Many human infections are polymicrobial in origin, and interactions among community inhabitants shape colonization patterns and pathogenic potential1. Periodontitis, which is the sixth most prevalent infectious disease worldwide2, ensues from the action of dysbiotic polymicrobial communities3. The keystone pathogen Porphyromonas gingivalis and the accessory pathogen Streptococcus gordonii interact to form communities in vitro and exhibit increased fitness in vivo3, 4. The mechanistic basis of this polymicrobial synergy, however, has not been fully elucidated. Here we show that streptococcal 4-aminobenzoate/para-amino benzoic acid (pABA) is required for maximal accumulation of P. gingivalis in dual species communities. Metabolomic and proteomic data showed that exogenous pABA is utilized for folate biosynthesis, and leads to decreased stress and elevated expression of fimbrial adhesins. Moreover, pABA increased the colonization and survival of P. gingivalis in a murine oral infection model. However, pABA also caused a reduction in virulence in vivo and suppressed extracellular polysaccharide production by P. gingivalis. Collectively, these data reveal a multidimensional aspect to P. gingivalis-S. gordonii interactions and establish pABA as a critical cue produced by a partner species that enhances fitness of P. gingivalis while diminishing virulence.
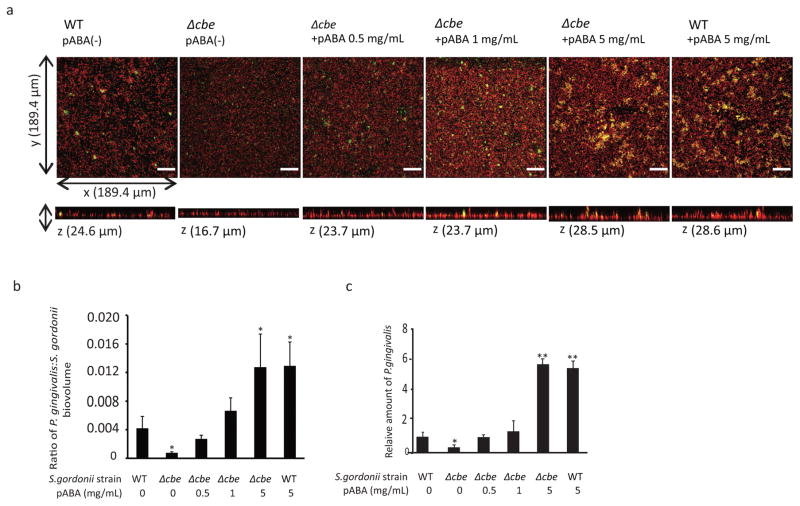
P. gingivalis accumulates into heterotypic communities with S. gordonii through an orchestrated series of developmental steps5. Initial perception of S. gordonii extracellular products initiates a gene expression program in P. gingivalis that prepares the organism for symbiotic community dwelling6. Mutation of the cbe gene (chorismate binding enzyme, SGO_0307) in S. gordonii reduces the amount of P. gingivalis that accumulates on a substratum of S. gordonii7; however, the nature of P. gingivalis responses to this interspecies recognition system, and the effect on P. gingivalis physiology and in vivo fitness are unknown. In many prokaryotes Cbe enzymes produce 4-aminobenzoate/para-amino benzoic acid (pABA), which is a precursor of folate synthesis and can be released extracellularly8. To investigate the role of pABA in metabolic crosstalk, the effect of exogenous pABA on the accumulation of P. gingivalis on substrata of S. gordonii was determined. At concentrations of 0.5 and 1 mg/mL, pABA restored P. gingivalis accretion on Δcbe mutant cells to levels observed with wild type S. gordonii, and 5 mg/mL pABA caused a further increase in P. gingivalis biovolume (Fig. 1a,b,c). Concentrations of 0.5 and 1 mg/mL are thus likely in the physiological range and were adopted in further studies. To verify that the effect of pABA is on P. gingivalis and not S. gordonii, we performed several additional controls. Treatment of only the S. gordonii parental or Δcbe strains with pABA did not affect amount of streptococcal substratum (Supplementary Fig. 1a,b), or the level of subsequent P. gingivalis accumulation (Supplementary Fig. 1c,d). In contrast, pretreatment of P. gingivalis with pABA increased the degree of association with S. gordonii substrata (Supplementary Fig. 2a,b), and with the glass support (Supplementary Fig. 2c,d). Collectively these results indicate that pABA is a cue recognized by P. gingivalis to stimulate accumulation with S. gordonii.
Figure 1.
Exogenous pABA enhances P. gingivalis community formation with S. gordonii. a, P. gingivalis (green) was reacted with a substratum of S. gordonii DL-1 parental strain (WT) or Δcbe mutant (red) for 18 h without or with pABA at the concentrations indicated. A series of 20–30 μm-deep optical fluorescent x-y sections were collected by confocal microscopy to create digitally reconstructed 3D images with Imaris software. Images are representative of 6 independent experiments. Bar is 25 μm. b, Ratio of P. gingivalis:S. gordonii biovolume in images represented in (a) measured with the Imaris Isosurface function. Data are representative of 6 independent experiments and are presented with mean with standard deviation of 5 random fields from one experiment. c, Numbers of P. gingivalis in dual species communities with S. gordonii under conditions described in (a) determined by qPCR. Results are means with standard deviation from one of three independent experiments performed in triplicate. For (b) and (c) * – P < 0.05, ** – P < 0.01 compared to WT without pABA using ANOVA with Tukey’s multiple comparison test.
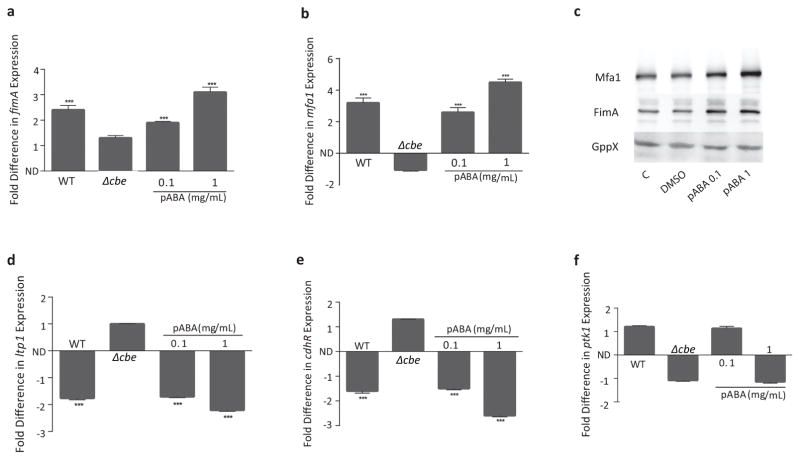
Attachment of P. gingivalis to S. gordonii is mediated by the FimA and Mfa1 component fimbriae5. Subsequently, signal transduction within P. gingivalis occurs through protein tyrosine (de)phosphorylaton circuitry5. Hence, we sought to determine the role of pABA on the expression of these effectors of community development. As presented in Fig. 2a,b, mRNA for both mfa1 and fimA was increased by pABA in a dose dependent manner. Upregulation of Mfa1 and FimA proteins was confirmed by western blotting with specific antibodies (Fig. 2c, and Supplementary Fig. 3a,b). In P. gingivalis expression of mfa1 is suppressed through a pathway that involves a tyrosine phosphatase (Ltp1) which dephosphorylates its cognate kinase (Ptk1) and indirectly upregulates the transcription factor CdhR, a negative regulator of mfa19. Both ltp1 and cdhR mRNA amounts were inhibited by pABA (Fig. 2d,e), indicating that pABA signaling occurs though the Ltp1-CdhR pathway. In contrast, Ptk1 which is regulated at the posttranslational level by phosphorylation status showed no statistical transcriptional change following pABA treatment (Fig. 2f). Expression of fimA continued to increase for up to 4 h after initial pABA treatment (Supplementary Fig. 4). These data are consistent with a model whereby pABA, produced and secreted by S. gordonii through the action of Cbe, is detected by P. gingivalis resulting in modulation of signal transduction pathways that control adhesin gene expression.
Figure 2.
pABA increases expression of P. gingivalis effectors of community development. a, b, d–f. qRT-PCR of mRNA extracted from P. gingivalis reacted for 2 h with pABA at the concentrations indicated or with secreted components of S. gordonii WT or Δcbe in a membrane insert transwell. 16S rRNA was used for normalization. Data are expressed as fold difference (or −1/ratio for negative fold change) relative to P. gingivalis incubated with vehicle (DMSO) alone (for pABA conditions) or P. gingivalis incubated with medium only in the transwell (for S. gordonii conditions). ND: no difference. *** – P < 0.001 using ANOVA with Tukey’s multiple comparison test. Data are means with standard deviation and representative of 3 biological replicates performed in triplicate. c, Immunoblots of cell lysates of P. gingivalis reacted with pABA for 18 h at the concentrations indicated, DMSO, or PBS alone (C) and probed with antibodies to Mfa1, FimA or GppX as a control. Data are representative of 3 independent experiments.
Previous studies have established that both P. gingivalis and S. gordonii adapt to the community environment on a global scale10, 11. Hence, we examined the impact of pABA on the proteome and metabolome of P. gingivalis. Qualitative detection of 1153 P. gingivalis proteins (55% of the total theoretical proteome) was observed. Based on quantitative spectral counting, 322 proteins were significantly reduced and 207 were significantly increased, compared to the control with vehicle alone (Supplementary Table S1). Distributions of the regulated proteins in functional categories is shown in Supplementary Fig. 5. Upregulation of P. gingivalis FimA and Mfa1 structural and accessory fimbrial proteins by pABA was observed in the proteome (Supplementary Fig. 6), and this was further verified by ELISA (Supplementary Fig. 7a), and functionally through increased adherence to gingival epithelial cells (Supplementary Fig. 7b). In addition to adhesive fimbriae, proteases, hemin uptake proteins, TonB-dependent proteins, and Type IX secretion system components were upregulated (Supplementary Fig. 6), indicating that P. gingivalis also utilizes pABA as a cue to adapt to the protein- and hemin-rich subgingival environment. An increase in gingipain protease enzyme activity was observed in whole-cell lysates of pABA-treated P. gingivalis, but not on the cell surface or in the culture supernatant, indicating that secretion and/or processing had become rate-limiting (Supplementary Fig. 8). Expression of a number of stress-related proteins was decreased by pABA (Supplementary Fig. 6), suggesting that P. gingivalis has adapted for community dwelling with S. gordonii.
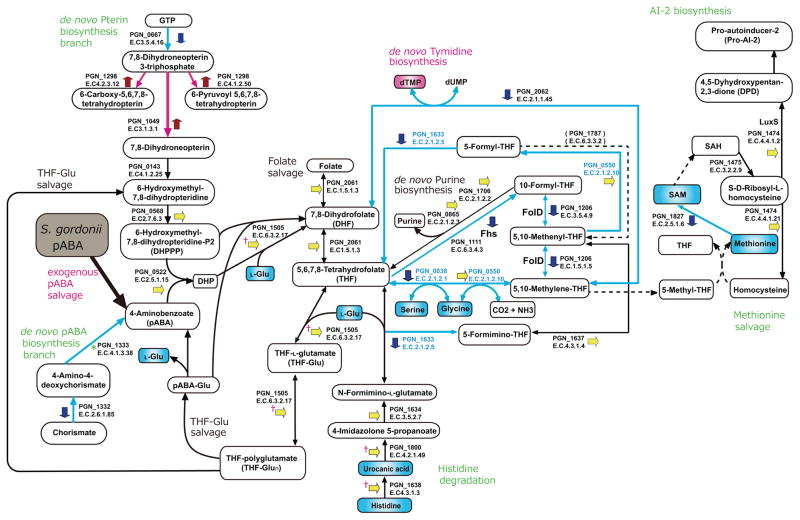
In bacterial metabolic pathways pABA is utilized in tetrahydrofolate (THF) metabolism12. Analysis of differentially regulated proteins, transcripts and metabolites (Fig. 3, Supplementary Fig. 9) established the role of exogenous pABA in THF physiology in P. gingivalis. THF and its one-carbon (C1) substituted derivatives, collectively termed folates, are cofactors for various C1 transfer reactions which produce essential metabolites such as purines, thymidylate, glycine, serine and methionine. THF is a tripartite molecule consisting of pterin, pABA, and glutamate moieties, and typically having a short, γ-linked polyglutamyl tail attached to the first glutamate13. Most bacteria, including P. gingivalis, make folates de novo, starting from GTP and chorismate12. Folates undergo spontaneous oxidative scission to yield pterin and p-aminobenzoylglutamate (pABAGlu) or its polyglutamyl forms (pABAGlun), which are re-used in folate synthesis through folate-salvage reactions13, 14. When P. gingivalis was treated with exogenous pABA, mRNA for PGN_1332 and PGN_1333 enzymes (PabB and PabC, pABA synthases) in the de novo pABA biosynthesis branch was reduced, and PGN_1332 was suppressed at the protein level (Fig. 3, Supplementary Fig. 9a), consistent with exogenous pABA salvage becoming predominant in the intermediate step of folate biosynthesis. Intracellularly, pABA and 2-Amino-4-hydroxy-6-hydroxymethyl-7,8-dihydropteridine-P2 (DHPPP) form 7,8-Dihydropteroate (DHP) mediated by PGN_0522 (FolP, dihydropteroate synthase), followed by enzymatic reactions leading to THF and THF-polyglutamate (THF-Glun) catalyzed by PGN_1505 (FolC, folylpolyglutamate synthase) which was transcriptionally upregulated (Fig. 3, Supplementary Fig. 9a). Conversion of THF to 5,10-Methylenyl-THF may be reduced as PGN_0038 (GlyA, serine hydroxymethyltransferase), PGN_1111 (formate-tetrahydrofolate ligase), PGN_1206 (methylenetetrahydrofolate dehydrogenase / methenyltetrahydrofolate cyclohydrolase), PGN_1633 (formiminotransferase-cyclodeaminase), and PGN_2062 (ThyA, thymidylate synthase) were downregulated by pABA (Supplementary Table 1 and Fig. 3). The upregulation of PGN_1505 (FolC), PGN_1638 (HutH, histidine ammonia-lyase), PGN_1800 (HutU, urocanic acid hydratase) mRNA (Supplementary Fig 9a), is suggestive of earlier activation of the THF-Glu and histidine degradation pathways. A metabolite set enrichment analysis (MSEA) showed that pABA significantly activated deoxyribonucleotide biosynthesis (Supplementary Table 2), accumulation of dTMP is likely due to the prior activation of thymidylate synthase (PGN_2062) in the early stage of pABA stimulation, although the protein amount was suppressed by 18 h (Supplementary Table 1). These results suggest that biosynthesis of THF and its derivatives in P. gingivalis is rapidly stimulated by pABA, and when internal THF-GLun amounts reach a sufficient level, THF metabolism becomes suppressed. Moreover, methionine salvage was significantly suppressed after pABA exposure (Fig. 3, Supplementary Tables 2, 3), thus regulation of folate derivative biosynthesis seems to be a prime determinant of metabolome alteration in pABA-treated P. gingivalis.
Figure 3.
Trans-omics of pABA effects of P. gingivalis on the tetrahydrofolate metabolic pathway. Red, blue and yellow short arrows represent proteins up-, down-, or non-regulated by pABA respectively (see Methods for statistical thresholds). * represents a non-detect in the proteome but down regulated at the transcriptional level. Ϯ represents a protein unchanged in the proteome but upregulated at the transcriptional level. Red and blue connecting arrows represent functionally predicted up- and down-regulation respectively. Blue font denotes PLP-dependent enzymes whose functions were suppressed as shown in the metabolome (Supplementary Table 4). Red and blue boxes represent increased and decreased metabolite production respectively.
Interestingly, many of the enzymes in the folate derivative biosynthesis pathways are pyridoxal 5-phosphate (PLP)-dependent. Our proteome and metabolome data suggest that pABA suppressed the biosynthesis of PLP and its derivatives by negative regulation of the key enzymes, transketolase (Tkt, PGN_1689) and pyridoxamine-phosphate oxidase (PdxH, PGN_0403) (Supplementary Fig. 9b and Supplementary Table 1). Reduction of transketolase resulted in a decrease of ribose 5-phosphate, and accumulation of fructose 6-phosphate and sedoheptulose 7-phosphate (Supplementary Fig. 9b). Strikingly, the metabolomic analyses showed that products of various PLP-dependent enzymes, such as serine, agmatine, beta-alanine, tryptophan, GABA, and 1-aminocyclopropane-1-carboxylic acid, were significantly decreased or not detected in pABA-treated P. gingivalis (Supplementary Table 4), suggestive of a global post-translational negative regulation of PLP-dependent enzymes by pABA. Furthermore, MSEA showed that Glycolysis/Glyconeogenesis pathways were significantly activated by pABA (Supplementary Table 2). As P. gingivalis is asaccharolytic, these pathways are likely utilized for processing or recycling of extracellular matrix components. Thus, while P. gingivalis is unable to utilize pABA as a sole nutritional source and pABA does not enhance the replication of P. gingivalis in complex medium, pABA nonetheless provides a metabolic cue that significantly alters physiology.
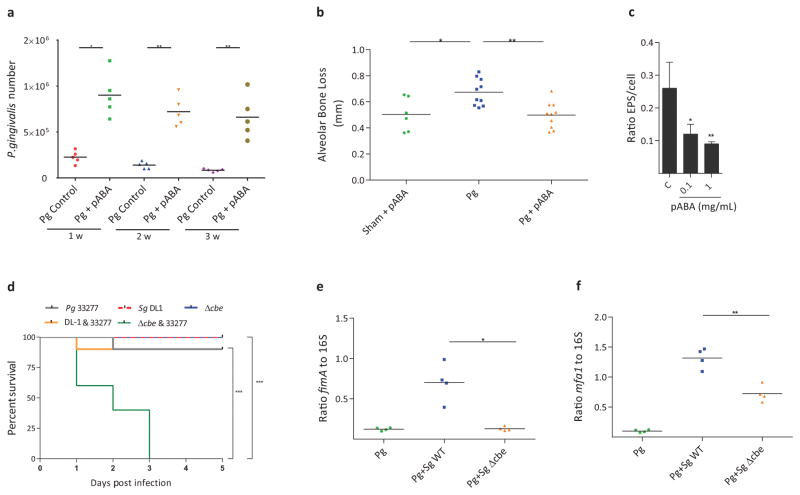
Next we sought to test the role of pABA on P. gingivalis in vivo. In a murine oral infection model, treatment of P. gingivalis with pABA resulted in recovery of around 106 P. gingivalis from each mouse mouth up to 3 weeks after infection, significantly higher than the control condition and previous studies15 (Fig. 4a), and consistent with increased expression of the fimbrial adhesins. Despite increased amounts, however, pABA-treated P. gingivalis induced significantly less alveolar bone loss compared to vehicle-treated bacteria (Fig. 4b), indicating that pABA dampens the pathogenic potential of P. gingivalis. As Ptk1 and Ltp1 regulate extracellular polysaccharide (EPS) production16, and EPS is a major virulence factor of P. gingivalis17, we examined the influence of pABA on EPS. Although not organized into a discrete capsule, strain 33277 produces EPS that can be detected by lectin binding16. Fig. 4c and Supplementary Fig. 10 show that pABA significantly reduced EPS production by P. gingivalis, which may provide the mechanistic underpinning for lower pathogenic potential in the murine bone loss model. These results also invited speculation that mixed infections of P. gingivalis and S. gordonii would be more pathogenic in the absence of streptococcal pABA production, and this was tested in the mouse abscess model. As shown in Fig. 4d, infection of mice with 5 x 109 total combined P. gingivalis WT and S. gordonii Δcbe resulted in the death of all the animals after 3 days. In contrast, 90% of animals survived in the P. gingivalis WT and S. gordonii WT combination, and in the P. gingivalis alone condition. All of the animals survived in the S. gordonii WT or Δcbe alone groups. To further investigate the involvement of pABA, the inoculum size was reduced to 2.5 x 109 total bacteria and abscess material was recovered. qRT-PCR (Fig. 4e,f) showed that in comparison to dual species P. gingivalis and S. gordonii WT infection, fimA and mfa1 expression was significantly reduced in the absence of pABA, consistent with the in vitro data (Fig. 2a–c). Collectively, these results reveal a multidimensional sensing-response system between P. gingivalis and S. gordonii. While S. gordonii-derived pABA provides a cue for P. gingivalis that enhances fimbrial expression and colonization, concomitantly virulence of P. gingivalis is constrained. The calming effect of pABA can be overcome when P. gingivalis aggregates develop on a S. gordonii substratum4, most likely due to spatial constraints in the accessibility of streptococcal pABA. When the influence of pABA is diminished there will be a subsequent increase in the pathogenicity of P. gingivalis-S. gordonii communities4.
Figure 4.
Effects of pABA on P. gingivalis in vivo and expression of extracellular polysaccharide. a, Colonization of the murine oral cavity. P. gingivalis (108) was reacted with pABA 1 mg/mL, or with DMSO (control) and inoculated into the oral cavity of Balb/c mice. One, 2 or 3 weeks (w) after the final inoculation, P. gingivalis was enumerated by qPCR. Each symbol represents an individual mouse and the short horizontal lines indicate the mean. One representative experiment of three is shown. b, pABA diminishes virulence of P. gingivalis in an alveolar bone loss model. P. gingivalis was inoculated orally as described in (a). Control mice were sham infected with pABA. Mice were given pABA (0.1 mg/mL) or vehicle only ad libitum in drinking water. Bone loss is expressed as mean distance from the cementoenamel junction (CEJ) to the alveolar bone crest (ABC) of 14 maxillary molar sites after 42 days. Each symbol represents an individual mouse and the short horizontal lines indicate the mean. One representative experiment of two is shown. c, P. gingivalis was reacted with pABA at the concentrations indicated for 18 h and extracellular polysaccharide was stained with FITC-labeled concanavalin A and wheat germ agglutinin. Bacterial cells were stained with Syto-17. The ratio of lectin binding (green) to whole cell staining (red) in confocal images was determined using Volocity software. Data are means with standard deviation of 5 random fields from one representative experiment of three. For a–c, data were analyzed with a two-tailed t-test. d, Kaplan-Meier plot showing survival of mice following subcutaneous inoculation of P. gingivalis, S. gordonii, or combinations of P. gingivalis with S. gordonii WT or Δcbe (5 x 109 total bacteria). n=10 mice per group and comparison between groups was by the log-rank test. One representative experiment of two is shown. e,f, qRT-PCR of fimA (e) or mfa1 (f) expression in abscess material from groups described in (d) (except with 2.5 x 109 total bacteria). Each symbol represents a single animal and the short horizontal lines indicate the mean. One representative experiment of two, performed in triplicate, is shown. Data were analyzed by a two-tailed t-test. For all panels * - P < 0.05, ** - P < 0.01, *** - P < 0.001.
The development of dental plaque biofilms follows a well-defined series of events where initial colonizers, predominantly streptococci, establish an environment conducive for later colonization by more overtly pathogenic species. While organisms such as S. gordonii are generally commensal in the oral cavity, they can increase the virulence of P. gingivalis in communities and hence have been designated as accessory pathogens17. P. gingivalis and S. gordonii are found in close proximity in vivo18, and S. gordonii can provide a substratum for the attachment and accretion of P. gingivalis cells7. Interactions between S. gordonii and P. gingivalis involve both direct contact19 and detection of soluble mediators such as pABA7. In this study we have begun the functional dissection of the P. gingivalis and S. gordonii interaction. pABA induced metabolic shifts within P. gingivalis and upregulation of pathways leading to the production of folylpolyglutamate along with gluconeogenesis and deoxyribonucleotide biosynthesis followed by nucleotide sugar biosynthesis. In contrast, pyridoxal 5′-phosphate was downregulated. Similar patterns in the periodontal metatranscriptome in vivo have also been reported20. The finding that pABA increased retention of P. gingivalis in the murine oral cavity while diminishing pathogenicity, shows that colonization and pathogenicity are functionally distinct. We thus propose that individual sensing and response systems can effectuate discrete outcomes, such as increased persistence, metabolic recalibration and regulation of pathogenicity, as part the overall process of synergy between the organisms.
Methods
Bacteria
P. gingivalis 33277 was cultured in trypticase soy broth supplemented with yeast extract (1 mg/mL), haemin (5 μg/mL) and menadione (1 μg/mL), anaerobically, at 37°C. S. gordonii DL1 (wild type, WT) and isogenic mutant Δcbe7 were cultured in brain–heart infusion broth containing 0.5% yeast extract, anaerobically at 37°C.
P. gingivalis-S. gordonii interactions
For image analysis, S. gordonii cells were stained with hexidium iodide (15 μg/mL; Invitrogen, Carlsbad, CA) and 2 × 108 cells were incubated on glass coverslips anaerobically for 16 h at 37°C. Mid-log cultures of P. gingivalis were stained with 5-(and-6)-carboxyfluorescein, succinimidyl ester (4 μg/mL, Invitrogen) and reacted with the S. gordonii substratum, or with the glass coverslip, anaerobically with rocking for 18 h at 37°C in pre-reduced PBS. For pABA involvement: S. gordonii substrata were incubated with pABA for 6 h, washed and reacted with or without P. gingivalis; P. gingivalis was incubated with pABA for 6 h, washed and reacted with substrata; or pABA was included in the buffer throughout the reaction. Bacterial accumulations were viewed with a Zeiss LSM 510 confocal laser scanning microscope (Carl Zeiss MicroImaging GmbH, Germany) and analyzed with Imaris 7.0.1 software (Bitplane AG, Zurich, Switzerland) as previously described21. To confirm the result of image analysis, qPCR was performed to quantify P. gingivalis cell numbers in the mixed species community. Briefly, 10-fold greater amounts of unstained mixed biofilms were similarly generated using 6-well microplates (Iwaki, Chiba, Japan), then washed gently twice with PBS and resuspended in 1.5 mL PBS after transferring with sterile cell scrapers. Bacteria harvested by centrifugation were immediately frozen and homogenized with zirconia beads, then total DNA from both S. gordonii and P. gingivalis was isolated using a Wizard genomic DNA purification kit (Promega, Madison, WI), and qPCR was performed using a P. gingivalis-specific TaqMan probe and primer set22. For gene expression, S. gordonii was cultured in the lower chamber of 0.4 μm transwell insert plates for 24 h. P. gingivalis cells were added to the upper chamber and reacted with the S. gordonii culture supernatant for 2 h.
Proteomics
Proteomics and data processing were performed as described23. In brief, bacteria were cultured to mid-log phase and reacted for 18 h with pABA (1 mg/mL) or vehicle control. Two technical replicates were measured for each of four independent biological replicates. For LC-MS/MS analysis, peptides were separated by reverse phase chromatography using an Acclaim C18 PepMap RSLC column (Dionex; Thermo Scientific, Waltham, MA) and reverse phase chromatography on a Dionex Ultimate 3000 RSLCnano UHPLC system (Thermo Scientific). Peptides were eluted using a 5–40% acetonitrile gradient over 90 min at 300 nL/min flow rate. Eluted peptides were directly injected into an Orbitrap Fusion mass spectrometer (Thermo Scientific) by nano-electrospray and subject to data-dependent tandem mass spectrometry, with full precursor ion scans (MS1) collected at 120,000 resolution. Monoisotopic precursor selection and charge-state screening were enabled, with ions of charge >+1 selected for collision-induced dissociation (CID). Fragmentation scans (MS2) were collected per MS1 using the topspeed mode with precursor priority set to most intense. Dynamic exclusion was active with 60 s exclusion for ions selected once within a 35 s window.
Spectra were searched against a P. gingivalis 33277 (NCBI NC_010729.1) protein sequence database and common contaminant proteins (ms-blender24 using tandem, comet, and MSGF+ search engines). Fully-tryptic peptides were considered, with up to two missed cleavages. Tolerances of 10 ppm (MS1) and 0.5 Da (MS2), carbamidomethylation of cysteine as static modification, and oxidized methionine as dynamic modification were used. High-confidence peptide-spectral matches (PSMs) were filtered at <1% false discovery rate.
To calculate protein abundance ratios, a normalization scheme was applied such that the total spectral counts for all P. gingivalis proteins in each condition were set equal for each comparison. For comparison, spectral counts for individual peptides were also calculated. Peptide spectral counts were normalized identically to the proteins and checked for consistency with the protein levels. A minimum of two peptide counts per protein were required for identification, and a pseudo-count of 1 was added to all proteins. Ratios were calculated for all combinations of repeats, for each normalized protein level, and these ratios were used in a t-test (using no change as the null hypothesis) to test for significance. The resultant p-value was then adjusted to correct for multi-hypothesis testing using the R package stats p-adjust function. Significant differential regulations was considered at greater than 1.5 fold regulation and an adjusted p-value of less than 0.01. Non-transformed spectral count data are presented in Supplementary Tables 7–9. To detect potential outliers the analysis was re-ran excluding the sample that correlated least with the others, as well testing for outliers on a per protein basis (assuming 30% outliers). Approximately 15% of proteins were flagged as outliers (Supplementary Table 10) and so this technical replicate was not included in the data presented in Supplementary Table 1 and Supplementary Figure 6.
Metabolomics
P. gingivalis cells were incubated anaerobically with PBS in the presence or absence of 1 mg/mL of pABA at 37°C for 2 h. Bacterial cells were collected and washed with Milli-Q water by centrifugation. Bacterial pellets (5×109 cfu) were immediately fixed by adding 5 μM internal standard-containing methanol25. Capillary electrophoresis time-of-flight mass spectrometry (CE-TOFMS) was performed using an Agilent CE-TOFMS System (Agilent Technologies, Santa Clara, CA) equipped with a fused silica capillary [50 μm (i.d.) × 80 cm]. The conditions for measurement of cationic/anionic metabolites were as follows. Run buffer: a solution composed of Cation Buffer Solution [H3301-1001; Human Metabolome Technologies (HMT), Tsuruoka, Japan] and Anion Buffer Solution (H3302-1021), CE voltage: +27 kV/+30 kV, MS ionization: ESI positive/ESI negative, MS capillary voltage: 4000 V/3500 V, MS scan range: m/z 50–1000, and sheath liquid: HMT Sheath Liquid (H3301-1020). Identification of metabolites and evaluation of the relative amounts were performed using Master Hands (version 2.1.0.1 and 2.9.0.9; Keio University, Tokyo, Japan) with the HMT metabolite database. The relative amount of each metabolite was calculated with reference to the internal standard material (HMT). A metabolite set enrichment analysis (MSEA) was performed to make biological inferences about statistically significant metabolites, which were selected by a statistical hypothesis test of the factor loading in principal component analysis26.
Quantitative RT-PCR
Total RNA was isolated from P. gingivalis and converted to cDNA with an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). qRT-PCR was performed with a StepOne plus (Applied Biosystems, Carlsbad, CA) by the ΔΔCt method using 16S rRNA as an internal control as described previously27. Primers are listed in Supplementary Table 6.
Western blotting
P. gingivalis cells lysates were separated by SDS-PAGE and electroblotted onto nitrocellulose membranes as described previously28. Blots were probed with polyclonal antibodies specific for FimA, Mfa1, or GppX29, 30. Antibodies were generated in our laboratory and screened for specificity by western blot with whole-cell lysates of P. gingivalis.
ELISA
P. gingivalis cells were pelleted by centrifugation, lysed in Bugbuster (Millipore, Billerica, MA) with protease inhibitor cocktail II (Sigma-Aldrich, St. Louis, MO). Cell lysates were diluted to 0.2 mg/mL in carbonate buffer (pH 9.6) and coated at 100 uL per well in microtiter plates overnight at 4C. The wells were blocked with 5% nonfat dry milk in TBS with 0.2% Tween-20 (TBSTM) for 1 h with constant agitation. Rabbit anti-FimA, FimD, Mfa1, Mfa3, Mfa4 and Ptk1 antisera were added to the appropriate wells in 5% TBSTM for 1 h, followed by three washes and the application of anti-rabbit IgG conjugated to HRP in TBSTM for 1 h. The plates were washed three times and binding was detected by the addition of 3,3′,5,5′-tetramethylbenzidine (TMB). The reaction was stopped with 0.2 M H2SO4 and measured at 405 nm.
Attachment to epithelial cells
Levels of P. gingivalis attached to the surface of gingival epithelial cells were measured as described previously31. Telomerase Immortalized Gingival Keratinocytes (TIGKs), derived from a primary gingival epithelial cell line32 were cultured in Lifeline DermaLife Keratinocyte Medium with supplements (Lifeline Cell Technology, Fredrick, MD) at 37 °C in 5% CO2. TIGKs express a cytokeratin profile consistent with basal cells of the oral junctional epithelium32. Mycoplasma-free cells were fixed with 5% paraformaldehyde, washed three times in PBS, and incubated with P. gingivalis MOI 10 for 1 h. After washing, P. gingivalis was quantified by reacting with P. gingivalis antibodies (1:5000) followed by HRP-conjugated secondary antibodies (1:10000) and TMB substrate. The reaction was stopped with 0.2 M H2SO4 and measured at 405 nm.
Extracellular polysaccharide detection
Exopolysaccharide production was determined with fluorescent lectins as described previously9. P. gingivalis cells were labeled with Syto-17 (Invitrogen) and deposited on glass coverslips. Polysaccharide was labeled with Concanavalin A-FITC and Wheat germ agglutinin-FITC (100 μg/mL) for 30 min at room temperature. After washing, images were collected and red (EPS)/green (bacterial cells) ratios determined by confocal microscopy and quantitative image analysis as described above.
Proteolytic activity
The amidolytic activity of arginine-specific (Rgp) and lysine-specific (Kgp) gingipains was assayed as described previously33. P. gingivalis was cultured to mid-log phase and reacted for 18 h with pABA (1 mg/mL) or vehicle control. Cells were separated from the culture supernatant fraction by centrifugation (5,000 × g, 20 min), washed and resuspended in PBS to the original volume, or lysed by Bubuster (Millipore). The chromogenic p-nitroanillide substrates N-Benzoyl-L-Arginine-pNA or toluenesulphonyl-glycyl-prolyl-L-lysine pNA (Sigma, St. Louis, MO) were used to detect RgpA/B and Kgp, respectively. Samples (50 μl) were preincubated in 200 mM Tris HCl, 5 mM CaCl2, 150 mM NaCl2, supplemented with 10 mM cysteine in 96-well plates for 10 min at 37°C and assayed with 0.5 mM substrate. The rate of substrate hydrolysis and the accumulation of p-nitroanilide were monitored spectrophotometrically at 405 nm over time in a Spectramax M5 reader (Molecular Devices, Sunnyvale, CA), and the activity of enzyme is given as mOD/min/μl.
Animal infection
All animal protocols were approved by the University of Louisville Institutional Animal Care and Use Committee. Group sizes were based on reported studies utilizing the same methodology. i) Oral inoculation. Female 10–12 week old BALB/c mice were initially treated with sulfamethoxazole (MP Biomedical, Solon, OH) at a final concentration of 800 μg/mL and trimethoprim (Sigma) at a final concentration of 400 μg/mL ad libitum in water for 10 days at 2-day intervals. P. gingivalis cells were reacted with 1 mg/mL pABA for 6 h or with 1% DMSO as a vehicle control prior to infection. There was no difference in viability of the bacteria between the pABA and DMSO conditions (laboratory observations). Four days after the last antibiotic treatment, the mice were orally infected five times with 108 P. gingivalis suspended in 1 mL of 2% carboxymethylcellulose (CMC) at 2-day intervals over a 10-day period. Mice were given pABA (0.1 mg/mL) or vehicle only control ad libitum in drinking water. To enumerate P. gingivalis colonization, oral samples were collected along the gingiva of the upper molars using a 15-cm sterile polyester-tipped applicator at 1, 2 and 3 weeks after the final bacterial infection. Total genomic DNA was purified using a Wizard Genomic DNA Purification Kit (Promega), and amplified by qPCR with primers to 16S rRNA (Supplementary Table S6). Numbers of P. gingivalis were calculated by comparison with a standard curve derived from known amounts of P. gingivalis. Forty two days after the last infection, mice were euthanized and skulls defleshed as described4. The cleaned skulls were stained with 1% methylene blue and bone loss was assessed by measuring the distance between the alveolar bone crest (ABC) and the cemento-enamel junction (CEJ) at 14 predetermined points on the maxillary molars. ii) Abscess model. Groups of female 8–12 week old BALB/c mice were inoculated subcutaneously with 100 μl containing either 2.5 x 109 or 5 x 109 total bacteria on the midpoint of the dorsal side. Groups were P. gingivalis WT, S. gordonii WT, S. gordonii Δcbe, S. gordonii WT with P. gingivalis WT, and S. gordonii Δcbe with P. gingivalis WT. Animals were monitored daily for general health status over 5 days, and moribund animals were euthanized and counted as non-surviving. Abscesses were harvested 2–3 days post infection by collection of the pus-filled sac, comprised of both host and bacterial cells, which was placed in RNALater and stored at −20°C. Total RNA was isolated using an RNeasy Mini kit (QIAGEN, Valencia, CA) with on-column DNase treatment and qRT-PCR performed as described above.
General statistical methods
Unless otherwise stated, statistical analysis of the data was performed with GraphPad Prism 6. For parametric tests, normal distribution of the data was examined with a D’Agostino-Pearson omnibus test, and an F-test was used to compare variance. Sample sizes were based on prior reports in the literature. For animal experiments, to minimize potential bias all mice were combined upon receipt and numerically labeled sequentially. Random integers were generated by the algorithm at random.org and mice selected for the experimental groups according to the integer. The investigator was blind for testing animal derived samples.
Supplementary Material
Acknowledgments
Supported by AMED-CREST, AMED, MEXT/JSPS KAKENHI Grant Numbers 15H05057, 15K20642 (MK), NIH grants DE012505, DE011111 (RJL), DE023193 (MW) DP1 OD009572 (EMM) and DE014372 (MH), Army Research Office Grant W911NF-12–1–0390 (EMM), and the Welch Foundation grant F1515 (EMM).
Footnotes
Data availability. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository34 http://www.ebi.ac.uk/pride with the dataset identifier PXD006153. Metabolic data have been deposited at http://www.metabolomicsworkbench.org/data/DRCCMetadata.php?Mode=Study&DataMode=AllData&StudyID=ST000384&StudyType=MS&ResultType=1 study accession number ST000384, project accession number PR000301.
Author Contributions MK, SAA, and AS performed metabolomics and community experiments. JRH, ELH, TW, and DACB performed proteomics experiments. QW and DPM performed PCR, blots, ELISAs, protease and attachment assays. QW, JAH and HW performed animal experiments. MK, MW, AA, HW, EMM, MH and RJL designed the study and interpreted data. MK, MW, AA, MH and RJL wrote the manuscript.
Competing Interests
The authors declare no competing financial interests.
Source proteomic spectral count data is available in Supplementary Tables 7–10. Additional data supporting the findings of this study are available from the corresponding author upon request.
References
- 1.Murray JL, Connell JL, Stacy A, Turner KH, Whiteley M. Mechanisms of synergy in polymicrobial infections. J Microbiol. 2014;52:188–199. doi: 10.1007/s12275-014-4067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kassebaum NJ, et al. Global Burden of Severe Periodontitis in 1990–2010: a systematic review and meta-regression. J Dent Res. 2014;93:1045–1053. doi: 10.1177/0022034514552491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamont RJ, Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med. 2015;21:172–183. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daep CA, Novak EA, Lamont RJ, Demuth DR. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect Immun. 2011;79:67–74. doi: 10.1128/IAI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright CJ, et al. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28:83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chawla A, et al. Community signalling between Streptococcus gordonii and Porphyromonas gingivalis is controlled by the transcriptional regulator CdhR. Mol Microbiol. 2010;78:1510–1522. doi: 10.1111/j.1365-2958.2010.07420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuboniwa M, et al. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol Microbiol. 2006;60:121–139. doi: 10.1111/j.1365-2958.2006.05099.x. [DOI] [PubMed] [Google Scholar]
- 8.Kubota T, et al. Production of para-aminobenzoate by genetically engineered Corynebacterium glutamicum and non-biological formation of an N-glucosyl byproduct. Metab Eng. 2016;38:322–330. doi: 10.1016/j.ymben.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 9.Maeda K, et al. A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol Microbiol. 2008;69:1153–1164. doi: 10.1111/j.1365-2958.2008.06338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuboniwa M, et al. Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiol. 2009;9:98. doi: 10.1186/1471-2180-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendrickson EL, et al. Proteomics of Streptococcus gordonii within a model developing oral microbial community. BMC Microbiol. 2012;12:211. doi: 10.1186/1471-2180-12-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wegkamp A, van Oorschot W, de Vos WM, Smid EJ. Characterization of the role of para-aminobenzoic acid biosynthesis in folate production by Lactococcus lactis. Appl Environ Microbiol. 2007;73:2673–2681. doi: 10.1128/AEM.02174-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orsomando G, et al. Evidence for folate-salvage reactions in plants. Plant J. 2006;46:426–435. doi: 10.1111/j.1365-313X.2006.02685.x. [DOI] [PubMed] [Google Scholar]
- 14.Orsomando G, et al. Plant gamma-glutamyl hydrolases and folate polyglutamates: characterization, compartmentation, and co-occurrence in vacuoles. J Biol Chem. 2005;280:28877–28884. doi: 10.1074/jbc.M504306200. [DOI] [PubMed] [Google Scholar]
- 15.Pathirana RD, O’Brien-Simpson NM, Brammar GC, Slakeski N, Reynolds EC. Kgp and RgpB, but not RgpA, are important for Porphyromonas gingivalis virulence in the murine periodontitis model. Infect Immun. 2007;75:1436–1442. doi: 10.1128/IAI.01627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wright CJ, et al. Characterization of a bacterial tyrosine kinase in Porphyromonas gingivalis involved in polymicrobial synergy. MicrobiologyOpen. 2014;3:383–394. doi: 10.1002/mbo3.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitmore SE, Lamont RJ. The pathogenic persona of community-associated oral streptococci. Mol Microbiol. 2011;81:305–314. doi: 10.1111/j.1365-2958.2011.07707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valm AM, et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc Natl Acad Sci U S A. 2011;108:4152–4157. doi: 10.1073/pnas.1101134108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamont RJ, et al. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiol. 2002;148:1627–1636. doi: 10.1099/00221287-148-6-1627. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, et al. Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients. ISME J. 2014;8:1879–1891. doi: 10.1038/ismej.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashino E, et al. Erythritol alters microstructure and metabolomic profiles of biofilm composed of Streptococcus gordonii and Porphyromonas gingivalis. Mol Oral Microbiol. 2013;28:435–451. doi: 10.1111/omi.12037. [DOI] [PubMed] [Google Scholar]
- 22.Kuboniwa M, et al. Quantitative detection of periodontal pathogens using real-time polymerase chain reaction with TaqMan probes. Oral Microbiol Immunol. 2004;19:168–176. doi: 10.1111/j.0902-0055.2004.00135.x. [DOI] [PubMed] [Google Scholar]
- 23.Houser JR, et al. Controlled measurement and comparative analysis of cellular components in E. coli reveals broad regulatory changes in response to glucose starvation. PLoS Comput Biol. 2015;11:e1004400. doi: 10.1371/journal.pcbi.1004400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon T, Choi H, Vogel C, Nesvizhskii AI, Marcotte EM. MSblender: A probabilistic approach for integrating peptide identifications from multiple database search engines. J Proteome Res. 2011;10:2949–2958. doi: 10.1021/pr2002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohashi Y, et al. Depiction of metabolome changes in histidine-starved Escherichia coli by CE-TOFMS. Mol bioSystems. 2008;4:135–147. doi: 10.1039/b714176a. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto H, et al. Statistical hypothesis testing of factor loading in principal component analysis and its application to metabolite set enrichment analysis. BMC Bioinformatics. 2014;15:51. doi: 10.1186/1471-2105-15-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirano T, Beck DA, Demuth DR, Hackett M, Lamont RJ. Deep sequencing of Porphyromonas gingivalis and comparative transcriptome analysis of a LuxS mutant. Frontiers Cell Infect Microbiol. 2012;2:79. doi: 10.3389/fcimb.2012.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, et al. FOXO responses to Porphyromonas gingivalis in epithelial cells. Cell Microbiol. 2015;17:1605–1617. doi: 10.1111/cmi.12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park Y, et al. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect Immun. 2005;73:3983–3989. doi: 10.1128/IAI.73.7.3983-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yilmaz O, Watanabe K, Lamont RJ. Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell Microbiol. 2002;4:305–314. doi: 10.1046/j.1462-5822.2002.00192.x. [DOI] [PubMed] [Google Scholar]
- 31.Capestany CA, Tribble GD, Maeda K, Demuth DR, Lamont RJ. Role of the Clp system in stress tolerance, biofilm formation, and intracellular invasion in Porphyromonas gingivalis. J Bacteriol. 2008;190:1436–1446. doi: 10.1128/JB.01632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moffatt-Jauregui CE, et al. Establishment and characterization of a telomerase immortalized human gingival epithelial cell line. J Periodontal Res. 2013;48:713–721. doi: 10.1111/jre.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sztukowska M, et al. The C-terminal domains of the gingipain K polyprotein are necessary for assembly of the active enzyme and expression of associated activities. Mol Microbiol. 2004;54:1393–1408. doi: 10.1111/j.1365-2958.2004.04357.x. [DOI] [PubMed] [Google Scholar]
- 34.Vizcaino JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:D447–456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.