Abstract
Background
MSC-NTF cells are Mesenchymal Stromal Cells (MSC) induced to express high levels of neurotrophic factors (NTFs) using a culture-medium based approach. MSC-NTF cells have been successfully studied in clinical trials for Amyotrophic Lateral Sclerosis (ALS) patients.
MicroRNAs (miRNA) are short non-coding RNA molecules that coordinate post-transcriptional regulation of multiple gene targets. The purpose of this study was to determine whether the miRNA profile could provide a tool for MSC-NTF cell characterization and to distinguish them from the matched MSC from which they are derived.
Methods
NTF secretion in the culture supernatant of MSC-NTF cells was evaluated by ELISA assays. The Agilent microarray miRNA platform was used for pairwise comparisons of MSC-NTF cells to MSC. The differentially expressed miRNAs and putative mRNA targets were validated using qPCR analyses.
Results
Principal component analysis revealed two distinct clusters based on cell type (MSC and MSC-NTFs). Nineteen miRNAs were found to be upregulated and 22 miRNAs were downregulated in MSC-NTF cells relative to the MSC cells of origin. Further validation of differentially expressed miRNAs confirmed that miR-3663 and miR-132 were increased 18.5- and 4.06-fold, respectively while hsa-miR-503 was reduced more than 15-fold, suggesting that miRNAs could form the basis of an MSC-NTF cell characterization assay. In an analysis of the miRNA mRNA targets, three mRNA targets of hsa-miR-132-3p (HN-1, RASA1 and KLH-L11) were found to be significantly downregulated.
Conclusions
We have demonstrated that MSC-NTF cells can be distinguished from their MSCs of origin by a unique miRNA expression profile.
Trial Registration
Clinicaltrial.gov identifier NCT01777646. Registered 12 December 2012.
Keywords: Mesenchymal Stromal Cells, Neurotrophic Factors, Amyotrophic Lateral Sclerosis, MicroRNAs
Background
Mesenchymal stromal cells (MSC) are multipotent cells capable of differentiating into cells of multiple lineages [1]. MSC are being evaluated as a potential therapy for a wide variety of degenerative and immunological disorders [2].
We have developed a culture-medium based method for inducing MSCs to secrete enhanced levels of multiple neurotrophic factors (NTFs) including glial-derived neurotrophic factor (GDNF), brain-derived neurotrophic factor (BDNF) vascular endothelial growth factor (VEGF) and hepatocyte growth factor (HGF) [3]. MSC-NTF cells (NurOwn®) have been successfully used in clinical trials for amyotrophic lateral sclerosis (ALS) patients [4].
NTFs are small, naturally occurring polypeptides that support the development and survival of neurons [5]. NTFs are potent survival factors for embryonic, neonatal, and adult neurons, and have been evaluated in various neurodegenerative disease clinical trials over the past 25 years [6, 7]. MSC-NTF cells that have been induced to enhanced secretion of NTFs, offer a novel method for simultaneously delivering multiple NTFs to patients with neurodegenerative diseases such as ALS, while leveraging the potential immunomodulatory therapeutic benefits of MSCs [8]. Furthermore, a recent study demonstrated potential therapeutic benefits of MSC-NTF cells in an animal model of autism [9].
ALS (also known as Lou Gehrig’s disease) is a rare, relentlessly progressive and lethal neurodegenerative disease. At the cellular level, ALS is characterized by the progressive degeneration of upper and lower motor neurons in the motor cortex, brainstem, and spinal cord leading to progressive functional impairment and ultimately death [10].
Three clinical studies with NurOwn® (the MSC-NTF cell therapy) in ALS patients have been completed. The first two open-label studies [4], confirmed that the treatment was safe and well tolerated either by the intrathecal (IT) or by the intramuscular (IM) route of administration as well as by combined IT and IM administration. These studies demonstrated preliminary indications of efficacy, by slowing the rate of disease progression, as measured by the ALS Functional Rating Scale-Revised (ALSFRS-R) score. The recently completed US phase 2 multicenter double-blind placebo-controlled study confirmed these preliminary findings and suggested that meaningful efficacy could be achieved following a single MSC-NTF cell transplantation in a subgroup of patients that excluded slow progressors (submitted for publication). These findings are to be confirmed in a larger repeat-dose multicenter US phase 3 program.
MicroRNAs (miRNAs) are short (17-24 nt), single-stranded, endogenous non-coding RNAs that regulate gene expression by post-transcriptional silencing and/or mRNA degradation and can play important regulatory roles in animals and plants by targeting mRNAs for cleavage or translational repression. miRNAs have been shown to play critical roles in several biological processes, including cell differentiation, cell development, cell growth and apoptosis, by regulating gene expression through either the inhibition of mRNA translation or the induction of mRNA degradation [11–13].
Single miRNA can target multiple mRNAs, overall targeting approximately 60% of human genes. They regulate multiple and diverse cellular pathways and processes in normal and disease situations leading to changes in cell phenotype.
miRNA act in concert with Argonaute proteins within the RISC complex to suppress translation in a manner which is dependent on incomplete Watson-Crick base pairing between the so-called ‘seed’ sequence of the miRNA with complementary sequences in the target gene, usually in the 3’-UTR region of the mature message.
This study aimed to characterize an MSC-NTF miRNA fingerprint by identifying miRNAs that are significantly differentially expressed in MSC-NTF cells compared to the donor-matched MSCs by selecting a panel of mi-RNAs to monitor cell differentiation and performance, to be used as release criteria, and as an in-vivo identification assay.
Methods
Cells
MSC were isolated from healthy volunteers (Lonza, Walkersville, MD, USA) and from ALS patients’ bone marrow and expanded in culture. ALS patients were consented in accordance with the Helsinki declaration in the context of the phase 2a clinical trial (Clinicaltrial.gov identifier NCT01777646). The study was approved by the ethics committee of the Hadassah Hebrew University Medical Center, Jerusalem, Israel, and by the Director General of the Israel Ministry of Health.
MSC-NTF cells were induced to differentiate from each of the MSC donors, using a culture medium-based approach as previously described [3]. Briefly, MSCs were induced to differentiate into MSC-NTF cells using a medium-based approach in which cells were incubated in medium containing 1 mM dibutyryl cyclic AMP (cAMP), 20 ng/ml human basic fibroblast growth factor (hbFGF), 5 ng/ml human platelet-derived growth factor (PDGF-AA), and 50 ng/ml human Heregulin β1.
NTF secretion
NTF secretion was evaluated by ELISA for GDNF (DuoSet, R&D Systems, Minneapolis, MN, USA) VEGF and HGF (Quantikine, R&D Systems) in cell culture supernatant before and after MSC differentiation into MSC-NTF cells.
Microarray profiling and validation
Total RNA was extracted from eight independent, matched donor bone marrow-derived MSC and derived MSC-NTF cells of healthy donors and ALS patients using the Cell & Plant miRCURY™ RNA isolation kit (Exiqon, Copenhagen, Denmark). All RNA samples had a RIN > 7.
Microarray analysis was performed on 100 ng total RNA using Agilent’s miRNA platform (SurePrint G3 Human v16 microRNA 8 × 60K microarray slides, Agilent Technologies, Cheadle, UK). Data pre-processing and normalization was carried out using the “AgiMicroRNA” package in Bioconductor (https://www.bioconductor.org/packages/devel/bioc/vignettes/AgiMicroRna/inst/doc/AgiMicroRna.pdf). miRNAs differentially expressed between the MSC-NTF and MSC cells were identified by fold change analysis (pFDR < 0.05, fold change > 1.5). Candidate miRNAs from microarray data for future normalization of quantitative reverse transcription (qRT)-PCR were identified using the two one-sided tests approach (pFDR < 0.05, fold change < 2.0).
Expression analysis of the differentially expressed mi-RNAs was carried out by qRT-PCR using miRCURY LNA™ Universal RT microRNA PCR (Exiqon) except for miR-3663 that was analyzed using a miScript assay (Qiagen, Hilden, Germany), and a Roche LightCycler 480 (Roche Diagnostics Ltd, Burgess Hill, UK). Identification of miRNAs for normalization of qRT-PCR was carried out using the GeNorm algorithm [14] as implemented in Biogazelle qbase + v2.5 (Biogazelle, Ghent, Belgium). Mean fold changes were determined between normalized relative expression values for MSC and MSC-NTF cells and tested for statistical significance using Student’s t test (p < 0.05).
Target mRNA validation
For validating the mRNA targets of the validated miRNAs, qPCR was carried out using RT2 mRNA PCR (Qiagen, Hilden, Germany) and a Roche LightCycler 480 on the RNA samples of MSC and MSC-NTF cells of eight different donors (six ALS patients and two healthy donors).
Results
NTF secretion
We induced MSCs to differentiate into MSC-NTF cells, using a culture medium-based process. The MSC-NTF cells maintained the MSC immunophenotype, whereby > 95% of the population expresses CD73, CD90 and CD105 as determined by flow cytometry analyses and secrete significantly higher levels of NTFs such as GDNF, VEGF and HGF versus naïve MSC (Fig. 1).
Fig. 1.
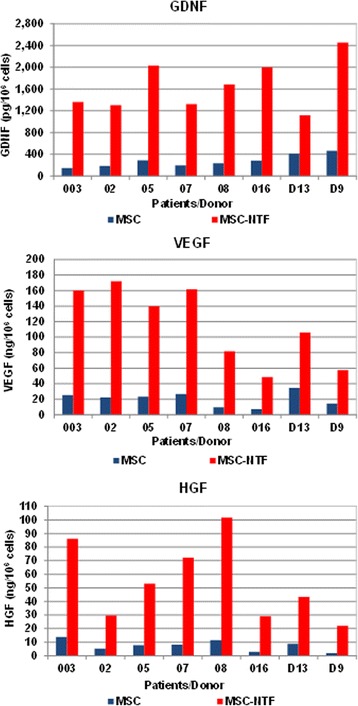
NTF secretion by MSC and MSC-NTF cells of the same patient/donor. MSC of six ALS patients and two healthy donors (D13 and D9) were induced to differentiate into MSC-NTF cells and secretion of neurotrophic factors GDNF, VEGF and HGF was measured in the culture supernatant by ELISA before and after differentiation. Average fold change MSC-NTF/MSC 6.56, 6.01, and 7.85 respectively (p < 0.005). GDNF glial-derived neurotrophic factor, HGF hepatocyte growth factor, MSC mesenchymal stromal cells, NTF neurotrophic factors, VEGF vascular endothelial growth factor
miRNA profiling
Matched MSC and MSC-NTF cells samples from four different ALS patients (patient ID 02, 03, 05, and 07) were analyzed using the Agilent miRNA platform. A total of 160 miRNAs were reliably detected across all the samples analyzed (present in at least one sample). An average of 199.75 ± 22.72 and 227.75 ± 14.48 (mean ± SD) miRNAs were identified in the MSC and MSC-NTF cell populations respectively.
To gain an overview of the donor-to-donor variability within each cell group and the relationships between the different cell groups, a visualization of the complete dataset was produced by PCA using all 160 detected miRNAs. The PCA plot represents the information content (variance) of each complete microRNA-ome dataset on the plot, as a single point in the principal component (PC) projection. The key point is the similar datasets cluster together.
This was initially done as a projection of the first three PCs (Fig. 2a). An alternative visualization of the expression patterns for the miRNAs in each sample and the sample relationships was generated using a heatmap based on agglomerative hierarchical clustering (Fig. 2b).
Fig. 2.
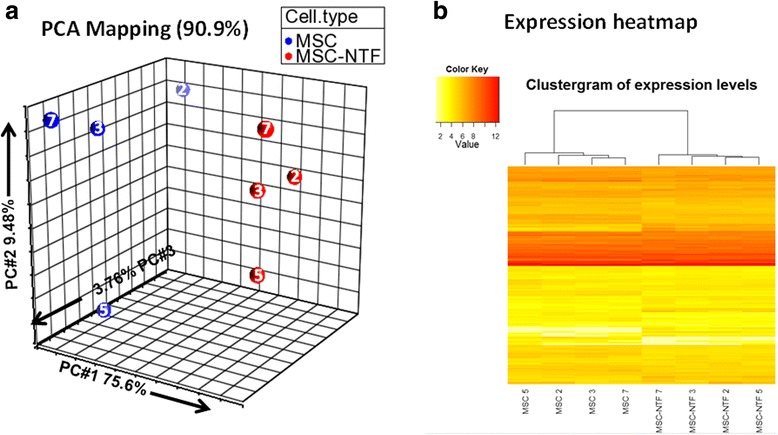
Comparison analysis of the MSC and MSC-NTF cell types based on all 160 detected miRNAs with cell type and donor ID indicated. a Representation of the eight cellular matched miRNA profiles of the four ALS patients in a 3D PCA projection, including donor ID (02, 03, 05, and 07); b Representation of the eight cellular miRNA profiles as a heatmap clustergram plot after hierarchical clustering, including donor ID. MSC mesenchymal stromal cells, NTF neurotrophic factors, PCA principal component analysis
The PCA and heatmap clustergram of MSC and MSC-NTF cells show that the sample set clearly separate, forming two distinct clusters based on cell type.
Analysis of miRNA
Forty-one different miRNAs were found to be differentially expressed in the MSC-NTF cells as compared to MSC. Eight miRNAs, equivalently expressed in the two cell types were identified, were used as candidate normalizers to support qPCR validation studies.
Statistical comparisons of the miRNA profiles of the differentially expressed miRNAs showed that 19 were upregulated in MSC-NTF versus MSC and 22 were downregulated in MSC-NTF versus MSC (Fig. 3).
Fig. 3.
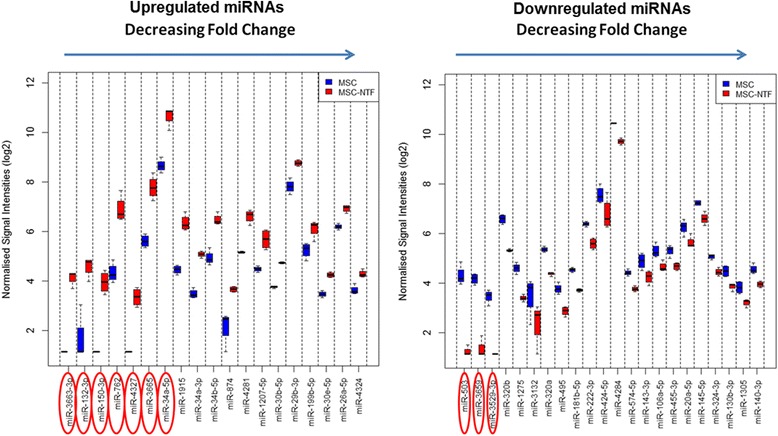
Differentially expressed miRNAs. Expression profiles of the 19 upregulated miRNAs (left panel) and the 22 downregulated miRNAs in MSC-NTF versus MSC on a log2 scale (right panel). The miRNAs most strongly down- and up-regulated in MSC-NTFs are highlighted with red ovals. When the expression of a miRNA was below the level of detection for the arrays, a nominal intensity value is given to these data points to avoid errors arising from non-computable mathematical operations during subsequent data analyses. MSC mesenchymal stromal cells, NTF neurotrophic factors
Gene ontology analysis of predicted target genes
The function of up- and downregulated miRNAs was studied using gene ontology analysis of predicted target genes. The analysis was performed on genes which were predicted to be miRNA targets by the miRNet database that integrates 11 different miRNA databases (http://www.mirnet.ca/faces/home.xhtml). To increase the level of reliability, only genes which were predicted to be targeted by at least three downregulated miRNAs or two upregulated miRNAs were included in the analysis (Table 1). While targets of upregulated miRNAs were found to be involved in promoting cell proliferation, targets of miRNAs which were downregulated were identified as genes which participate in neurogenesis, central nervous system development and cell cycle regulation (Table 1).
Table 1.
Gene ontology analysis
| GO biological process | No of genes | p value |
|---|---|---|
| Gene sets predicted to be upregulated following downregulation of at least three miRNAs (248 genes in analysis) | ||
| Central nervous system development | 31 | 5.15E-05 |
| Regulation of cellular protein metabolic process | 49 | 5.15E-05 |
| Regulation of growth | 25 | 5.15E-05 |
| Cell division | 24 | 5.15E-05 |
| Regulation of cell cycle | 34 | 5.15E-05 |
| Regulation of translation | 15 | 0.000127 |
| Negative regulation of cellular metabolic process | 48 | 0.000127 |
| Negative regulation of apoptotic process | 27 | 0.000127 |
| Regulation of protein metabolic process | 51 | 0.000127 |
| Neurogenesis | 27 | 0.000127 |
| Gene sets predicted to be targeted (downregulated) by at least two upregulated miRNAs (164 genes in analysis) | ||
| Gland development | 13 | 0.00222 |
| Positive regulation of cell proliferation | 20 | 0.00701 |
| Negative regulation of transcription from RNA polymerase II promoter | 15 | 0.00831 |
| Positive regulation of cell migration | 10 | 0.00831 |
| Regulation of chromosome organization | 7 | 0.00831 |
| Regulation of cell proliferation | 28 | 0.00831 |
| Negative regulation of apoptotic process | 17 | 0.00831 |
| Negative regulation of programmed cell death | 17 | 0.00831 |
| Positive regulation of developmental processes | 19 | 0.00831 |
| Positive regulation of cellular component organization | 15 | 0.00831 |
miRNA: microRNAs
Validation of differentially expressed miRNA
To validate the differentially expressed miRNAs, matched RNA samples of MSC and MSC-NTF cells of eight different donors (six ALS patients and two healthy donors) were analyzed. Of a panel of seven candidate-normalizing miRNAs tested, hsa-miR-19b-3p and hsa-miR-22-3p were selected as appropriate normalizers for this study.
The most differentially expressed miRNAs identified in the microarray analysis or those with a biologically relevant target gene function were chosen for validation by qRT-PCR (Table 2).
Table 2.
qPCR validation of differentially expressed miRNAs
| miRNA ID | Microarray fold change (MSC-NTF vs MSC) |
Function | qPCR validated (p ≤ 0.05) |
Fold change (MSC-NTF vs MSC) |
|---|---|---|---|---|
| hsa-miR-503-5p | −8.38 | Anti-angiogenic [17, 25] Promotes osteogenesis [26] (Originates from the same primary RNA transcript as miR-424 [27] | Yes | −30.5 |
| hsa-miR-320b | −2.41 | Involved in neural development, anti-angiogenic [28] | Yes | −1.36 |
| hsa-miR-320a | −1.97 | Targets VEGF A [16] | No | −1.28 |
| hsa-miR-222-3p | −1.74 | Negative modulator of angiogenesis Inhibition of osteogenic differentiation [29] | No | −1.25 |
| hsa-miR-424-5p | −1.72 | Anti-angiogenic [25] Targets VEGF A, VEGFR-2 and FGF2 [15] | Yes | −1.94 |
| hsa-miR-3663-3p | 8.03 | On/off signal. No validated mRNA target | Yes | 18.5 |
| hsa-miR-132-3p | 7.85 | Pro-angiogenic [30] Inhibits osteoblast differentiation [31] Regulation of synaptic structure [18] | Yes | 4.06 |
| hsa-miR-150-3p | 6.99 | On/off signal. Inhibits osteoblast differentiation downstream of TNF-α [32] | Not validated but significantly downregulated | −4.44 |
| hsa-miR-762 | 5.91 | Upregulated in neural precursors [33] | No | 1.47 |
| hsa-miR-34a-5p | 4.05 | Tumor suppressor: pro-apoptotic and anti-proliferative [34] TNF-α suppressor [35] Inhibits osteoblast differentiation [36] | Yes | 3.16 |
miRNA microRNAs, MSC mesenchymal stromal cells, NTF neurotrophic factors, VEGF(R) vascular endothelial growth factor (receptor), FGF fibroblast growth factor, TNF-α tumor necrosis factor alpha
qRT-PCR validation confirmed that six (hsa-miR-503, hsa-miR-320b, hsa-miR-424-5p, hsa-miR-34a-5p hsa-miR-132-3p and hsa-miR-3663-3p) of the ten miRNAs that were analyzed were found to be differentially expressed (p < 0.05), while the differential expression of hsa-miR-320a, hsa-miR-222-3p and hsa-miR-762) was just outside the significance cutoff p value (Table 2).
Of the differentially expressed miRNAs, hsa-miR-3663-3p was upregulated 18.5-fold and hsa-miR-503 was downregulated 30.5-fold (Fig. 4).
Fig. 4.
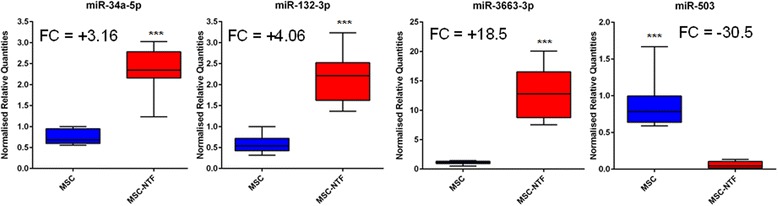
Validated differentially expressed miRNAs. Differential expression of miRNAs identified in the microarray was validated by qPCR analysis of MSC and MSC-NTF cells of eight different donors (six ALS patients and two healthy donors). ***p < 0.001, two-sided t test. FC fold change, MSC mesenchymal stromal cells, NTF neurotrophic factors
Non-specific amplification products were identified for miR-150-3p. miR-222-3p, miR-320a and 320b and miR-424-5p had low (and in some cases not significant) fold change between MSC and MSC-NTF cells limiting their diagnostic usefulness.
Target mRNA expression
To further investigate the potential effect of differentially expressed miRNAs we looked at the putative mRNA targets of the identified miRNAs.
Of the most significantly differentially expressed miRNAs that were validated by qPCR, miR-3663 has no known mRNA targets. We therefore further investigated the mRNA targets of hsa-miR-132-3p and hsa-miR-503, which were confirmed to be upregulated 4-fold and downregulated 30-fold, respectively.
Putative validated mRNA targets of hsa-miR-132-3p and miRNA hsa-miR-503-5p were identified by literature database searches.
Six mRNA targets of hsa-miR-132-3p (HN-1, RASA1, KLH-L11, MAPT, SPRED1 and ARHGAP32) and four mRNA targets of hsa-miR-503-5p (IGF1-R, FGF-R, L1CAM and CX3CL1) were chosen for further investigation based on their functional properties.
Of the known mRNA targets of hsa-miR-132-3p that were analyzed, HN-1, RASA1, and KLH-L11 were found to be significantly downregulated 11.86-fold, 3.73-fold, and 1.5-fold, respectively. These mRNA targets were significantly downregulated in MSC-NTF cells compared to MSCs (p < 0.05, Fig. 5) while MAPT and SPRED1 were not found to be differentially expressed between the two cell types (Fig. 5).
Fig. 5.
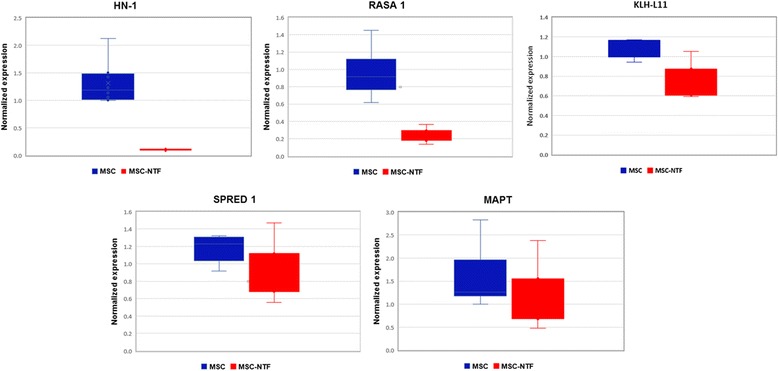
Differential expression of mRNAs targets of hsa-miR-132-3p. Expression of mRNA targets of miR-132-3p were compared in MSC and MSC-NTF cells of MSC and MSC-NTF cells of eight different donors (six ALS patients and two healthy donors) by qPCR analysis. HN-1, RASA1 and KLH-L11 were significantly downregulated (11.86 fold, 3.73 -fold and 1.5-fold respectively, p < 0.05). MSC mesenchymal stromal cells, NTF neurotrophic factors
Of the four mRNA targets of hsa-miR-503-5p, that were evaluated, expression of FGF-R and IGF1-R were found to be unchanged between MSC and MSC-NTF cells (data not shown). L1CAM and CX3CL1 were not expressed in either MSC nor in MSC-NTF cell samples (data not shown).
Discussion
The results of this study suggest that the differentiation process of MSC-NTF cells leads to both an increased secretion of NTFs and a characteristic miRNA profile. Furthermore, individual miRNAs biomarkers can be used to characterize MSC-NTF cells and distinguish them from the MSCs of origin. The changes in miRNA expression following MSC-NTF differentiation appear to lead to a repression of pro-proliferative target genes (inducing cell cycle arrest) and anti-apoptotic genes, while inducing neuronal-related genes providing the molecular basis for the functional phenotype and putative mechanism of action of the differentiated cells.
Of the 160 miRNAs identified by the microarray analysis, hsa-miR-3663-3p was found to be the most strongly upregulated (Fig. 4) and confirmed by qRT-PCR to be strongly induced (18.5-fold). Microarray analysis also identified hsa-miR-132-3p, which was confirmed to be upregulated 4-fold, while upregulation of hsa-miR-150-3p could not be confirmed by qPCR.
The putative mRNAs targets of the miRNAs that were identified to be characteristic of MSC-NTF cells were also studied. The most strongly induced miRNA, miR-3663, has no known mRNA targets. We therefore focused our investigation on known mRNA targets of miR-132-3p and miR-503-5p.
We found the mRNA targets of hsa-miR-132-3p, HN-1 RASA1 and KLH-L11, to be significantly downregulated in MSC-NTF cells compared to MSC samples from the same donor (p < 0.05), while mRNA targets of miR-503-5p FGF-R and IGF1-R were unchanged.
Importantly, miR-320a, miR-424-5p and miR-503 which were downregulated in MSC-NTF cells, were previously reported to target VEGF [15–17]. Indeed, downregulation of these three miRNAs is correlated with a significant increase in VEGF secretion in MSC-NTF cells (Fig. 1), suggesting that the observed change in these microRNAs may contribute to the increase in VEGF expression. Further confirmatory studies will be required.
miR-132-3p, upregulated in MSC-NTF cells, is an evolutionarily conserved and neuron-enriched miRNA [18, 19] that has been shown to be a positive regulator of developing neuron axon extension, acting through repression of Rasa1 mRNA (a Ras GTPase activator), in a mechanism that operates locally within the axon [20].
The Hematopoietic- and neurologic-expressed sequence 1 (Hn1) gene encodes a small protein that is highly conserved among species. Hn1 has previously identified as a gene associated with nervous system development and nerve regeneration [21]. MiR-132 has been found to downregulate HN1 expression leading to reduced cell proliferation [22]. Depletion of Hn1 has been shown to result in cell cycle arrest, which is consistent with the differentiated phenotype of MSC-NTF cells [23].
Very little is known about the Kelch-like family member 11 (KLH-L11); however, other members of the Kelch-like family of genes are involved in a wide range of processes that are relevant to LTP, including inter/intracellular communication, cell morphology, cytoskeletal organization and protein binding [24].
Conclusions
The results of this study suggest that a distinct miRNA signature may differentiate MSC-NTF cells from the MSCs of origin and potentially could be used as a sensitive criterion for identity -release testing in clinical trials. The upregulation of miR-132 supports a molecular mechanism for cell cycle arrest, neuronal differentiation and axonal extension, while downregulation of miR-320a, miR-424-5p and miR-503 are consistent with the observed increase in VEGF signaling. These observations support the putative mechanism of action of MSC-NTF cells and may have important implications to their application in ALS clinical studies and more broadly in a variety of neurodegenerative disorders that share common biology.
Acknowledgements
The authors wish to thank Sistemic UK for the miRNA analyses
Funding
Study was funded by Brainstorm Cell Therapeutics
Availability of supporting data
Some of the data used and/or analyzed during the current study are available from the corresponding author on reasonable request but restrictions may apply to the availability of some data, which were used under license for the current study, and so are not publicly available.
Authors’ contributions
YG contributed to conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. HK contributed to data analysis and interpretation, final approval of manuscript. NA contributed to collection and assembly of data, data analysis. RA contributed to conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. All authors read and approved the final manuscript.
Authors’ information
Not applicable.
Ethics approval and consent to participate
ALS patients were consented in accordance with the Helsinki declaration in the context of the phase 2a clinical trial
Consent for publication
Not applicable.
Competing interests
Authors are Brainstorm Cell Therapeutics employees
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yael Gothelf, Phone: ++972-3-9236384, Email: ygothelf@brainstorm-cell.com.
Haggai Kaspi, Email: hkaspi@brainstorm-cell.com.
Natalie Abramov, Email: nabramov@brainstorm-cell.com.
Revital Aricha, Email: raricha@brainstorm-cell.com.
References
- 1.Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 2.Bernardo ME, Fibbe WE. Mesenchymal stromal cells: sensors and switchers of inflammation. Cell Stem Cell. 2013;13:392–402. doi: 10.1016/j.stem.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Gothelf Y, Abramov N, Harel A, et al. Safety of repeated transplantations of neurotrophic factors-secreting human mesenchymal stromal stem cells. Clin Transl Med. 2014;3:21. doi: 10.1186/2001-1326-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrou P, Gothelf Y, Argov Z, et al. Safety and clinical effects of mesenchymal stem cells secreting neurotrophic factor transplantation in patients with amyotrophic lateral sclerosis: results of phase 1/2 and 2a clinical trials. JAMA Neurol. 2016;73:337–44. doi: 10.1001/jamaneurol.2015.4321. [DOI] [PubMed] [Google Scholar]
- 5.Henderson CE. Role of neurotrophic factors in neuronal development. Curr Opin Neurobiol. 1996;6:64–70. doi: 10.1016/S0959-4388(96)80010-9. [DOI] [PubMed] [Google Scholar]
- 6.Bartus RT, Johnson EM., Jr Clinical tests of neurotrophic factors for human neurodegenerative diseases, part 1: where have we been and what have we learned? Neurobiol Dis. 2017;97:156–68. doi: 10.1016/j.nbd.2016.03.027. [DOI] [PubMed] [Google Scholar]
- 7.Bartus RT, Johnson EM., Jr Clinical tests of neurotrophic factors for human neurodegenerative diseases, part 2: where do we stand and where must we go next? Neurobiol Dis. 2017;97:169–78. doi: 10.1016/j.nbd.2016.03.026. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Chen X, Cao W, et al. Plasticity of mesenchymal stem cells in immunomodulation: pathological and therapeutic implications. Nat Immunol. 2014;15:1009–16. doi: 10.1038/ni.3002. [DOI] [PubMed] [Google Scholar]
- 9.Perets N, Segal-Gavish H, Gothelf Y, et al. Long term beneficial effect of neurotrophic factors-secreting mesenchymal stem cells transplantation in the BTBR mouse model of autism. Behav Brain Res. 2017;331:254–60. doi: 10.1016/j.bbr.2017.03.047. [DOI] [PubMed] [Google Scholar]
- 10.Kiernan MC, Vucic S, Cheah BC, et al. Amyotrophic lateral sclerosis. Lancet. 2011;377:942–55. doi: 10.1016/S0140-6736(10)61156-7. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–24. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 13.Lim LP, Glasner ME, Yekta S, et al. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 14.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chamorro-Jorganes A, Araldi E, Penalva LO, et al. MicroRNA-16 and microRNA-424 regulate cell-autonomous angiogenic functions in endothelial cells via targeting vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1. Arterioscler Thromb Vasc Biol. 2011;31:2595–606. doi: 10.1161/ATVBAHA.111.236521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin Z, Zhao Y, Li H, et al. miR-320a mediates doxorubicin-induced cardiotoxicity by targeting VEGF signal pathway. Aging (Albany NY) 2016;8:192–207. doi: 10.18632/aging.100876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou B, Ma R, Si W, et al. MicroRNA-503 targets FGF2 and VEGFA and inhibits tumor angiogenesis and growth. Cancer Lett. 2013;333:159–69. doi: 10.1016/j.canlet.2013.01.028. [DOI] [PubMed] [Google Scholar]
- 18.Edbauer D, Neilson JR, Foster KA, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–84. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magill ST, Cambronne XA, Luikart BW, et al. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc Natl Acad Sci U S A. 2010;107:20382–7. doi: 10.1073/pnas.1015691107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hancock ML, Preitner N, Quan J, et al. MicroRNA-132 is enriched in developing axons, locally regulates Rasa1 mRNA, and promotes axon extension. J Neurosci. 2014;34:66–78. doi: 10.1523/JNEUROSCI.3371-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zujovic V, Luo D, Baker HV, et al. The facial motor nucleus transcriptional program in response to peripheral nerve injury identifies Hn1 as a regeneration-associated gene. J Neurosci Res. 2005;82:581–91. doi: 10.1002/jnr.20676. [DOI] [PubMed] [Google Scholar]
- 22.Zhang ZG, Chen WX, Wu YH, et al. MiR-132 prohibits proliferation, invasion, migration, and metastasis in breast cancer by targeting HN1. Biochem Biophys Res Commun. 2014;454:109–14. doi: 10.1016/j.bbrc.2014.10.049. [DOI] [PubMed] [Google Scholar]
- 23.Laughlin KM, Luo D, Liu C, et al. Hematopoietic- and neurologic-expressed sequence 1 (Hn1) depletion in B16.F10 melanoma cells promotes a differentiated phenotype that includes increased melanogenesis and cell cycle arrest. Differentiation. 2009;78:35–44. doi: 10.1016/j.diff.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhanoa BS, Cogliati T, Satish AG, et al. Update on the Kelch-like (KLHL) gene family. Hum Genomics. 2013;7:13. doi: 10.1186/1479-7364-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caporali A, Emanueli C. MicroRNA-503 and the extended microRNA-16 family in angiogenesis. Trends Cardiovasc Med. 2011;21:162–6. doi: 10.1016/j.tcm.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Y, Xu J, Xu L, et al. MiR-503 promotes bone formation in distraction osteogenesis through suppressing Smurf1 expression. Sci Rep. 2017;7:409. doi: 10.1038/s41598-017-00466-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forrest AR, Kanamori-Katayama M, Tomaru Y, et al. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia. 2010;24:460–6. doi: 10.1038/leu.2009.246. [DOI] [PubMed] [Google Scholar]
- 28.Somel M, Liu X, Tang L, et al. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011;9:e1001214. doi: 10.1371/journal.pbio.1001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshizuka M, Nakasa T, Kawanishi Y, et al. Inhibition of microRNA-222 expression accelerates bone healing with enhancement of osteogenesis, chondrogenesis, and angiogenesis in a rat refractory fracture model. J Orthop Sci. 2016;21:852–8. doi: 10.1016/j.jos.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 30.Anand S, Majeti BK, Acevedo LM, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16:909–14. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Z, Wang Y, Sun Z, et al. miRNA-132-3p inhibits osteoblast differentiation by targeting Ep300 in simulated microgravity. Sci Rep. 2015;5:18655. doi: 10.1038/srep18655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang N, Zhou Z, Wu T, et al. TNF-alpha-induced NF-kappaB activation upregulates microRNA-150-3p and inhibits osteogenesis of mesenchymal stem cells by targeting beta-catenin. Open Biol. 2016;6:150258. doi: 10.1098/rsob.150258. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Zhang D, Zhao T, Ang HS, et al. AMD1 is essential for ESC self-renewal and is translationally down-regulated on differentiation to neural precursor cells. Genes Dev. 2012;26:461–73. doi: 10.1101/gad.182998.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Farooqi AA, Fayyaz S, Shatynska-Mytsyk I, et al. Is miR-34a a Well-equipped swordsman to conquer temple of molecular oncology? Chem Biol Drug Des. 2016;87:321–34. doi: 10.1111/cbdd.12634. [DOI] [PubMed] [Google Scholar]
- 35.Guennewig B, Roos M, Dogar AM, et al. Synthetic pre-microRNAs reveal dual-strand activity of miR-34a on TNF-alpha. RNA. 2014;20:61–75. doi: 10.1261/rna.038968.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L, Holmstrom K, Qiu W, et al. MicroRNA-34a inhibits osteoblast differentiation and in vivo bone formation of human stromal stem cells. Stem Cells. 2014;32:902–12. doi: 10.1002/stem.1615. [DOI] [PubMed] [Google Scholar]


