Abstract
Objective
Prior hypothesis-driven studies identified immunophenotypic characteristics associated with the control of HIV replication without antiretroviral therapy (HIV controllers) as well as with the degree of CD4+ T cell recovery during ART. We hypothesized that an unbiased “discovery-based” approach might identify novel immunologic characteristics of these phenotypes.
Design
We performed immunophenotyping on four “aviremic” patient groups: (1) HIV controllers (n=98), (2) antiretroviral-treated immunologic non-responders (CD4 <350; n=59), (3) antiretroviral-treated immunologic responders (CD4 >350, n=142), and as a control group (4) HIV-negative adults (n=43). We measured levels of T cell maturation, activation, dysfunction, senescence, functionality and proliferation.
Methods
Supervised Learning assessed the relative importance of immune parameters in predicting clinical phenotypes (controller, immunologic responder or immunologic non-responder). Unsupervised learning clustered immune parameters and examined if these clusters corresponded to clinical phenotypes.
Results
HIV controllers were characterized by high percentages of HIV-specific T cell responses and decreased percentages of cells expressing HLA-DR in naïve, central memory and effector T cell subsets. Immunologic non-responders were characterized by higher percentages of CD4+ T cells that were TNF-α+ or INF-γ+, higher percentages of activated naïve and central memory T cells, and higher percentages of cells expressing PD-1. Unsupervised learning found two distinct clusters of controllers and two distinct clusters of immunologic non-responders, perhaps suggesting different mechanisms for the clinical outcomes.
Conclusions
Our discovery-based approach confirmed previously reported characteristics that distinguish aviremic individuals, but also identified novel immunologic phenotypes and distinct clinical sub-populations that should lead to more focused pathogenesis studies that might identify targets for novel therapeutic interventions.
Keywords: HIV, machine-learning, controllers, immunologic non-responders, immunophenotype, random forests, fuzzy forests
INTRODUCTION
The vast majority of HIV-infected adults who have access to combination antiretroviral therapy (ART) and are motivated to take these drugs can achieve and maintain undetectable plasma HIV RNA levels (viral load). As a consequence of treatment-mediated viral suppression, peripheral CD4+ T cell counts increase, and AIDS-related complications are largely prevented. Although most patients taking ART achieve virologic suppression, a substantial (approximately 15–30%) but poorly described subset of treated adults maintain abnormally low peripheral blood CD4+ T cell counts (“immunologic non-responders”) [1, 2]. We define an immunologic non-responder as an individual whose on-therapy CD4+ T cell count plateaus to a level < 350 cells/mm3 despite being aviremic. A low on-therapy CD4+ T cell count has been associated with increased risk of serious non-AIDS events, including heart disease and cancer, as well as an increased risk of mortality[3–5]. Understanding how immunologic responders and non-responders differ at a more granular level in terms of precise patterns of immune function will be critical in identifying targets for therapeutic interventions that may augment or optimize ART delivery.
In parallel with ongoing advances in ART agents and ART delivery strategies, there is a large and interest in identifying “curative” interventions [6]. Optimism that a cure might be feasible is driven in part by the fact that a small proportion of HIV-infected adults are able to control HIV indefinitely in the absence of ART. The majority of these so-called “HIV controllers” harbor replication-competent virus yet exhibit durable viral control, normal or very slowly declining CD4+ T cell counts, and have low risk for AIDS-related morbidity. Controllers – particularly those who also maintain low levels of immune activation – are now being studied in detail as they may provide clues as to how to effectively control HIV infection [7].
To more fully characterize immunologic features seen in varying states of “aviremia”—mediated by ART for those on treatment and presumably mediated by host mechanisms for those not on treatment—we assembled a cohort of four unique groups from patients enrolled in two longitudinal HIV patients cohorts at UCSF who have undetectable HIV RNA levels: (1) HIV controllers, (2) immunologic responders, (3) immunologic non-responders, and as a control group (4) HIV-uninfected individuals. T cell activation, senescence, maturation, and function were measured using well-validated assays generating over 514 Boolean subpopulations. Because there were many more parameters, p, than observations, n, and the parameters were highly correlated it was necessary reduce the parameter space in a systematically unbiased way. Recent advances in machine learning have been developed to deal with the so-called “p >> n problem”[8]. Recursive feature elimination Random Forests [9], conditional Random Forests[10, 11], and Fuzzy Forests have been shown to produce relatively unbiased variable importance lists in these types of settings. Using data-mining approaches and complex multi-parameter immunophenotypic data, we sought to identify immunologic signatures associated with each of the above four clinical phenotypes. We report here several novel correlates to each phenotype, which may have implications for future therapeutic strategies.
METHODS
Subjects
All participants were enrolled in one of two longitudinal cohorts based at the University of California, San Francisco (UCSF): the Options Project (which enrolls individuals diagnosed with acute/early HIV infection and at-risk HIV-uninfected controls) and the SCOPE cohort (which enrolls individuals with chronic HIV infection, and HIV-negative controls). Cryopreserved peripheral blood mononuclear cells (PBMCs) were stored and sent in batch to the UCSF Core Immunology Laboratory for advanced immunophenotyping (see below). All immunologic data were assembled in a single database. We selected four unique groups (1) HIV controllers: persons with documented HIV-1 infection for a t least 1 year, ART naïve and with a plasma HIV RNA level < 40–75 copies RNA/mL for all visits (n=98), (2) Immunologic responders: individuals with chronic HIV who achieved durable viral suppression (plasma HIV RNA < 40–75 copies/RNA, larger blips were allowed if they were flanked by undetectable values) on a stable ART regimen and who had high CD4+ T cell counts (> 350 cells/mm3, n=142), (3) Immunologic non-responders: individuals with chronic HIV who achieved durable viral suppression (plasma HIV RNA < 40–75 copies/RNA) on a stable ART regimen but whose CD4+ T cell counts plateaued to a level < 350 cells/mm3, n=59), and (4) HIV-negative controls (n=43).
Measurements
Immunophenotypic parameters of T cell maturation (CD45RA, CCR7, CD27, CD28), activation (CD38, HLA-DR, CCR5), dysfunction (PD-1), senescence (CD57), HIV and/or CMV (ex vivo) antigen-specificity (IFN-gamma, TNF-alpha, IL-2, CD107a) and proliferation (CFSE) were measured on (PBMCs) from all participants, using previously described methods [12, 13]. Digital Supplemental Table 1 lists all of the potential parameters.
Statistical Methods
Data on 514 unique immunologic profiles were generated on each individual subject. Due to the complex nature of the data, we employed several machine learning techniques to fulfill our objectives. Because variables such as CD4+ T cell count and viral load were used to define the groups, these variables were left out of all machine learning analyses.
We used a variant of Random Forests, Fuzzy Forests [14, 15], which are Random Forests (RF) with recursive feature elimination (RFE) and variable importance that are done on individual modules created using weighted gene co-expression network analysis. We briefly review Random Forests. RFs are a supervised learning method, meaning the outcome labels (in this case the clinical phenotypes) are used to classify the observations into categories [16]. RFs are useful in the so-called “large p, small n problem” wherein one has more potential predictors than observations. These methods have an advantage over other machine learning algorithms in that they incorporate high-order interactions, are largely unbiased even in the case of different scales among the predictor variables, have a natural variable importance measure, and are expedient even in very large datasets.
RFs are an ensemble of individual classification and regression trees. The data are bootstrapped and a tree is built on each bootstrap sample. To reduce the correlation between trees, at each node the number of variables chosen at each node is of size "mtry." For example, if mtry is 5, then 5 predictors are randomly selected from all possible predictors, p, at each node. The usual default for mtry is the square root of p. The best splitter in terms of the Gini index is chosen from this set to split the sample. Each tree is grown out and not pruned. The observations that are not in each individual tree (about 33% of the data) are called the Out-of-Bag (OOB) sample. The OOB predictive error can be estimated by predicting the outcome by a majority rules vote involving trees in the forest that did not have that observation in the training sample. Because of these features, there is low correlation among the individual trees. By averaging over a large number of trees, this learner has both low bias and low variance. This type of ensemble predictor is attractive because it is invariant to monotonic transformations, can handle large number of variables and is robust to outliers. These features are very amenable for flow-cytometry data in that it handles highly skewed variables without any need to symmetrize skewed variable distributions. The more trees in the forest, the more accurate the ensemble [16]. All analyses were conducted in R and C++. This approach has been previously used for gene filtering in microarrays [9, 17–20].
To select the approximately 50 most important parameters using Fuzzy Forests, we used the wff option in Fuzzy Forests. This option is used to account for the network structure among the predictors. First modules are constructed using Weighted Gene Co-expression network analysis. This creates modules that have higher correlation within module than between [21]. For each module, we then fit RFE-RF [22–28] and then ranked the predictors using permutation importance [10, 29–31]. After each iteration, we discarded the variables with the smallest variable importance retaining the set of parameters that has the smallest OOB rate. It is important to note that the OOB error rate for this part is used only for variable selection, and not to obtain an estimate of the error rate. This iterative procedure has been shown to yield good stability [9]. For each iteration, we grew 50,000 trees. The survivors of this process for each module were then pooled and another RFE-RF was performed. This was done until we had a set of approximately 50 features. We tried several values of “mtry” with similar results.
Unsupervised learning was performed using hierarchical clustering and a random forest dissimilarity matrix. In this analysis, the clinical phenotype (the clinical group the subject is in) as well as CD4+ T cell counts and viremia were excluded from the analysis.
RESULTS
We performed immunophenotyping on 342 individuals from four unique “aviremic” clinical phenotypes: (1) HIV controllers (n=98), (2) immunologic non-responders (n=59), (3) immunologic responders (n=142) and (4) HIV-negative controls (n=43). Most HIV-positive participants were men between 40–55 years old; HIV-negative controls were slightly younger (Table 1). While the median CD4+ T cell count was 235 cells/mm3 in the immunologic non-responders, the median CD4+ T cell count was well above 500 cells/mm3 for each of the other groups.
Table 1.
Demographic characteristics of the four aviremic groups. Median and Interquartile Range are given for continuous variables and N and percent are given for categorical variables.
| HIV Negative (N=43) | Controllers (N=98) | Chronic Responders (N=142) | Chronic Nonresponders (N=59) | |||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| Variable | Median | IQR | Median | IQR | Median | IQR | Median | IQR |
| Age | 40 | (33.6, 46) | 49 | (42.0, 53.5) | 47 | (43, 56.6) | 51 | (46.0, 56.9) |
| CD4 count | 863 | (765, 1164) | 740 | (606, 1012) | 640 | (586, 809) | 235 | (185, 292 |
| CD8 count | 531 | (397, 631 | 955 | (642, 1201) | 878 | (742, 1375) | 844 | (608, 1077) |
| Viral Load | ND | ND | 40 | (40,62) | 40 | (40, 52) | 40 | (40, 40) |
| Nadir CD4 | ND | ND | 560 | (400, 696) | 225 | (91, 317) | 70 | (19, 137) |
| Ethnicity | N | % | N | % | N | % | N | % |
|
|
|
|
|
|||||
| Asian | 1 | 2.33 | 3 | 3.06 | 9 | 6.34 | 1 | 1.69 |
| Black | 4 | 9.30 | 28 | 28.57 | 19 | 13.38 | 7 | 11.86 |
| Hispanic | 5 | 11.63 | 12 | 12.24 | 5 | 3.52 | 5 | 8.47 |
| Mixed | 1 | 2.33 | 10 | 10.20 | 4 | 2.82 | 3 | 5.08 |
| White | 30 | 69.77 | 42 | 42.86 | 104 | 73.24 | 43 | 72.88 |
| Other | 2 | 4.65 | 3 | 3.06 | 1 | 0.70 | 0 | 0.00 |
| Gender | ||||||||
| Female | 3 | 6.98 | 26 | 26.53 | 12 | 8.45 | 2 | 3.39 |
| Male | 40 | 93.02 | 69 | 70.41 | 127 | 89.44 | 56 | 94.92 |
| Male to Female | 0 | 0.00 | 3 | 3.06 | 3 | 2.11 | 1 | 1.69 |
We obtained data on the level of T cell maturation (CD45RA, CCR7, CD27, CD28), activation (CD38, HLA-DR, CCR5), dysfunction (PD-1), senescence (CD57), and functional capacity (cytokine production in response to HIV or CMV peptide pools) and T cell proliferation (CFSE). There were 514 measurements included in the analysis (Supplemental Table 1). Variables including absolute CD4+ and CD8+ T-cell counts, the CD4/CD8 ratio, nadir CD4 count, and viral load measures were excluded because they were used to define the four clinical phenotypes.
Identification of five unique immunologic clusters using unsupervised learning
We first used unsupervised hierarchical clustering using a Random Forest dissimilarity matrix to identify groups of HIV-infected participants that shared related immunologic signatures when the clinically-defined groups (controllers, immunologic responders/non-responders) were not specified a priori. CD4+ T cell counts, viral load and any group-defining characteristics were not included in the analysis. As shown in Figure 1, five unique clusters were identified (labeled A–E), two of which were highly enriched for HIV controllers and two that were highly enriched for immunologic non-responders. Immunologic responders clustered together and are identified by Cluster D. Variables that distinguished the clusters included Gag-specific CD4+ and CD8+ T-cells, HLA DR+ central memory T cells, resting CD4+ central memory cells, central memory activated CD8+ T-cells, perforin positive cells and PD-1.
Figure 1.
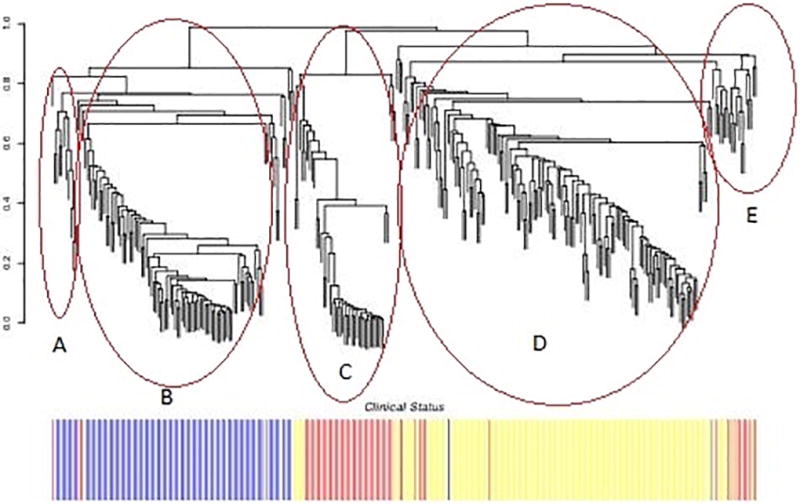
Hierarchical clustering plot using Unsupervised Learning shows three “aviremic” HIV-positive phenotypes (HIV controllers, immunologic responders, and immunologic non-responders) have several discrete subgroups. Each branch in the tree represents a patient. The bar graph below shows the phenotype membership of each subject. HIV controllers are shown in blue and labeled Clusters A and B, immunologic responders are shown in yellow and labeled Cluster D, and immunologic non-responders are shown in red and labeled Clusters C and E.
Overall HIV controllers had higher percentages of Gag-specific CD4+ and CD8+ T-cells relative to the other groups. Controllers also had much lower percentages of central memory CD4+ T-cells that express HLA-DR. There were two distinct clusters of controllers which we labeled Cluster A and Cluster B in Figure 1. Controller Cluster B had higher percentages of CD8+ T-cells that are CD28−CD57+ (19.7% versus 26.7% respectively, p=0.0078 by the t-test comparing cluster A and B) as well as higher percentages of CD8+HLA−DR+ cells (39% versus 48.9%, p=0.014) than controller cluster A. Controller Cluster A had higher percentages of CD8+ cells that were RA+ (61.9% versus 51.6%, p=0.0025) and higher percentages of CD8+HLADR+CCR7− that are CD38+ (55.3% versus 38.8, p=0.0068) compared to cluster B. Similarly Cluster A had higher percentages of CD8+ cells that were RA+CCR7+CD27+CD28+57− than Cluster B (26.4% versus 15.1%, p < 0.0001). Conversely, Cluster B had higher percentages of CD8+ cells that were RA+CCR7−CD27−CD28−57+ (7.7% versus 11.97%, p=0.0001). This perhaps suggests that the enrichment for RA+ cells appears to be driven mainly by naïve CD8 T cells and not TEMRAs (CD8+CCR7−CD45RA+) in Cluster A. Cluster B had slightly more subjects who self-reported black ethnicity, but this did not reach statistical significance.
Immunologic non-responders had higher levels of CD4+ activation (CD38+ HLA−DR+) as well as higher percentages of cells that expressed PD-1 compared to the other groups. There were also two distinct immunologic non-responder clusters, Clusters C and E. While both clusters of immunologic non-responders were characterized by high levels of both CD4+ and CD8+ T cell activation and PD1 expression, immunologic non-responder Cluster E appeared to be distinct from all of the other clusters and was characterized by lower percentages of activated CD8+ T-cells that are CCR5+ (60.7% versus 44.3, p=0.0068) and higher percentages of central memory CD4+ T-cells than Immunologic Non-Responder Cluster C (28.6% versus 33.9%, p=0.0013).
Correlates of HIV control using supervised learning
We next used fuzzy forests to identify the most important immunologic characteristics that predict each clinically defined group. We compared the HIV controllers to the immunologic responders, as both groups were distinguished by having a relatively intact immune system, with the groups differing primary based on the mechanism by which HIV was controlled (host-mediated control versus antiretroviral treatment).
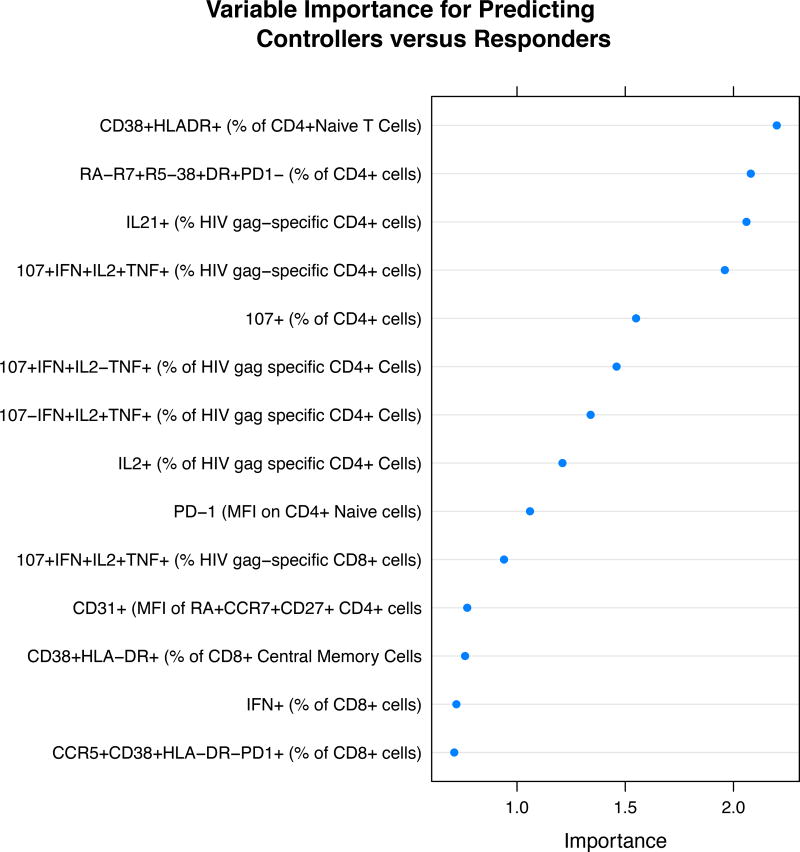
The best predictors are given in the variable importance plot for responders versus controllers (Figure 2a). The predictors are ranked with the best predictor at the top. As expected, we found that compared to the immunologic responders, HIV controllers had much higher levels of polyfunctional (ex vivo) Gag-specific CD4+ and CD8+ T cells than the other groups. This was particularly true for the frequency of polyfunctional (CD107+, IFN-gamma+, IL-2+, TNF-alpha+) CD4+ and CD8+ T cells. There were also marked differences in activation markers particularly HLA-DR and differences in central memory T cells (Table 2a lists post-hoc t-test p-values for the most important variables).
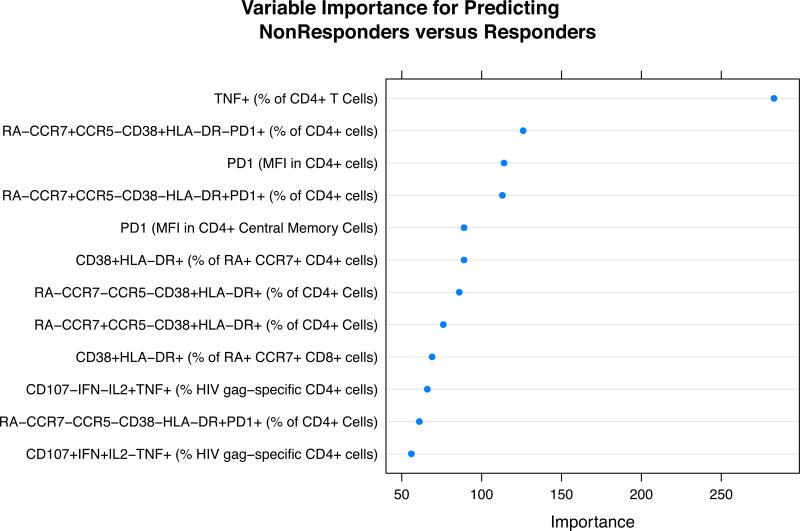
Figure 2.
a) Variable Importance Plots for the most important variables in predicting controllers from responders. b) Variable Importance Plots for the most important variables in predicting responders from immunologic non-responders. Variables are ranked in order of their importance, with the most important variable on top.
Table 2.
a) P-values obtained via the univariate t-test for the most important variables in predicting controllers versus responders as given by Fuzzy Forests. b) P-values obtained via the univariate t-test for the most important variables in predicting responders versus immunologic non-responders as given by Fuzzy Forests.
| Variable | t−value | P-value |
|---|---|---|
| CD38+HLADR+ (% of CD4+ naïve cells) | −6.428 | <0.0001 |
| RA−CCR7+CCR5−CD38+HLADR+PD1− (% of CD4+ cells) | 7.3834 | <0.0001 |
| IL21+ (% of HIV gag-specific CD4+ cells) | −3.1299 | 0.0023 |
| CD107+IFN+IL2+TNF+ (% of HIV gag-specific CD4+ cells) | −4.1031 | <0.0001 |
| CD107+ (% of CD4+ cells) | 5.9309 | <0.0001 |
| CD107−IFN+IL2+TNF+ (% of HIV gag-specific CD4+ cells) | −4.3022 | <0.0001 |
| CD107+IFN+IL2−TNF+ (% of HIV gag-specific CD4+ cells) | −1.805 | 0.07 |
| IL2+ (% of HIV gag-specific CD8+ cells) | −6.1202 | <0.0001 |
| CD107+IFN+IL2+TNF+ (% of HIV gag-specific CD8+ cells) | −5.5631 | <0.0001 |
| PD1+ (MFI CD4+ naïve cells) | −2.7265 | 0.007 |
| CCR5+38+HLADR−PD1+ (% of CD8+ cells) | 4.1607 | <0.0001 |
| IFN+ (% of CD8+ cells) | −1.9 | 0.06 |
| IFN+ (% of CD4+ cells) | −1.6378 | 0.1 |
| CD31+ (MFI of CD4+ naïve (RA+CCR7+CD27+) | 4.5962 | <0.0001 |
| CD107+IFN+IL2−TNF+ (% HIV gag-specific CD8+ cells) | −5.9161 | <0.0001 |
| CD38+HLADR+ (% of CD8+ Central Memory cells) | −6.3878 | <0.0001 |
| RA+CCR7+CCR5−CD38+HLADR+PD1− (% of CD4+ cells) | 8.441 | <0.0001 |
| CCR5+CD38+HLADR−PD1− (% of CD8+ cells) | 4.5843 | <0.0001 |
Red indicates that Controllers have higher average values
Black indicates that Controllers have lower average values
The percentage of “activated” (CD38+ HLA−DR+) CD8+ T cells was higher in controllers than in other groups (a median of 27% in controllers, versus 24% in non-responders, 21% in ART responders and 11% in HIV negatives), an observation that is consistent with prior studies from this and other cohorts [32–34]. Compared to responders controllers have lower percentages of CD4+ naïve cells that are HLA-DR positive (1.56% versus 3.73%, p< 0.0001), lower percentages of CD4+ central memory T cells that are DR positive (9.89% versus 15.51%, p < 0.0001) and lower percentages of CD4+ effector memory cells that are HLA−DR+ (15.43% versus 12.93%, p < 0.0001).
Correlates of immunologic response during ART using supervised learning
We also used the supervised learning approach to identify unique signatures associated with a low CD4+ T cell count while on ART (immunologic non-responder versus responder). Figure 2b shows the most important variables in predicting immunologic non-response and Table 2b lists the post-hoc p-values in comparing non-responders and responders on these variables. One of the most notable immunological differences that uniquely identified this group is the expression of PD-1 on CD4+ T cells and CD4+ T cell subsets, a finding consistent with prior observations from this cohort[26]. Immunologic non-responders had higher frequencies of TNF-α+ and INF-γ+ CD4 cells compared to responders. Non-responders also had higher percentages of CD4+ T cell activation in all subsets (naïve, central memory and effector memory) and lower frequencies of naïve CD4+ T cells than responders (see Table 2b).
Predictive capacity of the model
Overall, the forests had excellent predictive accuracy. Because using the OOB error rate on iterated sets can lead to biased estimates of the true error rate, we used the original 0.632 + bootstrap method to estimate the error rate. The overall OOB error rate was approximately 11% for each comparison meaning that, on average, 11% of the participants were misclassified. Moreover, the low OOB error rate suggests that the results may be generalizable to other cohorts.
Using Random Forests-type algorithms, we did not explicitly control for age or any other variable. Age was not listed as one of the top predictors and hence was not included as a potential predictor in the final Random Forest. Of note, HIV negative individuals were significantly younger than the other 3 groups. However the three HIV-infected groups did not differ significantly in age (P=0.1348 by the F test). When age was included into the top 50 variables and Random Forests were run and conditional variable importance results generated, age was again not selected as one of the most important variables in terms of classifying participants.
DISCUSSION
The overall objective of our analysis was to find, in an unbiased manner, the key immunologic signatures associated with three distinct clinical phenotypes: HIV controllers, immunologic non-responders, and immunologic responders. Using sophisticated machine-learning techniques designed to find novel observations within complex datasets where the number of parameters is much larger than the number of observations, we were able to meaningfully sort through the data in a systematic manner. The algorithm found a very strong and predictive signal, with an overall misclassification across the three HIV-seropositive groups and the HIV-seronegative group of approximately 11%. This unbiased approach confirmed a number of prior observations, and identified several unique associations that might lead to novel therapeutic approaches aimed at either controlling HIV in the absence of therapy (“functional” cure) or enhancing immune reconstitution during therapy. Importantly, these variable importance lists are not simply univariate tests of importance similar to a t-test, but rather allow for interaction of the variables. We also show excellent predictive capacity of the model. The low “Out of Bag” error rate suggests that these results may be generalizable to other cohorts.
This study confirmed a number of prior observations, specifically we found that HIV controllers have higher levels of HIV-specific T cells and higher levels of CD8+ T cell activation [35–40], while identifying unique immunophenotypes that differentiate these different HIV participant groups. For example, we identified a unique cluster of HIV controllers who maintained low CD8+ T cell activation levels despite robust HIV-specific T cell responsiveness. Non-responders clustered in to 2 groups. One of these groups (group E), in particular was distinct in that it had, on average, lower percentages of activated CD8+ T-cells that expressed CCR5 and higher percentages of central memory CD4+ T cells. These novel associations are the advantage of using a discovery-based approach to identify potential important parameters that distinguish these unique participant groups.
Interestingly, the unsupervised analysis identified a unique cluster of HIV controllers that maintained low CD8+ T cell activation despite robust Gag-specific T cell responses, representing a potentially attractive model for functional cure. In the CD4+ T cell compartment, we also found that although controllers and responders had similar percentages of CD4+ T cells that were activated (CD38+ HLADR+), controllers had lower percentages of CD4+ central memory T cells that were activated (HLA−DR+). It is interesting to note that in this respect HIV controllers are the group most similar to HIV negatives. Further we found that HIV controllers had the highest median percentage of CD4+ T cells that were resting (CD38− HLADR−) central memory cells and CCR5+.
Similar to other studies [41], we found a higher proportion of patients who reported black ethnicity as well as a higher proportion of women in the controller group. Elite Cluster B (see Figure 1) was enriched for the black ethnicity, the proportion not statistically different from cluster A. However, this could be due to sample size issues. Further larger studies are needed.
We then evaluated the immunological differences in patients on ART who failed to have sustained increases in CD4+ T cell counts. Consistent with previous studies [42], we found that most of the group-specific characteristics were in the CD4+ T cell compartment. One of most striking and consistent correlates was the expression of PD-1 on CD4+ T cells, including CD4+ central memory T cells [43]. Similarly, the percentage of CD4+ cells expressing TNF-α and IFN-γ was higher in immunologic non-responders. The expression of activation markers CD38 and HLA-DR was markedly elevated on all CD4+ T-cell subsets as well as naïve CD8+ T-cells, confirming prior observations [44–46]. Non-responders also had a lower percentage of naïve CD4+ T cells. These markers gave excellent prediction results in classifying patients who were non-responders. It is interesting to note that Non-responders clustered into two groups. One of these groups (group E), in particular was distinct in that it had, on average, lower percentages of activated CD8+ T-cells that expressed CCR5 and higher percentages of central memory CD4+ T cells. Collectively, these data suggest that ART-non-response might be due to a combination of factors, including failure to generate new cells and a shorter lifespan (consistent with the expression of activation markers). The markedly elevated levels of PD-1 suggest that many of the T cells are dysfunctional, which might contribute to persistent immunodeficiency and perhaps explain in part the excess morbidity and mortality in this group.
Three limitations of this analysis deserve comment. First, this is a cross-sectional study and has the well-recognized limitations of such a design. Specifically, it is difficult to disentangle causality. This is particularly true with regard to our analysis of immunologic non-responders. Although it is likely that the lack of naïve CD4+ T cells most likely causes immunologic non-response, one could also envision a model in which T cell activation causes immunologic non-response (e.g., by causing excess turnover, or by increasing PD-1 and anergy). A model can also be constructed in which immunologic non-response causes T cell activation (e.g., by persistent immunodeficiency and excess pathogen burden, or by stimulating release of homeostatic signals such as IL-7 that stimulate T cell proliferation/activation).
A second limitation is that we did not select our participants based on age, gender, or ethnicity; we observed group-to-group differences that might have been mitigated by matching or selection. Notably, the HIV-negative group was younger than the other groups, which may have biased any comparison to the normal state, as age is known to impact immune function. However, we were reassured by the fact that age did not emerge as a key predictor in Fuzzy Forests algorithms; based on this, age was not included as a potential predictor in the final analysis.
Third, our analysis explored multiple hypotheses, and although Random Forests theoretically do not overfit, and post-hoc comparisons of important variables are generally robust to a Bonferroni-type correction, our results should be interpreted in this light. P-values, in general, are not appropriate for machine learning-type applications. Instead, we used Out of Bag error rates. The estimated OOB error rates suggest that our results may be generalizable, although validation studies are needed.
In the current era of widely available, highly effective ART, there is intense interest in understanding immunophenotypes of individuals who achieve undetectable viral load, either naturally or by taking ART. Residual disease during otherwise successful ART is consistently predicted by and possibly caused by persistent inflammation and/or immunodeficiency. There is also intense interest in identifying interventions that will allow durable control of HIV in the absence of therapy. The dataset and approach outlined here clearly points to very distinct groups of “aviremic” individuals, with at least two distinct groups of immunologic non-responders and two distinct groups of controllers. It is hoped that further characterization of the heterogeneity within these groups will lead to novel approaches to improve health among those on therapy and perhaps to accelerate discovery of potentially curative interventions.
Supplementary Material
Acknowledgments
Funding: This work was supported by amfAR, the Delaney AIDS Research Enterprise (DARE; U19 AI0961090), NIAID (K24 AI069994), the UCSF/Gladstone Institute of Virology & Immunology CFAR (P30 AI027763), the UCSF Clinical and Translational Research Institute Clinical Research Center (UL1 RR024131), the Center for AIDS Prevention Studies (P30 MH62246), NCATS (UCSF-CTSI KL2TR000143), the UCLA Center for AIDS Research (CFAR) grant 5P30AI028697, Core H, NSF IIS 1251151 and the CFAR Network of Integrated Systems (R24 AI067039).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Authorship contribution: CMR, PWH and SGD served as the chief investigators, designed the study and developed the protocol. CMR is responsible for the statistical analysis plan, algorithm development and conception and data analysis. DC assisted with the statistical programming of the algorithms and performed some of the statistical analysis. ES and LE performed the flow cytometry analysis. SL, VJ, PYH, HH, FMH, SGD, JNM, and PWH contributed to the recruitment and selection of patients and obtaining funding for the primary immunologic measurements. CMR, PWH, SGD, HH, and JMM contributed to the primary interpretation of the analyses. CMR wrote the initial draft of the manuscript and all authors contributed to the editing of the manuscript.
References
- 1.Kelley CF, Kitchen CM, Hunt PW, Rodriguez B, Hecht FM, Kitahata M, et al. Incomplete Peripheral CD4(+) Cell Count Restoration in HIV-Infected Patients Receiving Long-Term Antiretroviral Treatment. Clin Infect Dis. 2009;48:787–794. doi: 10.1086/597093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore RD, Keruly JC. CD4+ cell count 6 years after commencement of highly active antiretroviral therapy in persons with sustained virologic suppression. Clin Infect Dis. 2007;44:441–446. doi: 10.1086/510746. [DOI] [PubMed] [Google Scholar]
- 3.Baker JV, Peng G, Rapkin J, Abrams DI, Silverberg MJ, MacArthur RD, et al. CD4+ count and risk of non-AIDS diseases following initial treatment for HIV infection. AIDS. 2008;22:841–848. doi: 10.1097/QAD.0b013e3282f7cb76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freiberg MS, Chang CC, Kuller LH, Skanderson M, Lowy E, Kraemer KL, et al. HIV Infection and the Risk of Acute Myocardial Infarction. JAMA internal medicine. 2013;173:614–622. doi: 10.1001/jamainternmed.2013.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Lelyveld SF, Gras L, Kesselring A, Zhang S, De Wolf F, Wensing AM, et al. Long-term complications in patients with poor immunological recovery despite virological successful HAART in Dutch ATHENA cohort. AIDS. 2012;26:465–474. doi: 10.1097/QAD.0b013e32834f32f8. [DOI] [PubMed] [Google Scholar]
- 6.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323:1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 7.Deeks SG, Autran B, Berkhout B, Benkirane M, Cairns S, Chomont N, et al. Towards an HIV cure: a global scientific strategy. Nature reviews. Immunology. 2012 doi: 10.1038/nri3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramirez Kitchen CM. Nonparametric variable selection using machine learning algorithms in high dimensional (largep, smalln) biomedical applications. In: Laskovski A, editor. Biomedical Engineering: Trends in electronics, communication and software. InTech; 2011. [Google Scholar]
- 9.Diaz-Uriarte R, Alvarez de Andes S. Gene selection and classification of microarray data using random forests. BMC Bioinformatics. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strobl C, Boulesteix A, Kneib T, Augustin T, Zeileis A. Conditional variable importance for random forests. BMC Bioinformatics. 2008;9:307. doi: 10.1186/1471-2105-9-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strobl C, Boulesteix A, Zeileis A, Hothorn T. Bias in random forest variable importance measures: illustrations, sources and a solution. BMC Bioinformatics. 2007;8:25. doi: 10.1186/1471-2105-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S, Sinclair E, Jain V, Huang Y, Epling L, Van Natta M, et al. Low proportions of CD28− CD8+ T cells expressing CD57 can be reversed by early ART initiation and predict mortality in treated HIV infection. J Infect Dis. 2014;210:374–382. doi: 10.1093/infdis/jiu109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatano H, Jain V, Hunt PW, Lee T, Sinclair E, Do T, et al. Cell-based measures of viral persistence are associated with immune activation and programmed cell death protein 1 (PD-1)-expressing CD4+ T cells. J Infect Dis. 2013;208:50–56. doi: 10.1093/infdis/jis630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conn D, Ramirez C. Random and Fuzzy Forests applied to feature selection in biomedical research. In: Alvarez R, editor. Computational Social Science: Discovery and Prediction. Oxford, UK: Cambridge University Press; 2015. In Press. [Google Scholar]
- 15.Conn D, Ngun T, Gang L, Ramirez C. Fuzzy Forests: Extending Random Forests for Correlated, High-Dimensional, Data. UCLA Biostatistics Working Paper Series. 2015 http://www.escholarship.org/uc/item/55h4h0w7.
- 16.Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 17.SR M, Chen C, Shit T, Horvath S, SF N, Reichardt J, Sawyers CL. IGFBP2 is a biomarker for PTEN status and PI3K/Akt pathway activation in glioblastoma and prostate cancer. Proc Natl Acad Sci USA. 2007;104:5563–5568. doi: 10.1073/pnas.0609139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng Y, Yang Q, Cuenco K, Cupples L, DeStefano A, Lunetta KL. Two-stage approach for identifying single-nucleotide polymorphisms associated with rheumatoid arthritis using random forests and Bayesian networks. BMC Proceedings. 2007;1:S56. doi: 10.1186/1753-6561-1-s1-s56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng Y, Yu Y, Adrienne Cupples L, Farrer L, Lunetta K. Performance of random forest when SNPs are in linkage disequilibrium. BMC Bioinformatics. 2009;10:78. doi: 10.1186/1471-2105-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicodemus K, Wang W, Shugart Y. Stability of variable importance scores and rankings using statistical learning tools on single nucleotide polymorphisms (SNPs) and risk factors involved in gene-gene and gene-environment interaction. BMC Proceedings. 2007;1:S58. doi: 10.1186/1753-6561-1-s1-s58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B, Horvath S. A general framework for Weighted Gene Co-expression Network Analysis. Statistical Applications in Genetics and Molecular Biology. 2005;4 doi: 10.2202/1544-6115.1128. Article 17. Epub 2005 Aug 2012. [DOI] [PubMed] [Google Scholar]
- 22.Deeks S, Kitchen C, Liu L, Guo H, Gasson R, Narvaez A, et al. Immune activation set point during HIV infection predicts subsequent T-cell changes independent of viral load. Blood. 2004;104:942–947. doi: 10.1182/blood-2003-09-3333. [DOI] [PubMed] [Google Scholar]
- 23.Deeks S, Martin JN, Sinclair E, Harris J, Neilands TB, Maecker HT, et al. Strong cell-mediated immune responses are associated with the maintenance of low-level viremia in antiretroviral-treated individuals with drug resistant human immunodeficiency virus type 1. J Infect Dis. 2004;189:312–321. doi: 10.1086/380098. [DOI] [PubMed] [Google Scholar]
- 24.Deeks SG. Virologic outcomes with protease inhibitor therapy in an urban AIDS clinic: relationship between baseline characteristics and response to both initial and salvage therapy. AIDS. 1999;13:F34–F44. doi: 10.1097/00002030-199904160-00001. [DOI] [PubMed] [Google Scholar]
- 25.Deeks SG, Barbour JD, Grant RM, Martin JN. Duration and predictors of CD4 T-cell gains in patients who continue combination therapy despite detectable plasma viremia. AIDS. 2002;16:201–207. doi: 10.1097/00002030-200201250-00009. [DOI] [PubMed] [Google Scholar]
- 26.Deeks SG, Barbour JD, Martin JN, Swanson MS, Grant R. Sustained CD4+ T-cell response after virologic failure of protease-based regimens in patients with HIV infection. J Infect Dis. 2000;181:946–953. doi: 10.1086/315334. [DOI] [PubMed] [Google Scholar]
- 27.Hatano H, Delwart E, Norris PJ, Tzong-Hae L, Dunn-Williams J, Hunt P, et al. Evidence for persistent low-level viremia in individuals who control human immunodeficiency virus in the absence of antiretroviral therapy. J Virol. 2009;83:329–335. doi: 10.1128/JVI.01763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunt P, Brenchley J, Sinclair E, McCune JM, Roland M, Page-Shafer K, et al. Relationship between T cell activation and CD4+ T cell count in HIV seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J Infect Dis. 2008;197:126–133. doi: 10.1086/524143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicodemus K, Malley J, Strobl C, Ziegler A. The behaviour of random forest permutation-based variable importance measures under predictor correlation. BMC Bioinformatics. 2010;11:110. doi: 10.1186/1471-2105-11-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hothorn T, Hornik K, Zeileis A. Unbiased recursive partitioning: a conditional inference frameork. Journal of Computational and Graphical Statistics. 2006;15:651–674. [Google Scholar]
- 31.Ishwaran H. Variable importance in binary regression trees and forests. Electronic Journal of Statistics. 2007;1:519–537. [Google Scholar]
- 32.Hunt PW, Brenchley J, Sinclair E, McCune JM, Roland M, Page-Shafer K, et al. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J Infect Dis. 2008;197:126–133. doi: 10.1086/524143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hunt PW, Landay AL, Sinclair E, Martinson JA, Hatano H, Emu B, et al. A low T regulatory cell response may contribute to both viral control and generalized immune activation in HIV controllers. PLoS ONE. 2011;6:e15924. doi: 10.1371/journal.pone.0015924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereyra F, Lo J, Triant VA, Wei J, Buzon MJ, Fitch KV, et al. Increased coronary atherosclerosis and immune activation in HIV-1 elite controllers. Aids. 2012;26:2409–2412. doi: 10.1097/QAD.0b013e32835a9950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emu B, Sinclair E, Favre D, Moretto WJ, Hsue P, Hoh R, et al. Phenotypic, Functional, and Kinetic Parameters Associated with Apparent T-Cell Control of Human Immunodeficiency Virus Replication in Individuals with and without Antiretroviral Treatment. J Virol. 2005;79:14169–14178. doi: 10.1128/JVI.79.22.14169-14178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hunt PW, Brenchley J, Sinclair E, McCune JM, Roland M, Page-Shafer K, et al. Relationship between T Cell Activation and CD4(+) T Cell Count in HIV-Seropositive Individuals with Undetectable Plasma HIV RNA Levels in the Absence of Therapy. J Infect Dis. 2008;197:126–133. doi: 10.1086/524143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams LD, Bansal A, Sabbaj S, Heath SL, Song W, Tang J, et al. Interleukin-21-producing HIV-1-specific CD8 T cells are preferentially seen in elite controllers. Journal of virology. 2011;85:2316–2324. doi: 10.1128/JVI.01476-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saez-Cirion A, Lacabaratz C, Lambotte O, Versmisse P, Urrutia A, Boufassa F, et al. HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc Natl Acad Sci U S A. 2007;104:6776–6781. doi: 10.1073/pnas.0611244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vingert B, Benati D, Lambotte O, de Truchis P, Slama L, Jeannin P, et al. HIV controllers maintain a population of highly efficient Th1 effector cells in contrast to patients treated in the long term. Journal of virology. 2012;86:10661–10674. doi: 10.1128/JVI.00056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crowell T, Gebo K, Blankson J, Korthuis P, Yehia B, Rutstein R, et al. Elite controllers are hospitalized more often than persons with mediclaly controlled HIV. J Infect Dis. 2015;211:1692–1702. doi: 10.1093/infdis/jiu809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teixeira L, Valdez H, McCune JM, Koup R, Badley A, Hellerstein MK, et al. Poor CD4 T cell restoration after suppression of HIV-1 replication may reflect lower thymic function. AIDS. 2001;15:1749–1756. doi: 10.1097/00002030-200109280-00002. [DOI] [PubMed] [Google Scholar]
- 43.Hatano H, Jain V, Hunt PW, Lee TH, Sinclair E, Do TD, et al. Cell-Based Measures of Viral Persistence Are Associated With Immune Activation and Programmed Cell Death Protein 1 (PD-1)-Expressing CD4+ T cells. The Journal of infectious diseases. 2012 doi: 10.1093/infdis/jis630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hunt PW, Martin JN, Sinclair E, Bredt B, Hagos E, Lampiris H, et al. T Cell Activation Is Associated with Lower CD4+ T Cell Gains in Human Immunodeficiency Virus-Infected Patients with Sustained Viral Suppression during Antiretroviral Therapy. J Infect Dis. 2003;187:1534–1543. doi: 10.1086/374786. [DOI] [PubMed] [Google Scholar]
- 45.Catalfamo M, Di Mascio M, Hu Z, Srinivasula S, Thaker V, Adelsberger J, et al. HIV infection-associated immune activation occurs by two distinct pathways that differentially affect CD4 and CD8 T cells. Proc Natl Acad Sci U S A. 2008;105:19851–19856. doi: 10.1073/pnas.0810032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lederman MM, Calabrese L, Funderburg NT, Clagett B, Medvik K, Bonilla H, et al. Immunologic failure despite suppressive antiretroviral therapy is related to activation and turnover of memory CD4 cells. The Journal of Infectious Diseases. 2011;204:1217–1226. doi: 10.1093/infdis/jir507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.