Abstract
Effective treatment of chronic myelogenous leukemia (CML) largely depends on eradication of CML leukemic stem cells (LSCs). We recently showed that CML LSCs depend on Tcf1 and Lef1 factors for self-renewal. Using a connectivity map we identified prostaglandin E1 (PGE1) as a small molecule that partly elicited the gene expression changes in LSCs caused by Tcf1/Lef1 deficiency. Although it has little impact on normal hematopoiesis, we found that PGE1 treatment impaired the persistence and activity of LSCs in a pre-clinical murine CML model and a xenograft model of transplanted CML patient CD34+ stem/progenitor cells. Mechanistically, PGE1 acted on the EP4 receptor and repressed Fosb and Fos AP-1 factors in a β-catenin-independent manner. Misoprostol, an FDA-approved EP4 agonist, conferred similar protection against CML. These findings suggest that activation of this PGE1-EP4 pathway specifically targets CML LSCs, and that combination of PGE1/misoprostol with conventional tyrosine-kinase inhibitors could provide effective therapy for CML.
ETOC summary
Xue and colleagues show that prostaglandin E1 (PGE1) inhibits the activity and self-renewal of human CML leukemic stem cells. Combination of PGE1 or an agonist for its receptor EP4 with conventional tyrosine kinase inhibitor treatment can effectively target CML leukemic stem cells and reduce leukemia growth.

Hematopoietic and leukemic stem cells (HSCs and LSCs, respectively) both have a capacity of self-renewal. Whereas HSCs give rise to all blood lineages during lifetime hematopoiesis, LSCs are responsible for initiation and propagation of leukemia, as well as drug resistance and disease relapse after treatment-induced remission (Visvader and Lindeman, 2012). Chronic myelogenous leukemia (CML) is a quintessential LSC-driven myeloproliferative disorder that results from transformation of HSCs by the BCR-ABL oncoprotein (Bhatia et al., 2003). BCR-ABL has constitutive tyrosine-kinase activity, and tyrosine-kinase inhibitors (TKIs), such as imatinib, induce remissions and improve survival in CML patients in the chronic phase (CP). CML LSCs do not, however, appear to depend on the BCR-ABL kinase activity for survival, and they are less sensitive to TKIs (Corbin et al., 2011). Failure to eliminate LSCs necessitates continuous TKI treatment to sustain remission (Mahon et al., 2010); when TKI resistance develops, CML relapses and/or progresses to an accelerated phase (AP) and/or blast crisis (BC) with features of aggressive, acute leukemia of the myeloid or lymphoid phenotype. Treatment options for AP or BC CML are limited, but CP represents a therapeutic window where eradication of LSCs may lead to a cure.
β-catenin, activated by Wnt ligands or prostaglandins, is implicated in HSC regulation (Castellone et al., 2005; Goessling et al., 2009; Malhotra and Kincade, 2009), and levels of β-catenin activation determine the impact on HSC activities (Luis et al., 2011). On the other hand, β-catenin is involved in many aspects of leukemogenesis, including development of LSCs in pre-clinical models of CML and acute myeloid leukemia (AML) (Jamieson et al., 2004; Wang et al., 2010; Zhao et al., 2007). β-catenin is also necessary for maintaining CML LSCs (Heidel et al., 2012), and is a contributing factor to TKI resistance (Hu et al., 2009) and progression to BC CML (Neviani et al., 2013; Scheller et al., 2013). Aberrant activation of β-catenin is a hallmark of tumor initiation, progression, and metastasis, making β-catenin a sought-after drug target in cancer therapy (Anastas and Moon, 2013). In a CML mouse model, blocking prostaglandin production diminishes β-catenin expression in CML LSCs and extends survival of CML mice in tertiary recipients (Heidel et al., 2012).
Upon activation, β-catenin translocates into the nucleus where it interacts with Tcf/Lef transcription factors to modulate gene expression (Staal et al., 2008; Xue and Zhao, 2012). Recently, we showed that two members of the Tcf/Lef family, Tcf1 and Lef1, are expressed in HSCs. Whereas HSCs require Tcf1/Lef1 for regenerative fitness, LSCs are more strongly dependent on both factors for self-renewal than HSCs (Yu et al., 2016). In the present study, we profiled Tcf1/Lef1 downstream genes in CML LSCs, and in search of small molecules that simulate gene expression changes caused by Tcf1/Lef1 deficiency using the Connectivity Map, we identified prostaglandin E1 (PGE1). In both pre-clinical and xenograft models, PGE1 treatment greatly diminished the activity and persistence of CML LSCs. The action of PGE1 is mechanistically distinct from PGE2 despite their structural similarity. Whereas PGE2 stimulates β-catenin accumulation, PGE1 acts through E-prostanoid receptor 4 (EP4) and represses AP-1 factors in LSCs in a β-catenin-independent manner. Therefore, activating the “EP4-AP-1 repression” pathway represents a different approach from inhibiting “PGE2-β-catenin activation” pathway to effectively subvert LSCs. PGE1 is an FDA-approved drug clinically known as alprostadil, and our study indicates that PGE1 can be repositioned in combination with TKIs for a more effective CML therapy, alleviating CML patients’ lifetime dependence on TKIs.
Results
Delineation of Tcf1/Lef1-dependent transcriptional programs in HSPCs and LSCs
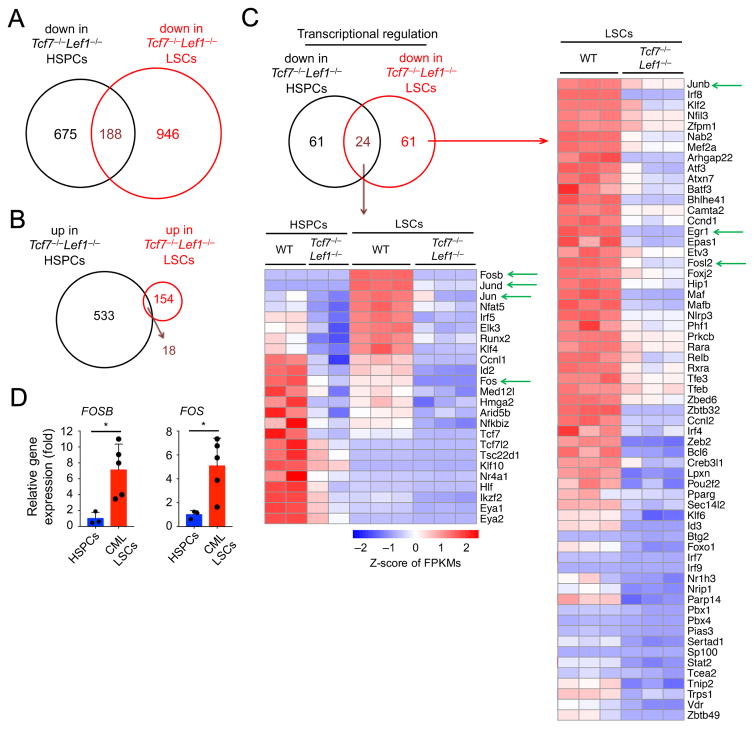
We recently demonstrated that CML LSCs are more strongly dependent on Tcf1 and Lef1 than hematopoietic stem/progenitor cells (HSPCs) for self-renewal (Yu et al., 2016). This observation suggests that Tcf1 and Lef1 are potential therapeutic targets to eliminate LSCs in CML without significantly affecting HSPCs. To explore this possibility, we performed RNA-Seq analyses comparing wild-type (WT) and Tcf1/Lef1-deficient HSPCs, as well as corresponding LSCs. HSPCs were sorted Flt3−Lin−Sca1+c-Kit+ (Flt3−LSK) cells from bone marrow (BM) cells of WT or Tcf7−/−Lef1−/− mice, and LSCs were sorted as GFP+ Lin−Sca1+c-Kit+ from WT or Tcf7−/−Lef1−/− BM cells after infection with bicistronic p210BCR-ABL-GFP retrovirus (Yu et al., 2012a). By cross-comparison of differentially expressed genes (DEGs) between Tcf7−/−Lef1−/− and WT HSPCs and those between Tcf7−/−Lef1−/− and WT LSCs, we found that Tcf1/Lef1 deficiency led to more genes downregulated, but fewer genes upregulated, in LSCs compared with HSPCs (Figure 1A–B). Tcf1/Lef1 appears to regulate a distinct transcriptional program in LSCs, which may account for the unique dependence of LSCs on these factors.
Figure 1. Tcf1/Lef1-dependent transcriptional program in HSPCs and LSCs.
A–B. Venn diagrams showing downregulated (A) and upregulated (B) genes resulting from comparing transcriptomes of Tcf7−/−Lef1−/− versus WT HSPCs and Tcf7−/−Lef1−/− versus WT LSCs.
C. Genes involved in transcriptional regulation that were downregulated in Tcf7−/−Lef1−/− LSCs, unique or common with those in Tcf7−/−Lef1−/− HSPCs (shown in heatmaps in right and lower panels, respectively). Green arrows mark genes of interest.
D. Elevated expression of FOSB and FOS in human CD34+ CML LSCs compared with human CD34+ HSPCs, as determined by quantitative RT-PCR. Each dot represents one healthy individual or one CML patient. *, p<0.05. See also Figure S1.
Functional annotation of the DEGs in Tcf7−/−Lef1−/− LSCs showed that a major group of genes is involved in transcriptional regulation (Figure 1C). Of particular interest are the AP-1 family members, with Fos, Fosb, Jun, and Jund as shared downregulated genes in Tcf7−/−Lef1−/− HSPCs and Tcf7−/−Lef1−/− LSCs, and Junb and Fosl2 downregulated in Tcf7−/−Lef1−/− LSCs alone. All but Fos showed induced expression in WT LSCs compared with WT HSPCs, with Fosb and Jund induced >60 fold (Figure 1C, S1A–B). Although some AP-1 genes showed reduced expression in Tcf7−/−Lef1−/− HSPCs, their reductions in Tcf7−/−Lef1−/− LSCs were more pronounced, with Fos and Fosb showing >40-fold reduction (Figure S1A–B). These data suggest a potential link of Tcf1/Lef1 with the AP-1 pathway in LSCs. By qRT-PCR, we found consistently elevated expression of FOSB and FOS in CML patient-derived CD34+ LSCs compared with human CD34+ HSPCs (Figure 1D). By gene set enrichment analysis, two gene sets, which are upregulated in quiescent human CML LSCs compared with human HSPCs (Graham et al., 2007), were enriched in WT LSCs, i.e., downregulated in Tcf7−/−Lef1−/− LSCs (Figure S1C–D). The transcriptomic analyses suggest that Tcf1/Lef1 deficiency at least partly impairs the transcriptional program that maintains CML LSCs in humans and the murine model.
Identification of PGE1 that confers protection from CML
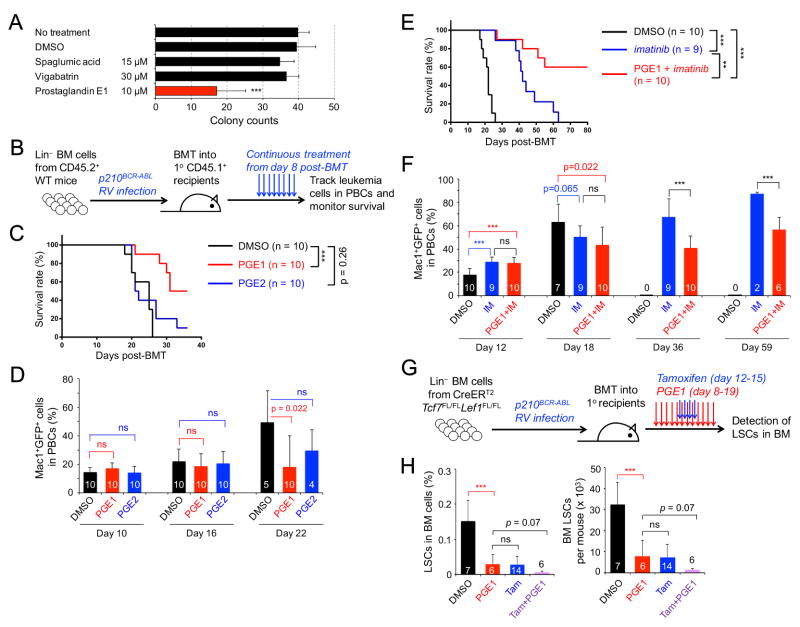
It is currently recognized that transcriptional programs can be druggable targets to eradicate LSCs (Ashton et al., 2012). The Connectivity Map (CMAP) database can be queried with a gene signature of interest to identify those compounds that induce desired gene expression changes (Lamb, 2007). From the DEGs of Tcf7−/−Lef1−/− LSCs, we extracted 4 sets of query signatures including: 1) transcription regulator signature (Figure 1C, S2A), 2) signal transduction signature (Figure S2B–C), 3) metabolism and chemotaxis signature (Figure S2D–E), and 4) 444 genes that were downregulated ≥ 2 fold and all 154 genes upregulated in Tcf7−/−Lef1−/− LSCs. We searched the CMAP with these gene signatures to identify small molecules that could simulate transcriptional changes caused by Tcf1/Lef1 deficiency in LSCs. Based on scores of “connectivity” and “enrichment”, we selected several candidate compounds, including vigabatrin, spaglumic acid, and prostaglandin E1 (PGE1). By in vitro colony formation assays, only PGE1 significantly reduced the LSC colonies (Figure 2A), whereas none strongly affected the colony formation capacity of normal HSPCs (Figure S3A).
Figure 2. PGE1 treatment targets CML LSCs and synergizes with imatinib therapy.
A. Effect of CMAP-selected chemicals on colony-forming capacity of LSCs. Data are means ± s.d. from ≥ 3 experiments.
B. Experimental design for establishing the murine CML model. Lin− BM cells from CD45.2+ WT mice were infected with p210BCR-ABL retrovirus (RV), followed by transplantation into irradiated CD45.1+ syngeneic mice as the primary (1°) recipients. Daily treatment was initiated on day 8 post-BMT and continued for the duration of observation.
C–D. PGE1 but not PGE2 prolongs survival of CML mice without affecting onset of CML. The 1° CML recipients were treated with 2.5 mg/kg body weight PGE1, PGE2, or the vehicle DMSO, and monitored for survival (C) and frequency of Mac1+GFP+ leukemic cells in PBCs (D).
E–F. PGE1 synergizes with imatinib therapy. The 1° CML recipients were treated with imatinib (IM) alone or together with 5 mg/kg body weight PGE1, and monitored for survival (E) and leukemic cells in PBCs (F). Data in C–F are representative from two independent experiments with similar results. Values in D and F indicate numbers of starting recipients (day 10 or 12) or surviving recipients observed.
G–H. PGE1 treatment and Tcf1/Lef1 ablation similarly diminished LSCs.
G. Experimental design. Lin− BM cells from CreERT2Tcf7FL/FLLef1FL/FL mice were used to establish CML in 1° recipients, followed by treatment with PGE1 and/or Tamoxifen (Tam) on indicated days.
H. Cumulative data on LSC frequency in whole BM (left) and LSC numbers (right), determined on day 20 post-BMT. DMSO-treated mice were analyzed on day 16 post-BMT because of uncurtailed leukemia progression. Data are ± s.d. from 2 experiments (n marked in the bars). ns, not statistically significant; **, p<0.01; ***, p<0.001 unless specified. See also Figure S2 and S3.
PGE1 and PGE2 are both structurally similar 20-carbon fatty acid derivatives, with PGE2 containing an extra carbon-carbon double bond at the C5 position. PGE1 is produced by cyclooxygenase (COX)-dependent oxygenation of dihomo-γ-linolenic acid (DGLA), and PGE2 production is also catalyzed by COX enzymes from arachidonic acid, a derivative of DGLA (Levin et al., 2002). To test their roles in controlling CML propagation in vivo, we transplanted p210BCR-ABL-transduced Lin− BM cells into irradiated congenic recipients (Figure 2B). Ectopic expression of BCR-ABL resulted in malignant transformation of HSPCs, and GFP+ Mac1+ myeloid leukemic cells were readily detectable in peripheral blood cells (PBCs) and BM. The CML recipients succumbed to the disease by day 30 post-BM transplantation (post-BMT; Figure 2C–2D, S3B–S3C). Neither PGE1 nor PGE2 treatment affected onset of CML as assessed by the frequency of GFP+Mac1+ leukemic cells in the PBCs on days 10 or 16 post-BMT (Figure 2D). On day 22 post-BMT, when approximately 50% of DMSO- or PGE2-treated recipients died of CML, all PGE1-treated animals were alive and exhibited reduced leukemic cell burden (Figure 2C–2D), highlighting a beneficial effect specific for PGE1.
The rapidly expanding CML leukemic blasts are better controlled by TKIs, and as expected, imatinib treatment extended survival of recipient mice to 60 days post-BMT (Figure 2E). We hypothesized that PGE1 acts on LSCs rather than leukemic blasts, which may account for its relatively modest survival benefit compared with imatinib treatment (compare Figure 2C with 2E). We tested this by combining PGE1 and imatinib therapy, which protected over 50% of the recipients for 80 days post-BMT (Figure 2E–2F). The recipients treated with imatinib alone exhibited a steady increase in peripheral leukemic cells; in contrast, the recipients treated with PGE1+imatinib had consistently lower leukemic cell burden for those having survived beyond day 30 post-BMT (Figure 2F). In parallel, we transplanted WT BM cells and treated the recipients with PGE1. After 70 days post-BMT, neither reconstitution of various blood lineages nor preservation of the HSC pool was detectably affected (Figure S3D–S3I). These findings demonstrate a synergistic therapeutic effect of imatinib and LSC-targeting PGE1, without affecting normal hematopoiesis.
To assess the impact of PGE1 on CML LSCs with direct comparison to genetic targeting of Tcf1/Lef1, we used Lin− BM cells from CreERT2Tcf7FL/FLLef1FL/FL mice to establish CML followed by treatment with PGE1 and/or Tamoxifen (Figure 2G). PGE1 showed a similar capacity of reducing BM LSCs as Tamoxifen-induced ablation of Tcf1/Lef1; interestingly, the combination of PGE1 and Tcf1/Lef1 ablation resulted in more consistent LSC reduction, albeit the difference with PGE1 treatment alone was not statistically significant (Figure 2H). These data further corroborate the notion that PGE1 simulates key gene expression changes caused by Tcf1/Lef1 deficiency, leading to LSC impairment.
PGE1 impairs LSC activities
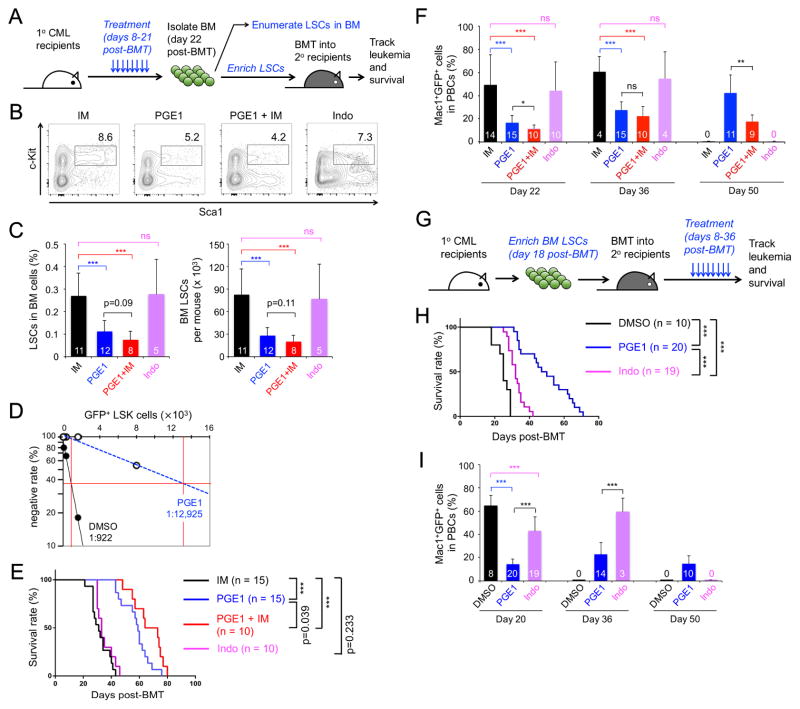
To further investigate the impact of PGE1 on LSCs and its synergy with imatinib in CML therapy, we treated primary CML recipients for a short-term and analyzed LSC number and functions (Figure 3A). Consistent with insensitivity of LSCs to TKIs (Hu et al., 2009), LSCs persisted in the BM of imatinib-treated mice; in contrast, PGE1 treatment greatly diminished LSCs and showed some additive effect when combined with imatinib (Figure 3B–3C). In addition to LSC enumeration based on phenotypic markers, we performed limiting dilution assays by transplanting graded numbers of BM cells from DMSO- or PGE1-treated primary recipients into secondary hosts (Jiang et al., 2003). By defining secondary hosts that contain <1% of CD45.2+GFP+ leukemic cells in PBCs as being negatively engrafted, we estimated that PGE1 treatment caused >10 fold reduction in functional LSCs (Figure 3D).
Figure 3. PGE1 impairs LSC activity.
A–F. Impact of PGE1 on primary LSCs.
A. Experimental design. CML was established in 1° recipients, followed by a 2-week treatment with imatinib (IM), PGE1, PGE1+IM, or indomethacin (Indo) and downstream analyses.
B. Detection of Sca1+c-Kit+ cells in Lin−GFP+ BM cells.
C. Cumulative data on frequency of LSCs (GFP+ LSK cells) in whole BM (left) and LSC numbers (right). The number of recipients is marked in the bars.
D. Detection of functional LSCs by limiting dilution assay. Graded numbers of BM cells from DMSO- or PGE1-treated 1° recipients (CD45.2+) were mixed with 2 × 105 protector BM cells (CD45.1+) and transplanted into secondary (2°) recipient mice (CD45.1+). Plotted are the percentages of 2° recipients containing < 1% CD45.2+GFP+ leukemia cells in the PBCs. Also marked is the frequency of functional LSCs calculated according to Poisson statistics.
E. LSCs were enriched from the BM of treated 1° recipients and transplanted into irradiated 2° recipients, which were monitored for survival.
F. Leukemia burden in the PBCs of 2° recipients. Values (in bars) indicate numbers of starting recipients (day 22) or surviving recipients observed.
G–I. Impact of PGE1 on serially transplanted LSCs.
G. Experimental design. LSCs were enriched from untreated 1° CML mice and transplanted into irradiated 2° recipients, followed by treatment with PGE1 or indomethacin.
H. Survival curve of the treated 2° recipients.
I. Leukemia burden in PBCs of treated 2° recipients. Values (in bars) indicate numbers of starting recipients (day 20) or surviving recipients observed. Data are means ± s.d. from 2–3 independent experiments. ns, not statistically significant; ***, p<0.001 unless specified.
PGE2 stabilizes β-catenin (Castellone et al., 2005). COX inhibitors, such as indomethacin, were used to block prostaglandin production and β-catenin activation, and showed some beneficial effects on serially transplanted LSCs (Heidel et al., 2012). In our experimental setting, treatment of the primary recipients with indomethacin did not result in significant reduction of LSCs (Figure 3B–C), highlighting a robust effect of PGE1 on LSCs, compared with the approach of blocking overall PGE production.
To determine if the short-term treatment exerted a lasting effect, we transplanted the same numbers of LSCs from the treated primary recipients into secondary hosts, and monitored their survival without additional treatments (Figure 3A). As expected, imatinib-pretreated LSCs propagated CML, leading to death of all secondary hosts by day ~40 post-BMT (Figure 3E). PGE1 pretreatment substantially extended survival of the secondary hosts, and the PGE1+imatinib regimen showed additional survival benefit (Figure 3E). In contrast, the effect of indomethacin pretreatment was similar as that of imatinib (Figure 3E). In addition, PGE1 but not indomethacin pretreatment reduced the leukemia burden in PBCs compared with imatinib at early time points, and PGE1+imatinib pretreatment appeared to be more effective than PGE1 alone in controlling leukemia (Figure 3F). These data suggest short-term exposure to PGE1 has lasting effects in inhibiting LSC self-renewal, in addition to directly reducing LSC numbers.
Tcf1/Lef1-deficient LSCs failed to propagate CML in secondary recipients (Yu et al., 2016), suggesting that LSCs are more sensitive to deregulation of Tcf1/Lef1-dependent target genes under regenerative stress. To determine if LSCs are more sensitive to PGE1-induced attrition when forced to self-renew, we established CML and isolated LSCs from untreated primary recipients, and after re-transplantation we treated the secondary recipients (Figure 3G). PGE1-treated mice showed reduced leukemia burden and survived substantially longer than DSMO-treated ones (Figure 3H–3I). Consistent with the previous report (Heidel et al., 2012), indomethacin exhibited an inhibitory effect on serially transplanted LSCs, albeit less potent than PGE1 (Figure 3H–3I). These beneficial effects by PGE1 were achieved without TKI treatment, further corroborating the notion that PGE1 potently inhibits LSC activities.
PGE1 targets Tcf1/Lef1-dependent AP-1 factors in LSCs
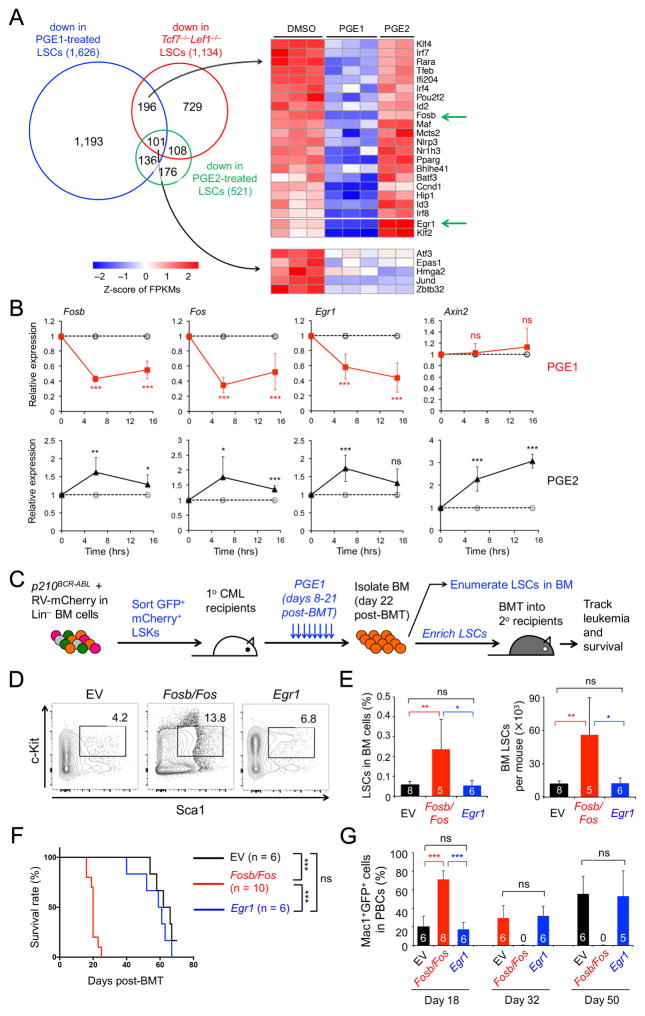
To investigate the mechanism(s) by which PGE1 impairs LSCs, we performed RNA-Seq on LSCs stimulated with PGE1 or PGE2 and found that PGE1 caused much broader transcriptomic changes than PGE2 (Figure 4A, S4A). Cross-comparison with transcriptomic changes caused by Tcf1/Lef1 deficiency showed that ~26% of downregulated genes in Tcf7−/−Lef1−/− LSCs were repressed by PGE1 stimulation, including Egr1 and AP-1 factors such as Fosb and Jund (compare Figure 4A with 1C). PGE1-mediated repression of Fosb and Egr1 was validated at 6 and 15 hrs after LSC stimulation (Figure 4B). Like Fosb, Fos expression was reduced by >40-fold in Tcf7−/−Lef1−/− LSCs (Figure S1A), and was also repressed by PGE1 in the gene-specific RT-PCR assay (Figure 4B). On the other hand, although PGE2-induced DEGs showed partial overlap with those in Tcf7−/−Lef1−/− LSCs (Figure 4A), PGE2 did not repress, but rather modestly induced Fosb, Fos and Egr1 along with Axin2, a known Wnt/β-catenin-responsive gene (Figure 4B). Similar gene expression patterns stimulated by PGE1 or PGE2 were also observed in human CML-derived K562 cells (Figure S4B). These data suggest that in spite of the structural similarity, PGE1 and PGE2 activate distinct pathways and downstream target genes in LSCs, which may underlie the specific effect of LSC subversion by PGE1.
Figure 4. PGE1 impairs LSC function by repressing Fosb and Fos expression.
A. Comparison of PGE1- and PGE2-induced transcriptome changes with those caused by Tcf1/Lef1 deficiency. LSCs were treated with 10 μM PGE1, PGE2, or DMSO for 6 hrs and analyzed by RNA-Seq. Venn diagram shows downregulated genes by PGE1, PGE2, or loss of Tcf1/Lef1. Genes involved in transcriptional regulation in indicated groups are shown in heatmaps.
B. Validation of PGE1- and PGE2-induced gene expression changes by quantitative RT-PCR. Data are means ± s.d. from 2 experiments.
C–G. Effect of forced expression of Fosb/Fos or Egr1 on LSC response to PGE1 therapy.
C. Experimental design. Lin− BM cells were infected with p210BCR-ABL-GFP and an empty vector (EV), Fosb/Fos-, or Egr1-retrovirus expressing mCherry. GFP+mCherry+ LSKs were sorted and transplanted into the 1° recipients, followed by PGE1 treatment and downstream analyses.
D. Detection of the frequency of Sca1+c-Kit+ cells in Lin− GFP+mCherry+ BM cells.
E. Cumulative data on frequency of GFP+mCherry+ LSCs in whole BM (left) and LSC numbers (right).
F. LSCs were enriched from the 1° recipients, and equal numbers of LSCs were transplanted into 2° recipients, which were monitored for survival.
G. Leukemia burden in the PBCs of 2° recipients. All data are from 2 independent experiments, and the numbers of recipients analyzed are marked in the bars. ns, not statistically significant; *, p<0.05; **, p<0.01; ***, p<0.001. See also Figure S4.
Because both genetic ablation of Tcf1/Lef1 and PGE1 stimulation reduced Fosb, Fos and Egr1 expression in LSCs, we next investigated how forced expression of these proteins altered LSCs’ response to PGE1 in vivo. We used bicistronic retroviruses to express genes of interest along with an mCherry indicator and co-infected Lin− BM cells together with the p210BCR-ABL-GFP retrovirus. We then sorted GFP+mCherry+ LSKs to establish CML, followed by PGE1 treatment (Figure 4C). After completion of the short-term PGE1 therapy, the empty vector (EV)-transduced LSCs remained sensitive to PGE1, and Egr1-transduced LSCs were detected at similar levels as the EV-transduced cells (Figure 4D–E, compare with 3B–C). In contrast, forced expression of Fosb and Fos resulted in death of 50% of the recipients before the completion of PGE1 treatment (Figure S4C), and the surviving recipients of Fosb/Fos-transduced cells showed LSC accumulation in the BM (Figure 4D–E). These data suggest that forced expression of Fosb and Fos renders LSCs less sensitive to PGE1 therapy.
To determine if these retrovirus-transduced LSCs were qualitatively different (in addition to numerical changes), we enriched LSCs from the PGE1-treated primary hosts and transplanted the cells into secondary recipients (Figure 4C). Whereas the recipients of EV- or Egr1-transduced LSCs showed prolonged survival, those of Fosb/Fos-transduced LSCs showed early onset of the leukemia and all succumbed to the disease at about 20 days post-BMT (Figure 4F, compare with 3E). The leukemic burden was substantially higher in the Fosb/Fos group (Figure 4G), indicating that LSCs with forced Fosb/Fos expression are refractory to PGE1 treatment. These data suggest that AP-1 factors are major targets for PGE1-induced impairment of LSCs.
PGE1 acts through EP4 receptor in LSCs
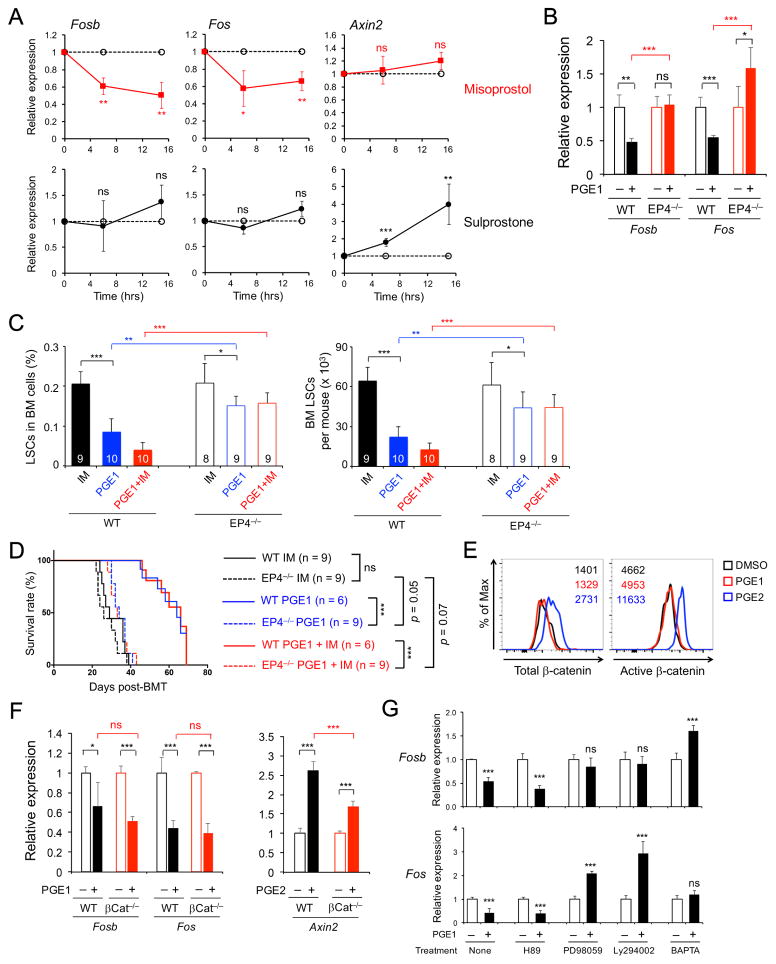
There are four E-prostanoid receptors, designated EP1, EP2, EP3 and EP4, and each has selective agonist(s) (Breyer et al., 2001; Sugimoto and Narumiya, 2007). Sulprostone preferentially acts on EP1 and EP3, and misoprostol is a strong agonist for EP3 and EP4 receptors. Misoprostol showed a remarkably similar effect as PGE1 on LSC gene expression, including repressing Fos and Fosb but without obvious impact on Axin2 (Figure 5A, compare with 4B). This was reproducible in K562 cells (Figure S4B). Sulprostone, however, did not affect Fosb and Fos expression but induced Axin2 (Figure 5A), partly simulating the PGE2 effect. By flow cytometry, EP1, EP3 and EP4 were all detected on LSCs (Figure S5A). We thus deduced that PGE1 most likely acts on EP4, whereas PGE2 may act through EP1.
Figure 5. PGE1 acts through EP4 receptor in LSCs but independent of β-catenin.
A. EP4 agonist misoprostol, but not EP1/3 agonist sulprostone, simulates gene expression changes induced by PGE1 in LSCs. LSCs were treated with misoprostol or sulprostone, and gene expression was analyzed as in Figure 4B.
B. PGE1-elicited gene expression changes depend on EP4. WT or EP4−/− LSCs were treated with DMSO or PGE1, and gene expression was analyzed. Data in A–B are means ± s.d. from 2 independent experiments.
C. EP4-deficient LSCs show reduced sensitivity to PGE1 treatment. Lin− BM cells from WT or EP4−/−mice were used to establish CML in 1° recipients, which were treated and analyzed as in Figure 3A. The frequency of LSCs in whole BM (left) and LSC numbers (right) in the 1° recipients are shown.
D. EP4-deficient LSCs rapidly propagate CML in spite of PGE1 treatment. WT or EP4−/−LSCs were enriched from the 1° recipients and transplanted into 2° recipients as in Figure 3A, which were monitored for survival. Data in C–D are from 2 independent experiments.
E. PGE2, but not PGE1, induces total and active β-catenin in LSCs. LSCs were stimulated and then intracellularly stained for total β-catenin (left) and active β-catenin (right). Representative data are from 2 experiments with values denoting geometric mean fluorescence intensity.
F. PGE1-mediated repression of Fosb and Fos is independent of β-catenin. WT or βCat−/−LSCs were treated with DMSO, PGE1, or PGE2, and gene expression was determined.
G. Multiple pathways contribute to PGE1-mediated repression of Fosb and Fos. Sorted WT LSCs were pre-incubated with indicated inhibitors and then stimulated with DMSO or 10 μM PGE1, and gene expression was analyzed. Data in F–G are means ± s.d. from 2 experiments. ns, not statistically significant; *, p<0.05; **, p<0.01; ***, p<0.001 unless specified. See also Figure S5.
To validate that EP4 mediates PGE1-induced LSC impairment, we generated Vav-Cre+EP4FL/FL (called EP4−/−) mice (Schneider et al., 2004) to delete EP4 in all hematopoietic cells. In EP4−/− LSCs, PGE1-induced repression of Fosb and Fos was abrogated (Figure 5B). We next used EP4−/− BM cells to establish CML in primary recipients and treated them with PGE1 as in Figure 3A. Whereas EP4-sufficient LSCs were substantially reduced by PGE1 (alone or combined with imatinib), a reduction of EP4−/− LSCs by PGE1 or PGE1+imatinib regimen was observed but markedly dampened (Figure 5C). We then transplanted the same numbers of enriched WT or EP4−/− LSCs into secondary hosts. The recipients of WT LSCs pretreated with PGE1 or PGE1+imatinib exhibited extended survival and reduced leukemic burden; in key contrast, those of EP4−/− LSCs (of any pretreatment) had elevated leukemic blasts in PBCs and died at an approximately similar rate as recipients of imatinib-pretreated WT LSCs (Figure 5D, S5B). It is of note that PGE1 did exhibit a marginal effect on EP4−/− LSCs (Figure 5C–D); nonetheless, our data suggest that EP4 is a major, if not the sole, receptor responsible for PGE1-induced effects on LSCs. Although EP4 was detected in BM stromal cells and endothelial cells (Figure S5A), the results from transplantation of EP4−/− LSCs demonstrate an intrinsic requirement for EP4. Without administration of exogenous PGE1, primary CML mice established using WT or EP4−/− Lin− BM cells showed similar survival rate and leukemia burden (Figure S5C), suggesting that the amount of endogenously produced PGE1 was not sufficient to confer protection.
We next probed the pathways elicited by PGE1-EP4 interaction. PGE2 stimulation induced accumulation of total β-catenin and the active, dephosphorylated form of β-catenin in LSCs (Figure 5E). We also generated Vav-Cre+β-cateninFL/FL (called βCat−/−) mice and found induction of Axin2 by PGE2 was diminished in βCat−/− LSCs (Figure 5F). In contrast, PGE1 did not cause β-catenin accumulation in LSCs, and PGE1-mediated repression of Fosb and Fos was not detectably affected in βCat−/− LSCs (Figure 5E–F). Further, PGE1 stimulation did not affect Tcf1, Lef1 or β-catenin gene expression in LSCs (Figure S5D). These data demonstrate that PGE1 acts through pathway(s) that are distinct from PGE2, independent of β-catenin and without directly affecting Tcf1/Lef1 expression per se.
Stimulation of EP receptors activates several other signaling cascades, including cAMP/PKA, Ras/MEK/Erk, PI3K/Akt pathways, and elevation of intracellular Ca2+ concentrations (Sugimoto and Narumiya, 2007; Wang and Dubois, 2010). We pretreated LSCs with PKA inhibitor H89, MEK inhibitor PD98059, PI3K inhibitor Ly294002, or BAPTA-AM to chelate intracellular Ca2+, and then stimulated with PGE1. PGE1-elicited Fos and Fosb repression was not affected by the PKA inhibitor, but was abrogated or even reversed by the MEK or PI3K inhibitor or the Ca2+ chelator (Figure 5G). These data suggest that PGE1 integrates multiple signaling cascades in LSCs to modulate downstream gene expression.
EP4 agonist misoprostol confers CML protection
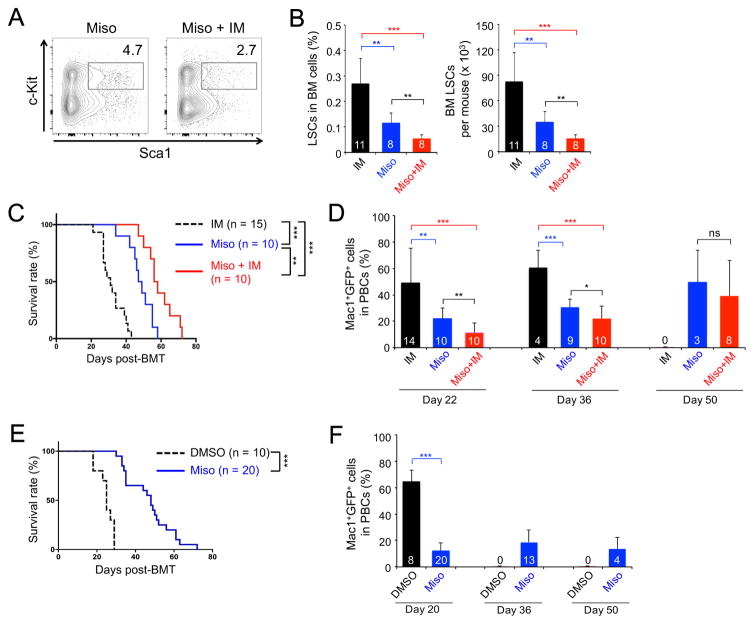
We next tested if misoprostol, another EP4 agonist, affected LSC self-renewal by treating primary CML recipients with misoprostol alone or in combination with imatinib as in Figure 3A. Misoprostol indeed diminished LSC frequency and numbers, and the combination of misoprostol+imatinib exhibited stronger effects (Figure 6A–B). We next enriched LSCs from the pretreated mice and transplanted the cells into secondary recipients. The recipients of misoprostol-pretreated LSCs survived longer and had diminished leukemia burden in PBCs (Figure 6C–6D), indicating a lasting inhibitory effect on LSCs in propagating CML. We also tested misoprostol on serially transplanted LSCs as in Figure 3F and found that misoprostol reduced leukemia burden and extended survival of secondary recipients (Figure 6E–6F). These data collectively indicate that specific stimulation of EP4 receptor compromises LSC activity.
Figure 6. Misoprostol, an EP4 agonist, impairs LSC activity.
All the experiments testing misoprostol (Miso), alone or with imatinib (IM), were performed together with those testing PGE1 as in Figure 3. Thus, the data on IM group are ‘re-used’ in panels of this figure for convenience of direct comparison.
A–B. Impact of misoprostol on maintaining LSCs. The 1° CML recipients were treated with misoprostol, and BM LSCs were analyzed as in Figure 3A. A. Representative data showing the frequency of Sca1+c-Kit+ cells in Lin−GFP+ BM cells. B. Cumulative data on LSC frequency in whole BM (left) and LSC numbers (right).
C–D. Impact of misoprostol on LSCs in propagating CML in secondary recipients. LSCs were enriched from BM of misoprostol-treated 1° recipients and transplanted into 2° recipients, as in Figure 3A. C. Survival curve of the 2° recipients. D. Leukemia burden in the PBCs of 2° recipients.
E–F. Impact of misoprostol on serially transplanted LSCs. LSCs were enriched from BM of untreated 1° recipients and transplanted into 2° recipients, which were treated with misoprostol as in Figure 3G. E. Survival curve of the treated 2° recipients. F. Leukemia burden in the treated 2° recipients. Data are from 2–3 independent experiments, and values (in bars in B, D, F) mark numbers of starting recipients or surviving recipients observed. ns, not statistically significant; *, p<0.05; **, p<0.01; ***, p<0.001.
PGE1 suppresses human CD34+ CML stem/progenitor cells in a murine xenograft model
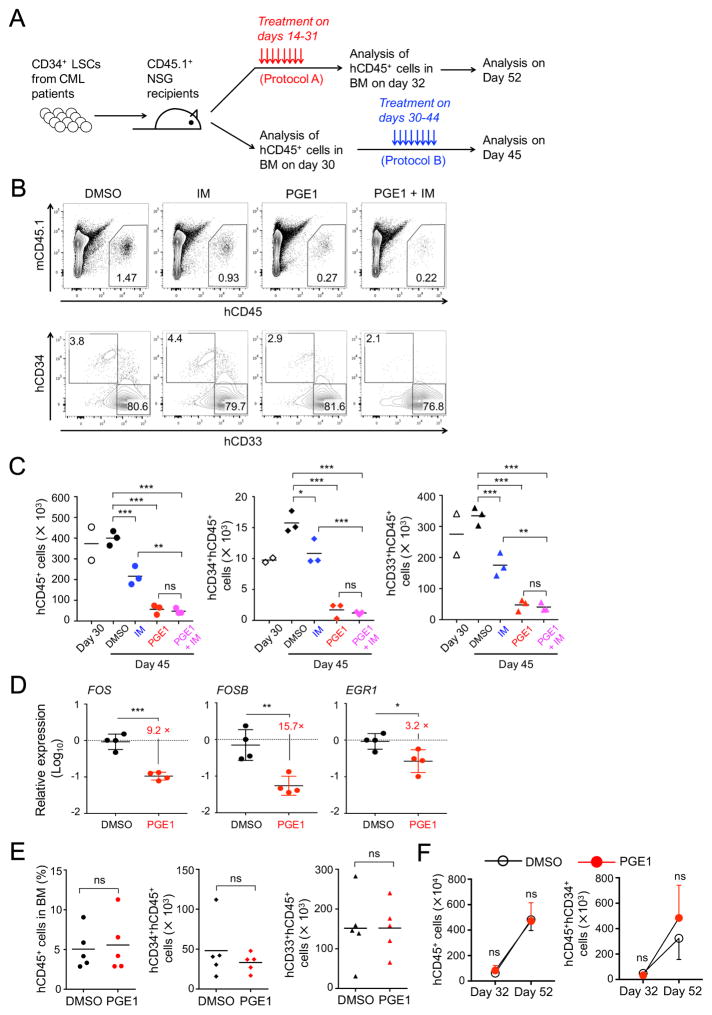
To directly investigate if PGE1 can target human CML LSCs as a clinical therapy, we employed an established, murine CML xenograft model system using non-obese diabetic (NOD)/SCID/IL2Rγnull (NSG) mice as recipients (Riether et al., 2015; Zhang et al., 2010). We enriched CD34+ CML stem/progenitor cells (called CML LSCs herein) from five newly diagnosed CML patients at the CP (Table S1) and transplanted the cells into sub-lethally irradiated NSG mice (Figure 7A). We adopted two protocols for delivering the PGE1 regimen. We initiated PGE1 treatment at an early stage of CML LSC engraftment in protocol A, so as to assess if the treatment had a lasting effect after withdrawal. In protocol B, we allowed longer time for CML LSC engraftment and then determined if late PGE1 therapy remained effective.
Figure 7. PGE1 treatment suppresses human CD34+ CML stem/progenitor cells in murine xenografts.
A. Experimental design for the murine xenograft model. CD34+ stem/progenitor cells from CML patients were transplanted into sub-lethally irradiated NSG mice. The recipients were treated, and grafted human CD45+ cells were analyzed in the BM following protocol A or B.
B–C. PGE1 suppresses activity of human CML LSCs. CML LSCs from Patient 4 were transplanted at 0.8×106 cells per NSG mouse, and the recipients treated with imatinib (IM) and/or PGE1 and analyzed following protocol B. B, detection of human CD45+ cells (upper panels) or CD34+ CML stem/progenitor cells and CD33+ CML myeloid cells (lower panels) in the BM of NSG recipients after completion of the treatment (day 45). C, cumulative numbers of human CD45+ (left), human CD45+CD34+ CML stem/progenitor cells (middle), and human CD45+ CD33+ CML myeloid cells (right) from multiple recipients. Also included is the analysis of two recipients on day 30 prior to the treatment. Horizontal bars denote means of the replicates.
D. PGE1 potently represses FOS and FOSB expression in human CML LSCs. CML LSCs from Patients 4 and 5 were transplanted into NSG mice, treated and analyzed following protocol B. On day 45, human CD45+CD34+ CML LSCs were sort-purified and analyzed for the expression of indicated genes.
E–F. PGE1 treatment does not affect HSPC activity. CD34+ human cord blood cells were transplanted into NSG mice (at 5×104 cells per mouse), which were treated and analyzed following protocol A. E, the impact of PGE1 right after the treatment. The frequency of all human CD45+ cells in the BM (left), the numbers of human CD45+CD34+ HSPCs (middle), and human CD45+CD33+ myeloid cells (right) are shown. F, the impact of PGE1 after withdrawal. The kinetic changes of all CD45+ cells (left) and human CD45+CD34+ HSPCs (right) from day 32 to day 52 post-transplantation are shown. Data are means ± s.d. in one experiment (n = 5 for all data points). ns, not statistically significant; *, p<0.05; **, p<0.01; ***, p<0.001. See also Figure S6, S7 and Table S1.
In protocol A, when analyzed in the BM of NSG recipient mice right after completion of an 18-day treatment, PGE1 greatly diminished the ability of CML LSCs (derived from Patient 1) to generate human CD45+ grafted cells (Figure S6A–B). In particular, the frequency and numbers of human CD34+ CML stem/progenitor cells and human CD45+CD33+ CML myeloid cells were substantially reduced by PGE1 (Figure S6A–S6B). Another cohort of NSG recipients was analyzed 20 days after the 18-day treatment, and both human CD45+ grafted cells and CD45+CD34+ CML stem/progenitor cells showed an evident increase on day 52 over day 32 post-transplantation in the DMSO-treated group, suggesting continued engraftment of the transplanted CML LSCs (Figure S6C–S6D). In contrast, in the PGE1-treated group, total human CD45+ cells were decreased, and CD45+CD34+ CML stem/progenitor cells showed only a minimal increase (Figure S6C–D). In testing CML LSCs from Patients 2 and 3, although the engraftment efficiency varied, PGE1 treatment invariably inhibited the engraftment of CD34+ CML stem/progenitor cells (Figure S6E–F). These data indicate that PGE1 treatment suppresses the activity of human CML LSCs, and the effect lasts after withdrawal.
In protocol B, we allowed 30-day engraftment by CML LSCs before a 15-day DMSO or PGE1 treatment (Figure 7A). After completion of the treatment, human CD45+ cells and CD45+CD34+ CML stem/progenitor cells generated from CML LSCs (derived from Patient 4) were greatly diminished in frequency and numbers by PGE1 (Figure 7B–7C). Compared with day 30 (prior to the treatment), the DMSO-treated group exhibited continuous engraftment, in particular the CD45+CD34+ CML stem/progenitor cells; in contrast, PGE1 treatment prevented the continuous engraftment and even substantially reduced already grafted cells (Figure 7C). Similar results were obtained when CML LSCs from Patient 5 were tested (Figure S6G). Clinical studies have shown that approximately 40% of TKI-treated patients maintained remission for 24–36 months after TKI discontinuation (Mahon et al., 2010; Ross et al., 2013), suggesting that TKI may have contributed to LSC erosion. We therefore tested imatinib alone or in combination with PGE1 in protocol B. Imatinib treatment alone modestly reduced grafted human CD45+ and human CD45+CD34+ CML stem/progenitor cells (Figure 7B–7C), consistent with reported observations (Zhang et al., 2010). The PGE1+imatinib regimen showed stronger impact than imatinib alone but did not improve over PGE1 single therapy (Figure 7B–7C), demonstrating a potent effect of PGE1 in inhibiting CML LSCs. To further determine the molecular targets of PGE1 in human CML LSCs, we sort-purified the engrafted human CD45+CD34+ CML stem/progenitor cells from the BM of NSG recipients of CML LSCs derived from Patient 4 and 5 after completion of the treatment in protocol B. Compared with DMSO treatment, PGE1 caused ~10-fold reduction in the expression of FOSB and FOS, and a modest reduction in EGR1 expression (Figure 7D). These data suggest that PGE1 acts through AP-1 repression to inhibit the engraftment and maintenance of human CML LSCs.
Patient 6 had bi-lineage blast crisis of CML (Table S1). We tested Patient 6-derived CML LSCs following protocol A, and found that PGE1 similarly reduced the engraftment of total human CD45+, CD34+ CML stem/progenitor cells and CD33+ CML myeloid cells in the BM of NSG recipients right after completion of the treatment (Figure S7A–B). Another cohort of NSG recipients was analyzed 20 days after the treatment; in the DMSO-treated group, human CD45+ and CD45+CD34+ CML stem/progenitor cells showed a substantial increase in the BM on day 52 over day 32 post-transplantation, and such increases were remarkably dampened in the PGE1-treated group (Figure S7C–D). Similar results were obtained when Patient 6-derived CML LSCs were tested using protocol B (Figure S7E). We also tested CML LSCs from Patient 7 in an accelerated phase of CML using Protocol B. Although the engraftment efficiency by Patient 7 cells was lower than others, PGE1 showed stronger inhibition of engraftment of human CD45+ cells than imatinib, and the combination of imatinib and PGE1 did not improve over PGE1 single therapy (Figure S7F). Molecularly, PGE1 treatment inhibited the expression of FOSB, FOS and EGR1 genes in human CD45+ cells derived from Patients 6 and 7 (Figure S7G). These data suggest that PGE1 remains effective in suppressing CML LSCs from patients at more advanced stages and that PGE1-mediated AP-1 repression is a conserved regulatory circuit in targeting CML LSCs.
The engraftment efficiency of human CML LSCs in this study appeared to be lower than previously reported (Li et al., 2012; Zhang et al., 2010). The other studies expanded human CML CD34+ cells in vitro with various cytokines; these cells engrafted NSG recipients with higher efficiency at 4-weeks post-transplantation, but the engraftment declined sharply thereafter. We transplanted freshly isolated or cryopreserved human CML LSCs without in vitro expansion. In this setting, although the engraftment efficiency was lower at approximately 4-weeks post-transplantation, the transplanted human CML LSCs showed continued engraftment over time (Figure 7C, S6D, S6G, S7D–F). We confirmed the origin of engrafted patient cells by RT-PCR detection of BCR-ABL transcripts (Figure S7H). The advantages of our approach lie in two aspects: one is to provide a wider window to observe the therapeutic effect of PGE1 in vivo without being limited to the first 4 weeks of transplantation; the other is to better simulate drug delivery in treating patients, as opposed to pre-treating human CML LSCs in vitro before transplantation.
Additionally, we tested PGE1 on the engraftment by normal CD34+ HSPCs from healthy donors using protocol A. We did not find a discernible impact on engraftment of human CD45+ cells and CD45+CD34+ stem/progenitor cells right after the treatment (Figure 7E and S7I), or after PGE1 withdrawal (Figure 7F). These data highlight a specific effect of PGE1 on LSCs, while sparing normal hematopoiesis.
Discussion
The advent of TKI treatment has greatly improved CML therapy. Because CML LSCs are less sensitive to TKIs, CML is usually controlled rather than cured, highlighting an unmet clinical need. Developed from our previous finding that CML LSCs are more dependent on Tcf1 and Lef1 transcription factors than normal HSCs for self-renewal (Yu et al., 2016), this study delves into the concept of using Tcf1/Lef1-dependent genes as a therapeutic target in LSCs (Ashton et al., 2012). Through CMAP data mining, we identified PGE1 and its analogue misoprostol as potent agents that simulate key gene expression changes, mainly downregulation of Fosb and Fos, caused by Tcf1/Lef1 deficiency in LSCs. PGE1 greatly suppressed LSC activity in a murine CML model and a xenograft model of transplanted human CML LSCs, but did not detectably affect normal hematopoiesis. The effect of Tcf1/Lef1 ablation and PGE1 treatment appeared to be specific to CML, because neither affected AML LSC self-renewal or AML progression in a murine MLL-AF9 model (data not shown). The synergistic effect of combining PGE1 and TKI regimens makes PGE1 an ideal therapeutic candidate for treating CML. PGE1 (clinically known as alprostadil) and misoprostol are FDA-approved drugs. PGE1 has a vasodilative effect, which is used to treat erectile dysfunction, pulmonary hypertension and peripheral artery occlusive disease (Murali et al., 1992; Weiss et al., 2002). Misoprostol is a gastric anti-secretory agent with protective effects on the gastrointestinal mucosa. In this study, PGE1 was used at 2.5–5 mg/kg body weight (equivalent to 7–14 μM), a concentration close to clinically relevant levels at 5–10 μM (Weiss et al., 2004). These considerations suggest PGE1 and misoprostol can be repositioned from their conventional use to CML treatment, allowing an expedited translational application (Ashburn and Thor, 2004).
PGE1 stimulation partly simulated gene expression changes caused by Tcf1/Lef1 deficiency, particularly downregulation of Fosb and Fos. Importantly, PGE1-mediated AP-1 repression was conserved in human CML LSCs in the xenograft model. AP-1 factors are known to regulate cell differentiation, stress response, and tumorigenesis (Lopez-Bergami et al., 2010); however, their roles in HSCs or LSCs have not been extensively investigated (Rossi et al., 2012). We discovered that several AP-1 genes showed strong upregulation in CML LSCs compared with HSPCs in the murine CML model and CML patient samples, and significantly, forced expression of Fosb and Fos in LSCs conferred resistance to PGE1 treatment in the murine CML model. In osteosarcoma and endometrical carcinoma, Fos expression is associated with high-grade lesions and adverse outcome (Bamberger et al., 2001; Gamberi et al., 1998). Identification of Fosb and Fos as key targets to subvert CML LSCs may lead to discovery of additional therapeutic options, and these factors may be utilized as new biomarkers in assessing therapeutic efficacy and prognosis in CML treatment.
In spite of their structural similarity, PGE1 and PGE2 have clearly distinct biological effects. PGE2 has been reported to enhance human cord blood stem cell xenotransplants (Goessling et al., 2011), partly owing to its ability to activate β-catenin (Goessling et al., 2009). Despite some contentions over the role of β-catenin in normal HSPCs, it is consistently found that β-catenin is required for development and maintenance of LSCs in several leukemias (Heidel et al., 2012; Jamieson et al., 2004; Wang et al., 2010; Yeung et al., 2010; Zhao et al., 2007) and represents a useful drug target in cancer therapy (Anastas and Moon, 2013). COX inhibitors (e.g., indomethacin) are used to block production of E prostanoids, in particular PGE2, and to prevent β-catenin stabilization in LSCs (Heidel et al., 2012). By direct comparison with PGE1, however, indomethacin showed less profound beneficial effects on CML. PGE1 treatment did not cause β-catenin accumulation or downregulation of Tcf1 or Lef1 expression per se, and PGE1-mediated repression of Fosb and Fos in LSCs was not dependent on β-catenin. Therefore, the action of PGE1 is merely a simulation of part of gene expression changes caused by genetic ablation of Tcf1 and Lef1, and is not necessarily directly linked to the Tcf/Lef-β-catenin pathway. Further mechanistic analysis showed that PGE1 mainly acted on EP4 receptor and integrated signals from MEK, PI3K, and intracellular Ca2+ to achieve repression of AP-1 factors. Although PGE1 and PGE2 are produced from the same precursors, PGE1 treatment represents a novel and potent approach to subvert CML LSCs, independent of blocking the PGE2-β-catenin pathway.
TKI discontinuation studies demonstrate that a portion of TKI-treated CML patients remain in remission for up to 3 years, albeit long-term risk assessment is still necessary (Bansal and Radich, 2016). The majority of CML patients depend on lifetime TKI treatment, and substantial efforts have been devoted to identifying LSC-targeting drugs for use together with TKIs, aiming for a deeper molecular response and improving the outcome after TKI discontinuation. Agonists of peroxisome proliferator-activated receptor γ (PPARγ), such as pioglitazone, erode CML LSCs, and the efficacy of TKI combination therapy has been demonstrated in phase II clinical trials (Prost et al., 2015). MEK inhibitors (such as trametinib), protein phosphatase 2A-activating drugs (such as FTY720), and a blocking anti-CD27 monoclonal antibody (to interrupt CD27-CD70 interaction) reportedly target CML LSCs and help overcome TKI resistance (Ma et al., 2014; Neviani et al., 2013; Riether et al., 2015); the latter two approaches are indirectly linked to diminishing β-catenin activation. A proteomics screen found that p53 and c-Myc pathways act in concert to maintain CML, and dual targeting of p53 and c-Myc is proposed to be a TKI replacement, especially for TKI-refractory CML patients (Abraham et al., 2016). Although CML is considered an “easily treatable” leukemia, most patients are committed to life-long TKI dependence, and the risks of progression to an advanced stage or blast crisis remain for TKI responders. In addition, the detailed molecular events underlying transformation to more ominous leukemia are still poorly understood. Because of heterogeneity in genetic background and responses to TKI therapy among the CML patients, identifying different pathways to target CML LSCs is thus a highly worthy effort to obtain complementary therapeutic options for CML at various stages. For new drugs, safety and specificity to LSCs remain top priorities. Our unbiased transcriptome-based approaches identified PGE1 (alprostadil) and its analogue, misoprostol, as potent suppressors of LSCs. In the murine CML model, p210BCR-ABL-transformed HSPCs give rise to rapidly developing, fatal myeloproliferative neoplasm-like disease or sometimes acute lymphoblastic leukemia, without a preceding chronic phase. In spite of these caveats, PGE1 and misoprostol remained effective in greatly reducing LSCs and extended recipient survival with lasting effects. The efficacy of PGE1 on human CD34+ CML stem/progenitor cells was consistent for all CML patients at CP, and was also evident for accelerated and BC CMLs. Importantly, the safety of PGE1 and misoprostol in clinical use has been demonstrated. This study thus exemplifies a precision-medicine strategy for targeting a transcriptional program that specifically affects LSCs, minimizing risks of perturbing critical factors/pathways utilized by normal hematopoietic cells. The same principle is potentially applicable to other types of malignancies, including AMLs and solid tumors.
STAR METHODS
CONTANCT FOR REAGENTS
Further information and requests for resources and reagents should be directed to and will be fulfilled by, the Lead Contact, Hai-Hui Xue (hai-hui-xue@uiowa.edu). Gene-targeted mouse strains generated in the Xue lab can be distributed upon establishing proper material transfer agreements.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Non-obese diabetic (NOD)/SCID/IL2Rγnull (NSG), B6.SJL, Vav-Cre, and β-catenin (gene name Ctnnb1)-floxed mice were from the Jackson Laboratories. Vav-Cre transgene was used to ablate floxed genes in all hematopoietic cells including HSPCs. Tcf1 germline-targeted mice (Tcf7−/−) were from Hans Clevers (Hubrecht Institute, the Netherland) (Verbeek et al., 1995), and Lef1 conditionally targeted (Lef1FL/FL) mice were previously described (Yu et al., 2012b). Vav-Cre mice were crossed to Tcf7−/− and Lef1FL/FL strains to obtain Vav-Cre+Tcf7−/−Lef1FL/FL (called Tcf7−/−Lef1−/−) mice, which were used for HSPC and LSC transcriptomic analysis. EP4 (gene name Ptger4)-floxed mice were from Matthew D. Breyer (Schneider et al., 2004). Vav-Cre+EP4FL/FL (called EP4−/−) and Vav-Cre+β-cateninFL/FL (called βCat−/−) were used to determine the receptor and signaling pathway(s) utilized by PGE1. Both male and female mice were used between 6–12 weeks of age for all experiments. All mouse experiments were performed under protocols approved by the Institutional Animal Use and Care Committee of the University of Iowa.
Human samples
Peripheral blood samples from CML patients were obtained from the University of Iowa Lymphoid and Myeloid Tissue Repository of the Lymphoma Molecular Epidemiological Resource (IRB No. 200002042), and facilitated by the University of Iowa Tissue Procurement Core. The specimens were obtained after written informed consent and were de-identified. The protocol was approved by the University of Iowa Institutional Review Board. Patient characteristics are listed in Table S1.
Hematopoietic stem/progenitor cell (HSPC) product was obtained from a healthy donor after HSPC mobilization and leukapheresis through the University of Iowa Tissue Procurement Core. Human core blood or bone marrow CD34+ HSPCs were obtained from StemCell Technologies.
Cell lines
K562 CML cell line, which was established from a female CML patient, was obtained from ATCC. The cells were cultured in Iscove’s Modified Dulbecco’s Medium, supplemented with 10% fetal bovine serum, 100 unit/ml penicillin and 0.1 mg/ml Streptomycin, at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
METHOD DETAILS
Flow cytometry and cell sorting
Peripheral blood was collected from mice through sub-mandibular puncture. To collect BM cells, long bones (femurs and humeri) were harvested from humanely euthanized mice and flushed with RPMI medium containing 10% of FBS. BM cells were filtered through a 40 μm strainer to obtain single cell suspension. The cells were surface-stained using fluorochrome-conjugated antibodies. For detection of EP receptors, the following rabbit polyclonal antibodies were used: EP1 (bs-6316R-A647) and EP3 (bs-1876R-A647, both from Bioss Antibodies), with rabbit IgG (IDA1E-A647, Cell Signaling Technologies) as an isotype control; EP2 (ab92755-PE) and EP4 (ab133716-PE), with rabbit IgG (ab37409-PE) as an isotype control (all from Abcam). For intracellular staining of total β-catenin while preserving GFP detection in LSCs, the surface-stained cells were fixed by 4% formaldehyde, followed by permeabilization using BD Cytoperm Permeabilization Buffer Plus and staining with an anti-β-catenin antibody (A647 conjugate, L54E2 clone, Cell Signaling Technologies). For detection of active β-catenin, the surface-stained and fixed cells were stained first with a primary antibody against dephosphorylated β-catenin (Clone 8e4, KP31715, Calbiochem) and then detected using an Alexa Flour 647 goat anti-mouse SFX kit (A31626, Invitrogen/Life Technologies). The stained cells were analyzed on an LSRII with Violet or a FACSVerse flow cytometer (BD Biosciences). Data were analyzed using FlowJo software (V10, TreeStar). For cell sorting, surface-stained cells were sorted on BD FACSAria II or FACSAria Fusion cell sorter.
RNA-Seq
For transcriptomic analysis of HSPCs, Flt3− Lin−Sca1+c-Kit+ (Flt3− LSK) cells were sorted from the BM cells of WT or Tcf7−/−Lef1−/− mice. Because Tcf1/Lef1 deficiency modestly diminished HSC quiescence, the LSK frequency in Tcf7−/−Lef1−/− mouse BM cells was increased by approximately 2.5 fold on average (Yu et al., 2016). In WT mice, LT-HSCs are detected at ~0.01% of whole BM cells in WT mice. To capture a larger spectrum of transcriptomic changes caused by loss of Tcf1 and Lef1, we chose to use HSPCs from Tcf7−/−Lef1−/− mice that showed LT-HSC frequency ≥ 0.025% of whole BM cells. For transcriptomic analysis of LSCs, Lin− BM cells from WT or Tcf7−/−Lef1−/− mice were transduced with p210BCR-ABL twice in two consecutive days, and GFP+ LSKs cells were sorted as LSCs. The sorted LSCs from WT mice were incubated with DMSO, 10 μM PGE1, or 10 μM PGE2 for 6 hrs.
Total RNA was extracted from the sorted cells using Trizol LS reagent (Invitrogen/Life Technologies). After chloroform extraction, the aqueous phase was mixed with 2 volumes of ethanol and loaded onto a purification column in RNeasy Mini Kit (QIAGEN) for further purification. RNA quality was assessed using the Agilent Model 2100 Bioanalyzer. cDNA synthesis and amplification were performed using SMARTer Ultra Low Input RNA Kit (Clontech) starting with 10 ng of total RNA per sample following manufacturer’s instructions. cDNA was fragmented with Q800R sonicator (Qsonica) and used as input for NEBNext Ultra DNA Library Preparation Kit (NEB). Libraries were sequenced on Illumina’s HiSeq2000 in single read mode with the read length of 50 nt producing 60–70 million reads per sample. Sequence data in fastq format were generated using CASAVA 1.8.2 processing pipeline from Illumina.
Transcriptome analysis and bioinformatics analysis
Raw reads from RNA-seq were mapped to the reference mouse genome (release mm9) using Tophat (Trapnell et al., 2009). Only unique reads with fewer than 2 mismatches were used for downstream analyses. Transcripts were assembled using Cufflinks using mapped fragments outputted by Tophat. Refseq (mm9) was used for the annotation of known transcripts. Normalized transcript abundance was computed using Cufflinks and expressed as FPKM (Fragments Per Kilobase of transcripts per Million mapped reads). Gene-level FPKM values were computed by summing up FPKM values of their corresponding transcripts. RNA-seq data reproducibility was assessed by computing Pearson correlation of the number of reads for a given gene measured in biological replicates. WT and Tcf7−/−Lef1−/− HSPCs, PGE2-stimulated LSCs were measured in duplicates, and WT and Tcf7−/−Lef1−/− LSCs, DMSO- and PGE1-stimulated LSCs were measured in triplicates. Pearson’s correlation coefficients between each set of duplicate samples or between any pairwise comparison among triplicate samples were all >0.99, indicating high degree of reproducibility. Genes with zero read counts in all biological replicates were excluded. The RNA-Seq data were then used in Gene Set Enrichment Analysis (GSEA, software.broadinstitute.org/gsea/index.jsp).
The R package GenomicRanges was used to summarize read counts for genes. EdgeR (Robinson et al., 2010) was used to detect differentially expressed genes. P-values were corrected for multiple-testing using the method of Benjamini-Hochberg. Genes were considered differentially expressed if the average FPKM changes were ≥1.5 fold and the corrected P-values were less than 0.05. Functional annotation of the differentially regulated genes were performed using the online DAVID bioinformatics resources (https://david.ncifcrf.gov). Heatmaps were generated using Package ‘pheatmap’ from the R Project.
Chemical selection using the Connectivity MAP (CMAP) 02 resource
The CMAP 02 resource (https://portals.broadinstitute.org/cmap/) was queried using gene expression signatures based on RNA-Seq analysis of WT and Tcf7−/−Lef1−/− LSCs. The first set of genes encodes transcription factors (15 up- and 96 down-regulated), set #2 genes encode signaling molecules including protein kinases and phosphatases (9 up- and 87 down-regulated), and set #3 genes encode molecules involved in metabolism and chemotaxis (33 up- and 70 down-regulated) (Figure 1C and S2). The 4th set includes 444 of 946 genes that were downregulated by ≥ 2 fold and all 154 upregulated genes in Tcf7−/−Lef1−/− LSCs independent of functional annotation. From the “detailed results” and “permuted results” of CMAP output, we were interested in chemicals with positive “connectivity” and “enrichment” scores, which indicate that the chemicals can simulate, but not reverse, gene expression changes caused by Tcf1/Lef1 deficiency. We selected recurring chemicals with high “connectivity scores” in the “detailed results” and high “enrichment scores” in the “permuted results”.
CML model, LSC secondary transplantation, and therapeutic treatment
To model CML in mice, Lin− BM cells were infected with p210BCR-ABL retrovirus, and the infected cells containing 3,000–6,000 GFP+ LSKs were transplanted into lethally irradiated congenic mice along with 2 ×105 protector BM cells (Yu et al., 2012a; Yu et al., 2016). In some experiments, RV-mCherry expressing Fosb/Fos or Egr1 was used in co-infection. For secondary transplantation of LSCs, primary recipients were sacrificed on days 18–22 post-BMT to harvest BM cells, and all lineage-positive cells (including GFP+Mac1+ leukemic cells) were depleted to enrich LSCs. The Lin− BM cells containing 1.2 × 104 GFP+ LSK cells were then transplanted into a secondary CD45.1+ syngeneic recipients along with 2 × 105 protector BM cells for CML propagation.
For therapeutic treatments of the primary or secondary recipients, imatinib (Novartis) was given at 100 mg/kg body weight by oral gavage twice a day. PGE1 and PGE2 (Sigma-Aldrich, at 2.5–5 mg/kg body weight), misoprostol (Sigma-Aldrich, 5 mg/kg body weight), and indomethacin (Sigma-Aldrich, 4 mg/kg body weight) were given via i.p. injection once a day. Tamoxifen was administered via oral gavage at 0.2 mg/g body weight for 4 consecutive days. The recipients were evaluated daily for lethargy, splenomegaly, and signs of morbidity. The GFP+ Mac1+ leukemic burden was assessed in the peripheral blood cells, and LSCs in the BM were enumerated in terminal experiments.
Colony formation assay and chemical treatment
WT Lin− BM cells were cultured for 24 hrs, retrovirally infected with BCR-ABL retrovirus and cultured for another 24 hrs. GFP+ LSKs were then sorted into 24-well plates containing complete methylcellulose M3434 medium (StemCell Technologies) and various chemicals at 200 cells/well. The colony numbers were counted after eight days. Prostaglandin E1 (alprostadil, 10 μM) was from Sigma-Aldrich (St. Louise, MO), Spaglumic acid (15 μM) and Vigabatrin (30 μM) were from Prestwick Chemical (France). To determine the effect of these chemicals on normal HSPCs, BM LSKs from WT C57BL/6 mice were sorted and assays under the same condition as above except that the colony numbers were counted after seven days.
Limiting dilution assays for LSCs
The primary CML recipients were established and treated with DMSO or PGE1 during days 8–17 post-BMT. The BM cells were harvested on day 18 post-BMT, where the early termination time point was used to avoid CML-caused death in the DMSO-treated group. Graded numbers of the BM cells from DMSO-treated mice (containing 8,000, 1,600, 320, and 64 CD45.2+ GFP+ LSK cells) were mixed with 2 × 105 CD45.1+ WT BM cells (as radio-protectors) and transplanted into irradiated CD45.1+ secondary recipients. The corresponding numbers of total BM cells from PGE1-treated mice were used in the assay without relying on the frequency of phenotypic CD45.2+ GFP+ LSK cells. On 14 days post-BMT, the frequency of CD45.2+GFP+ leukemic cells in the PBCs of secondary recipients was determined, and those containing <1% leukemic cells were considered negatively engrafted. Frequency of functional LSCs was calculated according to Poisson statistics using the L-Calc software (StemCell Technologies).
Isolation of human CML CD34+ stem/progenitor cells and murine xenografts
Peripheral blood from CML patients were layered on Ficoll-Paque PLUS (1.077 g/ml) for gradient centrifugation. The layer of mononuclear cells was collected, washed, and enriched for CD34+ CML stem/progenitor cells by positive selection using Human CD34 Positive Selection Kit (StemCell Technologies). Depending on the numbers of recovered cells, 7×104 to 1×106 CD34+ CML stem/progenitor cells were transplanted into sub-lethally irradiated (2.25–2.5 Gy) NSG mice via tail-vein injection. The NSG recipients were treated with DMSO, PGE1 and/or imatinib, monitored and analyzed for engraftment of human CD45+ cells.
Cell stimulation and quantitative RT-PCR
For analysis of murine LSCs, GFP+ LSKs were sorted from p210BCR-ABL retrovirus-infected Lin− BM cells from WT, EP4−/−, or β-catenin−/− mice. The sorted cells were stimulated with 10 μM PGE1, PGE2, sulprostone, or butaprost (all from Sigma-Aldrich), or 35 μM misoprostol (Sigma-Aldrich) for 6 or 15 hrs. In some experiments, the sorted LSCs were pre-incubated for 30 min before 6-hr PGE1 stimulation with the following signaling pathway inhibitors, 1.5 μM H89 (IC50 for PKA, 0.14 μM), 20 μM PD98059 (IC50 for MEK, 2–7 μM), and 15 μM Ly294002 (IC50 for PI-3K, 1.4 μM), where concentrations of inhibitors were used at approximately 10 fold of the reported IC50 values shown in parentheses. 1,2-bis-(o-aminophenoxy) ethane-tetra-acetic acid tetra-(acetoxymethyl) ester (BAPTA-AM) was used at 50 μM for the 30 min pre-incubation to chelate intracellular Ca2+. K562 CML cells (ATCC, CCL243) were treated with PGE1, PGE2, misoprostol, or 5 μM BIO (EMD Millipore) for 6 or 24 hrs. The treated cells were harvested for RNA extraction, reverse-transcription, and quantitative PCR on ABI 7300 Real Time PCR System (Applied Biosystems). For calculation of relative gene expression after chemical treatment, the expression of each gene was first normalized to Hprt in the same sample, and the chemical-treated sample was then normalized to that of DMSO-treated sample. The primers are listed in Table S2.
For comparison of gene expression in human HSPCs and CML LSCs, the expression of each gene of interest was first normalized to GAPDH, and its expression in human HSPCs was set to 1, then its relative expression in CML LSCs was normalized accordingly. For comparison of gene expression in human CD45+ or human CML LSCs after chemical treatment in vivo in NSG recipients, the expression of each gene of interest was first normalized to GAPDH in the same sample, and the chemical-treated sample was then normalized to that of DMSO-treated sample.
QUATIFICATION AND STATISTICAL ANALYSIS
For comparison between two experimental groups (different genotypes or different treatments), the Student’s t-test was used, with a two-tailed distribution assuming equal sample variance. For multi-group comparisons, one-way ANOVA was used to first determine whether any of the differences between the means are statistically significant, followed by unpaired Student’s t-test to determine the statistical significance for a specific pair. For survival of CML recipients, statistical significance between different genetic and/or treatment conditions was assessed using log-rank test using Prism6 software. Statistical parameters, including numbers of samples or recipient mice analyzed (n), descriptive statistics (means and standard deviation) are reported in the figures and figure legends. P values of no more than 0.05 are considered statistically significant, the following asterisk marks are used to indicate the level of significance: *, p<0.05; **, p<0.01; ***, p<0.001. P values equal to or more than 0.05 are considered not statistically significant (marked as ‘ns’ or in actual p values).
DATA AND SOFTWARE AVALABILITY
The RNA-Seq data have been deposited at the NCBI Gene Expression Omnibus (GEO). The data on WT and Tcf7−/−Lef1−/− HSPCs are under accession number GSE60587, and those on WT and Tcf7−/−Lef1−/− LSCs, DMSO-, PGE1-, or PGE2-treated LSCs are under GSE92361.
Supplementary Material
Highlights.
Signature-based query of connectivity maps identifies PGE1 as a CML LSC inhibitor
PGE1 targets AP-1 factors and impairs activity of CML LSCs in mice and humans
The synthetic EP4 receptor agonist misoprostol also impairs CML LSC activity
PGE1 enhances the efficacy of imatinib treatment in CML xenograft models
Acknowledgments
We thank Matthew D. Breyer for permission to use EP4FL/FL mice, Richard M. Breyer (Vanderbilt University) and Pamela Harding (Henry Ford Health System) for sharing the EP4FL/FL mice. We thank I. Antoshechkin (Millard and Muriel Jacobs Genetics and Genomics Laboratory at the Caltech) for RNA-Seq, the University of Iowa Flow Cytometry Core facility (J. Fishbaugh, H. Vignes and G. Rasmussen) for cell sorting, Radiation Core facility (A. Kalen) for mouse irradiation, the University of Iowa Tissue Procurement Core facility (J. Galbraith and M. Knudson) for acquiring HSPC products, and Jessica C. Parrott (Iowa State University) for proof-reading of the manuscript. The Flow Cytometry Core Facility is supported by the Carver College of Medicine/Holden Comprehensive Cancer Center (the University of Iowa), the Iowa City Veteran’s Administration Medical Center, and the National Center for Research Resources of the NIH (1S10 OD016199). The Tissue Procurement Core is supported by an NCI award (P30CA086862) and funding from the Carver College of Medicine (University of Iowa). This study is supported in-part by grants from the NIH (AI112579, AI115149, AI119160 and AI121080 to H.-H.X., HG006130 to K.T., P50 CA097274 to S.R.L), and the Veteran Affairs BLR&D Merit Review Program (BX002903A to H.-H.X). H.H.X. is the founder of Cure-it LifeSciences, and this capacity has no roles in experimental design and data interpretation in this study. All other authors have no competing financial interests.
Footnotes
Author contributions
F.L. performed the experiments and analyzed the data with help from S.Y.; R.R., S.R.L. and M.L.G coordinated the acquisition of CML patient samples; B.H. and X.M. analyzed the RNA-Seq data under supervision of K.T.; H.H.X. and C.Z. conceived and supervised the overall study and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham SA, Hopcroft LE, Carrick E, Drotar ME, Dunn K, Williamson AJ, Korfi K, Baquero P, Park LE, Scott MT, et al. Dual targeting of p53 and c-MYC selectively eliminates leukaemic stem cells. Nature. 2016;534:341–346. doi: 10.1038/nature18288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nature reviews Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- Ashburn TT, Thor KB. Drug repositioning: identifying and developing new uses for existing drugs. Nature reviews Drug discovery. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- Ashton JM, Balys M, Neering SJ, Hassane DC, Cowley G, Root DE, Miller PG, Ebert BL, McMurray HR, Land H, et al. Gene sets identified with oncogene cooperativity analysis regulate in vivo growth and survival of leukemia stem cells. Cell Stem Cell. 2012;11:359–372. doi: 10.1016/j.stem.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamberger AM, Milde-Langosch K, Rossing E, Goemann C, Loning T. Expression pattern of the AP-1 family in endometrial cancer: correlations with cell cycle regulators. J Cancer Res Clin Oncol. 2001;127:545–550. doi: 10.1007/s004320100255. [DOI] [PubMed] [Google Scholar]
- Bansal A, Radich J. Is cure for chronic myeloid leukemia possible in the tyrosine kinase inhibitors era? Current opinion in hematology. 2016;23:115–120. doi: 10.1097/MOH.0000000000000224. [DOI] [PubMed] [Google Scholar]
- Bhatia R, Holtz M, Niu N, Gray R, Snyder DS, Sawyers CL, Arber DA, Slovak ML, Forman SJ. Persistence of malignant hematopoietic progenitors in chronic myelogenous leukemia patients in complete cytogenetic remission following imatinib mesylate treatment. Blood. 2003;101:4701–4707. doi: 10.1182/blood-2002-09-2780. [DOI] [PubMed] [Google Scholar]
- Breyer RM, Bagdassarian CK, Myers SA, Breyer MD. Prostanoid receptors: subtypes and signaling. Annu Rev Pharmacol Toxicol. 2001;41:661–690. doi: 10.1146/annurev.pharmtox.41.1.661. [DOI] [PubMed] [Google Scholar]
- Castellone MD, Teramoto H, Williams BO, Druey KM, Gutkind JS. Prostaglandin E2 promotes colon cancer cell growth through a Gs-axin-beta-catenin signaling axis. Science. 2005;310:1504–1510. doi: 10.1126/science.1116221. [DOI] [PubMed] [Google Scholar]
- Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. The Journal of clinical investigation. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamberi G, Benassi MS, Bohling T, Ragazzini P, Molendini L, Sollazzo MR, Pompetti F, Merli M, Magagnoli G, Balladelli A, et al. C-myc and c-fos in human osteosarcoma: prognostic value of mRNA and protein expression. Oncology. 1998;55:556–563. doi: 10.1159/000011912. [DOI] [PubMed] [Google Scholar]
- Goessling W, Allen RS, Guan X, Jin P, Uchida N, Dovey M, Harris JM, Metzger ME, Bonifacino AC, Stroncek D, et al. Prostaglandin E2 enhances human cord blood stem cell xenotransplants and shows long-term safety in preclinical nonhuman primate transplant models. Cell Stem Cell. 2011;8:445–458. doi: 10.1016/j.stem.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goessling W, North TE, Loewer S, Lord AM, Lee S, Stoick-Cooper CL, Weidinger G, Puder M, Daley GQ, Moon RT, et al. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136:1136–1147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SM, Vass JK, Holyoake TL, Graham GJ. Transcriptional analysis of quiescent and proliferating CD34+ human hemopoietic cells from normal and chronic myeloid leukemia sources. Stem Cells. 2007;25:3111–3120. doi: 10.1634/stemcells.2007-0250. [DOI] [PubMed] [Google Scholar]
- Heidel FH, Bullinger L, Feng Z, Wang Z, Neff TA, Stein L, Kalaitzidis D, Lane SW, Armstrong SA. Genetic and pharmacologic inhibition of beta-catenin targets imatinib-resistant leukemia stem cells in CML. Cell Stem Cell. 2012;10:412–424. doi: 10.1016/j.stem.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Chen Y, Douglas L, Li S. beta-Catenin is essential for survival of leukemic stem cells insensitive to kinase inhibition in mice with BCR-ABL-induced chronic myeloid leukemia. Leukemia. 2009;23:109–116. doi: 10.1038/leu.2008.262. [DOI] [PubMed] [Google Scholar]
- Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating A, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. The New England journal of medicine. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- Jiang X, Stuible M, Chalandon Y, Li A, Chan WY, Eisterer W, Krystal G, Eaves A, Eaves C. Evidence for a positive role of SHIP in the BCR-ABL-mediated transformation of primitive murine hematopoietic cells and in human chronic myeloid leukemia. Blood. 2003;102:2976–2984. doi: 10.1182/blood-2003-05-1550. [DOI] [PubMed] [Google Scholar]
- Lamb J. The Connectivity Map: a new tool for biomedical research. Nature reviews Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- Levin G, Duffin KL, Obukowicz MG, Hummert SL, Fujiwara H, Needleman P, Raz A. Differential metabolism of dihomo-gamma-linolenic acid and arachidonic acid by cyclo-oxygenase-1 and cyclo-oxygenase-2: implications for cellular synthesis of prostaglandin E1 and prostaglandin E2. The Biochemical journal. 2002;365:489–496. doi: 10.1042/BJ20011798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang L, Li L, Wang Z, Ho Y, McDonald T, Holyoake TL, Chen W, Bhatia R. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer cell. 2012;21:266–281. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bergami P, Lau E, Ronai Z. Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nature reviews Cancer. 2010;10:65–76. doi: 10.1038/nrc2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis TC, Naber BA, Roozen PP, Brugman MH, de Haas EF, Ghazvini M, Fibbe WE, van Dongen JJ, Fodde R, Staal FJ. Canonical wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell Stem Cell. 2011;9:345–356. doi: 10.1016/j.stem.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Ma L, Shan Y, Bai R, Xue L, Eide CA, Ou J, Zhu LJ, Hutchinson L, Cerny J, Khoury HJ, et al. A therapeutically targetable mechanism of BCR-ABL-independent imatinib resistance in chronic myeloid leukemia. Sci Transl Med. 2014;6:252ra121. doi: 10.1126/scitranslmed.3009073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon FX, Rea D, Guilhot J, Guilhot F, Huguet F, Nicolini F, Legros L, Charbonnier A, Guerci A, Varet B, et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. The Lancet Oncology. 2010;11:1029–1035. doi: 10.1016/S1470-2045(10)70233-3. [DOI] [PubMed] [Google Scholar]
- Malhotra S, Kincade PW. Wnt-related molecules and signaling pathway equilibrium in hematopoiesis. Cell Stem Cell. 2009;4:27–36. doi: 10.1016/j.stem.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murali S, Uretsky BF, Armitage JM, Tokarczyk TR, Betschart AR, Kormos RL, Stein KL, Reddy PS, Hardesty RL, Griffith BP. Utility of prostaglandin E1 in the pretransplantation evaluation of heart failure patients with significant pulmonary hypertension. J Heart Lung Transplant. 1992;11:716–723. [PubMed] [Google Scholar]
- Neviani P, Harb JG, Oaks JJ, Santhanam R, Walker CJ, Ellis JJ, Ferenchak G, Dorrance AM, Paisie CA, Eiring AM, et al. PP2A-activating drugs selectively eradicate TKI-resistant chronic myeloid leukemic stem cells. The Journal of clinical investigation. 2013;123:4144–4157. doi: 10.1172/JCI68951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost S, Relouzat F, Spentchian M, Ouzegdouh Y, Saliba J, Massonnet G, Beressi JP, Verhoeyen E, Raggueneau V, Maneglier B, et al. Erosion of the chronic myeloid leukaemia stem cell pool by PPARgamma agonists. Nature. 2015;525:380–383. doi: 10.1038/nature15248. [DOI] [PubMed] [Google Scholar]
- Riether C, Schurch CM, Flury C, Hinterbrandner M, Druck L, Huguenin AL, Baerlocher GM, Radpour R, Ochsenbein AF. Tyrosine kinase inhibitor-induced CD70 expression mediates drug resistance in leukemia stem cells by activating Wnt signaling. Sci Transl Med. 2015;7:298ra119. doi: 10.1126/scitranslmed.aab1740. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross DM, Branford S, Seymour JF, Schwarer AP, Arthur C, Yeung DT, Dang P, Goyne JM, Slader C, Filshie RJ, et al. Safety and efficacy of imatinib cessation for CML patients with stable undetectable minimal residual disease: results from the TWISTER study. Blood. 2013;122:515–522. doi: 10.1182/blood-2013-02-483750. [DOI] [PubMed] [Google Scholar]
- Rossi L, Lin KK, Boles NC, Yang L, King KY, Jeong M, Mayle A, Goodell MA. Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell. 2012;11:302–317. doi: 10.1016/j.stem.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller M, Schonheit J, Zimmermann K, Leser U, Rosenbauer F, Leutz A. Cross talk between Wnt/beta-catenin and Irf8 in leukemia progression and drug resistance. J Exp Med. 2013;210:2239–2256. doi: 10.1084/jem.20130706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Guan Y, Zhang Y, Magnuson MA, Pettepher C, Loftin CD, Langenbach R, Breyer RM, Breyer MD. Generation of a conditional allele of the mouse prostaglandin EP4 receptor. Genesis. 2004;40:7–14. doi: 10.1002/gene.20048. [DOI] [PubMed] [Google Scholar]
- Staal FJ, Luis TC, Tiemessen MM. WNT signalling in the immune system: WNT is spreading its wings. Nature reviews Immunology. 2008;8:581–593. doi: 10.1038/nri2360. [DOI] [PubMed] [Google Scholar]
- Sugimoto Y, Narumiya S. Prostaglandin E receptors. The Journal of biological chemistry. 2007;282:11613–11617. doi: 10.1074/jbc.R600038200. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, Oosterwegel M, Wilson A, MacDonald HR, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Wang D, Dubois RN. Eicosanoids and cancer. Nature reviews Cancer. 2010;10:181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, Zon LI, Armstrong SA. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss T, Fischer D, Hausmann D, Weiss C. Endothelial function in patients with peripheral vascular disease: influence of prostaglandin E1. Prostaglandins Leukot Essent Fatty Acids. 2002;67:277–281. doi: 10.1054/plef.2002.0429. [DOI] [PubMed] [Google Scholar]
- Weiss TW, Mehrabi MR, Kaun C, Zorn G, Kastl SP, Speidl WS, Pfaffenberger S, Rega G, Glogar HD, Maurer G, et al. Prostaglandin E1 induces vascular endothelial growth factor-1 in human adult cardiac myocytes but not in human adult cardiac fibroblasts via a cAMP-dependent mechanism. J Mol Cell Cardiol. 2004;36:539–546. doi: 10.1016/j.yjmcc.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Xue HH, Zhao DM. Regulation of mature T cell responses by the Wnt signaling pathway. Ann N Y Acad Sci. 2012;1247:16–33. doi: 10.1111/j.1749-6632.2011.06302.x. [DOI] [PubMed] [Google Scholar]
- Yeung J, Esposito MT, Gandillet A, Zeisig BB, Griessinger E, Bonnet D, So CW. beta-Catenin mediates the establishment and drug resistance of MLL leukemic stem cells. Cancer cell. 2010;18:606–618. doi: 10.1016/j.ccr.2010.10.032. [DOI] [PubMed] [Google Scholar]
- Yu S, Jing X, Colgan JD, Zhao DM, Xue HH. Targeting tetramer-forming GABPbeta isoforms impairs self-renewal of hematopoietic and leukemic stem cells. Cell Stem Cell. 2012a;11:207–219. doi: 10.1016/j.stem.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Li F, Xing S, Zhao T, Peng W, Xue HH. Hematopoietic and Leukemic Stem Cells Have Distinct Dependence on Tcf1 and Lef1 Transcription Factors. The Journal of biological chemistry. 2016;291:11148–11160. doi: 10.1074/jbc.M116.717801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Zhou X, Steinke FC, Liu C, Chen SC, Zagorodna O, Jing X, Yokota Y, Meyerholz DK, Mullighan CG, et al. The TCF-1 and LEF-1 Transcription Factors Have Cooperative and Opposing Roles in T Cell Development and Malignancy. Immunity. 2012b;37:813–826. doi: 10.1016/j.immuni.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Strauss AC, Chu S, Li M, Ho Y, Shiang KD, Snyder DS, Huettner CS, Shultz L, Holyoake T, et al. Effective targeting of quiescent chronic myelogenous leukemia stem cells by histone deacetylase inhibitors in combination with imatinib mesylate. Cancer cell. 2010;17:427–442. doi: 10.1016/j.ccr.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Blum J, Chen A, Kwon HY, Jung SH, Cook JM, Lagoo A, Reya T. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer cell. 2007;12:528–541. doi: 10.1016/j.ccr.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.