Abstract
Aspergillus section Restricti together with sister section Aspergillus (formerly Eurotium) comprises xerophilic species, that are able to grow on substrates with low water activity and in extreme environments. We adressed the monophyly of both sections within subgenus Aspergillus and applied a multidisciplinary approach for definition of species boundaries in sect. Restricti. The monophyly of sections Aspergillus and Restricti was tested on a set of 102 isolates comprising all currently accepted species and was strongly supported by Maximum likelihood (ML) and Bayesian inferrence (BI) analysis based on β-tubulin (benA), calmodulin (CaM) and RNA polymerase II second largest subunit (RPB2) loci. More than 300 strains belonging to sect. Restricti from various isolation sources and four continents were characterized by DNA sequencing, and 193 isolates were selected for phylogenetic analyses and phenotypic studies. Species delimitation methods based on multispecies coalescent model were employed on DNA sequences from four loci, i.e., ID region of rDNA (ITS + 28S), CaM, benA and RPB2, and supported recognition of 21 species, including 14 new. All these species were also strongly supported in ML and BI analyses. All recognised species can be reliably identified by all four examined genetic loci. Phenotype analysis was performed to support the delimitation of new species and includes colony characteristics on seven cultivation media incubated at several temperatures, growth on an osmotic gradient (six media with NaCl concentration from 0 to 25 %) and analysis of morphology including scanning electron microscopy. The micromorphology of conidial heads, vesicle dimensions, temperature profiles and growth parameters in osmotic gradient were useful criteria for species identification.
The vast majority of species in sect. Restricti produce asperglaucide, asperphenamate or both in contrast to species in sect. Aspergillus. Mycophenolic acid was detected for the first time in at least six members of the section. The ascomata of A. halophilicus do not contain auroglaucin, epiheveadride or flavoglaucin which are common in sect. Aspergillus, but shares the echinulins with sect. Aspergillus.
Key words: Aspergillus restrictus, Aspergillus penicillioides, Eurotium, food spoilage, indoor fungi, linear discriminant analysis, multigene phylogeny, multispecies coalescent model, sick building syndrome, xerophilic fungi
Taxonomic novelties: Aspergillus canadensis Visagie, Yilmaz, F. Sklenar & Seifert; Aspergillus clavatophorus F. Sklenar, S.W. Peterson & Hubka; Aspergillus destruens Zalar, F. Sklenar, S.W. Peterson & Hubka; Aspergillus domesticus F. Sklenar, Houbraken, Zalar & Hubka; Aspergillus glabripes F. Sklenar, Ž. Jurjević & Hubka; Aspergillus hordei F. Sklenar, S.W. Peterson & Hubka; Aspergillus infrequens F. Sklenar, S.W. Peterson & Hubka; Aspergillus magnivesiculatus F. Sklenar, Zalar, Ž. Jurjević & Hubka; Aspergillus pachycaulis F. Sklenar, S.W. Peterson, Ž. Jurjević & Hubka; Aspergillus pseudogracilis F. Sklenar, Ž. Jurjević & Hubka; Aspergillus reticulatus F. Sklenar, Ž. Jurjević, S.W. Peterson & Hubka; Aspergillus salinicola Zalar, F. Sklenar, Visagie & Hubka; Aspergillus tardicrescens F. Sklenar, Houbraken, Zalar, & Hubka; Aspergillus villosus F. Sklenar, S.W. Peterson & Hubka
Introduction
Aspergillus section Restricti species occurs in environments with low water activity (aw). They are commonly found on building materials, in house dust (Kaarakainen et al., 2009, Visagie et al., 2014) and dried, salted or high sugar content foods (Pitt and Hocking, 2009, Frasz and Miller, 2015). Much attention is being paid to the indoor air quality (Kasuga, 2012, Flannigan et al., 2016) and these fungi are repeatedly reported to be present in this environment (Samson et al., 2002, Meklin et al., 2004, Meklin et al., 2007), where they are considered a potential agent responsible for sick building syndrome, respiratory problems and allergies (Terr, 2009, Saijo et al., 2011, Abe, 2012). Species may cause post-harvest rot in improperly dried commodities such as maize or wheat (Christensen & Kaufmann 1965), cotton goods are susceptible to A. restrictus rot (Smith 1931), while A. vitricola can even damage optical instruments (Ohtsuki 1962). Recently, fatal disseminated aspergillosis in an infant was proved to be caused by A. penicillioides (Gupta et al. 2016) and A. conicus was detected as the causal agent of an intraocular infection (Smith et al. 2013). While there are reports in the literature with infection cases from A. restrictus-like fungi, the patient nearly always has a documented underlying disease state, suggesting opportunistic infections (de Hoog et al. 2009).
Secondary metabolite production in these species has not been studied extensively. According to Micheluz et al. (2016), the homothallic species A. halophilicus produces many metabolites (chaetoviridin A, deoxybrevianamid E, pseurotin A, pseurotin D, rugulusovin, stachybotryamide and tryprostatin B) compared to anamorphic species producing a much narrower spectrum of substances with only asperglaucide detected in A. penicillioides and some unspecified metabolites in A. vitricola (Micheluz et al. 2016). There are no known mycotoxins produced by members of sect. Restricti and they do not pose a direct threat to consumers, but they cause significant losses for food and agricultural industries (Deschuyffeleer et al. 2015). More worrying is their potential for creating more favourable conditions for less xerophilic fungal species that may produce hazardous mycotoxins. They do this by producing metabolic water, thereby increasing water activity of the substrate. From a biotechnological perspective, polyextremophilic α-amylase produced by A. penicillioides has significant potential for use as a detergent (Ali et al. 2015).
The first species described from this section was A. penicillioides observed from Argentinian sugar cane (Spegazzini 1896). Aspergillus caesiellus was subsequently described from air in Tokyo (Saito 1904), A. gracilis from a Monilinia fructigena fruiting body and at the time thought to resemble A. fumigatus (Bainier 1907). Blochwitz described A. conicus from chalky soil (Dale 1914); Smith (1931) described A. restrictus causing degradation of cotton in the manufacturing proces; and finally, A. vitricola was described from binocular lens (Ohtsuki 1962). The taxonomy of Aspergillus was advanced greatly by the designation of type specimens and ex-type cultures by Samson & Gams (1985). Types for all aforementioned species in sect. Restricti were summarised or newly designated with the exception of A. vitricola (was not accepted by the authors). Pitt & Samson (1990) reduced the section to three species on the basis of physiology and morphology. Peterson (2000) used a phylogenetic analysis of LSU rDNA (28S) sequences to provide genetic evidence that there are seven species in the section, including A. halophilicus, which was described as Eurotium halophilicum from corn seeds by Christensen et al. (1959). Peterson (2008) used multilocus DNA sequence data, phylogenetic and concordance analysis to produce a statistically supported analysis of sect. Restricti containing seven species.
The morphology of sect. Restricti species is very simple and the number of taxonomically relevant morphological characters is low. Correct identification based solely on morphology is therefore challenging if not impossible and sequence comparisons represent the best method for fast and robust identifications. Phylogenetic analysis based on multiple loci has become an indispensable part of taxonomic studies. A polyphasic approach to species delimitation is currently standard in Aspergillus (Samson and Varga, 2009, Samson et al., 2014) with the phylogenetic component usually relying on the genealogical concordance phylogenetic species recognition (GCPSR) approach proposed by Taylor et al. (2000).
New and advanced multi-locus methods for species delimitation were introduced recently (Fujita et al., 2012, Tang et al., 2014, Fontaneto et al., 2015). The majority of new approaches and associated software is based on coalescent theory and multispecies coalescent model (Flot 2015). Simultaneously because of the possible incongruence between gene trees, the focus is also shifting from gene tree to species tree inference using methods, that take into account incomplete lineage sorting (ILS), the most common cause of locus incongruence (Edwards 2009). We followed the suggestion of Carstens et al. (2013) that unites the two mentioned tasks into one analysis, i.e., species delimitation and species tree estimation. Firstly, species are delimited from a set of individuals by several species delimitation methods, possibly based on different approaches (based on trees, genetic distances, haplowebs, etc.). Some methods or some loci may be more prone to over delimitation (i.e., the method splits the dataset into more potential species compared with other methods or loci) of species than others, so it is recommended to compare more methods with as much loci as possible (Carstens et al. 2013). The species tree is subsequently inferred with individuals assigned into putative species based on the results of species delimitation. These methods are currently being applied to many different groups of organisms (Flot 2015) but infrequently in fungi. In this study, we applied them for the first time to delimit species boundaries in Aspergillus.
During identifications of isolates of sect. Restricti from various substrates and locations, we encountered many isolates that could not credibly be placed in the seven species accepted by Peterson (2008). In order to substantiate the initial finding we assembled a set of 193 isolates from sect. Restricti including type isolates, conducted DNA sequencing of four genetic loci, coalescence analysis, physiological testing (temperature and aw gradients), micro- and macromorphology and SEM (scanning electron microscopy) in order to describe the biodiversity found in this section. Additionally, the monophyly of section Restricti within subgenus Aspergillus was addressed.
Materials and methods
Fungal strains
Strains used in this study were obtained from and deposited into various culture collections: 1) ATCC, American Type culture collection, Manassas, Virginia, USA (https://www.lgcstandards-atcc.org/en/Products/ATCC_Genuine_Cultures.aspx); 2) BCCM/IHEM, Biomedical Fungi and Yeasts Collection, Scientific Institute of Public Health, Brussels, Belgium (http://bccm.belspo.be/about-us/bccm-ihem); 3) CCF, Culture Collection of Fungi, Charles University, Prague, Czech Republic (https://www.natur.cuni.cz/biology/botany/structure/culture-collection-of-fungi-ccf); 4) CBS, culture collection of the Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands (http://www.westerdijkinstitute.nl/Collections); 5) DAOMC, Canadian Collection of Fungal Cultures, Agriculture and Agri-Food Canada, Ottawa, Canada; 6) DTO, working collection of the Applied and Industrial Mycology department housed at the Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands; 7) EMSL, EMSL Analytical Inc., New Jersey, USA (http://www.emsl.com); 8) EXF, Culture Collection of Extremophilic Fungi, University of Ljubljana, Slovenia (http://www.ex-genebank.com/index.php/en/fungi-2); 9) IBT, Culture Collection at Department of Biotechnology and beimedicine, Lyngby, Denmark; 10) IMI/CABI, International Mycological Institute, Kew, England (http://www.cabi.org/services/microbial-services); 11) KAS, fungal collection of Keith A. Seifert, Ottawa, Canada; 12) NRRL, Agricultural Research Service Culture Collection, Peoria, Illinois, USA (https://nrrl.ncaur.usda.gov); 13) UBOCC, Université de Bretagne Occidentale Culture Collection, Brest, France (https://www.univ-brest.fr/plateformes-technologiques/menu/nos-plates-formes/UBOCC); 14) MUT, Mycotheca dell'Università degli Studi di Torino, Turin, Italy, http://www.mut.unito.it/en/Collezione). Dried holotype and isotype specimens were deposited into the herbarium of the Mycological Department, National Museum, Prague, Czech Republic (PRM) or Canadian National Mycological Herbarium, Ottawa, Canada (DAOM).
Many strains were specifically isolated for the purposes of this study from the indoor environment in the USA, Bermuda, Puerto Rico, Trinidad and Tobago (Ž. Jurjević), house dust from Canada (C.M. Visagie), bakery products and deteriorated paintings in France (M. Coton, F. Déniel), deteriorated paintings from Slovenia (P. Zalar, D.D. Graf) and hypersaline water from Slovenia (P. Zalar, N. Gunde-Cimerman) and Israel (R. Tkavc).
Different isolation techniques were used for species isolation. Samples from indoor environments across the USA were collected using the following techniques: air samples were collected as detailed previously (Peterson & Jurjević 2013) using malt extract agar (MEA) as isolation medium. Dilution plates were used to isolate fungi that were taken by swabs as described previously (Jurjević et al. 2015), using MEA supplemented with chloramphenicol and dichloran-glycerol (DG18) agar as isolation medium. Sedimentation plate samples were taken for a one-hour exposure time using Potato Dextrose agar (PDA) as isolation medium. Isolations from Canadian dust were made using a modified dilution-to-extinction method (Collado et al. 2007) as described in (Visagie et al., 2014, Visagie et al., 2017) using DG18, Malt extract Yeast extract 10 % glucose 12 % NaCl (MY10-12) and Malt extract Yeast extract 50 % glucose agar (MY50G) as isolation media.
The samples from paintings in Slovenia were collected as follows. The sampling was performed in 2014 on old (from 300 to 400 years) oil canvas paintings originating from various Slovene churches and at the time of sampling stored in the Restoration Centre for the Protection of Cultural Heritage of Slovenia (ZVKDS). The sampled paintings were partly visibly overgrown with fungi, on the front painted side or on the back of the canvas. Samples were collected by rubbing overgrown areas with sterile cotton swabs soaked in physiological solution [0.9 % (w/v) NaCl]. Inoculum was subsequently spread onto plates containing DG18, MY10-12 or MY50G, all media amended with chloramphenicol (50 mg/l). The plates were incubated at 25 °C for up to 21 d. Pure cultures of the fungi were obtained from the primary isolation plates by further subculturing. Direct isolations were made from fungal growth on deteriorated paintings and bakery products in France using salt malt medium [5 % malt extract, 5 % NaCl, 1.5 % agar, (w/v)] as isolation media. Dilution series were sometimes also used. The paintings were visibly damaged and had been stored in the painting storage area at the Musée des beaux Arts in Brest, France. Plates were incubated at 25 °C for up to 21 d.
Several strains were isolated from different salterns in Slovenia and Israel using filtration of hypersaline water through membrane filters (pore diam 0.45 μm), followed by incubation of the filters on different cultivation media with lowered water activity, as reported by (Gunde-Cimerman et al. 2000).
Molecular studies
All isolates included in this study were identified using sequence data, but amplification of four genetic loci (see below) was performed only in 193 isolates selected for phylogenetic analyses (Table 1).
Table 1.
Provenance and GenBank accession numbers for Aspergillus sect. Restricti species used for phylogenetic analysis and comparative phenotypic studies.
| Species | Strain no.1,2 | Source | GenBank accession nos. |
|||
|---|---|---|---|---|---|---|
| ITS | benA | CaM | RPB2 | |||
| A. caesiellus | NRRL 5061T = CBS 470.65 = DTO 093-H3 = ATCC 11905 = IMI 172278 = CCF 5447 = IBT 34620 | Unknown, Japan | EF652044 | EF651884 | EF652030 | EF651981 |
| NRRL 25810 = CCF 5662 | Cloth, Panama | KY087751 | KY117814 | KY068301 | KY117992 | |
| CCF 5450 = EMSL No. 1614 | Air, outside, Delaware, USA | KY087598 | KY117667 | KY068151 | KY117844 | |
| CCF 5448 = EMSL No. 1383 = IBT 34621 | Air, home, Pennsylvania, USA | KY117665 | KY068149 | KY117842 | ||
| DTO 026-C7 | Indoor environment, Germany | KY087684 | KY117748 | KY068232 | KY117925 | |
| DTO 025-I4 = IBT 34538 | Indoor environment, Germany | KY087683 | KY117747 | KY068231 | KY117924 | |
| CCF 5451 = EMSL No. 1650 = IBT 34622 | Air, pineapple room, warehouse, Delaware, USA | KY087599 | KY117668 | KY068153 | KY117845 | |
| CCF 5449 = EMSL No. 1499 | Air, home, Delaware, USA | KY087597 | KY117666 | KY068150 | KY117843 | |
| NRRL 25861 | Unknown, Gorakpur, India | KY087766 | KY117829 | KY068316 | KY118007 | |
| NRRL 25815 = DTO 356-D1 = CCF 5663 | Hobnail shoes, Florida, USA | KY087753 | KY117816 | KY068303 | KY117994 | |
| A. canadensis | CCF 5548T = KAS 6194 = DTO 356-H9 = IBT 34520 = IBT 34642 = NRRL 66614 | House dust, Nova Scotia, Wolfville, Canada | KY087667 | KY117731 | KY068215 | KY117909 |
| KAS 7705 = DTO 357-A8 | House dust, Ottawa, Ontario, Canada | KY087672 | KY117736 | KY068220 | KY117914 | |
| KAS 7707 = DTO 357-B1 | House dust, Ottawa, Ontario, Canada | KY087673 | KY117737 | KY068221 | KY117915 | |
| KAS 7708 = DTO 357-B2 = CCF 5550 = IBT 34637 | House dust, Ottawa, Ontario, Canada | KY087674 | KY117738 | KY068222 | KY117916 | |
| KAS 7710 = DTO 357-B4 = CCF 5552 = IBT 34636 | House dust, Ottawa, Ontario, Canada | KY087675 | KY117739 | KY068223 | KY117917 | |
| KAS 7711 = DTO 357-B5 | House dust, Ottawa, Ontario, Canada | KY087676 | KY117740 | KY068224 | KY117918 | |
| KAS 7716 = DTO 357-B8 | House dust, Ottawa, Ontario, Canada | KY087677 | KY117741 | KY068225 | KY117919 | |
| KAS 7717 = DTO 357-B9 | House dust, Ottawa, Ontario, Canada | KY087678 | KY117742 | KY068226 | KY117920 | |
| KAS 7718 = DTO 357-C1 | House dust, Ottawa, Ontario, Canada | KY087679 | KY117743 | KY068227 | KY117921 | |
| KAS 7719 = DTO 357-C2 = CCF 5553 = IBT 34638 | House dust, Ottawa, Ontario, Canada | KY087680 | KY117744 | KY068228 | KY117922 | |
| KAS 7704 = DTO 357-A7 | House dust, Ottawa, Ontario, Canada | KY087671 | KY117735 | KY068219 | KY117913 | |
| KAS 7721 = DTO 357-C4 | House dust, Ottawa, Ontario, Canada | KY087681 | KY117745 | KY068229 | KY117923 | |
| A. clavatophorus | NRRL 25874T = CCF 5454 = IBT 34560 = IBT 34823 = DTO 356-D8 | Mouldy paper, Athens, Georgia, USA | KY087772 | KY117836 | KY068323 | KY118014 |
| NRRL 25873 = CCF 5453 = IBT 34632 | Mouldy paper, Athens, Georgia, USA | KY117835 | KY068322 | KY118013 | ||
| DTO 257-G5 = IBT 34561 = CCF 5669 | Puerh tea, China | KY087703 | KY117764 | KY068251 | KY117943 | |
| A. conicus | NRRL 149T = CBS 475.65 = IBT 33667 = DTO 096-H6 = ATCC 16908 = IMI 172281 = CCF 5456 | Unknown | EF652039 | EF651881 | EF652033 | EF651975 |
| CCF 5458 = EMSL No. 1490 | Air, home, California, USA | KY087601 | KY117670 | KY068155 | KY117846 | |
| CCF 5457 = EMSL No. 1318 = NRRL 62007 | Air, home, Idaho, USA | KY087600 | KY117669 | KY068154 | ||
| NRRL 25881 | Unknown, New York, USA | KY087775 | KY117839 | KY068326 | KY118017 | |
| CCF 5459 = EMSL No. 1649 | Air, pineapple room, warehouse, Delaware, USA | KY087602 | KY117671 | KY068156 | KY117847 | |
| CCF 5461 = EMSL No. 2549 | Air, office, Bayamon, Puerto Rico | KY087604 | KY117673 | KY068157 | KY117849 | |
| CCF 5460 = EMSL No. 2217 | Air, living room, West Chester, Pennsylvania, USA | KY087603 | KY117672 | KY068152 | KY117848 | |
| EXF-7663 = IBT 34267 = IBT 33574 | Oil painting on canvas, Ljubljana, Slovenia | KY087715 | KY117778 | KY068265 | KY117956 | |
| DTO 077-H5 | Indoor air, Witten, The Netherlands | KY087687 | KY117751 | KY068235 | ||
| IHEM 16531 | Wooden statue, Braine-l'Alleud, Belgium | KY087735 | KY117798 | KY068285 | KY117976 | |
| CCF 4042 | Kernel of Bertholletia excelsa, Czech Republic | KY087659 | KY117723 | KY068207 | KY117903 | |
| EXF-7660 = IBT 34263 = IBT 33577 | Oil painting on canvas, Ljubljana, Slovenia | KY087713 | KY117776 | KY068263 | KY117954 | |
| EXF-5015 = IBT 34273 = CCF 5650 | Microbial mat, Eliat, Israel | KY117774 | KY068261 | KY117952 | ||
| DTO 231-C3 | Museum piece, Zwartewaal, The Netherlands | KY087701 | KY117763 | KY068249 | KY117941 | |
| NRRL 25848 | Asphalt roof shingle, Chicago, Illinois, USA | KY087764 | KY117827 | KY068314 | KY118005 | |
| IHEM 20709 | Candy, Belgium | KY087736 | KY117799 | KY068286 | KY117977 | |
| DTO 110-C5 | Air in bathroom, near Copenhagen, Denmark | KY087690 | KY117754 | KY068238 | KY117930 | |
| DTO 110-F5 = IBT 34534 = CCF 5667 | Air in living room, near Copenhagen, Denmark | KY087691 | KY117755 | KY068239 | KY117931 | |
| DTO 017-B9 | Indoor air, Eindhoven, The Netherlands | KY087682 | KY117746 | KY068230 | ||
| A. destruens | NRRL 145T = CBS 593.91 = DTO 079-A8 = IMI 358691 = CCF 5462 = IBT 34818 | Maize seed, Maryland, USA | KY087748 | KY117811 | KY068298 | KY117989 |
| EXF-7699 = IBT 34262 | Oil painting on canvas, Ljubljana, Slovenia | KY087717 | KY117780 | KY068267 | KY117958 | |
| EXF-7651 = IBT 34258 = CCF 5653 | Oil painting on canvas, Ljubljana, Slovenia | KY087712 | KY117775 | KY068262 | KY117953 | |
| EXF-7661 = IBT 34271 = IBT 33573 | Oil painting on canvas, Ljubljana, Slovenia | KY087714 | KY117777 | KY068264 | KY117955 | |
| DTO 254-B2 = IBT 34522 | Air in villa, Utrecht, The Netherlands | KY087702 | KY068250 | KY117942 | ||
| EXF-10411 = IBT 34265 | Oil painting on canvas, Ljubljana, Slovenia | KY087724 | KY117787 | KY068274 | KY117965 | |
| EXF-7667 = IBT 34288 | Oil painting on canvas, Ljubljana, Slovenia | KY087716 | KY117779 | KY068266 | KY117957 | |
| DTO 220-B2 | Air in bakery, Tilburg, The Netherlands | KY087698 | KY117760 | KY068246 | KY117938 | |
| DTO 161-B7 = CCF 5671 | Surface of cheese, The Netherlands | KY087697 | KY117759 | KY068245 | KY117937 | |
| NRRL 157 = CCF 5463 | Unknown, USA | KY087749 | KY117812 | KY068299 | KY117990 | |
| EXF-10407 = IBT 34285 = CCF 5652 | Oil painting on canvas, Ljubljana, Slovenia | KY087723 | KY117786 | KY068273 | KY117964 | |
| EXF-7703 = IBT 34259 | Oil painting on canvas, Ljubljana, Slovenia | KY087718 | KY117781 | KY068268 | KY117959 | |
| DTO 147-E2 | Indoor air, Hungary | KY087696 | KY117758 | KY068244 | KY117936 | |
| DTO 113-E7 = CCF 5668 | Air in bakery, Tilburg, The Netherlands | KY087692 | KY117756 | KY068240 | KY117932 | |
| A. domesticus | DTO 079-F2T = CCF 5464 = NRRL 66616 = IBT 34814 | Wallpaper, Tiel, The Netherlands | KY087688 | KY117752 | KY068236 | KY117928 |
| DTO 231-C1 = NRRL 66617 = CCF 5665 | Museum piece, Zwartewaal, The Netherlands | KY087700 | KY117762 | KY068248 | KY117940 | |
| DTO 231-B9 | Museum piece (mouldy chair backrest), Zwartewaal, The Netherlands | KY087699 | KY117761 | KY068247 | KY117939 | |
| DTO 086-D1 = CCF 5670 | Archive material, Gorinchem, The Netherlands | KY087689 | KY117753 | KY068237 | KY117929 | |
| IHEM 6549 | Dust from mattress, Brussels, Belgium | KY087734 | KY117797 | KY068284 | KY117975 | |
| EXF-10012 = IBT 34274 | Statue made of wood, Textile and sea shells, Ljubljana, Slovenia | KY087719 | KY117782 | KY068269 | KY117960 | |
| UBOCC-A-116022 = CCF 5465 | Painting, Brittany, France | KY087605 | KY117674 | KY068158 | KY117850 | |
| CCF 5466 = EMSL No. 1316 | Air, home, Idaho, USA | KY087606 | KY117675 | KY068159 | KY117851 | |
| A. glabripes | CCF 5474T = EMSL No. 2462 = DTO 356-E8 = NRRL 66618 = IBT 34820 | Office folder, Macoya, Trinidad & Tobago | KY087614 | KY117683 | KY068166 | KY117859 |
| CCF 5473 = EMSL No. 2442 = IBT 34626 | Green fabric covered binders, import from China, New Jersey, USA | KY087613 | KY117682 | KY068165 | KY117858 | |
| CCF 5475 = EMSL No. 2463 | Office folder, Macoya, Trinidad & Tobago | KY087615 | KY117684 | KY068167 | KY117860 | |
| CCF 5476 = EMSL No. 2464 | Office folder, Macoya, Trinidad & Tobago | KY087616 | KY117685 | KY068168 | KY117861 | |
| CCF 5477 = EMSL No. 2465 | Office folder, Macoya, Trinidad & Tobago | KY087617 | KY117686 | KY068169 | KY117862 | |
| CCF 5469 = EMSL No. 1483 = DTO 356-E5 = IBT 34519 = IBT 34821 = NRRL 66619 | Air, home, California, USA | KY087609 | KY117678 | KY068161 | KY117854 | |
| EMSL No. 1812 = CCF 5470 | Front cover of log book, library, Louisiana, USA | KY087610 | KY117679 | KY068162 | KY117855 | |
| EMSL No. 1813 = CCF 5471 | Book, library, Louisiana, USA | KY087611 | KY117680 | KY068163 | KY117856 | |
| EMSL No. 1317 = CCF 5468 | Air, home, Idaho, USA | KY087608 | KY117677 | KY068160 | KY117853 | |
| EMSL No. 2305 = CCF 5472 | Air, kitchen, Summerville, South Carolina, USA | KY087612 | KY117681 | KY068164 | KY117857 | |
| UBOCC-A-116021 = CCF 5467 = IBT 34625 | Painting, Brittany, France | KY087607 | KY117676 | KY117852 | ||
| A. gracilis | NRRL 4962T = CBS 539.65 = DTO 351-H7 = CCF 5478 = ATCC 16906 = IMI 211393 = IBT 34817 | Gun-firing mechanism, South Pacific | EF652045 | EF651883 | EF652031 | EF651980 |
| CCF 5479 = EMSL No. 2775 = DTO 356-F4 = IBT 34559 | Child carrier, San Diego, California, USA | KY087618 | KY117687 | KY068170 | KY117863 | |
| CCF 5480 = EMSL No. 2920 | Child carrier, San Diego, California, USA | KY087619 | KY117688 | KY068171 | KY117864 | |
| CCF 5481 = EMSL No. 2922 | Child carrier, San Diego, California, USA | KY087620 | KY117689 | KY068172 | KY117865 | |
| CCF 5482 = EMSL No. 2923 = IBT 34623 | Child carrier, San Diego, California, USA | KY087621 | KY117690 | KY068173 | KY117866 | |
| A. halophilicus | NRRL 2739T = ATCC 16401 = CBS 122.62 = IMI 211802 = IBT 34878 = CCF 5687 | Dried corn, St. Paul, Minnesota, USA | EF652088 | EF651926 | EF652034 | EF651982 |
| DTO 271-F4 = CCF 5825 = IBT 34881 | Textile, imported into the Netherlands | KY087705 | KY117766 | KY068253 | KY117945 | |
| A. hordei | NRRL 25825T = CCF 5483 = DTO 356-D3 = IBT 34539 | Barley, St. Paul, Minnesota, USA | KY087759 | KY117822 | KY068309 | KY118000 |
| NRRL 25826 = CCF 5484 | Barley, St. Paul, Minnesota, USA | KY087760 | KY117823 | KY068310 | KY118001 | |
| NRRL 25830 = CCF 5485 = IBT 34631 | Insulating board, St. Paul, Minnesota, USA | KY087761 | KY117824 | KY068311 | KY118002 | |
| A. infrequens | NRRL 25868T = CCF 5486 = DTO 356-D6 = IBT 34524 | Wheat, Peoria, Illionois, USA | KY087770 | KY117833 | KY068320 | KY118011 |
| A. magnivesiculatus | NRRL 25866T = CCF 5488 = IBT 34816 | Katsuobushi, Tokyo, Japan | KY087768 | KY117831 | KY068318 | KY118009 |
| CCF 5491 = EMSL No. 2918 | Child carrier, San Diego, California, USA | KY087624 | KY117692 | KY068176 | KY117869 | |
| CCF 5489 = EMSL No. 1315 = DTO 356-E2 = IBT 34516 | Air, home, Idaho, USA | KY087622 | KY117691 | KY068174 | KY117867 | |
| CCF 5490 = EMSL No. 2741 | Child carrier, San Diego, California, USA | KY087623 | KY068175 | KY117868 | ||
| NRRL 25867 = CCF 5660 | Katsuobushi, Tokyo, Japan | KY087769 | KY117832 | KY068319 | KY118010 | |
| NRRL 25821 = CCF 5487 | Dried corn, St. Paul, Minnesota, USA | KY087756 | KY117819 | KY068306 | KY117997 | |
| EXF-10353 = IBT 34284 | Oil painting on canvas, Ljubljana, Slovenia | KY087720 | KY117783 | KY068270 | KY117961 | |
| KAS 5655 = DTO 356-G8 | House dust, Ottawa, Ontario, Canada | KY087661 | KY117725 | KY068209 | KY117905 | |
| KAS 5754 = DTO 356-G9 | House dust, Ottawa, Ontario, Canada | KY087662 | KY117726 | KY068210 | ||
| EXF-10377 | Oil painting on canvas, Ljubljana, Slovenia | KY087721 | KY117784 | KY068271 | KY117962 | |
| KAS 5623 = DTO 356-G7 | House dust, Stittsville, Ontario, Canada | KY087660 | KY117724 | KY068208 | KY117904 | |
| KAS 6089 = DTO 356-H3 | House dust, Wolfville, Nova Scotia, Canada | KY087664 | KY117728 | KY068212 | KY117907 | |
| A. pachycaulis | NRRL 25824T = CCF 5492 = DTO 356-D2 = IBT 34521 = IBT 34812 | Unknown, Washington, District of Columbia, USA | KY087758 | KY117821 | KY068308 | KY117999 |
| CCF 5493 = EMSL No. 2310 = DTO 356-E6 = IBT 34536 | Air, home, California, USA | KY087625 | KY117693 | KY068177 | KY117870 | |
| A. penicillioides | NRRL 4548T = CBS 540.65 = ATCC 16910 = IMI 211342 = DTO 207-I7 = CCF 5494 = IBT 34627 | Human skin, Recife, Brazil | EF652036 | EF651928 | EF652024 | EF651930 |
| CCF 5497 = EMSL No. 2430 = IBT 34628 | Green fabric covered binders, import from China, New Jersey, USA | KY087626 | KY117694 | KY068178 | KY117871 | |
| IHEM 2330 | Seeds of cereal, France | KY087730 | KY117793 | KY068280 | KY117971 | |
| IHEM 2476 | Indoor air, Brussels, Belgium | KY087732 | KY117795 | KY068282 | KY117973 | |
| DTO 281-A7 | Leather, imported into the Netherlands | KY087706 | KY117767 | KY068254 | KY117946 | |
| CCF 3282 | Sweet roll with chocolate glaze, Prague, Czech Republic | KY087657 | FR775347 | HE578103 | KY117902 | |
| CBS 140430 = UBOCC-A-115042 = DTO 334-E1 | French madeleines, France | KY087596 | KY117664 | KY068148 | KY117841 | |
| CCF 5500 = EMSL No. 2651 = IBT 34630 | Baseball gloves, store, O'fallon, Illinois, USA | KY087629 | KY117697 | KY068181 | KY117874 | |
| CCF 5503 = EMSL No. 2909 | Child carrier, San Diego, California, USA | KY087632 | KY117700 | KY068184 | KY117877 | |
| NRRL 25816 = CCF 5661 | Unknown, Durham, North Carolina, USA | KY087754 | KY117817 | KY068304 | KY117995 | |
| NRRL 25834 = CCF 5659 | Peas, St. Paul, Minnesota, USA | KY087762 | KY117825 | KY068312 | KY118003 | |
| NRRL 25835 | Wheat, St. Paul, Minnesota, USA | KY087763 | KY117826 | KY068313 | KY118004 | |
| KAS 7745 | House dust, Ottawa, Ontario, Canada | KY087746 | KY117809 | KY068296 | KY117987 | |
| KAS 7746 | House dust, Ottawa, Ontario, Canada | KY087747 | KY117810 | KY068297 | KY117988 | |
| DTO 267-A9 = CCF 5664 | House dust, Micronesia | KY087704 | KY117765 | KY068252 | KY117944 | |
| CCF 2666 | Leather shoe, Zlín, Czech republic | KY087655 | HE578081 | HE578102 | KY117900 | |
| CCF 5501 = EMSL No. 2749 = IBT 34629 | Child carrier, San Diego, California, USA | KY087630 | KY117698 | KY068182 | KY117875 | |
| CCF 5504 = EMSL No. 3264 | Archival cabinet, Bethesda, Maryland, USA | KY087633 | KY117701 | KY068185 | KY117878 | |
| NRRL 25870 | Unknown | KY087771 | KY117834 | KY068321 | KY118012 | |
| NRRL 25879 | Blood sample, New York, USA | KY087774 | KY117838 | KY068325 | KY118016 | |
| CCF 5499 = EMSL No. 2441 | Green fabric covered binders, import from China, New Jersey, USA | KY087628 | KY117696 | KY068180 | KY117873 | |
| NRRL 4550 = CCF 5495 | Human skin, Recife, Brazil | EF652037 | EF651929 | EF652025 | EF651931 | |
| NRRL 4553 = CCF 5496 | Human skin, Recife, Brazil | KY087750 | KY117813 | KY068300 | KY117991 | |
| CCF 5498 = EMSL No. 2440 = DTO 356-E7 = IBT 34815 | Green fabric covered binders, import from China, New Jersey, USA | KY087627 | KY117695 | KY068179 | KY117872 | |
| CCF 5502 = EMSL No. 2900 | Child carrier, San Diego, California, USA | KY087631 | KY117699 | KY068183 | KY117876 | |
| NRRL 25820 | Dried corn, St. Paul, Minnesota, USA | KY087755 | KY117818 | KY068305 | KY117996 | |
| NRRL 25822 | Dried corn, St. Paul, Minnesota, USA | KY087757 | KY117820 | KY068307 | KY117998 | |
| A. pseudogracilis | CCF 5505T = EMSL No. 2765 = DTO 356-F3 = NRRL 66620 = IBT 34813 | Child carrier, San Diego, California, USA | KY087634 | KY117702 | KY068186 | KY117879 |
| A. restrictus | NRRL 154T = CBS 117.33 = CBS 541.65 = DTO 079-B2 = ATCC 16912 = IHEM 3920 = IMI 16267 = IHEM 3920 = CCF 5506 = IBT 34615 | Cloth, United Kingdom | EF652042 | EF651880 | EF652029 | EF651978 |
| CCF 5511 = EMSL No. 1675 = IBT 34616 | Packing material, Maryland, USA | KY087639 | KY117707 | KY068191 | KY117884 | |
| NRRL 25862 | Culture contaminant, Peoria, Illinois, USA | KY087767 | KY117830 | KY068317 | KY118008 | |
| IHEM 2121 | Dust from mattress, Antwerp, Belgium | KY087729 | KY117792 | KY068279 | KY117970 | |
| CCF 5512 = EMSL No. 2206 = IBT 34617 | Air, auditorium, school, Sicklerville, New Jersey, USA | KY087640 | KY117708 | KY068192 | KY117885 | |
| CCF 5513 = EMSL No. 2429 | Green fabric covered binders, import from China, New Jersey, USA | KY087641 | KY117709 | KY068193 | KY117886 | |
| CCF 5509 = EMSL No. 1611 | Mattress cover, North Carolina, USA | KY087637 | KY117705 | KY068189 | KY117882 | |
| DTO 065-C7 = IBT 34541 | Corn kernels, Indonesia | KY087685 | KY117749 | KY068233 | KY117926 | |
| CCF 3364 = IBT 34619 | Sclerotium of fungus Corallocytostroma ornicopreoides imported from Australia, Prague, Czech Republic | KY087658 | FR775348 | HE578101 | HE578109 | |
| CCF 5514 = EMSL No. 2652 | Baseball gloves, store, O'fallon, Illinois, USA | KY087642 | KY117710 | KY068194 | KY117887 | |
| NRRL 25882 | Cattle feed, USA | KY087776 | KY117840 | KY068327 | KY118018 | |
| CCF 5515 = EMSL No. 2906 | Child carrier, San Diego, California, USA | KY087643 | KY117711 | KY068195 | KY117888 | |
| IHEM 818 | Indoor air, Estinnes-au-Mont, Belgium | KY087728 | KY117791 | KY068278 | KY117969 | |
| CCF 5510 = EMSL No. 1633 | Air, hospital, New Jersey, USA | KY087638 | KY117706 | KY068190 | KY117883 | |
| CCF 5508 = EMSL No. 1416 | Air, home, Alabama, USA | KY087636 | KY117704 | KY068188 | KY117881 | |
| CCF 5507 = EMSL No. 1379 = IBT 34618 | Air, home, Bermuda | KY087635 | KY117703 | KY068187 | KY117880 | |
| IHEM 2477 | Indoor air, Brussels, Belgium | KY087733 | KY117796 | KY068283 | KY117974 | |
| IHEM 2373 | Indoor air, Brussels, Belgium | KY087731 | KY117794 | KY068281 | KY117972 | |
| A. reticulatus | NRRL 25852T = CCF 5516 = DTO 356-D4 = IBT 34540 | Lung biopsy, Charleston, South Carolina, USA | KY087765 | KY117828 | KY068315 | KY118006 |
| CCF 5523 = EMSL No. 2526 | Air, administrative area, Bayamon, Puerto Rico | KY087649 | KY117717 | KY068201 | KY117894 | |
| CCF 5524 = EMSL No. 2548 = IBT 34637 | Air, office, Bayamon, Puerto Rico | KY087650 | KY117718 | KY068202 | KY117895 | |
| CCF 5518 = EMSL No. 1272 = NRRL 58903 = IBT 34819 | Air, home, Idaho, USA | KY087644 | KY117712 | KY068196 | KY117889 | |
| CCF 5519 = EMSL No. 1313 = NRRL 62004 | Air, home, Idaho, USA | KY087645 | KY117713 | KY068197 | KY117890 | |
| CCF 5520 = EMSL No. 1314 = NRRL 62005 | Air, home, Idaho, USA | KY087646 | KY117714 | KY068198 | KY117891 | |
| CCF 5521 = EMSL No. 1362 = DTO 356-E4 = IBT 34518 | Air, outside, Idaho, USA | KY087647 | KY117715 | KY068199 | KY117892 | |
| CCF 5525 = EMSL No. 885 = NRRL 58572 = IBT 34880 | Air, home, Florida, USA | KY087651 | KY117719 | KY068203 | KY117896 | |
| CCF 5522 = EMSL No. 2525 | Air, administrative area, Bayamon, Puerto Rico | KY087648 | KY117716 | KY068200 | KY117893 | |
| IHEM 22696 | Dust from carpet, Brussels, Belgium | KY087737 | KY117800 | KY068287 | KY117978 | |
| EXF-10429 = CCF 5656 | Oil painting on canvas, Ljubljana, Slovenia | KY087725 | KY117788 | KY068275 | KY117966 | |
| CCF 3112 = IBT 34634 = NRRL 62490 | Leather shoe, Zlín, Czech Republic | KY087656 | FR775323 | FR751451 | KY117901 | |
| NRRL 25878 = CCF 5517 | Lung biopsy, Chamblee, Georgia, USA | KY087773 | KY117837 | KY068324 | KY118015 | |
| A. salinicola | EXF-10401T = IBT 34266 = CCF 5526 = NRRL 66621 | Oil painting on canvas, Ljubljana, Slovenia | KY087722 | KY117785 | KY068272 | KY117963 |
| KAS 6054 | House dust, Wolfville, Nova Scotia, Canada | KY087738 | KY117801 | KY068288 | KY117979 | |
| UBOCC-A-116019 = CCF 5528 = IBT 34635 | Painting, Brittany, France | KY087652 | KY117720 | KY068204 | KY117897 | |
| EXF-226 = CCF 5527 = IBT 34277 = NRRL 66622 | Hypersaline water from salterns, Sečovlje salterns, Slovenia | KY087711 | KY117773 | KY068260 | ||
| A. tardicrescens | DTO 316-B5T = CCF 5529 = IBT 34558 = NRRL 66623 | Museum piece (measuring table), Alphen aan den Rijn, The Netherlands | KY087710 | KY117772 | KY068259 | KY117951 |
| DTO 316-A7 | Museum piece (dentist chair), Alphen aan den Rijn, The Netherlands | KY087707 | KY117768 | KY068255 | KY117947 | |
| DTO 316-A8 = IBT 34562 | Museum piece (rubber tyre of brancard), Alphen aan den Rijn, The Netherlands | KY087708 | KY117769 | KY068256 | KY117948 | |
| DTO 316-A9 | Museum piece (x-ray table), Alphen aan den Rijn, The Netherlands | KY117770 | KY068257 | KY117949 | ||
| DTO 316-B4 | Museum piece (vitrine), Alphen aan den Rijn, The Netherlands | KY087709 | KY117771 | KY068258 | KY117950 | |
| EXF-10456 = IBT 34286 | Air in depot of Conservation Centre of the Institute for the Protection of Cultrural Heritage of Slovenia, Ljubljana, Slovenia | KY087727 | KY117790 | KY068277 | KY117968 | |
| KAS 6252 = DTO 356-I5 | House dust, Wolfville, Nova Scotia, Canada | KY087669 | KY117733 | KY068217 | KY117911 | |
| EXF-10454 = IBT 34281 = CCF 5530 = NRRL 66624 | Oil painting on canvas, Ljubljana, Slovenia | KY087726 | KY117789 | KY068276 | KY117967 | |
| DTO 073-H6 | Moist wall of archive, Tilburg, The Netherlands | KY087686 | KY117750 | KY068234 | KY117927 | |
| A. villosus | NRRL 25813T = CCF 5531 = DTO 356-C9 = IBT 34822 | Unknown, Kirkhill, Scotland, United Kingdom | KY087752 | KY117815 | KY068302 | KY117993 |
| UBOCC-A-116020 = CCF 5532 = IBT 34624 | Painting, Brittany, France | KY087653 | KY117721 | KY068205 | KY117898 | |
| A. vitricola | NRRL 5125T = DTO 356-F7 = ATCC 16905 = ATCC 36505 = IMI 108298 = CCF 5533 = IBT 34530 | Binocular lens, Japan | EF652046 | EF651927 | EF652035 | EF651973 |
| KAS 6086 | House dust, Little Lepreau, New Brunswick, Canada | KY087739 | KY117802 | KY068289 | KY117980 | |
| KAS 6281 | House dust, Victoria, British Columbia, Canada | KY087744 | KY117807 | KY068294 | KY117985 | |
| KAS 6087 = DTO 356-H2 = IBT 34532 | House dust, Victoria, British Columbia, Canada | KY087663 | KY117727 | KY068211 | KY117906 | |
| KAS 6238 | House dust, Victoria, British Columbia, Canada | KY087743 | KY117806 | KY068293 | KY117984 | |
| KAS 6199 | House dust, Victoria, British Columbia, Canada | KY087742 | KY117805 | KY068292 | KY117983 | |
| KAS 6288 | House dust, Wolfville, Nova Scotia, Canada | KY087745 | KY117808 | KY068295 | KY117986 | |
| KAS 6237 = DTO 356-I2 | House dust, Victoria, British Columbia, Canada | KY087668 | KY117732 | KY068216 | KY117910 | |
| KAS 6133 = DAOMC 251500 | House dust, Little Lepreau, New Brunswick, Canada | KY087741 | KY117804 | KY068291 | KY117982 | |
| KAS 6093 | House dust, Victoria, British Columbia, Canada | KY087740 | KY117803 | KY068290 | KY117981 | |
| DTO 122-I4 | Archive material, Gorinchem, The Netherlands | KY087693 | KY117757 | KY068241 | KY117933 | |
| DTO 123-B2 | Archive material, Gorinchem, The Netherlands | KY087695 | KY068243 | KY117935 | ||
| DTO 122-I5 | Archive material, Gorinchem, The Netherlands | KY087694 | KY068242 | KY117934 | ||
| KAS 6278 = DTO 356-I8 | House dust, Wolfville, Nova Scotia, Canada | KY087670 | KY117734 | KY068218 | KY117912 | |
| KAS 6150 = DTO 356-H5 = IBT 34531 | House dust, Wolfville, Nova Scotia, Canada | KY087665 | KY117729 | KY068213 | ||
| KAS 6178 = DTO 356-H8 | House dust, Little Lepreau, New Brunswick, Canada | KY087666 | KY117730 | KY068214 | KY117908 | |
| CCF 5534 = EMSL No. 2785 | Child carrier, San Diego, California, USA | KY087654 | KY117722 | KY068206 | KY117899 | |
Acronyms of culture collections: ATCC, American Type culture collection, Manassas, Virginia, USA; CBS, culture collection of Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CCF, Culture Collection of Fungi, Charles University, Czech Republic; DAOMC, Canadian Collection of Fungal Cultures, Agriculture and Agri-Food Canada, Ottawa, Canada; DTO, working collection of the department of Applied and Industrial Mycology housed at CBS; EMSL, EMSL Analytical Inc., New Jersey, USA; EXF, Culture Collection of Extremophilic Fungi, University of Ljubljana, Slovenia; IBT, Culture Collection at Center for Microbial Biotechnology, Lyngby, Denmark; BCCM/IHEM, Biomedical Fungi and Yeasts Collection, Scientific Institute of Public Health, Brussels, Belgium; IMI/CABI, International Mycological Institute, Kew, England; KAS, fungal collection of Keith A. Seifert, Ottawa, Canada; NRRL, Agricultural Research Service Culture Collection, Peoria, Illinois, USA; UBOCC, Université de Bretagne Occidentale Culture Collection, Brest, France.
Ex-type strains are designated with superscript T.
ArchivePure DNA yeast and Gram2+ kit (5 PRIME Inc., Gaithersburg, MD) were used for DNA isolation from 14 d old cultures according to manufacturer instructions as updated by Hubka et al. (2015). Target genetic loci, including ITS + LSU rDNA (ID region), partial genes encoding calmodulin (CaM), β-tubulin (benA) and the second largest subunit of RNA polymerase II (RPB2), were amplified using primer combinations listed in Table 2. Amplification of RPB2 with the widely used primers (fRPB2-5F, fRPB2-7CR) designed by Liu et al. (1999) was problematic for many sect. Restricti isolates. Hence, new primers specific for section Restricti (fRPB2ResF100, fRPB2ResR950, Table 2) were designed based on the alignment of available sequences obtained with the Liu et al. (1999) primer set. Quality control (hairpin, self-dimer or hetero-dimer formation, melting temperature mismatch) was performed in OligoAnalyzer v. 3.1 (available online http://eu.idtdna.com/calc/analyzer). Standard and touchdown PCR protocols were described previously (Hubka et al., 2014, Hubka et al., 2016). PCR product purification followed Réblová et al. (2016). Automated sequencing was performed at Macrogen Sequencing Service (Amsterdam, The Netherlands) using both terminal primers. Sequences were deposited into GenBank with accession numbers shown in Table 1. All alignments are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.3t423.
Table 2.
Primers used for amplification and sequencing.
| Locus | Primer | Orientation | Sequence (from 5′ to 3′) | References |
|---|---|---|---|---|
| benA | Bt2a | Forward | GGTAACCAAATCGGTGCTGCTTTC | Glass & Donaldson (1995) |
| T10 | Forward | ACGATAGGTTCACCTCCAGAC | O'Donnell & Cigelnik (1997) | |
| Bt2b | Reverse | ACCCTCAGTGTAGTGACCCTTGGC | Glass & Donaldson (1995) | |
| CaM | CF1L | Forward | GCCGACTCTTTGACYGARGAR | Peterson (2008) |
| CF1M | Forward | AGGCCGAYTCTYTGACYGA | Peterson (2008) | |
| cmd5 | Forward | CCGAGTACAAGGAGGCCTTC | Hong et al. (2006) | |
| CF4 | Reverse | TTTYTGCATCATRAGYTGGAC | Peterson (2008) | |
| cmd6 | Reverse | TTTYTGCATCATRAGYTGGAC | Hong et al. (2006) | |
| RPB2 | fRPB2-5F | Forward | GAYGAYMGWGATCAYTTYGG | Liu et al. (1999) |
| fRPB2ResF100 | Forward | TGAARTAYGCICTTGCYAC | Newly designed | |
| fRPB2-7CR | Reverse | CCCATRGCTTGYTTRCCCAT | Liu et al. (1999) | |
| fRPB2ResR950 | Reverse | CARTGYGTCCADGTRTGKGC | Newly designed | |
| ITS | ITS1 | Forward | TCCGTAGGTGAACCTGCGG | White et al. (1990) |
| NL4 | Reverse | GGTCCGTGTTTCAAGACGG | O'Donnell (1993) | |
| ITS4 | Reverse | TCCTCCGCTTATTGATATGC | White et al. (1990) |
Phylogenetic analysis
Sequences were inspected in FinchTV (available online http://www.geospiza.com/Products/finchtv.shtml) and assembled in Bioedit v. 7.2.5 (Hall 1999). Alignments were performed using the G-INS-i strategy, as implemented in MAFFT v. 7 (Katoh & Standley 2013). The benA alignment contained 431 characters with 227 variable and 217 parsimony informative sites, CaM 652 characters with 288 variable and 268 parsimony informative sites, RPB2 819 characters with 272 variable and 249 parsimony informative sites, ID 1 123 characters with 231 variable and 175 parsimony informative sites. The concatenated alignment contained 3 025 characters, with 1 018 variable and 909 parsimony informative sites.
Phylogenetic trees based on the concatenated dataset were inferred with both Maximum likelihood (ML) and Bayesian inference (BI) analysis. Partitioning scheme and substitution models for analyses were selected using PartitionFinder v. 1.1.1 (Lanfear et al. 2012) with settings allowing introns, exons and codon positions to be independent datasets. Proposed partitioning schemes for each dataset are listed in Table 3. Hamigera avellanea NRRL 1938 was used as outgroup.
Table 3.
Partition-merging results and best substitution model for each partition according to Bayesian information criterion as proposed by PartitionFinder v1.1.1.
| Dataset | Phylogenetic method1 | Partitioning scheme (substitution model) |
|---|---|---|
| Sect. Restricti (ITS + benA + CaM + RPB2) | ML | benA + CaM introns (HKY+I+G); 1st codon positions of benA + CaM + RPB2 + 2nd codon positions of RPB2 (TrNef+I+G); ITS + LSU (TrNef+I+G); 2nd codon positions of benA + CaM + 3rd codon positions of benA + CaM (TIM+G); 3rd codon positions of RPB2 (HKY+G) |
| BI | benA + CaM introns (HKY+I+G); 1st codon positions of benA + CaM + 2nd codon positions of RPB2 (K80+I+G); 2nd codon positions of benA + CaM + 3rd codon positions of benA (SYM+G); 1st codon positions of RPB2 + 3rd codon positions of CaM + ITS + LSU (GTR+I+G); 3rd codon positions of RPB2 (HKY+G) | |
| Subg. Aspergillus (benA + CaM + RPB2) | ML | benA + CaM introns (HKY+I+G); 1st codon positions of benA (JC+I); 2nd codon positions of benA + CaM + RPB2 (F81+I); 3rd codon positions of benA + CaM (GTR+G); 1st codon positions of CaM + RPB2 (TrN+I+G); 3rd codon positions of RPB2 (TrNef+G) |
| BI | benA + CaM introns (HKY+I+G); 1st codon positions of benA (JC+I); 2nd codon positions of benA + CaM + RPB2 (F81+I); 3rd codon positions of benA + CaM (GTR+G); 1st codon positions of CaM + RPB2 (GTR+I+G); 3rd codon positions of RPB2 (HKY+G) |
ML, Maximum likelihood; BI, Bayesian inferrence.
The ML trees were constructed with IQ-TREE v. 1.4.4 (Nguyen et al. 2015) with branch support values obtained from 1 000 bootstrap replicates. Bayesian posterior probabilities (PP) were calculated using MrBayes v. 3.2.6 (Ronquist et al. 2012). Optimal partitioning scheme and substitution models were selected using PartitionFinder v. 1.1.1 as described above. The analyses ran for 5 × 106 generations, two parallel runs with four chains each were used, every 1 000th tree was retained, and the first 25 % of trees were discarded as burn-in.
For inferring relationships within subg. Aspergillus, phylogenies were calculated from a benA, CaM and RPB2 concatenated dataset containing 102 individuals and 1 902 characters, of which 929 characters were variable and 864 were parsimony informative. ML and BI analyses were inferred as described above. Suitable partitioning schemes selected using PartitionFinder v. 1.1.1 are listed in Table 3.
Species delimitation and species tree inference
Nucleotide substitution models for each locus were determined using jModeltest v. 2.1.7 (Posada 2008) based on the Bayesian information criterion (BIC) and are listed in Table 4.
Table 4.
Nucleotide substitution models selected by jModeltest 2.1.7 for each locus according to Bayesian information criterion.
| Clade | Locus | Selected substitution model |
|---|---|---|
| A. restrictus, A. conicus, A. vitricola clades | benA | SYM + G |
| CaM | TrNef + G | |
| RPB2 | K80 + G | |
| ITS + LSU | TrN + I | |
| A. penicillioides clade | benA | TrNef + G |
| CaM | TrNef + I | |
| RPB2 | TrNef + G | |
| ITS + LSU | SYM + I + G | |
| Whole dataset | benA | TrNef + G |
| CaM | TrNef + G | |
| RPB2 | TrNef + I + G | |
| ITS + LSU | TrNef + I + G |
To assign individuals into species, several species delimitation methods were employed. In all cases the alignment was split into two parts. The first part contained the A. restrictus, A. conicus and A. vitricola clades, the second part the A. penicillioides clade. The Bayesian version of the general mixed yule-coalescent model (bgmyc) was performed in R v. 3.3.1 (R Core Team 2015) with the bGMYC package (Reid & Carstens 2012). The general mixed yule-coalescent method (GMYC) was performed in R v. 3.3.1 using the splits package (Fujisawa & Barraclough 2013). Single-locus ultrametric trees created in BEAST v. 2.4.2 (Bouckaert et al. 2014) were used as an input for both methods. Chain length for each tree was 1 × 107 generations with 25 % burn-in. The highest credibility tree was used for the GMYC method and 100 trees sampled throughout the analysis were used for the bGMYC method. These trees were obtained by equal sampling of all the trees from the analysis after discarding the first 60 % of trees. The analysis according Poisson tree processes (PTP) model was performed on species delimitation server (Zhang et al. 2013). The method does not require an ultrametric tree, so the single-locus input trees were calculated using ML analysis in IQ-TREE web server (Trifinopoulos et al. 2016). Species delimitation using the Automatic barcode gap discovery (ABGD) was performed on ABGD web (Puillandre et al. 2012). Finally multilocus species delimitation (STACEY) was performed with the BEAST v. 2.4.2 add-on STACEY v. 1.2.2 (Jones 2017). The chain length was set to 5 × 108 generations, priors were set as follows: the species tree prior was set to the Yule model, growth rate prior was set to lognormal distribution (M = 5, S = 2), clock rate priors for all loci were set to lognormal distribution (M = 0, S = 1), PopPriorScale prior was set to lognormal distribution (M = -7, S = 2) and relativeDeathRate prior was set to beta distribution (α = 1, β = 1 000). The output was processed with SpeciesDelimitationAnalyzer (Jones 2017).
Species trees were inferred using *BEAST (Heled & Drummond 2010) implemented in BEAST v. 2.4.2. (Bouckaert et al. 2014). Individuals were assigned into species based on the consensual results from the above-mentioned species delimitation methods. The MCMC analysis was run for 1 × 108 of generations, 25 % of trees was discarded as burn-in. Strict molecular clock was chosen for all loci and population function was set as constant. Convergence was assessed by examining the likelihood plots in Tracer v. 1.6 (Rambaut et al. 2014).
Species delimitation hypotheses were tested by a coalescent-based approach implemented in BP&P v. 3.1 (Bayesian phylogenetics and phylogeography) (Yang & Rannala 2010). Species delimitation using rjMCMC (reversible jump MCMC algorithm allows to inspect different models with given species tree) was performed with similar isolates allocation to species as during the species tree inference and the tree topology created according to the results from *BEAST. We analysed three combinations on the prior distributions of the parameters ϑ (ancestral population size) and τ0 (root age) as proposed by Leaché & Fujita (2010), i.e. large ancestral population sizes and deep divergence: ϑ ∼ G (1, 10) and τ0 ∼ G (1, 10); small ancestral population sizes and shallow divergences among species: ϑ ∼ G (2, 2 000) and τ0 ∼ G (2, 2 000); large ancestral populations sizes and shallow divergences among species: ϑ ∼ G (1, 10) and τ0 ∼ G (2, 2 000).
Species boundaries were further validated by calculating the genealogical sorting index (GSI) (Cummings et al. 2008) which quantifies the degree of exclusive ancestry of hypothetical species. In order to perform the analysis, 100 trees inferred from each locus were created using RAxML (Stamatakis et al. 2008) with the bootstrap option. Calculation of gsi statistics was performed at http://www.molecularevolution.org, with 1 × 104 permutations for evaluation of statistical significance.
Comparison with 454 sequence data
Reference sequences generated in this study were compared to 454-pyrosequences obtained from house dust collected during a world-wide survey (Amend et al. 2010). Information with regards to dust collection and metagenomic analyses methods, readers are refered to Amend et al. (2010). For our comparisons, 454-sequences belonging to sect. Restricti were harvested by firstly doing a BLAST search of ITS barcodes from sect. Restricti against the main 454 database and retaining all sequences with at least 90 % similarity. This dataset was aligned in MAFFT v. 7 using the G-INS-i algorithm and subsequent neighbour-joining tree calculated in MEGA v. 7 (Kumar et al. 2016). This tree was used to remove all sequences that do not belong in sect. Restricti. The dataset was subsequently re-aligned and neighbour-joining tree calculated using Hamigera avellanea as outgroup.
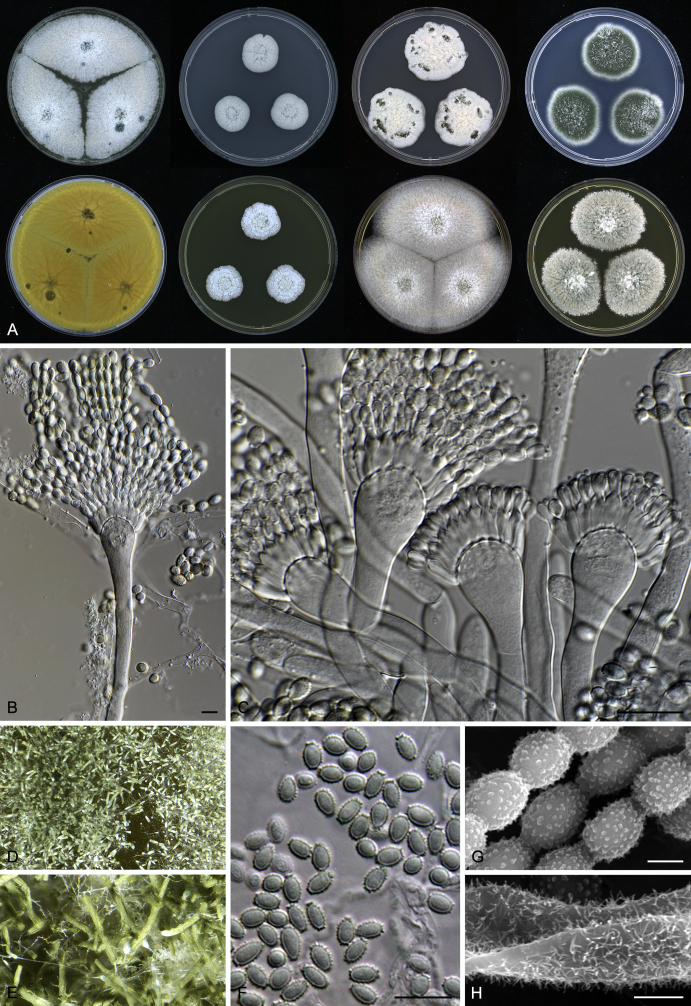
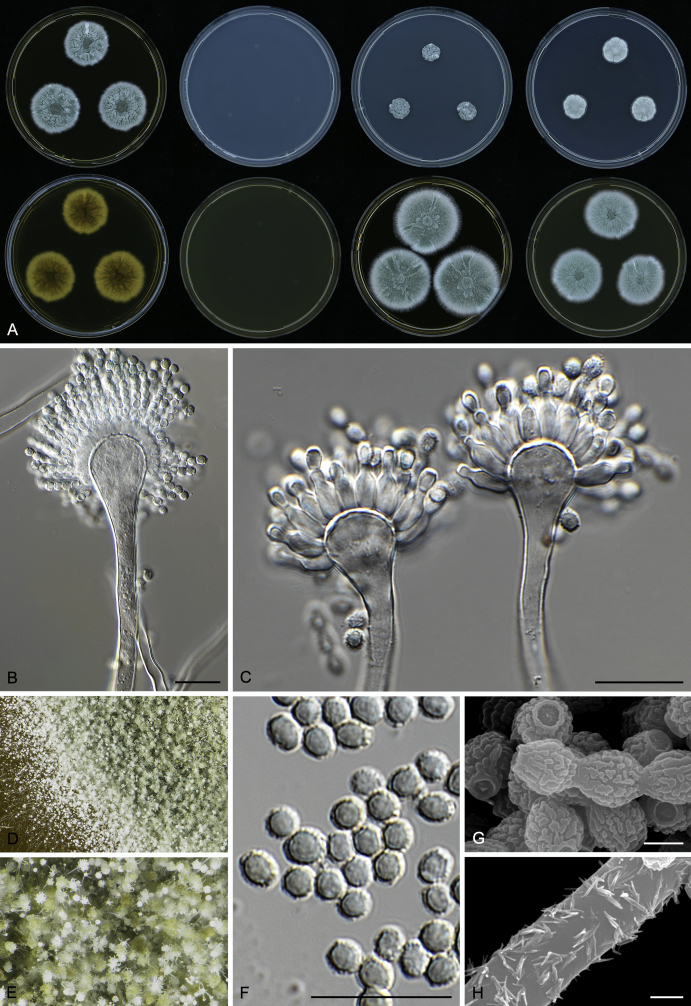
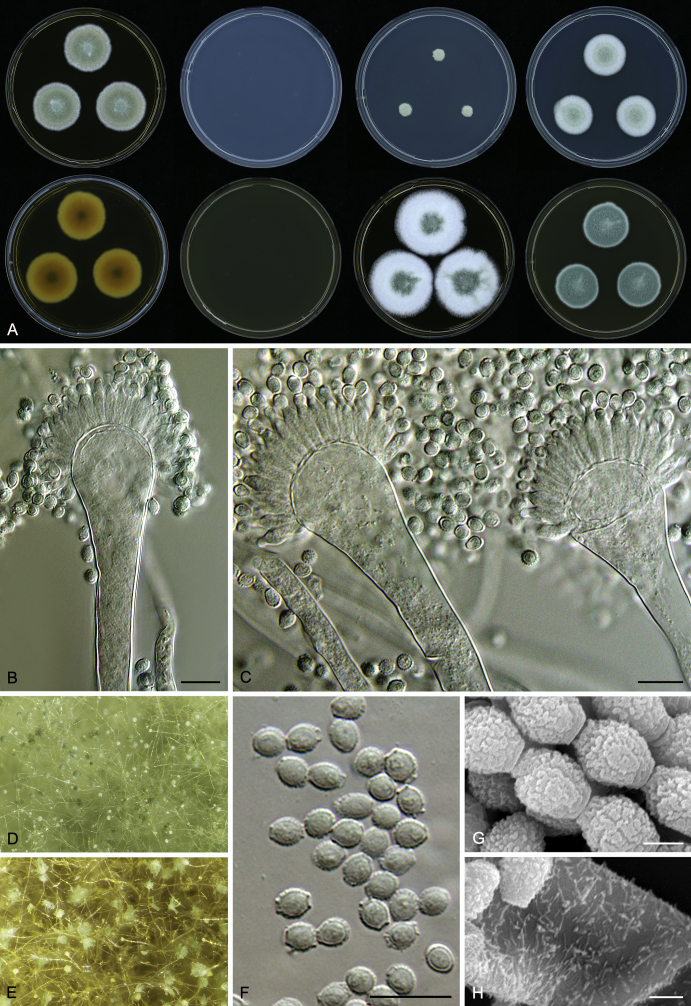
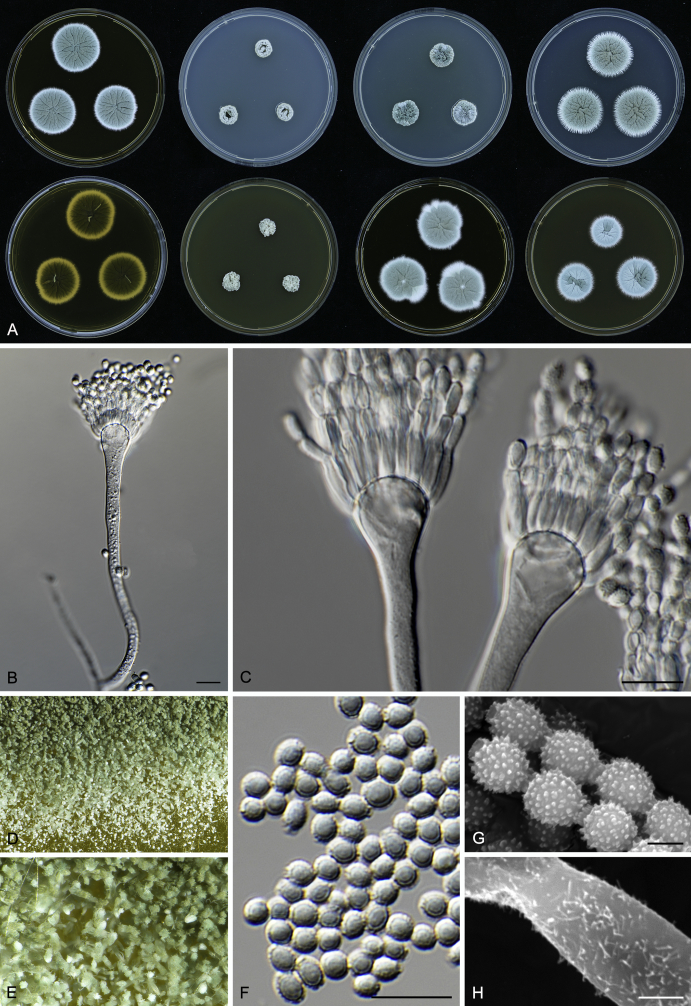
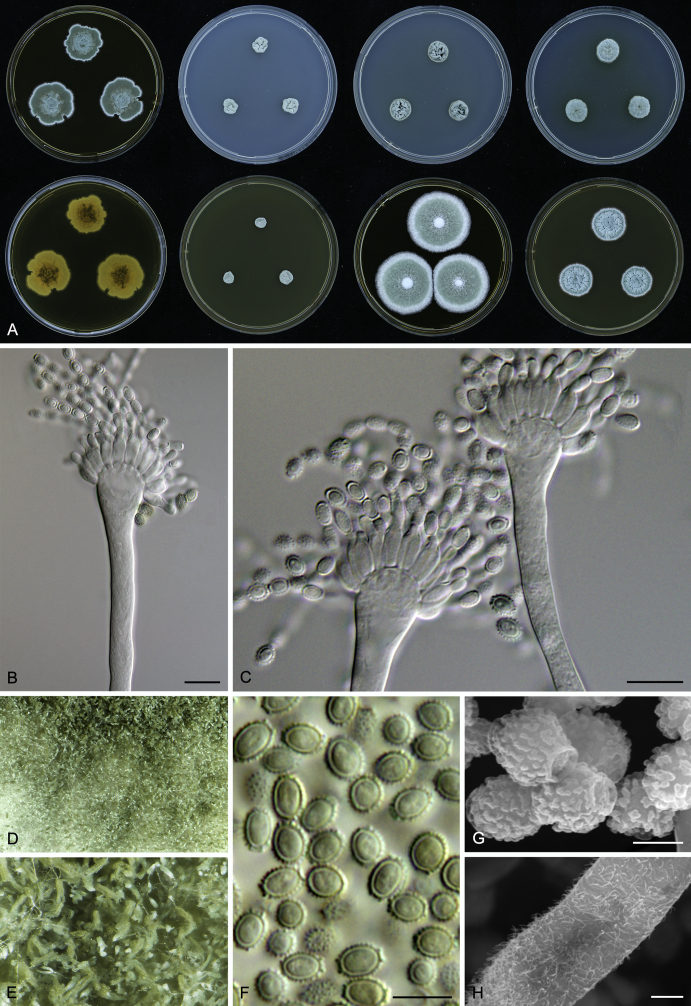
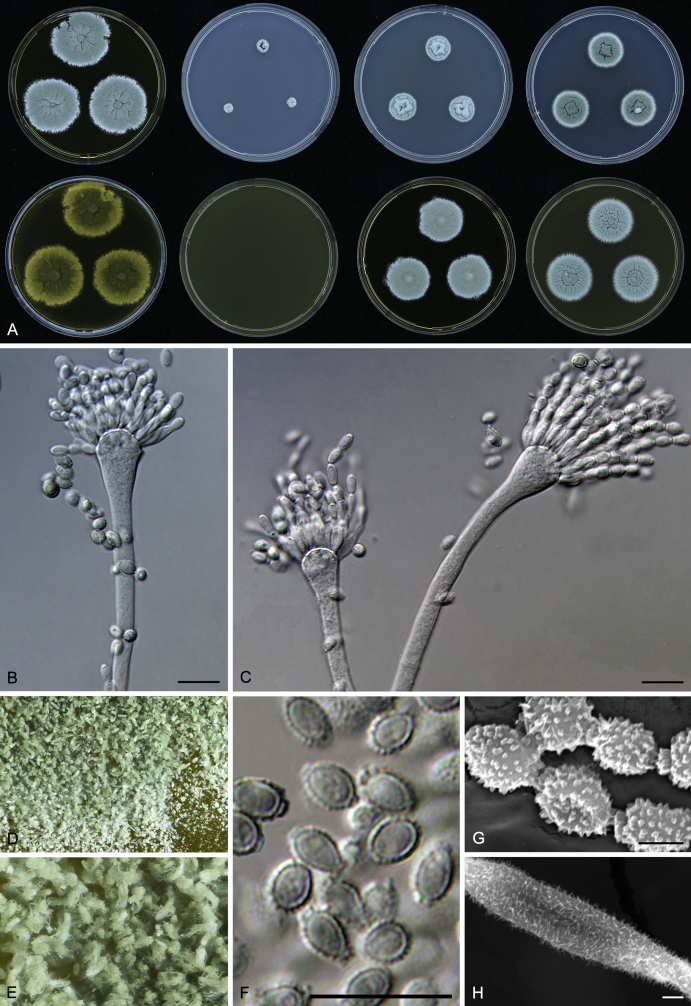
Morphology
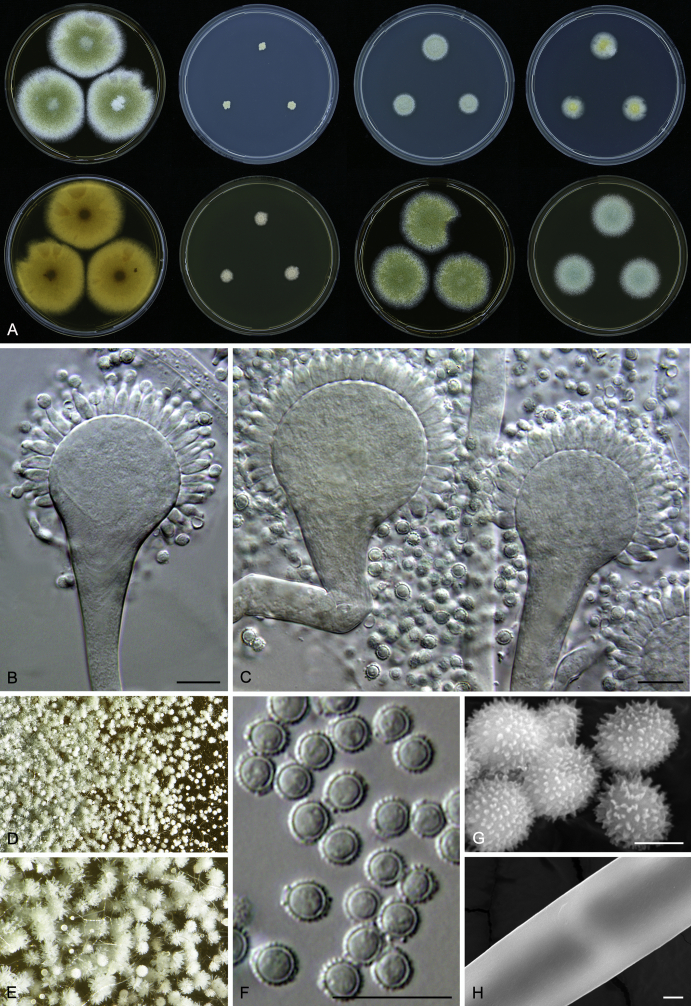
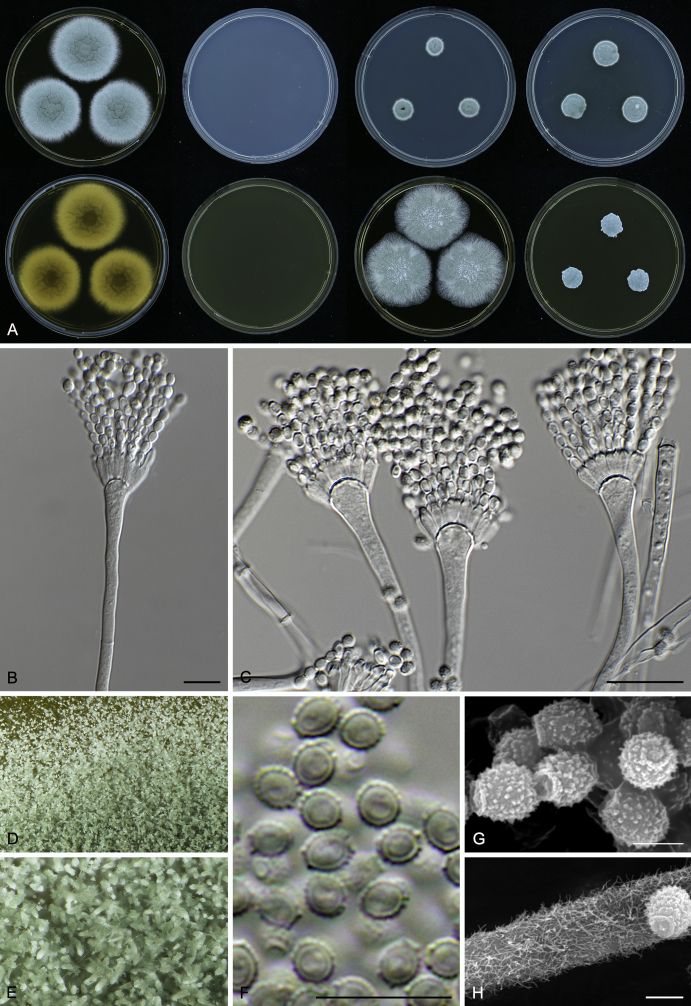
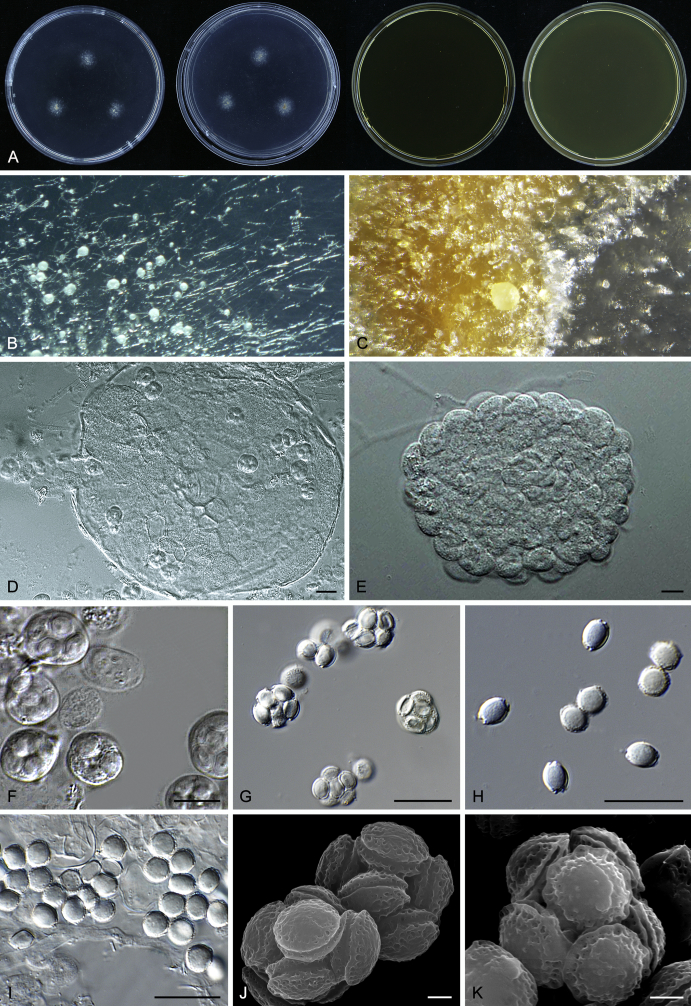
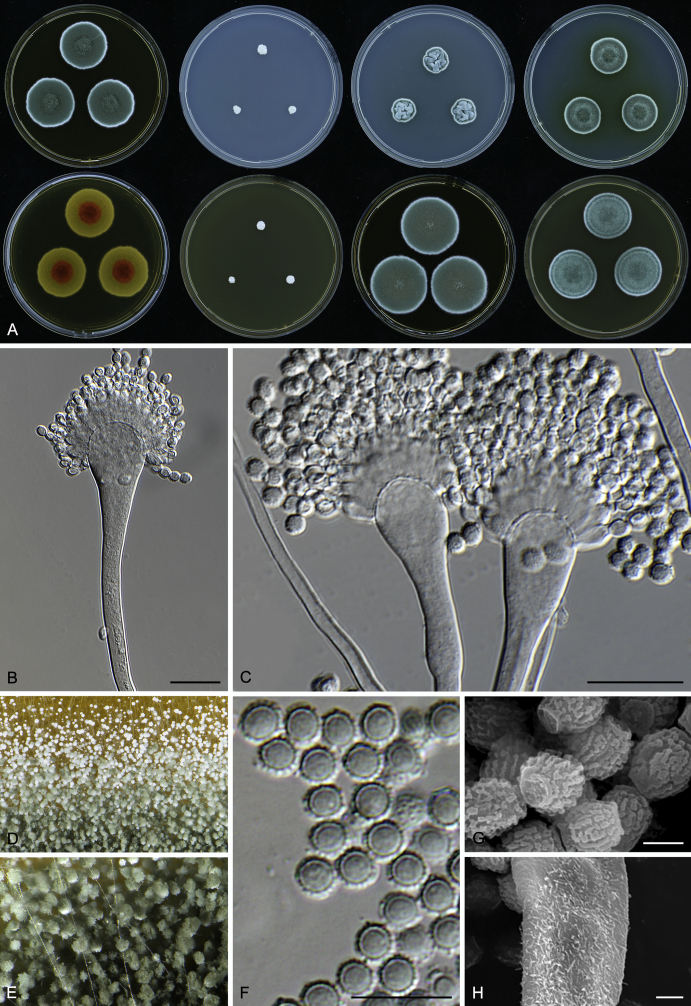
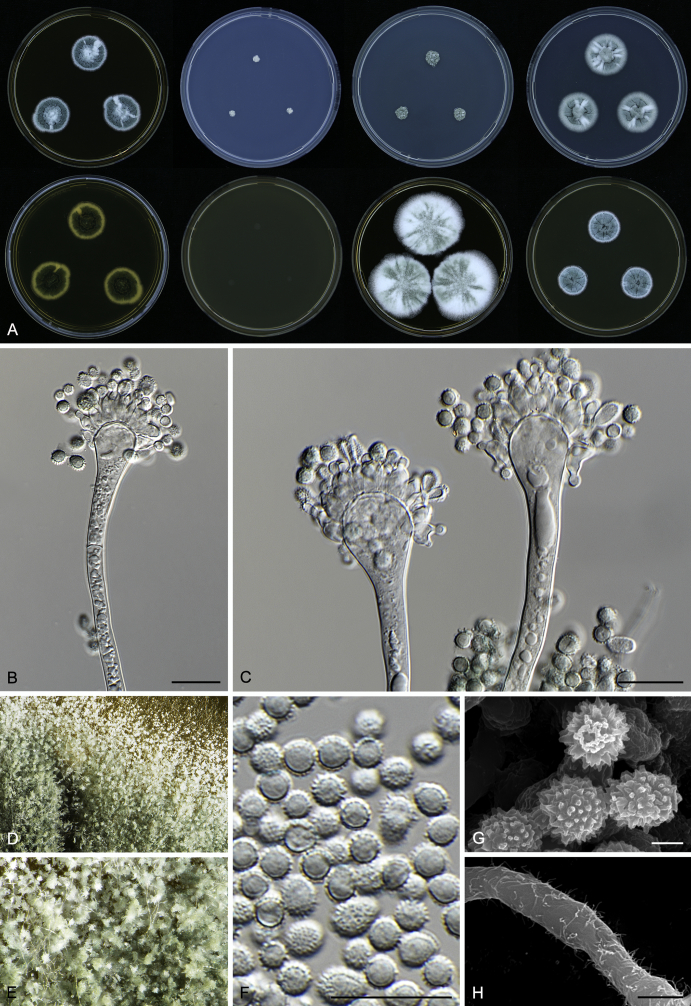
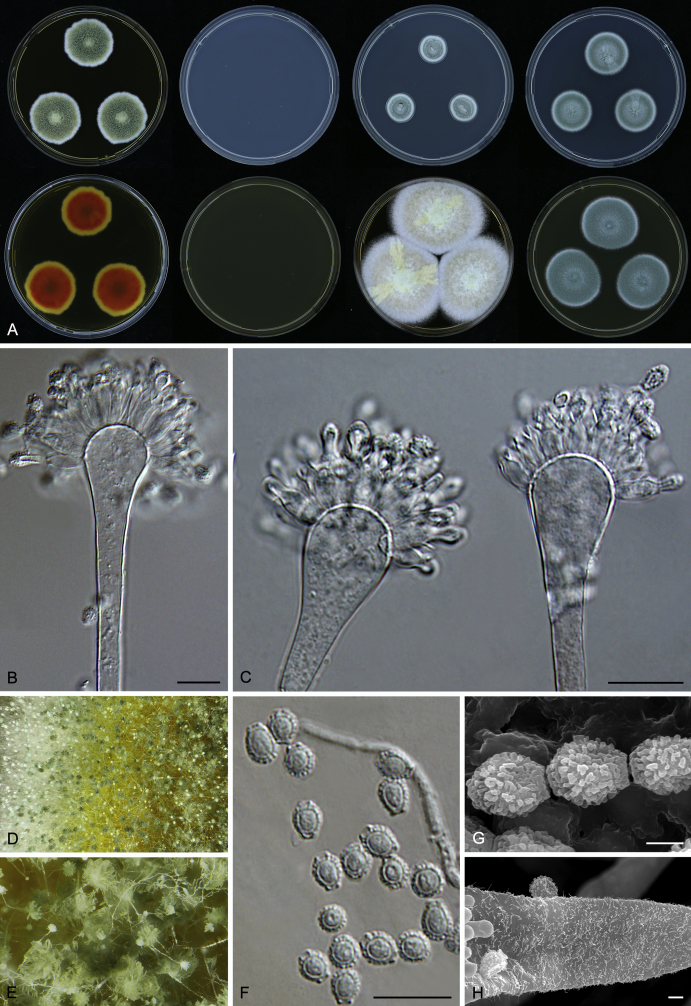
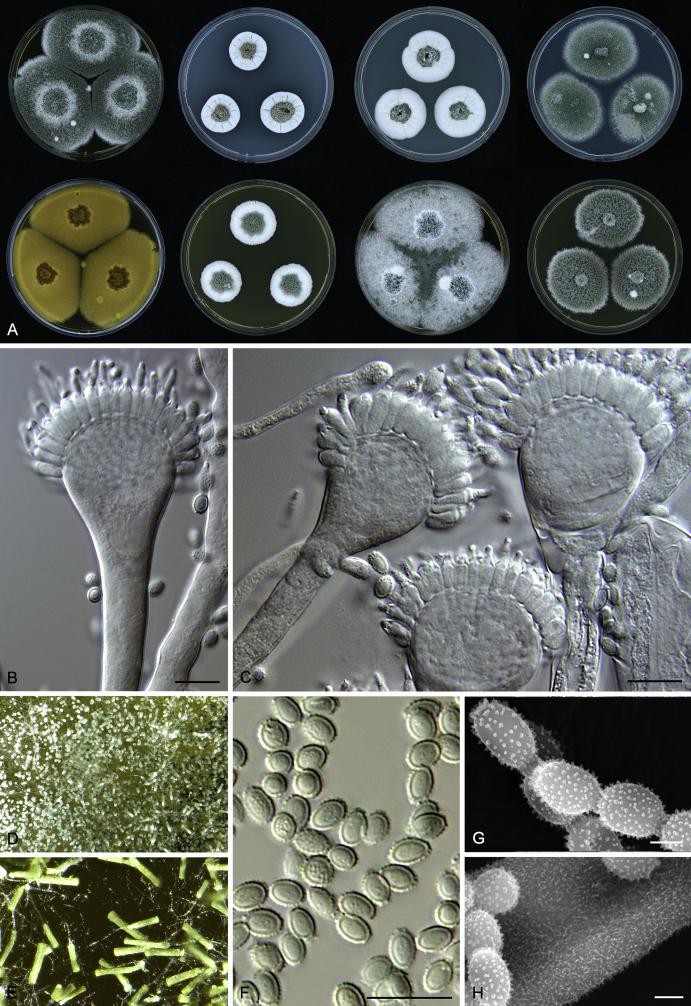
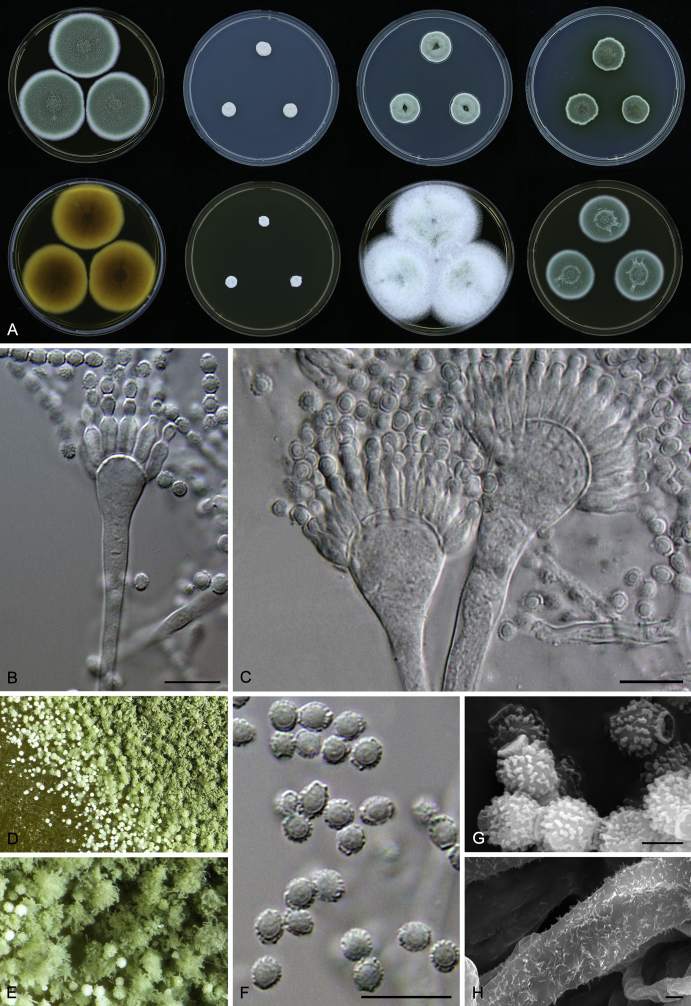
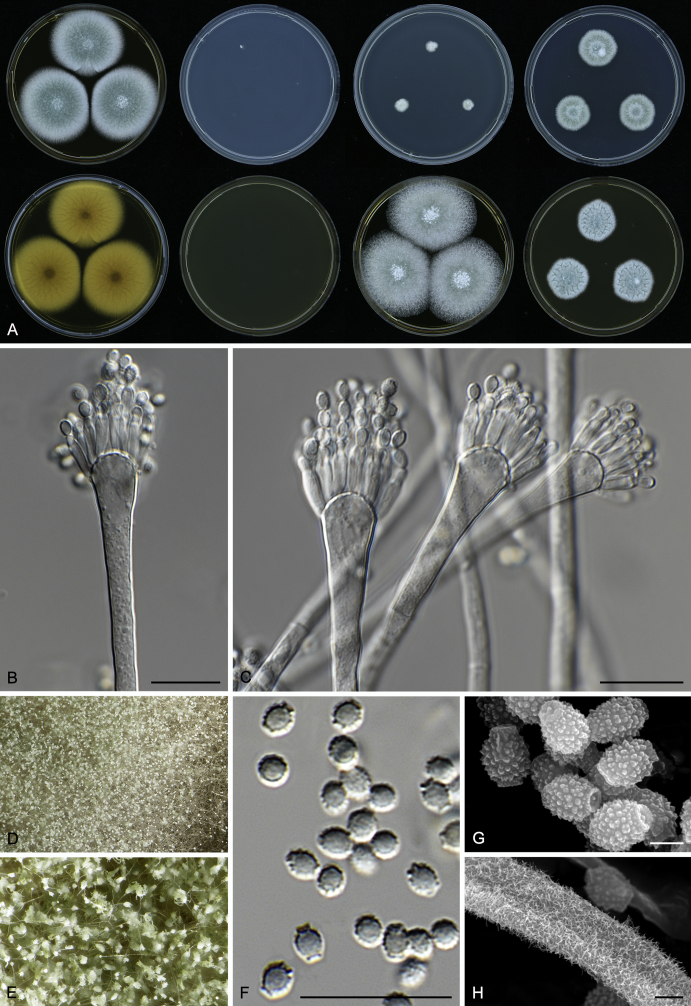
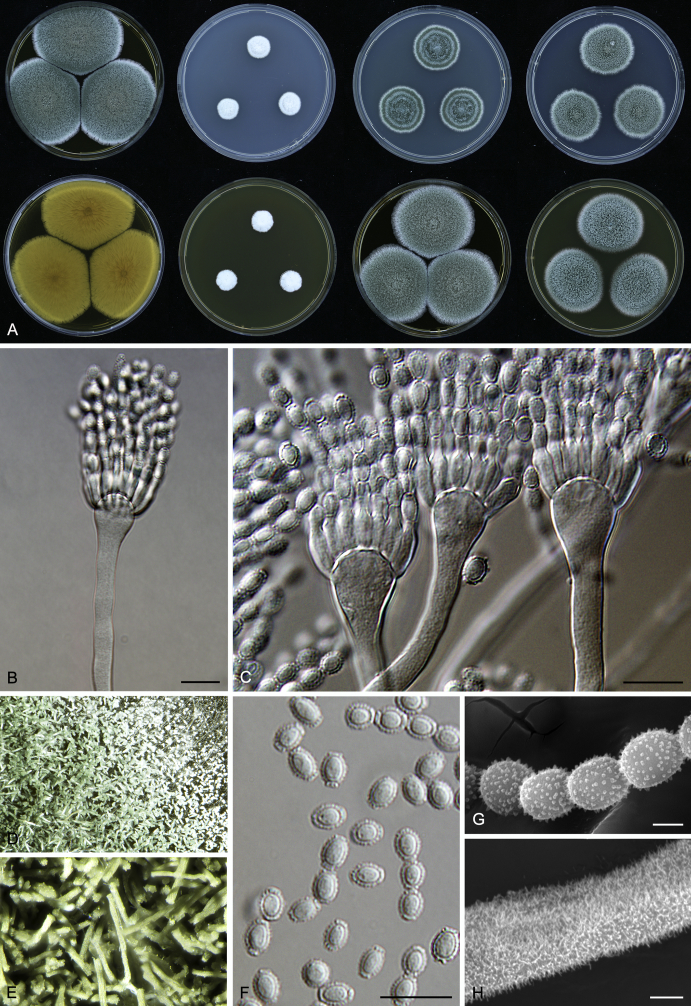
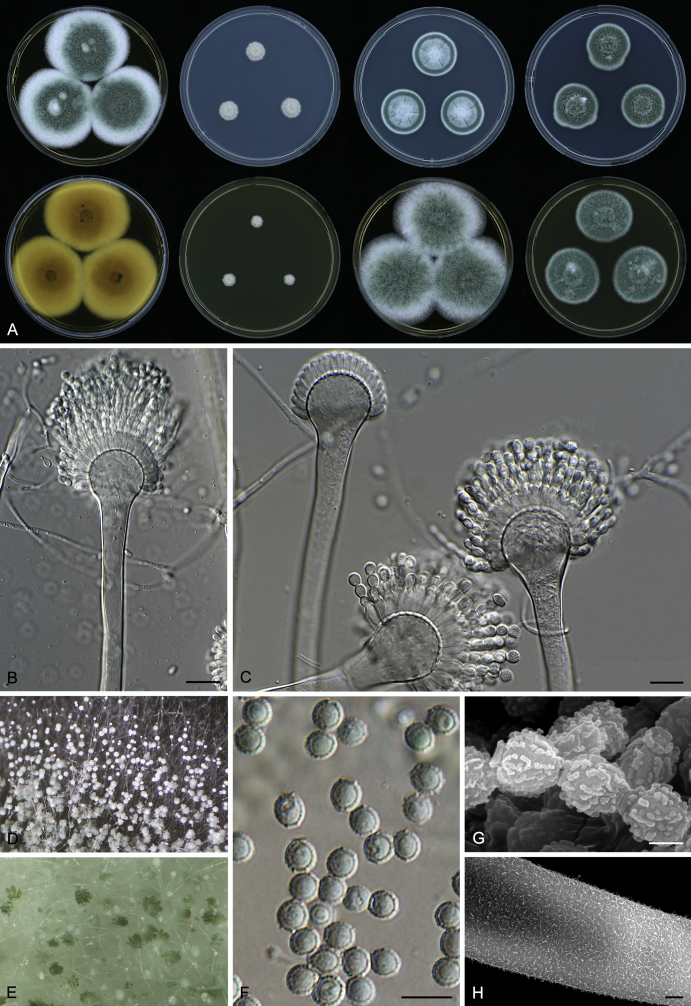
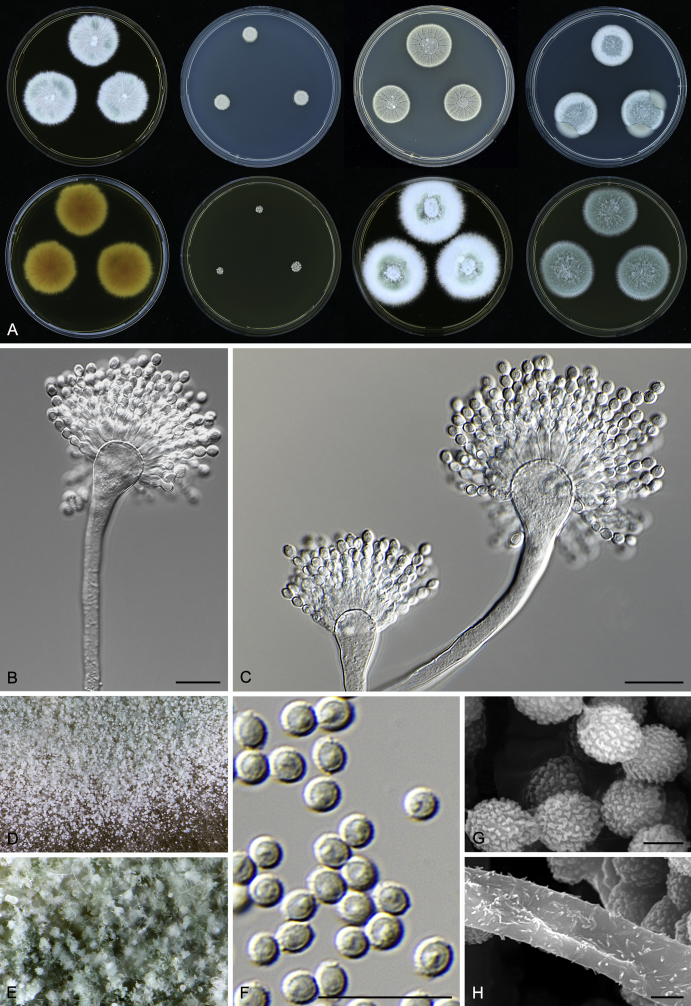
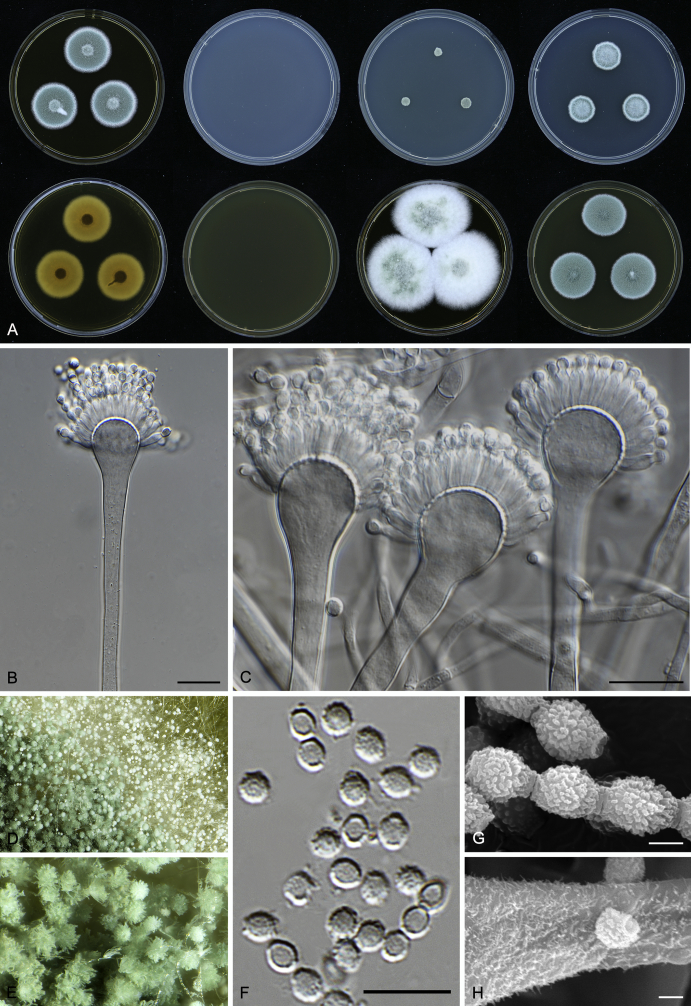
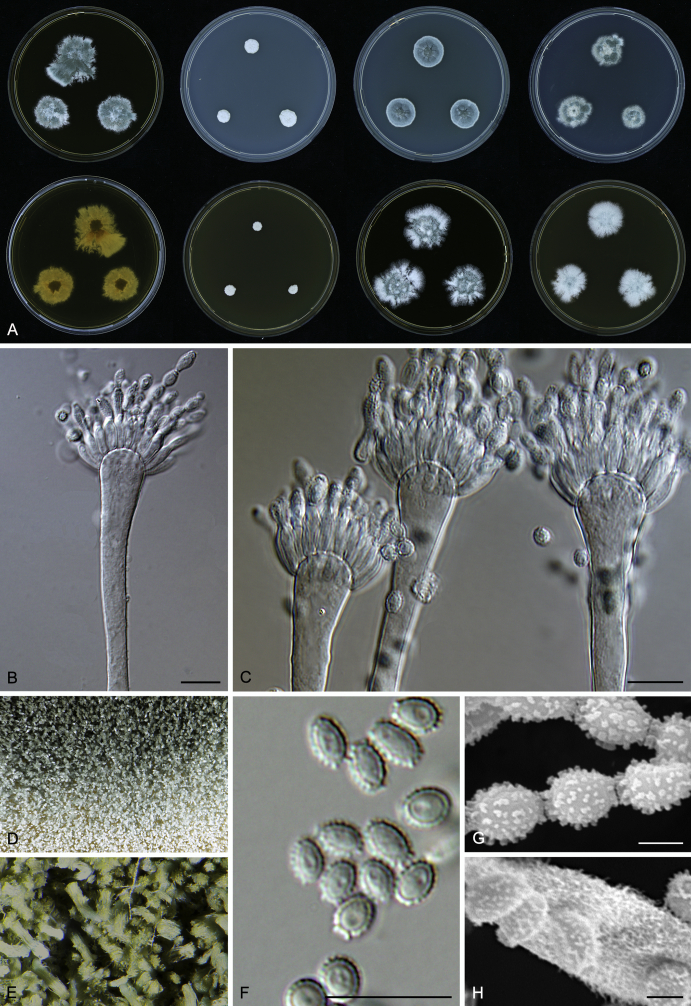
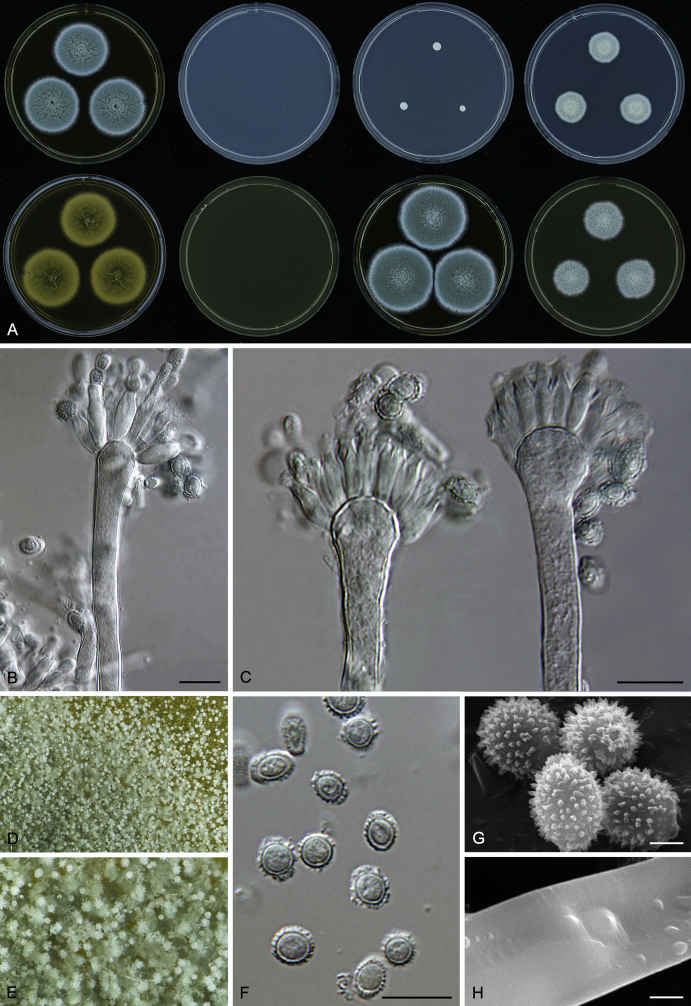
Macromorphological characters of colonies were observed on Harrold’s agar (M40Y) (Harrold 1950), Czapek yeast extract agar (CYA) (Pitt 1979), CYA supplemented with 20 % sucrose (CY20S) (Klich 2002), dichloran 18 % glycerol agar (DG18) (Hocking & Pitt 1980), malt extract agar (MEA; Oxoid) (Samson et al. 2010), Harrold's agar supplemented with 60 % Sucrose (M60Y) (Raper & Fennell 1965) and MEA supplemented with 10 % NaCl (MEA + 10 % NaCl). The isolates were inoculated in three points on 90 mm Petri dishes and incubated for 14 d at 25 °C in darkness. In addition, CY20S and M60Y plates were incubated at 30 °C and 37 °C. Colony diameters were measured after 14 d of incubation. The colony shape and texture, degree of sporulation, obverse and reverse colony colours, the production of soluble pigments and exudates were determined. The isolates of A. halophilicus were cultivated on Czapek–Dox agar (Thom & Church 1926) supplemented with 70 % sucrose (CZA70S) for 30 d. Colour names and codes used in descriptions refer to Kornerup & Wanscher (1967).
Light microscope preparations were made from 14 d old colonies grown on M40Y. Lactic acid (60 %) was used as mounting fluid and ethanol (96 %) used to remove excess conidia and prevent air bubble formation. Microphotography was done using an Olympus BX-51 microscope with an Olympus DP72 camera and Zeiss AX10 Imager A2 light microscope equipped with a Nikon DS-Ri2 camera. Macromorphology of the colonies was observed and captured on a Zeiss Stereo Discovery V20 dissecting microscope equipped with a Nikon DS-Ri2 camera. Pictures were processed and photographic plates prepared in Adobe Photoshop CS6. Micromorphological characters (length and width of conidia, width of stipes and vesicles and length of phialides) were measured from at least five isolates of each species (when available). Slides were prepared from both the colony centre and margins. Each character was recorded at least forty times for each character and isolate. Linear discriminant analysis was performed with measured data in R 3.3.1 (R Core Team 2015) with packages MASS (Venables & Ripley 2002) and ggplot2 (Wickham 2009). The isolates were assigned to groups based on the results of molecular phylogenetic analyses (see above).
Scanning electron microscopy (SEM) was performed using a JEOL-6380 LV microscope (JEOL Ltd. Tokyo, Japan) as described previously (Hubka et al. 2013).
In brief, plugs from colonies (5 × 5 mm) grown 14 d on M40Y containing conidiophores, and ascomata in the case of A. halophilicus (longer incubation on CZA70S was necessary) were fixed in osmium tetroxide vapours for 2 wk at 5–10 °C and gold-coated in a Bal-Tec SCD 050 sputter coater. The specimens were observed with spot size 40–42 μm and accelerating voltage 25 kV. Terminology of the surface ornamentation of the conidia was adopted from Kozakiewicz (1989).
Physiology
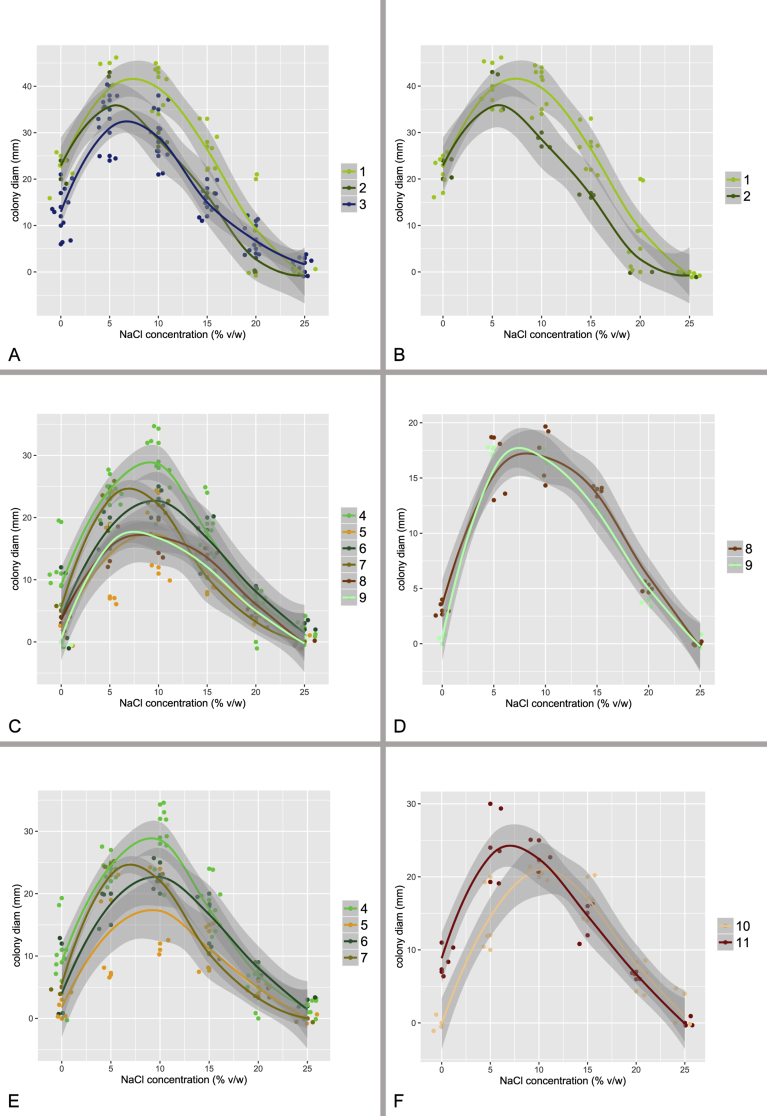
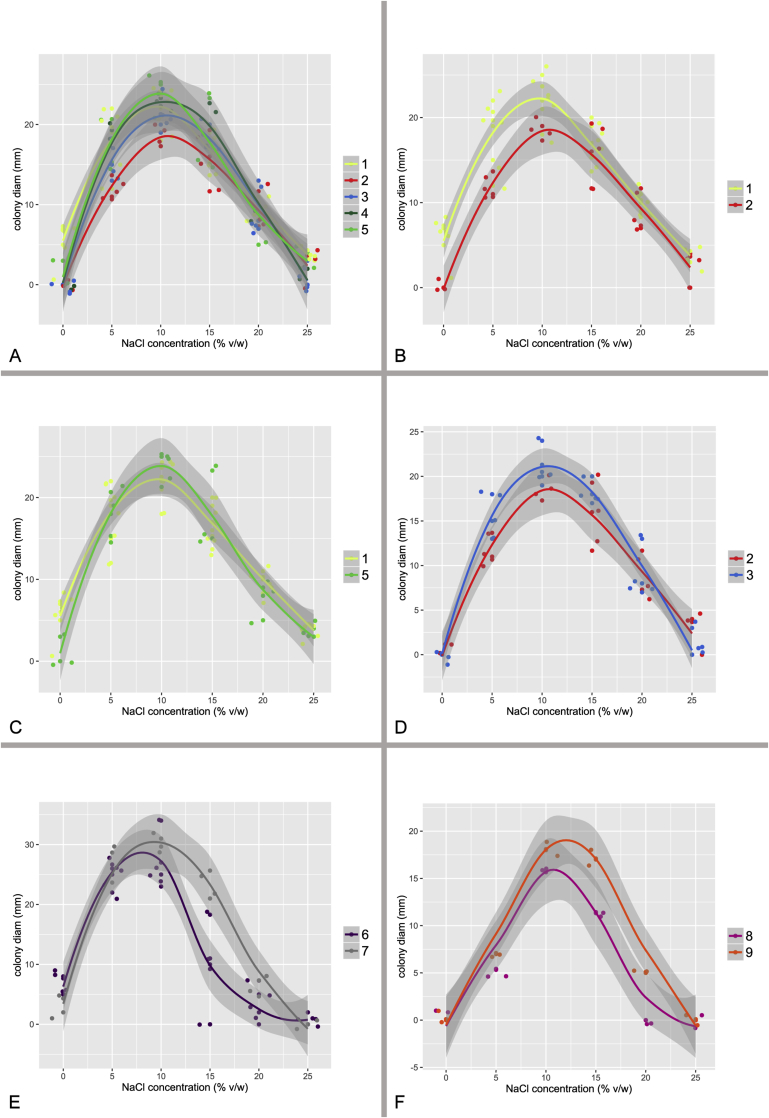
At least five isolates from each species, when available, were selected for determining species growth rates in an osmotic gradient. Each strain was cultivated at 25 °C on MEA with six different concentrations of NaCl ranging from 0 to 25 %. After 14 d, colony sizes were recorded and growth curves for each species calculated using local regression (LOESS) in R v. 3.3.1. (R Core Team 2015) using the ggplot2 package (Wickham 2009).
Exometabolite analysis
The isolates of Aspergillus sect. Restricti were incubated on DG18, CY20S and yeast extract sucrose agar (YES) agar, for 2 wk at 25 °C. Two agar plugs from each medium (6 plugs in total) were combined in one vial and extrolites extracted with ethylacetate / isopropanol (3:1) with added 1 % formic acid, and ultrasonication (50 min). In the case of A. halophilicus, six CZA70S plugs containing ascomata (ca 1–2 mo old colonies) were extracted. After ultrasonication, the plugs were removed and the organic solvent evaporated. The remaining extract was re-dissolved in methanol, centrifuged at 13 300 rpm and transferred to a small vial with a V-formed insert. HPLC analysis was done according to Frisvad & Thrane (1987) as modified by Nielsen et al. (2011).
Results
Phylogeny of subgenus Aspergillus
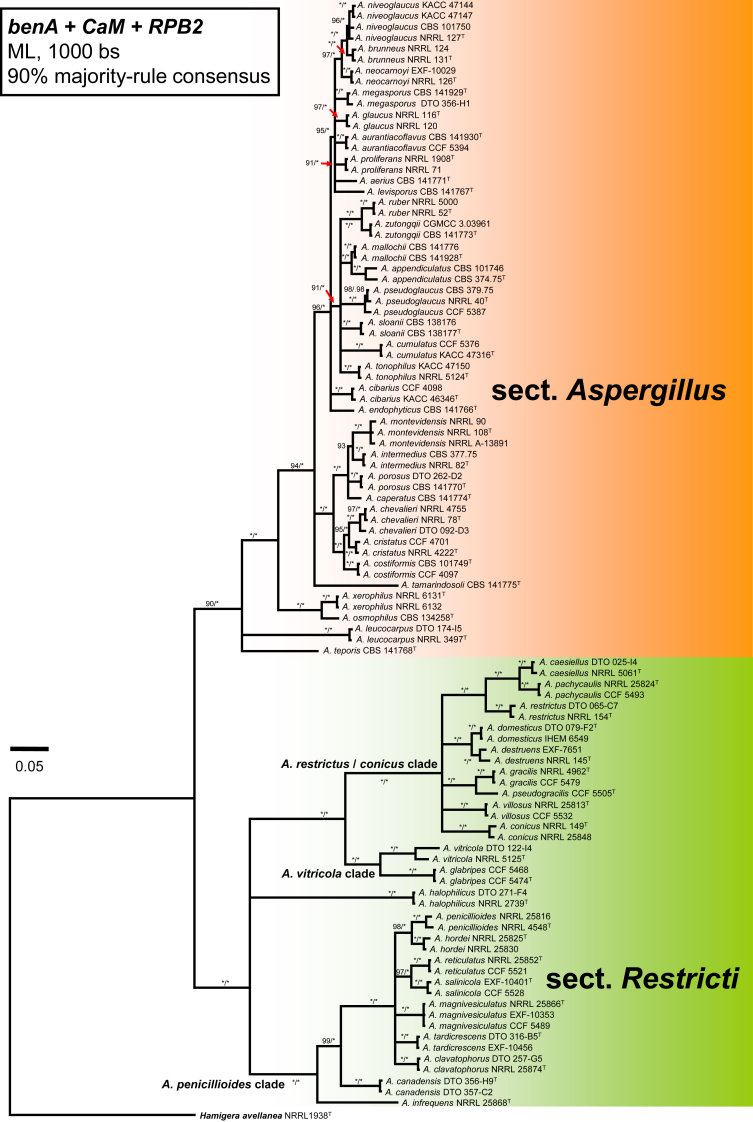
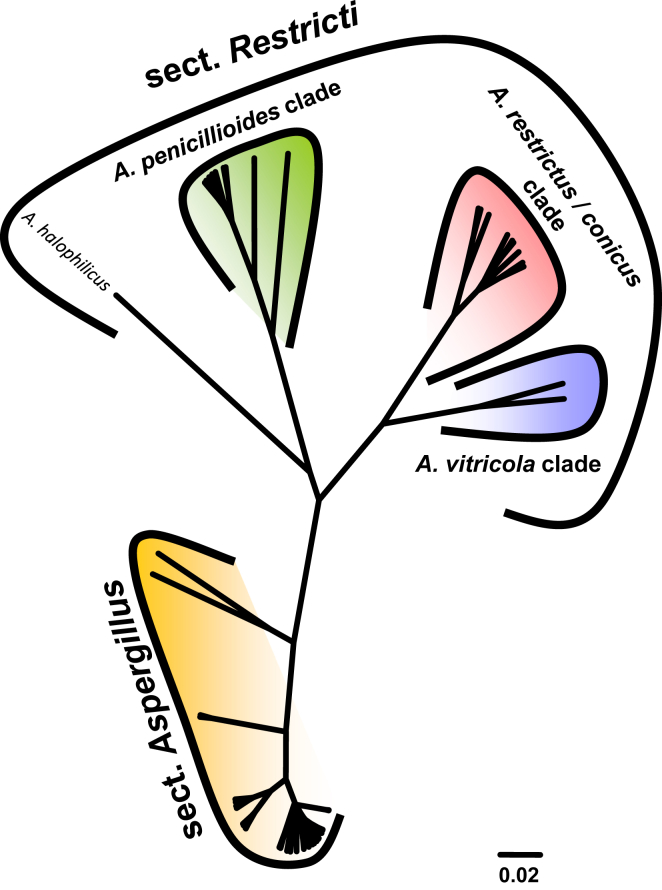
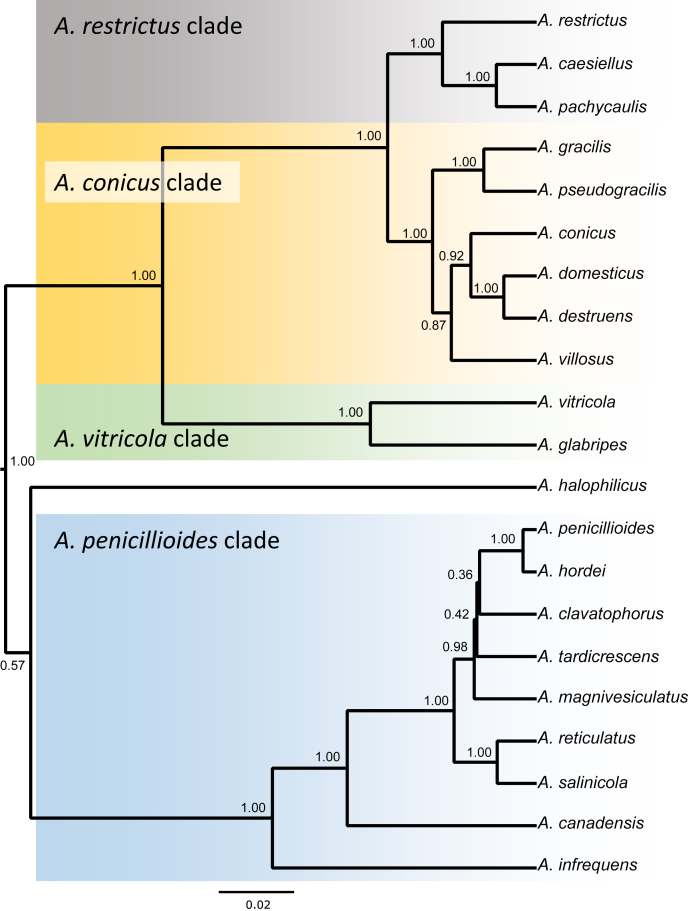
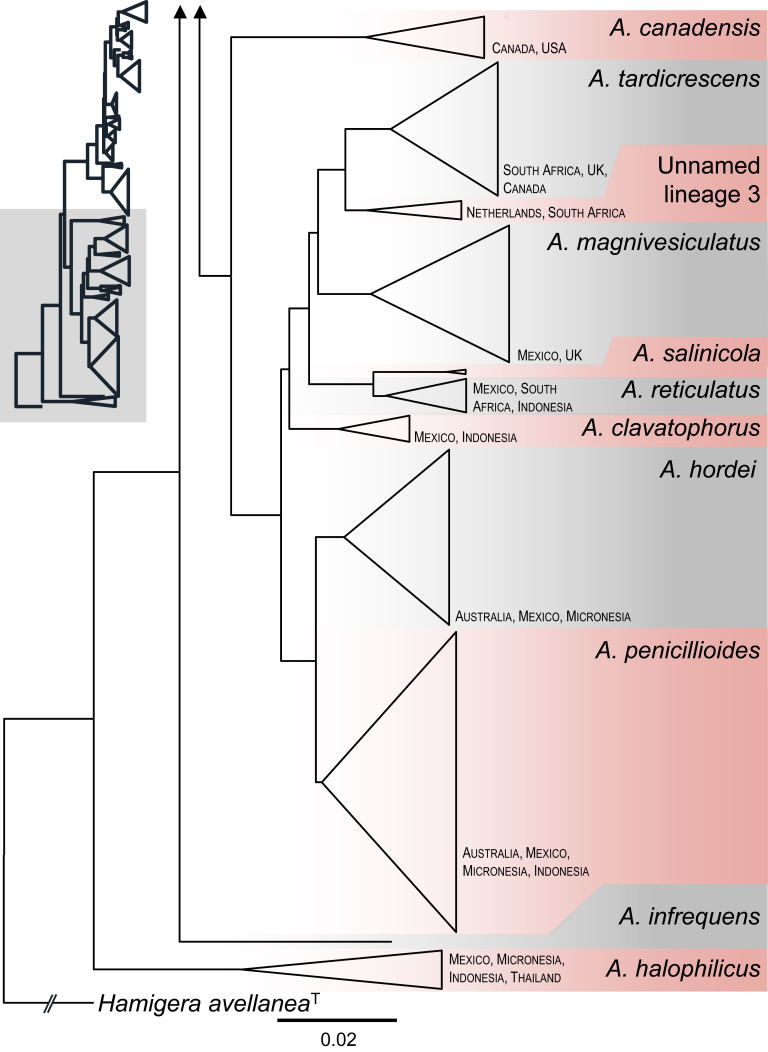
ML and BI analysis of 102 concatenated sequences of benA, CaM and RPB2 contained 31 species from sect. Aspergillus as recognised by Chen et al. (2017) and 21 species from sect. Restricti recognised here (see below). Tree topologies between ML and BI did not differ and the ML tree was used with both bootstrap and pp values included (Fig. 1, Fig. 2). Both analyses supported the monophyly of both sections. Each section contains several highly supported clades, but the exact position of species within the clades is often unresolved. Despite producing a eurotium-like sexual state common in sect. Aspergillus, A. halophilicus is resolved with high statistical support in sect. Restricti, but its exact position remains unclear. It is apparent from the radial representation of the tree (Fig. 2), that there are large genetic distances between the different clades of sect. Restricti, but these gaps may represent only hidden variability that has not been discovered during our study due to insufficient sampling or the use of inappropriate isolation media. The retaining of the current classification scheme of subg. Aspergilllus with two sections seems currently the best solution until more data on species diversity in sect. Restricti are collected. Additionally, sects. Restricti and Aspergillus are well supported by phenotypic data (see discussion).
Fig. 1.
A 90 % majority consensus tree of the subgenus Aspergillus inferred with Maximum likelihood analysis based on benA, CaM and RPB2 loci (partitioning scheme and substitution models are listed in Table 3). The data set contained 102 strains and 1902 characters, of which 929 characters were variable and 864 were parsimony informative. Support values represent maximum likelihood bootstrap/ Bayesian posterior probability values, 100 % bootstrap values and 1.00 posterior probability are designated by asterisk *. The ex-type isolates are designated by a superscript T. Hamigera avellanea (NRRL 1938) was used as outgroup.
Fig. 2.
Maximum likelihood phylogenetic tree of the subgenus Aspergillus inferred from partitioned analysis of concatenated dataset (benA, CaM and RPB2) with Maximum likelihood method and presented in radial format.
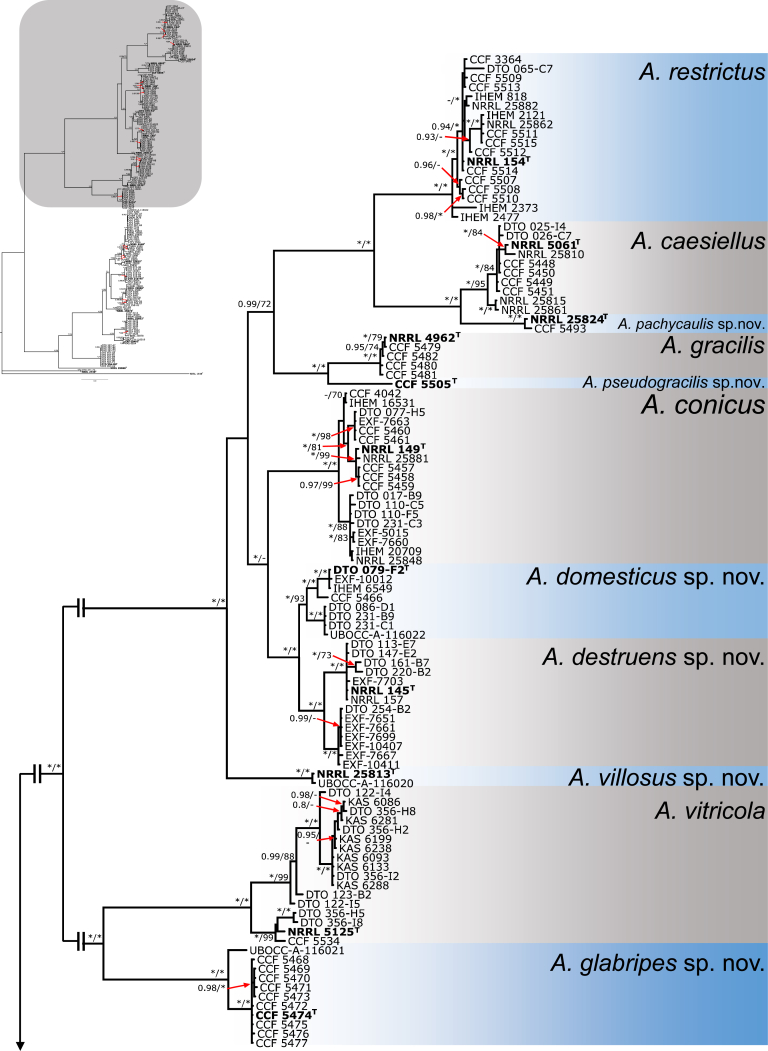
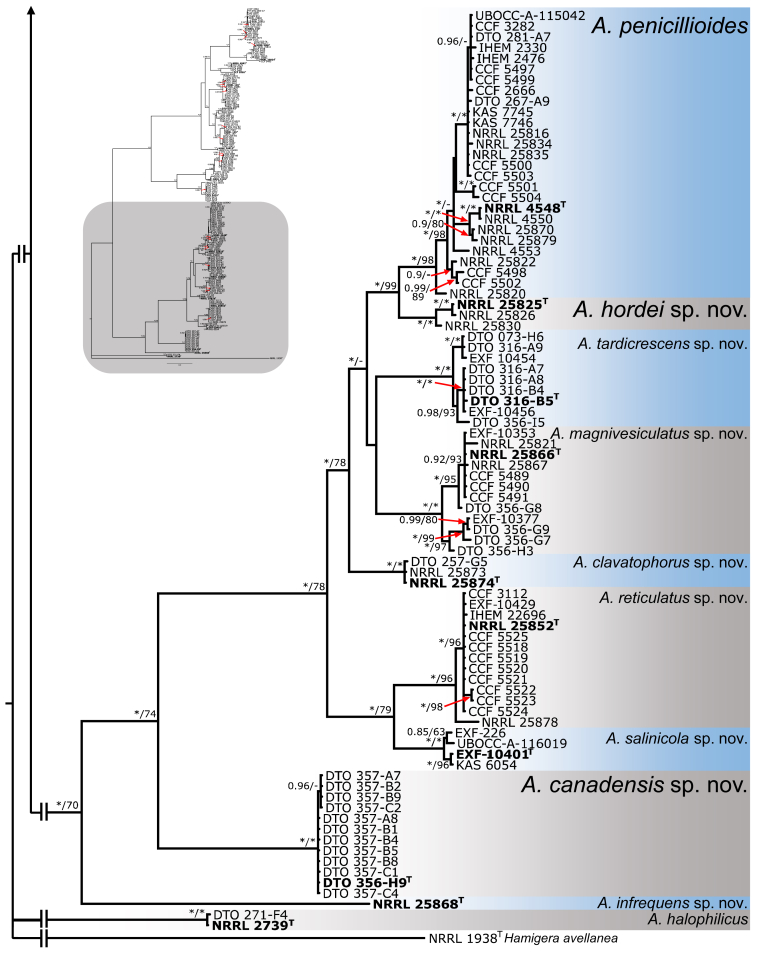
Species delimitation and validation in sect. Restricti
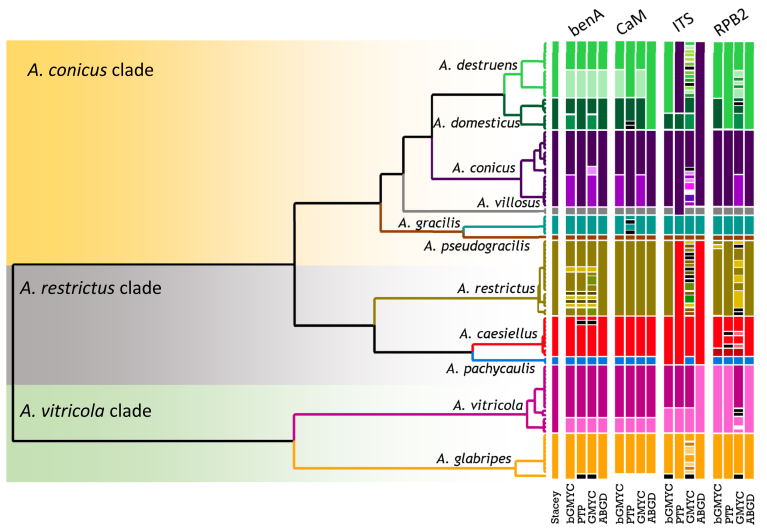
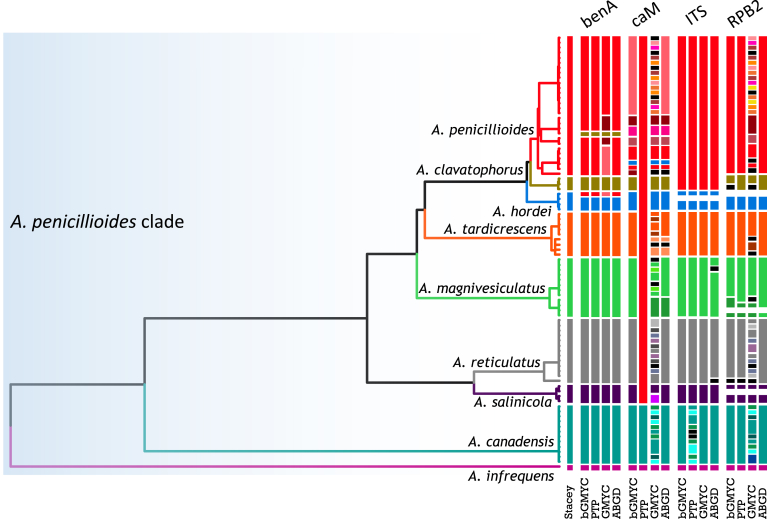
For species delimitation, the alignment was divided into two parts as discussed earlier. Eleven species were delimited within the first part that consisted of the A. restrictus, A. conicus and A. vitricola clades using STACEY and similarly, nine species were delimited within the second part of the data set (A. penicillioides clade). Aspergillus halophilicus was exluded from species delimitation analyses because it clearly represents a distantly related clade within the section. Results are summarised in Fig. 3, Fig. 4. Tree topologies in the Fig. 3, Fig. 4 were inferred in STACEY and used solely for the comprehensive presentation of the results from different methods; the evolutionary relationships in the section inferred by *BEAST are presented as the most robust (Fig. 5). In the first step, we compared results from four single-locus species delimitation methods with those derived from STACEY, that is currently one of the most advanced species delimitation methods because it processes multiple loci simultaneously during a single analysis (Jones 2017). Although the results vary across the methods and loci, the consensual results from single-locus species delimitation methods are generally in agreement with the results of STACEY. Single-locus method bGMYC was the most computationally intensive method among those used in this study and its results were most similar to STACEY. The method with greatest variability across the four loci was GMYC with 14 to 39 delimited species for the first part of the analysed data set (A. restrictus, A. conicus and A. vitricola clades) and eight to 38 species for A. penicillioides clade. A significant over delimitation was observed when analysing the ID region of the first part of the data set and also CaM and RPB2 loci using GMYC method (Fig. 3). A similar problem was observed in the case of CaM when analysing the second part of the data set and also in RPB2 locus when using GMYC method (Fig. 4). Although ABGD is a quite simplistic method compared to other used methods it yielded similar results to STACEY and bGMYC but over delimitation was observed when analysing the CaM locus in the A. penicillioides clade (Fig. 3, Fig. 4). The number of species delimited by PTP was slightly higher compared to bGMYC, but lower than in the case of GMYC. Single-locus methods often delimited additional species within A. conicus and A. vitricola, but the results were not consistent and in some cases even contradictory, suggesting recombination within the clade (Fig. 3). These tentative species had no or very limited phenotypic support, which is the reason for adopting a broader species concept. In contrast, the majority of single-locus methods did not support delimitation of A. clavatophorus and A. pachycaulis based on the ID region; the methods also did not support recognition of A. destruens and A. domesticus when analysing ID-region and RPB2 in contrast to benA and CaM. All these species were supported by STACEY and phenotype analysis, resulting in us proposing them as new species.
Fig. 3.
Schematic representation of results of species delimitation methods in A. restrictus, A. conicus and A. vitricola clades (108 isolates). The results of multilocus method (STACEY) are compared to results of single-locus methods (bGMYC, PTP, GMYC, ABGD). Results from different methods are depicted with coloured bars highlighting congruence across methods. Displayed tree comes from STACEY analysis and is used solely for the comprehensive presentation of the results from different methods.
Fig. 4.
Schematic representation of results of species delimitation methods in A. penicillioides clade (86 isolates). The results of multilocus method (STACEY) are compared to results of single-locus methods (bGMYC, PTP, GMYC, ABGD). Results from different methods are depicted with coloured bars highlighting congruence across methods. Displayed tree comes from STACEY analysis and is used only used solely for the comprehensive presentation of the results from different methods.
Fig. 5.
Bayesian species tree based on sequence data from four loci of 193 isolates inferred by *BEAST with posterior probabilities appended to nodes. Terminal branches represent delimited species (each comprises all isolates of respective species).
Based on consensus results of species delimitation methods and after reflection of phenotypic data in ambiguous species, we recognise 21 species within the sect. Restricti. This number comprises seven previously recognised and 14 new species proposed here (see section Taxonomy). All four loci have sufficient variability for reliable species identification and can be used as DNA barcodes. ID region has the lowest discriminative power but it is still sufficient for differentiation of all species. The locus with the highest ratio of variable positions to the sequence length was benA.
The species validation analysis results are listed in Table 5. All species were supported by the posterior probability 1.00 based on the analysis in BP&P v. 3.1 (Yang & Rannala 2010) under all three scenarios simulated by different prior distributions of parameters ϑ (ancestral population size) and τ0 (root age). The gsi calculations and significance testing performed using the genealogical sorting index software also confirmed that all delimited species can be considered separate evolutionary lineages. The ensemble statistic gsiT (weighted sum of gsi across genealogies) for each dataset (100 bootstrap trees for each locus and concensus trees) are listed in Table 5. Relatively low gsi value was obtained from the analysis of β-tubulin dataset in A. hordei and ID dataset in A. domesticus, however even in these cases the p-value of the permutation test is nearly 0. The gsi statistic was not calculated for A. infrequens and A. pseudogracilis because they were both represented by only one isolate.
Table 5.
Ensemble statistic (gsiT) and p-value of the genealogical sorting index calculation.
| Species |
benA |
CaM |
ITS |
RPB2 |
Consensus trees |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| gsiT | p-value | gsiT | p-value | gsiT | p-value | gsiT | p-value | gsiT | p-value | |
| A. restrictus clade | ||||||||||
| A. restrictus | 0.997 | 0.0001 | 0.989 | 0.0001 | 0.898 | 0.0001 | 0.999 | 0.0001 | 0.983 | 0.0001 |
| A. caesiellus | 1 | 0.0001 | 0.959 | 0.0001 | 0.947 | 0.0001 | 0.959 | 0.0001 | 1 | 0.0001 |
| A. pachycaulis | 1 | 0.0003 | 1 | 0.0003 | 0.942 | 0.0001 | 1 | 0.0009 | 1 | 0.0005 |
| A. conicus clade | ||||||||||
| A. conicus | 0.999 | 0.0001 | 0.994 | 0.0001 | 0.914 | 0.0001 | 0.979 | 0.0001 | 1 | 0.0001 |
| A. destruens | 0.997 | 0.0001 | 1 | 0.0001 | 0.894 | 0.0001 | 0.991 | 0.0001 | 1 | 0.0001 |
| A. domesticus | 0.98 | 0.0001 | 0.897 | 0.0001 | 0.707 | 0.0001 | 0.891 | 0.0001 | 0.933 | 0.0001 |
| A. gracilis | 1 | 0.0001 | 1 | 0.0001 | 0.946 | 0.0001 | 1 | 0.0001 | 1 | 0.0001 |
| A. villosus | 1 | 0.0005 | 1 | 0.0007 | 0.952 | 0.0001 | 1 | 0.0006 | 1 | 0.0007 |
| A. vitricola clade | ||||||||||
| A. vitricola | 0.942 | 0.0001 | 1 | 0.0001 | 0.958 | 0.0001 | 0.995 | 0.0001 | 0.981 | 0.0001 |
| A. glabripes | 0.999 | 0.0001 | 1 | 0.0001 | 0.913 | 0.0001 | 0.979 | 0.0001 | 0.975 | 0.0001 |
| A. penicillioides clade | ||||||||||
| A. penicillioides | 0.871 | 0.0001 | 0.983 | 0.0001 | 0.948 | 0.0001 | 0.995 | 0.0001 | 0.960 | 0.0001 |
| A. canadensis | 0.987 | 0.0001 | 1 | 0.0001 | 1 | 0.0001 | 1 | 0.0001 | 1 | 0.0001 |
| A. clavatophorus | 1 | 0.0001 | 0.980 | 0.0001 | 1 | 0.0005 | 1 | 0.0001 | 1 | 0.0003 |
| A. hordei | 0.675 | 0.0001 | 1 | 0.0001 | 0.948 | 0.0001 | 0.997 | 0.0001 | 0.915 | 0.0001 |
| A. magnivesiculatus | 0.997 | 0.0001 | 0.974 | 0.0001 | 0.898 | 0.0001 | 0.998 | 0.0001 | 1 | 0.0001 |
| A. reticulatus | 0.995 | 0.0001 | 0.995 | 0.0001 | 0.930 | 0.0001 | 1 | 0.0001 | 0.977 | 0.0001 |
| A. salinicola | 0.971 | 0.0001 | 0.997 | 0.0001 | 0.987 | 0.0001 | 1 | 0.0003 | 1 | 0.0001 |
| A. tardicrescens | 1 | 0.0001 | 0.933 | 0.0001 | 1 | 0.0001 | 0.985 | 0.0001 | 1 | 0.0001 |
Species tree
The species tree topology was inferred with *BEAST (Heled & Drummond 2010) and is depicted on Fig. 5, Fig. 6. It was used as a guide tree for species validation in BP&P but it also represents the most probable evolutionary relationships between the species. Four clusters of species designated as the A. restrictus, A. conicus, A. vitricola and A. penicillioides clades are denoted in Fig. 5. We observed only slight differences in the topology of the species tree and statistical support of some nodes inferred with *BEAST when compared to the results of ML and BI analysis of the concatenated and partitioned dataset (see below). Several nodes had only limited support in all types of analyses and there is definitely some degree of uncertainty. These uncertainties can be visualized in DensiTree (Bouckaert 2010) (Fig. 6), that displays all trees created during the analysis except burn-in phase and trees with one of the three most common topologies are differently coloured. The most obvious problem is the position of A. halophilicus. Based on the available sequence data it is not clear whether it is situated basally within sect. Restricti or if it is rather phylogenetically related to some clade within the section. In addition, *BEAST analysis supported (PP = 1.00) an A. conicus clade consisting of six species, A. conicus, A. domesticus, A. destruens, A. villosus, A. gracilis and A. pseudogracilis, however this arrangement was not supported by the concatenated dataset analysis (see Fig. 7) where A. gracilis and A. pseudogracilis formed a distinct clade from the remaining four species. The position of A. villosus within the clade is also not fully resolved and differs between phylogenetic methods (compare Figs 5 and 7). The topology of the A. penicillioides clade was almost identical when using all methods. The exception is an unresolved position of A. clavatophorus and A. tardicrescens. The respective nodes gained only limited support while all other nodes are suported by PP = 1.00 in *BEAST analysis.
Fig. 6.
Species tree inferred with *BEAST visualized by using DensiTree (Bouckaert 2010). All trees created in the analysis (except burn-in phase) are displayed. Trees with the most common topology are highlighted by blue colour, trees with the second most common topology by red colour, trees with the third most common topology by pale green and all other trees by dark green.
Fig. 7.
Bayesian tree depicting the relationship of species from Aspergillus section Restricti based on four loci (partitioning scheme and substitution models are listed in Table 3). Total length of the concatenated alignment was 2093 characters, with 837 variable and 668 parsimony informative sites. Support values represent Bayesian posterior probability/maximum likelihood bootstrap values, 100 % bootstrap values and 1.00 posterior probability are designated by asterisk *. The ex-type isolates are designated by bold font and superscript T. Hamigera avellanea (NRRL 1938) was used as outgroup.
Bayesian and Maximum likelihood analysis of the partitioned sequence data
All species delimited by methods based on coalescent model were clearly supported by BI and ML analyses (Fig. 7). All clades representing species were supported by PP = 1.00 in BI analysis and at least 93 % bootstrap support in ML analysis. Aspergillus halophilicus was in a polytomy on the base of the section in agreement with the *BEAST analysis. Slight differences were observed in the interspecies relationships between core species of A. penicillioides as mentioned above. The ML and BI analysis of partitioned dataset did not separate A. restrictus and A. conicus clades as did the *BEAST analysis (compare Figs 5 and 7).
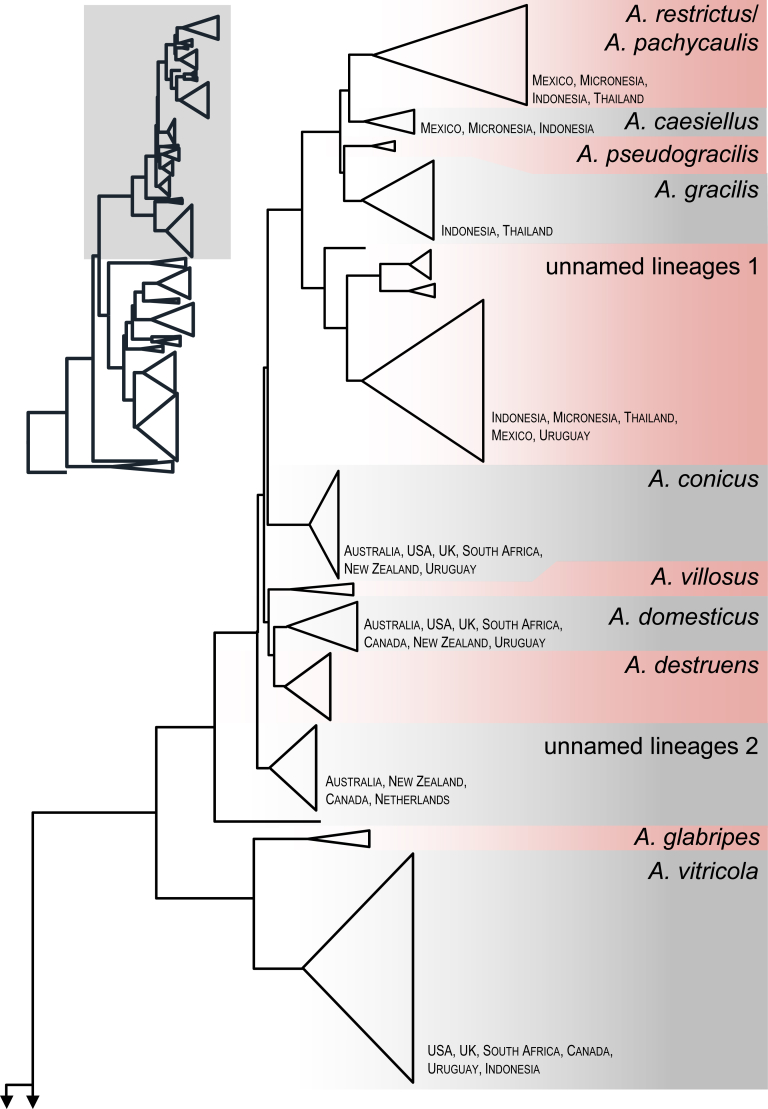
Comparisons with 454 data
A total of 1061 454-pyrosequences were found to belong to sect. Restricti and included in a dataset with 188 ITS barcodes generated during this study. The aligned dataset was 402 bp long and the calculated neighbour-joining tree is shown in Fig. 8. Many species were detected from dust samples, while A. pseudogracilis, A. villosus, A. destruens, A. glabripes, A. salinicola and A. infrequens were not detected. The phylogeny revealed 3–5 unnamed lineages from dust collected from Indonesia, Mexico, Micronesia, the Netherlands, South Africa, Thailand and Uruguay. Two singletons were also detected across the tree. Interestingly, each lineage is represented by OTU's from multiple countries, meaning they are probably common species.
Fig. 8.
Neighbour Joining tree created in MEGA 7 with 1000 bootstrap replicates based on ITS from 454-pyrosequences obtained from house dust samples (n = 1061; Amend et al. 2010) and from this study (n = 188). The tree is rooted with Hamigera avellanea. Monophyletic groups/species are collapsed and shown as proportional triangles. The information about country of origin pertains to and is given only for species found in house dust.
Phenotype analysis
Micromorphology
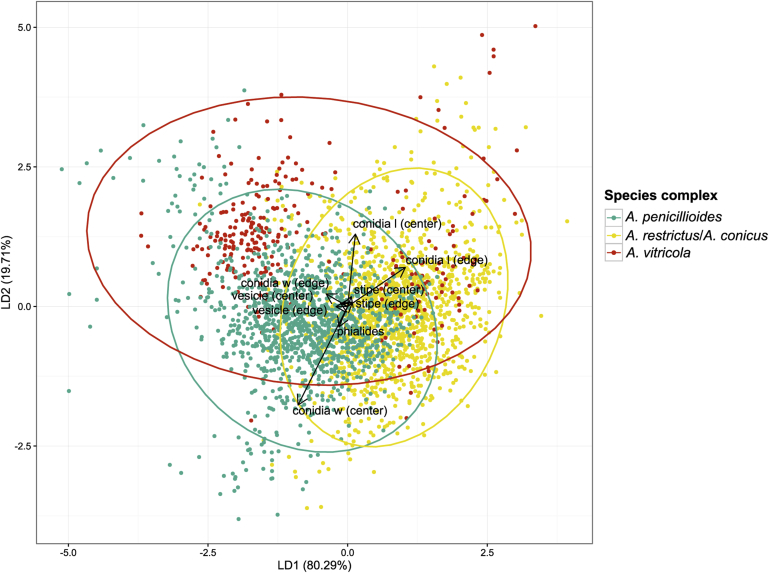
Results of linear discriminant analysis (LDA) based on measurements of micromorphological characters (length and width of conidia, width of stipe and vesicle from the centre and edge of the colony and length of phialides) are shown on Fig. 9, Fig. 10, Fig. 11. The values of individual phenotypic characters (average value ± standard deviation) for each species can be found in Table 6. The results of the analysis with individuals assigned to the species complexes are depicted on Fig. 9. While species from A. penicillioides complex and A. restrictcus/conicus complex form well-defined and relatively separated clusters, individuals from the A. vitricola complex formed two distinct clusters. These two clusters correspond to the two micromorphologically dissimilar species within the complex, i.e., A. vitricola and A. glabripes. It is also apparent from the analysis, that the most important character discriminating species complexes is the conidial size.
Fig. 9.
Results of linear discriminant analysis (LDA) performed in R 3.3.1 based on micromorphological measurements of all individuals assigned into species complexes. Ellipses represent 95 % confidence interval and arrows represent the contribution of each character to the axes. The length (l) and width (w) of conidia, diameter of stipes and vesicles were measured separately from the center and the edge of colonies.
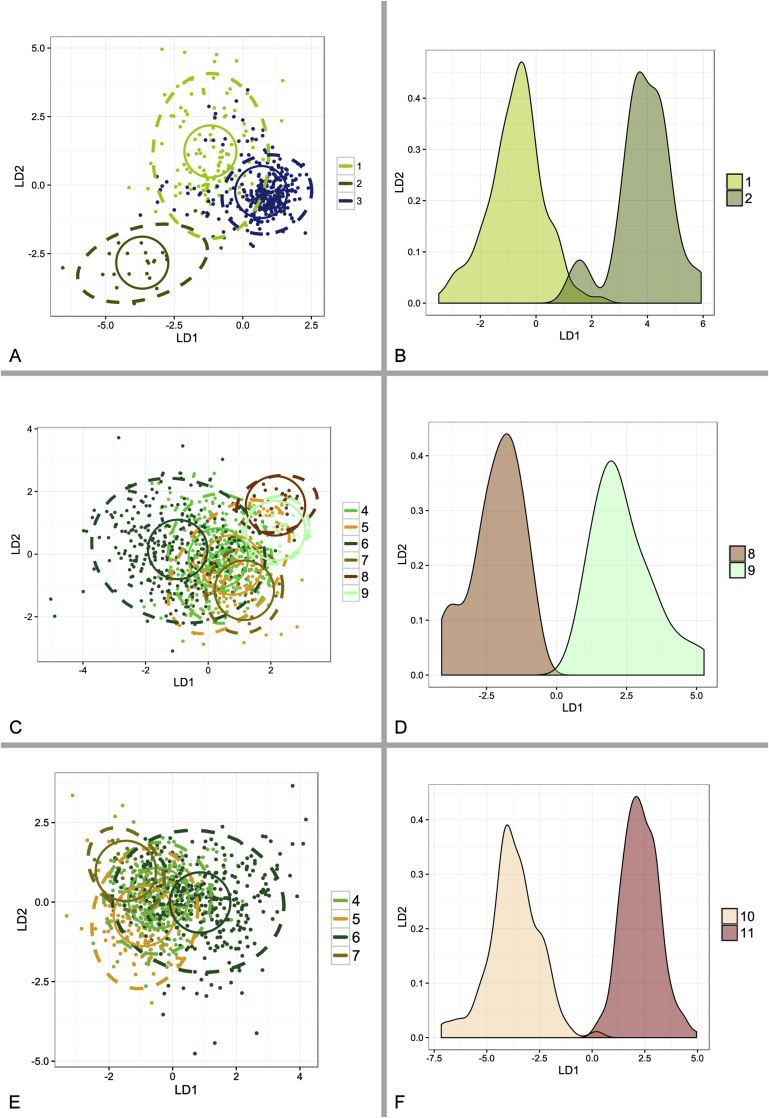
Fig. 10.
Results of linear discriminant analysis (LDA) performed in R 3.3.1 based on micromorphological measurements of species from A. restrictus, A. conicus and A. vitricola clade. Dashed ellipses represent 95 % confidence interval, full circles euclidean distances. Analyses involving only two species (B, D, F) are represented by probability density function. 1 – A. restrictus; 2 – A. caesiellus; 3 – A. pachycaulis; 4 – A. conicus; 5 – A. domesticus; 6 – A. destruens; 7 – A. villosus; 8 – A. gracilis; 9 – A. pseudogracilis; 10 – A. vitricola; 11 – A. glabripes.
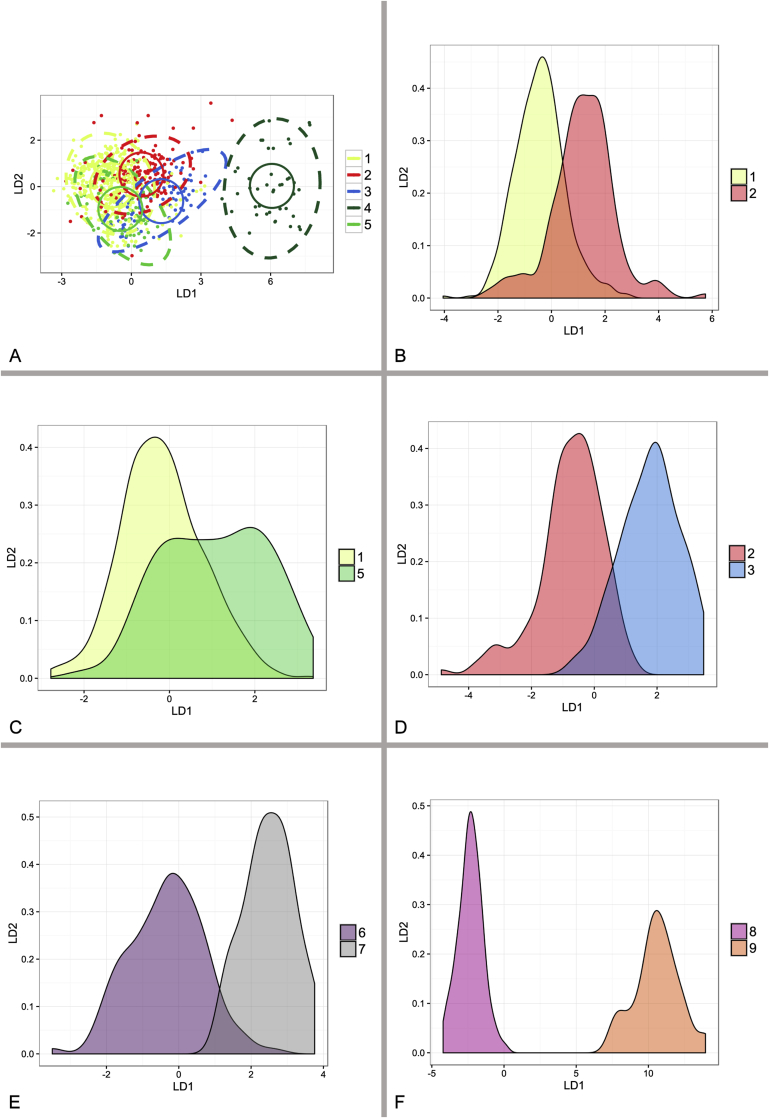
Fig. 11.
Results of linear discriminant analysis (LDA) performed in R 3.3.1 based on micromorphological measurements of species from A. penicillioides clade. Dashed ellipses represent 95 % confidence interval, full circles euclidean distances. Analyses involving only two species (B, D, F) are represented by probability density function. 1 – A. penicillioides; 2 – A. tardicrescens; 3 – A. clavatophorus; 4 – A. magnivesiculatus; 5 – A. hordei; 6 – A. reticulatus; 7 – A. salinicola; 8 – A. canadensis; 9 – A. infrequens.
Table 6.
Overview of conidia and conidiophore characteristics.
| Species | Conidia |
Conidiophores |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Length | Width | Ornamentation (SEM) | Mature conidia shape | Stipe width (μm) | Vesicle width (μm) | Phialides length (μm) | Stipe surface (SEM) | Shape of vesicle | |
| A. restrictus clade | |||||||||
| A. restrictus | 3.5–4.5 (4.1±0.4) | 2.5–3 (2.9±0.4) | Aculeate | Ellipsoidal to ovate | 4–5.5 (4.8±1) | 9–12 (11.5±2) | 8–9 (8.4±0.9) | Densely covered by long hairs | Pyriform to clavate |
| A. caesiellus | 4.5–6 (5±0.8) | 3–4.5 (3.6±0.5) | Aculeate | Ellipsoidal to ovate | 5–7.5 (5.5±0.8) | 11–16 (13.2±2) | 8–9 (8.2±2) | Densely covered by long hairs | Pyriform to clavate |
| A. pachycaulis | 4–5 (4.4±0.2) | 2.5–3.5 (3±0.2) | Aculeate | Ellipsoidal to ovate | 7–8 (7.3±0.8) | 16–20 (18.6±1.8) | 8–9 (8.7±0.5) | Densely covered by short hairs | Pyriform to subclavate |
| A. conicus clade | |||||||||
| A. conicus | 4–4.5 (4.1±0.4) | 2.5–3 (2.9±0.3) | Aculeate | Globose to subglobose or barrel-shaped | 4–5.5 (4.5±0.9) | 8–13 (10.5±2.5) | 8.5–10 (9.2±1.5) | Sparsely covered by short hairs | Pyriform, spatulate to clavate |
| A. destruens | 3.5–5.5 (4.3±0.6) | 2.5–3.5 (3.1±0.4) | Aculeate | Subglobose to ovate | 4–6 (4.2±0.9) | 8–14 (10.2±2.1) | 8.5–11.5 (8.6±1.7) | Densely covered by long hairs | Predominantly ellipsoidal to pyriform |
| A. domesticus | 3.5–4.5 (4.2±0.5) | 2.5–3.5 (2.8±0.3) | Aculeate | Ellipsoidal or barrel-shaped | 4–6 (4.8±0.8) | 7–13 (10.6±2.7) | 8.5–11 (9.7±1) | Densely covered by long hairs | Pyriform, spatulate to clavate |
| A. gracilis | 3–4 (3.4±0.2) | 2–3 (2.5±0.2) | Aculeate | Subglobose or barrel-shaped | 3–5 (4.1±0.5) | 6.5–9 (8±0.7) | 7.5–9 (8.7±0.6) | Densely covered by very long hairs | Spatulate to clavate |
| A. pseudogracilis | 3–4 (3.6±0.3) | 2–3 (2.7±0.2) | Aculeate | Subglobose or barrel-shaped | 4–6 (5±0.6) | 9–13 (10.3±1.6) | 9–10.5 (10±0.4) | Densely covered by long hairs | Spatulate to clavate |
| A. villosus | 3.5–4.5 (3.9±0.4) | 2.5–3.5 (2.9±0.2) | Tuberculate | Ellipsoidal or barrel-shaped | 5–6 (5.5±0.5) | 10–14 (12.6±1.4) | 9–12 (10.8±1.1) | Densely covered by long hairs | Pyriform, spatulate to clavate |
| A. vitricola clade | |||||||||
| A. vitricola | 4.5–5.5 (4.8±0.4) | 3–4 (3.4±0.3) | Aculeate | Subglobose to ellipsoidal | 4.5–6 (5.1±0.6) | 7–12 (9.6±1.7) | 8–10 (9.1±0.7) | Smooth | Spatulate, pyriform to clavate |
| A. glabripes | 3.5–4.5 (4.1±0.2) | 2–3 (2.9±0.2) | Aculeate | Subglobose | 6–8 (6.9±0.8) | 16–22 (19.9±2.3) | 7–9 (8.5±1) | Smooth | Globose, pyriform to subclavate |
| A. penicillioides clade | |||||||||
| A. penicillioides | 3.5–4.5 (4±0.6) | 2.5–3.5 (3±0.5) | Tuberculate | Subglobose or barrel-shaped | 4–6 (5.1±0.9) | 10–18 (13.5±2.3) | 8–10 (9.3±1.7) | Sparsely covered by short hairs | Pyriform, spatulate to clavate |
| A. canadensis | 3–4 (3.3±0.4) | 2.5–3 (2.8±0.4) | Lobate-reticulate | Subglobose or barrel-shaped | 3.5–5 (4.2±0.7) | (9–)13–16(–20) (13.9±2.5) | 6–8 (6.9±0.4) | Sparsely covered by bundles of long hairs | Pyriform to clavate |
| A. clavatophorus | 4–5 (4.8±0.2) | 3–4 (3.5±0.2) | Tuberculate | Subglobose, ovate or barrel-shaped | 6–9 (6.6±0.5) | 14–19 (16.5±1.3) | 8–10 (9.3±0.7) | Densely covered by short hairs | Spatulate to clavate |
| A. hordei | 3.5–5 (4.1±0.4) | 2.5–4 (3.2±0.4) | Lobate-reticulate | Globose or barrel-shaped | 4.5–6 (5.5±0.6) | 12–14.5 (13.2±1.4) | 8.5–11 (9.5±1) | Densely covered by short hairs | Spatulate, ellipsoidal, pyriform to subclavate |
| A. infrequens | 4–5 (4.8±0.3) | 3–4 (3.6±0.3) | Aculeate | Subglobose, ovate or barrel-shaped | 6–7 (6.7±0.8) | (10–)13–17(–20) (15.9±2.6) | 8–10 (9.5±1) | Sparsely covered by bundles of long hairs | Pyriform, spatulate to clavate |
| A. magnivesiculatus | 4.5–5.5 (5.2±0.3) | 3.5–4.5 (3.9±0.3) | Tuberculate | Ovate to barrel-shaped | 5–7 (5.9±1.2) | 17–22 (21.4±3.2) | 9–11 (9.6±0.8) | Densely covered by short hairs | Pyriform, subclavate to clavate |
| A. reticulatus | 3.5–4.5 (3.9±0.3) | 2.5–3.5 (3±0.3) | Tuberculate to lobate-reticulate | Globose or barrel-shaped | 5–7 (6±1) | 14–20 (16.3±3.2) | 9–11 (9.3±0.8) | Densely covered by short hairs | Globose or pyriform |
| A. salinicola | 3–4 (3.6±0.4) | 2–3 (2.6±0.2) | Tuberculate to lobate-reticulate | Globose to subglobose | 4–6 (3.6±0.4) | 11–14 (12.6±1.3) | 8–9 (8.7±0.4) | Sparsely covered by short hairs | Pyriform, spatulate to subclavate |
| A. tardicrescens | 3.5–4.5 (4±0.4) | 2.5–3.5 (2.9±0.3) | Tuberculate | Subglobose or barrel-shaped | 5–8 (6±1.2) | 14–19 (16.4±3.3) | 8–11 (8.9±1.2) | Densely covered by short hairs | Subglobose, spatulate, pyriform to subclavate |
Fig. 10, Fig. 11 show results of the analysis separately for particular species within species complexes/clades. The LDA clearly discriminated A. pachycaulis within the A. restrictus clade (Fig. 10A); the discrimation between A. restrictus and A. caesiellus was not so obvious because some isolates have similar micromorphologies. The discriminatory power of micromorphological characters within the A. conicus clade is low except for A. gracilis and A. pseudogracilis that were relatively well separated from other species (see Fig. 10C). The two species from the A. vitricola clade vary greatly in their micromorphological characters and consequently it resulted in clear separation of species in LDA.
The micromorphology of core species of the A. penicillioides clade is nearly identical, except A. magnivesiculatus that has much larger vesicles. It is more meaningful to compare micromorphology only in pairs of related species and even then some species are not well separated as in the case of A. penicillioides and A. hordei (see Fig. 11C). On the other hand two pairs of closely related species, distant from the core group (A. reticulatus – A. salinicola and A. canadensis – A. infrequens) can be distinguished very well by the LDA (Fig. 11E, F).
Growth in an osmotic gradient
The ability to grow in an osmotic gradient proved to be very consistent during repeated testing of the same isolate and similarly, there was only low infraspecific variability. In general, it can be considered a useful supplementary taxonomic character that can distinguish some species. Growth curves created from measurements of the extension rate after 14 d are displayed on Fig. 12, Fig. 13. The A. restrictus clade contains less xerophilic species compared to others and grow faster on media with higher water activity (MEA or MEA + 5 % NaCl); A. caesiellus differs from the two remaining species of A. restrictus clade by faster growth on MEA + 10 % NaCl and 15 % NaCl. Species from the A. conicus clade show relatively similar growth curves; A. domesticus, A. gracilis and A. pseudogracilis grow slower than the three remaining species on MEA + 5 % and 10 % NaCl; A. conicus grows fastest on MEA + 5 % and 10 % NaCl; A. villosus grows faster on MEA and MEA + 5 % NaCl but with decreasing water activity the curve quickly declines and the species grows poorly on MEA + 15 % and 20 % NaCl. Aspergillus vitricola and A. glabripes differ significantly in their growth on MEA and MEA + 5 % NaCl.
Fig. 12.
Osmotic gradient growth curves of species from A. restrictus, A. conicus and A. vitricola clades. Growth curves were created using local regression (LOESS) in R 3.3.1, grey zones represent 95 % confidence intervals. 1 – A. restrictus; 2 – A. caesiellus; 3 – A. pachycaulis; 4 – A. conicus; 5 – A. domesticus; 6 – A. destruens; 7 – A. villosus; 8 – A. gracilis; 9 – A. pseudogracilis; 10 – A. vitricola; 11 – A. glabripes.
Fig. 13.
Osmotic gradient growth curves of species from A. penicillioides clade. Growth curves were created using local regression (LOESS) in R 3.3.1, grey zones represent 95 % confidence intervals. 1 – A. penicillioides; 2 – A. tardicrescens; 3 – A. clavatophorus; 4 – A. magnivesiculatus; 5 – A. hordei; 6 – A. reticulatus; 7 – A. salinicola; 8 – A. canadensis; 9 – A. infrequens.
Growth curves of species belonging to the core group of the A. penicillioides clade are nearly identical but differences can be found when comparing some species pairs (see Fig. 13), e.g. A. penicillioides and A. tardicrescens (Fig. 13B). Aspergillus reticulatus and A. salinicola grow faster than all other species on MEA + 5 % NaCl and slower on MEA + 20 % NaCl. Aspergillus canadensis and A. infrequens grow slowest on MEA + 5 % NaCl. The growth curves of the last mentioned species pairs are also different (see Fig. 13F).
Other phenotypic characters
Some other phenotypic characters are taxonomically relevant and useful for differentiation of species complexes or particular species. An overview of recorded characters is given in Table 6, Table 7 and their usability for differentiation of species complexes is detailed in the Taxonomy section; possibilities of distinguishing species within complexes are detailed in Distinguishing characters that follows the descriptions of species. The shape of conidial heads can be used to distinguish between clades. Conidial heads of A. restrictus clade form compact columns, heads of A. conicus clade are loosely columnar, those of A. vitricola clade are radiate and A. penicillioides clade have mainly globose conidial heads.
Table 7.
Overview of colony characters on M40Y medium.
| Species | Sporulation colour | Reverse colour in the center | Shape of conidial heads |
|---|---|---|---|
| A. restrictus clade | |||
| A. restrictus | Greyish turquoise (24C4) | Cinnamon brown (6D6) | Columnar |
| A. caesiellus | Greyish turquoise (24E4) | Sunburn light brown (6D5) | Columnar |
| A. pachycaulis | Greyish turquoise (24C4) | Brownish yellow (5C8) | Columnar |
| A. conicus clade | |||
| A. conicus | Pale turquoise (24A3) | Olive grey (2F2) | Loosely columnar |
| A. destruens | Myrtle green (25F7) | Apricot greyish orange (5B6) | Loosely columnar |
| A. domesticus | Aquamarine greyish turquoise (24B3) | Olive grey (2F2) | Loosely columnar |
| A. gracilis | Aquamarine greyish turquoise (24B3) to turquoise white (24A2) | Olive grey (2F2) | Loosely columnar |
| A. pseudogracilis | Light turquoise (24A4) | Cinnamon brown (6D6) | Loosely columnar |
| A. villosus | Greyish green (25E7) to bluish green (25A6) | Olive green (2F6) | Loosely columnar |
| A. vitricola clade | |||
| A. vitricola | Greyish turquoise (water blue) (24C5) | Olive green (2F6) | Radiate |
| A. glabripes | Yellowish green (29C7) | Dark brown (6F7) | Radiate |
| A. penicillioides clade | |||
| A. penicillioides | Deep green (25E8) | Coffee brown (5F7) | First globose, later becoming loosely columnar |
| A. canadensis | Jungle green (25F5) to pale turquoise (24A3) | Olive grey (2F2) | Radiate |
| A. clavatophorus | Greyish green (28D6) | Coffee brown (5F7) | First globose, later becoming radiate |
| A. hordei | Dark turquoise (24F8) | Cuba reddish brown (9E8) | First globose, later becoming loosely columnar |
| A. infrequens | Pale turquoise (24A3) to dark turquoise (24F8) | Dark green (30F8) | Radiate |
| A. magnivesiculatus | Greyish green (26D5) | Mahogany red (8E7) | First globose, later becoming loosely columnar to radiate |
| A. reticulatus | Greyish turquoise (24E6) | Golden brown (5D7) | Globose to radiate |
| A. salinicola | Greyish green (25B5) | Golden brown (5D7) | First globose, later becoming loosely radiate |
| A. tardicrescens | Greyish turquoise (water blue) (24C5) | Olive brown (4F8) | First globose, later becoming radiate |
The extension rates of each species are given in Table 8. Aspergillus pachycaulis is the only member of the section Restricti that was able to grow on CY20S at 37 °C, but several species were able to grow at 37 °C on a more sucrose rich medium like M60Y. The extension rates of the species on optimal media (M40Y, M60Y, MEA + 10 % NaCl) was generally the same, but the species exhibit a greater degree of variability in more extreme conditions (high water activity, high temperature) and the ability or inability to grow on these suboptimal media can be used as taxonomic characters to distinguish between closely related species (e.g. A. conicus, A. domesticus and A. destruens).
Table 8.
Growth rate comparison after 14 d (mm).
| Species | CYA, 25 °C | MEA, 25 °C | MEA+10 % NaCl, 25 °C | DG18, 25 °C | M40Y, 25 °C | CY20S |
M60Y |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 25 °C | 30 °C | 37 °C | 25 °C | 30 °C | 37 °C | ||||||
| A. restrictus clade | |||||||||||
| A. restrictus | 12–16 | 5–13 | 33–40 | 28–33 | 42–53 | 24–29 | 12–25 | 0 | 38–50 | 45–55 | 20–40 |
| A. caesiellus | 11–22 | 17–22 | 36–45 | 32–38 | 58–60 | 21–35 | 12–40 | 0 | 48–60 | 45–60 | 32–55 |
| A. pachycaulis | 11–26 | 21–31 | 27–43 | 35–48 | 20–60 | 15–40 | 22–36 | (0) 8–10 | 25–60 | 36–60 | 38–60 |
| A. conicus clade | |||||||||||
| A. conicus | (0) 10–12 | (0) 8–10 | 16–36 | 18–31 | 32–40 | 5–18 | 0 | 0 | 26–40 | 26–37 | 0 |
| A. destruens | 5–10 | (0) 4–7 | 20–30 | 14–30 | 19–36 | 6–13 | 0 | 0 | 34–41 | 16–41 | 0 |
| A. domesticus | 0–8 | 0 | 23–31 | 20–24 | 36–38 | 5–17 | 0 | 0 | 29–38 | 15–30 | 0 |
| A. gracilis | 0–8 | 0 | 12–24 | 9–16 | 30–42 | 7–13 | 0 | 0 | 38–44 | 39–45 | 0 |
| A. pseudogracilis | 0 | 0 | 25–26 | 20–22 | 42–45 | 7–8 | 0 | 0 | 52–53 | 43–45 | 0 |
| A. villosus | 8–10 | 5–6 | 22–23 | 16–20 | 21–23 | 17–19 | 7–8 | 0 | 26–28 | 28–30 | 6–8 |
| A. vitricola clade | |||||||||||
| A. vitricola | 0 | 0 | 9–26 | 8–18 | 19–36 | (0) 4–7 | 0 | 0 | 25–42 | 16–42 | 0 |
| A. glabripes | 3–7 | 4–11 | 22–28 | 15–25 | 38–52 | 8–18 | 5–16 | 0 | 35–40 | 41–56 | (0) 7–10 |
| A. penicillioides clade | |||||||||||
| A. penicillioides | 7–9 | 5–9 | 24–37 | 17–36 | 27–43 | 11–20 | 11–15 | 0 | 28–59 | 40–60 | 17–45 |
| A. canadensis | 0 | 0 | 17–33 | 7–15 | 25–29 | (0) 10–11 | 0 | 0 | 23–42 | 2–20 | 0 |
| A. clavatophorus | 0 | 0 | 18–28 | 18–25 | 27–33 | 7–16 | 0 | 0 | 35–44 | 29–54 | (0) 5–17 |
| A. hordei | 0–8 | 0–5 | 26–29 | 13–26 | 25–29 | (0) 13–16 | (0)8–15 | 0 | 31–47 | 25–48 | 22–33 |
| A. infrequens | 4–5 | 0 | 18–19 | 25–26 | 21–22 | 7–8 | 0 | 0 | 40–43 | 8–10 | 0 |
| A. magnivesiculatus | 0 | 0 | 27–34 | 20–26 | 27–35 | 11–17 | 0 | 0 | 47–60 | 40–50 | 0 |
| A. reticulatus | 8–14 | 2–7 | 32–40 | 26–33 | 46–50 | 21–30 | 8–18 | 0 | 55–60 | 50–55 | 3–20 |
| A. salinicola | (0) 8–9 | 0–5 | 32–35 | 21–30 | 32–40 | 9–20 | 0–5 | 0 | 42–55 | 27–43 | 0 |
| A. tardicrescens | 0 | 0 | 19–29 | 12–17 | 17–29 | 0–5 | 0 | 0 | 31–50 | 15–35 | 0 |
Exometabolite analysis
The overview of metabolites produced by sect. Restricti species is given in Table 9 and their absorption maxima and bracketed retention indexes are listed in Table S1. Six species had isolates that could produce mycophenolic acid: A. glabripes, A. gracilis, A. halophilicus, A. pachycaulis, A. penicillioides and A. tardicrescens (Table 9). We detected asperglaucide, its precursor aurantiamide and/or asperphenamate in all species in sect. Restricti (Table 9). Other extrolites detected include antarone A, asperentins, clavatols, cyclopaldic acid, orthosporins, naphtho-gamma-pyrones and chrysogine. Several other extrolites were detected including indole alkaloids, phthalides, pyrones and some compounds with yellow chromophores (Table 9). Members of the echinulin/neoechinulin/isoechinulin/cristatine biosynthetic family were detected in A. halophilicus, A. hordei, A. magnivesiculatus, A. penicillioides and A. tardicrescens, indicating a strong chemical relationship with Aspergillus section Aspergillus. However, while nearly all isolates in species of section Aspergillus produce the yellow auroglaucin and flaviglaucin-type metabolites (colouring the ascomata), these yellow metabolites were not found in any species of section Restricti.
Table 9.
Exometabolites found in members of Aspergillus section Restricti.
| Species | Strain number1,2 | Exometabolites3 |
|---|---|---|
| A. caesiellus | NRRL 5061T = CBS 470.65 = DTO 093-H3 = ATCC 11905 = IMI 172278 = CCF 5447 = IBT 34620 | Asperphenamate, indole alkaloid “A” & “B”, CADA1, CADA2 |
| DTO 025-I4 = IBT 34538 | Asperphenamate, CAS1-CAS5 | |
| CCF 5448 = EMSL No. 1383 = IBT 34621 | Asperphenamate, indole alkaloid “A”, CA1, CA2, CAD1, CADA2, CAS1-CAS4 | |
| CCF 5451 = EMSL No. 1650 = IBT 34622 | Asperphenamate, indole alkaloid “A” & “B”, CA1, CAS1-CAS5 | |
| A. canadensis | CCF 5548T = KAS 6194 = DTO 356-H9 = IBT 34520 = IBT 34642 = NRRL 66614 | Asperglaucide, aurantiamide4, aurantiamide “B”, chrysogine, indole alkaloid “A”, CAN1 |
| KAS 7710 = DTO 357-B4 = CCF 5552 = IBT 34636 | Asperglaucide, aurantiamide, indole alkaloid “A” | |
| KAS 7708 = DTO 357-B2 = CCF 5550 = IBT 34637 | asperglaucide, indole alkaloid “A”, ASPHAM1 | |
| KAS 7719 = DTO 357-C2 = CCF 5553 = IBT 34638 | Asperglaucide, aurantiamide , indole alkaloid “A”, ASPHAM1, CANA1-CANA4 | |
| KAS 7706 = DTO 357-A9 = CCF 5549 = IBT 34639 = NRRL 66615 | Asperglaucide, aurantiamide , indole alkaloid “A”, ASPHAM1 | |
| KAS 7709 = DTO 357-B3 = CCF 5551 = IBT 34640 | Asperglaucide, asperphenamate, indole alkaloid “A” | |
| KAS 7720 = DTO 357-C3 = IBT 34641 | Asperglaucide, aurantiamide | |
| A. clavatophorus | NRRL 25874T = CCF 5454 = IBT 34560 = DTO 356-D8 = IBT 34823 | Asperglaucide, aurantiamide, indole alkaloid “A” |
| DTO 257-G5 = CCF 5669 = IBT 34561 | Asperglaucide, PRO1, Raistrick phenol “A” | |
| NRRL 25873 = CCF 5453 = IBT 34632 | Asperglaucide, aurantiamide, PRO1, Raistrick phenol “B” | |
| A. conicus | NRRL 149T = CBS 475.65 = IBT 33667 = DTO 096-H6 = ATCC 16908 = IMI 172281 = CCF 5456 | Asperglaucide, asperphenamate, indole alkaloid “A”, phthalide |
| DTO 110-F5 = CCF 5667 = IBT 34534 | Asperphenamate, phthalide, PRO2 | |
| EXF-7660 = IBT 34263 = IBT 33577 | Asperphenamate, phthalide, WOVI, PITAL | |
| EXF-7663 = IBT 34267 = IBT 33574 | Asperphenamate, phthalide, clavatol “A”, FOLS, indole alkaloid “B”, PITAL | |
| EXF-5015 = CCF 5650 = IBT 34273 | Asperphenamate | |
| EXF-10387 = CCF 5651 = IBT 34289 | Asperglaucide, PI-4 (fulvic acid biosynthetic family) | |
| A. destruens | NRRL 145T = IMI 358691 = CCF 5462 = CBS 593.91 = DTO 079-A8 = IBT 34818 | Asperglaucide, chromanol “X”, naphtho-gamma-pyrone “A”, YELLO |
| DTO 254-B2 = IBT 34522 | Asperphenamate, clavatol “B” & “C”, pyrone “A”, “B”, “C”, “D”, “E”, “F”, “G”, “H” & “I”, WOVI | |
| EXF-7657 = IBT 34264 | Asperphenamate, pyrone “A”, pyrone “G”, WOVI | |
| EXF-7703 = IBT 34259 | Asperphenamate, clavatol “D” | |
| EXF-7651 = CCF 5653 = IBT 34258 | Asperphenamate, pyrone “B”, “D” & “G”, clavatol “A” | |
| EXF-7699 = IBT 34262 | Asperphenamate, pyrone “A”, pyrone “B” & “G”, clavatol “A” | |
| EXF-7666 = IBT 34268 | Asperphenamate | |
| EXF-7661 = IBT 34271 = IBT 33573 | Asperphenamate, clavatol “A”, ASPHAM1, CAS1, CAS2, naphtho-gamma-pyrone “A” | |
| EXF-7667 = IBT 34288 | Asperphenamate, pyrone “G” | |
| EXF-10431 = IBT 34257 | Asperphenamate, pyrone “A”, “B” & “G” | |
| EXF-10411 = IBT 34265 | Asperphenamate, pyrone “A”, “B”, “E” & “G” | |
| EXF-10350 = IBT 34270 | Asperphenamate, WOVI | |
| EXF-10407 = CCF 5652 = IBT 34285 | Asperglaucide | |
| EXF-10017 = IBT 33576 | Asperphenamate, ASPHAM1, naphtho-gamma-pyrone “A”, WOVI | |
| EXF-10390 = IBT 34276 | Asperphenamate, clavatol “A”, SKIZ1, SKIZ2, CAS5 | |
| A. domesticus | DTO 079-F2T = CCF 5464 = NRRL 66616 = IBT 34814 | Asperphenamate, clavatol “D”, FOLS |
| CBS 233.90 = DTO 079-A7 = IBT 34515 | Asperphenamate, WOVI | |
| CBS 116419 = DTO 077-G6 = IBT 34535 | Asperphenamate, indole alkaloid “B”, FOLS | |
| EXF-10012 = IBT 34274 | Asperphenamate, clavatol “D”, FOLS, indole alkaloid “B” | |
| EXF-10583 = IBT 34275 | Asperphenamate, indole alkaloid “B” | |
| A. glabripes | CCF 5474T = DTO 356-E8 = EMSL No. 2462 = NRRL 66618 = IBT 34820 | SCUB, STALD1, STALD2, FJOS, indole alkaloid “A”, PRO1 |
| CCF 5469 = EMSL No. 1483 = DTO 356-E5 = IBT 34519 = IBT 34821 = NRRL 66619 | Mycophenolic acid, FAESI, GUOY, GUOY2, PRO1, SCUB, SPOJN1, SPOJN2, STALD1-STALD3, TERRIT1-TERRIT3, XANS | |
| UBOCC-A-116021 = CCF 5467 = IBT 34625 | Indole alkaloid “A”, PITAL | |
| CCF 5473 = EMSL No. 2442 = IBT 34626 | Asperphenamate, indole alkaloid “A”, PRO1, SCUB | |
| IBT 266035 | SCUB, STALD1, STALD2, FJOS, indole alkaloid “A”, PRO1 | |
| A. gracilis | NRRL 4962T = CBS 539.65 = DTO 351-H7 = CCF 5478 = ATCC 16906 = IMI 211393 = IBT 34817 | Asperphenamate, CHE1-CHE3, chromanol “X”, clavatol “D”, indole alkaloid “B”, FOL1, FOL2, FOLS, SCAB |
| CCF 5479 = EMSL No. 2775 = DTO 356-F4 = IBT 34559 | Asperphenamate, mycophenolic acid, FOL2, FOLS, indole alkaloid “B”, SCAB | |
| CCF 5482 = EMSL No. 2923 = IBT 34623 | Indole alkaloid “A”, SCAB | |
| A. halophilicus | NRRL 2739T = CCF 5687 = ATCC 16401 = CBS 12262 = IMI 211802 = IBT 34878 | Asperphenamate, echinulin, preechinulin, cristatin A, pyrone “R”, “U”, “V”, “W”, “X”, “Z”, cristatin “X”, DYLD1-DYLD4, PITAL |
| DTO 271-F4 = CCF 5825 = IBT 34881 | Asperphenamate, echinulin, preechinulin, cristatin A, trace of mycophenolic acid, pyrone “K”, “L”, “M”, “N”, “O”, “P”, “R”, “S” & “T”, indole alkaloid “A”, cristatine “X”, PITAL | |
| MUT 562 | Asperphenamate, cristatin A, echinulin, pyrone “K”, “N”, “U”, “V”, “W”, “X”, “R”, DYLD1-DYLD4, PITAL | |
| MUT 1305 | Asperphenamate, cristatin A, echinulin, pyrone “K”, “N”, “R”, “V”, “W”, “X”, DYLD1-DYLD4, PITAL | |
| MUT 798 | Asperphenamate, cristatin A, echinulin, pyrone “K”, “M”, “N”, “O”, “P”, “R”, DYLD1-DYLD4, PITAL | |
| MUT 1307 | Asperphenamate, cristatin A, echinulin, pyrone “K”, “N”, “R”, “U”, “V”, “W”, “X”, “Y”, “Z”, DYLD1-DYLD4, PITAL | |
| A. hordei | NRRL 25825T = CCF 5483 = DTO 356-D3 = IBT 34539 | Asperglaucide, aurantiamide, aurantiamide “B”, cristatine A, echinulin, indole alkaloid “C”, “D” & “E” |
| DTO 281-A4 = IBT 34517 | Asperglaucide, aurantiamide, chromanol “X”, cristatin A, echinulin, YEY | |
| NRRL 25830 = CCF 5485 = IBT 34631 | Asperglaucide, aurantiamide, aurantiamide “B”, echinulin, cristatin A, KOS3, PENIT | |
| A. infrequens | NRRL 25868T = CCF 5486 = DTO 356-D6 = IBT 34524 | Asperphenamate |
| A. magnivesiculatus | NRRL 25866T = CCF 5488 = IBT 34816 | Asperglaucide, cristatin A, echinulin, indole alkaloid “A”, KOS1-KOS3 |
| CCF 5489 = EMSL No. 1315 = DTO 356-E2 = IBT 34516 | Antarone A, asperglaucide, cristatin A, echinulin, FU1, FU2, KOS1-KOS3, FUSM1, FUSM2 | |
| EXF-10353 = IBT 34284 | Asperglaucide, pyrone “A”, “B”, “D” & “G” | |
| EXF-10356 = IBT 34279 | Asperglaucide, pyrone “G” & “I” | |
| EXF-10347 = IBT 34283 | Asperglaucide | |
| A. pachycaulis | NRRL 25824T = CCF 5492 = DTO 356-D2 = IBT 34521 = IBT 34812 | Asperphenamate, indole alkaloid “A” & “B”, phthalide, mycophenolic acid, ASPHAM2, CACCA, OSM1, OSM2, clavatol “D”, FIMUI, naphtho-gamma-pyrone “C” & “D” |
| CCF 5493 = EMSL No. 2310 = DTO 356-E6 = IBT 34536 | Asperphenamate, phthalide, indole alkaloid “A” and “B” | |
| A. penicillioides | NRRL 4548T = CBS 540.65 = ATCC 16910 = IMI 211342 = DTO 207-I7 = CCF 5494 = IBT 34627 | Asperglaucide, aurantiamide |
| DTO 267-A4 = IBT 34614 | Asperglaucide, cristatine A, trace of mycophenolic acid, YEY, echinulin | |
| CCF 5498 = EMSL No. 2440 = DTO 356-E7 = IBT 34815 | Asperglaucide | |
| CCF 5497 = EMSL No. 2430 = IBT 34628 | Asperglaucide, aurantiamide, cristatine A, YEY | |
| CCF 5501 = EMSL No. 2749 = IBT 34629 | Asperglaucide, YEY, echinulin, met k | |
| CCF 5500 = EMSL No. 2651 = IBT 34630 | Asperglaucide, YEY | |
| A. pseudogracilis | CCF 5505T = EMSL No. 2765 = DTO 356-F3 = NRRL 66620 = IBT 34813 | Asperphenamate, indole alkaloid “B”, FOLS |
| A. restrictus | NRRL 154T = CBS 117.33 = CBS 541.65 = DTO 079-B2 = ATCC 16912 = IHEM 3920 = IMI 16267 = CCF 5506 = IBT 34615 | Asperphenamate, CACCA, indole alkaloid “A” & “B” |
| DTO 065-C7 = IBT 34541 | Asperphenamate, indole alkaloid “B”, CRYPT1, CRYPT2, FNOVL1, FNOVL2 | |
| CCF 5511 = EMSL No. 1675 = IBT 34616 | Asperphenamate, FNOVL2, FOLS, indole alkaloid “B” | |
| CCF 3364 = IBT 34619 | Asperphenamate, CRYPT1, CRYPT2, CRYPT4, FNOVL1, FNOVL2, FOLS, indole alkaloid “B” | |
| CCF 5507 = EMSL No. 1379 = IBT 34618 | Asperphenamate, indole alkaloid “B”, FOLS | |
| CCF 5512 = EMSL No. 2206 = IBT 34617 | Asperphenamate, CRYPT1-CRYPT5, FNOVL1-FNOVL3, PALO1-PALO3 | |
| A. reticulatus | NRRL 25852T = CCF 5516 = DTO 356-D4 = IBT 34540 | Asperglaucide, aurantiamide, indole alkaloid “A”, clavatol “D” |
| CCF 5521 = EMSL No. 1362 = DTO 356-E4 = IBT 34518 | Asperglaucide, aurantiamide, indole alkaloid “A”, PITAL | |
| CCF 5524 = EMSL No. 2548 = IBT 34637 | Asperglaucide, aurantiamide, clavatol “D” | |
| CCF 3112 = IBT 34634 = NRRL 62490 | Asperglaucide, aurantiamide, indole alkaloid “A”, cyclopaldic acid chromanols | |
| CCF 5525 = NRRL 58572 = EMSL No. 885 = IBT 34880 | Asperglaucide, cyclopaldic acid, cyclopaldic acid chromanols | |
| CCF 5518 = NRRL 58903 = EMSL No. 1272 = IBT 34819 | Asperglaucide, aurantiamide, indole alkaloid “A”, cyclopaldic acid chromanols | |
| A. salinicola | EXF-10401T = IBT 34266 = NRRL 66621 = CCF 5526 | Antarone A, asperentin “A”, “B”, “C”, “D”, “E”, “F”, “G”, “H” & “I”, asperglaucide, aurantiamide, met k, SOSO |
| EXF-226 = CCF 5527 = IBT 34277 = NRRL 66622 | Asperglaucide, aurantiamide, indole alkaloid “A” | |
| UBOCC-A-116019 = CCF 5528 = IBT 34635 | Asperglaucide, aurantiamide | |
| A. tardicrescens | DTO 316-B5T = CCF 5529 = NRRL 66623 = IBT 34558 | Asperglaucide, mycophenolic acid, clavatol “D”, FOL2, indole alkaloid “A” |
| DTO 316-A8 = IBT 34562 | Asperglaucide, mycophenolic acid, FOL2, KOS3, indole alkaloid “A”, indole alkaloid “F” | |
| EXF-10454 = IBT 34281 = CCF 5530 = NRRL 66624 | Asperfuran, asperglaucide, echinulin, indole alkaloid “D”, FOL2, KOS3 | |
| EXF-10456 = IBT 34286 | Asperglaucide | |
| A. villosus | NRRL 25813T = CCF 5531 = DTO 356-C9 = IBT 34822 | Asperphenamate, orthosporin “A”, “A1”, “D”, “F”, “G”, “H”, “I”, “J”, “K”, “L”, “M”, “N”, “O”, “P”, “Q”, “R” & “S”, MOL |
| UBOCC-A-116020 = CCF 5532 = IBT 34624 | Indol alkaloid “A” | |
| IBT 22529, IBT 22530, IBT 225466 | Asperphenamate, orthosporin “A”, “F”, “G”, “O”, “R” & “S” | |
| A. vitricola | NRRL 5125T = DTO 356-F7 = ATCC 16905 = ATCC 36505 = IMI 108298 = CCF 5533 = IBT 34530 | No exometabolites detected |
| KAS 6150 = DTO 356-H5 = IBT 34531 | Orthosporin “A”, “B”, “C”, “D” & “E” | |
| KAS 6087 = DTO 356-H2 = IBT 34532 | Indole alkaloid “G” | |
| EXF-7700 = CCF 5657 = IBT 34282 = IBT 33575 | Asperglaucide, YELL | |
| EXF-10383 = CCF 5655 = IBT 34272 | Asperglaucide, Raistrick phenol “A” | |
| EXF-10301 = CCF 5654 = IBT 34290 | DOKU1, DOKU2 |
Acronyms of culture collections: ATCC, American Type Culture Collection, Manassas, Virginia, USA; CBS, culture collection of Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CCF, Culture Collection of Fungi, Charles University, Czech Republic; DTO, working collection of the department of Applied and Industrial Mycology housed at CBS; EMSL, EMSL Analytical Inc., New Jersey, USA; EXF, Culture Collection of Extremophilic Fungi, University of Ljubljana, Slovenia; IBT, Fungal Culture Collection at DTU-Bioengineering, Lyngby, Denmark; BCCM/IHEM, Biomedical Fungi and Yeasts Collection, Scientific Institute of Public Health, Brussels, Belgium; IMI/CABI, International Mycological Institute, Kew, England; KAS, fungal collection of Keith A. Seifert, Ottawa, Canada; MUT, Mycotheca dell'Università degli Studi di Torino, Turin, Italy; NRRL, Agricultural Research Service Culture Collection, Peoria, Illinois, USA; UBOCC, Université de Bretagne Occidentale Culture Collection, Brest, France.
Ex-type strains are designated with superscript T.
Absorption maxima and bracketed retention indexes of all detected exometabolites are listed in Table S1.
Aurantiamide = asperglucide = aurantiamide acetate = lyciumamide = suropeptate.
This isolate originated from mouldy rye bread, Denmark; it was identified solely based on its exometabolite spectrum.
All three isolates originated from whale, Greenland; they were identified solely based on their exometabolite spectra.
Taxonomy
Aspergillus section Restricti (Gams et al. 1985), Adv. Penicillium Aspergillus Syst.: 56.
Typus: Aspergillus restrictus G. Sm., J. Textile Inst. 22: T115. 1931.
The section includes 20 anamorphic species and one homothallic species. Xerophilic, growing optimally either on substrates containing high concentrations of sugar or salt or with low water content. Young conidial heads columnar (A. restrictus and A. conicus clade), radiate (A. vitricola clade) or globose (A. penicillioides clade). Conidiophores uniseriate with hyaline stipes, usually villose in SEM. Vesicles pyriform, subglobose or clavate, fertile over one to two thirds of the surface. Phialides flask-shaped, usually villose in SEM. Conidia ellipsoidal to ovate, barrel-shaped or subglobose, rough-walled; aculeate, tuberculate or lobate-reticulate in SEM. Ascomata present only in A. halophilicus, cleistothecial, hyaline to pale yellow, globose to subglobose. Asci globose to subglobose. Ascospores hyaline, lenticular with two equatorial crests, convex surface finely roughened in the area neighbouring the equatorial region, dimpled in SEM.
Aspergillus restrictus clade
Aspergillus restrictus, A. caesiellus and A. pachycaulis are distinguished from all other members of sect. Restricti by compact columnar conidial heads, different growth parameters in an osmotic gradient (compared to other clades, they grow faster on media with high water activity, e.g. MEA and MEA + 5 % NaCl and slower on MEA + 20 % NaCl and MEA + 25 % NaCl) and rapid growth and deep green sporulation on CY20S.
Accepted species:
Aspergillus caesiellus Saito, J. Fac. Sci. Coll. Imper. Univ. Tokyo 18: 49. 1904. [MB205025].
Aspergillus pachycaulis F. Sklenar, S.W. Peterson, Ž. Jurjević & Hubka, this study. [MB823048].
Aspergillus restrictus G. Sm., J. Textile Inst. 22: T115. 1931. [MB276290].
Aspergillus conicus clade
This clade contains six species (A. conicus, A. domesticus, A. destruens, A. villosus, A. gracilis and A. pseudogracilis). Members of the A. conicus clade have loosely columnar conidial heads, while species from the closely related A. vitricola clade have radiate heads. Several characters are useful for distinguishing between members of the A. conicus and A. penicillioides clade. The colour of sporulation of A. conicus clade species is usually paler green, colony reverse colour on M40Y is usually olive grey compared to the brown or red reverse of A. penicillioides clade species. Further, the members of A. penicillioides clade produce white cottony secondary mycelium on M60Y in contrast to A. conicus clade and conidial heads are usually globose. Conidia of A. conicus clade species in SEM are usually aculeate, whereas conidia of A. penicillioides clade species are tuberculate or lobate-reticulate. The differences from A. restrictus clade were described above (see distinguishing characters of A. restrictus).
Accepted species:
Aspergillus conicus Blochwitz in Dale, Ann. Mycol. 12: 38. 1914. [MB120214].
Aspergillus destruens Zalar, F. Sklenar, S.W. Peterson & Hubka, this study. [MB818930].
Aspergillus domesticus F. Sklenar, Houbraken, Zalar & Hubka, this study. [MB818931].
Aspergillus gracilis Bainier, Bull. Soc. Mycol. Fr. 23: 92. 1907. [MB167554].
Aspergillus pseudogracilis F. Sklenar, Ž. Jurjević & Hubka, this study. [MB818932].
Aspergillus villosus F. Sklenar, S.W. Peterson & Hubka, this study. [MB818933].
Aspergillus vitricola clade
This clade contains two species, A. vitricola and A. glabripes, and is defined by their radiate conidial heads, smooth stipes and phialides in SEM (unique through the whole section), and aculeate conidia in SEM.
Accepted species:
Aspergillus vitricola Ohtsuki, Bot. Mag. (Tokyo) 75: 436. 1962. [MB326665].
Aspergillus glabripes F. Sklenar, Ž. Jurjević & Hubka, this study. [MB818934].
Aspergillus penicillioides clade
This clade encompasses nine species (Fig. 7). Aspegillus tardicrescens, A. clavatophorus, A. magnivesiculatus and A. hordei are closely related to A. penicillioides, while the remaining four species are relatively distant. The core species can be considered the most xerophilic members of the section, and generally some of the most xerophilic fungi. Conidial heads of these species are always globose when young and sporulation colour is usually darker compared to species from other clades. Stipes are covered by short hairs observed with SEM and conidia are usually born as ellipsoidal (not cylindrical) and have tuberculate or lobate-reticulate ornamentation in SEM.
Accepted species:
Aspergillus canadensis Visagie, Yilmaz, F. Sklenar & Seifert, this study. [MB818935].
Aspergillus clavatophorus F. Sklenar, S.W. Peterson & Hubka, this study. [MB818936].
Aspergillus hordei F. Sklenar, S.W. Peterson & Hubka, this study. [MB818937].
Aspergillus infrequens F. Sklenar, S.W. Peterson & Hubka, this study. [MB818938].
Aspergillus magnivesiculatus F. Sklenar, Zalar, Ž. Jurjević & Hubka, this study. [MB818939].
Aspergillus reticulatus F. Sklenar, Ž. Jurjević, S.W. Peterson & Hubka, this study. [MB818940].
Aspergillus salinicola Zalar, F. Sklenar, Visagie & Hubka, this study. [MB818941].
Aspergillus tardicrescens F. Sklenar, Houbraken, Zalar & Hubka, this study. [MB818942].
Aspergillus penicillioides Speg., Revista Fac. Agron. Univ. Nac. La Plata 2: 245. 1896. [MB309234].
Aspergillus halophilicus clade
This clade contains only one species, Aspergillus halophilicus. It is easily recognisable, because it is the only species from the section that produces sexual reproductive state.
Accepted species:
Aspergillus halophilicus C.M. Chr., Papav. & C.R. Benj., Mycologia 51: 636. 1959. [MB326633].
Species descriptions
Aspergillus caesiellus Saito, J. Fac. Sci. Coll. Imper. Univ. Tokyo 18: 49. 1904, emended description. MycoBank MB205025. Fig. 14.
Fig. 14.
Aspergillus caesiellus. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Synonyms: Aspergillus gracilis var. sartoryi Bat., O.G. Lima & A.F. Vital, Mycopath. Mycol. Appl. 8: 96. 1957.
Aspergillus restrictus mut. eborinus G. Sm., Trans. Brit. Mycol. Soc. 44: 45. 1961.
Typus: Japan, unknown substrate, unknown year of isolation, isolated by K. Saito [neotype Herb. IMI 172278 designated by Samson & Gams (1985) and modified by Pitt & Samson (1993), culture ex-type NRRL 5061 = CBS 470.65 = DTO 093-H3 = ATCC 11905 = IMI 172278 = CCF 5447 = IBT 34620].
Colony diam, 14 d (mm): M40Y 58–60; CYA 11–22; CY20S 21–35; CY20S 30 °C 12–40; CY20S 37 °C No growth; DG18 32–38; MEA 17–22; M60Y 48–60; M60Y 30 °C 45–60; M60Y 37 °C 32–55; MEA + 10 % NaCl 36–45.
Colony characters: M40Y 25 °C, 14 d: flat; colony surface floccose; margin entire; mycelial areas white; sporulation greyish turquoise (24E4); soluble pigment absent; exudate absent; reverse centrally sunburn light brown (6D5) to greyish orange (5B4) in margins. CYA 25 °C, 14 d: with central depression; colony surface floccose, margin entire; mycelial areas white; sporulation sparse; soluble pigment absent; exudate absent; reverse centrally dull blue (23E4) to orange white (5A2) in margins. CY20S 25 °C, 14 d: raised; colony surface floccose; margin entire to undulate; mycelial areas white; sporulation jungle green (25F5) to dull green (25E4); soluble pigment absent; exudate absent; reverse centrally baby blue (bluish grey) (23B3) to orange grey (5B2) in margins. DG18 25 °C, 14 d: centrally raised; colony surface velvety to floccose in the centre, margin entire to delicately filiform; mycelial areas white; sporulation dull green (25D4); soluble pigment absent; exudate absent; reverse centrally birch grey (5C2) to orange grey (5B2) in margins. MEA 25 °C, 14 d: with central depression, irregulary wrinkled; colony surface floccose, margin delicately undulate; mycelial areas white; sporulation sparse; soluble pigment absent; exudate absent; reverse centrally dark blonde (5D4) to greyish orange (5B3) in margins. M60Y 25 °C, 14 d: flat; colony surface filamentous; margin filiform; mycelial areas white; sporulation dull green (27E3); soluble pigment absent; exudate absent; reverse pale yellow (3A3). MEA + 10 % NaCl 25 °C, 14 d: centrally raised; colony surface floccose; margin irregular; mycelial areas white; sporulation dull green (26D3); soluble pigment absent; exudate absent; reverse centrally light yellow (4A4) to pastel yellow (3A4) in margins.
Micromorphology: Conidial heads columnar, frequently sinuous. Conidiophores uniseriate. Stipes smooth, densely covered by long hairs in SEM, occasionally with septa, 5–7.5 μm wide. Vesicles pyriform to clavate, 11–16 μm in diameter. Phialides flask-shaped, densely covered by long hairs in SEM, 8–9 μm long, covering one third to one half of the vesicle. Conidia borne as smooth cylinders, at maturity ellipsoidal to ovate, connectives evident, echinulate, aculeate in SEM, 4.5–6 × 3–4.5 μm.
Distinguishing characters: See distinguishing characters of A. restrictus. The stipes of A. caesiellus are usually wider than those of A. restrictus and narrower than those of A. pachycaulis, vesicles of A. caesiellus are usually larger than those of A. restrictus and smaller than those of A. pachycaulis.
Taxonomic notes: The LSU region of the ex-type strain of A. gracilis var. sartoryi (NRRL 4745; GenBank U29642) is identical with the ex-type strain of A. caesiellus and confirms conclusions of Raper and Fennell (1965) based on morphology.
Authentic strain of A. restrictus mut. eborinus NRRL 4756 is conspecific with A. caesiellus based on molecular data.
Additional materials examined: Germany, air, unknown year of isolation, collector unknown, DTO 077-H6 = CBS 117330. Germany, indoor environment, 2006, collector unknown, DTO 026-C7, DTO 025-I4 = IBT 34538. India, Gorakpur, unknown substrate, before 1974, isolated by T. N. Bhargana, NRRL 25861. Indonesia, corn kernels, unknown year of isolation, collector unknown, DTO 065-D7. Panama, cloth, 1945, isolated by W. H. Weston, NRRL 25810 = CCF 5662. The Netherlands, wood of crate, imported from China, unknown year of isolation, isolated by J. Houbraken, DTO 060-H9. USA, Delaware, outside air, 2012, isolated by Ž. Jurjević, CCF 5450 = EMSL No. 1614. USA, Pennsylvania, home air, 2010, isolated by Ž. Jurjević, CCF 5448 = EMSL No. 1383 = IBT 34621. USA, Delaware, air, pineapple room, warehouse, 2012, isolated by Ž. Jurjević, CCF 5451 = EMSL No. 1650 = IBT 34622. USA, Delaware, home air, 2011, isolated by Ž. Jurjević, CCF 5449 = EMSL No. 1499. USA, Florida, hobnail shoes, 1944, isolated by W. H. Weston, NRRL 25815 = CCF 5663 = DTO 356-D1.
Aspergillus canadensis Visagie, Yilmaz, F. Sklenar & Seifert, sp. nov. Mycobank MB818935. Fig. 15.
Fig. 15.
Aspergillus canadensis. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Etymology: Named after country of origin.
Typus: Canada, Nova Scotia, Wolfville, house dust, 2015, collected by A. Walker, isolated by C.M. Visagie, (holotype DAOM 740109, culture ex-type CCF 5548 = KAS 6194 = DTO 356-H9 = IBT 34520 = IBT 34642 = NRRL 66614).
Colony diam, 14 d (mm): M40Y 25–29; CYA No growth; CY20S (0)10–11; CY20S 30 °C No growth; CY20S 37 °C No growth; DG18 7–15; MEA No growth; M60Y 23–42; M60Y 30 °C 2–20; M60Y 37 °C No growth; MEA + 10 % NaCl 17–33.
Colony characters: M40Y 25 °C, 14 d: Colonies umbonate, irregularly wrinkled, surface velutinous; margin delicately filiform; mycelial areas white; sporulation jungle green (25F5) to pale turquoise (24A3); soluble pigment absent; exudate absent; reverse centrally olive grey (2F2), corn greyish yellow (4B5) to champagne greyish yellow (4B4) in margins. CYA 25 °C, 14 d: no growth. CY20S 25 °C, 14 d: cerebriform, wrinkled; colony surface velutinous, margin undulate; mycelial areas white; sporulation pale turquoise (24A3); soluble pigment absent; exudate absent; reverse centrally dark green (27F7) to greenish grey (25D2), orange white (5A2) in margins. DG18 25 °C, 14 d: centrally raised, radially wrinkled, colony surface velutinous, margin undulate; mycelial areas white; sporulation turquoise grey (24B2); soluble pigment absent; exudate absent; reverse centrally orange grey (5B2) to orange white (5A2) in margins. MEA 25 °C, 14 d: no growth. M60Y 25 °C, 14 d: umbonate; colony surface floccose; margin delicately filiform; mycelial areas white; sporulation greyish green (25C5); soluble pigment absent; exudate absent; reverse centrally olive green (2F8) to mustard yellow (3B6), yellowish white (4A2) in the margins. MEA + 10 % NaCl 25 °C, 14 d: flat to umbonate, irregularly wrinkled; colony surface velutinous; margin entire to delicately filiform; mycelial areas white; sporulation dull green (25D4) to greyish green (25B4); soluble pigment absent; exudate absent; reverse centrally brownish grey (5F2) to mouse grey (5E3), champagne greyish yellow (4B4) in margins.
Micromorphology: Conidial heads radiate. Conidiophores uniseriate. Stipes smooth, sparsely covered by bundles of long hairs in SEM, 3.5–5 μm wide in the middle part, non-septate. Vesicles pyriform to clavate, (9–)13–16(–20) μm wide. Phialides flask-shaped, sparsely covered by long hairs in SEM, 6–8 μm long, covering two thirds of the vesicle. Conidia borne smooth and ellipsoidal, becoming rough-walled, subglobose or barrel-shaped at maturity, lobate-reticulate in SEM, commonly remaining in chains, connectives evident, 3–4 × 2.5–3 μm.
Distinguishing characters: Aspergillus canadensis and A. infrequens are phylogenetically distinct from the core species in the A. penicillioides clade. Aspergillus canadensis is unable to grow on CYA and MEA at 25 °C and grows much slower on M60Y at 30 °C than remaining species from the A. penicillioides clade. The phialides of this species are the shortest across the whole section.
Additional materials examined: Canada, Ontario, Ottawa, house dust, 2015, collected by J. Mack, isolated by C.M. Visagie, DAOMC 251505 = KAS 6095, DAOMC 251506 = KAS 6269, KAS 7704 = DTO 357-A7, KAS 7705 = DTO 357-A8, CCF 5549 = KAS 7706 = DTO 357-A9 = IBT 34639 = NRRL 66615, KAS 7707 = DTO 357-B1, CCF 5550 = KAS 7708 = DTO 357-B2 = IBT 34637, CCF 5551 = KAS 7709 = DTO 357-B3 = IBT 34640, CCF 5552 = KAS 7710 = DTO 357-B4 = IBT 34636, KAS 7711 = DTO 357-B5, KAS 7716 = DTO 357-B8, KAS 7717 = DTO 357-B9, KAS 7718 = DTO 357-C1, CCF 5553 = KAS 7719 = DTO 357-C2 = IBT 34638, KAS 7720 = DTO 357-C3 = IBT 34641, KAS 7721 = DTO 357-C4.
Note: The majority of strains of A. canadensis quickly degenerated in culture and changed significantly their phenotype (restricted growth, waxy colonies with poor or no sporulation).
Aspergillus clavatophorus F. Sklenar, S.W. Peterson & Hubka, sp. nov. MycoBank MB818936. Fig. 16.
Fig. 16.
Aspergillus clavatophorus. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Etymology: Refers to the clavate vesicles of the conidiophores
Typus: USA, Georgia, Athens, mouldy paper, 1987, isolated by R. T. Hanlin (holotype PRM 944440, isotype PRM 944441, culture ex-type NRRL 25874 = CCF 5454 = IBT 34560 = IBT 34823 = DTO 356-D8).
Colony diam, 14 d (mm): M40Y 27–33; CYA No growth; CY20S 7–16; CY20S 30 °C No growth; CY20S 37 °C No growth; DG18 18–25; MEA No growth; M60Y 35–44; M60Y 30 °C 29–54; M60Y 37 °C (0)5–17; MEA + 10 % NaCl 18–28.
Colony characters: M40Y 25 °C, 14 d: Colonies raised to umbonate, surface floccose to velutinous; margin entire to delicately filiform; mycelial areas white; sporulation greyish green (28D6); soluble pigment absent; exudate absent; reverse centrally coffee brown (5F7), golden brown (5D7) to pale yellow (4A3) in margins. CYA 25 °C, 14 d: no growth to microcolonies. CY20S 25 °C, 14 d: convex, radially wrinkled; colony surface floccose, margin undulate; mycelial areas white; sporulation rare, sand greyish yellow (4B3); soluble pigment absent; exudate absent; reverse centrally yellowish grey (4B2) to orange white (5A2) in margins. DG18 25 °C, 14 d: centrally raised; colony surface floccose to cottony, margin entire to delicately filiform; mycelial areas white; sporulation aquamarine greyish green (25B4) to pale green (27A3); soluble pigment absent; exudate absent; reverse centrally greyish turquoise (24C3) to orange white (5A2) in margins. MEA 25 °C, 14 d: no growth. M60Y 25 °C, 14 d: raised; colony surface floccose to lanose; margin entire to delicately filiform; mycelial areas white; sporulation greyish green (25D6); soluble pigment absent; exudate absent; reverse centrally dark brown (6F6) to golden brownish orange (5C6), pale yellow (4A3) in margins. MEA + 10 % NaCl 25 °C, 14 d: flat to umbonate; colony surface powdery to floccose; margin entire to delicately filiform; mycelial areas white; sporulation greyish turquoise (water blue) (24C5); soluble pigment absent; exudate absent; reverse centrally coffee brown (5F7), olive brown (4E5) to yellowish orange (4B7), cream pale yellow (4A3) in margins.
Micromorphology: Conidial heads at first globose, later becoming radiate. Conidiophores uniseriate. Stipes smooth, densely covered by short hairs in SEM, 6–9 μm wide in the middle part, widening gradually toward the vesicle, non-septate. Vesicles spatulate to clavate, 14–19 μm wide. Phialides flask-shaped, densely covered by short hairs in SEM, 8–10 μm long, covering two thirds to three quarters of the vesicle. Conidia borne smooth, ellipsoidal, becoming rough-walled, tuberculate in SEM, subglobose, ovate or barrel-shaped at maturity, connectives evident, 4–5 × 3–4 μm.
Distinguishing characters: See distinguishing characters of A. penicillioides. Isolates of A. clavatophorus have larger conidia (on average 4.8 × 3.5 μm) compared to A. tardicrescens (on average 4 × 2.9 μm) and smaller vesicles (on average 16.5 μm) compared to A. magnivesiculatus (on average 21.4 μm). The shape of the vesicles is usually spatulate to clavate in contrast to all other members of the A. penicillioides clade that have at least in part pyriform vesicles. Isolates of this species can grow on M60Y at 37 °C.
Additional materials examined: China, puerh tea, unknown year of isolation, isolated by J. Houbraken, DTO 257-G5 = CCF 5669 = IBT 34561. The Netherlands, unknown substrate, 2014, isolated by M. Meijer, DTO 316-B3. The Netherlands, painting, 2014, isolated by J. Houbraken, DTO 324-G2. USA, Georgia, Athens, mouldy paper, 1987, isolated by R. T. Hanlin, NRRL 25873 = CCF 5453 = IBT 34632. USA, Illinois, Peoria, mouldy cardboard, 1989, collector unknown, NRRL 25875 = CCF 5455.
Aspergillus conicus Blochwitz in Dale, Ann. Mycol. 12: 38. 1914, emended description. MycoBank MB120214. Fig. 17.
Fig. 17.
Aspergillus conicus. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Typus: Unknown locality of isolation, unknown substrate, 1924, isolated by P. Biourge [neotype Herb. IMI 172281 designated by Samson & Gams (1985), culture ex-type NRRL 149 = CBS 475.65 = IBT 33667 = DTO 096-H6 = ATCC 16908 = IMI 172281 = CCF 5456].
Colony diam, 14 d (mm): M40Y 32–40; CYA (0)10–12; CY20S 5–18; CY20S 30 °C No growth; CY20S 37 °C No growth; DG18 18–31; MEA (0)8–10; M60Y 26–40; M60Y 30 °C 26–37; M60Y 37 °C No growth; MEA + 10 % NaCl 16–36.
Colony characters: M40Y 25 °C, 14 d: Colonies flat, wrinkled, surface velutinous; margin filiform; mycelial areas white; sporulation pale turquoise (24A3); soluble pigment absent; exudate absent; reverse centrally olive grey (2F2) to champagne greyish yellow (4B4) in margins. CYA 25 °C, 14 d: crateriform, wrinkled; colony surface velutinous, margin undulate; mycelial areas white; sporulation sparse; soluble pigment absent; exudate absent; reverse centrally Persian blue (23A3) to yellowish white (4A2) in margins. CY20S 25 °C, 14 d: cerebriform; colony surface velutinous, margin undulate; mycelial areas white; sporulation light turquoise (24A4); soluble pigment absent; exudate absent; reverse dark turquoise (23F7) to greyish brown (7E3), orange white (5A2) in margins. DG18 25 °C, 14 d: flat, wrinkled; colony surface velutinous to filamentous, margin filiform; mycelial areas white; sporulation bluish grey (23B3); soluble pigment absent; exudate absent; reverse centrally greyish turquoise (24D3) to orange white (5A2) in margins. MEA 25 °C, 14 d: cerebriform; colony surface velutinous, margin undulate; mycelial areas white; sporulation Persian blue (23A3); soluble pigment absent; exudate absent; reverse centrally camel light brown (6D4) to sand greyish yellow (4B3) in margins. M60Y 25 °C, 14 d: flat; colony surface velutinous; margin filiform; mycelial areas white; sporulation pale turquoise (24A3); soluble pigment absent; exudate absent; reverse olive grey (2F2) to lead grey (2D2), champagne greyish yellow (4B4) in margins. MEA + 10 % NaCl 25 °C, 14 d: flat; colony surface velutinous; margin filiform; mycelial areas white; sporulation pale turquoise (24A3); soluble pigment absent; exudate absent; reverse centrally greenish grey (27D2) to sand greyish yellow (4B3) in margins.
Micromorphology: Conidial heads loosely columnar, frequently twisted. Conidiophores uniseriate. Stipes smooth, sparsely covered by short hairs in SEM, 4–5.5 μm wide in the middle part, widening gradually toward the vesicle, with occasional septa. Vesicles pyriform, spatulate to clavate, 8–13 μm wide. Phialides flask-shaped, sparsely covered by short hairs in SEM, 8.5–10 μm long, covering one third to one half of the vesicle. Conidia borne as smooth long cylinders, then becoming delicately rough to definitely aculeate in SEM, globose to subglobose or barrel-shaped, commonly remaining in chains, connectives evident, 4–4.5 × 2.5–3 μm.
Distinguishing characters: Aspergillus conicus is most closely related to A. domesticus and A. destruens. The colour of sporulating colonies on M40Y is pale turquoise in A. conicus, while myrtle green in A. destruens; reverse of A. conicus is olive grey in the centre, but apricot greyish orange in A. destruens. These three species share almost identical micromorphology, but differences can be found in their response to the osmotic stress. Aspergillus conicus grows fastest on MEA + 5 % NaCl and MEA + 10 % NaCl, while A. domesticus grows slowest (see Fig. 12E). Aspergillus gracilis and A. pseudogracilis differ from these three species by their inability to grow on MEA and CYA at 25 °C (several strains of A. gracilis exhibited very restricted growth on CYA) and more rapid growth on M60Y at 25 °C and 30 °C. In contrast to A. domesticus, isolates of A. conicus can grow on MEA and grow slightly faster on CYA. Aspergillus villosus can be distinguished from A. conicus, A. domesticus and A. destruens by its faster growth on CY20S, ability to grow on CY20S at 30 °C and on M60Y at 37 °C.
Additional materials examined: Belgium, candy, 2004, collector unknown, IHEM 20709. Belgium, Braine-l'Alleud, wooden statue, 2000, collector unknown, IHEM 16531. Czech Republic, kernel of Bertholletia excelsa (Brazil nut), unknown year of isolation, isolated by L. Marvanová, CCF 4042. Denmark, near Copenhagen, air in bathroom, 2009, isolated by J. C. Frisvad, DTO 110-C5. Denmark, near Copenhagen, air in living room, 2009, isolated by J. C. Frisvad, DTO 110-F5 = CCF 5667 = IBT 34534. France, French madeleines duchesse, 2013, isolated by F. Déniel, CBS 140429 = UBOCC-A-115041 = DTO 334-D9. Germany, Koln, air, unknown year of isolation, collector unknown, DTO 077-H2 = CBS 117332. Germany, indoor air, 2014, isolated by U. Hack, DTO 321-H2, DTO 321-H5. India, Tamil Nadu, Tiruchirapally, Musa acuminate, unknown year of isolation, DTO 079-A9 = CBS 122.453. Israel, Eliat, microbial mat, 2008, isolated by R. Tkavc, EXF-5015 = CCF 5650 = IBT 34273. Puerto Rico, Bayamon, office air, 2014, isolated by Ž. Jurjević, CCF 5461 = EMSL No. 2549. Slovenia, Ljubljana, oil painting on canvas, 2012, isolated by P. Zalar, EXF-7663 = IBT 34267 = IBT 33574, EXF-7660 = IBT 34263 = IBT 33577. Slovenia, Ljubljana, oil painting on canvas, 2015, isolated by P. Zalar and D.D. Graf, EXF-10387 = CCF 5651 = IBT 34289. The Netherlands, Witten, indoor air, unknown year of isolation, isolated by C. Trautmann and I. Dill, DTO 077-H5. The Netherlands, Zwartewaal, museum piece, 2012, isolated by M. Meijer, DTO 231-C2, DTO 231-C3 = CCF 5665. The Netherlands, Eindhoven, indoor air, 2006, isolated by J. Houbraken, DTO 017-B9. USA, Illinois, Chicago, asphalt roof shingle, 1959, isolated by H. G. Hedrick, NRRL 25848. USA, Delaware, air, pineapple room, warehouse, 2012, isolated by Ž. Jurjević, CCF 5459 = EMSL No. 1649. USA, California, home air, 2011, isolated by Ž. Jurjević, CCF 5458 = EMSL No. 1490. USA, Idaho, home air, 2010, isolated by Ž. Jurjević, CCF 5457 = NRRL 62007 = EMSL No. 1318. USA, New York, unknown substrate, 1975, isolated by I. Weitzman, NRRL 25881. USA, Pennsylvania, West Chester, living room air, 2013, isolated by Ž. Jurjević, CCF 5460 = EMSL No. 2217. USA, Georgia, Kennesaw, basement, 2015, isolated by Ž. Jurjević, EMSL No. 3245. USA, Missouri, Saint Louis, bedroom, wood base, 2015, isolated by Ž. Jurjević, EMSL No. 3209. Unknown locality of isolation, unknown substrate, 2007, isolated by T. van Doorn, DTO 039-G7.
Aspergillus destruens Zalar, F. Sklenar, S.W. Peterson & Hubka, sp. nov. MycoBank MB818930. Fig. 18.
Fig. 18.
Aspergillus destruens. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Etymology: Named after the common occurrence of this species on old deteriorated paintings.
Typus: USA, Maryland, maize seed, 1917, isolated by C. V. Piper (holotype PRM 944428, isotype PRM 944429, culture ex-type NRRL 145 = IMI 358691 = CCF 5462 = CBS 593.91 = DTO 079-A8 = IBT 34818).
Colony diam, 14 d (mm): M40Y 19–36; CYA 5–10; CY20S 6–13; CY20S 30 °C No growth; CY20S 37 °C No growth; DG18 14–30; MEA (0) 4–7; M60Y 34–41; M60Y 30 °C 16–41; M60Y 37 °C No growth; MEA + 10 % NaCl 20–30.
Colony characters: M40Y 25 °C, 14 d: Colonies centrally raised and winkled, surface velutinous; margin irregular and delicately filiform; mycelial areas white; sporulation myrtle green (25F7); soluble pigment absent; exudate absent; reverse centrally apricot greyish orange (5B6) to light yellow (4A4) in margins. CYA 25 °C, 14 d: cerebriform; colony surface velutinous, margin undulate; mycelial areas white; sporulation turquoise white (24A2); soluble pigment absent; exudate absent; reverse centrally sky grey (23B2) to cream pale yellow (4A3) in margins. CY20S 25 °C, 14 d: cerebriform to crateriform; colony surface velutinous, margin entire to delicately filiform; mycelial areas white; sporulation light turquoise (24A4); soluble pigment absent; exudate absent; reverse greyish turquoise (24E7) to pale orange (5A3) in margins. DG18 25 °C, 14 d: craterifiorm; colony surface floccose, margin entire to undulate; mycelial areas white; sporulation aquamarine greyish turquoise (23B3); soluble pigment greyish yellow (2B3); exudate absent; reverse centrally dull green (27E3) to yellowish white (4A2) in margins. MEA 25 °C, 14 d: cerebriform; colony surface velutinous, margin entire; sporulation greenish grey (25B2); soluble pigment absent; exudate absent; reverse centrally camel olive grey (2F2) to sand dark yellow (4C8) in margins. M60Y 25 °C, 14 d: umbonate; colony surface floccose to filamentous; margin entire to delicately filiform; mycelial areas white; sporulation pastel green (25B4); soluble pigment absent; exudate absent; reverse cinnamon brown (6D6) to champagne greyish yellow (4B4). MEA + 10 % NaCl 25 °C, 14 d: raised, irregularly wrinkled; colony surface velutinous; margin delicately filiform; mycelial areas white; sporulation bluish grey (23B3); soluble pigment absent; exudate absent; reverse centrally greenish grey (27D2) to cream pale yellow (4A3) in margins.
Micromorphology: Conidial heads loosely columnar. Conidiophores uniseriate. Stipes smooth, densely covered by long hairs in SEM, 4–6 μm wide in the middle part, widening gradually toward the vesicle, with occasional septa. Vesicles predominantly ellipsoidal to pyriform, less commonly spatulate to clavate, 8–14 μm wide. Phialides flask-shaped, densely covered by long hairs in SEM, 8.5–11.5 μm long, covering one to two thirds of the vesicle. Conidia borne as smooth, long cylinders, becoming delicately rough and definitely echinulate at maturity, aculeate in SEM, subglobose to ovate, commonly remaining in chains, connectives evident, 3.5–5.5 × 2.5–3.5 μm.
Distinguishing characters: See distinguishing characters of A. conicus. Aspergillus destruens is most closely related to A. conicus and A. domesticus, but even though the latter two are morphologically similar, they are different from A. destruens. The colour of sporulating colonies of A. destruens on M40Y and M60Y is darker, the reverse colour is apricot greyish orange in the centre in contrast to olive grey in the other two species.
Additional materials examined: Hungary, indoor air, 2009, collector unknown, DTO 147-E2. Slovenia, Ljubljana, oil painting on canvas, 2015, isolated by P. Zalar and D.D. Graf, EXF-10350 = IBT 34270, EXF-10407 = CCF 5652 = IBT 34285, EXF-10411 = IBT 34265, EXF-10390 = IBT 34276, EXF-10017 = IBT 33576. Slovenia, Ljubljana, oil painting on canvas, 2012, isolated by P. Zalar and D.D. Graf, EXF-7699 = IBT 34262, EXF-7651 = CCF 5653 = IBT 34258, EXF-7661 = IBT 34271 = IBT 33573, EXF-7667 = IBT 34288, EXF-7703 = IBT 34259, EXF-7657 = IBT 34264, EXF-10431 = IBT 34257, EXF-7666 = IBT 34268. The Netherlands, Utrecht, air in villa, unknown year of isolation, collector unknown, DTO 254-B2 = IBT 34522. The Netherlands, Tilburg, air in bakery, 2012, collector unknown, DTO 220-B2. The Netherlands, surface of cheese, 2011, isolated by J. Houbraken, DTO 161-B7 = CCF 5671. The Netherlands, Tilburg, air in bakery, 2009, isolated by J. Houbraken, DTO 113-E7 = CCF 5668. The Netherlands, Leerdam, unknown substrate, unknown year of isolation, isolated by M. Meijer, DTO 303-H2. USA, unknown substrate, 1939, isolated by C.W. Emmons, NRRL 157 = CCF 5463.
Aspergillus domesticus F. Sklenar, Houbraken, Zalar & Hubka, sp. nov. MycoBank MB818931. Fig. 19.
Fig. 19.
Aspergillus domesticus. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Etymology: Referring to the common occurrence of this species in indoor environments.
Typus: The Netherlands, Tiel, wallpaper, 2008, collector unknown (holotype PRM 944426, isotype PRM 944427, culture ex-type DTO 079-F2 = CCF 5464 = NRRL 66616 = IBT 34814).
Colony diam, 14 d (mm): M40Y 36–38; CYA 0–8; CY20S 5–17; CY20S 30 °C No growth; CY20S 37 °C No growth; DG18 20–24; MEA No growth; M60Y 29–38; M60Y 30 °C 15–30; M60Y 37 °C No growth; MEA + 10 % NaCl 23–31.
Colony characters: M40Y 25 °C, 14 d: Colonies flat, wrinkled in the centre, surface velutinous; margin filiform; mycelial areas white; sporulation aquamarine greyish turquoise (24B3); soluble pigment absent; exudate absent; reverse centrally olive grey (2F2) to champagne greyish yellow (4B4) in margins. CYA 25 °C, 14 d: crateriform; colony surface velutinous, margin undulate; mycelial areas white; sporulation turquoise white (24A2); soluble pigment absent; exudate absent; reverse centrally fog blue (23C3) to orange white (5A2) in margins. CY20S 25 °C, 14 d: cerebriform; colony surface velutinous, margin entire to delicately filiform; mycelial areas white; sporulation light turquoise (24A5) to turquoise white (24A2); soluble pigment absent; exudate absent; reverse dark turquoise (23F7) to fog blue (23C3), pale orange (5A3) in margins. DG18 25 °C, 14 d: umbonate, delicately wrinkled; colony surface powdery to velutinous, margin entire to delicately filiform; mycelial areas white; sporulation greyish turquoise (24C3); soluble pigment absent; exudate absent; reverse centrally peacock blue (24D7) to orange white (5A2) in margins. MEA 25 °C, 14 d: no growth. M60Y 25 °C, 14 d: flat; colony surface velutinous to powdery; margin filiform; mycelial areas white; sporulation turquoise grey (24B2); soluble pigment absent; exudate absent; reverse light blond greyish yellow (4C3). MEA + 10 % NaCl 25 °C, 14 d: flat, wrinkled; colony surface velutinous; margin entire to delicately filiform; mycelial areas white; sporulation light turquoise (24A4); soluble pigment absent; exudate absent; reverse centrally nougat greyish brown (5D3) to sand greyish yellow (4B3) in margins.
Micromorphology: Conidial heads loosely columnar, commonly twisted. Conidiophores uniseriate. Stipes smooth, densely covered by long hairs in SEM, 4–6 μm wide in the middle part, widening gradually toward the vesicle, with occasional septa. Vesicles pyriform, spatulate to clavate, 7–13 μm wide. Phialides flask-shaped, densely covered by long hairs in SEM, 8.5–11 μm long, covering the upper half of the vesicle. Conidia borne as smooth, long cylinders, becoming echinulate at maturity, aculeate in SEM, ellipsoidal or barrel-shaped, commonly remaining in chains, connectives evident, 3.5–4.5 × 2.5–3.5 μm.
Distinguishing characters: See distinguishing characters of A. conicus. Aspergillus domesticus is most closely related to A. conicus and A. destruens (Fig. 5). Unlike the two other species, A. domesticus does not grow on MEA and also grows slower on CYA.
Additional materials examined: Belgium, Brussels, dust from mattress, 1991, collector unknown, IHEM 6549. France, French almond madeleines, 2014, isolated by F. Déniel, UBOCC-A-115040 = CBS 140428 = DTO 334-D8, UBOCC-A-115039 = CBS 140427 = DTO 334-D7. France, air from the bakery oven room, 2014, isolated by F. Déniel, UBOCC-A-115038. France, French mini cakes, 2013, isolated by F. Déniel, UBOCC-A-115048 = CBS 140436 = DTO 334-E7. France, Brest, painting from Brittany (painter: E.J. Damery; painting title: Portrait de femme; painting technique: oil on canvas; and painter: J. Gamelin; painting title: Sainte en extase; painting technique: oil on canvas), 2015, isolated by F. Déniel, UBOCC-A-116022 = CCF 5465. Korea, Phragmidium sp. (rust) on Rubus coreanus, 2004, collected by H.D. Shin, isolated by P. Crous, CBS 116419 = DTO 077-G6 = IBT 34535. Slovenia, Ljubljana, oil painting on canvas, 2015, isolated by P. Zalar, EXF-10583 = IBT 34275. Slovenia, Ljubljana, statue made of wood, textile and sea shells, 2015, isolated by P. Zalar, EXF-10012 = IBT 34274. The Netherlands, Zwartewaal, museum piece, 2012, isolated by M. Meijer, DTO 231-C1 = CCF 5666 = NRRL 66617, DTO 231-B9. The Netherlands, Gorinchem, archive material, 2008, isolated by J. Houbraken and M. Meijer, DTO 086-D1 = CCF 5670. The Netherlands, Paleis het Loo, wallpaper, unknown year of isolation, isolated by J. Varga, CBS 233.90 = DTO 079-A7 = IBT 34515. USA, Idaho, home air, 2010, isolated by Ž. Jurjević, CCF 5466 = EMSL No. 1316. USA, New Jersey, Freehold, breakfast room, settle plates, 2015, isolated by Ž. Jurjević, EMSL No. 2780.
Aspergillus glabripes F. Sklenar, Ž. Jurjević & Hubka, sp. nov. MycoBank MB818934. Fig. 20.
Fig. 20.
Aspergillus glabripes. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Etymology: Referring to the absence of fine hairs on the stipe which are visible in majority of sect. Restricti species by SEM.
Typus: Trinidad & Tobago, Macoya, office folder, 2014, isolated by Ž. Jurjević (holotype PRM 944436, isotype PRM 944437, culture ex-type CCF 5474 = DTO 356-E8 = EMSL No. 2462 = NRRL 66618 = IBT 34820).
Colony diam, 14 d (mm): M40Y 38–52; CYA 3–7; CY20S 8–18; CY20S 30 °C 5–16; CY20S 37 °C No growth; DG18 15–25; MEA 4–11; M60Y 35–40; M60Y 30 °C 41–56; M60Y 37 °C (0)7–10; MEA + 10 % NaCl 22–28.
Colony characters: M40Y 25 °C, 14 d: Colonies raised, surface floccose; margin entire to delicately filiform; mycelial areas white; sporulation yellowish green (29C7); soluble pigment absent; exudate absent; reverse centrally dark brown (6F7) to champagne greyish yellow (4B4) in margins. CYA 25 °C, 14 d: cerebriform; colony surface floccose, margin undulate; mycelial areas white; sporulation sparse; soluble pigment absent; exudate absent; reverse pale orange (5A3). CY20S 25 °C, 14 d: centrally slightly raised; colony surface floccose, margin entire to delicately filiform; mycelial areas white; sporulation yellowish green (29B7); soluble pigment absent; exudate absent; reverse centrally yellowish grey (4D3) to yellowish white (4A2) in margins. DG18 25 °C, 14 d: umbonate; colony surface floccose, margin entire; mycelial areas yellowish white (4A2); sporulation pale green (29A3); soluble pigment absent; exudate absent; reverse centrally yellowish orange (4B7) to yellowish white (4A2) in margins. MEA 25 °C, 14 d: cerebriform; colony surface floccose, margin filiform; mycelial areas white; sporulation sparse; soluble pigment absent; exudate absent; reverse centrally clay brownish orange (5C5) to pale orange (5A3) in margins. M60Y 25 °C, 14 d: flat; colony surface floccose; margin entire to delicately filiform; mycelial areas white; sporulation deep green (26D8) to yellowish green (29A8); soluble pigment absent; exudate absent; reverse centrally olive brown (4E5) to corn greyish yellow (4B5) in margins. MEA + 10 % NaCl 25 °C, 14 d: flat; colony surface floccose, margin entire to delicately filiform; mycelial areas white; sporulation opal greyish green (25C6) to light green (28A5); soluble pigment absent; exudate absent; reverse centrally olive brown (2F6) to champagne greyish yellow (4B4) in margins.
Micromorphology: Conidial heads radiate. Conidiophores uniseriate. Stipes smooth in SEM, 6–8 μm wide in the middle part, widening gradually toward the vesicle, with occasional septa. Vesicles globose, pyriform to subclavate, 16–22 μm wide. Phialides flask-shaped, 7–9 μm long, covering three quarters of the vesicle. Conidia borne as smooth cylinders, echinulate at maturity, aculeate in SEM, subglobose, connectives evident, 3.5–4.5 × 2–3 μm.
Distinguishing characters: See distinguishing characters of A. vitricola. Aspergillus glabripes can easily be differentiated from A. vitricola by its large vesicles that are most commonly globose or pyriform. Most A. glabripes isolates have a yellowish green colour of sporulation on M40Y and M60Y.
Additional materials examined: Canada, New Brunswick, Little Lepreau, house dust, 2015, collected by A. Walker, isolated by C.M. Visagie, KAS 6253. France, Brest, painting from Brittany (painter: A. Nozal; painting title: Saint-Briac, grève à marée basse; painting technique: pastel; and painter: B.M. Agüro; painting title: La vision de Saint Antoine de Padoue; painting technique: oil on canvas), 2015, isolated by F. Déniel, UBOCC-A-116021 = CCF 5467 = IBT 34625. France, brownies, 2014, isolated by F. Déniel, UBOCC-A-115046. Trinidad & Tobago, Macoya, office folder, 2014, isolated by Ž. Jurjević, CCF 5475 = EMSL No. 2463, CCF 5476 = EMSL No. 2464, CCF 5477 = EMSL No. 2465. USA, New Jersey, green fabric covered binders, import from China, 2014, isolated by Ž. Jurjević, CCF 5473 = EMSL No. 2442 = IBT 34626. USA, California, home air, 2011, isolated by Ž. Jurjević, CCF 5469 = EMSL No. 1483 = DTO 356-E5 = IBT 34519 = IBT 34821 = NRRL 66619. USA, Louisiana, library, front cover of log book, 2012, isolated by Ž. Jurjević, CCF 5470 = EMSL No. 1812. USA, Louisiana, library, book, 2012, isolated by Ž. Jurjević, CCF 5471 = EMSL No. 1813. USA, Idaho, home air, 2010, isolated by Ž. Jurjević, CCF 5468 = EMSL No. 1317. USA, South Carolina, Summerville, kitchen air, 2014, isolated by Ž. Jurjević, CCF 5472 = EMSL No. 2305.
Aspergillus gracilis Bainier, Bull. Soc. Mycol. Fr. 23: 92. 1907, emended description. MycoBank MB167554. Fig. 21.
Fig. 21.
Aspergillus gracilis. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Synonyms: Aspergillus gracilis var. exiguus Bainier & Sartory, Bull. Soc. Mycol. Fr. 28: 47. 1912.
Typus: South Pacific, gun-firing mechanism, unknown year of isolation, R. Emerson (neotype Herb. IMI 211393 designated by Samson & Gams (1985), culture ex-type NRRL 4962 = CBS 539.65 = DTO 351-H7 = CCF 5478 = ATCC 16906 = IMI 211393 = IBT 34817).
Colony diam, 14 d (mm): M40Y 30–42; CYA 0–8; CY20S 7–13; CY20S 30 °C No growth; CY20S 37 °C No growth; DG18 9–16; MEA No growth; M60Y 38–44; M60Y 30 °C 39–45; M60Y 37 °C No growth; MEA + 10 % NaCl 12–24.
Colony characters: M40Y 25 °C, 14 d: Colonies flat, wrinkled in the centre, surface velutinous; margin entire to delicately filiform; mycelial areas white; sporulation aquamarine greyish turquoise (24B3) to turquoise white (24A2); soluble pigment absent; exudate absent; reverse centrally olive grey (2F2) to straw yellow (3B4), cream pale yellow (4A3) in margins. CYA 25 °C, 14 d: no growth to microcolonies. CY20S 25 °C, 14 d: crateriform; colony surface velutinous, margin filiform; mycelial areas white; sporulation turquoise grey (24C2); soluble pigment absent; exudate absent; reverse dull green (25D3) to pale orange (5A3) in margins. DG18 25 °C, 14 d: convex; colony surface floccose to velutinous, margin filiform; mycelial areas white; sporulation greyish turquoise (24D5); soluble pigment absent; exudate absent; reverse centrally yellowish grey (4B2) to orange white (5A2) in margins. MEA 25 °C, 14 d: no growth. M60Y 25 °C, 14 d: raised; colony surface granular to floccose in the centre; margin irregular, filiform; mycelial areas white; sporulation pale turquoise (24A3); soluble pigment absent; exudate absent; reverse centrally golden yellow (5B7) to champagne greyish yellow (4B4) in margins. MEA + 10 % NaCl 25 °C, 14 d: raised; colony surface velutinous, margin entire to irregular; mycelial areas white; sporulation pale turquoise (24A3); soluble pigment absent; exudate absent; reverse centrally cinnamon tan brown (6E6) to champagne greyish yellow (4B4) in margins.
Micromorphology: Conidial heads loosely columnar, frequently twisted. Conidiophores uniseriate. Stipes smooth, densely covered by very long hairs in SEM, 3–5 μm wide in the middle part, with occasional septa. Vesicles spatulate to clavate, 6.5–9 μm wide. Phialides flask-shaped, densely covered by very long hairs in SEM, 7.5–9 μm long, covering up to two thirds of the vesicle. Conidia borne as smooth cylinders, echinulate at maturity, aculeate in SEM, subglobose or barrel-shaped, connectives evident, 3–4 × 2–3 μm.
Distinguishing characters: Aspergillus gracilis is most closely related to A. pseudogracilis. These two species can be distinguished from other species belonging to the A. conicus clade by their inability to grow on MEA and CYA at 25 °C (several strains of A. gracilis exhibit very restricted growth on CYA) and more rapid growth on M60Y at 25 °C and 30 °C. Aspergillus pseudogracilis can be distinguished from A. gracilis by its wider stipes, larger vesicles and longer phialides. Growth parameters of both species in the osmotic gradient are identical (see Fig. 12D).
Notes: Aspergillus gracilis var. exiguus was proposed based on slight differences in physiological characters and treated as synonym of A. gracilis by Raper & Fennell (1965). We could not obtain a living culture and no herbarium material is extant, resulting in us following conclusions of Raper & Fennell (1965).
Additional materials examined: USA, California, San Diego, child carrier, 2015, isolated by Ž. Jurjević, CCF 5479 = EMSL No. 2775 = DTO 356-F4 = IBT 34559, CCF 5480 = EMSL No. 2920, CCF 5481 = EMSL No. 2922, CCF 5482 = EMSL No. 2923 = IBT 34623, EMSL No. 2763, EMSL No. 2774.
Aspergillus halophilicus C.M. Chr., Papav. & C.R. Benj., Mycologia 51: 636. 1959, emended description. MycoBank MB326633. Fig. 22.
Fig. 22.
Aspergillus halophilicus. A. Colonies after 14 d at 25 °C: left to right, obverse CZA70S, reverse CZA70S, M40Y and MEA + 10 % NaCl. B–E. Cleistothecia. F, G. Asci. H, I. Ascospores. J, K. SEM (ascospores). Scale bars: D–I = 10 μm; J, K = 2 μm.
Typus: USA, Minnesota, St. Paul, dried corn, 1957, isolated by C. M. Christensen [lectotype BPI 566153 designated by Samson & Gams (1985), culture ex-type NRRL 2739 = CCF 5687 = ATCC 16401 = CBS 12262 = IMI 211802 = IBT 34878].
Colony diam, 30 d (mm): CZA70S 10–12; no growth on all other tested media.
Colony characters: CZA70S 25 °C, 30 d: Colonies flat, surface floccose; margin filiform; mycelial areas hyaline to white; sporulation absent; ascomata produced after months of incubation, eurotium-like, hyaline to pale yellow; soluble pigment absent; exudate absent; reverse centrally greyish orange (6C6) to orange yellow (5A2) in margins.
Micromorphology: Ascomata maturing slowly within 1–2 mo on CZA70S at 25 °C, 100–150 μm diam, hyaline to pale yellow (3A3), asci globose to subglobose, 9–15 × 7–11 μm, ascospores hyaline, lenticular with two equatorial crests, convex surface finely roughened in the area neighbouring the equatorial region, with holes, 4.5–6.5 × 3.5–5 μm including crests.
Conidial state not observed in our cultures but (Christensen et al. 1959) observed it after 45 d on PDA + 20 % NaCl. Conidial heads at first globose, later becoming loosely radiate. Conidiophores uniseriate. Vesicles globose to subglobose or club-shaped, occasionally with one or two enlargements below the vesicle, 6–10 μm wide. Phialides flask-shaped, 6–14 μm long, covering only a portion of the top but occasionally up to three quarters of the vesicle, sometimes growing from the swellings below the vesicle. Conidia globose, elliptical, pyriform or irregular, spiny, hyaline to light blue-green, 6–11 × 4–6.5 μm.
Distinguishing characters: Aspergillus halophilicus can easily be distinguished from all other members of section Restricti by its eurotium-like sexual reproductive states and inability to grow on majority of tested media, including those with high sugar (M60Y) or salt (MEA + 10 % NaCl) content after 14 d.
Additional materials examined: Italy, Venice, book cover (swab), 2013, isolated by A. Micheluz, MUT 562. Italy, Lazio, Rome, Archivio Stampati del Vaticano, book cover (swab), 2010, isolated by M. Montanari, MUT 798. Italy, Marche, Loreto, Archivio della Casa Madre, book cover (swab), 2012, isolated by M. Montanari, MUT 1305. Italy, Piedmont, Torre Pellice, Archivio della Tavola Valdese, book cover (swab), 2012, isolated by M. Montanari, MUT 1307. The Netherlands, textile imported from unknown country, 2013, isolated by M. Meijer, DTO 271-F4 = CCF 5825 = IBT 34881.
Aspergillus hordei F. Sklenar, S.W. Peterson & Hubka, sp. nov. MycoBank MB818937. Fig. 23.
Fig. 23.
Aspergillus hordei. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Etymology: Named after the isolation of species from barley (Hordeum vulgare).
Typus: USA, Minnesota, St. Paul, barley, 1957, isolated by G. C. Papavizas (holotype PRM 944446, isotype PRM 944447, culture ex-type NRRL 25825 = CCF 5483 = DTO 356-D3 = IBT 34539).
Colony diam, 14 d (mm): M40Y 25–29; CYA 0–8; CY20S (0)13–16; CY20S 30 °C (0)8–15; CY20S 37 °C No growth; DG18 13–26; MEA 0–5; M60Y 31–47; M60Y 30 °C 25–48; M60Y 37 °C 22–33; MEA + 10 % NaCl 26–29.
Colony characters: M40Y 25 °C, 14 d: Colonies umbonate, surface velutinous; margin entire; mycelial areas white; sporulation dark turquoise (24F8); soluble pigment absent; exudate absent; reverse centrally Cuba reddish brown (9E8), less commonly brownish red (8C7) to olive yellow (3C7), pale yellow (4A3) in margins. CYA 25 °C, 14 d: cerebriform; colony surface floccose, margin undulate; mycelial areas white; sporulation sparse; soluble pigment absent; exudate absent; reverse centrally orange grey (5B2) to orange white (5A2) in margins. CY20S 25 °C, 14 d: cerebriform to crateriform; colony surface velutinous, margin entire to delicately filiform; mycelial areas white; sporulation aquamarine greyish turquoise (24B3) to greyish turquoise (24D4); soluble pigment absent; exudate absent; reverse centrally aquamarine turquoise grey (24E2) to turquoise grey (24C2), orange white (5A2) in margins. DG18 25 °C, 14 d: crateriform; colony surface velutinous, margin entire to delicately filiform; mycelial areas white; sporulation aquamarine dark turquoise (24F8); soluble pigment greyish yellow (2B3); exudate absent; reverse centrally yellowish brown (5E8) to olive grey (2E2), champagne greyish yellow (4B4) to yellowish white (4A2) in margins. MEA 25 °C, 14 d: no growth to microcolonies. M60Y 25 °C, 14 d: flat; colony surface velutinous; margin entire; mycelial areas white; sporulation dark turquoise (24F7); soluble pigment absent; exudate absent; reverse centrally cinnamon brown (6D6) to pale yellow (4A3), greyish yellow (4C5) to pale yellow (4A3) in margins. MEA + 10 % NaCl 25 °C, 14 d: flat to slightly centrally raised; colony surface floccose to velutinose; margin entire; mycelial areas white; sporulation deep turquoise (24E8) to greyish turquoise (24B4); soluble pigment absent; exudate absent; reverse centrally olive grey (2F2), greyish orange (5B4) to cream pale yellow (4A3) in margins.
Micromorphology: Conidial heads at first globose, later becoming loosely columnar. Conidiophores uniseriate. Stipes smooth, densely covered by short hairs in SEM, 4.5–6 μm wide in the middle part, with occasional septa. Vesicles spatulate, ellipsoidal, pyriform to subclavate, 12–14.5 μm wide. Phialides flask-shaped, densely covered by short hairs in SEM, 8.5–11 μm long, covering one to two thirds of the vesicle. Conidia borne smooth and ellipsoidal, becoming coarsely roughned, globose or barrel-shaped at maturity, lobate-reticulate in SEM, connectives evident, 3.5–5 × 2.5–4 μm.
Distinguishing characters: See distinguishing characters of A. penicillioides.
Additional materials examined: Panama, telescope reticle, 1945, collector unknown, NRRL 25812 = CCF 5658. The Netherlands, cigars, unknown year of isolation, isolated by M. Meijer, DTO 318-F5, DTO 318-F6, DTO 318-F7, DTO 321-F9, DTO 321-G1. The Netherlands, leather, imported from unknown country, 2013, isolated by J. Houbraken, DTO 281-A4 = IBT 34517. USA, Minnesota, St. Paul, barley, 1957, isolated by G. C. Papavizas, NRRL 25826 = CCF 5484. USA, Minnesota, St. Paul, insulating board, 1957, isolated by G. C. Papavizas, NRRL 25830 = CCF 5485 = IBT 34631.
Aspergillus infrequens F. Sklenar, S.W. Peterson & Hubka, sp. nov. Mycobank MB818938. Fig. 24.
Fig. 24.
Aspergillus infrequens. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Etymology: Referring to its rare occurrence.
Typus: USA, Illinois, Peoria, wheat, 1971, isolated by D. I. Fennell (holotype PRM 944449, isotype PRM 944450, culture ex-type NRRL 25868 = CCF 5486 = DTO 356-D6 = IBT 34524).
Colony diam, 14 d (mm): M40Y 21–22; CYA 4–5; CY20S 7–8; CY20S 30 °C No growth; CY20S 37 °C No growth; DG18 25–26; MEA No growth; M60Y 40–43; M60Y 30 °C 8–10; M60Y 37 °C No growth; MEA + 10 % NaCl 18–19.
Colony characters: M40Y 25 °C, 14 d: Colonies crateriform, wrinkled, surface floccose; margin filiform; mycelial areas white; sporulation pale turquoise (24A3) to dark turquoise (24F8); soluble pigment absent; exudate absent; reverse centrally dark green (30F8), olive grey (2F2) to dark green (30F8), champagne greyish yellow (4B4) in margins. CYA 25 °C, 14 d: no growth to microcolonies. CY20S 25 °C, 14 d: crateriform, wrinkled; colony surface floccose, margin undulate; mycelial areas white; sporulation pale green (26A3) to pastel green (25A4); soluble pigment absent; exudate absent; reverse centrally turquoise grey (24D2) to greyish orange (5B3), orange white (5A2) in margins. DG18 25 °C, 14 d: umbonate, wrinkled; colony surface velutinous, margin filiform; mycelial areas white; sporulation pastel green (25A4) to greyish turquoise (water blue) (24C5); soluble pigment absent; exudate absent; reverse centrally dark turquoise (24F8) to greyish turquoise (24D5), orange white (5A2) in margins. MEA 25 °C, 14 d: no growth. M60Y 25 °C, 14 d: raised; colony surface powdery to cottony; margin filiform; mycelial areas white; sporulation greyish green (26B4); soluble pigment absent; exudate absent; reverse centrally olive (2E7) to mustard yellow (3B6), cream pale yellow (4A3) in margins. MEA + 10 % NaCl 25 °C, 14 d: crateriform, wrinkled; colony surface velutinous; margin filiform; mycelial areas white; sporulation light turquoise (24A4); soluble pigment absent; exudate absent; reverse centrally slate grey (3F2) to olive (3E7), champagne greyish yellow (4B4) in margins.
Micromorphology: Conidial heads radiate. Conidiophores uniseriate. Stipes smooth, sparsely covered by bundles of long hairs in SEM, 6–7 μm wide in the middle part, non-septate. Vesicles pyriform, spatulate to clavate, (10–)13–17(–20) μm wide. Phialides flask-shaped, sparsely covered by long hairs in SEM, 8–10 μm long, covering one third to two thirds of the vesicle. Conidia borne smooth and ellipsoidal, becoming echinulate, subglobose, ovate or barrel-shaped at maturity, aculeate in SEM, commonly remaining in chains, connectives evident, 4–5 × 3–4 μm.
Distinguishing characters: Aspergillus infrequens is represented only by the ex-type isolate. It is phylogenetically distinct from other species in the A. penicillioides clade and resolves in the basal position. Although A. infrequens phenotypically resembles species from the A. penicillioides clade, the ornamentation of conidia in SEM is aculeate and atypical for the clade (this ornamentation is found in the A. restrictus, A. conicus and A. vitricola clades).
Aspergillus magnivesiculatus F. Sklenar, Zalar, Ž. Jurjević & Hubka, sp. nov. Mycobank MB818939. Fig. 25.
Fig. 25.
Aspergillus magnivesiculatus. A. Colonies: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Etymology: Referring to its large vesicles.
Typus: Japan, Tokyo, katsuobushi, 1967, isolated by K. Yamada (holotype PRM 944444, isotype PRM 944445, culture ex-type NRRL 25866 = CCF 5488 = IBT 34816).
Colony diam, 14 d (mm): M40Y 27–35; CYA No growth; CY20S 11–17; CY20S 30 °C No growth; CY20S 37 °C No growth; DG18 20–26; MEA No growth; M60Y 47–60; M60Y 30 °C 40–50; M60Y 37 °C No growth; MEA + 10 % NaCl 27–34.
Colony characters: M40Y 25 °C, 14 d: Colonies raised to umbonate, surface floccose; margin entire to delicately filiform; mycelial areas centrally yellow (3A6) to white in margins; sporulation greyish green (26D5); soluble pigment absent; exudate absent; reverse centrally mahogany red (8E7), yellowish red (8B8) to pale yellow (4A3) in margins. CYA 25 °C, 14 d: no growth. CY20S 25 °C, 14 d: crateriform; colony surface velutinous, margin entire; mycelial areas white; sporulation greyish green (25B4) to greyish turquoise (24B4); soluble pigment absent; exudate absent; reverse centrally aquamarine turquoise grey (24D2) to orange white (5A2) in margins. DG18 25 °C, 14 d: umbonate; colony surface velutinous to floccose, margin entire to delicately filiform; mycelial areas centrally yellowish white (4A2) to white in margins; sporulation aquamarine greenish white (27A2); soluble pigment absent; exudate absent; reverse centrally corn greyish yellow (4B5) to yellowish white (4A2) in margins. MEA 25 °C, 14 d: no growth. M60Y 25 °C, 14 d: raised; colony surface floccose to cottony; margin filiform; mycelial areas pastel yellow (3A4), pale yellow (4A3) or white; sporulation pale green (27A3) to white; soluble pigment absent; exudate absent; reverse centrally caramel brown (6C6), apricot yellow (5B6) to corn greyish yellow (4B5), pale yellow (4A3) in margins. MEA + 10 % NaCl 25 °C, 14 d: flat or centrally slightly raised; colony surface floccose to powdery; margin entire to delicately filiform; mycelial areas centraly light yellow (4A4) to white in margins; sporulation greyish green (26D5); soluble pigment absent; exudate absent; reverse centrally olive grey (2F2) to golden brown (5D7), cream pale yellow (4A3) in margins.
Micromorphology: Conidial heads at first globose, later becoming loosely columnar to radiate. Conidiophores uniseriate. Stipes smooth, densely covered by short hairs in SEM, 5–7 μm wide in the middle part, non-septate. Vesicles pyriform, subclavate to clavate, 17–22 μm wide. Phialides flask-shaped, densely covered by short hairs in SEM, 9–11 μm long, covering two thirds to three quarters of the vesicle. Conidia borne smooth, ellipsoidal, coarsely roughned, ovate to barrel-shaped at maturity, tuberculate in SEM, connectives evident, 4.5–5.5 × 3.5–4.5 μm.
Distinguishing characters: Isolates of A. magnivesiculatus have the largest vesicles among species of the A. penicillioides clade; reverse colour on M40Y is usually mahogany red. See also distinguishing characters of A. penicillioides.
Additional materials examined: Canada, Nova Scotia, Wolfville, house dust, 2015, collected by A. Walker, isolated by C.M. Visagie, KAS 6089 = DTO 356-H3. Canada, Ontario, Ottawa, house dust, 2015, collected and isolated by C. M. Visagie, KAS 5655 = DTO 356-G8, KAS 5754 = DTO 356-G9. Canada, Ontario, Stittsville, house dust, 2014, collected by K. A. Seifert, isolated by C. M. Visagie, KAS 5623 = DTO 356-G7. Japan, Tokyo, katsuobushi, 1967, isolated by K. Yamada, NRRL 25867 = CCF 5660. Slovenia, Ljubljana, oil painting on canvas, 2015, isolated by P. Zalar and D.D. Graf, EXF-10353 = IBT 34284. Slovenia, Ljubljana, oil painting on canvas, 2015, isolated by P. Zalar and D.D. Graf, EXF-10377, EXF-10347 = IBT 34283, EXF-10356 = IBT 34279. USA, California, San Diego, child carrier, 2014, isolated by Ž. Jurjević, CCF 5490 = EMSL No. 2741. USA, California, San Diego, child carrier, 2015, isolated by Ž. Jurjević, CCF 5491 = EMSL No. 2918. USA, Idaho, home air, 2010, isolated by Ž. Jurjević, CCF 5489 = EMSL No. 1315 = DTO 356-E2 = IBT 34516. USA, Minnesota, St. Paul, dried corn, 1954, isolated by C. M. Christensen, NRRL 25821 = CCF 5487.
Aspergillus pachycaulis F. Sklenar, S.W. Peterson, Ž. Jurjević & Hubka, sp. nov. MycoBank MB823048. Fig. 26.
Fig. 26.
Aspergillus pachycaulis. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Etymology: Referring to the wide conidiophore stipes, especially compared to the closely related A. caesiellus.
Typus: USA, District of Columbia, Washington, unknown substrate, 1956, isolated by H. E. Wheeler (holotype PRM 944432, isotype PRM 944433, culture ex-type NRRL 25824 = CCF 5492 = DTO 356-D2 = IBT 34521 = IBT 34812).
Colony diam, 14 d (mm): M40Y 20–60; CYA 11–26; CY20S 15–40; CY20S 30 °C 22–36; CY20S 37 °C (0)8–10; DG18 35–48; MEA 21–31; M60Y 25–60; M60Y 30 °C 36–60; M60Y 37 °C 38–60; MEA + 10 % NaCl 27–43.
Colony characters: M40Y 25 °C, 14 d: Colonies raised, surface powdery to floccose; margin entire to delicately filiform; mycelial areas white; sporulation greyish turquoise (24C4); soluble pigment absent; exudate absent; reverse centrally brownish yellow (5C8) to light yellow (4A4) in margins. CYA 25 °C, 14 d: centrally raised, radially wrinkled; colony surface lanose to floccose, margin entire; mycelial areas white; sporulation greyish green (25C3); soluble pigment absent; exudate absent; reverse centrally deep blue (21D8) to yellowish white (2A2) in margins. CY20S 25 °C, 14 d: with central depression; colony surface lanose to floccose; margin entire; mycelial areas white; sporulation dull green (25D3); soluble pigment absent; exudate absent; reverse centrally greyish brown nougat (5D3) to orange white (5A2) in margins. DG18 25 °C, 14 d: flat or umbonate; colony surface powdery to floccose, margin entire to delicately filiform; mycelial areas white; sporulation greyish turquoise (water blue) (24D5); soluble pigment absent; exudate absent; reverse milk white (1A2). MEA 25 °C, 14 d: centrally raised; colony surface lanose to floccose, margin entire; mycelial areas white; sporulation sparse; soluble pigment absent; exudate absent; reverse centrally reddish golden (6C7) to pale orange (5A3) in margins. M60Y 25 °C, 14 d: raised; colony surface lanose to filamentous; margin entire to delicately filiform; mycelial areas white; sporulation dull green (27D4); soluble pigment absent; exudate absent; reverse centrally Pompeian yellow (5C6) to pastel yellow (3A4) in margins. MEA + 10 % NaCl 25 °C, 14 d: umbonate; colony surface powdery; margin irregularly filiform; mycelial areas white; sporulation greyish blue (23C5); soluble pigment absent; exudate absent; reverse centrally greyish orange (5B4) to light yellow (4A4) in margins.
Micromorphology: Conidial heads columnar, frequently sinuous. Conidiophores uniseriate. Stipes smooth, densely covered by short hairs in SEM, occasionally with septa, 7–8 μm wide. Vesicles pyriform to subclavate, 16–20 μm wide. Phialides flask-shaped, densely covered by short hairs in SEM, 8–9 μm long, covering one to two thirds of the vesicle. Conidia borne as smooth cylinders, at maturity ellipsoidal to ovate, aculeate in SEM, connectives evident, 4–5 × 2.5–3.5 μm.
Distinguishing characters: See distinguishing characters of A. restrictus. Aspergillus pachycaulis has wider stipes and larger vesicles compared to A. caesiellus and A. restrictus. It also grows slightly faster on MEA and some isolates can grow on CY20S at 37 °C (exception throughout the whole sect. Restricti).
Additional materials examined: Canada, Ontario, Ottawa, unknown substrate, isolated by J. Bissett, DAOMC 226750 = KAS 7748. USA, California, home air, 2005, isolated by Ž. Jurjević, CCF 5493 = EMSL No. 2310 = DTO 356-E6 = IBT 34536. Unknown locality of isolation, unknown substrate, unknown year of isolation, collector unknown, DTO 066-I6.
Aspergillus penicillioides Speg., Revista Fac. Agron. Univ. Nac. La Plata 2: 245. 1896, emended description. MycoBank MB309234. Fig. 27.
Fig. 27.
Aspergillus penicillioides. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Typus: Brazil, Recife, isolated from human cutaneous disease, 1958, D. Borelli (neotype Herb. IMI 211392 designated by Samson & Gams (1985), culture ex-type NRRL 4548 = CBS 540.65 = ATCC 16910 = IMI 211342 = DTO 207-I7 = CCF 5494 = IBT 34627).
Colony diam, 14 d (mm): M40Y 27–43; CYA 7–9; CY20S 11–20; CY20S 30 °C 11–15; CY20S 37 °C No growth; DG18 17–36; MEA 5–9; M60Y 28–59; M60Y 30 °C 40–60; M60Y 37 °C 17–45; MEA + 10 % NaCl 24–37.
Colony characters: M40Y 25 °C, 14 d: Colonies raised to umbonate, surface powdery; margin entire to delicately filiform; mycelial areas white; sporulation deep green (25E8); soluble pigment absent; exudate absent; reverse centrally coffee brown (5F7) to yellowish brown (5E8), golden greyish yellow (4C6) to orange white (5A2) in margins. CYA 25 °C, 14 d: convex, wrinkled; colony surface velutinous, margin undulate; mycelial areas white; no sporulation; soluble pigment absent; exudate absent; reverse centrally greyish orange (5B3) to cream orange white (5A2) in margins. CY20S 25 °C, 14 d: crateriform; colony surface velutinous, margin filiform; mycelial areas white; sporulation pale green (25A3); soluble pigment absent; exudate absent; reverse centrally greyish yellow (2B5) to olive grey (2F2), greyish orange (5B3) to orange white (5A2) in margins. DG18 25 °C, 14 d: umbonate to convex; colony surface floccose, margin filiform; mycelial areas white; sporulation aquamarine light turquoise (24A4); soluble pigment greyish yellow (2B3); exudate absent; reverse centrally olive yellow (2D6) to yellowish white (4A2) in margins. MEA 25 °C, 14 d: crateriform; colony surface velutinous, margin entire; no sporulation; soluble pigment absent; exudate absent; reverse centrally caramel brown (6C6) to pale orange (5A3) in margins. M60Y 25 °C, 14 d: raised; colony surface cottony; margin entire to delicately filiform; mycelial areas white; sporulation greyish green (25D7); soluble pigment absent; exudate absent; reverse centrally dark brown (6F6) to golden greyish yellow (4C6), pale yellow (4A3) in margins. MEA + 10 % NaCl 25 °C, 14 d: centrally raised; colony surface floccose; margin entire to delicately filiform; mycelial areas white; sporulation greyish turquoise (24C6); soluble pigment absent; exudate absent; reverse centrally olive brown (4F5) to yellowish orange (4B7), cream pale yellow (4A3) in margins.
Micromorphology: Conidial heads at first globose, later becoming loosely columnar. Conidiophores uniseriate. Stipes smooth, sparsely covered by short hairs in SEM, 4–6 μm wide in the middle part, widening gradually toward the vesicle, non-septate. Vesicles pyriform, spatulate to clavate, 10–18 μm wide. Phialides flask-shaped, sparsely covered by short hairs in SEM, 8–10 μm long, covering upper half to two thirds of the vesicle. Conidia borne smooth and ellipsoidal, later becoming coarsely roughened, subglobose or barrel-shaped, tuberculate in SEM, commonly remaining in chains, connectives evident, 3.5–4.5 × 2.5–3.5 μm.
Distinguishing characters: All species belonging to the A. penicillioides clade are morphologically very similar and particularly A. penicillioides and A. hordei share almost identical morphologies. The latter two species can be distinguished from others in the clade by their ability to grow on CY20S at 30 °C, production of greyish yellow soluble pigment when cultivated on DG18, narrow stipes, small vesicles and loosely columnar rather then loosely radiate conidial heads. In contrast to most A. hordei isolates, A. penicillioides always grows on MEA and CYA. Conidia of A. penicillioides are tuberculate in SEM, but lobate-reticulate in A. hordei.
Additional materials examined: Belgium, Brussels, indoor air (air-conditioned office), 1985, collector unknown, IHEM 2476. Brazil, Recife, human cutaneous disease isolate, 1958, collector unknown, NRRL 4550 = CCF 5495, NRRL 4553 = CCF 5496. Canada, Ontario, Ottawa, house dust, 2015, collected by N. Yilmaz, isolated by C. M. Visagie, KAS 7745, KAS 7746. Czech Republic, Prague, sweet roll with chocolate glaze, 2002, isolated by H. Růžičková, CCF 3282. Czech Republic, Zlín, leather shoe, 1990, isolated by T. Skálová, CCF 2666. France, seeds of cereal, 1984, collector unknown, IHEM 2330. France, French madeleines, 2013, isolated by F. Déniel, UBOCC-A-115042 = CBS 140430 = DTO 334-E1. Germany, unknown substrate, unknown year of isolation, collector unknown, DTO 334-A9. Indonesia, peanuts, 2008, isolated by E.S. Rahayu and J. Houbraken, DTO 063-G8. Micronesia, house dust, unknown year of isolation, isolated by C. M. Visagie, DTO 267-A4 = IBT 34614. Micronesia, house dust, 2009, collected by W. Law, isolated by E. Whitfield and K. Mwange, DTO 267-A9 = CCF 5664. The Netherlands, leather imported from unknown country, 2013, isolated by J. Houbraken, DTO 281-A7. The Netherlands, Zwartewaal, air, 2012, isolated by M. Meijer, 231-A7. The Netherlands, flour imported from unknown country, 2005, isolated by J. Houbraken, DTO 011-C4. USA, Illinois, O'fallon, baseball gloves in store, 2014, isolated by Ž. Jurjević, CCF 5500 = EMSL No. 2651 = IBT 34630. USA, New Jersey, green fabric covered binders, import from China, 2014, isolated by Ž. Jurjević, CCF 5499 = EMSL No. 2441, CCF 5497 = EMSL No. 2430 = IBT 34628, CCF 5498 = EMSL No. 2440 = DTO 356-E7 = IBT 34815. USA, North Carolina, Durham, unknown substrate, 1952, isolated by J. L. Miranda, NRRL 25816 = CCF 5661. USA, Minnesota, St. Paul, peas, 1957, isolated by G. C. Papavizas, NRRL 25834 = CCF 5659. USA, Minnesota, St. Paul, wheat, 1957, isolated by G. C. Papavizas, NRRL 25835. USA, California, San Diego, child carrier, 2015, isolated by Ž. Jurjević, CCF 5501 = EMSL No. 2749 = IBT 34629, CCF 5502 = EMSL No. 2900, CCF 5503 = EMSL No. 2909, EMSL No. 2747, EMSL No. 2777, EMSL No. 2766, EMSL No. 2786, EMSL No. 2760, EMSL No. 2757, EMSL No. 2904, EMSL No. 2898. USA, Maryland, Bethesda, archival cabinet, 2015, isolated by Ž. Jurjević, CCF 5504 = EMSL No. 3264. USA, New York, blood sample, 1951, isolated by A. Howell, NRRL 25879. USA, Minnesota, St. Paul, dried corn, 1954, isolated by C. M. Christensen, NRRL 25820, NRRL 25822. Unknown locality of isolation, unknown substrate, unknown year of isolation, collector unknown, DTO 008-H4. Unknown locality of isolation, unknown substrate, unknown year of isolation, collector unknown, NRRL 25870.
Aspergillus pseudogracilis F. Sklenar, Ž. Jurjević & Hubka, sp. nov. MycoBank MB818932. Fig. 28.
Fig. 28.
Aspergillus pseudogracilis. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Etymology: Referring to its close phylogenetic and phenotypic relatedness to A. gracillis.
Typus: USA, California, San Diego, child carrier, 2015, isolated by Ž. Jurjević (holotype PRM 944434, isotype PRM 944435, culture ex-type CCF 5505 = EMSL No. 2765 = DTO 356-F3 = NRRL 66620 = IBT 34813).
Colony diam, 14 d (mm): M40Y 42–45; CYA No growth; CY20S 7–8; CY20S 30 °C No growth; CY20S 37 °C No growth; DG18 20–22; MEA No growth; M60Y 52–53; M60Y 30 °C 43–45; M60Y 37 °C No growth; MEA + 10 % NaCl 25–26.
Colony characters: M40Y 25 °C, 14 d: Colonies raised, surface floccose; margin entire to delicately filiform; mycelial areas white; sporulation light turquoise (24A4); soluble pigment absent; exudate absent; reverse centrally cinnamon brown (6D6) to champagne greyish yellow (4B4) in margins. CYA 25 °C, 14 d: no growth to microcolonies. CY20S 25 °C, 14 d: convex; colony surface floccose to cottony, margin entire to delicately filiform; mycelial areas white; sporulation sparse; soluble pigment absent; exudate absent; reverse centrally greyish orange (5B3) to yellowish white (4A2) in margins. DG18 25 °C, 14 d: centrally raised; colony surface floccose, margin entire to delicately filiform; mycelial areas white; sporulation greyish turquoise (24B4); soluble pigment absent; exudate absent; reverse centrally turquoise grey (24C2) to orange white (5A2) in margins. MEA 25 °C, 14 d: no growth. M60Y 25 °C, 14 d: raised; colony surface floccose; margin entire to delicately filiform; mycelial areas white; sporulation greyish turquoise (24B4); soluble pigment absent; exudate absent; reverse champagne greyish yellow (4B4). MEA + 10 % NaCl 25 °C, 14 d: flat, wrinkled; colony surface floccose, margin entire to delicately filiform; mycelial areas white; sporulation pale turquoise (24A3); soluble pigment absent; exudate absent; reverse centrally light brown (5D6) to champagne greyish yellow (4B4) in margins.
Micromorphology: Conidial heads loosely columnar, frequently twisted. Conidiophores uniseriate. Stipes smooth, densely covered by long hairs in SEM, 4–6 μm wide in the middle part, widening gradually toward the vesicle, septate. Vesicles spatulate to clavate, 9–13 μm wide. Phialides flask-shaped, densely covered by long hairs in SEM, 9–10.5 μm long, covering two thirds of the vesicle. Conidia borne as smooth cylinders, echinulate at maturity, aculeate in SEM, subglobose or barrel-shaped, connectives evident, 3–4 × 2–3 μm.
Distinguishing characters: See distinguishing characters of A. conicus and A. gracilis.
Aspergillus restrictus G. Sm., J. Textile Inst. 22: T115. 1931, emended description. MycoBank MB276290. Fig. 29.
Fig. 29.
Aspergillus restrictus. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Synonyms: Penicillium fuscoflavum S. Abe, J. Gen. Appl. Microbiol., Tokyo 2: 64. 1956.
Aspergillus restrictus var. B G. Sm., J. Textile Inst. 22: T115. 1931.
Typus: United Kingdom, cloth, 1928, isolated by G. Smith [neotype Herb. IMI 16267 designated by Samson & Gams (1985) and modified by Pitt & Samson (1993), culture ex-type NRRL 154 = CBS 117.33 = CBS 541.65 = DTO 079-B2 = ATCC 16912 = IHEM 3920 = IMI 16267= CCF 5506 = IBT 34615].
Colony diam, 14 d (mm): M40Y 42–53; CYA 12–16; CY20S 24–29; CY20S 30 °C 12–25; CY20S 37 °C No growth; DG18 28–33; MEA 5–13; M60Y 38–50; M60Y 30 °C 45–55; M60Y 37 °C 20–40; MEA + 10 % NaCl 33–40.
Colony characters: M40Y 25 °C, 14 d: Colonies flat, surface powdery, filamentous in margins; margin entire to delicately filiform; mycelial areas white; sporulation greyish turquoise (24C4); soluble pigment absent; exudate absent; reverse centrally cinnamon brown (6D6) to sand greyish yellow (4B3) in margins. CYA 25 °C, 14 d: centrally raised, irregulary wrinkled; colony surface floccose, margin entire; mycelial areas white; sporulation sparse; soluble pigment absent; exudate absent; reverse centrally dull blue (23E5) to orange white (5A2) in margins. CY20S 25 °C, 14 d: umbonate with concentric ring pattern; colony surface coarsely granular in the centre, velvety in margins; margin entire to delicately filiform; mycelial areas white; sporulation deep green (29E8) to light turquoise (25A4); soluble pigment absent; exudate absent; reverse centrally dull blue (23E5) to orange white (5A2) in margins. DG18 25 °C, 14 d: umbonate; colony surface powdery, margin entire to delicately filiform; mycelial areas white; sporulation greyish turquoise (24D4); soluble pigment absent; exudate absent; reverse centrally brownish orange (6C3) to orange white (5A2) in margins. MEA 25 °C, 14 d: convex, radially wrinkled; colony surface floccose, margin entire; mycelial areas white; sporulation sparse; soluble pigment absent; exudate absent; reverse centrally light brown (7D4) to flesh greyish orange (6B3) in margins. M60Y 25 °C, 14 d: flat; colony surface powdery; margin filiform; mycelial areas white; sporulation greyish turquoise (24C4); soluble pigment absent; exudate absent; reverse light yellow (4C4). MEA + 10 % NaCl 25 °C, 14 d: flat; colony surface powdery, filamentous in margins; margin entire to delicately filamentous; mycelial areas white; sporulation greyish turquoise (24C4); soluble pigment absent; exudate absent; reverse centrally brownish grey (8E2) to champagne greyish yellow (4B4) in margins.
Micromorphology: Conidial heads columnar, frequently sinuous. Conidiophores uniseriate. Stipes smooth, densely covered by long hairs in SEM, non-septate, 4–5.5 μm wide. Vesicles pyriform to clavate, 9–12 μm in diameter. Phialides flask-shaped, densely covered by long hairs in SEM, 8–9 μm long, covering one third to one half of the vesicle. Conidia borne as smooth cylinders in long appressed columns, at maturity ellipsoidal to ovate, commonly remaining in chains, connectives evident, coarsely roughned to echinulate, aculeate in SEM, 3.5–4.5 × 2.5–3 μm.
Distinguishing characters: Aspergillus restrictus can be distinguished from A. caesiellus and A. pachycaulis by smaller vesicles and narrower stipes, slower growth on M40Y and M60Y and no sporulation on CYA. Osmotic gradient growth curves of these three species differ mainly on MEA, where A. pachycaulis grows slowest on MEA + 10 % NaCl and MEA + 15 % NaCl where A. restrictus grows fastest (see Fig. 12A).
Taxonomic notes: Penicillium fuscoflavum (ex-type strain NRRL 4783) and A. restrictus var. B (represented by NRRL 154) are taxa conspecific with A. restrictus, based on phylogenetic analysis (Peterson 2008, this study – see Fig. 7).
Additional materials examined: Belgium, Antwerp, dust from mattress, 1984, collector unknown, IHEM 2121. Belgium, Estinnes-au-Mont, indoor air, 1981, collector unknown, IHEM 818. Belgium, Brussels, indoor air (air-conditioned office), 1985, collector unknown, IHEM 2477. Belgium, Brussels, indoor air (air-conditioned office), 1984, collector unknown, IHEM 2373. Bermuda, home air, 2010, isolated by Ž. Jurjević, CCF 5507 = EMSL No. 1379 = IBT 34618. Canada, Ontario, Oshawa, maple syrup, unknown year of isolation, isolated by Samantha Frasz, DAOMC 250090 = KAS 7744. Czech Republic, Prague, sclerotium of fungus Corallocytostroma ornicopreoides imported from Australia, 2003, isolated by A. Kubátová, CCF 3364 = IBT 34619. Indonesia, corn, unknown year of isolation, collector unknown, DTO 065-C7 = IBT 34541. Indonesia, dust, unknown year of isolation, collector unknown, DTO 236-I7, DTO 237-A2, DTO 236-I8. USA, Maryland, packing material, 2012, isolated by Ž. Jurjević, CCF 5511 = EMSL No. 1675 = IBT 34616. USA, Illinois, Peoria, culture contaminant, 1975, isolated by T. G. Pridham, NRRL 25862. USA, New Jersey, Sicklerville, air in school auditorium, 2013, isolated by Ž. Jurjević, CCF 5512 = EMSL No. 2206 = IBT 34617. USA, New Jersey, green fabric covered binders, import from China, 2014, isolated by Ž. Jurjević, CCF 5513 = EMSL No. 2429. USA, North Carolina, mattress cover, 2012, isolated by Ž. Jurjević, CCF 5509 = EMSL No. 1611. USA, Illinois, O'fallon, baseball gloves in store, 2014, isolated by Ž. Jurjević, CCF 5514 = EMSL No. 2652. USA, cattle feed, 1984, isolated by B. W. Horn, NRRL 25882. USA, New Jersey, hospital air, 2012, isolated by Ž. Jurjević, CCF 5510 = EMSL No. 1633. USA, Alabama, home air, 2010, isolated by Ž. Jurjević, CCF 5508 = EMSL No. 1416. USA, New York, Upton, work station, indoor air, 2015, isolated by Ž. Jurjević, EMSL No. 2892. USA, California, San Diego, child carrier, 2015, isolated by Ž. Jurjević, CCF 5515 = EMSL No. 2906, EMSL No. 2746, EMSL No. 2897, EMSL No. 2748.
Aspergillus reticulatus F. Sklenar, Ž. Jurjević, S.W. Peterson & Hubka, sp. nov. MycoBank MB818940. Fig. 30.
Fig. 30.
Aspergillus reticulatus. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Etymology: Referring to the lobate-reticulate ornamentation of the conidia.
Typus: USA, South Carolina, Charleston, lung biopsy, 1965, isolated by L. Jaynes (holotype PRM 944442, isotype PRM 944443, culture ex-type NRRL 25852 = CCF 5516 = DTO 356-D4 = IBT 34540).
Colony diam, 14 d (mm): M40Y 46–50; CYA 8–14; CY20S 21–30; CY20S 30 °C 8–18; CY20S 37 °C No growth; DG18 26–33; MEA 2–7; M60Y 55–60; M60Y 30 °C 50–55; M60Y 37 °C 3–20; MEA + 10 % NaCl 32–40.
Colony characters: M40Y 25 °C, 14 d: Colonies raised, surface floccose to lanose; margin entire to delicately filiform; mycelial areas white; sporulation greyish turquoise (24E6); soluble pigment absent; exudate absent; reverse centrally golden brown (5D7) to greyish yellow (4C4), pale yellow (4A3) in margins. CYA 25 °C, 14 d: crateriform, radially wrinkled; colony surface velutinous, margin undulate; mycelial areas white; sporulation sparse; soluble pigment absent; exudate absent; reverse centrally greyish orange (5B3) to orange white (5A2) in margins. CY20S 25 °C, 14 d: centrally raised; colony surface velutinous to floccose, margin entire to delicately filiform; mycelial areas white; sporulation pale turquoise (24A3) to deep turquoise (24E8); soluble pigment absent; exudate absent; reverse centrally aquamarine greyish turquoise (24B3) to yellowish white (4A2) in margins. DG18 25 °C, 14 d: umbonate; colony surface floccose, margin entire to delicately filiform; mycelial areas white; sporulation aquamarine deep green (25E8); soluble pigment absent; exudate absent; reverse centrally dull green (25D3) to orange grey (5B2), yellowish white (4A2) in margins. MEA 25 °C, 14 d: cerebriform; colony surface velutinous, margin undulate; mycelial areas white; sporulation sparse; soluble pigment absent; exudate absent; reverse centrally greyish orange (5C5) to orange white (5A2) in margins. M60Y 25 °C, 14 d: raised; colony surface floccose to lanose; margin entire to delicately filiform; mycelial areas white; sporulation greyish green (25E6); soluble pigment absent; exudate absent; reverse centrally cinnamon brown (6D6), pale yellow (4A3) to greyish yellow (4C5), pale yellow (4A3) in margins. MEA + 10 % NaCl 25 °C, 14 d: flat to umbonate; colony surface floccose; margin entire to delicately filiform; mycelial areas white; sporulation deep turquoise (24E8); soluble pigment absent; exudate absent; reverse centrally olive grey (2F2) to brownish orange (5C5), cream pale yellow (4A3) in margins.
Micromorphology: Conidial heads globose to radiate. Conidiophores uniseriate. Stipes smooth, densely covered by short hairs in SEM, 5–7 μm wide in the middle part, non-septate. Vesicles globose or pyriform, 14–20 μm wide. Phialides flask-shaped, densely covered by short hairs in SEM, 9–11 μm long, covering two thirds to three quarters of the vesicle. Conidia borne smooth, ellipsoidal, becoming rough-walled, globose or barrel-shaped at maturity, tuberculate to lobate-reticulate in SEM, sometimes remaining in chains, connectives evident, 3.5–4.5 × 2.5–3.5 μm.
Distinguishing characters: Aspergillus reticulatus is phylogenetically closely related to A. salinicola (Fig. 7). These two species are less xerophilic than species closely related to A. penicillioides, because they grow slightly faster in conditions with higher water activity and slower in conditions with lower water activity (higher concentration of salt). In contrast to A. salinicola, isolates of A. reticulatus can grow on M60Y at 37 °C, grow faster on CY20S at 25 °C and 30 °C and usually have wider stipes and larger vesicles. See also distinguishing characters of A. penicillioides.
Additional materials examined: Belgium, Brussels, dust from carpet, 2008, collector unknown, IHEM 22696. Czech Republic, Zlín, leather shoe, 1998, isolated by T. Skálová, CCF 3112 = IBT 34634 = NRRL 62490. France, French madeleines, 2013, F. Déniel, UBOCC-A-115044, UBOCC-A-115045. Puerto Rico, Bayamon, administrative area, air, 2014, isolated by Ž. Jurjević, CCF 5522 = EMSL No. 2525, CCF 5523 = EMSL No. 2526. Puerto Rico, Bayamon, office air, 2014, isolated by Ž. Jurjević, CCF 5524 = EMSL No. 2548 = IBT 34637. Slovenia, Ljubljana, oil painting on canvas, 2015, isolated by P. Zalar and D.D. Graf, EXF-10429 = CCF 5656. USA, Idaho, home air, 2009, isolated by Ž. Jurjević, CCF 5518 = NRRL 58903 = EMSL No. 1272 = IBT 34819. USA, Idaho, home air, 2010, isolated by Ž. Jurjević, CCF 5519 = NRRL 62004 = EMSL No. 1313, CCF 5520 = NRRL 62005 = EMSL No. 1314. USA, Idaho, outside air, 2010, isolated by Ž. Jurjević, CCF 5521 = EMSL No. 1362 = DTO 356-E4 = IBT 34518. USA, Florida, home air, 2008, isolated by Ž. Jurjević, CCF 5525 = NRRL 58572 = EMSL No. 885 = IBT 34880. USA, Georgia, Chamblee, lung biopsy, 1950, isolated by L. Ajello, NRRL 25878 = CCF 5517. USA, California, San Diego, child carrier, 2015, isolated by Ž. Jurjević, EMSL No. 2778, EMSL No. 2767, EMSL No. 2750, EMSL No. 2760, EMSL No. 2770, EMSL No. 2771, EMSL No. 2772, EMSL No. 2745, EMSL No. 2756. USA, Hawaii, Honolulu, drawer, unknown year of isolation, isolated by J. Varga, CBS 114325 = DTO 077-H3.
Aspergillus salinicola Zalar, F. Sklenar, Visagie & Hubka, sp. nov. MycoBank MB818941. Fig. 31.
Fig. 31.
Aspergillus salinicola. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Etymology: Referring to salterns, a source of isolation of some isolates.
Typus: Slovenia, Ljubljana, oil painting on canvas, 2015, isolated by P. Zalar (holotype PRM 944448, culture ex-type EXF-10401 = IBT 34266 = CCF 5526 = NRRL 66621).
Colony diam, 14 d (mm): M40Y 32–40; CYA (0)8–9; CY20S 9–20; CY20S 30 °C 0–5; CY20S 37 °C No growth; DG18 21–30; MEA 0–5; M60Y 42–55; M60Y 30 °C 27–43; M60Y 37 °C No growth; MEA + 10 % NaCl 32–35.
Colony characters: M40Y 25 °C, 14 d: Colonies flat to umbonate, surface velutinous; margin delicately filiform; mycelial areas pinkish white (7A2) to white; sporulation greyish green (25B5); soluble pigment absent; exudate absent; reverse centrally golden brown (5D7) to pale yellow (4A3) in margins. CYA 25 °C, 14 d: cerebriform; colony surface floccose, margin undulate; mycelial areas white; sporulation sparse; soluble pigment absent; exudate absent; reverse centrally greyish orange (5B3) to orange white (5A2) in margins. CY20S 25 °C, 14 d: centrally raised, radially wrinkled; colony surface velutinous, margin entire to delicately filiform; mycelial areas white; sporulation turquoise grey (24D2); soluble pigment absent; exudate absent; reverse centrally nougat dull yellow (3D4) to pale yellow (4A3) in margins. DG18 25 °C, 14 d: flat, slightly centrally raised; colony surface floccose, sometimes forming sectors, margin entire; mycelial areas white; sporulation greyish green (25D6); soluble pigment absent; exudate absent; reverse centrally greyish green (25C3) to greyish orange (5B3), orange white (5A2) in margins. MEA 25 °C, 14 d: no growth to microcolonies. M60Y 25 °C, 14 d: raised; colony surface floccose to lanose; margin filiform; mycelial areas white; sporulation greyish green (25D7); soluble pigment absent; exudate absent; reverse centrally olive green (2F6), pale yellow (4A3) to greyish yellow (4C5), pale yellow (4A3) in margins. MEA + 10 % NaCl 25 °C, 14 d: centrally raised; colony surface granular to filamentous, floccose in the centre; margin entire to delicately filiform; mycelial areas white; sporulation greyish turquoise (24C4); soluble pigment absent; exudate absent; reverse centrally light brown (5D6) to amber yellow (4D6), champagne greyish yellow (4B4) in margins.
Micromorphology: Conidial heads at first globose, later becoming loosely radiate. Conidiophores uniseriate. Stipes smooth, sparsely covered by short hairs in SEM, 4–6 μm wide in the middle part, non-septate. Vesicles pyriform, spatulate to subclavate, 11–14 μm wide. Phialides flask-shaped, sparsely covered by short hairs in SEM, 8–9 μm long, covering one third to one half of the vesicle. Conidia borne smooth and ellipsoidal, becoming irregularly rough-walled, globose to subglobose, tuberculate to lobate-reticulate in SEM, connectives evident, 3–4 × 2–3 μm.
Distinguishing characters: See distinguishing characters of A. reticulatus. A relatively unique character of A. salinicola is the pale colour of sporulation in comparison with other species belonging to the A. penicillioides clade.
Additional materials examined: Canada, Nova Scotia, Wolfville, house dust, 2015, collected by A. Walker, isolated by C.M. Visagie, KAS 6054. France, Brest, painting from Brittany (painter: J.M. Vien; painting title: Jésus et le fils du centenier de Capharnaüm; painting technique: oil on paper mounted on canvas), 2015, isolated by F. Déniel, UBOCC-A-116019 = CCF 5528 = IBT 34635. France, French savaroise, 2013, F. Déniel, UBOCC-A-115043 = CBS 140431 = DTO 334-E2. Germany, Essen, kindergarten, unknown year of isolation, isolated by J. Varga, CBS 117344 = DTO 077-I1. Slovenia, Sečovlje salterns, 1996, isolated by N. Gunde-Cimerman and P. Zalar, EXF-226 = CCF 5527 = IBT 34277 = NRRL 66622.
Aspergillus tardicrescens F. Sklenar, Houbraken, Zalar & Hubka, sp. nov. MycoBank MB818942. Fig. 32.
Fig. 32.
Aspergillus tardicrescens. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Etymology: Referring to the slow growth of the species.
Typus: The Netherlands, Alphen aan den Rijn, museum piece (measuring table), 2014, collector unknown (holotype PRM 944438, isotype PRM 944439, culture ex-type DTO 316-B5 = CCF 5529 = IBT 34558 = NRRL 66623).
Colony diam, 14 d (mm): M40Y 17–29; CYA No growth; CY20S 0–5; CY20S 30 °C No growth; CY20S 37 °C No growth; DG18 12–17; MEA No growth; M60Y 31–50; M60Y 30 °C 15–35; M60Y 37 °C No growth; MEA + 10 % NaCl 19–29.
Colony characters: M40Y 25 °C, 14 d: Colonies umbonate, surface powdery; margin entire to delicately filiform; mycelial areas white; sporulation greyish turquoise (water blue) (24C5); soluble pigment absent; exudate absent; reverse centrally olive brown (4F8) to greyish yellow (4C7), pale orange (5A3) in margins. CYA 25 °C, 14 d: no growth. CY20S 25 °C, 14 d: convex; colony surface velutinous, margin undulate; mycelial areas white; sporulation pale green (25A3); soluble pigment absent; exudate absent; reverse centrally greenish grey (25D2) to orange white (5A2) in margins. DG18 25 °C, 14 d: convex; colony surface floccose, margin entire; mycelial areas white; sporulation aquamarine turquoise blue (24A6); soluble pigment absent; exudate absent; reverse centrally turquoise grey (24C2) to orange white (5A2) in margins. MEA 25 °C, 14 d: no growth. M60Y 25 °C, 14 d: raised; colony surface floccose to cottony; margin entire to delicately filiform; mycelial areas white; sporulation aquamarine greyish turquoise (24B3) to pastel green (25A4); soluble pigment absent; exudate absent; reverse centrally brownish orange (5C5) to pale yellow (4A3) in margins. MEA + 10 % NaCl 25 °C, 14 d: flat to umbonate; colony surface powdery to floccose; margin entire to delicately filiform; mycelial areas white; sporulation greyish turquoise (24C3); soluble pigment absent; exudate absent; reverse centrally bronze brown (5E5) to greyish yellow (4C7), cream pale yellow (4A3) in margins.
Micromorphology: Conidial heads at first globose, later becoming radiate. Conidiophores uniseriate. Stipes smooth, densely covered by short hairs in SEM, 5–8 μm wide in the middle part, with occasional septa. Vesicles subglobose, spatulate, pyriform to subclavate, 14–19 μm wide. Phialides flask-shaped, densely covered by short hairs in SEM, 8–11 μm long, covering upper half to two thirds of the vesicle. Conidia borne irregularly rough and ellipsoidal, becoming coarsely rough-walled, subglobose or barrel-shaped at maturity, tuberculate in SEM, connectives evident, 3.5–4.5 × 2.5–3.5 μm.
Distinguishing characters: See distinguishing characters of A. penicillioides. Conidia of A. tardicrescens are smaller than those of A. clavatophorus and A. magnivesiculatus; its vesicles are smaller than of A. magnivesiculatus. The majority of isolates is not able to grow on CY20S at 30 °C or even at 25 °C.
Additional materials examined: Canada, Nova Scotia, Wolfville, house dust, 2015, collected by A. Walker, isolated by C. M. Visagie, KAS 6252 = DTO 356-I5. Slovenia, Ljubljana, oil painting on canvas, 2015, isolated by P. Zalar and D.D. Graf, EXF-10454 = IBT 34281 = CCF 5530 = NRRL 66624. Slovenia, Ljubljana, air in depot of Conservation Centre of the Institute for the Protection of Cultrural Heritage of Slovenia, 2015, isolated by P. Zalar and D.D. Graf, EXF-10456 = IBT 34286. The Netherlands, Alphen aan den Rijn, museum piece (dentist chair), 2014, collector unknown, DTO 316-A7. The Netherlands, Alphen aan den Rijn, museum piece (rubber tyre of brancard), 2014, collector unknown, DTO 316-A8 = IBT 34562. The Netherlands, Alphen aan den Rijn, museum piece (x-ray table), 2014, collector unknown, DTO 316-A9. The Netherlands, Alphen aan den Rijn, museum piece (vitrine), 2014, collector unknown, DTO 316-B4. The Netherlands, Tilburg, moist wall of archive, 2008, isolated by J. Houbraken, DTO 073-H6. The Netherlands, Zwartewaal, unknown substrate, 2012, isolated by M. Meijer, DTO 231-B8. The Netherlands, archive material, unknown year of isolation, isolated by J. Houbraken, DTO 014-A3.
Aspergillus villosus F. Sklenar, S.W. Peterson & Hubka, sp. nov. MycoBank MB818933. Fig. 33.
Fig. 33.
Aspergillus villosus. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Etymology: Referring to stipes and phialides, which are villose when observed using SEM.
Typus: United Kingdom, Scotland, Kirkhill, unknown substrate, 1945, isolated by D. Snow (holotype PRM 944430, isotype PRM 944431, culture ex-type NRRL 25813 = CCF 5531 = DTO 356-C9 = IBT 34822).
Colony diam, 14 d (mm): M40Y 21–23; CYA 8–10; CY20S 17–19; CY20S 30 °C 7–8; CY20S 37 °C No growth; DG18 16–20; MEA 5–6; M60Y 26–28; M60Y 30 °C 28–30; M60Y 37 °C 6–8; MEA + 10 % NaCl 22–23.
Colony characters: M40Y 25 °C, 14 d: Colonies flat, surface floccose; margin irregular, filiform; mycelial areas white; sporulation greyish green (25E7) to bluish green (25A6); soluble pigment absent; exudate absent; reverse centrally olive green (2F6) to champagne greyish yellow (4B4) in margins. CYA 25 °C, 14 d: convex; colony surface floccose, margin entire to undulate; mycelial areas white; sporulation sparse; soluble pigment absent; exudate absent; reverse centrally yellowish grey (4B2) to yellowish white (4A2) in margins. CY20S 25 °C, 14 d: umbonate, slightly radially wrinkled; colony surface velutinous to floccose, margin entire; mycelial areas white; sporulation light turquoise (24A4) to pastel blue (23A4); soluble pigment absent; exudate absent; reverse dull green (25D3) to pale orange (5A3) in margins. DG18 25 °C, 14 d: centrally raised; colony surface floccose, margin filiform; mycelial areas pinkish white (7A2); sporulation greyish turquoise (24D5); soluble pigment absent; exudate absent; reverse centrally orange white (5A2) to yellowish white (4A2) in margins. MEA 25 °C, 14 d: convex; colony surface floccose, margin undulate; mycelial areas white; sporulation sparse; soluble pigment absent; exudate absent; reverse centrally clay brownish orange (5C5) to pale orange (5A3) in margins. M60Y 25 °C, 14 d: raised; colony surface floccose; margin irregular, filiform; mycelial areas white; sporulation pastel green (25B4); soluble pigment absent; exudate absent; reverse centrally olive grey (2F2) to champagne greyish yellow (4B4) in margins. MEA + 10 % NaCl 25 °C, 14 d: flat; colony surface floccose, margin filiform; mycelial areas pinkish white (7A2); sporulation greyish turquoise (24D5); soluble pigment absent; exudate absent; reverse centrally cinnamon brown (6D6) to champagne greyish yellow (4B4) in margins.
Micromorphology: Conidial heads loosely columnar. Conidiophores uniseriate. Stipes smooth, densely covered by long hairs in SEM, 5–6 μm wide in the middle part, widening gradually toward the vesicle, with occasional septa. Vesicles pyriform, spatulate to clavate, 10–14 μm wide. Phialides flask-shaped, densely covered by long hairs in SEM, 9–12 μm long, covering the upper half of the vesicle. Conidia borne as smooth, long cylinders, echinulate at maturity, tuberculate in SEM, ellipsoidal or barrel-shaped, connectives evident, 3.5–4.5 × 2.5–3.5 μm.
Distinguishing characters: See the distinguishing characters of A. conicus. The phylogenetic position of A. villosus in the A. conicus clade is not fully resolved. It is a member of the A. conicus clade based on the *BEAST analysis, but the concatenated dataset analysis resolves it outside the clade with absolute support. Probably its closest relatives are A. conicus, A. domesticus and A. destruens. Aspergillus villosus can be distinguished by its faster growth on CY20S, ability to grow on CY20S at 30 °C and on M60Y at 37 °C. Additionally, A. villosus has rather spatulate vesicles in contrast to pyriform or ellipsoidal vesicles in mentioned relatives; conidia ornamentation in SEM is tuberculate, rather than aculeate common in the A. conicus clade.
Additional materials examined: France, Brest, painting from Brittany (painter: A.D. Hondius; painting title: Jésus à la piscine de Bethsaïda ou La guérison du paralytique; painting technique: oil on canvas), 2015, isolated by F. Déniel, UBOCC-A-116020 = CCF 5532 = IBT 34624.
Aspergillus vitricola Ohtsuki, Bot. Mag. (Tokyo) 75: 436. 1962, emended description. MycoBank MB326665. Fig. 34.
Fig. 34.
Aspergillus vitricola. A. Colonies after 14 d at 25 °C: top row left to right, obverse M40Y, CYA, CY20S and DG18; bottom row left to right, reverse M40Y, MEA, M60Y and MEA + 10 % NaCl. B, C. Conidiophores. D, E. Conidial heads. F. Conidia. G. Conidia in SEM. H. Stipe in SEM. Scale bars: B, C, F = 10 μm; G, H = 2 μm.
Typus: Japan, binocular lens, 1962, collector unknown (holotype in the Herbarium of the Nagao Institute, culture ex-type NRRL 5125 = DTO 356-F7 = ATCC 16905 = ATCC 36505 = IMI 108298 = CCF 5533 = IBT 34530).
Colony diam, 14 d (mm): M40Y 19–36; CYA No growth; CY20S (0)4–7; CY20S 30 °C No growth; CY20S 37 °C No growth; DG18 8–18; MEA No growth; M60Y 25–42; M60Y 30 °C 16–42; M60Y 37 °C No growth; MEA + 10 % NaCl 9–26.
Colony characters: M40Y 25 °C, 14 d: Colonies flat, wrinkled, surface velutinous; margin entire to delicately filiform; mycelial areas white; sporulation greyish turquoise (water blue) (24C5); soluble pigment absent; exudate absent; reverse centrally olive green (2F6) to olive yellow (3D6), champagne greyish yellow (4B4) in margins. CYA 25 °C, 14 d: no growth. CY20S 25 °C, 14 d: no growth to microcolonies. DG18 25 °C, 14 d: centrally raised; colony surface velutinous, margin entire to delicately filiform; mycelial areas white; sporulation sparse, turquoise white (24A2); soluble pigment absent; exudate absent; reverse centrally turquoise white (24D2) to orange white (5A2) in margins. MEA 25 °C, 14 d: no growth. M60Y 25 °C, 14 d: flat to slightly centrally raised; colony surface velutinous to powdery; margin entire to delicately filiform; mycelial areas white; sporulation deep turquoise (25E8); soluble pigment absent; exudate absent; reverse centrally dark green (29F3) to champagne greyish yellow (4B4) in margins. MEA + 10 % NaCl 25 °C, 14 d: flat, wrinkled; colony surface velutinous, margin entire to delicately filiform; mycelial areas white; sporulation aquamarine greyish turquoise (24B3); soluble pigment absent; exudate absent; reverse centrally cinnamon brown (6D6) to olive yellow (3D6), champagne greyish yellow (4B4) in margins.
Micromorphology: Conidial heads radiate. Conidiophores uniseriate. Stipes smooth in SEM, 4.5–6 μm wide in the middle part, widening gradually toward the vesicle, with occasional septa. Vesicles spatulate, pyriform to clavate, 7–12 μm wide. Phialides flask-shaped, 8–10 μm long, covering the upper half to two thirds of the vesicle. Conidia borne as rough-walled long cylinders, echinulate at maturity, aculeate in SEM, subglobose to ellipsoidal, connectives evident, commonly remaining in chains, 4.5–5.5 × 3–4 μm.
Distinguishing characters: Isolates of A. vitricola have larger conidia, narrower stipes and much smaller vesicles compared to A. glabripes. Aspergillus vitricola does not grow on MEA and CYA at 25 °C and CY20S at 30 °C and exhibit only very restricted growth on CY20S at 25 °C. Both species are also different micromorphologically and in growth parameters on most media.
Additional materials examined: Canada, British Columbia, Victoria, house dust, 2015, collected by B. Kendrick, isolated by C.M. Visagie, KAS 6093, KAS 6087 = DTO 356-H2 = IBT 34532, KAS 6199, KAS 6237 = DTO 356-I2, KAS 6238, KAS 6281. Canada, New Brunswick, Little Lepreau, house dust, 2015, collected by A. Walker, isolated by C.M. Visagie, DAOMC 251500 = KAS 6133, KAS 6086, KAS 6178 = DTO 356-H8, KAS 6279. Canada, Nova Scotia, Wolfville, house dust, 2015, collected by A. Walker, isolated by C.M. Visagie, KAS 6150 = DTO 356-H5 = IBT 34531, KAS 6235, KAS 6236, KAS 6250, KAS 6251, KAS 6263, KAS 6264, KAS 6278 = DTO 356-I8, KAS 6285, KAS 6287, KAS 6288, KAS 6290. Canada, Ontario, Stittsville, house dust, collected by K.A. Seifert, isolated by C.M. Visagie, KAS 5687. Slovenia, Ljubljana, mouldy hat of a national costume of a folklore group (stored in basement depot), 2015, isolated by P. Zalar, EXF-10301 = CCF 5654 = IBT 34290. Slovenia, Ljubljana, oil painting on canvas, 2015, isolated by P. Zalar and D.D. Graf, EXF-10383 = CCF 5655 = IBT 34272. Slovenia, Ljubljana, oil painting on canvas, 2012, isolated by P. Zalar, EXF-7700 = CCF 5657 = IBT 34282 = IBT 33575. France, raisin madeleines, 2014, isolated by F. Déniel, CBS 140435 = UBOCC-A-115047 = DTO 334-E6. The Netherlands, archive material, 2006, isolated by J. Houbraken, DTO 014-A4. The Netherlands, Noordwijk, carpet “zeegrastapijt” in house, 2015, J. Houbraken, DTO 331-D6. The Netherlands, Gorinchem, archive material, 2010, isolated by M. Meijer and O. Terhoeven, DTO 122-I4, DTO 123-B2, DTO 122-I5. USA, California, San Diego, child carrier, 2015, isolated by Ž. Jurjević, CCF 5534 = EMSL No. 2785.
Dichotomous key to species from section Restricti
-
1a)
Homothallic, no growth on M40Y at 25 °C …………………………………………….… A. halophilicus
-
1b)
Anamorphic, good growth on M40Y at 25 °C ………… 2
-
2a)
Conidial heads from colony edge radiate or globose when young ……………………………………………… 11
-
2b)
Conidial heads columnar or loosely columnar when young ……………………………………………………… 3
Aspergillus restrictus/conicus clades
-
3a)
No growth on CY20S at 30 °C and M60Y at 37 °C after 14 d ………………………………………………………. 4
-
3b)
Growth on CY20S at 30 °C and M60Y at 37 °C after 14 d ………………………………………………………. 8
-
4a)
Colonies on M60Y at 25 and 30 °C exceed 40 mm after 14 d ………………………………………………………. 5
-
4b)
Colonies on M60Y at 25 and 30 °C do not exceed 40 mm after 14 d ……………….………………………. 6
-
5a)
Stipes 3–5 μm wide, vesicles 6.5–9 μm wide, phialides 7.5–9 μm long, reverse colour on M40Y olive grey in the centre ……………………………………… A. gracilis
-
5b)
Stipes 4–6 μm wide, vesicles 9–13 μm wide, phialides 9–10.5 μm long, reverse colour on M40Y cinnamon brown in the centre …………………… A. pseudogracilis
-
6a)
Sporulation colour on M40Y myrtle green, reverse apricot greyish orange in the centre ……... A. destruens
-
6b)
Sporulation colour on M40Y turquoise, reverse olive grey in the centre ……………………………………..… 7
-
7a)
Sporulation colour on M40Y pale turquoise, conidia predominantly globose to subglobose (or barrel-shaped) ……………………………………………. A. conicus
-
7b)
Sporulation colour on M40Y greyish turquoise, conidia predominantly ellipsoidal (or barrel-shaped) ……………………………………………... A. domesticus
-
8a)
Length of phialides ≥ 9 μm, colonies on CY20S at 30 °C and CYA at 25 °C and M60Y at 37 °C ≤ 10 mm after 14 d, conidia tuberculate in SEM ……… A. villosus
-
8b)
Length of phialides ≤ 9 μm, colonies on CY20S at 30 °C and CYA at 25 °C and M60Y at 37 °C > 10 mm after 14 d, conidia aculeate in SEM …………………… 9
-
9a)
Diameter of vesicle ≥ 16 μm …………... A. pachycaulis
-
9a)
Diameter of vesicle ≤ 16 μm ………………………… 10
-
10a)
Conidia 3.5–4.5 × 2.5–3 μm, vesicle diameter 9–12 μm ……………………………………. A. restrictus
-
10b)
Conidia 4.5–6 × 3–4.5 μm, vesicle diameter 11–16 μm ……………………………………………….. A. caesiellus
Aspergillus penicillioides/vitricola clades
-
11a)
Average diameter of vesicles ≥ 19 μm ……………... 12
-
11b)
Average diameter of vesicles < 17 μm ………………. 13
-
12a)
Conidia 3.5–4.5 × 2–3 μm, length of phialides 7–9 μm ………………………………………………... A. glabripes
-
12b)
Conidia 4.5–5.5 × 3.5–4.5 μm, length of phialides 9–11 μm …………………………… A. magnivesiculatus
-
13a)
Length of phialides ≤ 8 μm, average length ∼ 7 μm …………………………………………..…. A. canadensis
-
13b)
Length of phialides ≥ 8 μm, average length ≥ 8.5 μm …………………………………………………………… 14
-
14a)
Conidia do not exceed 4 μm in long axis, average length ca 3.6 μm ……………………………. A. salinicola
-
14b)
Conidia commonly exceed 4 μm in long axis, average length ca 4–5 μm ……………………………………… 15
-
15a)
Colony diameter on CY20S and M40Y at 25 °C after 14 d > 20 and 45 mm, respectively ……… A. reticulatus
-
15b)
Colony diameter smaller on both media …………….. 16
-
16a)
Average width of stipes ≤ 5.5 μm ……………………………………………………………17
-
16b)
Average width of stipes frequently ≥ 6 μm ………………………………………………………..…. 19
-
17a)
No production of soluble pigment on DG18, average length of conidia > 4.5 μm, stipes smooth in SEM ………………………………………………….. A. vitricola
-
17b)
Greyish yellow soluble pigment on DG18, average length of conidia ca 4 μm, stipes covered by hairs in SEM …………………………………………………….. 18
-
18a)
Sporulation colour on M40Y deep green, conidia in SEM tuberculate ……………………….. A. penicillioides
-
18b)
Sporulation colour on M40Y dark turquoise, conidia in SEM lobate-reticulate …………………………. A. hordei
-
19a)
No growth on CYA at 25 °C after 14 d, colony reverse on M40Y in shades of yellow or brown, conidia tuberculate in SEM ………………………………………….. 20
-
19b)
Growth on CYA at 25 °C after 14 d, colony reverse on M40Y dark green, conidia aculeate in SEM ………………………………………………. A. infrequens
-
20a)
Sporulation colour on M40Y greyish green, average dimensions of conidia 4.8 × 3.5 μm, colony diameter on CY20S at 25 °C after 14 d > 6 mm ….. A. clavatophorus
-
20b)
Sporulation colour on M40Y greyish turquoise (water blue), average dimensions of conidia 4 × 2.9 μm, colony diameter on CY20S at 25 °C after 14 d < 6 mm …………………………………………... A. tardicrescens
Discussion
Past and current concept of section Restricti
Thom & Church (1926) were the first who studied the classification of the species of sect. Restricti. They placed A. penicilliodies, A. gracilis and A. conicus into a transitional group between A. glaucus and A. fumigatus. Thom & Raper (1945) included also A. restrictus and considered the members of sect. Restricti as a series within the A. glaucus group. The A. restrictus group was subsequently introduced by Raper & Fennell (1965) who added also A. caesiellus and treated A. vitricola as a synonym of A. penicillioides. Although the first described species was A. penicillioides, the authors decided to name the group after the species (A. restrictus) they considered most important. The authors also observed variability within A. restrictus and mentioned unique growth characterictics of isolate NRRL 145. This isolate is in the present study designated as the ex-type of a novel species A. destruens, a member of the A. conicus clade. This species resembles A. conicus rather than A. restrictus in almost all phenotypic characters.
The taxonomic rank of section was later introduced by Samson & Gams (1985). The number of species forming the section was subsequently reduced to three by Pitt & Samson (1990), rejecting A. conicus and A. gracilis. Even though the morphological variability within the section is rather small, at least the clades (A. restrictus, A. conicus, A. vitricola and A. penicillioides) can be clearly distinguished solely based on morphology.
Molecular studies conducted by Peterson (2000) confirmed that re-expansion of the section was necessary, supported recognition of at least six anamorphic species and placed A. halophilicus, which produces a homothallic teleomorph similar to those seen in sect. Aspergillus, into the sect. Restricti.
In this study, we performed a new revision of sect. Restricti that involved a large number of isolates and used both phenotypic and molecular genetic data for species delimitation. The number of accepted species was tripled compared to previous studies and 14 new species were described. The genetic distances between the species are greater (similarity between sequences of sister species within the clades varies from 95 % to 97 %, sequence similarity between the clades is only slightly above 90 %) than typically seen in other sections of the genus Aspergillus, including sister sect. Aspergillus. Our study could not sufficiently cover all common isolation sources of sect. Restricti members and description of new taxa in future can be expected if studies with extensive sampling and focused on particular substrates or environments are conducted. This assumption is supported by a comparison of our generated ITS barcodes with 1061 sect. Restricti 454-pyrosequences from Amend et al. (2010). A phylogenetic tree (Fig. 8) confirms that there is still uncovered species diversity within section Restricti and discovery of new species is expected in future. The data showed that the house dust conceals high number of species that belong to all species complexes recognized in this study including A. halophilicus. Aspergillus penicillioides, A. vitricola, A. hordei, A. tardicrescens and A. magnivesiculatus belonged to the most abundantly detected species. Several undescribed phylogenetic lineages are present in A. restrictus/conicus clade and one unnamed lineage sister to A. tardicrescens is present in A. penicillioides clade (Fig. 8).
Two of the 21 species accepted in this work were represented only by one isolate and three species by only two isolates. This situation, although not often discussed, is very common in the taxonomy of fungi (Lim et al. 2012). Even though working with species with no infraspecific variability is problematic, it is a general practice to include also these species into the analyses because the authors always want to cover all known variability (Ahrens et al. 2016). The results of some phylogenetic methods may be biased due to the presence of species with low or no variability, and population genetic methods cannot be used at all with datasets containing species without infraspecific variability. Since the aim of taxonomic revisions and monographs is to describe all current knowledge we included all species irrespective of the number of isolates and presence or absence of infraspecific variability. When we re-inspect the results of our phylogenetic analyses, it is common that the unresolved nodes are occupied by those species represented by a few isolates. Better resolution of phylogenetic relationships can be expected when the inevitable additional strains of rare species and discovery of novel taxa are made.
Phylogeny of subgenus Aspergillus
In contrast to sect. Restricti, sect. Aspergillus forms a compact clade. The phylogenetic analysis across species in subg. Aspergillus strongly supported the monophyly of both, sect. Restricti and sect. Aspergillus (Fig. 1, Fig. 2). This topology is in agreement with trees published by Peterson (2008) and Peterson et al. (2008), however, especially in the latter study the sect. Restricti received only limited statistical support. In the more recent studies of Houbraken and Samson, 2011, Houbraken et al., 2014 and Kocsubé et al. (2016), members of section Restricti did not form a separate monophyletic clade with respect to sister sect. Aspergillus. This was probably caused by the inadequate taxon sampling (low number of species included). Large genetic distances between particular clades within sect. Restricti could warrant proposal of several new sections (e.g., A. restrictus+A. conicus+A. vitricola clades = section Restricti s. str.; A. penicillioides clade; and A. halophilicus) solely based on phylogenetic distances (Fig. 1, Fig. 2) and their comparison with those between sections in other subgenera. Several factors prevent us doing so. Most importantly, the section forms a coherent group in terms of morphology (except A. halophilicus) and is strikingly different from homothallic species of sister sect. Aspergillus that produce abundant ascomata in vitro. When viewing the phylogenetic tree of the subgenus in the radial form (Fig. 2), the real relationships between sections and clades seem clearer. There are at least three clades in sect. Restricti separated by large genetic distances that are similar to distances of these clades from basal taxa of sect. Aspergillus. One could even argue that there are enough clades on the tree spread along the transition between sections Restricti and Aspergillus to merge both sections into one (Fig. 2). Another problem is the unresolved position of some taxa belonging to sect. Restricti, especially A. halophilicus. Considering the uniqueness of A. halophilicus in terms of morphology, physiology, reproductive strategy in addition to phylogenetic isolation, it might seem appropriate to exclude this species from sect. Restricti and create a separate, single-species section. However, such action would probably result in a paraphyletic sect. Restricti due to the unclear position of A. halophilicus. Isolation of additional taxa related to A. halophilicus could improve results of the phylogenetic analyses and provide new phenotypic data supporting or contradicting this scenario. Furthermore, the sexual reproductive strategy of anamorphic species from sect. Restricti is unknown but discovery of a sexual state would not be unexpected in light of recent developments in this field (Horn et al., 2009, O'Gorman et al., 2009, Horn et al., 2011, Nováková et al., 2014).
Exometabolites
All species in Aspergillus sect. Restricti produced asperglaucide, asperphenamate or both. This is in contrast to species of Aspergillus sect. Aspergillus that do not produce these compounds (Chen et al. 2017). On the other hand the species in both sections do produce some extrolites in common. Mycophenolic acid is produced by several species from both sections, similarly to echinulin and derivatives such as neoechinulin-type metabolites (Chen et al. 2017). In sect. Restricti no isolates were found to produce auroglaucins or epiheveadride, compounds which are very common in the ascomata of Aspergillus sect. Aspergillus. The ascomata of A. halophilicus do not contain the yellow auroglaucin or flavoglaucin compounds. It is interesting to note that antarone A and asperentins produced by A. salinicola are shared with a halotolerant Penicillium antarcticum (Vansteelandt et al., 2012, Geiger et al., 2013).
The asperglaucides and asperphenamate are simple phenylalanine derived compounds with anticancer activity (Bladt et al. 2013). These compounds have been found in many plants (Frisvad et al. 2013), and one can speculate that these medicinal compounds are not produced by the plants but rather by its endophytic fungi, including possibly endophytic isolates of Aspergillus sect. Restricti.
A very high chemical diversity was observed in section Restricti, but a large number of extrolites have until now only been found in this section. The chemoconsistency on the species level appeared to be less pronounced than in for example section Aspergillus, but the absence of metabolites may be caused by poor growth in some cases. For example A. villosus NRRL 25813T, IBT 22529, IBT 22530 and IBT 22546 produced a large number of orthosporins, that were not detected in the strain UBOCC-A-116020. In such cases other media than those used here may be more suitable for production of a broader spectrum of metabolites. Addition of a high concentration of salts or sugars strongly increased the growth rate of isolates from section Restricti, and also secondary metabolite production. More media, such as potato dextrose agar and malt agars may be more suitable for secondary metabolite production if they are supplemented with more salt or sugar. The overview of metabolites produced by species from section Restricti reported from previous studies is given in Table 10. Most secondary metabolites reported from A. halophilicus by Micheluz et al. (2016), such as chaetoviridin A, deoxybrevianamide E, pseurotin A and D, rugulosuvine, stachybotryamide, tenellin and tryprostatin B could not be recovered from any strain of A. halophilicus examined in this study. However these extrolites may have been detected because of contamination of the YES 15 % medium (YES agar with 15 % NaCl) they used. Several extrolites detected by Micheluz et al. (2016) are products of Aspergillus fumigatus, including pseurotins A, D, tenellin and tryprostatin (Frisvad and Larsen, 2016), and were only detected in one of three examined isolates. However, YES agar with 15 % NaCl has not been used in this study, so a final conclusion on the production of these A. fumigatus metabolites by A. halophilicus must await further testing.
Table 10.
Exometabolites reported from Aspergillus section Restricti isolates.
| Exometabolite | Producer | Reference |
|---|---|---|
| Arestrictin A and B | A. restrictus | Itabashi et al. (2006) |
| Arestrictin A and B | A. penicillioides | Itabashi et al. (2006) |
| Cristatin A | A. restrictus | Itabashi et al. (2006) |
| Neoechinulin A | A. halophilicus | Micheluz et al. (2016) |
| Asperglaucide (= aurantiamide acetate) | A. penicillioides | Isshiki et al., 2001, Micheluz et al., 2016 |
| Asperglaucide (= aurantiamide acetate) | A. restrictus | Micheluz et al. (2016) |
| Rugulusovin | A. halophilicus | Micheluz et al. (2016) |
| Citreorosein | A. penicillioides | Micheluz et al. (2016) |
| Deoxybrevianamide A | A. penicillioides | Micheluz et al. (2016) |
| Deoxybrevianamide E | A. halophilicus | Micheluz et al. (2016) |
| Exometabolites incorrectly ascribed to sect. Restricti species | ||
| Restrictocin | A. restrictus1 | Lamy and Davies, 1991, Arruda et al., 1992, Brandhorst and Kenealy, 1992, Yang and Kenealy, 1992, Brandhorst et al., 1996 |
| Chaetoviridin A | A. halophilicus | Micheluz et al. (2016) |
| Pseurotin A & D | A. halophilicus2 | Micheluz et al. (2016) |
| Tenellin | A. halophilicus2 | Micheluz et al. (2016) |
| Tryprostatin B | A. halophilicus2 | Micheluz et al. (2016) |
The isolates producing restrictocin have been re-identified as A. fumigatus (strains MDH 13462L = ATCC 34475 = NRRL 2869 and MDH 17070 = NRRL 3050 = ATCC 34506) (Arruda et al. 1992); see also Jong et al. (1996).
One of three isolates identified as A. halophilicus produced metabolites that are present in A. fumigatus (Frisvad et al., 2009, Frisvad and Larsen, 2016), so it was probably contaminated with A. fumigatus.
Ecology and pathogenicity
Species of sect. Restricti occur worldwide and they used to be most frequently isolated from food, feed and indoor environments. Pitt & Hocking (2009) summarised spoilage of different foods (e.g., flour, wheat, maize, rice, dried, cured or salted meat, dried fruit, spices, beans, nuts, coffee beans and salted dried fish) by A. restrictus and A. penicillioides. Aspergillus penicillioides is probably the most frequently isolated species from the section although it may be partly due to the broad concept adopted in the past. Members of sect. Restricti are commonly isolated from grain, such as corn, wheat and barley, and many species were found on grain during this study, namely A. penicillioides, A. restrictus, A. caesiellus, A. magnivesiculatus, A. hordei, A. halophilicus, A. infrequens. The isolates from food examined in this study originated mostly from candy and bakery products (A. penicillioides, A. restrictus, A. glabripes, A. destruens, A. domesticus, A. conicus, A. vitricola, A. salinicola), rarely from other foods such as cheese (A. destruens), nuts (A. penicillioides, A. conicus), tea (A. clavatophorus), katsuobushi (A. magnivesiculatus), etc. Some examples of spoiled foods examined in this study are shown in Fig. 35. The species are also commonly isolated from leather and textile goods, e.g. shoes, baseball gloves (see Fig. 36 G, H) or mattress covers (A. penicillioides, A. restrictus, A. reticulatus, A. caesiellus). The presence of these species in the indoor environment and house dust is well-known from past studies (Samson and Lustgraaf, 1978, Frisvad and Gravesen, 1994, Meklin et al., 2004, Kaarakainen et al., 2009, Amend et al., 2010, Visagie et al., 2014). Eleven species which were isolated from indoor air are included in our study, namely A. caesiellus, A. conicus, A. destruens, A. domesticus, A. glabripes, A. magnivesiculatus, A. pachycaulis, A. penicillioides, A. restrictus, A. reticulatus and A. tardicrescens. Examples of sect. Restricti members growing in various conditions, including indoor environment and textile can be found in Fig. 36. In a world-wide survey of fungi occurring in house dust using a portion of the ID locus as used here (Amend et al. 2010), A. penicillioides was a common OTU identified. In these analyses a 97 % cutoff is most commonly used, however, the low amount of sequence data available for sect. Restricti before this study makes the analysis of species diversity complicated. This is clear from our sect. Restricti phylogeny (Fig. 8) incorporating newly generated ITS barcodes and 454-pyrosequences from Amend et al. (2010), where we can now positively identify 14 of the OTU's down to species level, in contrast to the at most seven previously possible in theory. One of the main problems is the high sequence divergence between sect. Restricti species where culture independent methods had almost no chance of getting to a correct species name. In this study, we release more than 180 ITS rDNA barcode sequences covering both infra- and interspecies variation. This will greatly aid these modern techniques to identify common indoor fungi. A significant portion of strains and new species examined in this study was isolated from air, especially from indoor air of various regions in the USA. It seems that this enviroment might still be an important source of undiscovered genetic variability and new species. Many specimens examined in this work also come from house dust; for instance isolates of A. canadensis thus far originate exclusively from house dust in Canada.
Fig. 35.
Different kinds of food and commodities on which members of section Restricti are to be found. A–F. Various bakery products: A. Chocolate chip cookies (“A. penicillioides” by morphology – isolate not included in this study; isolated together with A. cibarius). B. Mouldy bread. C. Madeleines (isolate UBOCC-A-115045 – A. reticulatus). D. French savaroise (isolate UBOCC-A-115043 – A. salinicola). E. French mini cakes (isolate UBOCC-A-115048 – A. domesticus). F. Mouldy cake (mixed spoilage with Penicillium and Eurotium spp.). G. Grain containing diverse fungal populations similar to nuts, spices and seeds. H. Cigars (“A. penicillioides” by morphology – isolate not included in this study).
Fig. 36.
Overview of some substrates and habitats associated with members of section Restricti. A. Solar salterns at the Adriatic coast, Sečovlje, Slovenia (isolate EXF 226 – A. salinicola). B. Indoor environment, a source of numerous species: one of the sampling sites in Canada where house dust was collected. C. Water damaged wall surface, dry at the time of sampling (isolate DTO 073-H6 – A. tardicrescens). D. Mouldy chair backrest (isolate DTO 231-B9 – A. domesticus). E. Textiles (“A. penicillioides” by morphology – isolate not included in this study). F. Textile water hose imported from China (“A. penicillioides” and Cladosporium sp. by morphology – isolates not included in this study). G. Leather baseball gloves (isolate CCF 5500 – A. penicillioides and CCF 5514 – A. restrictus). H. Leather armrest of a dentist chair (isolate DTO 316-A7 – A. tardicrescens). I. Skeleton of Elephas maximus (Asian elephant) (“A. penicillioides” by morphology – isolate not included in this study). J. Mouldy sclerotium of Corallocytostroma ornicopreoides imported from Australia (isolate CCF 3364 – A. restrictus).
The species can also cause significant damage to specimens deposited in herbaria (Zhang et al. 2016) and also to works of art, especially paintings (Fig. 37). Examination of fungi isolated from deteriorated paintings from Musée des beaux Arts de Brest (France) and Institute for the Protection of Cultural Heritage of Slovenia during this study showed that species A. destruens, A. conicus, A. domesticus, A. vilosus, A. vitricola, A. glabripes, A. tardicrescens, A. reticulatus, A. magnivesiculatus and A. salinicola can deteriorate paintings and generate cultural and economic losses. Aspergillus clavatophorus was also isolated from a painting in the Netherlands, while A. halophilicus was recently identified as a causative agent of book biodeterioration in archives and libraries from Micheluz et al. (2015).
Fig. 37.
Deteriorated paintings and books from archives. A–H. Deteriorated painting from Capuchin monastery of St. Francis, Ajdovščina, Slovenia (A) and Musée des beaux Arts in Brest, France (B–H); isolated species: A. destruens, A. domesticus, A. conicus, A. glabripes, A. magnivesiculatus, A. reticulatus, A. salinicola, A. tardicrescens, A. villosus, A. vitricola. I–M. Deteriorated books from archives; isolated species: A. vitricola and A. domesticus.
Hypersaline water has proved to be a rich source of xerophilic fungi (Gunde-Cimerman et al. 2000), including members of sect. Restricti (Cantrell et al., 2006, Nazareth and Gonsalves, 2014). The novel species A. salinicola described in this study is a saltern inhabitant (Fig. 36. A).
Some species can be isolated from very uncommon substrates as in the case of A. vitricola that was isolated from glass in Japan and is able to damage optical lenses (Ohtsuki 1962). As we found here, this species is also very common in house dust in Canada.
The pathogenic potential of sect. Restricti seems to be low considering the small number of reported cases of opportunistic human infections. Several records of human infection caused by A. penicillioides and A. restrictus is summarised by de Hoog et al. (2009), but the identification of the pathogens was performed exclusively by morphological examination. It is very likely that some records represent confusion with A. fumigatus due to similar sporulation colour and micromorphology. However, a confirmed case of invasive aspergillosis in human has been described recently that was probably caused by an undescribed species from the A. penicillioides clade (ITS GenBank: KP316198) (Gupta et al. 2016). This case demonstrated that athough the species of the sect. Restricti are rarely found as pathogens, they have at least limited pathogenic potential.
Acknowledgements
This research was supported by the the project of the Charles University Grant Agency (GAUK 1434217) and the project BIOCEV (CZ.1.05/1.1.00/02.0109) provided by the Ministry of Education, Youth and Sports of CR and ERDF. Microbiological and structural investigations of biologically damaged textiles from Slovenian museums was financed by Slovenian Research Agency (J7-4208). Cobus M. Visagie, Neriman Yilmaz, Keith A. Seifert, Amanda Chen, Robert A, Samson and Jos Houbraken received financial support from the Alfred P. Sloan foundation Program on the Microbiology of the Built Environment. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer. We thank Milada Chudíčková and Alena Gabrielová for their invaluable assistance in the laboratory, CCF collection staff (Alena Kubátová – curator, Ivana Kelnarová and Adéla Kovaříčková) for deposition and lyophilization of the cultures, Alena Kubátová and Miroslav Hyliš for assistance with scanning electron microscopy, Dirk Stubbe for providing cultures from BCCM/IHEM collection, Martin Meijer for isolation and providing interesting fungal strains, Daniela Daša Graf for her work on isolation and characterisation of Aspergillus strains from oil paintings on canvas included in the manuscript. Vit Hubka is grateful for support from Czechoslovak Microscopy Society (CSMS scholarship 2016).
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.simyco.2017.09.002.
Contributor Information
S.W. Peterson, Email: Stephen.Peterson@ars.usda.gov.
V. Hubka, Email: hubka@biomed.cas.cz.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Abe K. Assessment of home environments with a fungal index using hydrophilic and xerophilic fungi as biologic sensors. Indoor Air. 2012;22:173–185. doi: 10.1111/j.1600-0668.2011.00752.x. [DOI] [PubMed] [Google Scholar]
- Ahrens D., Fujisawa T., Krammer H.J. Rarity and incomplete sampling in DNA-based species delimitation. Systematic Biology. 2016;65:478–494. doi: 10.1093/sysbio/syw002. [DOI] [PubMed] [Google Scholar]
- Ali I., Akbar A., Anwar M. Purification and characterization of a polyextremophilic α-Amylase from an obligate halophilic Aspergillus penicillioides isolate and its potential for souse with detergents. BioMed Research International. 2015;2015:245649. doi: 10.1155/2015/245649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amend A.S., Seifert K.A., Samson R. Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13748–13753. doi: 10.1073/pnas.1000454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruda L.K., Mann B.J., Chapman M.D. Selective expression of a major allergen and cytotoxin, Asp f I, in Aspergillus fumigatus – implications for the immunopathogenesis of Aspergillus-related diseases. Journal of Immunology. 1992;149:3354–3359. [PubMed] [Google Scholar]
- Bainier G. Mycothèque de l'Ecole de Pharmacie, XII-XVI. Bulletin de la Société Mycologique de France. 1907;23:90–93. [Google Scholar]
- Bladt T.T., Frisvad J.C., Knudsen P.B. Anticancer and antifungal compounds from Aspergillus, Penicillium and other filamentous fungi. Molecules. 2013;18:11338–11376. doi: 10.3390/molecules180911338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R., Heled J., Kuhnert D. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Computational Biology. 2014;10:6. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R.R. DensiTree: making sense of sets of phylogenetic trees. Bioinformatics. 2010;26:1372–1373. doi: 10.1093/bioinformatics/btq110. [DOI] [PubMed] [Google Scholar]
- Brandhorst T.T., Dowd P.F., Kenealy W.R. The ribosome-inactivating protein restrictocin deters insect feeding on Aspergillus restrictus. Microbiology. 1996;142:1551–1556. doi: 10.1099/13500872-142-6-1551. [DOI] [PubMed] [Google Scholar]
- Brandhorst T.T., Kenealy W.R. Production and localization of restrictocin in Aspergillus restrictus. Journal of General Microbiology. 1992;138:1429–1435. doi: 10.1099/00221287-138-7-1429. [DOI] [PubMed] [Google Scholar]
- Cantrell S.A., Casillas-Martínez L., Molina M. Characterization of fungi from hypersaline environments of solar salterns using morphological and molecular techniques. Mycological Research. 2006;110:962–970. doi: 10.1016/j.mycres.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Carstens B.C., Pelletier T.A., Reid N.M. How to fail at species delimitation. Molecular Ecology. 2013;22:4369–4383. doi: 10.1111/mec.12413. [DOI] [PubMed] [Google Scholar]
- Chen A.J., Hubka V., Frisvad J.C. Polyphasic taxonomy of Aspergillus section Aspergillus (formerly Eurotium) and its occurrence in indoor environment and food. Studies in Mycology. 2017;88:37–135. doi: 10.1016/j.simyco.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen C.M., Kaufmann H.H. Deterioration of stored grains by fungi. Annual Review of Phytopathology. 1965;3:69–84. [Google Scholar]
- Christensen C.M., Papavizas G.C., Benjamin C.R. A new halophilic species of Eurotium. Mycologia. 1959;51:636–640. [Google Scholar]
- Collado J., Platas G., Paulus B. High-throughput culturing of fungi from plant litter by a dilution-to-extinction technique. FEMS Microbiology Ecology. 2007;60:521–533. doi: 10.1111/j.1574-6941.2007.00294.x. [DOI] [PubMed] [Google Scholar]
- Cummings M.P., Neel M.C., Shaw K.L. A genealogical approach to quantifying lineage divergence. Evolution. 2008;62:2411–2422. doi: 10.1111/j.1558-5646.2008.00442.x. [DOI] [PubMed] [Google Scholar]
- Dale E. On the fungi of the soil. Part II. Fungi from chalky soil, uncultivated mountain peat, and the “black earth” of the reclaimed fenland. Annales Mycologici. 1914;12:38. [Google Scholar]
- de Hoog G., Guarro J., Gené J. 3rd CD-ROM ed. CBS-KNAW Fungal Biodiversity Centre; Utrecht, The Netherlands: 2009. Atlas of clinical fungi. [Google Scholar]
- Deschuyffeleer N., Vermeulen A., Daelman J. Modelling of the growth/no growth interface of Wallemia sebi and Eurotium herbariorum as a function of pH, aw and ethanol concentration. International Journal of Food Microbiology. 2015;192:77–85. doi: 10.1016/j.ijfoodmicro.2014.09.022. [DOI] [PubMed] [Google Scholar]
- Edwards S.V. Is a new and general theory of molecular systematics emerging? Evolution. 2009;63:1–19. doi: 10.1111/j.1558-5646.2008.00549.x. [DOI] [PubMed] [Google Scholar]
- Flannigan B., Samson R.A., Miller J.D. CRC Press; London, UK: 2016. Microorganisms in home and indoor work environments: diversity, health impacts, investigation and control. [Google Scholar]
- Flot J.F. Species delimitation's coming of age. Systematic Biology. 2015;64:897–899. doi: 10.1093/sysbio/syv071. [DOI] [PubMed] [Google Scholar]
- Fontaneto D., Flot J.F., Tang C.Q. Guidelines for DNA taxonomy, with a focus on the meiofauna. Marine Biodiversity. 2015;45:433–451. [Google Scholar]
- Frasz S.L., Miller J.D. Fungi in Ontario maple syrup & some factors that determine the presence of mold damage. International Journal of Food Microbiology. 2015;207:66–70. doi: 10.1016/j.ijfoodmicro.2015.04.038. [DOI] [PubMed] [Google Scholar]
- Frisvad J.C., Gravesen S. Penicillium and Aspergillus from Danish homes and working places with indoor air problems: identification and mycotoxin determination. In: Samson R.A., Flannigan B., Flanigan M.E., Verhoeff A.P., editors. Health implications of fungi in indoor environments: air quality monographs. Pergamon Press; Amsterdam, The Netherlands: 1994. pp. 281–290. [Google Scholar]
- Frisvad J.C., Houbraken J., Popma S. Two new Penicillium species Penicillium buchwaldii and Penicillium spathulatum, producing the anticancer compound asperphenamate. FEMS Microbiology Letters. 2013;339:77–92. doi: 10.1111/1574-6968.12054. [DOI] [PubMed] [Google Scholar]
- Frisvad J.C., Larsen T.O. Extrolites of Aspergillus fumigatus and other pathogenic species in Aspergillus section Fumigati. Frontiers in Microbiology. 2016;6:1485. doi: 10.3389/fmicb.2015.01485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad J.C., Rank C., Nielsen K.F. Metabolomics of Aspergillus fumigatus. Medical Mycology. 2009;47:S53–S71. doi: 10.1080/13693780802307720. [DOI] [PubMed] [Google Scholar]
- Frisvad J.C., Thrane U. Standardized high-performance liquid chromatography of 182 mycotoxins and other fungal metabolites based on alkylphenone retention indices and UV-VIS spectra (diode array detection) Journal of Chromatography A. 1987;404:195–214. doi: 10.1016/s0021-9673(01)86850-3. [DOI] [PubMed] [Google Scholar]
- Fujisawa T., Barraclough T.G. Delimiting species using single-locus data and the Generalized Mixed Yule Coalescent (GMYC) approach: a revised method and evaluation on simulated datasets. Systematic Biology. 2013;62:707–724. doi: 10.1093/sysbio/syt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M.K., Leaché A.D., Burbrink F.T. Coalescent-based species delimitation in an integrative taxonomy. Trends in Ecology & Evolution. 2012;27:480–488. doi: 10.1016/j.tree.2012.04.012. [DOI] [PubMed] [Google Scholar]
- Gams W., Christensen M., Onions A. Infrageneric taxa of Aspergillus. In: Samson R.A., Pitt J.I., editors. Advances in Penicillium and Aspergillus systematics. Vol. 102. Plenum Press; New York: 1985. pp. 55–62. (NATO ASI Series, Ser. A: Life Sciences). [Google Scholar]
- Geiger M., Guitton Y., Vansteelandt M. Cytotoxicity and mycotoxin production of shellfish-derived Penicillium spp., a risk for shellfish consumers. Letters in Applied Microbiology. 2013;57:385–392. doi: 10.1111/lam.12143. [DOI] [PubMed] [Google Scholar]
- Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunde-Cimerman N., Zalar P., de Hoog S. Hypersaline waters in salterns – natural ecological niches for halophilic black yeasts. FEMS Microbiology Ecology. 2000;32:235–240. doi: 10.1111/j.1574-6941.2000.tb00716.x. [DOI] [PubMed] [Google Scholar]
- Gupta K., Gupta P., Mathew J.L. Fatal disseminated Aspergillus penicillioides infection in a three-month-old infant with suspected cystic fibrosis: autopsy case with review of literature. Pediatric and Developmental Pathology. 2016;19:506–511. doi: 10.2350/15-10-1729-CR.1. [DOI] [PubMed] [Google Scholar]
- Hall T.A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series. [Google Scholar]
- Harrold C.E. Studies in the Genus Eremascus: I. The rediscovery of Eremascus albus Eidam and some new observations concerning its life-history and cytology. Annals of Botany. 1950;14:127–148. [Google Scholar]
- Heled J., Drummond A.J. Bayesian inference of species trees from multilocus data. Molecular Biology and Evolution. 2010;27:570–580. doi: 10.1093/molbev/msp274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hocking A.D., Pitt J.I. Dichloran-glycerol medium for enumeration of xerophillic fungi from low-moisture foods. Applied and Environmental Microbiology. 1980;39:488–492. doi: 10.1128/aem.39.3.488-492.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.-B., Cho H.-S., Shin H.-D. Novel Neosartorya species isolated from soil in Korea. International Journal of Systematic and Evolutionary Microbiology. 2006;56:477–486. doi: 10.1099/ijs.0.63980-0. [DOI] [PubMed] [Google Scholar]
- Horn B.W., Moore G.G., Carbone I. Sexual reproduction in Aspergillus flavus. Mycologia. 2009;101:423–429. doi: 10.3852/09-011. [DOI] [PubMed] [Google Scholar]
- Horn B.W., Moore G.G., Carbone I. Sexual reproduction in aflatoxin-producing Aspergillus nomius. Mycologia. 2011;103:174–183. doi: 10.3852/10-115. [DOI] [PubMed] [Google Scholar]
- Houbraken J., Samson R.A. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology. 2011;70:1–51. doi: 10.3114/sim.2011.70.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., de Vries R.P., Samson R.A. Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Advances in Applied Microbiology. 2014;86:199–249. doi: 10.1016/B978-0-12-800262-9.00004-4. [DOI] [PubMed] [Google Scholar]
- Hubka V., Lysková P., Frisvad J.C. Aspergillus pragensis sp. nov. discovered during molecular reidentification of clinical isolates belonging to Aspergillus section Candidi. Medical Mycology. 2014;52:565–576. doi: 10.1093/mmy/myu022. [DOI] [PubMed] [Google Scholar]
- Hubka V., Nováková A., Kolařík M. Revision of Aspergillus section Flavipedes: seven new species and proposal of section Jani sect. nov. Mycologia. 2015;107:169–208. doi: 10.3852/14-059. [DOI] [PubMed] [Google Scholar]
- Hubka V., Nováková A., Samson R.A. Aspergillus europaeus sp. nov., a widely distributed soil-borne species related to A. wentii (section Cremei) Plant Systematics and Evolution. 2016;302:641–650. [Google Scholar]
- Hubka V., Peterson S.W., Frisvad J.C. Aspergillus waksmanii sp. nov. and Aspergillus marvanovae sp. nov., two closely related species in section Fumigati. International Journal of Systematic and Evolutionary Microbiology. 2013;63:783–789. doi: 10.1099/ijs.0.047076-0. [DOI] [PubMed] [Google Scholar]
- Isshiki K., Asai Y., Tanaka S. Aurantiamide acetate, a selective cathepsin inhibitor, produced by Aspergillus penicilloides. Bioscience Biotechnology and Biochemistry. 2001;65:1195–1197. doi: 10.1271/bbb.65.1195. [DOI] [PubMed] [Google Scholar]
- Itabashi T., Matsuishi N., Hosoe T. Two new dioxopiperazine derivatives, arestrictins A and B, isolated from Aspergillus restrictus and Aspergillus penicilloides. Chemical & Pharmaceutical Bulletin. 2006;54:1639–1641. doi: 10.1248/cpb.54.1639. [DOI] [PubMed] [Google Scholar]
- Jones G. Algorithmic improvements to species delimitation and phylogeny estimation under the multispecies coalescent. Journal of Mathematical Biology. 2017;74:447–467. doi: 10.1007/s00285-016-1034-0. [DOI] [PubMed] [Google Scholar]
- Jong S., Dugan F., Edwards M. 19th edn. Catalogue; Rockville, Maryland, USA: 1996. ATCC filamentous fungi. [Google Scholar]
- Jurjević Ž., Kubátová A., Kolařík M. Taxonomy of Aspergillus section Petersonii sect. nov encompassing indoor and soil-borne species with predominant tropical distribution. Plant Systematics and Evolution. 2015;301:2441–2462. [Google Scholar]
- Kaarakainen P., Rintala H., Vepsalainen A. Microbial content of house dust samples determined with qPCR. Science of the Total Environment. 2009;407:4673–4680. doi: 10.1016/j.scitotenv.2009.04.046. [DOI] [PubMed] [Google Scholar]
- Kasuga H. Springer; Darmstadt, Germany: 2012. Indoor air quality. [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klich M.A. Centraalbureau voor schimmelcultures; Utrecht: 2002. Identification of common Aspergillus species. [Google Scholar]
- Kocsubé S., Perrone G., Magistà D. Aspergillus is monophyletic: evidence from multiple gene phylogenies and extrolites profiles. Studies in Mycology. 2016;85:199–213. doi: 10.1016/j.simyco.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornerup A., Wanscher J.H. Methuen; London: 1967. Methuen handbook of colour. [Google Scholar]
- Kozakiewicz Z. CAB International; Wallingford, UK: 1989. Aspergillus species on stored products. [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamy B., Davies J. Isolation and nucleotide-sequence of the Aspergillus restrictus gene coding for the ribonucleolytic toxin restrictocin and its expression in Aspergillus nidulans – the leader sequence protects producing strains from suicide. Nucleic Acids Research. 1991;19:1001–1006. doi: 10.1093/nar/19.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R., Calcott B., Ho S.Y. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- Leaché A.D., Fujita M.K. Bayesian species delimitation in West African forest geckos (Hemidactylus fasciatus) Proceedings of the Royal Society of London B: Biological Sciences. 2010;277:3071–3077. doi: 10.1098/rspb.2010.0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim G.S., Balke M., Meier R. Determining species boundaries in a world full of rarity: singletons, species delimitation methods. Systematic Biology. 2012;61:165–169. doi: 10.1093/sysbio/syr030. [DOI] [PubMed] [Google Scholar]
- Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Meklin T., Haugland R.A., Reponen T. Quantitative PCR analysis of house dust can reveal abnormal mold conditions. Journal of Environmental Monitoring. 2004;6:615–620. doi: 10.1039/b400250d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meklin T., Reponen T., McKinstry C. Comparison of mold concentrations quantified by MSQPCR in indoor and outdoor air sampled simultaneously. Science of the Total Environment. 2007;382:130–134. doi: 10.1016/j.scitotenv.2007.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheluz A., Manente S., Tigini V. The extreme environment of a library: xerophilic fungi inhabiting indoor niches. International Biodeterioration & Biodegradation. 2015;99:1–7. [Google Scholar]
- Micheluz A., Sulyok M., Manente S. Fungal secondary metabolite analysis applied to cultural heritage: the case of a contaminated library in Venice. World Mycotoxin Journal. 2016;9:397–407. [Google Scholar]
- Nazareth S., Gonsalves V. Aspergillus penicillioides-a true halophile existing in hypersaline and polyhaline econiches. Annals of Microbiology. 2014;64:397–402. [Google Scholar]
- Nguyen L.-T., Schmidt H.A., von Haeseler A. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K.F., Mansson M., Rank C. Dereplication of microbial natural products by LC-DAD-TOFMS. Journal of Natural Products. 2011;74:2338–2348. doi: 10.1021/np200254t. [DOI] [PubMed] [Google Scholar]
- Nováková A., Hubka V., Dudová Z. New species in Aspergillus section Fumigati from reclamation sites in Wyoming (USA) and revision of A. viridinutans complex. Fungal Diversity. 2014;64:253–274. [Google Scholar]
- O'Donnell K. Wallingford; England: 1993. Fusarium and its near relatives. The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. [Google Scholar]
- O'Donnell K., Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium ere nonorthologous. Molecular Phylogenetics and Evolution. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- O'Gorman C.M., Fuller H.T., Dyer P.S. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature. 2009;457:471–475. doi: 10.1038/nature07528. [DOI] [PubMed] [Google Scholar]
- Ohtsuki T. Studies on the glass mould. V. On two species of Aspergillus isolated from glass. The Botanical Magazine, Tokyo. 1962;75:436–442. [Google Scholar]
- Peterson S.W. Classification of Penicillium and Aspergillus: integration of modern taxonomic methods. Harwood Publishers; Reading, UK: 2000. Phylogenetic relationships in Aspergillus based upon rDNA sequence analysis; pp. 323–356. [Google Scholar]
- Peterson S.W. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia. 2008;100:205–226. doi: 10.3852/mycologia.100.2.205. [DOI] [PubMed] [Google Scholar]
- Peterson S.W., Jurjević Ž. Talaromyces columbinus sp. nov., and genealogical concordance analysis in Talaromyces clade 2a. PLoS One. 2013;8:11. doi: 10.1371/journal.pone.0078084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson S.W., Varga J., Frisvad J.C. Aspergillus in the genomic era. Wageningen Academic Pub; The Netherlands: 2008. Phylogeny and subgeneric taxonomy of Aspergillus; pp. 33–56. [Google Scholar]
- Pitt J.I. Academic Press Inc. Ltd.; London, UK: 1979. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. [Google Scholar]
- Pitt J.I., Hocking A.D. 3rd edn. Springer; London, UK: 2009. Fungi and food spoilage. [Google Scholar]
- Pitt J.I., Samson R.A. Taxonomy of Aspergillus section Restricta. In: Samson R.A., Pitt J.I., editors. Modern concepts in Penicillium and Aspergillus classification. Vol. 185. Springer; Boston, MA: 1990. pp. 249–257. (NATO ASI Series, Ser. A: Life Sciences). [Google Scholar]
- Pitt J.I., Samson R.A. Names in current use in the families Trichocomaceae, Cladoniaceae, Pinaceae and Lemnaceae. Koeltz Scientific Books; Koenigstein, Germany: 1993. Species names in current use in the Trichocomaceae (Fungi, Eurotiales) pp. 13–57. [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Puillandre N., Lambert A., Brouillet S. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Molecular Ecology. 2012;21:1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x. [DOI] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2015. R: A language and environment for statistical computing. [Google Scholar]
- Rambaut A., Suchard M., Xie D. 2014. Tracer v1.6.http://beast.bio.ed.ac.uk/Tracer Available online. [Google Scholar]
- Raper K.B., Fennell D.I. Williams & Wilkins; Baltimore, MD: 1965. The genus Aspergillus. [Google Scholar]
- Réblová M., Hubka V., Thureborn O. From the tunnels into the treetops: new lineages of black yeasts from biofilm in the Stockholm metro system and their relatives among ant-associated fungi in the Chaetothyriales. PLoS One. 2016;11:e0163396. doi: 10.1371/journal.pone.0163396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid N.M., Carstens B.C. Phylogenetic estimation error can decrease the accuracy of species delimitation: a Bayesian implementation of the general mixed Yule-coalescent model. BMC Evolutionary Biology. 2012;12:196. doi: 10.1186/1471-2148-12-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y., Kanazawa A., Araki A. Relationships between mite allergen levels, mold concentrations, and sick building syndrome symptoms in newly built dwellings in Japan. Indoor Air. 2011;21:253–263. doi: 10.1111/j.1600-0668.2010.00698.x. [DOI] [PubMed] [Google Scholar]
- Saito K. Untersuchungen über die atmosphärischen Pilzkeime. Journal of the College of Science, Imperial University of Tokyo. 1904;18:49–50. [Google Scholar]
- Samson R.A., Gams W. Advances in Penicillium and Aspergillus systematics. Springer; 1985. Typification of the species of Aspergillus and associated teleomorphs; pp. 31–54. [Google Scholar]
- Samson R.A., Houbraken J., Summerbell R.C. Microorganisms in home and indoor work environments: diversity, health impacts, investigation and control. CRC Press; Boca Raton, FL: 2002. Common and important species of fungi and actinomycetes in indoor environments; pp. 247–266. [Google Scholar]
- Samson R.A., Houbraken J., Thrane U. CBS-KNAW Fungal Biodiversity Centre; Utrecht, The Netherlands: 2010. Food and indoor fungi. [Google Scholar]
- Samson R.A., Lustgraaf B. Aspergillus penicilloides and Eurotium halophilicum in association with house-dust mites. Mycopathologia. 1978;64:13–16. doi: 10.1007/BF00443082. [DOI] [PubMed] [Google Scholar]
- Samson R.A., Varga J. What is a species in Aspergillus? Medical Mycology. 2009;47:13–20. doi: 10.1080/13693780802354011. [DOI] [PubMed] [Google Scholar]
- Samson R.A., Visagie C.M., Houbraken J. Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in Mycology. 2014;78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. The identification of fungi causing mildew in cotton goods: the genus Aspergillus-part II. Journal of the Textile Institute. 1931;22:110–116. [Google Scholar]
- Smith W.M., Fahle G., Nussenblatt R.B. A rare case of endogenous Aspergillus conicus endophthalmitis in an immunocompromised patient. Journal of Ophthalmic Inflammation and Infection. 2013;3:37. doi: 10.1186/1869-5760-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spegazzini C. Hongos de la cana de azucar. Revista de la Facultad de Agronomía y Veterinaria, Universidad Nacional de La Plata. 1896;2:227–258. [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Tang C.Q., Humphreys A.M., Fontaneto D. Effects of phylogenetic reconstruction method on the robustness of species delimitation using single-locus data. Methods in Ecology and Evolution. 2014;5:1086–1094. doi: 10.1111/2041-210X.12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.W., Jacobson D.J., Kroken S. Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- Terr A.I. Sick building syndrome: is mould the cause? Medical Mycology. 2009;47:217–222. doi: 10.1080/13693780802510216. [DOI] [PubMed] [Google Scholar]
- Thom C., Church M.B. Williams & Wilkins; Maryland, MD: 1926. The Aspergilli. [Google Scholar]
- Thom C., Raper K.B. Williams & Wilkins; Maryland, MD: 1945. A manual of the Aspergilli. [Google Scholar]
- Trifinopoulos J., Nguyen L.-T., von Haeseler A. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research. 2016;44:232–235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vansteelandt M., Kerzaon I., Blanchet E. Patulin and secondary metabolite production by marine-derived Penicillium strains. Fungal Biology. 2012;116:954–961. doi: 10.1016/j.funbio.2012.06.005. [DOI] [PubMed] [Google Scholar]
- Venables W.N., Ripley B.D. Springer; New York, USA: 2002. Modern applied statistics with S. [Google Scholar]
- Visagie C.M., Hirooka Y., Tanney J.B. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Studies in Mycology. 2014;78:63–139. doi: 10.1016/j.simyco.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visagie C.M., Yilmaz N., Renaud J.B. A survey of xerophilic Aspergillus from indoor environment, including descriptions of two new section Aspergillus species producing eurotium-like sexual states. MycoKeys. 2017;19:1–30. [Google Scholar]
- White T., Bruns T., Lee S. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Shinsky T.J., White T.J., editors. PCR protocols: a guide to methods and applications. Academic Press Inc.; New York, USA: 1990. pp. 315–322. [Google Scholar]
- Wickham H. Springer-Verlag; New York, USA: 2009. ggplot2: elegant graphics for data analysis. [Google Scholar]
- Yang R., Kenealy W.R. Regulation of restrictocin production in Aspergillus restrictus. Journal of General Microbiology. 1992;138:1421–1427. doi: 10.1099/00221287-138-7-1421. [DOI] [PubMed] [Google Scholar]
- Yang Z., Rannala B. Bayesian species delimitation using multilocus sequence data. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9264–9269. doi: 10.1073/pnas.0913022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kapli P., Pavlidis P. A general species delimitation method with applications to phylogenetic placements. Bioinformatics. 2013;29:2869–2876. doi: 10.1093/bioinformatics/btt499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang K., Sun M. Characterizations and analysis of the mold from animal specimens in Shenzhen Museum. Journal of Environmental Protection. 2016;7:1033–1040. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.