A major obstacle to an HIV cure in adult infection is a viral reservoir largely composed of CTL escape mutants. Leitman et al. demonstrate that in children, but not adults, escape variant–specific CTLs are generated that can successfully “corner” the virus.
Abstract
Recent studies have suggested greater HIV cure potential among infected children than adults. A major obstacle to HIV eradication in adults is that the viral reservoir is largely comprised of HIV-specific cytotoxic T lymphocyte (CTL) escape variants. We here evaluate the potential for CTL in HIV-infected slow-progressor children to play an effective role in “shock-and-kill” cure strategies. Two distinct subgroups of children were identified on the basis of viral load. Unexpectedly, in both groups, as in adults, HIV-specific CTL drove the selection of escape variants across a range of epitopes within the first weeks of infection. However, in HIV-infected children, but not adults, de novo autologous variant-specific CTL responses were generated, enabling the pediatric immune system to “corner” the virus. Thus, even when escape variants are selected in early infection, the capacity in children to generate variant-specific anti-HIV CTL responses maintains the potential for CTL to contribute to effective shock-and-kill cure strategies in pediatric HIV infection.
Introduction
In the absence of antiretroviral therapy (ART), children progress more rapidly to HIV disease than adults. In ART-naive adults, the median time to AIDS is 10 yr and to death 11 yr (Babiker et al., 2000), whereas in perinatally infected children, the median time to AIDS is 1 yr, and >50% HIV-infected ART-naive children die by 2 yr of age (Goulder et al., 2016). However, ∼5–10% of ART-naive HIV-infected children remain healthy and maintain normal-for-age CD4 counts throughout childhood (i.e., the first decade of life; Blanche et al., 1997; Paul et al., 2005; Mphatswe et al., 2007; Muenchhoff et al., 2016). In contrast to “elite controller” adults (Lefrère et al., 1999; Lambotte et al., 2005), in whom nonprogression is characterized by undetectable viremia (<50 HIV copies/ml) and the expression of “protective” HLA molecules such as HLA-B*57, HLA-B*58:01, and HLA-B*81:01, pediatric slow progressors (PSPs) typically have viral loads of ∼30,000 HIV copies/ml and generally do not express HLA class I molecules that are protective against adult disease progression (Adland et al., 2015; Muenchhoff et al., 2016). These children thus share some features of nonpathogenic SIV infection observed in natural nonhuman primate hosts such as the sooty mangabey, that is characterized by preservation of normal peripheral CD4 counts in the face of persistently high viral loads (Rey-Cuillé et al., 1998). In addition, the localization of the virus predominantly in the short-lived memory CD4+ T cells is a second aspect of immunodeficiency virus infection that is shared by HIV-infected pediatric nonprogressors and SIV-infected sooty mangabeys (Paiardini et al., 2011; Muenchhoff et al., 2016). In combination with the tolerogenic immune environment in early life that limits the establishment of the viral reservoir, these features together suggest a greater potential for HIV cure in children than adults (Goulder et al., 2016).
To further investigate HIV cure potential among PSPs, defined here as perinatally infected children who did not meet prevailing clinical (World Health Organization [WHO] stage III/IV clinical disease) or CD4 (maintained CD4 >350 cells/mm3) criteria for ART initiation by the age of 5 yr, we studied HIV-specific CD8+ T cell activity among these children, because, according to “shock-and-kill” strategies proposed (Deeks, 2012), these cells would play a critical role in clearance of HIV-infected cells after latency reversal in ART-treated individuals (Deng et al., 2015; Jones and Walker, 2016; Margolis et al., 2016). We hypothesized that the nature of the viral reservoir may differ between children and adults (Goulder et al., 2016), in that multiple escape mutants are rapidly selected early in the course of adult infection, and these escape viruses constitute the major part of the adult viral reservoir (Deng et al., 2015). In contrast, although HIV-specific CD8+ T cell responses are detectable early in life, viral setpoint is not established in infected children until 5 yr of age, even in those maintaining normal-for-age CD4 counts (Muenchhoff et al., 2016). Furthermore, studies of early pediatric progression have failed to demonstrate the selection of escape mutants, even when there are detectable CTL responses restricted by HLA alleles that are “protective” in adult infection, such as HLA-B*57/58:01/81:01 (Adland et al., 2015). This prompts the hypothesis that HIV-specific CD8+ T cells do not impose significant selection pressure on HIV in early life sufficient for the widespread selection of escape mutants and, therefore, that viral reservoirs in infected children on ART are populated largely by wild-type virus. However, studies of CD8+ T cell escape have not been undertaken to date in PSPs, with access to maternal samples and longitudinal pediatric samples in combination with next-generation sequencing, that together allow this issue to be interrogated.
To examine the impact of HIV-specific CD8+ T cell responses on circulating virus in PSPs, we determined the HIV-specific CD8+ T cell responses and ultra-deep sequenced virus within longitudinal samples from 11 PSPs from a historical cohort of perinatally infected children and their mothers followed from birth through the first decade of life, recruited in 2003–2005 to the Pediatric Early HAART and Strategic Treatment Interruption Study (Mphatswe et al., 2007; Prendergast et al., 2008). We hypothesized, based on previous studies in adults (Kiepiela et al., 2007), that the breadth, specificity and function, reflected by the selection of virus escape mutants, of HIV-specific CD8+ T cell responses would differ according to viral load in the PSPs. We report here that two distinct HIV-specific CD8+ T cell immune strategies can be discerned among PSPs: one is similar to that adopted by adult elite controllers and viremic controllers (VCs), and the other is reminiscent of that described among adult viremic non-controllers (VNCs; Kiepiela et al., 2007).
Results
PSPs categorized into high- and low-viremic groups
Of 75 perinatally infected South African infants followed longitudinally from birth (Mphatswe et al., 2007; Prendergast et al., 2008), before universal ART recommendation for pediatric HIV infection, 11 children had failed to meet prevailing clinical (WHO stage III/IV clinical disease) or CD4 criteria (absolute CD4 count of <350 cells/mm3) for ART initiation by 5 yr of age. These children we define here as PSPs (Table 1).
Table 1. Clinical characteristics of the PSPs.
| PID | DOB | Trans mission | Sex | C or M | HLA-A | HLA-A | HLA-B | HLA-B | HLA-C | HLA-C | Early ART duration | Early ART start (age) | Early ART stop (age) | ART (re)start age | Type of slow progression |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| yr | yr | yr | yr | ||||||||||||
| 133-C | 11/2003 | IU | f | C | 68:01 | 74:01 | 58:02 | 81:01 | 04:01 | 06:02 | none | n/a | n/a | left study at 9 yr, no ART | VC group |
| M | 03:01 | 68:01 | 42:01 | 58:02 | 06:02 | 12:04 | |||||||||
| 468-C | 12/2004 | IU | m | C | 02:02 | 68:01 | 57:03 | 58:01 | 06:02 | 07:01 | 1.5 | 0.1 | 1.6 | 6.1 | |
| M | 34:02 | 68:01 | 44:03 | 58:01 | 04:01 | 06:02 | |||||||||
| 349-C | 11/2004 | IU | f | C | 23:01 | 43:01 | 45:01 | 58:02 | 06:02 | 06:02 | none | n/a | n/a | 9 | |
| M | 23:01 | 30:02 | 45:01 | 57:02 | 06:02 | 07:01 | |||||||||
| 364-C | 09/2004 | IU | f | C | 29:02 | 30:02 | 41:02 | 44:03 | 07:01 | 17:01 | 1.5 | 0.2 | 1.7 | 8 | |
| M | 03:01 | 29:02 | 35:01 | 44:03 | 04:01 | 07:01 | |||||||||
| 385-C | 01/2005 | IP | f | C | 30:01 | 34:02 | 15:03 | 58:02 | 02:10 | 06:02 | 1.3 | 0.3 | 1.6 | left study at 8 yr, no ART | |
| M | 30:01 | 43:01 | 15:03 | 58:02 | 02:02 | 06:02 | |||||||||
| 517-C | 01/2005 | IP | m | C | 30:02 | 68:02 | 15:10 | 42:01 | 03:04 | 17:01 | none | n/a | n/a | never | VNC group |
| M | 30:02 | 30:04 | 42:01 | 58:02 | 06:02 | 17:01 | |||||||||
| 021-C | 09/2005 | IU | f | C | 02:05 | 29:02 | 42:01 | 44:03 | 02:10 | 17:01 | 1.0 | 0.1 | 1.1 | never | |
| M | 02:05 | 33:01 | 42:01 | 44:03 | 02:10 | 17:01 | |||||||||
| 114-C | 09/2003 | IP | f | C | 03:01 | 24:02 | 07:02 | 08:01 | 07:01 | 07:02 | 1.0 | 0.1 | 1.1 | left study at 7.3 yr, no ART | |
| M | 03:01 | 68:02 | 08:01 | 57:02 | 07:01 | 18:01 | |||||||||
| 559-C | 02/2005 | IU | m | C | 23:01 | 68:02 | 08:01 | 58:01 | 07:01 | 07:01 | 1.0 | 0.1 | 1.1 | 5.2 | |
| M | 23:01 | 30:02 | 08:01 | 58:02 | 06:02 | 07:01 | |||||||||
| 380-C | 10/2004 | IU | m | C | 23:01 | 68:02 | 15:10 | 15:10 | 03:04 | 08:01 | 1.1 | 0.1 | 1.2 | 7.6 | |
| M | 02:00 | 23:01 | 07:02 | 15:10 | 03:04 | 04:01 | |||||||||
| 081-C | 08/2003 | IU | f | C | 29:02 | 68:02 | 15:16 | 44:03 | 03:04 | 07:01 | 1.4 | 0.1 | 1.5 | 6.7 | |
| M | 29:02 | 30:01 | 42:02 | 44:03 | 07:01 | 17:01 |
For each subject, child’s HLA type is shown in the first row and maternal HLA is in the second row. C, child; DOB, date of birth; f, female; IP, intrapartum infection; IU, in utero infection; m, male; M, mother; n/a, not applicable; PID, participant identification.
Hypothesizing that, as in adults, the HIV-specific CD8+ T cell responses might differ with respect to specificity, breadth, and function, PSPs were subdivided into two subgroups according to the median viral load: one with persistently higher and one with persistently lower viral loads (Fig. 1 A). These we have termed VCs and VNCs. Although the viral loads overall were 0.87 log10 lower in the VNC group (P = 5 × 10−14), there was no significant difference between the groups in either absolute CD4 count or CD4% (Fig. 1, B and C), which remained healthy (median 735 cells/mm3 and 29.5%) and very similar in both groups, after the natural CD4 and CD4% decline over the first 5 yr that is observed in healthy uninfected children (Shearer et al., 2003; Lugada et al., 2004).
Figure 1.
Slow-progressor children categorized according to viremia into VC and VNC groups. (A) Viral loads for the 11 PSPs in the first decade of life for all time points off ART. (B and C) Absolute CD4 counts and CD4% for the 11 PSPs for all time points off ART. Each individual is shown in a unique symbol and linking line, colored according to VC (blue) or VNC (red) grouping. An estimate of best fit (LOWESS lines) is shown for each PSP group in a thick solid line and the corresponding 95% confidence interval is denoted by the blue or red shaded area. Likelihood-ratio test p-values are from linear mixed-effects modeling comparing PSP groups and take into account all time points.
Broad, high-magnitude non–Gag-specific CD8 responses among VNC PSPs in early life
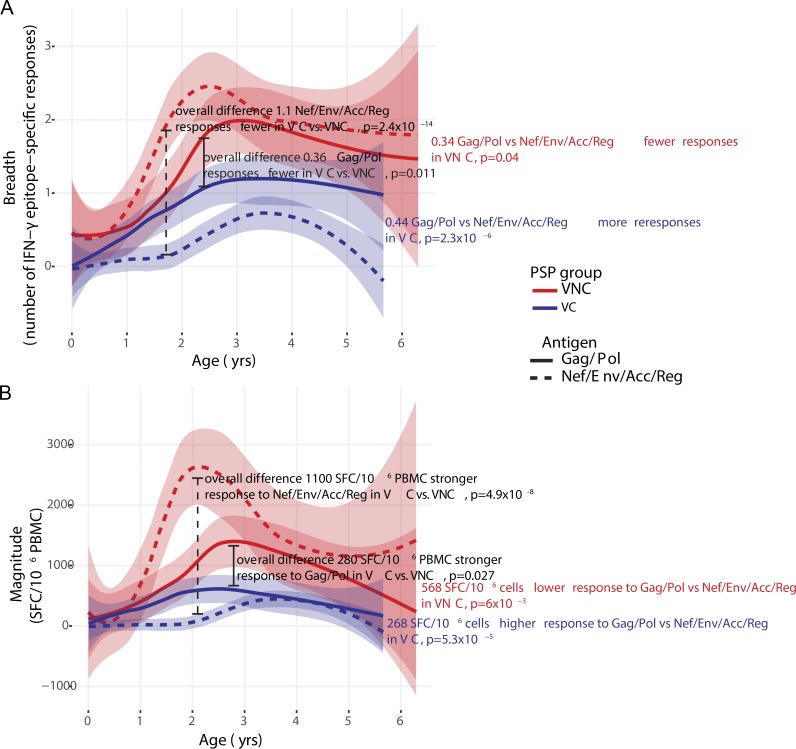
Specificity and breadth of the HIV-specific CD8+ T cell response has been related to viral load in previous studies of adult HIV infection, with broad Gag-specific responses associated with lower viral loads, and broad Env-, Nef- and accessory-regulatory protein (Vif/Vpr/Vpu/Tat/Rev)–specific responses associated with higher viral loads (Klein et al., 1995; Riviére et al., 1995; Ogg et al., 1998; Barouch et al., 2002, 2003; Edwards et al., 2002; Novitsky et al., 2003; Masemola et al., 2004; Zuñiga et al., 2006; Geldmacher et al., 2007; Kiepiela et al., 2007; Streeck et al., 2007; Janes et al., 2013). We therefore next investigated whether differential CD8+ T cell activity might explain the distinct viral loads among the two groups of PSPs. Using a panel of 410 overlapping 18-mer peptides spanning the clade C proteome, we determined in IFN-γ ELISPOT assays which individual peptides were recognized (Addo et al., 2003; Kiepiela et al., 2004). In both groups, HIV-specific CD8+ T cell responses significantly increased with age in breadth and magnitude (Fig. 2, A and B). However, total breadth was 1.5 responses higher in VNC than in VC PSPs (P = 3 × 10−9; Fig. 2 A). This difference in breadth was most significant in Nef-, Acc-Reg-, and Env-specific responses (P = 4 × 10−10, 4 × 10−5, and 4 × 10−4, respectively), but not in Gag (P = 0.07; Fig. 2 A). A similar pattern was observed for the magnitude of CD8+ T cell responses (Fig. 2 B). Overall, breadth and magnitude of non–Gag/Pol-specific responses were higher than breadth and magnitude of Gag/Pol-specific responses in the VNC children, whereas the reverse pattern was observed in the VC children (Fig. 3). These pediatric data are therefore consistent with adult studies indicating increased viral loads associated with non–Gag-specific responses, particularly those directed toward Env or Nef.
Figure 2.
Breadth and magnitude of HIV-specific IFN-γ ELISPOT responses among PSPs grouped according to antigen. (A) Breadth. (B) Magnitude. The magnitude of differences between five VC and six VNC children is shown in black text, and estimates of annual change shown in blue and red text, respectively. RTVVV, Rev/Tat/Vif/Vpu/Vpr/Tat/Rev. An estimate of best fit (LOWESS lines) is shown for each PSP group in a thick solid line, and the corresponding 95% confidence interval is denoted by the blue or red shaded area. P-values were determined by linear mixed-effects modeling that take into account all time points. Each ELISPOT measurement is the sum of two to three technical replicates, depending on cell availability.
Figure 3.
Gag/Pol versus Nef/Env/Accessory-Regulatory protein-specific IFN-γ ELISPOT responses in VC compared with VNC children. (A) Breadth. (B) Magnitude. Magnitude of differences between five VC and six VNC children is as shown on each panel, in black text. Magnitude of differences between Gag/Pol and Nef/Env/Acc/Reg targeted responses is shown in red (VNC) or blue (VC) bold text. An estimate of best fit (LOWESS lines) is shown for each PSP group and antigen group in a thick solid line (Gag/Pol) or thick dotted line (Nef/Env/Acc/Reg). Corresponding 95% confidence intervals for best fit lines are denoted by the blue or red shaded areas. For each of the 11 individuals the sum of the breadth, or magnitude of responses to the combination of antigens (Gag/Pol or Nef/Env/Acc/Reg) was calculated at each time point, and p-values were determined by linear mixed-effects modeling that take into account all time points. SFC, spot-forming cell.
Selection pressure in VC and VNC children targeting Gag-TL9
To investigate the impact on the virus of HIV-specific CD8+ T cells in these two distinct groups of PSPs, we next tracked the HIV-specific CD8+ T cell responses and kinetics of selection pressure imposed on HIV from birth through childhood in each child also having access to samples from the transmitting mother. To maximize the resolution of adaptive sequence changes, we deep sequenced the virus. Phylogenetic analysis of full-length consensus sequences for all 11 mother–child pairs confirmed the close relationship between virus in each mother–child pair (Fig. S1). In each child, we analyzed the regions of the virus encoding the CD8+ T cell epitopes restricted by any of the six HLA alleles expressed by that child using a panel of well-characterized epitopes (Goulder et al., 1997, 2000, 2001a; Honeyborne et al., 2006; Matthews et al., 2008, 2011, 2012; Ngumbela et al., 2008; Kløverpris et al., 2012a,b, 2013, 2014a,b; Llano et al., 2013) representing all the HIV proteins (see Materials and methods). For this analysis, variants shared with the mother expressing the same HLA class I molecule as the child were presumed to have been transmitted and not to have arisen de novo in the child. If the variant arose after the initial time point in the child, it was assumed that this variant was selected in the child and was not transmitted.
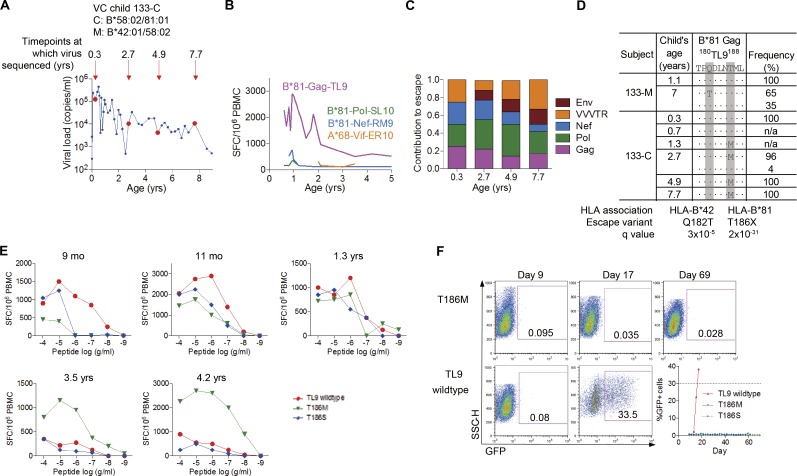
We first focused on the specific CD8+ T cell responses and viral sequence changes in three children, all of whom targeted the same immunodominant Gag-TL9 epitope (TPQDLNTML, Gag residues 180–188). These three pediatric subjects comprised one VC child, 133-C, and two VNC children, 517-C and 021-C.
The VC slow progressor 133-C made a dominant HLA-B*81:01–restricted response from the first time point tested (Figs. 4, A–C). The virus transmitted to this child encoded the wild-type TL9 epitope, and a T186M variant was selected that had almost reached fixation (96% frequency) by 2.7 yr of age (Fig. 4 D). Population sequencing showed that the T186M mutant arose between 0.7 and 1.3 yr (Thobakgale et al., 2009). A T186M variant–specific CD8+ T cell response was detectable by 1.3 yr of age and ultimately dominated the TL9-specific response (Fig. 4 E). The more common HLA-B*81:01-associated T186S mutation has previously been shown to substantially reduce viral replicative capacity (VRC; Wright et al., 2010), and analyses here of the T186M mutant demonstrated the same result (Fig. 4 F). Thus, the VC child 133-C achieved successful control of HIV (viral load of 520 copies/ml when leaving the study to relocate at 9 yr of age) via (1) an early, strong Gag-TL9–specific response driving the selection of an escape mutant that abolished viral growth, at least in vitro; and (2) the generation of a strong TL9-T186M variant–specific response.
Figure 4.
Escape in the immunodominant Gag-TL9 epitope develops in the first months of life in the VC child 133-C. (A) Virological profile of the child 133-C with deep-sequenced time points indicated by large circles and child’s age (years) indicated above the arrows. (B) ELISPOT CD8+ T cell responses in the VC child 133-C. (C) Contribution of different HIV proteins to HLA-associated escape across HIV proteome. VVVTR, Vif/Vpr/Vpu/Tat/Rev in 133-C. (D) Development of escape in Gag-TL9 epitope in the child 133-C. Q-values as per Carlson et al. (2012) and Carlson et al. (2014). (E) CD8+ T cell response to the Gag-TL9 wild-type epitope and its variants at five different time points in the child133-C. Responses were determined by IFN-γ ELISPOT assay. Assays repeated in triplicate. (F) Growth of viruses encoding TL9 wild-type and mutants T186M and T186S. Dotted line in the plot with viral growth curves indicates the threshold of 30% cells being infected at which time virus should be harvested. Each virus grown in triplicate; confirmed in two independent experiments. SFC, spot-forming cell.
In addition to the Gag-TL9–mediated selection pressure on the virus during the first months of life in 133-C, there was also evidence of broad, CD8+ T cell–driven escape selection (Table 2), including within Gag. The epitope Gag-SK10 (SILRGGKLDK, Gag residues 6–15) is restricted by HLA-A*74:01 (Matthews et al., 2011), an allele not shared with the mother 133-M. In previous studies of C clade–infected adults, the characteristic HLA-A*74:01–associated escape mutant is Gag K12N (at P7 in the epitope); the association between selection of K12N and expression of HLA-A*74:01 carries a q value of 6 × 10−17 (Carlson et al., 2014). The virus transmitted by the HLA-A*74:01–negative mother, as expected, encoded the wild-type HLA-A*74:01-Gag-SK10 epitope, and K12N was selected in 100% of sequences in the HLA-A*74:01–positive child by 3.6 mo of age. Similarly, in two HLA-B*81:01–restricted epitopes (HLA-B*81:01 was also not expressed in the mother 133-M), in Pol-SL10 (SPIETVPVKL, Pol residues 158–167) and Nef-RM9 (RPQVPLRPM, Nef residues 71–79), 100% of transmitted sequences encoded wild-type epitopes, yet escape mutations were selected in 133-C by 3.6 mo of age. These data demonstrate that CD8+ T cell responses in the VC slow progressor 133-C exerted selection pressure on the virus within the first months of life in Gag and non-Gag epitopes. In addition, variant-specific Gag-TL9 responses were generated, likely in the first year of life, effectively negating the benefit to the virus of the original escape mutant.
Table 2. HLA-associated mutations in the VC child 133-C that developed within the first months of life.
| Subject | Child’s age | A*74 Gag 6SK1015 | Frequency | B*81 Pol 158SL10167 | Frequency | A*74 Pol 423SR10432 | Frequency | B*81 Nef 71RM979 | Frequency |
|---|---|---|---|---|---|---|---|---|---|
| SILRGGKLDK | SPIETVPVKL | SQIYPGIKVR | RPQVPLRPM | ||||||
| yr | |||||||||
| 133-M | 1.1 | ·····E···· | 100% | nd | nd | nd | |||
| 7 | ·V···E···· | 100% | ·········· | 100% | ·······R·· | 81% | ········· | 100% | |
| ······VR·· | 13% | ||||||||
| ·····R·R·· | 6% | ||||||||
| 133-C | 0.3 | ·····EN··· | 100% | ·········· | 65% | ·····R·R·· | 100% | ·····V··· | 60% |
| ···D······ | 32% | ········· | 32% | ||||||
| ····P····· | 2% | ···A····· | 6% | ||||||
| ···K······ | 1% | K········ | 2% | ||||||
| 2.7 | ·····EN··· | 75% | ·S········ | 92% | ·······RIK | 55% | ·····I··· | 54% | |
| ·V···EN··· | 24% | ···K······ | 5% | ·······R·K | 42% | ·····V··· | 36% | ||
| ·········· | 3% | ·····R·R·· | 3% | K····V··· | 8% | ||||
| ········· | 2% | ||||||||
| 4.9 | ·V···EN··· | 99% | ·S········ | 100% | ·······RIK | 98.7% | ·····T··· | 89% | |
| ·······R·K | 1.3% | ·····V··· | 11% | ||||||
| 7.7 | ·····EN··· | 100% | ·S········ | 76% | ·····R·R·K | 81% | ·····V··· | 99% | |
| ·T········ | 24% | ·······RIK | 17% | ||||||
| ·······R·K | 2% | ||||||||
| HLA association | HLA-A*74 | HLA-B*81 | HLA-A*74 | HLA-B*81 | |||||
| HLA-B*81 | |||||||||
| Escape variant | K12N | P189S | R432K | L76X | |||||
| E191X | |||||||||
| q value | 6 × 10−17 | 3 × 10−39 | 7 × 10−3 | 2 × 10−38 | |||||
| 3 × 10−4 |
Mother 133-M: A*74:01−, B*81:01−. VC child 133-C: A*74:01+, B*81:01+. Footprints associated with the HLA class I allele are indicated below the table; known escape variants and q values indicating significance of an HLA-polymorphism association are shown (Carlson et al., 2014). 133-M indicates mother of the child 133-C. Frequency represents percentage at which a particular haplotype is present at the indicated time point.
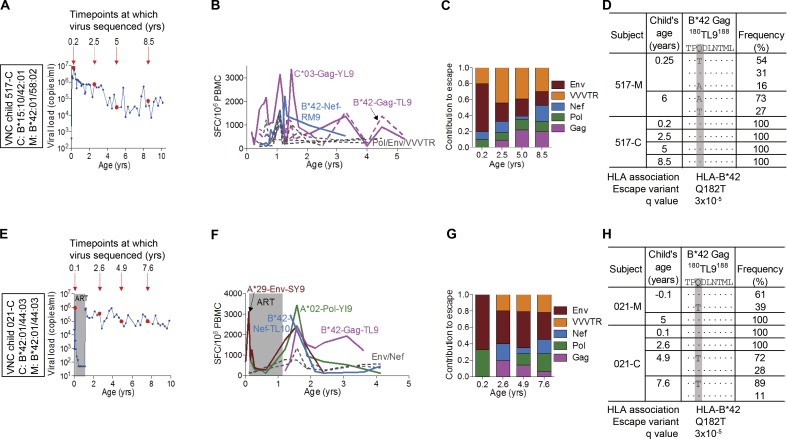
In both VNC slow progressors, 517-C and 021-C, the Gag-TL9 responses were restricted by HLA-B*42:01 as opposed to HLA-B*81:01, arose later in the course of infection (Fig. 5), and did not become immunodominant for several years. In 517-C, the dominant response initially was to the HLA-C*03–restricted Gag-YL9 epitope (YVDRFFKTL, Gag 286–304). Early Gag escape mutants were not observed in 517-C, and no Gag-TL9 escape mutants were observed at any time point studied through 8.5 yr (Fig. 5 D and Tables 3, S1, and S2). However, a broad array of non-Gag escape mutants were selected to fixation within 2.4 mo of birth (517-C was infected intrapartum and only became viremic at 28 d), for example within the HLA-B*15:10–restricted Rev epitope IL9 (IHSISERIL, Rev residues 52–60; Table 3). HLA-B*15:10 was not expressed in the mother 517-M and, as expected, the maternal virus transmitted encoded the wild-type Rev-IL9 epitope. Other epitopes in which escape to fixation appeared to have occurred by 2.4 mo of age in 517-C were in two HLA-B*42:01–restricted Env epitopes, RI10 (RPNNNTRKSI, Env residues 298–307) and IF9 (IPRRIRQGF, Env residues 843–851), and in two HLA-A*30:02–restricted Env epitopes, KY9 (KYLGSLVQY, Env residues 794–802) and IY9 (IVNRVRQGY, Env residues 704–712). In all four cases, the epitope mutant in the child’s viral population differed in 100% of viral sequences from the maternal sequence. The caveat here would be that both mother and child express HLA-A*30:02 and HLA-B*42:01 and that the samples sequenced were of plasma virus 2–3 mo after transmission. However, particularly in view of the clear-cut evidence of early escape within the HLA-B*15:10 Rev IL9 epitope, it would seem very likely that early escape is driven to fixation in these four Env epitopes, with additional evidence of early escape in almost 50% of sequences within the HLA-A*30:02–restricted Pol epitope AY9 (AQNPEIVIY, Pol residues 328–366) and in 60% of sequences within the HLA-B*15:10–restricted Vif epitope WI9 (WHLGHGVSI, Vif residues 79–87; Table 3).
Figure 5.
Selection pressure in Gag-TL9 epitope in the VNC children 517-C and 021-C. (A and E) Virological profiles of the children 517-C (A) and 021-C (E) with deep-sequenced time points indicated by large circles and child’s age (years) indicated above the arrows. (B and F) ELISPOT CD8+ T cell responses of the children 517-C (B) and 021-C (F). Assays repeated in triplicate. (C and G) Contribution of different HIV proteins to HLA-associated escape across HIV proteome in the children 517-C (C) and 021-C (G). VVVTR, Vif/Vpr/Vpu/Tat/Rev. (D and H) Sequences of the Gag-TL9 epitope in the first decade of life in 517-C (D) and 021-C (H). Q-values as per Carlson et al. (2012) and Carlson et al. (2014).
Table 3. Early escape in the VNC child 517-C develops in non-Gag epitopes.
| Subject | Child's age | C*03 Gag 296YL9304 | Frequencey | B*42 Nef 71RM979 | Frequency | A*30 Pol 328AY9366 | Frequency | A*30 Env 704IY9712 | Frequency | A*30 Env 794KY9802 | Frequency | B*42 Env 298RI10307 | Frequency | B*42 Env 843IF9851 | Frequency | B*15:10 Rev 52IL960 | Frequency | B*15:10 Vif 78WI987a | Frequency |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YVDRFFKTL | RPQVPLRPM | AQNPEIVIY | IVNRVRQGY | KYLGSLVQY | RPNNNTRKSI | IPRRIRQGF | IHSISERIL | DWHLGHGVSI | |||||||||||
| yr | |||||||||||||||||||
| 517-M | 0.25 | ········· | 100% | ········· | 100% | ········· | 100% | V·R······ | 68% | ········· | 100% | ·········· | 85% | L········ | 86% | ········· | 100% | ·········· | 100% |
| V·S······ | 22% | ·······R·· | 15% | L···V···· | 13% | ||||||||||||||
| V········ | 4% | ||||||||||||||||||
| I·S······ | 3% | ||||||||||||||||||
| 6 | ········· | 100% | ········· | 100% | ········· | 93% | V·R······ | 97% | ········· | 100% | ·········T | 55% | L········ | 36% | ········· | 100% | ·········· | 100% | |
| ·R······· | 4% | V·RK····· | 2% | ·········V | 29% | L···L···· | 29% | ||||||||||||
| ·K······· | 2% | ··S······V | 13% | L···F···· | 29% | ||||||||||||||
| ··S······I | 1.6% | L···V···· | 5% | ||||||||||||||||
| 517-C | 0.2 | ········· | 100% | ········· | 100% | ········· | 53% | ··K······ | 100% | ····N··L· | 100% | ·TG····R·· | 100% | ········L | 100% | ·RA······ | 94% | ·········· | 40% |
| ········C | 47% | ·RA·····I | 6% | ·······A·· | 36% | ||||||||||||||
| E········· | 18% | ||||||||||||||||||
| ··Q······· | 2% | ||||||||||||||||||
| ··L······· | 2% | ||||||||||||||||||
| ·······I·· | 1.1% | ||||||||||||||||||
| 2.5 | ········· | 98% | ········· | 100% | ········· | 88% | V·R······ | 98.6% | ········· | 91% | ··G····R·· | 51% | L········ | 100% | ·N······· | 76% | E········· | 61% | |
| ······R·· | 2% | ·····L··· | 12% | V·RK····· | 1.2% | ····N···· | 9% | ··G······· | 40% | ·RA······ | 12% | ·······A·· | 30% | ||||||
| ··S······· | 9% | ·H······· | 10% | E······A·· | 5% | ||||||||||||||
| LN······· | 1.3% | ·········· | 4% | ||||||||||||||||
| 5 | ········· | 58% | ········· | 100% | ·····L··· | 82% | V·R······ | 86% | ········· | 91% | ··G····R·· | 82% | L········ | 96% | ·N······· | 59% | E········· | 70% | |
| ·······V· | 39% | ········· | 16% | ··R······ | 13% | ····N···· | 9% | ··G····R·M | 14% | L···V···· | 3% | ·N·L····· | 17% | ·······A·· | 25% | ||||
| ·······A· | 3% | T····L··· | 2% | ··S····R·· | 2% | LN······· | 11% | E······A·· | 5% | ||||||||||
| ·H······· | 7% | ||||||||||||||||||
| ·RA······ | 4% | ||||||||||||||||||
| VN······· | 2% | ||||||||||||||||||
| 8.5 | ·······V· | 75% | ········· | 100% | ·····L··· | 55% | V·R······ | 97% | ········· | 98% | ··G····R·T | 69% | L········ | 49% | LN······· | 71% | E········· | 61% | |
| ········· | 22% | ········· | 41% | V·RK····· | 1.5% | ····N···· | 2% | ··G····R·· | 31% | L·T······ | 21% | ·N·L····· | 21% | E······A·· | 22% | ||||
| ······R·· | 1.6% | T····L··· | 2% | ··R······ | 1.2% | L···L···· | 17% | ·N······· | 7% | ·······A·· | 17% | ||||||||
| ·······A· | 1.5% | L···F···· | 14% | ||||||||||||||||
| HLA association | HLA-C*03 | HLA-B*42 | |||||||||||||||||
| Escape variant | Y303X | R71X | |||||||||||||||||
| q value | 4 × 10-18 |
Mother 517-M: A*30:02+ B*42+/B*15:10− C*03-ve. Child 517-C: A*30:02+ B*42+/B*15:10+ C*03+. Footprints associated with the HLA class I allele are indicated below the table; known escape variants and q values indicating significance of an HLA-polymorphism association are shown (Carlson et al., 2014). 517-M indicates mother of the child 517-C. Frequency represents percentage at which a particular haplotype is present at the indicated time point.
HLA-B*15:10–restricted Vif epitope is WI9 (WHLGHGVSI, residues 79–87; in bold), but HLA-B*15:10–associated polymorphisms are at the residue 85 (V85A) within the epitope and at the residue 78 (D78E) upstream of the epitope.
In the other VNC child, 021-C, escape in the Gag-TL9 epitope was observed, but only relatively late, with no epitope variants being evident at 2.6 yr (Fig. 5 H). Again, early escape within 2 mo of birth was evident in non-Gag epitopes. HLA-A*29:02 was not expressed in the mother (Table S2). This low-frequency variant was ultimately replaced by the characteristic HLA-A*29:02 escape footprint in this epitope (Carlson et al., 2012). Another instance of early escape within 2 mo of birth was evident again at low frequency within the HLA-A*02–restricted Pol-VL9 (VIYQYMDDL, Pol residues 334–342). HLA-A*02 was not shared with the mother (Table S2). However, this variant did not persist and reverted back to wild-type in 100% sequences. There were no other instances of early escape identified in this child’s viral sequences, although many escape mutants were transmitted, and in this mother–child pair, five of the six HLA class I molecules were shared.
Thus, in the VC child 133-C, there was evidence of strong selection pressure within the first 3 mo of life, with broad Gag-specific CD8+ T cell responses generated (to HLA-A*74-Gag-SK10 and HLA-B*81-Gag-TL9) and escape mutants within Gag, Pol, and Nef, all reaching fixation in that time. In contrast, although escape mutants were selected within the same time period in the two VNC children (517-C and 021-C), these were mostly in Env, and none were in Gag. These data are consistent with the notion of control of broad Gag-specific CD8+ T cell responses contributing to lower viremia in early pediatric infection, as previously shown in adult infection (Klein et al., 1995; Riviére et al., 1995; Ogg et al., 1998; Edwards et al., 2002; Novitsky et al., 2003; Masemola et al., 2004; Zuñiga et al., 2006; Geldmacher et al., 2007; Kiepiela et al., 2007; Streeck et al., 2007).
Preferential recognition of autologous Gag-TL9 variant in children, but not adults
The observation that preferential recognition of the autologous variant is more common in HIV-infected children than adults was previously made with respect to the HLA-B*57/58:01–restricted Gag-TW10 epitope (Feeney et al., 2005). To investigate whether this might apply more generally, we here investigated whether preferential recognition of the autologous Gag-TL9 variant is also observed more commonly in pediatric than adult infection. We identified 11 children and 16 adults from Southern Africa infected with C clade virus who expressed either HLA-B*42:01 and/or HLA-B*81:01 and in whom autologous virus encoded a Gag-TL9 variant (Table S3). In two of the HIV-infected children, population sequencing of autologous virus initially demonstrated the presence of a mixture of variants at Gag-TL9 variants at Gag-182 (position 3 in the epitope; Fig. 6, A and B), suggesting recent selection of escape (Goonetilleke et al., 2009). Preferential recognition of the single autologous Gag-TL9 variant that subsequently emerged was demonstrated in both of these two children (Fig. 6, A and B). Overall, in the 11 children studied, preferential recognition of the autologous Gag-TL9 variant was observed in all but one (91%), whereas in the 16 adults studied, preferential recognition of the autologous variant was observed in only 5 (31%; P = 0.0047; Fig. 6, C and D).
Figure 6.
Preferential recognition of autologous TL9 variants by HIV-infected children compared with adults. (A) Development of preferential recognition of autologous TL9-Q182G variant by pediatric subject K-004-C between age 8.2 yr, at which time autologous virus encoded a mix of Q182P/Q182G variants, and 12.8 yr, at which time autologous virus encoded only the Q182G variant, by population sequencing of virus. HLA type and autologous sequence of the mother K-004-M are unknown. ELISPOT assays repeated in triplicate. (B) Development of preferential recognition of autologous TL9-Q182G variant by pediatric subject K-044-C between age 4.5 yr, at which time autologous virus encoded a mix of Q182P/Q182T variants, and 8.7 yr, at which time autologous virus encoded only the Q182G variant, by population sequencing of virus. The mother (K-044-M) was HLA-B*42:01/81:01 negative and autologous virus when sequenced at child’s age 4.5 yr encoded wild-type TL9, suggesting that wild-type TL9 was transmitted to the child. ELISPOT assays repeated in triplicate. (C) The magnitude of the IFN-g ELISPOT response to wild-type TL9 is lower than to the autologous variant among children (n = 11; mean 531 vs. 1,010 spot-forming cells per million PBMCs; P = 0.02, paired t test; P = 0.002, Wilcoxon signed-rank test). Among adults, the IFN-γ ELISPOT response to wild-type TL9 tended to be higher than to the autologous TL9 variant, although this did not reach statistical significance (n = 16; median 1,597 vs, 882 spot-forming cells per million PBMCs; P = 0.25, paired t test; P = 0.40, Wilcoxon signed-rank test). (D) Frequency of subjects showing preferential recognition of autologous variant is significantly higher in pediatric subjects than adult subjects (10/11 = 91%; vs. 5/16 = 31%; P = 0.0047, Fisher’s exact test). SFC, spot-forming cell.
The caveats here (discussed further below) are that the transmitted virus in all of the adults and some of the children were unknown. However, in 3 of the children, an evolving preferential recognition of the autologous Gag-TL9 variant was demonstrated (Figs. 4 and 6), and in 4 more children, maternal virus encoded wild-type Gag-TL9 and/or the mother was HLA-B*42:01/81:01 negative. Even if one restricts the analysis to these 7 children, the observed preferential recognition of autologous Gag-TL9 variants remains significantly greater in children than adults (P = 0.0045, Fisher’s exact test).
Contrasting selection pressure in VC and VNC children
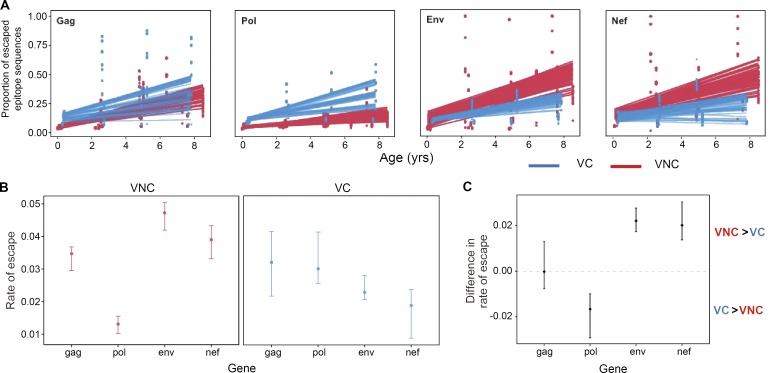
To examine selection pressure more broadly and compare the differences between VNC and VC children in the kinetics of accumulation of escape mutants, we calculated the proportion of epitopes within the most immunogenic proteins (Gag, Pol, Nef, and Env) restricted by the HLA alleles expressed in each subject that contained at least one nonsynonymous mutation for each time point. The rates of escape accumulation were calculated by linear regression modeling, and the statistical uncertainty of these rates was assessed by generating 50 bootstrapped datasets for each patient. This revealed that there was significantly greater selection for HLA-associated escape in Env and Nef and less selection for escape in Pol in the VNC group compared with VC children (Fig. 7, A and B). Gag-specific escape did not differ significantly between the two groups. The interquartile range (IQR) of the difference in the rate of escape between VNC and VC children (calculated by bootstrapping viral sequence data from each subject) was estimated in order to assess the statistical robustness of these patterns (Fig. 7 C). These analyses confirm the robustness of the specificity differences of CD8+ T cell selection pressure observed in favor of Nef and Env among VNC children and in favor of Pol among VC children.
Figure 7.
Differential patterns of CD8+ T cell–mediated escape in VC versus VNC children. (A) Mean proportion of escaped variants over time in CD8+ T cell-restricted epitopes in gag, pol, env, and nef for the VNC and VC children. The points represent the proportion of escaped variants at a particular CD8+ T cell–restricted epitope from 50 bootstrapped replicates of the patients’ sequence data. The lines show the bootstrap distribution of regression lines of the mean rate of escape variants accumulated over time for each gene in VNC and VC children. (B) Rate of escape across genes in VNC and VC children: the error bars depict the interquartile range of the mean rate of escaped variants accumulated in the CD8+ T cell-restricted epitopes over time for VNC and VC children. The points represent the median, whereas the top and bottom edges represent the upper and lower quartiles. This was estimated from the slopes of the linear regression model based for each bootstrap replicate (n = 50). In VNC children, the rate of escape in env and nef is significantly greater than in pol and to lesser extent than in gag. Conversely, the opposite pattern is observed in VC children, where the rate of escape is significantly greater in gag and pol than in env and nef. (C) Difference in the rate of escaped variants accumulating in the different gene regions over time in VNC and VC children. This was estimated by calculating the difference in the slopes of the linear regression for VNC and VC children for each bootstrap replicate (n = 50). The interquartile range of this statistic is plotted to indicate that VNC children have greater accumulation of escaped variants in env and nef and lower accumulation of escaped variants in pol than VC children. There is statistically no difference in the rate of escape in gag-specific restricted CD8+ T cell epitopes. (A–C) Linear regression model.
Collectively, these data indicate that CD8+ T cells among the slow-progressor children studied exert HLA-associated pressure on the virus from the very first weeks of infection, with unequivocal evidence of escape selection within the first months of life and escape mutants accumulating with age. The patterns of CD8+ T cell activity in the PSPs differ such that in the VNC group, CD8+ T cells preferentially target non-Gag/Pol epitopes and exert greater HLA-associated selection pressure in Nef/Env epitopes and vice versa in the VC children. These distinct patterns of CD8+ T cell activity, together with the observed differences in viral loads in the VC and VNC groups, are consistent with previous studies linking non-Gag/Pol specificity of CD8+ T cell responses with high viremia (Klein et al., 1995; Riviére et al., 1995; Ogg et al., 1998; Barouch et al., 2002, 2003; Edwards et al., 2002; Novitsky et al., 2003; Masemola et al., 2004; Zuñiga et al., 2006; Geldmacher et al., 2007; Kiepiela et al., 2007; Streeck et al., 2007; Janes et al., 2013), as discussed further below.
Discussion
In this study, we sought to investigate the potential for HIV-specific CD8+ T lymphocyte activity to play an effective role in viral eradication of pediatric HIV infection, because CD8+ T cells play a key role in shock-and-kill HIV cure strategies (Deeks, 2012), and because previous studies have indicated that the potential for HIV cure may be higher among infected children than adults (Goulder et al., 2016). Studying ART-naive PSPs followed through childhood, we hypothesized that the breadth, specificity, and function (reflected by the selection of CTL escape mutants) of HIV-specific CD8+ T cells would differ according to viremia. We divided these children into low and viremia groups (termed VCs and VNCs, respectively) that did not differ significantly by absolute CD4 count or CD4%. The HIV-specific CD8+ T cell responses in VCs were of lower breadth and magnitude than those of VNCs but focused predominantly on Gag and Pol specificities compared with Nef and other accessory/regulatory proteins (Nef/Acc-Reg) and Env, whereas the reverse applied among VNCs. Unexpectedly, in both groups, escape mutations were selected from early infancy; however, in VCs, the selection pressure on the virus focused more on Gag/Pol than on Nef/Acc-Reg/Env, and the reverse was the case in VNCs. Finally, preferential recognition of autologous epitope variants in children, but not adults, was shown with respect to Gag-TL9, the immunodominant epitope for approximately one third of HIV-infected individuals in sub-Saharan Africa, also demonstrating the potential for HIV-specific CTL in children to corner the virus.
This escape-cornering strategy has long been proposed as a theoretical immune tactic against HIV (O’Connor et al., 2001; Altfeld and Allen, 2006; Allen and Altfeld, 2008) and has been mathematically modeled (Ferguson et al., 2013). That such a strategy can be effective is demonstrated for the first time here. In the current study, we observe the HLA-B*81:01 TL9-Gag response first drives the selection of an escape variant such as T186M, which substantially reduces viral replicative capacity (Fig. 4); then, by the generation of a variant-specific CTL response, the virus is down an evolutionary cul-de-sac with apparently no further viable escape options. These findings suggest the potential for HIV-specific CD8+ T cells to play an effective role in shock-and-kill HIV-cure strategies among slow-progressor children, because after latency reversal, HIV-specific CD8+ T cells will be required to kill virus-infected cells expressing escape variants in both pediatric and adult infection (Deng et al., 2015).
However, the success of an escape-cornering strategy depends on the ability of the immune system to not only generate variant-specific immune responses as the virus escapes but also drive the selection of escape mutants that significantly diminish viral replicative capacity and limit further escape options. The ability of the immune system to generate sequential immune responses against viral variants and yet fail to corner HIV is especially well described in the context of neutralizing antibody activity against HIV, in which the virus is always one step ahead of contemporaneous antibody (Wei et al., 2003). Env is highly variable and sequence changes appear to have little impact on viral replicative capacity (Troyer et al., 2009). VNC children whose HIV-specific CD8+ T cell responses are not typically targeting conserved epitopes in Gag and Pol may therefore lack an escape-cornering facility, irrespective of their ability to generate variant-specific CTL responses. To induce such responses may require immunotherapeutic vaccination, ideally using a mosaic insert encoding the common escape variants (Fischer et al., 2007; Barouch et al., 2010), in those conserved epitopes (Hayton et al., 2014; Ondondo et al., 2016). A precedent for the effectiveness of such an approach in redirecting the specificity of the CTL response toward more conserved regions of the proteome has been described in the Phambili trial of the MRKAd5 Gag/Pol/Nef T cell vaccine, in which induction of higher Gag-specific responses among HLA-B*58:02-positive subjects who became HIV-infected was associated with lower viral loads than in the HLA-B*58:02-positive placebo controls who later became infected (Leitman et al., 2016). In natural HIV infection, HLA-B*58:02 is associated with dominant Env-specific CD8+ T cell responses and high viral loads (Ngumbela et al., 2008). Thus, eradication of the viral reservoir likely requires the ability of HIV-specific CD8+ T cell activity to both recognize autologous variants and operate effectively in killing virus-infected cells, which in part is related to specificity of the response.
The greater ability of infected children compared with adults to generate de novo variant-specific CD8+ T cell responses is described here in relation to the Gag-TL9 epitope that is immunodominant among HIV-individuals expressing HLA-B*42:01 or HLA-B*81:01 and some other HLA class I molecules in the B7 superfamily, such as HLA-B*39:10 (Leslie et al., 2006). These are highly prevalent in the sub-Saharan African populations worst affected by the HIV epidemic, and one or more of these HLA class I molecules are expressed in approximately one third of these individuals. We previously described the same ability of children and not adults to generate autologous variant-specific responses in a study of HLA-B*57/58:01–restricted Gag-TW10 responses, where 73% of children mounted de novo variant-specific responses compared with 9% of adults (P = 0.0004; Feeney et al., 2005). Thus, although the generation of de novo responses to variants is not unique to HIV-infected children (Allen et al., 2005), this phenomenon appears on the basis of this and the current study to be much more common among children. Caveats to this analysis, as mentioned above, include the fact that the viral sequence transmitted by the mother was not known in all cases; however, restricting the analysis to the children in whom either evolution of the variant-specific response could be shown via longitudinal analyses, or in whom maternal virus encoded wild-type Gag-TL9, the preferential recognition of the autologous variant remained significantly more common among children than adults (P = 0.0045). Furthermore, our finding that 5 of 16 (31%) adults had Gag-TL9 variant–specific responses may overestimate the number generating these responses subsequent to a wild-type–specific response, because it is likely that in a proportion of these adults, the virus transmitted encoded a Gag-TL9 variant. Although wild-type virus tends to be preferentially transmitted (Carlson et al., 2016), even when it is present at lower frequency than the escape variant, as illustrated in the current study (for example, Fig. 5 D), as many as 18% of HLA-B*42:01/81:01–negative individuals carry Gag-TL9 variants (Payne et al., 2014), presumably that were selected before transmission.
Why adults might be less able to mount variant-specific responses than children is unknown, but one may speculate that this difference is related to the respective numbers of naive and memory T cells in children and adults. In adults, it may be more likely that a memory CD8+ T cell response would cross-react weakly with a novel CD8+ T cell escape variant; hence, no de novo response would be initiated, giving rise to a situation akin to “original antigenic sin” (Klenerman and Zinkernagel, 1998) among adults. This phenomenon has been proposed to explain disease after dengue reinfection by a distinct subtype of virus from the virus that caused primary infection (Mongkolsapaya et al., 2003).
The observations made here of distinct CD8+ T cell specificities among VC and VNC children are consistent with several studies indicating that Gag- and, to a lesser extent, Pol-specific CD8+ T cell responses are more effective at controlling viremia than non-Gag/Pol specificities (Klein et al., 1995; Riviére et al., 1995; Ogg et al., 1998; Barouch et al., 2002, 2003; Edwards et al., 2002; Novitsky et al., 2003; Masemola et al., 2004; Zuñiga et al., 2006; Geldmacher et al., 2007; Kiepiela et al., 2007; Sacha et al., 2007a,b; Streeck et al., 2007; Kawashima et al., 2009, 2010; Payne et al., 2010; Goulder and Walker, 2012; Janes et al., 2013). The mechanisms for these protein-specific differences remain unclear but may be related to the ability of Gag and Pol epitopes in particular to be presented on the surface of infected cells before de novo HIV protein synthesis and concomitant HLA down-regulation (Sacha et al., 2007a,b) and substantially earlier than Nef or Env specificities. However, it is evident that factors other than protein specificity, including functional avidity and polyfunctionality (Almeida et al., 2007, 2009), also play an important role in the ability of a particular CD8+ T cell response to control HIV replication. Factors contributing to the differences in specificity of the HIV-specific CD8+ T cell responses in the VNC and VC groups of children are likely to include those shown to be important in adult infection, namely the HLA class I molecules expressed (Kiepiela et al., 2004) and the viral sequence transmitted (Goepfert et al., 2008; Carlson et al., 2016). These factors have previously been shown also to have a strong influence on specificity of the CD8+ T cell response in infected infants (Goulder et al., 2001b; Thobakgale et al., 2007). The impact of transmission of a virus preadapted to HLA in the child will be especially marked, because the mother shares at least half the class I molecules expressed in the child, and in such cases, the transmitted virus may be highly preadapted (Goulder et al., 2001b; Thobakgale et al., 2007; Goepfert et al., 2008; Carlson et al., 2016). Notwithstanding these differences in the protein specificity of the CD8+ T cell response and in the viral loads of the two subgroups of PSPs studied here, these were not reflected in CD4 count differences (Fig. 1), underlining the lack of a significant correlation typically observed between viral load and absolute CD4 count in pediatric infection (Ssewanyana et al., 2007; Muenchhoff et al., 2016).
The observation of high rates of CD8+ T cell escape in Nef and Env in the VNC pediatric subjects is consistent with the dominant Env- and Nef-specific CD8+ T cell responses detected. Previous studies in adults have shown that rate of escape is related to the magnitude of the response, its relative immunodominance compared with the other responses generated, and epitope entropy (Ferrari et al., 2011; Liu et al., 2013). The rate and location of escape mutations is to a surprisingly large degree predictable, independent of the CD8+ T cell response actually generated (Barton et al., 2016), based on knowledge of epitopes, the transmitted viral sequence, and entropy. Although the mean entropies of Gag and Pol proteins are substantially less than those of Env and Nef (Yusim et al., 2002), and much of the analysis here has focused on protein-specific CD8+ T cell responses and immune escape, it is important to note that there are also relatively conserved regions within Nef and Env and that the impact of escape mutations on viral replicative capacity may differ greatly between epitopes within the same protein. Indeed, the impact of an escape mutation may be vastly different, depending on which particular mutation is selected within a single epitope. This is well illustrated by the Gag-TL9 epitope studied here and previously (Fig. 4; Wright et al., 2010, 2012; Tsai et al., 2016), in which the T186S and T186M mutants dramatically decrease viral replicative capacity, whereas mutants such as Q182S have relatively little impact.
The observation here of widespread early HIV-specific CD8+ T cell escape in pediatric infection was unexpected given previous studies that have shown a striking lack of efficacy of anti-HIV CD8+ T cell responses in infected children, even involving the same specificities that are believed to underlie HLA class I–mediated control of viremia in adult infection. For example, the HLA-B*57/58:01–restricted Gag TW10 epitope (Gag residues 240–248) is one of the earliest HIV-specific CD8+ T cell responses generated in acute infection (Altfeld et al., 2001, 2006; Leslie et al., 2004; Brumme et al., 2008) and characteristically drives the selection of escape mutants such as T242N that reduce viral replicative capacity (Martinez-Picado et al., 2006) or, more rarely, other variants such as A248D that substantially cripple the virus (Miura et al., 2009). However, even though a high-frequency HLA-B*57/58:01–restricted TW10 response can be evident at birth among in utero–infected infants, progression is observed without any selection of escape mutants, suggesting lack of antiviral efficacy in this age group (Adland et al., 2015). In contrast, among HLA-B*57/58:01 adults with disease progression, TW10 escape is almost universal (Martinez-Picado et al., 2006). Furthermore, the absence of HLA-B*57/58:01/81:01–mediated protection against disease progression in pediatric infection, contrasting with the strong influence of these alleles in adult infection, is additional indirect evidence that HIV-specific CD8+ T cell responses play an insignificant role in pediatric slow progression (Adland et al., 2015). Finally, more recent data directly measuring magnitude and breadth of virus-specific CD8+ T cell activity further indicate that these responses do not contribute significantly to progression rates in pediatric infection (Muenchhoff et al., 2016). Indeed, a progressively more active CD8+ T cell response that is observed through childhood (Luzuriaga et al., 1995; Scott et al., 2001; Thobakgale et al., 2007) can be associated with higher levels of immune activation and accelerated CD4 decline in these children (Muenchhoff et al., 2016).
The current studies demonstrate the benefit of next-generation deep sequencing (NGS) compared with previous approaches involving population sequencing and/or sequencing of limited numbers of viral clones to address the question of early escape in pediatric infection have (Leslie et al., 2004; Pillay et al., 2005; Sanchez-Merino et al., 2005). Here, we have been able to demonstrate via NGS the absence of rare variants in the transmitting mother. This is important, because even if the mother does not carry the HLA allele driving selection pressure in the child, it is not uncommon for the virus that is transmitted to the mother to have come from the father of the child and hence to include variants within epitopes that are restricted by HLA only shared by the father and the child (Pillay et al., 2005). The combination of NGS showing no variant in the transmitting mother, and 100% selection of a variant within an epitope restricted by an HLA allele only expressed in the child, as illustrated by the HLA-A*74–associated variant K12N (Table 2), is compelling evidence of early selection pressure on the virus driven by CD8+ T cell responses among these infants.
The significance of these findings for slow-progressor children who have gone onto ART as per the guidelines recommending universal ART for all HIV-infected children, irrespective of health of CD4 count, has been discussed above. HIV-specific CTL responses in VC children have the potential to make an effective contribution to eradication of viral reservoir after latency reversal, both because of the capacity to recognize autologous variants and because the CTL specificities are generally Gag/Pol specific and effective at killing virus-infected cells. HIV-specific CTL responses in VNC children may require redirection toward these more conserved Gag/Pol specificities via immunotherapeutic immunization in order to be effective. For the HIV-infected children not studied here in whom ART was initiated early, either prophylactically to prevent immune decline or therapeutically to allow immune reconstitution, it is possible that the viral reservoir may not be comprised so extensively with escape variants because of lack of selection pressure as described above. However, it is likely that immunotherapeutic immunization via a T cell vaccine inducing broad, conserved Gag/Pol specificities in these children also would increase the likelihood of HIV-specific CTL making an effective impact on the viral reservoir.
In considering how the situation might differ between HIV-infected adults and children, several factors may contribute. In HIV-infected adults, widespread CD8+ T cell escape typically occurs early in infection (Goulder and Walker, 1999; Brumme et al., 2008; Goonetilleke et al., 2009; Boutwell et al., 2013), and most of the latent reservoir virus expresses escape mutants (Deng et al., 2015). Thus, the relative inability of the adult immune system to mount autologous variant-specific CD8+ T cell responses may represent a major obstacle to effective clearance of the adult viral reservoir. Immune reconstitution is both more rapid and more complete in children than in adults on ART (Franco et al., 2000; Gibb et al., 2000; Douek et al., 2003; Feeney et al., 2003; Walker et al., 2004; Sabin et al., 2008), and therefore, the immunogenicity of a T cell vaccine may be substantially greater in ART-treated children (Swadling et al., 2016). Immunotherapeutic vaccination using a mosaic insert encoding the common escape variants (Fischer et al., 2007; Barouch et al., 2010) might be expected to induce responses to the autologous variants more effectively in children than in adults. In all these respects, therefore, the HIV-specific CD8+ T cell responses in infected children may be more likely than those in adults to play an effective part in eradication of the viral reservoir. If CD8+ T cell responses against “low fitness” variants can be induced either naturally or via vaccination in children, the prospects for HIV “remission” after ART (Persaud et al., 2013; Sáez-Cirión et al., 2013; Frange et al., 2016) may be greater in pediatric than adult infection.
Materials and methods
Study subjects
Eleven children studied here were from the previously described Pediatric Early HAART and Strategic Treatment Interruption Study (Mphatswe et al., 2007; Thobakgale et al., 2007, 2009; Prendergast et al., 2008). This feasibility study enrolled 75 HIV-infected infants born to HIV-positive mothers at St. Mary’s and Prince Mshiyeni Hospitals in KwaZulu-Natal, South Africa, in 2003–2005. Antenatal mothers were recruited in the third trimester of pregnancy; mothers and infants received a single dose of nevirapine at delivery (the only available regimen for prevention of mother-to-child transmission at the time; infant ART was not available at government hospitals). Infants were randomized to one of three study arms: deferred treatment, immediate uninterrupted treatment for 12 mo, or immediate 18-mo treatment with structured interruptions (Mphatswe et al., 2007; Prendergast et al., 2008). Diagnosis of HIV infection in infants and HLA class I typing were performed as previously described (Kiepiela et al., 2004; Mphatswe et al., 2007; Thobakgale et al., 2007; Prendergast et al., 2008). CD4+ T cell counts were determined by flow cytometry using standard clinical protocols. HIV plasma viral load measurements were done using the Roche Amplicor Monitor assay according to the manufacturer’s instructions. Eleven children from this cohort were termed PSPs, defined here as children infected via mother-to-child transmission who before universal ART recommendations for pediatric HIV infection had not met ART initiation criteria by the age of 5 yr (CD4 >350 cells/mm3 or WHO stage III or IV; Table 1).
The study was approved by the Biomedical Research Ethics Committee of the University of KwaZulu-Natal and the Institutional Review Boards of the Massachusetts General Hospital and the University of Oxford. The mothers gave written informed consent for participation of their children.
Additionally, for the analysis of CD8+ T cell responses to the wild-type versus autologous variant-specific epitopes (Fig. 6), we used PBMCs from treatment-naive HIV C clade–infected subjects from the following previously described cohorts: pediatric Kimberley cohort from Kimberley, South Africa (Adland et al., 2015); adult Gateway cohort from Durban, South Africa (Payne et al., 2014); and adult Thames Valley Cohort from Northampton General Hospital, Northampton, UK, and Wycombe Hospital, High Wycombe, UK (Payne et al., 2010). All were C clade–infected adults with chronic HIV infection. Among the Kimberley pediatric subjects, the mean age at enrolment was 6.5 yr, mean CD4 count was 876 cells/mm3, mean CD4% was 31.4%, and median viral load was 33,000 copies/ml. Among the adults, the mean CD4 count was 531 cells/mm3, (IQR, 429–576 cells/mm3), and the median viral load was 8,700 HIV copies/ml plasma (IQR, 5,090–27,814). All participants provided written informed consent. Ethics approval was given by the University of KwaZulu-Natal Biomedical Research Board and the University of Oxford Ethics Committee.
IFN-γ ELISPOT assays
Ex vivo HIV-specific CD8+ T cell responses were screened in IFN-γ ELISPOT assays using a panel of 410 overlapping 18mer peptides, spanning the entire HIV proteome, based on 2001 C clade consensus sequence, as described previously (Addo et al., 2003; Kiepiela et al., 2004; Thobakgale et al., 2007). Spots were counted using an automated ELISPOT reader (AID ELISPOT v4.0; Autoimmun Diagnostika) and manually checked. Background (>3 SDs above the mean of the 4 wells containing PBMCs without a peptide) was subtracted from values of all wells, and the final result was expressed as number of spot-forming cells per 106 PBMC. Positive responses were considered >100 spot-forming cells/106 PBMCs after background subtraction. In IFN-γ ELISPOT assays to determine recognition of wild-type TL9 and autologous Gag-TL9 variants, where adequate cell numbers were available, assays were performed in triplicate at peptide concentrations of 10−10 M to 10−4 M (Fig. 6, A and B). In subjects where cell numbers were limiting, peptide concentrations of 10−6 M to 10−4 M were used. Data shown in Fig. 6 (C and D) were determined by calculating the mean of the responses to TL9 wild-type and autologous TL9 variant peptides at concentrations of 10−6 M to 10−4 M for each subject.
Viral RNA extraction
One-milliliter plasma aliquots were thawed at 37°C in a water bath and centrifuged for 1.5 h at 15,000 rpm; 860 µl supernatant was removed, and viral RNA was isolated from the remaining 140 µl using QIAmp Viral RNA Mini kit (QIAGEN) following the manufacturer’s instructions. Samples with a viral load <2,000 copies/ml were concentrated by putting two or three aliquots of plasma (if available) through the same QIAmp column.
Population sequencing of gag
Genomic DNA was extracted from whole blood via the QIAamp DNA Blood Mini kit (QIAGEN). HIV gag sequences were amplified by nested PCR to obtain population sequences using the primers 5′-CTCTAGCAGTGGCGCCCGAA-3′ and 5′-TCCTTTCCACATTTCCAACAGCC-3′ for the first round and 5′-CAATTTCTGGCTATGTGCCC-3′ and 5′-ACTCGGCTTGCTGAAGTGC-3′ for the second round. Viral RNA was isolated from plasma by use of a QIAamp Viral RNA Mini kit (QIAGEN). The Gag-Protease region was amplified by RT-PCR from plasma HIV-1 RNA using Superscript III One-Step Reverse transcription kit (Invitrogen) and the following Gag-protease-specific primers: 5′-CACTGCTTAAGCCTCAATAAAGCTTGCC-3′ (HXB2 nucleotides 512–539) and 5′-TTTAACCCTGCTGGGTGTGGTATTCCT-3′ (nucleotides 2,851–2,825). Second-round PCR was performed using forward primer (5′-GACTCGGCTTGCTGAAGCGCGCACGGCAAGAGGCGAGGGGCGACTGGTGAGTACGCCAAAAATTTTGACTAGCGGAGGCTAGAAGGAGAGAGATGGG-3′, 695–794) and reverse primer (5′-GGCCCAATTTTTGAAATTTTTCCTTCCTTTTCCATTTCTGTACAAATTTCTACTAATGCTTTTATTTTTTCTGTCAATGGCCATTGTTTAACTTTTG-3′, 2,646–2,547). All PCR products were purified using a QIAquick PCR purification kit (QIAGEN) according to manufacturer’s instructions. Sequencing was undertaken using the Big Dye Ready Reaction Terminator Mix (V3; Applied Biosystems) analyzed using Sequencher v4.8 (Gene Codes Corporation) and manually aligned using Se_Al software.
Amplification of full-length viral RNA for ultra-deep sequencing
Amplification of the full HIV genome in four overlapping fragments was performed using SuperScript III One-Step RT-PCR System with Platinum Taq High Fidelity (Invitrogen), as previously described (Gall et al., 2012). To confirm amplification, PCR products were run on 1% agarose gel with 240 V, 300 mA for 1.5 h, and bands were visualized under UV light. Amplicons were then combined into one pool (5 µl of amplicon 1 and 10 µl each of amplicons 2, 3, and 4) and ultra-deep sequenced at the Wellcome Trust Sanger Institute (Cambridge, UK).
Ultra-deep sequencing and de novo assembly of viral genomes
Libraries were prepared from 50–1,000 ng DNA as described previously (Quail et al., 2008, 2011) using one of 96 multiplex adaptors for each pool of amplicons. Paired-end sequencing with a read length of 300 bp was performed using the Illumina MiSeq instrument as described previously (Gall et al., 2014). Consensus sequences were generated by de novo assembly (i.e., without a reference sequence) using Iterative Virus Assembler (Hunt et al., 2015).
Epitope haplotype calling from ultra-deep sequencing data
Gaps in the de novo assembled genomes were filled using maternal or a reference sequence. The reads were then mapped to the assemblies using MOSAIK (Lee et al., 2014) with default parameters. A subtype C reference genome was annotated with the location of the epitopes of interest. The de novo assemblies were aligned using MUSCLE (Edgar, 2004) to the annotated reference to determine the location of the epitopes in each assembly. V-Phaser 2 with default parameters was used to call variants (Yang et al., 2013) and V-Profiler to call haplotypes (Henn et al., 2012) for the HLA-associated epitopes. Only haplotypes with greater than 1% frequency were included in the analysis.
Phylogenetic analysis of HIV genomes
HIV genomes from 11 PSP mother–child pairs were ultra-deep sequenced. Maternal time points included closest to delivery, if available, and 5–7 yr after delivery, if available; children’s time points included earliest available, ∼2.5, ∼5, and ∼7 yr of age or only the earliest and 5 yr of age time points (43 full genomes and seven partial genomes were generated). To confirm the close relationship between viral sequences from mother–child pairs, a maximum-likelihood phylogenetic tree was constructed using the General Time Reversible models of nucleotide substitution, from the 43 full-genome sequences and eight clade-specific reference sequences with 1,000 bootstrap replicates in MEGA version 6.06-mac software (Tamura et al., 2013). The tree was visualized in FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) and Adobe Illustrator CS4 v14.0.0. HIV subtype (C) was confirmed with REGA tool, v3.0 (de Oliveira et al., 2005).
Calculating the contribution to escape in VC and VNC children targeting Gag-TL9
The Gene Cutter tool (http://www.hiv.lanl.gov/content/sequence/GENE_CUTTER/cutter.html) was used to extract sequences of the nine HIV proteins from the de novo assembled consensus sequences from mother–child pairs; protein-encoding sequences were then translated in Geneious 6.1.8 and aligned in Mega6.06.
A panel of well-characterized epitopes (Table S4) across full HIV proteome restricted by the six HLA-A/B/C alleles expressed among the 11 PSPs was compiled. To examine escape in HLA-relevant CD8+ T cell epitopes, for each child, regions of the virus encoding the CD8+ T cell epitopes restricted by any of the six HLA alleles expressed by that child were analyzed. Only polymorphisms that developed independently in the child (i.e., had not been transmitted by the mother) were included and confirmed using epitope minor variant data.
The extent of HLA-driven escape was quantified as the proportion of HLA-associated sites that were adapted to that allele (i.e., polymorphisms that are enriched in the presence of an HLA allele or polymorphisms that differ from the polymorphisms that are less frequently observed in the presence of that HLA allele) based on previously defined HLA-associated escape sites (Carlson et al., 2012, 2014). The number of HLA-associated adaptations observed across the HIV proteome in each child was divided by the total number of HLA-associated epitopes relevant to that child’s HLA repertoire (Payne et al., 2014).
Calculating the rate of escape in HLA-restricted CD8+ T cell epitopes
We have defined escape as a nonsynonymous mutation that has arisen in the HLA-restricted CD8+ T cell epitope sequence since the founder virus, which we infer from the consensus sequence of the first time point. A panel of well-characterized epitopes (Table S4) across full HIV proteome restricted by six HLA-A/B/C alleles expressed among the 11 PSPs was compiled. To examine the accumulation of escape mutations in different gene regions, the proportion of sequences that accumulated at least one escape mutation was calculated for each time point for known restricted CD8+ T cell epitopes per patient. Furthermore, we ensured that there were at least 100 sequences with 95% coverage of the epitope region to estimate the proportion of “escaped” sequences. The mean accumulation of escaped variants per gene was obtained by fitting a linear regression model to the proportion of escaped variants over time. We also explored a logistic regression model, although the sum of squared errors and Akaike information criterion indicated that the linear regression model provided a significantly better fit to the data. The statistical uncertainty of these rates was assessed by generating 50 bootstrapped datasets for each patient. The difference in the rates of escape was estimated by calculating the difference in the slope of the linear regression model (per bootstrap replicate) between VNC and VC children for each gene. These analyses were performed using custom-made python scripts (code available at https://github.com/jnarag/DeepGenomeAnalysis).
Site-directed mutagenesis of T186M
T186M mutation of HIV-1 Gag sequence was introduced into SK-254(M), a modified version of a patient-derived subtype C HIV-1 Gag-protease sequence (SK-254, GenBank accession no. HM593258) with p24 Gag identical to consensus C inserted in pNL4-3. The mutation was engineered by using QuikChange Lightning site-directed mutagenesis kit (Agilent Technologies) along with custom-designed mutagenesis forward and reversed primers: 5′-CACCCCACAAGATTTAAACATGATGTTAAATACAGTGGGGGG-3′ and 5′-CCCCCCACTGTATTTAACATCATGTTTAAATCTTGTGGGGTG-3′. The mutation was confirmed by sequencing after the mutagenesis.
Virus propagation
Plasmid preparation was performed according to manufacturer’s instructions (HiSpeed plasmid Maxi kit; QIAGEN). To generate mutant viruses, the T186M Gag-Pro amplified and purified PCR products and BstE II linearized pNL4-3Δgag-protease (New England Biolabs) were transfected into the GFP-reporter GXR cell line via electroporation in a Bio-Rad GenePulsar II using 0.4-cm cuvettes at 300 V, 500 µF and infinite resistance as previously described (Miura et al., 2009). Virus propagation was monitored by flow cytometry to detect infected GFP-positive cells. Virus culture supernatants were harvested when 30% of cells were GFP-positive. Viruses were aliquoted and stored at −80°C until use.
Statistical analysis
Statistical analyses were performed in Prism (v5.0c; GraphPad) or R (v3.3.1; The R Foundation for Statistical Computing). Linear mixed effect regression analyses were performed using the lme4 package in R, with the lmer (for continuous outcomes) or glm (for binomial outcomes such as PSP group) functions, in each case we allowed for a random effect attributable to the age in days at which each donor was sampled, and solved for the (fixed) effect estimate caused by PSP group. When estimating the change in immune response over time (as in Fig. 2), we modeled age as a fixed effect and allowed for a random effect caused by donor. We calculated likelihood ratio p-values using the ANOVA function in R to compare two nested models (i.e., a model with the predictor variable of interest versus a null model omitting only that variable) that were fit under a maximum-likelihood scenario. The p-values reported are not adjusted for multiple comparisons. Data plots were made using the ggplot2 package, and the default span (0.75) was used for the LOWESS smoothed lines shown in the figures. The code used for analyses is available from the authors. The response variables of interest were left untransformed in the case of the absolute CD4 count, CD4+ T cell percentage, IFN-γ ELISPOT breadth and IFN-γ ELISPOT magnitude, but the HIV viral load was modeled after log10 transformation.
For comparisons between two groups at a single time point, or of mean measures, data were analyzed in Prism. Data were first confirmed to have parametric or nonparametric distribution (D’Agostino and Pearson omnibus normality test). For two-group analyses t test (parametric) or Mann–Whitney U test (nonparametric) was performed; for ≥three-group analyses ANOVA (parametric) or Kruskal–Wallis (nonparametric) followed by post-hoc test was performed. Strength of association between two variables was analyzed by Pearson (nonparametric) or Spearman (parametric) correlation. P-values < 0.05 were considered significant.
Online supplemental material
Fig. S1 shows phylogenetic tree from 43 full-genome sequences of the 11 PSP and their mothers. Table S1 shows HLA-associated escape mutations that developed in the VNC child 517-C. Table S2 shows HLA-associated escape mutations that developed in the VNC child 021-C. Table S3 shows 11 HLA-B*42:01–positive and/or HLA-B*81:01–positive children from Southern Africa in whom autologous virus encoded a Gag-TL9 variant. Table S4 lists epitopes used in the analysis of pediatric deep sequence data. Tables S1–S4 are provided as Excel files.
Supplementary Material
Acknowledgments
This work was supported by the Wellcome Trust (WT 104748MA to P.J.R. Goulder, 098051 to P. Kellam and A. Gall, and 093768/Z/15/X to A.J. Prendergast); the Clarendon Fund (E.M. Leitman); National Institute for Health Research research capability funding (P.C. Matthews); Linacre College, University of Oxford (J. Hemelaar); and the Mochida Memorial Foundation for Medical and Pharmaceutical Research (M. Mori).
The authors declare no competing financial interests.
Author contributions: E.M. Leitman designed the study, performed experiments, analyzed data, and wrote the manuscript. C.F. Thobakgale and E. Adland performed experiments and analyzed data. M.A. Ansari analyzed data. J. Raghwani and O.G. Pybus analyzed data and contributed to manuscript writing. A.J. Prendergast, G. Tudor-Willliams, P. Kiepiela, J. Hemelaar, P. Jooste, T. Ndung’u, L. Riddell, and G. Luzzi managed cohort recruitment and reviewed the manuscript. J. Brener, M.-H. Tsai, and M. Mori contributed to experiment performance and reviewed the manuscript. B.D. Walker contributed to study design and reviewed the manuscript. P. Kellam and A. Gall performed some of the experiments, contributed to data analyses, and reviewed the manuscript. V. Naranbhai analyzed data and contributed to manuscript revision. P.C. Matthews designed and supervised the study, contributed to data analyses, and wrote and reviewed the manuscript. P.J.R. Goulder conceived, designed, and supervised the study, contributed to data analyses, found funding for the study, and wrote the manuscript.
Footnotes
Abbreviations used:
- ART
- antiretroviral therapy
- IQR
- interquartile range
- NGS
- next-generation deep sequencing
- PSP
- pediatric slow progressor
- VC
- viremic controller
- VNC
- viremic non-controller
References
- Addo M.M., Yu X.G., Rathod A., Cohen D., Eldridge R.L., Strick D., Johnston M.N., Corcoran C., Wurcel A.G., Fitzpatrick C.A., et al. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081–2092. 10.1128/JVI.77.3.2081-2092.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adland E., Paioni P., Thobakgale C., Laker L., Mori L., Muenchhoff M., Csala A., Clapson M., Flynn J., Novelli V., et al. 2015. Discordant Impact of HLA on Viral Replicative Capacity and Disease Progression in Pediatric and Adult HIV Infection. PLoS Pathog. 11:e1004954 10.1371/journal.ppat.1004954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen T.M., and Altfeld M.. 2008. Crippling HIV one mutation at a time. J. Exp. Med. 205:1003–1007. 10.1084/jem.20080569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen T.M., Yu X.G., Kalife E.T., Reyor L.L., Lichterfeld M., John M., Cheng M., Allgaier R.L., Mui S., Frahm N., et al. 2005. De novo generation of escape variant-specific CD8+ T-cell responses following cytotoxic T-lymphocyte escape in chronic human immunodeficiency virus type 1 infection. J. Virol. 79:12952–12960. 10.1128/JVI.79.20.12952-12960.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida J.R., Price D.A., Papagno L., Arkoub Z.A., Sauce D., Bornstein E., Asher T.E., Samri A., Schnuriger A., Theodorou I., et al. 2007. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 204:2473–2485. 10.1084/jem.20070784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida J.R., Sauce D., Price D.A., Papagno L., Shin S.Y., Moris A., Larsen M., Pancino G., Douek D.C., Autran B., et al. 2009. Antigen sensitivity is a major determinant of CD8+ T-cell polyfunctionality and HIV-suppressive activity. Blood. 113:6351–6360. 10.1182/blood-2009-02-206557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altfeld M., and Allen T.M.. 2006. Hitting HIV where it hurts: an alternative approach to HIV vaccine design. Trends Immunol. 27:504–510. 10.1016/j.it.2006.09.007 [DOI] [PubMed] [Google Scholar]
- Altfeld M., Rosenberg E.S., Shankarappa R., Mukherjee J.S., Hecht F.M., Eldridge R.L., Addo M.M., Poon S.H., Phillips M.N., Robbins G.K., et al. 2001. Cellular immune responses and viral diversity in individuals treated during acute and early HIV-1 infection. J. Exp. Med. 193:169–180. 10.1084/jem.193.2.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altfeld M., Kalife E.T., Qi Y., Streeck H., Lichterfeld M., Johnston M.N., Burgett N., Swartz M.E., Yang A., Alter G., et al. 2006. HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8(+) T Cell Response against HIV-1. PLoS Med. 3:e403 10.1371/journal.pmed.0030403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiker A., Darby S., De Angelis D., Ewart D., and Porter K.. 2000. Collaborative Group on AIDS Incubation and HIV Survival including the CASCADE EU Concerted Action. Concerted Action on SeroConversion to AIDS and Death in Europe. Time from HIV-1 seroconversion to AIDS and death before widespread use of highly-active antiretroviral therapy: a collaborative re-analysis. Lancet. 355:1131–1137. 10.1016/S0140-6736(00)02061-4 [DOI] [PubMed] [Google Scholar]
- Barouch D.H., Kunstman J., Kuroda M.J., Schmitz J.E., Santra S., Peyerl F.W., Krivulka G.R., Beaudry K., Lifton M.A., Gorgone D.A., et al. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature. 415:335–339. 10.1038/415335a [DOI] [PubMed] [Google Scholar]
- Barouch D.H., Kunstman J., Glowczwskie J., Kunstman K.J., Egan M.A., Peyerl F.W., Santra S., Kuroda M.J., Schmitz J.E., Beaudry K., et al. 2003. Viral escape from dominant simian immunodeficiency virus epitope-specific cytotoxic T lymphocytes in DNA-vaccinated rhesus monkeys. J. Virol. 77:7367–7375. 10.1128/JVI.77.13.7367-7375.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch D.H., O’Brien K.L., Simmons N.L., King S.L., Abbink P., Maxfield L.F., Sun Y.H., La Porte A., Riggs A.M., Lynch D.M., et al. 2010. Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat. Med. 16:319–323. 10.1038/nm.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton J.P., Goonetilleke N., Butler T.C., Walker B.D., McMichael A.J., and Chakraborty A.K.. 2016. Relative rate and location of intra-host HIV evolution to evade cellular immunity are predictable. Nat. Commun. 7:11660 10.1038/ncomms11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanche S., Newell M.L., Mayaux M.J., Dunn D.T., Teglas J.P., Rouzioux C., and Peckham C.S.. 1997. Morbidity and mortality in European children vertically infected by HIV-1. The French Pediatric HIV Infection Study Group and European Collaborative Study. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 14:442–450. 10.1097/00042560-199704150-00008 [DOI] [PubMed] [Google Scholar]
- Boutwell C.L., Carlson J.M., Lin T.H., Seese A., Power K.A., Peng J., Tang Y., Brumme Z.L., Heckerman D., Schneidewind A., and Allen T.M.. 2013. Frequent and variable cytotoxic-T-lymphocyte escape-associated fitness costs in the human immunodeficiency virus type 1 subtype B Gag proteins. J. Virol. 87:3952–3965. 10.1128/JVI.03233-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumme Z.L., Brumme C.J., Carlson J., Streeck H., John M., Eichbaum Q., Block B.L., Baker B., Kadie C., Markowitz M., et al. 2008. Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J. Virol. 82:9216–9227. 10.1128/JVI.01041-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson J.M., Brumme C.J., Martin E., Listgarten J., Brockman M.A., Le A.Q., Chui C.K., Cotton L.A., Knapp D.J., Riddler S.A., et al. International HIV Adaptation Collaborative . 2012. Correlates of protective cellular immunity revealed by analysis of population-level immune escape pathways in HIV-1. J. Virol. 86:13202–13216. 10.1128/JVI.01998-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson J.M., Schaefer M., Monaco D.C., Batorsky R., Claiborne D.T., Prince J., Deymier M.J., Ende Z.S., Klatt N.R., DeZiel C.E., et al. 2014. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science. 345:1254031 10.1126/science.1254031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson J.M., Du V.Y., Pfeifer N., Bansal A., Tan V.Y., Power K., Brumme C.J., Kreimer A., DeZiel C.E., Fusi N., et al. 2016. Impact of pre-adapted HIV transmission. Nat. Med. 22:606–613. 10.1038/nm.4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks S.G. 2012. HIV: Shock and kill. Nature. 487:439–440. 10.1038/487439a [DOI] [PubMed] [Google Scholar]
- Deng K., Pertea M., Rongvaux A., Wang L., Durand C.M., Ghiaur G., Lai J., McHugh H.L., Hao H., Zhang H., et al. 2015. Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature. 517:381–385. 10.1038/nature14053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira T., Deforche K., Cassol S., Salminen M., Paraskevis D., Seebregts C., Snoeck J., van Rensburg E.J., Wensing A.M., van de Vijver D.A., et al. 2005. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics. 21:3797–3800. 10.1093/bioinformatics/bti607 [DOI] [PubMed] [Google Scholar]
- Douek D.C., Picker L.J., and Koup R.A.. 2003. T cell dynamics in HIV-1 infection. Annu. Rev. Immunol. 21:265–304. 10.1146/annurev.immunol.21.120601.141053 [DOI] [PubMed] [Google Scholar]
- Edgar R.C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards B.H., Bansal A., Sabbaj S., Bakari J., Mulligan M.J., and Goepfert P.A.. 2002. Magnitude of functional CD8+ T-cell responses to the gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J. Virol. 76:2298–2305. 10.1128/jvi.76.5.2298-2305.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney M.E., Draenert R., Roosevelt K.A., Pelton S.I., McIntosh K., Burchett S.K., Mao C., Walker B.D., and Goulder P.J.. 2003. Reconstitution of virus-specific CD4 proliferative responses in pediatric HIV-1 infection. J. Immunol. 171:6968–6975. 10.4049/jimmunol.171.12.6968 [DOI] [PubMed] [Google Scholar]
- Feeney M.E., Tang Y., Pfafferott K., Roosevelt K.A., Draenert R., Trocha A., Yu X.G., Verrill C., Allen T., Moore C., et al. 2005. HIV-1 viral escape in infancy followed by emergence of a variant-specific CTL response. J. Immunol. 174:7524–7530. 10.4049/jimmunol.174.12.7524 [DOI] [PubMed] [Google Scholar]
- Ferguson A.L., Mann J.K., Omarjee S., Ndung’u T., Walker B.D., and Chakraborty A.K.. 2013. Translating HIV sequences into quantitative fitness landscapes predicts viral vulnerabilities for rational immunogen design. Immunity. 38:606–617. 10.1016/j.immuni.2012.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari G., Pollara J., Kozink D., Harms T., Drinker M., Freel S., Moody M.A., Alam S.M., Tomaras G.D., Ochsenbauer C., et al. 2011. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J. Virol. 85:7029–7036. 10.1128/JVI.00171-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W., Perkins S., Theiler J., Bhattacharya T., Yusim K., Funkhouser R., Kuiken C., Haynes B., Letvin N.L., Walker B.D., et al. 2007. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat. Med. 13:100–106. 10.1038/nm1461 [DOI] [PubMed] [Google Scholar]
- Franco J.M., León-Leal J.A., Leal M., Cano-Rodriguez A., Pineda J.A., Macías J., Rubio A., Rey C., Sanchez B., and Lissen E.. 2000. CD4+ and CD8+ T lymphocyte regeneration after anti-retroviral therapy in HIV-1-infected children and adult patients. Clin. Exp. Immunol. 119:493–498. 10.1046/j.1365-2249.2000.01152.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frange P., Faye A., Avettand-Fenoël V., Bellaton E., Descamps D., Angin M., David A., Caillat-Zucman S., Peytavin G., Dollfus C., et al. ANRS EPF-CO10 Pediatric Cohort and the ANRS EP47 VISCONTI study group . 2016. HIV-1 virological remission lasting more than 12 years after interruption of early antiretroviral therapy in a perinatally infected teenager enrolled in the French ANRS EPF-CO10 paediatric cohort: a case report. Lancet HIV. 3:e49–e54. 10.1016/S2352-3018(15)00232-5 [DOI] [PubMed] [Google Scholar]
- Gall A., Ferns B., Morris C., Watson S., Cotten M., Robinson M., Berry N., Pillay D., and Kellam P.. 2012. Universal amplification, next-generation sequencing, and assembly of HIV-1 genomes. J. Clin. Microbiol. 50:3838–3844. 10.1128/JCM.01516-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall A., Morris C., Kellam P., and Berry N.. 2014. Complete Genome Sequence of the WHO International Standard for HIV-1 RNA Determined by Deep Sequencing. Genome Announc. 2:e01254-13 10.1128/genomeA.01254-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldmacher C., Currier J.R., Herrmann E., Haule A., Kuta E., McCutchan F., Njovu L., Geis S., Hoffmann O., Maboko L., et al. 2007. CD8 T-cell recognition of multiple epitopes within specific Gag regions is associated with maintenance of a low steady-state viremia in human immunodeficiency virus type 1-seropositive patients. J. Virol. 81:2440–2448. 10.1128/JVI.01847-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb D.M., Newberry A., Klein N., de Rossi A., Grosch-Woerner I., and Babiker A.. Paediatric European Network for Treatment of AIDS (PENTA) Steering Committee . 2000. Immune repopulation after HAART in previously untreated HIV-1-infected children. Lancet. 355:1331–1332. 10.1016/S0140-6736(00)02117-6 [DOI] [PubMed] [Google Scholar]
- Goepfert P.A., Lumm W., Farmer P., Matthews P., Prendergast A., Carlson J.M., Derdeyn C.A., Tang J., Kaslow R.A., Bansal A., et al. 2008. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J. Exp. Med. 205:1009–1017. 10.1084/jem.20072457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goonetilleke N., Liu M.K., Salazar-Gonzalez J.F., Ferrari G., Giorgi E., Ganusov V.V., Keele B.F., Learn G.H., Turnbull E.L., Salazar M.G., et al. CHAVI Clinical Core B . 2009. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 206:1253–1272. 10.1084/jem.20090365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder P.J., and Walker B.D.. 1999. The great escape - AIDS viruses and immune control. Nat. Med. 5:1233–1235. 10.1038/15184 [DOI] [PubMed] [Google Scholar]
- Goulder P.J., and Walker B.D.. 2012. HIV and HLA class I: an evolving relationship. Immunity. 37:426–440. 10.1016/j.immuni.2012.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder P.J., Edwards A., Phillips R.E., and McMichael A.J.. 1997. Identification of a novel HLA-A24-restricted cytotoxic T-lymphocyte epitope within HIV-1 Nef. AIDS. 11:1883–1884. 10.1097/00002030-199715000-00015 [DOI] [PubMed] [Google Scholar]
- Goulder P.J., Brander C., Annamalai K., Mngqundaniso N., Govender U., Tang Y., He S., Hartman K.E., O’Callaghan C.A., Ogg G.S., et al. 2000. Differential narrow focusing of immunodominant human immunodeficiency virus gag-specific cytotoxic T-lymphocyte responses in infected African and caucasoid adults and children. J. Virol. 74:5679–5690. 10.1128/JVI.74.12.5679-5690.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder P.J., Addo M.M., Altfeld M.A., Rosenberg E.S., Tang Y., Govender U., Mngqundaniso N., Annamalai K., Vogel T.U., Hammond M., et al. 2001a Rapid definition of five novel HLA-A*3002-restricted human immunodeficiency virus-specific cytotoxic T-lymphocyte epitopes by elispot and intracellular cytokine staining assays. J. Virol. 75:1339–1347. 10.1128/JVI.75.3.1339-1347.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder P.J., Brander C., Tang Y., Tremblay C., Colbert R.A., Addo M.M., Rosenberg E.S., Nguyen T., Allen R., Trocha A., et al. 2001b Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 412:334–338. 10.1038/35085576 [DOI] [PubMed] [Google Scholar]
- Goulder P.J., Lewin S.R., and Leitman E.M.. 2016. Paediatric HIV infection: the potential for cure. Nat. Rev. Immunol. 16:259–271. 10.1038/nri.2016.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayton E.J., Rose A., Ibrahimsa U., Del Sorbo M., Capone S., Crook A., Black A.P., Dorrell L., and Hanke T.. 2014. Safety and tolerability of conserved region vaccines vectored by plasmid DNA, simian adenovirus and modified vaccinia virus ankara administered to human immunodeficiency virus type 1-uninfected adults in a randomized, single-blind phase I trial. PLoS One. 9:e101591 10.1371/journal.pone.0101591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn M.R., Boutwell C.L., Charlebois P., Lennon N.J., Power K.A., Macalalad A.R., Berlin A.M., Malboeuf C.M., Ryan E.M., Gnerre S., et al. 2012. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog. 8:e1002529 10.1371/journal.ppat.1002529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeyborne I., Rathod A., Buchli R., Ramduth D., Moodley E., Rathnavalu P., Chetty S., Day C., Brander C., Hildebrand W., et al. 2006. Motif inference reveals optimal CTL epitopes presented by HLA class I alleles highly prevalent in southern Africa. J. Immunol. 176:4699–4705. 10.4049/jimmunol.176.8.4699 [DOI] [PubMed] [Google Scholar]
- Hunt M., Gall A., Ong S.H., Brener J., Ferns B., Goulder P., Nastouli E., Keane J.A., Kellam P., and Otto T.D.. 2015. IVA: accurate de novo assembly of RNA virus genomes. Bioinformatics. 31:2374–2376. 10.1093/bioinformatics/btv120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes H., Friedrich D.P., Krambrink A., Smith R.J., Kallas E.G., Horton H., Casimiro D.R., Carrington M., Geraghty D.E., Gilbert P.B., et al. 2013. Vaccine-induced gag-specific T cells are associated with reduced viremia after HIV-1 infection. J. Infect. Dis. 208:1231–1239. 10.1093/infdis/jit322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.B., and Walker B.D.. 2016. HIV-specific CD8+ T cells and HIV eradication. J. Clin. Invest. 126:455–463. 10.1172/JCI80566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima Y., Pfafferott K., Frater J., Matthews P., Payne R., Addo M., Gatanaga H., Fujiwara M., Hachiya A., Koizumi H., et al. 2009. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 458:641–645. 10.1038/nature07746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima Y., Kuse N., Gatanaga H., Naruto T., Fujiwara M., Dohki S., Akahoshi T., Maenaka K., Goulder P., Oka S., and Takiguchi M.. 2010. Long-term control of HIV-1 in hemophiliacs carrying slow-progressing allele HLA-B*5101. J. Virol. 84:7151–7160. 10.1128/JVI.00171-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiepiela P., Leslie A.J., Honeyborne I., Ramduth D., Thobakgale C., Chetty S., Rathnavalu P., Moore C., Pfafferott K.J., Hilton L., et al. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 432:769–775. 10.1038/nature03113 [DOI] [PubMed] [Google Scholar]
- Kiepiela P., Ngumbela K., Thobakgale C., Ramduth D., Honeyborne I., Moodley E., Reddy S., de Pierres C., Mncube Z., Mkhwanazi N., et al. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46–53. 10.1038/nm1520 [DOI] [PubMed] [Google Scholar]
- Klein M.R., van Baalen C.A., Holwerda A.M., Kerkhof Garde S.R., Bende R.J., Keet I.P., Eeftinck-Schattenkerk J.K., Osterhaus A.D., Schuitemaker H., and Miedema F.. 1995. Kinetics of Gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J. Exp. Med. 181:1365–1372. 10.1084/jem.181.4.1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenerman P., and Zinkernagel R.M.. 1998. Original antigenic sin impairs cytotoxic T lymphocyte responses to viruses bearing variant epitopes. Nature. 394:482–485. 10.1038/28860 [DOI] [PubMed] [Google Scholar]
- Kløverpris H.N., Harndahl M., Leslie A.J., Carlson J.M., Ismail N., van der Stok M., Huang K.H., Chen F., Riddell L., Steyn D., et al. 2012a HIV control through a single nucleotide on the HLA-B locus. J. Virol. 86:11493–11500. 10.1128/JVI.01020-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kløverpris H.N., Stryhn A., Harndahl M., van der Stok M., Payne R.P., Matthews P.C., Chen F., Riddell L., Walker B.D., Ndung’u T., et al. 2012b HLA-B*57 Micropolymorphism shapes HLA allele-specific epitope immunogenicity, selection pressure, and HIV immune control. J. Virol. 86:919–929. 10.1128/JVI.06150-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kløverpris H.N., Stryhn A., Harndahl M., Carlson J.M., Leslie A.J., Chen F., Riddell L., Mulenga J., Walker B.D., Ndung’u T., et al. 2013. HLA-A*68:02-restricted Gag-specific cytotoxic T lymphocyte responses can drive selection pressure on HIV but are subdominant and ineffective. AIDS. 27:1717–1723. 10.1097/QAD.0b013e32836146cd [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kløverpris H.N., Adland E., Koyanagi M., Stryhn A., Harndahl M., Matthews P.C., Shapiro R., Walker B.D., Ndung’u T., Brander C., et al. 2014a HIV subtype influences HLA-B*07:02-associated HIV disease outcome. AIDS Res. Hum. Retroviruses. 30:468–475. 10.1089/aid.2013.0197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kløverpris H.N., McGregor R., McLaren J.E., Ladell K., Stryhn A., Koofhethile C., Brener J., Chen F., Riddell L., Graziano L., et al. 2014b Programmed death-1 expression on HIV-1-specific CD8+ T cells is shaped by epitope specificity, T-cell receptor clonotype usage and antigen load. AIDS. 28:2007–2021. 10.1097/QAD.0000000000000362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambotte O., Boufassa F., Madec Y., Nguyen A., Goujard C., Meyer L., Rouzioux C., Venet A., and Delfraissy J.F.. SEROCO-HEMOCO Study Group . 2005. HIV controllers: a homogeneous group of HIV-1-infected patients with spontaneous control of viral replication. Clin. Infect. Dis. 41:1053–1056. 10.1086/433188 [DOI] [PubMed] [Google Scholar]
- Lee W.P., Stromberg M.P., Ward A., Stewart C., Garrison E.P., and Marth G.T.. 2014. MOSAIK: a hash-based algorithm for accurate next-generation sequencing short-read mapping. PLoS One. 9:e90581 10.1371/journal.pone.0090581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrère J.J., Mariotti M., Morand-Joubert L., Thauvin M., and Roudot-Thoraval F.. 1999. Plasma human immunodeficiency virus RNA below 40 Copies/mL is rare in untreated persons even in the first years of infection. J. Infect. Dis. 180:526–529. 10.1086/314906 [DOI] [PubMed] [Google Scholar]
- Leitman E.M., Hurst J., Mori M., Kublin J., Ndung’u T., Walker B.D., Carlson J., Gray G.E., Matthews P.C., Frahm N., and Goulder P.J.. 2016. Lower Viral Loads and Slower CD4+ T-Cell Count Decline in MRKAd5 HIV-1 Vaccinees Expressing Disease-Susceptible HLA-B*58:02. J. Infect. Dis. 214:379–389. 10.1093/infdis/jiw093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A.J., Pfafferott K.J., Chetty P., Draenert R., Addo M.M., Feeney M., Tang Y., Holmes E.C., Allen T., Prado J.G., et al. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282–289. 10.1038/nm992 [DOI] [PubMed] [Google Scholar]
- Leslie A., Price D.A., Mkhize P., Bishop K., Rathod A., Day C., Crawford H., Honeyborne I., Asher T.E., Luzzi G., et al. 2006. Differential selection pressure exerted on HIV by CTL targeting identical epitopes but restricted by distinct HLA alleles from the same HLA supertype. J. Immunol. 177:4699–4708. 10.4049/jimmunol.177.7.4699 [DOI] [PubMed] [Google Scholar]
- Liu M.K., Hawkins N., Ritchie A.J., Ganusov V.V., Whale V., Brackenridge S., Li H., Pavlicek J.W., Cai F., Rose-Abrahams M.; CHAVI Core B, et al. 2013. Vertical T cell immunodominance and epitope entropy determine HIV-1 escape. J. Clin. Invest. 123:380–393. 10.1172/JCI65330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano A., Williams A., Overa A., Silva-Arrieta A., and Brander C.. 2013. Best-characterized HIV-1 CTL epitopes: the 2013 update. In HIV Molecular Immunology 2013. Yusim K., Korber B., Brander C., Barouch D., de Boer R., Haynes B.F., Koup R., Moore J.P., and Walker B.D., editors. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM: 3–19. [Google Scholar]
- Lugada E.S., Mermin J., Kaharuza F., Ulvestad E., Were W., Langeland N., Asjo B., Malamba S., and Downing R.. 2004. Population-based hematologic and immunologic reference values for a healthy Ugandan population. Clin. Diagn. Lab. Immunol. 11:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzuriaga K., Holmes D., Hereema A., Wong J., Panicali D.L., and Sullivan J.L.. 1995. HIV-1-specific cytotoxic T lymphocyte responses in the first year of life. J. Immunol. 154:433–443. [PubMed] [Google Scholar]
- Margolis D.M., Garcia J.V., Hazuda D.J., and Haynes B.F.. 2016. Latency reversal and viral clearance to cure HIV-1. Science. 353:aaf6517 10.1126/science.aaf6517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Picado J., Prado J.G., Fry E.E., Pfafferott K., Leslie A., Chetty S., Thobakgale C., Honeyborne I., Crawford H., Matthews P., et al. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 80:3617–3623. 10.1128/JVI.80.7.3617-3623.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masemola A., Mashishi T., Khoury G., Mohube P., Mokgotho P., Vardas E., Colvin M., Zijenah L., Katzenstein D., Musonda R., et al. HIVNET 028 Study Team . 2004. Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. J. Virol. 78:3233–3243. 10.1128/JVI.78.7.3233-3243.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews P.C., Prendergast A., Leslie A., Crawford H., Payne R., Rousseau C., Rolland M., Honeyborne I., Carlson J., Kadie C., et al. 2008. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J. Virol. 82:8548–8559. 10.1128/JVI.00580-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews P.C., Adland E., Listgarten J., Leslie A., Mkhwanazi N., Carlson J.M., Harndahl M., Stryhn A., Payne R.P., Ogwu A., et al. 2011. HLA-A*7401-mediated control of HIV viremia is independent of its linkage disequilibrium with HLA-B*5703. J. Immunol. 186:5675–5686. 10.4049/jimmunol.1003711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews P.C., Koyanagi M., Kløverpris H.N., Harndahl M., Stryhn A., Akahoshi T., Gatanaga H., Oka S., Juarez Molina C., Valenzuela Ponce H., et al. 2012. Differential clade-specific HLA-B*3501 association with HIV-1 disease outcome is linked to immunogenicity of a single Gag epitope. J. Virol. 86:12643–12654. 10.1128/JVI.01381-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T., Brockman M.A., Schneidewind A., Lobritz M., Pereyra F., Rathod A., Block B.L., Brumme Z.L., Brumme C.J., Baker B., et al. 2009. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte [corrected] recognition. J. Virol. 83:2743–2755. 10.1128/JVI.02265-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsapaya J., Dejnirattisai W., Xu X.N., Vasanawathana S., Tangthawornchaikul N., Chairunsri A., Sawasdivorn S., Duangchinda T., Dong T., Rowland-Jones S., et al. 2003. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat. Med. 9:921–927. 10.1038/nm887 [DOI] [PubMed] [Google Scholar]
- Mphatswe W., Blanckenberg N., Tudor-Williams G., Prendergast A., Thobakgale C., Mkhwanazi N., McCarthy N., Walker B.D., Kiepiela P., and Goulder P.. 2007. High frequency of rapid immunological progression in African infants infected in the era of perinatal HIV prophylaxis. AIDS. 21:1253–1261. 10.1097/QAD.0b013e3281a3bec2 [DOI] [PubMed] [Google Scholar]
- Muenchhoff M., Adland E., Karimanzira O., Crowther C., Pace M., Csala A., Leitman E., Moonsamy A., McGregor C., Hurst J., et al. 2016. Nonprogressing HIV-infected children share fundamental immunological features of nonpathogenic SIV infection. Sci. Transl. Med. 8:358ra125 10.1126/scitranslmed.aag1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngumbela K.C., Day C.L., Mncube Z., Nair K., Ramduth D., Thobakgale C., Moodley E., Reddy S., de Pierres C., Mkhwanazi N., et al. 2008. Targeting of a CD8 T cell env epitope presented by HLA-B*5802 is associated with markers of HIV disease progression and lack of selection pressure. AIDS Res. Hum. Retroviruses. 24:72–82. 10.1089/aid.2007.0124 [DOI] [PubMed] [Google Scholar]
- Novitsky V., Gilbert P., Peter T., McLane M.F., Gaolekwe S., Rybak N., Thior I., Ndung’u T., Marlink R., Lee T.H., and Essex M.. 2003. Association between virus-specific T-cell responses and plasma viral load in human immunodeficiency virus type 1 subtype C infection. J. Virol. 77:882–890. 10.1128/JVI.77.2.882-890.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor D., Friedrich T., Hughes A., Allen T.M., and Watkins D.. 2001. Understanding cytotoxic T-lymphocyte escape during simian immunodeficiency virus infection. Immunol. Rev. 183:115–126. 10.1034/j.1600-065x.2001.1830110.x [DOI] [PubMed] [Google Scholar]
- Ogg G.S., Jin X., Bonhoeffer S., Dunbar P.R., Nowak M.A., Monard S., Segal J.P., Cao Y., Rowland-Jones S.L., Cerundolo V., et al. 1998. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 279:2103–2106. 10.1126/science.279.5359.2103 [DOI] [PubMed] [Google Scholar]
- Ondondo B., Murakoshi H., Clutton G., Abdul-Jawad S., Wee E.G., Gatanaga H., Oka S., McMichael A.J., Takiguchi M., Korber B., and Hanke T.. 2016. Novel Conserved-region T-cell Mosaic Vaccine With High Global HIV-1 Coverage Is Recognized by Protective Responses in Untreated Infection. Mol. Ther. 24:832–842. 10.1038/mt.2016.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiardini M., Cervasi B., Reyes-Aviles E., Micci L., Ortiz A.M., Chahroudi A., Vinton C., Gordon S.N., Bosinger S.E., Francella N., et al. 2011. Low levels of SIV infection in sooty mangabey central memory CD4+ T cells are associated with limited CCR5 expression. Nat. Med. 17:830–836. 10.1038/nm.2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul M.E., Mao C., Charurat M., Serchuck L., Foca M., Hayani K., Handelsman E.L., Diaz C., McIntosh K., and Shearer W.T.. Women and Infants Transmission Study . 2005. Predictors of immunologic long-term nonprogression in HIV-infected children: implications for initiating therapy. J. Allergy Clin. Immunol. 115:848–855. 10.1016/j.jaci.2004.11.054 [DOI] [PubMed] [Google Scholar]
- Payne R.P., Kløverpris H., Sacha J.B., Brumme Z., Brumme C., Buus S., Sims S., Hickling S., Riddell L., Chen F., et al. 2010. Efficacious early antiviral activity of HIV Gag- and Pol-specific HLA-B 2705-restricted CD8+ T cells. J. Virol. 84:10543–10557. 10.1128/JVI.00793-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne R., Muenchhoff M., Mann J., Roberts H.E., Matthews P., Adland E., Hempenstall A., Huang K.H., Brockman M., Brumme Z., et al. 2014. Impact of HLA-driven HIV adaptation on virulence in populations of high HIV seroprevalence. Proc. Natl. Acad. Sci. USA. 111:E5393–E5400. 10.1073/pnas.1413339111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persaud D., Gay H., Ziemniak C., Chen Y.H., Piatak M. Jr., Chun T.W., Strain M., Richman D., and Luzuriaga K.. 2013. Absence of detectable HIV-1 viremia after treatment cessation in an infant. N. Engl. J. Med. 369:1828–1835. 10.1056/NEJMoa1302976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay T., Zhang H.T., Drijfhout J.W., Robinson N., Brown H., Khan M., Moodley J., Adhikari M., Pfafferott K., Feeney M.E., et al. 2005. Unique acquisition of cytotoxic T-lymphocyte escape mutants in infant human immunodeficiency virus type 1 infection. J. Virol. 79:12100–12105. 10.1128/JVI.79.18.12100-12105.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast A., Mphatswe W., Tudor-Williams G., Rakgotho M., Pillay V., Thobakgale C., McCarthy N., Morris L., Walker B.D., and Goulder P.. 2008. Early virological suppression with three-class antiretroviral therapy in HIV-infected African infants. AIDS. 22:1333–1343. 10.1097/QAD.0b013e32830437df [DOI] [PubMed] [Google Scholar]
- Quail M.A., Kozarewa I., Smith F., Scally A., Stephens P.J., Durbin R., Swerdlow H., and Turner D.J.. 2008. A large genome center’s improvements to the Illumina sequencing system. Nat. Methods. 5:1005–1010. 10.1038/nmeth.1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail M.A., Otto T.D., Gu Y., Harris S.R., Skelly T.F., McQuillan J.A., Swerdlow H.P., and Oyola S.O.. 2011. Optimal enzymes for amplifying sequencing libraries. Nat. Methods. 9:10–11. 10.1038/nmeth.1814 [DOI] [PubMed] [Google Scholar]
- Rey-Cuillé M.A., Berthier J.L., Bomsel-Demontoy M.C., Chaduc Y., Montagnier L., Hovanessian A.G., and Chakrabarti L.A.. 1998. Simian immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease. J. Virol. 72:3872–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviére Y., McChesney M.B., Porrot F., Tanneau-Salvadori F., Sansonetti P., Lopez O., Pialoux G., Feuillie V., Mollereau M., Chamaret S., et al. 1995. Gag-specific cytotoxic responses to HIV type 1 are associated with a decreased risk of progression to AIDS-related complex or AIDS. AIDS Res. Hum. Retroviruses. 11:903–907. 10.1089/aid.1995.11.903 [DOI] [PubMed] [Google Scholar]
- Sabin C.A., Smith C.J., d’Arminio Monforte A., Battegay M., Gabiano C., Galli L., Geelen S., Gibb D., Guiguet M., Judd A., et al. Collaboration of Observational HIV Epidemiological Research Europe (COHERE) Study Group . 2008. Response to combination antiretroviral therapy: variation by age. AIDS. 22:1463–1473. 10.1097/QAD.0b013e3282f88d02 [DOI] [PubMed] [Google Scholar]
- Sacha J.B., Chung C., Rakasz E.G., Spencer S.P., Jonas A.K., Bean A.T., Lee W., Burwitz B.J., Stephany J.J., Loffredo J.T., et al. 2007a Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression. J. Immunol. 178:2746–2754. 10.4049/jimmunol.178.5.2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacha J.B., Chung C., Reed J., Jonas A.K., Bean A.T., Spencer S.P., Lee W., Vojnov L., Rudersdorf R., Friedrich T.C., et al. 2007b Pol-specific CD8+ T cells recognize simian immunodeficiency virus-infected cells prior to Nef-mediated major histocompatibility complex class I downregulation. J. Virol. 81:11703–11712. 10.1128/JVI.00926-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez-Cirión A., Bacchus C., Hocqueloux L., Avettand-Fenoel V., Girault I., Lecuroux C., Potard V., Versmisse P., Melard A., Prazuck T., et al. ANRS VISCONTI Study Group . 2013. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog. 9:e1003211 10.1371/journal.ppat.1003211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Merino V., Nie S., and Luzuriaga K.. 2005. HIV-1-specific CD8+ T cell responses and viral evolution in women and infants. J. Immunol. 175:6976–6986. 10.4049/jimmunol.175.10.6976 [DOI] [PubMed] [Google Scholar]
- Scott Z.A., Chadwick E.G., Gibson L.L., Catalina M.D., McManus M.M., Yogev R., Palumbo P., Sullivan J.L., Britto P., Gay H., and Luzuriaga K.. PACTG(Pediatric AIDS Clinical Trial Group) 345 Investigators . 2001. Infrequent detection of HIV-1-specific, but not cytomegalovirus-specific, CD8(+) T cell responses in young HIV-1-infected infants. J. Immunol. 167:7134–7140. 10.4049/jimmunol.167.12.7134 [DOI] [PubMed] [Google Scholar]
- Shearer W.T., Rosenblatt H.M., Gelman R.S., Oyomopito R., Plaeger S., Stiehm E.R., Wara D.W., Douglas S.D., Luzuriaga K., McFarland E.J., et al. Pediatric AIDS Clinical Trials Group . 2003. Lymphocyte subsets in healthy children from birth through 18 years of age: the Pediatric AIDS Clinical Trials Group P1009 study. J. Allergy Clin. Immunol. 112:973–980. 10.1016/j.jaci.2003.07.003 [DOI] [PubMed] [Google Scholar]
- Ssewanyana I., Elrefaei M., Dorsey G., Ruel T., Jones N.G., Gasasira A., Kamya M., Nakiwala J., Achan J., Charlebois E., et al. 2007. Profile of T cell immune responses in HIV-infected children from Uganda. J. Infect. Dis. 196:1667–1670. 10.1086/522013 [DOI] [PubMed] [Google Scholar]
- Streeck H., Lichterfeld M., Alter G., Meier A., Teigen N., Yassine-Diab B., Sidhu H.K., Little S., Kelleher A., Routy J.P., et al. 2007. Recognition of a defined region within p24 gag by CD8+ T cells during primary human immunodeficiency virus type 1 infection in individuals expressing protective HLA class I alleles. J. Virol. 81:7725–7731. 10.1128/JVI.00708-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swadling L., Halliday J., Kelly C., Brown A., Capone S., Ansari M.A., Bonsall D., Richardson R., Hartnell F., Collier J., et al. 2016. Highly-Immunogenic Virally-Vectored T-cell Vaccines Cannot Overcome Subversion of the T-cell Response by HCV during Chronic Infection. Vaccines (Basel). 4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S.. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30:2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thobakgale C.F., Ramduth D., Reddy S., Mkhwanazi N., de Pierres C., Moodley E., Mphatswe W., Blanckenberg N., Cengimbo A., Prendergast A., et al. 2007. Human immunodeficiency virus-specific CD8+ T-cell activity is detectable from birth in the majority of in utero-infected infants. J. Virol. 81:12775–12784. 10.1128/JVI.00624-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thobakgale C.F., Prendergast A., Crawford H., Mkhwanazi N., Ramduth D., Reddy S., Molina C., Mncube Z., Leslie A., Prado J., et al. 2009. Impact of HLA in mother and child on disease progression of pediatric human immunodeficiency virus type 1 infection. J. Virol. 83:10234–10244. 10.1128/JVI.00921-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer R.M., McNevin J., Liu Y., Zhang S.C., Krizan R.W., Abraha A., Tebit D.M., Zhao H., Avila S., Lobritz M.A., et al. 2009. Variable fitness impact of HIV-1 escape mutations to cytotoxic T lymphocyte (CTL) response. PLoS Pathog. 5:e1000365 10.1371/journal.ppat.1000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.H., Muenchhoff M., Adland E., Carlqvist A., Roider J., Cole D.K., Sewell A.K., Carlson J., Ndung’u T., and Goulder P.J.. 2016. Paediatric non-progression following grandmother-to-child HIV transmission. Retrovirology. 13:65 10.1186/s12977-016-0300-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker A.S., Doerholt K., Sharland M., Gibb D.M., and Collaborative H.I.V.P.S.S.C.. Collaborative HIV Paediatric Study (CHIPS) Steering Committee . 2004. Response to highly active antiretroviral therapy varies with age: the UK and Ireland Collaborative HIV Paediatric Study. AIDS. 18:1915–1924. 10.1097/00002030-200409240-00007 [DOI] [PubMed] [Google Scholar]
- Wei X., Decker J.M., Wang S., Hui H., Kappes J.C., Wu X., Salazar-Gonzalez J.F., Salazar M.G., Kilby J.M., Saag M.S., et al. 2003. Antibody neutralization and escape by HIV-1. Nature. 422:307–312. 10.1038/nature01470 [DOI] [PubMed] [Google Scholar]
- Wright J.K., Brumme Z.L., Carlson J.M., Heckerman D., Kadie C.M., Brumme C.J., Wang B., Losina E., Miura T., Chonco F., et al. 2010. Gag-protease-mediated replication capacity in HIV-1 subtype C chronic infection: associations with HLA type and clinical parameters. J. Virol. 84:10820–10831. 10.1128/JVI.01084-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J.K., Naidoo V.L., Brumme Z.L., Prince J.L., Claiborne D.T., Goulder P.J., Brockman M.A., Hunter E., and Ndung’u T.. 2012. Impact of HLA-B*81-associated mutations in HIV-1 Gag on viral replication capacity. J. Virol. 86:3193–3199. 10.1128/JVI.06682-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Yorke E., Hancock G., Clutton G., Sande N., Angus B., Smyth R., Mak J., and Dorrell L.. 2013. Improved quantification of HIV-1-infected CD4+ T cells using an optimised method of intracellular HIV-1 gag p24 antigen detection. J. Immunol. Methods. 391:174–178. 10.1016/j.jim.2013.03.001 [DOI] [PubMed] [Google Scholar]
- Yusim K., Kesmir C., Gaschen B., Addo M.M., Altfeld M., Brunak S., Chigaev A., Detours V., and Korber B.T.. 2002. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J. Virol. 76:8757–8768. 10.1128/JVI.76.17.8757-8768.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuñiga R., Lucchetti A., Galvan P., Sanchez S., Sanchez C., Hernandez A., Sanchez H., Frahm N., Linde C.H., Hewitt H.S., et al. 2006. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J. Virol. 80:3122–3125. 10.1128/JVI.80.6.3122-3125.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.