Abstract
Nitric oxide (NO) is a signaling molecule with pleiotropic physiological roles in normal cells and pathophysiological roles in cancer. NO synthetase expression and NO synthesis are linked to altered metabolism, neoplasticity, invasiveness, chemoresistance, immune evasion, and ultimately to poor prognosis of cancer patients. Exogenous NO in the microenvironment facilitates paracrine signaling, mediates immune responses, and triggers angiogenesis. NO regulates posttranslational protein modifications, S-nitrosation, and genome-wide epigenetic modifications that can have both tumor-promoting and tumor-suppressing effects. We review mechanisms that link NO to cancer hallmarks, with a perspective of co-targeting NO metabolism with first-line therapies for improved outcome. We highlight the need for quantitative flux analysis to study NO in tumors.
Keywords: Nitric oxide, Cancer metabolism, Tumor microenvironment, Epigenetics, S-nitrosation, NO flux analysis
Dissecting the links between cancer hallmarks and nitric oxide
The uncontrolled proliferation of cancer cells requires a significant shift in metabolism. To support a higher growth rate, cancer cells redirect nutrients into anabolic pathways to maintain biomass production. However, to survive in a harsh tumor microenvironment (TME) (Glossary), tumors need to strike a balance between anabolic demands and catabolic energy production. Rewired energy metabolism is, therefore, a ubiquitous hallmark of cancer. Rapid formation of solid tumors is accompanied with poor vasculature leading to limited supply of nutrients and oxygen, which further contributes to the altered metabolism in tumors [1]. Therefore, there is a complex interplay between (i) cell-autonomous metabolic alterations, (ii) intercellular metabolic crosstalk, and (iii) extracellular stimuli. Nitric oxide (NO) is a metabolic product of the nitric oxide synthase (NOS) reaction that catalyzes the conversion of arginine into citrulline. NOS enzyme exists in three isoforms: neuronal NOS (nNOS or NOS1), inducible NOS (iNOS or NOS2), and endothelial NOS (eNOS or NOS3) (Text Box 1). Historically, researchers have focused on the functions of NO as a signaling molecule, but have overlooked the ubiquitous interplay between NO synthesis and tumor metabolism, and the role NO plays in the TME.
Box 1. Biochemistry of Nitric Oxide and Nitric Oxide Synthetase.
Nitric oxide (NO) is a lipophilic, highly diffusible, and short-lived molecule. These characteristics make it an ideal physiological messenger capable of regulating intercellular and extracellular signaling pathways. NO can be endogenously synthesized by the enzyme nitric oxide synthase (NOS) from the guanido nitrogen of L-arginine. It is known to regulate a variety of important cellular functions such as vasodilation, respiration, neurotransmission, cell migration, immune response, apoptosis, and metabolism [77]. The wide range of biological actions has implicated its role in pathophysiological actions, especially in cancer.
The enzyme responsible for its synthesis exists in three isoforms enzymes that are different in structure and function: neuronal NOS (nNOS or NOS1), inducible NOS (iNOS or NOS2), and endothelial NOS (eNOS or NOS3) [78]. NOS1 and NOS3 are constitutive isoforms that are modulated by calcium-calmodium concentrations. These isoforms have a lower capacity of producing NO than the inducible isoform, NOS2 [79]. NOS2 activity is independent of calcium concentrations but can be stimulated by cytokines in all cell types (Fig. 1, Key Figure). NOS2 induction requires two signals, one from interferon gamma (IFNγ) and another trigger such as the endotoxin, tumor necrosis factor alpha (TNFα). The activation of NOS2 by TNFα occurs via stimulation of the transcription factor NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) which bonds to a κB element in NOS promoter [80]. In tumors with chronic hypoxia due to the lack of proper vasculature, hypoxia inducible factor-1α (HIF-1α) interacts with IFNγ and induces NOS2 expression. All isoforms convert arginine to citrulline with the release of NO. This two-step reaction catalyzed by NOS is dependent on oxygen and nicotinamide-adenine-dinucleotide phosphate (NADPH). Therefore, this reaction directly affects arginine utilization and redox homeostasis in cells. In the context of tumor formation, major advances in the investigation of NO biology have been witnessed. NO acts like a double-edged sword, where its level of expression and duration of NO exposure determine the often-contradictory cellular outcomes. Higher concentrations (more than 200nM) induce apoptosis while low levels (less than 200nM), have been associated with tumor progression (Fig. 1, Key Figure).
Metabolic traits that make cancer cells distinct from healthy tissues present opportunities for therapy [2]. However, cancer cells share many metabolic features with healthy proliferating cells, making the search for metabolic targets that selectively attack cancer cells challenging. Moreover, due to redundancies in metabolic pathways, cancer cells can activate compensatory pathways that perform similar functions as the metabolic pathways being targeted by drugs. This enables cancer cells to acquire resistance to metabolic drugs. Despite advancements in developing small-molecule metabolic drugs, their efficacy as anticancer drugs has been underwhelming in the clinic. NO plays strategic roles in signaling and metabolic pathways, making NO metabolism a hub that control pathways responsible for supporting tumorigenesis or suppress tumor growth altogether (Fig. 1, Key Figure). NO metabolism, thus, presents a viable therapeutic target as it has a wide range of control over tumorigenic functions. Treatments targeting key regulators, such as NO, can be more effective when combined with conventional therapeutic strategies to achieve synergistic effects. Combination therapies target multiple pathways that contribute to complementary aspects of tumor pathology. Such an approach reduces the probability of tumors acquiring drug resistance by activating compensatory pathways [3]. This review enumerates the mechanisms of tumor progression that are initiated and supported by NO metabolism, which may be targetable using small-molecule drugs. More importantly, we present a novel perspective of co-targeting NO metabolism with conventional targets to achieve a synthetic lethal effect. Additionally, we highlight the importance of metabolic flux analysis (Text Box 2) of endogenous NO synthesis in providing a quantitative approach to help design therapies that effectively target NO metabolism and maximize the therapeutic window.
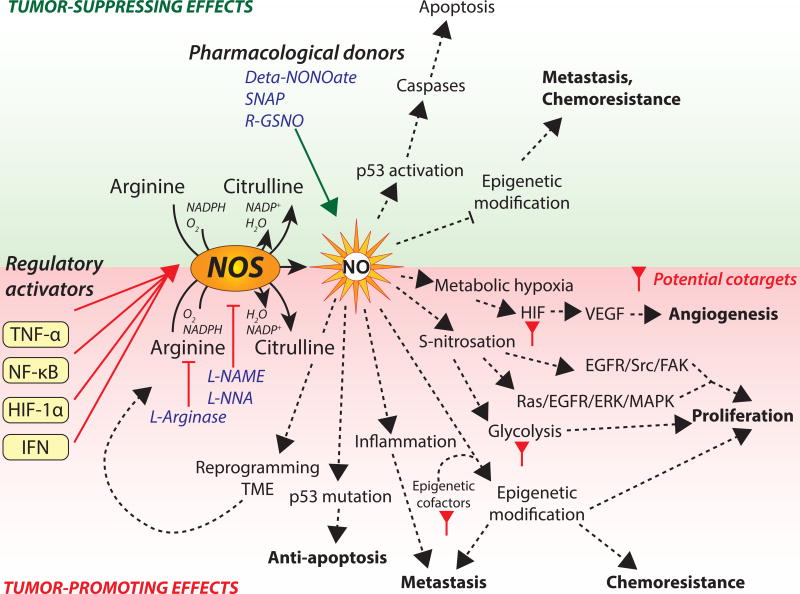
Figure 1, Key Figure. Pleotropic effects of nitric oxide in cancer.
NO and NOS activate anti-oncogenic pathways that can suppress tumor progression in certain types of cancers by activating p53 or suppressing epigenetic modifications (green panel). NO donors diethylenetriamine NONOate (deta-NONOate), S-Nitroso-N-Acetyl-D,L-Penicillamine (SNAP), and S-Nitrosoglutathiones (R-GSNO) provide exogenous NO to activate or amplify tumor-suppressing effects.
NO synthesis and NOS expression promote tumorigenic properties via induction of metabolic hypoxia, aberrant S-nitrosation, irregular epigenetic modifications, increased inflammation, exploiting p53 mutations and reprogramming TME metabolism (red panel). NO synthesis or NOS is targeted for anticancer treatment using NOS inhibitors L-NG-Nitroarginine methyl ester (L-NAME), L-NG-Nitroarginine (L-NNA), or arginine depletion drug, L-asparaginase. Key intermediate pathways induced by NO or NOS are proposed as co-targets for combination therapy to improve therapeutic efficacy.
Box 2. Metabolic Flux Analysis: Measuring Endogenous NO Synthesis.
NO production in cells has a wide spectrum of effects, depending on local NO concentration and duration of exposure. Quantification of cellular NO production is important, especially when NO metabolism is being targeted for therapeutic purposes. Direct methods are techniques that rely on physical sensors to measure NO secreted by cells, as opposed to indirect methods that measure NO-derived molecules such as nitrates or nitrites.
The most recent platform was designed based on a tunable diode laser absorption spectroscopy (TDLAS) sensor that used a continuous wave quantum cascade laser. The setup was used to measure NO emissions from ovarian cancer cells with detection limits as low as 124 ppt [81]. A previous technique relied on reconversion of nitrate enzymatically into nitrite, and nitrite into NO in an acidic iodide solution. The NO is released stoichiometrically, which is detected using an amperometric electrode. However, this setup does not allow recording of real-time measurements [82]. Another method based on electrochemical principles using a microcoaxial electrode can, however, measure local NO concentrations with high spatial and temporal resolutions allowing real-time measurements in vitro [83]. Indirect approaches to quantify NO produced via the NOS pathway in vivo measure nitrate and nitrite species formed by the unstable NO. The most widely used spectrophotometric technique measures nitrite produced from diazotization of NO using Griess reagents [84, 85]. This assay has been applied to a variety of biological samples in liquid matrices and can detect nitrite down to 2.5 µM. Like direct NO detection techniques, this method cannot resolve between endogenously produced and exogenously acquired NO.
To address this drawback, stable-isotope tracer-based techniques employed for quantifying carbon fluxes in cellular metabolism have been adapted to determine de novo NO synthesis. NOS-derived NO requires nitrogen from arginine, therefore, naturally occurring arginine is replaced with isotope-labeled L-[guanidino-15N2] arginine. Upon oxidation by NOS, 15NO is produced and is subsequently converted into [15N] nitrite and [15N] nitrate species that are detectable using a gas-chromatograph mass spectrometer. This method has been employed both, in vitro and in vivo, since it relies on measuring stable species. To measure NO synthesis in patients, stable-isotope tracers are infused intravenously. Blood and plasma samples are obtained from tracer-infused patients to analyze for enrichment of 15N in nitrate and nitrite pools. The rate of tracer infusion and enrichment of 15N within blood and plasma can help quantify the rate of NO synthesis [86–88]. A study on NO-mediated hypertension in pregnant women designed a tracer experiment to resolve NO production by NOS enzyme and production of its precursor arginine. They infused patients with a combination of [guanidino-15N2] arginine and [5,5-2H2] citrulline or [15N] citrulline alone. This combination of labeling was used to estimate NO synthesis from arginine, arginine production from citrulline in the urea cycle, and arginine from dietary sources [89]. These techniques provide researchers with a powerful arsenal to understand NO metabolism quantitatively and performing dose-response analysis of NOS inhibitors or NO-donors to design effective therapeutic treatments.
NO: modulator of the TME
Cancers develop within a complex TME that provides support for sustained growth, invasion, and metastasis. Non-malignant cells in TME often have tumor-promoting functions. NO secreted by cancer cells (Fig. 2) reprograms stromal cells to support tumor progression. For example, cancer cell-derived NO induces chronic inflammation in the TME of melanoma to promote drug resistance [4]; and elevated levels of NO in the microenvironment contribute to increased migration of breast cancer via upregulation of caveolin-1 (Cav-1) expression [5]. Similarly, cell-autonomous induction of NOS in stromal cells also contributes to tumor progression. For example, in cancer associated fibroblasts (CAFs) expressing chemokine ligand (CXCL14), NOS1 expression is essential for CAF-supported growth of breast and prostate cancer cells. Furthermore, suppressing NOS1 expression disrupted pro-tumorigenic functions of CXCL14-expressing CAFs and reduced tumor growth in mice. Since NOS1 expression did not increase extracellular NO, NOS1-derived NO supported CXCL14 activity within CAFs [6] (Fig. 2). Increase in exogenous NO in the TME is also associated with tumorigenic functions in colon cancer patients. Colon cancer patients with high NOS2 expression have increased incidences of lymph node metastasis; and elevated NOS2 expression observed in the upper colon in colitis patients indicates higher risk of developing colon cancer [7, 8]. In addition to regulating metastasis and tumor initiation, NO is also a key regulator of angiogenesis. Increased NO in the TME has been observed to upregulate vascular endothelial growth factor (VEGF) in glioblastoma and hepatoma cells [9]. Furthermore, influences of NO on angiogenesis has been exploited to sensitize glioma tumors in mice to radiotherapy by inhibiting NOS1 expression. Suppressing NO production in glioma tumors leads to normalization of tumor vasculature, leading to oxygenation of tumors that supports radiation treatment [10].
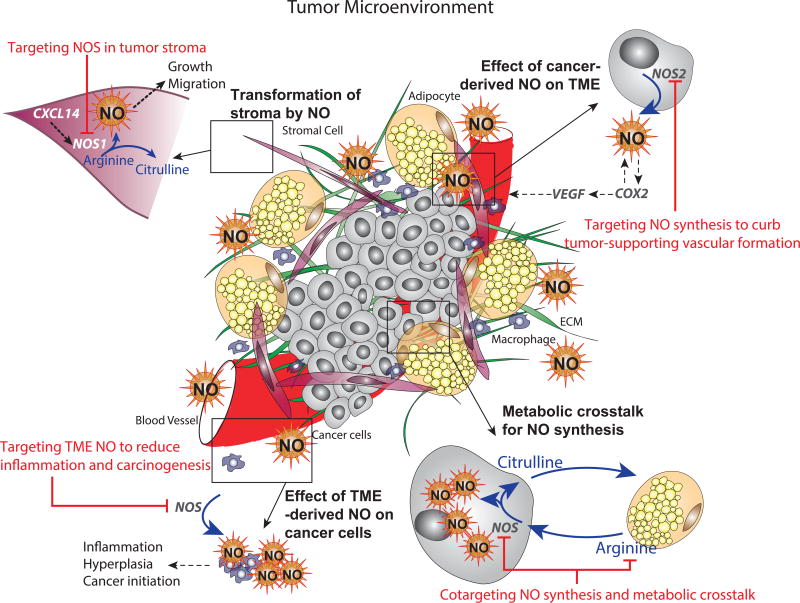
Figure 2. Targeting NOS and NO-mediated pathways in the TME.
Increased NOS expression or NO production in non-malignant stromal compartments are induced by upregulation of chemokine ligands. NO produced by stromal cells contributes to tumor growth and increases migration potential of tumors. Targeting stromal NOS diminishes tumorigenic properties (top left panel).
Cancer cells expressing NOS2 produce NO to induce angiogenesis in tumors by upregulating VEGF. Targeting NOS in cancer cells prevents formation of tumor-supporting blood vessels (top right panel).
O-ASCs in the TME secrete arginine that is utilized by ovarian cancer cells to produce NO via NOS. Citrulline regenerated by cancer cells is utilized by O-ASCs to supplement arginine production. Cotargeting NOS in cancer cells and depleting arginine effectively eliminates metabolic crosstalk in the TME and reduce cancer cell viability (bottom right panel).
Exogenous NO in the TME produced by stromal cells induces inflammation, hyperplasia and initiates tumorigenesis in colorectal cancers. Inhibiting stromal NOS in early stage patients can curb tumor progression (bottom left panel).
NOS, nitric oxide synthase; VEGF, Vascular endothelial growth factor; COX2, Cytochrome oxidase-2; TME, tumor microenvironment; O-ASCs, Omental adipose stellate cells
The dependence of several tumors on NO and successful use of NOS inhibitors as described in these studies, presents a novel therapeutic strategy. NO synthesis can be targeted in combination with first line therapies that target cancer cells to improve the therapeutic outcome (Fig. 2). The synergistic effect of NOS inhibitor, L-NG-Nitroarginine methyl ester (L-NAME), and carboplatin has been demonstrated in preclinical studies [11]. Combination therapy improved the survival of mice with mutant p53 and KRAS non-small cell lung cancers, as compared to the mono carboplatin arm. Incidentally, the anti-tumor effect of L-NAME was due to inhibition of NOS in the stromal cells, since cancer cells did not express NOS [11]. In concordance with these results, our lab has observed that combining L-NAME with L-asparaginase (which depletes extracellular arginine) significantly reduces cell viability of ovarian cancer cells, as compared to individual treatments (Fig. 2) [12]. Next generation drugs that deplete arginine have shown significant antitumor activity in melanoma and hepatocellular carcinoma during Phase I and II clinical trials [13]. These studies suggest that both NOS inhibition and arginine depletion are viable strategies to eliminate tumor-promoting NO in patients.
In contrast to supporting tumorigenic functions in cancer cells, stroma-derived NO has also been observed to have tumor suppressive effects. This has been demonstrated in NOS2-deficient mouse embryonic fibroblasts (MEFs). Loss of NOS2 leads to higher COX2 expression in these MEFs, as compared to wild-type MEFs. Treating these cells with the NO-donor, S-nitrosoglutathione, reverses pro-tumorigenic effects of COX2 and causes growth arrest in tumors (Fig. 2) [14]. These studies are preamble to the divergent properties of NO in the TME on tumors. The context-dependent role of NO has been demonstrated in the immune response, disease progression and metabolic interactions in tumors.
NO and immune response
The effects of NO in cancer gained attention after the observation of activated macrophages metabolizing arginine to generate NO, an effector molecule that induced cytotoxicity in hepatoma cells [15]. NO has since been observed to have a diverse and multifaceted role in antitumor immune response. For instance, an anticancer drug, OM-174 a Toll-like receptor (TLR) 4 agonist, has been found to induce NOS2 expression in mouse breast cancer models. Inhibiting NOS2 expression diminishes the antitumor effect of OM-174 indicating that NO synthesis is essential to the tumor-suppressive properties of the TLR agonist [16]. Similarly, a study on T-cell immunotherapy in lymphoma tumor-bearing mice also discovered that NO production in the TME was essential for anti-tumor activity of CD8+ T-cells [17]. The NO required to activate CD8+ T-cells is synthesized by NOS2-positive tumor infiltrating myeloid cells.
However, recent studies show that the function of NO is not limited to activating innate and adaptive responses of the immune system against tumors; it can also facilitate pathways that aid cancer cells in evading antitumor immune responses [18]. Several studies have demonstrated that suppressing NO production in tumors using NOS inhibitors can enhance antitumor immune response in animal models [19–21]. For example, a TLR7 agonist, Imiquimod, has been shown to be more effective in reducing tumor growth in lymphoma-bearing mice that do not express NOS2, as compared to wild-type mice [20, 22]. This suggests that NOS2 expression is crucial to tumor growth and resistance to Imiquimod. The significant difference in these two studies was the localization of NO production. NO generated by NOS2-expressing tumor cells impairs the anti-tumor immunity induced by TLR agonists; whereas, NO produced by NOS2-expressing myeloid cells acts in conjunction with CD8+ T-cells to eradicate tumor cells. Therefore, identifying the duality of NO’s role in tumor immune response is essential for developing immunotherapies that cotarget NO metabolism.
NO in cancer cells
The contextual role of NO in the tumor stroma extends to cancer cells, where it can have pro- and anti-tumorigenic effects. The effect of NO has been found to be linked to disease progression and hypoxic status in tumors. The influence of disease stage on the action of NO is exemplified by the high NOS2 expression observed in colonic mucosa of patients with inflammatory bowel disease [23]. NOS2 expression in these patients greatly increases their risk of developing colon cancer [24]. Additionally, colorectal cancer stem cells are dependent on NO synthesis and NOS expression for tumor initiation [25]. However, early stage colon cancer patients have been shown to have low NOS2 in their colonic epithelium. This suggests that NO is essential for tumor initiation but not essential during the early stages of tumor progression. Furthermore, hypoxic status in solid tumors also plays a role in determining the effect of NO on apoptosis in cancer cells. The accumulation of NO caused by anticancer NO-donor, Poly-SNO-HSA, induces cGMP-dependent apoptosis in colon cancer cells under oxygen-replete conditions. However, the same cells under hypoxia upregulate PDE5 to inhibit cyclic GMP and subsequently limit NO-induced apoptosis [26]. Combination therapy of Poly-SNO-HSA and PDE5 inhibitor proves to be an effective strategy to induce growth arrest in normoxic and hypoxic regions of tumors in colon cancer-bearing mice [26]. In contrast, increased NO production can also enable apoptotic evasion in colorectal carcinomas that harbor p53 mutations. NO induces apoptosis in healthy colonic epithelial cells by stimulating proapoptotic miRNA in a p53-dependent manner. However, cells with p53 mutations can evade this apoptotic mechanism. Therefore, elevated NO concentration in tumors selectively kills healthy cells and allows cancer cells to survive, thereby contributing to tumor progression [27]. A correlative study in oral squamous cell carcinoma patients made a similar observation, where tumors in advanced stages of the disease showed higher NOS expression and lower p53 expression [28].
NO and metabolism
Metabolic crosstalk between cancer cells and stromal cells within the TME is well-documented and considered an indispensable interaction [12, 29–31]. NO is a byproduct of cellular metabolism and plays a vital role in this metabolic interaction. NO inhibits enzyme activity by reacting with the metal centers of these proteins to form a complex. For example, NO inhibits prolyl hydroxylase (PHD), which is responsible for degradation of hypoxia-inducible factor 1 (HIF-1α) to actively elicit a hypoxic response, even in the presence of oxygen [32]. NO also reacts with Complex IV of the electron transport chain to compete with oxygen for electrons, thereby inhibiting oxidative phosphorylation and mitochondrial respiration [33]. This results in reduced consumption of cellular oxygen leading to a condition known as metabolic hypoxia. Conversely, the activation of nuclear factor, NF-κB and hypoxia-inducible factor, HIF-1α leads to the induction of NOS2 in several cell types establishing a positive feedback mechanism between NO synthesis and HIF-1α stabilization [34]. Furthermore, NO-mediated induction of hypoxia contributes to several tumor-promoting functions such as the Warburg effect. Cellular hypoxic response typically involves upregulation of glycolytic enzymes and inhibition of mitochondrial function [33, 35]. Caneba et al. have shown that NO generation increases with disease progression and invasive capacity of ovarian tumors [36]. Caneba et al. have observed that NO positively regulates glycolysis in highly invasive ovarian cancers. Furthermore, NO also inhibits mitochondrial respiration, increases glutamine consumption in the TCA cycle and ultimately increases tumor growth and confers chemoresistance. However, this link between NO and the Warburg effect is not observed in less invasive ovarian cancers, indicating that the effect of NO varies as the disease progresses [36]. In highly invasive ovarian cancers, the tumor-promoting effects of NO can be reversed and mitochondrial oxidation can be restored by inhibiting NO synthesis using NOS inhibitors or depleting arginine. In addition to the cell-autonomous regulation of metabolism by NO, it also facilitates metabolic interactions between ovarian cancers and stromal cells. Salimian et al. have revealed a bidirectional metabolic link between omental adipose stromal cells (O-ASCs) and ovarian or endometrial cancer cells [12]. They have shown that O-ASC-secreted arginine when uptaken and metabolized by cancer cells generates NO, which subsequently modulates their metabolism and proliferation [12]. Furthermore, the citrulline secreted by cancer cells during NO synthesis is utilized by O-ASCs to complete the cross-talk between cancer cells and O-ASCs (Fig. 2, bottom right panel). Moreover, via controlled differentiation of O-ASCs it has been shown that L-citrulline significantly increases the lipid storage. This indicates the presence of another metabolic interaction, wherein fat deposits released by stromal cells are utilized by tumor cells for energy production [37, 38].
As such, there is sufficient evidence to suggest that considerable advantage could be gained by targeting the enhanced NOS expression in cancer cells. It could help distinguish between healthy and malignant tissues. Importantly, the mechanisms of action that lead to the pro-neoplastic activity of NO could provide unexplored avenues for developing novel cancer therapies [39]. The study by Salimian et al. established a cotargeting strategy, where NOS in the cancer cells and arginine-producing enzymes ASS or ASL in the stromal compartments can be inhibited. Such novel strategies that cotarget cancer and stromal components have shown promise in disrupting glutamine-mediated crosstalk in orthotopic ovarian cancer mouse models [31].
S-nitrosation – an unexplored Achilles heel of cancer metabolism
S-nitrosation has emerged as a ubiquitous mechanism of posttranslational protein modification mediated by NO. Proteins are S-nitrosated when the thiol moiety of cysteine residues on peptides or proteins reversibly bind with NO to generate an S-nitrosothiols. This form of post-translational modification (PTM) is highly conserved and occurs in proteins in all biological systems [40]. S-nitrosation can control a diverse set of biological functions by regulating protein activity, altering protein localization, and mediating protein-protein interactions [41–43]. Particularly, S-nitrosation is involved in critical biological processes such as immune response, transcriptional regulation, DNA repair, and apoptosis. Not surprisingly, several pathophysiological traits in cancers have been associated with deregulated S-nitrosation [44]. Accumulating evidence suggests that deregulated S-nitrosation is a key event in tumor initiation that may considerably increase cancer risk [45, 46]. In normal cells, S-nitrosation predominantly occurs at sites where NOS isoforms are expressed. However, NOS isoforms are ubiquitously expressed in several types of cancers and macrophages, predisposing them to effects of S-nitrosation [47, 48]. Surprisingly, S-nitrosation in some cancers has been observed to be independent of NOS expression. For example, cellular nitrite reserves can supply NO to induce S-nitrosation. This is observed in the case of S-nitrosation of caspase-3 in endothelial cells under hypoxia [49]. Further, hypoxic conditions in the TME of solid tumors can induce interleukin-6 (IL-6), interferon gamma (IFNγ), and prostaglandin E2 (PGE2) to promote NOS2 expression [50–52]. Incidentally, Sonveaux et al. observed in erythrocytes, exposure to NO can cause S-nitrosation of oxygenated haemoglobin to form S-nitrosohemoglobin. S-nitrosohemoglobin was found to mediate reoxygenation of hypoxic tumor tissues via controlled release of NO and oxygen in low oxygen regions. This property of S-nitrosohemoglobin can be exploited to improve tumor blood flow and drug delivery [53]. This suggests that S-nitrosation has opposing effects on redox status of tumors that depends on which cells within the TME are affected by NO.
Typically, higher NOS expression and NO production increase the likelihood of S-nitrosation of cellular proteins that can promote tumorigenesis. NO-induced S-nitrosation activates oncogenic signaling cascades such as the EGFR-Src-c-Myc/Akt and Ras-EGFR-ERK1/2-MAP kinases in breast cancers [54–56]. Further, S-nitrosation can modulate cancer metabolism via these signaling pathways or directly alter enzyme activity (Fig. 3). As such, NO-mediated S-nitrosation can regulate cellular bioenergetics [57–60]. The strongest evidence of systematic control of energy metabolism has been provided by a novel mass spectrometry-based proteomics approach. A study revealed that S-nitrosation can alter activity of enzymes involved in glycolysis, gluconeogenesis, tricarboxylic acid cycle (TCA cycle), and oxidative phosphorylation in distinct types of tissue in mice (Fig. 3) [57]. Thus, elevated levels of NO found in the TME of many cancers can significantly alter tumor metabolism. For example, S-nitrosation of the rate limiting lipid synthesis enzyme fatty acid synthase (FASN), triggers differentiation of adipocyte stem cells into adipocytes (Fig. 3) [58]. This process is known as adipogenesis and occurs when adipocytes are required to store lipids and maintain energy homeostasis in tissues. Aberrant S-nitrosation due to high NO levels in tumors can enhance adipogenesis, leading to increase in adipocytes that support tumor growth by providing stored lipids [37, 58].
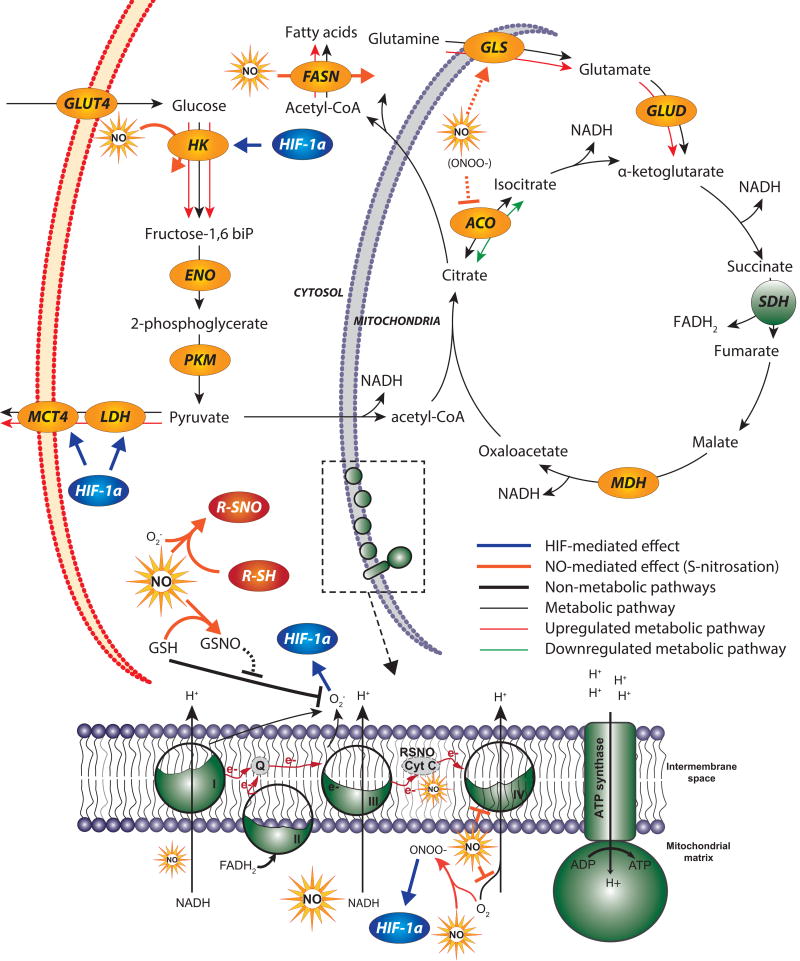
Figure 3. NO-mediated effects on cellular metabolism via S-nitrosation and signaling pathways.
NO, in the presence of oxygen postranslationally modifies proteins via S-nitrosation. S-nitrosation can lead to either enhanced or repressed metabolic pathway activity. Excess NO inactivates antioxidant GSH, which leads to build up of ROS and subsequently induces HIF-1α. HIF-1α initiates a signaling cascade that regulates glycolytic enzymes such as HK, LDH and MCT. NO inhibits the electron transport chain activity and subsequently mitochondrial respiration by interacting with complex IV. NO enhances lipid metabolism by inhibiting ACO and upregulating FASN to redirect citrate into lipogenic acetyl-CoA. NO also affects TCA cycle fluxes by increasing GLS activity, which enhances glutamine utilization by the TCA cycle.
ROS, Reactive oxygen species; GSH, glutathione; GSSG, reduced glutathione; HIF, hypoxia inducible factor; GLUT4, Glucose transporter type 2; HK, Hexokinase; ENO, Enolase; PKM, pyruvate kinase; LDH, Lactate dehydrogenase; MCT, Monocarboxylate transporters; GLS, glutaminase; GLUD, glutamate dehydrogenase; ACO: aconitase; SDH, succinate dehydrogenase; MDH, malate dehydrogenase
In contrast to tumor-promoting metabolic adaptations induced by S-nitrosation, S-nitrosation can have tumor-suppressing properties in highly glycolytic or hypoxic cancer cells. Dysfunctional mitochondrial oxidation is common in hypoxic and highly glycolytic cells, which leads to production of reactive oxygen species (ROS). Elevation in ROS level affects the redox homeostasis and can initiate apoptosis. Under these conditions, the cells divert glucose to the pentose phosphate pathway (PPP) to produce NADPH required for the regeneration of the antioxidant, glutathione (GSH) (Fig. 3) [59]. GSH is the key antioxidant that prevents ROS accumulation. However, GSH can be S-nitrosated to a biologically inactive complex, S-nitrosoglutathione. This leads to diminished GSH levels that can hinder the ability of cancer cells to prevent oxidative stress caused by ROS accumulation, ultimately leading to apoptosis [60].
Clearly, S-nitrosation is one of the most important functions of NO, as it can comprehensively affect metabolism of healthy and cancer cells alike. Therefore, a focused approach is required to understand which S-nitrosation events in tumors lead to pro- or antitumorigenic traits. Further, novel proteomic techniques [57, 61] and NO flux analysis (Text Box 2) are essential to understand how NO controls S-nitrosation events.
The role of NO in epigenetics
NO has been long known to be a driver of epigenetic changes and is responsible for genome-wide epigenetic regulation via several mechanisms, including: (i) interaction with heme proteins to alter their catalytic activity, (ii) generation of higher oxides of nitrogen under sufficient oxygen availability that form protein adducts (see S-nitrosation) to post translationally modify proteins, and (iii) inducing histone posttranslational modifications. Targeting NO metabolism to reverse these epigenetic changes presents a viable therapeutic approach. However, the strategy of targeting NO metabolism is highly dependent on the epigenetic mechanism responsible for tumor progression. Much like the double-edged effects of NO on tumor progression, its epigenetic effects are contradictory and context dependent.
One of the earliest studies investigating the carcinogenic properties of NO done using yeast p53 functional assays showed that NO treatment could preferentially enhance C:G→T:A transversions leading to higher mutation rates in the p53 genes. Although this is not a canonical epigenetic modification, it is initiated by enhanced methylation of cytosine that leads to its deamination into thymine [62] (Fig. 4). Since then, several studies have described NO-driven epigenetic modifications in a wide range of organisms, which control normal biological development and mediate tumorigenesis [63]. In oral squamous cell carcinoma patients, it is common to find histone hyperacetylation that promotes tumor progression. A study found that NO-mediated overexpression of NPMI1 and GAPDH was responsible for acetylation of p300 and subsequently, histone hyperacetylation. p300 histone acetylase (HAT) activity was dependent on endogenously generated NO and inhibiting p300 HAT restricted tumor growth [64]. The dependence of p300 HAT activity on NOS2-derived NO suggests that a NOS inhibitor could prove equally effective in treating the tumors (Fig. 4). NO has also been found to be a mediator of gastric cancer initiation induced by H. pylori infection. Elevated NO production via NOS2 causes aberrant DNA methylation of the E-cad gene, leading to E-cad repression, an early event in gastric cancer development [65, 66]. Patients infected with H. pylori may be treated with NOS inhibitors to reduce risk of disease development [67]. A recent study showed that NO could, in fact, directly affect the histone posttranslational modifications (PTMs) in breast cancer cells. Treating breast cancer cells with NO-donors caused differential expression of over 6500 genes and the pattern of PTMs correlated with an oncogenic signature [68] (Fig. 4). In contradiction to these cases, NO has also been found to inhibit KDM3A, a histone demethylase, in a HIF1-α independent manner [69]. KDM3A is known to positively regulate cancer cell invasion, chemoresistance, and metastasis in breast and ovarian cancer cells [70, 71]. In such cases, increasing NO levels instead of inhibiting NO production, could potentially reverse tumorigenic properties that were induced by KDM3A expression (Fig. 4).
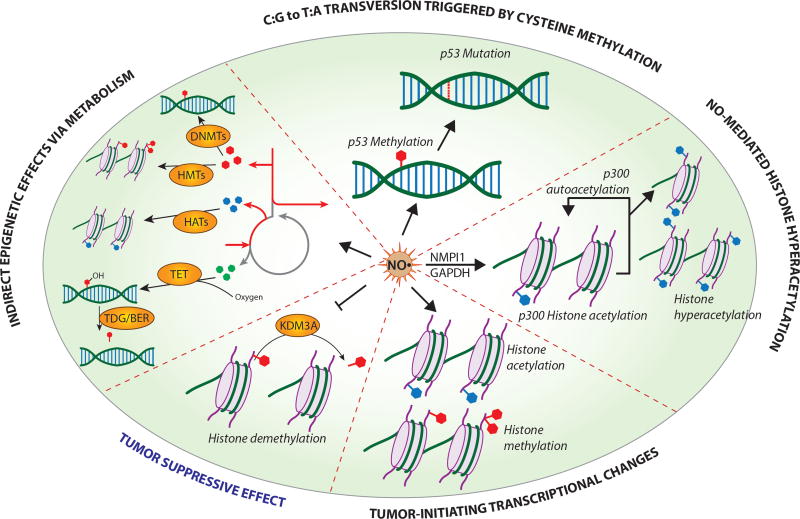
Figure 4. NO regulates epigenetic modifications in tumors.
NO regulates epigenetic modifications via various mechanisms to support tumor progression, or inhibits epigenetic changes that lead to tumorigenesis. NO induces p53 methylation that can lead up to tumor-supporting p53 mutations. NO upregulates NMP1 and GAPDH expression to promote p300 autoacetylation and histone hyperacetylation that is associated with tumor initiation. NO is responsible for genome-wide posttranslational modifications that can lead to oncogenic signatures in breast epithelial cells. NO is speculated to indirectly regulate epigenetics by affecting cellular metabolism that generates cofactors required for epigenetic modifications. Cotargeting NO synthesis and cofactor-producing metabolic reprogramming can provide novel therapeutic approaches. NO suppresses tumor progression by inhibiting KDM3A to reduce histone demethylation known to support tumor growth.
NMP1, Neucleophosmin; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; KDM3A, Lysine demethylase.
Since the tumor-promoting epigenetic effects of NO are localized within the tumor, inhibiting NOS or depleting arginine will have potentially fewer off-target effects and thus, a reduced risk of side-effects. Furthermore, the reversible nature of most epigenetic modifications means that targeting NO could systematically reverse tumor-supporting epigenetic modifications, thereby proving to be an effective therapeutic strategy. The mechanistic effects of NO on tumorigenesis are not restricted to direct modulation of epigenetic modifications, but may also be prompted indirectly via NO-induced metabolic alterations. Caneba et al. have shown that NO is responsible for inducing the Warburg effect, enhancing glutamine consumption and reductive carboxylation of glutamine, and regulating TCA metabolite levels [36]. These effects can have major influence on epigenetic modifications, because histone acetylation is dependent on acetyl-CoA availability, which can be regulated by reductive carboxylation of glutamine. Furthermore, histone deacetylation via sirtuin 1 (SIRT1) is dependent on NAD+ levels, which are controlled by TCA cycle activity. Even methylation of histones catalyzed by Jumonji C Domain-containing demethylases (JmjC) is dependent on glutamine-derived α-ketoglutarate [72] (Fig. 4). Methylation of DNA and histones requires the cofactor S-Adenosyl methionine (SAM) and acetylation requires acetyl-CoA. Both cofactors are inextricably linked to the central carbon metabolism of cells; therefore, any metabolic perturbations can affect their synthesis. SAM is produced in the coupled SAM and folate cycles, which are driven by the conversion of serine to glycine. Changes in glycolysis pathway can hinder the supply of serine thereby maintaining control over SAM-availability for epigenetic modifications [73]. On the other hand, acetyl-CoA is required for several functions and can be synthesized from glucose-derived citrate, glutamine-derived citrate, and fatty acid oxidation. All three pathways are known to be altered in many cancers and studies have linked aerobic glycolysis to support histone acetylation [74]. However, no studies have shown mechanistic link between metabolism-induced epigenetic changes and NO. Uncovering these links may present an opportunity of cotargeting NOS2 and complementary metabolic enzymes that control tumor-promoting epigenetic modifications.
Concluding remarks
It is generally observed that constitutive exposure of cancer cells to NO promotes tumor growth; however, NO is associated with both tumor-promoting and tumor-suppressive functions. Although, the role NO plays in tumors is difficult to predict, the evidence suggests that it depends on the organ of primary tumor [6, 14], stage of disease progression [7, 8] and the types of cancer-associated stromal cells within the TME [16, 20]. Sufficient interest has been generated in developing anticancer drugs that target NO metabolism, but progress towards clinical applications of NOS inhibitors or NO-donors has been limited. Several studies have exemplified the compatibility of NO inhibitors with drugs that target complementary pathways to achieve improved therapeutic outcomes [3, 11, 12]. However, the limitations of effective treatment are attributed to unresolved questions surrounding the effect of NOS inhibitors and NO-donors in vivo. In parallel to clinical trials for novel drugs that selectively inhibit NOS, researchers have looked to FDA-approved off-target drugs, such as L-asparaginase, which can reduce NOS activity by depleting arginine. However, there is a lack of evidence of their efficacy in selectively inhibiting NO synthesis using L-asparaginase (see Outstanding Questions). Similarly, the feasibility of using anticancer NO-donors has been demonstrated in scientific publications and patents, but none have progressed to clinical trials. It is also difficult to predict the pharmacological outcome of NO-donors due to the distinct biochemical properties of different NO-donors. For instance, NO release kinetics or TME conditions can greatly influence the concentration of NO in tumors induced by NO-donors [75]; and biological functions of NO can switch between tumor-promoting and tumor suppressing based on NO concentrations [76]. The systematic consequences of drug-induced off-target effects on NO-induced S-nitrosation, epigenetic modifications and metabolic alterations have also not been investigated (see Outstanding Questions). Therefore, characterization of therapeutic effects of modulating NO metabolism requires well-designed studies with quantifiable parameters. Metabolic flux analysis techniques designed specifically to quantify dynamic changes in NO synthesis in distinct conditions will provide further insight into the effects of NO on tumors (Text Box 2). A formalized approach that involves flux analysis techniques will help design a rubric for the use of NOS inhibitors, arginine depletion drugs, NO donors, or combination drugs to treat cancer of distinct types and at various stages of the disease.
Outstanding Questions.
Is arginine metabolism always perturbed in tumors with dysregulated NO metabolism?
In cases where NO acts as a tumor-suppressor, what are the prospects and potential off-target effects of inducing NO synthesis in a clinical setting?
Do NOS inhibitors have a systemic effect on S-nitrosation and epigenetic modifications?
Is metabolic alteration another mechanism through which NO regulates epigenetics?
Trends Box.
NO is a key messenger in the TME with pro- and anti-tumorigenic roles.
NO is emerging as a key regulator of cancer metabolism via S-nitrosation of enzymes.
NO has a wide range of control over gene expression in tumors via NO-mediated epigenetic modifications
Quantified approach to studying effects of NO synthesis in cancers should guide the design of therapies targeting NO – from in vitro experiments to clinical trials.
Acknowledgments
We apologize for the omission of any primary citations. This work was supported by NIH Research Project Grant (R01CA204969). We thank Luciana Pinheiro, Nithya Ramnath and Frank Weinberg for critical reading of this manuscript.
Glossary
- Tumor microenvironment
tumors are populations of cancer cells and non-neoplastic stroma cells including fibroblasts, vascular cells, immune cells, bone marrow-derived inflammatory cells, lymphocytes, and the extracellular matrix (ECM). The vicinity in which malignant cells thrive by dynamically interacting with the non-malignant components is known as the TME.
- Cancer-associated fibroblasts
are a subpopulation of cells within the tumor that are transformed fibroblasts, but share properties of myofibroblasts that are found during the process of wound healing.
- Toll-like receptors
these protein receptors are characterized by their ability to respond to invading pathogens by recognizing conserved molecular structures. TLRs are primarily expressed by immune cells such as monocytes, macrophages, mast cells and dendritic cells. Upregulated TLR expression has been found in almost all types of cancer cells and has been linked to oncogenesis and cancer progression. TLR agonists are emerging as anti-tumor agents and are exploited to enhance the immunogenicity of current chemotherapeutic regimens.
- S-nitrosation
is the process of protein post-translational modification, where NO is attached to the thiol group of a cysteine residue. It’s can cause repression or enhance protein activity depending on the protein and location of the active thiol group.
- Epigenetic modifications
environmental factors can lead to modifications in the DNA strands or histones around which DNA strands are wrapped. These are in the form of reversible attachment of methyl and/or acetyl groups on segments of the DNA or on histone tails.
- Therapeutic window
is the range of drug dose that targets cancer cells effectively but avoids adversely affecting healthy cells to minimize side-effects.
- Co-targets
a novel concept where genes or metabolic enzymes that are involved in a strong interplay are both targeted to achieve a synergistic therapeutic effect.
- Flux analysis
a computational technique that uses mass balance principles to estimate intracellular metabolic rates (or fluxes) from empirically measurable metabolic parameters. Fluxes are the closest representation of metabolic pathway activity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tannock IF, Rotin D. Acid Ph in Tumors and Its Potential for Therapeutic Exploitation. Cancer Research. 1989;49(16):4373–4384. [PubMed] [Google Scholar]
- 2.Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology. Cell. 2017;168(4):657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li F, Zhao C, Wang L. Molecular-targeted agents combination therapy for cancer: developments and potentials. Int J Cancer. 2014;134(6):1257–69. doi: 10.1002/ijc.28261. [DOI] [PubMed] [Google Scholar]
- 4.Tanese K, Grimm EA, Ekmekcioglu S. The role of melanoma tumor-derived nitric oxide in the tumor inflammatory microenvironment: its impact on the chemokine expression profile, including suppression of CXCL10. Int J Cancer. 2012;131(4):891–901. doi: 10.1002/ijc.26451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanuphan A, et al. Long-term nitric oxide exposure enhances lung cancer cell migration. Biomed Res Int. 2013;2013:186972. doi: 10.1155/2013/186972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Augsten M, et al. Cancer-associated fibroblasts expressing CXCL14 rely upon NOS1-derived nitric oxide signaling for their tumor-supporting properties. Cancer Res. 2014;74(11):2999–3010. doi: 10.1158/0008-5472.CAN-13-2740. [DOI] [PubMed] [Google Scholar]
- 7.de Oliveira GA, et al. Inducible Nitric Oxide Synthase in the Carcinogenesis of Gastrointestinal Cancers. Antioxid Redox Signal. 2016 doi: 10.1089/ars.2016.6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erdman SE, et al. Nitric oxide and TNF-alpha trigger colonic inflammation and carcinogenesis in Helicobacter hepaticus-infected, Rag2-deficient mice. Proc Natl Acad Sci U S A. 2009;106(4):1027–32. doi: 10.1073/pnas.0812347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kimura H, et al. Hypoxia response element of the human vascular endothelial growth factor gene mediates transcriptional regulation by nitric oxide: control of hypoxia-inducible factor-1 activity by nitric oxide. Blood. 2000;95(1):189–197. [PubMed] [Google Scholar]
- 10.Kashiwagi S, et al. Perivascular nitric oxide gradients normalize tumor vasculature. Nat Med. 2008;14(3):255–7. doi: 10.1038/nm1730. [DOI] [PubMed] [Google Scholar]
- 11.Pershing NL, et al. Treatment with the nitric oxide synthase inhibitor L-NAME provides a survival advantage in a mouse model of Kras mutation-positive, non-small cell lung cancer. Oncotarget. 2016;7(27):42385–42392. doi: 10.18632/oncotarget.9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salimian Rizi B, et al. Nitric oxide mediates metabolic coupling of omentum-derived adipose stroma to ovarian and endometrial cancer cells. Cancer Res. 2015;75(2):456–71. doi: 10.1158/0008-5472.CAN-14-1337. [DOI] [PubMed] [Google Scholar]
- 13.Feun L, et al. Arginine deprivation as a targeted therapy for cancer. Current Pharmaceutical Design. 2008;14(11):1049–1057. doi: 10.2174/138161208784246199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wada S, et al. Loss of p53 in stromal fibroblasts enhances tumor cell proliferation through nitric-oxide-mediated cyclooxygenase 2 activation. Free Radical Research. 2015;49(3):269–278. doi: 10.3109/10715762.2014.997230. [DOI] [PubMed] [Google Scholar]
- 15.Hibbs JB, Jr, et al. Nitric oxide: a cytotoxic activated macrophage effector molecule. Biochem Biophys Res Commun. 1988;157(1):87–94. doi: 10.1016/s0006-291x(88)80015-9. [DOI] [PubMed] [Google Scholar]
- 16.Lamrani M, et al. TLR4/IFN gamma pathways induce tumor regression via NOS II-dependent NO and ROS production in murine breast cancer models. Oncoimmunology. 2016;5(5) doi: 10.1080/2162402X.2015.1123369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marigo I, et al. T Cell Cancer Therapy Requires CD40-CD40L Activation of Tumor Necrosis Factor and Inducible Nitric-Oxide-Synthase-Producing Dendritic Cells. Cancer Cell. 2016;30(3):377–390. doi: 10.1016/j.ccell.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogdan C. Nitric oxide synthase in innate and adaptive immunity: an update. Trends Immunol. 2015;36(3):161–78. doi: 10.1016/j.it.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Hegardt P, et al. Nitric oxide synthase inhibitor and IL-18 enhance the anti-tumor immune response of rats carrying an intrahepatic colon carcinoma. Cancer Immunol Immunother. 2001;50(9):491–501. doi: 10.1007/s002620100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito H, et al. Inhibition of induced nitric oxide synthase enhances the anti-tumor effects on cancer immunotherapy using TLR7 agonist in mice. Cancer Immunol Immunother. 2015;64(4):429–36. doi: 10.1007/s00262-014-1644-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ridnour LA, et al. NOS Inhibition Modulates Immune Polarization and Improves Radiation-Induced Tumor Growth Delay. Cancer Res. 2015;75(14):2788–99. doi: 10.1158/0008-5472.CAN-14-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozato K, Tsujimura H, Tamura T. Toll-like receptor signaling and regulation of cytokine gene expression in the immune system. Biotechniques. 2002:66-+. [PubMed] [Google Scholar]
- 23.Dhillon SS, et al. Higher activity of the inducible nitric oxide synthase contributes to very early onset inflammatory bowel disease. Clin Transl Gastroenterol. 2014;5:e46. doi: 10.1038/ctg.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freeman HJ. Colorectal cancer risk in Crohn's disease. World J Gastroenterol. 2008;14(12):1810–1. doi: 10.3748/wjg.14.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puglisi MA, et al. High nitric oxide production, secondary to inducible nitric oxide synthase expression, is essential for regulation of the tumour-initiating properties of colon cancer stem cells. J Pathol. 2015;236(4):479–90. doi: 10.1002/path.4545. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda M, et al. Apoptosis induction of poly-S-nitrosated human serum albumin in resistant solid tumor under hypoxia can be restored by phosphodiesterase 5 inhibition. Nitric Oxide. 2017 doi: 10.1016/j.niox.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Li WW, et al. P53-dependent miRNAs mediate nitric oxide-induced apoptosis in colonic carcinogenesis. Free Radical Biology and Medicine. 2015;85:105–113. doi: 10.1016/j.freeradbiomed.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 28.Yang L, et al. The Expression and Correlation of iNOS and p53 in Oral Squamous Cell Carcinoma. Biomed Res Int. 2015;2015:637853. doi: 10.1155/2015/637853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Achreja A, et al. Exo-MFA - A 13C metabolic flux analysis framework to dissect tumor microenvironment-secreted exosome contributions towards cancer cell metabolism. Metab Eng. 2017 doi: 10.1016/j.ymben.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao H, et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife. 2016;5:e10250. doi: 10.7554/eLife.10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L, et al. Targeting Stromal Glutamine Synthetase in Tumors Disrupts Tumor Microenvironment-Regulated Cancer Cell Growth. Cell Metab. 2016;24(5):685–700. doi: 10.1016/j.cmet.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berchner-Pfannschmidt U, et al. Nitric oxide modulates oxygen sensing by hypoxia-inducible factor 1-dependent induction of prolyl hydroxylase 2. J Biol Chem. 2007;282(3):1788–96. doi: 10.1074/jbc.M607065200. [DOI] [PubMed] [Google Scholar]
- 33.Nisoli E, Carruba MO. Nitric oxide and mitochondrial biogenesis. Journal of Cell Science. 2006;119(14):2855–2862. doi: 10.1242/jcs.03062. [DOI] [PubMed] [Google Scholar]
- 34.Palmer LA, Johns RA. Hypoxia upregulates inducible (Type II) nitric oxide synthase in an HIF-1 dependent manner in rat pulmonary microvascular but not aortic smooth muscle cells. Chest. 1998;114(1):33s–34s. doi: 10.1378/chest.114.1_supplement.33s. [DOI] [PubMed] [Google Scholar]
- 35.Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate hypoxic signaling. Current Opinion in Cell Biology. 2009;21(6):894–899. doi: 10.1016/j.ceb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caneba CA, et al. Nitric oxide is a positive regulator of the Warburg effect in ovarian cancer cells. Cell Death & Disease. 2014;5 doi: 10.1038/cddis.2014.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nieman KM, et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17(11):1498–503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheng X, Mittelman SD. The role of adipose tissue and obesity in causing treatment resistance of acute lymphoblastic leukemia. Front Pediatr. 2014;2:53. doi: 10.3389/fped.2014.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ridnour LA, et al. Molecular mechanisms for discrete nitric oxide levels in cancer. Nitric Oxide. 2008;19(2):73–6. doi: 10.1016/j.niox.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen YJ, et al. dbSNO 2.0: a resource for exploring structural environment, functional and disease association and regulatory network of protein S-nitrosylation. Nucleic Acids Res. 2015;43(Database issue):D503–11. doi: 10.1093/nar/gku1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seth D, Stamler JS. The SNO-proteome: causation and classifications. Curr Opin Chem Biol. 2011;15(1):129–36. doi: 10.1016/j.cbpa.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Broniowska KA, Hogg N. The chemical biology of S-nitrosothiols. Antioxid Redox Signal. 2012;17(7):969–80. doi: 10.1089/ars.2012.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marozkina NV, Gaston B. S-Nitrosylation signaling regulates cellular protein interactions. Biochim Biophys Acta. 2012;1820(6):722–9. doi: 10.1016/j.bbagen.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z. Protein S-nitrosylation and cancer. Cancer Lett. 2012;320(2):123–9. doi: 10.1016/j.canlet.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 45.Fukumura D, Kashiwagi S, Jain RK. The role of nitric oxide in tumour progression. Nat Rev Cancer. 2006;6(7):521–34. doi: 10.1038/nrc1910. [DOI] [PubMed] [Google Scholar]
- 46.Jadeski LC, Chakraborty C, Lala PK. Role of nitric oxide in tumour progression with special reference to a murine breast cancer model. Can J Physiol Pharmacol. 2002;80(2):125–35. doi: 10.1139/y02-007. [DOI] [PubMed] [Google Scholar]
- 47.Forstermann U, Sessa WC. Nitric oxide synthases: regulation and function. Eur Heart J. 2012;33(7):829–37. 837a–837d. doi: 10.1093/eurheartj/ehr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dioh A, Hennen G. Nitric oxide. Current findings on the synthesis and genetics of nitric oxide synthases, regulation of hypothalamic and vascular function. Rev Med Liege. 1995;50(11):480–6. [PubMed] [Google Scholar]
- 49.Lai YC, et al. Nitrite-mediated S-nitrosylation of caspase-3 prevents hypoxia-induced endothelial barrier dysfunction. Circ Res. 2011;109(12):1375–86. doi: 10.1161/CIRCRESAHA.111.256479. [DOI] [PubMed] [Google Scholar]
- 50.Heinecke JL, et al. Tumor microenvironment-based feed-forward regulation of NOS2 in breast cancer progression. Proc Natl Acad Sci U S A. 2014;111(17):6323–8. doi: 10.1073/pnas.1401799111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Timoshenko AV, Lala PK, Chakraborty C. PGE2-mediated upregulation of iNOS in murine breast cancer cells through the activation of EP4 receptors. Int J Cancer. 2004;108(3):384–9. doi: 10.1002/ijc.11575. [DOI] [PubMed] [Google Scholar]
- 52.Qian D, et al. Normoxic induction of the hypoxic-inducible factor-1 alpha by interleukin-1 beta involves the extracellular signal-regulated kinase 1/2 pathway in normal human cytotrophoblast cells. Biol Reprod. 2004;70(6):1822–7. doi: 10.1095/biolreprod.103.025031. [DOI] [PubMed] [Google Scholar]
- 53.Sonveaux P, et al. Oxygen regulation of tumor perfusion by S-nitrosohemoglobin reveals a pressor activity of nitric oxide. Circ Res. 2005;96(10):1119–26. doi: 10.1161/01.RES.0000168740.04986.a7. [DOI] [PubMed] [Google Scholar]
- 54.Monteiro HP, et al. Nitric oxide: Protein tyrosine phosphorylation and protein S-nitrosylation in cancer. Biomed J. 2015;38(5):380–8. doi: 10.4103/2319-4170.158624. [DOI] [PubMed] [Google Scholar]
- 55.Switzer CH, et al. S-nitrosylation of EGFR and Src activates an oncogenic signaling network in human basal-like breast cancer. Mol Cancer Res. 2012;10(9):1203–15. doi: 10.1158/1541-7786.MCR-12-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murillo-Carretero M, et al. S-Nitrosylation of the epidermal growth factor receptor: a regulatory mechanism of receptor tyrosine kinase activity. Free Radic Biol Med. 2009;46(4):471–9. doi: 10.1016/j.freeradbiomed.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 57.Doulias PT, et al. Nitric oxide regulates mitochondrial fatty acid metabolism through reversible protein S-nitrosylation. Sci Signal. 2013;6(256):rs1. doi: 10.1126/scisignal.2003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi MS, et al. S-nitrosylation of fatty acid synthase regulates its activity through dimerization. J Lipid Res. 2016;57(4):607–15. doi: 10.1194/jlr.M065805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rizi BS, Nagrath D. Linking omentum and ovarian cancer: NO. Oncoscience. 2015;2(10):797–8. doi: 10.18632/oncoscience.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marozkina NV, et al. S-nitrosoglutathione reductase in human lung cancer. Am J Respir Cell Mol Biol. 2012;46(1):63–70. doi: 10.1165/rcmb.2011-0147OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gould N, et al. Regulation of protein function and signaling by reversible cysteine S-nitrosylation. J Biol Chem. 2013;288(37):26473–9. doi: 10.1074/jbc.R113.460261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murata J-I, et al. Nitric oxide as a carcinogen: analysis by yeast functional assay of inactivating p53 mutations induced by nitric oxide. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 1997;379:211–218. doi: 10.1016/s0027-5107(97)00149-8. [DOI] [PubMed] [Google Scholar]
- 63.Vasudevan D, Bovee RC, Thomas DD. Nitric oxide, the new architect of epigenetic landscapes. Nitric Oxide. 2016;59:54–62. doi: 10.1016/j.niox.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 64.Arif M, et al. Nitric Oxide-Mediated Histone Hyperacetylation in Oral Cancer: Target for a Water-Soluble HAT Inhibitor, CTK7A. Chemistry & Biology. 2010;17:903–913. doi: 10.1016/j.chembiol.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 65.Attwood J, Richardson B. Relative quantitation of DNA methyltransferase mRNA by real-time RT-PCR assay. Methods in Molecular Biology (Clifton, N.J.) 2004;287:273–283. doi: 10.1385/1-59259-828-5:273. [DOI] [PubMed] [Google Scholar]
- 66.Huang F-Y, et al. Helicobacter pylori induces promoter methylation of E-cadherin via interleukin-1β activation of nitric oxide production in gastric cancer cells. Cancer. 2012;118:4969–4980. doi: 10.1002/cncr.27519. [DOI] [PubMed] [Google Scholar]
- 67.Huang F-Y, et al. Interleukin-1β increases the risk of gastric cancer through induction of aberrant DNA methylation in a mouse model. Oncology Letters. 2016;11:2919–2924. doi: 10.3892/ol.2016.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vasudevan D, et al. Nitric Oxide Regulates Gene Expression in Cancers by Controlling Histone Posttranslational Modifications. Cancer Research. 2015;75:5299–5308. doi: 10.1158/0008-5472.CAN-15-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hickok JR, et al. Nitric Oxide Modifies Global Histone Methylation by Inhibiting Jumonji C Domain-containing Demethylases. Journal of Biological Chemistry. 2013;288:16004–16015. doi: 10.1074/jbc.M112.432294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramadoss S, Guo G, Wang C-Y. Lysine demethylase KDM3A regulates breast cancer cell invasion and apoptosis by targeting histone and the non-histone protein p53. Oncogene. 2017;36:47–59. doi: 10.1038/onc.2016.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramadoss S, et al. Lysine-specific demethylase KDM3A regulates ovarian cancer stemness and chemoresistance. Oncogene. 2016 doi: 10.1038/onc.2016.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaelin William G, Jr, McKnight Steven L. Influence of Metabolism on Epigenetics and Disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kottakis F, et al. LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature. 2016;539(7629):390–395. doi: 10.1038/nature20132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peng M, et al. Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science. 2016;354(6311):481–484. doi: 10.1126/science.aaf6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hickok JR, Thomas DD. Nitric oxide and cancer therapy: the emperor has NO clothes. Curr Pharm Des. 2010;16(4):381–91. doi: 10.2174/138161210790232149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thomas DD, et al. Hypoxic inducible factor 1 alpha, extracellular signal-regulated kinase, and p53 are regulated by distinct threshold concentrations of nitric oxide. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(24):8894–8899. doi: 10.1073/pnas.0400453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moncada S, et al. Nitric Oxide and the Cell: Proliferation, Differentiation, and Death. Princeton University Press; 2017. [Google Scholar]
- 78.Gorren ACF, Mayer B. The versatile and complex enzymology of nitric oxide synthase. Biochemistry-Moscow. 1998;63(7):734–743. [PubMed] [Google Scholar]
- 79.Vannini F, Kashfi K, Nath N. The dual role of iNOS in cancer. Redox Biol. 2015;6:334–43. doi: 10.1016/j.redox.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saura M, et al. Interaction of interferon regulatory factor-1 and nuclear factor kappa B during activation of inducible nitric oxide synthase transcription. Journal of Molecular Biology. 1999;289(3):459–471. doi: 10.1006/jmbi.1999.2752. [DOI] [PubMed] [Google Scholar]
- 81.Köhring M, et al. QCL-based TDLAS sensor for detection of NO toward emission measurements from ovarian cancer cells. Applied Physics B. 2014:1–7. [Google Scholar]
- 82.Berkels R, Purol-Schnabel S, Roesen R. Measurement of Nitric Oxide by Reconversion of Nitrate/Nitrite to NO. In: Hassid A, editor. Nitric Oxide Protocols. Humana Press; 2004. pp. 1–8. [DOI] [PubMed] [Google Scholar]
- 83.Kitamura Y, et al. In Vivo Nitric Oxide Measurements Using a Microcoaxial Electrode. In: Hassid A, editor. Nitric Oxide Protocols. Humana Press; 2004. pp. 35–43. [DOI] [PubMed] [Google Scholar]
- 84.Granger DL, et al. Measurement of nitrate and nitrite in biological samples using nitrate reductase and Griess reaction. Methods in Enzymology. 1996;268:142–151. doi: 10.1016/s0076-6879(96)68016-1. [DOI] [PubMed] [Google Scholar]
- 85.Griess P. Bemerkungen zu der Abhandlung der HH. Weselsky und Benedikt „Ueber einige Azoverbindungen”. Berichte der deutschen chemischen Gesellschaft. 1879;12:426–428. [Google Scholar]
- 86.Jahoor F, et al. Arginine flux and intravascular nitric oxide synthesis in severe childhood undernutrition. The American Journal of Clinical Nutrition. 2007;86:1024–1031. doi: 10.1093/ajcn/86.4.1024. [DOI] [PubMed] [Google Scholar]
- 87.Tsikas D. Measurement of Nitric Oxide Synthase Activity In Vivo and In Vitro by Gas Chromatography-Mass Spectrometry. In: Hassid A, editor. Nitric Oxide Protocols. Humana Press; 2004. pp. 81–103. [DOI] [PubMed] [Google Scholar]
- 88.Villalpando S, et al. In vivo arginine production and intravascular nitric oxide synthesis in hypotensive sepsis. The American Journal of Clinical Nutrition. 2006;84:197–203. doi: 10.1093/ajcn/84.1.197. [DOI] [PubMed] [Google Scholar]
- 89.Thame MM, et al. Arginine Flux, but Not Nitric Oxide Synthesis, Decreases in Adolescent Girls Compared with Adult Women during Pregnancy. The Journal of Nutrition. 2011;141:71–74. doi: 10.3945/jn.110.129403. [DOI] [PubMed] [Google Scholar]