Cancer. 2016;122:20–33. doi: 10.1002/cncr.29706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodeur GM, Bagatell R. Mechanisms of neuroblastoma regression. Nat Rev Clin Oncol. 2014;11:704–713. doi: 10.1038/nrclinonc.2014.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hero B, Simon T, Spitz R, Ernestus K, Gnekow AK, Scheel-Walter HG, Schwabe D, Schilling FH, Benz-Bohm G, Berthold F. Localized infant neuroblastomas often show spontaneous regression: results of the prospective trials NB95-S and NB97. J Clin Oncol. 2008;26:1504–1510. doi: 10.1200/JCO.2007.12.3349. [DOI] [PubMed] [Google Scholar]
- 5.Brodeur GM, Pritchard J, Berthold F, Carlsen NL, Castel V, Castelberry RP, De Bernardi B, Evans AE, Favrot M, Hedborg F. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J Clin Oncol. 1993;11:1466–1477. doi: 10.1200/JCO.1993.11.8.1466. [DOI] [PubMed] [Google Scholar]
- 6.Cohn SL, Pearson AD, London WB, Monclair T, Ambros PF, Brodeur GM, Faldum A, Hero B, Iehara T, Machin D. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol. 2009;27:289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.London WB, Castleberry RP, Matthay KK, Look AT, Seeger RC, Shimada H, Thorner P, Brodeur G, Maris JM, Reynolds CP. Evidence for an age cutoff greater than 365 days for neuroblastoma risk group stratification in the Children's Oncology Group. J Clin Oncol. 2005;23:6459–6465. doi: 10.1200/JCO.2005.05.571. [DOI] [PubMed] [Google Scholar]
- 8.Monclair T, Brodeur GM, Ambros PF, Brisse HJ, Cecchetto G, Holmes K, Kaneko M, London WB, Matthay KK, Nuchtern JG. The International Neuroblastoma Risk Group (INRG) staging system: an INRG Task Force report. J Clin Oncol. 2009;27:298–303. doi: 10.1200/JCO.2008.16.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 10.Attiyeh EF, London WB, Mosse YP, Wang Q, Winter C, Khazi D, McGrady PW, Seeger RC, Look AT, Shimada H. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353:2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- 11.Simon T, Spitz R, Faldum A, Hero B, Berthold F. New definition of low-risk neuroblastoma using stage, age, and 1p and MYCN status. J Pediatr Hematol Oncol. 2004;26:791–796. [PubMed] [Google Scholar]
- 12.Pinto NR, Applebaum MA, Volchenboum SL, Matthay KK, London WB, Ambros PF, Nakagawara A, Berthold F, Schleiermacher G, Park JR. Advances in Risk Classification and Treatment Strategies for Neuroblastoma. J Clin Oncol. 2015;33:3008–3017. doi: 10.1200/JCO.2014.59.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maris JM. The biologic basis for neuroblastoma heterogeneity and risk stratification. Curr Opin Pediatr. 2005;17:7–13. doi: 10.1097/01.mop.0000150631.60571.89. [DOI] [PubMed] [Google Scholar]
- 14.Janoueix-Lerosey I, Schleiermacher G, Michels E, Mosseri V, Ribeiro A, Lequin D, Vermeulen J, Couturier J, Peuchmaur M, Valent A. Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol. 2009;27:1026–1033. doi: 10.1200/JCO.2008.16.0630. [DOI] [PubMed] [Google Scholar]
- 15.Tomioka N, Oba S, Ohira M, Misra A, Fridlyand J, Ishii S, Nakamura Y, Isogai E, Hirata T, Yoshida Y. Novel risk stratification of patients with neuroblastoma by genomic signature, which is independent of molecular signature. Oncogene. 2008;27:441–449. doi: 10.1038/sj.onc.1210661. [DOI] [PubMed] [Google Scholar]
- 16.Bresler SC, Weiser DA, Huwe PJ, Park JH, Krytska K, Ryles H, Laudenslager M, Rappaport EF, Wood AC, McGrady PW. ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell. 2014;26:682–694. doi: 10.1016/j.ccell.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulte JH, Bachmann HS, Brockmeyer B, Depreter K, Oberthur A, Ackermann S, Kahlert Y, Pajtler K, Theissen J, Westermann F. High ALK receptor tyrosine kinase expression supersedes ALK mutation as a determining factor of an unfavorable phenotype in primary neuroblastoma. Clin Cancer Res. 2011;17:5082–5092. doi: 10.1158/1078-0432.CCR-10-2809. [DOI] [PubMed] [Google Scholar]
- 18.Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, Laquaglia MJ, Sennett R, Lynch JE, Perri P. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 20.Cheung NK, Zhang J, Lu C, Parker M, Bahrami A, Tickoo SK, Heguy A, Pappo AS, Federico S, Dalton J. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA. 2012;307:1062–1071. doi: 10.1001/jama.2012.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, Kramer A, Roncaioli JL, Sand F, Heuckmann JM. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526:700–704. doi: 10.1038/nature14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valentijn LJ, Koster J, Zwijnenburg DA, Hasselt NE, van Sluis P, Volckmann R, van Noesel MM, George RE, Tytgat GA, Molenaar JJ. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet. 2015;47:1411–1414. doi: 10.1038/ng.3438. [DOI] [PubMed] [Google Scholar]
- 23.Oberthuer A, Juraeva D, Hero B, Volland R, Sterz C, Schmidt R, Faldum A, Kahlert Y, Engesser A, Asgharzadeh S. Revised risk estimation and treatment stratification of low- and intermediate-risk neuroblastoma patients by integrating clinical and molecular prognostic markers. Clin Cancer Res. 2015;21:1904–1915. doi: 10.1158/1078-0432.CCR-14-0817. [DOI] [PubMed] [Google Scholar]
- 24.Oberthuer A, Hero B, Berthold F, Juraeva D, Faldum A, Kahlert Y, Asgharzadeh S, Seeger R, Scaruffi P, Tonini GP. Prognostic impact of gene expression-based classification for neuroblastoma. J Clin Oncol. 2010;28:3506–3515. doi: 10.1200/JCO.2009.27.3367. [DOI] [PubMed] [Google Scholar]
- 25.De Preter K, Vermeulen J, Brors B, Delattre O, Eggert A, Fischer M, Janoueix-Lerosey I, Lavarino C, Maris JM, Mora J. Accurate outcome prediction in neuroblastoma across independent data sets using a multigene signature. Clin Cancer Res. 2010;16:1532–1541. doi: 10.1158/1078-0432.CCR-09-2607. [DOI] [PubMed] [Google Scholar]
- 26.Ohira M, Oba S, Nakamura Y, Isogai E, Kaneko S, Nakagawa A, Hirata T, Kubo H, Goto T, Yamada S. Expression profiling using a tumor-specific cDNA microarray predicts the prognosis of intermediate risk neuroblastomas. Cancer Cell. 2005;7:337–350. doi: 10.1016/j.ccr.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 27.Vermeulen J, De Preter K, Naranjo A, Vercruysse L, Van Roy N, Hellemans J, Swerts K, Bravo S, Scaruffi P, Tonini GP. Predicting outcomes for children with neuroblastoma using a multigene-expression signature: a retrospective SIOPEN/COG/GPOH study. Lancet Oncol. 2009;10:663–671. doi: 10.1016/S1470-2045(09)70154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mosse YP, Deyell RJ, Berthold F, Nagakawara A, Ambros PF, Monclair T, Cohn SL, Pearson AD, London WB, Matthay KK. Neuroblastoma in older children, adolescents and young adults: a report from the International Neuroblastoma Risk Group project. Pediatr Blood Cancer. 2014;61:627–635. doi: 10.1002/pbc.24777. [DOI] [PubMed] [Google Scholar]
- 29.Castel V, Villamon E, Canete A, Navarro S, Ruiz A, Melero C, Herrero A, Yanez Y, Noguera R. Neuroblastoma in adolescents: genetic and clinical characterisation. Clin Transl Oncol. 2010;12:49–54. doi: 10.1007/s12094-010-0466-z. [DOI] [PubMed] [Google Scholar]
- 30.Mazzocco K, Defferrari R, Sementa AR, Garaventa A, Longo L, De Mariano M, Esposito MR, Negri F, Ircolo D, Viscardi E. Genetic abnormalities in adolescents and young adults with neuroblastoma: A report from the Italian Neuroblastoma group. Pediatr Blood Cancer. 2015;62:1725–1732. doi: 10.1002/pbc.25552. [DOI] [PubMed] [Google Scholar]
- 31.Gui J, Li H. Penalized Cox regression analysis in the high-dimensional and low-sample size settings, with applications to microarray gene expression data. Bioinformatics. 2005;21:3001–3008. doi: 10.1093/bioinformatics/bti422. [DOI] [PubMed] [Google Scholar]
- 32.Theissen J, Oberthuer A, Hombach A, Volland R, Hertwig F, Fischer M, Spitz R, Zapatka M, Brors B, Ortmann M. Chromosome 17/17q gain and unaltered profiles in high resolution array-CGH are prognostically informative in neuroblastoma. Genes Chromosomes Cancer. 2014;53:639–649. doi: 10.1002/gcc.22174. [DOI] [PubMed] [Google Scholar]
- 33.Spitz R, Hero B, Simon T, Berthold F. Loss in chromosome 11q identifies tumors with increased risk for metastatic relapses in localized and 4S neuroblastoma. Clin Cancer Res. 2006;12:3368–3373. doi: 10.1158/1078-0432.CCR-05-2495. [DOI] [PubMed] [Google Scholar]
- 34.Miyashita M, Ohnishi H, Okazawa H, Tomonaga H, Hayashi A, Fujimoto TT, Furuya N, Matozaki T. Promotion of neurite and filopodium formation by CD47: roles of integrins, Rac, and Cdc42. Mol Biol Cell. 2004;15:3950–3963. doi: 10.1091/mbc.E04-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Decock A, Ongenaert M, Hoebeeck J, De Preter K, Van Peer G, Van Criekinge W, Ladenstein R, Schulte JH, Noguera R, Stallings RL. Genome-wide promoter methylation analysis in neuroblastoma identifies prognostic methylation biomarkers. Genome Biol. 2012;13:R95. doi: 10.1186/gb-2012-13-10-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer M, Oberthuer A, Brors B, Kahlert Y, Skowron M, Voth H, Warnat P, Ernestus K, Hero B, Berthold F. Differential expression of neuronal genes defines subtypes of disseminated neuroblastoma with favorable and unfavorable outcome. Clin Cancer Res. 2006;12:5118–5128. doi: 10.1158/1078-0432.CCR-06-0985. [DOI] [PubMed] [Google Scholar]
- 37.De Bernardi B, Gerrard M, Boni L, Rubie H, Canete A, Di Cataldo A, Castel V, Forjaz de Lacerda A, Ladenstein R, Ruud E. Excellent outcome with reduced treatment for infants with disseminated neuroblastoma without MYCN gene amplification. J Clin Oncol. 2009;27:1034–1040. doi: 10.1200/JCO.2008.17.5877. [DOI] [PubMed] [Google Scholar]
- 38.Bown N, Cotterill S, Lastowska M, O'Neill S, Pearson AD, Plantaz D, Meddeb M, Danglot G, Brinkschmidt C, Christiansen H. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med. 1999;340:1954–1961. doi: 10.1056/NEJM199906243402504. [DOI] [PubMed] [Google Scholar]
- 39.Collett D. Modelling Survival Data in Medical Research. Chapman & Hall; London: 1994. Strategy for model selection; pp. 78–83. [Google Scholar]
- 40.Goeman JJ. L1 penalized estimation in the Cox proportional hazards model. Biom J. 2010;52:70–84. doi: 10.1002/bimj.200900028. [DOI] [PubMed] [Google Scholar]
- 41.Zhang W, Yu Y, Hertwig F, Thierry-Mieg J, Zhang W, Thierry-Mieg D, Wang J, Furlanello C, Devanarayan V, Cheng J. Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol. 2015;16:133. doi: 10.1186/s13059-015-0694-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su Z, Fang H, Hong H, Shi L, Zhang W, Zhang W, Zhang Y, Dong Z, Lancashire LJ, Bessarabova M. An investigation of biomarkers derived from legacy microarray data for their utility in the RNA-seq era. Genome Biol. 2014;15:523. doi: 10.1186/s13059-014-0523-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Annotations of All Probes of the Two Gene Expression–Based Classifiers NB-th44 and NB-th24
Supplementary material