Promyelocytic leukemia (PML) nuclear bodies modulate several processes, including senescence or apoptosis. Niwa-Kawakita et al. demonstrate that PML regulates reactive oxygen species (ROS) homeostasis in vivo by coupling ROS to p53 signaling to enforce basal ROS protection and mediate their acute toxicity.
Abstract
Promyelocytic leukemia (PML) nuclear bodies (NBs) recruit partner proteins, including p53 and its regulators, thereby controlling their abundance or function. Investigating arsenic sensitivity of acute promyelocytic leukemia, we proposed that PML oxidation promotes NB biogenesis. However, physiological links between PML and oxidative stress response in vivo remain unexplored. Here, we identify PML as a reactive oxygen species (ROS) sensor. Pml−/− cells accumulate ROS, whereas PML expression decreases ROS levels. Unexpectedly, Pml−/− embryos survive acute glutathione depletion. Moreover, Pml−/− animals are resistant to acetaminophen hepatotoxicity or fasting-induced steatosis. Molecularly, Pml−/− animals fail to properly activate oxidative stress–responsive p53 targets, whereas the NRF2 response is amplified and accelerated. Finally, in an oxidative stress–prone background, Pml−/− animals display a longevity phenotype, likely reflecting decreased basal p53 activation. Thus, similar to p53, PML exerts basal antioxidant properties but also drives oxidative stress–induced changes in cell survival/proliferation or metabolism in vivo. Through NB biogenesis, PML therefore couples ROS sensing to p53 responses, shedding a new light on the role of PML in senescence or stem cell biology.
Introduction
Promyelocytic leukemia (PML) organizes nuclear bodies (NBs), domains recruiting a variety of unrelated proteins and modulating their posttranslational modifications (Lallemand-Breitenbach and de Thé, 2010). PML has antiproliferative functions at least in part mediated by PML’s ability to enhance p53 function (Guo et al., 2000; Pearson et al., 2000). p53 is a key regulator of survival and proliferation, but other functions were more recently assigned to p53, notably control of metabolism, oxidative stress, autophagy, and stem cell self-renewal (Berkers et al., 2013). Exactly how PML controls p53 activity remains disputed. p53 and most of its regulators (ARF, HIPK2, CBP, MDM2 SIRT1, or MOZ) traffic through PML NBs, suggesting that these domains may represent privileged sites to fine-tune p53 posttranslational modifications and ultimately determine its activation status (Garcia and Attardi, 2014). Although PML is required for efficient induction of p53 targets like Bax or CDKN1/p21 upon p53 or PML overexpression or PML knockdown ex vivo (Fogal et al., 2000; Guo et al., 2000; Pearson et al., 2000), in vivo transcriptomics has yet failed to identify a defective p53 signature in Pml−/− mice.
Arsenic, a strong oxidant, cures acute promyelocytic leukemia (APL) by initiating PML/RARA oncoprotein degradation. Studies on the biochemical basis of arsenic therapeutic action have revealed that both PML/RARA and PML proteins are oxidation prone (Jeanne et al., 2010). Arsenic induces serial changes in PML state, including oxidation, assembly into insoluble nuclear matrix–associated shells, sumoylation, and ultimately allowing recruitment of protein partners into NBs (Lallemand-Breitenbach et al., 2001; Sahin et al., 2014b). The conjunction of arsenic-induced PML/RARA degradation and PML NB biogenesis activates a prosenescent p53 program driving APL cure (Ablain et al., 2014).
In addition to transcriptional activation by NRF2, the response to oxidative stress involves a subset of p53 targets (Desaint et al., 2004; Gambino et al., 2013). p53 exerts dual effects on oxidative stress: protection at basal levels and senescence/apoptosis at high reactive oxygen species (ROS) levels. Importantly, p53 antioxidant functions were directly linked to its antitumor effects (Sablina et al., 2005). In addition to its tumor-suppressive effects, p53 also drives aging in activated p53 mutant mice (Matheu et al., 2007; Gambino et al., 2013). Here, we show that similar to p53, PML controls basal intracellular ROS levels and is required for oxidative stress response in vivo. Our studies identify PML as a critical ROS sensor coupling NB formation to p53 activation.
Results and discussion
PML protects against ROS
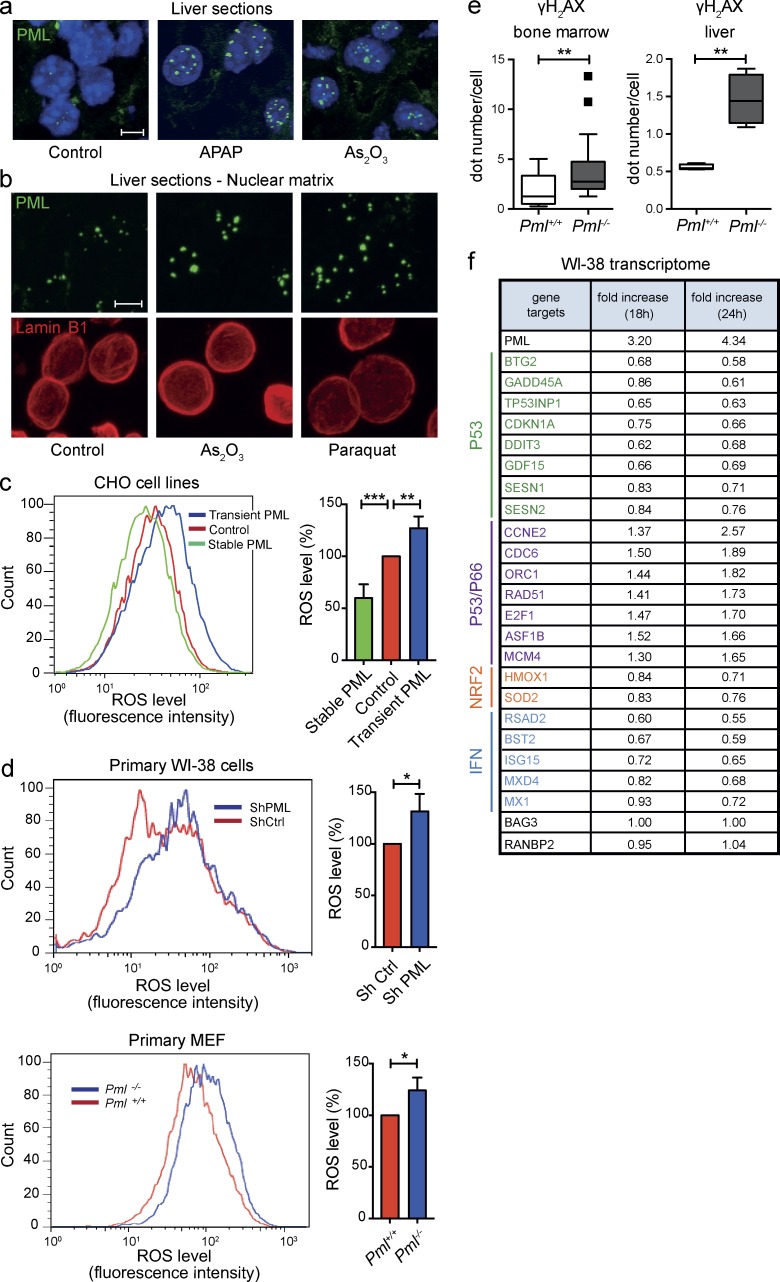
Arsenic triggers changes in PML state ex vivo, inducing a switch from diffuse nuclear PML proteins to insoluble multimers assembled into NBs (Zhu et al., 1997; Lallemand-Breitenbach et al., 2001; Jeanne et al., 2010; Sahin et al., 2014a; Ribet et al., 2017). Exploring NB biogenesis in vivo, we confirmed that NB formation is sharply enhanced in vivo by acetaminophen (APAP) and arsenic trioxide (Fig. 1 a). We then directly assessed endogenous PML solubility by extracting in situ the nuclear matrix from liver sections. Importantly, PML immunostaining of the insoluble matrix sharply increased 1 h after injection of prooxidant stimuli (Fig. 1 b). Our results identify ROS, produced by oxidants other than arsenic, as key regulators of PML NB aggregation and matrix association in vivo.
Figure 1.
PML controls ROS levels. (a) Immunofluorescent staining for PML in response to oxidative stress in vivo. Mice were i.p. injected with arsenic trioxide or APAP and sacrificed 1 h (arsenic) or 2 h (APAP) after injection. PML NBs (green) were analyzed on liver cryosections. DAPI is shown (blue). Data are representative of at least three independent experiments (n = 3) with two mice per condition. Bar, 5 µm. (b) Immunofluorescence analysis of insoluble PML (green) associated with the nuclear matrix prepared from liver cryosections. Livers were collected 1 h after i.p. injection with arsenic or Paraquat or from untreated mice. Lamin B1 (red) was used as a positive control and DAPI as a negative control (not depicted) for in situ nuclear matrix preparation. Bar, 5 µm. (c and d) FACS analysis of ROS levels in the indicated cells using a fluorescent CellROX probe (left), and graphs representing mean fluorescent intensity in indicated cells, normalized to control cells (right). Mean fluorescence intensity was set to 100% in control cells. Data in c and d are representative (left) or quantitative (right) of n = 5 and n = 2 independent experiments, respectively. Error bars represent SEM. (c) CHO cells were transiently or stably transfected with expression vectors encoding PML III isoform or empty vector control. **, P < 0.01; ***, P < 0.001 (t test). (d) Primary WI-38 human fibroblasts were analyzed for ROS content 24 h after transduction with PML or control shRNA lentiviral vector (top). ROS levels were assessed in MEFs freshly extracted from Pml−/− embryos or wild-type littermates (bottom). *, P < 0.05 (t test). (e) Box and whisker plot (Tukey) representing γH2AX dots counts from 100 nuclei assessed by immunofluorescence on the indicated tissues (two mice each/tissue). Mann–Whitney test is indicated (**, P < 0.01). (f) Table extracted from transcriptomic analysis of primary WI-38 cells 18 h and 24 h after transduction with lentivirus encoding the PML IV isoform or with control empty virus. RNA levels relative to control for oxidative stress–induced p53 target genes, some IFNs, p53/p66, or NRF2 targets are indicated. Two unaffected genes are shown as control (black). Data are the mean of n = 2 replicates.
Transient overexpression of PML III rapidly increases ROS levels (Fig. 1 c), whereas the arsenic-insensitive C212A mutant does not (not depicted), suggesting that PML has intrinsic redox activity. Conversely, stable PML III expression in CHO cells significantly decreased basal ROS levels (Fig. 1 c), whereas down-regulation of all endogenous PML isoforms by shRNA increased ROS levels in primary WI-38 human fibroblasts; Fig. 1 d). Similarly, Pml genetic inactivation led to ROS accumulation in MEFs (Fig. 1 d) or mouse hematopoietic progenitors (not depicted), whereas stable reexpression of PML isoforms I, III, or IV in primary Pml−/− MEFs restored basal low ROS levels (unpublished data). Consistent with ROS accumulation, we observed higher levels of phosphorylated histone H2AX (γH2AX) foci in Pml−/− hematopoietic progenitors or hepatocytes (Fig. 1 e and Fig. S1; Zhong et al., 1999).
We then explored PML-regulated gene expression in WI-38 fibroblasts infected with a PML IV–expressing lentivirus (Ivanschitz et al., 2015). A three- to fourfold enhancement of PML expression down-regulated p53 targets known to be up-regulated by oxidative stress, consistent with a direct or indirect antioxidant function of PML (Fig. 1 f; Desaint et al., 2004). Enhanced PML expression also repressed canonical NRF2 targets such as HMOX1 and PML up-regulated cell-cycle targets known to be repressed by p53 upon oxidative stress (Fig. 1 f; Gambino et al., 2013). Finally, PML induction also down-regulated IFN signaling (Fig. 1 f; Choi et al., 2006). Collectively, PML induction down-regulates intracellular ROS, blunting oxidation-responsive signaling, demonstrating that PML is a redox-sensitive protein that controls basal ROS levels.
The acute oxidative stress response is altered in Pml−/− animals
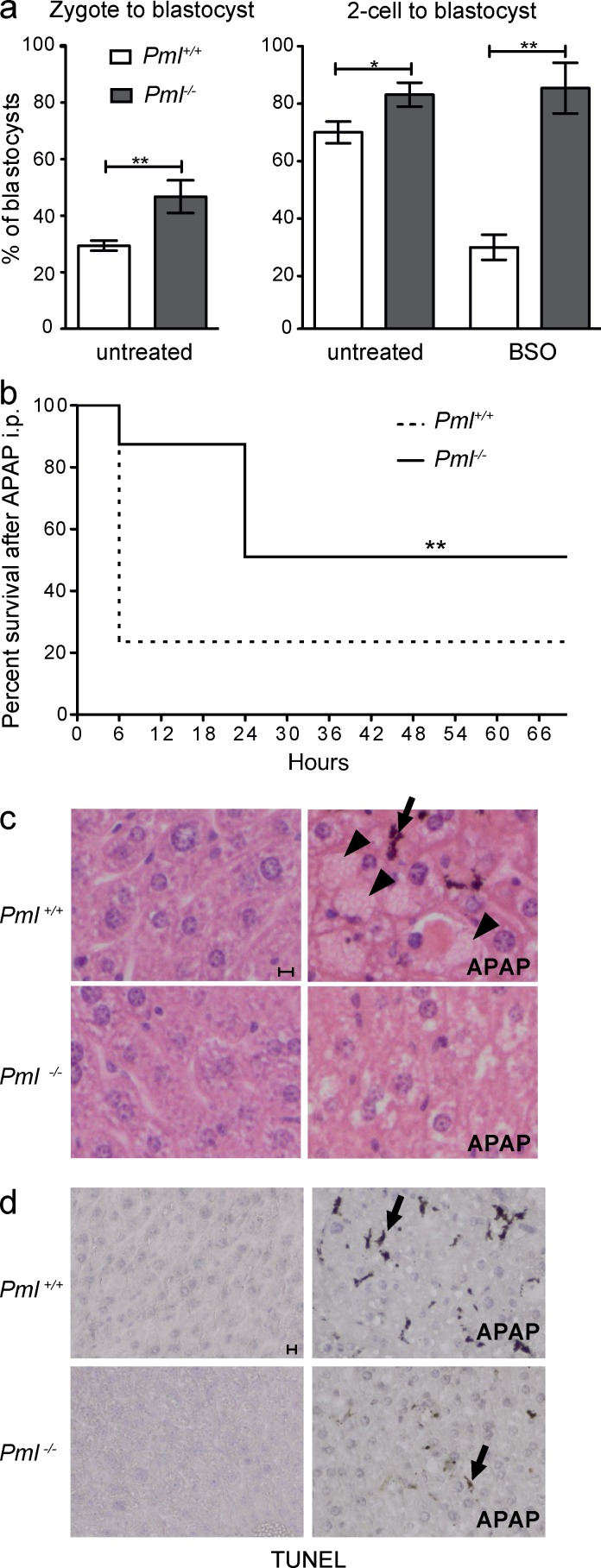
Such reciprocal link between PML and ROS levels prompted us to investigate the role of Pml in the biological response to oxidative stresses. We first explored ex vivo survival and development of preimplantation embryos, some of the most oxidative stress–sensitive cells known. No difference in the litter number was observed, suggesting that Pml+/+ and Pml−/− embryo survival is similar in vivo. Yet, in culture (a condition of sharp oxidative stress), Pml−/− zygotes progressed up to the blastocyst stage significantly more often than wild-type ones (Fig. 2 a, left). The same difference in spontaneous survival at blastocyst stage was observed with ex vivo two-cell-stage embryos (Fig. 2 a, right). This difference was exacerbated in the presence of buthionine sulfoximine (BSO), an inhibitor of glutathione synthesis, which yields acute oxidative stress. Indeed, Pml−/− embryos were insensitive to BSO, whereas survival of Pml+/+ embryos was drastically decreased (Fig. 2 a, right). BSO promoted aggregation of PML bodies (not depicted).
Figure 2.
Pml−/− embryos and mice are resistant to oxidative stress. (a) Graphs representing percentage of blastocysts derived from Pml+/+ and Pml−/− preimplantation embryos. Embryos were obtained from n = 4 (untreated) and n = 3 (BSO) independent experiments involving 15–45 embryos each; error bars represent SEM (Fisher’s exact test: *, P < 0.05; **, P < 0.01). The total number of embryos was as follows: zygotes: Pml+/+, 123; Pml−/−, 106; 2-cell embryos untreated: Pml+/+, 108; Pml−/−, 140; 2-cell embryos BSO treated: Pml+/+, 45; Pml−/−, 65. (b) Survival (percentage) of Pml−/− and Pml+/+ mice after i.p. injection with APAP (time 0). A total of n = 4 independent experiments were performed (with n = 4–6 mice for each group). **, P < 0.01 (log-rank test or Gehan–Breslow–Wilcoxon tests). (c) Hematoxylin and eosin staining of liver sections from mice treated or not with APAP and sacrificed 6 h later. Note hemorrhages (arrowheads) and an apoptotic cell (arrow). (d) TUNEL performed on liver cryosections from c. Bars, 10 µm.
Acute oxidative stress induced by APAP overdose is highly hepatotoxic (Fig. 2, b–d; Woolbright and Jaeschke, 2017). We thus investigated the effects of Pml inactivation on response to a high dose of APAP in vivo. Unexpectedly, despite their higher basal ROS levels, APAP-treated Pml−/− animals survived better than Pml+/+ ones in two genetic backgrounds (Fig. 2 b and Fig. S2). Histologically, 6 h after APAP injections, only moderate cellular damage was observed in Pml−/− animals, whereas massive hemorrhage and apoptosis were evident in Pml+/+ livers (Fig. 2, c and d). These data demonstrate that Pml mediates liver apoptosis upon acute oxidative stress. Thus, in different ex or in vivo models, Pml inactivation significantly increases survival upon oxidative stress, supporting the idea that apart from its regulation of basal ROS levels, PML also senses, conveys, and/or amplifies the biological response to ROS.
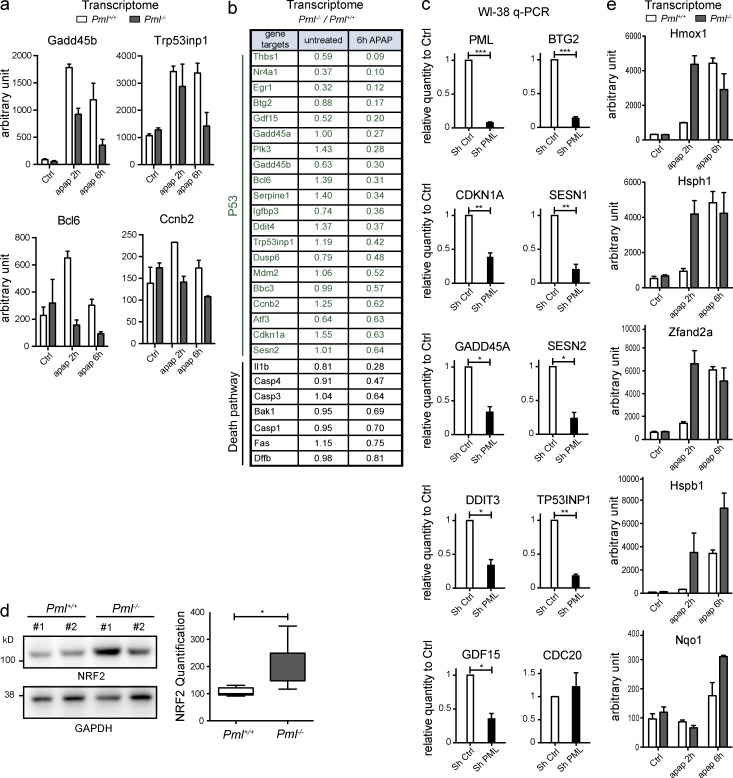
To obtain mechanistic insights into the role of Pml in ROS response, we profiled mRNA expression in livers from untreated or APAP-treated Pml−/− and Pml+/+ animals. In Pml+/+ mice, p53 signaling was activated after APAP injection, although p53 protein was not stabilized (Fig. 3 a and not depicted). Importantly, activation of oxidative stress–responsive p53 targets was blunted in Pml−/− animals (Fig. 3, a and b). Some targets, such as Trp53inp1 or Sesn2, have antioxidant properties. Their basal expression levels were decreased in Pml−/− animals, which may explain higher ROS levels of Pml−/− cells (Fig. 3 b). We also explored the p53 signature in WI-38 primary fibroblasts upon PML knockdown. Again, PML was required for basal activation of p53 targets (Fig. 3 c). Genes controlling fatty acid oxidation were also differentially expressed between Pml−/− and Pml+/+ hepatocytes upon APAP treatment in vivo (not depicted; Carracedo et al., 2012). Finally, consistent with higher basal ROS levels in Pml−/− cells, basal NRF2 protein expression was significantly increased in Pml−/− livers and activation of Nrf2-target genes was sharply accelerated upon APAP treatment (Fig. 3, d and e). Collectively, these data reveal that in Pml−/− animals, both basal and oxidative stress–induced p53 signaling is defective, likely explaining higher basal ROS levels and priming of an adaptive NRF2 response.
Figure 3.
Pml−/− mice fail to activate p53 upon oxidative stress. (a) Graphs representing transcript levels for the indicated p53 target genes determined by transcriptomic arrays of Pml−/− and Pml+/+ mouse livers 2 and 6 h after APAP or vehicle injections. Mean values from two mice per condition are shown. Error bars represent SEM. (b) Table from the transcriptomic analysis in a indicating p53 targets and death regulators and the mRNA ratio in Pml−/− versus Pml+/+ mice. (c) Transcripts from WI-38 fibroblasts transduced with PML or control shRNA were quantified by quantitative PCR. Error bars represent SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (d) Western blot analysis of NRF2 protein from Pml−/− and Pml+/+ mouse livers (left; two representative untreated mice). Box and whisker plot (Tukey) showing NRF2 protein quantification from six mice (right; Mann–Whitney test: *, P < 0.05). (e) Levels of NRF2 target mRNAs (transcriptomic analysis in b) 2 h or 6 h after APAP treatment. Error bars represent SEM.
PML is required for fasting-induced p53 activation
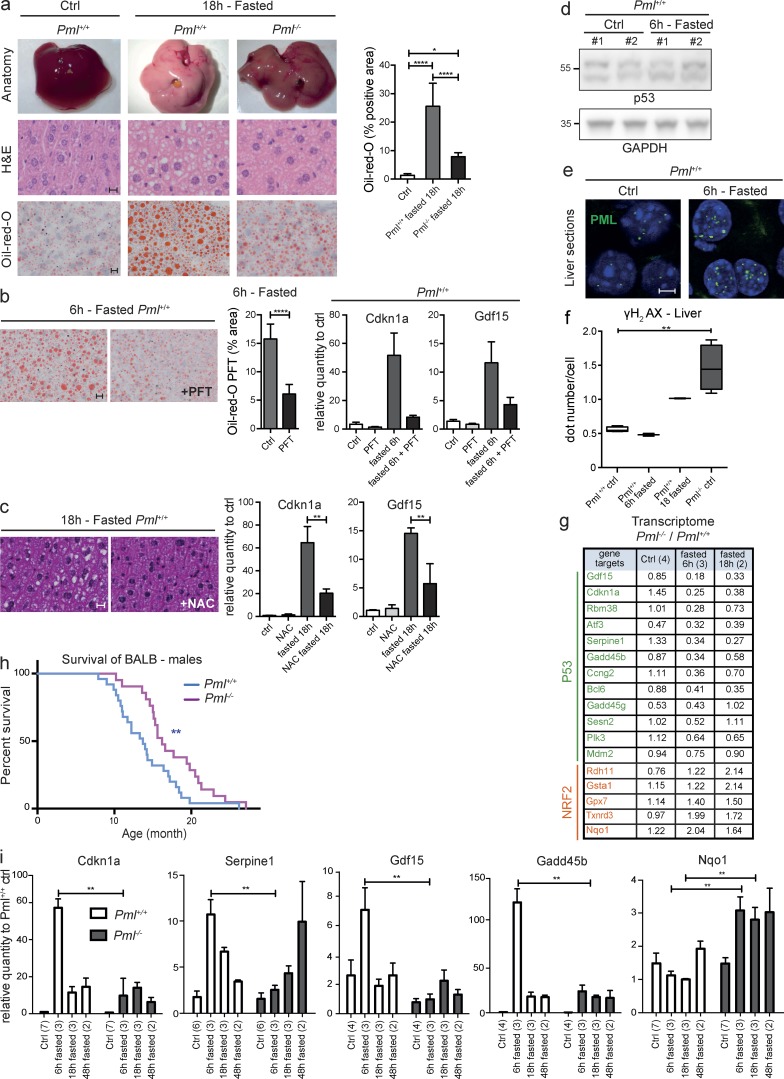
We investigated fasting, which triggers profound metabolic changes, notably through glucose deprivation–induced p53 activation (Berkers et al., 2013). We used BALB/cByJ mice, which are exquisitely sensitive to fasting because of a mutation of the Acads gene encoding a mitochondrial enzyme of fatty acid β-oxidation. Loss of this enzyme results in fasting-enhanced ROS production (Wood et al., 1989) and fatty acid accumulation in the liver (Fig. 4 a). Fasting-induced steatosis and activation of oxidative stress–sensitive p53 targets were both antagonized by either the p53 inhibitor pifithrin α or the ROS scavenger N-acetyl cysteine in Pml+/+ animals (Fig. 4, b and c). Thus, in BALB/cByJ mice, fasting-induced ROS trigger p53-driven liver steatosis. Interestingly, although p53 protein levels in the liver remained unchanged upon fasting, NB formation was consistently increased and PML abundance was occasionally enhanced (Fig. 4, d and e; and Fig. S2; Scaglioni et al., 2012).
Figure 4.
PML is required for the fasting-activated p53 response. (a) Livers from fed or 18-h–fasted Pml−/− and Pml+/+ mice (top), hematoxylin, phloxine, and saffron staining (middle; bar, 10 µm), and oil red O staining (bottom; bar, 10 µm). Quantification of oil red staining from n = 5 mice. Error bars represent SEM. *, P < 0.05; ****, P < 0.0001 (t test). (b) Image (left; bar, 10 µm) and quantification (middle; ****, P < 0.0001 [t test]; error bars represent SEM) of oil red O staining and quantitative PCR analysis of the indicated p53 targets (right; error bars represent SEM) from fasted or fed mice treated or not with pifithrin-α. (c) Hematoxylin, phloxine, and saffron staining (left; bar, 10 µm) and quantitative PCR analysis of indicated p53 targets (right) from fasted mice treated or not with N-acetylcysteine (NAC). Error bars represent SEM. **, P < 0.01. (d) Western blot analysis of p53 and GAPDH from livers of indicated mice. Representative experiment with two mice per condition (fed and 6-h fasted) from n = 2 replicates. (e) Immunofluorescent PML staining (green) upon fasting in liver cryosections (blue, DAPI). Bar, 5 µm. (f) Box and whisker plot (Tukey) representing γH2AX foci on liver sections of Pml−/− and Pml+/+ littermates. Dots were counted from 100 nuclei of four fed mice or two mice fasted for 6 h or 18 h. **, P < 0.01 (Mann–Whitney test). (g) mRNA levels (transcriptomes) from the p53 (top) and NRF2 (bottom) signature in Pml−/− livers relative to their control littermate from fed or fasted mice. (h) Survival of Pml−/− and Pml+/+ littermates. **, P < 0.01 (Gehan–Breslow–Wilcoxon test). (i) Graphs representing quantitative PCR measurement of indicated p53 and NRF2 targets. Mean values with SEM from the indicated number of mice (in parentheses) are shown. **, P < 0.01.
Critically, fasting-induced steatosis was dramatically dampened in Pml−/− animals compared with their wild-type counterparts (Fig. 4 a, right) and activation of oxidative stress–sensitive p53 targets was sharply delayed in Pml−/− animals (Fig. 4 g). Again, the NRF2 response to fasting-induced ROS was accelerated in Pml−/− mice (Fig. 4 g). These data were confirmed by qPCR in independent animals (Fig. 4 h). In this background, Pml inactivation was also associated with high basal ROS and/or γH2AX levels (Fig. 1 e and 4 f and Fig. S1). Fatty acid β-oxidation is transcriptionally regulated by p53 (Berkers et al., 2013; Bieging et al., 2014) and was not efficiently activated in fasted Pml−/− mice (not depicted). Altogether, these data establish that fasting-induced oxidative stress triggers Pml-dependent p53 activation, extending the results obtained with drug-induced oxidative stress.
PML may be solely an indirect global enhancer of p53 responses initiated by other stress-induced pathways, such as DNA damage (Bernardi and Pandolfi, 2007; Brady et al., 2011). We thus examined γH2AX foci in hepatocytes upon fasting. Importantly, γH2AX foci number increased only after 18 h (Fig. 4 f), whereas the p53 transcriptional response was already evident at 6 h (Fig. 4, g and i). Thus, PML is not a mere amplifier of DNA-damage–induced p53 transcriptional response. Collectively, our data unravel PML as a broad oxidative stress sensor conveying the ROS response, at least in part through activation of p53 signaling.
Longevity of BALB Pml−/− mice despite chronic oxidative stress
Chronic oxidative stress accelerates aging. We thus examined the survival of BALB.Pml−/− and BALB.Pml+/+ mice derived from heterozygote crossings. A significant survival advantage was observed for Pml−/− males relative to Pml+/+ males (Fig. 4 h). A similar difference of borderline significance was observed in females, which live significantly longer irrespective of their Pml status. Pathological analyses did not reveal major differences between Pml-proficient or deficient males (Fig. S3). The apparent causes of death were thrombosis and heart disease. Delayed aging may reflect defective p53-driven senescence, although we cannot exclude a contribution of other Pml-dependent pathways (HIF1a and PPAR; Carracedo et al., 2012). Thus, in the context of this oxidative stress–prone strain, the absence of Pml delays aging, despite high basal ROS levels and DNA damage.
PML is an oxidation-sensitive protein (Jeanne et al., 2010; Sahin et al., 2014b; Ribet et al., 2017; Fig. 1, a and b). Our current data show that PML acts as a broad ROS sensor that regulates ROS homeostasis at least in part by enhancing p53 responses (Fig. 5). Similar to p53, PML has a dual role, enforcing basal protection against ROS (Figs. 2 and 5) and facilitating ROS-triggered apoptosis (Figs. 3 and 5). Mechanistically, multiple feedback loops and cross talk exist among ROS, PML, and p53. For example, p53 transcriptionally induces PML expression (de Stanchina et al., 2004), enhancing NB recruitment of p53 and its activating kinases/acetylases (Pearson et al., 2000; Rokudai et al., 2013). In addition to promoting antioxidant responses, PML may also directly behave as a ROS scavenger because of its intrinsic ROS sensitivity (Fig. 1). PML was proposed to interfere with mitochondrial complex II, explaining basal ROS production and enhanced NFR2 activity (Guo et al., 2014). Should defective mitochondrial function be the primum movens explaining ROS production in Pml−/− animals, high ROS and DNA damage would be expected to sensitize both NRF2 and p53 responses. PML was also proposed to favor NRF2 degradation (Malloy et al., 2013), but loss of this PML negative-feedback mechanism should yield low basal ROS levels. We propose that, at least in vivo, the primary target of PML is p53 and that NRF2 activation is a compensatory effect resulting from insufficient p53-driven antioxidant signaling (Fig. 5). PML NBs may also serve as a molecular switch activating p53 targets at the expense of NRF2 ones. Our studies highlight how PML couples ROS and stress-induced changes in nuclear organization to control of gene expression. Using a specific, stress-prone genetic background (Fig. 4), we unraveled an unexpected role for PML in aging. Although the later may be connected to p53 pathway, this deserves further investigation. Defective ROS homeostasis in Pml−/− animals may explain the multiplicity of PML-sensitive biological pathways, including PTEN/AKT or HIF1A (Bernardi et al., 2006; Trotman et al., 2006; Song et al., 2008). Significantly, PML’s control of the ROS response and cellular redox status could be particularly important in senescent or stem cells, which express high levels of PML and are exquisitely ROS and PML sensitive.
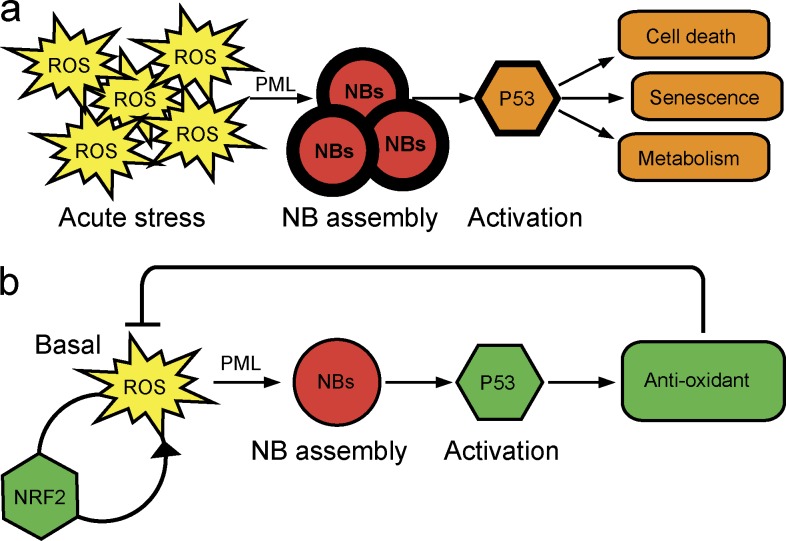
Figure 5.
Model for PML function in oxidative stress response in vivo. (a) Upon acute oxidative stress, high ROS levels increase NB formation, leading to p53 activation, mediating toxic effects. (b) Basal ROS levels induce PML NB assembly, recruiting p53 and its regulators and inducing antioxidant responses that ultimately decrease ROS levels. With active PML/p53 antioxidant signaling, NRF2 responses are blunted.
Materials and methods
Antibodies, Western blot, and immunofluorescence
Western blot analysis was performed using rabbit anti–NRF2 antibody (12721; Cell Signaling), mouse monoclonal anti–mouse PML, clone 36.1–104 (MAB3738; Merck Millipore), rabbit anti–phospho-H2AX antibody (2577; Cell Signaling), rabbit anti-p53, CM5 (NCL-p53-CM5p; Novocastra), or rabbit anti-GAPDH antibody (G9545; Sigma).
Total cell extracts were obtained from mouse livers conserved in RNAlater using Tissue Extraction Reagent I (ThermoFisher, Life Technologies), homogenized using lyser LT (Qiagen), and lysed in Laemmli buffer or from fresh cultured cells by direct lysis in Laemmli buffer. These extracts were run on 4–12% gradient SDS-PAGE (Bolt; Life Technologies), and immunoblots were performed as described previously (Lallemand-Breitenbach et al., 2008).
For immunofluorescence, 10-µm frozen liver sections were fixed for 10 min in acetone, permeabilized in PBS 0.2% Triton for 30 min at 4°C, and blocked for 1 h with PBS 0.05% Triton-1% BSA. Immunostaining was then processed as previously described (Lallemand-Breitenbach et al., 2008) using anti–mouse PML directly coupled to FITC fluorochrome (homemade) or detected by Alexa Fluor 488–coupled secondary antibodies (from Jackson ImmunoResearch). Confocal analysis was performed with a LSM510 Meta laser microscope (Carl Zeiss Micro-Imaging, Inc.) with a Plan-Apochromat 63× NA 1.4 oil-immersion objective. In situ nuclear Matrix preparation was performed as described previously (Sahin et al., 2014b) on liver cryosections (directly on slides) before immunostaining.
ROS level detection
Total ROS levels were measured using CellROX probes (C10422; Molecular Probes). Cells were incubated with CellROX Deep red reagent for 30 min and washed, and probe signals were measured by flow cytometry on a FACSCalibur flow cytometer and analyzed using CellQuest software (BD Biosciences).
Primary cell culture, transfection, and shRNA
Primary MEFs were prepared from littermate embryos. MEFs were pooled from at least three different embryos with the same genotype and from two mothers. MEFs, WI-38 cells, and CHO cells were cultivated in DMEM, 10% FCS. MEFs were analyzed during the first five passages. WI-38 transduction using lentivirus expressing shPML or control scrambled shRNA was performed as described previously (Ivanschitz et al., 2015). Transient transfections of CHO cells with pSG5-PMLIII vector using Effecten reagent (Qiagen) was performed as described elsewhere (Lallemand-Breitenbach et al., 2008).
Animal experiments
Animals were handled according to the guidelines of institutional animal care committees using protocols approved by the Comité d’Ethique Experimentation Animal Paris-nord (no. 121). Mice were maintained in a 12-h light–dark cycle animal facility under specific pathogen–free conditions with free access to water and food (A03: SAFE; Institut Universitaire d’Hématologie Institute, Paris, France). Pml−/− mice (129sv background) were provided by P.P. Pandolfi (Harvard Medical School, Boston, MA) and maintained in the facility. The BALB/cByJ congenic strain is deficient for acyl-coenzyme A dehydrogenase, short chain (Acads−/−). The BALB congenic strain of Pml−/− (BALB.Pml−/−) was generated by backcrossing 129sv.Pml−/− mice to the BALB/cByJ inbred strain (Charles River) for at least 10 generations. Mice were genotyped by multiplex PCR using following primers: Pml PCR (5′-AAGCCATACAGGAGGAATTTCA-3′; 5′-GTGGTTGGTATTGGAGCAGAA-3′; 5′-ATCAGGATGATCTGGACGAAG-3′) yields fragments of 443 bp (Pml+/+) or ∼660 bp (Pml−/−), and Acads PCR (5′-AGTTCAAGCTGGCAGACATGG-3′ and 5′-TAAAGAGGCAGCCAAGCTCAG-3′) yields fragments of 815 bp (Acads wt) or 536 bp (Acads mut).
9–12-wk-old age-, sex-, and body weight–matched Pml−/− mice or Pml+/+ mice were used for the following treatments. APAP (N-acetyl-para-amino-phenol; Sigma-Aldrich) was dissolved in warm PBS and i.p. injected at a dose of 500–800 mg/kg for the survival analysis or 300 mg/kg for the histological and array analysis. Arsenic was injected at 5 µg/g for the indicated time. For short-term fasting, food was removed for the indicated time. 2.2 mg/kg pifithrin-α (P4359; Sigma-Aldrich) or control vehicle was i.p. injected when the food was removed. 10 mg/ml N-acetylcysteine (Sigma-Aldrich) was used in drinking water and changed once per week during a 4-wk period before fasting. Mice were sacrificed by cervical dislocation.
Lifespan
The Pml−/− and Pml+/+ littermates derived from BALB/cByJ congenic Pml heterozygote mouse intercrosses were used for the lifespan experiments.
Preimplantation embryos
Preimplantation embryos were collected from oviducts either in the morning of copulation plug (zygote) or the day after (two-cell stage). The embryos were cultured in M16 medium (Sigma). Before incubation or after 24 h of culture for zygotes, the number of two-cell-stage embryos was counted (as total embryos). The embryos were observed daily to evaluate the developmental stage. The number of embryos that developed up to the blastocyst stage were counted during 7-d (zygote) or 6-d (two-cell stage) culture. Culture media was changed every 2 d. After an initial 24 h of culture (four-cell stage), 5 mM BSO (19176; Sigma) treatment was performed for 48 h.
Histological analysis
Tissue samples were fixed in alcohol-formaldehyde-acetic acid reagent (Carlo Erba) or immediately frozen in liquid nitrogen. The fixed samples were embedded in paraffin, and 5-µm sections were stained with hematoxylin and eosin or hematoxylin, phloxine, and saffron. TUNEL was performed using the In Situ Cell Death Detection kit (Roche). For oil red O staining (Sigma-Aldrich), 10-µm frozen liver sections were dried, fixed with 10% formalin, and stained using standard protocols.
TaqMan quantitative PCR analysis and Affymetrix array
Total RNA was extracted from tissues with RNeasy Plus Universal kit or miRNeasy kit (Qiagen). Tissues were previously stabilized with RNAlater (ThermoFisher) and lysate homogenized in Tissue Lyser LT (Qiagen). First-strand cDNA was synthesized using a reverse transcription kit (4368813; ThermoFisher). qPCR was performed using TaqMan probes with the 7500 Fast Real-Time PCR system (ThermoFisher). Gapdh was used as an endogenous control. The mean value of two replicates for each sample was used. Transcriptomic analyses were performed at the Curie Institute. Samples were hybridized on Affymetrix Mouse Gene 1.1 ST or 2.1 ST arrays and normalized with GcRMA.
Statistical analysis
All statistical analyses and the number of replicates are indicated in the figures or figure legends.
Online supplemental material
Fig. S1 compares γH2AX dots in primary cells from Pml−/− and Pml+/+ animals, as well as untreated mouse liver (3.5 mo) showing no pathological differences. Fig. S2 compares survival upon APAP administration in Pml-proficient or deficient 129 mice, as well as Pml stabilization upon fasting in BALB/cByJ mice. Fig. S3 summarizes pathological analysis of old Pml−/− and Pml+/+ mice.
Supplementary Material
Acknowledgments
The authors thank N. Setterblad and Institut Universitaire d’Hématologie technology platforms for imaging; L. Peres, L. Huynh, and E. Gelabale for histology; D. Gentien (Institut Curie) for arrays; V. Parietti for the animal house; laboratory members; and A. Carrier, F. Mechta-Grigoriou, A. Bazarbachi, and U. Sahin for critical reading of the manuscript.
Work in the laboratory is supported by Institut National du Cancer, the Association pour la Recherche Contre le Cancer (Prix Griffuel), the European Research Council (senior grant 268729 – STEMAPL to H. de Thé), and the French National Research Agency (Investissements d’Avenir program grants ANR-11-PHUC-002 and ANR-10-IHUB-0002 and grant ANR-13-JSV2-0005-01).
The authors declare no competing financial interests.
Author contributions: M. Niwa-Kawakita performed and analyzed in vivo experiments and transcriptomic data. O. Ferhi performed in vivo and ex vivo experiments and prepared the figures. H. Soilihi performed in vivo experiments. M. Le Bras performed experiments in WI-38 cells. V. Lallemand-Breitenbach and H. de Thé designed the study, analyzed data, and wrote the manuscript, which was approved by all authors.
Footnotes
Abbreviations used:
- APAP
- acetaminophen
- BSO
- buthionine sulfoximine
- γH2AX
- phosphorylated histone H2AX
- NB
- nuclear body
- PML
- promyelocytic leukemia
- ROS
- reactive oxygen species
References
- Ablain J., Rice K., Soilihi H., de Reynies A., Minucci S., and de Thé H.. 2014. Activation of a promyelocytic leukemia-tumor protein 53 axis underlies acute promyelocytic leukemia cure. Nat. Med. 20:167–174. 10.1038/nm.3441 [DOI] [PubMed] [Google Scholar]
- Berkers C.R., Maddocks O.D., Cheung E.C., Mor I., and Vousden K.H.. 2013. Metabolic regulation by p53 family members. Cell Metab. 18:617–633. 10.1016/j.cmet.2013.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R., and Pandolfi P.P.. 2007. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 8:1006–1016. 10.1038/nrm2277 [DOI] [PubMed] [Google Scholar]
- Bernardi R., Guernah I., Jin D., Grisendi S., Alimonti A., Teruya-Feldstein J., Cordon-Cardo C., Simon M.C., Rafii S., and Pandolfi P.P.. 2006. PML inhibits HIF-1alpha translation and neoangiogenesis through repression of mTOR. Nature. 442:779–785. 10.1038/nature05029 [DOI] [PubMed] [Google Scholar]
- Bieging K.T., Mello S.S., and Attardi L.D.. 2014. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer. 14:359–370. 10.1038/nrc3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady C.A., Jiang D., Mello S.S., Johnson T.M., Jarvis L.A., Kozak M.M., Kenzelmann Broz D., Basak S., Park E.J., McLaughlin M.E., et al. . 2011. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 145:571–583. 10.1016/j.cell.2011.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carracedo A., Weiss D., Leliaert A.K., Bhasin M., de Boer V.C., Laurent G., Adams A.C., Sundvall M., Song S.J., Ito K., et al. . 2012. A metabolic prosurvival role for PML in breast cancer. J. Clin. Invest. 122:3088–3100. 10.1172/JCI62129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y.H., Bernardi R., Pandolfi P.P., and Benveniste E.N.. 2006. The promyelocytic leukemia protein functions as a negative regulator of IFN-gamma signaling. Proc. Natl. Acad. Sci. USA. 103:18715–18720. 10.1073/pnas.0604800103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desaint S., Luriau S., Aude J.C., Rousselet G., and Toledano M.B.. 2004. Mammalian antioxidant defenses are not inducible by H2O2. J. Biol. Chem. 279:31157–31163. 10.1074/jbc.M401888200 [DOI] [PubMed] [Google Scholar]
- de Stanchina E., Querido E., Narita M., Davuluri R.V., Pandolfi P.P., Ferbeyre G., and Lowe S.W.. 2004. PML is a direct p53 target that modulates p53 effector functions. Mol. Cell. 13:523–535. 10.1016/S1097-2765(04)00062-0 [DOI] [PubMed] [Google Scholar]
- Fogal V., Gostissa M., Sandy P., Zacchi P., Sternsdorf T., Jensen K., Pandolfi P.P., Will H., Schneider C., and Del Sal G.. 2000. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 19:6185–6195. 10.1093/emboj/19.22.6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambino V., De Michele G., Venezia O., Migliaccio P., Dall’Olio V., Bernard L., Minardi S.P., Della Fazia M.A., Bartoli D., Servillo G., et al. . 2013. Oxidative stress activates a specific p53 transcriptional response that regulates cellular senescence and aging. Aging Cell. 12:435–445. 10.1111/acel.12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia P.B., and Attardi L.D.. 2014. Illuminating p53 function in cancer with genetically engineered mouse models. Semin. Cell Dev. Biol. 27:74–85. 10.1016/j.semcdb.2013.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A., Salomoni P., Luo J., Shih A., Zhong S., Gu W., and Pandolfi P.P.. 2000. The function of PML in p53-dependent apoptosis. Nat. Cell Biol. 2:730–736. 10.1038/35036365 [DOI] [PubMed] [Google Scholar]
- Guo S., Cheng X., Lim J.H., Liu Y., and Kao H.Y.. 2014. Control of antioxidative response by the tumor suppressor protein PML through regulating Nrf2 activity. Mol. Biol. Cell. 25:2485–2498. 10.1091/mbc.E13-11-0692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanschitz L., Takahashi Y., Jollivet F., Ayrault O., Le Bras M., and de Thé H.. 2015. PML IV/ARF interaction enhances p53 SUMO-1 conjugation, activation, and senescence. Proc. Natl. Acad. Sci. USA. 112:14278–14283. 10.1073/pnas.1507540112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanne M., Lallemand-Breitenbach V., Ferhi O., Koken M., Le Bras M., Duffort S., Peres L., Berthier C., Soilihi H., Raught B., and de Thé H.. 2010. PML/RARA oxidation and arsenic binding initiate the antileukemia response of As2O3. Cancer Cell. 18:88–98. 10.1016/j.ccr.2010.06.003 [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., and de Thé H.. 2010. PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2:a000661 10.1101/cshperspect.a000661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., Zhu J., Puvion F., Koken M., Honoré N., Doubeikovsky A., Duprez E., Pandolfi P.P., Puvion E., Freemont P., and de Thé H.. 2001. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 193:1361–1372. 10.1084/jem.193.12.1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., and de Thé H.. 2008. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 10:547–555. 10.1038/ncb1717 [DOI] [PubMed] [Google Scholar]
- Malloy M.T., McIntosh D.J., Walters T.S., Flores A., Goodwin J.S., and Arinze I.J.. 2013. Trafficking of the transcription factor Nrf2 to promyelocytic leukemia-nuclear bodies: implications for degradation of NRF2 in the nucleus. J. Biol. Chem. 288:14569–14583. 10.1074/jbc.M112.437392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu A., Maraver A., Klatt P., Flores I., Garcia-Cao I., Borras C., Flores J.M., Viña J., Blasco M.A., and Serrano M.. 2007. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 448:375–379. 10.1038/nature05949 [DOI] [PubMed] [Google Scholar]
- Pearson M., Carbone R., Sebastiani C., Cioce M., Fagioli M., Saito S., Higashimoto Y., Appella E., Minucci S., Pandolfi P.P., and Pelicci P.G.. 2000. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature. 406:207–210. 10.1038/35021000 [DOI] [PubMed] [Google Scholar]
- Ribet D., Lallemand-Breitenbach V., Ferhi O., Nahori M.A., Varet H., de Thé H., and Cossart P.. 2017. Promyelocytic leukemia protein (PML) controls Listeria monocytogenes infection. MBio. 8:e02179-16 10.1128/mBio.02179-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokudai S., Laptenko O., Arnal S.M., Taya Y., Kitabayashi I., and Prives C.. 2013. MOZ increases p53 acetylation and premature senescence through its complex formation with PML. Proc. Natl. Acad. Sci. USA. 110:3895–3900. 10.1073/pnas.1300490110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablina A.A., Budanov A.V., Ilyinskaya G.V., Agapova L.S., Kravchenko J.E., and Chumakov P.M.. 2005. The antioxidant function of the p53 tumor suppressor. Nat. Med. 11:1306–1313. 10.1038/nm1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., de Thé H., and Lallemand-Breitenbach V.. 2014a PML nuclear bodies: assembly and oxidative stress-sensitive sumoylation. Nucleus. 5:499–507. 10.4161/19491034.2014.970104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Ferhi O., Jeanne M., Benhenda S., Berthier C., Jollivet F., Niwa-Kawakita M., Faklaris O., Setterblad N., de Thé H., and Lallemand-Breitenbach V.. 2014b Oxidative stress-induced assembly of PML nuclear bodies controls sumoylation of partner proteins. J. Cell Biol. 204:931–945. 10.1083/jcb.201305148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaglioni P.P., Rabellino A., Yung T.M., Bernardi R., Choi S., Konstantinidou G., Nardella C., Cheng K., and Pandolfi P.P.. 2012. Translation-dependent mechanisms lead to PML upregulation and mediate oncogenic K-RAS-induced cellular senescence. EMBO Mol. Med. 4:594–602. 10.1002/emmm.201200233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M.S., Salmena L., Carracedo A., Egia A., Lo-Coco F., Teruya-Feldstein J., and Pandolfi P.P.. 2008. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 455:813–817. 10.1038/nature07290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman L.C., Alimonti A., Scaglioni P.P., Koutcher J.A., Cordon-Cardo C., and Pandolfi P.P.. 2006. Identification of a tumour suppressor network opposing nuclear Akt function. Nature. 441:523–527. 10.1038/nature04809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood P.A., Amendt B.A., Rhead W.J., Millington D.S., Inoue F., and Armstrong D.. 1989. Short-chain acyl-coenzyme A dehydrogenase deficiency in mice. Pediatr. Res. 25:38–43. 10.1203/00006450-198901000-00010 [DOI] [PubMed] [Google Scholar]
- Woolbright B.L., and Jaeschke H.. 2017. Role of the inflammasome in acetaminophen-induced liver injury and acute liver failure. J. Hepatol. 66:836–848. 10.1016/j.jhep.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Hu P., Ye T.Z., Stan R., Ellis N.A., and Pandolfi P.P.. 1999. A role for PML and the nuclear body in genomic stability. Oncogene. 18:7941–7947. 10.1038/sj.onc.1203367 [DOI] [PubMed] [Google Scholar]
- Zhu J., Koken M.H.M., Quignon F., Chelbi-Alix M.K., Degos L., Wang Z.Y., Chen Z., and de Thé H.. 1997. Arsenic-induced PML targeting onto nuclear bodies: implications for the treatment of acute promyelocytic leukemia. Proc. Natl. Acad. Sci. USA. 94:3978–3983. 10.1073/pnas.94.8.3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.