Abstract
Protein kinase-like domains that lack conserved residues known to catalyse phosphoryl transfer, termed pseudokinases, have emerged as important signalling domains across all kingdoms of life. Although predicted to function principally as catalysis-independent protein-interaction modules, several pseudokinase domains have been attributed unexpected catalytic functions, often amid controversy. We established a thermal-shift assay as a benchmark technique to define the nucleotide-binding properties of kinase-like domains. Unlike in vitro kinase assays, this assay is insensitive to the presence of minor quantities of contaminating kinases that may otherwise lead to incorrect attribution of catalytic functions to pseudokinases. We demonstrated the utility of this method by classifying 31 diverse pseudokinase domains into four groups: devoid of detectable nucleotide or cation binding; cation-independent nucleotide binding; cation binding; and nucleotide binding enhanced by cations. Whereas nine pseudokinases bound ATP in a divalent cation-dependent manner, over half of those examined did not detectably bind nucleotides, illustrating that pseudokinase domains predominantly function as non-catalytic protein-interaction modules within signalling networks and that only a small subset is potentially catalytically active. We propose that henceforth the thermal-shift assay be adopted as the standard technique for establishing the nucleotide-binding and catalytic potential of kinase-like domains.
Keywords: nucleotide binding, non-catalytic protein-interaction domain, protein kinase, pseudoenzyme, pseudokinase
INTRODUCTION
Although first described in sea urchins [1], examples of pseudokinase domains are now known across all kingdoms of life. In previous years, essential functions in signal transduction have been attributed to these protein kinase-like domains, principally as modulators of the catalytic activities of bona fide protein kinases or as scaffolding proteins that nucleate the assembly of signalling complexes [2–14]. Often amid controversy, several pseudokinase domains have been reported to exhibit a low catalytic activity, yet the importance of this enzymatic activity to the protein’s biological function and the ubiquity of this phenomenon across all pseudokinases remain unclear.
Pseudokinase domains were originally predicted to be devoid of any catalytic activity owing to the absence of one or more of the three crucial residues known to catalyse phosphoryl transfer in active protein kinases (Figure 1A). These residues are located within highly conserved motifs: (i) the VAIK motif, where the lysine residue in the β3 strand of the N-lobe positions the ATP α- and β-phosphates for catalysis; (ii) the HRD motif in the catalytic loop, which crucially contributes the catalytic aspartic acid residue; and (iii) the DFG motif within the activation loop contributes an aspartic acid residue involved in binding Mg2+ that stabilizes the bound ATP [15,16]. More recently, further consideration has been given to the integrity of the [GSA]xGxx[GSA] motif (where x is any amino acid) in the glycine-rich loop (‘G-loop’ or ‘P-loop’) that connects the β1 and β2 strands of the kinase domain N-lobe, since the flexibility of this loop is a key factor in promoting ATP binding [17]. Despite the absence of one or more of the three canonical catalytic residues, previous studies have reported nucleotide binding and enzymatic activities for a number of pseudokinases [14,18–21], raising the prospect that it might be a more general phenomenon of pseudokinases.
Figure 1. Pseudokinase domains selected for characterization of nucleotide binding.
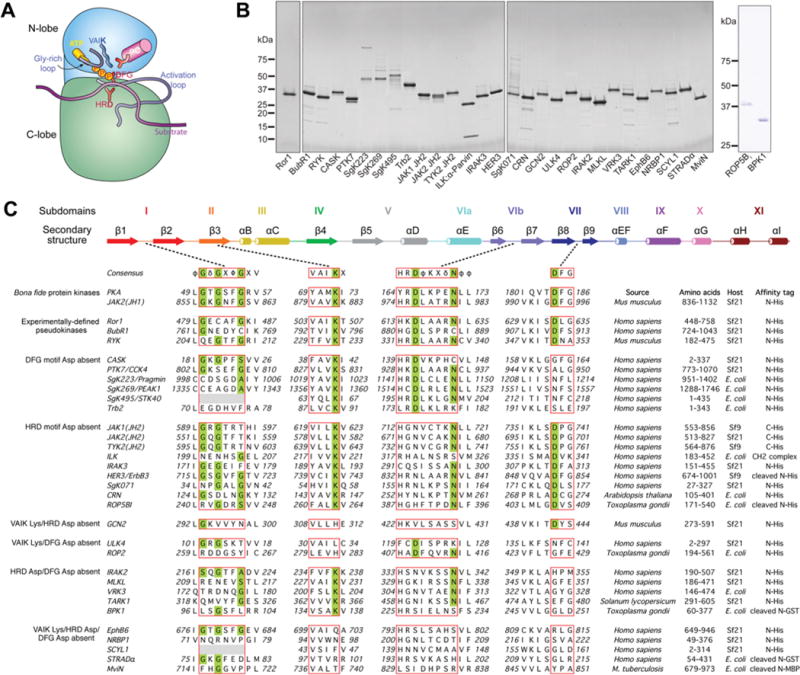
(A) Schematic cartoon of the eukaryotic protein kinase domain illustrating the amino acid motifs thought to be required for phosphoryl transfer that are usually absent from pseudokinase domains (depicted in the same style as [71]). (B) Purified pseudokinase domains (∼1 μg) were resolved by reducing SDS/PAGE and then were Coomassie Blue-stained. Molecular mass markers are indicated on the left-hand side. In each case, the predominant species present in the preparation is the pseudokinase domain. (C) Multiple sequence alignment of canonical catalytic motifs in PKA (protein kinase A), a model serine/threonine protein kinase, and JAK2(JH1), a model tyrosine kinase, with the 31 pseudokinase domains examined in the present study. Consensus sequences of the catalytic motifs and the protein kinase secondary structure are depicted as described previously ([72] and [73]). φ, hydrophobic; δ, hydrophilic; Φ, large hydrophobic; X, any amino acid. Subdomains are labelled according to the nomenclature proposed in [15]. The G-loop and the VAIK, HRD and DFG motifs of catalytically active protein kinases and their pseudokinase counterparts are boxed in red, and residues thought to be essential for robust catalytic activity are shaded in green. Details of the species of origin, domain boundaries and expression strategy are given for each protein studied, with further details presented in the Experimental section and Supplementary Table S1 (at http://www.biochemj.org/bj/457/bj4570323add.htm). It should be noted that equivalent results were obtained for BubR1 with the domain boundaries of residues 720–1043 (results not shown).
A prerequisite underlying whether a pseudokinase might exhibit catalytic activity is to define whether it binds to nucleotides in the presence of divalent cations. Having extensively reviewed the methods available to characterize the nucleotide-binding properties of pseudokinases [22], we used the most robust technique, the thermal-shift assay, to examine nucleotide and divalent cation binding by pseudokinase domains in vitro. This assay obviates the need for specific fluorescently tagged nucleotides, such as TNP-ATP [2(3)-O-(2,4,6-trinitrophenyl) adenosine 5-triphosphate], which have been reported to bind targets with affinities several hundred-fold greater than ATP in some cases [23] or non-specifically fluoresce in the presence of BSA (P.A. Eyers and D. Ungureanu, unpublished work). Additionally, the assay monitors the thermal denaturation of the major protein species, so any minor contaminants, including co-purified protein kinases, that would lead to misleading results in radiometric in vitro kinase assays do not contribute to the signal. Using this assay, we experimentally defined the nucleotide- and divalent cation-binding properties of a collection of 31 diverse pseudokinase domains, including almost half of the human pseudokinome, and found that the majority of examined pseudokinases did not detectably bind nucleotides. The findings of the present study are consistent with the idea that these domains predominantly serve non-enzymatic functions in cellular signalling networks.
EXPERIMENTAL
Recombinant protein expression and purification
The sources of cDNA templates used in the preparation of the expression constructs are shown in Supplementary Table S1 (at http://www.biochemj.org/bj/457/bj4570323add.htm). Insect cell expression was performed using Sf21 cells [except for JAK1(JH2) (where JAK is Janus kinase and JH2 is JAK homology 1), TYK2(JH2) (where TYK2 is tyrosine kinase 2) and HER3 (human epidermal growth factor receptor 3)/ErbB3 (v-erb-b2 avian erythroblastic leukaemia viral oncogene homologue 3), which used Sf9 cells] as described previously [20] starting from the pFastBac HTb or pFastBac1 vectors (LifeTechnologies) or a pAceBac1-derived vector (ATG Biosynthetics) to generate His6−tagged proteins. Bacterial expression constructs were prepared analogously to incorporate N-terminal His6 tags. VRK3 (vaccinia-related kinase 3) and CRN (also known as CORYNE) were cloned into pProEX HTb (LifeTechnologies); SgK495 [sugen kinase 495; also known as STK40 (serine/threonine kinase 40)] and TRB2 [also known as TRIB2 (tribbles pseudokinase 2)] were cloned into pET30 Ek/LIC (Novagen); SgK223 and SgK269 [also known as PEAK1 (pseudopodium-enriched atypical kinase 1)] were cloned into pCOLD IV (Clontech); and ROP2 (rhoptry protein kinase 2) and BPK1 (bradyzoite pseudokinase 1) were cloned into pET28a (Clontech), and expression was performed according to established protocols [24]. Typically, proteins were purified using a standardized procedure [20], before Superdex-200 gel-filtration chromatography (GE Healthcare) in 200 mM NaCl, 20 mM Tris/HCl, 10% (v/v) glycerol and 0.5 mM TCEP [tris-(2-carboxyethyl)phosphine] (pH 8.0).
The expression and purification of JAK2(JH1) [25,26], ILK (integrin-linked kinase)–α-parvin CH2 (calponin homology 2) domain complex [27], HER3/ErbB3 [4], MviN [28], ROP5BI [29], JAK2(JH2) [20] and STRADα (STE20-related kinase adaptor α) [30] were performed as described in their respective references. Typically, proteins were concentrated to 2 mg/ml or higher, snapfrozen in liquid N2 and stored at −80◦C until required. Protein concentrations were estimated on the basis of absorbance at 280 nm and the calculated molar absorption coefficients.
Thermal-shift assay for nucleotide binding
Thermal-shift assays were performed using a Corbett Real Time PCR machine with proteins diluted in 150 mM NaCl, 20 mM Tris/HCl (pH 8.0) and 1 mM DTT to 2–5 μM and assayed with the appropriate concentration of ligand in a total reaction volume of 25 μl. SYPRO Orange (Molecular Probes) was used as a probe with fluorescence detected at 530 nm. The temperature was raised in 1◦C per min steps from 25◦C to 95◦C and fluorescence readings were taken at each interval. Nucleotide- or cation-binding experiments were assessed relative to a buffer control. Two generic inhibitors, DAP {N′2′-(4-aminomethyl-phenyl)-5-fluoro-N′4′-phenyl-pyrimidine-2,4-diamine [31]; supplied by SYNthesis Med Chem} and VI16832 [32], were assessed relative to a 2% (v/v) DMSO control. With the exception of the titration experiments, nucleotide concentrations of 0.2 mM, divalent cation concentrations of 1 mM, and DAP or VI16832 concentrations of 40 μM were used in each experiment. For each well, sample fluorescence was plotted as a function of increasing temperature. The melting temperature (Tm) corresponding to the midpoint for the protein unfolding transition was calculated by fitting the sigmoidal melt curve to the Boltzmann equation using GraphPad Prism, with R2 values of >0.99. Data points after the fluorescence intensity maximum were excluded from fitting. Changes in the unfolding transition temperature compared with the control curve (ΔTm) were calculated for each ligand (nucleotides cations). A positive ΔTm value indicates that the ligand stabilizes the protein from denaturation, and therefore binds the protein. A minimum of two independent assays was performed for each protein and representative data are shown for each.
ITC
ITC (isothermal titration calorimetry) was performed using a MicroCal instrument (GE Healthcare). ATP (0.5 mM with no added divalent cations) was titrated into a solution of 50 μM human MLKL (mixed lineage kinase domain-like) pseudokinase domain at 25◦C. Both samples were prepared in 200 mM NaCl, 20 mM Hepes and 5% (v/v) glycerol (pH 7.5).
RESULTS
Selection and preparation of target pseudokinase domains
A diverse group of pseudokinase domains were selected for expression, purification and nucleotide-binding studies to represent a cross-section of domains lacking different combinations of catalytic residues from the VAIK, HRD and DFG motifs (Figure 1). We also examined the proteins Ror1 (receptor tyrosine kinase-like orphan receptor 1), BubR1 [also known as BUB1B (BUB1 mitotic checkpoint serine/threonine kinase B)] and RYK (receptor-like tyrosine kinase), which despite containing the key residues of the VAIK, HRD and DFG motifs did not exhibit catalytic activity in earlier studies [33–36]. In addition to the 23 human and two mouse pseudokinases chosen for examination, we also analysed the bacterial pseudokinase MviN [28], two plant pseudokinases, CRN (CORYNE) [37] and TARK1 (tomato atypical receptor-like kinase 1) [38], and three Toxoplasma gondii pseudokinases, ROP5BI [29], ROP2 [39] and BPK1 [40].
In most cases, pseudokinases were overexpressed and purified from insect cells (Figures 1B and 1C). All were prepared either with a His6 tag or proteolytically cleaved to remove globular affinity tags, such as GST and MBP (maltose-binding protein), since these domains will themselves undergo denaturation in the thermal-shift assay and may obscure detection of ligand binding to pseudokinase domains. Because some pseudokinase domains are unstable in their apo forms, we sought to test whether the assay was amenable to examining ligand binding within protein complexes by examining ILK co-expressed and purified in complex with the CH2 domain of α-parvin [27,41]. Coomassie Blue-stained SDS/PAGE gels of the recombinant proteins examined in the present study are shown in Figure 1(B).
A thermal-shift assay for examining nucleotide and cation binding
Initially, we established an assay to evaluate nucleotide and cation binding to pseudokinase domains. The assay, based on an earlier method [42], monitors the thermal denaturation of a test protein by measuring the fluorescence arising from binding of the dye SYPRO Orange to hydrophobic patches that become exposed during denaturation. Ligand binding to a protein is known to enhance a protein’s thermal stability, which manifests in an elevated melting temperature (Tm) that can be used to measure ligand binding. Relative to many other methods that have previously been explored to characterize nucleotide binding to kinase-like domains [22], the thermal-shift assay offers the following advantages: (i) modest quantities of recombinant protein (2–5 μg per condition) can be used; (ii) an RT-PCR (real-time PCR) instrument, a common piece of laboratory equipment, can be used; (iii) depending upon the RT-PCR instrument, 96 or 384 conditions can be tested simultaneously; (iv) the binding of any ligand, even a simple divalent cation such as Mg2+, can be assessed without fluorescent tagging of the ligand; (v) the assay is extremely sensitive, being able to detect interactions with affinities in the 10−4 M range; and (vi) the presence of minor quantities of contaminating kinases will not affect the assay, since the denaturation measurement is dominated by the major species.
We validated our assay using human MLKL, whose mouse orthologue had already shown ATP binding in this assay [43]. We measured ATP binding to human MLKL by both thermal-shift assay and ITC, which yielded comparable Kd values (∼15 μM; Supplementary Figures S1A and S1B at http://www.biochemj.org/bj/457/bj4570323add.htm). These studies, coupled with those reported previously for other kinases [44], provide important validation that the magnitudes of the thermal shifts correlate with ligand-binding affinities and thus that the thermal-shift assay provides an accurate representation of ligand binding. To gauge the sensitivity of the assay, we examined whether the assay could detect ADP–Mg2+ binding to the active tyrosine kinase domain of JAK2 (the ‘JH1’ domain), a domain we have previously shown to bind ADP–Mg2+ with a Kd value of ∼100 μM [45]. As shown in Supplementary Figures S1(C) and S1(D), we could indeed detect binding of JAK2(JH1) to a variety of nucleotides in the presence of Mg2+, including ADP. On the basis of these preliminary studies, we estimated that this assay can detect ligand binding down to sub-millimolar Kd values. Indeed, a recent study using an alternative technique demonstrated that EphB6 (Ephrin type-B receptor 6) binds GTP–Mg2+ with a Kd value of >500 μM [46], and this interaction was also successfully detected in our assay (Figure 3C).
Figure 3. Thermal denaturation curves and ΔTm analyses of Class 2 (nucleotide binding) and Class 3 (cation binding) pseudokinases.
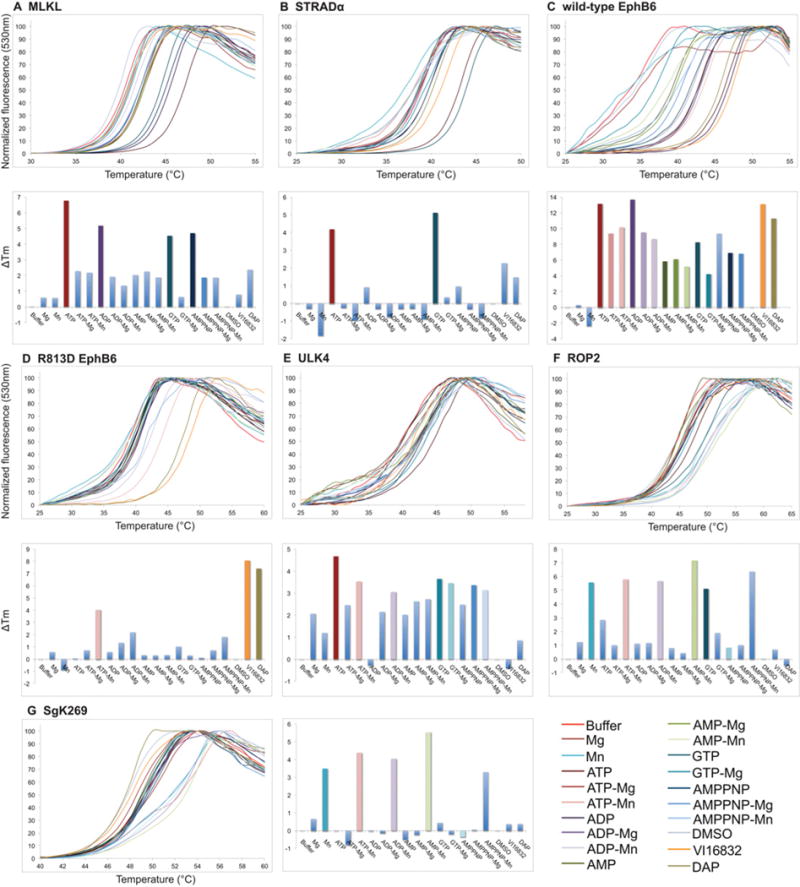
Thermal denaturation data of the Class 2 pseudokinases, MLKL (A), STRADα (B), wild-type EphB6 (C) and ULK4 (E), presented as described in the legend to Figure 2. (D) The R813D mutation converts EphB6 from a Class 2 into Class 4 pseudokinase. Note that wild-type EphB6 (C) and ULK4 (E) are classified as Class 2 (nucleotide-binding) as the presence of Mn2+ and Mg2+ does not enhance ATP binding. Data for the Class 3 (cation-binding) pseudokinases ROP2 and SgK269 are presented in (F) and (G) respectively. Note that ROP2 and SgK269 are classified as Class 3 (cation-binding) pseudokinases because the presence of nucleotides does not confer further thermal stability above and beyond that of cation alone.
Classification of pseudokinases
We proceeded to qualitatively assess the propensity of each of 31 recombinant pseudokinase domains (Figure 1) to bind divalent cations (Mg2+ or Mn2+), nucleotides {AMP, ADP, ATP, AMP-PNP (adenosine 5′-[β,γ-imido]triphosphate) and GTP}or both cation and nucleotide together. We also examined binding to two promiscuous ATP-competitive inhibitors, DAP [31] and VI16832 [32] (structures shown in Supplementary Figures S1E and S1F), as a means of evaluating whether the proteins contained an accessible, potentially druggable, pocket equivalent to the nucleotide-binding site of a catalytically active kinase. On the basis of our experience using this technique to screen for small molecule kinase inhibitors, we deemed a ΔTm value of ⩾3◦C to be a robust indicator of ligand binding.
On the basis of their ligand-binding properties in the thermal-shift assay, we categorized the 31 pseudokinases into four classes: (i) devoid of detectable nucleotide or cation binding; (ii) nucleotide binding; (iii) cation binding; and (iv) nucleotide and cation binding. As summarized in Table 1, more than half (16) of the pseudokinases did not detectably bind nucleotides or cations in our assay (Figure 2 and Supplementary Figure S2 at http://www.biochemj.org/bj/457/bj4570323add.htm). Interestingly, although we did not detect nucleotide or cation binding for TYK2(JH2) and IRAK3 (interleukin-1-receptor-associated kinase 3) (Figures 2E and 2F), both proteins were capable of binding the inhibitors DAP and VI16832, indicating the presence of an intact ATP-binding cleft and raising the possibility that these domains may bind nucleotides at affinities below the sensitivity of the assay. As shown in Figure 3, four proteins bound nucleotide alone [MLKL, EphB6, STRADα and ULK4 (unc-51 like kinase 4); Class 2] and two bound cations alone (SgK269 and ROP2; Class 3). It should be noted that the thermal shifts observed for Class 3 pseudokinases in the presence of nucleotides and cations can be accounted for by the presence of Mn2+ alone (Figures 3F and 3G). Fewer than one-third of the pseudokinase domains tested (9 of 31) bound nucleotide and cation: JAK1(JH2), JAK2(JH2), ILK, HER3/ErbB3, SgK071, CRN, ROP5BI, IRAK2 and TARK1 (Figure 4 and Supplementary Figure S3 at http://www.biochemj.org/bj/457/bj4570323add.htm). Of note, our assay was amenable to examining the nucleotide and cation binding of pseudokinases within complexes with other protein-interaction domains. We were able to assay ILK (Figure 4C and Supplementary Figure S3C), even though its stability relies on being in complex with the α-Parvin CH2 domain.
Table 1. Summary of nucleotide, divalent cation and inhibitor binding by pseudokinases.
The G-loop was defined as degraded if two of three positions do not conform to the sequence [GSA]xGxx[GSA] where x is any amino acid.
| (a) Class 1 (devoid of nucleotide- or cation-binding) | ||||||
|---|---|---|---|---|---|---|
|
| ||||||
| Pseudokinase | Degraded G-loop | VAIK motif lysine residue present | HRD motif aspartic acid residue present | DFG motif aspartic acid residue present | DAP or VI16832 binding | |
| Ror1 | No | + | + | + | − | |
| BubR1 | Yes | + (TFVK)* | + | + (DNA)* | − | |
| RYK | No | + | + | + (DNA)* | − (DAP) | |
| RYK TFVK>VAVK | No | + | + | + | + | |
| CASK | No | + | + | − | − | |
| PTK7/CCK4 | No | + | + | − | − | |
| SgK223 | Yes | + | + | − | − | |
| SgK495/STK40 | Yes | + | + | − | − | |
| TRB2 | Yes | + | + | − | − | |
| TYK2(JH2) | No | + | − | + | + | |
| IRAK3 | No | + | − | + | + | |
| GCN2 | Yes | − | − | + | − | |
| VRK3 | Yes | + | − | − | − | |
| BPK1 | Yes | + | − | − | − | |
| NRBP1 | Yes | − | − | − | − | |
| SCYL1 | Yes | − | − | − | − | |
| MviN | Yes | − | − | − | − | |
|
| ||||||
| (b) Class 2 (nucleotide-binding) | ||||||
|
| ||||||
| Pseudokinase | Degraded G-loop | VAIK motif lysine residue present | HRD motif aspartic acid residue present | DFG motif aspartic acid residue present | DAP or VI16832 binding | |
|
| ||||||
| ULK4 | No | − | + (FCD)* | − | − | |
| MLKL | Yes | + | − | − | − | |
| EphB6 | No | − (VTVR)* | − | − | + | |
| STRADα | No | − | − | − | − | |
|
| ||||||
| (c) Class 3 (cation-binding) | ||||||
|
| ||||||
| Pseudokinase | Degraded G-loop | VAIK motif lysine residue present | HRD motif aspartic acid residue present | DFG motif aspartic acid residue present | DAP or VI16832 binding | |
|
| ||||||
| SgK269 | Yes | + | + | − | − | |
| ROP2 | Yes | − | + | − | − | |
|
| ||||||
| (d) Class 4 (nucleotide- and cation-binding) | ||||||
|
| ||||||
| Pseudokinase | Degraded G-loop | VAIK motif lysine residue present | HRD motif aspartic acid residue present | DFG motif aspartic acid residue present | DAP or VI16832 binding | Catalytic activity? |
|
| ||||||
| JAK1(JH2) | No | + | − | + | + (VI16832) | No [70] |
| JAK2(JH2) | No | + | − | + (DVK)* | + | Yes [20,53] |
| ILK | Yes | + | − | + | − | No [27,41] |
| HER3/ErbB3 | No | + | − | + | − | Yes [19] |
| SgK071 | No | + | − | + | − | Unknown |
| CRN | No | + | − | + | + | No [37] |
| ROP5B I | No | + | − | + | − (VI16832) | No [29] |
| IRAK2 | No | + | − | − (EFG)* | + (VI16832) | Unlikely [66,67] |
| TARK1 | Yes | + | − | − | + | No [38] |
| EphB6 R813D | No | − | − | + | + | Unknown |
Motif sequence noted due to similarity/difference from conventional motif
Figure 2. Thermal denaturation curves for selected Class 1 (non-binding) pseudokinase domains.
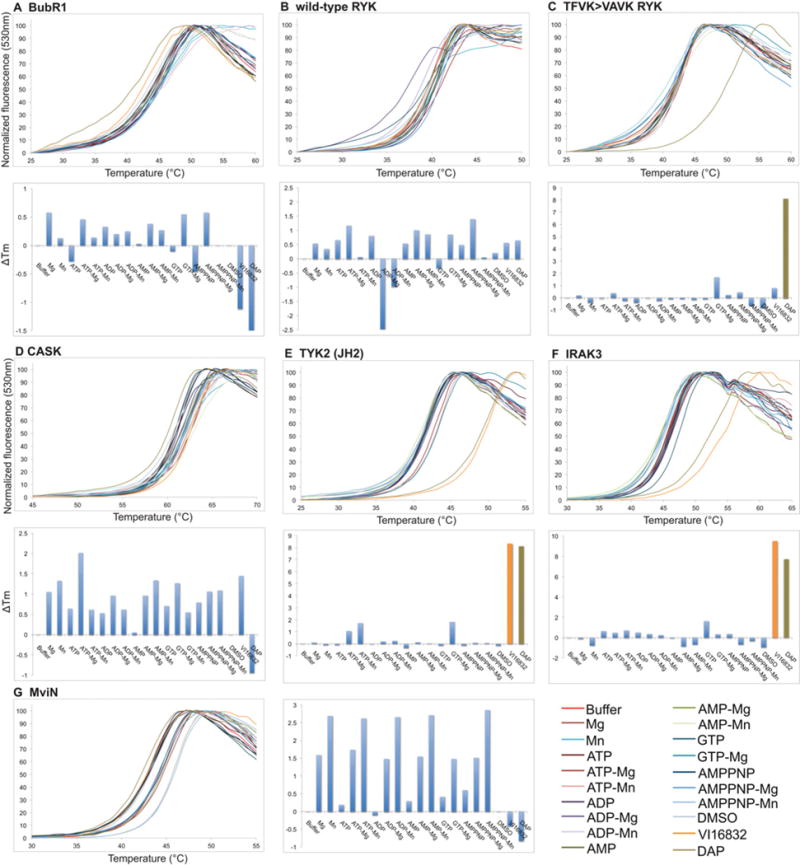
Thermal denaturation curves of the Class 1 pseudokinases, BubR1 (A), wild-type RYK (B), CASK (D) and MviN (G) (upper panels) with the corresponding histograms depicting the ΔTm values in each condition (lower panels), as derived from analysis of upper panel curves. Although thermal denaturation of TFVK>VAVK RYK (C) and IRAK3 (F) was not affected by nucleotides or cations, TYK2(JH2) (E) exhibited modest, but consistent, thermal shifts between experiments in the presence of ATP–Mg2+, ATP–Mn2+ or GTP–Mg2+ . Thermal shifts for all three of these proteins were detected in the presence of the inhibitor VI16832 and/or DAP. A colour key for the curve labelling is to the right-hand side of the curves. In the histograms, bars are coloured according to the colour key when ΔTm>3◦C and pale blue otherwise. Thermal denaturation curves for the other Class 1 pseudokinases, Ror1, PTK7/CCK4, SgK223, SgK495, TRB2, GCN2 [also known as EIF2AK4 (eukaryotic translation initiation factor 2-α kinase 4)], VRK3, BPK1, NRBP1 and SCYL1 (SCY1-like 1), are shown in Supplementary Figure S2 (at http://www.biochemj.org/bj/457/bj4570323add.htm).
Figure 4. ΔTm analyses of Class 4 (nucleotide- and cation-binding) pseudokinases.
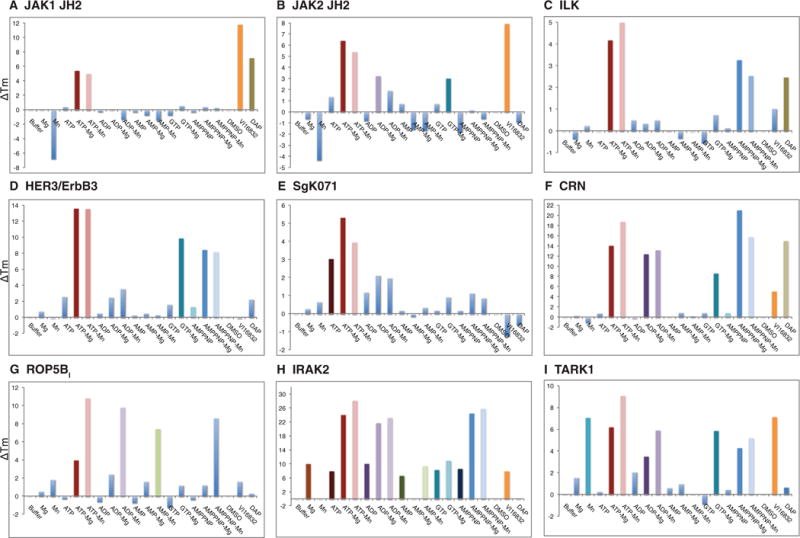
Histograms depicting the ΔTm value in each condition derived from thermal denaturation curves presented in Supplementary Figure S3 (at http://www.biochemj.org/bj/457/bj4570323add.htm) for JAK1(JH2) (A), JAK2(JH2) (B), ILK (C), HER3/ErbB3 (D), SgK071 (E), CRN (F), ROP5BI (G), IRAK2 (H) and TARK1 (I). Note that some conditions are destabilizing, leading to negative ΔTm values. TARK1 (I) was categorized as Class 4 owing to the observation of ATP–Mg2+, ADP–Mg2+, AMP-PNP–Mg2+ and GTP–Mg2+ binding. Interestingly TARK1 also appears to bind Mn2+ in the absence of nucleotides, leading to thermal-stability shifts under all Mn2+ -containing conditions.
RYK contains each of the three canonical residues present in active protein kinases, but is known to be catalytically inactive [36] and did not detectably bind nucleotides or cations in our assays. To test whether the highly conserved phenylalanine residue within the TFVK sequence, the counterpart of a conventional kinase’s VAIK motif, might occupy the adenine-binding pocket and prevent nucleotide binding, we examined a TFVK>VAVK mutant RYK by thermal shift. Although this mutation did not result in detectable nucleotide binding, the mutant was able to engage the promiscuous ATP-competitive inhibitor DAP, whereas wild-type RYK did not (Figures 2B and 2C). ATP binding by a second protein, EphB6, was not enhanced by the presence of divalent cations (Figure 3C), leading us to query whether the R813LG sequence in place of the canonical DFG motif might serve a role in binding the γ-phosphate of ATP, effectively supplanting the conventional Mg2+ . Introduction of the R813D mutation abrogated the direct binding of ATP by EphB6 in the absence of cations, but conferred an unexpected ability to bind ATP in the presence of Mn2+ (Figure 3D).
It is surprising that BubR1, which contains the catalytic residues typical of an active protein kinase, did not bind nucleotides in the presence or absence of divalent cations (Figure 2A). To ensure that this was not a consequence of the domain boundaries (residues 724–1043) we used initially, we also characterized a longer construct (residues 720–1043), similar to that used in [35], which contains the kinase extension domain that is critical for catalytic activity of the related bona fide protein kinase BUB1 [47]. We expressed BubR1 in both Sf21 and Escherichia coli cells and found that all preparations behaved equivalently in the thermal-shift assay (results not shown), confirming the inability of BubR1 to detectably bind to nucleotides or divalent cations.
DISCUSSION
The importance of pseudokinases to cellular signalling has only recently begun to be appreciated. Fuelled by the discovery that mutations within the pseudokinase domain of JAK2 underlie human blood cell malignancies [48–52], pseudokinases have emerged as an unexplored family of therapeutic targets. Previous studies have attributed unexpected catalytic functions to several pseudokinase domains, including that of JAK2 [20], ErbB3 [19] and KSR2 (kinase suppressor of Ras 2) [14], although whether nucleotide-binding and, relatedly, phosphoryl transfer activity is a widespread property within this domain family has remained unclear. We have addressed this issue by profiling the nucleotide-binding properties of a collection of 31 diverse pseudokinase domains using a robust thermal-shift assay.
Thermal-shift assay data are supported by prior structural studies
Although we have performed a comprehensive examination of ATP, ADP, AMP, AMP-PNP and GTP binding in the presence or absence of Mg2+ or Mn2+ in the present study, structural and biophysical studies have previously explored the ATP-binding capacities of some pseudokinases included in the present study [4,17–19,22,27–30,39,41,53,54]. With the exception of CASK (calcium/calmodulin-dependent serine protein kinase), all of these prior findings are supported by the present study, providing a sound foundation for the results reported in the present paper for formerly uncharacterized pseudokinases. The absence of ATP binding to CASK using the thermal-shift assay is unsurprising (Figure 2D), since our interpretation of the prior CASK ATP-binding data [18] suggests a Kd(ATP) value in the millimolar range, an affinity beyond the range of detection using most biophysical techniques. Accordingly, we were unable to detect nucleotide binding to CASK prepared from E. coli (results not shown) or insect cells (Figure 2D). The structure of the TYK2(JH2) domain bound to an ATP-mimetic inhibitor was recently solved (PDB code 3ZON), confirming the presence of an accessible active site: a finding consistent with the binding to the small-molecule kinase inhibitors DAP and VI16832 observed in the present study (Figure 2E). Although the structure of the Ror1 pseudokinase domain has not yet been solved, an absence of ATP binding in the present study can be rationalized by inspection of the structures of the related Ror2 (PDB codes 3ZZW and 4GT4) [55], where the adenine-binding pocket is partly occupied by the side chain of the highly conserved Tyr555.
Pseudokinases function principally as catalysis-independent protein-interaction modules
The largest class of pseudokinases observed in the present study did not detectably bind nucleotides or divalent cations (Class 1; Figure 2 and Table 1, and Supplementary Figure S2), providing compelling support for the hypothesis that pseudokinase domains serve predominantly non-catalytic functions within signalling networks in Nature. Of these, Ror1 [33,34], RYK [36], CCK4 (colon carcinoma kinase 4)/PTK7 (protein tyrosine kinase 7) [56], BubR1 [35], IRAK3/IRAK-M [57] and MviN [28] were not catalytically active in vitro in previously published studies, consistent with our observations that they do not bind ATP.
Nucleotide-binding (Class 2) pseudokinases
Prior studies of the nucleotide-binding-only proteins STRADα and EphB6 did not reveal kinase activity in vitro [30,58,59], in keeping with our premise that both ATP and cation binding are necessary for catalytic activity. Human MLKL has previously been attributed a catalytic function based on in vitro kinase assays with protein immunoprecipitated from HEK (human embryonic kidney)-293T cells [60]. However, on the basis of our previous characterization of mouse MLKL [43], and the analogous cation-independent nucleotide-binding properties observed for human MLKL in the present study, it is probable that the catalytic activity previously attributed to MLKL is likely to arise from the catalytic activity of a contaminating protein. A fourth protein, ULK4, has not been characterized biochemically, though cation-independent nucleotide binding indicates that ULK4 will similarly serve non-catalytic signalling roles.
A common feature of STRADα, MLKL, EphB6 and ULK4 is divergent sequences in place of the conventional cation-binding DFG motif. In the case of STRADα and EphB6, the DFG motif is replaced by GLR and RLG respectively. Since the GLR sequence was shown to play a key role in mediating ATP binding by STRADα [30], we examined whether the R813LG counterpart in EphB6 might play a similar role. R813D mutant EphB6 exhibited a predictable loss of nucleotide binding in the absence of divalent cations in the thermal-shift assay, but an unexpected gain of Mn2+ -dependent ATP binding (Figure 3D). This finding is intriguing, since the RLG>DLG mutation confers nucleotide-binding properties on EphB6 that are synonymous with a conventional protein kinase, although in the absence of a catalytic residue in the catalytic loop, we anticipate that R813D EphB6 will not be enzymatically active. An examination of orthologous sequences revealed that the arginine residue of the RLG motif is conserved almost universally as one of two basic residues (arginine or histidine) in mammalian EphB6 orthologues, whereas other orthologues, including those in fish (including lamprey), chicken, zebra finch and turkey, contain a DFG sequence in addition to intact VAIK (or VAVK) and HRD motifs. These observations indicate that mammalian EphB6 orthologues evolved from a catalytically active ancestral protein kinase to serve non-catalytic nucleotide-binding functions in cell signalling, presumably as means of modulating protein conformation. Remarkably, the evolution of the RLG sequence in place of the canonical DFG motif has coevolved with substitutions within the VAIK and HRD motifs. Our findings demonstrate an important role for the RLG sequence in facilitating nucleotide binding in mammalian EphB6 orthologues that is likely to have evolved to compensate for the concomitant substitution of the VAIK motif to VAIQ. The existence of the Class 2 pseudokinases provides important support for the emerging idea that nucleotide binding among pseudokinases may have evolved as a non-catalytic conformational switch, as proposed for STRADα [30,54] and MLKL [43]. An exciting possibility is that conformational regulation by nucleotide binding, rather than catalytic functions, may similarly underlie the functions of Class 4 pseudokinases as protein-interaction modules within signalling networks.
The Class 2 pseudokinases also offer important mechanistic insights into the contributions of the VAIK motif lysine residue to ATP binding in both pseudokinases and catalytically active kinases. Consistent with a prior study of the bona fide protein kinase Lck (lymphocyte-specific protein tyrosine kinase) [61], where the VAIK motif lysine residue was shown to serve a catalytic rather than ATP-binding function, we observed that ATP binding was still accomplished by EphB6 and ULK4 (Figures 3C and 3E), two pseudokinases containing VAIQ and VAIL respectively in place of the canonical VAIK motif. In some pseudokinase domains, however, it appears that the VAIK motif lysine residue has evolved an essential role in ATP binding, as illustrated by mutational analyses of mouse [43] and human [62] MLKL, both Class 2 pseudokinases, and HSER (heat-stable enterotoxin receptor)/GC-C (guanylate cyclase C) [63]. As a result, it appears that in the absence of other key ATP- and/or divalent cation-binding residues, some pseudokinases have evolved a greater reliance on the VAIK motif lysine residue to mediate ATP binding. Furthermore, these findings raise the intriguing possibility that the VAIK motif lysine residue may mediate ATP binding, rather than have a catalytic function, in a subset of catalytically active protein kinases.
Cation-binding (Class 3) pseudokinases
Pseudokinases that bound divalent cations alone include SgK269/PEAK1 and the T. gondii protein ROP2 (Figures 3F and 3G and Table 1). Consistent with the original study of ROP2 [39], we observed no ATP binding in our assays although, curiously, we did detect GTP binding in a cation-independent manner. This observation is most consistent with ROP2 containing an intact nucleotide-binding pocket that allows ROP2 to retain either a Mn2+ - or GTP-binding capacity, albeit in the absence of catalytic activity. By comparison, SgK269 had previously been attributed a catalytic activity [64,65], although in our hands it did not detectably bind ATP: a finding more consistent with the highly degraded G-loop and the absence of the conserved aspartic acid residue in the DFG motif. Interestingly, SgK223, a protein closely related to SgK269, likewise did not bind nucleotides in our assays, but, in contrast with SgK269, did not bind Mn2+ ions (Supplementary Figure S2B). It should be noted that the bacterial pseudokinase MviN showed evidence of Mn2+ binding, but because the ΔTm value in the presence of Mn2+ was <3◦C, it did not meet our criteria to be categorized as cation binding (Figure 2G).
Nucleotide- and cation-binding (Class 4) pseudokinases
The prevalence of Class 1 pseudokinases and the identification of Classes 2 and 3 demonstrate that the catalytic activity attributed to a subset of pseudokinase domains is not a widespread feature. We deemed nine of the 31 pseudokinases examined in the present study to be potentially catalytically active on the basis of the premise that both nucleotide and cation must be bound simultaneously for efficient phosphoryl transfer. Of these Class 4 pseudokinases (Figure 4), catalytic activities have been reported for purified recombinant JAK2(JH2) [20] and ErbB3/HER3 [19], indicating that in some pseudokinases asparagine residues can substitute (albeit poorly) for the catalytic aspartic acid of the canonical HRD motif. IRAK2 immunoprecipitated from macrophages was previously attributed a catalytic function [66], although this has been recently queried [67]. In addition, on the basis of our JAK2(JH2) data, we anticipated that the related proteins, JAK1(JH2) and TYK2(JH2), would behave similarly to JAK2(JH2). Unexpectedly, we observed that TYK2(JH2) exhibited only modest, albeit consistent, thermal shifts in the presence of ATP–Mg2+, ATP–Mn2+ or GTP–Mg2+ (Figure 2E), indicating that TYK2(JH2) may bind nucleotides in a cation-dependent manner, but with affinities below the sensitivity of our assay. In contrast, recent comprehensive structure/function studies have demonstrated that the pseudokinase domain of ILK is completely devoid of catalytic activity owing to a severely degraded catalytic core, and catalysis-independent functions have been elegantly established using knockin studies of ILK kinase-dead mutants in mice [68]. In addition, CRN, ROP5BI and TARK1 did not exhibit catalytic activity in published studies [29,37,38], supporting the idea that nucleotide binding by pseudokinase domains predominantly serves a role in modulating protein conformation, rather than catalytic functions, in cell signalling.
Can nucleotide-binding properties be predicted from pseudokinase amino acid sequences?
Although the nucleotide- and cation-binding (Class 4) pseudokinases constitute less than one-third of those profiled in the present paper, these domains were considered the most likely to be catalytically active and, as a result, we discuss some trends among this group that might aid their prediction in the future. First, combined G-loop degradation and the loss of the VAIK, DFG and HRD motifs thwarts nucleotide and cation binding and, accordingly, such sequence divergence is predictive of a catalytically inactive pseudokinase domain. Secondly, the DFG motif aspartic acid residue is present in nearly all pseudokinases that bind VI16832, DAP or nucleotide and cation (Table 1). Finally, among pseudokinase domains that bind nucleotide and cation (Class 4), the VAIK motif lysine residue (or, in some cases, arginine, such as in KSR2 [14]) and the DFG motif aspartic acid [or, in some cases, glutamic acid, such as in TARK1 (the present study)] are almost entirely conserved, with loss of only the catalytic aspartic acid from the HRD motif evident throughout Class 4. The sole loss of the catalytic aspartic acid residue in the HRD motif is not deleterious to nucleotide binding, since this catalytic residue is typically peripheral to the bound nucleotide and divalent cation in conventional protein kinases. Although not essential for nucleotide and cation engagement, the loss of the catalytic aspartic acid residue from the HRD motifs in the Class 4 pseudokinases is likely to severely limit their capacity to catalyse the phosphoryl transfer reaction. However, as noted previously [69], it remains unclear at this point how much catalytic activity is required in vivo for pseudokinases to play a significant role in modulating signal transduction and, as discussed above, whether ATP binding might principally serve as a conformational switch that governs the capacities of pseudokinases to serve as protein-interaction domains.
Another trend that we observed is that, in general, Class 4 pseudokinases were less thermostable than Class 1. Although the Tm values of Class 1 pseudokinases were typically >40◦C, with Tm>50◦C observed for PTK7, BPK1, NRBP1 (nuclear receptor-binding protein 1) and CASK, Class 4 proteins typically exhibited lower Tm values (<40◦C), suggesting a high degree of flexibility between the N- and C-lobes corresponding to a more ‘open’ ATP-binding cleft in the absence of ligand. Exceptions to this generalization, such as the high thermal stability of the Class 4 protein ROP5BI (Supplementary Figure S3G), prevent thermal stability itself serving diagnostically as an indicator of the nucleotide-binding propensity. Nonetheless these data raise the interesting possibility that, for Class 2 and 4 pseudokinases, the significant stabilization induced by ATP binding may be central to their ability to act as conformational switches or rigid scaffolds for intermolecular interactions.
Although a comprehensive understanding of this important family of domains awaits further biochemical, structural and biological characterization, the methodology and data of the present study provide a foundation for identifying pseudokinase domains that have retained a functional nucleotide-binding pocket. This subset of pseudokinase domains, including those known to be mutated in human cancers, such as JAK2(JH2) and HER3/ErbB3, are potentially druggable by ATP-competitive small molecules that could either inhibit weak catalytic activities or modulate the conformations of pseudokinases that serve as molecular switches in signalling pathways [30,43,54]. Consequently, these pseudokinases represent an exciting emergent class of drug targets. To this end, in future, we envisage that the methodology described in the present study will be utilized to screen for small-molecule lead compounds from which specific therapeutic modulators of pseudokinase functions could be developed.
Supplementary Material
Acknowledgments
We thank Professor Dario Alessi (University of Dundee, Dundee, U.K.) for providing the GST–STRADα expression construct; Professor Seung-Taek Lee (Yonsei University, Seoul, Korea) for the PTK7/CCK4 template DNA; Dr Alessandra Gentile (Institute for Cancer Research and Treatment, Candiolo, Italy) for the Ror1 template cDNA; Dr Elton Zeqiraj (Tanenbaum-Lunenfeld Research Institute, Toronto, Canada) for assistance with the secondary-structure cartoon in Figure 1(C); and the Monash University Protein Production Unit for access to the Corbett RT-PCR instrument used for all of the thermal-shift assays.
FUNDING
This work was supported by the National Health and Medical Research Council (NHMRC) [grant numbers 637342 and 1011804]; the Australian Research Council (ARC) [fellowships FT100100100 and FT110100169 (to J.M.M. and J.J.B.)]; the Leukaemia Foundation and the Australian Stem Cell Centre via scholarships to L.N.V.; the National Institutes of Health (NIH) [grant numbers R01 HL58758 (to J.Q.) and AI73756 (to M.L.R.)]; the Medical Research Council of Academy of Finland, the Sigrid Juselius Foundation, the Medical Research Fund of Tampere University Hospital, the Finnish Cancer Foundation and the Tampere Tuberculosis Foundation (to O.S.); the American Heart Association [grant number 11BGIA7440051 (to N.J.)]; and the National Science Foundation [grant number IOS-0821801 (to M.B.M.)]; with additional support from the Victorian State Government Operational Infrastructure Support and the NHMRC Independent Research Institutes Infrastructure Support Scheme (IRIISS) [grant number 361646].
Abbreviations
- AMP-PNP
adenosine 5′-[β,γ-imido]triphosphate
- BPK1
bradyzoite pseudokinase 1
- CASK
calcium/calmodulin-dependent serine protein kinase
- CCK4
colon carcinoma kinase 4
- CH2
calponin homology 2
- CRN
CORYNE
- DAP
N′2′ -(4-aminomethyl-phenyl)-5-fluoro-N′4′ -phenyl-pyrimidine-2,4-diamine
- EphB6
Ephrin type-B receptor 6
- ErbB3
v-erb-b2 avian erythroblastic leukaemia viral oncogene homologue 3
- HER3
human epidermal growth factor receptor 3
- ILK
integrin-linked kinase
- IRAK
interleukin-1-receptor-associated kinase
- ITC
isothermal titration calorimetry
- JAK
Janus kinase
- JH
JAK homology
- KSR2
kinase suppressor of Ras 2
- MLKL
mixed lineage kinase domain-like
- NRBP1
nuclear receptor-binding protein 1
- PEAK1
pseudopodium-enriched atypical kinase 1
- PTK7
protein tyrosine kinase 7
- ROP
rhoptry protein kinase
- Ror1
receptor tyrosine kinase-like orphan receptor
- RT-PCR
real-time PCR
- RYK
receptor-like tyrosine kinase
- SgK
sugen kinase
- STRADα
STE20-related kinase adaptor α
- TARK1
tomato atypical receptor-like kinase 1
- TRB2
tribbles pseudokinase 2
- TYK2
tyrosine kinase 2
- ULK4
unc-51 like kinase 4
- VRK3
vaccinia-related kinase 3
Footnotes
AUTHOR CONTRIBUTION
The present study was designed and supervised by James Murphy and Isabelle Lucet. James Murphy, Qingwei Zhang, Samuel Young, Leila Varghese, Kelan Chen, Anne Tripaydonis, Jeffrey Babon and Isabelle Lucet performed the experiments. Michael Reese, Fiona Bailey, Patrick Eyers, Daniela Ungureanu, Henrik Hammaren, Olli Silvennoinen, Natalia Jura, Koichi Fukuda, Jun Qin, Zachary Nimchuk, Mary Beth Mudgett, Sabine Elowe, Christine Gee, Ling Liu and Roger Daly contributed vital reagents. James Murphy, Gerard Manning, Jeffrey Babon and Isabelle Lucet analysed the data with input from all of the other authors. James Murphy, Jeffrey Babon and Isabelle Lucet wrote the paper with input from all of the other authors.
References
- 1.Singh S, Lowe DG, Thorpe DS, Rodriguez H, Kuang WJ, Dangott LJ, Chinkers M, Goeddel DV, Garbers DL. Membrane guanylate cyclase is a cell-surface receptor with homology to protein kinases. Nature. 1988;334:708–712. doi: 10.1038/334708a0. [DOI] [PubMed] [Google Scholar]
- 2.Baas AF, Boudeau J, Sapkota GP, Smit L, Medema R, Morrice NA, Alessi DR, Clevers HC. Activation of the tumour suppressor kinase LKB1 by the STE20-like pseudokinase STRAD. EMBO J. 2003;22:3062–3072. doi: 10.1093/emboj/cdg292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16:443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Jura N, Shan Y, Cao X, Shaw DE, Kuriyan J. Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc Natl Acad Sci USA. 2009;106:21608–21613. doi: 10.1073/pnas.0912101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keeshan K, Bailis W, Dedhia PH, Vega ME, Shestova O, Xu L, Toscano K, Uljon SN, Blacklow SC, Pear WS. Transformation by Tribbles homolog 2 (Trib2) requires both the Trib2 kinase domain and COP1 binding. Blood. 2010;116:4948–4957. doi: 10.1182/blood-2009-10-247361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saharinen P, Takaluoma K, Silvennoinen O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol Cell Biol. 2000;20:3387–3395. doi: 10.1128/mcb.20.10.3387-3395.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu H, He K, Li L, Sun L, Tang F, Li R, Ning W, Jin Y. Deletion of Stk40 in the mouse causes respiratory failure and death at birth. J Biol Chem. 2013;288:5342–5352. doi: 10.1074/jbc.M112.409433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burman JL, Bourbonniere L, Philie J, Stroh T, Dejgaard SY, Presley JF, McPherson PS. Scyl1, mutated in a recessive form of spinocerebellar neurodegeneration, regulates COPI-mediated retrograde traffic. J Biol Chem. 2008;283:22774–22786. doi: 10.1074/jbc.M801869200. [DOI] [PubMed] [Google Scholar]
- 9.Fleckenstein MC, Reese ML, Konen-Waisman S, Boothroyd JC, Howard JC, Steinfeldt T. A Toxoplasma gondii pseudokinase inhibits host IRG resistance proteins. PLoS Biol. 2012;10:e1001358. doi: 10.1371/journal.pbio.1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson CH, Crombie C, van der Weyden L, Poulogiannis G, Rust AG, Pardo M, Gracia T, Yu L, Choudhary J, Poulin GB, et al. Nuclear receptor binding protein 1 regulates intestinal progenitor cell homeostasis and tumour formation. EMBO J. 2012;31:2486–2497. doi: 10.1038/emboj.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Croucher DR, Hochgrafe F, Zhang L, Liu L, Lyons RJ, Rickwood D, Tactacan CM, Browne BC, Ali N, Chan H, et al. A novel signaling pathway in basal breast cancer involving Lyn and the atypical kinase SgK269/PEAK1. Cancer Res. 2013;73:1969–1980. doi: 10.1158/0008-5472.CAN-12-1472. [DOI] [PubMed] [Google Scholar]
- 12.Zheng Y, Zhang C, Croucher DR, Soliman MA, St-Denis N, Pasculescu A, Taylor L, Tate SA, Hardy WR, Colwill K, et al. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature. 2013;499:166–171. doi: 10.1038/nature12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eyers PA, Murphy JM. Dawn of the dead: protein pseudokinases signal new adventures in cell biology. Biochem Soc Trans. 2013;41:969–974. doi: 10.1042/BST20130115. [DOI] [PubMed] [Google Scholar]
- 14.Brennan DF, Dar AC, Hertz NT, Chao WC, Burlingame AL, Shokat KM, Barford D. A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature. 2011;472:366–369. doi: 10.1038/nature09860. [DOI] [PubMed] [Google Scholar]
- 15.Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 16.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 17.Scheeff ED, Eswaran J, Bunkoczi G, Knapp S, Manning G. Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Structure. 2009;17:128–138. doi: 10.1016/j.str.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee K, Sharma M, Urlaub H, Bourenkov GP, Jahn R, Sudhof TC, Wahl MC. CASK functions as a Mg2+ -independent neurexin kinase. Cell. 2008;133:328–339. doi: 10.1016/j.cell.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi F, Telesco SE, Liu Y, Radhakrishnan R, Lemmon MA. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc Natl Acad Sci USA. 2010;107:7692–7697. doi: 10.1073/pnas.1002753107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ungureanu D, Wu J, Pekkala T, Niranjan Y, Young C, Jensen ON, Xu CF, Neubert TA, Skoda RC, Hubbard SR, Silvennoinen O. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat Struct Mol Biol. 2011;18:971–976. doi: 10.1038/nsmb.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida-Moriguchi T, Willer T, Anderson ME, Venzke D, Whyte T, Muntoni F, Lee H, Nelson SF, Yu L, Campbell KP. SGK196 is a glycosylation-specific O-mannose kinase required for dystroglycan function. Science. 2013;341:896–899. doi: 10.1126/science.1239951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lucet IS, Babon JJ, Murphy JM. Techniques to examine nucleotide binding by pseudokinases. Biochem Soc Trans. 2013;41:974–980. doi: 10.1042/BST20130075. [DOI] [PubMed] [Google Scholar]
- 23.Stewart RC, VanBruggen R, Ellefson DD, Wolfe AJ. TNP-ATP and TNP-ADP as probes of the nucleotide binding site of CheA, the histidine protein kinase in the chemotaxis signal transduction pathway of Escherichia coli. Biochemistry. 1998;37:12269–12279. doi: 10.1021/bi980970n. [DOI] [PubMed] [Google Scholar]
- 24.Murphy JM, Metcalf D, Young IG, Hilton DJ. A convenient method for preparation of an engineered mouse interleukin-3 analog with high solubility and wild-type bioactivity. Growth Factors. 2010;28:104–110. doi: 10.3109/08977190903443048. [DOI] [PubMed] [Google Scholar]
- 25.Babon JJ, Murphy JM. In vitro JAK kinase activity and inhibition assays. Methods Mol Biol. 2013;967:39–55. doi: 10.1007/978-1-62703-242-1_3. [DOI] [PubMed] [Google Scholar]
- 26.Lucet IS, Bamert R. Production and crystallization of recombinant JAK proteins. Methods Mol Biol. 2013;967:275–300. doi: 10.1007/978-1-62703-242-1_20. [DOI] [PubMed] [Google Scholar]
- 27.Fukuda K, Gupta S, Chen K, Wu C, Qin J. The pseudoactive site of ILK is essential for its binding to α-Parvin and localization to focal adhesions. Mol Cell. 2009;36:819–830. doi: 10.1016/j.molcel.2009.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gee CL, Papavinasasundaram KG, Blair SR, Baer CE, Falick AM, King DS, Griffin JE, Venghatakrishnan H, Zukauskas A, Wei JR, et al. A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci Signaling. 2012;5:ra7. doi: 10.1126/scisignal.2002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reese ML, Boothroyd JC. A conserved non-canonical motif in the pseudoactive site of the ROP5 pseudokinase domain mediates its effect on Toxoplasma virulence. J Biol Chem. 2011;286:29366–29375. doi: 10.1074/jbc.M111.253435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeqiraj E, Filippi BM, Goldie S, Navratilova I, Boudeau J, Deak M, Alessi DR, van Aalten DM. ATP and MO25α regulate the conformational state of the STRADα pseudokinase and activation of the LKB1 tumour suppressor. PLoS Biol. 2009;7:e1000126. doi: 10.1371/journal.pbio.1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 32.Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Bicocca VT, Chang BH, Masouleh BK, Muschen M, Loriaux MM, Druker BJ, Tyner JW. Crosstalk between ROR1 and the pre-B cell receptor promotes survival of t(1;19) acute lymphoblastic leukemia. Cancer Cell. 2012;22:656–667. doi: 10.1016/j.ccr.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gentile A, Lazzari L, Benvenuti S, Trusolino L, Comoglio PM. Ror1 is a pseudokinase that is crucial for Met-driven tumorigenesis. Cancer Res. 2011;71:3132–3141. doi: 10.1158/0008-5472.CAN-10-2662. [DOI] [PubMed] [Google Scholar]
- 35.Suijkerbuijk SJ, van Dam TJ, Karagoz GE, von Castelmur E, Hubner NC, Duarte AM, Vleugel M, Perrakis A, Rudiger SG, Snel B, Kops GJ. The vertebrate mitotic checkpoint protein BUBR1 is an unusual pseudokinase. Dev Cell. 2012;22:1321–1329. doi: 10.1016/j.devcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 36.Hovens CM, Stacker SA, Andres AC, Harpur AG, Ziemiecki A, Wilks AF. RYK, a receptor tyrosine kinase-related molecule with unusual kinase domain motifs. Proc Natl Acad Sci USA. 1992;89:11818–11822. doi: 10.1073/pnas.89.24.11818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nimchuk ZL, Tarr PT, Meyerowitz EM. An evolutionarily conserved pseudokinase mediates stem cell production in plants. Plant Cell. 2011;23:851–854. doi: 10.1105/tpc.110.075622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim JG, Li X, Roden JA, Taylor KW, Aakre CD, Su B, Lalonde S, Kirik A, Chen Y, Baranage G, et al. Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell. 2009;21:1305–1323. doi: 10.1105/tpc.108.063123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Labesse G, Gelin M, Bessin Y, Lebrun M, Papoin J, Cerdan R, Arold ST, Dubremetz JF. ROP2 from Toxoplasma gondii: a virulence factor with a protein-kinase fold and no enzymatic activity. Structure. 2009;17:139–146. doi: 10.1016/j.str.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 40.Buchholz KR, Bowyer PW, Boothroyd JC. Bradyzoite pseudokinase 1 is crucial for efficient oral infectivity of the Toxoplasma tissue cyst. Eukaryotic Cell. 2013;12:399–410. doi: 10.1128/EC.00343-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukuda K, Knight JD, Piszczek G, Kothary R, Qin J. Biochemical, proteomic, structural, and thermodynamic characterizations of integrin-linked kinase (ILK): cross-validation of the pseudokinase. J Biol Chem. 2011;286:21886–21895. doi: 10.1074/jbc.M111.240093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lo MC, Aulabaugh A, Jin G, Cowling R, Bard J, Malamas M, Ellestad G. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem. 2004;332:153–159. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 43.Murphy JM, Czabotar PE, Hildebrand JM, Lucet IS, Zhang JG, Alvarez-Diaz S, Lewis R, Lalaoui N, Metcalf D, Webb AI, et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39:443–453. doi: 10.1016/j.immuni.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 44.Bullock AN, Debreczeni JE, Fedorov OY, Nelson A, Marsden BD, Knapp S. Structural basis of inhibitor specificity of the human protooncogene proviral insertion site in Moloney murine leukemia virus (PIM-1) kinase. J Med Chem. 2005;48:7604–7614. doi: 10.1021/jm0504858. [DOI] [PubMed] [Google Scholar]
- 45.Babon JJ, Kershaw NJ, Murphy JM, Varghese LN, Laktyushin A, Young SN, Lucet IS, Norton RS, Nicola NA. Suppression of cytokine signaling by SOCS3: characterization of the mode of inhibition and the basis of its specificity. Immunity. 2012;36:239–250. doi: 10.1016/j.immuni.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becher I, Savitski MM, Savitski MF, Hopf C, Bantscheff M, Drewes G. Affinity profiling of the cellular kinome for the nucleotide cofactors ATP, ADP, and GTP. ACS Chem Biol. 2013;8:599–607. doi: 10.1021/cb3005879. [DOI] [PubMed] [Google Scholar]
- 47.Kang J, Yang M, Li B, Qi W, Zhang C, Shokat KM, Tomchick DR, Machius M, Yu H. Structure and substrate recruitment of the human spindle checkpoint kinase Bub1. Mol Cell. 2008;32:394–405. doi: 10.1016/j.molcel.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 49.James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, Garcon L, Raslova H, Berger R, Bennaceur-Griscelli A, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 50.Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M, Skoda RC. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 51.Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 52.Zhao R, Xing S, Li Z, Fu X, Li Q, Krantz SB, Zhao ZJ. Identification of an acquired JAK2 mutation in polycythemia vera. J Biol Chem. 2005;280:22788–22792. doi: 10.1074/jbc.C500138200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bandaranayake RM, Ungureanu D, Shan Y, Shaw DE, Silvennoinen O, Hubbard SR. Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F. Nat Struct Mol Biol. 2012;19:754–759. doi: 10.1038/nsmb.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeqiraj E, Filippi BM, Deak M, Alessi DR, van Aalten DM. Structure of the LKB1–STRAD–MO25 complex reveals an allosteric mechanism of kinase activation. Science. 2009;326:1707–1711. doi: 10.1126/science.1178377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Artim SC, Mendrola JM, Lemmon MA. Assessing the range of kinase autoinhibition mechanisms in the insulin receptor family. Biochem J. 2012;448:213–220. doi: 10.1042/BJ20121365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jung JW, Shin WS, Song J, Lee ST. Cloning and characterization of the full-length mouse Ptk7 cDNA encoding a defective receptor protein tyrosine kinase. Gene. 2004;328:75–84. doi: 10.1016/j.gene.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 57.Wesche H, Gao X, Li X, Kirschning CJ, Stark GR, Cao Z. IRAK-M is a novel member of the Pelle/interleukin-1 receptor-associated kinase (IRAK) family. J Biol Chem. 1999;274:19403–19410. doi: 10.1074/jbc.274.27.19403. [DOI] [PubMed] [Google Scholar]
- 58.Gurniak CB, Berg LJ. A new member of the Eph family of receptors that lacks protein tyrosine kinase activity. Oncogene. 1996;13:777–786. [PubMed] [Google Scholar]
- 59.Matsuoka H, Iwata N, Ito M, Shimoyama M, Nagata A, Chihara K, Takai S, Matsui T. Expression of a kinase-defective Eph-like receptor in the normal human brain. Biochem Biophys Res Commun. 1997;235:487–492. doi: 10.1006/bbrc.1997.6812. [DOI] [PubMed] [Google Scholar]
- 60.Zhao J, Jitkaew S, Cai Z, Choksi S, Li Q, Luo J, Liu ZG. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc Natl Acad Sci USA. 2012;109:5322–5327. doi: 10.1073/pnas.1200012109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carrera AC, Alexandrov K, Roberts TM. The conserved lysine of the catalytic domain of protein kinases is actively involved in the phosphotransfer reaction and not required for anchoring ATP. Proc Natl Acad Sci USA. 1993;90:442–446. doi: 10.1073/pnas.90.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murphy JM, Lucet IS, Hildebrand JM, Tanzer MC, Young SN, Sharma P, Lessene G, Alexander WS, Babon JJ, Silke J, Czabotar PE. Insights into the evolution of divergent nucleotide-binding mechanisms among pseudokinases revealed by crystal structures of human and mouse MLKL. Biochem J. 2013 doi: 10.1042/BJ20131270. [DOI] [PubMed] [Google Scholar]
- 63.Jaleel M, Saha S, Shenoy AR, Visweswariah SS. The kinase homology domain of receptor guanylyl cyclase C: ATP binding and identification of an adenine nucleotide sensitive site. Biochemistry. 2006;45:1888–1898. doi: 10.1021/bi052089x. [DOI] [PubMed] [Google Scholar]
- 64.Kelber JA, Reno T, Kaushal S, Metildi C, Wright T, Stoletov K, Weems JM, Park FD, Mose E, Wang Y, et al. KRas induces a Src/PEAK1/ErbB2 kinase amplification loop that drives metastatic growth and therapy resistance in pancreatic cancer. Cancer Res. 2012;72:2554–2564. doi: 10.1158/0008-5472.CAN-11-3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Y, Kelber JA, Tran Cao HS, Cantin GT, Lin R, Wang W, Kaushal S, Bristow JM, Edgington TS, Hoffman RM, et al. Pseudopodium-enriched atypical kinase 1 regulates the cytoskeleton and cancer progression. Proc Natl Acad Sci USA. 2010;107:10920–10925. doi: 10.1073/pnas.0914776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kawagoe T, Sato S, Matsushita K, Kato H, Matsui K, Kumagai Y, Saitoh T, Kawai T, Takeuchi O, Akira S. Sequential control of Toll-like receptor-dependent responses by IRAK1 and IRAK2. Nat Immunol. 2008;9:684–691. doi: 10.1038/ni.1606. [DOI] [PubMed] [Google Scholar]
- 67.Pauls E, Nanda SK, Smith H, Toth R, Arthur JS, Cohen P. Two phases of inflammatory mediator production defined by the study of IRAK2 and IRAK1 knock-in mice. J Immunol. 2013;191:2717–2730. doi: 10.4049/jimmunol.1203268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lange A, Wickstrom SA, Jakobson M, Zent R, Sainio K, Fassler R. Integrin-linked kinase is an adaptor with essential functions during mouse development. Nature. 2009;461:1002–1006. doi: 10.1038/nature08468. [DOI] [PubMed] [Google Scholar]
- 69.Mendrola JM, Shi F, Park JH, Lemmon MA. Receptor tyrosine kinases with intracellular pseudokinase domains. Biochem Soc Trans. 2013;41:1029–1036. doi: 10.1042/BST20130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toms AV, Deshpande A, McNally R, Jeong Y, Rogers JM, Kim CU, Gruner SM, Ficarro SB, Marto JA, Sattler M, et al. Structure of a pseudokinase-domain switch that controls oncogenic activation of Jak kinases. Nat Struct Mol Biol. 2013;20:1221–1223. doi: 10.1038/nsmb.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 72.Zeqiraj E, van Aalten DM. Pseudokinases: remnants of evolution or key allosteric regulators? Curr Opin Struct Biol. 2010;20:772–781. doi: 10.1016/j.sbi.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G. Structural and functional diversity of the microbial kinome. PLoS Biol. 2007;5:e17. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


