Abstract
Bats are unique mammals that are reservoirs of high levels of virus diversity. Although several of these viruses are zoonotic, the majority are not. Astroviruses, transmitted fecal-orally, are commonly detected in a wide diversity of bat species, are prevalent at high rates and are not thought to directly infect humans. These features make astroviruses useful in examining virus evolutionary history, epidemiology in the host, and temporal shedding trends. Our study screened for the presence of astroviruses in bats in Singapore, reconstructed the phylogenetic relations of the polymerase genes and tested for population characteristics associated with infection. Of the seven species screened, astroviruses were detected in Rhinolophus lepidus and Eonycteris spelaea. The R. lepidus sequences grouped with other Rhinolophus astrovirus sequences from China and Laos, while the Eoncyteris sequences formed a distinct clade with astroviruses from Rousettus spp. in Laos and Pteropus giganteus in Bangladesh, but not with other E. spelaea sequences. Longitudinal collections of Eonycteris feces demonstrated variable shedding. Juvenile status of bats was a risk factor for astroviruses. This study highlights the diversity of astroviruses in nectivorous and insectivorous bats in Singapore and provides a predictive framework for understanding astrovirus infection in these bats. It also suggests that in addition to host phylogenetic relatedness, host ecology, such as roosting behavior, may drive co-infections, virus maintenance and spillover.
Keywords: Bat borne virus, Eonycteris spelaea, Rhinolophus lepidus, Epidemiology, Phylogenetics, Pooling prevalence, Southeast Asia
1. Introduction
Bats are the second most speciose group of mammals behind rodents [1], their species diversity provides opportunity for virus diversity through co-evolution [2]. The intensive surveillance in bats for zoonotic viruses has provided biological samples that are screened for other virus families that may not be of human public health importance or pathogenic to the reservoir, but may be informative of evolutionary processes of bat viruses due to their high prevalence [3], [4]. In addition to zoonotic viruses, such as Ebola and SARS-CoV, there are a number of commonly detected viruses that demonstrate varying degrees of host specificity at genus or species level [5], [6], [7].
The Astroviridae, a family of single stranded, positive sense non-enveloped RNA viruses [8], is a favorable candidate for studying evolutionary history because they infect a wide variety of species and recombine [9], [10], [11]. In addition, the phylogeny of the astrovirus RdRp segment indicates that cross-species transmission has driven the evolution of this virus family [12]. A number of these viruses have public health and economic importance [12], [13]. Transmission is fecal-oral and diarrhea is the primary clinical symptom in most hosts, but astroviruses can also cause kidney-related ailments in chickens and humans and encephalitis in humans and cows [14], [15], [16]. Astrovirus outbreaks in commercial cow, pig, mink, turkey, duck, goose, chicken, and guinea fowl production facilities cause major economic losses [17], [18], [19], [20], [21], [22], [23]. In wildlife, avastroviruses have been detected in wild ducks, wading birds, passerines, pigeons, and penguins [24], [25], [26], [27]. When infected, bats have been hypothesized notto become sick or display signs due to their unique immune response [28].
Singapore, a predominantly urban island that has lost the great majority of its original habitats and suffered a number of local species extinctions [29], harbors an estimated 26 resident bat species. Certain species of bats in Singapore exist in urbanized sites, while others reside in small forest and grassland fragments. As part of efforts to characterize the virus communities of Singapore bats, including the dynamics of infection, we caught and screened bats and then tested the impact of bat species, age class (juvenile vs. adult), sex, and body condition on the presence of astroviruses while also monitoring the temporal shedding of these viruses.
2. Materials and methods
2.1. Sample collection
Animal ethics approval for the study was given by the National University of Singapore - International Animal Care & Use Committee (Permit # B01/12) and bats were sampled with permission from the National Parks Board, Singapore (NP/RP11-011). Bats were trapped at seven different sites in Singapore from April 2011 through March 2014 as previously described [30]. Briefly, rectal and oral swabs were taken and age (classified as juvenile or adult), gender, forearm length, and weight were recorded from each individual. In addition, longitudinal fecal collections were taken from a colony of cave nectar bats (Eonycteris spelaea) from April 2011 until June 2015.
2.2. RNA extraction, PCR, sequencing and phylogenetic analysis
Fecal samples were vortexed for 15 s and then centrifuged at 4000 rpm for 1 min. Oral and rectal swabs were vortexed and then pooled for each individual. Samples collected between 12 April 2011 to 2 March 2012 were pooled by date (5 individual samples per pool) to test for pooled prevalence. RNA extraction cDNA synthesis, PCR amplification, cloning and sequencing were performed as previously described [31], [32]. 5′ and 3′ RACE was attempted to generate cDNA from the ends (Life Technologies, Carlsbad, CA), but neither approach was successful. RACE generally requires high amounts and high quality of target nucleic acids and these fecal samples may have relatively few astrovirus reads compared to the host and bacterial background. Over 700 global astrovirus RdRp sequences were downloaded from the NCBI GenBank and analyzed with 38 bat astrovirus sequences generated from this study (GenBank accession numbers MF983815-MF983852) using Geneious 7.1.7 software [33]. The final dataset was reduced to a total of 449 astrovirus sequences after the removal of short sequences and phylogenies were reconstructed using RAxML v8.0.14 [34] as previously described [30].
2.3. Epidemiological analysis
Individual prevalence from pools was estimated using a frequentist approach that assumed a fixed pool size and a perfect detection and a Bayesian approach with a Gibbs sampler model with AusVet EpiTools [35]. Details of the criteria used in the analysis are available in the supplementary materials. These were compared to individual prevalence rates on four dates within the sampling period. Multiple logistic regression (MLogR) was used to examine the effects of species, catch/roost location, sex, age, and bat body condition [36] on astrovirus infection. The scale of each predictor was determined by performing exploratory data analysis. A variance inflation factor (VIF) was calculated to assess multi-collinearity (inter-relatedness of the covariates). Hierarchical logistic models were tested to investigate significant predictors of astrovirus infection. Likelihood ratio tests (LRT) were generated to compare full, extended MLogR to more constrained models and assess whether the inclusion of interaction terms within a larger model were significant. Akaike Information Criterion (AIC) values were used to rank models and to determine the optimal set of predictors. Odds ratios were computed for the explanatory variables in the resulting model.
Forward and backward stepwise selection based on AIC and a 5% p-value cut-off were utilized as an aid to choose predictors for the final model. All covariates were considered for elimination. Based upon the statistical output of the stepwise processes, likelihood ratio tests outcomes, the variance inflation factor procedure, confounding assessments, cross tabulations of the outcome and variables, and prior knowledge regarding the subject, the final model was fit. The final model included 1) bat age and 2) bat species. The Pearson's chi-squared goodness-of-fit test (H0: the model M0 fits) was used to determine the overall fit of the final model. The p-value was computed at P: 0.27, indicating a well-fit model. All analyses were carried out using Stata version 12.1 [37].
3. Results
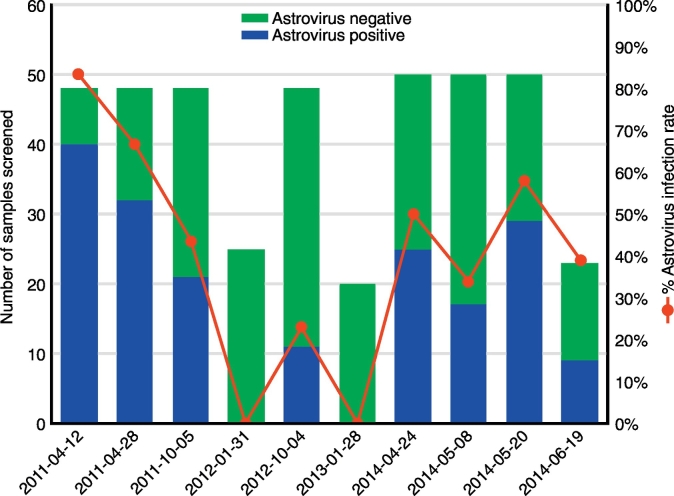
A total of 431 bats from 7 species were trapped and sampled (Table 1). Astroviruses were only detected in Eonycteris spelaea (30/169, 17.8%) and Rhinolophus lepidus (13/36, 36.1%), with an overall detection rate of 9.9% for the pooled oral-rectal swabs (Fig. 1). A total of 410 E. spelaea fecal samples were screened, with 184 (44.9%) astrovirus positives detected. However, the number of infected fecal samples varied by week with a high of 83% positive (2011-04-12) to a low of 0% positive (2012-01-31 and 2013-01-28) (Fig. 2). A total of 144 pools out of 221 (1105 individual samples) made from 19 sampling occasions were positive for astroviruses (65.16%) (Table 2). Minimum infection rates of individual samples throughout the sampling period ranged from 6%–20%. Frequentist analysis predicted individual sample prevalence ranged from 6.89%–36.9% across the sampling period with an estimated overall prevalence of 19.01% (95% Confidence Interval: 16.1%–22.2%), while the Bayesian analysis calculated an estimated mean prevalence of 23.75% (95% Credible Interval: 18.9%–34.0%) (Supplementary data 1). Both the frequentist confidence interval and the Bayesian credible interval produced similar values supporting the estimated prevalence.
Table 1.
Bat baseline descriptive characteristics by outcome group.
| Bat characteristics | Astrovirus + ve |
Astrovirus − ve |
Total |
P-value⁎ |
|---|---|---|---|---|
| N = 43 (9.98%) | N = 388 (90.02%) | N = 431 | ||
| Bat species | < 0.001 | |||
| Eonycteris spelaea | 30 (69.8%) | 139 (35.8%) | 169 (39.2%) | |
| Cynopterus brachyotis | 0 (0%) | 144 (37.11%) | 144 (33.4%) | |
| Penthetor lucasi | 0 (0%) | 79 (20.4%) | 79 (18.3%) | |
| Rhinolophus lepidus | 13 (30.2%) | 23 (5.9%) | 36 (8.4%) | |
| Macroglossus minimus | 0 (0%) | 1 (0.3%) | 1 (0.2%) | |
| Myotis sp. | 0 (0%) | 1 (0.3%) | 1 (0.2%) | |
| Pipstrelle stenopterus | 0 (0%) | 1 (0.3%) | 1 (0.2%) | |
| Sex | 0.17 | |||
| Female | 15 (34.9%) | 179 (46.1%) | 194 (45%) | |
| Male | 28 (62.8%) | 203 (52.6%) | 231 (53.6%) | |
| Missing dataa | 1 (2.3%) | 5 (1.3%) | 6 (1.4%) | |
| Age | 0.02 | |||
| Juvenile | 14 (31.8%) | 81 (20.9%) | 95 (22%) | |
| Sub-adult/adult | 24 (54.6%) | 299 (77.3%) | 323 (74.9%) | |
| Missing dataa | 6 (13.6%) | 7 (1.8%) | 13 (3%) | |
| Roost location | < 0.001 | |||
| Bukit Timah | 13 (30.2%) | 100 (25.8%) | 113 (26.2%) | |
| Kent Ridge | 1 (2.3%) | 94 (24.2%) | 95 (22%) | |
| Pulau Ubin | 0 (0%) | 3 (0.8%) | 3 (0.7%) | |
| Rifle Range Road | 29 (67.4%) | 149 (38.4%) | 178 (41.3%) | |
| Dairy Farm | 0 (0%) | 23 (5.9%) | 23 (5.3%) | |
| Sian Tan Avenue | 0 (0%) | 1 (0.3%) | 1 (0.2%) | |
| Telok Blangah | 0 (0%) | 18 (4.6%) | 18 (4.2%) | |
| Body condition index | < 0.001 | |||
| Mean | 0.491 | 0.608 | 0.597 | |
| Std. dev. | 0.274 | 0.199 | 0.210 | |
| Median | 0.541 | 0.592 | 0.590 | |
| Missing dataa | 5 (11.4%) | 20 (5.2%) | 25 (5.8%) |
In categorical variables, the first characterization under the variable heading is the reference category.
Information in parentheses (%).
Categorical variables' p-value based on Chi-squared test (≥ 5 per cell) or Fisher's exact test (< 5 per cell). Continuous variable p-value based on Wald Z-test.
Missing data for this variable; statistical analyses based on available data.
Fig. 1.
Photographs of Rhinolophus lepidus (Blyth's horseshoe bat) and Eonycteris spelaea (Cave nectar bat).
Fig. 2.
Temporal variation in the infection rate of astrovirus detected in the feces collected from an Eonycteris spelaea colony using a hemi-nested PCR detecting the RNA-dependent reverse polymerase gene.
Table 2.
Pooled prevalence estimates using frequentist and Bayesian methods with a Gibbs sampler compared to individual prevalence rates.
| Collection date | Number of Pools | Number of positive pools | Minimum infection rate | Estimated individual prevalence (% CL: 2.5–97.5) | Standard error | Individual samples screened | Individual samples PCR positive | Individual sample prevalence | |
|---|---|---|---|---|---|---|---|---|---|
| Frequentist estimation of pooled prevalence with fixed pool size and perfect test | 12-Apr-11 | 35 | 15 | 8.6% | 10.6% (0.059;0.170) | 0.026 | 48 | 40 | 83.3% |
| 28-Apr-11 | 16 | 11 | 13.8% | 20.8% (0.101;0.357) | 0.059 | 48 | 32 | 66.7% | |
| 12-May-11 | 10 | 6 | 12% | 16.7% (0.059; 0.344) | 0.064 | – | – | – | |
| 25-May-11 | 10 | 6 | 12% | 16.7% (0.059; 0.344) | 0.064 | – | – | – | |
| 13-Jun-11 | 10 | 3 | 6% | 6.9% (0.014; 0.191) | 0.039 | – | – | – | |
| 1-Jul-11 | 10 | 5 | 10% | 12.9% (0.041; 0.285) | 0.055 | – | – | – | |
| 14-Jul-11 | 10 | 10 | 20% | – | – | – | – | – | |
| 27-Jul-11 | 10 | 10 | 20% | – | – | – | – | – | |
| 8-Aug-11 | 10 | 8 | 16% | 27.5% (0.111; 0.521) | 0.092 | – | – | – | |
| 22-Aug-11 | 10 | 9 | 18% | 36.9% (0.149; 0.698) | 0.120 | – | – | – | |
| 9-Sep-11 | 10 | 10 | 20% | – | – | – | – | – | |
| 21-Sep-11 | 10 | 9 | 18% | 36.9% (0.149; 0.698) | 0.120 | – | – | – | |
| 5-Oct-11 | 10 | 6 | 12% | 16.7% (0.059; 0.344) | 0.064 | 48 | 21 | 43.8% | |
| 21-Oct-11 | 10 | 9 | 18% | 36.9% (0.149; 0.698) | 0.120 | – | – | – | |
| 2-Nov-11 | 10 | 4 | 8% | 9.7% (0.0256; 0.235) | 0.047 | – | – | – | |
| 30-Nov-11 | 10 | 6 | 12% | 16.7% (0.059; 0.344) | 0.064 | – | – | – | |
| 1-Jan-12 | 10 | 8 | 16% | 27.5% (0.111; 0.521) | 0.092 | – | – | – | |
| 31-Jan-12 | 10 | 5 | 10% | 12.9% (0.041; 0.285) | 0.055 | 25 | 0 | 0% | |
| 2-Mar-12 | 10 | 4 | 8% | 9.7% (0.0256; 0.235) | 0.047 | – | – | – | |
| Total | 221 | 144 | 13% | 19% (0.161; 0.222) | 0.015 | 169 | 93 | 55% | |
| Pooled prevalence with a Bayesian approach and a Gibbs sampler | Minimum Prevalence | Estimated Mean Prevalence | Standard deviation | ||||||
| Total | 221 | 144 | 15.3% | 23.8% (0.189; 0.340) | 0.040 | ||||
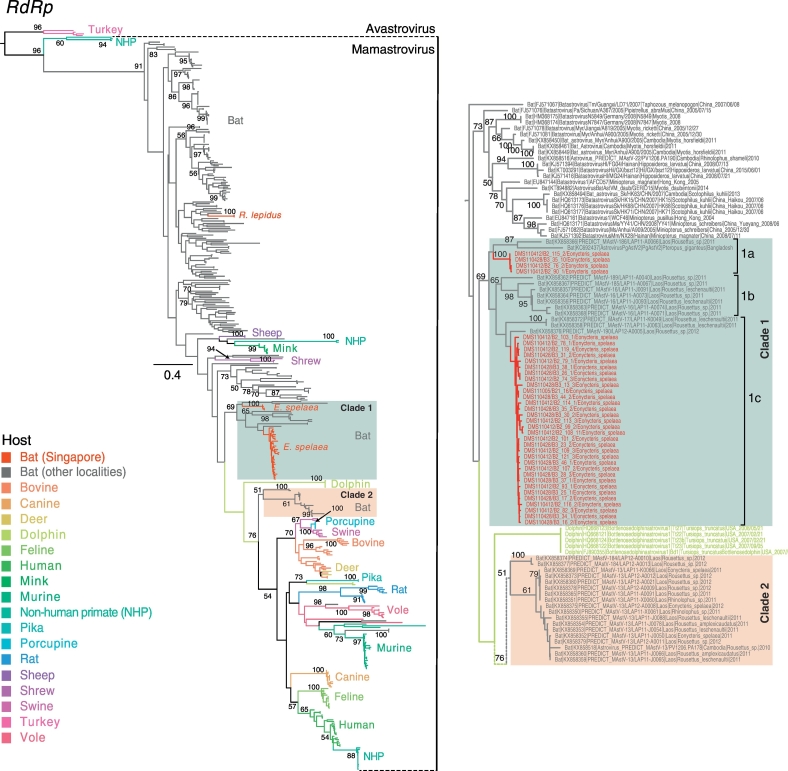
The astroviruses phylogeny showed that the majority of bat astroviruses (indicated by grey branches) were basal to other mammalian astroviruses, but with a general lack of statistical support (Fig. 3). The 38 novel bat astroviruses sequences collected in Singapore (denoted by red branches in Fig. 3) fell into 2 groups. Two bat astrovirus sequences identified from R. lepidus formed a monophyletic clade (bootstrap [BS] = 100%) that clustered within other Rhinolophus astroviruses from China and Laos (BS < 60%). Similarly, 36 astroviruses sequences from E. spelaea formed a monophyletic lineage (BS = 69%) with two distinct sub-clades, and this lineage was closely related to other pteropodid bat astroviruses from Bangladesh and Laos. Furthermore, the clustering of all E. spelaea astrovirus sequences, despite being collected in different years, suggests sustained transmission in this host. The astroviruses from R. lepidus and E. spelaea from Singapore were genetically dissimilar (nucleotide sequence identity 54.0–57.2%) even though their sampling locations and collection periods were similar.
Fig. 3.
Maximum likelihood phylogeny of the RNA-dependent RNA polymerase (RdRp) region of global astrovirues. Red branches denote novel bat astrovirus generated from this study, whereas the grey branches represent bat astrovirus collected from other geographical regions. The coloured branches denote different host species. Bootstrap values > 50% are indicated at major nodes. The scale bar represents the nucleotide substitutions per site. The insert on the right shows the details of astrovirus strains in Clade 1 and 2. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Interestingly, we revealed phylogenetically diverse astroviruses among E. spelaea bats, with at least two distinctive groups (Clade 1 and 2). Clade 1 (BS = 69%) is further composed of three smaller sub-clades (designated as a-c). Clade 1a (nucleotide identity 60.8–99.2%) includes 4 E. spelaea astroviruses from Singapore (BS = 100%) whereas the other comprises one Pteropus giganteus astrovirus from Bangladesh and a Rousettus sp. astrovirus from Laos. Clade 1b (BS = 65%; nucleotide identity 64.8–100%) is composed of Rousettus astroviruses from Laos. Clade 1c (nucleotide identity: 66.8–99.2%) includes astroviruses from E. spelaea (Singapore) and Rousettus leschenaultia (Laos). Both Singapore E. spelaea astrovirus clades were collected in the same sampling period. Clade 2 (BS = 51%; nucleotide identity 65.3–99.2%) contains two E. spelaea astroviruses, one Rhinolophus astrovirus and numerous Rousettus astroviruses, all from Laos. This suggests that E. spelaea astroviruses from Singapore are closely related to those of Laos Rousettus spp., yet are distantly related to Laos E. spelaea astroviruses. Clade 2 further displays the close phylogenetic relationship between E. spelaea and Rousettus spp. astroviruses (BS = 61%), all occurring in Laos. It is likely that the co-roosting of these bat species facilitated the interspecies virus transmission.
The most predictive model for astrovirus infection included bat age and bat species: log(p/1 − p) = βo + β1(Bat Age) + β2(Bat Species). In the crude analysis, bat age was found to be a significant risk factor for astrovirus, (OR: 0.44, P: 0.02, 95% CI: 0.22–0.90) providing evidence that astroviruses were less likely to be detected in adults. Thus we estimate the odds of astrovirus infection to be reduced by 56% given an adult bat. Similarly, unadjusted bat species was a strong indicator of astrovirus, (OR: 2.62, P: 0.02, 95% CI: 1.19–5.75); thus, it is estimated that the odds of contracting astrovirus infection are ~ 2.6 × greater among Rhinolophus lepidus bats as compared to the referent group Eonycteris spelaea bat. The five remaining species were automatically eliminated from the model, as none of them predicted astrovirus (astrovirus detection rate = 0%). However, adjusting for bat age in the larger model displayed more modest evidence for a bat age effect (OR: 0.59, P: 0.20, 95% CI: 0.26–1.34). Although, bat species did not appear to be an important confounder in the bat age/astrovirus relationship (determined by the adjusted bat age odds ratio lying between the 95% CI in the unadjusted model). Hence, the evidence in these data suggest, but are insufficient to conclude, that bat age has an independent effect on astrovirus outcome beyond that of bat species (Table 3).
Table 3.
Risk of astrovirus infection according to bat age and bat species.
| Unadjusted |
Adjusted |
|||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | |
| Bat age | ||||||
| Juvenile | 1 [Reference] | 1 [Reference] | ||||
| Adult | 0.46 | 0.22–0.90 | 0.02 | 0.59 | 0.26–1.34 | 0.20 |
| Bat speciesa | ||||||
| Eonycteris spelaea | 1 [Reference] | 1 [Reference] | ||||
| Rhinolophus lepidus | 2.61 | 1.19–5.75 | 0.02 | 3.41 | 1.35–8.60 | 0.01 |
Abbreviations: OR, Odds Ratio; 95% CI, 95% Confidence Interval; P, P-value.
Remaining bat species categories dropped due to perfect prediction of failure.
4. Discussion
This is the first report of bat astroviruses from Singapore and the first report of astrovirus infection in Rhinolophus lepidus. Previous surveillance efforts have demonstrated variable rates of astrovirus infection from diverse bat species, including those tested here [38]. Astroviruses have been detected from ten other species of Rhinolophus bats in previous studies [39], [40], [41], [42], [43]. A recent study in Cambodia and Laos screened six members of the family Pteropodidae and astroviruses were only detected in Eoncyteris spelaea (3.8%) and Rousettus spp. (7.2%) [44]. For longitudinal monitoring, the volume of samples may hinder processing, making pooling a feasible option. However, if the number of positive samples is low, the sensitivity of the test may present false negatives and if all the pools are positive, as occurred on three sampling occasions in this study, one is unable to estimate individual prevalence. In cases where we screened individual samples, the individual detection rate was greater than the estimated prevalence. This may result from the pooling of the samples lowering the relative quantity of virus in comparison to the background material.
As Astroviruses replicate in the gastro-intestinal tract and are fecally-orally transmitted, the relative viral load may be higher in feces compared to swabs. This may explain our higher detection rate from E. spelaea feces, but as bat viruses may be shed differentially over time, we cannot exclude changes in viral load affecting our results. We found variable rates of shedding across our sampling period. In Singapore, temperatures are not variable month to month, but there are two monsoon seasons, which may be associated with bat borne virus shedding [43]. A study on Myotis myotis in Germany demonstrated distinct astrovirus amplification peaks that correlated with the formation of a suitable roost size and also a post-parturition period [45]. In the tropics, where there is lower seasonal variation and an absence of torpor, the reproductive behavior of bats may drive virus persistence [46]. This may not always be the case as astrovirus and coronavirus shedding in Borneo was not associated with reproductive status [43].
We analyzed the astrovirus RdRp sequences from a broad range of hosts including bats and non-human primates [44], [47], [48]. Consistent with previous studies, our results indicate that bats harbor a genetically diverse group of astroviruses that are typically polyphyletic and predominantly situated in a basal position of the phylogenetic tree. Interestingly, we detected diverse lineages of bat astroviruses in our samples that were collected from a small geographic area. We demonstrated that the astroviruses from Rhinolophus lepidus and Eonycteris spelaea in Singapore were phylogenetically distinct and clearly segregated into two distantly-related lineages. This is reflected by the low percentage (55.3%–56.9%) of nucleotide sequence identity between Rhinolophus and Eonycteris RdRp sequences. We also observed that the E. spelaea astroviruses from Singapore and Laos exhibited high levels of sequence variation and at least two clades were identified. Clade 1 is genetically more diverse than Clade 2, and further split into smaller sub-groups. This includes all E. spelaea astroviruses collected from Singapore, although these are not monophyletic. The majority of E. spelaea astroviruses detected in Singapore are more closely related with Rousettus astroviruses from Laos. As Rousettus and Eonycteris are commonly encountered in the same roosting environment, and there is evidence that Rousettus bat coronaviruses can infect other genera of bats [49], it is unsurprising that closely related astroviruses were detected in these species. In addition, we observed that some astroviruses of E. spelaea in Singapore are closely related with Pteropus giganteus astroviruses detected in Bangladesh. The close genetic relationship of these viruses from Bangladesh, Laos and Singapore indicates an important role for bats in the dispersal of astroviruses over a wide geographical range, consistent with previous observations for lineage D betacoronaviruses [30].
Clade 2 is composed of astroviruses from several bat species from Cambodia and Laos that are most closely related to astroviruses from other mammalian species. Notably, E. spelaea astroviruses from Singapore (Clade 1) are only distantly related to E. spelaea astroviruses from Laos (Clade 2), indicating these astroviruses have arisen from different origins. Specifically, the Clade 2 bat astroviruses are most closely related to viruses from other mammals, indicating that the may have arisen by interspecies transmission from a non-bat host. This is in contrast to previously observed relationships of coronaviruses and astroviruses, where bats generally harbor virus lineages that are ancestral to viruses found in other mammalian hosts [50], [51], [52]. However, this could also reflect sampling biases. Furthermore, some terrestrial mammalian astroviruses in Clade 2 were monophyletic (e.g. dogs and rats) that may reflect host restrictions, while others (e.g. those from humans and non-human primates and swine) were not monophyletic, indicating that the interspecies transmission among these hosts is less restricted. More sequence data from different host species are necessary to better understand the underlying factors influencing the cross-species transmission and persistence of the virus.
The epidemiological models indicate that bat age is the most predictive characteristic beyond that of species, displaying an overall viral burden bias towards juveniles. The overall effect was largely driven by the larger E. spelaea sample size, especially the larger juvenile cohort, however, the proportion of juveniles positive for astrovirus in both species tested were not significantly different. Astroviruses predominantly infect juveniles [53], [54], [55], [56], [57] and the later an animal in life is infected, the shorter the period of detection, possibly reflecting a reduced period of viral shedding [58]. Our results did not yield a significant effect of bat gender on astrovirus infection, and while bat body condition and roost location were significant in the crude analyses, these two variables were not included in our main model due to extreme collinearity with other predictors. This was mirrored in a study on insectivorous bats in Borneo, where poor body condition was slightly significant for astrovirus detection while gender was not [35].
Bats have several traits that make them ideal reservoirs, including being long lived, the ability to fly, and living in large roosts [59]. Contact rates drive infectious disease transmission and larger colony sizes may play a role in transmission [60], [61] through periodic introductions of immunologically naïve individuals [46]. Differences in roosting behaviors between the three pteropodid bats in Singapore may be a reason why astroviruses were only detected in E. spelaea. This species roosts in large colonies and the study site has over 3000 individuals, while Cynopterus bat species roost in small colonies, one study observing that 87.5% of colonies contained fewer than 15 individuals [62]. The Penthetor lucasi colony in Singapore roost in a single-species, artificial cave and unless the founding individuals introduced astroviruses, the colony may be uninfected.
The developed model provides a predictive framework for understanding the burden of astrovirus in Singapore bats. Periodic sampling of tagged bats may help to determine if reproductive status plays a role in female bats' infection status, and further, if there are periods within the life cycle that coincide with higher pulses of virus [63]. Dichotomizing female sex by reproduction status not only in the study design phase, but also in the study analysis phase, may shed light on the effects of pregnancy and lactation, which may be currently hidden within the female variable. For example, Heideman and Utzurrum [64] showed that there are two distinct periods of births in E. spelaea – February to mid-May and from July to mid-October. Sampling of adults within these two periods as well as outside would be useful for the detection patterns in astrovirus infection. Serological testing would have revealed if older bats had been exposed to astrovirus at a younger age. Performing a longitudinal study with both PCR testing and serological surveys would help to characterize the distribution of astrovirus infection as well as determine temporal patterns of immunity in the bats.
The following are the supplementary data related to this article.
Supplementary Fig. 1.
Maximum likelihood phylogeny of the RNA-dependent RNA polymerase (RdRp) region of global astrovirues. Red branches denote novel bat astrovirus generated from this study, whereas the gray branches represent bat astrovirus collected from other geographical regions. The coloured branches denote different host species. Bootstrap values greater than 50% are indicated at major nodes. The scale bar represents the nucleotide substitutions per site. The insert on the right shows the details of astrovirus strains in Clade 1 and 2.
Astrovirus frequentist and Bayesian pooled prevalence estimate parameters and results
Acknowledgments
Acknowledgements
This study was supported by the Duke-NUS Signature Research Program funded by the Ministry of Health, Singapore, the National Medical Research Council (NMRC/BNIG/2005/2013) and the NUS-Global Asia Institute grant NIHA-2011-1-005. BPYH Lee was supported by a National Parks Board Postgraduate Scholarship and the Wildlife Reserves Singapore Conservation Fund. We would like to thank Dr. Vijaykrishna Dhanasekaran for his assistance with preliminary phylogenetic analysis.
Contributor Information
Ian H. Mendenhall, Email: ian.mendenhall@duke-nus.edu.sg.
Gavin J.D. Smith, Email: gavin.smith@duke-nus.edu.sg.
References
- 1.IUCN . International Union for Conservation of Nature; 2016. Status Category Summary by Major Taxonomic Groups (Animals) [Google Scholar]
- 2.Nunn C.L., Altizer S., Sechrest W., Jones K.E., Barton R.A., Gittleman J.L. Parasites and the evolutionary diversification of primate clades. Am. Nat. 2004;164:S90–S103. doi: 10.1086/424608. [DOI] [PubMed] [Google Scholar]
- 3.Morse S.S., Mazet J.A., Woolhouse M., Parrish C.R., Carroll D., Karesh W.B., Zambrana-Torrelio C., Lipkin W.I., Daszak P. Prediction and prevention of the next pandemic zoonosis. Lancet. 2012;380:1956–1965. doi: 10.1016/S0140-6736(12)61684-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabaa M.A., Tue N.T., Phuc T.M., Carrique-Mas J., Saylors K., Cotten M., Bryant J.E., Nghia H.D., Cuong N.V., Pham H.A., Berto A., Phat V.V., Dung T.T., Bao L.H., Hoa N.T., Wertheim H., Nadjm B., Monagin C., van Doorn H.R., Rahman M., Tra M.P., Campbell J.I., Boni M.F., Tam P.T., van der Hoek L., Simmonds P., Rambaut A., Toan T.K., Van Vinh Chau N., Hien T.T., Wolfe N., Farrar J.J., Thwaites G., Kellam P., Woolhouse M.E., Baker S. The Vietnam initiative on zoonotic infections (VIZIONS): a strategic approach to studying emerging zoonotic infectious diseases. EcoHealth. 2015;12:726–735. doi: 10.1007/s10393-015-1061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Longdon B., Hadfield J.D., Webster C.L., Obbard D.J., Jiggins F.M. Host phylogeny determines viral persistence and replication in novel hosts. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Webby R., Hoffmann E., Webster R. Molecular constraints to interspecies transmission of viral pathogens. Nat. Med. 2004;10:S77–81. doi: 10.1038/nm1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parrish C.R., Holmes E.C., Morens D.M., Park E.C., Burke D.S., Calisher C.H., Laughlin C.A., Saif L.J., Daszak P. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 2008;72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Méndez E., Murillo A., Velázquez R., Burnham A., Arias C.F. Astrovirus Research. Springer; 2013. Replication cycle of astroviruses; pp. 19–45. [Google Scholar]
- 9.Bidin M., Bidin Z., Majnaric D., Tisljar M., Lojkic I. Circulation and phylogenetic relationship of chicken and turkey-origin astroviruses detected in domestic ducks (Anas platyrhynchos domesticus) Avian. Pathol. 2012;41:555–562. doi: 10.1080/03079457.2012.733340. [DOI] [PubMed] [Google Scholar]
- 10.Pantin-Jackwood M.J., Spackman E., Woolcock P.R. Phylogenetic analysis of Turkey astroviruses reveals evidence of recombination. Virus Genes. 2006;32:187–192. doi: 10.1007/s11262-005-6875-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cattoli G., Chu D.K.W., Peiris M. Springer; 2013. Astrovirus Infections in Animal Mammalian Species, Astrovirus Research; pp. 135–149. [Google Scholar]
- 12.Benedictis P., Schultz-Cherry S., Burnham A., Cattoli G. Astrovirus infections in humans and animals - molecular biology, genetic diversity, and interspecies transmissions. Infect. Genet. Evol. 2011;11 doi: 10.1016/j.meegid.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koci M.D., Schultz-Cherry S. Avian astroviruses. Avian Pathol. 2002;31:213–227. doi: 10.1080/03079450220136521. [DOI] [PubMed] [Google Scholar]
- 14.Clark B., McKendrick M. A review of viral gastroenteritis. Curr. Opin. Infect. Dis. 2004;17:461–469. doi: 10.1097/00001432-200410000-00011. [DOI] [PubMed] [Google Scholar]
- 15.Li L., Diab S., McGraw S., Barr B., Traslavina R., Higgins R., Talbot T., Blanchard P., Rimoldi G., Fahsbender E., Page B., Phan T.G., Wang C., Deng X., Pesavento P., Delwart E. Divergent astrovirus associated with neurologic disease in cattle. Emerg. Infect. Dis. 2013;19:1385–1392. doi: 10.3201/eid1909.130682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlottau K., Schulze C., Bilk S., Hanke D., Hoper D., Beer M., Hoffmann B. Detection of a novel bovine astrovirus in a cow with encephalitis. Transbound. Emerg. Dis. 2016;63:253–259. doi: 10.1111/tbed.12493. [DOI] [PubMed] [Google Scholar]
- 17.Sun N., Yang Y., Wang G.S., Shao X.Q., Zhang S.Q., Wang F.X., Tan B., Tian F.L., Cheng S.P., Wen Y.J. Detection and characterization of avastrovirus associated with diarrhea isolated from minks in China. Food Environ. Virol. 2014;6:169–174. doi: 10.1007/s12560-014-9155-3. [DOI] [PubMed] [Google Scholar]
- 18.Da Silva S.E., Bonetti A.M., Petrocelli A.T., Ferrari H.F., Luvizotto M.C., Cardoso T.C. Detection of Turkey astrovirus in young poults affected with poult enteritis complex in Brazil. J. Vet. Med. Sci. 2008;70:629–631. doi: 10.1292/jvms.70.629. [DOI] [PubMed] [Google Scholar]
- 19.Cattoli G., Toffan A., De Battisti C., Salviato A., Terregino C., Capua I. Astroviruses found in the intestinal contents of guinea fowl suffering from enteritis. Vet. Rec. 2005;156:220. [PubMed] [Google Scholar]
- 20.Mor S.K., Chander Y., Marthaler D., Patnayak D.P., Goyal S.M. Detection and molecular characterization of porcine astrovirus strains associated with swine diarrhea. J. Vet. Diagn. Investig. 2012;24:1064–1067. doi: 10.1177/1040638712458781. [DOI] [PubMed] [Google Scholar]
- 21.Bidin M., Lojkic I., Tisljar M., Bidin Z., Majnaric D. Astroviruses associated with stunting and pre-hatching mortality in duck and goose embryos. Avian Pathol. 2012;41:91–97. doi: 10.1080/03079457.2011.642796. [DOI] [PubMed] [Google Scholar]
- 22.Bulbule N.R., Mandakhalikar K.D., Kapgate S.S., Deshmukh V.V., Schat K.A., Chawak M.M. Role of chicken astrovirus as a causative agent of gout in commercial broilers in India. Avian Pathol. 2013;42:464–473. doi: 10.1080/03079457.2013.828194. [DOI] [PubMed] [Google Scholar]
- 23.Candido M., Alencar A.L., Almeida-Queiroz S.R., Buzinaro Mda G., Munin F.S., de Godoy S.H., Livonesi M.C., Fernandes A.M., de Sousa R.L. Molecular detection and phylogenetic analysis of bovine astrovirus in Brazil. Arch. Virol. 2015;160:1519–1525. doi: 10.1007/s00705-015-2400-8. [DOI] [PubMed] [Google Scholar]
- 24.Phan T.G., Vo N.P., Boros A., Pankovics P., Reuter G., Li O.T., Wang C., Deng X., Poon L.L., Delwart E. The viruses of wild pigeon droppings. PLoS One. 2013;8 doi: 10.1371/journal.pone.0072787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu D.K., Leung C.Y., Perera H.K., Ng E.M., Gilbert M., Joyner P.H., Grioni A., Ades G., Guan Y., Peiris J.S., Poon L.L. A novel group of avian astroviruses in wild aquatic birds. J. Virol. 2012;86:13772–13778. doi: 10.1128/JVI.02105-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grimaldi W.W., Hall R.J., White D.D., Wang J., Massaro M., Tompkins D.M. First report of a feather loss condition in Adelie penguins (Pygoscelis adeliae) on Ross Island, Antarctica, and a preliminary investigation of its cause. Emu. 2015;115:185–189. [Google Scholar]
- 27.Mendenhall I.H., Yaung K.N., Joyner P.H., Keatts L., Borthwick S., Neves E.S., San S., Gilbert M., Smith G.J. Detection of a novel astrovirus from a black-naped monarch (Hypothymis azurea) in Cambodia. Virol. J. 2015;12:182. doi: 10.1186/s12985-015-0413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou P., Tachedjian M., Wynne J.W., Boyd V., Cui J., Smith I., Cowled C., Ng J.H., Mok L., Michalski W.P., Mendenhall I.H., Tachedjian G., Wang L.F., Baker M.L. Contraction of the type I IFN locus and unusual constitutive expression of IFN-alpha in bats. PNAS. 2016;113:2696–2701. doi: 10.1073/pnas.1518240113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sodhi N.S., Koh L.P., Brook B.W., Ng P.K.L. Southeast Asian biodiversity: an impending disaster. Trends Ecol. Evol. 2004;19:654–660. doi: 10.1016/j.tree.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Mendenhall I.H., Borthwick S., Neves E.S., Low D., Linster M., Liang B., Skiles M., Jayakumar J., Han H., Gunalan V., Lee B.P., Okahara K., Wang L.F., Maurer-Stroh S., Su Y.C., Smith G.J. Identification of a lineage D Betacoronavirus in cave nectar bats (Eonycteris Spelaea) in Singapore and an overview of lineage D reservoir ecology in SE Asian bats. Transbound. Emerg. Dis. 2016 doi: 10.1111/tbed.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendenhall I.H., Low D., Neves E.S., Anwar A., Oh S., Su Y.C., Smith G.J. One Health; 2016. Evidence of Canine Parvovirus Transmission to a Civet Cat (Paradoxurus musangus) in Singapore. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu D.K., Poon L.L., Guan Y., Peiris J.S. Novel astroviruses in insectivorous bats. J. Virol. 2008;82:9107–9114. doi: 10.1128/JVI.00857-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Meintjes P., Drummond A. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stamatakis A., Ludwig T., Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- 35.Sergeant E.S.G. Ausvet Pty Ltd; 2017. Epitools Epidemiological Calculators. [Google Scholar]
- 36.Green A.J. Mass/length residuals: measures of body condition or generators of spurious results? Ecology. 2001;82:1473–1483. [Google Scholar]
- 37.Hamilton L.C. Cengage Learning; 2012. Statistics With Stata: version 12. [Google Scholar]
- 38.Fischer K., Pinho Dos Reis V., Balkema-Buschmann A. Bat astroviruses: towards understanding the transmission dynamics of a neglected virus family. Viruses. 2017;9 doi: 10.3390/v9020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu B., Chmura A.A., Li J., Zhu G., Desmond J.S., Zhang Y., Zhang W., Epstein J.H., Daszak P., Shi Z. Detection of diverse novel astroviruses from small mammals in China. J. Gen. Virol. 2014;95:2442–2449. doi: 10.1099/vir.0.067686-0. [DOI] [PubMed] [Google Scholar]
- 40.Wu Z., Ren X., Yang L., Hu Y., Yang J., He G., Zhang J., Dong J., Sun L., Du J., Liu L., Xue Y., Wang J., Yang F., Zhang S., Jin Q. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J. Virol. 2012;86:10999–11012. doi: 10.1128/JVI.01394-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu H.C., Chu D.K., Liu W., Dong B.Q., Zhang S.Y., Zhang J.X., Li L.F., Vijaykrishna D., Smith G.J., Chen H.L., Poon L.L., Peiris J.S., Guan Y. Detection of diverse astroviruses from bats in China. J. Gen. Virol. 2009;90:883–887. doi: 10.1099/vir.0.007732-0. [DOI] [PubMed] [Google Scholar]
- 42.Dufkova L., Strakova P., Sirmarova J., Salat J., Moutelikova R., Chrudimsky T., Bartonicka T., Nowotny N., Ruzek D. Detection of diverse novel bat Astrovirus sequences in the Czech Republic. Vector Borne Zoonot. Dis. 2015;15:518–521. doi: 10.1089/vbz.2015.1813. [DOI] [PubMed] [Google Scholar]
- 43.Seltmann A., Corman V.M., Rasche A., Drosten C., Czirjak G.A., Bernard H., Struebig M.J., Voigt C.C. Seasonal fluctuations of astrovirus, but not coronavirus shedding in bats inhabiting human-modified tropical forests. EcoHealth. 2017;14:272–284. doi: 10.1007/s10393-017-1245-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lacroix A., Duong V., Hul V., San S., Davun H., Omaliss K., Chea S., Hassanin A., Theppangna W., Silithammavong S., Khammavong K., Singhalath S., Afelt A., Greatorex Z., Fine A.E., Goldstein T., Olson S., Joly D.O., Keatts L., Dussart P., Frutos R., Buchy P. Diversity of bat astroviruses in Lao PDR and Cambodia. Infect. Genet. Evol. 2017;47:41–50. doi: 10.1016/j.meegid.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drexler J.F., Corman V.M., Wegner T., Tateno A.F., Zerbinati R.M., Gloza-Rausch F., Seebens A., Muller M.A., Drosten C. Amplification of emerging viruses in a bat colony. Emerg. Infect. Dis. 2011;17:449–456. doi: 10.3201/eid1703.100526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayman D.T. Biannual birth pulses allow filoviruses to persist in bat populations. Proc. R. Soc. Lond. Biol. 2015;282:2014–2591. doi: 10.1098/rspb.2014.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karlsson E.A., Small C.T., Freiden P., Feeroz M.M., Matsen F.A.t., San S., Hasan M.K., Wang D., Jones-Engel L., Schultz-Cherry S. Non-human primates harbor diverse mammalian and avian astroviruses including those associated with human infections. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fischer K., Zeus V., Kwasnitschka L., Kerth G., Haase M., Groschup M.H., Balkema-Buschmann A. Insectivorous bats carry host specific astroviruses and coronaviruses across different regions in Germany. Infect. Genet. Evol. 2016;37:108–116. doi: 10.1016/j.meegid.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lau S.K., Li K.S., Tsang A.K., Shek C.T., Wang M., Choi G.K., Guo R., Wong B.H., Poon R.W., Lam C.S., Wang S.Y., Fan R.Y., Chan K.H., Zheng B.J., Woo P.C., Yuen K.Y. Recent transmission of a novel alphacoronavirus, bat coronavirus HKU10, from Leschenault's rousettes to pomona leaf-nosed bats: first evidence of interspecies transmission of coronavirus between bats of different suborders. J. Virol. 2012;86:11906–11918. doi: 10.1128/JVI.01305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mendenhall I.H., Smith G.J., Dhanasekaran V. Ecological drivers of virus evolution: astrovirus as a case study. J. Virol. 2015;(JVI):02971–02974. doi: 10.1128/JVI.02971-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vijaykrishna D., Smith G.J., Zhang J.X., Peiris J.S., Chen H., Guan Y. Evolutionary insights into the ecology of coronaviruses. J. Virol. 2007:4012–4020. doi: 10.1128/JVI.02605-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu B., Ge X., Wang L.F., Shi Z. Bat origin of human coronaviruses. Virol. J. 2015;12:221. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guix S., Caballero S., Villena C., Bartolome R., Latorre C., Rabella N., Simo M., Bosch A., Pinto R.M. Molecular epidemiology of astrovirus infection in Barcelona, Spain. J. Clin. Microbiol. 2002;40:133–139. doi: 10.1128/JCM.40.1.133-139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kofstad T., Jonassen C.M. Screening of feral and wood pigeons for viruses harboring a conserved mobile viral element: characterization of novel Astroviruses and Picornaviruses. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larsen P.F., Møller S., Clausen T., Hammer A. Wageningen Academic Pub; 2012. Proceedings of the Xth International Scientific Congress in Fur Animal Production. [Google Scholar]
- 56.Grellet A., De Battisti C., Feugier A., Pantile M., Marciano S., Grandjean D., Cattoli G. Prevalence and risk factors of astrovirus infection in puppies from French breeding kennels. Vet. Microbiol. 2012;157:214–219. doi: 10.1016/j.vetmic.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 57.Fu Y., Pan M., Wang X., Xu Y., Xie X., Knowles N.J., Yang H., Zhang D. Complete sequence of a duck astrovirus associated with fatal hepatitis in ducklings. J. Gen. Virol. 2009;90:1104–1108. doi: 10.1099/vir.0.008599-0. [DOI] [PubMed] [Google Scholar]
- 58.Awe O.O., Kang K.I., Ibrahim M., Ali A., Elaish M., Saif Y.M., Lee C.W. Age-related susceptibility of turkeys to enteric viruses. Avian Dis. 2015;59:207–212. doi: 10.1637/10907-071514-Reg. [DOI] [PubMed] [Google Scholar]
- 59.Calisher C.H., Childs J.E., Field H.E., Holmes K.V., Schountz T. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006;19:531–545. doi: 10.1128/CMR.00017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farrington C.P., Whitaker H.J. Estimation of effective reproduction numbers for infectious diseases using serological survey data. Biostatistics. 2003;4:621–632. doi: 10.1093/biostatistics/4.4.621. [DOI] [PubMed] [Google Scholar]
- 61.Nunn C.L., Jordan F., McCabe C.M., Verdolin J.L., Fewell J.H. Infectious disease and group size: more than just a numbers game. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2015;370 doi: 10.1098/rstb.2014.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan K., Akbar Z., Kunz T. Roost selection and social organisation in Cynopterus horsfieldi (Chiroptera: Pteropodidae) Malay. Nat. J. 1999;53:295–298. [Google Scholar]
- 63.Hayman D.T., Bowen R.A., Cryan P.M., McCracken G.F., O'Shea T.J., Peel A.J., Gilbert A., Webb C.T., Wood J.L. Ecology of zoonotic infectious diseases in bats: current knowledge and future directions. Zoonoses Public Health. 2013;60:2–21. doi: 10.1111/zph.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heideman P.D., Utzurrum R.C. Seasonality and synchrony of reproduction in three species of nectarivorous Philippines bats. BMC Ecol. 2003;3:1–14. doi: 10.1186/1472-6785-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Astrovirus frequentist and Bayesian pooled prevalence estimate parameters and results