Abstract
Elongation factor, RNA polymerase II, 2 (ELL2) is an RNA Pol II elongation factor with functional properties similar to ELL that can interact with the prostate tumor suppressor EAF2. In the prostate, ELL2 is an androgen response gene that is up-regulated in benign prostatic hyperplasia (BPH). We recently showed that ELL2 loss could enhance prostate cancer cell proliferation and migration, and that ELL2 gene expression was down-regulated in high Gleason score prostate cancer specimens. Here, prostate-specific deletion of ELL2 in a mouse model revealed a potential role for ELL2 as a prostate tumor suppressor in vivo. Ell2 knockout mice exhibited prostatic defects including increased epithelial proliferation, vascularity and PIN lesions similar to the previously determined prostate phenotype in Eaf2 knockout mice. Microarray analysis of prostates from Ell2 knockout and wild-type mice on a C57BL/6J background at age 3 mos and qPCR validation at 17 mos of age revealed a number of differentially expressed genes associated with proliferation, cellular motility and epithelial and neural differentiation. OncoPrint analysis identified combined down-regulation or deletion in prostate adenocarcinoma cases from the Cancer Genome Atlas (TCGA) data portal. These results suggest that ELL2 and its pathway genes likely play an important role in the development and progression of prostate cancer.
Keywords: ELL2, prostatic intraepithelial neoplasia, prostate cancer, EAF2, HIF1α
Introduction
ELL2 (elongation factor, RNA polymerase II, 2; previously eleven-nineteen lysine-rich leukemia 2) is an RNA Pol II elongation factor with functional properties similar to ELL and ELL3 (Shilatifard, et al. 1997; Miller, et al. 2000). ELL2 suppresses transient pausing of RNA polymerase II activity along the DNA strand and facilitates the transcription process (Shilatifard et al. 1997). The ELL family proteins are components of the super elongation complex (SEC) which regulate HOX gene expression in MLL-based hematological malignancies by controlling genes involved in early development and in immediate early gene transcription (Lin, et al. 2010; Lin, et al. 2011; Smith, et al. 2011; Takahashi, et al. 2011). ELL was recently identified as a component in the little elongation complex (LEC), which is involved in RNA polymerase II transcription of small nuclear RNA (snRNA) genes (Smith et al. 2011). ELL and EAF proteins also bind to MED26, a component of the human mediator that plays a key role in transcriptional activation (Takahashi et al. 2011). ELL2 was also reported to direct immunoglobulin secretion in plasma cells by stimulating alternative RNA processing associated with histone methylations (Martincic, et al. 2009; Milcarek, et al. 2011).
ELL is frequently translocated with the MLL gene on chromosome 11q23 in acute myeloid leukemia (Thirman, et al. 1994; Mitani, et al. 1995); and homozygous deletion of ELL is embryonic lethal in the mouse as well as in Drosophila (Mitani, et al. 2000; Eissenberg, et al. 2002). ELL and ELL2 interact with ELL-associated factors 1 and 2 (EAF1 and EAF2) (Simone, et al. 2003) resulting in enhanced ELL elongation activity (Kong, et al. 2005). ELL and ELL2 expression ratios vary in different human tissues, suggesting tissue-specific roles for these genes (Shilatifard et al. 1997). Since high expression of ELL2 has been reported in the prostate (Uhlen, et al. 2005), ELL2 may be important for prostate homeostasis. ELL2 was also found to be an androgen response gene (Nelson, et al. 2002; Bolton, et al. 2007) that is up-regulated in response to chronic prostatic inflammation in rats (Funahashi, et al. 2015) and was up-regulated in human benign prostatic hyperplasia (BPH) (O’Malley, et al. 2009). The specific role of ELL2 in the prostate has not been fully elucidated; however, transfected ELL2 protein has been shown to interact with the potential prostate tumor suppressor gene ELL-associated factor 2 (EAF2) (Simone et al. 2003). In this study, Simone et al., showed that endogenous EAF2 coimmunoprecipitated with transfected ELL2 in 293 cells and that like the MLL-ELL fusion protein (Lavau, et al. 2000), MLL-EAF2 could immortalize hematopoietic progenitor cells in vitro. Recently, eaf-1 and ell-1, worm orthologs of EAF1, EAF2 and ELL1 and ELL2, were shown to have overlapping function in the regulation of fertility, survival and cuticle formation in C. elegans (Cai, et al. 2011). In advanced prostate cancer, EAF2 protein was down-regulated (Xiao, et al. 2003; Ai, et al. 2013; Pascal, et al. 2013); and, overexpression of EAF2 in prostate cancer cell lines induced apoptosis and inhibited the growth of xenograft tumors (Xiao et al. 2003). Eaf2 knockout mice developed high-grade murine prostatic intraepithelial neoplasia and increased vascularity in several murine strains (mPIN) (Xiao, et al. 2008; Pascal et al. 2013). Recently, we reported that siRNA knockdown of ELL2 in combination with RB enhanced proliferation, migration and invasion in prostate cancer cell lines LNCaP, C4-2 and 22RV1, while ELL2 knockdown alone enhanced migration and invasion but did not induce a statistically significant increased proliferation (Qiu, et al. 2017). ELL2 expression was down-regulated in high Gleason score prostate cancer specimens. Cumulatively, these studies suggest that ELL2 may play a significant role in maintaining prostate homeostasis similar to EAF2.
In the current study, the potential role of ELL2 in the prostate was explored in a murine knockout model. Conditional prostate epithelial cell-specific Ell2 knockout mice were generated and examined for histologic defects. Microarray analysis of mouse prostates was performed to identify potential target genes of ELL2. Differentially expressed genes identified by microarray analysis in animals at 3 mos of age were validated by qPCR in animals at 17–20 mos of age.
Materials and Methods
In situ Hybridization
Before hybridization, murine prostate tissue cryosections (ProbeOn, Fisher Biotech, Pittsburgh, PA) were washed with PBS, fixed in 4% paraformaldehyde, digested with proteinase K at 20 μg/ml in PBS, refixed in 4% paraformaldehyde, rewashed in PBS, and then acetylated in 0.25% acetic anhydride in 0.1 M triethanolamine, pH 8.0. Full-length mouse ELL2 cDNA was inserted into the EcoRI and XhoI site between T3 and T7 promoters in pBluescript II SK plasmid vector. The plasmid was purified by CsCl double banding, linearized with EcoRI or XhoI, and proteinase K-treated. Purified linear DNA templates were used in the synthesis of both sense and antisense digoxygenin-labeled riboprobes using either T3 or T7 RNA polymerase (Promega Corp., Madison, WI) as previously described (Cyriac, et al. 2002). Riboprobe size was reduced to approximately 250 bp using limited alkaline hydrolysis.
For hybridization, the probe was diluted in hybridization solution (5x SSC, 1x Denhardt’s, 100 μg/ml salmon testis DNA, 50% formamide, and 250 μg/ml yeast transfer RNA), and slides were hybridized overnight at 67°C in a sealed chamber humidified with 5x SSC/50% formamide. Coverslips were removed, and slides were washed in 0.2x SSC at 72°C for 1 h. After washing in buffer (0.1 M Tris (pH 7.6), 0.15 M NaCl), slides were blocked in 10% horse serum at room temperature for 1 h. Slides were then incubated overnight at 4°C with antidigoxigenin-AP Fab fragments (1:2000, Boehringer Mannheim, Mannheim, Germany) in 1% horse serum. Slides were washed and then developed with nitro blue tetrazolium (2.25 μl/ml) and 5-bromo-4-chloro-3-indolyl-phosphate, 4-toluidine salt (0.6 μg/ml) in alkaline phosphatase buffer (0.1 M Tris (pH 9.5), 0.05 M MgCl2, 0.1 M NaCl).
Generation of prostate-specific ELL2 deletion mice
Mice with prostate specific deletion of the Ell2 gene expression were generated by cross-breeding the Ell2loxp/loxp mice with probasin-Cre4 (Wu, et al. 2001) mice. Ell2loxp/loxp mice were cloned by homologous recombination between Ell2loxp/loxp targeting vector and the Ell2 genomic locus (Park, et al. 2014). Briefly, a conditional targeting vector in which Ell2 exon 3 was flanked by a single upstream loxP site and a downstream <FRT/neomycin resistance/FRT/loxP> cassette was constructed. Correctly targeted embryonic stem cells contained the cko allele with an 8.6 Sacl band, in addition to a 14 kb wild-type band, following hybridization with the 5′ probe. These cko clones also contained a 13.9 kb EcoRI-targeted band as well as a 12 kb wild-type band, following hybridization with the 3′ probe. Mice carrying the Ell2-cko allele were maintained on a C57BL/6J background and cross-bred with probasin-Cre4 mice (PBCre4), which provide prostate-specific expression of Cre recombinase, to generate mice with prostate epithelial cell specific deletion of Ell2-cko (Ell2loxp/loxpPBCre4).
Experimental cohorts were wild-type (WT) and homozygous Ell2-cko male littermates; all mice were maintained identically. All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Pittsburgh and were conducted in strict accordance with the standards for humane animal care and use as set by the Animal Welfare Act and the National Institutes of Health guidelines for the use of laboratory animals under Animal Welfare Assurance number A3187-01. Genotyping was determined by PCR analysis of mouse tail genomic DNA at 21 days of age and confirmed on muscle DNA when animals were euthanized at 3 mos of age (n=12) or 17–20 mos of age (n=29). Primer pairs included Ell2ko-cSalI-38915, 5′-ATGCATCGTCGAACAGGAGTTCAAGGT-3′ and ELL2ko-SaclI-5′-CTGATACCGCGGGTGGAAATCACTCC-3′ (reverse), and Cre-upstream, 5′-TTGCCTGCATTACCGGTCGATG-3′ and Cre-downstream, 5′-TCCAGCCACCAGCTTGCATG-3′ (forward) and 5′-CGGTGCCCGGTGGTAAGATC-3′ (reverse). Prostate tissue necropsy was performed and organs were cleaned of excess fat and membrane with phosphate-buffered saline; mass of each prostate lobe was determined after blotting with filtration paper to remove excess water.
Histopathologic analysis
Samples were fixed in 10% formalin for at least 24 hrs, then embedded in paraffin, sectioned at 5 μm, and stained with hematoxylin and eosin. All tissues were examined by a board-certified animal pathologist in a blinded fashion (LHR, V.M.D). Lesions were identified as epithelial hyperplasia, stromal hyperplasia, and mPIN per the criteria published by Shappell, et al. commonly used to score prostate lesions in transgenic mouse models (Shappell, et al. 2004). Epithelial hyperplasia was recognized as an increase in epithelial cells within normal-appearing gland profiles, reflected by stratification of epithelial cells (Shappell et al. 2004). Stromal hyperplasia was identified as a non-neoplastic increase in the cellularity of the stromal component of the prostate compared with age-matched controls (Shappell et al. 2004). mPIN ranged from low grade, characterized by glands lined by 1 to 3 layers of epithelial cells displaying minimal pleomorphism or hyperchromasia, slight nuclear enlargement with little atypia, infrequent mitosis, and essentially normal glandular profiles with only occasional hints of papillary epithelial proliferation to high grade, characterized by extensive intraglandular epithelial proliferation, formation of papillary or cribriform structures consisting of epithelial cells displaying significant nuclear atypia and hyperchromasia, cellular pleomorphism, and increased frequency of mitoses (Shappell et al. 2004).
Tissue preparation and microarray hybridization
All lobes of the prostates from 6 wild-type and 6 Ell2-cko virgin male mice littermates at 3 mos of age were microdissected from the urogenital tract in phosphate-buffered saline with the aid of a dissecting Carl Zeiss Stemi 2000 Stereomicroscope (Zeiss) and stored in 1 ml of RNA later at −80 °C. Expression profiling experiments were performed by the High Throughput Genome Center at the Department of Pathology, University of Pittsburgh. Total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. RNA quantity and quality was measured by obtaining A260 and an A260/A280 wavelength ratio of 1.8 – 2.1 via Agilent 2100 bioanalyzer (Agilent, Santa Clara, CA). Microarray hybridization was performed with a pre-equilibrated Mouse Genome 430 2.0 Genechip array (Affymetrix Inc., Santa Clara, CA). For each sample, 8 μg total RNA was used for retro-transcription, and 15 μg cRNA to the chip for hybridization. Hybridization data were normalized to an average target intensity of 500 per chip using Affymetrix GeneChip® Operating Software (GCOS 1.4), and were converted to a Microsoft Excel spreadsheet text file.
Bioinformatics data analysis
The differential expression result was achieved fitting a lognormal distribution to all 6 Affymetrix intensity signals of each group, wild-type (WT) and Ell2-cko and asking what is the probability that Ell2-cko > WT (and conversely WT > Ell2-cko). Along with probabilistic support, the magnitude of the effect was also considered taking into account the average fold-changes. The final gene list considered was obtained from filtering the gene list at two simultaneous thresholds: (i) log2-ratios greater than two-fold and (ii) probability Pr(Ell2-cko > WT) > 95%, for up-regulated, or Pr(WT > Ell2-cko) > 95%, for down-regulated. Functional and ontology enrichment analysis was performed using the DAVID web-based tool (Dennis, et al. 2003) and Ingenuity Pathways Analysis (IPA) 5.0 (Ingenuity Systems, Redwood City, CA) as described in Haram et al. (Haram, et al. 2008).
Gene expression validation
The anterior prostates of an independent set of WT (n=5) or Ell2-cko male mice (n=3) at 17 mos of age were used for total RNA isolation using the RNeasy minikit (Qiagen, Germantown, MD). Animal tissues were homogenized with a Kontes pellet pestle for 30 sec twice (Fisher Scientific, Fair View, NJ). RNA quality was analyzed by bleach gel electrophoresis (Aranda, et al. 2012). qPCR verified expression scored by cDNA arrays of ventral prostate tissue and expression levels in anterior prostate tissue (SYBR Green/ROX, Thermo Scientific, Waltham, MA, USA). PCR amplification was carried out using Applied Biosystems StepOne™Plus™ Real-Time PCR Systems (Applied Biosystems CA, USA). PCR amplification of various genes was normalized to the housekeeping gene Gapdh using the comparative CT method (Schmittgen and Livak 2008). Primer sequences are listed in Table 1. GAPDH was chosen as an internal control because there was no difference in Gapdh expression between wild-type and Ell2-cko prostate in the microarray data. Also Gapdh has been used as a normalization control in murine prostate research (Ai, et al. 2009). Each experimental sample was assayed in triplicate from a minimum of 3 animals per group.
Table 1.
Primers for qPCR
| Gene | HUGO* Gene Name | Forward | Reverse | Species |
|---|---|---|---|---|
| ARSK | arylsulfatase family member K | GAGATGTTGCATTCTTGCTCCG | GCCATTCGATTGCTTTGTCTGT | mouse |
| ARSK | arylsulfatase family member K | GTGAGCGACTCCTTCGATGG | GGAGAGTTTGTGTAGGCATTCA | human |
| CTSE | cathepsin E | GACATCAGTCCGTTCGGAAGA | AGGGGTTCATTGACACTCGAATA | mouse |
| CXCL10 | C-X-C motif chemokine ligand 10 | CCAAGTGCTGCCGTCATTTTC | GGCTCGCAGGGATGATTTCAA | mouse |
| ELL2 | elongation factor for RNA polymerase II 2 | CGCTGGAGACTTACCAGAGC | CATTGAAGGGATCATTTTTGG | human |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase | AGGTCGGTGTGAACGGATTTG | GTAGACCATGTAGTTGAGGTCA | mouse |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase | CGACCACTTTGTCAAGCTCA | AGGGGAGATTCAGTGTGGTG | human |
| GBP2 | guanylate binding protein 2 | CTGCACTATGTGACGGAGCTA | GAGTCCACACAAAGGTTGGAAA | mouse |
| HIF1A | hypoxia inducible factor 1 alpha subunit | ACCTTCATCGGAAACTCCAAAG | CTGTTAGGCTGGGAAAAGTTAGG | mouse |
| MUC4 | mucin 4, cell surface associated | GGAACTTGGAGTATCCTGTTG | CCTCCTCTTGCTACCTGATGC | mouse |
| OLIG2 | oligodendrocyte transcription factor 2 | TCCCCAGAACCCGATGATCTT | CGTGGACGAGGACACAGTC | mouse |
| PBSN | probasin | GCATGTGCTAGGCGTCTCC | GTTCTCAATGGTGAGCCTTCAT | mouse |
| PDS5A | PDS5 cohesin associated factor A | TTGGGAAACTGATGACCATAGC | ACACAAACGTCAGCCTGCTT | mouse |
| PDS5A | PDS5 cohesin associated factor A | AGATCGCTTACCCTCCGGG | ACTACCATCTTCAGGCGTTTGA | human |
| RHOBTB3 | Rho related BTB domain containing 3 | CTGAGGCATCACACTATCACTCC | AGAGAACGATCTTGTGGGCTT | mouse |
| RHOBTB3 | Rho related BTB domain containing | GGGGCTTATCCGCACTTACC | TGGCCTGATACTCGGTGAACA | human |
| RTN4 | reticulon 4 | TGCCTTCATTGTTTGTCGGG | TTCCTAGCTGCTGATAGGCGA | mouse |
| S1PR1 | sphingosine-1-phosphate receptor 1 | ATGGTGTCCACTAGCATCCC | CGATGTTCAACTTGCCTGTGTAG | mouse |
| S1PR3 | sphingosine-1-phosphate receptor 3 | ACTCTCCGGGAACATTACGAT | CAAGACGATGAAGCTACAGGTG | mouse |
| SFI1 | SFI1 centrin binding protein | TTGGGGAGCAGCAGTTAGAGA | CGGACCAGGAACATTCGGC | mouse |
| SNAI3 | snail family transcriptional repressor 3 | GGTCCCCAACTACGGGAAAC | CTGTAGGGGGTCACTGGGATT | mouse |
| VEGF | vascular endothelial growth factor | GCACATAGAGAGAATGAGCTTCC | CTCCGCTCTGAACAAGGCT | mouse |
HUGO Gene Nomenclature Committee at the European Bioinformatics Institute www.genenames.org
Cell culture experiments
Human C4-2 prostate cancer cells (kind gift from Leland K. Chung) were maintained in RPMI 1640 (10-040-CV, Corning cellgro) supplemented with 10% fetal bovine serum (FBS) (S11150, Atlanta Biologicals) and 5% antibiotics. LNCaP prostate cancer cells were purchased from ATCC and maintained in RPMI 1640. ELL2 response to androgen was analyzed by culturing cells over a concentration range of R1881 (0–2 nM). After treatment, cells were lysed in modified radioimmune precipitation assay buffer [50 mm Tris (pH 7.4), 1% Igepal CA-630, 0.25% Na-deoxycholate, 150 mm NaCl, 1 mm EDTA (pH 8.0), 1 mm NaF, 2 mm phenylmethylsulfonylfluoride, 1 mm Na3VO4, and protease inhibitor cocktail (Sigma)]. Western blot analysis of ELL2, HIF1α and GAPDH was as described previously (Saporita, et al. 2007). Antibodies used were rabbit polyclonal anti-ELL2 (1:1000, A302-505A, Bethyl Laboratories, Inc., Montgomery, TX, USA), HIF1α (1:1000, 54/HIF-1α, 610959, BD Transduction Laboratories, San Jose, CA, USA) and rabbit polyclonal anti-GAPDH (1:6000, FL-335, sc25778, Santa Cruz Biotechnology, Dallas, TX, USA). All experiments were performed in triplicate.
The effects of ELL2 knockdown were determined in C4-2 cells. Small interfering RNAs (siRNAs) targeting ELL2 (siELL2, 5′-AUUUACAAUCUGAGGAGGAUGUGAGAU, 3′-TAAAUGUUAGACUCCUCCUACACUC) and EAF2 (siEAF2,5′-GGAGAAUGUCGGCUAGAAATT, 3′-GACCUCUUACAGCCGAUCUUU) were purchased from Integrated DNA Technologies (Coralville, IA, USA), and negative control (siSCR) siRNAs were purchased from Santa Cruz (Control siRNA-A:sc-37007). Cells were transfected with siRNA for 48 hours using DharmaFECT™ Transfection Reagents (GE Healthcare) following the manufacturer’s instructions. Experiments were performed in triplicate.
Immunohistochemistry
A group of mice including 10 wild-type and 19 Ell2-cko mice was generated and examined for histological defects in the prostate at age 17–20 mos. Immunohistochemical stains were performed on five-micron sections of paraffin-embedded murine tissue specimens as described previously (Pascal, et al. 2011). Briefly, sections were deparaffinized and rehydrated through a graded series of ethanol. Heat-induced epitope retrieval was performed using a decloaker, followed by rinsing in TBS buffer for 5 minutes. Primary antibodies for immunostaining of murine tissue sections were rabbit polyclonal anti-ELL2 (bs-6993R, Bioss Antibodies, Woburn, MA, USA), rat monoclonal anti-Ki-67 (TEC-3, M7249, Dako, Carpinteria, CA, USA) and rat monoclonal anti-CD31 (MEC 13.3, 550274, BD Biosciences, San Jose, CA, USA) (working dilution 1:100 for ELL2, and 1:400 for Ki-67 and CD31). Slides were then counterstained in hematoxylin and coverslipped. Immunostained sections were imaged with a Leica DM LB microscope (Leica Microsystems Inc., Bannockburn, IL, USA) equipped with a QImaging MicroPublisher 3.3 RTV digital camera (QImaging, Surry, BC, Canada).
For murine tissues, Ki-67-positive cell density was determined by analysis of sections from at least 6 independent mice from each genotype. Slides stained with Ki-67 were scanned and digitized using the Aperio ScanScope CS scanner (Aperio, Vista, CA) to capture digital whole slide images (WSI) using the x 20 objective lens at a spatial sampling period of 0.47 μm per pixel. The digital WSI were analyzed using Aperio ImageScope software (http://www.aperio.com/pathology-services/image-scope-slide-viewing-software.asp). The manufacturer’s (Aperio Technologies, Inc.) algorithms were used to quantify nuclear staining. Composite images were constructed with Photoshop CS (Adobe Systems, San Jose, CA). Assessment of microvessel density was determined based on CD31-positive blood vessel count as previously described (Pascal et al. 2011). Briefly, microvessel density was determined from at least 20 fields imaged at 10X magnification for prostate with no overlap and identified by evaluating histological sections, and CD31-positive vessels were counted to determine the average vessel numbers per field for each section.
Data analysis using cBioPortal
The cBioPortal was utilized to determine the percentage of alterations and co-occurrence between ELL2 its potential target genes in two data sets of prostate cancers. In cBioPortal, OncoPrints are generated for visualizing gene and pathway alterations across a set of cases. Individual genes are represented as rows, and individual cases or patients are represented as columns. We studied the alterations in a set of genes potentially regulated by ELL2 identified by microarray analysis of murine Ell2-cko prostate at age 3 mos and validated by qPCR analysis of murine prostate at age 17 mos. The cBioPortal for Cancer Genomics site (http://cbioportal.org) provides a Web resource for exploring, visualizing, and analyzing multidimensional cancer genomics data and is subjected to scheduled updates (Cerami, et al. 2012; Gao, et al. 2013).
Statistical analysis
For non-microarray data, straightforward established biostatistics analyses were used. Comparisons between groups were calculated using the Student’s t-test or two-tailed Fisher exact test as appropriate. A p-value of p < 0.05 was considered significant. GraphPad Prism version 4 was used for graphics (GraphPad Software, San Diego, CA, USA). Values are expressed as means ± S.E.M.
Results
Conditional deletion of ELL2 in the murine prostate epithelial cell results in murine prostatic intraepithelial neoplasia (mPIN)
The cell type specific expression of Ell2 mRNA in the wild-type murine prostate was determined by in situ hybridization. Ell2 expression was localized to prostate epithelial cells of all lobes of the murine prostate (Figure 1). Expression of Ell2 was also evident in smooth muscle cells and fibroblasts in the stroma. In order to determine the function of Ell2 in the murine prostate epithelial cell, mice with conditional deletion of Ell2 were generated by crossing PB-Cre4 mice to mice harboring a floxed Ell2 allele on a pure C57BL6/J background. Genotyping to confirm Ell2-cko was determined by performing PCR analysis of mouse tail genomic DNA at 21 days of age and confirmed on muscle DNA when animals were euthanized (Figure 2A). A group of 19 male Ell2-cko mice were generated and examined at 17–20 mos of age for prostatic defects. Since EAF2 knockout mice on a pure C57BL/6J background did not develop epithelial hyperplasia and neoplasia until ~20 mos (PMID 24260246), we examined Ell2-cko mice at a similar timepoint. Loss of ELL2 protein in prostate epithelial cells was verified by immunostaining (Figure 2B). Nuclear and cytoplasmic immunoexpression of ELL2 was evident in the epithelial cells of the wild-type murine prostate. The prostate epithelial cells of Ell2-cko mice only had background staining. Ell2-cko mice at 17–20 mos displayed a statistically significant increased incidence in epithelial hyperplasia, stromal hyperplasia and murine prostatic intraepithelial neoplasia (mPIN) compared to wild-type controls (Table 2, Figure 2C). No wild-type animals displayed mPIN lesions.
Figure 1.
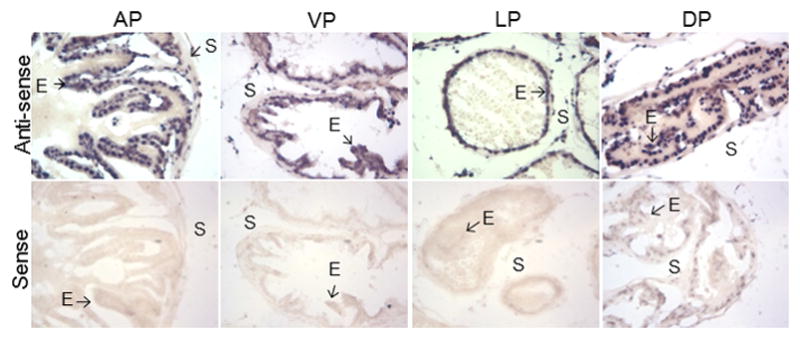
In situ hybridization analysis of ELL2 mRNA in murine prostate tissue. Both anti-sense (upper panels) and sense (lower panels) ELL2 RNA probes were labeled with DIG and visualized with alkaline phosphatase-conjugated anti-DIG antibody. Epithelial (E) and stromal (S) cells are indicated by arrows. AP, anterior prostate, DP, dorsal prostate, LP, lateral prostate, VP, ventral prostate.
Figure 2.
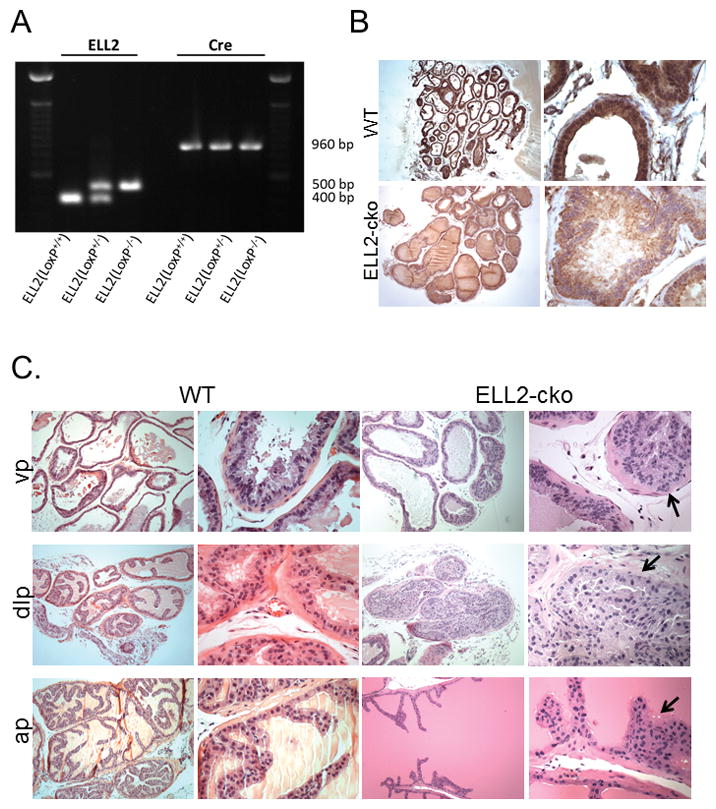
Generation of ELL2 conditional knockout mouse. A. Genotyping PCR analysis of mouse tail or skeletal muscle genomic DNA. PCR primers amplified ELL2 wild-type at 400 bp and mutant at 500 bp; and probasin-Cre at 960 bp. B. Immunoexpression of ELL2 in wild-type (WT) and Ell2-cko mouse prostate at 17–20 mos of age. C. Ell2-cko mice at 17–20 mos of age on a C57BL/6J background displayed increased mPIN (solid black arrows) compared to WT controls in the ventral (vp), dorsal-lateral (dlp) and anterior (ap) prostate lobes. Original magnification 10X, inset 40X.
Table 2.
Distribution of mice studied for prostatic defects
| Genotype | Number of animals analyzed | Animals with mPIN (%) | Fisher’s exact p-value | Animals with epithelial hyperplasia (%) | Fisher’s exact p-value | Animals with stromal hyperplasia (%) | Fisher’s exact p-value |
|---|---|---|---|---|---|---|---|
| WT | 10 | 0 (0%) | 0 (0%) | 0 (0%) | |||
| Ell2-cko | 19 | 8 (42%) | 0.0265 | 12 (63%) | 0.0012 | 8 (42%) | 0.0265 |
In addition to histologic defects, the prostates of Ell2-cko mice had increased mass in all lobes of the prostate compared to age-matched wild-type controls, further suggesting that Ell2 loss could induce increased epithelial proliferation (Figure 3A). The proliferative marker Ki-67 was used to detect dividing cells in the prostates of Ell2-cko and wild-type mice. In agreement with the increased mass, the number of Ki-67-positive epithelial cells was significantly increased in all lobes of the Ell2-cko mice compared to wild-type controls (Figure 3B,C). These results suggest that ELL2 loss in the murine model could induce epithelial proliferation, contributing to the development of mPIN lesions. ELL2 was previously identified as androgen responsive gene in the prostate cancer cell line LNCaP [17]. We also recently showed that knockdown of ELL2 in C4-2 and LNCaP cells significantly enhanced invasion and migration and induced a slight increase in BrdU incorporation (Qiu et al. 2017). Here, knockdown of ELL2 in LNCaP prostate cancer cells induced an increase in cellular proliferation (Figure 3D). In the prostate cancer cell line C4-2, ELL2 expression following exposure to increasing concentrations of R1881 was determined by western blot (Figure 3E). As expected, ELL2 protein levels were increased by androgens in a dose-dependent manner. Cumulatively, these results suggest that ELL2 loss could contribute to an increase in prostate cancer proliferation, invasion and migration.
Figure 3.
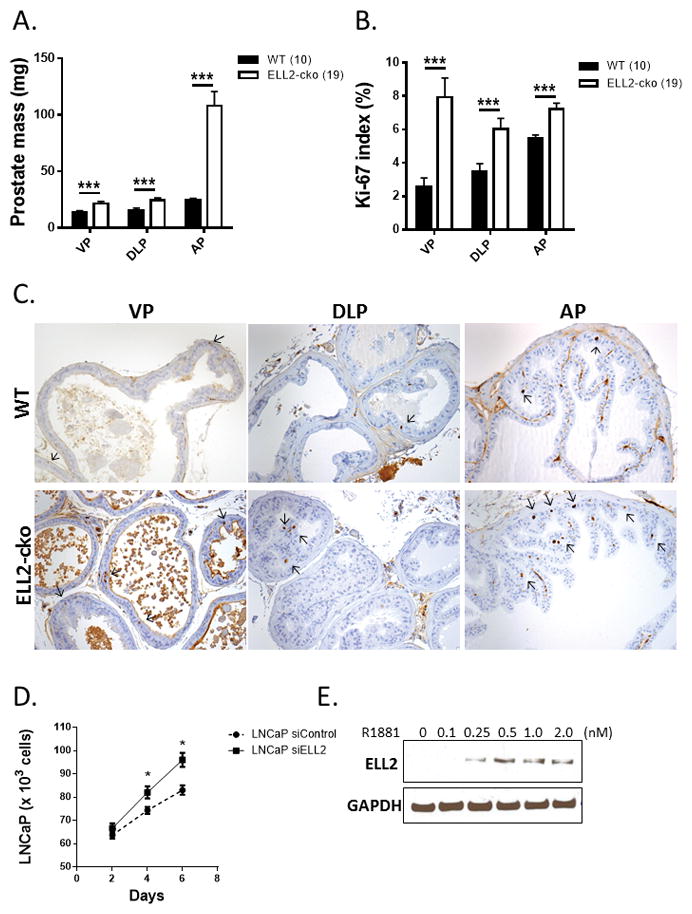
Effects of ELL2 loss on epithelial proliferation in the C57BL/6J mouse prostate at age of 17–20 mos. A. Mass of murine prostate in Ell2-cko mice compared to age-matched wild-type (WT) controls at age 17–20 mos. B. Percentage of Ki-67 positive epithelial cells in the ventral (vp), dorsal-lateral (dlp), and anterior (ap) prostate of wild-type (WT) and Ell2-cko mice at age 17–20 mos. C. Ki-67 immunostaining in transverse sections of vp, dlp and ap lobes from WT and Ell2-cko C57BL/6J mice at age 17–20 mos. Ki-67 positive cells identified with black arrows. Original magnification 20X. Number of animals in each group is indicated in parenthesis. D. Growth curve analysis of LNCaP cells transfected with ELL2 siRNA (siELL2) or control (siControl). LNCaP cells were cultured in the presence of 1 nM R1881. E. Western blot analysis of ELL2 protein expression in C4-2 cells treated with R1881. LNCaP cells were cultured in the presence of 1 nM R1881, and results are representative of at least 3 experimental replicates. (*p<0.05, ***p<0.0001).
ELL2 gene deletion is associated with increased vascular density in the prostate
To investigate the effects of ELL2 loss on microvessel density in the prostate, we examined the number of CD31-positive blood vessels by immunostaining in a subset of animals (Figure 4). The normal prostate is characterized by prominent stromal vasculature and rarely intraductal vessels, whereas there is a noticeable migration of vessels into the prostatic duct within PIN lesions (Huss, et al. 2001). In agreement with the increased incidence of mPIN lesions, the number of CD31-positive intraductal vessels as well as total microvessel density increased significantly in all lobes of the Ell2-cko prostate as compared to wild-type control animals (Figure 4A, B, C). Since EAF and ELL proteins have overlapping functions, and both EAF2 (Xiao, et al. 2009; Chen, et al. 2014) and ELL (Liu, et al. 2010) interact with HIF1α, we examined its expression in response to ELL2 loss. Hif1α gene expression was significantly increased in Ell2-cko mice compared to wild-type controls (Figure 4D), while VEGF levels were not significantly different (see Figure 5). siRNA knockdown of ELL2 in C4-2 cells also induced an increase in HIF1α protein levels were increased compared to siSCR (Figure 4E). ELL2 could regulate prostate vascularity through the HIF1α pathway directly or through its interaction with EAF2 protein.
Figure 4.
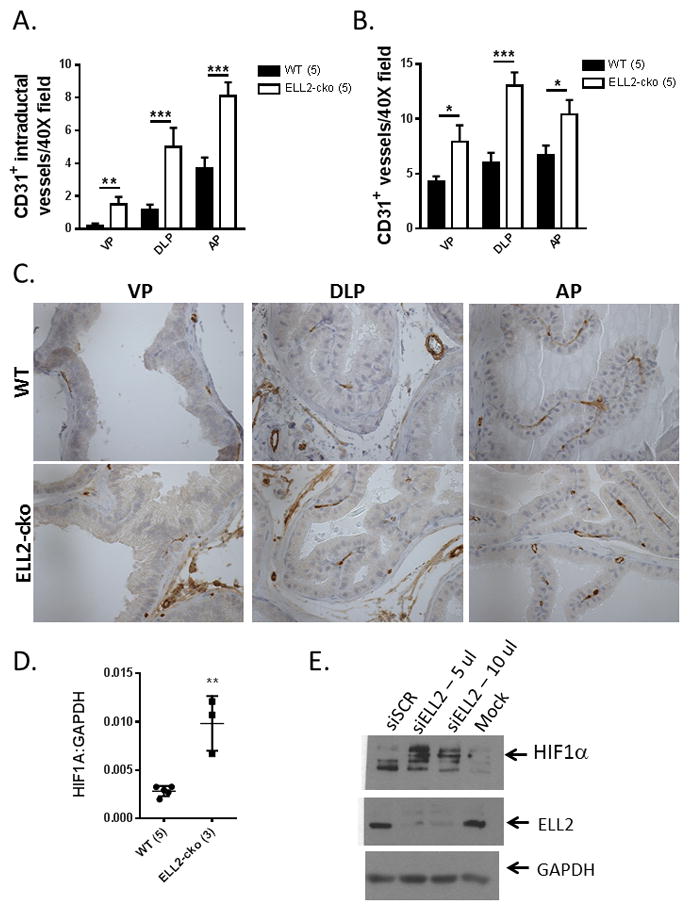
CD31-positive microvessel density in Ell2-cko mice at age 17–20 mos. A. Quantification of CD31-positive intraductal microvessels in Ell2-cko mice vs wild-type (WT) controls. B. Quantification of total CD31-positive microvessels Ell2-cko mice vs wild-type (WT) controls. C. Immunostaining analysis of EAF2 and CD31-positive microvessels in prostate tissues. Original magnification 20X. D. Expression of HIF1α mRNA in Ell2-cko and WT mice relative to GAPDH using the comparative CT method. E. Expression of HIF1α protein in C4-2 cells treated with siELL2 or siSCR. GAPDH served as loading control, and results are representative of 3 separate experiments. (*p<0.05, **p<0.001, ***p<0.0001). Number of animals in each group is indicated in parenthesis.
Figure 5.

qPCR data results for validated gene expression in mouse prostate at 17 mos of age. ELL2 knockout and wild-type mice were analyzed for mRNA expression of genes identified by microarray. Data were normalized to GAPDH using the comparative CT method. (*p<0.05, **p<0.001, ***p<0.0001). Number of animals in each group is indicated in parenthesis.
Microarray analysis and qPCR validation of genes differentially expressed in the prostate of ELL2 knockout mouse
To identify target genes of ELL2 prior to the development of histological defects, we performed cDNA microarray analysis of the prostate from 6 wild-type and 6 Ell2-cko mice at 3 mos of age. The most differentially regulated genes are listed in Supplemental Table S1. The most up-regulated genes identified in the ELL2 knockout mice included Sfi1, Muc4, Cd209b, Zfp786, Armc9, Plcd3 and Ctse. The most down-regulated genes identified included L3mbtl3, Rbm41, S1pr1, Hps1, Scarb1, Mepce, Atl2, and Lrpap1.
qPCR analysis of anterior prostates isolated from WT (n = 5) and Ell2-cko (n=3) mice at 17 mos of age was used to validate several genes identified by cDNA microarray as up- or down-regulated by ELL2 knockout in the mouse prostate at 3 mos of age. Insufficient samples for qPCR analyses were available for mice at 3 mos of age, therefore genes that were altered at both 3 mos of age as well as at 17 mos of age were considered. Fifteen genes were chosen randomly or because they encode products that are important in tumorigenesis for qPCR validation. In ELL2 knockout prostates at age 17 mos, Sfi1 and Cxcl10 were significantly up-regulated compared to wild-type controls, while Pds5a, Rtn4, Olig2, Rhobtb3 and Gbp2 were significantly decreased in agreement with the cDNA microarray results from mice at age 3 mos (Figure 5). Additionally, Pbsn and Vegf were not significantly altered, also in agreement with the microarray data. Several of the genes analyzed by qPCR in mice at 17 mos of age did not agree with the microarray results at 3 mos of age. These included Muc4, Ctse and S1pr1, which were not differentially expressed; as well as, Arsk, S1pr3 and Snai3, which were down-regulated in mice at age 3 mos, but up-regulated in the mice at 17 mos of age (Figure 6A). ELL2 knockdown in C4-2 cells induced a statistically significant decrease in PDS5A, while the effects of ELL2 knockdown RHOBTB3 and ARSK were not significant (Figure 6B). Fold changes from the microarray platforms were listed in Supplemental Table 1.
Figure 6.
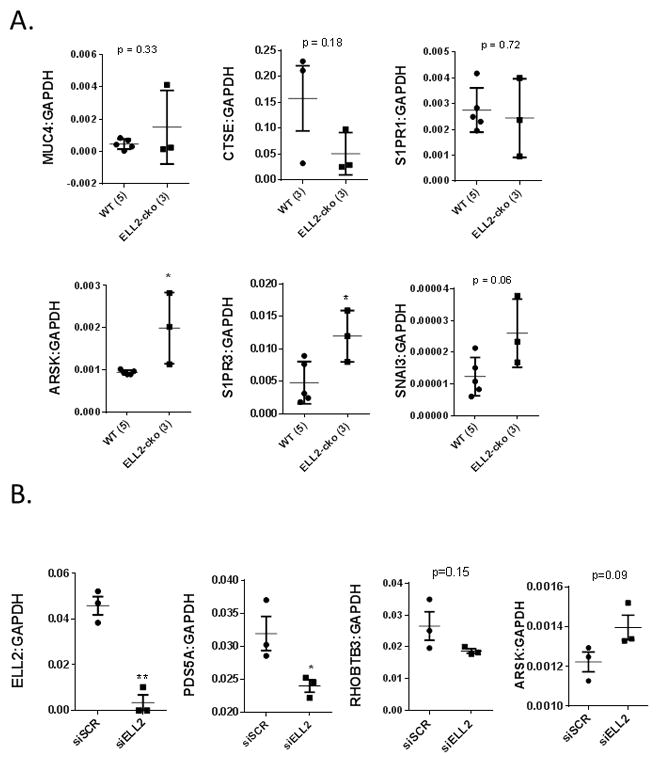
qPCR data results for gene expression in mouse prostate at 17 mos of age. A. Expression of genes in mice at 17 mos of age which differed from microarray results at 3 mos of age. ELL2 knockout and wild-type mice were analyzed for mRNA expression of genes identified by microarray. B. Expression of genes in C4-2 cells treated with siELL2 knockdown or siSCR control. Data were normalized to GAPDH using the comparative CT method. (*p<0.05, **p<0.001, ***p<0.0001). Number of animals in each group is indicated in parenthesis.
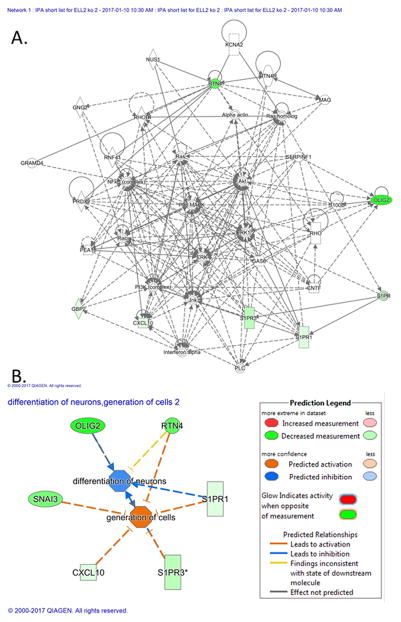
Functional classification of differentially expressed genes was analyzed for significant enrichment using the DAVID annotation tool (Dennis et al. 2003) (http://david.abcc.ncifcrf.gov/). Gene ontology-based analysis performed using DAVID identified 65 Annotation Clusters with gene count >2 (Supplemental Table S2). Annotation Clusters 1, 2 and 3 had enrichment scores >1.4 and included terms involving golgi apparatus, intracellular transport, protein transport, protein localization, protein targeting, organelle membrane, and mitochondrion. Ingenuity Pathway Analysis (IPA) (http://www.ingenuity.com) was also used to identify networks of interacting genes. The most significant network was associated with differentiation of neurons and the generation of cells (Figure 7A, B).
Figure 7.
A. Highest scoring interactive network generated by mapping differentially expressed genes to Ingenuity Pathway Knowledge Base information. Node color intensity indicates degree of upregulation (red) or downregulation (green) in Ell2-cko mice compared to wild-type mice at 3 mos of age. B. Schematic pathways activated by down-regulation of several genes. Down-regulation of genes (green) leading to activation of generation of cells (orange) and inhibited differentiation of neurons (blue).
The expression of genes associated with ELL2 loss was also examined in several large-scale genomics data sets available through the cBioPortal for Cancer Genomics (Cerami et al. 2012; Grasso, et al. 2012). Interestingly, several of the genes identified by microarray analysis of the Ell2 knockout mouse as potentially regulated by ELL2 were frequently altered in patients with ELL2 changes in two prostate cancer data sets. In the data set Neuroendocrine Prostate Cancer (Trento/Cornell/Broad 2016), ELL2 was amplified or up-regulated in 17% (13 of 77 sequenced) of patients (Figure 8A). Genes PDS5A, RHOBTB3, CTSE, ARSK, S1PR3, HIF1A and EAF2 were most frequently amplified or up-regulated in conjunction with ELL2. This dataset included whole exome and RNA Seq data of castration resistant adenocarcinoma and castration resistant neuroendocrine prostate cancer (somatic mutations and copy number aberrations) (Beltran, et al. 2016). Conversely, the TCGA, Provisional dataset generated by the TCGA Research Network: http://cancergenome.nih.gov/ identified several patients with gene down-regulation coinciding with deep deletion or mRNA upregulation of ELL2. In this dataset, 9% (43 of 492 sequenced cases) of patients had ELL2 alterations. RHOBTB3, GBP2 and ARSK were most frequently altered in combination with ELL2 (Figure 8B). One missense mutation was identified in the occluding homology domain of one patient (1 out of 112 sequenced cases) in the dataset Prostate Adenocarcinoma (Broad/Cornell 2012) (Barbieri, et al. 2012). These results suggest that ELL2 alterations in prostate cancer are most commonly copy number alteration or mRNA dysregulation rather than somatic mutations.
Figure 8.
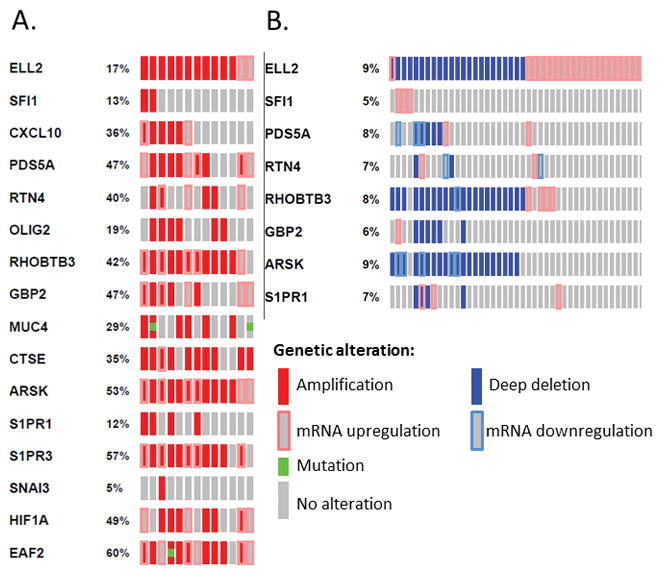
Oncoprint data represents samples from two published sets from the cBioPortal for cancer genomics (Cerami, et al. 2012; Gao, et al. 2013). A. The queried samples represent ELL2 enriched tumors from neuroendocrine prostate cancer (107 samples, whole exome and RNA Seq data of castration resistant adenocarcinoma and castration resistant neuroendocrine prostate cancer (somatic mutations and copy number aberrations)) (Beltran, et al. 2016). B. ELL2 enriched tumors from prostate adenocarcinoma tumor samples with sequencing and CNA data (492 samples, TCGA Research Network: http://cancergenome.nih.gov/). Individual genes are represented as rows, and individual cases or patients are represented as columns. The types of genetic alteration included in both sets are: amplification, deep deletion, mRNA upregulation, mRNA downregulation and missense mutation. cBioPortal data is subjected to scheduled updates.
Discussion
ELL and EAF family proteins play important roles in both development and tumorigenesis. ELL2 is a binding partner of EAF2, which is androgen responsive and acts as a prostate tumor suppressor (Xiao et al. 2003; Xiao et al. 2008). EAF2 knockout mice have increased incidence in neoplastic PIN lesions, increased epithelial proliferation and increased vascularity (Xiao et al. 2008; Pascal et al. 2013). Here, prostate-specific deletion of ELL2 in the murine prostate also induced an increased incidence in epithelial proliferation and mPIN lesions in older mice. EAF2 can induce increased HIF1α through the stabilization of VHL protein (Xiao et al. 2009) and can decrease expression of anti-angiogenic TSP1 (Su, et al. 2010). ELL has also been shown to influence the HIF1α pathway by modulating expression of VEGF and Glut-1, and knockdown of ELL increased HIF1α protein expression (Liu et al. 2010). Here we show that Ell2 loss in the murine prostate was also associated with an increase in microvessel density and recruitment of intraductal vessels to mPIN lesions compared to wild-type controls (Figures 2 and 3). The prostates of Ell2 knockout mice had increased mRNA expression of Hif1α (see Figure 4). Knockdown of ELL2 in C4-2 prostate cancer cells also resulted in an increase in HIF1 α protein. Furthermore, Rhobtb3 mRNA was identified by cDNA microarray as one of the most differentially down-regulated genes in ELL2 knockout mouse prostate in mice at 3 mos of age (see Table S1) as well as in the anterior prostates of mice at 18 mos of age (see Figure 5). RHOBTB3 was recently shown to promote the hydroxylation, ubiquitination and degradation of HIFα (Zhang, et al. 2015). Increased expression of HIF1α has been reported as a potential early ‘angiogenic event’ in the development of prostate cancer in the TRAMP model (Huss et al. 2001) and is negatively regulated by EAF2 (Chen et al. 2014). ELL2 and EAF2 appear to similarly regulate prostate angiogenesis in part through modulation of HIF1α. Future studies will be required to determine if ELL2 and EAF2 interaction is critical for regulation of prostate vascularity.
Microarray analysis identified several other differentially expressed genes in the Ell2-cko knockout mouse at 3 mos of age in addition to Rhobtb3. Functional classification of these genes utilizing DAVID and Ingenuity Pathways Analysis revealed alteration of genes involved in golgi apparatus, intracellular transport and cell-cell signaling, and cancer. qPCR validation confirmed the persistent altered expression of several of these genes in animals at 17 mos of age, including Sfi1, Cxcl10, Pds5a, Rtn4, Olig2, Rhobtb3, and Gbp2 suggesting that these genes and their associated pathways might contribute to the development of the histological defects identified in the prostates of aged animals with ELL2 loss. SFI1 is a key regulator of normal endocrine steroidogenesis, which is up-regulated in castration resistant prostate cancer (Lewis, et al. 2014). PDS5A, also known as SCC-112, was recently reported as a translocation partner of MLL (Put, et al. 2012) and was initially characterized as a cell cycle regulator and promoter of apoptosis (Kumar, et al. 2004). Persistent alteration of these genes likely contributes to the proliferative and highly vascularized preneoplastic phenotype observed in the prostates of Ell2 knockout mice at age 17–20 mos.
RTN4, OLIG2, S1PR1 and S1PR3 interact with several pathways previously associated with prostate carcinogenesis and are involved in neural differentiation (see Figure 7). Eaf2 loss was previously shown to activate the ERK pathway (Su, et al. 2013). RTN4 is a neurite outgrowth inhibitor (Spillmann, et al. 1998) that was shown to inhibit proliferation and promote apoptosis when transfected into human hepatocellular carcinoma cells (Chen, et al. 2005). OLIG2 is a neural stem cell transcription factor that regulates differentiation of oligodendrocytes (Zhou and Anderson 2002). S1PR1 and S1PR3 are involved in dendritic remodeling (Willems, et al. 2016). Since these genes were also frequently co-up-regulated with ELL2 in the neuroendocrine prostate cancer dataset (see Figure 8A), ELL2 may play a role in the differentiation and proliferation of neuroendocrine cells through the regulation of RTN4, OLIG2, S1PR1 and S1PR3. Overexpression of ELL2 and its target genes was associated with prostate cancers with a neuroendocrine phenotype, whereas downregulation of ELL2 and ELL2 target genes was associated with prostate adenocarcinoma. Future studies will be required to fully elucidate the role of ELL2 interaction with these genes in prostate carcinogenesis.
Cumulatively, these results show that similar to EAF2 loss, ELL2 loss in the prostate induced increased epithelial proliferation, increased microvessel density and preneoplastic lesions. Several pathways were identified that are potentially regulated by ELL2 in the prostate, and many of these genes were found to be altered in conjunction with ELL2 in patient tumors in the cBioPortal database. Interestingly, ELL2 and its regulated genes were up-regulated in neuroendocrine prostate tumors, and down-regulated in prostate adenocarcinoma. These findings provide a strong foundation for further studies to elucidate the mechanisms by which ELL2 loss promotes prostate carcinogenesis.
Supplementary Material
Acknowledgments
We are grateful to Marie Acquafondata, Megan Lambert, Katie Leschak, and Aiyuan Zhang for technical support and Karen Seisek for reading the manuscript. This work was funded in part by NIH grants R01CA186780, P50 CA090386, T32 DK007774, and 1R50 CA211242 and scholarships from the Tippins Foundation (LEP) and the Mellam Foundation (KZM). The microarray work was performed in the Genomics Research Core at the University of Pittsburgh. This project used the UPCI Animal Facility and was supported in part by award P30CA047904.
Footnotes
Disclosure statement: The authors have nothing to disclose.
Contributions:
LEP, KZM, JL and ZW designed experiments. LEP, KZM, JL, XQ, QS, YW, YZ, TY, and YW contributed reagents, animals, tissue specimens and/or performed research. LHR analyzed murine prostate pathology. LEP, UC, LMC and RZV analyzed microarray data. LEP wrote the manuscript with help from KZM, YL, JZ and ZW. All authors reviewed and edited the manuscript.
References
- Ai J, Pascal LE, O’Malley KJ, Dar JA, Isharwal S, Qiao Z, Ren B, Rigatti LH, Dhir R, Xiao W, et al. Concomitant loss of EAF2/U19 and Pten synergistically promotes prostate carcinogenesis in the mouse model. Oncogene. 2013 doi: 10.1038/onc.2013.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai J, Wang Y, Dar JA, Liu J, Liu L, Nelson JB, Wang Z. HDAC6 regulates androgen receptor hypersensitivity and nuclear localization via modulating Hsp90 acetylation in castration-resistant prostate cancer. Mol Endocrinol. 2009;23:1963–1972. doi: 10.1210/me.2009-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda PS, LaJoie DM, Jorcyk CL. Bleach gel: a simple agarose gel for analyzing RNA quality. Electrophoresis. 2012;33:366–369. doi: 10.1002/elps.201100335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV, Varambally S, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21:2005–2017. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Phong BL, Fisher AL, Wang Z. Regulation of fertility, survival, and cuticle collagen function by the Caenorhabditis elegans eaf-1 and ell-1 genes. J Biol Chem. 2011;28(6):35915–35921. doi: 10.1074/jbc.M111.270454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Lu DD, Cao XR, Zhang XR. RTN4-C gene expression in hepatocellular carcinoma and its influence on SMMC7721 cell growth and apoptosis. Yi Chuan Xue Bao. 2005;32:891–897. [PubMed] [Google Scholar]
- Chen Z, Liu X, Mei Z, Wang Z, Xiao W. EAF2 suppresses hypoxia-induced factor 1alpha transcriptional activity by disrupting its interaction with coactivator CBP/p300. Mol Cell Biol. 2014;34:1085–1099. doi: 10.1128/MCB.00718-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyriac J, Haleem R, Cai X, Wang Z. Androgen regulation of spermidine synthase expression in the rat prostate. Prostate. 2002;50:252–261. doi: 10.1002/pros.10052. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Eissenberg JC, Ma J, Gerber MA, Christensen A, Kennison JA, Shilatifard A. dELL is an essential RNA polymerase II elongation factor with a general role in development. Proc Natl Acad Sci U S A. 2002;99:9894–9899. doi: 10.1073/pnas.152193699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funahashi Y, Wang Z, O’Malley KJ, Tyagi P, DeFranco DB, Gingrich JR, Takahashi R, Majima T, Gotoh M, Yoshimura N. Influence of E. coli-induced prostatic inflammation on expression of androgen-responsive genes and transforming growth factor beta 1 cascade genes in rats. Prostate. 2015;75:381–389. doi: 10.1002/pros.22924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haram KM, Peltier HJ, Lu B, Bhasin M, Otu HH, Choy B, Regan M, Libermann TA, Latham GJ, Sanda MG, et al. Gene expression profile of mouse prostate tumors reveals dysregulations in major biological processes and identifies potential murine targets for preclinical development of human prostate cancer therapy. Prostate. 2008;68:1517–1530. doi: 10.1002/pros.20803. [DOI] [PubMed] [Google Scholar]
- Huss WJ, Hanrahan CF, Barrios RJ, Simons JW, Greenberg NM. Angiogenesis and prostate cancer: identification of a molecular progression switch. Cancer Res. 2001;61:2736–2743. [PubMed] [Google Scholar]
- Kong SE, Banks CA, Shilatifard A, Conaway JW, Conaway RC. ELL-associated factors 1 and 2 are positive regulators of RNA polymerase II elongation factor ELL. Proc Natl Acad Sci U S A. 2005;102:10094–10098. doi: 10.1073/pnas.0503017102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Sakabe I, Patel S, Zhang Y, Ahmad I, Gehan EA, Whiteside TL, Kasid U. SCC-112, a novel cell cycle-regulated molecule, exhibits reduced expression in human renal carcinomas. Gene. 2004;328:187–196. doi: 10.1016/j.gene.2003.12.013. [DOI] [PubMed] [Google Scholar]
- Lavau C, Luo RT, Du C, Thirman MJ. Retrovirus-mediated gene transfer of MLL-ELL transforms primary myeloid progenitors and causes acute myeloid leukemias in mice. Proc Natl Acad Sci U S A. 2000;97:10984–10989. doi: 10.1073/pnas.190167297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SR, Hedman CJ, Ziegler T, Ricke WA, Jorgensen JS. Steroidogenic factor 1 promotes aggressive growth of castration-resistant prostate cancer cells by stimulating steroid synthesis and cell proliferation. Endocrinology. 2014;155:358–369. doi: 10.1210/en.2013-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, Shilatifard A. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC) Genes Dev. 2011;25:1486–1498. doi: 10.1101/gad.2059211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Ai J, Xiao W, Liu J, Wang Y, Xin D, He Z, Guo Y, Wang Z. ELL is an HIF-1alpha partner that regulates and responds to hypoxia response in PC3 cells. Prostate. 2010;70:797–805. doi: 10.1002/pros.21113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincic K, Alkan SA, Cheatle A, Borghesi L, Milcarek C. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol. 2009;10:1102–1109. doi: 10.1038/ni.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milcarek C, Albring M, Langer C, Park KS. The eleven-nineteen lysine-rich leukemia gene (ELL2) influences the histone H3 protein modifications accompanying the shift to secretory immunoglobulin heavy chain mRNA production. J Biol Chem. 2011;286:33795–33803. doi: 10.1074/jbc.M111.272096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Williams K, Johnstone RW, Shilatifard A. Identification, cloning, expression, and biochemical characterization of the testis-specific RNA polymerase II elongation factor ELL3. J Biol Chem. 2000;275:32052–32056. doi: 10.1074/jbc.M005175200. [DOI] [PubMed] [Google Scholar]
- Mitani K, Kanda Y, Ogawa S, Tanaka T, Inazawa J, Yazaki Y, Hirai H. Cloning of several species of MLL/MEN chimeric cDNAs in myeloid leukemia with t(11;19)(q23;p13.1) translocation. Blood. 1995;85:2017–2024. [PubMed] [Google Scholar]
- Mitani K, Yamagata T, Iida C, Oda H, Maki K, Ichikawa M, Asai T, Honda H, Kurokawa M, Hirai H. Nonredundant roles of the elongation factor MEN in postimplantation development. Biochem Biophys Res Commun. 2000;279:563–567. doi: 10.1006/bbrc.2000.3970. [DOI] [PubMed] [Google Scholar]
- Nelson PS, Clegg N, Arnold H, Ferguson C, Bonham M, White J, Hood L, Lin B. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc Natl Acad Sci U S A. 2002;99:11890–11895. doi: 10.1073/pnas.182376299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley KJ, Dhir R, Nelson JB, Bost J, Lin Y, Wang Z. The expression of androgen-responsive genes is up-regulated in the epithelia of benign prostatic hyperplasia. Prostate. 2009;69:1716–1723. doi: 10.1002/pros.21034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Bayles I, Szlachta-McGinn A, Paul J, Boiko J, Santos P, Liu J, Wang Z, Borghesi L, Milcarek C. Transcription elongation factor ELL2 drives Ig secretory-specific mRNA production and the unfolded protein response. J Immunol. 2014;193:4663–4674. doi: 10.4049/jimmunol.1401608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal LE, Ai J, Masoodi KZ, Wang Y, Wang D, Eisermann K, Rigatti LH, O’Malley KJ, Ma HM, Wang X, et al. Development of a reactive stroma associated with prostatic intraepithelial neoplasia in EAF2 deficient mice. PLoS One. 2013;8:e79542. doi: 10.1371/journal.pone.0079542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal LE, Ai J, Rigatti LH, Lipton AK, Xiao W, Gnarra JR, Wang Z. EAF2 loss enhances angiogenic effects of Von Hippel-Lindau heterozygosity on the murine liver and prostate. Angiogenesis. 2011;14:331–343. doi: 10.1007/s10456-011-9217-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Put N, Van Roosbroeck K, Vande Broek I, Michaux L, Vandenberghe P. PDS5A, a novel translocation partner of MLL in acute myeloid leukemia. Leuk Res. 2012;36:e87–89. doi: 10.1016/j.leukres.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Qiu X, Pascal LE, Song Q, Zang Y, Ai J, O’Malley KJ, Nelson JB, Wang Z. Physical and Functional Interactions between ELL2 and RB in the Suppression of Prostate Cancer Cell Proliferation, Migration, and Invasion. Neoplasia. 2017;19:207–215. doi: 10.1016/j.neo.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saporita AJ, Ai J, Wang Z. The Hsp90 inhibitor, 17-AAG, prevents the ligand-independent nuclear localization of androgen receptor in refractory prostate cancer cells. Prostate. 2007;67:509–520. doi: 10.1002/pros.20541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Shappell SB, Thomas GV, Roberts RL, Herbert R, Ittmann MM, Rubin MA, Humphrey PA, Sundberg JP, Rozengurt N, Barrios R, et al. Prostate pathology of genetically engineered mice: definitions and classification. The consensus report from the Bar Harbor meeting of the Mouse Models of Human Cancer Consortium Prostate Pathology Committee. Cancer Res. 2004;64:2270–2305. doi: 10.1158/0008-5472.can-03-0946. [DOI] [PubMed] [Google Scholar]
- Shilatifard A, Duan DR, Haque D, Florence C, Schubach WH, Conaway JW, Conaway RC. ELL2, a new member of an ELL family of RNA polymerase II elongation factors. Proc Natl Acad Sci U S A. 1997;94:3639–3643. doi: 10.1073/pnas.94.8.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone F, Luo RT, Polak PE, Kaberlein JJ, Thirman MJ. ELL-associated factor 2 (EAF2), a functional homolog of EAF1 with alternative ELL binding properties. Blood. 2003;101:2355–2362. doi: 10.1182/blood-2002-06-1664. [DOI] [PubMed] [Google Scholar]
- Smith ER, Lin C, Garrett AS, Thornton J, Mohaghegh N, Hu D, Jackson J, Saraf A, Swanson SK, Seidel C, et al. The little elongation complex regulates small nuclear RNA transcription. Mol Cell. 2011;44:954–965. doi: 10.1016/j.molcel.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillmann AA, Bandtlow CE, Lottspeich F, Keller F, Schwab ME. Identification and characterization of a bovine neurite growth inhibitor (bNI-220) J Biol Chem. 1998;273:19283–19293. doi: 10.1074/jbc.273.30.19283. [DOI] [PubMed] [Google Scholar]
- Su F, Correa BR, Luo J, Vencio RZ, Pascal LE, Wang Z. Gene Expression Profiling Reveals Regulation of ERK Phosphorylation by Androgen-Induced Tumor Suppressor U19/EAF2 in the Mouse Prostate. Cancer Microenviron. 2013;6:247–261. doi: 10.1007/s12307-013-0132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su F, Pascal LE, Xiao W, Wang Z. Tumor suppressor U19/EAF2 regulates thrombospondin-1 expression via p53. Oncogene. 2010;29:421–431. doi: 10.1038/onc.2009.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, et al. 2011 Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirman MJ, Levitan DA, Kobayashi H, Simon MC, Rowley JD. Cloning of ELL, a gene that fuses to MLL in a t(11;19)(q23;p13.1) in acute myeloid leukemia. Proc Natl Acad Sci U S A. 1994;91:12110–12114. doi: 10.1073/pnas.91.25.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- Willems LM, Zahn N, Ferreiros N, Scholich K, Maggio N, Deller T, Vlachos A. Sphingosine-1-phosphate receptor inhibition prevents denervation-induced dendritic atrophy. Acta Neuropathol Commun. 2016;4:28. doi: 10.1186/s40478-016-0303-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wu J, Huang J, Powell WC, Zhang J, Matusik RJ, Sangiorgi FO, Maxson RE, Sucov HM, Roy-Burman P. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech Dev. 2001;101:61–69. doi: 10.1016/s0925-4773(00)00551-7. [DOI] [PubMed] [Google Scholar]
- Xiao W, Ai J, Habermacher G, Volpert O, Yang X, Zhang AY, Hahn J, Cai X, Wang Z. U19/Eaf2 binds to and stabilizes von hippel-lindau protein. Cancer Res. 2009;69:2599–2606. doi: 10.1158/0008-5472.CAN-08-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Zhang Q, Habermacher G, Yang X, Zhang AY, Cai X, Hahn J, Liu J, Pins M, Doglio L, et al. U19/Eaf2 knockout causes lung adenocarcinoma, B-cell lymphoma, hepatocellular carcinoma and prostatic intraepithelial neoplasia. Oncogene. 2008;27:1536–1544. doi: 10.1038/sj.onc.1210786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Zhang Q, Jiang F, Pins M, Kozlowski JM, Wang Z. Suppression of prostate tumor growth by U19, a novel testosterone-regulated apoptosis inducer. Cancer Res. 2003;63:4698–4704. [PubMed] [Google Scholar]
- Zhang CS, Liu Q, Li M, Lin SY, Peng Y, Wu D, Li TY, Fu Q, Jia W, Wang X, et al. RHOBTB3 promotes proteasomal degradation of HIFalpha through facilitating hydroxylation and suppresses the Warburg effect. Cell Res. 2015;25:1025–1042. doi: 10.1038/cr.2015.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.