Abstract
High‐mobility‐group protein 2 (HMGB2) expression is up‐regulated in human liver cancer; however, little is known about its regulatory function. Here, we establish HMGB2 as a new modulator of the pluripotency of mouse embryonic stem cells. Similar to octamer‐binding transcription factor 4 (OCT4) and sex‐determining region Y‐box 2 (SOX2), HMGB2 protein is highly expressed in undifferentiated CGR8 cells, whereas it undergoes rapid decline during embryonic body formation. HMGB2 interacts with OCT4, increases protein expression of OCT4 and SOX2, and enhances their transcriptional activities. We also show that microRNA (miRNA)‐127 is a translational repressor of HMGB2 protein expression by targeting its 3′ untranslated region. We further elucidate a transcriptional mechanism controlling HMGB2 messenger RNA expression by the nuclear receptor small heterodimer partner (SHP) and transcription factor E2F1. Diminishing HMGB2 expression by ectopic expression of miR‐127 or SHP or treatment with the small molecule inhibitor inflachromene decreases OCT4 and SOX2 expression and facilitates CGR8 differentiation. In addition, HMGB2 is markedly induced in liver tumor initiating cells. Diminishing HMGB2 expression by short hairpin RNA for HMGB2 (shHMGB2), miR‐127, or SHP impairs spheroid formation. Importantly, HMGB2 expression is elevated in various human cancers. Conclusion: HMGB2 acts upstream of OCT4/SOX2 signaling to control embryonic stem cell pluripotency. Diminishing HMGB2 expression by miR‐127 or SHP may provide a potential means to decrease the pluripotency of tumor initiating cells. (Hepatology Communications 2017;1:816–830)
Abbreviations
- AP
alkaline phosphatase
- DDC
3,5‐diethoxycarbonyl‐1,4‐dihydrocollidine
- EB
embryonic body
- EID1
EP300 interacting inhibitor of differentiation 1
- ESC
embryonic stem cell
- FGF
fibroblast growth factor
- GFP
green fluorescent protein
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HMGB2
high‐mobility‐group protein 2
- ICM
inflachromene
- IP
immunoprecipitation
- LIF
leukemia inhibitory factor
- luc
luciferase
- miR, miRNA
micro RNA
- SHP
small heterodimer partner
- TIC
tumor initiating cell
- UTR
untranslated region
- WT
wild type
Introduction
High‐mobility‐group protein 2 (HMGB2) belongs to the HMG protein family, a group of nonhistone nuclear proteins that contain HMG‐box motifs. It has been proposed that HMGB proteins can act as architectural facilitators in the assembly of nucleoprotein complexes by bending DNA.1 HMGB2 interacts with several proteins, including casein kinase 1α2 and tumor suppressor p53.3 These interactions eventually lead to the recruitment of HMGB to specific sites of the genome where it locally modulates the association of transcription factors to their cognate DNA‐binding sites. HMGB2 expression is increased in hepatocellular carcinoma (HCC), which is associated with tumor aggressiveness and prognosis of HCC.4 Recent studies have shown that HMGB2 plays an important role in mesenchymal stem cell differentiation5 and erythroid differentiation.6 A small molecule inhibitor of HMGB, inflachromene (ICM), was identified to inhibit microglia‐mediated neuroinflammation by perturbing its posttranslational modification.7 Despite these studies, the regulatory mechanisms that control HMGB2 expression and function remain largely unexplored, particularly in the context of embryonic stem cells (ESCs).
The microRNAs (miRNA, miR) are small noncoding RNA transcripts and critical regulators of gene expression at the translational and transcriptional level.8 miR‐127 forms a cluster with miR‐433,9 and both miRNAs are commonly regulated by nuclear receptors.10, 11 miR‐127 was down‐regulated in liver cancer12 and breast cancer,13 implicating its role in carcinogenesis. miR‐127 promotes mesendoderm differentiation of mouse ESCs.14 However, additional target genes of miR‐127 remain to be identified to better understand its regulatory function in stem cells.
The small heterodimer partner (SHP, NROB2) is a unique orphan nuclear receptor lacking the DNA‐binding domain15 but exerts its transrepressive effect through physical interaction with its regulatory partners,16, 17 including liver receptor homolog 1,18 hepatocyte nuclear factor 4 alpha,15 estrogen‐related receptor gamma,11 and EP300 interacting inhibitor of differentiation 1 (EID1).19 Acting as a transcriptional repressor, SHP plays important roles in liver metabolic disease,20 cellular proliferation,21 liver carcinogenesis,22 and monocytic differentiation.23
ESCs can be derived from the inner cell mass of the preimplantation embryo and are characterized by their unlimited capacity for self‐renewal and pluripotency.24 The undifferentiated state of ESCs can be characterized by a high level of expression of alkaline phosphatase (AP). A group of transcription factors are essential for the establishment and maintenance of the pluripotent state.25 Among these, OCT4 is central to the machinery governing pluripotency, which is highly expressed in pluripotent embryonic cells as well as cells of the germline, and rapidly decreases on differentiation.26 In the pluripotent state, OCT4, protein kinase B (PKB/Akt), and HMGB2 participate in a regulatory feedback loop.27
In this study, we explored the relationship between HMGB2 and mouse germ layer differentiation. We found that HMGB2 was enriched in undifferentiated stem cells but significantly decreased during embryonic body (EB) formation. Furthermore, we identified HMGB2 as a novel target of miR‐127, which was up‐regulated on differentiation. In addition, we demonstrated that nuclear receptor SHP was a new modulator of HMGB2 by suppressing its promoter activity with EID1. Hence, miR‐127‐ and SHP‐mediated down‐regulation of HMGB2 at both posttranscriptional and transcriptional levels, respectively, provide a new mechanism in modulating stem cell pluripotency.
Materials and Methods
ANIMALS
C57BL/6 (wild‐type [WT]), Shp –/–, and E2f1 –/– mice have been described and were maintained on a pure C57BL/6 background.18, 19 Mice were fed a standard rodent chow diet (Harlan No. 2018) with free access to water and maintained on a 12‐hour light/dark cycle (light on 6 am to 6 pm) in a temperature‐controlled (23 °C) and pathogen‐free facility. The treatment of mice with 3,5‐diethoxycarbonyl‐1,4‐dihydrocollidine (DDC)‐supplemented diet has been described.19 In vivo experiments were performed on male mice at the age of 8 weeks unless stated otherwise (n = 5 mice/group). Protocols for animal use were approved by the Institutional Animal Care and Use Committee at the University of Connecticut. All methods were performed in accordance with the relevant guidelines and regulations.
HUMAN CANCER ANALYSIS
Human cancer data were extracted from the starBase Pan‐Cancer Analysis Platform (http://starbase.sysu.edu.cn/panCancer.php). The clinical and expression profiles of different cancer types were originally from the Cancer Genome Atlas data portal. Box plots of HMGB2 or miR‐127 expression (log2 read per million) were generated from RNA‐seq or miRNA‐seq data sets, respectively. The fold change, P value, and sample size were provided in each chart.
MAMMALIAN EXPRESSION PLASMIDS
Expression plasmids for epitope‐tagged E2F1, SHP, and EID1 have been described.15, 19 The human HMGB2 3′ untranslated region (UTR) containing the predicted miR‐127 binding site was cloned into pMIR‐Reporter (Invitrogen) using primers 5′‐TCAGACTAGTCAAGTGCAGCTCAATACT‐3′ and 5′‐ACCAAGCTTCACCTGAGGAACAATT TA‐3′. Mutations in pMIR‐Reporter‐HMGB2‐3′UTR were generated with a SiteMutant kit using primers 5′‐TGAATTCAAGTGCAGCTCAATACTA GCTAGTGTATAAAAA
CTGTACAGATTTTTGTATAG‐3′ and 5′‐CT ATACAAAAATCTGTACAGTTTTTATACACT AGCTA
GTATTGAGCTGCACTTGAATTCA‐3′. The plasmids were confirmed by sequencing. The expression plasmid pTarget‐miR‐127 and pTarget‐miR‐433 were generated as described.11 Plasmids pCAG‐Myc‐Oct3/4‐IP and pCAG‐HA‐Sox2‐IP were gifts from Dr. Shinya Yamanaka. pCDNA3.1 Flag mHMGB2 and reporter gene plasmid 6xO/S luciferase (luc) were gifts from Dr. Yasuhiko Kawakam and Dr. Lisa Dailey, respectively.
CELL CULTURE AND TRANSFECTION
HEK293T, HeLa, Hep3B, and MHCC97H cells were cultured and transfected using X‐tremeGene HP DNA transfection reagent (Roche) as described.28, 29 Mouse ESCs (CGR8; Sigma) were maintained on 0.1% gelatin‐coated tissue‐culture dishes in knockout Dulbecco's modified Eagle's medium containing 15% ESCs qualified fetal bovine serum, 2 mM L‐glutamin, 0.1 mM nonessential amino acids, 0.1 mM β‐mercaptoethanol, 50 μg/mL penicillin–streptomycin, and 10 ng/mL leukemia inhibitory factor (LIF). Primary mouse embryonic fibroblasts were isolated and cultured as described.21 Mouse liver tumor initiating cells (TICs) were originally isolated by fluorescence‐activated cell sorting based on surface expression of CD133 and CD49 from liver tumors developed in hepatitis C virus (HCV) nonstructural protein 5A transgenic mice fed alcohol according to the described protocol.30 They were cultured and maintained as a cell line in Dulbecco's modified Eagle's medium (F‐12 media with 10% fetal bovine serum, 20 ng/mL murine epidermal growth factor (EGF), EmbryoMax Nucleosides, 100 nM dexamethasone, and an antibiotic–antimycotic agent). TICs are able to form tumors in immunodeficient mice after subcutaneous transplantation and immunocompetent mice fed with an alcohol diet after orthotopic transplantation.
EMBRYONIC BODY DIFFERENTIATION
CGR8 cells were trypsinized into single cells and cultured by seeding 900 cells in 30 μL ESCs hanging droplets of ESCs medium devoid of LIF. Two days later, embryonic bodies (EBs) were transferred to an ultralow attachment plate for another 2‐day suspension culture. EBs were then plated onto gelatin‐coated tissue‐culture plates for a further 6‐day spontaneous differentiation.
TICs SPHEROID ASSAY
We suspended 1,000 cells in serum‐free TIC medium with 1% methylcellulose supplemented with B27, 20 ng/mL EGF, and 20 ng/mL fibroblast growth factor (FGF) in each well of a 96‐well ultralow attachment plate. The cells were transduced with the indicated viruses overnight, followed by the spheroid assay for 7 days.
ADENOVIRAL AND LENTIVIRAL TRANSDUCTION
Recombinant adenovirus vectors for expression of green fluorescent protein (GFP)‐mouse SHP (mSHP) and GFP control were prepared as described.31, 32 Adenoviral supernatants were generated by standard methods at the University of Pennsylvania Penn Vector Core facility. For lentivirus production of pSico‐GFP‐miR‐127, pSico‐GFP, and Cre‐IRES‐PuroR, HEK293T cells were transfected with pSico‐GFP‐miR‐127, pSico‐GFP, and Cre together with lentiviral packaging plasmids pRSV‐REV, pMDLG/PRRE, and pVSVG (Addgene).28 After 48 hours of culture, lentiviruses were collected from the medium, purified by centrifugation, and then used to infect host cells. CGR8 cells were cultured on a gelatin‐coated dish and infected with viral supernatant at different multiplicities of infection for 6 hours. Virus‐containing media were removed, and cells were continuously cultured.
IMMUNOPRECIPITATION AND WESTERN BLOTTING
Immunoprecipitation was carried out as described.33 CGR8 cell lysates were harvested by Myc lysis buffer (138 mM NaCl, 20 mM Tris‐HCl (pH 8.0), 1% Nonidet P‐40), and anti‐OCT4 antibody or rabbit immunoglobulin G were used to immunoprecipitate OCT4 by incubating with protein A/G‐Magnetic Beads (Thermo Scientific) at 4 °C for 4 hours. After extensive washes, immunoprecipitated proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred onto polyvinylidene fluoride membrane, immunostained with anti‐HMGB2 antibody, and finally detected by horseradish peroxidase‐conjugated secondary antibodies and visualized by chemiluminescence (Pierce). For whole‐cell lysate analysis, cells were resuspended in radio immunoprecipitation assay lysis buffer and total protein was quantified by the BCA protein assay kit (Pierce). Western blotting analysis was conducted with antibodies as follows: rabbit anti‐OCT4 (Abcam); mouse anti‐SOX2 (Abcam); rabbit anti‐HMGB2 (Cell Signaling Technology); rabbit anti‐HMGB1 (Cell Signaling Technology); mouse anti‐β‐ACTIN (Sigma); rabbit anti‐cyclic adenosine monophosphate response element‐binding protein (Cell Signaling Technology); mouse anti‐LamA/C (Santa Cruz); and mouse anti‐α‐Tubulin (Santa Cruz).
ALKALINE PHOSPHATASE STAINING ASSAY
CGR8 cells treated with compounds, adenovirus, or lentivirus for 48 hours were cultured in medium containing LIF for another 5 days. The medium was changed every other day. AP activity was measured with the Alkaline Phosphatase Detection Kit (EMD Millipore).
RNA ISOLATION, REVERSE TRANSCRIPTION, AND QUANTITATIVE REAL‐TIME REVERSE‐TRANSCRIPTION POLYMERASE CHAIN REACTION
Total RNAs were extracted using TRIzol reagent (Invitrogen) as described.34 One microgram of total RNA was reverse transcribed to complementary DNA using High‐Capacity cDNA Reverse Transcription kits (Applied Biosystems). Quantitative real‐time reverse‐transcription polymerase chain reaction was performed using SYBR Green (Applied Biosystems) on a CFX384 Real‐Time system (Bio‐Rad). Hprt and U6 were used as reference genes for normalization of messenger RNA (mRNA) and miRNA levels. Samples were done in triplicate, and data were analyzed using the 2‐ΔΔCT method. The primers used for specific mouse genes are provided in http://onlinelibrary.wiley.com/doi/10.1002/hep4.1086/suppinfo.
LUCIFERASE REPORTER ASSAY
Forty‐eight hours after transfection with luciferase reporter plasmid and Renilla plasmid, cells were harvested and analyzed with the Dual‐Luciferase reporter assay system (Promega) as described.35 All assays were done in triplicate, and all values were normalized for transfection efficiency against Renilla luciferase activities.
STATISTICAL ANALYSIS
Data are shown as the mean ± SEM and are representative of at least three independent experiments from triplicate assays. Statistical analysis was carried out using the Student t test between two groups and one way analysis of variance among multiple groups. P < 0.05 was considered statistically significant.
Results
EXPRESSION OF OCT4, SOX2, AND NANOG WERE DECREASED DURING EB FORMATION
To investigate the functional role of HMGB2 in ESCs and germ layer differentiation, we used CGR8 cells, a feeder‐free ESC line and a well‐established model of ESC differentiation into germ layer lineages during EB formation. EB mimics normal mouse embryonic development and differentiation over the course of a few days into both embryonic and extraembryonic cell types. Undifferentiated CGR8 cells (Fig. 1A) were tightly packed on the surface of a gelatinized dish in the presence of LIF, and these disappeared on LIF withdrawal for 2 days. Balloon‐like cysts were observed on day 2 (2D) of the EB formation, indicating ESCs in these three‐dimensional cell aggregates underwent spontaneous differentiation and cell specification along the three germ lineages. Transferring D4 EBs to a gelatinized dish allowed them to complete their further development. To validate the pluripotency of CGR8 cells, we measured the expression of several pluripotent genes and a number of germ layer‐specific genes. Pluripotency genes (Oct4 and Nanog) were highly expressed in CGR8 cells, while lineage markers (Cxcl12, Brachyury/T, BMP5, Eomes, Gata4, Cdx2, Foxa2) were not expressed (Fig. 1B), suggesting that CGR8 cells were well maintained in the undifferentiated state. As expected, during EB differentiation from 0 to 10 days, the expression of Oct4, Sox2, and Nanog were significantly down‐regulated whereas germ layer‐specific genes representing ectoderm (Cxcl12), mesoderm (Brachyury/T, BMP5), endoderm (Foxa2, Gata4), and trophectoderm (Cdx2, Eomes) were markedly elevated to various extents (Fig. 1C).
Figure 1.
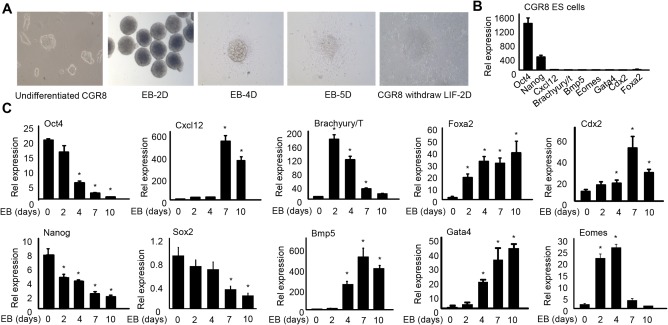
Establishing mouse ES cell line CGR8 cultures and embryonic body differentiation. (A) Microscopic images of mouse ES cell line CGR8 and EB outgrowths. Undifferentiated CGR8 cells were maintained in ES cell medium containing LIF. EB‐2D shows EBs transferred to an ultralow attachment plate by the hanging drop method on EB formation protocol day 2. Aggregates were round and had a well‐defined border. EB‐4D and 5D show cellular outgrowth and migration on day 4 and day 5 from attached EBs after being transferred to a gelatin‐coated plate on day 4. CGR8 withdraw LIF‐2D shows CGR8 cells cultured in ES cell medium devoid of LIF for 2 days. (B) qPCR of mRNA expression of pluripotency and three germ layer markers in undifferentiated CGR8 cells. (C) qPCR of mRNA expression of pluripotency and three‐germ layer markers during EB formation on day 0 (ESCs), 2, 4, 7, and 10 (EB2D, 4D, 7D, 10D). Data are shown as mean ± SEM (triplicate assays). *P < 0.05 versus EB day 0. Abbreviation: qPCR, quantitative polymerase chain reaction.
HMGB2 EXPRESSION WAS DECREASED CONCOMITANTLY WITH OCT4, SOX2, AND NANOG AND ENHANCED THEIR TRANSCRIPTIONAL ACTIVITY
As expected, OCT4, SOX2, and NANOG proteins were highly expressed in undifferentiated CGR8 cells, but their levels underwent a rapid decline during EB formation (Fig. 2A). In contrast, none of the three proteins could be detected in completely differentiated primary mouse embryonic fibroblasts. When we examined the HMGB2 protein, it exhibited a similar expression pattern to that of OCT4, SOX2, and NANOG. HMGB1 is a close relative of HMGB2 and was detected in undifferentiated CGR8 cells, but its expression remained little changed during the first 7 days of differentiation, which was distinct from the HMGB2 protein.
Figure 2.
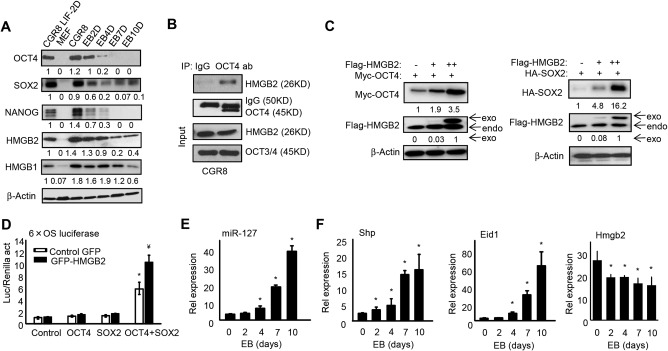
HMGB2 is highly expressed in ESCs to enhance OCT4 and SOX2 expression and transcriptional activity. (A) Western blot to determine protein expression in CGR8 cells devoid of LIF (CGR8LIF‐2D), MEF cells, and EB formation on day 0, 2, 4, 7, and 10 from undifferentiated CGR8 cells. (B) Co‐IP followed by western blot. CGR8 cells were harvested and cell lysates were immunoprecipitated with anti‐OCT4 antibody or control IgG. The co‐IP complexes and the inputs were analyzed by western blot with the indicated antibodies. (C) Left, western blot to determine HMGB2 and OCT4 proteins in HEK293T cells that were transfected with 0.5 μg Myc‐OCT4 expression plasmid (+) and 0 (–), 1 (+), or 2 (++) μg Flag‐HMGB2 expression plasmid. Right, western blot to determine HMGB2 and SOX2 proteins in HEK293T cells transfected with 0.5 μg HA‐SOX2 expression plasmid and 0, 1, or 2 μg Flag‐HMGB2 expression plasmid. (D) Luciferase reporter assays. HepG2 cells were transduced with AAV8‐GFP or AAV8‐GFP‐HMGB2 for 24 hours before being transfected with the 6xO/S luc reporter together with OCT4 and SOX2 expression plasmids alone or in combination. Data are shown as mean ± SEM (triplicate assays). *P < 0.05 OCT4+SOX2 versus OCT4 or SOX2; ¥ P < 0.05 HMGB2 versus GFP. (E) qPCR of miR‐127 expression and (F) qPCR of mRNA expression during EB formation. Data are shown as mean ± SEM (triplicate assays). *P < 0.05 versus EB day 0. Abbreviations: IgG, immunoglobulin G; MEF, mouse embryonic fibroblasts; qPCR, quantitative polymerase chain reaction.
To explore the mechanistic action of HMGB2 during ESCs differentiation, we tested the interaction between endogenous HMGB2 and OCT4 or HMGB2 and SOX2 in CGR8 cells by performing an endogenous immunoprecipitation (IP) experiment. HMGB2 was retrieved by anti‐OCT4 IP but not by control immunoglobulin G (Fig. 2B). However, we failed to detect the HMGB2 and SOX2 protein association (not shown). We next determined the effect of HMGB2 on static levels of OCT4 or SOX2 proteins. Equal amounts of Oct4 or Sox2 plasmids were cotransfected with increased doses of HMGB2 in HEK293T cells. The levels of OCT4 (Fig. 2C, left) and SOX2 proteins (Fig. 2C, right) were increased by HMGB2 in a dosage‐dependent manner.
Because the heterodimer interaction of OCT4 and SOX2 induces gene transcription by binding to promoters of target genes, such as Nanog and FGF4, we examined the effect of HMGB2 on Oct4/Sox2 transactivation. The promoter reporter 6xO/S luc contained six tandem copies of the composite Oct4/Sox2 binding site of the FGF4 enhancer, which was used for determining the transcriptional activity of Oct4/Sox2 complexes in HepG2 cells. Co‐expression of OCT4 and SOX2 markedly increased 6xO/S luc reporter activity compared with OCT4 or SOX2 alone (Fig. 2D). Notably, transient expression of HMGB2 enhanced the transcriptional activity of Oct4/Sox2 to a significantly higher level. Overexpression of HMGB2 alone did not increase the 6xO/S luc reporter activity, suggesting its effect is through regulating OCT4/SOX2 expression. The results suggest that HMGB2 overexpression leads to enhanced transcriptional activity of the OCT4/SOX2 heterodimer.
Because miRNA‐127 was reported to promote ESC differentiation,14 we examined its expression and observed a progressive induction during EB formation (Fig. 2E). As our laboratory has been working on the SHP function over the past decade, we also examined
SHP and its co‐repressor EID1 and observed a similar induction during ESC differentiation (Fig. 2F). This finding is interesting as SHP has not been reported to be involved in ESC function. In contrast, Hmgb2 mRNA exhibited a negative expression correlation with SHP. Taken together, the results demonstrated that the expression of HMGB2 was inversely correlated with the differentiation status of ESCs as well as with miR‐127 and Shp levels.
miR‐127 INHIBITED HMGB2 PROTEIN EXPRESSION TO MODULATE STEM CELL PLURIPOTENCY
As we were interested in the relationship between miR‐127 and HMGB2 based on the above results, we used TargetScan and miRanda, which predicted a conserved seed region for miR‐127 in the HMBG2 3′UTR of human, mouse, and rat (Fig. 3A). To verify whether HMGB2 is targeted by miR‐127, human HMBG2 3′UTR was cloned into pMIR‐reporter. Luciferase reporter assay showed that the HMGB2 3′UTR reporter activity was decreased in a dose‐dependent fashion by ectopic expression of miR‐127 in HeLa cells (Fig. 3B, left); this was relieved when the miR‐127 seed region was mutated (Fig. 3B, middle). In addition, a specific miR‐127 antagomir, anti‐miR‐127, led to an increase in HMGB2 3′UTR activity (Fig. 3B, right). Ectopic expression of miR‐127 reduced HMGB2 protein in a dose‐dependent fashion (Fig. 3C). In contrast, the expression of HMGB2 was not altered by miR‐433, an miRNA that was expressed in the same cluster as miR‐127,9 confirming the specific inhibition by miR‐127. Therefore, although the transcription of miR‐127 and miR‐433 genes shares common genomic regions and regulation by nuclear receptors,10 both miRNAs likely exert a differential function through targeting distinct genes.
Figure 3.
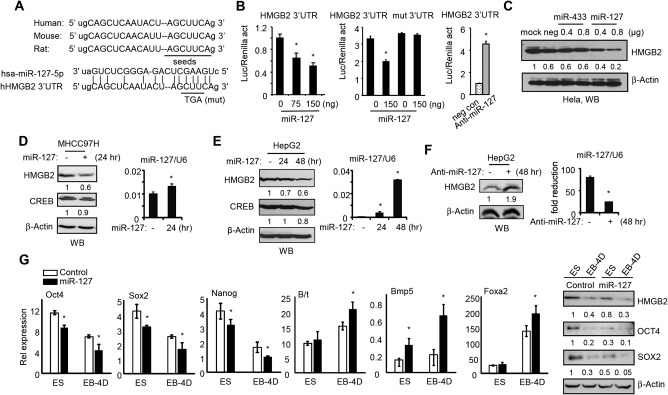
miR‐127 represses HMGB2 protein expression to facilitate EB body formation. (A) Sequence alignment of miR‐127 and the 3'UTR of HMGB2 in human, mouse, and rat. The seed‐match region is underlined. (B) Transient transfection assays. Left, HeLa cells were transfected with HMGB2 3'UTR luciferase reporter along with miR‐127 expression plasmid. Middle, HeLa cells were transfected with WT or mutant HMGB2 3'UTR with or without miR‐127. Right, HeLa cells were transfected with HMGB2 3'UTR with or without miR‐127 antagomir. Luciferase reporter activity was normalized to Renilla activity. (C) Western blot to determine endogenous HMGB2 protein in HeLa cells that were transfected with pTarget, pTarget‐miR‐433, or pTarget‐miR‐127. (D) Western blot to determine HMGB2 and CREB proteins (left) and qPCR to determine miR‐127 expression (right) in MHCC97H cells transfected with (+) or without (–) miR‐127. (E) Western blot to determine HMGB2 and CREB proteins (left) and qPCR to determine miR‐127 expression (right) in HepG2 cells transfected with or without miR‐127. (F) Western blot to determine HMGB2 protein (left) and qPCR to determine miR‐127 expression (right) in HepG2 cells transfected with or without miR‐127 antagomir (anti‐miR‐127). (G) Left, qPCR of mRNA expression of pluripotency markers and mesoderm markers in undifferentiated CGR8 cells and EB‐4D after transduction with GFP control or GFP‐miR‐127 lentivirus. Data are shown as mean ± SEM (triplicate assays). *P < 0.05 versus corresponding control. Right, western blot of protein expression. Abbreviations: CREB, cyclic adenosine monophosphate response element‐binding protein; mut, mutant; hr, hours; qPCR, quantitative polymerase chain reaction.
We further determined miR‐127 inhibition of HMGB2 in hepatic cells, including MHCC97H and HepG2 cells. Similar to HeLa cells, miR‐127 decreased HMGB2 protein levels in both cell types (Fig. 3D,E). The cyclic adenosine monophosphate response element‐binding protein, a known miR‐433 target,29 showed minimal changes. In addition, transient expression of anti‐miR‐127 increased HMGB2 protein in HepG2 cells (Fig. 3F). Taken together, the results suggest that HMGB2 is a novel target of miR‐127, as demonstrated by using multiple cell types.
To investigate whether miR‐127‐mediated down‐regulation of HMGB2 affected stem cell pluripotency, we generated a lentivirus containing scramble control or miR‐127 (http://onlinelibrary.wiley.com/doi/10.1002/hep4.1086/suppinfo) to obtain efficient expression in CGR8 cells. CGR8 cells were transduced with lenti‐control or lenti‐miR‐127 for 48 hours, followed by inducing EB differentiation. miR‐127 decreased the expression of Oct4 and Sox2 on day 0 (ESCs) and day 4 EB (EB‐4D), whereas mesodermal markers B/t, BMP5, and Foxa2 were significantly increased by miR‐127 (Fig. 3G). Protein levels of HMGB2, OCT4, and SOX2 were reduced by miR‐127 as well. These results were consistent with the low expression of miR‐127 in undifferentiated ESCs but with dramatically increased levels during EB formation (Fig. 2E).
HMGB2 GENE EXPRESSION WAS ACTIVATED BY E2F1 AND REPRESSED BY SHP AND EID1
Our previous studies demonstrated that SHP functioned as a transcriptional repressor.36 Because Shp is a circadian clock‐controlled gene37 and is also part of the liver clock machinery,38 we analyzed HMGB2 protein expression in WT and Shp –/– mouse livers that were collected over a 24‐hour light/dark cycle. HMGB2 proteins were highly elevated in Shp –/– versus WT livers and also exhibited a cyclic expression (Fig. 4A, left). This expression pattern was in contrast to that of Shp expression, as reported in our previous studies.39 Hmgb2 mRNA was also induced in Shp –/– livers (Fig. 4A, right). The results suggested SHP as a potential inhibitor of HMGB2 gene transcription.
Figure 4.
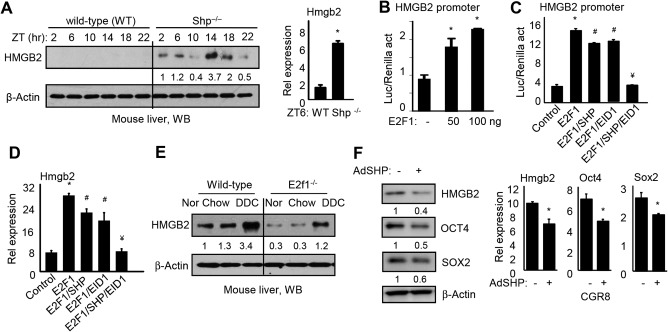
HMGB2 is transcriptionally regulated by the E2F1/SHP/EID1 network. (A) Left, western blot to determine HMGB2 protein in WT and Shp –/– mouse livers collected at ZT 2, 6, 10, 14, 18, and 22. Right, qPCR of HMGB2 mRNA in WT and Shp –/– mouse livers at ZT6. *P < 0.05 Shp –/– versus WT. (B) Transient transfection assay. HEK293T cells were transfected with HMGB2 promoter reporter with increasing doses of E2F1 expression plasmid. Luciferase reporter activity was normalized to Renilla activity. *P < 0.05 E2F1 versus empty vector (–). (C) HEK293T cells were transfected with HMGB2 promoter reporter together with E2F1, SHP, and EID expression plasmids alone or in combination. Luciferase reporter activity was normalized to Renilla activity. *P < 0.05 E2F1 versus control; # P < 0.05 E2F1/SHP or E2F1/EID1 versus E2F1 alone; ¥ P < 0.05 E2F1/SHP/EID1 versus E2F1/SHP or E2F1/EID1. (D) qPCR of HMGB2 mRNA in HepG2 cells transfected with E2F1, SHP, and EID1 expression plasmids alone or in combination. *P < 0.05 E2F1 versus control; # P < 0.05 E2F1/SHP or E2F1/EID1 versus E2F1 alone; ¥ P < 0.05 E2F1/SHP/EID1 versus E2F1/SHP or E2F1/EID1. (E) Western blot to determine HMGB2 protein in WT or E2f1 –/– mouse livers fed with normal chow, chow (DDC control), or DDC diet. (F) Western blot of HMGB2, OCT4, and SOX2 proteins (left) and qPCR of Hmgb2, Oct4, and Sox2 mRNAs in CGR8 cells treated with AdGFP or AdSHP for 48 hours. *P < 0.05 AdSHP (+) versus AdGFP (–). Data in all bar graphs are shown as mean ± SEM (triplicate assays). Abbreviations: ad, ; Nor, normal; qPCR, quantitative polymerase chain reaction; Ad, adenovirus; ZT, Zeitgeber time.
SHP lacks a DNA binding domain and is known to repress its target gene through interaction with other transcription factors.40 Promoter analysis identified a putative E2F1 binding site within the Hmgb2 gene 5' flanking region (http://onlinelibrary.wiley.com/doi/10.1002/hep4.1086/suppinfo). Our previous study showed that SHP interacted with E2F1 to repress E2F1 activity through the co‐repressor EID1.19 Thus, we hypothesized that SHP may repress Hmgb2 through E2F1. As expected, ectopic expression of E2F1 activated the Hmgb2 promoter in a dose‐dependent manner (Fig. 4B). Forced expression of SHP or its co‐repressor EID1 alone showed inhibition of E2F1 activity, whereas co‐expression of both genes completely diminished E2F1 activity (Fig. 4C). Consistently, Hmgb2 mRNA was induced by E2F1 but inhibited by SHP and EID1 in HepG2 cells (Fig. 4D). We next determined HMGB2 proteins in WT and E2f1 –/– mouse livers under normal (regulator chow), chow (DDC control), or DDC‐supplemented diet; the latter was known to activate E2F1.19 HMGB2 proteins were significantly down‐regulated in E2f1 –/– livers (Fig. 4E).
We next determined whether SHP affected stem cell pluripotency by overexpressing it in CGR8 cells using adenoviral transduction.32 The protein and mRNA of Hmgb2 along with Oct4 and Sox2 were down‐regulated by SHP compared to the GFP control (–) (Fig. 4F). Thus, SHP suppressed HMGB2 expression, which in turn facilitated EB differentiation.
INHIBITION OF HMGB2 EXPRESSION BY A SMALL MOLECULE INHIBITOR, miR‐127, OR SHP ATTENUATED ESC PLURIPOTENCY
A recent study identified a small molecule inhibitor of HMGB2, namely ICM.7 To investigate the pluripotent state of stem cells affected by ICM, we determined EB formation after treatment of CGR8 cells with ICM for 48 hours. The total HMGB2 protein (Fig. 5A, left) was decreased by ICM in CGR8 cells, which was in agreement with the report that ICM modified HMGB2 protein stability but not mRNA expression.7 Interestingly, the nuclear HMGB2 protein was most significantly degraded by ICM (Fig. 5A, right). OCT4 protein was only detected in the nucleus, and its level was markedly decreased by ICM. In addition, both cytosolic and nuclear levels of SOX2 proteins were reduced by ICM. As expected, Hmgb2 mRNA remained unaltered by ICM (Fig. 5B). Interestingly, the mRNA expression of Oct4, Sox2, and Nanog were diminished by ICM treatment (Fig. 5B), suggesting that the diminished HMGB2 protein function attenuated pluripotency. Morphologically, ICM‐treated CGR8 exhibited scattered clones, and the EB lost a round and well‐defined border (Fig. 5C). AP activity has been shown to be up‐regulated in pluripotent stem cells, including undifferentiated ESCs. The AP staining assay showed that ICM, miR‐127, and SHP decreased AP expression in CGR8 cells, indicating an induction of differentiation (http://onlinelibrary.wiley.com/doi/10.1002/hep4.1086/suppinfo). Taken together, inhibition of HMGB2 function by ICM, miR‐127, or SHP promoted mouse stem cell differentiation.
Figure 5.
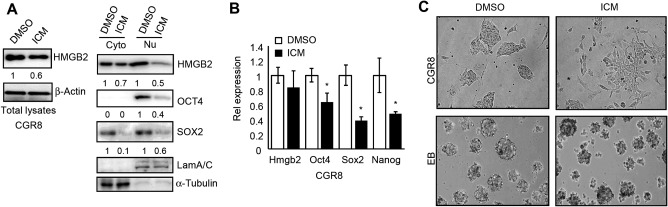
A small molecule inhibitor of HMGB2 disrupts stem cell differentiation. (A) Western blot to determine HMGB2, OCT4, and SOX2 proteins in CGR8 cells treated with 5 μM ICM or DMSO for 48 hours. Proteins were detected in total cell lysates, cytoplasmic, or nuclear fractions; Lamin A/C, nuclear fraction; α‐Tubulin, cytoplasmic fraction. (B) qPCR of Hmgb2, Oct4, Sox2, and Nanog mRNAs in CGR8 cells treated with 5 μM ICM or DMSO for 48 hours. Data are shown as mean ± SEM (triplicate assays). *P < 0.05 ICM versus DMSO. (C) Microscopic images of the CGR8 cells and EB formation. CGR8 cells were treated with 5 μM ICM or DMSO for 48 hours (upper). EB formation from CGR8 cells pretreated with ICM or DMSO for 48 hours was induced by the hanging drop method in ESCs medium removal of LIF but containing 5 μM ICM or DMSO for 4 days. Abbreviations: Cyto, cytoplasmic; DMSO, dimethyl sulfoxide; Nu, nuclear; qPCR, quantitative polymerase chain reaction.
HMGB2 WAS MARKEDLY ELEVATED IN LIVER TICs TO FACILITATE SPHEROID FORMATION
The important function of HMGB2 in ESC differentiation as elucidated above suggested that it may play a potential role in cancer stem cells. TICs are malignant cells that share important similarities with undifferentiated ESCs.41 Like ESCs, TICs have the ability to unlimited self‐renewal and to create differentiated progenies. Thus, we extended our study on TICs isolated from liver tumors developed in alcohol‐fed HCV Ns5a transgenic mice.30 We observed a striking induction of HMGB2 protein in TICs compared to the control cells (Fig. 6A). In addition, ectopic expression of miR‐127 or SHP decreased HMGB2 protein in TICs (Fig. 6B). Diminishing HMGB2 expression using short hairpin RNA for HMGB2 (shHMGB2) (http://onlinelibrary.wiley.com/doi/10.1002/hep4.1086/suppinfo) repressed TIC spheroid formation (Fig. 6C). Similarly, overexpression of miR‐127 or SHP reduced spheroid formation, and the inhibitory effects of miR‐127 and SHP were rescued by overexpression of HMGB2 (Fig. 6D).
Figure 6.
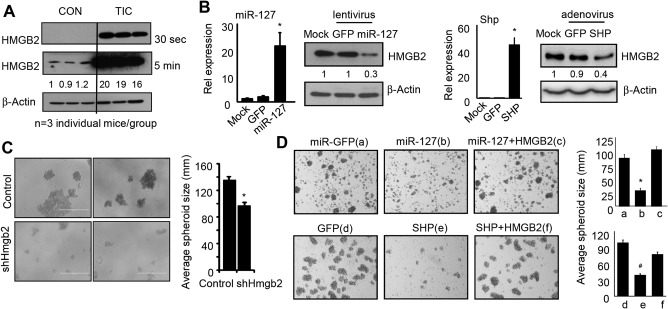
HMGB2 protein is markedly induced in liver TICs, and inhibition of HMGB2 expression reduces spheroid formation. (A) Western blot of HMGB2 protein in control and TICs, which were isolated from three individual HCV Ns5a Tg mice per group. (B) Western blot to determine HMGB2 proteins and qPCR to determine miR‐127 and Shp mRNAs in TICs. Data are shown as mean ± SEM (triplicate assays). *P < 0.05 versus GFP control. (C) Left, microscopic images of spheroid formation from TICs transduced with EV or shHmgb2 lentivirus. Right, analysis of average size of spheroids. *P < 0.01 shHmgb2 versus EV. (D) Left, microscopic images of spheroid formation from TICs transduced with GFP, miR‐127, or miR‐127 plus HMGB2 (top) or GFP, SHP, SHP plus HMGB2 (bottom) for 7 days. Right, analysis of average size of spheroids. *P < 0.01 miR‐127 versus miR‐GFP. # P < 0.01 SHP versus GFP. Abbreviations: CON, control; EV, empty vector; qPCR, quantitative polymerase chain reaction.
We also treated TICs with ICM or two other compounds, namely ICM‐Linker and ICM‐H (http://onlinelibrary.wiley.com/doi/10.1002/hep4.1086/suppinfo). ICM‐Linker decreased cell proliferation and HMGB2 expression, which served as a positive control. ICM‐H showed no effect on HMGB2 protein expression, which served as a negative control. The present ICM concentration, although relatively high, showed no cytotoxicity. Although future studies are needed to further understand the function of HMGB2 in cancer stem cells, the results implied an important oncogenic potential of HMGB2 in tumor formation.
HMGB2 WAS UP‐REGULATED IN HUMAN CANCERS AND WAS ASSOCIATED WITH PATIENT SURVIVAL
To establish the potential association of HMGB2 with human cancers, we analyzed an existing database (http://starbase.sysu.edu.cn/) to retrieve Hmgb2 expression profiles from multiple types of human cancer (http://onlinelibrary.wiley.com/doi/10.1002/hep4.1086/suppinfo). The analysis revealed a significant induction of Hmgb2 expression in several human cancers that negatively correlated with miR‐127 down‐regulation, including colon adenocarcinoma (Fig. 7A), head and neck squamous cell carcinoma (Fig. 7B), and uterine corpus endometrial carcinoma (Fig. 7C). Shp expression profiles were not available from the same data sets, thus we could not conduct a comparable analysis. Nonetheless, Shp expression was markedly down‐regulated in metastatic HCC and gastric cancers using data derived from ONCOMINE.42 Importantly, a higher expression of Hmgb2 was also found in breast cancers and was associated with lower patient survival (http://kmplot.com/analysis/index.php?p=service-start=1) (http://onlinelibrary.wiley.com/doi/10.1002/hep4.1086/suppinfo). Taken together, the results suggested that HMGB2 activation was positively associated with human cancer development.
Figure 7.
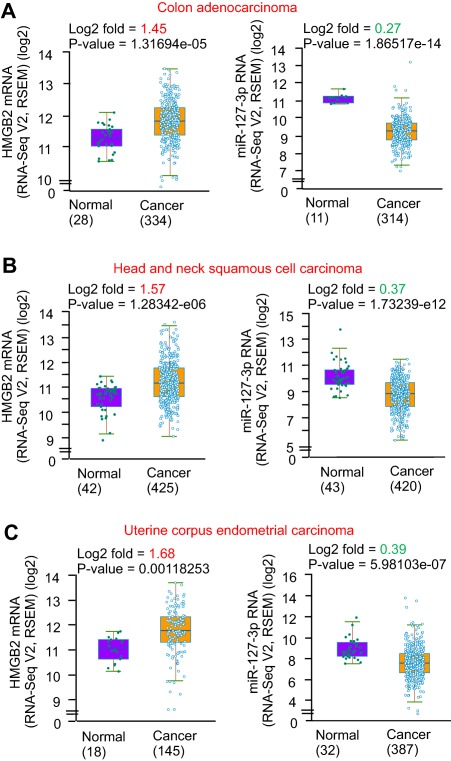
HMGB2 is markedly up‐regulated in human cancers, which correlates with miR‐127 down‐regulation. The expression of HMGB2 (left) and miR‐127 (right) in (A) human cancers, including colon adenocarcinoma, (B) head and neck squamous cell carcinoma, and (C) uterine corpus endometrial carcinoma. The data were derived from http://starbase.sysu.edu.cn/. The log2 of read per million was calculated from RNA‐seq or miRNA‐seq data. Statistical analysis was carried out using the Student t test. Abbreviation: RSEM, RNA‐Seq by Expectation Maximization.
Discussion
HMGB2 has been implicated in multiple cellular and physiologic processes, including gene transcription and DNA repair. However, limited information is available regarding the molecular mechanisms and pathways that control its expression and how changes in HMGB2 expression affect its regulatory function. In this study, we demonstrated that HMGB2 is highly expressed in undifferentiated mouse ESCs, and its expression is down‐regulated during germ layer differentiation. HMGB2 increases the transcriptional activity of the OCT4/SOX2 complex, and diminishing HMGB2 expression destabilizes OCT4 and SOX2 proteins and their associated ESC pluripotency. Thus, we identified HMGB2 as a critical upstream regulator of OCT4 and SOX2 and a gatekeeper of ESC pluripotency (Fig. 8).
Figure 8.
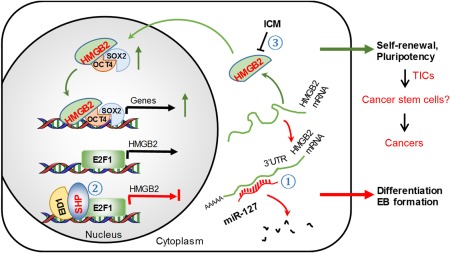
Schematics summarizing major findings in the present study. HMGB2 is critical to maintain the pluripotent state of mouse ESCs. HMGB2 enhances OCT4/SOX2 transcriptional activity by binding to OCT4 and increasing OCT4 and SOX2 protein expression. Down‐regulation of HMGB2 protein by ICM disrupts stem cell polarity and impacts normal EB formation. miR‐127 functions as a new repressor of HMGB2 protein expression by inhibiting HMGB2 3'UTR activity and HMGB2 mRNA translation. SHP serves as a new transcriptional repressor of HMGB2 mRNA expression by inhibiting the promotor activity of HMGB2 through E2F1. Overexpression of miR‐127 or SHP facilitates germ layer differentiation by diminishing HMGB2 expression. The induction of HMGB2 in TICs as well as in various human cancers suggests its potential oncogenic function. This effect could be mediated through regulating cancer stem cells, which will be elucidated in future studies (question mark). Overall, our study unravels a crosstalk between HMGB2 and miR‐127/SHP to control mouse ESC differentiation.
We are promoted to determine the mechanisms that cause HMGB2 down‐regulation during ESC differentiation. Synthesis and degradation are two important aspects that determine the quantity of a protein in cells. Because miRNAs are important posttranscriptional regulators,28 we are interested in identifying miRNA‐mediated control of HMGB2 protein expression. Multiple miRNAs were predicted to bind HMGB2 3′UTR, and miR‐127 was validated to inhibit HMGB2 protein expression by targeting HMGB2 3′UTR. The result is further supported by the inverse expression correlation between miR‐127 and HMGB2 during early embryonic development. To the best of our knowledge, no other miRNAs have been reported to repress HMGB2 protein expression, and this is the first study that establishes HMGB2 expression inhibition by an miRNA.
At the transcriptional level, we identified SHP as a repressor of HMGB2 mRNA expression. We showed that SHP directly inhibits HMGB2 promoter activity by repressing E2F1 transactivation. Interestingly, the effect of SHP is augmented in the presence of its co‐repressor EID1, which is also observed in early growth response‐1 gene promoter.19 However, such synergy is not observed in the neuronal periodic acid–Schiff domain protein 2(NPAS2) promoter,38 suggesting a cellular context and promoter‐specific effect. The expression of Shp and Eid1 is progressively up‐regulated during EB formation, suggesting their physiologic roles in ESC differentiation. Indeed, Shp expression is low in the early embryonic liver but is increased during liver maturation,32 emphasizing the potential importance of SHP in hepatocyte differentiation. Because SHP is most highly expressed in mature livers in which HMGB2 is barely detectable, it is postulated that the increased HMGB2 expression in Shp –/– livers may induce the activation of liver progenitor cells. Future studies are necessary to elucidate the crosstalk between HMGB2 and SHP in liver function and diseases.
It is intriguing that a short‐term treatment of CGR8 cells with a small molecule inhibitor of HMGB2 ICM results in rapid degradation of HMGB2 as well as OCT4 and SOX2 proteins, which subsequently alters the morphology and pluripotency of ESCs. Constant treatment of ICM during the differentiation process disrupts proper EB formation. It is interesting to note that ICM was reported to bind both HMGB2 and HMGB1 proteins in microglia.7 However, the involvement of HMGB1 in ESC could be minimal based on its expression pattern during ESC differentiation. In addition, both miR‐127 and SHP were not predicted to regulate HMGB1 expression. Therefore, considering the emerging importance of HMGB2 in mesenchymal stem cell differentiation5 and miR‐127 in germ layer specification,19 our studies may have significant relevance for better characterizing the underlying molecular mechanisms.
TICs are not only responsible for resistance to chemotherapy and associated with metastatic HCC but also are endowed with stem/progenitor cell properties, including the ability of self‐renewal and the generation of mature cells of a particular lineage through differentiation. The stem cell‐like property and their limited number within the bulk of the tumor account for their capability to escape conventional therapies. Inhibiting self‐renewal or promoting TIC differentiation to a less stemness state would make them more vulnerable to current therapies. Overexpression of miR‐127 or SHP decreased spheroid formation, which was contributed by the reduction of HMGB2. Our study published in Hepatology demonstrated that SHP inhibited hepatocyte cell proliferation.21 Our study published in PLOS ONE also demonstrated that miR‐127 inhibited tumor growth.28 The present study not only supported our prior findings but also provides a new regulatory pathway by identifying HMGB2 as a target of miR‐127 and SHP. As TICs were generated from the liver of alcohol‐fed HCV Ns5a Tg mice, an interesting question is whether the treatment of ICM of alcohol‐fed HCV Ns5a Tg mice will lead to the reduction of TIC number in this model. Due to the important role of HMGB2 and its high expression in TICs as well as in other human cancers, it would be of great interest to explore the function of HMGB2 in cancer stem cells in future studies.
Author names in bold designate shared co‐first authorship.
Supporting information
Additional Supporting Information may be found at http://onlinelibrary.wiley.com/doi/10.1002/hep4.1086/full.
Supporting Information Table 1
Supporting Information Figure 1
Supporting Information Figure 2
Supporting Information Figure 3
Supporting Information Figure 4
Supporting Information Figure 5
Supporting Information Figure 6
Supporting Information Figure 7
Acknowledgment
We thank Drs. Shinya Yamanaka (Kyoto University), Yasuhiko Kawakami (University of Minnesota), and Lisa Dailey (New York University School of Medicine) for generously providing the plasmids.
Potential conflict of interest: Nothing to report.
Supported by the National Institutes of Health (R01DK104656, R01DK080440, R01ES025909, R21AA022482, R21AA024935 to L.W.); VA Merit Award (1I01BX002634 to L.W.); and the National Natural Scientific Foundation of China (Grant No. 81572443 to L.W.).
Funding for open access charge from the National Institutes of Health.
REFERENCES
- 1. Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev 2005;15:496‐506. [DOI] [PubMed] [Google Scholar]
- 2. Dubois T, Howell S, ZemLickova E, Aitken A. Identification of casein kinase Ialpha interacting protein partners. FEBS Lett 2002;517:167‐171. [DOI] [PubMed] [Google Scholar]
- 3. Stros M, Ozaki T, Bacikova A, Kageyama H, Nakagawara A. HMGB1 and HMGB2 cell‐specifically down‐regulate the p53‐ and p73‐dependent sequence‐specific transactivation from the human Bax gene promoter. J Biol Chem 2002;277:7157‐7164. [DOI] [PubMed] [Google Scholar]
- 4. Kwon JH, Kim J, Park JY, Hong SM, Park CW, Hong SJ, et al. Overexpression of high‐mobility group box 2 is associated with tumor aggressiveness and prognosis of hepatocellular carcinoma. Clin Cancer Res 2010;16:5511‐5521. [DOI] [PubMed] [Google Scholar]
- 5. Taniguchi N, Carames B, Hsu E, Cherqui S, Kawakami Y, Lotz M. Expression patterns and function of chromatin protein HMGB2 during mesenchymal stem cell differentiation. J Biol Chem 2011;286:41489‐41498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Laurent B, Randrianarison‐Huetz V, Marechal V, Mayeux P, Dusanter‐Fourt I, Dumenil D. High‐mobility group protein HMGB2 regulates human erythroid differentiation through trans‐activation of GFI1B transcription. Blood 2010;115:687‐695. [DOI] [PubMed] [Google Scholar]
- 7. Lee S, Nam Y, Koo JY, Lim D, Park J, Ock J, et al. A small molecule binding HMGB1 and HMGB2 inhibits microglia‐mediated neuroinflammation. Nat Chem Biol 2014;10:1055‐1060. [DOI] [PubMed] [Google Scholar]
- 8. Yang Z, Wang L. Regulation of microRNA expression and function by nuclear receptor signaling. Cell Biosci 2011;1:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Song G, Wang L. MiR‐433 and miR‐127 arise from independent overlapping primary transcripts encoded by the miR‐433‐127 locus. PLoS One 2008;3:e3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Song G, Wang L. A conserved gene structure and expression regulation of miR‐433 and miR‐127 in mammals. PLoS One 2009;4:e7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Song G, Wang L. Transcriptional mechanism for the paired miR‐433 and miR‐127 genes by nuclear receptors SHP and ERRgamma. Nucleic Acids Res 2008;36:5727‐5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tryndyak VP, Ross SA, Beland FA, Pogribny IP. Down‐regulation of the microRNAs miR‐34a, miR‐127, and miR‐200b in rat liver during hepatocarcinogenesis induced by a methyl‐deficient diet. Mol Carcinog 2009;48:479‐487. [DOI] [PubMed] [Google Scholar]
- 13. Wang S, Li H, Wang J, Wang D, Yao A, Li Q. Prognostic and biological significance of microRNA‐127 expression in human breast cancer. Dis Markers 2014;2014:401986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ma H, Lin Y, Zhao ZA, Lu X, Yu Y, Zhang X, et al. MicroRNA‐127 promotes mesendoderm differentiation of mouse embryonic stem cells by targeting left‐right determination factor 2. J Biol Chem 2016;291:12126‐12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou T, Zhang Y, Macchiarulo A, Yang Z, Cellanetti M, Coto E, et al. Novel polymorphisms of nuclear receptor SHP associated with functional and structural changes. J Biol Chem 2010;285:24871‐24881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rudraiah S, Zhang X, Wang L. Nuclear receptors as therapeutic targets in liver disease: are we there yet? Annu Rev Pharmacol Toxicol 2016;56:605‐626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Y, Hagedorn CH, Wang L. Role of nuclear receptor SHP in metabolism and cancer. Biochim Biophys Acta 2011;1812:893‐908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Y, Soto J, Park K, Viswanath G, Kuwada S, Abel ED, et al. Nuclear receptor SHP, a death receptor that targets mitochondria, induces apoptosis and inhibits tumor growth. Mol Cell Biol 2010;30:1341‐1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y, Xu N, Xu J, Kong B, Copple B, Guo GL, et al. E2F1 is a novel fibrogenic gene that regulates cholestatic liver fibrosis through the Egr‐1/SHP/EID1 network. Hepatology 2014;60:919‐930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Smalling RL, Delker DA, Zhang Y, Nieto N, McGuiness MS, Liu S, et al. Genome‐wide transcriptome analysis identifies novel gene signatures implicated in human chronic liver disease. Am J Physiol Gastrointest Liver Physiol 2013;305:G364‐G374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y, Xu P, Park K, Choi Y, Moore DD, Wang L. Orphan receptor small heterodimer partner suppresses tumorigenesis by modulating cyclin D1 expression and cellular proliferation. Hepatology 2008;48:289‐298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. He N, Park K, Zhang Y, Huang J, Lu S, Wang L. Epigenetic inhibition of nuclear receptor small heterodimer partner is associated with and regulates hepatocellular carcinoma growth. Gastroenterology 2008;134:793‐802. [DOI] [PubMed] [Google Scholar]
- 23. Choi YH, Park MJ, Kim KW, Lee HC, Cheong J. The orphan nuclear receptor SHP is involved in monocytic differentiation, and its expression is increased by c‐Jun. J Leukoc Biol 2004;76:1082‐1088. [DOI] [PubMed] [Google Scholar]
- 24. Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A 1981;78:7634‐7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hammachi F, Morrison GM, Sharov AA, Livigni A, Narayan S, Papapetrou EP, et al. Transcriptional activation by Oct4 is sufficient for the maintenance and induction of pluripotency. Cell Rep 2012;1:99‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scholer HR, Ruppert S, Suzuki N, Chowdhury K, Gruss P. New type of POU domain in germ line‐specific protein Oct‐4. Nature 1990;344:435‐439. [DOI] [PubMed] [Google Scholar]
- 27. Campbell PA, Rudnicki MA. Oct4 interaction with Hmgb2 regulates Akt signaling and pluripotency. Stem Cells 2013;31:1107‐1120. [DOI] [PubMed] [Google Scholar]
- 28. Yang Z, Zhang Y, Wang L. A feedback inhibition between miRNA‐127 and TGFbeta/c‐Jun cascade in HCC cell migration via MMP13. PLoS One 2013;8:e65256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang Z, Tsuchiya H, Zhang Y, Hartnett ME, Wang L. MicroRNA‐433 inhibits liver cancer cell migration by repressing the protein expression and function of cAMP response element‐binding protein. J Biol Chem 2013;288:28893‐28899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Machida K, Tsukamoto H, Mkrtchyan H, Duan L, Dynnyk A, Liu HM, et al. Toll‐like receptor 4 mediates synergism between alcohol and HCV in hepatic oncogenesis involving stem cell marker Nanog. Proc Natl Acad Sci U S A 2009;106:1548‐1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang L, Liu J, Saha P, Huang J, Chan L, Spiegelman B, et al. The orphan nuclear receptor SHP regulates PGC‐1alpha expression and energy production in brown adipocytes. Cell Metab 2005;2:227‐238. [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y, Liu C, Barbier O, Smalling R, Tsuchiya H, Lee S, et al. Bcl2 is a critical regulator of bile acid homeostasis by dictating Shp and lncRNA H19 function. Sci Rep 2016;6:20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhao Y, Xiao M, Sun B, Zhang Z, Shen T, Duan X, et al. C‐terminal domain (CTD) small phosphatase‐like 2 modulates the canonical bone morphogenetic protein (BMP) signaling and mesenchymal differentiation via Smad dephosphorylation. J Biol Chem 2014;289:26441‐26450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tsuchiya H. Membrane interactions of phytochemicals as their molecular mechanism applicable to the discovery of drug leads from plants. Molecules 2015;20:18923‐18966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang X, Wu J, Choiniere J, Yang Z, Huang Y, Bennett J, et al. Arsenic silences hepatic PDK4 expression through activation of histone H3K9 methylatransferase G9a. Toxicol Appl Pharmacol 2016;304:42‐47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Y, Bonzo JA, Gonzalez FJ, Wang L. Diurnal regulation of the early growth response 1 (Egr‐1) protein expression by hepatocyte nuclear factor 4alpha (HNF4alpha) and small heterodimer partner (SHP) cross‐talk in liver fibrosis. J Biol Chem 2011;286:29635‐29643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pan X, Zhang Y, Wang L, Hussain MM. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab 2010;12:174‐186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee SM, Zhang Y, Tsuchiya H, Smalling R, Jetten AM, Wang L. Small heterodimer partner/neuronal PAS domain protein 2 axis regulates the oscillation of liver lipid metabolism. Hepatology 2015;61:497‐505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsuchiya H, da Costa KA, Lee S, Renga B, Jaeschke H, Yang Z, et al. Interactions between nuclear receptor SHP and FOXA1 maintain oscillatory homocysteine homeostasis in mice. Gastroenterology 2015;148:1012‐1023.e1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Y, Wang L. Nuclear receptor SHP inhibition of Dnmt1 expression via ERRgamma. FEBS Lett 2011;585:1269‐1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem‐cell biology to cancer. Nat Rev Cancer 2003;3:895‐902. [DOI] [PubMed] [Google Scholar]
- 42. Yang Z, Koehler AN, Wang L. A novel small molecule activator of nuclear receptor SHP inhibits HCC cell migration via suppressing Ccl2. Mol Cancer Ther 2016;15:2294‐2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found at http://onlinelibrary.wiley.com/doi/10.1002/hep4.1086/full.
Supporting Information Table 1
Supporting Information Figure 1
Supporting Information Figure 2
Supporting Information Figure 3
Supporting Information Figure 4
Supporting Information Figure 5
Supporting Information Figure 6
Supporting Information Figure 7


