Abstract
Multiple myeloma (MM) is a hematological malignancy resulting from the uncontrolled proliferation of antibody-producing plasma cells in the bone marrow. At diagnosis, independent plasma cell tumors are found throughout the skeleton. The recirculation of mutant plasma cells from the initial lesion and their recolonization of distant marrow sites are thought to occur by a process similar to solid tumor metastasis. However, the efficiency of this bone marrow homing process and the proportion of disseminated cells that actively divide and contribute to new tumor growth in MM are both unknown. We used the C57BL/KaLwRij mouse model of myeloma, lentiviral-mediated DNA barcoding of 5TGM1 myeloma cells, and next-generation sequencing to investigate the relative efficiency of plasma cell migration to, and growth within, the bone marrow. This approach revealed three major findings: firstly, establishment of metastasis within the bone marrow was extremely inefficient, with approximately 0.01% of circulating myeloma cells becoming resident long term in the bone marrow of each long bone; secondly, the individual cells of each metastasis exhibited marked differences in their proliferative fates, with the majority of final tumor burden within a bone being attributable to the progeny of between 1 and 8 cells; and, thirdly, the proliferative fate of individual clonal plasma cells differed at each bone marrow site in which the cells “landed.” These findings suggest that individual myeloma plasma cells are subjected to vastly different selection pressures within the bone marrow microenvironment, highlighting the importance of niche-driven factors, which determine the disease course and outcome.
Abbreviations: MM, multiple myeloma; MGUS, monoclonal gammopathy of undetermined significance; SMM, smoldering multiple myeloma; NGS, next-generation sequencing; PC, plasma cell; GFP, green fluorescent protein; tdT, tandem dimer tomato; PCR, polymerase chain reaction
Background
Multiple myeloma (MM) is a hematological malignancy caused by the clonal proliferation of antibody-producing plasma cells (PCs) within the bone marrow [1]. MM is the second most common hematological malignancy with an incidence of 6.6 per 100,000 in the United States [2]. Despite recent therapeutic advances, MM remains a largely incurable cancer, relapse is common, and the 5-year relative survival rate is approximately 50% [2]. Symptomatic MM is characterized by >10% clonal PCs within the bone marrow and the presentation of end organ damage (CRAB symptoms: hypercalcemia, renal insufficiency, anemia, or bone lesions) or other myeloma-defining events [3]. The disease is characterized by multiple sites of tumor growth throughout the axial and appendicular skeleton. It is usually preceded by the asymptomatic condition, termed monoclonal gammopathy of undetermined significance (MGUS), which is characterized by a lower tumor burden and the absence of end organ damage [3], [4]. Despite having multiple sites of tumor growth within the bone marrow, patients with MGUS transition to MM at a rate of 1% per annum. What triggers this transition remains to be determined, but studies suggest that it is a combination of intrinsic changes, such as an increase in the mutational load of the clonal PCs [5], and extrinsic cellular changes within the bone marrow microenvironment [6] involving stromal cells [7], macrophages [8], and osteoclasts [9], [10]. The development of both MGUS and MM relies on the systemic recirculation and continued bone marrow metastasis of myeloma PCs from the site of the initial lesion to distal sites [11]. As the mechanisms of myeloma PC metastasis remain to be fully elucidated, a more complete understanding of these processes would assist with the design of strategies to prevent posttherapy disease relapse and prevent the reestablishment of mutant PCs at new bone sites.
To date, the in vivo models used to investigate the migration of and dissemination of myeloma PCs have relied on the MM1.S xenogeneic transplant model [12] or the 5TMM series of transplantable C57BL/KaLwRij-derived tumors [13]. Using in vivo microscopy, small numbers of fluorescently labeled MM1.S cells have been shown to rapidly migrate from the peripheral circulation to the bone marrow [14], [15], [16]. However, there remains little understanding of the number of MM1.S cells, of the initial migrating pool, that enter the bone marrow and become long-term resident MM cells. Furthermore, these studies were limited to the examination of the bones of the calvaria, which, while more accessible, have a different ontogeny and physiology to the long bones of the appendicular skeleton. Interestingly, studies using radioactively labeled 5T2MM or 5T33MM cells in C57BL/KaLwRij mice suggest that approximately 10% to 15% of intravenously injected cells home to the skeleton within 18 hours [17]. We have recently used a membrane-label retention system and longitudinal intravital microscopy to assess the fates of 5TGM1 and 5T2MM PCs following intravenous injection in C57BL/KaLwRij mice. These studies highlight that the 5TGM1 and 5T2MM PCs that migrate to the bone marrow following intravenous injection have different fates, with the majority remaining dormant, while only a small number proliferate rapidly [9]. Again, enumeration of the absolute numbers of these colonizing cells was not possible, and the nature of any ancestral relationships between the dormant cells and the proliferative cells could not be measured.
Cellular DNA “barcoding” and deep sequencing [18] have been previously combined to examine lineage differentiation of hematopoietic stem cells/progenitors [19], [20]and mammary epithelial cells [21] in vivo, and to track the clonal origins of hepatocyte and Bcr-Abl leukemia cell engraftment [22], [23] and Cbx7-induced leukemias [24]. In the present study, we used functionally neutral, heritable DNA tags to uniquely label each myeloma PC. As each daughter cell will harbor the same DNA barcode as their parental cell, this enables the tracking of the progeny of individual injected MM PC. Furthermore, it makes it possible to measure the precise number of 5TGM1 cells that migrate and become resident long term in the bone marrow of the long bones of C57BL/KaLwRij mice. We propose that the relative frequency of each barcode in an established MM tumor could provide a direct measure of the number of proliferative versus dormant myeloma PC clones within the bone marrow. Using this approach, we show, for the first time, that large numbers of distinct, unrelated MM PCs become stably resident in the bone marrow but that the progeny of only a small number of these myeloma PCs contribute to the majority of the tumor burden in each long bone, while the rest remain dormant.
Methods
Library Construction
Oligo duplexes encoding a partially degenerate DNA “barcode” sequence [19], [21] were generated by reannealing of the following two oligonucleotides: 5′-CATNNTAANNATCNNGATSSAAANNGGTNNAACNNTGTGC-3′ and 5′-GGCCGCACANNGTTNNACCNNTTTSSATCNNGATNNTTANNATGGGCC-3′.
In vitro phosphorylated duplex barcodes were subsequently cloned into the pLeGO-T2 lentiviral vector [25] (gift from Boris Fehse, Addgene plasmid # 27342) using ApaI and NotI, replacing one of the LoxP sites in the original vector. The ligated barcode-lentiviral library was transformed into MegaX DH10B T1R electrocompetent cells (Invitrogen) by electroporation at 2.0 kV, 200 Ω, and 25 μF with a Gene Pulser (Bio-Rad). An aliquot of the transformed Escherichia coli was plated onto agar overnight and gave an estimated CFU frequency of ~3 × 106 for the whole library. Concurrently, a DNA preparation of the whole barcode-lentiviral library was generated from 400 ml of overnight culture using the Pure Link HiPure Maxiprep kit (Invitrogen). Screening by ApaI and XmaI double digestion of plasmid DNA isolated from 48 colonies showed an oligo duplex insertion frequency of 100% (with only 4 colonies showing bands indicative of tandem oligo duplex insertion). The following two primers were used in 30 cycles of polymerase chain reactions (PCRs) to amplify the “degenerate barcode” from genomic DNA isolated from whole bone marrow using PfuTurbo polymerase (Agilent Technologies):
CS1.LeGO.T2.F-5′ ACA CTG ACG ACA TGG TTC TACA ACA GGG ACA GCA GAG ATC CAG 3′
CS2.LeGO.T2.R-5′ TAC GGT AGC AGA GAC TTG GTCT CGT TAC TGC AGC TAG CTT GCC 3′
PCR products were then purified following agarose gel electrophoresis using an Ultraclean Gelspin DNA extraction kit (Mo Bio Laboratories). Ten nanograms of purified PCR product was amplified for a further 10 cycles using High Fidelity Master Mix (Roche) and 384 Barcode Access Array primers (Fluidigm). Final amplicons were purified with Ampure XP beads (Beckman Coulter) and quantitated with Qubit. Libraries were spiked with PhiX prior to running on the MiSeq (Illumina). A v2 500-cycle kit was used to perform 2 × 180-bp reads plus 10-bp index reads.
Sequencing Metrics and Bioinformatics Analysis
Sequencing was performed on the Illumina MiSeq and NextSeq instruments using barcodes with a minimum Hamming distance of 3. Base calling was performed by Illumina RTA version 1.18.54 for the MiSeq and CASAVA 1.8.2 for the NextSeq with default parameters. The FastQ files were trimmed using Cutadapt v1.3 [26] using the front adapter ACAGGGACAGCAGAGATCCAGTTTGGTTAGTACCGGGCCC and back adapter GCGGCCGCTGAAAGACCCCACCTGTAGGTTTGGCAAGCTAGCTGCAGTAACG for the first of pair reads (R1) and swapped and reverse complimented for the second of pair reads (R2). A 10% error rate was allowed, and only reads trimmed for both adapters were kept for further analysis.
Trimmed reads were matched against the barcode pattern ATNNTAANNATCNNGATSSAAANNGGTNNAACNNTGT where N can be any base and S is G or C, and then unique sequences conforming to the pattern were counted. Barcodes from the initial barcode library pools were kept and used to filter remaining experiments. Source code is hosted online at https://bitbucket.org/sacgf/17hewetttumorbarcodes. Library complexity (number of unique barcodes) was estimated from PCR amplification and deep sequencing of the maxiprep. Four independent PCRs were performed on the library and sequenced. A “catalogue” of verified barcodes was generated from barcode sequences that were detected in two out of the four replicate PCRs. The size of this catalogue of verified barcodes was slightly higher than that obtained from CFU estimation (4.2 × 106).
Cell Lines
Mouse 5TGM1 myeloma cells [27] were maintained in Iscove's modified Dulbecco's medium (SIGMA) supplemented with 20% fetal calf serum (FCS), 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, and 10 mM hydroxyethyl piperazineethanesulfonic acid (Life Technologies). We have previously modified the 5TGM1 cells [28] with a trimodality retroviral NES-TGL construct [29], [30] encoding thymidine kinase, green fluorescent protein (GFP), and firefly luciferase [27], and a new clonal subline was established that exhibits consistent bone tropism [27], [31].
In Vivo Myeloma Model
Viral particles containing pLeGO-T2 barcode library were generated following Lipofectamine 2000 (Invitrogen) transfection of HEK293T cells concomitantly with psPAX2 (gift from Didier Trono, Addgene plasmid #12260) and pECO (Clontech) packaging constructs. 5TGM1 cells were infected with lentiviral particles by centrifugation at 1000g for 1 hour with 8 μg/ml polybrene. Forty hours later, tandem dimer tomato (tdT)–positive 5TGM1 cells that had been successfully transduced with the lentiviral-barcode library were isolated by flow cytometry. Single infected cells were also isolated by flow cytometry. Clonal cell lines established from these single cells were screened for barcode-lentivirus insertion frequencies following PCR and Sanger sequencing. After brief recovery at 37°C, the barcoded cells were injected via the tail vein into 6- to 8-week-old C57BL/KaLwRij mice as previously described [27]. Briefly, 5TGM1 cells were resuspended at 5 × 106 cells per ml in phosphate-buffered saline (PBS), and then 5 × 105 cells were injected into the tail vein of 6- to 8-week-old C57BL/KaLwRij mice. Growth of tumors was monitored by whole animal bioluminescent imaging (BLI) using a Xenogen IVIS 100 Imaging System (Caliper Life Sciences) after intraperitoneal injection of 150 mg/kg of D-Luciferin (BIOSYNTH). Tumor burden was quantitated using Living Image software. Long bones with the highest tumor burden were dissected, and the bone marrow was harvested into PBS, 2% FCS, and 2 mM EDTA (PFE). Genomic DNA was extracted from bone marrow cells using DNeasy Blood and Tissue Kit (QIAGEN) according to manufacturer's instructions.
Results
DNA Barcoding of 5TGM1 Cells In Vitro and In Vivo
The 5TGM1-C57BL/KaLwRij mouse model of myeloma faithfully replicates many of the features of aggressive myeloma [27], [31], [32], [33]. In this model, 5 × 105 5TGM1 cells are routinely injected via the tail vein, after which an unknown number of these cells home to and then proliferate at multiple sites within the apical and appendicular skeleton, leading to very high levels of tumor burden within 4 weeks [27], [31]. In this study, each 5TGM1 PC was uniquely marked with a permanent viral-integrated DNA barcode, allowing assessment of the fate of each clone and its progeny. The read counts generated for each tag/barcode, following deep sequencing, allowed for the direct quantitation of the contribution of the progeny of each individually tagged cell with respect to final tumor burden within each long bone (see Figure 1 for experimental plan). To achieve this, a partially degenerate oligonucleotide duplex was cloned into a lentiviral vector containing a tdT fluorescence reporter. The degenerate oligonucleotide was a longer version of a previously used barcode sequence [19], [21] and is predicted to yield 6.7 × 107 potential unique nucleotide combinations. Estimation of the complexity of the resulting barcode-lentiviral library by enumeration of the colony forming units and by deep sequencing revealed that the constructed library had between 3 × 106 and 4 × 106 barcodes. 5TGM1 cells were infected with this library, and transduced cells were isolated based on their GFP+tdT+ phenotype. In order to limit the number of 5TGM1 cells harboring more than one DNA barcode [18], the viral particles were titered to maintain a 5TGM1 transduction efficiency of only 10%. Analysis of 5TGM1 subclones derived from single cells post lentiviral infection showed that more than 70% of the GFP+tdT+5TGM1 cells contained a single DNA barcode (data not shown).
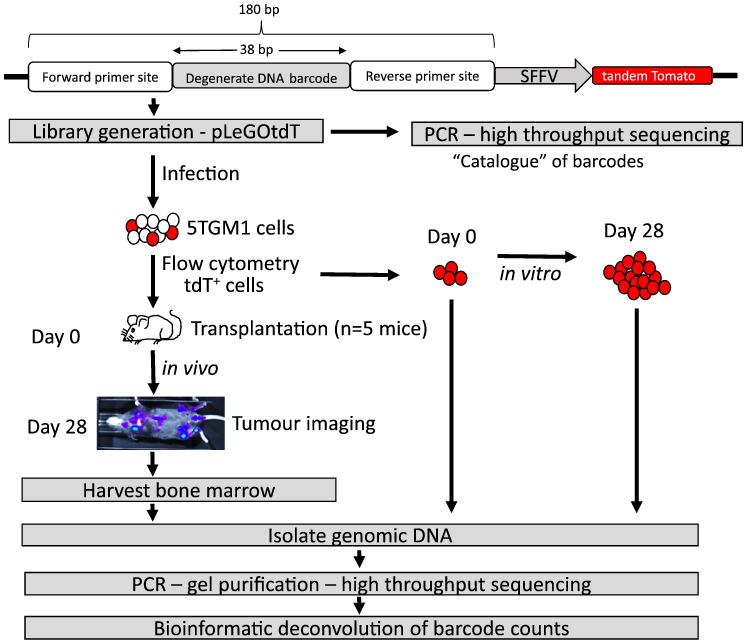
Figure 1.
DNA-barcoding experimental workflow. The strategy for tracking uniquely labeled 5TGM1 cells in vitro and in vivo using a DNA-barcode lentiviral library is shown.
In order to assess whether there were intrinsic differences in the proliferation rates of individual 5TGM1 cells within our clonally derived cell line, the barcoded 5TGM1 cells (5 × 105 cells) were initially used to seed a culture that was maintained in vitro for 28 days. As anticipated, the day 0 and day 28 in vitro samples contained large numbers of different barcodes (75,812 and 21,504, respectively) with no high-frequency “common” barcodes present at >5% of the total reads (Figure 2, Supplementary Table S1). Indeed, only 4 barcodes out of a combined 97,316 in vitro barcode sequences individually constituted >0.1% of the total reads, consistent with all the barcoded cells proliferating at very similar rates. Given the random nature of the lentiviral integration, occasional insertion-mediated inactivation of a tumor suppressor gene or activation of a proto-oncogene, which would provide an intrinsic growth advantages to some of the barcoded cells, cannot be actively avoided. However, based on the data presented here, we saw no evidence of such events, as no highly proliferative barcoded cells were present in the in vitro culture.
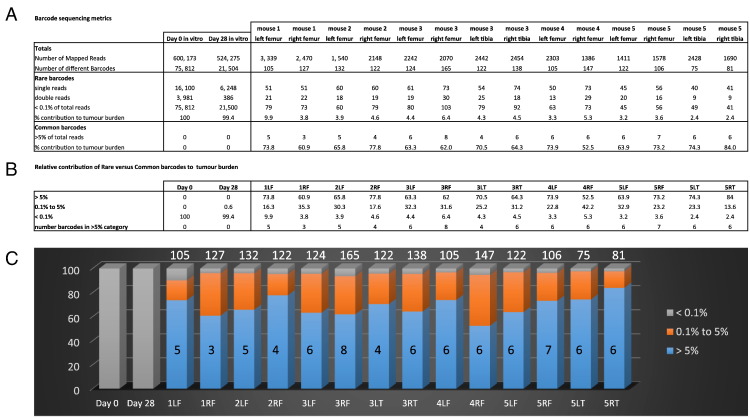
Figure 2.
Growth heterogeneity of myeloma cells in vivo compared to in vitro as revealed by DNA barcoding. (A) Barcode read count metrics are shown for PCR products generated from in vitro (day 0, day 28) 5TGM1 cultures versus in vivo 5TGM1 skeletal tumors (day 28). The numbers of barcodes falling into the “rare” (<0.1% of total reads for tumor) and “common” (>5% of total read for tumor) frequency subclasses of barcodes are indicated. (B) Summaries of the percentages of each tumor mass (i.e., total number of read counts) that was attributable to each frequency subclass of barcode. The numbers of individual barcodes that were of the common subclass in each tumor are also shown. (C) Graphical representation of the growth heterogeneity that is seen in vivo compared to in vitro. The proportion of each sample that is attributable to barcodes of each frequency subclass is shown by the different colors. For the in vivo tumor samples, the absolute numbers of common (>5%) barcodes and the total numbers of individual barcodes detected are superimposed over and above the histogram bars respectively. Key: R/L = left/right; F/T = femur/tibia.
DNA Barcode Quantitation
To investigate whether the mapped read counts provide an accurate measure of the relative frequencies of low-frequency clones, we also performed deep sequencing on admixtures of two 5TGM1 subclones each harboring a single barcode. Clone #23 and clone #31 were mixed in the following three ratios: 99 to 1, 99.9 to 0.1, and 99.99 to 0.01. Each sample had 190–200,000 mapped reads. The observed ratios from barcode read depth were 98.30 to 1.02, 99.17 to 0.09, and 99.37 to 0.01 (Supplementary Table S2). This remarkable alignment to expected ratios highlights that barcode-read counting of rare cells is quantitative. It also indicates the sensitivity of the approach, with 1 in 1000 cells being readily detectable.
In order to investigate the potential cross-contamination between samples and the degree of nonspecific background noise in the barcoding system as a whole, these single-barcode controls detailed above were sequenced alongside the experimental tumor samples detailed later (Supplementary Table S2). As expected, the vast majority of the sequences in the two control samples were the predicted barcode (99.37% for clone #23, 99.61% for clone #31). However, each sample did show a large number of additional low-frequency barcodes (n = 121 barcodes in clone #23; n = 132 in clone #31). Careful examination of the sequence of each of these additional barcodes revealed that a proportion of these barcodes were likely to be the result of sequencing errors rather than contamination. As such, 22.7% to 46.3% of these barcodes were only 1 or 2 mismatches away from the expected sequence (n = 56 barcodes in clone #23, and n = 30 barcodes for clone #31). Scoring the single and double mismatches reveals an error rate of 0.006 per base for clone#23 and 0.002 per base for clone#31. These findings are in line with previous estimates of Miseq error rates ranging from 0.001 to 0.01 per base [34], [35], [36]. For clone #23, 13/15 of the next most frequent barcode sequences (after the correct sequence) all differed by 1 to 2 mismatches. For clone #31, 6/9 of the next most frequent barcode sequences (after the correct sequence) all differed by 1 to 2 mismatches (Supplementary Table S2). The second type of low-frequency barcodes was unrelated to the cloned barcode and is likely intersample contaminants. Many of these were subsequently seen to be common barcodes from other tumor samples or single clones. The three most frequent contaminants of this type, seen in clone #23, were present at read counts of 25, 12, and 9 (in a total read count of 193,101) representing 0.013%, 0.006%, and 0.005% of the totals, respectively. These findings suggest that even if mismatch errors are excluded, it is challenging to confidently call the exact numbers of low-frequency clones (i.e., those contributing to <0.02% of the total reads for each sample).
5TGM1 Growth Kinetics In Vivo
In order to investigate the growth of 5TGM1 tumors in vivo, two independent experiments were carried out in which 5 × 105 cells were injected into each of the 5 C57BL/KaLwRij mice, and tumors were allowed to establish for 28 days (Figure 1). As the cells were previously infected with a lentiviral construct harboring a luciferase reporter [29], [32], tumor growth could be monitored by whole animal BLI at weekly intervals. At day 28, bone marrow was collected from representative long bones in which tumor growth was detected by BLI. Deep sequencing and deconvolution of barcode frequencies were carried out, and the barcodes were then grouped according to their relative abundance within each long bone and separated into three classes: 1) “common” barcodes constituting >5% of the total tumor mass (i.e., total number of mapped reads), 2) barcodes constituting 0.1% to 5% of the total tumor mass, and 3) “rare” barcodes constituting <0.1% of the total tumor mass.
In the initial proof-of-concept experiment, a total of 14 different long bones were isolated from 5 tumor-bearing mice, and barcode frequency was assessed (Figure 2, Supplementary Table S1). In contrast to the in vitro findings, the in vivo samples exhibited a remarkable skewing, with the bulk of the reads within each tumor derived from a very small number of barcodes. Thus, there was a median of just 6 “common” (>5%) barcodes per tumor (range: 3-8; n = 14 tumors) which made up an average of 68.6% of each tumor (range: 52.5%-84.0%). Each of the 78 common barcodes seen across the tumor set was unique (Supplementary Table S3). In contrast, a large number of “rare” (<0.1%) barcodes were evident in each tumor sample, but their combined contribution to the overall tumor mass was very small. Thus, the tumors recovered from all mice had a median of 73 “rare” barcodes each (range: 41-103), with these rare barcodes making up an average of 4.4% of each total tumor (range: 2.4%-9.9%). Together, these results suggest that only a small number of injected 5TGM1 PCs home to, and persist within, the bone marrow and that very few of the clones actively proliferate to generate the majority of the tumor burden.
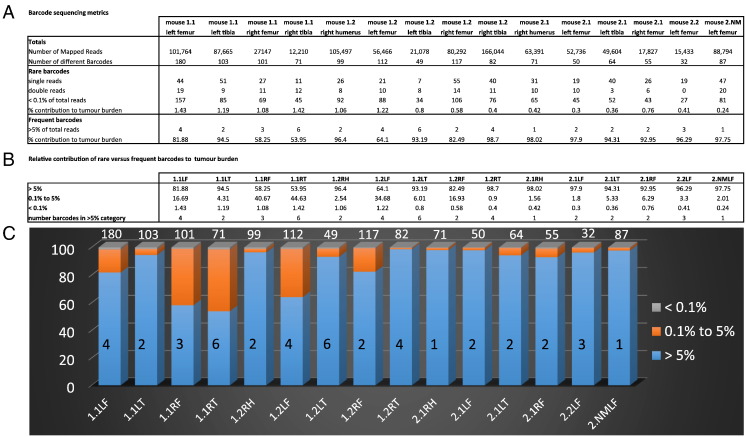
To more accurately evaluate the consistent clonal dominance observed in vivo, a second experiment was performed with much higher sequencing depth. In an effort to eliminate any false-positive barcodes, particularly infrequent barcode sequences that may have been introduced by PCR or Illumina sequencing errors, the following precautions were observed, in keeping with the recommendations of Naik et al. [18]: 1) each tumor DNA sample was independently amplified in separate PCRs, and only those barcodes that were detectable by next-generation sequencing (NGS) from both PCRs were retained; 2) only barcodes that were present in both the tumor DNA sample and the reference catalogue of barcodes from the original lentiviral barcode library were included (with the exception of the common subclass of barcodes); 3) barcodes which differed by only 1 to 2 mismatches from barcodes with >10,000 reads within each tumor were discounted as PCR/Miseq errors; and 4) to increase the accuracy of the barcode enumeration, only samples with a final mapped read depth of at least 10,000 reads were analyzed, leaving assessable data from 15 tumors which were recovered from 5 different mice. As described above, the barcodes were partitioned according to their frequency into three subclasses (Figure 3, Supplementary Table S4). As with the first in vivo experiment, a large number of rare barcodes (median: 69, range: 27-157) were present in each bone, and the majority of the tumor mass resulted from the expansion of a very small number of “common” barcodes (median: 2, range: 1-6). The number of “common” barcodes per long bone ranged from single barcodes, which constituted 98% of the tumor mass of both the right humerus of mouse 2.1 and the left femur of mouse 2.NM, to 6 barcodes, which made up 92% of the tumor mass in left tibia of mouse 1.2.The common barcodes identified across all tumors and all mice are shown in Supplementary Table S5. Reassuringly, all of the 44 common barcodes were only ranked as common in either the bone or the mouse from which they were derived (consistent with what was seen in the first in vivo experiment). There were 35 different sequences within these 44 common barcodes, with 7 of these barcodes belonging to the common subclass at more than one tumor site within the same mouse (Supplementary Table S5). These findings are consistent with the occasional metastasis of myeloma cells from one bone to another, a phenomenon which we next examined in more detail.
Figure 3.
DNA barcoding shows the growth heterogeneity of myeloma cells in a second in vivo experiment. (A) Barcode read count metrics are shown for PCR products generated from in vivo 5TGM1 skeletal tumors (day 28). The numbers of barcodes falling into the rare (<0.1%) and common (>5%) frequency subclasses of barcodes are indicated. (B) Summaries of the percentages of each tumor mass that was attributable to each frequency subclass of barcode. The numbers of individual barcodes that were of the common subclass in each tumor are also shown. (C) Graphical representation of the growth heterogeneity that is seen in vivo. The proportion of each sample that is attributable to barcodes of each frequency subclass is shown by the different colors. The absolute numbers of common (>5%) barcodes and the total numbers of individual barcodes detected are superimposed over and above the histogram bars respectively. Key: R/L = left/right; F/T = femur/tibia.
Evidence of Metastasis
The ability to tag myeloma PCs with heritable DNA barcodes should also allow for the assessment of the direct relationship between individual myeloma cells present at distant sites within the same mouse. If cells within tumors from different long bones show a common ancestry, this would imply metastasis between different tumor sites — a feature for which there were limited data in this mouse model.
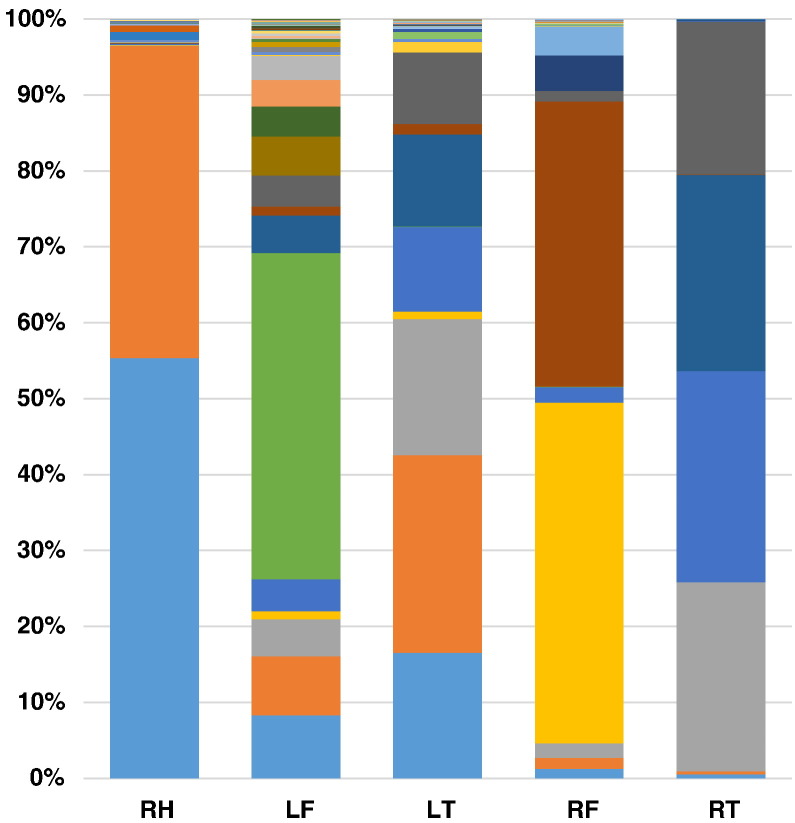
However, the ability to detect metastases in this model was limited by the potential for cross-contamination between samples (as was seen when examining the single clones earlier). Some potential cross-contamination between samples was evident with a number of individual barcodes observed in many bones from different mice, the most common pattern relating to a very high frequency barcode in one sample being seen at very low frequency in several other samples (Supplementary Table S4). When limiting the analysis to barcodes unique to each mouse, there were still many examples of apparent metastases among the read counts for the rare (<0.1%) subclass of barcodes — but in these cases, it was impossible to truly differentiate metastasis of a very high frequency barcode from an intersample contamination error. This issue points to the limitations of the barcoding approach to accurately enumerate metastasis. Despite these caveats, there were clearer examples of metastases observed in this model. Mouse 1.2 was the most apparent example which exhibited evidence of multiple metastases, where six of the “common” (>5%) barcodes showed evidence of being in the “common” subclass of barcode in more than one long bone (Figure 4, Supplementary Table S5). These barcodes showed the following differential read counts across two to three different bones within the same mouse: (barcode #1) 58381, 4663, and 3491; (barcode #2) 43315, 5474, and 4399; (barcode #3) 46155 and 2354; (barcode #4) 42937 and 2548; (barcode #5) 41333 and 3788; and (barcode #6) 33453 and 1987.
Figure 4.
Evidence of shared barcodes between different bones. The relative frequencies (as percentage of total reads) and identities (colors) of barcodes enumerated in 5 different long bones from mouse 1.2 are shown. Key: R/L = left/right; F/T = femur/tibia.
To further address the impact that the intersample contamination could have on enumeration of the rare subclass of barcodes, we evaluated whether each of the 1273 individual barcodes was unique to each bone or to each mouse. Almost half (45.2%) of the rare (<0.1%) subclass of barcode sequences was unique to each tumor or mouse (Supplementary Table S6). Even after removing all of the “rare” barcodes that could result from potential contamination issues, “rare” barcodes still greatly outnumbered the “common” barcodes present in each tumor. As such, the “rare” barcodes were usually observed at least one order of magnitude more frequently than the common barcodes (mean 16.4-fold ± SD 12.5). As such, these studies suggest that typically only 1 out of every 16 cells that become a long-term bone marrow PC will become highly proliferative (Supplementary Table S6).
Discussion
MM is characterized by the presence of multiple clonal PC tumors which are disseminated throughout the skeleton [1]. Myeloma can be viewed as the archetype of a cancer that has arisen as a result of metastases of circulating PCs to multiple distal sites from the initial site of growth [11]. As such, an understanding of the mechanisms and characteristics of the myeloma PC trafficking and metastasis is crucial to understanding disease initiation, progression, and relapse following therapy.
Using DNA barcoding of 5TGM1 PCs, prior to administration to C57BL/KaLwRij mice, we have been able to use cell-lineage tracking to examine the fates and net tumor contribution of individual MM PCs in vivo. This approach revealed that only a small proportion (approximately 0.01%) of the myeloma PCs injected migrate to, and are retained within, the bone marrow of each long bone. A more striking finding was that the progeny of only a small number of PCs (always fewer than nine cells and often less than three cells) contributed to the majority of the tumor burden in each long bone at day 28. To this end, we estimated that for each proliferative myeloma cell that makes a substantial contribution to tumor burden, 16 times as many PCs remain dormant. This skewing of the growth of individual cells was not seen in in vitro cultures of the same duration, suggesting that there are strong selection pressures in vivo. Similar examples of clonal growth heterogeneity have previously been observed using DNA barcoding in xenogeneic transplants. The HCT-116 colorectal cancer cell line was barcoded using uninduced shRNAs as neutral DNA tags and then grown subcutaneously in immunodeficient mice [37]. After 12 days of growth, deep sequencing of the xenograft revealed that only 6% of the injected cells contributed to 95% of the total aggregate cell number, while 75% of the injected cells had undergone less than 2 cell divisions. By contrast, DNA barcode complexity was maintained in vitro, with 95% of descendant barcoded cells being derived from 80% of the initially tagged clones. Clonal growth heterogeneity in vivo has also been demonstrated using other cell lines [37]. By contrast, xenografts of a barcoded HCC827 small cell lung cancer cell line exhibit less population skewing in vivo when compared with growth in vitro [38]. The more pronounced skewing of the growth trajectories of individual PCs in our model may be the result of using an orthotopic and syngeneic model, whereby a relatively small number of cells (rather than a large bolus of cells) end up being dispersed throughout their native bone marrow environment where they can be subjected to differential gradients of growth-supportive/-suppressive factors. This is consistent with a limited number of very strong and very localized microenvironmental selection pressures within the bone marrow. Very recent findings using xenografts of barcoded primary B-ALL leukemic cells also showed clonal dominance in vivo [39]. To this end, patient-derived xenografts in the bone marrow were highly polyclonal with tens to hundreds of barcodes being detectable following injection of 250,000 cells. Comparison of different skeletal sites in the same recipient mouse showed that often >80% of the barcodes per tumor site were contributed by only 3 to 7 barcodes. In contrast, this clear clonal dominance was not seen in the bone marrow of mice xenografted with a barcoded SupB15 leukemic cell line, but there were still significantly fewer clones compared to the same cells cultured in vitro [43]. Interestingly, the authors also noted a degree of “metastasis” of their patient xenografts, with the same group of barcodes often seen in more than one skeletal site and the relative frequencies of each of these barcodes at each site being moderately different [39]. Our studies showed even more marked spatial asymmetry — where often the predominant clones/barcodes in one bone are barely detectable above background levels in other bones.
The findings presented here are consistent with our previous studies examining longitudinal tracking of 5TGM1 myeloma cell fate determination using intravital imaging and dye labeling to differentiate dormant from actively dividing myeloma cells [9]. Large numbers of individual myeloma cells were shown to migrate to the endosteal bone surfaces of femurs by day 7 post cell injection. At these sites, the cells were held in a long-term dormant state, in part by soluble factors produced by osteoblasts. By contrast, only a limited number of regions of active myeloma cell growth were observed, peaking with ~15 colonies per femur at day 14 post tumor cell administration. At all of the time points examined, the numbers of isolated, individual myeloma cells seen in the bone marrow were always significantly more than the number of myeloma cell colonies. At day 7, the number of individual cells observed at this point was approximately 50 compared with 2 to 3 colonies of active cell growth. These findings bear striking similarity to our DNA-barcoding estimates from day 28, where there were 16 relatively quiescent MM cells for each highly proliferative MM cell. Our ability to track the fate of individual MM cells with heritable tags also suggests that the areas of focal cell growth described in Lawson et al. [9] are likely to be clonal in origin and that at least some of the individual MM cells observed at sites distant to these colonies are likely to be independently migrated cells rather than cells that have been “shed” by the growing colonies. Both these studies highlight the fact that unrestricted cell growth and tumor formation are not the inevitable outcome for a myeloma PC that has successfully migrated to and become resident in the bone marrow. The bone marrow is a complex microenvironment with distinct niches, each of which has a different cellular composition and function, such as the well-characterized endosteal/osteoblastic and vascular niches that support HSC (haematopoietic stem cell) maintenance and dissemination [40], [41], [42]. Myeloma PCs have well-characterized interactions with cellular elements of the bone marrow, including osteoclasts, osteoblasts, stromal cells, macrophages, and endothelial cells [11], [43], many of which produce stimulatory and antiapoptotic factors. It is likely to be the balance between prostimulatory (e.g., stromal cell–produced IL-6 [44]) and growth-inhibiting (e.g. osteoblast-produced decorin [45], or host-derived matrix metalloproteinase-7 [46]) factors that will determine the growth rate of a mutated PC.
MM is almost invariably preceded by the premalignant condition of monoclonal gammopathy of undetermined significance (MGUS) characterized by a lower mutant PC burden and lack of any end organ damage [4]. MM can also be preceded by an intermediate asymptomatic state known as smoldering MM (SMM) [47]. It has been traditionally viewed that the transition from asymptomatic MGUS/SMM to MM is associated with an increased mutational burden in the clonal PC, resulting in increased activation of tumorigenic pathways [5]. Indeed, the average number of NS-SNV mutations in MGUS PCs does seem to be lower than that seen in MM PCs [48], [49]. However, examination of matched samples taken from patients before and after malignant transformation has shown that MGUS/SMM PCs can be genetically complex [6] and that transition is often associated with the accumulation of very few new mutations [49], [50]. Furthermore, recent xenograft studies have shown that MGUS PCs have similar tumorigenic potential to MM PCs [51]. These findings suggest that extrinsic factors play an important role in malignant transformation, with disease progression being triggered by cues from the microenvironment [43]. For instance, MGUS patients with increased bone resorption have been seen to be more likely to develop overt myeloma [52], and induction of bone resorption can increase tumor growth in a mouse model of myeloma [53].
Using DNA barcoding to track myeloma PC fate has proved to have its own technical challenges. The problem of closely related barcodes “piggy backing” as sequence errors when particular barcodes become very abundant has previously been raised as a potential concern with this technology [54]. We would recommend that barcodes with up to two mismatches or in\dels from these abundant barcodes be excluded from further analysis. Despite the inclusion of technical replicates, intersample contaminants that required exclusion were also evident. The modest frequency of double viral-integration events (i.e., 29% of 5TGM1 cells tagged with two barcodes) evident from the sequence analysis of single sorted cells (data not shown) also means that we cannot definitively discount the possibility that, in some individual long bones, we may have overestimated the number of “seed cells” whose progeny went on to contribute to the tumors at day 28. With these recommendations in mind, DNA barcoding would be a useful tool for tracking cell fate in cancers that metastasize to bone, particularly breast and prostate cancer [55], [56]. It would also prove interesting to examine whether stimulation of bone remodeling (e.g., following sRANKL exposure) would alter the proportions of frequent and rare barcodes in our model system, as osteoclast activation has previously been associated with a release of MM cells from dormancy [9].
Conclusion
We have used heritable DNA tags or barcodes to label a clonal population of myeloma cells and enumerate the numbers of cells which become long-term resident in the bone marrow in vivo. The myeloma cells displayed striking differences in their growth trajectories, with the progeny of less than nine myeloma cells (often less than three cells) contributing to the bulk of the tumor in each long bone by disease end point. Such skewed growth was not observed in vitro and highlights the strong selection pressures to which myeloma cells are subjected to in the bone marrow microenvironment. We have termed this phenomenon “habitual clonal dominance,” and it is consistent with the previous studies demonstrating that the majority of myeloma cells are relatively quiescent. Multiple myeloma is almost invariably preceded by an asymptomatic condition with a lower tumor cell burden, and our results suggest that extrinsic (bone marrow–derived) factors could play an important part in expanding PC numbers and triggering transition from asymptomatic to diagnostic MM. Our findings also suggest that targeting such niche components, be they activators of proliferation or quiescence of myeloma cells, will be an increasingly important therapeutic strategy.
The following are the supplementary data related to this article.
In vitro and in vivo barcode data from pilot experiments. The sequences and read counts of all the barcodes detected in day 0 and day 28 in vitro 5TGM1 cultures, and tumors from the individual long bones of 5 mice on day 28 are shown. Frequencies of the rare (<0.1%) and common (>5%) subclasses of barcodes are also shown. Key: L/R = left/right. F/T = femur/tibia.
Deep sequencing of single barcode control cell lines. Read counts, mismatch data, and frequency distributions are shown for NGS of PCR products from 5TGM1 clones #23 and #31, each of which contains a single DNA barcode (shaded yellow and green, respectively). Read counts are also shown for deep sequencing of PCR products derived from admixtures of the two cell lines at the ratios indicated. The red shaded barcode is an example of a low-frequency barcode that was detected in all 15 tumors and controls that were analyzed.
Common subclasses of barcodes in pilot in vivo experiment. The sequences and read counts for all 78 barcodes that were classified as common (each representing at least 5% of the bulk of their respective tumors) in individual long bones from n = 5 different mice. Key: L/R = left/right. F/T = femur/tibia.
Tumor barcode data from second in vivo experiment. The sequences and read counts are shown for all barcodes detected in both replicate PCR products generated from tumors in the indicated long bones. Only those barcodes seen in the reference library of barcodes are shown. Sequences with less than 3 mismatches from barcodes with >10,000 read counts have been removed. Overall classification of barcodes into rare (<0.1%) or common (>5%) subclasses is also shown. Barcodes shaded in green are unique to either the sample or the mouse as shown.
Uniqueness of the common subclass of barcodes. The read counts for each common (>5%) barcode are shown together with their bone of origin and whether they were detectable within the common subclass in only one bone and/or one mouse. Barcodes whose frequency subclass was common in more than one long bone in the same mouse are indicated by the same color shading.
Filtering rare barcodes for sample uniqueness. The uniqueness of all the barcodes of the rare (<0.1%) subclass is shown. A comparison to the numbers of common (>5%) subclass of barcodes per sample (from Figure 3) is shown.
Acknowledgments
Acknowledgements
This work was supported by the National Health and Medical Research Council (Grant ID: 1104031). Dr. Kate Vandyke is the recipient of a Mary Overton Early Career Research Fellowship from the Royal Adelaide Hospital.
References
- 1.Kyle RA, Rajkumar SV. Multiple myeloma. Blood. 2008;111:2962–2972. doi: 10.1182/blood-2007-10-078022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Howlader NNA, Krapcho M, Miller D, Bishop K, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA, editors. SEER Cancer Statistics Review, 1975–2014. National Cancer Institute; Bethesda, MD: 2017. [Google Scholar]
- 3.Rajkumar SV, Dimopoulos MA, Palumbo A, Blade J, Merlini G, Mateos MV, Kumar S, Hillengass J, Kastritis E, Richardson P. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014;15:e538–e548. doi: 10.1016/S1470-2045(14)70442-5. [DOI] [PubMed] [Google Scholar]
- 4.Landgren O, Kyle RA, Pfeiffer RM, Katzmann JA, Caporaso NE, Hayes RB, Dispenzieri A, Kumar S, Clark RJ, Baris D. Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: a prospective study. Blood. 2009;113:5412–5417. doi: 10.1182/blood-2008-12-194241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12:335–348. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
- 6.Dhodapkar MV. MGUS to myeloma: a mysterious gammopathy of underexplored significance. Blood. 2016;128:2599–2606. doi: 10.1182/blood-2016-09-692954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noll JE, Williams SA, Tong CM, Wang H, Quach JM, Purton LE, Pilkington K, To LB, Evdokiou A, Gronthos S. Myeloma plasma cells alter the bone marrow microenvironment by stimulating the proliferation of mesenchymal stromal cells. Haematologica. 2014;99:163–171. doi: 10.3324/haematol.2013.090977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng Y, Cai Z, Wang S, Zhang X, Qian J, Hong S, Li H, Wang M, Yang J, Yi Q. Macrophages are an abundant component of myeloma microenvironment and protect myeloma cells from chemotherapy drug-induced apoptosis. Blood. 2009;114:3625–3628. doi: 10.1182/blood-2009-05-220285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawson MA, McDonald MM, Kovacic N, Hua Khoo W, Terry RL, Down J, Kaplan W, Paton-Hough J, Fellows C, Pettitt JA. Osteoclasts control reactivation of dormant myeloma cells by remodelling the endosteal niche. Nat Commun. 2015;6:8983–8997. doi: 10.1038/ncomms9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yaccoby S, Wezeman MJ, Henderson A, Cottler-Fox M, Yi Q, Barlogie B, Epstein J. Cancer and the microenvironment: myeloma-osteoclast interactions as a model. Cancer Res. 2004;64:2016–2023. doi: 10.1158/0008-5472.can-03-1131. [DOI] [PubMed] [Google Scholar]
- 11.Ghobrial IM. Myeloma as a model for the process of metastasis: implications for therapy. Blood. 2012;120:20–30. doi: 10.1182/blood-2012-01-379024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenstein S, Krett NL, Kurosawa Y, Ma C, Chauhan D, Hideshima T, Anderson KC, Rosen ST. Characterization of the MM.1 human multiple myeloma (MM) cell lines: a model system to elucidate the characteristics, behavior, and signaling of steroid-sensitive and -resistant MM cells. Exp Hematol. 2003;31:271–282. doi: 10.1016/s0301-472x(03)00023-7. [DOI] [PubMed] [Google Scholar]
- 13.Vanderkerken K, Asosingh K, Croucher P, Van Camp B. Multiple myeloma biology: lessons from the 5TMM models. Immunol Rev. 2003;194:196–206. doi: 10.1034/j.1600-065x.2003.00035.x. [DOI] [PubMed] [Google Scholar]
- 14.Alsayed Y, Ngo H, Runnels J, Leleu X, Singha UK, Pitsillides CM, Spencer JA, Kimlinger T, Ghobrial JM, Jia X. Mechanisms of regulation of CXCR4/SDF-1 (CXCL12)-dependent migration and homing in multiple myeloma. Blood. 2007;109:2708–2717. doi: 10.1182/blood-2006-07-035857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glavey SV, Manier S, Natoni A, Sacco A, Moschetta M, Reagan MR, Murillo LS, Sahin I, Wu P, Mishima Y. The sialyltransferase ST3GAL6 influences homing and survival in multiple myeloma. Blood. 2014;124:1765–1776. doi: 10.1182/blood-2014-03-560862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azab AK, Hu J, Quang P, Azab F, Pitsillides C, Awwad R, Thompson B, Maiso P, Sun JD, Hart CP. Hypoxia promotes dissemination of multiple myeloma through acquisition of epithelial to mesenchymal transition-like features. Blood. 2012;119:5782–5794. doi: 10.1182/blood-2011-09-380410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Asosingh K, De Raeve H, Croucher P, Goes E, Van Riet I, Van Camp B, Vanderkerken K. In vivo homing and differentiation characteristics of mature (CD45-) and immature (CD45+) 5T multiple myeloma cells. Exp Hematol. 2001;29:77–84. doi: 10.1016/s0301-472x(00)00625-1. [DOI] [PubMed] [Google Scholar]
- 18.Naik SH, Schumacher TN, Perie L. Cellular barcoding: a technical appraisal. Exp Hematol. 2014;42:598–608. doi: 10.1016/j.exphem.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Cheung AM, Nguyen LV, Carles A, Beer P, Miller PH, Knapp DJ, Dhillon K, Hirst M, Eaves CJ. Analysis of the clonal growth and differentiation dynamics of primitive barcoded human cord blood cells in NSG mice. Blood. 2013;122:3129–3137. doi: 10.1182/blood-2013-06-508432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naik SH, Perie L, Swart E, Gerlach C, van Rooij N, de Boer RJ, Schumacher TN. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature. 2013;496:229–232. doi: 10.1038/nature12013. [DOI] [PubMed] [Google Scholar]
- 21.Nguyen LV, Makarem M, Carles A, Moksa M, Kannan N, Pandoh P, Eirew P, Osako T, Kardel M, Cheung AM. Clonal analysis via barcoding reveals diverse growth and differentiation of transplanted mouse and human mammary stem cells. Cell Stem Cell. 2014;14:253–263. doi: 10.1016/j.stem.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Cornils K, Thielecke L, Huser S, Forgber M, Thomaschewski M, Kleist N, Hussein K, Riecken K, Volz T, Gerdes S. Multiplexing clonality: combining RGB marking and genetic barcoding. Nucleic Acids Res. 2014;42:e56. doi: 10.1093/nar/gku081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cornils K, Thielecke L, Winkelmann D, Aranyossy T, Lesche M, Dahl A, Roeder I, Fehse B, Glauche I. Clonal competition in BcrAbl-driven leukemia: how transplantations can accelerate clonal conversion. Mol Cancer. 2017;16:120–131. doi: 10.1186/s12943-017-0668-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klauke K, Broekhuis MJ, Weersing E, Dethmers-Ausema A, Ritsema M, Gonzalez MV, Zwart E, Bystrykh LV, de Haan G. Tracing dynamics and clonal heterogeneity of Cbx7-induced leukemic stem cells by cellular barcoding. Stem Cell Rep. 2015;4:74–89. doi: 10.1016/j.stemcr.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber K, Bartsch U, Stocking C, Fehse B. A multicolor panel of novel lentiviral "gene ontology" (LeGO) vectors for functional gene analysis. Mol Ther. 2008;16:698–706. doi: 10.1038/mt.2008.6. [DOI] [PubMed] [Google Scholar]
- 26.Martin M. vol. 17. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads; pp. 10–12. [Google Scholar]
- 27.Noll JE, Hewett DR, Williams SA, Vandyke K, Kok C, To LB, Zannettino AC. SAMSN1 is a tumor suppressor gene in multiple myeloma. Neoplasia. 2014;16:572–585. doi: 10.1016/j.neo.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dallas SL, Garrett IR, Oyajobi BO, Dallas MR, Boyce BF, Bauss F, Radl J, Mundy GR. Ibandronate reduces osteolytic lesions but not tumor burden in a murine model of myeloma bone disease. Blood. 1999;93:1697–1706. [PubMed] [Google Scholar]
- 29.Diamond P, Labrinidis A, Martin SK, Farrugia AN, Gronthos S, To LB, Fujii N, O'Loughlin PD, Evdokiou A, Zannettino AC. Targeted disruption of the CXCL12/CXCR4 axis inhibits osteolysis in a murine model of myeloma-associated bone loss. J Bone Miner Res. 2009;24:1150–1161. doi: 10.1359/jbmr.090210. [DOI] [PubMed] [Google Scholar]
- 30.Ponomarev V, Doubrovin M, Serganova I, Vider J, Shavrin A, Beresten T, Ivanova A, Ageyeva L, Tourkova V, Balatoni J. A novel triple-modality reporter gene for whole-body fluorescent, bioluminescent, and nuclear noninvasive imaging. Eur J Nucl Med Mol Imaging. 2004;31:740–751. doi: 10.1007/s00259-003-1441-5. [DOI] [PubMed] [Google Scholar]
- 31.Noll JE, Vandyke K, Hewett DR, Mrozik KM, Bala RJ, Williams SA, Kok CH, Zannettino AC. PTTG1 expression is associated with hyperproliferative disease and poor prognosis in multiple myeloma. J Hematol Oncol. 2015;8:106–121. doi: 10.1186/s13045-015-0209-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheong CM, Chow AW, Fitter S, Hewett DR, Martin SK, Williams SA, Toh LB, Zannettino AC, Vandyke K. Tetraspanin 7 (TSPAN7) expression is upregulated in multiple myeloma patients and inhibits myeloma tumour development in vivo. Exp Cell Res. 2015;332:24–38. doi: 10.1016/j.yexcr.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 33.Oyajobi BO, Munoz S, Kakonen R, Williams PJ, Gupta A, Wideman CL, Story B, Grubbs B, Armstrong A, Dougall WC. Detection of myeloma in skeleton of mice by whole-body optical fluorescence imaging. Mol Cancer Ther. 2007;6:1701–1708. doi: 10.1158/1535-7163.MCT-07-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30:434–439. doi: 10.1038/nbt.2198. [DOI] [PubMed] [Google Scholar]
- 35.Meacham F, Boffelli D, Dhahbi J, Martin DI, Singer M, Pachter L. Identification and correction of systematic error in high-throughput sequence data. BMC Bioinformatics. 2011;12:451–461. doi: 10.1186/1471-2105-12-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lou DI, Hussmann JA, McBee RM, Acevedo A, Andino R, Press WH, Sawyer SL. High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc Natl Acad Sci U S A. 2013;110:19872–19877. doi: 10.1073/pnas.1319590110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nolan-Stevaux O, Tedesco D, Ragan S, Makhanov M, Chenchik A, Ruefli-Brasse A, Quon K, Kassner PD. Measurement of cancer cell growth heterogeneity through lentiviral barcoding identifies clonal dominance as a characteristic of in vivo tumor engraftment. PLoS One. 2013;8:e67316. doi: 10.1371/journal.pone.0067316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porter SN, Baker LC, Mittelman D, Porteus MH. Lentiviral and targeted cellular barcoding reveals ongoing clonal dynamics of cell lines in vitro and in vivo. Genome Biol. 2014;15:R75. doi: 10.1186/gb-2014-15-5-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belderbos ME, Koster T, Ausema B, Jacobs S, Sowdagar S, Zwart E, de Bont E, de Haan G, Bystrykh LV. Clonal selection and asymmetric distribution of human leukemia in murine xenografts revealed by cellular barcoding. Blood. 2017;129:3210–3220. doi: 10.1182/blood-2016-12-758250. [DOI] [PubMed] [Google Scholar]
- 40.Sipkins DA, Wei X, Wu JW, Runnels JM, Cote D, Means TK, Luster AD, Scadden DT, Lin CP. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature. 2005;435:969–973. doi: 10.1038/nature03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin T, Li L. The stem cell niches in bone. J Clin Invest. 2006;116:1195–1201. doi: 10.1172/JCI28568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Croucher PI, McDonald MM, Martin TJ. Bone metastasis: the importance of the neighbourhood. Nat Rev Cancer. 2016;16:373–386. doi: 10.1038/nrc.2016.44. [DOI] [PubMed] [Google Scholar]
- 43.Manier S, Kawano Y, Bianchi G, Roccaro AM, Ghobrial IM. Cell autonomous and microenvironmental regulation of tumor progression in precursor states of multiple myeloma. Curr Opin Hematol. 2016;23:426–433. doi: 10.1097/MOH.0000000000000259. [DOI] [PubMed] [Google Scholar]
- 44.Uchiyama H, Barut BA, Mohrbacher AF, Chauhan D, Anderson KC. Adhesion of human myeloma-derived cell lines to bone marrow stromal cells stimulates interleukin-6 secretion. Blood. 1993;82:3712–3720. [PubMed] [Google Scholar]
- 45.Li X, Pennisi A, Yaccoby S. Role of decorin in the antimyeloma effects of osteoblasts. Blood. 2008;112:159–168. doi: 10.1182/blood-2007-11-124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lwin ST, Fowler JA, Drake MT, Edwards JR, Lynch CC, Edwards CM. A loss of host-derived MMP-7 promotes myeloma growth and osteolytic bone disease in vivo. Mol Cancer. 2017;16:49–56. doi: 10.1186/s12943-017-0616-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kyle RA, Durie BG, Rajkumar SV, Landgren O, Blade J, Merlini G, Kroger N, Einsele H, Vesole DH, Dimopoulos M. Monoclonal gammopathy of undetermined significance (MGUS) and smoldering (asymptomatic) multiple myeloma: IMWG consensus perspectives risk factors for progression and guidelines for monitoring and management. Leukemia. 2010;24:1121–1127. doi: 10.1038/leu.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mikulasova A, Wardell CP, Murison A, Boyle EM, Jackson GH, Smetana J, Kufova Z, Pour L, Sandecka V, Almasi M. Somatic mutation spectrum in monoclonal gammopathy of undetermined significance indicates a less complex genomic landscape compared to multiple myeloma. Haematologica. 2017;102:1617–1625. doi: 10.3324/haematol.2017.163766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker BA, Wardell CP, Melchor L, Brioli A, Johnson DC, Kaiser MF, Mirabella F, Lopez-Corral L, Humphray S, Murray L. Intraclonal heterogeneity is a critical early event in the development of myeloma and precedes the development of clinical symptoms. Leukemia. 2014;28:384–390. doi: 10.1038/leu.2013.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao S, Choi M, Heuck C, Mane S, Barlogie B, Lifton RP, Dhodapkar MV. Serial exome analysis of disease progression in premalignant gammopathies. Leukemia. 2014;28:1548–1552. doi: 10.1038/leu.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das R, Strowig T, Verma R, Koduru S, Hafemann A, Hopf S, Kocoglu MH, Borsotti C, Zhang L, Branagan A. Microenvironment-dependent growth of preneoplastic and malignant plasma cells in humanized mice. Nat Med. 2016;22:1351–1357. doi: 10.1038/nm.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bataille R, Chappard D, Basle MF. Quantifiable excess of bone resorption in monoclonal gammopathy is an early symptom of malignancy: a prospective study of 87 bone biopsies. Blood. 1996;87:4762–4769. [PubMed] [Google Scholar]
- 53.Libouban H, Moreau MF, Basle MF, Bataille R, Chappard D. Increased bone remodeling due to ovariectomy dramatically increases tumoral growth in the 5T2 multiple myeloma mouse model. Bone. 2003;33:283–292. doi: 10.1016/s8756-3282(03)00196-0. [DOI] [PubMed] [Google Scholar]
- 54.Blundell JR, Levy SF. Beyond genome sequencing: lineage tracking with barcodes to study the dynamics of evolution, infection, and cancer. Genomics. 2014;104:417–430. doi: 10.1016/j.ygeno.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 55.Roodman GD. Mechanisms of bone metastasis. Discov Med. 2004;4:144–148. [PubMed] [Google Scholar]
- 56.de Groot AE, Roy S, Brown JS, Pienta KJ, Amend SR. Revisiting seed and soil: examining the primary tumor and cancer cell foraging in metastasis. Mol Cancer Res. 2017;15:361–370. doi: 10.1158/1541-7786.MCR-16-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In vitro and in vivo barcode data from pilot experiments. The sequences and read counts of all the barcodes detected in day 0 and day 28 in vitro 5TGM1 cultures, and tumors from the individual long bones of 5 mice on day 28 are shown. Frequencies of the rare (<0.1%) and common (>5%) subclasses of barcodes are also shown. Key: L/R = left/right. F/T = femur/tibia.
Deep sequencing of single barcode control cell lines. Read counts, mismatch data, and frequency distributions are shown for NGS of PCR products from 5TGM1 clones #23 and #31, each of which contains a single DNA barcode (shaded yellow and green, respectively). Read counts are also shown for deep sequencing of PCR products derived from admixtures of the two cell lines at the ratios indicated. The red shaded barcode is an example of a low-frequency barcode that was detected in all 15 tumors and controls that were analyzed.
Common subclasses of barcodes in pilot in vivo experiment. The sequences and read counts for all 78 barcodes that were classified as common (each representing at least 5% of the bulk of their respective tumors) in individual long bones from n = 5 different mice. Key: L/R = left/right. F/T = femur/tibia.
Tumor barcode data from second in vivo experiment. The sequences and read counts are shown for all barcodes detected in both replicate PCR products generated from tumors in the indicated long bones. Only those barcodes seen in the reference library of barcodes are shown. Sequences with less than 3 mismatches from barcodes with >10,000 read counts have been removed. Overall classification of barcodes into rare (<0.1%) or common (>5%) subclasses is also shown. Barcodes shaded in green are unique to either the sample or the mouse as shown.
Uniqueness of the common subclass of barcodes. The read counts for each common (>5%) barcode are shown together with their bone of origin and whether they were detectable within the common subclass in only one bone and/or one mouse. Barcodes whose frequency subclass was common in more than one long bone in the same mouse are indicated by the same color shading.
Filtering rare barcodes for sample uniqueness. The uniqueness of all the barcodes of the rare (<0.1%) subclass is shown. A comparison to the numbers of common (>5%) subclass of barcodes per sample (from Figure 3) is shown.