Abstract
Molecular docking is a standard computational approach to predict binding modes of protein-ligand complexes, by exploring alternative orientations and conformations of the ligand (i.e., by exploring ligand flexibility). Docking tools are largely used for virtual screening of small drug-like molecules, but their accuracy and efficiency greatly decays for ligands with more than 10 flexible bonds. This prevents a broader use of these tools to dock larger ligands such as peptides, which are molecules of growing interest in cancer research. To overcome this limitation, our group has previously proposed a meta-docking strategy, called DINC, to predict binding modes of large ligands. By incrementally docking overlapping fragments of a ligand, DINC allowed predicting binding modes of peptide-based inhibitors of transcription factors involved in cancer. Here we describe DINC 2.0, a revamped version of the DINC webserver with enhanced capabilities and a more user-friendly interface. DINC 2.0 allows docking ligands that were previously too challenging for DINC, such as peptides with more than 25 flexible bonds. The webserver is freely accessible at http://dinc.kavrakilab.org, together with additional documentation and video tutorials. Our team will provide continuous support for this tool and is working on extending its applicability to other challenging fields, such as personalized immunotherapy against cancer.
Keywords: DINC webserver, incremental algorithm, molecular docking, protein-peptide complexes
Introduction
Given the importance of the structure-function relationship in proteins, knowledge of the structure of a protein or a macromolecular complex can provide insights on how to inhibit a protein target or disrupt a complex, which can be key to preventing a pathological outcome [1]. In this context, computational methods have been widely applied to the virtual screening of small molecules with potential use as inhibitors (or agonists), which constitutes one of the initial steps for drug discovery and rational drug design [2, 3]. Molecular docking methods, for instance, can predict both the geometry of a protein-ligand complex and its corresponding binding energy [4]. These methods are known to be generally accurate for small drug-like ligands with up to 10 flexible bonds, or degrees of freedom (DoFs) [5]. In recent years, there has been a growing interest in the use of peptides as protein inhibitors, given their natural role as binders and regulators in a variety of pathways [6,7]. However, even small peptides and peptidomimetics (i.e., synthetic peptide-based inhibitors [8]) are too large and too flexible for most available molecular docking methods [9]. Note that peptide-docking is a growing field of research and that other tools have been recently proposed [7]. However, a more detailed review of these methods goes beyond the scope of this paper.
To address the challenge of docking large ligands, our group has previously developed a meta-docking approach called DINC (Docking INCrementally). DINC was evaluated on a dataset of large ligands, by trying to reproduce 73 complexes reported in the Protein Data Bank (PDB) [5]. DINC was shown to be much faster than the standard tool AutoDock 4 (AD4) [10] and to provide binding modes that were consistent with experimental data [5]. However, this experiment also highlighted the limitations of the original implementation: indeed, good reproductions were only observed for drug-like ligands with up to 16 DoFs. In another study, our group investigated the use of peptidomimetics as inhibitors of the Src Homology 2 (SH2) [11] domain of STAT3, a transcription factor that was found to be constitutively activated in a number of human cancers [8]. Since no structural information was available on potential SH2 inhibitors, we used DINC to predict the binding modes of a group of previously experimentally-identified peptidomimetic inhibitors with up to 22 DoFs. Besides predicting binding modes that were in agreement with data from previous studies, we were able to differentiate between strong and weak binders using predicted free-energies and conformational analysis through molecular dynamics simulations. In addition, DINC predicted a yet unknown alternative binding mode to SH2, leading to new opportunities for developing stronger inhibitors [8].
Incremental docking of overlapping fragments
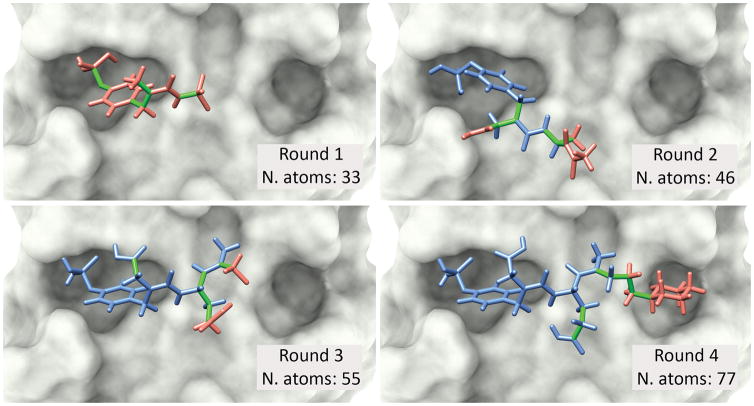
DINC is a parallelized meta-docking method for the incremental docking of large ligands (Figure 1). Instead of docking the whole ligand by exploring all its DoFs at once, DINC reduces the problem complexity by incrementally docking larger and larger overlapping fragments of the ligand. DINC starts by selecting a small fragment of the ligand and calling a standard molecular docking software (currently AD4 [10]) to dock this fragment, in a first round of sampling and scoring. AD4 uses a Lamarckian genetic algorithm for sampling, and a semi-empirical free energy force-field for scoring, based on pair-wise evaluations (V) with pre-defined weights (Wi) [10]:
Figure 1. Incremental docking of a modified peptide.
The proto-oncogene tyrosine protein kinase Src (c-Src) has been associated with breast cancer and osteoporosis [11]. A known peptidomimetic inhibitor, capable of binding to the Src SH2 domain, has 77 atoms and 17 DoFs. This modified peptide provides a good example of the use of DINC’s incremental approach for binding mode prediction. DINC starts by selecting a small fragment of the ligand (top left), with only 6 DoFs (depicted in green), and using it as input for the first round of docking to the SH2 domain (depicted in grey). The best binding modes are selected across multiple parallel docking runs, and the corresponding fragments are expanded by adding a small number of atoms. These extended fragments are used as input for the second round of docking, in which a new subset of 6 flexible DoFs is explored. These flexible DoFs involve some of the “new” atoms (red) and some of the “old” atoms (blue), as depicted in the best binding mode obtained in round 2 (top right). This process continues until the entire ligand has been reconstructed (bottom left and right). In this example, the root mean square deviation (RMSD) between the obtained model and the corresponding crystal structure of the same complex (PDB code 1SKJ) is only 1.97 Å.
The best binding modes obtained in the first round of docking are then selected, and the corresponding fragment is expanded by adding atoms of the ligand to it. Then, the next round of docking is performed. This process of expanding and docking is repeated incrementally, until the whole ligand is reconstructed and docked (Figure 1). By default, the initial fragment is defined to have only 6 DoFs. At each docking round, 3 new DoFs are added and only 3 of the previously-explored DoFs are kept flexible. This way, each docking round considers only 6 internal DoFs of the ligand (in addition to its rotation and translation), regardless of fragment size.
Note that, in the context of DINC, the meaning of “fragment” differs from its most common use in fragment-based drug discovery (FBDD). FBDD involves creating new drugs or drug-like ligands using libraries of fragments, which are very small molecules with no more than two functional groups [12]. A related strategy, anchor-and-grow, was implemented in the docking software DOCK [13]. DINC was inspired by these developments, but provides a higher-level method that leverages and enhances traditional docking software (such as AD4 [10]), therefore improving efficiency for large ligands.
A new user-friendly webserver for docking peptides
Aiming to dock even larger ligands, such as peptides, we upgraded the DINC algorithm and created a whole new webserver: DINC 2.0. New heuristics were introduced to select the initial fragment and expand the subsequent fragments, based on maximizing the potential for hydrogen bonds with the receptor. DINC 2.0 also features an improved management of the parallelization process, which is crucial to docking large peptides; users can increase the number of top conformations selected at each round, therefore expanding the sampling without increasing the running time. Finally, our team improved several technical components of the original software; we are continuously updating the algorithm to improve its efficiency and allow for the processing of very challenging complexes.
The main page of DINC 2.0 is clear and user-friendly, making docking accessible to both new and experienced users. To run a docking job, users have to provide the structures of the ligand and receptor of interest. While the original DINC [5] required specific preprocessing of the ligands, DINC 2.0 accepts standard PDB files as input. Then, users have to provide information on the “grid box,” a volume that should be large enough to contain the ligand and the receptor’s binding cleft. The docking search is based on the position of this box and the points of this three-dimensional grid are used for the energy pre-computation (scoring).
In a re-docking experiment (i.e., when trying to reproduce a known protein-ligand complex), users can easily define the grid box location and size based on the input ligand (see Video 1). In other docking experiments, when the ligand coordinates do not correspond to the receptor’s binding site, users must provide specific values for “grid center” and “grid size” (see Video 2). To find appropriate values, users can use various graphical tools (see our “HELP” page). Users can also copy the values from previous experiments with AD4 [10] or AutoDock Vina [14] (see Video 2). The interface automatically converts all values to angstroms, facilitating the comparison to grid boxes defined in other docking software.
Additional parameters that directly impact the search can be accessed in the “Advanced options” menu. Increasing the values of these parameters can improve the likelihood of finding accurate binding modes, but can increase running time. Further information can be found in the “HELP” page, which also contains video tutorials, links to related resources and tools, as well as examples of the expected output files for three large ligands; one of them is an 8-mer peptide with 26 DoFs, involved in a complex that DINC 2.0 reproduced with an accuracy of 1.61 Å.
Finally, another new feature in DINC 2.0 is a “Results” page that summarizes the input parameters and provides the best alternative binding modes, selected based on binding energy ranking or RMSD clustering. Using the embedded visualization tool, users can quickly inspect the output and compare the predicted binding modes, which can also be downloaded for further analysis (see Video 1). We encourage users to further evaluate the complexes produced by DINC 2.0 using other methods, such as molecular dynamics, especially when predicting complexes for which no crystal structure is available.
Future directions and challenges in cancer immunotherapy
Following a meta-docking approach, we envision to extend our methods to other popular docking tools, to investigate the benefits of alternative scoring functions and consensus docking. Regarding future applications, one of the greatest challenges in peptide docking relates to the structural prediction of Human Leukocyte Antigen (HLA) complexes [5]. HLAs are key molecules for the cellular immune response because they can bind peptides derived from intracellular proteins and display them at the cell surface for recognition by circulating cytotoxic T-cells [15]. Given their role in the immunity against tumors [15], predicting the structure of such peptide-HLA complexes is becoming a major need for T-cell-based immunotherapy. The challenge, however, is the size of these peptides, which are typically 8 to 12 amino acids long and present more than 35 DoFs. Our group is now focused on further expanding the capabilities of DINC 2.0, specifically aiming at applications towards personalized T-cell-based immunotherapy against cancer.
Acknowledgments
Financial support: Work on this project by D.A. Antunes, L.E. Kavraki, M. Moll, G. Lizée and K.R. Jackson was supported by NIH R21CA209941-01 (co-PIs: L.E. Kavraki and G. Lizée) through the Informatics Technology for Cancer Research (ITCR) initiative of the National Cancer Institute (NCI). Work on this project by D. Devaurs was supported by a training fellowship from the Gulf Coast Consortia, on the Computational Cancer Biology Training Program (CPRIT Grant No. RP170593, PI: M. Pettitt).
We thank Sujay Tadwalkar, Angela Hoch, and Sarah Hall-Swan for their contributions to this project.
Footnotes
Disclosure statement: The authors declare no potential conflicts of interest.
References
- 1.Cordomi A, Fourmy D, Tikhonova IG. Gut hormone GPCRs: structure, function, drug discovery. Curr Opin Pharmacol. 2016;31:63–67. doi: 10.1016/j.coph.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 2.Sliwoski G, Kothiwale S, Meiler J, Lowe EW. Computational methods in drug discovery. Pharmacol Rev. 2014;66:334–395. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haga JH, Ichikawa K, Date S. Virtual screening techniques and current computational infrastructures. Curr Pharm Des. 2016;22:3576–3584. doi: 10.2174/1381612822666160414142530. [DOI] [PubMed] [Google Scholar]
- 4.Antunes DA, Devaurs D, Kavraki LE. Understanding the challenges of protein flexibility in drug design. Expert Opin Drug Discov. 2015;10:1301–1313. doi: 10.1517/17460441.2015.1094458. [DOI] [PubMed] [Google Scholar]
- 5.Dhanik A, McMurray JS, Kavraki LE. DINC: a new AutoDock-based protocol for docking large ligands. BMC Struct Biol. 2013;13:S11. doi: 10.1186/1472-6807-13-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craik DJ, Fairlie DP, Liras S, Price D. The future of peptide-based drugs. Chem Biol Drug Des. 2013;81:136–147. doi: 10.1111/cbdd.12055. [DOI] [PubMed] [Google Scholar]
- 7.Kilburg D, Gallicchio E. Recent advances in computational models for the study of protein-peptide interactions. Adv Protein Chem Struct Biol. 2016;105:27–57. doi: 10.1016/bs.apcsb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 8.Dhanik A, McMurray JS, Kavraki LE. Binding modes of peptidomimetics designed to inhibit STAT3. PLoS One. 2012;7:e51603. doi: 10.1371/journal.pone.0051603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rentzsch R, Renard BY. Docking small peptides remains a great challenge: an assessment using AutoDock Vina. Brief Bioinform. 2015;16:1045–1056. doi: 10.1093/bib/bbv008. [DOI] [PubMed] [Google Scholar]
- 10.Morris GM, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plummer MS, et al. Design, synthesis, and cocrystal structure of a nonpeptide Src SH2 domain ligand. J Med Chem. 1997;40:3719–3725. doi: 10.1021/jm970402q. [DOI] [PubMed] [Google Scholar]
- 12.Erlanson DA, Fesik SW, Hubbard RE, Jahnke W, Jhoti H. Twenty years on: the impact of fragments on drug discovery. Nat Rev Drug Discov. 2016;15:605–619. doi: 10.1038/nrd.2016.109. [DOI] [PubMed] [Google Scholar]
- 13.Ewing TJ, Makino S, Skillman AG, Kuntz ID. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des. 2001;15:411–428. doi: 10.1023/A:1011115820450. [DOI] [PubMed] [Google Scholar]
- 14.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lizée G, et al. Harnessing the power of the immune system to target cancer. Annu Rev Med. 2013;64:71–90. doi: 10.1146/annurev-med-112311-083918. [DOI] [PubMed] [Google Scholar]