Chen et al. demonstrate that human MHC selects a larger human TCR repertoire than mouse MHC. They show how humans optimized TCR diversity and suggest that CDR3 length adjusts for different V segment–MHC affinity.
Abstract
For thymic selection and responses to pathogens, T cells interact through their αβ T cell receptor (TCR) with peptide–major histocompatibility complex (MHC) molecules on antigen-presenting cells. How the diverse TCRs interact with a multitude of MHC molecules is unresolved. It is also unclear how humans generate larger TCR repertoires than mice do. We compared the TCR repertoire of CD4 T cells selected from a single mouse or human MHC class II (MHC II) in mice containing the human TCR gene loci. Human MHC II yielded greater thymic output and a more diverse TCR repertoire. The complementarity determining region 3 (CDR3) length adjusted for different inherent V-segment affinities to MHC II. Humans evolved with greater nontemplate-encoded CDR3 diversity than did mice. Our data, which demonstrate human TCR–MHC coevolution after divergence from rodents, explain the greater T cell diversity in humans and suggest a mechanism for ensuring that any V–J gene combination can be selected by a single MHC II.
Introduction
A key event in αβ T cell–mediated interactions is the binding of the TCR to its ligand in the form of short peptides, which are bound to MHC molecules on the surface of APCs. To accommodate the vast amount of antigens presented by various MHC molecules, T cells must generate a diverse αβ TCR repertoire. T cells achieve that task by recombining each one of the multiple germline-encoded variable (V), diversity (D), and joining (J) gene segments; nontemplate additions/deletions of nucleotides in the V(D)J junctional region; and random αβ chain pairing (Davis and Bjorkman, 1988). Each T cell expresses a unique TCR. Upon encountering antigens, TCRs also undergo conformational adjustments, a so-called induced-fit binding, to ensure specific recognition of respective peptide-MHCs (pMHCs; Krogsgaard and Davis, 2005).
An old question is how T cells, with such TCR diversity (theoretically ∼1015 clonotypes) and TCR plasticity, react almost exclusively in a MHC-restricted fashion and can react to almost any MHC molecule, considering the great polymorphism of MHC genes (∼15,000 variants in humans; Robinson et al., 2003). Positive selection during T cell development in the thymus imposes self-MHC restriction on T cells because only αβ T cells that bind to self-pMHC complexes with low affinity receive a survival signal (Davis and Bjorkman, 1988; Jameson et al., 1995). Approximately 15% of thymocytes induce signaling for thymic selection; of which, half are negatively selected, likely because of too great an affinity for self-pMHC and cross-reactivity (Merkenschlager et al., 1997; McDonald et al., 2015). The relatively high proportion of MHC-reactive T cells in the preselection pool (∼5–20%) or the fact that ∼10% of the peripheral T cells are MHC alloreactive indicates an intrinsic affinity of TCRs toward MHC (Blackman et al., 1986; Zerrahn et al., 1997; Suchin et al., 2001; Blattman et al., 2002). Namely, the germline-encoded complementarity determining region (CDR) 1 and CDR2 of the Vα and Vβ segments are evolutionarily conserved to react with MHC molecules, which was termed TCR germline bias (Huseby et al., 2005; Marrack et al., 2008; Garcia et al., 2009).
Compelling evidence for this hypothesis resulted from structural and mutational analysis, showing that single amino acid substitutions in a mouse Vβ CDR2, e.g., Tyr48, Tyr50, and Glu54, decreased positive selection in a TCR transgenic mouse model (Dai et al., 2008; Scott-Browne et al., 2009). Furthermore, some Vβ genes of jawed vertebrates (frog, shark, trout, and lizard), which diverged from mammals ∼400 million years ago, share sequences in the CDR2 region of mouse Vβ8.2 but otherwise exhibit little similarity. T cells with chimeric TCRs, containing such Vβ genes, e.g., derived from frogs, were positively selected in mice (Scott-Browne et al., 2011). Further evidence is mounting from the growing database of TCR–pMHC ternary, crystallographic structures (Rossjohn et al., 2015). With few exceptions (Beringer et al., 2015; Rossjohn et al., 2015), many of the solved TCR–pMHC structures to date have adapted a diagonal docking topology atop the pMHC complex. Namely, the CDR1 and CDR2 domains of TCRα or β chains fix over the α2 and α1 helix of MHC class I (MHC I) or β and α helix of MHC II, whereas the CDR3α and the CDR3β are mainly in contact with the presented peptide, respectively (Rossjohn et al., 2015; Adams et al., 2016).
However, not all V gene segments share conserved residues in CDR1 and CDR2. Therefore, it was suggested that each V segment engages to its cognate MHC through a menu of structurally coded recognition motifs that have arisen evolutionarily (Feng et al., 2007; Marrack et al., 2008; Garcia et al., 2009), a comprehensive hypothesis, which, however, is difficult to address experimentally. A number of similarly convincing studies, including the demonstration of antibody-like T cells that developed in coreceptor and MHC-deficient mice and some structural analyses of TCR–pMHC complexes did not support the TCR germline bias for MHC. Hence, it is not generally accepted that TCR and MHC coevolved (Tynan et al., 2005; Gras et al., 2010; Sethi et al., 2011; Tikhonova et al., 2012; Van Laethem et al., 2013; Beringer et al., 2015), which is not surprising, given the complex and flexible interactions that TCR and MHC can undergo.
Structural and mutational analysis of TCR–pMHC complexes depicts only a few of the billions of different possible combinations. Therefore, we wished to address the problem differently, based on several assumptions. We reasoned that thymic selection is the most sensitive readout to detect subtle differences in affinity between a defined MHC molecule and any given TCR. Even though mouse TCRs can be selected on human MHC (Kievits et al., 1987; Ito et al., 1996) and human TCRs can be selected on mouse MHC (Li et al., 2010), we assumed that mouse and human TCR and MHC gene loci further coevolved after their divergence ∼75 million years ago (Waterston et al., 2002), resulting in changes in thymic selection of a polyclonal repertoire, depending on whether the TCR–pMHC interaction was specific for inter- or intraspecies. Therefore, we employed mice with a polyclonal human αβ TCR repertoire, which were deficient for mouse αβ TCRs and expressed either a single human MHC II (HLA-DRA/HLA-DRB1*0401; HLA-DR4, hereafter) or a single-mouse MHC II gene (I-Ab). TCR deep-sequencing of peripheral CD4 T cells from both mouse lines revealed distinct differences in their repertoire, compatible with coevolution of TCR and MHC.
Results
Human TCR gene loci transgenic mice with mouse or human MHC II gene
ABabDII and ABabDR4 mice, which both contain complete human TCRα and TCRβ gene loci and a single MHC II, mouse I-Ab, or human HLA-DR4, were employed in this study. Both strains are deficient for mouse αβ TCRs. ABabDII mice contain the HLA-A*0201 gene and are deficient for mouse MHC I expression (β2m- and Db-deficient), whereas ABabDR4 mice contain two mouse MHC I genes (Kb and Db). HLA-A*0201 and HLA-DR4 are both chimeric molecules allowing mouse CD8 and CD4 coreceptor binding, respectively. The α1 and β1 regions of I-Ab share 56 and 61% homology with the human HLA-DR4 molecule at the amino acid level. Even though I-Ab and HLA-DR4 are not the closest homologues to each other, the TCR repertoire selected by either molecule can be compared, assuming that different MHC II alleles have similar ability to select a diverse repertoire. The cortical and medullary thymic epithelial cells, as well as thymic DCs, which are critical for positive and negative selection of T cells (Klein et al., 2014), expressed comparable levels of MHC II in the two mouse strains (Fig. S1).
Reduced thymic selection by mouse MHC II compared with human MHC II
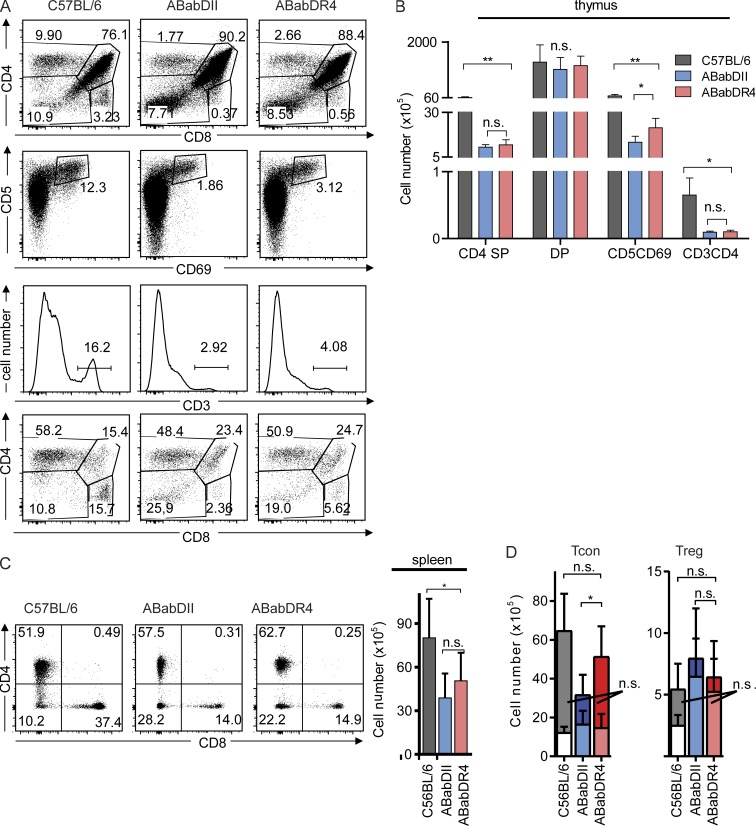
ABabDII and ABabDR4 mice contained comparable levels of double-positive, CD4 single-positive, and CD8 single-positive cells, as well as CD3+ thymocytes (Fig. 1, A and B). However, ABabDR4 mice contained more CD5/CD69-positive thymocytes (2.106 ± 0.5 × 106 cells) than ABabDII mice did (1.3 × 106 ± 0.3 × 106 cells), indicating that more T cells received a positive/negative selection signal (Fig. 1, A and B). However, thymocyte development in ABabDII and ABabDR4 mice was less efficient than it was in C57BL/6 mice (Fig. 1).
Figure 1.
T cell development in C57BL/6, ABabDII, and ABabDR4 mice. (A) Thymocyte staining. First lane, CD4, CD8 double- and single-positive cells; second lane, CD5 and CD69 staining; third lane, CD3 staining, gated on living lymphocytes; and fourth lane, CD4 and CD8 double- and single-positive cells gated on CD3+ cells. One representative FACS plot from each of C57BL/6 (n = 3), ABabDII (n = 6), and ABabDR4 (n = 6) mice. Percentage of the gated cells for the whole population is indicated inside or above the gates. (B) Numbers of CD4 single-positive, CD5+CD69+, CD4CD8 double-positive, and CD3+CD4+ thymocytes. (C, left) Representative FACS plots of CD4 and CD8 staining from splenocytes, gated on CD3+ living lymphocytes. (C, right) Absolute numbers of CD4 T cells from spleen of C57BL/6, ABabDII, and ABabDR4 mice. The percentage of positive cells is indicated in both A and C. (D) Tcon (left) and Treg (right) cell numbers from spleen. Bars with lighter color indicate the CD44+ portion of cells from each subgroup. Summarized data (B, C, and D) from C57BL/6 (n = 3), ABabDII (n = 6), and ABabDR4 (n = 6) mice, shown as means ± SD. **, 0.001 ≤ P < 0.05; *, 0.05 ≤ P < 0.1; n.s., not significant (Mann-Whitney test, two-tailed). In addition, Fig. S2 shows FoxP3 thymic and spleen staining.
In the periphery, ABabDR4 had more CD4 T cells than ABabDII mice had, which, however, did not reach statistical significance (Fig. 1 C). However, conventional CD4 T cell (Tcon; FoxP3−CD4+CD3+) numbers were significantly greater in the periphery in ABabDR4 compared with ABabDII mice (Fig. 1 D). Regulatory T cell (Treg; FoxP3+CD4+CD3+) numbers in both the thymus and spleen in ABabDII mice were comparable to those in ABabDR4 mice, but the frequency of Treg within the CD4 T cells was substantially greater in ABabDII (19.9 ± 4.4%) compared with ABabDR4 mice (10.8 ± 4.0%; Fig. 1 D and Fig. S2, A and B).
Similar numbers and frequencies of Treg cells expressed high levels of CD44, a homeostatic proliferation marker for naive T cells, although greater frequency of Tcon cells in ABabDII mice were CD44hi (51.7 ± 6.1%) compared with ABabDR4 mice (30.0 ± 15.1%; Fig. 1 D and Fig. S2, A and C). Treg cells and Tcon cells had similar Vβ usages in both ABabDII and ABabDR4 mice, based on staining with 24 human Vβ antibodies (Fig. S2 D).
Because ABabDII and ABabDR4 mice had comparable frequencies of peripheral CD8 T cells, it is unlikely that the different MHC I molecules shaped the development of CD4 T cells (Fig. 1 C). Collectively, these data showed that the development of CD4 T cells with human TCRs differed depending on whether they were selected by mouse or human MHC II.
Diverse but nonrandom V-J usage in both mice and humans
We compared the αβ TCR repertoire of CD4 T cells of ABabDII and ABabDR4 mice by quantitative deep sequencing. The CD8 T cell repertoires were not sequenced because ABabDR4 possess two MHC I alleles, whereas ABabDII mice have only one human MHC I. We included similar numbers of naive (CD62L+/CD45RO−) CD4 T cells from three human donors as a case of TCR selection on multiple MHC II alleles. All CD4 T cells were isolated by FACS sort with purities >97%. We wish to point out, however, that certain parameters, such as V(D)J usage frequency or CDR3 length, but not repertoire diversity, can be compared between the mice and humans because humans contain a set of six different MHC II alleles by which the T cells were selected. Genomic DNA from ∼2.5 × 105 purified CD4 T cells from ABabDII and ABabDR4 mice and ∼1.8 × 105 from human were submitted for sequencing. Between 0.6 × 107 and 1.6 × 107 valid reads were obtained (Table 1).
Table 1. In-frame and out-of-frame TCRα/β diversity.
| TCR | Source | In-frame nt clonotypes | In-frame amino acid clonotypes | In-frame amino acid clonotypes (%) | Out-of-frame clonotypes | Out-of-frame clonotypes (%) | Summary read |
|---|---|---|---|---|---|---|---|
| TCRα | ABabDII | 8 × 104 | 6.8 × 104 | 0.70 | 10 × 104 | 0.54 | 1.5 × 107 |
| 9 × 104 | 7.6 × 104 | 0.68 | 12 × 104 | 0.54 | 1.0 × 107 | ||
| 9 × 104 | 7.6 × 104 | 0.68 | 13 × 104 | 0.56 | 1.3 × 107 | ||
| 10 × 104 | 8.2 × 104 | 0.68 | 14 × 104 | 0.56 | 1.2 × 107 | ||
| 8 × 104 | 7.0 × 104 | 0.70 | 11 × 104 | 0.56 | 1.1 × 107 | ||
| ABabDR4 | 14 × 104 | 10.8 × 104 | 0.65 | 20 × 104 | 0.58 | 1.6 × 107 | |
| 12 × 104 | 9.5 × 104 | 0.66 | 1.8 × 104 | 0.59 | 1.0 × 107 | ||
| 11 × 104 | 8.7 × 104 | 0.68 | 1.7 × 104 | 0.59 | 1.0 × 107 | ||
| 13 × 104 | 10.7 × 104 | 0.65 | 20 × 104 | 0.59 | 1.4 × 107 | ||
| 13 × 104 | 10.4 × 104 | 0.65 | 18 × 104 | 0.56 | 1.3 × 107 | ||
| Humans | 21 × 104 | 18.6 × 104 | 0.77 | 21 × 104 | 0.49 | 1.3 × 107 | |
| 18 × 104 | 15.2 × 104 | 0.78 | 15 × 104 | 0.45 | 1.0 × 107 | ||
| 12 × 104 | 10.4 × 104 | 0.80 | 8.6 × 104 | 0.41 | 1.0 × 107 | ||
| TCRβ | ABabDII | 10 × 104 | 9.0 × 104 | 0.78 | 1.1 × 104 | 0.50 | 0.7 × 107 |
| 8 × 104 | 7.5 × 104 | 0.79 | 0.9 × 104 | 0.49 | 0.6 × 107 | ||
| 9 × 104 | 8.4 × 104 | 0.79 | 0.9 × 104 | 0.48 | 0.7 × 107 | ||
| 11 × 104 | 10.2 × 104 | 0.79 | 1.0 × 104 | 0.46 | 0.7 × 107 | ||
| 10 × 104 | 8.7 × 104 | 0.79 | 1.0 × 104 | 0.50 | 0.7 × 107 | ||
| ABabDR4 | 12 × 104 | 11.0 × 104 | 0.77 | 1.4 × 104 | 0.51 | 0.8 × 107 | |
| 12 × 104 | 11.1 × 104 | 0.77 | 1.4 × 104 | 0.51 | 0.9 × 107 | ||
| 13 × 104 | 11.7 × 104 | 0.76 | 1.5 × 104 | 0.51 | 0.9 × 107 | ||
| 16 × 104 | 14.1 × 104 | 0.76 | 1.8 × 104 | 0.51 | 0.9 × 107 | ||
| 17 × 104 | 15.0 × 104 | 0.75 | 1.9 × 104 | 0.50 | 1.0 × 107 | ||
| Humans | 13 × 104 | 12.3 × 104 | 0.92 | 3 × 104 | 0.17 | 0.6 × 107 | |
| 14 × 104 | 13.2 × 104 | 0.91 | 2 × 104 | 0.14 | 0.7 × 107 | ||
| 10 × 104 | 9.6 × 104 | 0.92 | 2 × 104 | 0.18 | 0.7 × 107 |
nt, nucleotide.
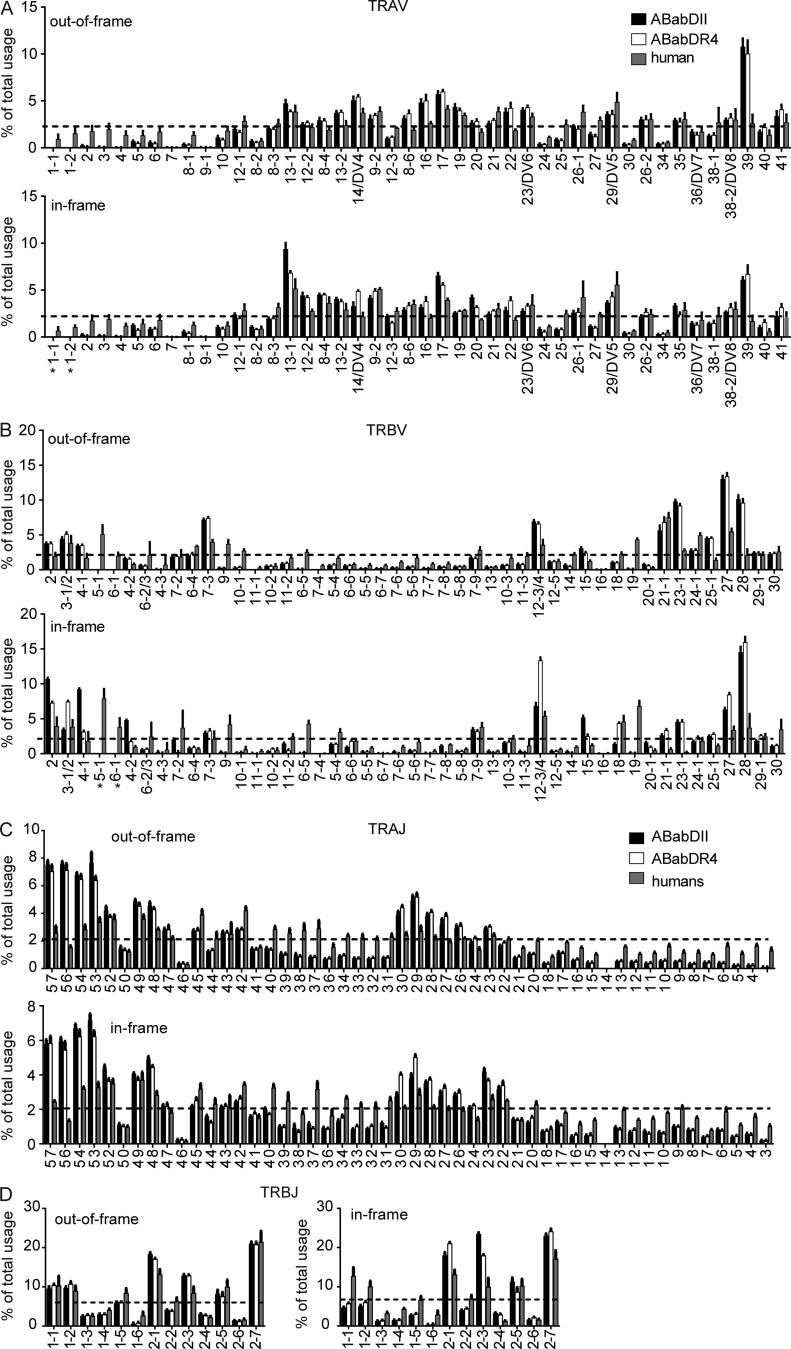
We analyzed V and J gene usages from both in-frame and out-of-frame TCRs, where the out-of-frame TCRs approximated the preselection pool (Zvyagin et al., 2014). Even though many T cells with functional TCR rearrangement are not positively selected and, thus, are part of the preselection pool (McDonald et al., 2015), the out-of-frame TCRs in T cells selected by their second functional TCR represents an unbiased estimate of V–J usage frequency. Both, the Vα and Vβ gene usage in the preselection repertoire did not differ between the two mouse groups because ABabDII and ABabDR4 mice shared the same TCR transgene loci and employed the same TCR recombination enzymes for rearrangement (i.e., RAG proteins).
Most Vα and Vβ genes were found to be rearranged, except for TRBV5-1, TRBV6-1, and TRAV1, which were previously reported to be missing or not expressed in the ABab transgenic mice (Fig. 2, A and B; Li et al., 2010). Humans showed a similar out-of-frame V gene usage. However, some Vα and Vβ genes were either more frequently (e.g., TRAV16, TRAV39, TRBV23-1, TRBV25-1, TRBV27, TRBV28) or less frequently (e.g., some of the most 5′ located Vα genes, TRBV9, TRBV10-1, TRBV6-5, TRBV19) used in the two mouse lines compared with humans. The reason for these differences is not clear but could be related, in some cases, to polymorphisms. For example, in the promoter region of TRAV39, a deletion of five nucleotides (TTTTC; available from GenBank under accession no. NC_000014, positions 22 and 125–22,130) was detected in the mouse samples, compared with the TRAV39 gene in the three human donors. A similar polymorphism (TTTTC deletion) has been observed in the CD4 promoter, which was associated with lower promoter activity (Kristiansen et al., 2004).
Figure 2.
V and J gene usage frequencies of out-of-frame and in-frame TCR clonotypes. Frequencies include TCRα (A) and TCRβ (B) V gene or TCRα (C) and TCRβ (D) J gene usage of unique TCR clonotypes in CD4 T cells of ABabDII mice (n = 5), ABabDR4 (n = 5) mice and human donors (n = 3). The gene segments were arranged on the x axis according to their position on the human chromosome from 5′ to 3′. TRAV18 usage was not detected, most likely because of PCR bias, and was excluded from the graph. Dashed line indicates frequency for random V gene usage (TRAV, 2.3%; TRBV, 2.1%). All data are shown as means ± SD. *, V genes whose expression is missing in ABab transgenic mice.
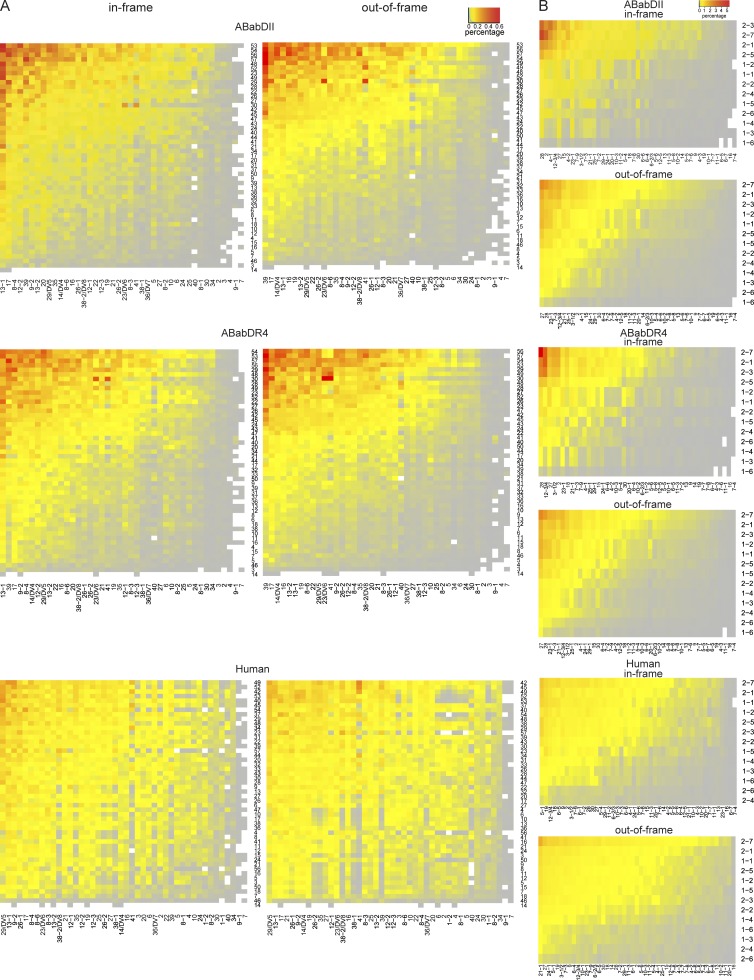
Both Vα and Vβ gene usage was nonrandom (P < 0.0001, χ2 test: actual frequencies to the random gene frequency usage; Fig. 2). The preference for Vα and Vβ gene usage appeared to be different. Vα genes that were closer to the 5′ region of the gene locus were underrepresented. Vβ genes located in the 5′ and 3′ region were preferentially rearranged similarly in mice and humans (i.e., TRBV12-3/4, TRBV21-1, and TRBV27; Fig. 2 B). Jα and Jβ usage was also nonrandom and similar between the transgenic mice and humans (Fig. 2, C and D), although the four Jα segments located closest to the 5′ end were used more frequently in mice than in humans. Most Vα–Jα and Vβ–Jβ gene combinations were detected in both the in-frame and the out-of-frame repertoire (Fig. 3). In general, the postselection repertoire for both the α and the β chains mirrored the usage pattern of the preselection repertoire similarly in ABabDII and ABabDR4 mice (Figs. 2 and 3). However, we also observed changes in the pre- versus postselection repertoire for some V genes and V–J pairing (see I-Ab and HLA-DR4 have distinct imprint in TCR selection).
Figure 3.
Mean frequencies of V–J pairing of TCR clonotypes. Frequencies include TCRα (A) and TCRβ (B) V–J pairing of unique TCR clonotypes in ABabDII mice (n = 5), ABabDR4 (n = 5) mice, and human donors (n = 3). The heat maps were arranged by frequency from greatest (red) to the least (gray; top → bottom, J segments; left → right, V segments). (Left) In-frame; (Right) Out-of-frame. TRAV1, TRBV5-1, and TRBV6-1 were excluded from the analysis for ABabDII and ABabDR4 mice; TRAV18 was excluded for all three groups.
Larger CD4 T cell repertoire in ABabDR4 compared with ABabDII mice
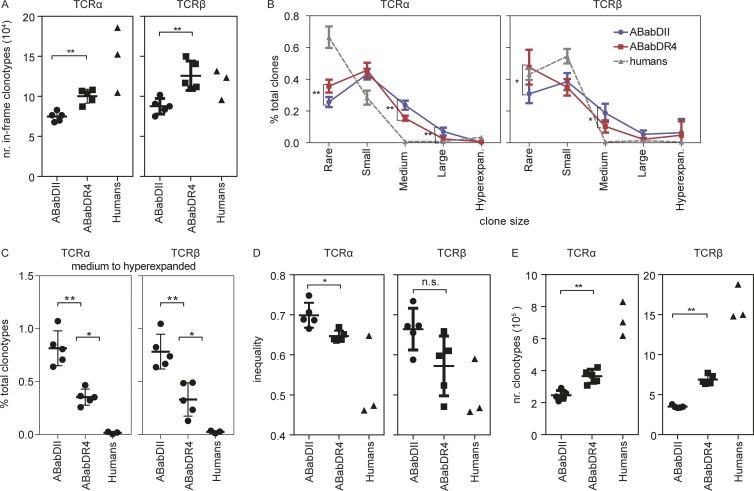
ABabDII mice were able to select a diverse human TCR repertoire with a mean of 7.5 × 104 ± 0.60 × 104 in-frame TCRα and 8.8 × 104 ± 0.97 × 104 TCRβ amino acid clonotypes from the CD4 T cells submitted for sequencing (Fig. 4 A and Table 1). Thus, I-Ab molecules can positively select all human Vα–Jα and Vβ–Jβ combinations (Fig. 3). However, HLA-DR4 selected significantly more functional TCRα and TCRβ clonotypes compared with I-Ab from the same number of T cells (10.0 × 104 ± 0.87 × 104 and 12.6 × 104 ± 1.83 × 104 TCRα and TCRβ clonotypes, respectively; Fig. 4 A and Table 1). Humans had the most TCRα clonotypes (14.8 × 104 ± 4.08 × 104), likely because of the effect of positive selection by multiple MHC II molecules but, surprisingly, similar numbers of TCRβ clonotypes (11.7 × 104 ± 1.89 × 104) compared with ABabDR4 mice.
Figure 4.
Comparison of TCRα and TCRβ repertoire among ABabDII mice, ABabDR4 mice, and human donors. (A) Absolute numbers of unique TCR amino acid clonotypes within 2.5 × 105 mouse or 1.8 × 105 human CD4 T cells. (B) Distribution of TCR amino acid clonotypes of different sizes: rare, 0 < x ≤ 0.001%; small, 0.001% < x ≤ 0.01%; mediumm 0.01% < x ≤ 0.1%; large, 0.1% < x < 1%; hyperexpanded (hyperexpan.), 1% < x < 10%. Human donor 3 was omitted from this analysis. Mann–Whitney test was used to compare clonotype sizes between ABabDII and ABabDR4 mice. (C) Percentage of medium to hyperexpanded TCR amino acid clonotypes of the total clonotypes. (D) Gini index indicating the equality of distribution of TCRα and TCRβ clonotypes (inequality). (E) Numbers of unique TCRα and TCRβ calculated using the iCHAO1 estimator. Data are from ABabDII (n = 5); ABabDR4 (n = 5) mice, and humans (n = 2 for B; n = 3 for A, C, and D). Mann–Whitney test (two-tailed) was used to compare values between ABabDII and ABabDR4 mice. **, 0.001 ≤ P < 0.05; *, 0.05 ≤ P < 0.1; n.s., not significant (Mann–Whitney test). Data from ABabDII and ABabDR4 mice are shown as means ± SD. Means and SD for humans are not depicted. In addition, Table 1 provides a summary of sequencing data and Fig. S2 provides further Treg and Tcon analysis.
There were more medium-to-large and fewer rare-to-small TCRα and TCRβ clonotypes in ABabDII mice than in ABabDR4 mice (Fig. 4, B and C). Most TCRα and TCRβ clonotypes were rare to small in the two younger human donors. The third donor, aged 60 yr, had some hyperexpanded TCR clones and was not included in the clone-size comparison. In general, ABabDR4 mice had a more homogenous TCRα and TCRβ distributions than ABabDII mice had (a mean inequality score of 0.65 ± 0.01 vs. 0.70 ± 0.03 for TCRα and 0.57 ± 0.07 vs. 0.66 ± 0.05 for TCRβ; Fig. 4 D). Two of the three human donors had the lowest inequality scores of all three groups.
Because only ∼0.5% of the total CD4 T cell repertoire from each mouse was sequenced, we applied a computational approach to determine the total TCR repertoire in CD4 T cells from the mice and humans. A lower-bound estimation on the TCR repertoire size was calculated with the acquired number of productive TCR sequences and the number of their templates detected in the sequencing samples using iCHAO1 estimator (Chiu et al., 2014). HLA-DR4 selected significantly more functional TCRα and TCRβ clonotypes (3.7 × 105 ± 0.4 × 105 and 6.9 × 105 ± 0.6 × 105 TCRα and TCRβ clonotypes, respectively) compared with I-Ab (2.4 × 105 ± 0.3 × 105 TCRα and 3.5 × 105 ± 0.2 × 105 TCRβ clonotypes; Fig. 4 E). Humans had the most of both TCRα (7.2 × 105 ± 1.105) and β clonotypes (16.2 × 105 ± 2.2 × 105), likely because of the effect of positive selection by multiple MHC II molecules.
Endogenous superantigens, such as the mouse mammary tumor virus (MMTV) superantigens, could alter CD4 T cell selection, e.g., deletion of T cells with certain Vβ segments, as observed in HLA-DR4 transgenic mice with a mouse TCR repertoire (Ito et al., 1996). We co-cultured purified CellTrace-labeled CD4 T cells from C57BL/6, ABabDII, ABabDR4 and DR4 mice with purified CD19+ cells from ABabDII, ABabDR4 or DR4 mice for 84 h. Compatible with superantigen recognition, C57BL/6 CD4 T cells responded to stimulation with B cells from DR4 and ABabDR4 mice, however, ABabDII and ABabDR4 mice, both expressing the human TCR repertoire, did not proliferate to the B cells of any mouse line (Fig. S3). Thus, we assume that human TCRs, unlike mouse TCRs, only weakly (or not at all) interact with MMTV and conclude that endogenous mouse superantigens did not obscure our results.
I-Ab and HLA-DR4 have distinct imprint in TCR selection
In general, after thymic selection the usage pattern remained similar for both Vα and Vβ to what it was before selection. However, we also observed changes for some V genes. For example, TRBV4-1 was strongly preferred by I-Ab, but not HLA-DR4, molecules (Fig. 2). Interestingly, TRBV4-1 is evolutionarily related to a mouse TCRβ chain (Vβ8.2) in the CDR2 region (Scott-Browne et al., 2011). Similarly, TRAV13-1 and TRBV2 were preferentially selected by I-Ab molecules. Conversely, some V genes were preferentially selected by HLA-DR4 but not I-Ab molecules, i.e., TRBV3-1/2 and TRBV12-3/4.
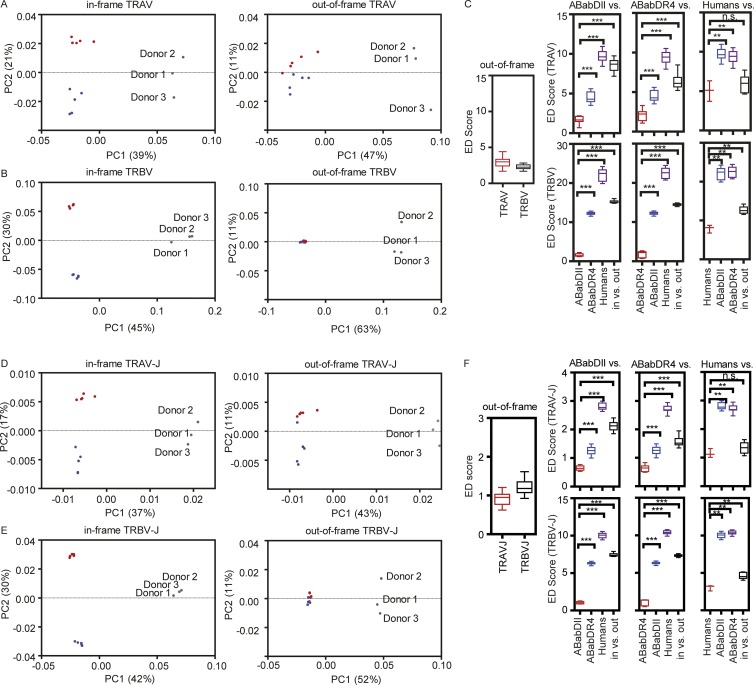
To analyze the V gene usage pattern more globally, an unsupervised method, principle component analysis (PCA), was performed on the V gene usage profiles of the three groups. PCA separated the human samples on the dominant axis (PC1) but did not do so for the two mouse groups for both in-frame and out-of-frame TCRs, which was also reflected by their closed Euclidean distance (Vα: 3.0 and Vβ: 2.3; Fig. 5, A–C). Before selection, the only variable among the three groups may have been solely species difference. Only after thymic selection did ABabDII and ABabDR4 mice and humans separate from each other and cluster on the PC2 axis, suggesting that different MHC II and number of MHC II molecules influenced the V gene usage and similarly for Vα (Fig. 5 A) and Vβ genes (Fig. 5 B). Indeed, in-frame V gene usage was rather similar within groups (ED score ∼1–2) but significantly different between groups (Fig. 5 C). Based on that observation, we grouped the V genes whose usage frequency changed significantly compared with out-of-frame TCR for the two mouse strains into “overrepresented,” “unchanged,” and “underrepresented” (Fig. 6 A). The same distribution pattern and clustering of the three groups in the post- versus preselection repertoire was observed for Vα–Jα and Vβ–Jβ combinatorial events (Fig. 5, D and E) and, surprisingly, also for Jα and Jβ usage frequency (Fig. S4).
Figure 5.
PCA correlations among mice and humans for V and V–J usage. PCA shows the correlation between ABabDII mice (blue dots), ABabDR4 mice (red dots), and human donors (gray dots) for their TRAV usage (A) and TRBV usage (B) of unique TCR clonotypes. (Left) PCA of in-frame sequences; (right) PCA for out-of-frame sequences. TRAV1-1, TRAV1-2, TRBV5-1, and TRBV6-1 were excluded. Each dot represents an individual of ABabDII mice, ABabDR4 mice, or humans. Proportions of variance (PC1 and PC2) are indicated at the axis. (C) Comparisons of V segment usages restricted to the same or different MHC alleles. (Left) Similarity comparisons of out-of-frame V gene usages in ABabDII and ABabDR4. (Right, top and bottom lanes) Similarity comparisons of in frame V segment usages within and between groups of ABabDII mice, ABabDR4 mice, and humans and to the out-of-frame sequences. (Top) TCRα (TRAV) V segments; (bottom) TCRβ (TRBV) V segments. (D and E) PCA of TRAV-J (D) and TRBV-J (E) pairing usage of unique TCR clonotypes, arranged the same as in A and B. (F) Comparisons of V–J pairing usages restricted to the same or different MHC allele(s), arranged the same as in C. ***, P ≤ 0.001; **, 0.001 ≤ P < 0.05; and *, 0.05 ≤ P < 0.1; n.s., not significant (Mann–Whitney test, two-tailed). ED, Euclidean distance. Data are from ABabDII mice (n = 5), ABabDR4 mice (n = 5), and humans (n = 3). Fig. S4 provides PCA analysis of J segment usages.
Figure 6.
Grouped V gene analysis. (A) Heat maps of TCR V gene usage changes after thymic selection. Red represents the V genes that were overpresented after thymic selection, yellow were the ones unchanged, and gray to blue were ones that were underrepresented. Values were calculated as log2 of the ratio of Frequency Vin-frame/Frequency Vout-of-frame. (B) V genes were grouped into overrepresented, unchanged, or underrepresented, and their CDR3 length distributions are shown in bars (mean ± SD). Gaussian distributions were assumed and are shown in lines for all groups. Data are from ABabDII mice (n = 5), ABabDR4 mice (n = 5), and humans (n = 3).
A skewed V–J pairing was observed for functional TCRα and β chains compared with the preselection pool (Fig. 3, A and B). ABabDII and ABabDR4 had almost the same out-of-frame V–J pairing patterns, and the TRAV13-1-TRAJ-54/53 and TRBV28-TRBJ2-3/7 were the most prominently selected in both strains. Despite those similarities, the two MHC II molecules also had their own features, e.g., TRBV12-3/4-TRBJ2-1 was the second most selected in ABabDR4 mice, whereas TRBV2-TRBJ2-7 was the one in ABabDII mice (Fig. 3 B). The distinctive patterns of V–J pairing between the two mouse strains, which are imposed by mouse or human MHC II, could also be seen in the PCA analysis (Fig. 5, D and E) and, by comparison, in their Euclidean distance (Fig. 5 F).
HLA-DR4 selects a longer TCRβ CDR3 compared with I-Ab
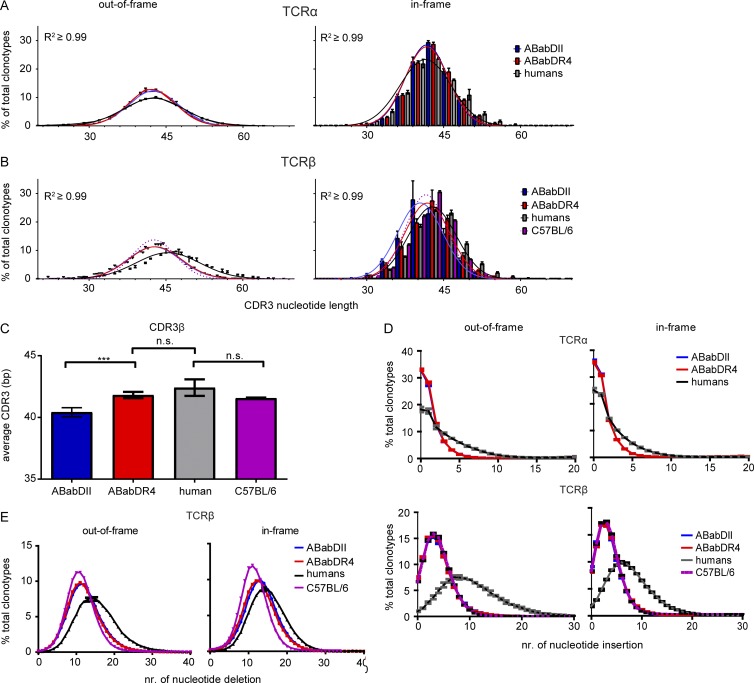
The V(D)J junctional region (CDR3) generates most diversity within the TCRs and is the major region for antigen contact and recognition. ABabDII and ABabDR4 mice showed rather similar mean TCRα CDR3 length (∼42 bp) in the preselection as well as the postselection pool (Figs. 6 B and 7 A). After selection, the CDR3 length distribution narrowed, but ABabDR4 mice contained a wider range of CDR3 length than ABabDII did. The TCRα CDR3 length distribution in humans was quite similar to that in the mice, yet slightly broader before or after selection (Fig. 7 A).
Figure 7.
CDR3 region analysis. CDR3 length distribution of TCRα (A) and TCRβ (B) clonotypes. For the in-frame clonotypes, the frequencies of different CDR3 lengths for ABabDII mice, ABabDR4 mice, humans, and C57BL/6 mice (only TCRβ) are shown in bar diagrams. For the out-of-frame clonotypes, only the mean frequency and SD for each CDR3 length are shown for each group at the corresponding length. Gaussian distributions were assumed and are shown as lines for all groups, including the C57BL/6 CDR3β (purple, dotted lines; R2 ≥ 0.99 for all samples). (C) Comparison of means of CDR3β lengths among ABabDII, ABabDR4, and C57BL/6 mice and humans. (D) Frequency of TCRs with the same number of CDR3 nucleotide insertions for TCRβ chains; insertions in C57BL/6 mice were included (purple dotted lines). (E) Frequency of TCRs with the same number of CDR3 nucleotide deletion of TCRβ chains from ABabDII mice, ABabDR4 mice, humans, and C57BL/6 mice. (D and E, left) Out-of-frame clonotypes from TCRα (top)/β (bottom). (D and E, right) In-frame. For all panels, **, 0.001 ≤ P < 0.05; n.s., not significant (Mann–Whitney test, two-tailed). Data are shown as means ± SD. Data are from ABabDII (n = 5), ABabDR4 (n = 5), and C57BL/6 (n = 3) mice and humans (n = 3).
The TCRβ CDR3 length distribution also did not differ in the preselection pool between the two mouse strains; however, humans generated, on average, longer and broader CDR3 region (Fig. 7, B and C). After selection, the CDR3 length distribution narrowed as seen for TCRα. The peak of CDR3 length remained the same in ABabDR4 mice postselection (42 bp), approaching that observed in humans, but decreased in ABabDII mice to 39 bp (Fig. 7, B and C). Most TCRβ chains selected by HLA-DR4 molecules had, on average, one amino acid longer CDR3 compared with those selected by I-Ab molecules (Fig. 7, B and C). On average, CDR3 was longer in humans, compared with the two mouse strains in both the pre- and postselection repertoire, which was more apparent for the TCRβ chain, likely because of two recombinatorial events (V–D–J; Fig. 7 C).
To determine whether the human MHCII molecule is imprinted to select for longer CDR3, we deep-sequenced the mouse TCRβ repertoire from CD4 T cells isolated from three C57BL/6 mice. Mouse TCRβ selected on the mouse I-Ab molecule showed a rather similar mean CDR3 length to ABabDR4 mice and human postselection (Fig. 7, B and C). Thus, selection of TCRβ chains with shorter CDR3 was a specific feature of ABabDII mice.
CDR3 length is the net result of exonuclease and terminal deoxynucleotidyl transferase (TdT) activity. ABabDII, ABabDR4, and C57BL/6 mice had similar exonuclease and TdT activity, reflected by their almost identical number of bp deletions and insertions in the CDR3 region of TCRα (Fig. 7 D) and β chains (Fig. 7, D and E). Compared with mice, humans revealed, on average, more deletions and insertions and, thus, had more exonuclease and TdT activity. When analyzing the CDR3 length in groups according to their V gene usage frequencies, the underrepresented Vβ genes in ABabDII mice had, on average, the shortest CDR3 length, whereas in ABabDR4 mice, they had, on average, the longest CDR3 length (Fig. 6 B).
Shared TCRα and β clonotypes
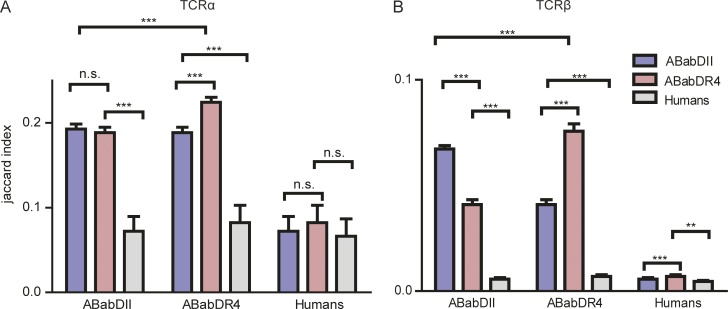
The number of shared clonotypes (based on amino acid sequences) after selection within and between the two mouse strains and humans was strikingly greater than if the repertoire is created randomly (Fig. S5, A and B; Robins et al., 2010). More shared clonotypes were detected within, compared with between, groups (Fig. 8), and ABabDR4 shared more clonotypes among each other (Jaccard index: 0.224 ± 0.006 for TCRα and 0.076 ± 0.003 for TCRβ) than ABabDII mice did (0.193 ± 0.006 for TCRα and 0.067 ± 0.002 for TCRβ).
Figure 8.
TCR shared repertoire analysis from ABabDII mice, ABabDR4 mice, and humans. Jaccard index scores within and between groups of ABabDII mice, ABabDR4 mice, and human samples for both TCRα (A) and TCRβ (B). **, 0.001 ≤ P < 0.05; ***, P ≤ 0.001 (Mann–Whitney test, two-tailed). The data shown are means ± SD from ABabDII mice (n = 5), ABabDR4 mice (n = 5), and humans (n = 3). Additional information in Fig. S5.
The number of shared clonotypes beyond MHC restriction was surprisingly high, and the number of shared TCRα clonotypes (0.189 ± 0.006) was significantly greater than shared TCRβ (0.041 ± 0.002) between ABabDII and ABabDR4 mice, likely because of one compared with two somatic recombination events. ABabDII and ABabDR4 generated more shared TCRα and β than they shared with humans (ABabDII with humans: TCRα 0.072 ± 0.017 and TCRβ 0.0057 ± 0.001; ABabDR4: TCRα 0.082 ± 0.020 and TCRβ 0.0070 ± 0.001), most likely because of the higher genetic similarity between the two mouse strains. Notably, random recombination would yield virtually no shared clonotypes within 2.5 × 105 CD4 T cells. The shared clonotypes within and among groups cumulated linearly with the increase in total clonotypes (Fig. S5 B).
Stronger CD4 T cell responses in ABabDR4 compared with DR4 mice
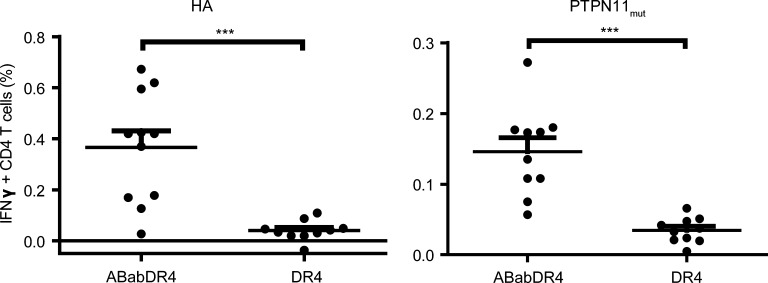
We investigated whether ABabDR4 mice could more efficiently respond to immunization than did DR4 mice expressing a mouse TCR repertoire selected on HLA-DR4. Therefore, we immunized ABabDR4 and DR4 mice with the two HLA-DR4–presented peptides derived from hemagglutinin (HA307–319) or PTPN11mut. PTPN11mut is a somatic mutation in cancer with a single amino acid substitution but is otherwise identical to the mouse homologue. The percentage of IFN-γ+ CD4 T cells in response to both peptides was significantly higher in ABabDR4 compared with DR4 mice (0.37% ± 0.22% vs. 0.04% ± 0.04% for HA307–319 and 0.15% ± 0.06% vs. 0.03% ± 0.02% for PTPN11mut peptide, respectively; Fig. 9). The data suggest that ABabDR4 have more HA307–319 and PTPN11mut CD4 T cell precursors than DR4 mice.
Figure 9.
Comparison of CD4 T cell responses against two HLA-DR4 epitopes between DR4 and ABabDR4 mice. ABabDR4 and DR4 mice were immunized twice with hemagglutinin peptide 307–319 (HA) or PTPN11G503A peptide 492–506 (PTPN11mut). At 2 wk after the second immunization, CD4 T cell responses were detected with peptide restimulation of draining LN cells, followed by intracellular IFN-γ staining. Percentages of IFN-γ+ CD4 T cells were measured by flow cytometry. Gates were set on live CD3+ lymphocytes. Each dot represents one mouse. Results are from two experiments combined and shown as mean ± SD. ***, P < 0.001 (Mann–Whitney test, two-tailed).
Discussion
We felt that the controversial discussion about whether and how the TCR and MHC coevolved reached a dead end with good arguments for either site. This is because, in most cases, single TCR–pMHC interactions were analyzed, either by resolving crystallographic structures or by analyzing mutations in the germline-encoded CDR1 and CDR2 (Marrack et al., 2008; Rossjohn et al., 2015). Although the data provided convincing evidence for TCR–MHC coevolution, inherent in the immune system, there are usually exceptions to that rule, so that it is not always easy to distinguish what is the exception and what is the rule. For example, a TCR germline bias (Garcia et al., 2009) is difficult to reconcile with the observation that 85% of the thymocytes do not receive a selection signal and, therefore, apparently lack a sufficient affinity for self-pMHC and that only 7.5% of the thymocytes are MHC or pMHC cross-reactive (Blackman et al., 1986; Merkenschlager et al., 1997; Zerrahn et al., 1997; Huseby et al., 2005; McDonald et al., 2015). Therefore, we addressed the issue differently, based on the assumption that evolving differences in the inherent affinity between TCR and MHC in mice and humans are subtle and that thymic selection is the most sensitive read-out to detect such differences. We investigated the human αβ TCR repertoire because previous studies focused mainly on mouse TCR–MHC interactions, and we analyzed the polyclonal repertoire to encompass the ability of a single MHC allele to select T cells with any possible, functionally rearranged TCR. By comparing the pre- and postselection repertoire selected on a single mouse or human MHC II allele, we indirectly addressed the inherent germline-encoded affinity for any human Vα (Jα) or Vβ (Jβ) segment, which evolved during the some 70-million-year divergence of rodents and humans. Our conclusions became apparent only through massive parallel TCR deep sequencing.
Both TCRα and TCRβ V–J gene usage in CD4 T cells as well as V–J combinatorial frequencies are highly biased, dramatically limiting the theoretically possible T cell repertoire, which was known for the TCRβ repertoire (Robins et al., 2010; Rubelt et al., 2016). The nonrandom usage is hardwired in the human TCR gene loci. It is imprinted in the postselection repertoire but shaped by the respective selecting MHC II molecule, shown by the PCA, and the higher number of shared clonotypes within than between the two mouse lines. Surprisingly, many TCRβ clonotypes were shared, and even more TCRα clonotypes were shared between humans and human MHC II-expressing mice. ABabDR4 mice shared more TCR clonotypes (11% ± 0.3% TCRα and 1.3 ± 0.1% TCRβ chains) with humans than ABabDII mice did (9.6% ± 0.2% TCRα and 1.1 ± 0.2% TCRβ). The abundance of shared TCRα or TCRβ single chains between different species and, independent of the MHC II profile, suggests that αβ chain combinatorial pairing has a larger role for creating diversity than previously thought (Arstila et al., 1999). We could not detect more shared clonotypes between ABabDR4 mice and the only HLA-DR4+ human, which is not surprising because humans bear six MHC II alleles, and which TCR is restricted to which MHC II allele is not known.
Mouse MHC II molecules almost perfectly select a human TCR repertoire, but only, almost. Basically, all human TCRα V-J and TCRβ V–D–J gene combinations were detected in the postselection repertoire of ABabDII mice, demonstrating that “structurally coded recognition motifs for MHC” (Marrack et al., 2008; Garcia et al., 2009) have been selected and fixed in most human V genes before mouse–human divergence. However, ABabDII mice had reduced thymic output and a greater clonality. The difference in I-Ab and HLA-DR4 in selecting a human TCR repertoire became clearly visible in the global comparison when ABabDR4 mice generated 30% more of both TCRα and TCRβ unique clonotypes (amino acids) than did ABabDII mice. This provides a strong hint that mouse MHC II molecules, at least I-Ab, do not select as efficiently a diverse, human αβ TCR repertoire as human MHC II molecules.
The increased TCR repertoire selected by HLA-DR4 compared with I-Ab molecules is likely due to a slightly increased inherent affinity, likely in the CDR1 and CDR2 regions (Marrack et al., 2008), of many human Vα genes for HLA-DR4. The increased TCRβ repertoire in ABabDR4, compared with ABabDII, mice is directly reflected in the CDR3 length. ABabDII and ABabDR4 mice revealed similar CDR3β length distribution in the preselection repertoire. After selection, the mean length of CDR3β selected by human MHC II in ABabDR4 mice and humans was one amino acid longer than that selected by mouse MHC II. The mean, shorter CDR3β was not seen in C57BL/6 mice, where the TCR and MHC were species compatible. Thus, the most likely explanation is that species-specific TCRs evolved to have an optimal intrinsic affinity for their own MHC or vice versa. Assuming that the intrinsic affinity is not optimal between many human TCRs and mouse MHC II, the peripheral CD4 T cell repertoire in ABabDII mice had to adopt a shorter CDR3 domain to become positively selected (Gilfillan et al., 1995; Marten et al., 1999; Yassai et al., 2002). Shorter CDR3 domains increase the risk that T cells will be cross-reactive (Gavin and Bevan, 1995; Huseby et al., 2008). In line with that finding, ABabDR4 mice had more peripheral Tcon cells, compared with ABabDII mice and, interestingly, the frequency of Treg within the CD4 T cell population was substantially higher in ABabDII (19.9 ± 4.4%) compared with ABabDR4 mice (10.8 ± 4.0%). Thus, we assume that CD4 T cells selected for short CDR3 of human TCRs by mouse MHC II are at higher risk of ending as Tregs because of cross-reactivity. Collectively, the interspecies incompatibility between TCR and MHC further supports TCR–MHC coevolution after divergence of the two species. However, the TCR repertoires selected by the closest homologues, e.g., I-Ab and HLA-DQ, need to be analyzed.
In ABabDII mice, underrepresented V genes seem to have suboptimal affinity for the I-Ab molecule because their CDR3 were, on average, the shortest. In contrast, the underrepresented V genes selected by HLA-DR4 had, on average, the longest CDR3. Thus, the underrepresented human V genes in ABabDII mice may have retained or gained affinity for HLA-DR4 but lost it for I-Ab. On the other hand, the underrepresented V genes in ABabDR4 mice may have a too-high, inherent affinity for HLA-DR4, assuming that longer CDR3 decrease the affinity. The picture may change with different MHC II alleles; each of which, may have a set of preferred and nonpreferred V genes (Sharon et al., 2016). We hypothesize that different, inherent affinity of any V segment for any MHC allele is adjusted by CDR3 length, ensuring that T cells with any V segment can be positively selected by any MHC allele, which is supported by the diverse human TCR repertoire in ABabDII mice. HLA-DR4 is capable, but less efficient, in selecting a mouse TCR repertoire than I-Ab because CD4 T cell responses were less efficient in HLA-DR4 transgenic, compared with ABabDR4, mice.
Mice generate, on average, a shorter TCRβ CDR3 region than do humans, which can be seen in the preselection repertoire. Surprisingly, the human recombination machinery evolved to both excise and add more nucleotides in the TCRβ V–D and D–J junctional regions, which is executed by combined exonuclease and TdT activity. Both enzymatic activities might increase TCR diversity. This evolutionary process provides a reasonable explanation for the larger T cell repertoire in humans, which was estimated to be 20-fold higher than that of mice (Arstila et al., 1999; Casrouge et al., 2000; Nikolich-Žugich et al., 2004; Vrisekoop et al., 2014).
In conclusion, our findings suggest that human TCRα and TCRβ V genes acquired an inherent affinity for MHC II before separation of rodents and humans. Afterward, human MHC and TCR gene loci further coevolved to maintain inherent affinity, and in order to compensate for nonrandom V(D)J usage, to increase T cell diversity by focusing on larger nontemplate-encoded CDR3 diversity. Our data also suggest that CDR3 length adjusts for different inherent V segment–MHC affinity and that T cells with shorter CDR3β are at increased risk of becoming Tregs.
Materials and methods
Mice
All mouse studies were performed in accordance with institutional, state, and federal (Landesamt für Arbeitsschutz, Gesundheitsschutz und technische Sicherheit, Berlin, Germany) guidelines. C57BL/6 and HLA-DR4 mice were purchased from The Jackson Laboratory and Taconic, respectively. ABabDII transgenic mice have been previously described (Li et al., 2010). ABabDR4 mice were established by crossing ABab transgenic mice (Li et al., 2010) to HLA-DR4 mice (Ito et al., 1996) and selecting mice homozygous for human TCRα and TCRβ gene loci, HLA-DR4 transgene, as well as mouse TCRα, TCRβ, I-Eα, and I-Aβ deficiency. The genotype of the mice was confirmed by PCR. Mice were bred in the Max-Delbrück-Center animal facility under specific pathogen–free condition and were on a mixed 129SV, C57BL/6, and BALB/c genetic background. Mice aged between 8 and 16 wk were used in this study.
Human donors
Three healthy human donors, aged 30, 48, and 60 yr at the time of blood collection, volunteered to donate blood with informed consent. Blood collecting and processing was performed according to human experimental guidelines under license EA4/046/10 (Ethikkommission). The MHC II profiles were determined by genotyping (Zentrum für Humangenetik und Laboratoriumsdiagnostik, Martinsried, Germany). Details are as follows: donor 1: HLA-DRB1 08:01, 11:12; HLA-DRB3 02; HLA-DQB1 03:01, 04:02; HLA-DPB1 03:01, 04:02; donor 2: HLA-DRB1 04:01, 15:01; HLA-DRB4 01:03; HLA-DRB5 01:01; HLA-DQA1 01:02, 03:01; HLA-DQA1 01:02, 03:01; HLA-DQB1 03:02, 06:02; HLA-DPA1 01:03; HLA-DPB1 04:01; and donor 3: HLA-DRB1 01:01, 13:01; HLA-DRB3 02:02; HLA-DQA1 01:01, 01:03; HLA-DQB1 05:01, 06:03; HLA-DPA1 01:03; HLA-DPB1 04:01.
Flow cytometry
Fluorochrome-conjugated antibodies specific for mouse CD4 (GK1.5), CD8a (53–6.7), CD5 (53–7.3, isotype: rat IgG2a, κ), IFN-γ (XMG1.2), CD45 (30-F11, isotype: rat IgG2b, κ), CD326 (EpCAM, G8.8, isotype: rat IgG2α, κ), I-Ab (AF6-120.1), CD11c (N418), and FoxP3 (MF-14), and human CD3 (HIT3a), CD8a (HIT8a), CD4 (OKT4), CD45RA (Hl100), CD45RO (UCHL1), and CD62L (DREG-56) were obtained from BioLegend. Mouse CD3ε (145-2C11), CD69 (H1.2F3, isotype: Armenian hamster IgG), Ly51 (6C3, isotype: rat IgG2a, κ), and HLA-DR (L243) specific antibodies were purchased from BD. The TCR Vβ repertoire kit (IOTest Beta Mark) was purchased from Beckman Coulter. UEA I lectin was obtained from GeneTex. Thymus, spleen, and LNs from 1–2-mo-old C57BL/6, ABabDII and ABabDR4 mice were isolated. Cells were obtained by mashing the organs through a 0.45-µm cell strainer. Isolation of thymic DCs and epithelial cells was performed as published (Xing and Hogquist, 2014). In brief, thymic lobes were digested in enzyme solution (RPMI-1640 medium with 0.05% Liberase TH and 100 U/ml of DNase I) at 37°C for 20 min. Single cells were then stained with antibodies specified in the respective figure legends and analyzed by flow cytometry (FACSCanto II; BD). FoxP3 staining was performed with True-Nuclear transcription factor buffer set from BioLegend.
FACS sorting
For mouse CD4 T cells, pooled cells from mouse spleen and LNs were collected. For human naive (CD45RO− and CD62L+) CD4 T cell isolation, ∼50 ml fresh blood from human donors was collected, and PBMCs were isolated by the Ficoll density centrifugation method. The cells were sorted by a FACS sorter (FACSARIA III; BD), and the purity for all samples was >95%.
Assay for detection of MMTV
Mouse CD4 T cells and CD19 cells from spleens of C57BL/6, DR4, ABabDII, and ABabDR4 mice were purified using mouse CD4+ T cell isolation kit and CD19 MicroBeads (Miltenyi Biotec). Subsequently, the purified CD4 T cells were labeled with CellTrace Violet (Thermo Fisher Scientific). 3 × 105 labeled CD4 T cells were co-cultured with 1.5 × 106 CD19 cells from different mouse strains at a ratio of 1:5 in a 96-well, round-bottom plate for 84 h at 37°C, with 5% CO2. Measurement of the proliferated CD4 T cells was accessed by flow cytometry.
Genomic DNA isolation and TCR deep sequencing
Genomic DNA was extracted with the QIAGEN blood and tissue kit, quantified with a NanoDrop 1000 (Thermo Fisher Scientific), and stored at −80°C. TCRα and TCRβ deep sequencing and quantification was performed on an immunoSEQ platform (Adaptive Biotechnologies). The technique has a sensitivity of 1 in 200,000 T cells and was optimized to minimize the effect of PCR bias introduced in the first multiplex PCR step (Robins et al., 2009; Carlson et al., 2013). 1.2 µg of genomic DNA, which corresponds to ∼2.5 × 105 mouse and ∼1.8 × 105 human CD4 T cells, was sequenced for each sample. TCR sequences were delineated according to the definition established by the International ImMunoGeneTics Information System collaboration.
Immunization
ABabDR4 and DR4 mice were immunized twice at an interval of 4 wk. 80 µg hemagglutinin peptide HA307–319 (KYVKQNTLKLATG) or mutant peptide PTPN11492–506 (KTIQMVRSQRSMVQ; G503A mutation), mixed with 100 µl IFA, and 50 µg CpG oligonucleotides were injected s.c. on both sides of the tail base of each mouse. 14 d after the second immunization, the draining LNs were isolated, single cells were restimulated in vitro for 12 h with the respective peptides, and the IFN-γ production by CD4 T cells was measured intracellularly using the kit and protocol from BD (Cytofix/Cytoperm kit).
Data analysis
Data analysis and statistics were performed in Excel (Microsoft), R (R Foundation for Statistical Computing), and Prism (GraphPad Software).
Gene segment (V, J and V-J pairing) frequencies
Calculation of random distribution frequencies was estimated by the reciprocal of the total number of functional TCR genes (V, J, or V–J pairing).
iCHAO1 estimator
The TCRα and β repertoire sizes of ABabDII and ABabDR4 mice and humans were estimated based on the deep-sequenced samples using iCHAO1 estimator provided with the immunoSEQ platform (Chiu et al., 2014).
Inequality Score
Inequality (Gini index) analysis on the in-frame TCR amino acid clonotypes was based on the Lorenz curve.
PCA
PCA was performed based on V, J, or V–J combinatorial frequencies from ABabDII and ABabDR4 mice and humans with the “prcomp” function in R software without data normalization (centralizing data). TRAV1, TRBV5-1, and TRBV6-1 were excluded from the analysis because they were known to be missing in the transgenic mouse repertoire.
Euclidean distance
The Euclidean distance (ED score) was calculated, as shown in Eq. 1, to evaluate the similarities of V gene or V–J pairing usage frequencies within and between different groups:
| (1) |
Estimation of Gaussian CDR3 length distribution
All CDR3 lengths from ABabDII and ABabDR4 mice and humans were assumed to have a Gaussian distribution (R2 > 0.99). We used the variance (square of the SD) of the predicted Gaussian curve to depict the width of the CDR3 distributions.
Absolute number of shared clonotypes
The number of shared clonotypes from any two of the reshaped samples was calculated using the “intersect” function in R software, package tcR (Nazarov et al., 2015).
Jaccard index for similarity analysis
The αβ TCR similarities between any two samples were evaluated with the Jaccard index, which uses the number of shared TCR clonotypes by the number of total clonotypes from the two samples, as shown in Eq. 2:
| (2) |
where A and B represent TCRα or β repertoires from any two samples.
Sample size
No specified effect size was used to determine sample sizes.
Data availability
TCR sequencing data underlying this study can be analyzed and downloaded from the Adaptive Biotechnologies immuneACCESS site at https://doi.org/10.21417/B7ZD0D.
Online supplemental material
Fig. S1 includes MHC II staining of thymic APCs from C57BL/6, ABabDII, and ABabDR4 mice. Fig. S2 includes additional data related to Fig. 1, showing FoxP3 and CD44 frequencies in C57BL/6, ABabDII, and ABabDR4 mice, including one representative staining and total columns for Treg and Tcon Vβ usages in ABabDII and ABabDR4 mice. Fig. S3 shows CD4 T cell responses to MMTV superantigen-presenting CD19 cells from C57BL/6, DR4, ABabDII, and ABabDR4 mice. Fig. S4 includes additional data related to Fig. 5, showing PCAs of TRAJ/BJ usages in ABabDII and ABabDR4 mice and human donors. Fig. S5 includes additional data related to Fig. 8, showing the absolute TCRα/β clonotypes shared either in total or from the most- to the least-abundant clonotypes among ABabDII and ABabDR4 mice and human donors.
Supplementary Material
Acknowledgments
We thank M. Obenaus for suggestions in data interpretation, bioinformatic support, and critical reading of this manuscript; M. Manzke and C. Westen for genotyping the mice; and A. Dhamodaran for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft through SFB-TR36.
The authors declare no competing financial interests.
Author contributions: X. Chen developed the concept, performed experiments, analyzed the data, and wrote the manuscript; L. Poncette performed the immunization experiments, analyzed the data, and revised the manuscript; and T. Blankenstein developed the concept, analyzed the data, and wrote the manuscript.
Footnotes
Abbreviations used:
- HA
- hemagglutinin
- MMTV
- mouse mammary tumor virus
- PCA
- principle component analysis
- pMHC
- peptide-MHC
- Tcon
- conventional CD4 T cell
- TdT
- terminal deoxynucleotidyl transferase
- Treg
- regulatory T cell
References
- Adams J.J., Narayanan S., Birnbaum M.E., Sidhu S.S., Blevins S.J., Gee M.H., Sibener L.V., Baker B.M., Kranz D.M., and Garcia K.C.. 2016. Structural interplay between germline interactions and adaptive recognition determines the bandwidth of TCRpeptide-MHC cross-reactivity. Nat. Immunol. 17:87–94. 10.1038/ni.3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arstila T.P., Casrouge A., Baron V., Even J., Kanellopoulos J., and Kourilsky P.. 1999. A direct estimate of the human αβ T cell receptor diversity. Science. 286:958–961. 10.1126/science.286.5441.958 [DOI] [PubMed] [Google Scholar]
- Beringer D.X., Kleijwegt F.S., Wiede F., van der Slik A.R., Loh K.L., Petersen J., Dudek N.L., Duinkerken G., Laban S., Joosten A., et al. 2015. T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex. Nat. Immunol. 16:1153–1161. 10.1038/ni.3271 [DOI] [PubMed] [Google Scholar]
- Blackman M., Yagüe J., Kubo R., Gay D., Coleclough C., Palmer E., Kappler J., and Marrack P.. 1986. The T cell repertoire may be biased in favor of MHC recognition. Cell. 47:349–357. 10.1016/0092-8674(86)90591-X [DOI] [PubMed] [Google Scholar]
- Blattman J.N., Antia R., Sourdive D.J., Wang X., Kaech S.M., Murali-Krishna K., Altman J.D., and Ahmed R.. 2002. Estimating the precursor frequency of naive antigen-specific CD8 T cells. J. Exp. Med. 195:657–664. 10.1084/jem.20001021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson C.S., Emerson R.O., Sherwood A.M., Desmarais C., Chung M.-W., Parsons J.M., Steen M.S., LaMadrid-Herrmannsfeldt M.A., Williamson D.W., Livingston R.J., et al. 2013. Using synthetic templates to design an unbiased multiplex PCR assay. Nat. Commun. 4:2680 10.1038/ncomms3680 [DOI] [PubMed] [Google Scholar]
- Casrouge A., Beaudoing E., Dalle S., Pannetier C., Kanellopoulos J., and Kourilsky P.. 2000. Size estimate of the α β TCR repertoire of naive mouse splenocytes. J. Immunol. 164:5782–5787. 10.4049/jimmunol.164.11.5782 [DOI] [PubMed] [Google Scholar]
- Chiu C.-H., Wang Y.-T., Walther B.A., and Chao A.. 2014. An improved nonparametric lower bound of species richness via a modified good-turing frequency formula. Biometrics. 70:671–682. 10.1111/biom.12200 [DOI] [PubMed] [Google Scholar]
- Dai S., Huseby E.S., Rubtsova K., Scott-Browne J., Crawford F., Macdonald W.A., Marrack P., and Kappler J.W.. 2008. Crossreactive T Cells spotlight the germline rules for αβ T cell-receptor interactions with MHC molecules. Immunity. 28:324–334. 10.1016/j.immuni.2008.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.M., and Bjorkman P.J.. 1988. T-cell antigen receptor genes and T-cell recognition. Nature. 334:395–402. 10.1038/334395a0 [DOI] [PubMed] [Google Scholar]
- Feng D., Bond C.J., Ely L.K., Maynard J., and Garcia K.C.. 2007. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat. Immunol. 8:975–983. 10.1038/ni1502 [DOI] [PubMed] [Google Scholar]
- Garcia K.C., Adams J.J., Feng D., and Ely L.K.. 2009. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat. Immunol. 10:143–147. 10.1038/ni.f.219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin M.A., and Bevan M.J.. 1995. Increased peptide promiscuity provides a rationale for the lack of N regions in the neonatal T cell repertoire. Immunity. 3:793–800. 10.1016/1074-7613(95)90068-3 [DOI] [PubMed] [Google Scholar]
- Gilfillan S., Benoist C., and Mathis D.. 1995. Mice lacking terminal deoxynucleotidyl transferase: adult mice with a fetal antigen receptor repertoire. Immunol. Rev. 148:201–219. 10.1111/j.1600-065X.1995.tb00099.x [DOI] [PubMed] [Google Scholar]
- Gras S., Chen Z., Miles J.J., Liu Y.C., Bell M.J., Sullivan L.C., Kjer-Nielsen L., Brennan R.M., Burrows J.M., Neller M.A., et al. 2010. Allelic polymorphism in the T cell receptor and its impact on immune responses. J. Exp. Med. 207:1555–1567. 10.1084/jem.20100603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huseby E.S., White J., Crawford F., Vass T., Becker D., Pinilla C., Marrack P., and Kappler J.W.. 2005. How the T cell repertoire becomes peptide and MHC specific. Cell. 122:247–260. 10.1016/j.cell.2005.05.013 [DOI] [PubMed] [Google Scholar]
- Huseby E.S., Kappler J.W., and Marrack P.. 2008. Thymic selection stifles TCR reactivity with the main chain structure of MHC and forces interactions with the peptide side chains. Mol. Immunol. 45:599–606. 10.1016/j.molimm.2006.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Bian H.J., Molina M., Han J., Magram J., Saar E., Belunis C., Bolin D.R., Arceo R., Campbell R., et al. 1996. HLA-DR4-IE chimeric class II transgenic, murine class II-deficient mice are susceptible to experimental allergic encephalomyelitis. J. Exp. Med. 183:2635–2644. 10.1084/jem.183.6.2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson S.C., Hogquist K.A., and Bevan M.J.. 1995. Positive selection of thymocytes. Annu. Rev. Immunol. 13:93–126. 10.1146/annurev.iy.13.040195.000521 [DOI] [PubMed] [Google Scholar]
- Kievits F., Ivanyi P., Krimpenfort P., Berns A., and Ploegh H.L.. 1987. HLA-restricted recognition of viral antigens in HLA transgenic mice. Nature. 329:447–449. 10.1038/329447a0 [DOI] [PubMed] [Google Scholar]
- Klein L., Kyewski B., Allen P.M., and Hogquist K.A.. 2014. Positive and negative selection of the T cell repertoire: What thymocytes see (and don’t see). Nat. Rev. Immunol. 14:377–391. 10.1038/nri3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen O.P., Karlsen A.E., Larsen Z.M., Johannesen J., Pociot F., and Mandrup-Poulsen T.. Danish IDDM Epidemiology and Genetics Group and Danish Study Group of IDDM in Childhood . 2004. Identification of a type 1 diabetes-associated CD4 promoter haplotype with high constitutive activity. Scand. J. Immunol. 59:582–591. 10.1111/j.1365-3083.2004.01444.x [DOI] [PubMed] [Google Scholar]
- Krogsgaard M., and Davis M.M.. 2005. How T cells ‘see’ antigen. Nat. Immunol. 6:239–245. 10.1038/ni1173 [DOI] [PubMed] [Google Scholar]
- Li L.P., Lampert J.C., Chen X., Leitao C., Popović J., Müller W., and Blankenstein T.. 2010. Transgenic mice with a diverse human T cell antigen receptor repertoire. Nat. Med. 16:1029–1034. 10.1038/nm.2197 [DOI] [PubMed] [Google Scholar]
- Marrack P., Scott-Browne J.P., Dai S., Gapin L., and Kappler J.W.. 2008. Evolutionarily conserved amino acids that control TCRMHC interaction. Annu. Rev. Immunol. 26:171–203. 10.1146/annurev.immunol.26.021607.090421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marten N.W., Stohlman S.A., Smith-Begolka W., Miller S.D., Dimacali E., Yao Q., Stohl S., Goverman J., and Bergmann C.C.. 1999. Selection of CD8+ T cells with highly focused specificity during viral persistence in the central nervous system. J. Immunol. 162:3905–3914. [PubMed] [Google Scholar]
- McDonald B.D., Bunker J.J., Erickson S.A., Oh-Hora M., Bendelac A., Mcdonald B.D., Bunker J.J., Erickson S.A., Oh-hora M., and Bendelac A.. 2015. Crossreactive αβ T Cell receptors are the predominant targets of thymocyte negative selection. Immunity. 43:859–869. 10.1016/j.immuni.2015.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M., Graf D., Lovatt M., Bommhardt U., Zamoyska R., and Fisher A.G.. 1997. How many thymocytes audition for selection? J. Exp. Med. 186:1149–1158. 10.1084/jem.186.7.1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarov V.I., Pogorelyy M.V., Komech E.A., Zvyagin I.V., Bolotin D.A., Shugay M., Chudakov D.M., Lebedev Y.B., and Mamedov I.Z.. 2015. tcR: an R package for T cell receptor repertoire advanced data analysis. BMC Bioinformatics. 16:175 10.1186/s12859-015-0613-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolich-Žugich J., Slifka M.K., and Messaoudi I.. 2004. The many important facets of T-cell repertoire diversity. Nat. Rev. Immunol. 4:123–132. 10.1038/nri1292 [DOI] [PubMed] [Google Scholar]
- Robins H.S., Campregher P.V., Srivastava S.K., Wacher A., Turtle C.J., Kahsai O., Riddell S.R., Warren E.H., and Carlson C.S.. 2009. Comprehensive assessment of T-cell receptor β-chain diversity in αβ T cells. Blood. 114:4099–4107. 10.1182/blood-2009-04-217604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins H.S., Srivastava S.K., Campregher P.V., Turtle C.J., Andriesen J., Riddell S.R., Carlson C.S., and Warren E.H.. 2010. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci. Transl. Med. 2:47ra64 10.1126/scitranslmed.3001442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J., Waller M.J., Parham P., de Groot N., Bontrop R., Kennedy L.J., Stoehr P., and Marsh S.G.E.. 2003. IMGT/HLA and IMGT/MHC: Sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 31:311–314. 10.1093/nar/gkg070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossjohn J., Gras S., Miles J.J., Turner S.J., Godfrey D.I., and McCluskey J.. 2015. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33:169–200. 10.1146/annurev-immunol-032414-112334 [DOI] [PubMed] [Google Scholar]
- Rubelt F., Bolen C.R., McGuire H.M., Vander Heiden J.A., Gadala-Maria D., Levin M., Euskirchen G.M., Mamedov M.R., Swan G.E., Dekker C.L., et al. 2016. Individual heritable differences result in unique cell lymphocyte receptor repertoires of naïve and antigen-experienced cells. Nat. Commun. 7:11112 10.1038/ncomms11112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Browne J.P., White J., Kappler J.W., Gapin L., and Marrack P.. 2009. Germline-encoded amino acids in the αβ T-cell receptor control thymic selection. Nature. 458:1043–1046. 10.1038/nature07812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Browne J.P., Crawford F., Young M.H., Kappler J.W., Marrack P., and Gapin L.. 2011. Evolutionarily conserved features contribute to αβ T cell receptor specificity. Immunity. 35:526–535. 10.1016/j.immuni.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi D.K., Schubert D.A., Anders A.K., Heroux A., Bonsor D.A., Thomas C.P., Sundberg E.J., Pyrdol J., and Wucherpfennig K.W.. 2011. A highly tilted binding mode by a self-reactive T cell receptor results in altered engagement of peptide and MHC. J. Exp. Med. 208:91–102. 10.1084/jem.20100725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon E., Sibener L.V., Battle A., Fraser H.B., Garcia K.C., and Pritchard J.K.. 2016. Genetic variation in MHC proteins is associated with T cell receptor expression biases. Nat. Genet. 48:995–1002. 10.1038/ng.3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchin E.J., Langmuir P.B., Palmer E., Sayegh M.H., Wells A.D., and Turka L.A.. 2001. Quantifying the frequency of alloreactive T cells in vivo: New answers to an old question. J. Immunol. 166:973–981. 10.4049/jimmunol.166.2.973 [DOI] [PubMed] [Google Scholar]
- Tikhonova A.N., Van Laethem F., Hanada K., Lu J., Pobezinsky L.A., Hong C., Guinter T.I., Jeurling S.K., Bernhardt G., Park J.-H., et al. 2012. αβ T cell receptors that do not undergo major histocompatibility complex-specific thymic selection possess antibody-like recognition specificities. Immunity. 36:79–91. 10.1016/j.immuni.2011.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynan F.E., Burrows S.R., Buckle A.M., Clements C.S., Borg N.A., Miles J.J., Beddoe T., Whisstock J.C., Wilce M.C., Silins S.L., et al. 2005. T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat. Immunol. 6:1114–1122. 10.1038/ni1257 [DOI] [PubMed] [Google Scholar]
- Van Laethem F., Tikhonova A.N., Pobezinsky L.A., Tai X., Kimura M.Y., Le Saout C., Guinter T.I., Adams A., Sharrow S.O., Bernhardt G., et al. 2013. Lck availability during thymic selection determines the recognition specificity of the T cell repertoire. Cell. 154:1326–1341. 10.1016/j.cell.2013.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrisekoop N., Monteiro J.P., Mandl J.N., Germain R.N., Vrisekoop N., Monteiro J.P., Mandl J.N., and Germain R.N.. 2014. Revisiting thymic positive selection and the mature T cell repertoire for antigen. Immunity. 41:181–190. 10.1016/j.immuni.2014.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Mouse Genome Sequencing Consortium . 2002. Initial sequencing and comparative analysis of the mouse genome. Nature. 420:520–562. 10.1038/nature01262 [DOI] [PubMed] [Google Scholar]
- Xing Y., and Hogquist K.A.. 2014. Isolation, identification, and purification of murine thymic epithelial cells. J. Vis. Exp. (90):e51780 10.3791/51780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassai M., Ammon K., Goverman J., Marrack P., Naumov Y., and Gorski J.. 2002. A molecular marker for thymocyte-positive selection: selection of CD4 single-positive thymocytes with shorter TCRB CDR3 during T cell development. J. Immunol. 168:3801–3807. 10.4049/jimmunol.168.8.3801 [DOI] [PubMed] [Google Scholar]
- Zerrahn J., Held W., and Raulet D.H.. 1997. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell. 88:627–636. 10.1016/S0092-8674(00)81905-4 [DOI] [PubMed] [Google Scholar]
- Zvyagin I.V., Pogorelyy M.V., Ivanova M.E., Komech E.A., Shugay M., Bolotin D.A., Shelenkov A.A., Kurnosov A.A., Staroverov D.B., Chudakov D.M., et al. 2014. Distinctive properties of identical twins’ TCR repertoires revealed by high-throughput sequencing. Proc. Natl. Acad. Sci. USA. 111:5980–5985. 10.1073/pnas.1319389111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TCR sequencing data underlying this study can be analyzed and downloaded from the Adaptive Biotechnologies immuneACCESS site at https://doi.org/10.21417/B7ZD0D.