Abstract
Manual review of aligned reads for confirmation and interpretation of variant calls is an important step in many variant calling pipelines for next-generation sequencing (NGS) data. Visual inspection can greatly increase the confidence in calls, reduce the risk of false positives, and help characterize complex events. The Integrative Genomics Viewer (IGV) was one of the first tools to provide NGS data visualization, and it currently provides a rich set of tools for inspection, validation, and interpretation of NGS datasets, as well as other types of genomic data. Here we present a short overview of IGV’s variant review features for both single nucleotide variants (SNV) and structural variants (SV), with examples from both cancer and germline datasets. IGV is freely available at https://www.igv.org.
Keywords (from list): New software for data analysis, Mutation detection methods, Chromosomal translocations: genomic aspects
Keywords (user-defined): IGV, Visualization, NGS
Introduction
Visual inspection of read alignments has been an important component of variant detection since the advent of next-generation sequencing (NGS). Although the identification of putative variants has been largely automated with tools such as the Genome Analysis Toolkit (GATK) [1], visual review remains critical to the verification and interpretation of these putative variants in many applications.
The Integrative Genomics Viewer (IGV) [2, 3] was one of the first tools to provide NGS data visualization. In May 2009, we introduced support for viewing short-read sequence alignment datasets in the, then nascent, SAM/BAM file format [4]. IGV remains in active development and is used extensively for variant inspection in research and clinical settings. It supports a rich set of features for quick identification of sequencing and analysis artifacts leading to errant single nucleotide variant (SNV) calls, as well as support for viewing large scale structural variants (SV) detected by paired-end read technology. More recently we have developed a suite of specialized features to support third-generation long-read sequencing technologies. Here we present a short overview of IGV variant visualization capabilities illustrated with examples drawn from cancer and germline datasets.
Single Nucleotide Variants
Currently, the most common application of next-generation sequencing technology in a clinical setting is the detection of SNVs and small insertions and deletions with respect to a reference genome. Output from the sequencer is typically run through a standard analysis pipeline, such as the GATK, to produce a list of putative variants (“calls”). Finally, manual review of aligned reads is often performed to reduce the risk of false positives and incorrect results [5–9].
A number of IGV features have been developed specifically to aid this manual review step, specifically: (1) highlighting mismatched bases in individual reads in color, to aid detection of unusual patterns and mis-mapped alignments; (2) highlighting ambiguously mapped reads (mapping quality = 0), indicative of high reference sequence homology, as such regions are known to produce many false positives; (3) shading of mismatched bases by read base quality, as clusters of bases of low quality can be indicative of sequencing errors; and (4) sorting, grouping, and coloring alignments by alignment, sequencing, and platform meta-data, which can be useful for detecting systematic errors upstream of read alignment. These features are described in more detail in the IGV user guide (http://www.broadinstitute.org/igv). Here we review a selection of these features for single nucleotide variant review focused on detection of possible false positives and mis-characterization of variants.
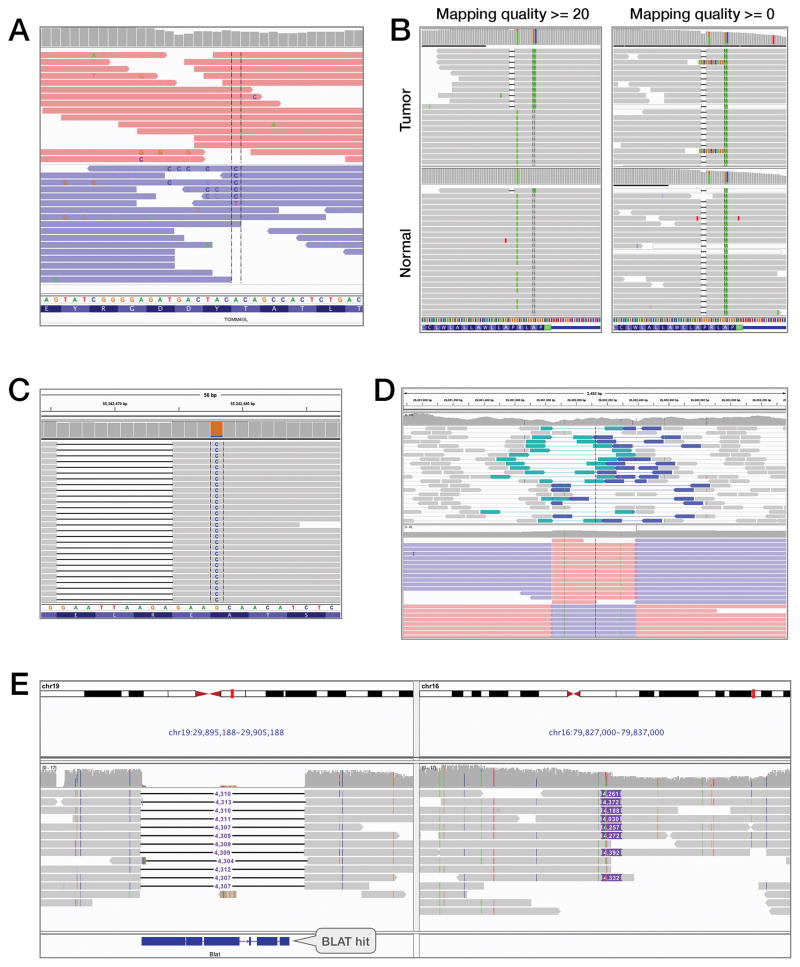
Figure 1A and the video in Supplementary Data illustrate a false positive potentially caused by a sequencing error. The locus was originally called an A→C SNV due to a variant base in 6 of 23 reads. However, close examination reveals an abnormally high number of variant “C” base calls on reads containing the putative variant, many of low base quality, which are indicated in IGV by shading of the bases. These “C” variants appear to be distributed randomly and are not well supported by other reads. This is an unusual pattern and indicative of possible sequencing errors. Importantly, sorting and coloring the alignments reveals that all variant bases at this locus are on the same read strand. The expected strand distribution for the protocol used for the sequencing of this sample is 50–50. Thus, this is likely to be a false positive.
Figure 1.
This figure illustrates five examples of how IGV was used to visually highlight anomalies in aligned sequencing data to aid the investigator in their interpretation.
A. Example of a false positive in a GC-rich region. Bases that do not match the reference genome are drawn with the letter of the called base (A, C, G, or T). Sorting and coloring alignments by strand reveal a strong strand bias indicative of a sequencing artifact. Pink and blue reads were aligned to the forward and reverse strand, respectively.
B. A germline mutation mischaracterized as a somatic NOTCH2 mutation in a clinical cancer sample. The mutation caller used a mapping quality filter with threshold set to 20, removing most alignments that support the variant from the normal sample and resulting in a reference call. The short horizontal black lines in the alignments represent 2-nucleotide deletions, and the colored bars in the grey alignments represent bases that did not match the reference sequence. In particular, the green bars represent bases that were called as A’s.
C. A clinically actionable multi-nucleotide deletion-insertion event (L747_A750delinsP) initially misclassified as two independent events. The horizontal black lines in the grey alignments represent deletions in alignments that otherwise match the reference sequence. The blue column of C’s indicates a large number of mismatches against the G at that locus of the reference sequence.
D. Comparison of NGS paired-end short reads (top panel) and third-generation long reads (bottom panel) spanning a 500 base pair inversion. The short reads are colored by pair orientation. The two reads of each pair were expected to point inward toward each other when aligned to the reference sequence. The teal color represents pairs where both reads unexpectedly aligned in the “right” direction, and the dark blue represents pairs where both aligned in the “left” direction. Breakpoints can be inferred from the short reads by careful examination of the outer read extents and dip in coverage. The long reads are split into multiple alignments and each alignment color-coded by strand. Pink and blue segments were aligned to the forward and reverse strand, respectively. The breakpoints are directly visible as the boundaries between alternating strands.
E. PacBio long reads support a 4.3 kb translocation from chr19 to chr16 in the SK-BR-3 breast cancer cell line. A black line represents a deletion, and a purple box is an insertion. Deletions and insertions are labeled with the event size.
In the clinical setting, false positive mutation calls may direct patients to wrong therapy decisions either by identifying a false actionable mutation or by inflating tumor mutation burden which has become a marker for immunotherapy trials. As a result, manual review of mutation calls from clinical samples is essential for proper patient management. An example of a false positive somatic mutation from a clinical cancer sample is presented in Figure 1B. The left panel presents an IGV screenshot of alignments from the tumor and normal samples as seen by the mutation caller, which had set a mapping quality threshold to filter out all alignments with mapping quality lower than 20. To simulate this, we set the IGV mapping quality to 20 to filter alignments below this threshold from view. The SNV was called a somatic mutation due to absence of a non-reference allele in the normal sample. However, visual inspection reveals the presence of a single read in the normal sample supporting the mutation, also in phase with a nearby deletion as seen in the tumor sample. Removing the mapping quality filter from IGV reveals a more complex region (right panel). The normal sample appears to harbor this mutation, albeit in lower mapping quality reads that are not filtered out in this view. The presence of non-unique mappings in the region, indicated by semi-transparent alignments in the IGV view, is evidence that the locus is homologous with other regions of the genome. As a result, we cannot conclude whether the mutation is germline or originates in the regions of the genome homologous to this locus. While the frequency of this type of false positive call may change from gene to gene, they occur frequently enough that we recommend manually reviewing all mutation calls passing pipeline filters.
Misclassification of variants is another source of errors. In a cancer genomics setting, variant misclassification can lead to actionable mutations being classified as novel or variants of unknown significance. This is a particular problem with complex multi-nucleotide variants. The majority of variant callers used in practice today cannot correctly identify these events. An example of a complex mutation from a cancer sample is presented in Figure 1C. The EGFR–L747_A750delinP mutation is a common EGFR event that is clinically actionable. This event was initially classified as two independent events E746_R748del and A750P. Visual inspection by an analyst confirms that the events are in phase, and somatic, and should be merged.
Structural Variants
While short-read NGS sequencing can directly detect SNVs and short indels, discovery of larger structural variants relies on interpretation of unexpected alignments of paired reads from the ends of the sequenced molecule. Evidence can include anomalies in mapped distance between the alignments and in their orientation with respect to each other. IGV supports visualization of discrepant pairs through color-coding, revealing distinctive patterns that can be used to distinguish between variant classes such as inversions, duplications, and translocations.
“Third-generation” long-read sequencing technologies produce average read lengths of 10,000 base pairs or more, making it possible to directly sequence and visualize many structural variants that are undetectable or poorly resolved by second-generation paired-end approaches [10]. IGV 3.0 includes new features to support these technologies, including: (1) support for linking split alignments that are common with longer reads; (2) a “consensus mode” that removes random single base and small indel errors from view; (3) a “quick-phasing” operation for grouping reads by base call at a selected locus; (4) new options for labeling indels and soft clipped regions; (5) the ability to expand and view inserted sequences in place; and (6) support for searching the reference genome for sequences similar to the sequence of an insertion, through an external BLAT server [11].
Figure 1D illustrates an inversion sequenced with both Illumina short-read paired-end alignments (top panel) and PacBio® SMRT® [12] long-read sequencing (bottom panel). Individual reads in the short-read data are of insufficient length to span the inversion, however paired reads from each end of the same molecule can be used to detect it. Pairs that conform to the expected orientation are colored grey, other pairs are assigned a color according to orientation. In an inversion, pairs spanning the left breakpoint will point “right-right”, while pairs spanning the right breakpoint point “left-left”. The approximate location of breakpoints can be ascertained by careful examination of the extent of the 3′ ends of the outside alignment of each discrepant pair. Dips in coverage are further evidence of breakpoints as individual reads will in general not align across the breakpoint.
By contrast, the long reads represented in the bottom panel are of sufficient length to completely span the inversion. Aligners tuned for long reads are able to split reads that span the breakpoint into multiple alignments as required to obtain the highest overall mapping score for the read. In this example, IGV options have been used to re-connect split alignments from the same read and to color-code each constitutive alignment by its strand. The breakpoints are clearly visible at the junctions of contrasting strand.
Figure 1E illustrates PacBio SMRT long reads supporting a 4.3 kilobase translocation from chromosome 19 to chromosome 16 in the SK-BR-3 breast cancer cell line [13]. In this view, new IGV features for base consensus and small indel filtering are enabled, suppressing random noise to reveal the putative rearrangement as a matched deletion and insertion event. The deleted sequence in chromosome 19 is represented by a black line labeled with the size of the deleted sequence. This sequence appears as an insertion in chromosome 16 of the same approximate size. IGV’s split-screen view is used to display both regions side by side. Right-clicking the insertion in chromosome 16 and performing a BLAT search on its sequence reveals a strong hit in the region of the deletion in chromosome 19.
Availability
IGV is freely available at https://www.igv.org under an MIT open-source license.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by the National Cancer Institute (NIH/NCI), grants R01CA157304 and U24CA210004; and the Starr Cancer Consortium, grant I5-A500.
Thank you to Michael Schatz for releasing the SK-BR-3 PacBio sequence data under the Toronto Agreement. We gratefully acknowledge the members of the Molecular Diagnostics Service in the Department of Pathology and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology. We also acknowledge the patients and their families for their participation.
Footnotes
Disclosure of Potential Conflicts of Interest
A.M.W. is an employee and shareholder of Pacific Biosciences, a company commercializing DNA sequencing technologies. The other authors declare no potential conflicts of interest.
References
- 1.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strom SP. Current practices and guidelines for clinical next-generation sequencing oncology testing. Cancer Biol Med. 2016;13:3–11. doi: 10.28092/j.issn.2095-3941.2016.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanchi KL, Johnson KJ, Lu C, McLellan MD, Leiserson MD, Wendl MC, et al. Integrated analysis of germline and somatic variants in ovarian cancer. Nat Commun. 2014;5:3156. doi: 10.1038/ncomms4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cazier JB, Rao SR, McLean CM, Walker AK, Wright BJ, Jaeger EE, et al. Whole-genome sequencing of bladder cancers reveals somatic CDKN1A mutations and clinicopathological associations with mutation burden. Nat Commun. 2014;5:3756. doi: 10.1038/ncomms4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maxwell KN, Wubbenhorst B, D’Andrea K, Garman B, Long JM, Powers J, et al. Prevalence of mutations in a panel of breast cancer susceptibility genes in BRCA1/2-negative patients with early-onset breast cancer. Genet Med. 2015;17:630–8. doi: 10.1038/gim.2014.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh RR, Patel KP, Routbort MJ, Aldape K, Lu X, Manekia J, et al. Clinical massively parallel next-generation sequencing analysis of 409 cancer-related genes for mutations and copy number variations in solid tumours. Br J Cancer. 2014;111:2014–23. doi: 10.1038/bjc.2014.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaisson MJ, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517:608–11. doi: 10.1038/nature13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kent WJ. BLAT--the BLAST-like alignment tool. Genome Res. 2002;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–8. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 13.Nattestad M, Chin CS, Schatz MC. Ribbon: Visualizing complex genome alignments and structural variation. 2016:082123. bioRxiv. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.