Loss of Krit1 disables an angiogenic checkpoint by reducing TSP1 expression, thereby enabling cerebral cavernous malformation (CCM) formation. Lopez-Ramirez et al. propose that replacing TSP1 by TSP1 fragments or angiogenesis inhibitors may provide an approach to inhibit CCM development.
Abstract
KRIT1 mutations are the most common cause of cerebral cavernous malformation (CCM). Acute Krit1 gene inactivation in mouse brain microvascular endothelial cells (BMECs) changes expression of multiple genes involved in vascular development. These changes include suppression of Thbs1, which encodes thrombospondin1 (TSP1) and has been ascribed to KLF2- and KLF4-mediated repression of Thbs1. In vitro reconstitution of TSP1 with either full-length TSP1 or 3TSR, an anti-angiogenic TSP1 fragment, suppresses heightened vascular endothelial growth factor signaling and preserves BMEC tight junctions. Furthermore, administration of 3TSR prevents the development of lesions in a mouse model of CCM1 (Krit1ECKO) as judged by histology and quantitative micro-computed tomography. Conversely, reduced TSP1 expression contributes to the pathogenesis of CCM, because inactivation of one or two copies of Thbs1 exacerbated CCM formation. Thus, loss of Krit1 function disables an angiogenic checkpoint to enable CCM formation. These results suggest that 3TSR, or other angiogenesis inhibitors, can be repurposed for TSP1 replacement therapy for CCMs.
Introduction
Cerebral cavernous malformations (CCMs) are central nervous system vascular anomalies that lead to significant morbidity and mortality (Golden et al., 2015). CCMs affect ∼1 of 200 humans and cause a lifelong risk of stroke and other neurological sequelae for which there is no pharmacologic therapy. Loss-of-function mutations of three genes (KRTI1, CCM2, and PDCD10) are associated with development of venous capillary dysplasias with disrupted junctions, leading to hemorrhage and increased vascular permeability (Mikati et al., 2015) characteristic of CCM (D’Angelo et al., 2011; McDonald et al., 2014). The KRIT1+/− genotype is the most common cause of the familial form of CCM (Tanriover et al., 2013). In mice, timed endothelial-specific inactivation of Krit1 results in cerebellar and retinal vascular lesions that are similar to those in CCM patients (Boulday et al., 2011; Maddaluno et al., 2013; Cuttano et al., 2016). These mouse studies, in combination with the finding of a “second hit” on the normal KRIT1 allele in human CCM endothelial cells (McDonald et al., 2011) indicate that a complete loss of KRIT1 function causes endothelial cell–autonomous CCM formation.
The consequences of loss of endothelial KRIT1 include abnormal angiogenesis (Whitehead et al., 2004; Boulday et al., 2011), dysregulation of endothelial metalloproteinases (Zhou et al., 2015), increased expression of the transcription factors KLF2 and KLF4 (Maddaluno et al., 2013; Renz et al., 2015; Zhou et al., 2015), and alterations in signaling pathways such as Notch (Wüstehube et al., 2010), vascular endothelial growth factor (VEGF; DiStefano et al., 2014), and Rho/Rho kinase (Whitehead et al., 2009; Stockton et al., 2010). In addition, increased cell migration caused by disruption of endothelial apical-basal polarity (Lampugnani et al., 2010) and endothelial-to-mesenchymal transition (Maddaluno et al., 2013) have been recently reported to be features of CCMs. Changes in gene expression associated with these phenotypic changes include increased expression of the transcription factors KLF2 and KLF4 (Renz et al., 2015; Zhou et al., 2015) and mesenchymal genes (Maddaluno et al., 2013; Cuttano et al., 2016); however, a detailed picture of the early changes in gene expression that follow loss of KRIT1 has been lacking. Here we perform genome-wide transcriptome analysis of brain microvascular endothelial cells (BMECs) after acute Krit1 inactivation and describe a signature of mRNA changes primarily affecting genes involved in cardiovascular development. A striking finding was the dramatic suppression of a potent endogenous angiogenesis inhibitor, TSP1, which we ascribe to KLF2- and KLF4-mediated repression of the gene encoding TSP1, Thbs1. Reduced TSP1 expression contributes to the pathogenesis of CCMs, because loss of one or two copies of Thbs1 exacerbated CCM formation. Replenishing TSP1 in vitro with either full-length TSP1 or 3TSR, an anti-angiogenic TSP1 fragment, prevented the disruption of BMEC tight junctions, a phenotypic result of Krit1 inactivation. Administration of 3TSR prevented disruption of intercellular junctions in Krit1ECKO mice in part because of the capacity of 3TSR to inhibit VEGF signaling. Administration of 3TSR inhibited formation of CCMs in Krit1ECKO mice. Thus, loss of expression of TSP1, a key angiogenic checkpoint, plays an important role in pathogenesis of CCM, and repurposing 3TSR or other angiogenesis inhibitors may provide a new approach to inhibit CCM development.
Results
Genetic inactivation of Krit1 inhibits expression of TSP1
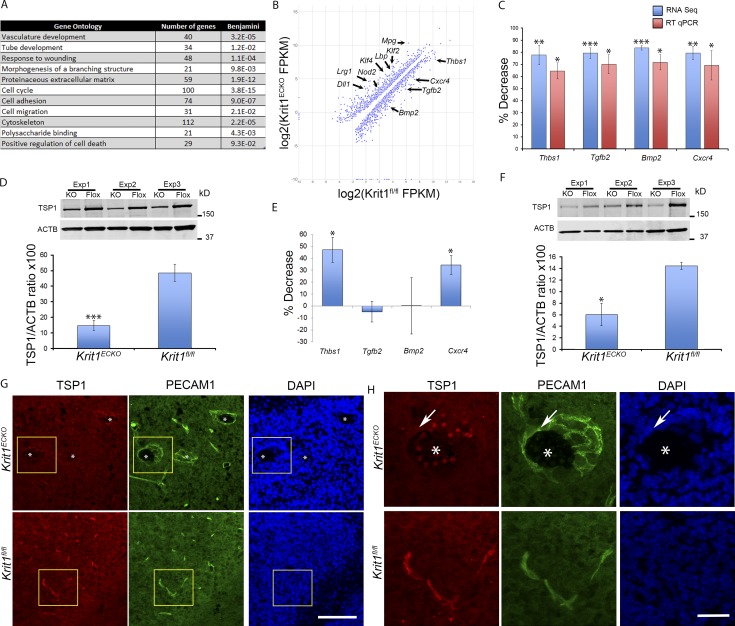
To elucidate the pathogenesis of CCM, we used genome-wide RNA sequencing (RNA-seq) to characterize the transcriptome of primary BMECs after acute genetic inactivation of Krit1. BMECs were isolated from mice bearing floxed alleles of Krit1 (Krit1fl/fl) and an endothelial-specific tamoxifen-regulated Cre recombinase (Pdgfb-iCreERT2; Claxton et al., 2008). Treatment of Krit1fl/fl-Pdgfb-iCreERT2 BMECs with 5 µM-hydroxy-tamoxifen deleted Krit1 (Krit1ECKO) and reduced KRIT1 mRNA and protein by >90% within 5 d compared with hydroxy-tamoxifen–treated Krit1fl/fl BMECs. Deep sequencing of cDNA from Krit1ECKO and Krit1fl/fl BMECs (Fig. S1) revealed that acute loss of brain endothelial KRIT1 caused a dramatic change in gene expression in BMECs. We identified 334 genes differentially expressed between Krit1ECKO and Krit1fl/fl BMECs (corrected P < 0.05, ≥2.5-fold change). Gene ontology analysis of the differentially expressed genes indicated significant enrichment for terms related to cardiovascular development (P < 3.2 × 10−5; Fig. 1 A). Among genes known to be important in cardiovascular development were up-regulation of Klf2 and Klf4, two transcription factors recently implicated in CCM pathogenesis (Maddaluno et al., 2013; Renz et al., 2015; Zhou et al., 2015; Cuttano et al., 2016; Fig. 1 B). Among the most notable changes were an ∼75% decrease in expression of genes encoding secreted proteins or receptors that regulate angiogenesis, including Thbs1 (whose protein product is TSP1), Cxcr4, Bmp2, and Tgfb2 (Fig. 1, B and C). The changes in TSP1 mRNA levels were associated with reduced TSP1 protein expression (∼3-fold decrease) in Krit1ECKO BMECs (Fig. 1 D).
Figure 1.
Loss of KRIT1 inhibits the expression of TSP1. (A) Genome-wide RNA-seq from three independent biological replicates followed by gene ontology analysis of genes differentially expressed in Krit1ECKO BMECs compared with Krit1fl/fl BMECs. Each term listed was the top term in a cluster of related terms, and the corrected p-values were calculated according to Benjamini’s method (Huang et al., 2009). (B) Expression levels of differentially expressed genes represented on a scatter plot; fragments per kilobase of transcript per million mapped reads (FPKM) of individual transcripts are represented on a log2 scale. A few of the most highly suppressed and up-regulated genes are labeled. (C) RT-qPCR confirmation of RNA-seq–identified marked decrease in mRNA of extracellular regulators of angiogenesis in Krit1ECKO BMECs compared with Krit1fl/fl BMECs (SEM, n = 3). (D) Quantification of TSP1 protein from three independent biological replicates in Krit1ECKO (KO) and in Krit1fl/fl (Flox) BMECs (SEM, n = 3). (E) RT-qPCR analysis of isolated brain microvasculature in Krit1ECKO compared with Krit1fl/fl littermate controls (SEM, n = 3). (F) Quantification of TSP1 protein from freshly isolated brain microvasculature in Krit1ECKO (KO) compared with Krit1fl/fl (Flox) littermate controls (SEM, n = 3). (G) Confocal microscopy of cerebellar cortex stained for TSP1 (red) and endothelial specific marker PECAM1 (green); DAPI staining (blue) was used to reveal nuclei (n = 3). (H) Higher-magnification images of boxed areas in G. TSP1 protein expression was decreased in CCM from Krit1ECKO mice (arrows). Asterisks, vascular lumen of CCM lesions. Bars: (G) 100 µm; (H) 25 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; determined by Student’s t test. Data in A–H are representative of three independent experiments.
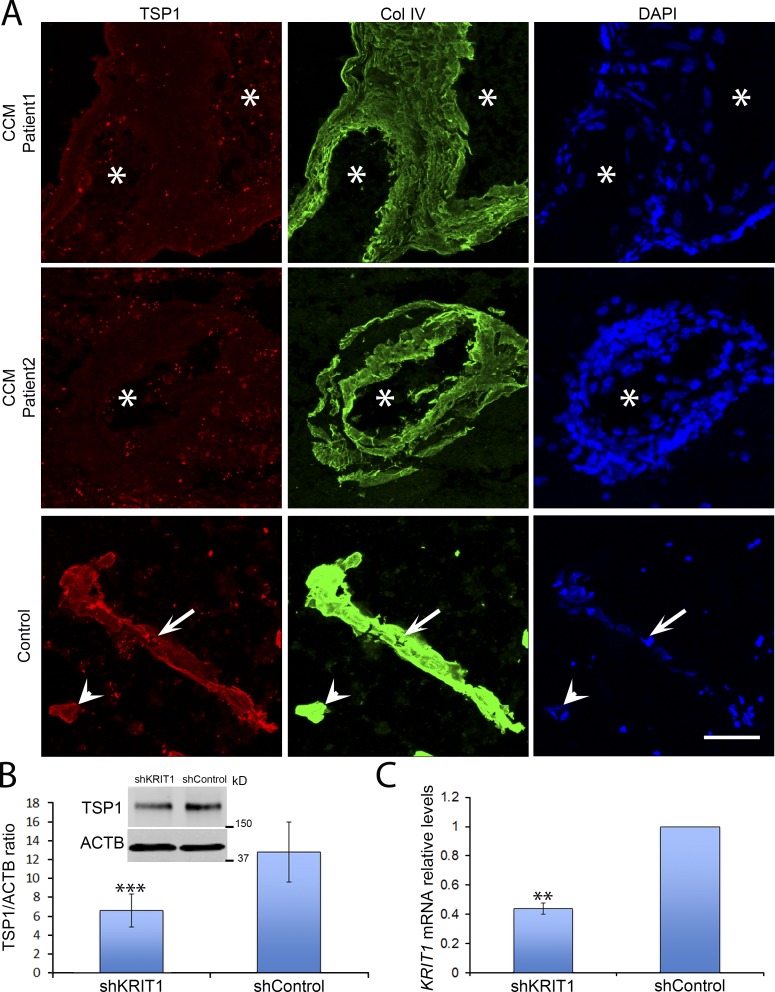
To confirm these reductions in potential extracellular regulators of angiogenesis in vivo, we isolated brain microvasculature from tamoxifen-treated neonatal Krit1ECKO or Krit1fl/fl mice and quantified mRNA with real-time quantitative PCR (RT-qPCR). Consistent with results observed in vitro, TSP1 and CXCR4 mRNA levels in freshly isolated brain microvasculature were reduced in postnatal day 7 (P7) Krit1ECKO mice (Fig. 1 E). In sharp contrast, we did not observe significant changes in BMP2 or TGFβ2 mRNA abundance (Fig. 1 E). We focused on TSP1 because reduced TSP1 mRNA expression was confirmed in vivo, TSP1 is an endogenous anti-angiogenic protein, and TSP1 fragments and analogues have been developed as potential cancer therapeutic agents (Zhang and Lawler, 2007; Lawler and Lawler, 2012). We confirmed that the reduction in mRNA was reflected in reduced TSP1 protein abundance in freshly isolated brain microvasculature after genetic inactivation of endothelial Krit1 (Fig. 1 F). Furthermore, there was a dramatic reduction in in situ TSP1 protein staining in CD31-positive endothelial cells in lesions of Krit1ECKO mice in comparison to Krit1fl/fl littermates (Fig. 1, G and H). Loss of TSP1 expression also occurs during the pathogenesis of human CCM, because there was a dramatic decrease in endothelial TSP1 staining in human CCM lesions in comparison to a lesion-free brain tissue (Fig. 2 A), and silencing KRIT1 in human endothelial cells led to reduced TSP1 expression in HUVECs (Fig. 2, B and C). Thus, inactivation of Krit1 in BMECs leads to early reductions in TSP1 mRNA and protein expression in vitro and in vivo, suggesting a role for the reduction of this secreted anti-angiogenic protein in the pathogenesis of CCM.
Figure 2.
Decreased human endothelial TSP1. (A) Immunofluorescent staining of TSP1 (red) and collagen IV (green) of human CCM and lesion-free brain tissue. Arrowheads, capillary; arrows, venule; asterisks, vascular lumen of CCM lesions. Bar, 100 µm (n = 2). (B and C) HUVECs were transduced with shKrit1 or shControl using lentivirus. (B) KRIT1-depleted HUVECs expressed ∼50% as much TSP1 protein as control cells (SEM, n = 3). (C) KRIT1 shRNA 55% decrease in KRIT1 mRNA as determined by RT-qPCR (SEM, n = 4). **, P < 0.01; ***, P < 0.001; determined by Student’s t test. Data in A is from two independent experiments, and data in B and C are representative of three or four independent experiments.
Altered tight junctions are an early event that follows loss of KRIT1
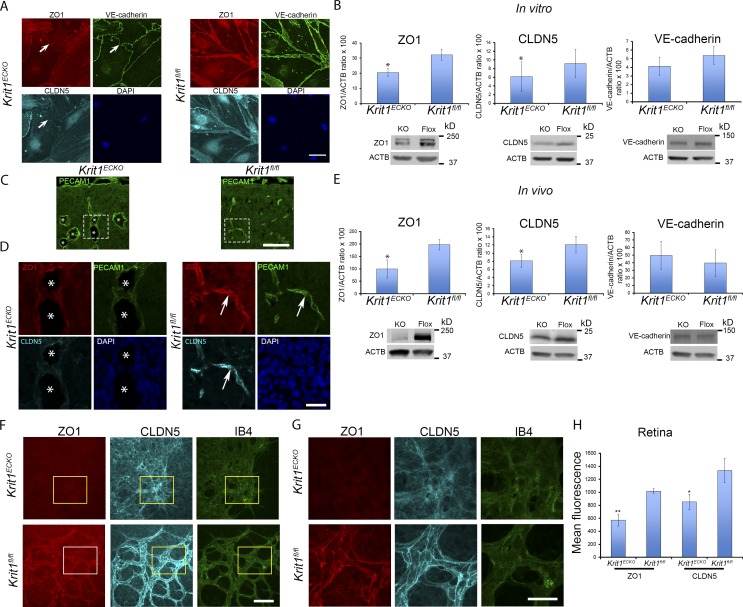
Because TSP1 is a large (∼450 kD) glycoprotein, we sought an in vitro intermediate phenotype to assess the effect of TSP1 reconstitution. Disruption of cell–cell junctions and increased vascular permeability are prominent features of CCM in humans (Tanriover et al., 2013; Mikati et al., 2015), and silencing of KRIT1 leads to disruption of intercellular junctions in HUVECs (Glading et al., 2007). We examined the time course of altered cell–cell junctions in cultured BMECs after acute genetic inactivation of Krit1. There was striking loss of tight junction proteins ZO-1 and claudin-5 from BMEC junctions within 5 d of 4-hydroxy-tamoxifen treatment, an early time after the concentration of KRIT1 protein and mRNA were decreased (Fig. 3 A and Fig. S2). Furthermore, immunoblotting revealed a 40–50% reduction in ZO-1 and claudin-5 protein abundance after inactivation of Krit1 in BMECs (Fig. 3 B). In contrast, at this early time point, the small decreases in vascular endothelial cadherin (VE-cadherin) protein abundance and distribution were not statistically significant (Fig. 3, A and B). To assess whether similar changes were observed at early times in lesion evolution in vivo, we examined the endothelial distribution of these junctional proteins in the brains of P7 Krit1ECKO mice. ZO-1 and claudin-5 staining were markedly reduced in the dilated early CCM in these mice, whereas both tight junction proteins were abundant in normal vessels from Krit1ECKO and Krit1fl/fl littermates (Fig. 3, C and D). Furthermore, there were reduced levels of ZO-1 and claudin-5 in microvasculature isolated from the cerebellum of P7–P10 Krit1ECKO mice as assessed by immunoblotting (Fig. 3 E). We did not observe significant changes in VE-cadherin protein levels in early lesions in Krit1ECKO (P7–P10) mice. Similar changes were seen in Krit1ECKO mouse retina (Fig. 3, F–H), a tissue that enables precise temporal and spatial assessment of vascular development (Pitulescu et al., 2010). Collectively, these data show that changes in tight junctions, which are features of CCM, are early abnormalities that follow loss of KRIT1 in BMECs. Such changes could therefore represent an intermediate phenotype to assess potential interventions in CCM pathogenesis.
Figure 3.
Altered tight junctions are an early phenotypic consequence of Krit1 inactivation. (A) Representative confocal images of ZO-1 (red), claudin5 (CLDN5; turquoise), and VE-cadherin (green) staining in primary BMEC Krit1ECKO or control Krit1fl/fl BMECs. Nuclei were counterstained with DAPI (blue; n = 4). Arrows indicate loss of tight junctions but not adherence junctions. (B) Quantification of brain endothelial ZO-1, claudin5, and VE-cadherin protein as assessed by Western blot analysis in Krit1ECKO compared with Krit1fl/fl BMEC controls (SEM, n = 3 or 4). (C) Confocal microscopy of cerebellar cortex at P7 stained with anti-PECAM1 (green). (D) Higher-magnification images of boxed areas in C stained for ZO-1 (red), claudin5 (turquoise), and PECAM1 (green). Arrows, staining of tight junction proteins ZO-1 and claudin5; asterisks, vascular lumen of CCM (n = 3). (E) Quantification of brain endothelial ZO-1, claudin5, and VE-cadherin protein abundance in freshly isolated cerebellar microvasculature in Krit1ECKO compared with Krit1fl/fl littermate controls (SEM, n = 3 or 4). (F) Maximum-intensity projection of whole-mount P7 retinal vasculature at the angiogenic growth front stained for ZO-1 (red), claudin5 (turquoise), and an endothelial marker, isolectin B4 (green). (G) Higher-magnification images of boxed areas in F show staining for ZO-1 (red), claudin5 (turquoise), and isolectin B4 (green). (H) Quantification of ZO-1 and claudin5 protein expression in retinal vasculature at the angiogenic front in Krit1ECKO compared with Krit1fl/fl littermate controls (SEM, n = 6 mice per group). Bars: (A) 50 µm; (C) 100 µm; (D, F, and G) 25 µm. *, P < 0.05; **, P < 0.01; determined by Student’s t test. Data in A–H are representative of at least three independent experiments.
Reconstitution of TSP1 prevents the loss of tight junctions that follows inactivation of Krit1
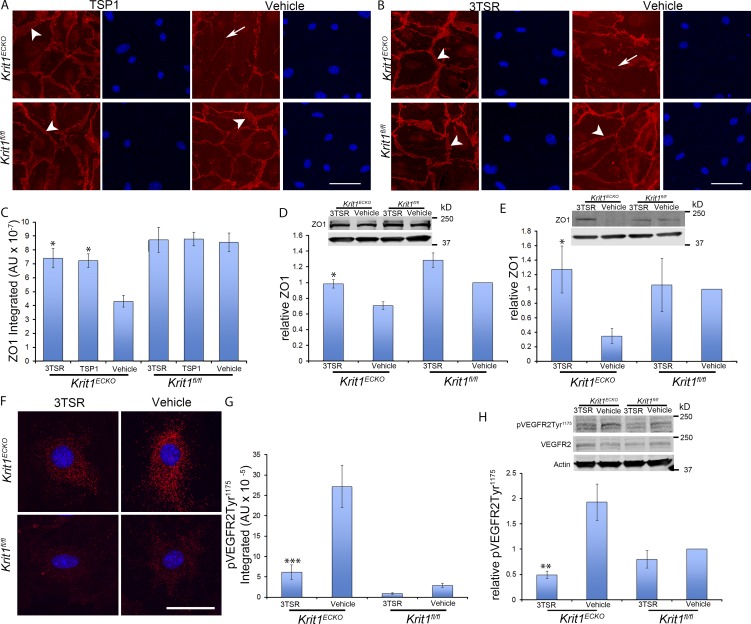
Having shown that altered tight junctions are a result of loss of KRIT1 both in vitro and in vivo, we assessed the effect of the addition of exogenous mouse TSP1 on this phenotype in vitro. Addition of 5 nM TSP1 prevented the loss of ZO-1 from BMEC tight junctions after Krit1 inactivation (Fig. 4 A). In contrast, prior studies showed that much higher doses of TSP-1 (∼200 nM) could destabilize junctions (Garg et al., 2011). TSP1 is a modular protein and a recombinant fragment containing three type I repeats (3TSR) that accounts for much of the anti-angiogenic activity through its capacity to engage both CD36 and integrins (Short et al., 2005; Lawler and Lawler, 2012). Treatment of BMEC with 20 nM recombinant 3TSR also prevented loss of ZO-1 from cell–cell junctions (Fig. 4 B) and the reduction in ZO-1 protein abundance that followed deletion of Krit1 (Fig. 4, C and D). These effects of 3TSR were also seen in vivo. Treatment of Krit1ECKO mice with 1.6 mg/kg 3TSR on two succeeding days increased ZO-1 expression in the cerebellum (Fig. 4 E). These data show that replacement of TSP1 can prevent the effects of Krit1 deletion on the distribution and abundance of ZO-1 in brain endothelium and that an anti-angiogenic domain of TSP1 (3TSR) is sufficient for this activity.
Figure 4.
Reconstitution of TSP1 prevents the tight junction loss that follows inactivation of Krit1. (A) 5 nM mouse recombinant TSP1 (mrTSP1) was added to cultured Krit1ECKO BMEC 72 h after initial treatment with 4-hydroxy-tamoxifen. Note continuous ZO-1 junctional staining in TSP1-treated Krit1ECKO BMEC (arrowheads), resembling the appearance of Krit1fl/fl BMEC. In sharp contrast, ZO-1 staining was reduced and discontinuous (arrows) in vehicle-treated Krit1ECKO. (B) Treatment with an anti-angiogenic domain (20 nM), 3TSR. The arrows in Krit1ECKO BMEC indicate immunostaining for ZO-1 that is punctate and reduced at the cell–cell contacts. Treatment with 3TSR prevented loss of ZO-1 protein from tight junctions in Krit1ECKO BMEC (arrowheads). (C) Quantification of ZO-1 protein expression in BMEC. Krit1ECKO or control Krit1fl/fl BMEC treated with TSP1, 3TSR, or vehicle as indicated (SEM, n = 3). (D) ZO-1 protein levels as determined by Western blot analysis in Krit1ECKO and Krit1fl/fl BMEC in the presence or absence of 20 nM 3TSR (SEM, n = 3). Bottom row is actin loading control. (E) Quantification of ZO-1 protein expression in cerebellar tissue in Krit1ECKO and control Krit1fl/fl mice treated with 3TSR or vehicle (SEM, n = 4 mice in each group). Bottom row is actin loading control. (F) VEGFR2-Tyr1175 phosphorylation in primary BMEC Krit1ECKO or control Krit1fl/fl BMECs treated with 3TSR or vehicle. Nuclei were counterstained with DAPI (blue). (G) Quantification of VEGFR2-Tyr1175 phosphorylation in BMECs is shown as integrated density in Krit1ECKO and Krit1fl/fl controls in the presence or absence of 20 nM 3TSR (SEM, n ≥ 47 cells). (H) Quantification of VEGFR2-Tyr1175 phosphorylation in cerebellar tissue in Krit1ECKO and control Krit1fl/fl treated with 3TSR or vehicle 30 min after VEGF treatment (75 µg/Kg), as assessed by Western blot analysis (SEM, n = 4 mice in each group). Bar: (A and B) 50 µm; (F) 25 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001 versus vehicle-treated Krit1ECKO ; determined by one-way ANOVA followed by Tukey’s post hoc test (C and G) or Student’s t test (D, E, and H). Data in A–H are representative of at least three independent experiments.
TSP1 replacement with 3TSR antagonizes increased VEGFR2 phosphorylation that follows inactivation of Krit1
VEGF signaling is enhanced in KRIT1-depleted endothelial cells (DiStefano et al., 2014) and can contribute to the disruption of interendothelial junctions (Morin-Brureau et al., 2011; Argaw et al., 2012) and capillary dilatation that occur in CCM (Jung et al., 2003). Therefore, we assessed the effect of loss of brain endothelial KRIT1 on VEGFR2 phosphorylation as an indicator of VEGFR2 signaling. Immunocytochemistry revealed elevated levels of VEGFR2-Tyr1175 phosphorylation in Krit1ECKO BMECs (Fig. 4, F and G). Treatment of BMECs with 3TSR prevented the increased VEGFR2-Tyr1175 phosphorylation that followed Krit1 deletion (Fig. 4, F and G). Furthermore, silencing KRIT1 in human endothelial cells increased VEGFR2 phosphorylation; this was prevented by 3TSR (Fig. S3), and a VEGFR2 antagonist ameliorated the effects of KRIT1 deletion on tight junctions (Fig. S3). 3TSR also prevented increased VEGFR2-Tyr1175 phosphorylation in the brains of Krit1ECKO mice (Fig. 4 H). 3TSR can also promote TGF-β activation; however, we noted no effect of 3TSR on expression of TGF-β–regulated genes or in SMAD3 phosphorylation in Krit1ECKO BMECs (Fig. S4). Thus, 3TSR limits the increased VEGFR2 signaling that follows loss of endothelial KRIT1, an effect that can account for both stabilization of tight junctions and prevention of capillary dilation in CCM.
TSP1 replacement with 3TSR prevents CCM
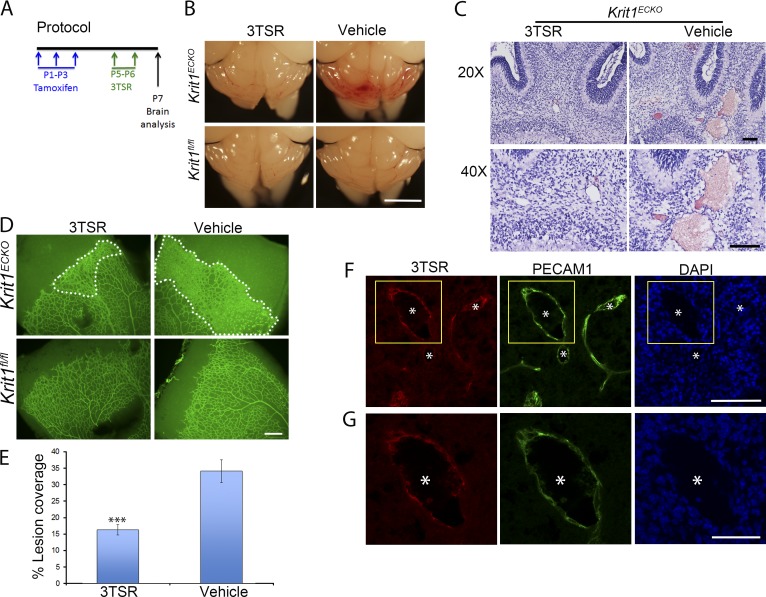
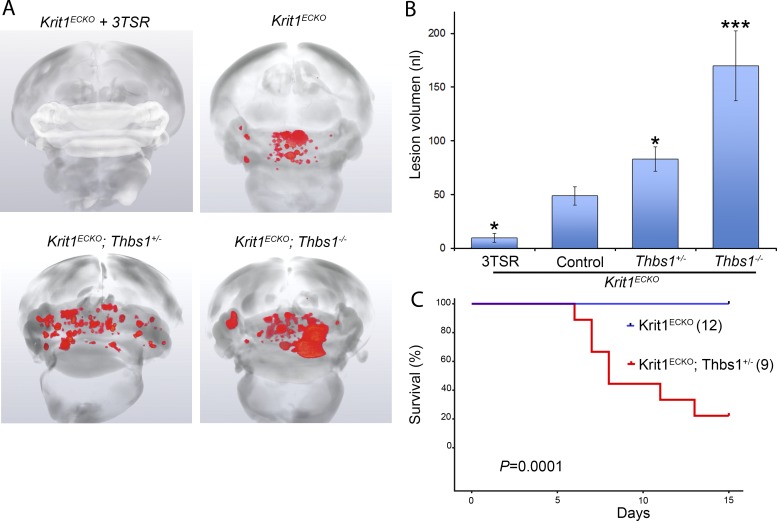
Visual inspection of the hindbrains of neonatal 3TSR-treated Krit1ECKO mice compared with vehicle-treated littermate Krit1ECKO controls (Fig. 5 A) revealed a notable reduction in the number and size of vascular lesions (Fig. 5 B). A similar marked reduction in histologically typical CCMs was observed in 3TSR-treated Krit1ECKO mice (Fig. 5 C), and a similar reduction was noted in retinal lesions (Fig. 5, D and E). Importantly, 3TSR administered into the retroorbital venous plexus was able to reach the CCM (Fig. 5, F and G), suggesting that it could act locally during CCM development. To quantify CCM formation, we imaged P7 hindbrains using contrast-enhanced, high-resolution x-ray micro-computed tomography (micro-CT), and measured lesion volumes using semiautomated software. 3TSR-treated Krit1ECKO mouse hindbrains exhibited near-complete prevention of CCM compared with vehicle-treated Krit1ECKO littermates, as assessed by hindbrain micro-CT imaging (Fig. 6 A). Blinded measurement of total CCM lesion volume (Fig. 6 B) confirmed the dramatic reduction in CCM as a consequence of 3TSR administration. Thus, replacement of TSP1 with 3TSR inhibited CCM formation.
Figure 5.
TSP1 derivative, 3TSR, prevents CCMs and retinal vascular lesions in Krit1ECKO mice. (A) Experimental protocol: vehicle or 3TSR (1.6 mg/Kg) was administered by retroorbital plexus injection at P5 and P6, and brains and retinas were analyzed at P7. (B) Prominent lesions are present in the cerebellum of Krit1ECKO mice, whereas administration of 3TSR suppressed lesion formation. (C) Hematoxylin and eosin staining of cerebellar sections from Krit1ECKO mice after treatment with 3TSR or vehicle (n = 4 mice in each group). (D) Representative image of whole-mount P7 retinal vasculature at the angiogenic growth front. The marked area in Krit1ECKO whole-mount retina shows decreased areas of condensed vascular plexus in Krit1ECKO treated with 3TSR compared with vehicle-treated Krit1ECKO littermates (SEM, n = 8 mice in each group). (E) Quantification of lesion coverage in Krit1ECKO mice treated with 3TSR or Vehicle (SEM, n = 8 mice in each group). (F) Administered 3TSR is present in CCM. 3TSR was injected retroorbitally into a Krit1ECKO;Thbs1−/− mouse. After 30 min, the mouse was killed, and its cerebellar cortex was stained for 3TSR (red, using anti-TSP1 antibodies) and endothelial marker PECAM1 (green); DAPI staining (blue) was used to reveal nuclei. 3TSR is observed in CCM. (G) Higher-magnification images of boxed areas in F. (F and G) Asterisks, vascular lumen of CCM. Bars: (B) 1 mm; (C and F) 100 µm; (D) 200 µm; (G) 50 µm. ***, P < 0.001 versus vehicle-treated Krit1ECKO mice; determined by Student’s t test.
Figure 6.
TSP1 limits CCM formation in Krit1ECKO mice. (A) Prominent lesions are present in the cerebellum of Krit1ECKO mice, whereas administration of 3TSR suppressed lesion formation. Increased CCM lesions were observed in Krit1ECKO;Thbs1+/− and Krit1ECKO;Thbs1−/− mice. (B) Quantification of lesion volumes by micro-CT analysis from mice in experiment depicted in A. Controls were either vehicle-treated mice or untreated mice that had similar lesion volumes. All groups were compared with control Krit1ECKO mice (SEM, n > 16 mice in each group, except Krit1ECKO;Thbs1−/− mice (n = 3). (C) Survival of Krit1ECKO and Krit1ECKO;Thbs1+/− mice. The number of mice in each group is in parentheses. Statistical significance was analyzed by log-rank test for comparing survival rates. *, P < 0.05, *** P < 0.001; determined by one-way ANOVA followed by Tukey’s post hoc test.
To assess the role of endogenous TSP1 in limiting CCM pathogenesis, we examined the impact of genetic inactivation of Thbs1 in Krit1ECKO mice. Micro-CT analysis of Krit1ECKO;Thbs1+/− mice revealed an ∼80% increase in the volume of CCM lesions relative to Krit1ECKO;Thbs1+/+ littermates, whereas the surviving Krit1ECKO;Thbs1−/− mice exhibited an ∼200% increase in CCM lesion volume (Fig. 6, A and B). Furthermore, there was a statistically significant reduction in survival of Krit1ECKO;Thbs1+/− versus Krit1ECKO;Thbs1+/+ mice (Fig. 6 C). These data show that TSP1 limits the formation of CCMs and that replacement of the loss of TSP1 with an anti-angiogenic fragment can prevent CCMs. Thus, the reduced expression of TSP1 that follows Krit1 inactivation contributes to CCM lesion pathogenesis.
KLF2 and KLF4 regulate expression of TSP1
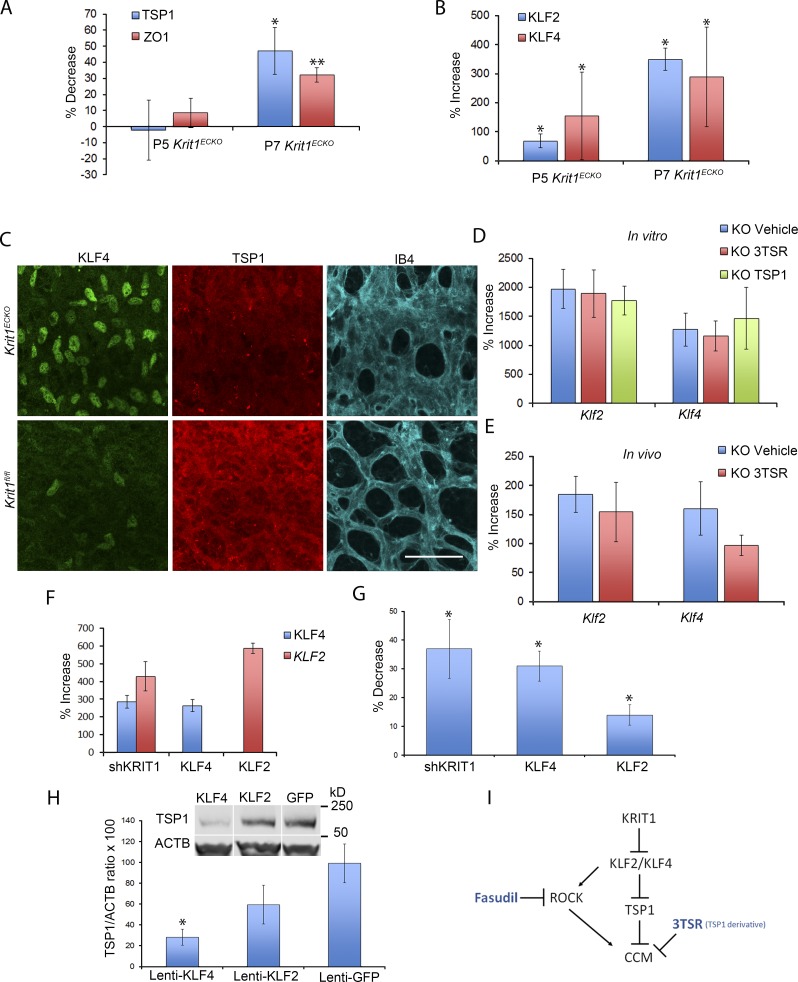
Recent studies established the importance of elevated expression of KLF2 and KLF4 transcription factors in the cardiovascular effects of loss of KRIT1 expression (Maddaluno et al., 2013; Renz et al., 2015; Zhou et al., 2015, 2016; Cuttano et al., 2016). Furthermore, elevation of KLF2 and KLF4 expression precedes an increase in Wnt-β-catenin signaling or endothelial-to-mesenchymal transition (Zhou et al., 2016). We noted that expression of both KLF2 and KLF4 was increased in freshly isolated brain microvasculature of Krit1ECKO mice at a time that roughly coincided with the decrease in TSP1 and ZO-1 mRNA levels (Fig. 7, A and B). Moreover, in retinas, there was marked up-regulation of nuclear KLF4 at areas of condensed peripheral vascular plexus (Maddaluno et al., 2013) that showed pronounced reduction in TSP1 immunostaining at P7 (Fig. 7 C). Because KLF2 and KLF4 are central transcriptional drivers of flow-mediated athero- and thrombo-protective vascular responses (Lin et al., 2005; Zhou et al., 2012), and loss of these transcription factors in endothelial cells is lethal in adult mice (Sangwung et al., 2017), we examined the effect of TSP1 and 3TSR on expression of these transcription factors. Neither TSP1 nor 3TSR prevented the increase in KLF2 or KLF4 mRNA after Krit1 inactivation in vitro (Fig. 7 D), and 3TSR did not do so in vivo (Fig. 7 E). Thus, exogenous addition of TSP1 or its anti-angiogenic domain, 3TSR, can block vascular effects that follow loss of KRIT1 in spite of maintained elevation of KLF2 and KLF4 expression.
Figure 7.
TSP1 replacement does not suppress the rise in KLF2 and KLF4 after loss of KRIT1. (A and B) Analysis of TSP1, ZO-1, KLF2, and KLF4 mRNA levels by RT-qPCR in freshly isolated microvasculature from mice at P5 and P7 as indicated. Krit1fl/fl littermate controls, at each developmental stage, were used to calculate percentage increase or decrease in Krit1ECKO mice using the following formulas: % increase = 100 × (X − F)/F and % decrease = 100 × ABS[(F − X)/F], where X and F = mRNA abundance in Krit1ECKO and Krit1fl/fl BMECs, respectively (SEM, n = 4 or 6). (C) Representative confocal images of retinal vasculature stained for KLF4 (green), TSP1 (red), or isolectin B4 (turquoise). TSP1 is decreased and KLF4 is increased at areas of condensed vasculature (n = 5 or 6 mice in each group). Bar, 25 µm. (D and E) Analysis of levels of KLF2 and KLF4 mRNA by RT-qPCR from Krit1ECKO BMEC (D) or cerebellar tissue from Krit1ECKO mice (E) treated with 3TSR, TSP1, or vehicle compared with Krit1fl/fl BMEC or Krit1fl/fl controls. Data are expressed as percentage increase or decrease in Krit1ECKO using the same formulas as in A and B (SEM, n = 3 or 4 in each group). (F) HUVECs were transduced with lentivirus encoding shKrit1, KLF2, or KLF4, and the increase in KLF2 or KLF4 mRNA relative to cells transduced with lentivirus encoding GFP was measured by RT-qPCR (SEM, n = 4). (G) HUVECs were transduced with lentivirus encoding ShKrit1, KLF2, or KLF4 as described in F, and the decrease of TSP1 mRNA levels was measured relative to cells transduced with EGFP control lentivirus (SEM, n = 4 or 5). (H) Analysis of TSP1 protein levels in HUVECs transduced with lentivirus encoding KLF2 or KLF4 as assessed by Western blot analysis; lentivirus encoding GFP was used as a control (SEM, n = 4). White lines indicate intervening lanes have been spliced out. (I) Loss of endothelial KRIT1 increases expression of KLF2 and KLF4 transcription factors, contributing to CCM formation by downstream effects including suppressed TSP1 expression. 3TSR (TSP1 derivative) reduces CCM lesion formation by replacing functions of TSP1 such as blocking VEGF signaling. Loss of KRIT1 also leads to ROCK activation in a KLF2-dependent manner, and blocking ROCK can also ameliorate CCMs. Thus, blockade of these and other downstream targets of KLF2 and KLF4 may offer a general strategy to reduce CCM formation in humans. *, P < 0.05; ** P < 0.01; determined by Student’s t test.
To test whether increased KLF2 or KLF4 was sufficient for suppression of TSP1 expression, we used lentivirus-mediated transduction to ectopically express KLF2 and KLF4 in human endothelial cells at levels similar to those that followed KRIT1 silencing (Fig. 7 F). Overexpression of KLF2 or KLF4 resulted in ∼15% or ∼30% decreases in TSP1 mRNA, respectively (Fig. 7 G). Moreover, ectopic expression of KLF4 induced an ∼3.5-fold decrease in TSP1 protein levels in human endothelial cells (Fig. 7 H), whereas the KLF2-induced ∼1.5-fold decrease in TSP1 protein abundance was not statistically significant (Fig. 7 H). Collectively, these data suggest that the suppression of TSP1 expression is an important downstream effect of the elevation in KLF2 and KLF4 that follows loss of KRIT1 (Fig. 7 I). Loss of the angiogenic checkpoint protein, TSP1, then leads to enhanced VEGFR2 signaling that contributes to the pathogenesis of CCM.
Discussion
Here, we show that acute Krit1 inactivation in BMECs causes rapid changes in expression of genes involved in cardiovascular development. Most notable is the dramatic suppression of TSP1, a potent endogenous angiogenesis inhibitor; this suppression is also seen in human CCM and follows the increase in expression of transcription factors KLF2 and KLF4. Replenishing TSP1 with either full-length TSP1 or 3TSR, an anti-angiogenic TSP1 fragment, prevents the disruption of BMEC tight junctions, an early phenotypic consequence of loss of KRIT1. Rescue of tight junctions is ascribable to the capacity of 3TSR to prevent increased VEGFR2 phosphorylation in Krit1ECKO BMECs and mice. Administration of 3TSR prevented the development of CCMs in Krit1ECKO mice as judged histologically and by quantitative micro-CT. Conversely, reduced Thbs1 gene dosage in Krit1ECKO mice increased the CCM lesion burden, demonstrating that endogenous TSP1 limits the pathogenesis of CCM. These studies reveal a critical mechanism in the pathogenesis of CCM and point to the possibility of repurposing 3TSR, or other angiogenesis inhibitors, for therapy of CCM.
Inactivation of brain microvascular endothelial Krit1 induced a rapid change in expression of genes that regulate cardiovascular development. We used primary BMECs and a conditional Cre recombinase to precisely control the time of deletion and analyzed gene expression at a time point when KRIT1 mRNA had just fallen to <90% of initial levels. Our data provide the first genome-wide view of the acute effects of loss of KRIT1 in the target cell for CCM formation. The dramatic changes in genes tied to the cell cycle and extracellular matrix provide a molecular signature that explains the observed increased proliferation and extracellular matrix seen in lesions from CCM patients (Shenkar et al., 2007). We noted dramatic up-regulation of KLF2 and KLF4, transcription factors recently implicated in development of CCM lesions (Maddaluno et al., 2013; Renz et al., 2015, 2016; Cuttano et al., 2016). Furthermore, confirming recent findings (Zhou et al., 2016), we found few early changes in genes involved in endothelial-to-mesenchymal transition, a result explained by a recent report that these markers are elevated ∼15 d after Krit1 inactivation, downstream of the elevation of KLF4 (Cuttano et al., 2016). We also found several KRIT1-regulated genes encoding secreted proteins and receptors (e.g., TSP1, CXCR4, BMP2, TGFb2, LRG1, and Dll1) that could modulate previously reported angiogenic remodeling (Whitehead et al., 2004, 2009; Boulday et al., 2011; Maddaluno et al., 2013) and inflammation (Shenkar et al., 2007; e.g., TSP1, LBP, ADAM8, NOD2). Thus, this analysis provides new molecular clues about the pathogenesis of CCM.
A striking finding was an ∼75% reduction in TSP1 mRNA and an ∼70% reduction in TSP1 protein. TSP1 is among the most potent and best-characterized endogenous inhibitors of angiogenesis. Up-regulation of expression of TSP1 during angiogenesis limits vascular density, thus serving as an angiogenic checkpoint to prevent neovascularization (Jiménez et al., 2000; Rodriguez-Manzaneque et al., 2001). Strikingly, in TSP1-null mice, angiogenic vessels are dramatically dilated within tumors (Rodriguez-Manzaneque et al., 2001; Velasco et al., 2009), thereby resembling early CCMs. Although loss of KRIT1 can destabilize adherens junctions (Glading and Ginsberg, 2010; Maddaluno et al., 2013), we find that loss of tight junctions occurs before loss of adherens junctions, thus mirroring the alterations in tight junctions in human CCM lesions (Schneider et al., 2011; Stamatovic et al., 2015). Replacement of TSP1 could prevent the loss of brain endothelial tight junctions that follows inactivation of Krit1 in vitro, indicating that disabling the TSP1 angiogenic checkpoint has a pathogenic role in the increased vascular permeability that characterizes CCMs (Tanriover et al., 2013; Mikati et al., 2015). Indeed, loss of TSP1 from brain endothelium alters VEGF signaling (Rodriguez-Manzaneque et al., 2001; Isenberg et al., 2009; Lawler and Lawler, 2012), which can contribute to disassembly of brain endothelial tight junctions (Morin-Brureau et al., 2011; Argaw et al., 2012) and cerebrovascular dysfunction (Whitehead et al., 2009; Argaw et al., 2012). Our finding that loss of brain endothelial KRIT1 led to increased levels of VEGFR2 phosphorylation and that VEGF inhibition preserves morphological tight junctions in Krit1ECKO mice are consistent with the findings that loss of KRIT1 increases VEGF signaling, and that VEGFR2 inhibitors prevent the resulting increase in endothelial paracellular permeability (DiStefano et al., 2014). Thus, 3TSR prevention of increased VEGFR2 signaling in Krit1ECKO endothelial cells provides a cogent explanation for the capacity of 3TSR to reduce vascular dysmorphology in Krit1ECKO mice.
We previously showed that loss of KRIT1 leads to activation of Rho kinase (ROCK), thereby increasing vascular leak (Stockton et al., 2010) and that blocking ROCK with inhibitors that are well tolerated in humans could ameliorate CCM (McDonald et al., 2012). Recent data suggest that ROCK activation during CCM formation is downstream of KLF2 elevation (Zhou et al., 2016). As shown here, KLF2 and KLF4 suppress TSP1 expression, and 3TSR (Lawler and Lawler, 2012), which was relatively nontoxic in preclinical studies (Russell et al., 2015), prevents CCM without suppressing KLF2 and KLF4. Moreover, ABT-510, a TSP-derived anti-angiogenic peptide, was well tolerated in phase I studies in humans (Hoekstra et al., 2005; Gietema et al., 2006). Importantly, 3TSR and ABT-510 lack the TSP1 epidermal growth factor (EGF) repeats that can disrupt cell–cell junctions (Garg et al., 2011). Loss of endothelial KLF2 and KLF4 is lethal (Sangwung et al., 2017), as is inactivation of MEKK3 (Parmar et al., 2006; Hamik et al., 2007; Ohnesorge et al., 2010), which is upstream of the elevation of KLF2 (Zhou et al., 2016). Thus, we propose that identification of the downstream targets of KLF2 and KLF4 that mediate CCM formation, such as ROCK activation and TSP1 suppression, can serve as a general strategy for discovery of agents that might act alone or synergistically to prevent these common vascular malformations (Fig. 7 I).
Materials and methods
Genetically modified mice
Endothelial-specific conditional Krit1-null mice were generated by breeding transgenic mice expressing endothelial-specific, Pdgfb promoter-driven, tamoxifen-regulated Cre recombinase, iCreERT2 (Claxton et al., 2008), in combination with loxP-flanked Krit1 exon 5 (Krit1fl/fl, a gift of Douglas A. Marchuk, Duke University, Durham, NC; Pdgfb-iCreERT2; Krit1fl/fl mice). All experiments were performed using aged matched Krit1fl/fl littermates on the same C57BL/6 background. TSP1-null (Thbs1tm1Hyn/J, Jackson Laboratory) mice were crossed with Pdgfb-iCreERT2; Krit1fl/fl mice to generate Pdgfb-iCreERT2; Krit1fl/fl; TSP1+/− mice. Mice were administered 50 µg tamoxifen (T5648; Sigma-Aldrich) by intragastric injection on P1, P2, and P3, inducing Cre activity and endothelial Krit1 gene inactivation in the littermates bearing the iCreERT2 (Krit1ECKO). These mice and control Krit1fl/fl mice were killed by decapitation for phenotypic analysis on the indicated postnatal days.
For 3TSR treatment, mice were randomized (by flipping a coin) to TSR (1.6 mg/kg) or vehicle treatment. 10 μl 3TSR was administered by retroorbital injection on P5 and P6 using a 28-gauge, 0.36 × 13-mm needle mounted on an insulin syringe (Ultra-Fine II; BD). 3TSR was prepared as described previously (Miao et al., 2001). All animal experiments were performed in compliance with animal procedure protocols approved by the University of California, San Diego Institutional Animal Care and Use Committee.
Isolation and purification of primary BMECs
Adult 2–4-mo-old mice were killed, and their brains were removed and dropped into ice-cold buffer A (10 mM Hepes, 1× penicillin-streptomycin, and 0.5% BSA in DMEM). Meninges and choroid plexus were removed, and brain tissue was minced with scissors. Tissue suspensions were centrifuged at 700 g for 5 min at 4°C, and the brain tissue pellet was digested with collagenase and dispase solution (DMEM containing 1 mg/ml collagenase/dispase [10269638001; Sigma-Aldrich], 20 units/ml DNase I [11284932001; Sigma-Aldrich], and 0.150 tosyl-lysine-chloromethyl-ketone [T7254; Sigma-Aldrich]) at 37°C for 1 h. After incubation, the brain tissue was triturated with Pasteur pipettes using different sized tips until a homogeneous suspension was obtained. This suspension was centrifuged (700 g for 5 min at 4°C), and the pellet was resuspended in ice-cold buffer B (10 mM Hepes, 1× penicillin streptomycin, and 25% BSA in DMEM). The suspension was centrifuged at 1000 g for 20 min at 4°C. The pellet containing microvascular fragments (heavier phase) underwent a second collagenase and dispase digestion for 30 min and was clarified by passage through a 70-µm mesh filter. BMECs were seeded onto collagen-coated plates and cultured in EBM-2 medium supplemented with complements (hereafter termed EGM-2-BMEC medium) obtained from the manufacturer (Lonza) at the following concentrations: 0.025% (vol/vol) recombinant human EGF, 0.1% (vol/vol) insulin-like growth factor, 0.1% (vol/vol) gentamicin, 0.04% (vol/vol) ascorbic acid, 0.04% (vol/vol) hydrocortisone, and 20% (vol/vol) FBS. BMECs were cultured for 2 d in 10 µg/ml puromycin and maintained in 2 µg/ml puromycin for 6 d. Purified primary BMECs were routinely characterized for morphology and formation of adherens and tight junctions by immunofluorescence, for the presence of mRNA from endothelial cell–specific genes, and for the absence of leukocyte, glia, and smooth muscle cell marker mRNAs.
Isolation of brain microvasculature
P5–P10 mice were killed. Brain and cerebellum were removed into ice-cold buffer A. Four to five brains from tamoxifen-injected Krit1ECKO were pooled, and comparisons were performed with similar pools from littermate tamoxifen-injected Krit1fl/f l mice. Brain tissue was minced with scissors and centrifuged at 700 g for 5 min at 4°C. The pellet was resuspended and incubated in ACK lysing buffer (10-548E; Lonza) for 5 min at room temperature to eliminate erythrocytes. The resulting suspension was sedimented by centrifugation (700 g, 5 min), and the pellet was digested with collagenase and dispase solution for 1 h at 37°C. The suspension was triturated and centrifuged at 700 g for 5 min at 4°C, and microvasculature was passed through a 70-µm mesh filter. Depletion of blood cell contaminants was performed using Dynabeads Untouched Mouse T cell kit (11413D; Thermo Fisher Scientific) according to the manufacturer’s protocol. For brains at P7 to P10, the tissue suspension was centrifuged at 700 g for 10 min at 4°C, and the pellet was resuspended in ice-cold buffer B and centrifuged at 1,000 g for 20 min at 4°C. Density-dependent centrifugation in BSA separates capillary fragments (heavier density) from myelin, neurons, astrocyte, and other brain-resident contaminants (lighter density). The capillary fragment phase was subjected to Percoll gradient as previously reported (Lopez-Ramirez et al., 2014). For brains at P5, the tissue suspension was centrifuged at 700 g for 10 min at 4°C, and the pellet was resuspended in 100 µl isolation buffer (PBS, pH 7.2, 0.5% BSA, and 2 mM EDTA) containing rat polyclonal antibody anti-mouse CD31 microbeads (130-097-418; Miltenyi Biotec). After incubation, the cell suspension was washed using 7 ml isolation buffer and centrifuged at 1,000 g for 10 min. The supernatant was removed, and the cell pellet was resuspended in 500 µl isolation buffer. CD31+ cells were sorted by applying cell suspension onto an LS column placed into a magnetic field according to the manufacturer’s protocol (130-042-401; Miltenyi Biotec). Endothelial cell identity was confirmed by RT-qPCR of mRNA from endothelial cell–specific genes and minor levels of leukocyte, glia, and smooth muscle cell marker mRNAs.
Genetic inactivation of KRIT1 in BMECs
BMECs at passage 1–3 were maintained at 37°C in 95% air and 5% CO2, grown to 85% confluence, and treated for 48 h with 5 µM 4-hydroxy-tamoxifen (H7904; Sigma-Aldrich), after which the medium was replaced with medium lacking 4-hydroxy-tamoxifen, and cells were harvested after 72 h in culture. For TSP1 and 3TSR addition experiments, after 24 h in EGM-2-BMEC medium without serum containing 0.5% BSA, 5 nM mrTSP1 (7859-TH; R&D Systems), 20 nM 3TSR, or vehicle was added. After 48 h, a second dose of TSP1, 3TSR, or vehicle was added, and cells were harvested after an additional 24 h.
Immunofluorescence microscopy
BMECs were grown to confluence on collagen-coated cover glass (12-545-81; Thermo Fisher Scientific), and cells were fixed for 10 min at room temperature with 4% PFA in PBS, pH 7.4, and permeabilized with 0.5% Triton X-100 in PBS for 5 min. The slides were blocked with 0.5% BSA for 30 min and incubated with rabbit polyclonal antibody anti–ZO-1 (1:80, 61-7300; Thermo Fisher Scientific), mouse monoclonal antibody anti-CLDN5 (1:50, 35-2500; Thermo Fisher Scientific), and rat polyclonal antibody anti–VE-cadherin (1:100, 550548; BD Pharmingen) overnight at room temperature. For VEGFR2 phosphorylation, BMECs were grown to subconfluence on collagen-coated cover glass and fixed in methanol for 30 min at 4°C, followed by cold acetone (maintained at −20°C before use) for 1 min at room temperature, and incubated with anti-pVEGFR2Tyr1175 antibody (1:100; Cell Signaling Technology). Cells were washed four times in PBS and incubated for 1 h at room temperature with a suitable Alexa Fluor–coupled secondary antibody (1:300; Thermo Fisher Scientific) in PBS. Cell nuclei were stained with DAPI and mounted with Fluoromount-G mounting medium (SouthernBiotech).
RNA extraction and qRT-PCR
Primary BMECs, HUVECs, and freshly isolated brain microvasculature total RNA were isolated using TRIzol reagent, according to the manufacturer’s protocol (Thermo Fisher Scientific). For gene expression analysis, single-stranded cDNA was produced from 10 ng total RNA of BMECs using SuperScript III First-Strand synthesis and random primers according to the manufacturer’s protocol (Thermo Fisher Scientific). Kapa SybrFast qPCR kit (Kapa Biosystems) and thermal cycler (CFX96 Real-Time System; Bio-Rad) were used to determine the relative levels of the genes analyzed (primer sequences are shown in Table S1) according to the manufacturer’s protocol. Actin mRNA levels were used as internal control, and the 2-ΔΔCT method was used for analysis of the data. Each control value (Krit1fl/fl) was normalized to 1, and Krit1ECKO values were relative to control.
Genome-wide RNA sequencing
The quantity (ND-1000 spectrophotometer; NanoDrop Technologies) and quality (TapeStation; Agilent) of total RNA were analyzed. RNA libraries were generated using Illumina’s TruSeq Stranded mRNA Sample Prep kit using 400 ng RNA. RNA libraries were multiplexed and sequenced with 100-bp paired single-end reads (SR100) to a depth of ∼30 million reads per sample on an Illumina HiSeq2500. Fastq files from RNA-seq experiments were mapped to individual genomes for the mouse strain of origin using STAR with default parameters (mm10 for C57BL/6J; Dobin et al., 2013). Reproducibility between samples was analyzed using the irreproducibility discovery rate tool (Li et al., 2011). Homer (Heinz et al., 2010) was used for further analysis. To measure gene expression, analyze repeats (with the option RNA and condense genes) was used along with the default parameters. Subset-specific expression was defined with a fourfold difference in expression between two experiments. Genes with fewer than 16 tag counts were defined as not expressed. To map subset-specific peaks to gene expression, only expressed genes were considered. Differential expression was defined by a fold change of at least 1.5-fold averaging over replicates (P < 0.05) and used for gene ontology annotation analysis with David Bioinformatics (Huang et al., 2009; Eichenfield et al., 2016). Sequencing data have been deposited in Gene Expression Omnibus (GEO) under accession no. GSE85657.
Whole-mount retinal staining
Eyes were harvested and fixed in PFA for 20 min at room temperature. Eyes were washed four times with PBS, and retinal whole-mount preparations were permeabilized, blocked using blocking buffer (PBS, 1% BSA, and 0.5% Triton X-100), and incubated at 4°C overnight for tight junction and 2 d for KLF4 staining. For tight junctions and adherens junction staining, whole-mount preparations were incubated with rabbit polyclonal antibody anti–ZO-1 (1:50), mouse monoclonal antibody anti-CLDN5 (1:80), and rat polyclonal antibody anti–VE-cadherin (1:100) in PBS overnight at room temperature. For KLF4 and TSP1 staining, whole-mount preparations were incubated with goat polyclonal antibody anti-KLF4 (1:100, AF3158; R&D Systems) and rabbit polyclonal antibody anti-TSP1 (1:500; Zhang and Lawler, 2007; Lawler and Lawler, 2012) in PBS at room temperature for 1 d followed by 4°C for 2 d. Preparations were washed three times in PBS and three times in Pblec buffer (PBS, 1 mM CaCl2, 1 mM MgCl2, 0.1 mM MnCl2, and 1% Triton X-100) and incubated with isolectin B4 FITC (1:80, L2895; Sigma-Aldrich) or Alexa Fluor 647 (1:80, I32450; Thermo Fisher Scientific) conjugated, as indicated, in Pblec buffer at 4°C overnight. Preparations were incubated at room temperature for 2 h with suitable secondary anti-rabbit Alexa Fluor 594, anti-goat Alexa Fluor 488, and anti-mouse Alexa Fluor 647 antibodies (1:250; Thermo Fisher Scientific) in PBS. Retinal whole-mount preparations were washed five times in PBS and flat-mounted using Fluoromount-G (SouthernBiotech).
Immunohistochemistry
P7 Krit1ECKO and littermate control Krit1fl/fl mice were perfused with HBSS containing 0.5% BSA to rinse out the blood, and brains were isolated and fixed in 4% PFA at 4°C overnight. After cryoprotection in sucrose and freezing, 12-µm sections of cerebellar tissue were cut onto Superfrost Plus slides (12-550-15; VWR International). The preparation was blocked and permeabilized using permeabilization buffer (PBS, 5% goat serum, 0.5% Triton X-100, and 0.5% BSA) for 2 h and incubated with rabbit polyclonal antibody anti-TSP1 (1:1000; Lawler et al., 1998), rat polyclonal antibody anti-PECAM1 (1:100, 553370; BD PharMingen), rabbit polyclonal antibody anti–ZO-1 (1:120), mouse monoclonal antibody anti-CLDN5 (1:100), or rat polyclonal antibody anti–VE-cadherin (1:100) in PBS. All antibodies were incubated at room temperature overnight in a humidified box. Human tissue was obtained after informed consent from patients undergoing lesion resection, under protocol #10-295-A, approved by the University of Chicago Institutional Review Board. Human tissue, CCM1 lesions, and lesion-free brain tissues were snap-frozen and sectioning using a cryostat (Leica). Specimens were fixed in 4% PFA at room temperature for 15 min and washed three times in PBS. The specimens were blocked and permeabilized using permeabilization buffer for 3 h and incubated with rabbit polyclonal antibody anti-TSP1 (1:1,000) and goat polyclonal antibody anti–collagen IV (1:100, AB769; Millipore) in PBS at room temperature overnight. Preparations were washed four times in PBS and incubated at room temperature for 1 h with suitable secondary anti-rabbit Alexa Fluor 594, anti-goat Alexa Fluor 488, anti-rat Alexa Fluor 488, and anti-mouse Alexa Fluor 647 antibodies (1:300; Thermo Fisher Scientific) in PBS. Cell nuclei were stained with DAPI (SouthernBiotech).
Image acquisition and quantitative analysis
The slides were viewed with a high-resolution SP5 confocal microscope (Leica Microsystems), and the images were captured with Leica application suite software (Leica Microsystems). For BMEC confocal microscopy, five images in z-stacks up to 4.5 µm in depth were acquired with a 60× oil-immersion objective and projected onto one image. For cerebellar tissue and retina confocal microscopy, six images in z-stacks up to 8 µm (brain) or 6 µm (retina) in depth were acquired with a 20× objective or six images to 6 µm in depth were acquired with a 60× oil-immersion objective and projected onto one image. The quantification analysis was performed using Volocity software on high-resolution confocal images.
Western blot analysis
BMECs or HUVECs as indicated were grown to confluence on collagen-coated six-well plates, whereas cerebellar frozen tissue was immersed in liquid nitrogen and pulverized with a tissue crusher. Cells and tissue were lysed using lysis solution containing 100 µl RIPA buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, and 0.1% SDS) and a mixture of inhibitors (Roche) and 1 mM sodium orthovanadate. Protein concentration was determined using a Micro BCA protein assay kit (Pierce). Cell lysates were diluted in Laemmli buffer solution at 95°C for 5 min. 25 µg total protein was resolved on 4–20% polyacrylamide gels (Thermo Fisher Scientific) in SDS-PAGE buffer and transferred onto nitrocellulose membranes (Amersham) using a wet transfer system (Bio-Rad). Membranes were blocked with blocking WB solution (PBS, 10% nonfat milk, and 0.05% Tween-20) for 1 h and incubated in the presence of rabbit polyclonal antibody anti-TSP1 (1:500, ab85762; Abcam), anti–ZO-1 (1:150), rat polyclonal antibody anti–VE-cadherin (1:75), rabbit polyclonal antibody anti–claudin-5 (1:170, 34-1600; Thermo Fisher Scientific), rabbit polyclonal anti-VEGFR2 (1:200; Cell Signaling Technology; 1:200; Santa Cruz Biotechnology), and rabbit monoclonal anti-VEGFR2pTyr1175 (1:120; Cell Signaling Technology) at 4°C overnight. After several washes, membranes were incubated with the appropriate IRDye/Alexa Fluor–coupled secondary antibody (1:10,000; Li-COR) and imaged using an infrared imaging system (Odyssey; Li-COR). Blots were processed using Image Studio Lite software (Li-COR). As a control for protein loading, mouse antibodies against actin (1:5,000, A1978; Sigma-Aldrich) were used.
Micro-CT scan image
P7 Krit1ECKO, Krit1ECKO;TSP1+/−, Krit1ECKO;TSP1−/−, and littermate control Krit1fl/fl, Krit1fl/fl;TSP1+/−, Krit1fl/fl;TSP1−/− mice were killed, and brains were removed and dropped into 10% neutral buffered formalin (Sigma-Aldrich). Brains were soaked in 50 ml of 1.25% Lugol iodine (Thermo Fisher Scientific) for 96 h. Imaging data acquisition was performed using the Phoenix v|tome|x s 180/240 micro-CT scanner system (General Electric), and hyperdense CCM lesions were computationally segmented with the AMIRA 5.5 software platform (FEI; Girard et al., 2016).
Expression constructs and shRNAs
To generate the KLF2 and KLF4 constructs, a plasmid template encoding KLF2 (50786; Addgene) and KLF4 (19764; Addgene) were amplified by PCR to place them downstream of mouse PGK promoter and fuse to IRES-puromycin resistance gene into pLVX vector (Clontech). KLF2- and KLF4-expressing lentiviral particles were prepared by cotransfection of pLVX;PGK-KLF2 or pLVX;PGK-KLF4 with pMDLg/pRRE, pRSV-Rev, and pMD2.G in HEK293T cells. Oligos for ShKRIT1 (clone TRCN0000072879, target sequence 5′-CGGGTAGATAAAGTGGTAATA-3′) is based on the public TRC (RNAi Consortium; Broad Institute) library and cloned into pLKO.1 using EcoRI and Age1 restriction sites. For KLF2, KLF4, or shRNA delivery, HUVECs were grown to 80% confluence on gelatin-coated six-well plates and transduced with lentiviral particles. 72 h after infection, HUVECs were prepared for RNA or protein analysis (described above).
Statistical analysis
Data are expressed as means ± SEM. For all experiments, the number of independent experiments (n) is indicated. Analyses of brain and retina experiments were performed blinded. The sample sizes were estimated with two-sample t test (two-tailed). The pooled standard deviation was used to account for unequal variances in the two groups (vehicle or 3TSR). Two-tailed unpaired Student’s t test was used to determine statistical significance. For multiple comparisons, one-way ANOVA followed by Tukey’s post hoc test was used. For survival rate analysis, long-rank test was performed using SAS Software.
Online supplemental material
Fig. S1 shows RNA-seq analysis of BMEC transcriptome after acute genetic inactivation of Krit1. Fig. S2 shows acute loss of KRIT1 decreases tight junctions in BMECs. Fig. S3 (related to Fig. 4) shows that 3TSR prevents VEGFR2-Tyr1175 phosphorylation in human endothelial cells. Fig. S4 shows that 3TSR does not activate TGF-β signaling in Krit1ECKO. Table S1, included as an Excel file, shows primers used for RT-qPCR.
Supplementary Material
Acknowledgments
We thank Mark Duquette for helping to prepare 3TSR and Nicholas Hobson, Rhoda Lightle, and Prof. Zhe-Xi Luo for assistance in the phenotypic assessment of mouse brains and micro-CT technique. We are grateful to Jennifer Santini for microscopy technical assistance and the UCSD School of Medicine Microscopy Core. We also thank Preston Hale and Wilma McLaughlin for technical assistance.
This work was supported by a CAO Pilot Grant from the Beth Israel Deaconess Medical Center (to J. Lawler), NIH grants HL078784 (to M.H. Ginsberg) and NS092521 (to M.H. Ginsberg and I.A. Awad), and the University of California, San Diego School of Medicine Microscopy Core (P30 NS047101).
The authors declare no competing financial interests.
Author contributions: M.A. Lopez-Ramirez designed and performed the experiments, performed data analysis and interpretation, and wrote the paper; M.A. Lopez-Ramirez, G. Fonseca, C.K. Glass, and M.H. Ginsberg performed analysis and interpretation of the RNA-seq data; H.A. Zeineddine, R. Girard, T. Moore, and I.A. Awad performed analysis and interpretation of micro-CT in mice; A. Pham performed histological analysis of experiments in conditional knockout mice; R. Shenkar and I.A. Awad provided human CCM samples; Y. Cao performed statistical analysis of micro-CT; M.A. Lopez-Ramirez, B.-J. de Kreuk, and F. Lagarrigue performed analysis and interpretation of experiments in HUVECs; J. Lawler provided reagents and help in interpretation of data and M.H. Ginsberg handled overall study design, analysis and interpretation of the data, generation of figures, and writing and final editing of the manuscript.
Footnotes
Abbreviations used:
- BMEC
- brain microvascular endothelial cell
- CCM
- cerebral cavernous malformation
- micro-CT
- micro-computed tomography
- RNA-seq
- RNA sequencing
- ROCK
- Rho kinase
- RT-qPCR
- real-time quantitative PCR
- VE-cadherin
- vascular endothelial cadherin
- VEGF
- vascular endothelial growth factor
References
- Argaw A.T., Asp L., Zhang J., Navrazhina K., Pham T., Mariani J.N., Mahase S., Dutta D.J., Seto J., Kramer E.G., et al. . 2012. Astrocyte-derived VEGF-A drives blood-brain barrier disruption in CNS inflammatory disease. J. Clin. Invest. 122:2454–2468. 10.1172/JCI60842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulday G., Rudini N., Maddaluno L., Blécon A., Arnould M., Gaudric A., Chapon F., Adams R.H., Dejana E., and Tournier-Lasserve E.. 2011. Developmental timing of CCM2 loss influences cerebral cavernous malformations in mice. J. Exp. Med. 208:1835–1847. 10.1084/jem.20110571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claxton S., Kostourou V., Jadeja S., Chambon P., Hodivala-Dilke K., and Fruttiger M.. 2008. Efficient, inducible Cre-recombinase activation in vascular endothelium. Genesis. 46:74–80. 10.1002/dvg.20367 [DOI] [PubMed] [Google Scholar]
- Cuttano R., Rudini N., Bravi L., Corada M., Giampietro C., Papa E., Morini M.F., Maddaluno L., Baeyens N., Adams R.H., et al. . 2016. KLF4 is a key determinant in the development and progression of cerebral cavernous malformations. EMBO Mol. Med. 8:6–24. 10.15252/emmm.201505433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo R., Marini V., Rinaldi C., Origone P., Dorcaratto A., Avolio M., Goitre L., Forni M., Capra V., Alafaci C., et al. . 2011. Mutation analysis of CCM1, CCM2 and CCM3 genes in a cohort of Italian patients with cerebral cavernous malformation. Brain Pathol. 21:215–224. 10.1111/j.1750-3639.2010.00441.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiStefano P.V., Kuebel J.M., Sarelius I.H., and Glading A.J.. 2014. KRIT1 protein depletion modifies endothelial cell behavior via increased vascular endothelial growth factor (VEGF) signaling. J. Biol. Chem. 289:33054–33065. 10.1074/jbc.M114.582304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenfield D.Z., Troutman T.D., Link V.M., Lam M.T., Cho H., Gosselin D., Spann N.J., Lesch H.P., Tao J., Muto J., et al. . 2016. Tissue damage drives co-localization of NF-κB, Smad3, and Nrf2 to direct Rev-erb sensitive wound repair in mouse macrophages. eLife. 5:e13024 10.7554/eLife.13024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg P., Yang S., Liu A., Pallero M.A., Buchsbaum D.J., Mosher D.F., Murphy-Ullrich J.E., and Goldblum S.E.. 2011. Thrombospondin-1 opens the paracellular pathway in pulmonary microvascular endothelia through EGFR/ErbB2 activation. Am. J. Physiol. Lung Cell. Mol. Physiol. 301:L79–L90. 10.1152/ajplung.00287.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietema J.A., Hoekstra R., de Vos F.Y., Uges D.R., van der Gaast A., Groen H.J., Loos W.J., Knight R.A., Carr R.A., Humerickhouse R.A., and Eskens F.A.. 2006. A phase I study assessing the safety and pharmacokinetics of the thrombospondin-1-mimetic angiogenesis inhibitor ABT-510 with gemcitabine and cisplatin in patients with solid tumors. Ann. Oncol. 17:1320–1327. 10.1093/annonc/mdl102 [DOI] [PubMed] [Google Scholar]
- Girard R., Zeineddine H.A., Orsbon C., Tan H., Moore T., Hobson N., Shenkar R., Lightle R., Shi C., Fam M.D., et al. . 2016. Micro-computed tomography in murine models of cerebral cavernous malformations as a paradigm for brain disease. J. Neurosci. Methods. 271:14–24. 10.1016/j.jneumeth.2016.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glading A.J., and Ginsberg M.H.. 2010. Rap1 and its effector KRIT1/CCM1 regulate beta-catenin signaling. Dis. Model. Mech. 3:73–83. 10.1242/dmm.003293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glading A., Han J., Stockton R.A., and Ginsberg M.H.. 2007. KRIT-1/CCM1 is a Rap1 effector that regulates endothelial cell–cell junctions. J. Cell Biol. 179:247–254. 10.1083/jcb.200705175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden M.J., Morrison L.A., Kim H., and Hart B.L.. 2015. Increased number of white matter lesions in patients with familial cerebral cavernous malformations. AJNR Am. J. Neuroradiol. 36:899–903. 10.3174/ajnr.A4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamik A., Lin Z., Kumar A., Balcells M., Sinha S., Katz J., Feinberg M.W., Gerzsten R.E., Edelman E.R., and Jain M.K.. 2007. Kruppel-like factor 4 regulates endothelial inflammation. J. Biol. Chem. 282:13769–13779. 10.1074/jbc.M700078200 [DOI] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra R., de Vos F.Y., Eskens F.A., Gietema J.A., van der Gaast A., Groen H.J., Knight R.A., Carr R.A., Humerickhouse R.A., Verweij J., and de Vries E.G.. 2005. Phase I safety, pharmacokinetic, and pharmacodynamic study of the thrombospondin-1-mimetic angiogenesis inhibitor ABT-510 in patients with advanced cancer. J. Clin. Oncol. 23:5188–5197. 10.1200/JCO.2005.05.013 [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., and Lempicki R.A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Isenberg J.S., Martin-Manso G., Maxhimer J.B., and Roberts D.D.. 2009. Regulation of nitric oxide signalling by thrombospondin 1: implications for anti-angiogenic therapies. Nat. Rev. Cancer. 9:182–194. 10.1038/nrc2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez B., Volpert O.V., Crawford S.E., Febbraio M., Silverstein R.L., and Bouck N.. 2000. Signals leading to apoptosis-dependent inhibition of neovascularization by thrombospondin-1. Nat. Med. 6:41–48. 10.1038/71517 [DOI] [PubMed] [Google Scholar]
- Jung K.H., Chu K., Jeong S.W., Park H.K., Bae H.J., and Yoon B.W.. 2003. Cerebral cavernous malformations with dynamic and progressive course: correlation study with vascular endothelial growth factor. Arch. Neurol. 60:1613–1618. 10.1001/archneur.60.11.1613 [DOI] [PubMed] [Google Scholar]
- Lampugnani M.G., Orsenigo F., Rudini N., Maddaluno L., Boulday G., Chapon F., and Dejana E.. 2010. CCM1 regulates vascular-lumen organization by inducing endothelial polarity. J. Cell Sci. 123:1073–1080. 10.1242/jcs.059329 [DOI] [PubMed] [Google Scholar]
- Lawler P.R., and Lawler J.. 2012. Molecular basis for the regulation of angiogenesis by thrombospondin-1 and -2. Cold Spring Harb. Perspect. Med. 2:a006627 10.1101/cshperspect.a006627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler J., Sunday M., Thibert V., Duquette M., George E.L., Rayburn H., and Hynes R.O.. 1998. Thrombospondin-1 is required for normal murine pulmonary homeostasis and its absence causes pneumonia. J. Clin. Invest. 101:982–992. 10.1172/JCI1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.H., Brown J.B., Huang H.Y., and Bickel P.J.. 2011. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 5:1752–1779. 10.1214/11-AOAS466 [DOI] [Google Scholar]
- Lin Z., Kumar A., SenBanerjee S., Staniszewski K., Parmar K., Vaughan D.E., Gimbrone M.A. Jr., Balasubramanian V., García-Cardeña G., and Jain M.K.. 2005. Kruppel-like factor 2 (KLF2) regulates endothelial thrombotic function. Circ. Res. 96:e48–e57. 10.1161/01.RES.0000159707.05637.a1 [DOI] [PubMed] [Google Scholar]
- Lopez-Ramirez M.A., Wu D., Pryce G., Simpson J.E., Reijerkerk A., King-Robson J., Kay O., de Vries H.E., Hirst M.C., Sharrack B., et al. . 2014. MicroRNA-155 negatively affects blood-brain barrier function during neuroinflammation. FASEB J. 28:2551–2565. 10.1096/fj.13-248880 [DOI] [PubMed] [Google Scholar]
- Maddaluno L., Rudini N., Cuttano R., Bravi L., Giampietro C., Corada M., Ferrarini L., Orsenigo F., Papa E., Boulday G., et al. . 2013. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature. 498:492–496. 10.1038/nature12207 [DOI] [PubMed] [Google Scholar]
- McDonald D.A., Shenkar R., Shi C., Stockton R.A., Akers A.L., Kucherlapati M.H., Kucherlapati R., Brainer J., Ginsberg M.H., Awad I.A., and Marchuk D.A.. 2011. A novel mouse model of cerebral cavernous malformations based on the two-hit mutation hypothesis recapitulates the human disease. Hum. Mol. Genet. 20:211–222. 10.1093/hmg/ddq433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D.A., Shi C., Shenkar R., Stockton R.A., Liu F., Ginsberg M.H., Marchuk D.A., and Awad I.A.. 2012. Fasudil decreases lesion burden in a murine model of cerebral cavernous malformation disease. Stroke. 43:571–574. 10.1161/STROKEAHA.111.625467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D.A., Shi C., Shenkar R., Gallione C.J., Akers A.L., Li S., De Castro N., Berg M.J., Corcoran D.L., Awad I.A., and Marchuk D.A.. 2014. Lesions from patients with sporadic cerebral cavernous malformations harbor somatic mutations in the CCM genes: Evidence for a common biochemical pathway for CCM pathogenesis. Hum. Mol. Genet. 23:4357–4370. 10.1093/hmg/ddu153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao W.M., Seng W.L., Duquette M., Lawler P., Laus C., and Lawler J.. 2001. Thrombospondin-1 type 1 repeat recombinant proteins inhibit tumor growth through transforming growth factor-beta-dependent and -independent mechanisms. Cancer Res. 61:7830–7839. [PubMed] [Google Scholar]
- Mikati A.G., Khanna O., Zhang L., Girard R., Shenkar R., Guo X., Shah A., Larsson H.B., Tan H., Li L., et al. . 2015. Vascular permeability in cerebral cavernous malformations. J. Cereb. Blood Flow Metab. 35:1632–1639. 10.1038/jcbfm.2015.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin-Brureau M., Lebrun A., Rousset M.C., Fagni L., Bockaert J., de Bock F., and Lerner-Natoli M.. 2011. Epileptiform activity induces vascular remodeling and zonula occludens 1 downregulation in organotypic hippocampal cultures: Role of VEGF signaling pathways. J. Neurosci. 31:10677–10688. 10.1523/JNEUROSCI.5692-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnesorge N., Viemann D., Schmidt N., Czymai T., Spiering D., Schmolke M., Ludwig S., Roth J., Goebeler M., and Schmidt M.. 2010. Erk5 activation elicits a vasoprotective endothelial phenotype via induction of Kruppel-like factor 4 (KLF4). J. Biol. Chem. 285:26199–26210. 10.1074/jbc.M110.103127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmar K.M., Larman H.B., Dai G., Zhang Y., Wang E.T., Moorthy S.N., Kratz J.R., Lin Z., Jain M.K., Gimbrone M.A. Jr., and García-Cardeña G.. 2006. Integration of flow-dependent endothelial phenotypes by Kruppel-like factor 2. J. Clin. Invest. 116:49–58. 10.1172/JCI24787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitulescu M.E., Schmidt I., Benedito R., and Adams R.H.. 2010. Inducible gene targeting in the neonatal vasculature and analysis of retinal angiogenesis in mice. Nat. Protoc. 5:1518–1534. 10.1038/nprot.2010.113 [DOI] [PubMed] [Google Scholar]
- Renz M., Otten C., Faurobert E., Rudolph F., Zhu Y., Boulday G., Duchene J., Mickoleit M., Dietrich A.C., Ramspacher C., et al. . 2015. Regulation of β1 integrin-Klf2-mediated angiogenesis by CCM proteins. Dev. Cell. 32:181–190. 10.1016/j.devcel.2014.12.016 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Manzaneque J.C., Lane T.F., Ortega M.A., Hynes R.O., Lawler J., and Iruela-Arispe M.L.. 2001. Thrombospondin-1 suppresses spontaneous tumor growth and inhibits activation of matrix metalloproteinase-9 and mobilization of vascular endothelial growth factor. Proc. Natl. Acad. Sci. USA. 98:12485–12490. 10.1073/pnas.171460498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S., Duquette M., Liu J., Drapkin R., Lawler J., and Petrik J.. 2015. Combined therapy with thrombospondin-1 type I repeats (3TSR) and chemotherapy induces regression and significantly improves survival in a preclinical model of advanced stage epithelial ovarian cancer. FASEB J. 29:576–588. 10.1096/fj.14-261636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwung P., Zhou G., Nayak L., Chan E.R., Kumar S., Kang D.W., Zhang R., Liao X., Lu Y., Sugi K., et al. . 2017. KLF2 and KLF4 control endothelial identity and vascular integrity. JCI Insight. 2:e91700 10.1172/jci.insight.91700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H., Errede M., Ulrich N.H., Virgintino D., Frei K., and Bertalanffy H.. 2011. Impairment of tight junctions and glucose transport in endothelial cells of human cerebral cavernous malformations. J. Neuropathol. Exp. Neurol. 70:417–429. 10.1097/NEN.0b013e31821bc40e [DOI] [PubMed] [Google Scholar]
- Shenkar R., Shi C., Check I.J., Lipton H.L., and Awad I.A.. 2007. Concepts and hypotheses: Inflammatory hypothesis in the pathogenesis of cerebral cavernous malformations. Neurosurgery. 61:693–703. 10.1227/01.NEU.0000298897.38979.07 [DOI] [PubMed] [Google Scholar]
- Short S.M., Derrien A., Narsimhan R.P., Lawler J., Ingber D.E., and Zetter B.R.. 2005. Inhibition of endothelial cell migration by thrombospondin-1 type-1 repeats is mediated by β1 integrins. J. Cell Biol. 168:643–653. 10.1083/jcb.200407060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatovic S.M., Sladojevic N., Keep R.F., and Andjelkovic A.V.. 2015. PDCD10 (CCM3) regulates brain endothelial barrier integrity in cerebral cavernous malformation type 3: Role of CCM3-ERK1/2-cortactin cross-talk. Acta Neuropathol. 130:731–750. 10.1007/s00401-015-1479-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockton R.A., Shenkar R., Awad I.A., and Ginsberg M.H.. 2010. Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J. Exp. Med. 207:881–896. 10.1084/jem.20091258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanriover G., Sozen B., Seker A., Kilic T., Gunel M., and Demir N.. 2013. Ultrastructural analysis of vascular features in cerebral cavernous malformations. Clin. Neurol. Neurosurg. 115:438–444. 10.1016/j.clineuro.2012.06.023 [DOI] [PubMed] [Google Scholar]
- Velasco P., Huegel R., Brasch J., Schröder J.M., Weichenthal M., Stockfleth E., Schwarz T., Lawler J., Detmar M., and Lange-Asschenfeldt B.. 2009. The angiogenesis inhibitor thrombospondin-1 inhibits acute cutaneous hypersensitivity reactions. J. Invest. Dermatol. 129:2022–2030. 10.1038/jid.2008.447 [DOI] [PubMed] [Google Scholar]
- Whitehead K.J., Plummer N.W., Adams J.A., Marchuk D.A., and Li D.Y.. 2004. Ccm1 is required for arterial morphogenesis: Implications for the etiology of human cavernous malformations. Development. 131:1437–1448. 10.1242/dev.01036 [DOI] [PubMed] [Google Scholar]
- Whitehead K.J., Chan A.C., Navankasattusas S., Koh W., London N.R., Ling J., Mayo A.H., Drakos S.G., Jones C.A., Zhu W., et al. . 2009. The cerebral cavernous malformation signaling pathway promotes vascular integrity via Rho GTPases. Nat. Med. 15:177–184. 10.1038/nm.1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüstehube J., Bartol A., Liebler S.S., Brütsch R., Zhu Y., Felbor U., Sure U., Augustin H.G., and Fischer A.. 2010. Cerebral cavernous malformation protein CCM1 inhibits sprouting angiogenesis by activating DELTA-NOTCH signaling. Proc. Natl. Acad. Sci. USA. 107:12640–12645. 10.1073/pnas.1000132107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., and Lawler J.. 2007. Thrombospondin-based antiangiogenic therapy. Microvasc. Res. 74:90–99. 10.1016/j.mvr.2007.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Hamik A., Nayak L., Tian H., Shi H., Lu Y., Sharma N., Liao X., Hale A., Boerboom L., et al. . 2012. Endothelial Kruppel-like factor 4 protects against atherothrombosis in mice. J. Clin. Invest. 122:4727–4731. 10.1172/JCI66056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Rawnsley D.R., Goddard L.M., Pan W., Cao X.J., Jakus Z., Zheng H., Yang J., Arthur J.S., Whitehead K.J., et al. . 2015. The cerebral cavernous malformation pathway controls cardiac development via regulation of endocardial MEKK3 signaling and KLF expression. Dev. Cell. 32:168–180. 10.1016/j.devcel.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Tang A.T., Wong W.Y., Bamezai S., Goddard L.M., Shenkar R., Zhou S., Yang J., Wright A.C., Foley M., et al. . 2016. Cerebral cavernous malformations arise from endothelial gain of MEKK3-KLF2/4 signalling. Nature. 532:122–126. 10.1038/nature17178 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.