Abstract
Quantitative measurement of transcription rates in live cells is important for revealing mechanisms of transcriptional regulation. This is particularly challenging for measuring the activity of RNA polymerase III (Pol III), which transcribes growth-promoting small RNAs. To address this, we developed Corn, a genetically encoded fluorescent RNA reporter suitable for quantifying RNA transcription in cells. Corn binds and induces fluorescence of 3,5-difluoro-4-hydroxybenzylidene-imidazolinone-2-oxime, which resembles the fluorophore found in red fluorescent protein (RFP). Notably, Corn shows high photostability, enabling quantitative fluorescence imaging of mTOR-dependent Pol III transcription. Unlike actinomycin D, we found that mTOR inhibitors resulted in heterogeneous transcription suppression in individual cells. Quantitative imaging of Corn-tagged Pol III transcript levels revealed distinct Pol III transcription “trajectories” elicited by mTOR inhibition. Together, these studies provide an approach for quantitative measurements of Pol III transcription by direct imaging of Pol III transcripts containing a photostable RNA-fluorophore complex.
RNA Pol III accounts for nearly 15% of the total RNA transcription in the cell, and synthesizes small noncoding RNA transcripts that coordinate cell growth and proliferation1. These include tRNAs needed for protein synthesis, small nucleolar RNAs and 5S ribosomal RNA for ribosome biogenesis, as well as small nuclear RNAs such as U6 that are needed for mRNA processing1. By controlling the levels of these RNAs needed for translation and mRNA processing, the rate of Pol III transcription could potentially determine the translational capacity of the cell1.
Consistent with this function, Pol III activity is regulated by pathways linked to cell growth and proliferation2–4. Pol III activity is upregulated by oncogenes such as c-myc, and downregulated by tumor suppressors, such as p53 and RB5. Regulation of Pol III transcription occurs, at least in part, through mTOR. mTOR phosphorylates and inactivates Maf1, an inhibitor of Pol III6,7. mTOR inhibitors lead to Maf1 dephosphorylation and reduce Pol III activity, which has been proposed to contribute to the anti-proliferative effects of these drugs6.
Monitoring Pol III transcription dynamics and how Pol III transcription is linked to signaling pathways is significantly more difficult than analysis of Pol II transcription, which produces mRNAs. mRNAs are capped and polyadenylated, and can be modified to contain reporter proteins such as GFP to reveal transcriptional dynamics in living cells8. In contrast. Pol III transcripts lack the 7-methylguanosine cap and poly(A) tail needed for translation9, so they cannot be modified to contain reporter proteins. Therefore, Northern blotting is typically used to infer changes in Pol III promoter activity. As a result, the temporal dynamics of Pol III transcription in the same cell over time, or among individual cells in a population cannot readily be measured.
An alternative approach to image Pol III promoter activity in living cells could be to directly quantify the transcript using a reporter RNA, rather than an encoded reporter protein. However, current RNA imaging tags are not suitable for quantitative measurements in living cells. These tags comprise RNA aptamers and cognate fluorophores that become fluorescent upon binding the aptamer10–13. These aptamers include the green fluorescent Spinach, Spinach2 and Broccoli aptamers, which bind 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI (1))10–12, an otherwise nonfluorescent small molecule fluorophore. However, RNA-bound DFHBI readily photobleaches due to light-induced isomerization of DFHBI from the cis to the trans form, which terminates fluorescence14,15. Although these tags provide qualitative detection of RNA in cells, they fail to provide quantitative measurements of the levels of a reporter RNA labeled with these imaging tags due to the loss of signal caused by photobleaching.
Here we describe an RNA mimic of red fluorescent protein that exhibits marked photostability and enables quantitative transcript level imaging in live cells. Since aptamers that bind DFHBI are photolabile, we designed a new fluorophore, DFHO (2), based on the naturally occurring fluorophore in DsRed and other red fluorescent proteins. Similar to DFHBI, DFHO exhibits negligible fluorescence in solution or when incubated with cells. We developed a novel RNA aptamer, Corn, which binds DFHO and converts it to a yellow fluorescent species. Notably, Corn exhibits considerably improved photostability compared to Spinach and Broccoli, enabling quantitative measurements of RNA levels in live cells. We quantified the fluorescence of Pol III transcripts tagged with Corn to determine how mTOR inhibitors suppress Pol III transcription in live cells. We find that mTOR inhibitors induce specific patterns of Pol III transcriptional inhibition “trajectories” over time. These data demonstrate the ability of these photostable RNA-fluorophore complexes to reveal patterns of Pol III transcriptional activity in live cells.
RESULTS
DFHO: A fluorophore mimic of red fluorescent proteins
Spinach-DFHBI complexes undergo rapid reversible photobleaching14,15, which complicates the use of this tag for quantitative measurements of RNA levels in live cells. Subsequent screens for DFHBI-binding aptamers resulted in the generation of Broccoli which also exhibits photobleaching12. We therefore sought to develop a different fluorophore, and determine if aptamers that activate this fluorophore would exhibit photostability.
Fluorogenic RNA imaging tags rely on fluorophores such as DFHBI, which exhibit essentially undetectable fluorescence when applied to cells10. Thus, fluorescence seen in DFHBI-treated cells can be specifically assigned to Broccoli-DFHBI or Spinach-DFHBI complexes10. This contrasts with most dyes, such as malachite green and thiazole orange, which exhibit minimal fluorescence in buffer but become highly fluorescent upon incubation with cells due to interactions with membranes or nucleic acids10.
In order to identify a new cell-compatible conditionally fluorescent molecule, we considered molecules that contained the same structural elements of DFHBI, since the hydroxybenzylidene imidazolinone structure of DFHBI might be responsible for low cell-induced background16. Since the photobleaching of Spinach-DFHBI reflects light-induced isomerization of the fluorophore14,15, we considered fluorophores that might have additional “handles” that could interact with the RNA and restrict the cis-trans isomerization that leads to photobleaching. Structural studies of Spinach-DFHBI complexes showed that the imidazolinone exhibits few contacts with RNA, especially at the 2′ position the fluorophore17,18 (Fig. 1a). Conceivably, the lack of contacts may enable the imidazolinone to isomerize while bound to the Spinach aptamer.
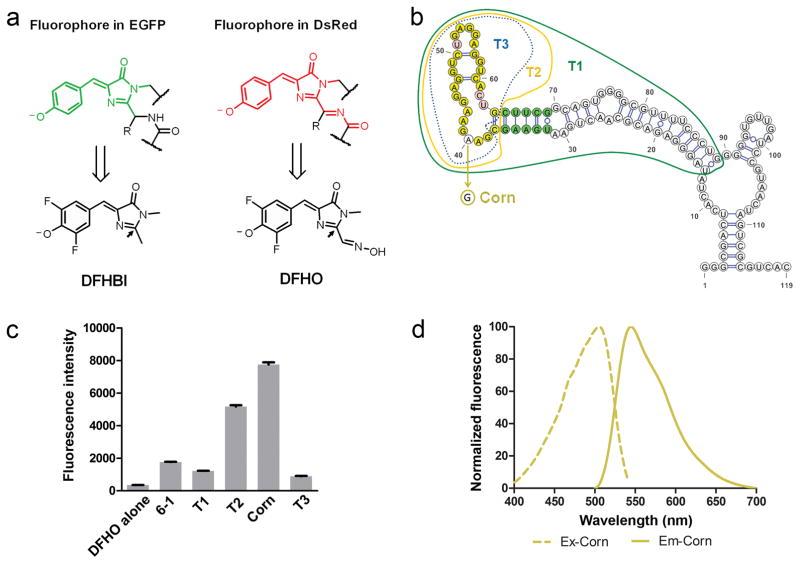
Figure 1. The Corn aptamer activates the fluorescence of the DFHO fluorophore.
(a) Structures of the GFP fluorophore (green) and DsRed fluorophore (red) (top row) and structures of the respective bio-mimicking synthetic compounds: DFHBI and DFHO, at pH 7.4 (bottom row). The 2′ position (indicated with an arrow) in DFHO contains additional recognition elements compared to DFHBI that may enable RNA-mediated photostability.
(b) Truncation and mutation analysis of the 6-1 aptamer identifies a core domain responsible for fluorescence activation of DFHO. The mFold-predicted secondary structure of 6-1 is presented. The borders of three truncations (T1, T2 and T3) are indicated. Green indicates the bases which form a stem required to maintain the fluorescence of 6-1, T1, and T2. Yellow indicates base positions which are not tolerant to mutations. Pink indicates base positions which tolerate mutations. A single A→G mutation at position 40 converts T2 into a brighter aptamer termed Corn.
(c) Quantification of fluorescence induced by Corn, full length 6-1, its truncation mutants from panel (b). “T2 - 1nt” is the T2 aptamer missing a single nucleotide: the 5′ C residue. The markedly reduced fluorescence of the T2 - 1nt suggests that T2 is the minimal aptamer sequence required for inducing DFHO fluorescence. All the aptamers tested, except 6-1 and T1, were expressed fused to the tRNA scaffold to maintain proper folding. Error bars indicate standard deviations (n=3).
(d) Excitation and emission spectra of Corn. Spectra were measured using 20 μM RNA and 2 μM DFHO.
We considered the fluorophore found in red fluorescent proteins (RFPs). DsRed utilizes a fluorophore that is similar to hydroxybenzylidene imidazolinone in GFP19–21. The difference is that it is modified with an acylimine substituent on the 2-position of the imidazolinone ring, which extends the π-electron conjugation19–22 (Fig. 1a). Importantly, the GFP family of proteins is conditionally fluorescent: when the protein is denatured, its fluorophore becomes nonfluorescent23. Thus the fluorophore is switched to a fluorescent form due to its interactions with the protein matrix surrounding the fluorophore in the folded structure. Based on this, we hypothesized that RFP-like fluorophores might exhibit low fluorescence in cells and might be activatable by RNA aptamers.
As an RFP fluorophore mimic, we synthesized DFHO (3,5-difluoro-4-hydroxybenzylidene imidazolinone-2-oxime) (Fig. 1a). We designed DFHO to contain a stable version of the characteristic N-acyl imine found in RFP. The N-acyl imine was mimicked in DFHO with an N-hydroxyl imine (i.e., oxime) substituent. Notably, DFHO is stable (Supplementary Results, Supplementary Fig. 1c and 2), in contrast to the N-acyl imine substituent of RFP which is readily hydrolyzed19. The fluorophore in RFP exists in a primarily negatively charged phenolate form, which increases its extinction coefficient and fluorophore brightness19–21. Therefore, we added two fluorine atoms on the hydroxybenzylidene ring of the fluorophore to reduce the pKa of the phenol and obtain the negatively charged fluorophore in neutral pH (Supplementary Fig. 1d).
We next tested whether DFHO fluorescence is nonspecifically activated by cellular constituents. Incubation of DFHO with human embryonic kidney (HEK293T) cellular RNA did not induce DFHO fluorescence (Supplementary Fig. 3a). Application of 10 μM DFHO to cultured HEK293T cells resulted in low level background fluorescence, while control compounds, malachite green and thiazole orange, produced robust cellular fluorescence (Supplementary Fig. 3b). Incubation of HEK293T cells with 10 μM DFHO did not induce cell death after irradiation on the microscope, while the control compound malachite green, produced substantial cytotoxicity (Supplementary Fig. 3c). Taken together, these data indicate that like DFHBI, DFHO exhibits low background fluorescence and cytotoxicity, making it a potentially suitable fluorophore for activation by RNA aptamers and for imaging experiments in living cells.
The Corn aptamer switches on the fluorescence of DFHO
We next generated aptamers that bind DFHO using a library of ~1014 random RNA sequences in the SELEX approach24. DFHO with aminohexyl linkers was synthesized for attachment to solid support. Boc-protected DFHO linker (3) was synthesized followed by the removal of the Boc protecting group to generate DFHO linker (4). DFHO-binding aptamers were recovered on DFHO-derivatized agarose beads. After 8 rounds of SELEX, we identified a single 119 nt-long aptamer (Aptamer 6-1) that induced the fluorescence of DFHO, with an excitation maximum (ExMax) of 505 nm and an emission maximum (EmMax) of 545 nm. This aptamer was truncated to a 76 nt-long aptamer (T1) that retained the ability to induce DFHO fluorescence (Fig. 1b,c).
We next used directed evolution to generate improved variants of T1. In this approach, a randomized DNA library was prepared12 based on the sequence of T1, so that each library member encodes a mutant T1 sequence. The library contains T1 mutants with every possible single, double, triple, up to seven-nucleotide mutation combination12. Aptamers were transcribed from the library and selected on DFHO-agarose to enrich for mutants that retain the ability to bind DFHO. DFHO-binding aptamers were then cloned into a bacterial expression library, and subjected to fluorescence-activated cell sorting (FACS) in PBS with no added Mg2+ to identify mutants that are less dependent on magnesium for folding. These experiments resulted in an aptamer that exhibited increased fluorescence due to a single A→G mutation (Fig. 1b and Supplementary Fig. 4a, b). We then performed further truncation analysis, resulting in the generation of T2, a 28-nt long aptamer that retained the ability to induce DFHO fluorescence (Fig. 1b,c). Introduction of the A→G mutation into T2 resulted in a 1.5-fold increase in fluorescence relative to T2 (Fig. 1b,c). Thus, the A→G T2 mutant (Fig. 1b and Supplementary Fig. 4c) was chosen for further characterization.
We next measured the spectral properties of this RNA-fluorophore complex (Table 1, Fig. 1d). The quantum yield (0.25) and extinction coefficient is similar to the initially reported coral RFPs25 (Table 1). Because of the yellow fluorescence of this RNA-fluorophore complex, this aptamer was designated Corn.
Table 1.
| Excitation maximum(nm) | Emission maximum (nm) | Extinction coefficient(M−1cm−1)a | Fluorescence quantum yield | Brightnessb | KD (nM)c | EC50 Mg2+(mM) | |
|---|---|---|---|---|---|---|---|
| DFHO alone | 473 | 561 | 19,800 | 0.0006 | 0.16 | --- | --- |
| Corn-DFHO | 505 | 545 | 29,000 | 0.25 | 100 | 70 | 0.45 |
| OrangeBroccoli-DFHO | 513 | 562 | 34,000 | 0.28 | 131 | 230 | 0.15 |
| Red Broccoli-DFHO | 518 | 582 | 35,000 | 0.34 | 164 | 206 | 0.18 |
Extinction coefficients for DFHO alone were measured at 505 nm in pH 7.4 buffer. RNA-DFHO complex extinction coefficients were all measured at maximum excitation wavelength in pH 7.4 buffer.
Brightness (extinction coefficient × quantum yield) is reported relative to Corn-DFHO complex.
KD based measured by fluorescence.
Corn-DFHO exhibits high fluorescence and efficient folding
We next asked if Corn exhibits properties needed to serve as a transcriptional reporter in live cells. For the biochemical characterizations, we incorporated Corn into the tRNALys scaffold, which improves the folding efficiency of aptamers26 (Supplementary Fig. 4c). Indeed, the folding of Corn was maintained in the presence of diverse flanking sequences (Supplementary Fig. 5a). Corn shows low dependence on magnesium for folding (Supplementary Fig. 5b), consistent with the fact that the directed evolution was performed in low-magnesium media.
Corn exhibits high thermal stability (Supplementary Fig. 5c), which helps to maximize aptamer fluorescence in mammalian cells11. Corn binds DFHO with high-nanomolar affinity (Table 1, Supplementary Fig. 5d and 6), similar to the affinity of Spinach and Broccoli for DFHBI10–12. This allows efficient formation of Corn-DFHO complexes when low concentrations of DFHO are added to cells. This is useful since unbound DFHO, has low but measureable fluorescence (Table 1) that would cause background fluorescence interference if applied at high concentrations.
Lastly, we sought to understand the structural basis for Corn-DFHO fluorescence. Spinach contains a G-quadruplex that interacts with DFHBI and enforces a planar conformation of the fluorophore17,18. Corn shows potassium-dependent folding (Supplementary Fig. 7a), suggesting that Corn contains a G-quadruplex. To test this, we used thioflavin T and thiazole orange, both of which show increased fluorescence upon binding to diverse G-quadruplex RNAs, including Broccoli27. As with Broccoli, both these dyes showed increased fluorescence upon interaction with Corn, but not a random RNA sequence (Supplementary Fig. 7b). These results, along with structural data28 indicates that Corn uses a G-quadruplex to activate DFHO fluorescence.
Another structural feature of Corn is that it forms a dimer in a manner that does not require DFHO28. The dimeric form of Corn is the only form that binds and activates the fluorescence of DFHO (Supplementary Fig. 8 and 9).
Corn-DFHO exhibits improved photostability in cells
We next asked if Corn can be used to encode yellow fluorescence in cells. We first tested Corn in E. coli. Minimal fluorescence was seen in DFHO-treated cells (20 μM) transformed with the empty vector. However robust yellow fluorescence was seen in E. coli expressing Corn (Supplementary Fig. 10).
We next imaged Corn-DFHO complexes in mammalian cells. Since the U6 promoter is one of the three major Pol III promoters, we expressed Corn from the U6 promoter and replaced the majority of the U6 sequence with Corn, leaving a minimal 5′ capping element leader sequence29. The majority of the cells expressed diffuse yellow fluorescence, most prominently in the cytoplasm (Fig. 2a).
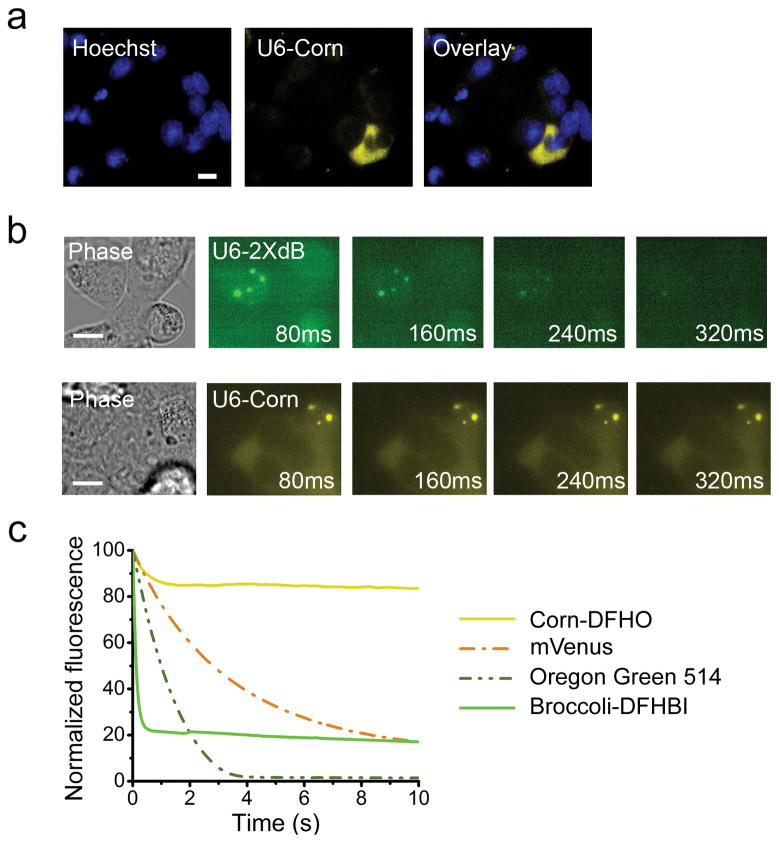
Figure 2. Corn exhibits remarkable photostability in vivo and in vitro.
(a) Corn expressed off of the U6 promoter shows robust fluorescence in HEK293T cells. U6-Corn was detected throughout the cell, with occasional puncta of U6-Corn seen in the cytoplasm. Images were acquired using a 60X objective (NA 1.40) and YFP filter cube for Corn (ex 500 ± 12 nm, em 542 ± 13.5 nm). Exposure times: 1000 ms for Corn, 200 ms for Hoechst. Scale bar, 10 μm.
(b) Corn-tagged RNA exhibits markedly improved photostability in living cells. In vivo photostability of Corn in comparison with Broccoli was assessed by imaging cells expressing Corn or Broccoli off the U6 promoter. Four consecutive images were acquired, each using an 80 ms exposure time. Cells with clear cytoplasmic puncta were selected for photostability experiments. Broccoli-tagged RNAs lost fluorescence by 240–320 ms, while the fluorescence of Corn-tagged RNA was maintained throughout imaging.
(c) Corn-DFHO exhibits markedly enhanced photostability in vitro, compared to Broccoli-DFHBI, small organic dye or a fluorescent protein. To measure the photostability of aptamer-fluorophore complexes, we prepared solutions containing ~20 μM Corn-DFHO and Broccoli-DFHBI by incubating 90 μM RNA aptamer and 20 μM DFHO (or DFHBI). Control solutions were the fluorescent protein mVenus (20 μM) or the control dye Oregon Green 514 (20 μM) in PBS. These solutions were emulsified in microscope immersion oil and three to five fluorescent aqueous droplets were repeatedly imaged under constant full-power light illumination in an epifluorescence microscope. Normalized bleaching curves of all droplets were averaged and plotted using Origin software. Exposure times were 20 ms.
We noticed that Corn fluorescence did not quickly photobleach, unlike Broccoli or Spinach. This was seen in all cells, but most easily quantified in a small subset of cells (~1%) where the fluorescence appeared as puncta in the cells rather than diffuse cytosolic labeling (Fig. 2b). In cells expressing U6-Corn, both the diffuse cytoplasmic labeling as well as the U6-Corn RNA puncta were readily detectable throughout the 320 ms imaging time, while Broccoli fluorescence was markedly reduced by 160 ms.
Constant irradiation of a solution of Broccoli-DFHBI showed rapid photobleaching, with >50% of the fluorescence lost following 200 ms (Fig. 2c). However, Corn-DFHO exhibited remarkable photostability, with minimal loss of fluorescence seen after 10 s of irradiation. Corn-DFHO showed higher photostability than a control dye Oregon Green 514 (20 μM), and mVenus (20 μM), a fluorescent protein with similar excitation and emission wavelengths as Corn30 (Fig. 2c).
We considered the possibility that the increased photostability of Corn-DFHO is due to rapid unbinding of photobleached DFHO followed by rapid rebinding of fresh DFHO. However, comparison of kunbind for Broccoli-DFHBI and Corn-DFHO showed that DFHO stays bound longer to Corn (0.018 ± 0.002 sec−1 and 0.008 ± 0.002 sec−1, respectively) (Supplementary Fig. 11). Furthermore, the association rate of DFHO on Corn is slower than DFHBI on Broccoli (kbind, 23,000 ± 3,000 M−1 sec−1 and 54,000 ± 4,000 M−1 sec−1, respectively). Thus, a higher DFHO recycling rate is unlikely to explain the improved photophysical properties of Corn-DFHO.
Notably, we noticed a small rapid loss of fluorescence for a small fraction of Corn-DFHO after in vitro irradiation (Fig. 2c). This may reflect a small subpopulation of DFHO bound to Corn in a conformation that is not photostable. The maintenance of Broccoli-DFHBI fluorescence at ~20% of its initial levels may reflect fluorescence recovery due to unbinding of isomerized DFHBI and rebinding of a nonisomerized DFHBI fluorophore.
Orange and Red Broccoli-DFHO complexes lack photostability
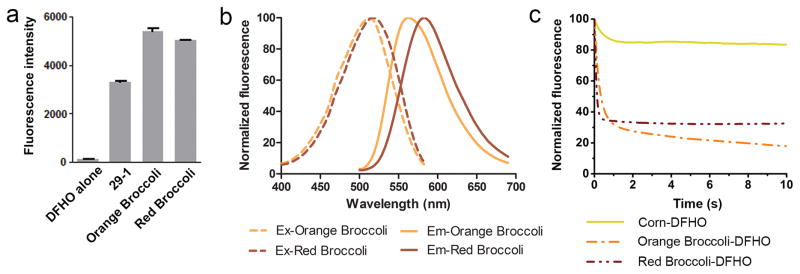
We next asked if the photostability seen with Corn-DFHO complexes reflects intrinsic photostability of DFHO. In this case, any aptamer that binds and activates DFHO should show photostability. To generate additional DFHO-binding aptamers, we considered the possibility that DFHO might bind Broccoli. Indeed, significant red fluorescence (ExMax = 515 nm, EmMax = 575 nm) was observed following incubation of 29-1, the parental aptamer of Broccoli12, with DFHO (Fig. 3a).
Figure 3. Orange Broccoli and Red Broccoli activate and tune the fluorescence of the DFHO fluorophore.
(a) Comparison of the molecular brightness of 29-1, Orange Broccoli and Red Broccoli. The fluorescence was recorded using 20 μM RNA and 1 μM DFHO at the respective excitation and emission maxima. Error bars indicate standard deviations (n=3).
(b) Excitation and emission spectra of Orange Broccoli and Red Broccoli. Spectra were measured using 20 μM RNA and 1 μM DFHO.
(c) Corn tunes the photostability of DFHO. Solutions containing ~20 μM Corn-DFHO, Orange Broccoli-DFHO or Red Broccoli-DFHO were prepared by incubating 90 μM RNA aptamer and 20 μM DFHO. These solutions were emulsified in microscope immersion oil and aqueous droplets were repeatedly imaged under constant full-power light illumination in an epifluorescence microscope. Unlike fluorescent dyes and fluorescent proteins, the fluorophore for these aptamers exhibits reversible binding and unbinding. As a result, the photobleaching of an RNA-fluorophore complex can be reversed by rebinding of a different fluorophore. This likely contributes to the lack of complete photobleaching seen for the Red/Orange Broccoli-fluorophore complexes. Images were acquired using a 60X objective (NA 1.40) and a YFP filter cube for Corn (ex 500 ± 12 nm, em 542 ± 13.5 nm), an orange fluorescence filter cube for Orange Broccoli (ex 512 ± 12.5 nm, em 532 nm (long pass)), a red fluorescence filter cube Red Broccoli (ex 535 ± 25 nm, em 580 nm (long pass)). Exposure times were 20 ms.
Directed evolution of 29-1 and DFHO resulted in a group of aptamers with red-shifted fluorescence (EmMax =582 nm) and another group with orange fluorescence (EmMax =562 nm). Because these clones are derived from the Broccoli precursor aptamer 29-1, the brightest aptamers were designated Orange Broccoli and Red Broccoli (Supplementary Fig. 12).
Characterization of the Orange Broccoli and Red Broccoli-DFHO complexes showed that both exhibited red-shifted excitation and emission peaks relative to Corn-DFHO with similar quantum yields (Fig. 3b, Table 1, Supplementary Fig. 5a–d).
We next asked if Orange Broccoli-DFHO and Red Broccoli-DFHO show that same photostability as Corn-DFHO. In both cases, Orange and Red Broccoli showed rapid photobleaching, similar to Broccoli-DFHBI complexes (Fig. 3c). Thus, DFHO is not intrinsically photostable—its photostability is selectively induced by its binding to Corn.
Imaging transcription from each Pol III promoter subclass
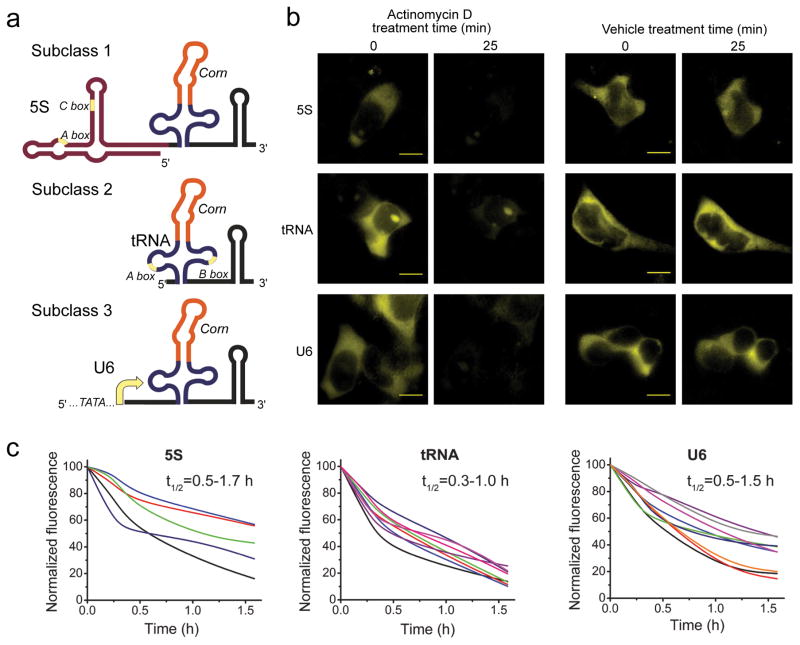
Because of the photostability of Corn during imaging, we reasoned that Corn could be used for quantitative measurements of cellular Pol III promoter activity in living cells. In these experiments, Pol III promoters are engineered so that they express Corn in place of their normal small RNA product. Corn serves as a direct reporter of transcript levels. We inserted Corn in the tRNALys folding scaffold downstream of the three main subclasses of Pol III promoters. The prototypical RNAs transcribed from each promoter are the 5S RNA, tRNA, and U6 RNA (subclasses 1, 2, and 3, respectively)31. In the case of subclasses 1 and 2, control sequences that direct transcription are not upstream of the encoded transcript, but are instead found internally within the portion of the gene that encodes the transcript. Thus, sequence elements needed for transcription are also transcribed. In contrast, subclass 3 utilizes upstream TATA-element control elements and the encoded RNA contains no promoter elements31. We thus fused the 5S promoter32 to Corn scaffolded in tRNALys. The tRNA promoter, which requires specific elements within the tRNA sequence, was modified to contain the scaffolded Corn in the anticodon loop. The U6 promoter was left unchanged. The U6 transcribed region was removed except for a 27-nucleotide stem that promotes capping32. tRNALys-Corn replaced the remaining sequence (Fig. 4a).
Figure 4. Imaging Pol III activity in single cells using Corn.
(a) Schematic of the Pol III reporter constructs. Corn in a tRNA scaffold was expressed off 5S, tRNA and U6 promoters. These promoters belong to three distinct subclasses of Pol III-dependent promoters.
(b) Pol III transcription inhibition with actinomycin D reduces Corn fluorescence in mammalian cells. HEK293T cells were preincubated with 10 μM DFHO and then treated with 5 μg/ml actinomycin D or vehicle for 25 min. The Corn signal rapidly disappears in actinomycin D-treated cells during the 25 min incubation, indicating that this signal can be attributed to RNA transcription. Exposure time 1000 ms. All images are contrast enhanced the same way to guarantee comparability. Scale bar, 10 μm.
(c) Corn fluorescence induced by each Pol III subclass promoter exhibits rapid turnover. The Corn reporter needs to turnover rapidly in order for changes in Pol III activity to cause a change in overall cellular Corn fluorescence. To determine turnover rates, we monitored Corn fluorescence after treatment with actinomycin D in individual cells. Cell trajectories were generated using the adjacent averaging algorithm in the Origin software package (see Online Methods). Each trajectory indicates the total cellular Corn fluorescence normalized to the cell area at each time point for an individual cell. The value at time zero is set to 100 and the background fluorescence is set to 0. Indicated in the upper right corner of each plot is the range of RNA half-life as calculated based on the single-cell trajectories. Cells were randomly selected from three separate experiments for each reporter.
Transfection of the reporter plasmids into HEK293T cells led to yellow fluorescence after addition of DFHO. To confirm that the yellow fluorescence reflects transcription of the Corn reporter, cells were treated with actinomycin D, an inhibitor of RNA polymerases33. In each case, substantial reduction of yellow fluorescence was seen in 25 min (Fig. 4b).
We validated that the reporters produce Corn reporter transcripts of the expected size by Northern blot analysis (Supplementary Fig. 13). No evidence for internally initiated reporter transcripts was detected.
To further characterize Corn reporter of Pol III dynamics, we monitored the levels of Corn fluorescence in individual cells over time. In each case, actinomycin D caused a reduction in Corn fluorescence in each cell (Fig. 4c), indicating that Corn turns over rapidly in cells, enabling Corn levels to indicate dynamic changes in Pol III promoter activity over time courses of several hours.
We noticed that a subset of cells lacked Corn fluorescence even though they expressed a cotransfection marker, blue fluorescent protein (BFP) (Supplementary Fig. 14a). In some cases, these cells started to spontaneously exhibit Corn fluorescence (Supplementary Fig. 14b). Thus, these initially Corn-negative cells may have been in low-transcription states, which spontaneously reactivated during imaging.
mTOR inhibitors partially inhibit Pol III transcription
We next monitored Pol III transcriptional dynamics in response to mTOR inhibition in cells expressing the 5S, tRNA, and U6 reporters. To quantify the expression in the population, Corn fluorescence was measured by flow cytometry. In the case of vehicle treatment, numerous fluorescent cells were detected; however, actinomycin D caused the majority of cells to lose fluorescence after 2 h. Following treatment with the clinically utilized rapamycin derivative, temsirolimus (also known as CCI-779)34, Corn fluorescence was reduced, but remained higher than in actinomycin D-treated cells after 2 hours (Fig. 5a and Supplementary Fig. 15). Thus, mTOR pathway inhibition and actinomycin D appear to display distinct effects of Pol III transcription.
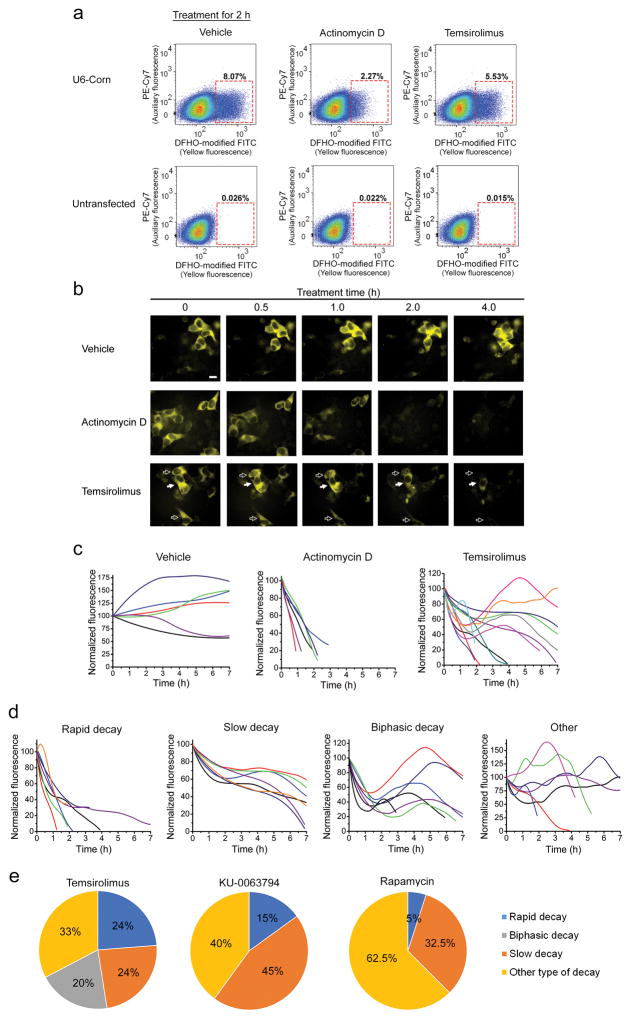
Figure 5. The mTOR inhibitor temsirolimus shows a variety of Pol III activity trajectories.
(a) Flow cytometry analysis of whole-population repression of Pol III activity after 5 μg/ml actinomycin D or 20 μM temsirolimus treatment. HEK293T cells were transfected with reporter plasmids expressing Corn from the 5S, tRNA, and U6 promoters (5S and tRNA reporters shown in Supplementary Fig. 15). Fluorescence was collected in two channels: yellow (modified FITC, ex 488 nm, em 545 ± 17.5 nm) and auxiliary far-red (PE-Cy7, ex 561 nm, em 780 ± 30) to measure cellular autofluorescence.
(b) Single-cell imaging reveals heterogeneous responses to temsirolimus. Temsirolimus causes some cells to show a rapid decline in the U6 reporter signal (open arrows), while other cells showed a delayed reduction in Corn signal (filled arrow). Images were acquired using a 40X objective (NA 0.75) and YFP filter cube for Corn (ex 500 ± 12 nm, em 542 ± 13.5 nm). Exposure time 1000 ms. Scale bar, 10 μm.
(c) Fluorescence imaging of the Corn reporter in cells reveals single-cell trajectories of Pol III activity in vehicle-, actinomycin D- and temsirolimus-treated cells. Trajectories were based on fluorescence measurements in individual cells using nine hourly time points (vehicle), 5–11 time points (actinomycin D), or 22 time points (temsirolimus) (see Online Methods).
(d) Examples of major types of temsirolimus-induced Pol III activity trajectories.
(e) Pie chart showing the prevalence each type of Pol III activity trajectory after mTOR inhibitor treatment. Temsirolimus (46 cells from three separate experiments), KU-0063794 and rapamycin (40 cells from three separate experiments).
In order to understand the basis for the partially reduced Pol III promoter activity following temsirolimus treatment, we imaged Pol III promoter activity in individual HEK293T cells over time. In vehicle-treated cells, fluorescence was largely stable (Fig. 5b). In actinomycin D-treated cells, the fluorescence of all cells was markedly reduced by 2 h and nearly undetectable at 4 h. However, in temsirolimus-treated cells, heterogeneous responses were evident, with some cells showing rapid reductions in fluorescence, while others showed notably lower reductions (Fig. 5b). During the entire time course, the reporter maintained its short half-life (Supplementary Fig. 16a,b), suggesting that the observed effects are not due to an unintended stabilization of the reporter RNA.
To assess the cell-to-cell variation in these cells, we generated Pol III “trajectories” for individual cells. Each trajectory indicates the Corn fluorescence in a single cell, normalized to the cell area at each time point. In vehicle-treated cells, the 7 h Pol III trajectory showed cells exhibiting steady increases or decreases in the Corn reporter (Fig. 5c). In actinomycin D-treated cells, the cell trajectories were homogeneous, with most cells showing a complete loss of Corn fluorescence by 2 h.
In contrast, temsirolimus induced marked alterations in Pol III promoter activity, with different HEK293T cells showing different trajectories (Fig. 5c). Many cells showed a rapid decay in Corn expression (Fig. 5d), while others showed a much slower rate of decay of Corn fluorescence. Many cells showed a biphasic pattern with an initial rapid decay, followed by increased Corn expression. Many of these cells remained fluorescent at the end of the 7 h time course (Fig. 5e). In addition, other trajectories were seen, including rare cells that exhibited increased Corn expression or no significant change in Corn expression throughout the time course (Fig. 5d).
We also tested two other mTOR inhibitors, rapamycin and KU-0063794. Rapamycin primarily affects mTORC135 while KU-0063794 is a potent dual inhibitor of mTORC1 and mTORC235. Single cell Pol III trajectories obtained following KU-0063794 treatment or rapamycin did not show the biphasic decay patterns seen with temsirolimus (Fig. 5e). Instead, KU-0063794 caused predominantly slow decay (Fig. 5e).
Together, these experiments confirm that mTOR inhibitors reduce Pol III activity; however, the cellular response shows cell-to-cell heterogeneity and different responses by different mTOR inhibitors (Fig. 5e and Supplementary Fig. 17). The heterogeneous responses likely account for the incomplete inhibition seen in the flow cytometry analysis (Supplementary Fig. 18).
We confirmed the imaging results by northern blotting for an endogenous Pol III tRNA transcript (Supplementary Fig. 19) and measuring the expression of the reporter RNA by native PAGE analysis (Supplementary Fig. 20). We also examined whether the heterogeneous Pol III transcriptional responses elicited by mTOR inhibition were seen in another cell line, COS7 cells. Trajectory plots of single COS7 cells expressing the U6-Corn reporter showed marked heterogeneity, although the fraction of each decay pattern was different than HEK-293 cells (Fig. 5e and 21b,c). Together, these results indicate that different cell lines exhibit unique effects on Pol III transcription in response to mTOR inhibition.
DISCUSSION
Although Pol III transcribes small RNAs with critical roles in cell growth and proliferation, quantification of Pol III transcriptional dynamics at cellular resolution has not been possible. Traditional transcriptional reporters are sequences that encode reporter proteins such as GFP; however, Pol III transcripts are not translatable, making this approach not possible for Pol III transcription. To overcome this, we developed Corn, a markedly photostable RNA-fluorophore complex that enables direct quantitative measurement of RNA levels when used as a RNA-based reporter for transcription. This reporter utilizes DFHO, an RFP-like fluorophore that exhibits minimal nonspecific fluorescence in cells. Upon binding Corn, DFHO is selectively activated and the resulting complex provides a photostable fluorescent reporter that enables Pol III transcriptional dynamics to be measured in living cells in real time.
Early studies on mTOR showed that mTOR promotes cell growth, in part, by increasing synthesis of ribosomal RNAs, 5S RNA, tRNAs, and snRNAs36. mTOR inhibitors are thought to reduce cell growth capacity, in part, by suppressing the synthesis of these Pol III transcripts37. Using Corn as a transcriptional reporter, we validated these early findings by showing a general drop in Pol III transcription in response to mTOR inhibition. However, the cellular responses were highly heterogeneous, with many cells maintaining Pol III transcription, while others showing rapid Pol III transcription inhibition similar to actinomycin D treatment. The inhibitory effects were also affected by the type of mTOR inhibitor that was used. Overall, these findings suggest that more effective mTOR inhibitors could be identified by screening for compounds based on Pol III inhibition.
DFHO is likely to have low background for the same reason as hydroxybenzylidene imidazolinone, which is the GFP fluorophore. In solution, hydroxybenzylidene imidazolinone loses absorbed energy by undergoing a highly efficient volume-conserving cis-trans isomerization upon excitation with light38. Corn likely suppresses this non-radiative decay pathway of the excited state DFHO fluorophore. Thus, Corn switches DFHO to a fluorescent form, enabling RNA imaging in cells.
Unlike previous RNA-fluorophore complexes, Corn-DFHO shows markedly increased photostability. Notably, Orange and Red Broccoli bind and activate fluorescence of DFHO but do not convert DFHO to a photostable fluorophore. Thus, photostability of DFHO in Corn reflects the ability of Corn to “tune” the photophysical properties of DFHO, similar to the spectral tuning of DFHO fluorescence emission by the different aptamers. The photostability of Corn allows longer exposure times, which is typically required for quantitative measurements of RNAs. Although Corn can be used for quantitative measurements, its dimerization makes it unsuitable for tagging and imaging mRNAs in cells. For this application, a photostable monomeric RNA-fluorophore complex would be needed.
Although the design of DFHO was inspired by the fluorophore found in DsRed and related RFPs, additional fluorophore structures are found in other RFPs39. These fluorophores contain diverse substituents on the imidazolinone ring that extend the π-conjugation of the fluorophore39. If these fluorophores exhibit low background fluorescence and cytotoxicity, they may be the starting point for other biomimetic and photostable RNA-fluorophore complexes.
METHODS
Reagents and equipment
Commercially available reagents were used without further purification. DFHBI fluorophores were obtained from Lucerna Technologies (New York, NY) or were synthesized as described previously10,22. Absorbance spectra were recorded with a Thermo Scientific NanoDrop 2000 spectrophotometer with cuvette capability. ChemiDoc MP imager (Bio-Rad) was used to record bacterial colony fluorescence on agar plates as described previously12. Fluorescence was measured on FluoroMax-4 spectrofluorometer (Horiba Scientific), Safire II or Genios Pro plate readers (Tecan). FACS experiments were performed using FACSAria II instrument (BD Biosciences). Fluorescence imaging experiments were performed using an Eclipse TE2000-E microscope (Nikon).
Preparation of affinity matrix
Amine-functionalized DFHO was first dissolved in DMSO at a concentration of 40 mM and then diluted into 100 mM HEPES buffer pH 7.5 with a final concentration of 5% DMSO and 2 mM amine-functionalized DFHO. This fluorophore solution was then added to NHS-activated Sepharose (GE Life Sciences), which had been preequilibrated with 2 volumes of ice-cold buffer. The resin was then incubated with amine-functionalized DFHO solution overnight at 4°C in the dark. The resin was washed with reaction buffer and incubated with 100 mM Tris pH 8.0 for 2 h at 25°C to react with any remaining NHS-activated sites. After thorough washing, the resin was stored in 1:1 ethanol:100 mM sodium acetate pH 5.4 at 4°C. The efficiency of sepharose coupling was monitored by measuring the absorbance at 400 nm of free DFHO in the flow-through. Using this approach, we estimate that the resin contains approximately 5 μmol of fluorophore per ml.
SELEX procedure
The random library used for our SELEX was generated before and previously used to isolate the Spinach and 29-1 aptamers12. In brief, this library contained two 26-base random stretches separated by a 12-base fixed sequence and flanked from 5′ and 3′ ends with constant regions used for PCR amplification and in vitro transcription. Doped libraries were described in detail previously12. In brief, these libraries were created in a way that each encoded aptamer resembles the parent aptamer, except that there are on average seven mutations per sequence. In order to obtain this library every position is chemically synthesized with a phosphoramidite nucleosides mixture that contains primarily the nucleotide that is found at that position in the parent aptamer, but also contains each of the other nucleotides at a lower concentration. dsDNA encoding doped libraries were designed with 14% mutagenesis efficiency and were ordered from Protein and Nucleic Acid Facility, Stanford University Medical Center.
1 × 1014 different sequences of double stranded DNA were transcribed in a 250 μl T7 RNA polymerase transcription reaction using the AmpliScribe T7-Flash Transcription Kit (Epicentre Biotechnologies). After treatment with DNase (Epicentre Biotechnologies) for 1 h, RNAs were purified using RNeasy Mini Kit (Qiagen) following manufacturer’s recommendations.
The SELEX procedure for random and doped libraries was conducted essentially as described previously12. Briefly, during the first step RNA species capable of binding to the DFHO-sepharose matrix were removed by incubation with “mock” resin. The resulting RNA solution was then incubated with DFHO-coupled matrix. RNA bound to DFHO resin was then washed with 3×0.5 ml of selection buffer during rounds 1–2, 4×0.5 ml during rounds 3–6 and 6×0.5 ml during round 7. Finally, specifically bound RNA was eluted with free DFHO. The doped library SELEX was conducted essentially the same way. The magnesium concentration in the selection buffer was decreased from 1 mM to 0.1 mM beginning at the second round of SELEX and maintained at this low concentration during the third round as well.
The eluted RNAs were then ethanol precipitated, reverse-transcribed, PCR amplified and in vitro transcribed to yield the pool for the next round. The presence of fluorescent RNA species in each pool was assessed by mixing 20 μM RNA and 10 μM DFHO and measuring fluorescence emission of this solution on a fluorometer in comparison with the fluorophore alone. At this point, RNAs were cloned into bacterial expression plasmids for FACS-based screening.
Bacterial library generation and FACS sorting
RNA libraries, expressed from the pBAD E plasmid12 were analyzed in LMG194 E. coli (ATCC). LMG194 cells then were grown in LB media overnight in presence of 0.002% arabinose and then collected for sorting. Typical bacterial libraries contain 5–30×106 individual members. Cells were preincubated with 40 μM DFHO and then sorted on a FACSAria II instrument (BD Biosciences). The sample compartment of the sorter was maintained at 37°C to facilitate sorting of cells expressing the most thermostable aptamers. To isolate yellow fluorescent events, cells were excited with the 488 nm laser and their emission was collected using 545±17.5 emission filter. Orange and red fluorescent cells were collected with 561 nm excitation and 585±21 nm emission filter. Typically the top one thousand brightest cells were sorted into 1 ml SOC media and cultured at 37°C for 1 h. The cells were then plated on LB-agar supplemented with carbenicillin, 0.002% arabinose and 20 μM DFHO.
The next day, the colonies on the LB-agar plate were imaged on a ChemiDoc MP imager (Bio-Rad). Yellow fluorescence was collected in a channel with 470±15 nm excitation and 532±14 nm emission. Orange and red fluorescent colonies were imaged with 530±15 nm excitation and 607±25 nm emission. The Cy5 channel (630±15 nm excitation and 697±22.5 nm emission) was used to collect autofluorescence signal from bacterial colonies, which allows normalization for colony size. Images were processed and normalized in ImageJ software (NIH) to identify colonies expressing the brightest aptamers.
In vitro characterization of aptamers
dsDNA from the brightest bacterial colonies was PCR amplified from the purified plasmids. Truncation, deletion and point mutation mutants were generated from dsDNAs that were PCR amplified from the appropriate ssDNA templates. Where indicated, dsDNA, and thus the encoded RNA, also contained the tRNA scaffold sequence. PCR products were then purified with PCR purification columns (Qiagen) and in vitro transcribed utilizing an AmpliScribe T7-Flash Transcription Kit (Epicenter). RNA was purified using Bio-Spin columns (Bio-Rad), and quantified using both absorbance values and the Quant-iT RiboGreen RNA Assay Kit (Life Technologies).
Absorption, excitation and emission spectra were measured for solutions using “excess RNA” conditions and limiting amount of fluorophore to ensure that no free fluorophore contributes to the absorbance or fluorescence signal11. The RNA concentration was 20 μM (for the fluorescence measurements) and 50 μM (for the absorbance measurements) while the DFHO concentration was 2 μM and 5 μM respectively.
Measuring in vitro photostability of Corn, Orange Broccoli and Red Broccoli
To measure aptamer photostability, 90 μM of Corn, Orange Broccoli or Red Broccoli RNA was mixed with 20 μM DFHO in buffer containing 40 mM HEPES pH 7.4, 100 mM KCl, and 5 mM MgCl2. 90 μM Broccoli and 20 μM DFHBI solution was used as a control. mVenus fluorescent protein or Oregon Green 514 dye (both 20 μM in PBS) served as additional controls. These solutions were emulsified in the microscope immersion oil and imaged using a Nikon T-2000E epifluorescence microscope. Normally, a group of three to five fluorescent aqueous droplets in one optical field were imaged with consecutive 20 ms exposures using constant full-power light illumination (i.e., neutral density setting 1). The fluorescence intensities of each droplet at different time point were quantified in NIS-Elements software (Nikon) and normalized to the intensity of the respective droplet at time 0. Finally, normalized bleaching curves of all droplets were averaged and plotted using GraphPad software. Images were acquired using a 60X objective (NA 1.40) and the YFP filter cube for Corn, mVenus or Oregon Green 514 (ex 500±12 nm, em 542±13.5 nm), an orange fluorescence filter cube for Orange Broccoli (ex 512±12.5 nm, em 532 nm (long pass)), a red fluorescence filter cube Red Broccoli (ex 535±25 nm, em 580 nm (long pass)), or a FITC filter cube for Broccoli (470±20 nm; emission filter 525±25 nm). Exposure times were 20 ms.
Quantum yield and extinction coefficient measurements
The extinction coefficient was calculated based on the absorbance spectrum and the Beer-Lambert-Bouguer law. All quantum yields were determined by comparing the integral of the corrected emission spectra for each fluorophore or RNA-fluorophore complex with the corresponding integral obtained from a solution of Rhodamine 123. Integrals at various concentrations were then plotted against the absorbance obtained at the wavelength corresponding to the excitation wavelength and the slope of the curve was compared to the slope of the curves found for reference fluorophores. All measurements for RNA-fluorophore complexes were taken in the presence of excess RNA to avoid interference from unbound fluorophore. The absolute quantum yields for reference fluorophores Rhodamine 123 was 0.9.
Native PAGE analysis
Native PAGE conditions were prepared by casting 10 × 10 cm 10% PAGE gels (29:1 acrylamide:bisacrylamide; Sigma) using using native PAGE buffer, i.e. the same final buffer composition as used for all the in vitro assays (40 mM HEPES, pH 7.5, 100 mM KCl, 5 mM MgCl2). After polymerization, the samples were run at 40 V for at least 2 h in a water-cooled PAGE chamber to keep the buffer temperature at ~25°C.
For staining, gels were incubated in buffer containing 10 μM DFHO for 15 min. Gels were then imaged to detect DFHO-Corn complexes. DFHO-stained gels were analyzed on a ChemiDoc MP imaging station (BioRad) at 470/30 nm excitation and 530/28 nm emission (green channel) and 530/28 nm excitation and 605/50 nm emission (red channel). Next, counterstaining with 1X SYBR Gold was performed for 15 min followed by gel imaging to detect all RNA species in each lane. SYBR Gold-stained bands were imaged using UV excitation (302 nm) and 590/110 nm emission on the ChemiDoc MP imaging station.
mVenus expression and purification
The pProEx-Htb-mVenus plasmid was transformed into BL21-AI One Shot Chemically Competent E. coli cells (Thermo Fisher Scientific) and colonies grew overnight. The next morning one colony was inoculated and grown during the day. In the evening, the cells were diluted 1/100 into fresh LB with ampicillin and 1 mM IPTG and the expression proceeded overnight. The next morning the cells were pelleted and lysed with B-PER bacterial protein extraction reagent (Thermo Fisher Scientific) following the manufacture’s protocol. Then the protein was purified using TALON His-Tag Purification Resin (Clontech). The resin was washed with 300 mM NaCl/PBS and the protein was finally eluted with 250 mM imidazole in 300 mM NaCl/PBS. The protein was then concentrated using Amicon Ultra-15 centrifugal filters and the buffer was changed to PBS. Finally, the concentration of the protein was calculated based on the measured absorbance at 515 nm and extinction coefficient of 92 mM−1 cm−1 (ref. 40).
Cloning of the plasmids used for expressing aptamer-tagged RNA in mammalian cells
U6-Corn was expressed from pAV-U6+27, which expresses the 27 nt-leader sequence of the U6 small nucleolar RNA from the U6 promoter32. 5S-Corn was expressed from pAV-5S, which expresses full length human 5S RNA from its endogenous promoter. The Corn reporter was fused to the 3′ end of either 27 nt leader sequence of U6 or 5S as the tRNALys-Corn chimera to facilitate better folding in vivo. To clone tRNALys-Corn into pAV-U6+27 or pAV-5S these plasmids were cut with XbaI and SalI. To express tRNALys-Corn off the tRNA promoter only, the pAV-U6+27-tCorn plasmid was digested with BamHI and SalI and then the cleaved sites were filled in using Klenow fragment and the vector backbone was self-ligated.
Cell culture conditions
Cell lines were obtained directly from the American Type Culture Collection (ATCC) for all experiments. HEK293T (ATCC-CRL-11268) and COS-7 (ATCC-CRL-1651) were grown according to ATCC instructions. Cells were screened for mycoplasma contamination before passaging using Hoechst 33258, according to ATCC recommendations.
Mammalian cell flow cytometry
HEK293T cells were transfected with the 5S, tRNA or U6 Pol III reporter (pAV-5S-tCorn, pAV-tCorn or pAV-U6+27-tCorn) using FuGENE HD. Untransfected cells were used as a negative control. After 48 h, the cells were treated with the indicated drug or vehicle (0.1% DMSO) for the indicated time. Then, cells were washed with 1X PBS once, harvested using TrypLE Express Enzyme (Thermo fisher scientific #12604013), resuspended in the 4% FBS/1X PBS solution containing 5 μM DFHO and 5 mM MgSO4, and kept on ice until analysis on the FACSAria II instrument (BD Biosciences). Transfected cells were analyzed in two channels: yellow (modified FITC, ex=488 nm, em= 545±17.5 nm) and auxiliary far-red channel (PE-Cy7, ex=561 nm, em=780±30). The auxiliary far-red channel measures cellular autofluorescence and facilitates presentation of the flow cytometry results in a scatter plot. Processing and analysis of the data was performed in the FlowJo program (Tree Star)
Imaging of Corn in mammalian cells
HEK293T cells were plated on poly-D-lysine- and mouse laminin-coated glass-bottom 24-well plates (MatTek) or 24-well ibiTreat μ-Plate (ibidi GmbH). The next day, cells were transfected with the 5S, tRNA or U6 Pol III reporter (pAV-5S-tCorn, pAV-tCorn or pAV-U6+27-tCorn) using FuGENE HD reagent (Promega). Cell culture media was changed to phenol red-free DMEM (Thermo Fisher Scientific #31053028) supplemented with 10% FBS and 1X GlutaMax-I (Thermo Fisher Scientific) 16 h before imaging. Next, HEK293T cells were pretreated with DFHO to a final concentration of 10 μM using 10X imaging buffer (100 μM DFHO in 500 mM HEPES pH 7.4 and 50 mM MgSO4) 30 min before imaging. Then cells were incubated with 5 μg/ml actinomycin D (Research Products International), 20 μM temsirolimus (Selleckchem), 1 μM KU-0063794 (Adipogen), 100 nM rapamycin (Adipogen) or 0.1% DMSO (New England Biolabs) as indicated. For the experiment using Hoechst (Fig. 2a), 5 μg/mL of Hoechst 33342 was used.
In the case of COS-7 cells, cells were grown on ECL cell attachment matrix (EMD Millipore)-coated 24-well ibiTreat μ-Plate (ibidi GmbH). Cell were transfected with the U6 Pol III reporter (pAV-U6+27-tCorn) using FuGENE HD reagent (Promega) or electroporated using a Nucleofector 2b Device (Lonza) following the manufacturers’ protocols. The cell culture media was changed to fresh phenol-free DMEM plus 10% FBS, 2X GlutaMax-I and sodium pyruvate (Thermo Fisher Scientific) 16 h before imaging. COS-7 cells were pretreated with DFHO (20 μM) using 10X imaging buffer (200 μM DFHO in 500 mM HEPES pH 7.4 and 100 mM MgSO4) at least 1 h before imaging. Cells were incubated with 2.5 μg/ml actinomycin D, 20 μM temsirolimus, 0.5 μM KU-0063794 or 0.1% DMSO (vehicle) as indicated.
Live fluorescence images were taken with a CoolSnap HQ2 CCD camera through a 40X air objective (NA 0.75) or 60X oil objective (NA 1.40) mounted on a Nikon Eclipse TE2000-E microscope and analyzed with the NIS-Elements software. For Corn detection we used the YFP filter cube (Semrock): excitation filter 500±12 nm, dichroic mirror 520 nm (long pass), and emission filter 542±13.5 nm. Hoechst-stained nuclei or blue fluorescent protein (BFP) were imaged with 350±25 nm excitation filter, 400 nm (long pass) dichroic mirror and 460±25 nm emission filter (Chroma Technology). Exposure times: 1000 ms for Corn, 200 ms for Hoechst, and 400 ms for BFP.
Plotting Pol III activity trajectories
To prepare the Pol III activity trajectory plot, individual cells in a captured field were manually traced as Regions of Interest (ROIs) and then tracked throughout the time series using the ImageJ software (NIH). To image N random fields of view, a well in 24-well plate was roughly divided into N squares and the center of each square was imaged. Fluorescence intensity for each time point was then calculated by dividing the total cellular fluorescence by the area for each ROI (cell), which resulted in the mean fluorescence intensity per unit area. The mean intensity of each cell at time zero is defined as 100% and background fluorescence intensity is defined as 0%. The mean fluorescence intensity at any other time point is normalized to the value at time 0. Normalized mean fluorescence of each cell were plotted as a function of time and then the resulting points were fitted with a smoothed curve to generate the Pol III activity trajectory. Dead cells were excluded from analysis based on morphological features (e.g. cell detachment and cell shrinkage).
All trajectories were generated using the adjacent averaging algorithm of Origin software. Five time points were used for the actinomycin D treatment experiment (Fig. 4c): images were taken at 0, 15, 25, 65, 95 min after treatment. To plot the curves for the vehicle-treated cell in Fig. 5c we collected nine time points approximately every hour. To plot the curves for the actinomycin D-treated cells in Fig. 5c, 5 to 11 time points were collected (for different curves) with intervals between time points ranging from 2 min to 20 min. To plot the curves for temsirolimus-treated cells (Figs. 5c and 5d), up to 22 points were used for each trajectory. These time points were collected at 15–20 min time intervals. 17–25 time points were collected every 15–30 min to plot the curves of the vehicle-treated HEK293T cells in Supplementary Fig. 17b. To plot the curves for the KU-0063794- or rapamycin-treated HEK293T cells (Supplementary Fig. 17b), time points were collected every 15 min, and 20–25 time points were used. To plot the curves for the vehicle-, temsirolimus- and KU-0063794-treated COS-7 cells (Supplementary Fig. 20b), time points were collected every 15 min, and 20–25 points were used. To plot the curves for the actinomycin D-treated COS7 cells (Supplementary Fig. 20b), 13–25 time points were collected every 15 min.
Based on their trajectories, cells were classified into four subtypes: rapid decay, slow decay, biphasic decay and “other,” which comprised trajectories that could not be assigned into the other three subtypes. Rapid decay trajectories were defined as showing substantial fluorescence loss by 3 h of treatment. Slow decay trajectories were the ones with major fluorescence loss happening after 5 h of treatment. Biphasic decay is defined as trajectories showing fluorescence loss during 1–3 h after treatment followed by fluorescence increase during the next 1–3 h after which fluorescence finally starts to decline again. Other trajectories showed diverse behaviors including gradual fluorescence increase or no substantial fluorescence decrease. Importantly, the specific patterns and frequency of each cell type trajectory, especially biphasic decay, are highly dependent on cell density, nutrient status, growth factors in the media, and potentially other factors. Therefore, it is important to maintain cells in as similar conditions as possible when comparing different drug treatments.
DATA AVAILABILITY
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files.
Supplementary Material
Acknowledgments
We thank members of the Jaffrey lab for helpful comments and suggestions. We thank J. S. Paige for early contributions to this project, M. You, R. S. Strack, and J. Litke for useful comments and suggestions, K. D. Warner and A. Ferré-D’Amaré (NIH) for communication of unpublished results, J. McCormick (Weill Cornell) for help with flow cytometry and J. Chiaravalli for guidance and support developing the fluorescence polarization assay at the High Throughput and Spectroscopy Resource Center at The Rockefeller University. This work was supported by NIH grants to S.R.J. (R01 NS064516 and R01 EB010249), Kwanjeong Educational Foundation to H.K, and the DFG to M.H.
Footnotes
AUTHOR CONTRIBUTIONS
W.S., G.S.F., H.K., M.H. and S.R.J. designed experiments, analyzed the data and wrote the manuscript. W.S. devised the synthetic methodologies, developed the aptamers, and characterized their binding properties in vitro and performed live imaging trajectory analysis in living cells. G.S.F. developed and imaged the Pol III reporters using FACS and using microscopy. H.K. performed imaging with different mTOR inhibitors in living cells and studies correlating reporter expression to RNA expression. M.H. performed anisotropy, melting, and affinity measurements. J.D.M. performed studies of reporter dynamics and half-life. X.L. performed studies of fluorophore stability in diverse conditions.
Competing financial interests
S.R.J. is the co-founder of Lucerna Technologies and has equity in this company. Lucerna has licensed commercialization of technology related to Spinach and other RNA-fluorophore complexes.
References
- 1.White RJ. RNA polymerase III transcription and cancer. Oncogene. 2004;23:3208–3216. doi: 10.1038/sj.onc.1207547. [DOI] [PubMed] [Google Scholar]
- 2.White RJ, Gottlieb TM, Downes CS, Jackson SP. Mitotic regulation of a TATA-binding-protein-containing complex. Mol Cell Biol. 1995;15:1983–1992. doi: 10.1128/mcb.15.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Felton-Edkins ZA, Fairley JA, Graham EL, Johnston IM, White RJ, Scott PH. The mitogen-activated protein (MAP) kinase ERK induces tRNA synthesis by phosphorylating TFIIIB. EMBO J. 2003;22:2422–2432. doi: 10.1093/emboj/cdg240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moir RD, Willis IM. Regulation of pol III transcription by nutrient and stress signaling pathways. Biochim Biophys Acta. 2013;1829:361–375. doi: 10.1016/j.bbagrm.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felton-Edkins ZA, Kenneth NS, Brown TR, Daly NL, Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct regulation of RNA polymerase III transcription by RB, p53 and c-Myc. Cell Cycle. 2003;2:181–184. [PubMed] [Google Scholar]
- 6.Shor B, Wu J, Shakey Q, Toral-Barza L, Shi C, Follettie M, Yu K. Requirement of the mTOR kinase for the regulation of Maf1 phosphorylation and control of RNA polymerase III-dependent transcription in cancer cells. J Biol Chem. 2010;285:15380–15392. doi: 10.1074/jbc.M109.071639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ. mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc Natl Acad Sci U S A. 2010;107:11823–11828. doi: 10.1073/pnas.1005188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR. Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- 9.Sisodia SS, Sollner-Webb B, Cleveland DW. Specificity of RNA maturation pathways: RNAs transcribed by RNA polymerase III are not substrates for splicing or polyadenylation. Mol Cell Biol. 1987;7:3602–3612. doi: 10.1128/mcb.7.10.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strack RL, Disney MD, Jaffrey SR. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat RNA. Nature Methods. 2013;10:1219–1224. doi: 10.1038/nmeth.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filonov GS, Moon JD, Svensen N, Jaffrey SR. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J Am Chem Soc. 2014;136:16299–16308. doi: 10.1021/ja508478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolgosheina EV, Jeng SC, Panchapakesan SS, Cojocaru R, Chen PS, Wilson PD, Hawkins N, Wiggins PA, Unrau PJ. RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem Biol. 2014;9:2412–2420. doi: 10.1021/cb500499x. [DOI] [PubMed] [Google Scholar]
- 14.Wang PC, Querard J, Maurin S, Nath SS, Le Saux T, Gautier A, Jullien L. Photochemical properties of Spinach and its use in selective imaging. Chem Sci. 2013;4:2865–2873. [Google Scholar]
- 15.Han KY, Leslie BJ, Fei JY, Zhang JC, Ha T. Understanding the Photophysics of the Spinach-DFHBI RNA Aptamer-Fluorogen Complex To Improve Live-Cell RNA Imaging. Journal of the American Chemical Society. 2013;135:19033–19038. doi: 10.1021/ja411060p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You M, Jaffrey SR. Structure and Mechanism of RNA Mimics of Green Fluorescent Protein. Annual review of biophysics. 2015;44:187–206. doi: 10.1146/annurev-biophys-060414-033954. [DOI] [PubMed] [Google Scholar]
- 17.Warner KD, Chen MC, Song W, Strack RL, Thorn A, Jaffrey SR, Ferre-D’Amare AR. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat Struct Mol Biol. 2014;21:658–663. doi: 10.1038/nsmb.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang H, Suslov NB, Li NS, Shelke SA, Evans ME, Koldobskaya Y, Rice PA, Piccirilli JA. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat Chem Biol. 2014;10:686–691. doi: 10.1038/nchembio.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci U S A. 2000;97:11990–11995. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wall MA, Socolich M, Ranganathan R. The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nat Struct Biol. 2000;7:1133–1138. doi: 10.1038/81992. [DOI] [PubMed] [Google Scholar]
- 21.Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-A resolution. Proc Natl Acad Sci U S A. 2001;98:462–467. doi: 10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song W, Strack RL, Svensen N, Jaffrey SR. Plug-and-play fluorophores extend the spectral properties of Spinach. J Am Chem Soc. 2014;136:1198–1201. doi: 10.1021/ja410819x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chudakov DM, Matz MV, Lukyanov S, Lukyanov KA. Fluorescent Proteins and Their Applications in Imaging Living Cells and Tissues. Physiol Rev. 2010;90:1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 24.Stoltenburg R, Reinemann C, Strehlitz B. SELEX--a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 26.Ponchon L, Dardel F. Recombinant RNA technology: the tRNA scaffold. Nat Methods. 2007;4:571–576. doi: 10.1038/nmeth1058. [DOI] [PubMed] [Google Scholar]
- 27.Filonov GS, Kam CW, Song W, Jaffrey SR. In-gel imaging of RNA processing using Broccoli reveals optimal aptamer expression strategies. Chem Biol. 2015;22:649–660. doi: 10.1016/j.chembiol.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warner KD, Sjekloća L, Song W, Filonov GS, Jaffrey SR, Ferre-D’Amare AR. A homodimer interface without basepairs in an RNA mimic of red fluorescent protein. Nature Chemical Biology. doi: 10.1038/nchembio.2475. This issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Good PD, Krikos AJ, Li SX, Bertrand E, Lee NS, Giver L, Ellington A, Zaia JA, Rossi JJ, Engelke DR. Expression of small, therapeutic RNAs in human cell nuclei. Gene Ther. 1997;4:45–54. doi: 10.1038/sj.gt.3300354. [DOI] [PubMed] [Google Scholar]
- 30.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nature biotechnology. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 31.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Paul CP, Good PD, Li SX, Kleihauer A, Rossi JJ, Engelke DR. Localized expression of small RNA inhibitors in human cells. Mol Ther. 2003;7:237–247. doi: 10.1016/s1525-0016(02)00038-2. [DOI] [PubMed] [Google Scholar]
- 33.Perry RP, Kelley DE. Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J Cell Physiol. 1970;76:127–139. doi: 10.1002/jcp.1040760202. [DOI] [PubMed] [Google Scholar]
- 34.Neshat MS, Mellinghoff IK, Tran C, Stiles B, Thomas G, Petersen R, Frost P, Gibbons JJ, Wu H, Sawyers CL. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc Natl Acad Sci U S A. 2001;98:10314–10319. doi: 10.1073/pnas.171076798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Martinez JM, Moran J, Clarke RG, Gray A, Cosulich SC, Chresta CM, Alessi DR. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem J. 2009;421:29–42. doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 37.Bonhoure N, Byrnes A, Moir RD, Hodroj W, Preitner F, Praz V, Marcelin G, Chua SC, Jr, Martinez-Lopez N, Singh R, Moullan N, Auwerx J, Willemin G, Shah H, Hartil K, Vaitheesvaran B, Kurland I, Hernandez N, Willis IM. Loss of the RNA polymerase III repressor MAF1 confers obesity resistance. Genes Dev. 2015;29:934–947. doi: 10.1101/gad.258350.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong J, Abulwerdi F, Baldridge A, Kowalik J, Solntsev KM, Tolbert LM. Isomerization in fluorescent protein chromophores involves addition/elimination. Journal of the American Chemical Society. 2008;130:14096–14098. doi: 10.1021/ja803416h. [DOI] [PubMed] [Google Scholar]
- 39.Subach FV, Verkhusha VV. Chromophore transformations in red fluorescent proteins. Chemical reviews. 2012;112:4308–4327. doi: 10.1021/cr2001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, Davidson MW, Wang J. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nature methods. 2013;10:407–409. doi: 10.1038/nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files.