Abstract
MicroRNAs (miRNAs) are small, non-coding RNA species essential for the post-translational regulation of gene expression. Several miRNA have been proposed to contribute to Human immunodeficiency virus-1 (HIV-1) infection establishment, progression and latency. Among them, miR-29a seems to be of particular interest. The aim of this study was to investigate the association between miR-29a expression and immunologic and virologic markers of HIV infection progression in long-term antiretroviral-treated individuals. In a homogenous group of 165 young adults, with chronic HIV infection, parenterally acquired during childhood, the expression level of miR-29a was found to be inversely correlated with HIV viral load and the degree of immunosuppression, expressed by both CD4 cell count and the CD4/ CD8 ratio. There was a significant difference in miR-29a expression according to the patient’s response to treatment, with the lowest levels expressed by patients with treatment failure, defined as detectable viremia and CD4 <350 cells/mm3. No significant correlation was found between miRNA level and the nadir CD4 count or zenith HIV viral load. This study establishes the association between miR-29a expression and markers of HIV infection in long-term survivors, treatment-experienced patients, suggesting its potential use as an indicator for the on-treatment disease evolution.
Keywords: microRNA, miR-29a, HIV, CD4/ CD8, antiretroviral treatment
INTRODUCTION
MicroRNAs (miRNAs) are species of small, non-coding RNA that play a crucial role in post-translational regulation of gene expression. More than 60% of human protein-coding genes seem to be influenced by these small molecules [Selbach et al., 2008]. Since their discovery some 20 years ago, miRNAs have been involved in all normal processes of the cellular cycle [Cannell et al., 2008] but also have emerged as important players in oncogenesis, neurological disorders and, more recently, in infectious diseases [Li and Kowdley, 2012].
The discovery of the pivotal contribution of miR-122 to the development and progression of hepatitis C [Jangra et al., 2010; Dubin et al., 2014] and the subsequent manufacture of Miravirsen [Janssen et al., 2013], the first anti-miR molecule to be used in the treatment of a viral infection, has opened a new path for the control of viral diseases. Viral replication seems to be affected by cellular miRNA expression and in turn, production of viral proteins seem to change the normal cellular miRNA profile [Gottwein and Cullen, 2008]. Recent studies established a link between the host microRNA machinery and the pathophysiologic processes occurring during infection with hepatitis B virus [Mosca et al., 2014], enteroviruses [Ho et al., 2016], or viral encephalitis [Chen et al., 2016].
Although the role of miRNA during HIV infection has not been fully elucidated, several of these small, host-encoded regulators have been involved in the disease establishment, progression, and latency. Among them, miR-29 family appears to be one of the strongest candidates for viral replication regulation [Chiang et al., 2012]. miR-29a, a highly conserved cellular miRNA, expressed in human peripheral blood mononuclear cells, is targeting the 3′UTR region of the HIV nef gene, efficiently inhibiting viral replication [Ahluwalia et al., 2008; Nathans et al., 2009]. Most published studies focused on the role of miRNA in recent HIV infected, treatment-naïve patients, or in vitro models of acute HIV infection, some proposed miR-29a as an important factor in viral latency establishment [Patel et al., 2014]. The present study, examines the expression levels of miR-29a, in a homogenous cohort of long-term infected, heavily treated patients. The objective was to investigate the potential association between miR-29a expression and markers of the on-treatment HIV infection.
MATERIALS AND METHODS
Study Population
Peripheral blood samples were collected from 165 participants with chronic HIV infection. All HIV participants belong to the Romanian cohort of long-term survivors parenterally infected in childhood with HIV-1 subtype F1 [Kozinetz et al., 2000].
Written informed consent was obtained from all participants and the study was approved by the ethics committee of the Stefan S Nicolau Institute of Virology.
Measurement of HIV-1 RNA
HIV viral load was determined with a commercial nucleic acid amplification test (COBAS TaqMan HIV-1 Test Version 2.0, Roche Molecular Systems, Branchburg, NJ), with a lower detection limit of 20 copies of HIV RNA/ml and a linear range between 34 and 10,000,000 copies HIV RNA/ml. Values ≤34 copies/ml were classified as undetectable for analysis.
CD4+ and CD8+ T Lymphocyte Count
CD4+ and CD8+ T lymphocyte counts were performed by flow cytometry (BD Tritest CD4/CD8/CD3, Becton Dickinson, San Jose, CA).
PBMC Isolation
Peripheral blood mononuclear cells (PBMC) were isolated from fresh blood by density gradient centrifugation (Biochrom AG, Berlin, Germany) and dry pellets of 106 PBMC were stored at −80°C.
TaqMan-Based Real Time PCR Technique for miRNAs
miRNA was quantified by a real-time PCR Taq-Man® Assay (hsa-miRNA-29a, Applied Biosystems, Foster City, CA). Briefly, total RNA was isolated from PBMCs using the PureLink® RNA Mini Kit (Ambion, Life Technologies, Darmstadt, Germany) according to manufacturer’s protocol. Cellular RNA purity was evaluated spectrophotometrically at 230, 260, and 280 nm (Eppendorf BioSpectrometer®, Eppendorf, Hamburg, Germany). Reverse transcription was performed with TaqMan miRNA Reverse Transcription Kit, (Applied Biosystems) and real-time PCR was carried out on an Applied Biosystems 7,300 cycler (Applied Biosystems). The constitutively expressed RNU43 (Applied Biosystems) a small nucleolar RNA, member of the large C/D box family, thought to direct 2′-O-ribose methylation in ribosome biogenesis [Kiss, 2002], was used as an endogenous control (RNU43, Applied Biosystems).
miR-29a expression levels were estimated by single normalization 2(−ΔCT), where ΔCT =CT of miR-29a −CT of RNU43.
Statistical Analysis
All statistical analysis was performed using SPSS v.20.0.0 for Windows.
Spearman’s rho coefficient was calculated to assess correlations between non normally distributed continuous variables. Differences between groups were evaluated using Mann–Whitney (for two groups) or Kruskal–Wallis (when more than two groups were compared) tests. Univariate and multivariate regression was used to assess the independent prediction factors for miR-29a expression level. The independent relationship between selected variables—miR-29a expression, exposure time to antiretroviral drugs, nadir CD4 count, zenith HIV viral load, CDC clinical classification—and HIV treatment outcome—assessed by current CD4 count and HIV viral load—was estimated through odds ratios (OR) with 95% confidence interval (95% CI) obtained from multinomial logistic regression. A P-value less than 0.05 was considered statistically significant.
RESULTS
Characteristics of Study Participants
The demographic and clinical characteristics of study participants are presented in Table I. The median current plasma viral load was 2.29 log10 (interquartile range [IQR]: 0–3.98) HIV RNA copies/ml, and the current median CD4 count was 476 (IQR: 263–698) cells/mm3, although the median CD4 nadir was low 85 (IQR: 23–183.5) cells/mm3. All participants had similar age, duration of HIV infection, as well as similar exposure to previous antiretroviral treatments.
TABLE I.
Demographic and Clinical Characteristics of HIV-1 Infected Participants
| HIV-1 infected participants (n =165) | |
|---|---|
| Males (n, %) | 74 (44.8) |
| Age (median, IQR) | 24 (23.5–25) |
| Current HIV RNA | |
| Undetectable (<=34 copies/ml) | 100 (60.6) |
| 35–1000 copies/ml | 25 (15.2) |
| >1000 copies/ml | 40 (24.2) |
| Zenith HIV RNA log10, copies/ml | 5.06 (4.3–5.6) |
| Nadir CD4 count, cells/mm3 (median, IQR) | 85 (23–183.5) |
| Current CD4 count (n, %) | |
| <350 cells/mm3 | 49 (29.7) |
| >350 cells/mm3 | 116 (70.3) |
| Current CD8 count, cells/mm3(median, IQR) | 743 (535–1015.5) |
| CD4/CD8 ratio (n, %) | |
| Q1 0–25% | 38 (23) |
| Q2 25–50% | 43 (26.1) |
| Q3 50–75% | 40 (24.2) |
| Q4 75–100% | 42 (25.5) |
| Time since HIV diagnosis, years (median, IQR) | 23.98 (22.9–24.7) |
| CDC clinical staging (n, %) | |
| A | 3 (1.8) |
| B | 81 (49.1) |
| C | 81 (49.1) |
| Antiretroviral treatment duration, months (median, IQR) | 143.51 (98.7–173) |
Values are presented as number (%) or median (interquartile range).
Currently, 63.3% of participants receive an antiretroviral regimen consisting of two nucleoside reverse transcriptase inhibitors and a protease inhibitor (2 NRTI +1 PI), 12.9% receive an antiretroviral regimen consisting of two nucleoside reverse transcriptase inhibitors and a non-nucleoside reverse transcriptase inhibitors (2 NRTI +1 NNRTI), 17.7% a three class regimen (1 NRTI +1 NNRTI +1 PI), and 6.1% of participants receive other therapeutic regimens. 49.7% of participants had a clinical C class event (AIDS defining disease) in the past but at the moment of the study no opportunistic infections were recorded in the group. The rate of HCV co-infection was very low (2.4%), 31.5% participants were HBsAg chronic carriers, but only 5 (3.03%) subjects had signs of active HBV replication.
Correlations Between miR-29a Expression and Clinical Markers of HIV Infection
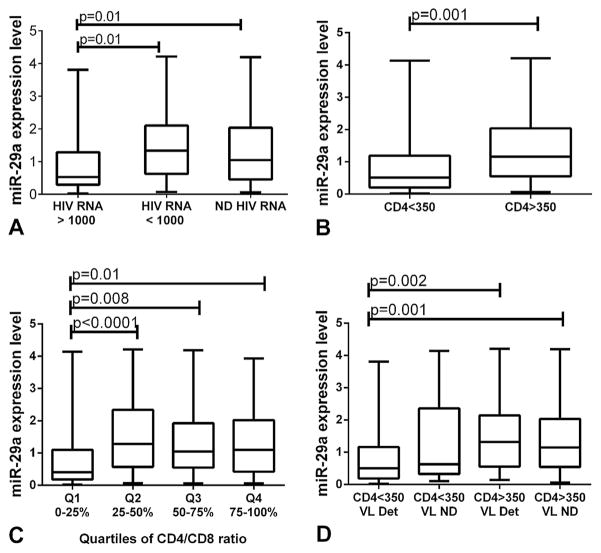
miR-29a expression was inversely correlated with HIV replication level, higher miR-29a level being associated with low viral load (P =0.002, rho =−0.36). There was a significant difference in miR-29a expression levels between participants with suppressed viral replication (undetectable HIV RNA) and those with high HIV viral load (>1000 copies HIV RNA/ml): median miR-29a expression level: 1.04, IQR: 0.45–2.03 versus 0.52, IQR: 0.28–1.29; P =0.01, as well as between participants with low (<1000 copies HIV RNA/ml) and high HIV viremia: median miR-29a expression level: 1.34, IQR: 0.62–2.10 versus 0.52, IQR: 0.28–1.29; P =0.01 (Fig. 1A).
Fig. 1.
Impact of miR-29a expression levels on viro-immunological markers of HIV infection. A: Differences in miR-29a expression levels between particular HIV viral load ranges. miR-29a levels were lower in patients with high levels of HIV RNA, compared to patients with not detectable (ND) (P =0.01) or low HIV RNA levels (P =0.01). B: Differences in miR-29a expression levels between CD4 categories. miR-29a levels were lower in patients with immunosuppression (CD4 <350) compared to patients with immune restoration (CD4 >350) (P =0.001). C: Differences in miR-29a expression levels between quartiles of CD4/CD8. miR-29a levels were lower in patients from Q1 (0–25%) compared with patients from Q2 (25–50%) P <0.0001, Q3 (50–75%) P =0.008, and Q4 (75–100%) P =0.01, respectively. D: Differences in miR-29a expression levels between HIV treatment outcome groups. miR-29a levels were lowest in patients with treatment failure—detectable HIV viral load (VL Det) and CD4 <350 and highest in patients with treatment success—not detectable HIV viral load (VL ND) and CD4 >350. The expression level of miR-29a differed significantly between the two classes of patients (P =0.001) and between patients with treatment failure and patients with immune reconstitution but virologic failure—CD4 >350 and detectable HIV viral load (P =0.002).
Significant correlations were found between miR-29a expression levels and the degree of immunosuppression, measured by CD4 cells number (P =0.003, rho =0.23) and the CD4/CD8 ratio (P =0.02, rho =0.17) but not with CD8 count (P =0.61, rho =0.04). Significant differences in miR-29a expression levels were recorded in participants with more than 350 CD4 cells/mm3, versus those with less than 350 cells/mm3: median miR-29a expression level: 1.15, IQR: 0.54–2.03 versus 0.51, IQR: 0.20–1.18; P =0.001 (Fig. 1B).
CD4/CD8 ratio was split into quartiles (Q1 =lowest quartile, 0–25%; Q2 =lower-middle quartile, 25–50%; Q3 =upper-middle quartile, 50–75%; Q4 =highest quartile, 75–100%) and Kruskal–Wallis test showed significant differences in miRNA expression levels across quartiles of CD4/CD8 (P =0.004) (Fig. 1C).
No correlation was found between miR-29a expression and nadir CD4, zenith viral load, or with the CDC clinical stage. miR-29a expression levels were neither correlated with gender, age nor the HBsAg status.
Linear regression was used to better assess the relation between miR-29a expression and HIV clinical markers—CD4 cells number, CD4/CD8 quartiles, and HIV viral load. In univariate models, miR-29a expression was significantly correlated with HIV viremia (OR: −0.12, 95% CI: −0.23, −0.02, P =0.02), and CD4 cell count (OR: 0.001, 95% CI: 0, 0.001, P =0.05) but not with the ordinal CD4/CD8 ratio (OR: 0.10, 95% CI: −0.04, 0.25, P =0.16). In a multivariate analysis including CD4 cells count, quartiles of CD4/CD8 ratio, and HIV viremia, only CD4 count remained independently significantly associated with miR-29a expression levels (OR: 0.29, 95% CI: 0, 0.002, P =0.05) while HIV RNA (OR: −0.19, 95% CI: −0.24, 0.03, P =0.13) and quartiles of CD4/CD8 ratio (OR: −0.17, 95% CI: −0.49, 0.14, P =0.28) have lost their significance.
miR-29a Association With the Response to HIV Treatment
Participants were divided into four groups according to their response to treatment: group 1—subjects with overall treatment failure (CD4 <350; detectable HIV plasma viral load), group 2—subjects with virologic success, but without immune restoration (CD4 <350; not detectable HIV viral load), group 3—subjects with immune restoration, but virologic failure (CD4 >350; detectable HIV plasma viral load), and group 4—subjects with overall treatment success (CD4 >350; not detectable HIV plasma viral load).
There was a significant difference in miR-29a expression between the four groups (P =0.005), participants with overall treatment failure (group 1) expressing the lowest levels of miR-29a (median miR-29a expression level: 0.15, IQR: 0.06–0.24), followed by those without immune restoration (group 2) (median miR-29a expression level: 0.25, IQR: 0.16–0.48), with immune restoration (group 3) (median miR-29a expression level: 0.65, IQR: 0.40–0.96), and participants with overall treatment success (group 4) (median miR-29a expression level: 0.90, IQR: 0.62–1.21) (Fig. 1D).
A multinomial logistic regression analysis was performed to test the association between the response to treatment and: miR-29a expression, the total time of exposure to antiretroviral drugs, the CDC clinical staging, nadir CD4 cells count and zenith HIV viral load. Treatment success (CD4 >350, ND HIV RNA) was used as the reference category. Overall the model fit was significant (χ2 =45.13; P <0.0001) and miR-29a expression (P =0.01), the total exposure time to ARV therapy (P =0.007) and nadir CD4 cells count (P =0.001) significantly contributed to the model but the CDC clinical staging (P =0.94), or zenith viral load (P =0.95) were not significant (Table II). Increased miR-29a expression levels were linked with higher odds of treatment success, one fold increase in miR-29a expression levels being associated with a 0.44 decrease in the relative risk of being in group 1 (overall treatment failure) versus group 4 (overall treatment success), 95% CI: 0.24–0.78, P =0.005.
TABLE II.
The Multinomial Logistic Regression Model
| Group 1 | Group 2 | Group 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | |
| miR-29a expression levels | 0.44 | 0.24–0.78 | 0.005 | 0.9 | 0.51–1.59 | 0.72 | 1.05 | 0.70–1.58 | 0.78 |
| Total exposure time to ARV (months) | 1.01 | 1–1.02 | 0.05 | 0.98 | 0.97–0.99 | 0.03 | 1 | 0.99–1.01 | 0.48 |
| Nadir CD4 cells count (cells/mm3) | 0.99 | 0.98–0.99 | 0.005 | 0.99 | 0.98–1 | 0.19 | 1 | 0.99–1 | 0.21 |
| Zenith HIV viral load (copies/ml plasma) | 0.96 | 0.56–1.65 | 0.9 | 0.88 | 0.53–1.45 | 0.62 | 1.05 | 0.65–1.69 | 0.83 |
| CDC clinical stage | |||||||||
| A | 0 | 0 | 0.99 | 0 | 0 | N/A | 1.28 | 0.05–31.7 | 0.87 |
| B | 0.56 | 0.21–1.47 | 0.24 | 0.92 | 0.28–2.98 | 0.89 | 0.86 | 0.35–2.11 | 0.74 |
| C | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
OR, odds ratio; ARVs, antiretrovirals.
DISCUSSION
The present study demonstrates a significant association between miR-29a expression levels and the virological and immunological markers of HIV-1 infection (plasma HIV viral load, CD4 cell count, and CD4/CD8 ratio) in antiretroviral treatment experienced patients. The strength of the study is the highly homogenous study group: all subjects being parenterally infected as children, with a particular and rather rarely encountered HIV subtype-F1, without significant comorbidities, and very similar in terms of age, length of HIV infection and of exposure to antiretroviral therapy.
We found a negative correlation between miR-29a expression level and HIV plasma viral load, with participants maintaining active viral replication having the lowest miR-29a expression levels. These find-ings are supported by a series of in vitro studies showing that inhibition of miR-29a expression enhances HIV-1 replication and infectivity [Hariharan et al., 2005; Nathans et al., 2009] as well as HIV activation from latent reservoirs [Patel et al., 2014] and some in vivo studies finding that low miR-29a expression associates with high viral load [Monteleone et al., 2015] in HIV-infected, treatment-naive individuals. Thus, miR-29a might be involved directly in controlling HIV replication, as has been hypothesized by a recent study that demonstrated enhanced HIV replication in human monocytic or lymphocytic cells only by inhibiting miR-29a expression without the need of additional cells activation [Patel et al., 2014].
miR-29a is part of the miR-29 family alongside miR-29b and miR-29c. All three miRNAs share an identical seed sequence and are highly similar so it is likely that all family members would exert some effect on HIV and the infection clinical markers.
Although miR-29b and miR-29c were recently connected with impaired HIV replication [Chiang et al., 2012; Adoro et al., 2015] or immunosuppression [Liu et al., 2015], miR-29a was reported to have the highest expression level both in monocytes and CD4 lymphocytes [Monteleone et al., 2015] probable exerting the strongest effect during HIV infection. Another study found evidence that HIV proteins reduce the cellular level of several anti-HIV miRNA, miR-29a and miR-29b among them, by incorporating them in exosomes secreted from the infected cells, and limiting the effects of RNA interference on viral replication [Aqil et al., 2014].
In the present study, miR-29a expression was directly correlated with markers of immunosuppres-sion: CD4 cells count that remained an independent predictor for miR-29a expression in the multivariate regression analysis and CD4/CD8 ratio, a marker associated by recent studies with high mortality and morbidity even in participants with apparent immune recovery [Mussini et al., 2015].
The connection between miR-29a expression level and the CD4 cell count was also found in previous studies [Witwer et al., 2012]. In addition to these independent correlations, miR-29a expression was found to be associated with the response to antiretroviral therapy—participants with treatment success (not detectable HIV viral load, >350 CD4 cells) having the highest miRNA expression. Moreover, a multinomial regression analysis that included miR-29a expression level, total exposure time to ARV, nadir CD4 cell count, CDC clinical stage and zenith viral load showed that high levels of miR-29a expression increase the chances of therapeutic success marking this miRNA as a possible additional marker for the treatment response.
A recent study reported higher levels of miR-29a in HIV-infected but asymptomatic participants (detectable viral load, and high CD4 count) compared with those with symptomatic disease (high viral load associated with CD4 cells depletion) in treatment naïve patients [Patel et al., 2014]. This association was also demonstrated during the natural progression of HIV infection, with down-regulation of miR-29a in immunosuppressed participants with high viral load [Houzet et al., 2008].
The role of miR-29 in HIV infection seems to be a complex one. Further studies could lead to a better understanding of the subtle mechanisms of host—virus interplay and if upregulation of this molecule could control the natural and on-treatment course of the HIV infection.
Acknowledgments
The authors want to thank Dr Gratiela Tardei for technical support in patients’ monitoring and Dr Camelia Sultana for critically reading the manuscript.
References
- Adoro S, Cubillos-Ruiz JR, Chen X, Deruaz M, Vrbanac VD, Song M, Park S, Murooka TT, Dudek TE, Luster AD, Tager AM, Streeck H, Bowman B, Walker BD, Kwon DS, Lazarevic V, Glimcher LH. IL-21 induces antiviral micro-RNA-29 in CD4T cells to limit HIV-1 infection. Nat Commun. 2015;6:7562. doi: 10.1038/ncomms8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahluwalia JK, Khan SZ, Soni K, Rawat P, Gupta A, Hariharan M, Scaria V, Lalwani M, Pillai B, Mitra D, Brahmachari SK. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology. 2008;5:117. doi: 10.1186/1742-4690-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aqil M, Naqvi AR, Mallik S, Bandyopadhyay S, Maulik U, Jameel S. The HIV Nef protein modulates cellular and exosomal miRNA profiles in human monocytic cells. J Extracell Vesicles. 2014:3. doi: 10.3402/jev.v3.23129. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannell IG, Kong YW, Bushell M. How do microRNAs regulate gene expression? Biochem Soc Trans. 2008;36:1224–1231. doi: 10.1042/BST0361224. [DOI] [PubMed] [Google Scholar]
- Chen Z, Ye J, Ashraf U, Li Y, Wei S, Wan S, Zohaib A, Song Y, Chen H, Cao S. MicroRNA-33a-5p modulates Japanese encephalitis virus replication by targeting eukaryotic translation elongation factor 1A1. J Virol. 2016;90:3722–3734. doi: 10.1128/JVI.03242-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang K, Sung TL, Rice AP. Regulation of cyclin T1 and HIV-1 replication by microRNAs in resting CD4+ T lymphocytes. J Virol. 2012;86:3244–3252. doi: 10.1128/JVI.05065-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin PH, Yuan H, Devine RK, Hynan LS, Jain MK, Lee WM. Micro-RNA-122 levels in acute liver failure and chronic hepatitis C. J Medical Virol. 2014;86:1507–1514. doi: 10.1002/jmv.23987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–387. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan M, Scaria V, Pillai B, Brahmachari SK. Targets for human encoded microRNAs in HIV genes. Biochem Biophys Res Commun. 2005;337:1214–1218. doi: 10.1016/j.bbrc.2005.09.183. [DOI] [PubMed] [Google Scholar]
- Ho BC, Yang PC, Yu SL. MicroRNA and pathogenesis of enterovirus infection. Viruses. 2016;8:11. doi: 10.3390/v8010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzet L, Yeung ML, de Lame V, Desai D, Smith SM, Jeang KT. MicroRNA profile changes in human immunodeficiency virus type 1 (HIV-1) seropositive individuals. Retrovirology. 2008;5:118. doi: 10.1186/1742-4690-5-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangra RK, Yi M, Lemon SM. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol. 2010;84:6615–6625. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- Kiss T. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- Kozinetz C, Matusa R, Cazazu A. The changing epidemic of pediatric hiv infection in romania. Ann Epidemiol. 2000;10:474–475. doi: 10.1016/s1047-2797(00)00082-x. [DOI] [PubMed] [Google Scholar]
- Li Y, Kowdley KV. MicroRNAs in common human diseases. Genomics Proteomics Bioinformatics. 2012;10:246–253. doi: 10.1016/j.gpb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MQ, Zhao M, Kong WH, Peng JS, Wang F, Qiu HY, Zhu ZR, Tang L, Sang M, Wu JG, Ho WZ, Zhou W. Antiretroviral therapy fails to restore levels of HIV-1 restriction miRNAs in PBMCs of HIV-1-infected MSM. Medicine. 2015;94:e2116. doi: 10.1097/MD.0000000000002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteleone K, Selvaggi C, Cacciotti G, Falasca F, Mezzaroma I, D’Ettorre G, Turriziani O, Vullo V, Antonelli G, Scagnolari C. MicroRNA-29 family expression and its relation to antiviral immune response and viro-immunological markers in HIV-1-infected patients. BMC Infect Dis. 2015;15:51. doi: 10.1186/s12879-015-0768-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca N, Castiello F, Coppola N, Trotta MC, Sagnelli C, Pisaturo M, Sagnelli E, Russo A, Potenza N. Functional interplay between hepatitis B virus X protein and human miR-125a in HBV infection. Biochem Biophys Res Commun. 2014;449:141–145. doi: 10.1016/j.bbrc.2014.05.009. [DOI] [PubMed] [Google Scholar]
- Mussini C, Lorenzini P, Cozzi-Lepri A, Lapadula G, Marchetti G, Nicastri E, Cingolani A, Lichtner M, Antinori A, Gori A, d’Arminio Monforte A. CD4/CD8 ratio normalisation and non-AIDS-related events in individuals with HIV who achieve viral load suppression with antiretroviral therapy: An observational cohort study. Lancet HIV. 2015;2:e98–e106. doi: 10.1016/S2352-3018(15)00006-5. [DOI] [PubMed] [Google Scholar]
- Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell. 2009;34:696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel P, Ansari MY, Bapat S, Thakar M, Gangakhedkar R, Jameel S. The microRNA miR-29a is associated with human immunodeficiency virus latency. Retrovirology. 2014;11:108. doi: 10.1186/s12977-014-0108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Witwer KW, Watson AK, Blankson JN, Clements JE. Relationships of PBMC microRNA expression, plasma viral load, and CD4+ T-cell count in HIV-1-infected elite suppressors and viremic patients. Retrovirology. 2012;9:5. doi: 10.1186/1742-4690-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]