Abstract
Red-shifted bioluminescence reporters are desirable for biological imaging. We describe the development of red-shifted luciferins based on synthetic coelenterazine analogs and corresponding mutants of NanoLuc that enable bright bioluminescence. One pair in particular shows superior sensitivity over other commonly used bioluminescence reporters in vitro and in vivo. This pair was adapted to develop a bioluminescence resonance energy-based Antares reporter called Antares2, which offers improved signal from deep tissues.
There is enormous interest in harnessing bioluminescence for ultrasensitive bioassays, drug screening, and optical imaging1–3. The photon fluxes of bioluminescent reporters are determined by the quantum efficiencies and catalytic rates of bioluminescence reactions. The slow catalysis of common luciferases often results in bioluminescence emission several orders of magnitude lower than that of fluorescence4. A recent advance was the development of NanoLuc luciferase, which, when paired with a synthetic furimazine substrate (Supplementary Fig. 1), generates intense blue bioluminescence much higher than other popular reporters5. However, the in vivo performance of NanoLuc, particularly in deep tissues, is poor since photons shorter than 600 nm interact strongly with mammalian tissues6. Because a significant portion of Firefly luciferase (FLuc) emission is longer than 600 nm, FLuc/d-luciferin remains among the top choices for deep-tissue bioluminescence imaging (BLI)7. Although both FLuc and d-luciferin have recently been engineered for longer wavelength emission, this desirable spectral shift is often offset by a substantially reduced total intensity7. Only a very few examples, such as the d-luciferin analogues CycLuc1 and AkaLumine-HCl8, 9, can modestly enhance FLuc bioluminescence at limited substrate concentration ranges.
It can be reasoned that an improved bioluminescence reporter may be derived by red-shifting the emission of NanoLuc. Early studies reported red-shifted coelentarazine (CTZ) analogs for Oplophorus luciferase, the ancestor of NanoLuc10, 11; however, their intensities were low and it is unclear whether the spectral shift could be extended to NanoLuc. Recent studies directly tested NanoLuc with several synthetic CTZ analogs but no significant red-shift was noted (Supplementary Note 1)12–14. Herein, we present novel luciferase-luciferin pairs based on other CTZ analogs and correspondingly re-engineered NanoLuc mutants for substantially brighter and red-shifted bioluminescence.
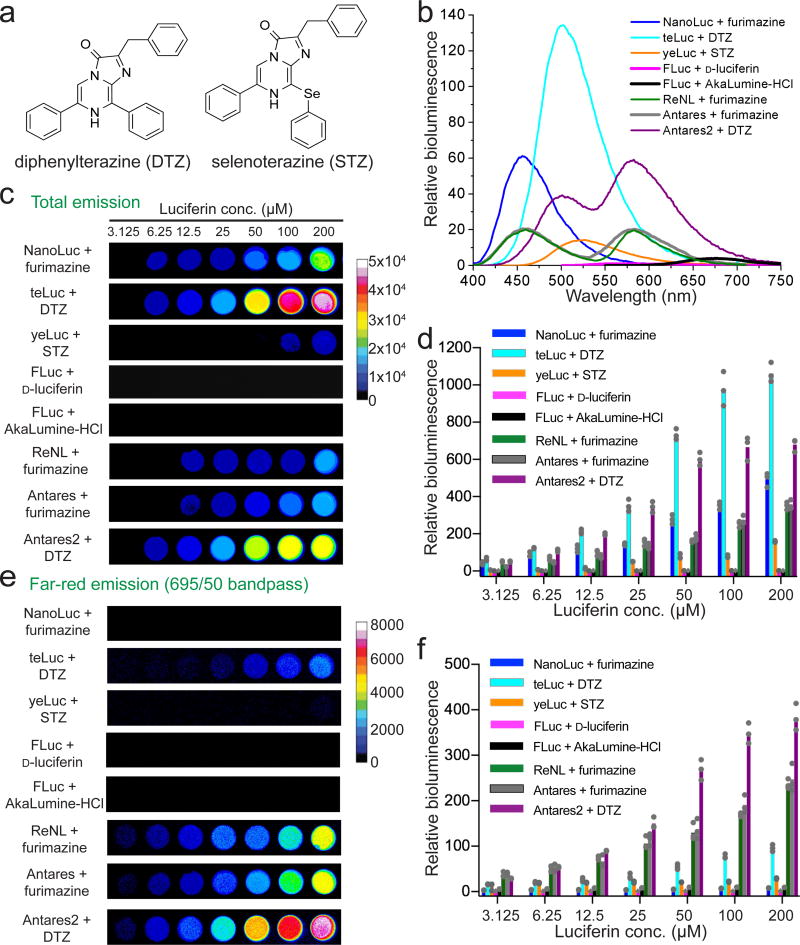
We prepared several CTZ analogs, including diphenylterazine (DTZ), which extends the CTZ conjugation system through an aromatic ring at C-8, and selenoterazine (STZ), which has a selenium heteroatom at C-8 (Fig. 1a, Supplementary Fig. 1, and Supplementary Notes 2,3). When assayed with NanoLuc, DTZ and STZ caused 44- and 71-nm red-shifts from furimazine, respectively, although with reduced brightness (Supplementary Table 1).
Figure 1. Bioluminescent reporters based on synthetic substrates and re-engineered luciferases.
(a) Chemical structures of diphenylterazine (DTZ) and selenoterazine (STZ). (b) Bioluminescence emission of purified luciferases (1 nM) with their corresponding luciferin substrates (30 µM). The spectra were normalized to peak emission of FLuc/d-luciferin. (c–f) Representative pseudocolored images (c,e) and quantifications (d,f) of luciferase-expressing HEK 293T cells in the presence of various luciferins. Images were acquired without a filter (c) or with a 695±25 nm NIR emission filter (e). Panels d and f are quantification results for Panels c and e, respectively. All values were normalized to the intensities of FLuc/d-luciferin (50 µM) under the same imaging conditions. The graphs show mean values and individual data points of three independent measurements.
We next engineered NanoLuc for enhanced activity toward either DTZ or STZ by simultaneously randomizing residues I44, I54, and I138 (Supplementary Fig. 2), since these positions have been reported to modulate the substrate preference of NanoLuc15. However, screening this library did not yield any useful mutants. We subsequently screened another library with full randomization at residues L18, D19, R162, and C164 based on their proximity to a putative substrate-binding site (Supplementary Fig. 3 and Supplementary Fig. 4)16. We also introduced random mutations across the gene using error-prone PCR. From this, we identified a NanoLuc mutant (NanoLuc-D19S/D85N/C164H) with a 5.7-fold enhancement of DTZ bioluminescence, which was designated “teLuc” for its teal emission peak at 502 nm (Table 1, Fig. 1b, and Supplementary Fig. 2). Additionally, we identified another NanoLuc mutant (yeLuc0.8 or NanoLuc-L18Q/D19A/S28T/C164S) displaying enhanced activity toward STZ. Further evolving yeLuc0.8 with three additional rounds of error-prone PCR and bacterial colony-based screening resulted in a variant with six additional mutations (yeLuc or yeLuc0.8-F1L/A14D/V27L/Q69R/R112Q/L142R). This new mutant shows an 11.5-fold overall enhancement of STZ bioluminescence over NanoLuc (Table 1, Fig. 1b, and Supplementary Table 1). teLuc/DTZ displayed sustained bioluminescence with an ~40 min half-life, whereas the fast decay of yeLuc/STZ emission has an ~5 min half-life (Supplementary Fig. 5). The apparent Michaelis constants (Km) for teLuc/DTZ and yeLuc/STZ were similar to those of NanoLuc/furimazine (Supplementary Fig. 6), while the quantum yield of teLuc/DTZ doubled from that of NanoLuc/furimazine (Supplementary Table 1). Compared to FLuc/d-luciferin, teLuc/DTZ emitted 13 times more photons at wavelengths longer than 600 nm (Fig. 1b and Table 1).
Table 1.
Photoluminescence properties of various luciferase-luciferin pairs.
| λmax (nm) |
Reporter size (kDa) |
Relative intensity a | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Proteinb | HEK 293T Cellsc | Mice | ||||||||
| Total | > 600 nm |
Intact (Total) |
Intact (695/50 nm) |
Lysate | Subcutaneously injected cellsd |
Hydrodynamic transfectione |
||||
| 0.3 µmol |
3.3 µmol | |||||||||
| NanoLuc + furimazine | 456 | 19 | 43.5 | 0.7 | 277 | 5 | 167 | 7.2 | NDf | NDf |
| teLuc + DTZ | 502 | 19 | 113 | 13 | 733 | 55 | 317 | 54 | 52 | 133 |
| yeLuc + STZ | 527 | 19 | 13 | 3.7 | 78 | 21 | 6.5 | 1.8 | 3.5 | NDf |
| FLuc + d-luciferin | 563 | 61 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4.2g |
| FLuc + AkaLumine-HCl | 677 | 61 | 3.4 | 8.7 | 2.3 | 10 | 11 | 1.3 | NDf | 9.3 |
| ReNL + furimazine | 459, 583 | 71.8 | 26 | 13 | 159 | 130 | 100 | NDf | NDf | NDf |
| Antares + furimazine | 456, 583 | 70.5 | 30 | 17 | 180 | 137 | 112 | 26 | 64 | 105 |
| Antares2 + DTZ | 501, 583 | 70.5 | 79 | 65 | 601 | 268 | 252 | 57 | 97 | 182 |
Intensity values normalized to FLuc/d-luciferin under comparable experimental conditions;
30 µM substrates and 100 pM proteins. Values are based on intensities integrated over the first 10 min post-substrate injection;
50 µM substrates and 2000 cells with an average transfection efficiency of ~ 70%;
Subcutaneous injection of two million HEK 293T cells and 100 µL of 100 µM of each substrate;
Intraperitoneal injection of each substrate. All intensity values are normalized to FLuc and 0.3 µmol d-luciferin;
Not determined.
The relative intensity increased to 11.7 when 10 µmol d-luciferin was intraperitoneally injected.
Recently, NanoLuc was fused to fluorescent proteins for BRET-based reporters, resulting in the two most red-shifted variants, Antares and ReNL17, 18. In particular, the absorbance of CyOFP1 in Antares overlaps better with teLuc/DTZ emission than with NanoLuc/furimazine (Supplementary Fig. 7). Replacing NanoLuc with teLuc in Antares derived a BRET-based Antares2 reporter emitting 3.8 times more photons above 600 nm than Antares and 65 times more than FLuc/d-luciferin (Fig. 1b and Table 1).
We further evaluated the bioluminescence of our new reporters using transiently transfected Human Embryonic Kidney (HEK) 293T cells. Under all conditions, the brightness of DTZ with teLuc or Antares2 was two to three orders of magnitude higher than that of FLuc/d-luciferin (Fig. 1c–f, Table 1, and Supplementary Figs. 8,9). These enhancements were more dramatic than the results observed in protein assays because of the high levels of teLuc and Antares2 in live cells (Supplementary Fig. 10) and better cell permeability of our synthetic substrates. teLuc and Antares2 also displayed improvements over NanoLuc, Antares, and ReNL (Table 1). teLuc emitted sustained bioluminescence with a half-life of >2 h in both intact cells and cell lysates, whereas yeLuc showed flash-type kinetics (Supplementary Figs. 11). Moreover, DTZ alone yielded very little background, leading to excellent signal-to-background ratios (Supplementary Fig. 12). Furthermore, DTZ elicited minimal cell toxicity at millimolar concentrations (Supplementary Fig. 13). In contrast, AkaLumine-HCl, furimazine, and STZ induced cell death within the tested substrate concentration range.
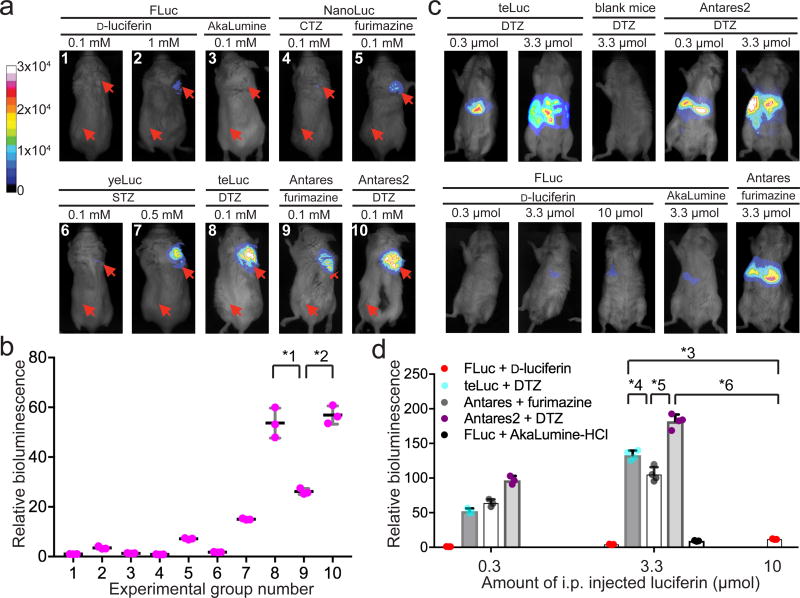
We next explored the use of our new reporters for in vivo BLI. To compare bioluminescence at superficial sites, we subcutaneously injected HEK 293T cells expressing individual luciferases near the right, dorsolateral trapezius region of unshaved BALB/c mice. For comparison, we also subcutaneously administered cells transfected with an empty vehicle to the same mice at the left, dorsolateral thoracolumbar region. These injections were followed up with an additional subcutaneous luciferin injection. At a 0.1 mM substrate concentration, teLuc/DTZ was ~54-fold brighter than FLuc/d-luciferin, and ~7.5-fold brighter than NanoLuc/furimazine (Fig. 2ab and Table 1). The brightness of Antares2 was comparable to that of teLuc, and no background bioluminescence was observed from sites injected with vehicle-transfected cells. To further evaluate the reporters for imaging deep-tissue targets, we utilized hydrodynamic transfection to express luciferase genes within the internal organs of mice. After intraperitoneal injections of 0.3 µmol individual luciferin substrates, teLuc/DTZ generated ~52-fold higher emission than FLuc/d-luciferin (Fig. 2cd and Table 1). With 3.3 µmol of each substrate (a dose recommended for AkaLumine-HCl9), teLuc/DTZ was ~32-fold brighter than FLuc/d-luciferin and ~15-fold brighter than FLuc/AkaLumine-HCl. Moreover, teLuc with 3.3 µmol DTZ was still ~12-fold brighter than FLuc with 10 µmol d-luciferin (equivalent to the standard ~150 mg/kg FLuc imaging condition). DTZ injections into untransfected BALB/c mice did not yield any background emission (Fig. 2c). When used for deep-tissue imaging, Antares2 shows an additional 35–90% signal increase over teLuc. Furthermore, the bioluminescence resulting from intraperitoneally injected DTZ displayed extended kinetics (Supplementary Fig. 14), suggesting that both teLuc and Antares2 are suitable for time-lapse BLI. To further demonstrate the use of these new bioluminescent reporters to detect low-abundant targets, we intravenously injected luciferase-expressing cells into unshaved BALB/c mice. Despite that ~5×105 FLuc-expressing cells were only marginally detectable, ~1×105 teLuc- or Antares2-labeled cells generated signals well above the background (Supplementary Fig. 15).
Figure 2. Bioluminescence imaging of luciferase-luciferin pairs at superficial sites and in deep tissues of live mice.
(a,b) Representative bioluminescence images (a) and quantitative analysis (b) of BALB/c mice with subcutaneously injected luciferase-expressing HEK 293T cells and 100 µL luciferin substrates at the indicated concentrations. The group numbers in panel b are aligned with those in panel a. Two injection sites (one for luciferase-expressing cells and one for empty vehicle controls) for each mouse are illustrated with red arrows. Intensity values were normalized to the intensity of FLuc/d-luciferin (0.1 mM) acquired under the same condition. The graphs show mean values and individual data points of three independent measurements. (c,d) Representative bioluminescence images (c) and quantitative analysis (d) of BALB/c mice, to which luciferase-coding plasmids were hydrodynamically delivered through tail vein injection, and luciferase substrates were intraperitoneally injected at 12 h post-plasmid injection. Intensity values were normalized to the intensity of FLuc/d-luciferin (0.3 µmol). Data are shown as individual data points and mean with s.d. (n = 4 for teLuc, Antares, and Antares2 with 3.3 µmol substrates, and n = 3 for all other groups). Unpaired two-tailed t-tests were used to compare teLuc/DTZ and Antares2/DTZ with FLuc/d-luciferin or Antares/furimazine (*1: P=0.0015; *2: P=0.0002; *3: P<0.0001; *4: P=0.0042; *5: P<0.0001; *6: P<0.0001), indicating the existence of a significant enhancement by teLuc and Antares2.
In summary, we have synthesized CTZ analogues with modifications at the C-8 position and re-engineered NanoLuc luciferase to catalyze these new substrates, thereby leading to brighter and more red-shifted bioluminescence. Although the water solubility of these CTZ analogs is worse than that of d-luciferin, the corresponding bioluminescence reactions with low amounts of the CTZ analogs are much brighter than FLuc with d-luciferin at saturated concentrations. In particular, teLuc/DTZ is one of the brightest bioluminescent systems showing robust performance in vitro, in cellulo, and in live mice. This will streamline a variety of applications to afford high sensitivity and reproducibility, and expanded the scope of BLI by allowing the use of less demanding instrumentation to track less abundant targets with higher spatiotemporal resolution. Moreover, our study demonstrates the general feasibility of co-engineering CTZ-utilizing luciferases and substrates for improved bioluminescence. Increased detection sensitivity is not the only advantage of teLuc/DTZ. The small sizes of NanoLuc and its derived teLuc and yeLuc (19 kDa), compared to FLuc (61 kDa) and some other luciferases, facilitate their use as fusion reporters, and to assemble viral vectors with limited packaging capacities. Moreover, NanoLuc forms a β-barrel structure that is amenable to various genetic and structural manipulations, such as split and fragment complementation19. Furthermore, their bioluminescence is independent of Mg2+, Ca2+, and ATP 5. All these suggest that teLuc may be an excellent scaffold for the development of bioluminescent biosensors compatible to other popular optogenetic tools20. We fused teLuc with CyOFP1 to derive Antares2, which further improved bioluminescence detection in deep tissues. Antares2 is an excellent bioluminescent reporter when the molecular size of the reporter is not important and when CyOFP1 does not caused spectral crosstalk. Overall, our work provides several robust bioluminescent reporters, including teLuc and Antares2, which are expected to have broad applications.
Editor’s summary
Red-shifted luciferins and corresponding mutants of NanoLuc enable brighter bioluminescence imaging in vitro, in cells, and in deep tissues of living mice alone and in the context of the newly developed Antares2 BRET reporter.
ONLINE METHODS
Materials and general methods
Synthetic DNA oligonucleotides were purchased from Integrated DNA Technologies. Restriction endonucleases were purchased from Thermo Scientific Fermentas. Accura high-fidelity DNA polymerase and EconoTaq DNA polymerase were purchased from Lucigen. Products of PCR and restriction digestion were purified by gel electrophoresis and Syd Laboratories Gel Extraction columns. Plasmid DNA was purified using Syd Laboratories Miniprep columns. DNA sequences were analyzed by Retrogen. d-luciferin was purchased from Thermo Fisher Scientific. Coelenterazine was purchased from Gold Biotechnology. Furimazine was purchased from Promega. AkaLumine was purchased from Wako Chemicals USA and acidified with one equivalent of HCl to derive AkaLumine-HCl. All other chemicals were purchased from Sigma-Aldrich, Fisher Scientific, or VWR, and used without further purification. Varian Inova 500 with a 5-mm triple resonance (1H/ 13C/ 15N) triple axis gradient probe at the UCR ACIF NMR Facility was used to record all NMR spectra. Chemical shift (δ) is given in parts per million relative to 1H (7.24 ppm) and 13C (77.23 ppm) for CDCl3; and 1H (2.50 ppm) and 13C (39.5 ppm) for DMSO-d6. Splitting patterns are reported as s (singlet), bs (broad singlet), d (doublet), t (triplet), dd (doublet of doublets), m (multiplet). Coupling constant (J) is given in Hz. ESI-MS was run on an Agilent LC-TOF system by direct infusion. A Gilson PLC 2020 Purification System coupled with an Agela Venusil XBP C18 HPLC Column (10 µM, 100 Å, 10×150mm) was used for preparative reverse-phase HPLC purifications. BALB/c mice obtained from the Jackson Laboratory (Cat. # 000651) were used for in vivo experiments. Animals were maintained and treated in standard conditions complied with all relevant ethical regulations and all animal procedures were approved by the UCR Institutional Animal Care and Use Committee. Images were analyzed using the Fiji image analysis software21, Microsoft Excel and GraphPad Prism were used to analyze data and prepare figures. Other general information is available from “Life Sciences Reporting Summary”.
Synthesis of DTZ and STZ
Methods for preparing DTZ and STZ are described in the Supplementary Information.
Construction of plasmids and libraries
Polymerase chain reaction (PCR) was used to amplify all genetic elements using various synthetic oligonucleotide pairs (see Supplementary Table 2). The gene for NanoLuc was purchased from Integrated DNA Technologies as a gBlock, and further amplified with oligos XhoI-NL-F and NL-R-HindIII. The product was digested with XhoI and HindIII restriction enzymes, and then ligated into a predigested, compatible pBAD/His B plasmid (Life Technologies). To create a gene library with randomization at residues 44, 54, and 138, oligo pairs XhoI-NL-F and I44I54-NNK-R, I44I54-NNK-F and I138-NNK-R, and I138-NNK-F and NL-R-HindIII were utilized to amplify three individual fragments from NanoLuc. The resultant three fragments were purified by agarose gel electrophoresis and utilized as templates for assembly in subsequent PCR reactions by using oligos XhoI-NL-F and NL-R-HindIII. The assembled full-length fragment was subsequently digested with XhoI and HindIII restriction enzymes and ligated into pBAD/His B. To create a library with randomization at residues 18, 19, 162, and 164, a similar multi-step overlap PCR strategy was used. XhoI-NL-F and L18D19-NNK-R, L18D19-NNK-F and R162C164-NNK-R were utilized to create two fragments, which were next assembled by using XhoI-NL-F and NL-R-HindIII. The resultant gene fragment was also treated with XhoI and HindIII, and ligated into a predigested pBAD/His B plasmid. To introduce random mutations across the NanoLuc gene, Taq DNA polymerase was used in all reactions with 0.2 mM MnCl2 added to promote amplification errors. To create mammalian expression plasmids, HindIII-NL-F-Koz and NL-R-XhoI (or NL-R-164H and NL-R-164S) were used to amplify NanoLuc and NanoLuc mutants. The products were treated with HindIII and XhoI restriction enzymes and ligated into a predigested compatible pcDNA3 plasmid (Life Technologies). The Firefly luciferase (FLuc) gene and the FLuc2 gene were amplified from a pGL2-GAL4-UAS-Luc plasmid (Addgene Cat # 33020) and pGL4.17 (Promega), respectively, by using FLuc-F and FLuc-R or Luc2-F-HindIII-Kozak and Luc2-Myc-R, and inserted into pcDNA3 between HindIII and XhoI sites. Ant-HindIII-F-koz and Ant-XhoI-R were used to amplify a fragment contains Antares gene from pNCS-Antares (Addgene Cat # 74279). The product was digested with HindIII and XhoI and then ligated into a predigested pcDNA3 plasmid as mentioned above. To replace the NanoLuc fragment in Antares with teLuc, oligo pairs Ant-HindIII-F-koz and Te19DtoS_R, Te19DtoS_F and Te85DtoN_R, Te85DtoN_F and Te164CtoH_R, Te164CtoH_F and Antares_R_HindIII were utilized to amplify four individual fragments from pNCS-Antares. The resultant four fragments were used as templates and assembled via PCR reactions by using oligo pairs Ant-HindIII-F-koz and Ant-XhoI-R. The product was digested with HindIII and XhoI, purified by agarose gel electrophoresis, and ligated into a predigested pcDNA3 plasmid to give pcDNA3-Antares2. To construct a bacterial expression plasmid for Antares2, oligos Antares_F_XhoI and Antares_R_HindIII were used to amplify the whole gene from pcDNA3-Antares2, which was subsequently digested with XhoI and HindIII and inserted into a compatible, predigested pBAD/HisB plasmid. To construct mammalian expression plasmids with C-terminal Myc tags, the aforementioned forward primers for the construction of pcDNA3 plasmids were individually paired with NL-Myc-R, teLuc-Myc-R, yeLuc-Myc-R, FLuc-Myc-R, Luc2-Myc-R, or Antares-Myc-R to amplify the corresponding luciferase genes, which were further extended using Myc-R-XhoI paired with the corresponding forward primers. To build bicistronic plasmids containing a self-cleaving porcine teschovirus-1 P2A peptide (GSGATNFSLLKQAGDVEENPGP), oligos P2A-FLuc-F and FLuc-R were used to amplify the FLuc gene, and teLuc-P2A-R, NLuc-P2A-R, and P2A-ext-R were paired with the corresponding forward primers to amplify teLuc or NanoLuc. These fragments were further assembled using overlap PCR to create NanoLuc-P2A-FLuc or teLuc-P2A-FLuc. The products were digested with HindIII and XhoI, purified by agarose gel electrophoresis, and ligated into a predigested pcDNA3 plasmid. All ligation products were used to transform Escherichia coli DH10B electrocompetent cells, which were next plated on LB agar plates supplemented with ampicillin (100 µg/mL). Additional L-arabinose (0.02%, w/v%) was supplemented to induce protein expression for direct bioluminescence imaging of bacterial colonies.
Library screening
DH10B cells containing NanoLuc mutants were plated on LB agar plates supplemented with ampicillin (100 µg/mL) and L-arabinose (0.02%, w/v%) and incubated at 37°C overnight to form bacterial colonies. Agar plates were left at room temperature for another 6 h, followed by bioluminescence imaging using a luminescence dark box (Stanford Photonics) equipped with a Pixis 1024B CCD camera (Princeton Instruments). Digital images were acquired after spraying ~100 µL of 200 µM substrates to each agar plate, and next processed with the Fiji image analysis software21 to derive bioluminescence intensities of individual colonies. For each compound, the brightest fifty colonies from a total of ~20,000 colonies were chosen and inoculated in 5 mL liquid LB broth containing ampicillin (100 µg/mL) and L-arabinose (0.02%, w/v%). After overnight growth at 37°C and 250 rpm, the cultures were moved onto a shaker at room temperature for another 6 h. Cells were next diluted with assay buffer (1 mM CDTA, 0.5% Tergitol NP-40, 0.05% Antifoam 204, 150 mM KCl, 100 mM MES, pH 6.0, 1 mM DTT, and 35 mM thiourea) to OD600 = 0.1. Next, bioluminescence activities of individual samples were measured in white 96-well plates (Costar 3912) on a Synergy Mx Microplate Reader (BioTek) after directly injecting substrates (final concentration of 30 µM). Kinetics were followed for 1-s signal integration every 60 s for a total of 40 min. Mutants showing exceptionally high bioluminescence activities and extended kinetics were chosen for sequencing, protein preparation, and other additional characterization.
Luciferase expression and purification
Luciferases were expressed and purified as His6-tagged fusion proteins. DH10B cells containing corresponding pBAD plasmids were grown in 5 mL LB starter cultures containing ampicillin (100 µg/mL) at 37°C and 250 rpm overnight. Each saturated overnight culture was diluted 100-fold into 2YT media containing the appropriate antibiotics and grown under the same conditions. When OD600 reached 0.7–0.9, the expression culture was induced with L-arabinose (0.2%, w/v%) and incubated at room temperature with shaking at 250 rpm for another 16 h. Cells were harvested by centrifugation at 4700 rpm for 15 min and lysed by sonication. The resulting cell lysate was clarified by centrifugation at 18,000 rpm for 30 min at 4 °C. The supernatant was incubated with Ni-NTA agarose beads (Pierce) at 4°C for 2 h. Agarose beads loaded to a plastic column were sequentially washed with 10 mL of wash buffer 1 (pH 8.0, 50 mM Tris HCl, 20 mM Imidazole, 300 mM NaCl, 1 mM DTT) and 10 mL of wash buffer 2 (pH 8.0, 50 mM Tris HCl, 50 mM Imidazole, 300 mM NaCl, 1 mM DTT), followed by elution with elution buffer (pH 8.0, 50 mM Tris HCl, 300 mM Imidazole, 300 mM NaCl, 1 mM DTT). Proteins were buffer-exchanged into Tris-HCl (50 mM, pH 7.4) containing 1 mM DTT using Thermo Scientific Snakeskin dialysis tubing, and next concentrated using 3-kDa Amicon Ultra Centrifugal Filters (EMD Millipore). Protein concentrations were determined using the Pierce Coomassie Bradford Protein Assay Kit (Thermo Fisher). For storage, glycerol was added to a final concentration of 50% (v/v) and the resultant mixtures were kept at −20°C.
In vitro bioluminescence characterization
A Synergy Mx Microplate Reader (BioTek) was used for all in vitro bioluminescence characterizations. For kinetics measurements, no emission filter or monochromator was used. 50 µL of luciferin substrates in the assay buffer (1 mM CDTA, 0.5% Tergitol NP-40, 0.05% Antifoam 204, 150 mM KCl, 100 mM MES pH 6.0, 1 mM DTT, and 35 mM thiourea) was injected into the wells of white 96-well plates containing 50 µL of pure enzymes in the same assay buffer. The final concentrations of all enzymes and substrates were 100 pM and 30 µM, respectively. Measurements were taken every 60 s post-injection (1-s integration and 10-s shaking during intervals). FLuc bioluminescence assays were performed similarly, except the assay buffer contained 30 mM MOPS, pH 7.0, 1.5 mM ATP, and 5 mM MgSO4. To derive values for apparent Michaelis constants (Km), substrate concentrations varied from 0.78 to 50 µM and peak bioluminescence intensities at individual substrate concentrations were used to fit the Michaelis-Menten equation. To convert relative arbitrary unit (RLU) to the number of photons, the instrument was calibrated by determining the chemiluminescence of 50–800 nM luminol (QY = 1.23%)22 in the presence of 100 nM horseradish peroxidase and 2 mM hydrogen peroxide in 0.1 M K2CO3 aqueous solution for a total volume of 200 µL. To determine the quantum yields of bioluminescent reactions, 0.01 nmol of each luciferin in 50 µL PBS was injected into 150 µL PBS containing 0.5 nmol of the corresponding luciferase. 1.5 mM ATP and 5 mM Mg were supplemented for the reaction between FLuc and d-luciferin. Signals were integrated until the substrates were completely consumed. Integrated total photos were divided by the total numbers of substrate molecules to derive the quantum yields of bioluminescence reactions. The validation of our results was confirmed by measuring the quantum yields of the Renilla luciferase mutant RLuc8 in the presence of CTZ (QY = 6.9%)23 and FLuc in the presence of d-luciferin (QY = 41±7.4%)24. kcat values for individual enzymes were determined using the equation: kcat = Imax/(QY*[E]), where Imax is the maximal luminescence intensity from the fitting of the Michaelis-Menten equation and [E] is the enzyme concentration. A Tecan M1000 Pro Plate Reader was used to record emission spectra. 50 µL of individual substrates (60 µM) in assay buffers were injected into 50 µL of 2 nM pure enzymes, and the bioluminescence spectra were collected with 0.1-s integration and 1-nm increments from 400 to 750 nm.
Mammalian cell culture and transfection
HEK 293T (purchased from ATCC and tested for mycoplasma by PCR), which is one of the most widely used and readily transfectable cell lines, was used for all tissue culture transfections. HEK 293T cells were cultured at 37°C with 5% CO2 in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Transfection mixtures were prepared with 3 µg of plasmid DNA and 9 µg of PEI (polyethylenimine, linear, MW 25 kDa) in DMEM and incubated for 20 min at room temperature. The medium was first aspirated, and subsequently, the transfection mixtures were added to cells at 70% confluency on 35-mm culture dishes seeded one day prior to transfection. Incubation lasted for 3 h at 37°C. Fresh DMEM containing 10% FBS was next utilized to replace the transfection mixtures. After incubation for another 24 h at 37°C in a CO2 incubator, the medium was removed and cells were collected and resuspended in Dulbecco's phosphate-buffered saline (DPBS).
Evaluation of the cytotoxicity of luciferins
Cell viability was determined using RealTime-Glo™ MT Cell Viability Assay (Promega) after incubation of HEK 293T cells with individual luciferin substrates for 24 h at 37°C. Cell morphology was further evaluated using microscopy. Briefly, HEK 293T cells in 35-mm culture dishes were washed twice with 400 µL DPBS pre-warmed to 37°C, and further incubated in 500 µL DPBS containing 20 µg/mL propidium iodide (PI), a fluorescent stain for dead cells, at room temperature for 15 min under light-shielded conditions. Cells were further rinsed with pre-warmed 300 µL DPBS three times to remove residual PI. Cells in 300 µL DPBS were next analyzed on a Motic AE31 microscope equipped with a 100 W Short-Arc Mercury lamp, a 540/25 nm excitation filter, a 565 nm longpass dichroic mirror, and a 605/55 nm emission filter.
Bioluminescence measurements in HEK 293T cells and cell lysates
The number and density of cells in DPBS suspension were determined using a hemocytometer. Cells were next diluted in DPBS to gain the needed numbers in each 100 µL solution. To use the luminescence dark box to directly image cells, we added luciferase-expressing HEK293T cells (2000 cells per well with ~70% transfection efficiency) and the corresponding luciferin substrates into wells of a white-wall, 96-well plate. Bioluminescence was imaged using a Pixis 1024B cooled CCD camera equipped with a 50-mm f/0.95 lens at one min post-substrate addition. The camera exposure time was 1 s and the field of view was 6×6 inches. A 695BP50 (Omega Optical) filter was utilized to acquire far-red emission. All images were analyzed using the Fiji image analysis software21. Cell lysates were prepared by sonication. Without further separation, luciferin substrates in 100 µL assay buffers were added to initiate bioluminescence reactions.
Western Blot
To examine the protein levels of tested luciferases in mammalian cells, we transfected HEK 293T cells with mammalian expression plasmids harboring C-terminal, c-Myc tagged luciferase genes. Cell lysates were prepared using CelLytic M Cell Lysis Buffer (Sigma-Aldrich) supplemented with cOmplete™ Protease Inhibitor Cocktail (Promega). Cell lysates were clarified by centrifugation at 14,000× g for 10 mins. The Bradford assay was used to determine total protein concentrations in the lysates. 10 µg of total proteins were loaded to each lane and resolved on a 12% SDS-PAGE gel. Proteins were next transferred to a nitrocellulose membrane at 40 V for 3 h. The membrane was blocked with 5% bovine serum albumin (BSA) in 1× phosphate buffered saline with Tween® 20 (PBST) for 1 h, and then incubated with either an anti-c-Myc (Santa Cruz Biotechnology, sc-40; 1:3000 dilution) or an anti-β-actin antibody (ThermoFisher Scientific, PA1-183; 1:10000 dilution) in PBST containing 3% BSA at 4°C overnight. After washing four times with PBST, the corresponding horseradish peroxidase (HRP) conjugated secondary antibodies (goat anti-mouse IgG, ThermoFisher Scientific, 31430; or goat anti-rabbit IgG, Sigma-Aldrich, A0545; both at 1:10000 dilution) were added and incubated for 1 h at room temperature. Membranes were visualized with Amersham ECL Western Blotting Detection Reagent (GE Healthcare) on a Bio-Rad Gel Doc XR System.
Bioluminescence imaging of HEK 293T cells at superficial sites on live mice
BALB/c mice on a 37°C electronic heat pad were anesthetized using 2% isoflurane in 100% oxygen with a flow of 0.5 L/min. We subcutaneously injected 2 million HEK 293T cells transfected with luciferase genes and resuspended in 100 µL PBS near the right, dorsolateral trapezius region of unshaved BALB/c mice; and another 2 million cells transfected with an empty vehicle vector and also resuspended in 100 µL PBS to the left, dorsolateral thoracolumbar region. After cells were settled for 5 min, the corresponding luciferase substrates with indicated concentrations in 100 µL PBS were also subcutaneously injected to each site. Mice were subsequently imaged with a 30-s exposure per frame for a total of 5 min using a luminescence dark box (Stanford Photonics) equipped with a Pixis 1024B cooled CCD camera. The Fiji image analysis software21 was used to analyze images and integrate bioluminescence intensities over common regions of interest encompassing all injected cells.
Bioluminescence imaging in deep tissues of live mice
Hydrodynamic transfections were performed on BALB/c mice as described elsewhere25. Briefly, 20 µg of each luciferase-expressing plasmid in sterilized saline (volume equivalent to 9% bodyweight of the treated mouse) was injected into restrained mice via the tail vein over 4–8 sec. Mice were allowed to recover on heat pads and were monitored until their breathing rate returned to normal. Bioluminescence images were acquired at 12 h post-injection. d-luciferin or AkaLumine-HCl at the indicated dose was dissolved in 100 µL PBS and intraperitoneally injected into FLuc-transfected mice. Prior to intraperitoneal injections of CTZ analogs to teLuc, Antares, or Antares2 transfected mice, the indicated dose of DTZ or furimazine was dissolved in a 100 µL solution containing 8% glycerol, 10% ethanol, 10% hydroxypropyl-β-cyclodextrin, and 35% PEG 400 in water. To inject 3.3 µmol DTZ or furimazine, the total volumes were increased to 500 µL. The luminescence dark box (Stanford Photonics) equipped with a Pixis 1024B cooled CCD camera was again used to image anesthetized mice with a 1-min exposure per frame over a course of 10 min. The Fiji image analysis software21 was used to process images and derive integrated intensities.
Bioluminescence imaging of intravenously injected HEK 293T cells
At 48 h after transfection, one million HEK 293T cells expressing luciferases were trypsinized, pelleted, and resuspended in 100 µL PBS. Cells expressing teLuc, Antares, or Antares2 were combined with the same number of cells expressing FLuc and injected into the tail vein of BALB/c mice placed in a restrainer. Mice were recovered for 5 h, anaesthetized, intraperitoneally injected with 3.3 µmol d-luciferin, and immediately imaged with a 1-min exposure per frame over a course of 20 min. Mice were kept under isoflurane anesthesia on a heat pad during imaging. After the diminishing of the FLuc bioluminescence, 0.3 µmol DTZ or furimazine was intraperitoneally injected. Again, mice were imaged with a 1-min exposure per frame over a course of 20 min. The images were processed using the Fiji image analysis software21 and the frames with highest signals in individual experiments were used for comparison.
Statistical analysis
Unpaired two-tailed t-tests were used to determine all P values. No statistical methods were used to pre-determine the sample size. No sample was excluded from data analysis, and no blinding was employed. Animals were randomly assigned to receive various treatments. Unless otherwise indicated, data are shown as mean ± s.d., and error bars in figures represent s.d.
Data availability
The gene sequences for teLuc, yeLuc, and Antares2 have been deposited to GenBank under the accession numbers KX963378, KX963379, and KY474379, respectively. The plasmids for teLuc (Plasmid #100026) and Antares2 (Plasmid #100027) have been deposited to Addgene. Materials, associated protocols, and other supporting data are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
pNCS-Antares and pcDNA3-ReNL were gifts from M. Lin and T. Nagai (Addgene plasmids # 74279 and # 85203), respectively. We thank the University of California-Riverside, the National Institutes of Health (R01GM118675 and R21EB021651), and the National Science Foundation (CHE-1351933) for financial support. We thank T. M. Truong for reading and revising the manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
H.A. conceived and supervised the entire project. H.A. and M.M.M. supervised the mouse imaging experiments. H.W.Y. performed synthetic, protein engineering, and in vitro and in cellulo characterization experiments. H.W.Y. and O.K. performed the mouse imaging experiments. A.J. assisted H.W.Y. in completing compound synthesis. D.C. assisted in the mouse imaging experiments. H.W.Y. and H.A. analyzed the data and wrote the manuscript.
Supplementary Information accompanies this paper at http://www.nature.com/******
Competing financial interests: The authors declare competing financial interests. UC Riverside has filed a provisional patent application in the USA that is partially based on results described in the manuscript. H.A. and H.W.Y. are listed as co-inventors.
References
- 1.Negrin RS, Contag CH. Nat. Rev. Immunol. 2006;6:484–490. doi: 10.1038/nri1879. [DOI] [PubMed] [Google Scholar]
- 2.Arranz A, Ripoll J. Front. Pharmacol. 2015;6:189. doi: 10.3389/fphar.2015.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inglese J, et al. Nat. Chem. Biol. 2007;3:466–479. doi: 10.1038/nchembio.2007.17. [DOI] [PubMed] [Google Scholar]
- 4.Saito K, et al. Nat. Commun. 2012;3:1262. doi: 10.1038/ncomms2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall MP, et al. ACS Chem. Biol. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stacer AC, et al. Mol. Imaging. 2013;12:1–13. [PMC free article] [PubMed] [Google Scholar]
- 7.Adams ST, Jr, Miller SC. Curr. Opin. Chem. Biol. 2014;21c:112–120. doi: 10.1016/j.cbpa.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans MS, et al. Nat. Methods. 2014;11:393–395. doi: 10.1038/nmeth.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuchimaru T, et al. Nat. Commun. 2016;7:11856. doi: 10.1038/ncomms11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inouye S, Shimomura O. Biochem. Biophys. Res. Commun. 1997;233:349–353. doi: 10.1006/bbrc.1997.6452. [DOI] [PubMed] [Google Scholar]
- 11.Wu C, Nakamura H, Murai A, Shimomura O. Tetrahedron Lett. 2001;42:2997–3000. [Google Scholar]
- 12.Inouye S, et al. Biochem. Biophys. Res. Commun. 2013;437:23–28. doi: 10.1016/j.bbrc.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 13.Jiang T, Du L, Li M. Photochem. Photobiol. Sci. 2016;15:466–480. doi: 10.1039/c5pp00456j. [DOI] [PubMed] [Google Scholar]
- 14.Nishihara R, et al. Chem. Commun. 2015;51:391–394. doi: 10.1039/c4cc06886f. [DOI] [PubMed] [Google Scholar]
- 15.Inouye S, Sato J, Sahara-Miura Y, Yoshida S, Hosoya T. Biochem. Biophys. Res. Commun. 2014;445:157–162. doi: 10.1016/j.bbrc.2014.01.133. [DOI] [PubMed] [Google Scholar]
- 16.Tomabechi Y, et al. Biochem. Biophys. Res. Commun. 2016;470:88–93. doi: 10.1016/j.bbrc.2015.12.123. [DOI] [PubMed] [Google Scholar]
- 17.Chu J, et al. Nat. Biotechnol. 2016;34:760–767. doi: 10.1038/nbt.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki K, et al. Nat. Commun. 2016;7:13718. doi: 10.1038/ncomms13718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon AS, et al. ACS Chem. Biol. 2016;11:400–408. doi: 10.1021/acschembio.5b00753. [DOI] [PubMed] [Google Scholar]
- 20.Yang J, et al. Nat. Commun. 2016;7:13268. doi: 10.1038/ncomms13268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schindelin J, et al. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ando Y, et al. Photochem. Photobiol. 2007;83:1205–1210. doi: 10.1111/j.1751-1097.2007.00140.x. [DOI] [PubMed] [Google Scholar]
- 23.Loening AM, Dragulescu-Andrasi A, Gambhir SS. Nat. Methods. 2010;7:5–6. doi: 10.1038/nmeth0110-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ando Y, et al. Nat Photon. 2008;2:44–47. [Google Scholar]
- 25.Liu F, Song Y, Liu D. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gene sequences for teLuc, yeLuc, and Antares2 have been deposited to GenBank under the accession numbers KX963378, KX963379, and KY474379, respectively. The plasmids for teLuc (Plasmid #100026) and Antares2 (Plasmid #100027) have been deposited to Addgene. Materials, associated protocols, and other supporting data are available from the corresponding author upon request.