Abstract
Acyl-Coenzyme As (acyl-CoAs) are a group of activated fatty acid molecules participating in multiple cellular processes including lipid synthesis, oxidative metabolism of fatty acids to produce ATP, transcriptional regulation and protein posttranslational modification. Quantification of cellular acyl-CoAs is challenging due to their instability in aqueous solutions and lack of blank matrices. Here we demonstrate an LC-MS/MS analytical method which allows for absolute quantitation with broad coverage of cellular acyl-CoAs. This assay was applied to profile endogenous acyl-CoAs under the challenge of a variety of dietary fatty acids in prostate and hepatic cells. Additionally, this approach allowed for detection of multiple fatty acid metabolic processes including the biogenesis of acyl-CoAs, and their elongation, degradation, and desaturation. Hierarchical clustering in the remodeling of acyl-CoA profiles revealed a fatty acid-specific pattern across all tested cell lines, which provides a valuable reference for making predictions in other cell models. Individual acyl-CoAs were identified which were altered differentially by exogenous fatty acids in divergent tumorigenicity states of cells. These findings demonstrate the power of acyl-CoA profiling toward understanding the mechanisms for the progression of tumors or other diseases in response to fatty acids.
For Table of Contents Only
Fatty acids play critical roles in the malignancy of cancer cells, including generating cellular building blocks for proliferation, regulating membrane structures for coordination of signal transduction and motility (e.g. lipid rafts),1 and synthesizing a variety of protumorigenic signaling molecules. A high-fat diet is a contributing factor in multiple types of cancer due to its central role in obesity.1,2 The major composition of dietary fats, ranging from vegetable to animal fats, are long-chain fatty acids with chain lengths ranging from C13 to C21 (Table 1). In addition to exogenous sources, levels of cellular fatty acids are also susceptible to metabolic abnormalities during cancer pathogenesis. For example, the elevation of monoacylglycerol lipase (MAGL) in aggressive cancers can increase the levels of free fatty acids in cancer cells.3 The current evidence to support this hypothesis was summarized in a review supporting limiting fatty acid availability as a therapeutic strategy to control cancer cell proliferation.3
Table 1.
Common dietary fatty acids and corresponding acyl-CoAs
| Fatty acids | Corresponding acyl-CoA |
|---|---|
| Decanoic acid, C10:0 | Decanoyl CoA, C10:0 CoA |
| Lauric acid, C12:0 | Lauroyl CoA, C12:0 CoA |
| Myristic acid, C14:0 | Myristoyl CoA, C14:0 CoA |
| Palmitic acid, C16:0 | Palmitoyl CoA, C16:0 CoA |
| Stearic acid, C18:0 | Stearoyl CoA, C18:0 CoA |
| Oleic acid, cis-9-C18:1 | Oleoyl CoA, cis-9-C18:1 CoA |
| Elaidic acid, trans-9-C18:1 | Elaidoyl CoA, trans-9-C18:1 CoA |
| Arachidic acid, C20:0 | Arachidoyl CoA, C20:0 CoA |
Exogenous and endogenous fatty acids require the formation of fatty acyl-coenzyme As (acyl-CoAs) before entry into bioactive lipid networks and participation in various metabolic pathways. Long-chain fatty acids are activated into acyl-CoAs by one of 13 acyl-CoA synthetase (ACS) isoforms, and are preferred substrates of long-chain acyl CoA synthetases (ACSL) including ACSL 1, 3, 4, 5 and 6. Acyl-CoAs are key transcriptional ligands and regulate critical cellular processes by complex lipid synthesis and posttranslational modification of proteins (acylation).4 The involvement of various long-chain acyl-CoAs in the development of cancer is increasingly appreciated. For example, myristoyl CoA modifies and activates a variety of oncogenic tyrosine kinases including the Src kinase family, and myristoylation of c-Src is increased in a number of human cancers, in conjunction with an upregulation of N-myristoyltransferase (NMT),5 Palmitoylation regulates a wide range of cancer-associated proteins involved with sustained proliferative signaling,6,7 resistance to cell death,8 invasion and metastasis,9 angiogenesis,10 and induction of inflammation.11
Considering the important roles of acyl-CoAs and related enzymes in physiological and pathological pathways, especially in cancer, it’s necessary to have an analytical method to profile acyl-CoAs in cells. In addition, to elucidate how a high-fat diet or enzyme abnormality-induced elevation of fatty acids correlates with tumorigenesis and progression, it would be meaningful to make a comprehensive assessment of alterations to the acyl-CoA profile following stress from the addition of different fatty acids. Given enzymatic activities including acyl-CoA synthesis, degradation and elongation, we reasoned that remodeling of the acyl-CoA profile would occur under such conditions. High performance liquid chromatography tandem mass spectrometry (LC-MS/MS) proved to be a reliable analytical technology for determining acyl-CoAs. Considerable efforts have been devoted to develop LC-MS/MS methods for acyl-CoAs profiling in bacteria,12 cancer cells,13, 14 and animal tissues.14, 15 However, most methods only determined relative levels of acyl-CoAs, and there is no method aimed at absolute determination of long-chain acyl-CoAs. More noteworthy, until now, there are no quantitative data reporting the alterations of acyl-CoAs in the presence of fatty acids, or a comparison of acyl-CoA synthesis between normal and cancer cells.
In this paper, we therefore established a new method, with protein precipitation for sample preparation, reversed phase chromatography for separation, followed by multiple reaction monitoring (MRM) mass spectrometry to simultaneously quantitate a variety of acyl-CoAs converted from dietary fatty acids in mammalian cells. Hepatic cells are the major element in the liver for fatty acid metabolism. Additionally, increasing evidence suggests an association of dietary fatty acids with prostate cancer initiation and progression.16,17,18,19 This analytical approach reveals a signature of these cells in multiple fatty acid metabolic processes in addition to metabolic modification after tumorigenic alteration.
EXPERIMENTAL PROCEDURES
Reagents and Materials
Various acyl coenzyme As, ammonium salts (acyl-CoAs), including C10:0 CoA, C12:0 CoA, C14:0 CoA, C15:0 CoA, C16:0 CoA, C18:0 CoA, C18:1 (n9) CoA, C18:2 (n6) CoA, C18:3 (n3) CoA, C20:0 CoA, C20:4 CoA, were purchased from Avanti Polar Lipids (Alabaster, AL). Ammonium acetate, LC-MS grade acetonitrile, methanol and water were purchased from Sigma–Aldrich (St. Louis, MO).
Instrumentation
An Agilent 1100 binary pump HPLC system (Santa Clara, CA) interfaced to a Waters Micromass Quattro Micro triple quadrupole mass spectrometer with an ESI source (Milford, MA) was operated for LC–MS/MS analysis. Masslynx 4.0 software by Waters (Beverly, MA) was applied for data processing. A Labconco CentriVap Complete Vacuum Concentrator (Kansas City, MO) was used to evaporate samples.
LC-MS/MS conditions
A Luna® C18(2) 100 Å LC column (100 × 2 mm, 3 µm) coupled with a Phenomenex SecurityGuard C-18 guard column (4.0 mm × 2.0 mm) was used to separate the analytes. The column temperature was controlled at 32 °C. The mobile phase A was 10 mM ammonium acetate (pH 6.8), and mobile phase B was acetonitrile. The injection volume was 30 µL. Analytes were separated using a gradient method, with a 0.2 mL/min flow rate, (time/minute, % mobile phase B): (0, 20), (15, 100), (22.5, 100), (22.51, 20), and the run time for each injection was 30 min. The LC system was interfaced by a six-port divert valve to the mass spectrometer, introducing eluates from 1 to 15 min to the ion source. The autosampler injection needle was washed with methanol after each injection. Samples were analyzed by the mass spectrometer in positive ion ESI mode. MS parameters were optimized by direct infusion of 5 µM acyl-CoAs dissolved in 50% acetonitrile at 30 µL/min into the instrument. The capillary voltage was 3.20 kV, the cone voltage was 45 V, the extractor voltage was 0 and RF lens voltage was 3.0 V. Nitrogen was used as the desolvation gas at a flow rate of 500 L/h. The desolvation temperature was 500 °C and the source temperature was 120 °C. Argon was used as the collision gas, the collision cell pressure was 3.5 × 10−3 mbar. A multiple reaction monitoring (MRM) function was applied for the simultaneous detection of all analytes. Ion transitions, retention times and collision energies for all acyl-CoAs are listed in Table 2.
Table 2.
Acyl-CoAs in this analytical assay.
| Q1 (m/z) | Q3 (m/z) | CE (eV) | RT (min) | LOD (pmol)a | LOQ (pmol)a | |
|---|---|---|---|---|---|---|
| C10:0 CoA | 922 | 415 | 32 | 7.30 | 0.14 | 0.47 |
| C12:0 CoA | 950 | 443 | 36 | 8.26 | 0.11 | 0.35 |
| C14:0 CoA | 978 | 471 | 40 | 9.14 | 0.12 | 0.39 |
| C15:0 CoA | 992 | 485 | 40 | 9.63 | 0.09 | 0.31 |
| C16:0 CoA | 1006 | 499 | 40 | 10.07 | 0.07 | 0.25 |
| C18:0 CoA | 1034 | 527 | 42 | 11.04 | 0.07 | 0.24 |
| C18:1 CoA | 1032 | 525 | 42 | 10.25 | 0.11 | 0.36 |
| C18:2 CoA | 1030 | 523 | 42 | 9.69 | 0.12 | 0.38 |
| C18:3 CoA | 1028 | 521 | 42 | 9.26 | 0.14 | 0.46 |
| C20:0 CoA | 1062 | 555 | 44 | 12.23 | 0.07 | 0.25 |
| C20:4 CoA | 1054 | 547 | 44 | 9.66 | 0.10 | 0.33 |
CE: collision energy, RT: retention times, LOD: limit of detection, LOQ: limit of quantitation.
Amount in a single injection.
Cell Culture
Human prostate cell lines (PNT2 and DU145) and hepatic cell lines (HepG2 and Hep3B) were purchased from American Type Culture Collection (ATCC). Cells were maintained in a humidified incubator under 5% CO2 at 37°C and grown in the recommended media supplemented with 5% fetal bovine serum (FBS) and 1% penicillin/streptomycin. When cells reached ~90% confluence, bovine serum albumin (BSA)-conjugated fatty acids including decanoic acid (C10:0), lauric acid (C12:0), myristic acid (C14:0), palmitic acid (C16:0), stearic acid (C18:0), oleic acid (cis-9-C18:1), elaidic acid (trans-9-C18:1) and arachidic acid (C20:0), were individually introduced to each cell line (n = 3) at a concentration of 400 µM and incubated for 24 h.
Sample Preparation
Cell culture media were removed and the cells were washed with phosphate-buffered saline (PBS) twice, incubated with 2 mL methanol and 15 µL 10 µM 15:0 CoA (internal standard, ISTD) at −80 °C for 15 min. The cells lysate was scraped from the culture plate and centrifuged at 15000 × g at 5 °C for 5 min. The supernatant was transferred to a glass tube, mixed with 1 mL of acetonitrile and evaporated in a vacuum concentrator at 55 °C for 1.5h. The sample was reconstituted with 150 µL methanol, vortexed and centrifuged at 15000 × g at 5 °C for 10 min. 100 µL supernatant was transferred into an autosampler vial for LC-MS/MS analysis.
Determination of Total Cellular Protein
Precipitated protein was solubilized using a RIPA lysis buffer containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM EDTA, 10% glycerol, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate and protease inhibitor cocktail. Protein concentration was determined using the Bio-Rad DC protein assay kit.
Determination of mRNAs for Long-chain Acyl-CoA Synthetase Isoforms
The total RNA was isolated from different cell lines using the Qiagen RNeasy Mini Kit (Valencia, CA). 1 µg of the total RNA was used for first-strand cDNA synthesis as recommended by the manufacturer Quanta Biosciences (Beverly, MA). Real time PCR was performed using SYBR Green as an indicator on the ABI StepOne Plus Real Time PCR system. The final reaction mixture contained 10 ng of cDNA, 100 nM of each primer, 5 µL of 2× SYBR Green PCR Master Mix (from Quanta Biosciences) and RNase-free water, to complete the reaction at a volume of 12.5 µL. PCR was carried out for 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Fluorescence was read during the reaction, allowing a continuous monitoring of the amount of PCR product. The data were normalized to internal control glyceralde hyde-3-phosphate dehydrogenase (GAPDH) mRNA. The sequences of primers are as shown in Table S1.
Quantitation of Acyl-CoAs in Cells
For each cell line, calibration samples (n = 3) were prepared by mixing 2.4, 6, 12, 24, 60, 120, 240 and 600 pmol acyl-CoAs with 8 × 105 cells prior to sample preparation. 11 acyl-CoA compounds were included in the calibration samples, and a calibration curve for each analyte was plotted using the peak area versus the amount of supplemented acyl-CoA, with 1/x weighted linear regression. Slopes (k) of the linear regression of the analyte’s calibration curve and the internal standard (ISTD) calibration curve were calculated, and the ratio of slopes (R) = kanalyte / kISTD were calculated. In a real sample spiked with 150 pmol ISTD, the amount (pmol) of an analyte was calculated when its signal-to-noise ratio > 10, using the formula:
To correct for potential differences in the cell number among samples, the analyte amount was normalized to per mg of total cellular protein, and the final unit for quantitation of acyl-CoAs in cells is pmol/mg (about 3 × 106 cells).
Statistical Analysis
For an analyte with a signal-to-noise ratio > 10, the level is regarded as above the limit of quantitation and is included for further data analysis. Calculation of peak areas and the linear regression were performed with Masslynx 4.0 software. Data were presented as mean ± SD. Significant differences are based on a student’s t-test (two tailed) with P < 0.05. Heat map and hierarchical clustering were done using the software Gene-e (Broad Institue, http://www.broadinstitute.org/cancer/software/GENE-E/download.html).
RESULTS AND DISCUSSION
Method Development
C15:0 CoA (pentadecanoyl CoA) is an odd-chain acyl-CoA undetectable in human cells. C15:0 CoA and other odd-chain acyl-CoAs have been reported to work as an ISTD for acyl-CoAs in other analytical assays.13,20 C15:0 CoA is selected as the ISTD for this assay because it has a median length of carbon chain among all acyl-CoAs to be determined.
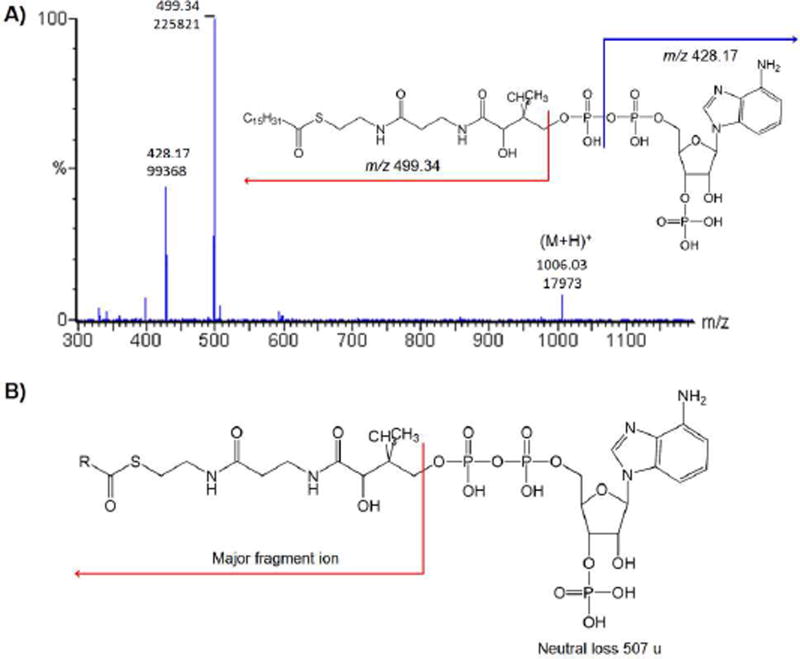
The autotune mode imbedded in Masslynx 4.0 software by Waters was utilized to optimize MS parameters to increase signal response, accompanied by a direct infusion of a standard solution containing all acyl-CoAs (5 µM in 50% acetonitrile) at 30 µL/min into the instrument. Product ion mass spectra for acyl-CoA were acquired to determine the precursor and production pairs to monitor. For all acyl-CoAs, the most abundant fragment ion was the one formed by a neutral loss of the phosphorylate ADP moiety (M-507) from the precursor ion (Figure 1). Ion transitions for detection of acyl-CoAs are listed in Table 2.
Figure 1.
Fragment ions of acyl-CoAs. (A) Fragment ions of C16:0 CoA. (B) The major fragmentation mechanism for acyl-CoAs: a neutral loss of 507.
Various acyl-CoAs share similar physicochemical properties, allowing one sample preparation method to extract all of them. Acyl-CoAs are soluble in water and methanol. 2 mL methanol and 2 mL 80% methanol/20% water were tested to lyse cells and extract acyl-CoAs. The former was finally selected as the extraction solvent because it took 1.5 h to completely evaporate the methanol extract at 55 °C in the vacuum concentrator, while it took the latter 4 h under the same conditions and presented a poorer signal for analytes. This is probably because of a higher level of water in the latter extract, which likely increased during evaporation, facilitated the hydrolysis of acyl-CoAs. 1 mL of acetonitrile was mixed with the extract before evaporation, to decrease the ratio of water derived from cell lysis and to facilitate evaporation.
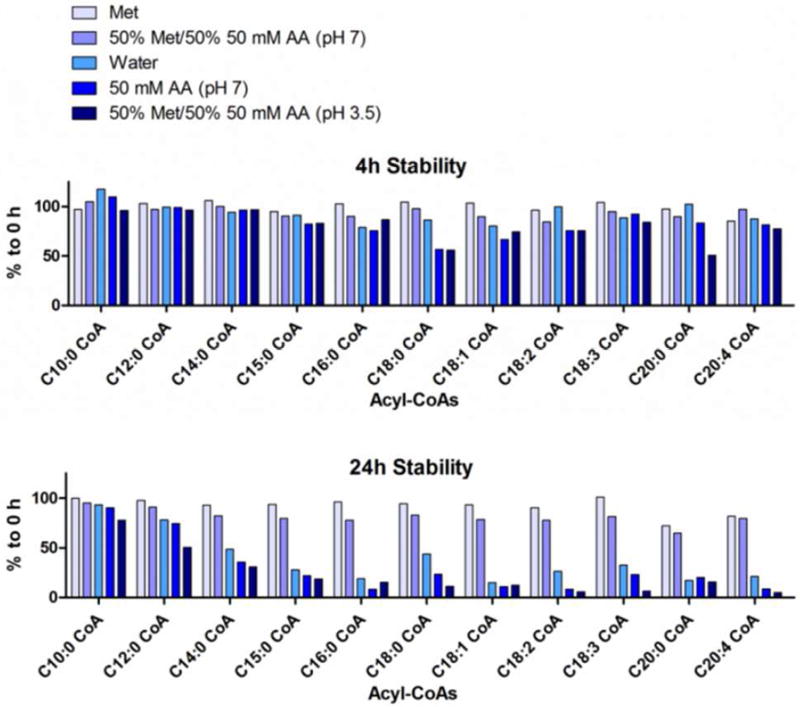
Acyl-CoAs are prone to hydrolysis, and are unstable in aqueous, especially alkaline and strongly acidic solutions. The stability of acyl-CoAs was tested in a series of solutions, to determine the reconstitution solution for dry samples after evaporation. Tested solutions include methanol, 50% methanol/50% 50 mM ammonium acetate (pH 7), water, 50 mM ammonium acetate (pH 7) and 50% methanol/50% 50 mM ammonium acetate (pH 3.5, adjusted with acetic acid). 500 nM acyl-CoAs in the above solutions were placed on the autosampler, and each sample was analyzed at time-zero, 4 h and 24h. The stability of each acyl-CoA was demonstrated as the percentage relative to the time-zero sample (Figure 2). Methanol was selected to reconstitute samples, because it provided the best stability for analytes over the tested periods of time. Another interesting observation was the instability of acyl-CoAs in aqueous solutions, except for 50% methanol/50% 50 mM ammonium acetate (pH 7), which caused deterioration that increased with the length of fatty acid chain especially from 10:0 CoA to 16:0 CoA. This is possibility due to a longer fatty acid chain facilitating the hydrolysis of acyl-CoAs.
Figure 2.
Stability of acyl-CoAs in solutions for reconstitution.
Separation of acyl-CoAs
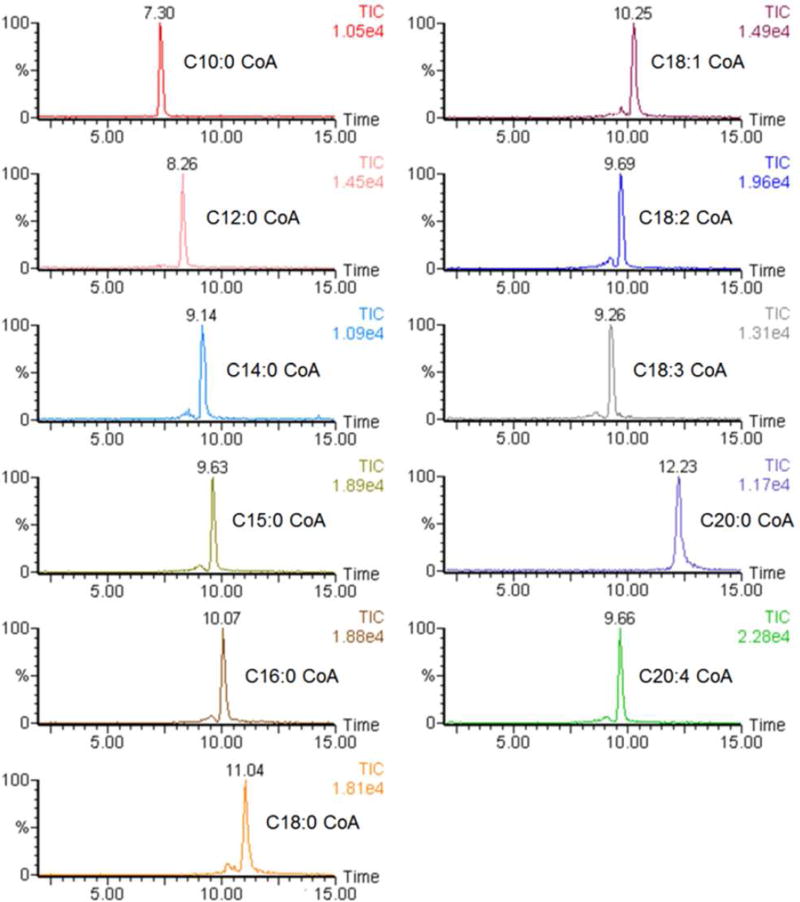
Chromatographic separation is important for analyzing acyl-CoAs when using LC-MS/MS, to reduce the competition for ionization between co-eluting analytes and other endogenous species, which would otherwise lead to serious ion suppression. A good separation was achieved with the LC method, with little co-elution of analytes. As shown in Figure 3, 11 acyl-CoA standard compounds, 150 pmol each, was retained on the column and yielded retention times between 7 and 13 min. The retention time increases with the length of fatty acid chain of the analyte, and decreases with the number of double bonds in the fatty acid chain.
Figure 3.
Extracted ion chromatograms of 150 pmol acyl-CoA standards.
Detection of acyl-CoAs in cells
Calibration curves (n = 3) for acyl-CoAs in PNT2, DU145, HepG2 and Hep3B cell lines were prepared as described above. The coefficient of determination is over 0.98 for either analyte in either cell line, suggesting a strong linear relationship between the amount of the supplemented acyl-CoA and the response. Slopes (k) of the linear regression of the analyte’s calibration curves and the ISTD calibration curve were calculated, and the ratio of slopes (R) = kanalyte / kISTD are shown in Table 3. The ratios are all between 0.6 and 1.6, but are slightly different for the same analyte in different cell lines, suggesting similar but different matrix effects from different cells.
Table 3.
Ratio of slopes between the analyte and the ISTD in the cell line from the calibration curve (n = 3).
| PNT2 | DU145 | HepG2 | Hep3B | |
|---|---|---|---|---|
| C10:0 CoA | 0.66 ± 0.03 | 0.61 ± 0.00 | 0.69 ± 0.00 | 0.74 ± 0.03 |
| C12:0 CoA | 0.77 ± 0.05 | 0.86 ± 0.06 | 0.98 ± 0.02 | 1.09 ± 0.05 |
| C14:0 CoA | 1.16 ± 0.07 | 0.64 ± 0.02 | 0.77 ± 0.01 | 0.70 ± 0.05 |
| C16:0 CoA | 1.60 ± 0.08 | 1.05 ± 0.02 | 1.28 ± 0.08 | 1.17 ± 0.07 |
| C18:0 CoA | 1.45 ± 0.05 | 1.51 ± 0.07 | 1.01 ± 0.05 | 1.37 ± 0.14 |
| C18:1 CoA | 0.78 ± 0.03 | 0.84 ± 0.12 | 1.02 ± 0.18 | 0.80 ± 0.06 |
| C18:2 CoA | 0.67 ± 0.02 | 0.84 ± 0.05 | 0.91 ± 0.03 | 0.92 ± 0.06 |
| C18:3 CoA | 0.57 ± 0.03 | 0.67 ± 0.05 | 0.77 ± 0.00 | 0.78 ± 0.06 |
| C20:0 CoA | 1.48 ± 0.04 | 1.40 ± 0.11 | 0.98 ± 0.08 | 1.36 ± 0.06 |
| C20:4 CoA | 0.79 ± 0.05 | 0.92 ± 0.02 | 1.11 ± 0.04 | 1.03 ± 0.07 |
The limit of detection (LOD) and the limit of quantitation (LOQ) are two key parameters for evaluating the sensitivity of an analytical assay. They were defined at a signal-to-noise (S/N) of 3, and at an S/N of 10, respectively. All analytes demonstrated a signal-to-noise ratio above 10 except C18:3 CoA in PNT2 and DU145 cells. The values for the ISTD were calculated from cells (n=3) spiked with 2.4 pmol 15:0 CoA standard, for a single injection (30 µL of the 150 µL final reconstituted sample) into the system. The LOD and LOQ for all analytes in each cell line were calculated using the values of the ISTD and their response ratio to the ISTD in Table 3, and the average values among tested cell lines are listed in Table 2.
Differential levels of endogenous acyl-CoA metabolites
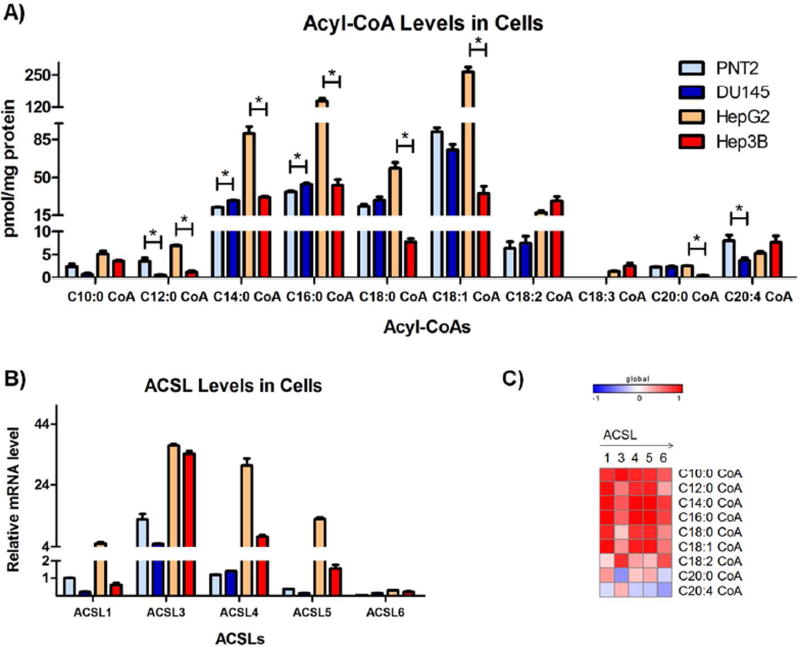
Examples for chromatograms of acyl-CoA compounds in cells (HepG2) were shown in Figure S1. The levels of acyl-CoAs including C10, C12, C14, C16, C18:0, C18:1, C18:2 and C18:3 CoA were higher in HepG2 cells (normal hepatic cells) than PNT2 cells (normal prostate cells) (Figure 4A and Table S2). Interestingly, the levels of acyl-CoAs were correlated with the mRNA levels of ACSL genes (Figure4B and 4C), indicating a higher activity of fatty acid metabolism in hepatic cells. Acyl-CoAs including C14, C16, C18:0 CoA and C18:1 CoA were predominant in the tested cells. The levels of these acyl-CoAs together with 12:0 CoA and 20:0 CoA were less expressed in Hep3B cells in comparison with HepG2 cells (P = 0.001, 0.001, 0.01, 0.0003, 0.0001, 0.00008, respectively), suggesting a loss of function for fatty acid metabolism in tumorigenic cells. The decreased levels of these acyl-CoAs were also correlated with the lower mRNA levels of ACSLs in Hep3B cells in comparison with HepG2 cells (Figure 4A and B). In two prostate cells lines, DU145 cells (prostate tumorigenic cells) had significantly lower levels of C12:0 CoA and C20:4 CoA (P = 0.04, 0.03, respectively), and significantly higher levels of C14:0 CoA and C16:0 CoA (P = 0.0002, 0.01, respectively) than PNT2 cells (nontumorigenic prostate cells). Collectively, the acyl-CoA profiles reflect the specificity in hepatic function and its alteration in fatty acid metabolism after tumorigenic change.
Figure 4.
The Acyl-CoA levels and the mRNA expression of ACSL genes in prostate and hepatic cells. The prostate cells, PNT2 (normal prostate cells) and DU145 (prostate cancer cells), and hepatic cells, HepG2 (normal hepatic cells) and Hep3B (hepatic tumorigenic cells) were grown in the media recommended by ATCC. A) Cellular levels of acyl-CoAs measured by LC-MS/MS. “*” represents significant differences between nontumorigenic and tumorigenic cells (P < 0.05). B) Expression levels of ACSL1, 3, 4, 5 and 6 measured by RT-PCR. C) The spearman correlation between the level of individual acyl-CoA and the levels of each ACSL gene.
Acyl-CoA synthesis from exogenous fatty acids
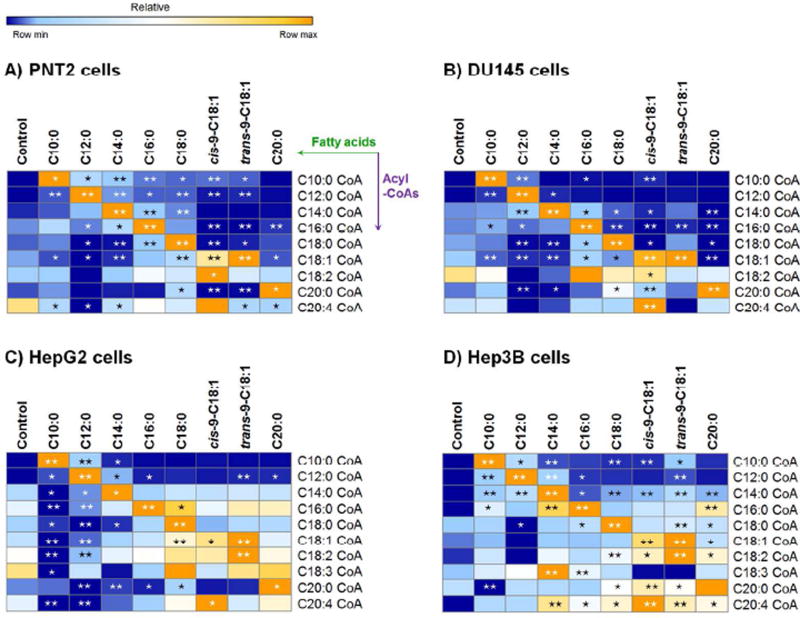
The fatty acids decanoic acid (C10:0), lauric acid (C12:0), myristic acid (C14:0), palmitic acid (C16:0), stearic acid (C18:0), oleic acid (cis-9-C18:1), elaidic acid (trans-9-C18:1) and aracidic acid (C20:0) are typical dietary fatty acids in high fat diets for animals and humans. Heat maps were generated to provide a graphical representation of acyl-CoA levels, particularly to reflect deviations from baseline levels when under the stress of exogenous fatty acids (Figure 5 and Table S2). The alteration of intracellular acyl-CoAs reflected fatty acid metabolism during the biosynthesis, degradation, elongation, and desaturation of acyl-CoAs, and different metabolic rates for the C18:1 isomers. While the heat map for each cell line was unique, some general rules are identifiable:
Biosynthesis of acyl-CoAs: The most noticeable elevation of an acyl-CoA always occurred after cells were treated with the fatty acid of the same length and degree of saturation, which corresponds to the process of fatty acid activation by acyl-CoA synthetases.
Degradation of acyl-CoAs: Treatment with an n-carbon saturated fatty acid (Cn:0) commonly resulted in a great increase of the C(n-2):0 CoA and smaller increases of other shorter acyl-CoAs, especially when n = 10~16. For example, palmitic acid (C16:0) significantly/very significantly increased the levels of C14:0 CoA, C12:0 CoA and C10:0 CoA in PNT2 cells, C14:0 CoA and C10:0 CoA in DU145 cells, C12:0 CoA in HepG2 cells, and C14:0 CoA and C12:0 CoA in Hep3B cells. The increases are possibly due to β-oxidation of the synthesized acyl-CoA inside the mitochondria, which yields an acetyl-CoA and a fatty acyl-CoA two carbons shorter after each cycle.
Elongation of acyl-CoAs: Likewise, treatment with an Cn:0 resulted in increases of the C(n+2):0 CoA in most cell lines. For example, treatment of C10:0 significantly increased the levels of C12:0 CoA in all cell lines; C12:0 increased C14:0 CoA in DU145, HepG2, and Hep3B cells; and C16:0 increased C18:0 CoA in PNT2, DU145, and Hep3B cells. The increase of C18:0 CoA in the presence of C16:0 is a reflection of fatty acid elongation in the endoplasmic reticulum, catalyzed by the very long chain fatty acid synthase complex, where C16:0 CoA is the substrate.21 Increases of shorter acyl-CoAs can be mediated by the fatty acid synthase complex in the cytoplasm, through the addition of two carbons from malonyl-CoA. This process is initiated by the sequential transfer of an acyl-CoA to the acyltransferase domain, to an acyl carrier protein (ACP) domain, and finally to the β-ketoacyl synthase domain. Multiple enzymes involved in fatty acid synthesis have broad specificity which ensures efficient elongation all the way up to C16:0 CoA.22,23
Competitive inhibition of ACSL: Decreases of C(n+m):0 CoA (m > 2) after the treatment with Cn:0 were also observed, such as significant decreases of C18:0 CoA in almost all cell lines following the metabolism of lauric acid (C12:0) and myristic acid (C14:0). The observation can be interpreted as the exogenous fatty acid inhibiting the synthesis of other acyl-CoAs from endogenous fatty acids, by competing for the same binding site on ACSLs. In addition, the decreases were frequently observed in Hep G2 cells, including C14:0 CoA and C16:0 CoA in the presence of decanoic acid (C10:0), C14:0 CoA and C16:0 CoA in the presence of lauric acid (C12:0). This might be due to the higher abundance of ACSLs in HepG2 cells, which the ACSL-mediated biosynthesis is more active for the generation of acyl-CoAs (Figure 4B). The decreases observed for C(n+m):0 CoA (m > 2) in cells cultured with Cn:0, suggest that the contribution from elongation would decline after multiple cycles, therefore being insufficient to offset other factors which may decrease C(n+m):0 CoA.
Desaturation of acyl-CoAs: C18:1 CoA and C18:2 CoA were significantly increased in some cells cultured with C18:0, cis-9-C18:1 or trans-9-C18:1. The desaturation process can be mediated either by fatty acid desaturases, a major pathway for unsaturated fatty acid synthesis., or by acyl-CoA dehydrogenase, which catalyzes the dehydrogenation process of acyl-CoAs in fatty acid β-oxidation.
Different metabolic rates for C18:1 fatty acid isomers: The metabolism of trans-9-C18:1 was similar to cis-9-C18:1 in terms of intracellular alterations of acyl-CoAs, however the levels of C18:1 CoA were slightly higher in the presence of trans-9-C18:1 than cis-9-C18:1. The difference is in agreement with previous reports indicating a slower degradation of trans-9-C18:1 CoA than cis-9-C18:1 CoA in rat heart mitochondria.24 Trans-unsaturated fatty acids mainly originate from the food industry. It was reported that the degradation of the trans-fatty acid (elaidic acid, trans-9-C18:1) was effective but compromised when compared to its cis isomer (oleic acid, cis-9-C18:1) or its saturated form (stearic acid, C18:0), because the β-oxidation intermediate 5-trans-tetradecenoyl-CoA (trans-5-C14:1CoA) is a poorer substrate for long-chain acyl-CoA dehydrogenase (LCAD).25 As a result, the difference could lead to alterations in its accumulation, hydrolysis and conversion into an acylcarnitine, and subsequently a deviation from the metabolic process in the mitochondria.25
Figure 5.
The acyl-CoA profile in prostate and hepatic cells. PNT2 (A), DU145 (B), HepG2 (C) and Hep3B (D) cells were cultured in media with and without 400 µM of individual fatty acids including decanoic acid (C10:0), lauric acid (C12:0), myristic acid (C14:0), palmitic acid (C16:0), stearic acid (C18:0), oleic acid (cis-9-C18:1), elaidic acid (trans-9-C18:1) and arachidic acid (C20:0) for 24 hours (n =3). Significant changes for each acyl-CoA between the control (the baseline level) and the treatment of individual fatty acid in respective group were labeled. *: p < 0.05, **: p < 0.01.
Next, exogenous cis-9-C18:1 led to higher levels of C10:0 CoA than trans-9-C18:1. The increases could be explained by the β-oxidation process. Because of the location of the double bond, after three cycles of β-oxidation, elaidoyl CoA and oleoyl CoA would respectively generate trans-Δ3-dodecenoyl-CoA and cis-Δ3-dodecenoyl-CoA, which can be catalyzed by enoyl CoA isomerase to trans-Δ2-dodecenoyl-CoA, a substrate of β-oxidation and subsequently be degraded to C10:0 CoA.26,27 Additionally, the increase of C12:0 CoA was possibly due to elongation from C10:0 CoA, and changes of C14:0 CoA, 16:0 CoA and 18:0 CoA resulted from a mixture of competition for ACSLs (decreases) and elongation from 10:0 CoA (increases).
Unique signature of individual fatty acid metabolism
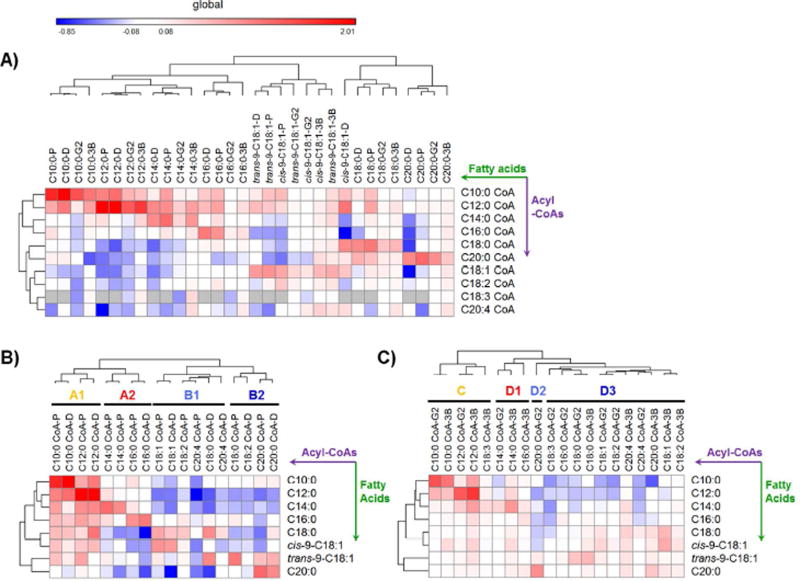
To analyze if the metabolism of individual fatty acids has a metabolic signature, hierarchical clustering on the levels of acyl-CoA metabolites was applied to the tested cell lines under the stress of a variety of fatty acids. The analysis was conducted based on the log ratio values due to a wide range of acyl-CoA abundance (0.5 pmol/mg ~ over 500 pmol/mg) in different cell lines and treatments. Clearly, cells cultured with the same saturated fatty acid were clustered despite the difference of cell types (Figure 6A), demonstrating that the metabolism of individual fatty acids has its signature from remodeling the intracellular acyl-CoA profile. This finding allows for a prediction on the alterations of the acyl-CoA profiles in other cells under the stress of the same saturated fatty acids. In addition, C18:1 trans and cis fatty acid isomers were clustered together (except cis-9-C18:1 in DU145 cells), demonstrating a similar impact on acyl-CoA remodeling from C18:1 isomers. Conversely, a different remodeling by cis-9-C18:1 in DU145 cells suggested a distinct alteration of the metabolic pathways for this unsaturated fatty acid in this prostate cell line.
Figure 6.
Remodeling of the acyl-CoA profile across cell lines after treatment with various fatty acids. A) Hierarchical clustering of cells with a variety of fatty acid treatments based on remodeling of the acyl-CoA profile. B–C) Hierarchical clustering of acyl-CoA changes (log ratio) in B) prostate cells and C) hepatic cells in response to fatty acids. The log ratio of the measure is defined as the average amount of an acyl-CoA in a treatment group over the amount in a control group. The color code for log ratio 0.08 indicates a log ratio ≥ 0.08 (log 1.2) and for −0.08 indicates log ratio ≤ −0.08. (P: PNT2 cells, D: DU145 cells, G2: HepG2 cells and 3B: Hep3B cells)
Tumorigenesis-associated fatty acid metabolism
To discover tumorigenesis-associated abnormalities in acyl-CoA synthesis and metabolism, we merged the changes (log ratio) of acyl-CoAs in tumorigenic and nontumorigenic cells from the cell type. Hierarchical clustering was performed to cluster cellular acyl-CoAs with similar changes under a panel of fatty acids (Fig 6 B and C). This analysis revealed four groups (A1, A2, B1 and B2) in prostate cells, and four groups (C, D1, D2 and D3) in hepatic cells. The same length acyl-CoAs were clustered in the same group, despite the disparity in tumorigenecity, such as C10:0 CoA, C12:0 CoA, C14:0 CoA, C16:0 CoA and 20:0 CoA in prostate cells. However, C18:0 CoA and 18:2 CoA were segregated into different groups between normal and tumorigenic prostate cells. C16:0 CoA, C18:3 CoA and C20:0 CoA were also clustered into different groups in normal or tumorigenic hepatic cells. The segregations suggest metabolic alterations between tumorigenic and normal cells under the challenge of exogenous fatty acids in acyl-CoA synthesis. The mechanisms for the alterations are not yet clear, but could be a result of dysregulation of multiple enzymes in the neoplasm, including ACSLs,28, 29 acyltransferases,30 desaturases,28 or fatty acid synthases30.
CONCLUSION
We developed a sensitive LC-MS/MS analytical method to quantitate acyl-CoAs in prostate and hepatic cell lines. Calibration curves were made from serially supplemented acyl-CoAs, and pentadecanoyl CoA (C15:0 CoA) was used as the internal standard. For the first time, how a variety of dietary fatty acids affect the pool of cellular acyl-CoAs was determined quantitatively, and interpretation of data reflected the involvement of multiple pathways and enzymes in acyl-CoA synthesis and metabolism. Hierarchical clustering in the remodeling of acyl-CoA profiles revealed a fatty acid-specific pattern across all cell lines. Individual acyl-CoAs which corresponded differently to fatty acids in cells at divergent states of tumorigenicity were identified. Altogether, we believe this method provides a valuable tool for studying the role of fatty acids and subsequent remodeling of the acyl-CoA profile, in the progression of tumors and other diseases associated with nutrition and/or metabolic abnormalities.
Supplementary Material
Acknowledgments
This research was supported by NIH (R01CA172495) and DOD (W81XWH-15-1-0507).
Footnotes
ASSOCIATED CONTENT
Figure S1 shows Representative chromatograms of acyl-CoA analytes in HepG2 cells. Table S1 shows primer sets for real-time RT-PCR analysis of expression of ACSL genes. Table S2 shows amounts of acyl-CoAs (pmol/mg protein) in PNT2, DU145, HepG2 and Hep3B cells (n = 3) incubated with 400 µM fatty acids for 24 h.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
References
- 1.Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Cell. 2010;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allott EH, Hursting SD. Endocr.-Relat. Cancer. 2015;22:R365–R386. doi: 10.1530/ERC-15-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Currie E, Schulze A, Zechner R, Walther TC, Farese RV. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grevengoed TJ, Klett EL, Coleman RA. In: Annual Review of Nutrition. Cousins RJ, editor. Vol. 34. Annual Reviews; Palo Alto: 2014. pp. 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selvakumar P, Lakshmikuttyamma A, Shrivastav A, Das SB, Dimmock JR, Sharma RK. Prog. Lipid Res. 2007;46:1–36. doi: 10.1016/j.plipres.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Yeste-Velasco M, Linder ME, Lu Y. J. Biochim. Biophys. Acta-Rev. Cancer. 2015;1856:107–120. doi: 10.1016/j.bbcan.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Schmick M, Kraemer A, Bastiaens PIH. Trends Cell Biol. 2015;25:190–197. doi: 10.1016/j.tcb.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Chakrabandhu K, Herincs Z, Huault S, Dost B, Peng L, Conchonaud F, Marguet D, He HT, Hueber AO. Embo J. 2007;26:209–220. doi: 10.1038/sj.emboj.7601456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou B, Liu L, Reddivari M, Zhang XA. Cancer Res. 2004;64:7455–7463. doi: 10.1158/0008-5472.CAN-04-1574. [DOI] [PubMed] [Google Scholar]
- 10.Hemler ME. Nat. Rev. Cancer. 2014;14:49–60. doi: 10.1038/nrc3640. [DOI] [PubMed] [Google Scholar]
- 11.Chow A, Zhou WY, Liu L, Fong MY, Champer J, Van Haute D, Chin AR, Ren XB, Gugiu BG, Meng ZP, Huang WD, Ngo V, Kortylewski M, Wang SE. Sci Rep. 2014;4:11. doi: 10.1038/srep05750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zimmermann M, Thormann V, Sauer U, Zamboni N. Anal. Chem. 2013;85:8284–8290. doi: 10.1021/ac401555n. [DOI] [PubMed] [Google Scholar]
- 13.Haynes CA, Allegood JC, Sims K, Wang EW, Sullards MC, Merrill AH. J. Lipid Res. 2008;49:1113–1125. doi: 10.1194/jlr.D800001-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu XJ, Sadhukhan S, Sun SY, Wagner GR, Hirschey MD, Qi L, Lin HN, Locasale JW. Mol. Cell. Proteomics. 2015;14:1489–1500. doi: 10.1074/mcp.M114.044859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao L, Chiou W, Tang H, Cheng XH, Camp HS, Burns DJ. J. Chromatogr. B. 2007;853:303–313. doi: 10.1016/j.jchromb.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 16.Crowe FL, Allen NE, Appleby PN, Overvad K, Aardestrup IV, Johnsen NF, Tjonneland A, Linseisen J, Kaaks R, Boeing H, Kroger J, Trichopoulou A, Zavitsanou A, Trichopoulos D, Sacerdote C, Palli D, Tumino R, Agnoli C, Kiemeney LA, Bueno-de-Mesquita HB, Chirlaque MD, Ardanaz E, Larranaga N, Quiros JR, Sanchez MJ, Gonzalez CA, Stattin P, Hallmans G, Bingham S, Khaw KT, Rinaldi S, Slimani N, Jenab M, Riboli E, Key TJ. Am. J. Clin. Nutr. 2008;88:1353–1363. doi: 10.3945/ajcn.2008.26369. [DOI] [PubMed] [Google Scholar]
- 17.Strom SS, Yamamura Y, Forman MR, Pettaway CA, Barrera SL, DiGiovanni J. Int. J. Cancer. 2008;122:2581–2585. doi: 10.1002/ijc.23414. [DOI] [PubMed] [Google Scholar]
- 18.Epstein MM, Kasperzyk JL, Mucci LA, Giovannucci E, Price A, Wolk A, Hakansson N, Fall K, Andersson SO, Andren O. Am. J. Epidemiol. 2012;176:240–252. doi: 10.1093/aje/kwr520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iordanescu G, Brendler C, Crawford SE, Wyrwicz AM, Venkatasubramanian PN, Doll JA. J. Magn. Reson. Imaging. 2015;42:651–657. doi: 10.1002/jmri.24824. [DOI] [PubMed] [Google Scholar]
- 20.Mauriala T, Herzig KH, Heinonen M, Idziak J, Auriola S. J. Chromatogr. B. 2004;808:263–268. doi: 10.1016/j.jchromb.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 21.Riezman H. Cell. 2007;130:587–588. doi: 10.1016/j.cell.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Jakobsson A, Westerberg R, Jacobsson A. Prog. Lipid Res. 2006;45:237–249. doi: 10.1016/j.plipres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Smith S, Witkowski A, Joshi AK. Prog. Lipid Res. 2003;42:289–317. doi: 10.1016/s0163-7827(02)00067-x. [DOI] [PubMed] [Google Scholar]
- 24.Lawson LD, Kummerow FA. Biochimica et biophysica acta. 1979;573:245–254. doi: 10.1016/0005-2760(79)90058-4. [DOI] [PubMed] [Google Scholar]
- 25.Yu WF, Liang XQ, Ensenauer RE, Vockley J, Sweetman L, Schulz H. J. Biol. Chem. 2004;279:52160–52167. doi: 10.1074/jbc.M409640200. [DOI] [PubMed] [Google Scholar]
- 26.Mursula AM, van Aalten DMF, Hiltunen JK, Wierenga RK. J. Mol. Biol. 2001;309:845–853. doi: 10.1006/jmbi.2001.4671. [DOI] [PubMed] [Google Scholar]
- 27.Janssen U, Fink T, Lichter P, Stoffel W. Genomics. 1994;23:223–228. doi: 10.1006/geno.1994.1480. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez-Martinez R, Cruz-Gil S, de Cedron MG, Alvarez-Fernandez M, Vargas T, Molina S, Garcia B, Herranz J, Moreno-Rubio J, Reglero G, Perez-Moreno M, Feliu J, Malumbres M, de Molina AR. Oncotarget. 2015;6:38719–38736. doi: 10.18632/oncotarget.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaisa NT, Reinartz A, Schneider U, Klaus C, Heidenreich A, Jakse G, Kaemmerer E, Klinkhammer BM, Knuechel R, Gassler N. Histol. Histopath. 2013;28:353–364. doi: 10.14670/HH-28.353. [DOI] [PubMed] [Google Scholar]
- 30.Wakamiya T, Suzuki SO, Hamasaki H, Honda H, Mizoguchi M, Yoshimoto K, Iwaki T. Neuropathology. 2014;34:465–474. doi: 10.1111/neup.12132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.