SUMMARY
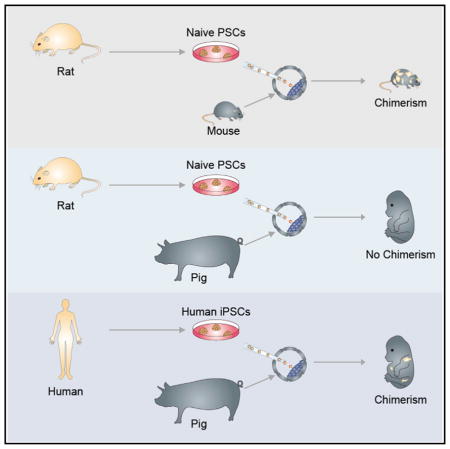
Interspecies blastocyst complementation enables organ-specific enrichment of xenogenic pluripotent stem cell (PSC) derivatives. Here, we establish a versatile blastocyst complementation platform based on CRISPR-Cas9-mediated zygote genome editing and show enrichment of rat PSC-derivatives in several tissues of gene-edited organogenesis-disabled mice. Besides gaining insights into species evolution, embryogenesis, and human disease, interspecies blastocyst complementation might allow human organ generation in animals whose organ size, anatomy, and physiology are closer to humans. To date, however, whether human PSCs (hPSCs) can contribute to chimera formation in non-rodent species remains unknown. We systematically evaluate the chimeric competency of several types of hPSCs using a more diversified clade of mammals, the ungulates. We find that naïve hPSCs robustly engraft in both pig and cattle pre-implantation blastocysts but show limited contribution to post-implantation pig embryos. Instead, an intermediate hPSC type exhibits higher degree of chimerism and is able to generate differentiated progenies in post-implantation pig embryos.
In Brief
Human pluripotent stem cells robustly engraft into both cattle and pig pre-implantation blastocysts, but show limited chimeric contribution to post-implantation pig embryos.
INTRODUCTION
Embryonic pluripotency has been captured in vitro at a spectrum of different states, ranging from the naive state, which reflects unbiased developmental potential, to the primed state, in which cells are poised for lineage differentiation (Weinberger et al., 2016; Wu and Izpisua Belmonte, 2016). When attempting to introduce cultured pluripotent stem cells (PSCs) into a developing embryo of the same species, recent studies demonstrated that matching developmental timing is critical for successful chimera formation. For example, naive mouse embryonic stem cells (mESCs) contribute to chimera formation when injected into a blastocyst, whereas primed mouse epiblast stem cells (mEpiSCs) efficiently engraft into mouse gastrula-stage embryos, but not vice versa (Huang et al., 2012; Wu et al., 2015). Live rodent interspecies chimeras have also been generated using naive PSCs (Isotani et al., 2011; Kobayashi et al., 2010; Xiang et al., 2008). However, it remains unclear whether naive PSCs can be used to generate chimeras between more distantly related species.
The successful derivation of human PSCs (hPSCs), including ESCs from pre-implantation human embryos (Reubinoff et al., 2000; Thomson et al., 1998), as well as the generation of induced pluripotent stem cells (iPSCs) from somatic cells through cellular reprograming (Takahashi et al., 2007; Park et al., 2008; Wernig et al., 2007; Yu et al., 2007; Aasen et al., 2008), has revolutionized the way we study human development and is heralding a new age of regenerative medicine. Several lines of evidence indicate that conventional hPSCs are in the primed pluripotent state, similar to mEpiSCs (Tesar et al., 2007; Wu et al., 2015). A number of recent studies have also reported the generation of putative naive hPSCs that molecularly resemble mESCs (Gafni et al., 2013; Takashima et al., 2014; Theunissen et al., 2014). These naive hPSCs have already provided practical and experimental advantages, including high single-cell cloning efficiency and facile genome editing (Gafni et al., 2013). Despite these advances, it remains unclear how the putative higher developmental potential of naive hPSCs can be used to better understand human embryogenesis and to develop regenerative therapies for treating patients.
Like naive rodent PSCs, naive hPSCs can potentially be used to generate interspecies chimeras for studying human development and disease, and producing functional human tissues via interspecies blastocyst complementation. To date, however, all reported attempts on generating hPSC-derived interspecies chimeras have used the mouse as the host animal, and the results obtained suggest that this process is rather inefficient (Gafni et al., 2013; Theunissen et al., 2014, 2016). Although the mouse is one of the most important experimental models for stem cell research, there are considerable differences between humans and mice (e.g., early post-implantation development, embryo size, gestational length, and developmental speed), which may hinder not only the efficiency but also the usefulness of human-mouse chimeric studies. Thus, expanding the repertoire of host species may complement this incipient but promising area of research in the field of regenerative medicine. In particular, interspecies chimera research of hPSCs using ungulates, e.g., pigs, cattle, and sheep, could lead to improved research models, as well as novel in vivo strategies for (1) generating human organs and tissues, (2) designing new drug screening methodologies, and (3) developing new human disease models (Wu and Izpisua Belmonte, 2015). Experiments to empirically test and evaluate the chimeric contribution of various types of hPSCs in the ungulates are thus imperative, but currently lacking. To start filling this void, we tested different types of hPSCs for their chimeric contribution potential in two ungulate species, pigs and cattle.
RESULTS
Naive Rat PSCs Robustly Contribute to Rat-Mouse Interspecies Chimera Formation
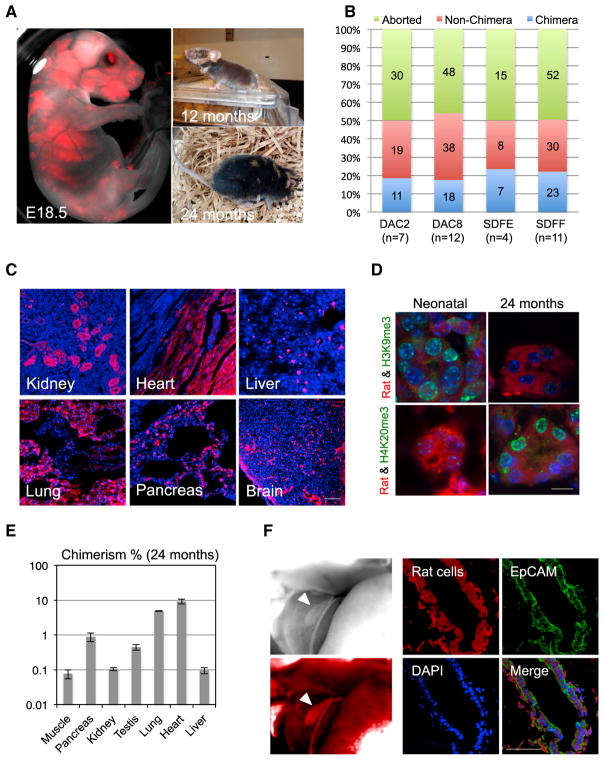
We first used rodent models to gain a better understanding of the factors and caveats underlying interspecies chimerism with PSCs. To this end, we used two chimeric-competent rat ESC lines, DAC2 and DAC8 (Li et al., 2008). We labeled both lines with a fluorescent marker, humanized kusabira orange (hKO), for cell tracking and injected them into mouse blastocysts. Following embryo transfer (ET) into surrogate mouse mothers, both DAC2 and DAC8 lines gave rise to live rat-mouse chimeras (Figures 1A and S1A). Many of the chimeras developed into adulthood, and one chimera reached 2 years of age (Figure 1A), indicating that the xenogeneic rat cells sustained the physiological requirements of the mouse host without compromising its life span. We also generated two rat iPSC lines (SDFE and SDFF) from tail tip fibroblasts (TTFs) isolated from a neonatal Sprague-Dawley rat and used them to generate rat-mouse chimeras. Similar to rat ESCs, rat iPSCs could also robustly contribute to chimera formation in mice (Figure S1B). Overall, the chimera forming efficiencies of all rat PSC lines tested were ~20%, consistent with a previous report (Figure 1B) (Kobayashi et al., 2010).
Figure 1. Interspecies Rat-Mouse Chimeras Derived from Rat PSCs.
(A) Rat-mouse chimeras generated by rat ESCs (DAC2). Left, an E18.5 rat-mouse chimeric fetus. Red, hKO-labeled rat cells. Right, a 12-month-old (top) and 24-month-old (bottom) rat-mouse chimera.
(B) Chimera forming efficiencies with rat ESC lines (DAC2 and DAC8) and rat iPSC lines (SDFE and SDFF). n, number of embryo transfers.
(C) Representative fluorescence images showing hKO-labeled rat ESCs (DAC2) contributed to different tissues in the 24-month-old rat-mouse chimera. Red, hKO-labeled rat cells. Blue, DAPI. Scale bar, 100 μm.
(D) Representative immunofluorescence images showing the expression of aging-related histone marks, including H3K9me3 and H4K20me3, in the kidney tissue of neonatal and 24-month-old chimeras. Scale bar, 10 μm.
(E) Levels of chimerism of rat ESCs (DAC2) in different tissues of the 24-month-old rat-mouse chimera. Error bars indicate SD.
(F) Rat iPSCs (SDFE) contributed to the neonatal mouse gall bladder. Left, bright-field (top) and fluorescence (bottom) images showing a neonatal mouse gallbladder contained cells derived from rat iPSCs. White arrowheads indicate the gallbladder. Right, representative immunofluorescence images showing the expression of a gallbladder epithelium marker (EpCAM) by rat cells. Red, hKO-labeled rat cells; blue, DAPI. Scale bar, 50 μm.
We observed contribution of rat cells to a wide range of tissues and organs in both neonatal and aged rat-mouse chimeras (Figures 1C, S1A, and S1B). We examined aging-related histone marks in both neonatal and aged chimeras and found that the 2-year-old chimera exhibited histone signatures characteristic of aging (Figure 1D). We quantified the degree of chimerism in different organs of the aged chimera via quantitative qPCR analysis of genomic DNA using a rat-specific primer (Table S2). We found that different tissues contained different percentages of rat cells, with the highest contribution observed in the heart (~10%) (Figure 1E).
One anatomical difference between mice and rats is that rats lack a gallbladder. In agreement with a previous report (Kobayashi et al., 2010), we also observed the presence of gallbladders in rat-mouse chimeras (chimeras derived from injecting rat PSCs into a mouse blastocyst). Interestingly, rat cells contributed to the chimeric gallbladder and expressed the gallbladder epithelium marker EpCAM (Figures 1F and S1C), which suggests that the mouse embryonic microenvironment was able to unlock a gallbladder developmental program in rat PSCs that is normally suppressed during rat development.
A Versatile CRISPR-Cas9-Mediated Interspecies Blastocyst Complementation System
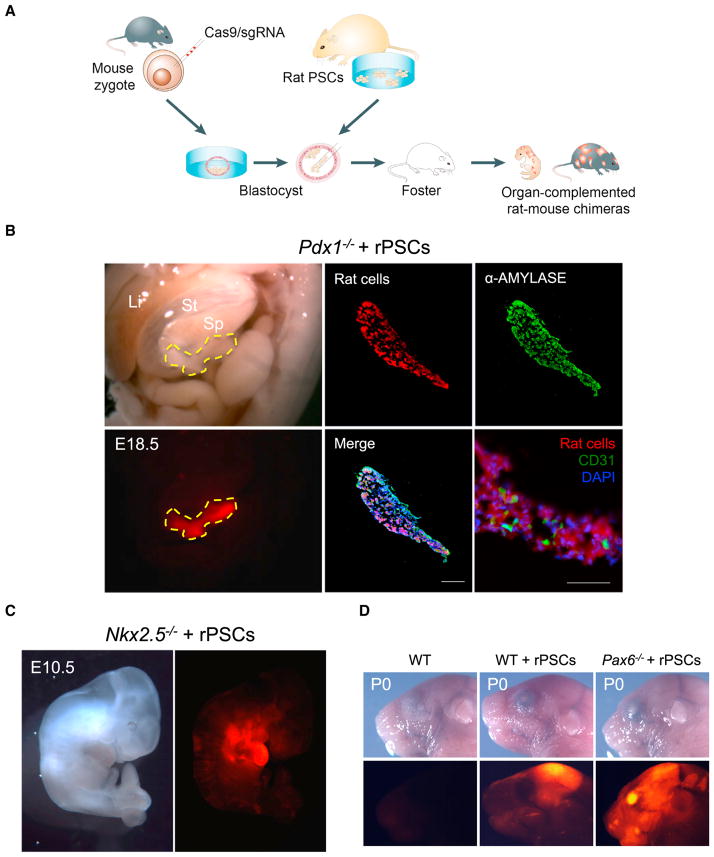
Chimeric contribution of PSCs is random and varies among different host blastocysts and donor cell lines used. To selectively enrich chimerism in a specific organ, a strategy called blastocyst complementation has been developed where the host blastocysts are obtained from a mutant mouse strain in which a gene critical for the development of a particular lineage is disabled (Chen et al., 1993; Kobayashi et al., 2010; Wu and Izpisua Belmonte, 2015). Mutant blastocysts used for complementation experiments were previously obtained from existing lines of knockout mice, which were generated by gene targeting in germ-line-competent mouse ESCs—a time-consuming process. To relieve the dependence on existing mutant strains, we developed a blastocyst complementation platform based on targeted genome editing in zygotes. We chose to use the CRISPR-Cas9 system, which has been harnessed for the efficient generation of knockout mouse models (Wang et al., 2013) (Figure 2A).
Figure 2. Interspecies Blastocyst Complementation via CRISPR-Cas9-Mediated Zygote Genome Editing.
(A) Schematic of the CRISPR-Cas9 mediated rat-mouse blastocyst complementation strategy.
(B) Left, bright-field (top) and fluorescence (bottom) images showing the enrichment of rat cells in the pancreas of an E18.5 Pdx1−/− mouse. Li, liver; St, stomach; Sp, spleen. Yellow-dotted line encircles the pancreas. Red, hKO-labeled rat cells. Middle and right (top), representative immunofluorescence images showing rat cells expressed α-amylase in the Pdx1−/− mouse pancreas. Blue, DAPI. Right (bottom), a representative immunofluorescence image showing that some pancreatic endothelial cells, as marked by a CD31 antibody, were not derived from rat PSCs. Scale bar, 100 μm.
(C) Bright field (left) and fluorescence (right) images showing the enrichment of rat cells in the heart of an E10.5 Nkx2.5−/− mouse. Red, hKO-labeled rat cells.
(D) Bright field (top) and fluorescence (bottom) images showing the enrichment of rat cells in the eye of a neonatal Pax6−/− mouse. Red, hKO-labeled rat cells. WT, mouse control; WT+rPSCs, control rat-mouse chimera without Cas9/sgRNA injection.
For proof-of-concept, we knocked out the Pdx1 gene in mouse by co-injecting Cas9 mRNA and Pdx1 single-guide RNA (sgRNA) into mouse zygotes. During mouse development, Pdx1 expression is restricted to the developing pancreatic anlagen and is a key player in pancreatic development. Mice homozygous for a targeted mutation in Pdx1 lack a pancreas and die within a few days after birth (Jonsson et al., 1994; Offield et al., 1996). Similarly, Pdx1−/− mice generated by the zygotic co-injection of Cas9 mRNA and Pdx1 sgRNA were apancreatic, whereas other internal organs appeared normal (Figure S2A). These mice survived only a few days after birth. We observed the efficiency for obtaining Pdx1−/− mouse via CRISPR-Cas9 zygote genome editing was ~60% (Figure S2F). Next, we combined zygotic co-injection of Cas9/sgRNA with blastocyst injection of rat PSCs, and found that rat PSC-derivatives were enriched in the neonatal pancreas of Pdx1 −/− mice and expressed α-AMYLAYSE, a pancreatic enzyme that helps digest carbohydrates (Figures 2B and S2B). Of note is that in these animals the pancreatic endothelial cells were still mostly of mouse origin, as revealed by staining with an anti-CD31 antibody (Figure 2B). Importantly, pancreas enriched with rat cells supported the successful development of Pdx1−/− mouse host into adulthood (>7 months), and maintained normal serum glucose levels in response to glucose loading, as determined using the glucose tolerance test (GTT) (Figure S2C).
Taking advantage of the flexibility of the CRISPR-Cas9 zygotic genome editing, we next sought to enrich xenogenic rat cells toward other lineages. Nkx2.5 plays a critical role in early stages of cardiogenesis, and its deficiency leads to severe growth retardation with abnormal cardiac looping morphogenesis, an important process that leads to chamber and valve formation (Lyons et al., 1995; Tanaka et al., 1999). Mice lacking Nkx2.5 typically die around E10.5 (Lyons et al., 1995; Tanaka et al., 1999). Consistent with previous observations, CRISPR-Cas9 mediated inactivation of Nkx2.5 resulted in marked growth-retardation and severe malformation of the heart at E10.5 (Figure S2D). In contrast, when complemented with rat PSCs, the resultant Nkx2.5−/− mouse hearts were enriched with rat cells and displayed a normal morphology, and the embryo size was restored to normal (Figures 2C and S2D). Of note is that although rat PSCs rescued embryo growth and cardiac formation in E10.5 Nkx2.5−/− mouse embryos, to date we still have not obtained a live rescued chimera (n = 12, where n is the number of ETs). Pax6 is a transcription factor that plays key roles in development of the eye, nose and brain. Mice homozygous for a Pax6 loss-of-function mutation lack eyes, nasal cavities, and olfactory bulbs, and exhibit abnormal cortical plate formation, among other phenotypes (Gehring and Ikeo, 1999). Pax6 is best known for its conserved function in eye development across all species examined (Gehring and Ikeo, 1999). In agreement with the published work, CRISPR-Cas9 mediated Pax6 inactivation disrupted eye formation in the E15.5 mouse embryo (Figure S2E). When complemented with rat PSCs, we observed the formation of chimeric eyes enriched with rat cells in Pax6−/− mouse neonate (Figures 2D and S2E). Similar to Pdx1−/−, we observed efficient generation of homozygous Nkx2.5−/− and Pax6−/− mouse embryos via zygotic co-injection of Cas9 mRNA and sgRNAs (Figure S2F). All DNA sequencing results of CRISPR-Cas9 mediated gene knockouts and gRNA sequences are summarized in Tables S1 and S2, respectively.
In sum, for the pancreas, heart, and eye, as well as several other organs (data not shown), we successfully generated chimerized organs that were enriched with rat cells, demonstrating the efficacy and versatility of the CRISPR-Cas9 mediated inter-species blastocyst complementation platform.
Naive Rodent PSCs Do Not Contribute to Chimera Formation in Pigs
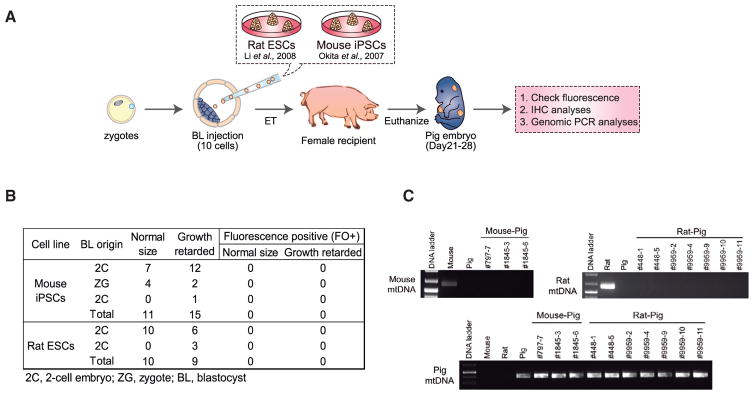
It is commonly accepted that the key functional feature of naive PSCs is their ability to generate intraspecies germline chimeras (Nichols and Smith, 2009). Studies in rodents also support the notion that attaining the naive pluripotent state is the key step in enabling chimera formation across species boundaries (Xiang et al., 2008; Isotani et al., 2011; Kobayashi et al., 2010). However, it has not yet been tested whether naive rodent PSCs can contribute to chimera formation when using a non-rodent host. To further examine the relationship between naive PSCs and interspecies chimerism, we injected rat ESCs into pig blastocysts followed by ET to recipient sows. In addition to rat ESCs, we also used a germline competent mouse iPSC line (Okita et al., 2007). Several criteria were used to determine the chimeric contribution of rodent cells in pig embryos, namely, (1) detection of fluorescence (hKO) signal, (2) immunohistochemical (IHC) labeling of embryo sections with an anti-hKO antibody, and (3) genomic PCR with mouse- or rat-specific primers targeting mitochondrial DNA (mtDNA) (Figure 3A). We terminated the pregnancy between day 21–28 of pig development and collected embryos derived from the injection of mouse iPSCs or rat ESCs into pig blastocyst (26 and 19 embryos, respectively) (Figure 3B; Table S3). We failed to detect any hKO signal in both normal size and growth retarded embryos (Figure 3B). We next sectioned the pig embryos and stained them with an antibody against hKO. Similarly, we did not detect any hKO-positive cells in the embryonic sections examined (data not shown). Finally, we employed a more sensitive test, using genomic PCR to amplify rat- or mouse-specific mtDNA sequences (pig-specific mtDNA primers served as the loading control) (Table S2). Consistently, genomic PCR analyses did not detect any rodent contribution to the pig embryos (Figure 3C). Taken together, although naive rodent PSCs can robustly contribute to rodent-specific interspecies chimeras, our results show that these cells are incapable of contributing to normal embryonic development in pigs.
Figure 3. Naive Rodent PSCs Fail to Contribute to Chimera Formation in Pigs.
(A) Schematic of the generation and analyses of post-implantation pig embryos derived from blastocyst injection of naive rodent PSCs.
(B) Summary of the pig embryos recovered between day 21–28 of pregnancy.
(C) Genomic PCR analyses of pig embryos derived from blastocyst injection of mouse iPSCs or rat ESCs. Mouse- and rat- specific mtDNA primers were used for the detection of chimeric contribution from mouse iPSCs and rat ESCs, respectively. Pig-specific mtDNA primers were used for the control.
Generation of Naive, Intermediate, and Primed hiPSCs
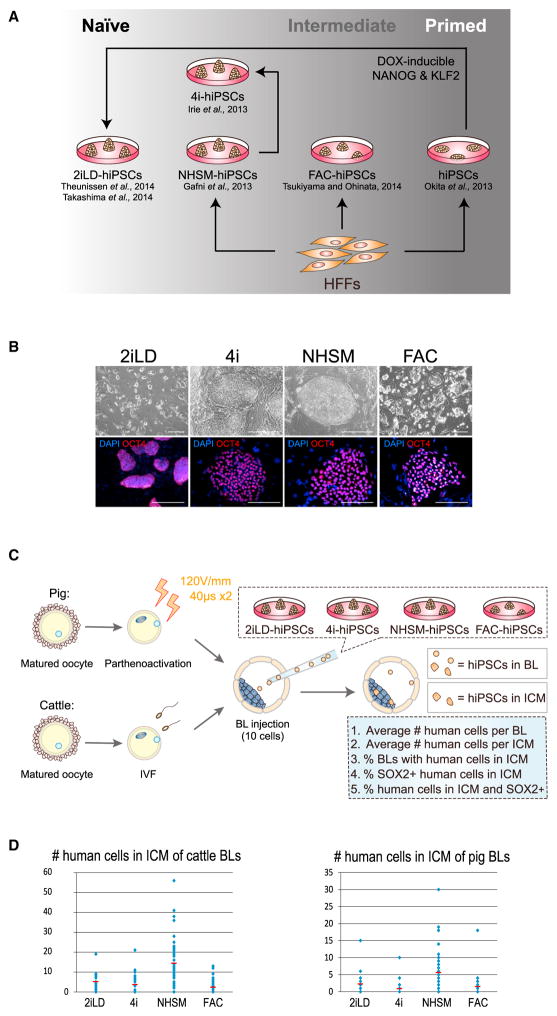
Next, we sought to systematically evaluate the chimeric competency of hPSCs in ungulate embryos. We generated hiPSCs using several reported naive PSC culture methods, a culture protocol supporting a putative intermediate pluripotent state between naive mESCs and primed mEpiSCs (Tsukiyama and Ohinata, 2014), and a primed culture condition (Figure 4A). Mouse ground state culture condition (2iL) induces the differentiation of primed hPSCs. However, when combined with the forced expression of NANOG and KLF2 (NK2), transcription factors that help to maintain murine naive pluripotency, 2iL culture can stabilize hPSCs in an immature state (Takashima et al., 2014; Theunissen et al., 2014). We generated doxycycline (DOX)-inducible NK2-expressing naive hiPSCs cultured in 2iL medium from primed hiPSCs (2iLD-hiPSCs). Transgene-free primed hiPSCs were reprogramed from human foreskin fibroblasts (HFFs) using episomal vectors (Okita et al., 2011). For comparison, we also generated naive hiPSCs from HFFs using the NHSM culture condition (Gafni et al., 2013) (NHSM-hiPSCs). It has been shown that cells grown in 4i medium, a simplified version of NHSM, have a significant potential for germ cell induction, a distinguishing feature between naive mESCs and primed mEpiSCs (Irie et al., 2015). Thus, we also culture-adapted NHSM-hiPSCs in 4i medium (4i-hiPSCs), resulting in stable 4i-hiPSCs with similar morphological and molecular characteristics to parental NHSM-hiPSCs (Figure 4B). In addition, we generated another type of hiPSC by direct reprogramming of HFFs in a modified mEpiSC medium containing bFGF, Activin-A, and CHIR99021 (FAC; Figure 4A). mEpiSCs cultured in FAC medium exhibited features characteristic of both naive mESCs and primed mEpiSCs, supporting an intermediate pluripotent state (Tsukiyama and Ohinata, 2014). hiPSCs generated and cultured in FAC medium (FAC-hiPSCs) displayed a colony morphology intermediate between that of 2iLD- and primed hiPSCs, with less defined borders (Figure 4B). 2iLD-hiPSCs, NHSM-hiPSCs, 4i-hiPSCs, and FAC-hiPSCs could all be stably maintained long term in culture, preserving normal karyotypes and the homogeneous, nuclear localization of OCT4 protein (Figure 4B; data not shown). Notably, similar to hiPSCs grown in naive cultures (2iLD-hiPSCs, NHSM-hiPSCs, 4i-hiPSCs), FAC-hiPSCs could also be efficiently propagated by single-cell dissociation without using a ROCK kinase inhibitor. After injecting cells into the kidney capsule of immunodeficient NSG mice, all of these hiPSCs formed teratomas that consisted of tissues from all three germ layers: endoderm, mesoderm, and ectoderm (Figure S3A). To facilitate the identification of human cells in subsequent chimera experiments, we labeled hiPSCs with either green fluorescence protein (GFP) or hKO fluorescence markers.
Figure 4. Generation and Interspecies ICM Incorporation of Different Types of hiPSCs.
(A) Schematic of the strategy for generating naive, intermediate, and primed hiPSCs.
(B) (Top) Representative bright-field images showing the colony morphologies of naive (2iLD-, 4i-, and NHSM-hiPSCs) and intermediate (FAC-hiPSCs) hiPSCs. Bottom, representative immunofluorescence images of naive and intermediate hiPSCs stained with an anti-OCT4 antibody. Red, OCT4; blue, DAPI. Scale bar, 100 μm.
(C) Schematic of the experimental procedures for producing cattle and pig blastocysts obtained from in vitro fertilization (IVF) and parthenoactivation, respectively. Blastocysts were subsequently used for laser-assisted blastocyst injection of hiPSCs. After hiPSC injection, blastocysts were cultured in vitro for 2 days before fixation and analyzed by immunostaining with an anti-HuNu and an anti-SOX2 antibodies. Criteria to evaluate the survival of human cells, as well as the degree and efficiency of ICM incorporation are shown in the blue box.
(D) Number of hiPSCs that integrated into the cattle (left) and pig (right) ICMs after ten hiPSCs were injected into the blastocyst followed by 2 days of in vitro culture. Red line, the average number of ICM-incorporated hiPSCs. Blue dot, the number of ICM-incorporated hiPSCs in each blastocyst.
Chimeric Contribution of hiPSCs to Pig and Cattle Blastocysts
The ability to integrate into the inner cell mass (ICM) of a blastocyst is informative for evaluating whether hiPSCs are compatible with pre-implantation epiblasts of the ungulate species. This is also one of the earliest indicators of chimeric capability. We therefore evaluated interspecies chimeric ICM formation by injecting hiPSCs into blastocysts from two ungulate species, pig and cattle.
Cattle-assisted reproductive technologies, such as in vitro embryo production, are well established given the commercial benefits of improving the genetics of these animals. Cattle also serve as a research model because of several similarities to human pre-implantation development (Hansen, 2014; Hasler, 2014). Using techniques for producing cattle embryos in vitro, we developed a system for testing the ability and efficiency of hiPSCs to survive in the blastocyst environment and to integrate into the cattle ICM (Figure 4C). Cattle embryos were obtained by in vitro fertilization (IVF) using in vitro matured oocytes collected from ovaries obtained from a local slaughterhouse. The tightly connected cells of the blastocyst trophectoderm from large livestock species, such as pig and cattle, form a barrier that complicates cell microinjection into the blastocoel. Thus, microinjection often results in embryo collapse and the inability to deposit the cells into the embryo. To facilitate cell injection we employed a laser-assisted approach, using the laser to perforate the zona pellucida and to induce damage to a limited number of trophectoderm cells. This allowed for easy access into the blastocyst cavity for transferring the human cells (Figure S3B). Furthermore, the zona ablation and trophectoderm access allowed use a blunt-end pipette for cell transfer, thus minimizing further embryo damage. This method resulted in a nearly 100% injection effectiveness and >90% embryo survival.
To determine whether hiPSCs could engraft into the cattle ICM, we injected ten cells from each condition into cattle blastocysts collected 7 days after fertilization. After injection, we cultured these blastocysts for additional 2 days before analysis. We used several criteria to evaluate the chimeric contribution of hiPSCs to cattle blastocysts: (1) average number of human cells in each blastocyst, (2) average number of human cells in each ICM, (3) percentage of blastocysts with the presence of human cells in the ICM, (4) percentage of SOX2+ human cells in the ICM, and (5) percentage of human cells in the ICM that are SOX2+ (Figure 4C). Our results indicated that both naive and intermediate (but not primed) hiPSCs could survive and integrate into cattle ICMs, albeit with variable efficiencies (Figures 4D and S3C–S3E; Table S4). Compared with other cell types, 4i-hiPSCs exhibited the best survival (22/23 blastocysts contained human cells), but the majority of these cells lost SOX2 expression (only 13.6% of human cells remained SOX2+). On average, 3.64 4i-hiPSCs were incorporated into the ICM. NHSM-hiPSCs were detected in 46 of 59 injected blastocysts, with 14.41 cells per ICM. Of these, 89.7% remained SOX2+. For 2iLD-hiPSCs, 40 of 52 injected blastocysts contained human cells, with 5.11 cells per ICM, and 69.9% of the ICM-incorporated human cells remained SOX2+. FAC-hiPSCs exhibited moderate survival rate (65/101) and ICM incorporation efficiency (39/101), with an average of 2.31 cells incorporated into the ICM, and 89.3% remaining SOX2+.
We also performed ICM incorporation assays by injecting hiPSCs into pig blastocysts. Because certain complications are frequently associated with pig IVF (Abeydeera, 2002; Grupen, 2014) (e.g., high levels of polyspermic fertilization), we used a parthenogenetic activation model, which enabled us to efficiently produce embryos that developed into blastocysts (King et al., 2002). Pig oocytes were obtained from ovaries collected at a local slaughterhouse. Once the oocytes were matured in vitro, we removed the cumulus cells and artificially activated the oocytes using electrical stimulation. They were then cultured to blastocyst stage (Figure 4C). We injected ten hiPSCs into each pig parthenogenetic blastocyst and evaluated their chimeric contribution after 2 days of in vitro culture (Figures 4C and S3C–S3E; Table S4). Similar to the results in cattle, we found that hiPSCs cultured in 4i and NHSM media survived better and yielded a higher percentage of blastocysts harboring human cells (28/35 and 37/44, respectively). Also, among all blastocysts containing human cells, we observed an average of 9.5 cells per blastocyst for 4i-hiPSCs and 9.97 cells for NHSM-hiPSCs. For NHSM-hiPSCs, 19/44 blastocysts had human cells incorporated into the ICM. In contrast, only 6/35 blastocysts had 4i-hiPSCs localized to the ICM. For 2iLD-hiPSCs, we observed an average of 5.7 cells per blastocyst, with 2.25 human cells localized to the ICM. For FAC-hiPSCs, an average of 3.96 and 1.62 human cells were found in the blastocyst and ICM, respectively. Once incorporated into the ICM, 82.2%, 72%, 60.9%, and 40% of 2iLD-, 4i-, NHSM-, and FAC-hiPSCs, respectively, stained positive for the pluripotency marker SOX2. These results indicate that both naive and intermediate hiPSCs seem to perform better when injected into cattle than pig blastocysts. This suggests a different in vivo blastocyst environment in pig and cattle, with the cattle blastocysts providing an environment that is more permissive for hiPSC integration and survival.
Chimeric Contribution of hiPSCs to Post-implantation Pig Embryos
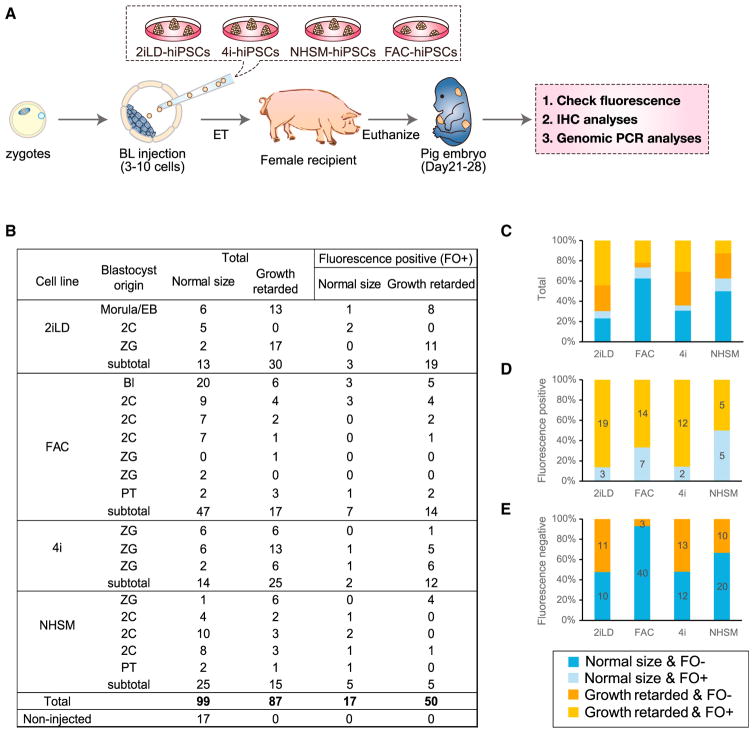
Although ICM incorporation of hiPSCs is the necessary first step to contribute to the embryo proper of host animals, it has limited predictive value for post-implantation chimera formation, as other factors are involved. Next, we investigated if any of the naive and intermediate hiPSCs that we generated, which showed robust ICM incorporation in pre-implantation blastocysts, could contribute to post-implantation development following ET. The pig has certain advantages over cattle for experiments involving post-implantation embryos, as they are a polytocus species, and are commonly used as a translational model given their similarities to humans concerning organ physiology, size, and anatomy. We thus chose the pig for these experiments. Since there was little to no contribution of primed hiPSCs, even at the pre-implantation blastocyst stage, we excluded these cells from the ET experiments. Pig embryos were derived in vivo or through parthenogenesis. A total of 167 embryo donors were used in this study, from which we collected 1,298 zygotes, 1,004 two-cell embryos and 91 morulae (Table S5). Embryos were cultured in vitro until they reached the blastocyst stage (Figures S4AA and S4B). Overall, 2,181 good quality blastocysts with a well-defined ICM were selected for subsequent blastocyst injections, of which 1,052 were derived from zygotes, 897 from two-cell embryos, 91 from morulae, and 141 from parthenogenetic activation (Table S5). We injected 3–10 hiPSCs into the blastocoel of each of these blastocysts (Figures 5A, S4A, and S4C; Table S6). After in vitro embryo culture, a total of 2,075 embryos (1,466 for hiPSCs; Table S6; 477 for rodent PSCs; Table S3) that retained good quality were transferred to surrogate sows. A total of 41 surrogate sows received 30–50 embryos each, resulting in 18 pregnancies (Table S6). Collection of embryos between day 21–28 of development resulted in the harvesting of 186 embryos: 43 from 2iLD-hiPSCs, 64 from FAC-hiPSCs, 39 from 4i-hiPSCs, and 40 from NHSM-hiPSCs (Figures 5B, S4A, S4D, and S4F). In addition, 17 control embryos were collected from an artificially inseminated sow (Figure 5B).
Figure 5. Generation of Post-implantation Human-Pig Chimeric Embryos.
(A) Schematic of the experimental procedures for the generation and analyses of post-implantation pig embryos derived from blastocyst injection of naive and intermediate hiPSCs.
(B) Summary of the pig embryos recovered between day 21–28 of pregnancy.
(C) Bar graph showing proportions of normal size and growth retarded embryos, as well as the proportion of fluorescence-positive and -negative embryos, generated from different types of hiPSCs.
(D) Bar graph showing the proportion of normal size and growth-retarded embryos (among those exhibiting a fluorescence signal) generated from different types of hiPSCs.
(E) Bar graph showing the proportion of normal-sized and growth-retarded embryos (among those without exhibiting a fluorescence signal) generated from different types of hiPSCs.
Following evaluating the developmental status of the obtained embryos, more than half showed retarded growth and were smaller than control embryos (Figures 5B and S4B), as was seen when pig blastocysts were injected with rodent PSCs (Figure 3B). Among different hiPSCs, embryos injected with FAC-hiPSCs were more frequently found to be normal size (Figure 5C). From the recovered embryos, and based on fluorescence imaging (GFP for 2iLD-hiPSCs and FAC-hiPSCs; hKO for 4i-hiPSCs and NHSM-hiPSCs), we observed positive fluorescence signal (FO+) in 67 embryos among which 17 showed a normal size and morphology, whereas the rest were morphologically under-developed (Figures 5B). In contrast, among fluorescence negative embryos we found the majority (82/119) appeared normal size (Figure 5E), suggesting contribution of hiPSCs might have interfered with normal pig development. Closer examination of the underdeveloped embryos revealed that 50 out of 87 were FO+ (Figures 5B). Among all the FO+ embryos the distribution of normal size versus growth retarded embryos for each cell lines was: 3:19 for 2iLD-hiPSCs, 7:14 for FAC-hiPSCs, 2:12 for 4i-hiPSCs, and 5:5 for NHSM-hiPSCs (Figure 5D). Among normal size embryos we found 3/13 from 2iLD-hiPSCs, 7/47 from FAC-hiPSCs, 2/14 from 4i-hiPSCs, and 5/25 from NHSM-hiPSCs that were FO+ (Figure 5B). All normal size FO+ embryos derived from 2iLD-hiPSCs, 4i-hiPSCs, or NHSM-hiPSCs showed a very limited fluorescence signal (Figure S5A). In contrast, normal size FO+ FAC-hiPSC-derived embryos typically exhibited a more robust fluorescence signal (Figures 6A and S5A).
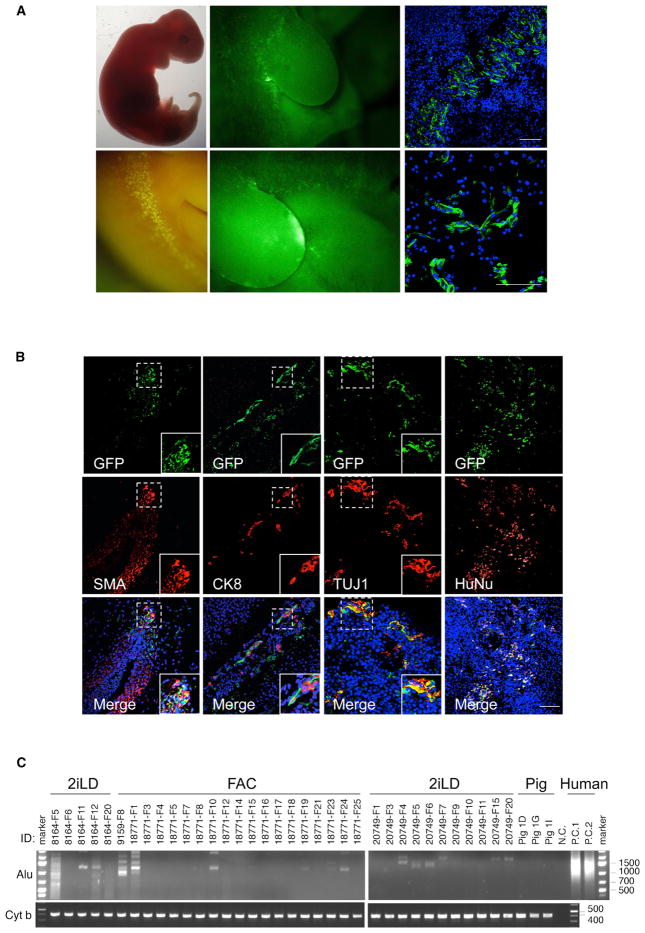
Figure 6. Chimeric Contribution of hiPSCs to Post-implantation Pig Embryos.
(A) Representative bright field (left top) fluorescence (left bottom and middle) and immunofluorescence (right) images of GFP-labeled FAC-hiPSCs derivatives in a normal size day 28 pig embryo (FAC #1). Scale bar, 100 μm.
(B) Representative immunofluorescence images showing chimeric contribution and differentiation of FAC-hiPSCs in a normal size, day 28 pig embryo (FAC #1). FAC-hiPSC derivatives are visualized by antibodies against GFP (top), TUJ1, SMA, CK8 and HuNu (middle). (Bottom) Merged images with DAPI. Insets are higher magnification images of boxed regions. Scale bar, 100 μm.
(C) Representative gel images showing genomic PCR analyses of pig embryos derived from blastocyst injection of 2iLD-iPSCs (surrogates #8164 and #20749) and FAC-hiPSCs (surrogates #9159 and #18771) using a human specific Alu primer. A pig specific primer Cyt b was used for loading control. nc, negative control with no genomic DNA loaded. pc, positive controls with human cells. Pig 1D, 1G, and 1I, pig controls. ID, surrogate and pig embryos.
Detecting fluorescence signal alone is insufficient to claim chimeric contribution of donor hiPSCs to these embryos, as auto-fluorescence from certain tissues and apoptotic cells can yield false positives, especially when chimerism is low. We thus sectioned all normal size embryos deemed positive based on the presence of fluorescence signal and subjected them to IHC analyses with antibodies detecting GFP or hKO. For 2iLD-hiPSC-, 4i-hiPSC-, and NHSM-hiPSC-derived embryos, in agreement with fluorescence signals observed in whole-embryo analysis, we detected only a few hKO- or GFP-positive cells in limited number of sections (Figure S5A). This precluded us from conducting further IHC analysis using lineage markers. For FAC-hiPSC-derived embryos, we confirmed via IHC analysis (using an anti-GFP antibody) that they contained more human cells (Figures 6A, S5A, and S5B). We then stained additional sections using antibodies against TUJ1, EPCAM, SMA, CK8, and HNF3β (Figures 6B and S5C) and observed differentiation of FAC-hiPSCs into different cell lineages. In addition, these cells were found negative for OCT4, a pluripotency marker (data not shown). Moreover, the presence of human cells was further verified with a human-specific HuNu antibody staining (Figure 6B) and a sensitive genomic PCR assay using a human specific Alu sequence primer (Figure 6C; Table S2). Together, these results indicate that naive hiPSCs injected into pig blastocysts inefficiently contribute to chimera formation, and are only rarely detected in post-implantation pig embryos. An intermediate hPSC type (FAC-hiPSCs) showed better chimeric contribution and differentiated to several cell types in post-implantation human-pig chimeric embryos. It should be noted that the levels of chimerism from all hiPSCs, including the FAC-hiPSCs, in pig embryos were much lower when compare to rat-mouse chimeras (Figures 1C, 1E, S1A, and 1B), which may reflect the larger evolutionary distance between human-pig than between rat-mouse.
DISCUSSION
Our study confirms that live rat-mouse chimeras with extensive contribution from naive rat PSCs can be generated. This is in contrast to earlier work in which rat ICMs were injected into mouse blastocysts (Gardner and Johnson, 1973). One possible explanation for this discrepancy is that cultured PSCs acquire artificial features that make them more proliferative and/or better able to survive than embryonic ICM cells, which in turn leads to their more robust xeno-engraftment capability in a mouse host. Rat-mouse chimeras generated by injecting donor rat PSCs into a mouse host were mouse-sized and developed into adulthood with apparently normal appearance and physiology. We further show in this study that a rat-mouse chimera could live a full mouse lifespan (about 2 years) and exhibit molecular signatures characteristic of aged cells. This demonstrates that cells from two different species, which diverged ~18 million years ago, can live in a symbiotic environment and are able to support normal organismal aging. The fact that rat PSCs were able to contribute to the mouse gallbladder, an organ that is absent in the rat, highlights the importance of embryonic niches in orchestrating the specification, proliferation, and morphogenesis of tissues and organs during organismal development and evolutionary speciation (Izpisúa-Belmonte et al., 1992).
Previous interspecies blastocyst complementation experiments generated host embryos by crossing heterozygous mutant mouse strains, which were themselves generated through targeted gene disruption in germline competent ESCs. These experiments are labor intensive and time consuming. Moreover, only ~25% of blastocysts derived from genetic crosses are homozygous mutants, posing a limitation for efficient complementation. CRISPR-Cas9 mediated zygote genome editing offers a faster and more efficient one-step process for generating mice carrying homozygous mutations, thereby providing a robust interspecies blastocyst complementation platform. Additionally, the multiplexing capability of CRISPR-Cas9 (Cong et al., 2013; Yang et al., 2015) could potentially be harnessed for multi-lineage complementation. For example, in the case of the pancreas, one might hope to eliminate both the pancreatic parenchyma and vasculature of the host to generate a more complete xenogeneic pancreas. Despite the advantages, there are several technical limitations of the CRISPR-Cas9 blastocyst complementation system that need to be overcome before unlocking its full potential. First, gene inactivation relies on the error-prone, non-homologous end joining (NHEJ) pathway, which is often unpredictable. In-frame mutations and mosaicism are among the factors that may affect outcomes. A more predictable targeted gene inactivation strategy that utilizes homologous recombination (HR) is still inefficient in the zygote. Second, each embryo must be injected twice when using this system and embryos must be cultured in vitro for several days before ET, thereby compromising embryo quality. Technical advancements that include a more robust gene-disruption strategy (e.g., targeted generation of frameshift mutations via homology independent targeted integration [Suzuki et al., 2016]), alternative CRISPR/Cas9 delivery methods, and improved culture conditions for manipulated embryos will likely help improve and optimize the generation of organogenesis-disabled hosts.
We observed a slower clearance of an intraperitoneally injected glucose load for Pdx1−/− than Pdx1+/− rat-mouse chimeras, while both were slower than wild-type mouse controls (Figure S2C). While this result may seem to contradict a previous report (Kobayashi et al., 2010), the discrepancy is likely due to the development of autoimmune type inflammation that is often observed in adult rat-mouse (chimeras made by injection of rat PSCs into mouse blastocyst, data not shown) (>7 months, this study) and mouse-rat chimeras (chimeras made by injection of mouse PSCs into rat blastocyst; H. Nakauchi, personal communication), which is less evident in young chimeras (~8 weeks; Kobayashi et al. 2010). Interestingly though, we did observe a similarly slower clearance of glucose load in wild-type rats, although the initial spike was much lower in rats compared to mice or chimeras (Figure S2C). Thus, the rat cellular origin might also have played a role in the different GTT responses observed.
Rodent ESCs/iPSCs, considered as the gold standard cells for defining naive pluripotency, can robustly contribute to intra- and inter-species chimeras within rodent species. These and other results have led to the assumption that naive PSCs are the cells of choice when attempting to generate interspecies chimeras involving more disparate species. Here, we show that rodent PSCs fail to contribute to chimera formation when injected into pig blastocysts. This highlights the importance of other contributing factors underlying interspecies chimerism that may include, but not limited to, species-specific differences in epiblast and trophectoderm development, developmental kinetics, and maternal microenvironment.
To date, and taking into consideration all published studies that have used the mouse as the host species, it is probably appropriate to conclude that interspecies chimera formation involving hPSCs is inefficient (De Los Angeles et al., 2015). It has been argued that this apparent inefficiency results from species-specific differences between human and mouse embryogenesis. Therefore, studies utilizing other animal hosts would help address this important question. Here we focused on two species, pig and cattle, from a more diverse clade of mammals and found that naive and intermediate, but not primed, hiPSCs could robustly incorporate into pre-implantation host ICMs. Following ET, we observed, in general and similar to the mouse studies, low chimera forming efficiencies for all hiPSCs tested. Interestingly, injected hiPSCs seemed to negatively affect normal pig development as evidenced by the high proportion of growth retarded embryos. Nonetheless, we observed that FAC-hiPSCs, a putative intermediate PSC type between naive and primed pluripotent states, displayed a higher level of chimerism in post-implantation pig embryos. IHC analyses revealed that FAC-hiPSCs integrated and subsequently differentiated in host pig embryos (as shown by the expression of different lineage markers, and the lack of expression of the pluripotency marker OCT4). Whether the degree of chimerism conferred by FAC-hiPSCs could be sufficient for eliciting a successful inter-species human-pig blastocyst complementation, as demonstrated herein between rats and mice, remains to be demonstrated. Studies and approaches to improve the efficiency and level of hPSC interspecies chimerism (Wu et al., 2016), such as matching developmental timing, providing a selective advantage for donor hPSCs, generating diverse hPSCs with a higher chimeric potential and selecting a species evolutionarily closer to humans, among others parameters, will be needed.
The procedures and observations reported here on the capability of human pluripotent stem cells to integrate and differentiate in a ungulate embryo, albeit at a low level and efficiency, when optimized, may constitute a first step towards realizing the potential of interspecies blastocyst complementation with hPSCs. In particular, they may provide a better understanding of human embryogenesis, facilitate the development and implementation of humanized animal drug test platforms, as well as offer new insights on the onset and progression of human diseases in an in vivo setting. Ultimately, these observations also raise the possibility of xeno-generating transplantable human tissues and organs towards addressing the worldwide shortage of organ donors.
STAR ★ METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-SOX2 | BioGenex | Cat# NU579-UC |
| Mouse anti-HuNu (Clone 235-1) | Millipore | Cat# MAB1281; RRID: AB_11212527 |
| Rabbit anti-monomeric Kusabira-Orange 2 | MBL | Code# PM051M |
| Rabbit anti-GFP | MBL | Code# 598 |
| Rat monoclonal anti-cytokeratin 8/18 (TROMA-I) | DSHB | RRID: AB_531826 |
| Mouse monoclonal anti-epithelial antigen | DAKO | Cat# M0804; RRID: AB_2335685 |
| Mouse monoclonal anti-Ep-CAM | Santa Cruz | Cat# sc-25308; RRID: AB_627531 |
| Mouse monoclonal anti-actin α-smooth muscle (Clone 1A4) | Sigma-Aldrich | Cat# A5228; RRID: AB_262054 |
| Rabbit polyclonal anti-Histone H3 (tri methyl K9) | Abcam | Cat# ab8898; RRID: AB_306848 |
| Rabbit polyclonal anti-Histone H4 (tri methyl K20) | Abcam | Cat# ab9053; RRID: AB_306969 |
| Rat anti-Mouse CD31 (Clone MEC13.3) | BD PharMingen | Cat# 553370; RRID: AB_394816 |
| Mouse monoclonal anti-Oct3/4 (C-10) | Santa Cruz | Cat# sc-5279; RRID: AB_628051 |
| Mouse anti-Tubulin β3 (Clone TUJ1) | BioLegend | Cat# 801202; RRID: AB_10063408 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Y-27632 dihydrochloride | Torcris | Cat# 1254 |
| Recombinant human FGF-basic (FGF2) | Peprotech | Cat# 100-18B |
| Recombinant human LIF | Peprotech | Cat# 300-05 |
| CHIR99021 | Selleckchem | Cat# S2924 |
| PD035901 | Selleckchem | Cat# S1036 |
| L-ascorbic acid 2-phosphate | Sigma-Aldrich | Cat# A8960 |
| Recombinant human IGF-I LR3 | Peprotech | Cat# 100-11R3 |
| Recombinant human TGF-β1 | Peprotech | Cat# 100-21C |
| SB203580 | Selleckchem | Cat# S1076 |
| SP600125 | Selleckchem | Cat# S1460 |
| LDN193189 | Selleckchem | Cat# S2618 |
| Recombinant human/murine/rat Activin A | Peprotech | Cat# 120-14P |
| Doxycycline hyclate (dox) | Stemgent | Cat# 04-0016 |
| Critical Commercial Assays | ||
| MEGAshortscript T7 Transcription Kit | Invitrogen | Cat# AM1354 |
| MEGAclear Transcription Clean-up Kit | Ambion | Cat# AM1908 |
| Cas9 mRNA | Sigma-Aldrich | Cat# CAS9MRNA-1EA |
| P2 Primary Cell 4D-Nucleofector kit | Lonza | Cat# V4XP-2024 |
| NucleoSpin Gel and PCR Clean-up | MACHEREY-NAGEL | Cat# 740609 |
| DNeasy Blood and Tissue kit | QIAGEN | Cat# 69506 |
| PicoPure DNA Extraction Kit | Thermo Fisher Scientific | Cat# KIT0103 |
| SYBR Green PCR Master Mix | Thermo Fisher Scientific | SKU# 4309155 |
| Experimental Models: Cell Lines | ||
| DAC2 | Li et al., 2008 | N/A |
| DAC8 | Li et al., 2008 | N/A |
| SDFE | This paper | N/A |
| SDFF | This paper | N/A |
| iPS-MEF-Ng-20D-17 | RIKEN BRC | Cat# APS001 |
| HFF hiPSCs | This paper | N/A |
| NHSM-hiPSCs | This paper | N/A |
| 2iLD-hiPSCs | This paper | N/A |
| 4i-hiPSCs | This paper | N/A |
| FAC-hiPSCs | This paper | N/A |
| Human foreskin fibroblast (HFF) | ATCC | CRL-2429 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | Stock No: 000664 |
| Mouse: B6D2F1/J | The Jackson Laboratory | Stock No: 100006 |
| Mouse: ICR (CD1) | Envigo | Order code: 030 |
| Mouse: NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ | The Jackson Laboratory | Stock No: 005557 |
| Rat: Sprague Dawley | Envigo | Order code: 002 |
| Pig: Large-White × Landrace | Murcia, Spain | N/A |
| Pig: Pietrain | Murcia, Spain | N/A |
| Pig: Yorkshire | University of California Davis | N/A |
| Pig: Yorkshire cross | University of California Davis | N/A |
| Recombinant DNA | ||
| pCXLE-hOCT3.4-shp53-F | Okita et al., 2011 | Addgene Plasmid #27077 |
| pCXLE-EGFP | Okita et al., 2011 | Addgene Plasmid #27082 |
| pCXLE-hSK | Okita et al., 2011 | Addgene Plasmid #27078 |
| pCXLE-hUL | Okita et al., 2011 | Addgene Plasmid #27080 |
| pFUW-tetO-hKlf2 | This paper | N/A |
| pFUW-tetO-hNanog | This paper | N/A |
| pCAG-IP-humanized Kusabira Orange | This paper | N/A |
| pmKO1-MC1 | MBL | Code No. AM-V0052M |
| LV-tetO-FU-Oct3/4 | Hockemeyer et al., 2008 | Addgene Plasmid #20726 |
| LV-tetO-FU-Sox2 | Hockemeyer et al., 2008 | Addgene Plasmid #20724 |
| LV-tetO-FU-KLF4 | Hockemeyer et al., 2008 | Addgene Plasmid #20725 |
| LV-tetO-FU-Myc | Hockemeyer et al., 2008 | Addgene Plasmid #20723 |
| LV-FUW-M2rtTA | Hockemeyer et al., 2008 | Addgene Plasmid #20342 |
| pEGIP | Zou et al., 2009 | Addgene Plasmid#26777 |
| pMDLg/pRRE | Dull et al., 1998 | Addgene Plasmid #12251 |
| pRSV-Rev | Dull et al., 1998 | Addgene Plasmid #12253 |
| pMD2.G | pMD2.G was a gift from Didler Trono | Addgene Plasmid #12259 |
| Sequence-Based Reagents | ||
| sgRNA sequences, see Table S2 | This paper | N/A |
| Genomic PCR primers, see Table S2 | This paper | N/A |
| DNA sequencing primers, see Table S2 | This paper | N/A |
| Genomic qPCR primers, see Table S2 | This paper | N/A |
| Software and Algorithms | ||
| CRISPR Design | MIT | http://crispr.mit.edu |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to and will be fulfilled by Lead Contact Juan Carlos Izpisua Belmonte (belmonte@salk.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Rodents
All the rodent experiments were performed under the ethical guidelines of the Salk Institute, and animal protocols were reviewed and approved by the Salk Institute Institutional Animal Care and Use Committee (Protocol #12-00021). All mice (C57BL/6J, B6D2F1/J, ICR and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) and rats (Sprague Dawley) used in this study were purchased from Envigo (Harlan) or The Jackson Laboratory, and bred in the animal facility of the Salk Institute.
Pigs
The pig experiments at University of Murcia, Spain, were performed in accordance with Directive 2010/63/EU EEC for animal experiments and were reviewed and approved by the Ethical Committee for Experimentation with Animals (code: 69/2014), the Research Ethics Committee (code: 1086/2015), and the Biosafety Committee (code:) of the University of Murcia, Spain; and by the Murcia Regional Ministry of Agriculture and Water (code: 273.705), and the Murcia Regional Ministry of Health (code: 061015), Spain. Day 21–28 gestation stages were selected to analyze the chimeric contribution of hiPSC to the pig post implantation embryo. Stopping development at this stage will also prevent that embryos with potentially high hiPSC contribution develop a mature central nervous system. The pig experiments in Spain were conducted in two commercial pig farms located in Southeastern Spain (Murcia, Spain) and in the pig experimental unit of the University of Murcia (Murcia, Spain). Weaned crossbred sows (Landrace × Large-White) from the same genetic line (2–6 parity) were used as embryo donors and recipients. The sows were kept individually in crates in a mechanically ventilated confinement facility. The semen donors were sexually mature Pietrain boars (2–3 years of age) housed in climate- controlled individual pens (20–25°C) at a commercial insemination station in Murcia (Spain). Animals had ad libitum access to water and were fed commercial diets according to their nutritional requirements.
Pig experiments performed at the University of California Davis, were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC# 18158) at the University of California, Davis. Experiments involving hiPSCs at the University of California Davis were reviewed and approved by the Stem Cell Research Oversight Committee (SCRO# 1127) and Biological Use Authorization (BUA# R1627). Day 28 or earlier gestation stages were selected to analyze the contribution of hiPSC to the pig post implantation embryo. Stopping development at this stage will also prevent that embryos with potentially high hiPSC contribution develop a mature central nervous system. For pig experiments performed at the University of California Davis, Yorkshire or Yorkshire cross females at 6–8 months of age were used as embryo recipients. The animals were kept in group pens with ad libitum access to water and were fed commercial diets according to their nutritional requirements.
Human iPSC Culture Media
Conventional primed hiPSCs were either cultured on Matrigel (BD Biosciences) coated plates in mTeSR1 medium (StemCell Technologies) or on MEFs in CDF12 medium: DMEM/F12 (Life Technologies, 11330-032), 20% Knockout Serum Replacement (KSR, Life Technologies, 10828), 2mM Glutamax (Life Technolgies, 35050-061), 0.1mM NEAA (Life Technologies, 11140-050), 0.1mM β-mercaptoethanol (GIBCO, 21985) and 4ng/ml FGF2 (Peprotech). 2iL medium contains: N2B27 medium supplemented with human LIF (10ng/ml, Peprotech), 3 μM CHIR99021 (Selleckchem) and 1 μM PD035901 (Selleckchem). To prepare 500ml of NHSM medium we added 500ml KnockOut DMEM (Invitrogen), 5ml Pen-strep (GIBCO), 5ml GlutaMax (GIBCO), 5ml NEAA (GIBCO), 5g AlbumaxI (Invitrogen), 5ml N2 supplement (Invitrogen; 17502048), L-ascorbic acid 2-phosphate (Sigma), 20ng/ml human LIF (Peprotech), 20ng/ml human LR3-IGF1 (Peprotech), 8g/ml FGF2 (Peprotech), 2ng/ml TGFβ1 (Peprotech), 3 μM CHIR99021 (Selleckchem), 1 μM PD0325901 (Selleckchem), 5 μM SB203580 (Selleckchem), 5 μM SP600125 (Selleckchem), 5 μM Y27632 (Torcris) and 0.4 μM LDN193189 (Selleckchem). 4i medium is based on NHSM medium with only the following differences: L-ascorbic acid 2-phosphate, Human LR3-IGF1 and LDN193189 were excluded from 4i medium. TGFβ1 was used at 1 ng/ml and Y27632 at 5 μM. FAC medium contains: DMEM/F12 (Invitrogen, 11320) and Neurobasal medium (Invitrogen; 21103) were mixed at 1:1 ratio; N2 supplement (Invitrogen; 17502048); 1 × B27 supplement (Invitrogen; 17504044); 2 mM GlutaMax; 1% NEAA; 0.1mM β-mercaptoethanol (Sigma); 1 × Pen-Strep; 50 μg/ml BSA (Sigma); 12 ng/ml FGF2, 10 ng/ml Activin-A (Peprotech) and 3 μM CHIR99021.
Culture and maintenance of rat ESCs/iPSCs and mouse iPSCs
Dr. Qilong Ying, University of Southern California, provided rat ESC lines DAC2 and DAC8. Rat iPSCs, SDFE and SDFF, were generated in house with tail tip fibroblasts derived from Sprague Dawley rat (Envigo). Rat ESCs/iPSCs were cultured either on MEFs or FBS-coated plates in rat ESC medium: N2B27 basal medium supplemented with human LIF (10ng/ml, Peprotech), 1.5uM or 3uM CHIR99021 (Selleckchem) and 1uM PD035901 (Selleckchem). Rat ESCs/iPSCs were passaged every 4–5 days at a split ratio of 1:10. The male mouse iPSCs (MEF-Ng-20D-17) were obtained from RIKEN BRC and were cultured either on MEFs or Matrigel-coated plates in mouse ESC medium: N2B27 basal medium supplemented with human LIF (10ng/ml, Peprotech), 3uM CHIR99021 (Selleckchem) and 1uM PD035901 (Selleckchem). Mouse iPSCs were passaged every 3–4 days at a split ratio of 1:20.
METHOD DETAILS
Chemicals Unless Otherwise Indicated, Chemicals Were Obtained from Sigma-Aldrich
Plasmids Construction
For pFUW-tetO-hNanog and pFUW-tetO-hKlf2, PCR-amplified ORF of human NANOG and KLF2 from human H9 ESCs was sequenced and subcloned into the EcoRI site of pFUW-tetO inducible lentivirus vector. To generate pCAG-IP-humanized Kusabira Orange (hKO), as described previously (Hishida et al., 2011), the ORF was amplified from pmKO1-MC1 (MBL) by PCR, sequenced and subcloned into the XhoI/NotI sites of the pCAG-IP.
Rat iPSC Generation
Rat iPSC reprograming was performed as descried previously. Briefly, LV-tetO-FU-Oct3/4 (addgene#20726), LV-tetO-FU-Sox2 (addgene#20724), LV-tetO-FU-KLF4 (addgene#20725), LV-tetO-FU-Myc (addgene#20723) or LV-FUW-M2rtTA (addgene#20342) was transfected into 293FT cells with packaging plasmids. Forty-eight hours after transfection, virus-containing supernatant was collected from transfected 293FT cells and filtered through a 0.45 μm filer. Rat tail-tip fibroblasts were infected with a mixture of collected viruses in the presence of 8 μg/ml of polybrene. One day after infection, culture medium was switched to 2 μg/ml Dox-containing medium to induce the expression of reprogramming factors. The cells were passaged onto mitotically inactivated MEFs at day 4 after infection, and, on the following day, the cells were cultured with Dox-containing rat ESC culture medium. Colonies started to appear at day 9 after infection. Dox was withdrawn at day 13 and individual colonies were picked and transferred to newly prepared MEF plates and further expanded into stable rat iPSC lines.
Human iPSC Generation
Primed hiPSCs, NHSM-hiPSCs and FAC-hiPSCs were generated by reprogramming of human foreskin fibroblasts (HFF, ATCC, CRL-2429) with episomal vectors. Episomal plasmids pCXLE-EGFP (addgene#27082), pCXLE-hOCT3.4-shp53-F (addgene#27077), pCXLE-hSK (addgene#27078), pCXLE-hUL (addgene#27080) were obtained from Addgene. HFFs (2×106) were nucleofected with the episomal vectors using a 4D-Nucleofector (Program EN150, Lonza) with the P2 Primary Cell 4D-Nucleofector kit (Lonza, V4XP-2024). Five days post-nucleofection, HFFs were re-plated onto mitotically inactivated MEFs. The next day the medium was changed to conventional hPSC culture medium (CDF12), NHSM medium or FAC medium. Putative iPSC colonies were picked between day 20 and 30 and transferred to newly prepared MEFs for further cultivation. For the generation of 2iLD-hiPSCs, primed hiPSCs grown on Matrigel in mTeSR1 medium were pre-treated with 10 μM Y-27632 (Torcris) for 24 hr and then dissociated using TrypLE (Invitrogen). Lentivirus was prepared as follows: pFUW-tetO-hNanog and pFUW-tetO-hKlf2 were transfected into 293FT cells with packaging plasmids: pMDLg/pRRE (addgene#12251), pRSV-Rev (addgene#12253) and pMD2.G (addgene#12259). Forty-eight hours after transfection, virus-containing supernatant was collected from transfected 293FT cells and filtered through a 0.45 μm filer. Primed hiPSCs were transduced in suspension with lentivirus in the presence of 10 μM Y-27632 and 6 μg/ml polybrene for 1 hr. After brief centrifugation, the cells were plated on irradiated DR4 MEF feeders (ATCC) in 2iL media containing 10 μM Y-27632 and 2 μg/ml DOX (Stemgent). Three days after transduction, puromycin (1 μg/ml; Invitrogen) was added to the medium. After 7–14 days, dome shaped colonies were manually picked onto fresh MEF feeders and expanded as 2ILD-hiPSCs. Conversion of NHSM-hiPSCs to 4i-hiPSCs was done by simply changing culture medium from NHSM to 4i. Converted cells were passaged for more than 8 times before further analysis and used for subsequent experiments.
Generation of Fluorescently Labeled Rat PSCs and hiPSCs
We used pCAG-IP-humanized Kusabira Orange (hKO) for labeling rat PSCs, mouse iPSCs and hiPSCs (NHSM-hiPSCs), Briefly, 1–2 μg of pCAG-IP-hKO were transfected into 1–2 million of dissociated PSCs using an Amaxia 4D-nucleofector following the protocol recommended by the manufacture. 0.5–1 μg/ml of puromycin (Invitrogen) was added to the culture 2–3 days post-transfection. Drug-resistant colonies were manually picked between 7–14 days and further expanded clonally. To label 2iLD-hiPSCs and FAC-hiPSCs, briefly, pEGIP (addgene#26777) was co-transfected with pMDLg/pRRE, pRSV-Rev and pMD2.G, and packaged and purified as lentiviral vectors according to a published protocol (Kutner et al., 2009). hiPSCs were individualized with Accumax (Innovative Cell Technologies). Cells were transduced in suspension with lentiviral pEGIP vector in the presence of 4 ug/ml polybrene for 1 hr. After brief centrifugation to remove any residual lentiviral vector, the cells were seeded on irradiated DR4 MEF feeders (ATCC) in 2iLD and FAC media. Three days after transduction, 1 ug/ml puromycin (Invitrogen) was added to the medium. After 2 weeks, Drug-resistant colonies were manually picked onto fresh MEF feeders and expanded as EGFP-labeled hiPSC lines.
Mouse Embryo Collection
C57BL/6 female mice (8–10 weeks old) were superovulated by intraperitoneal injection with 5 IU of PMSG (Millipore), followed by injection of 5 IU of hCG (Millipore) 48 hr later. After mating with BDF1 male mice, zygotes were collected the next day at E0.5 in mKSOM-HEPES medium from oviduct and cultured in the mKSOMaa until Cas9/sgRNA microinjection. mKSOMaa medium: NaCl (95 mM), KCl (2.5 mM), KH2PO4 (0.35 mM), MgSO4 (0.2 mM), NaHCO3 (25 mM), CaCl2 (1.71 mM), Na2-EDTA (0.01 mM), L-glutamine (1.0 mM), Na lactate (10 mM), Na pyruvate (0.2 mM), glucose (5.56 mM), essential amino acid (EAA; 10.0 ml/l), non-essential amino acid (NEAA; 5.0ml/l), penicillin (0.06 g/l), streptomycin (0.05 g/l), and BSA (4 g/l). mKSOM-HEPES medium: same amount of chemicals as KSOMaa, but include decreased amount of NaHCO3 (5 mM), HEPES-Na (20 mM), does not include EAA, NEAA, and BSA substituted by PVA (0.1 g/l).
sgRNA Design and In Vitro Transcription
We used the online software (MIT CRISPR Design Tool: http://crispr.mit.edu) to design sgRNAs. The sgRNAs containing T7 promoter were amplified by PCR with the following primers (5′-TAATACGACTCACTATA-G-[19bp sgRNA target sequence]-GTTTTAGAGCTA GAAATAGC-3′ and 5′-AAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCT CTAAAAC-3′) using Q5 High-Fidelity DNA Polymerase (NEB). The PCR product was purified using NucleoSpin Gel and PCR Clean-up (MACHEREY-NAGEL). To prepare sgRNA mRNA, the purified PCR product was in vitro transcribed by MEGAshortscript T7 Transcription Kit (Invitrogen) following the manufacturer’s instruction. Prepared RNAs were purified using MEGAclearTM Kit Purification for Large Scale Transcription Reactions (Ambion) and dissolved in water for embryo transfer (Sigma).
Microinjection of Cas9 mRNA and sgRNAs to Mouse Zygotes
The zygotes show clear two pronuclei were selected and transferred into a 40 μL drop of KSOM-HEPES and placed on an inverted microscope (Olympus, Tokyo, Japan) fitted with micromanipulators (Narishige, Tokyo, Japan). Mixture of Cas9 mRNA (Sigma-Aldrich, 25–100u/g/ml) and sgRNA (25–50 ug/ml) was loaded to a blunt-end micropipette (Sutter Instrument, CA) of 2–3 μm internal diameter (ID) connected to a manual hydraulic oil microinjector (Eppendorf, Hamburg, Germany). Then, the zygotes were secured by a holding pipette and Piezo Micro Manipulator (Prime Tech Ltd, Japan) was used to create a hole in the zona pellucida. The zygote membranes were broken with a single piezo pulse in the lowest intensity and the injection of the mixture RNA was confirmed by the bulge of membrane. Groups of 10–12 zygotes were manipulated simultaneously and each session was limited to 10 min. After microinjection, the zygotes were cultured in the 40 μL droplet of mKSOMaa for 3days in a humidified atmosphere of 5% CO2 in air at 37.0°C.
Microinjection of Rat PSCs to Mouse Blastocysts
The embryos that have obvious blastocoel at E3.5 were defined as blastocysts. Single cell suspensions were added to a 40 μL drop of KSOM-HEPES containing the blastocysts to be injected. Individual cells were collected into a 20 μm ID of micropipette. Six cells were introduced into the blastocoel near the ICM. Groups of 10–12 blastocysts were manipulated simultaneously and each session was limited to 30 min. After microinjection, the blastocysts were cultured in mKSOMaa for at least 1 hr until the embryo transfer.
Mouse Embryo Transfer
ICR female mice as surrogates (8 weeks old) in the estrus were mated with vasectomized ICR male mice to induce pseudopregnancy Embryo transfer to the surrogate at E2.5 was performed surgically under the anesthesia with ketamine (Putney) and xylazine (Akorn). Injected blastocysts at E3.5 were loaded to the pipette with air bubble and transferred to the uterine horn, which were previously punctured with a 27G needle connected to a 1.5 mL syringe. 14–22 blastocysts were transferred and performed within 20–30 min per surrogate.
Genomic PCR
Genomic PCR was carried out for the detection of rat, mouse or human cells in pig fetuses. Genomic DNAs were extracted using DNeasy Blood and Tissue kit (QIAGEN) or PicoPure DNA Extraction Kit (Thermo Fisher Scientific). Genomic PCRs were performed using PrimeSTAR GXL DNA polymerase (Takara). Primer sequences are provided in Table S2.
Quantitative Genomic PCR
Quantitative PCR (qPCR) for quantifying rat contribution in rat-mouse chimera was performed using SYBR Green PCR Master Mix (Applied Biosystems) and total genomic DNAs isolated from different tissues of the chimera, mouse tail tip and DAC2 rat ESCs. The data were analyzed using the ΔΔCT method, which were first normalized to the values of the mouse specific primers. A rat specific primer was used for detecting rat cells. The levels of chimerism were determined based on the values generated from serial dilutions of rat:mouse genomic DNA. The primers used for genomic qPCR are listed in Table S2.
Genotyping and DNA Sequencing
To determine genotypes of gene modified mouse fetuses or neonates, yolk sac (fetus) or tail tip (neonates) were used for genomic DNA extraction using DNeasy Blood and Tissue kit (QIAGEN) or PicoPure DNA Extraction Kit (Thermo Fisher Scientific). The genomic DNA sequences including target site were amplified with PrimeSTAR GXL DNA Polymerase. Amplicons were sequenced using an ABI 3730xl sequencer (Applied Biosystems). Primer sequences are provided in Table S2.
Glucose Tolerance Test
Mice and Rats were fasted overnight for approximately 16 hr before the test. Fasted blood glucose levels were measured and used as the baseline glucose levels. A solution of glucose is administered by intra-peritoneal (IP) injection (2g of glucose/kg body weight). Subsequently, the blood glucose levels were measured at 15, 30, 60 and 120 min after glucose injection, by placing a small drop of blood on a on the test strip of the blood glucose meter.
Cattle In Vitro Embryo Production
Oocyte Recovery and In Vitro Maturation
Ovaries were collected from a slaughterhouse (Cargill Meat Solutions, Fresno, CA, USA) and transported to the laboratory in insulated container filled with pre-warmed saline solution at ~32°C. The ovaries were washed several times and placed in a water bath at (37°C) in saline solution for oocyte aspiration. Oocytes were aspirated from 2 to 6 mm antral follicles using a 21 G butterfly needle connected to a vacuum pump. Cumulus-oocyte complexes (COCs) containing compact and complete cumulus cell layers were selected and matured in groups of 50 COCs in 400 μl of M199 medium supplemented with ALA-glutamine (0.1 mM), Na pyruvate (0.2 mM), gentamicin (5 μg/ml), EGF (50 ng/ml), oFSH (50 ng/ml; National Hormone and Peptide Program), bLH (1 ug/ml; National Hormone and Peptide Program), cysteamine (0.1 mM), and 10% fetal bovine serum (FBS; Hyclone, South Logan, UT, USA). In vitro maturation (IVM) was performed for 22–24 hr in a humidified atmosphere of 5% CO2 in air at 38.5°C.
In Vitro Fertilization
Fertilization (Day 0) was carried out using frozen-thawed semen. Straws were thawed at 37°C for 45 s and then semen layered onto a 90%/45% Percoll discontinuous density gradient for centrifugation (700 × g for 15 min) at room temperature. A second centrifugation (300 × g for 5 min) was performed after discarding the supernatant and re-suspending the spermatozoa pellet in TALP-Sperm (pH = 7.4, 295 mOsm) (Parrish et al., 1988; Parrish et al., 1986). Matured groups of 15–20 COCs were washed twice and placed in 50 μL of fertilization medium. The final sperm concentration was adjusted to 1×106 sperm/ml using a hemocytometer. The fertilization medium was supplemented with BSA (essentially fatty acid free, 6 mg/ml), fructose (90 μg/ml), penicillamine (3 μg/ml), hypotaurine (11 μg/ml) and heparin (20 μg/ml). Oocytes were co-incubated with spermatozoa at 38.5°C in humidified atmosphere of 5% CO2 in air. Embryo culture (IVC): Presumptive zygotes were mechanically denuded by vortexing for 3–5 min in a 1.5mL tube and 100 uL of SOF-HEPES medium (Holm et al., 1999) and cultured in groups of 15–20 in 50 μL drops of potassium simplex optimized medium supplemented with amino acids and 4 mg/mL of BSA (KSOMaa, pH = 7.4, 275 mOsm) (Evolve ZEBV-100, Zenith Biotech, Guilford, CT, USA) for 7 days. On Day 3, 5% FBS was added. Culture conditions were 38.5°C in a humidified atmosphere of 5% CO2, 5% O2, and 90% N2. On Days 4 and 7 mourlae and blastocysts, respectively, were selected for cell injection.
Pig Parthenogenetic Embryo Production
Oocyte Collection and IVM
Oocytes were aspirated from antral follicles (2–4 diameters) of ovaries from prepubertal gilt ovaries collected at a local slaughterhouse (Olson Meat Company, Orland, CA, USA). COCs were washed in TCM-199 (GIBCO) containing 0.1% polyvinyl alcohol (PVA), and incubated at 38°C and 5% CO2 for 48 hr in TCM-199 containing 0.1% PVA, 3.05 mM D-glucose, 0.91 mM sodium pyruvate, 0.5 μg/ml oFSH, 0.5 μg/ml bLH, 10 ng/ml EGF, 10 μg/ml gentamicin (GIBCO) and 10% porcine follicle fluid.
Parthenogenetic Activation
After IVM, maturated oocytes were stripped of their cumulus cells by incubation in 1 mg/ml hyaluronidase and gentle pipetting. Denuded oocytes were washed with MEM containing 25 mM HEPES (GIBCO) and electrically activated with two pulses of 120 V/mm for 40 μs, delivered by a BTX Electro Cell Manipulator 2001 (BTX, San Diego, CA, USA) in a 0.5 mm chamber containing 0.3 M mannitol, 0.05 mM CaCl2, 0.1 mM MgSO4 and 0.1% bovine serum albumin (BSA). After washing with PZM-5 (Yoshioka et al., 2012), the oocytes were incubated in the presence of 5 μg/ml cytochalasin B in PZM-5 for 3 hr to prevent second polar body extrusion and thus generate diploid parthenogenetic embryos.
Embryo Culture
After activation, pig zygotes were cultured in the 500 μL of PZM-5 (Yoshioka et al., 2012) containing 0.3% BSA for 3–5 days. After 4 days of culture, the culture medium was supplemented with 10% FBS (Gemini Bio-Product, CA, USA) at 38.5°C in a humidified atmosphere of 5% CO2, 5% O2, and 90% N2.
Microinjection of PSCs to Cattle and Pig Blastocysts and Embryo Culture
For morula injections, embryos with more than eight blastomeres and before compaction were selected on days 3–4 for pig and day 4 for cattle embryos. For blastocyst injections, embryos with an obvious blastocoel on days 5–6 for the pig and 6–7 for cattle were used. Single cell suspensions were added to a 50 μL drop of cell culture medium containing the embryos to be injected, and placed on an inverted microscope fitted with micromanipulators. Individual cells were collected into a blunt-end micropipette of 20–30 μm internal diameter (ID) connected to a manual hydraulic oil microinjector. Then, the embryo was secured by a holding pipette and a laser system (Saturn 5 Active, Research Instruments, UK) was used to create a whole in the zona pellucida of the embryo. For blastocysts, another laser pulse was applied to the trophectoderm in order to allow access to the blastocele. Then, the micropipette containing the cells was advanced into the embryo and 10 cells deposited in the blastocoel or periviteline space, for blastocysts and morulae, respectively. Groups of 10–20 embryos were manipulated simultaneously and each session was limited to 40 min. Following cell injection, morulae and blastocysts were cultured in the respective cell culture medium for 4 hr, followed by culture for 20 hr in mix medium (1:1) of each cell culture medium and PZM-5 containing 10% FBS (for the porcine embryos) or KSOMaa containing 4 mg/ml BSA (for the cattle embryos). Then, embryos were cultured in PZM-5 containing 10% FBS or KSOMaa containing 4 mg/ml BSA, for pig and cattle embryos respectively, for another 24 hr, except injected cattle morulae that were cultured for 96 hr. At the end of the culture period, GFP or hKO signals were observed using an inverted fluorescence microscope (Nikon, Tokyo, Japan) and then embryos were fixed for immunostaining.
Pig and Cattle Blastocyst Immunostaining
Only blastocyst stage embryos at time of collection were processed for immunostaining as previously described (Ross et al., 2008). Embryos were washed with phosphate buffered saline (PBS; GIBCO) containing 1 mg/mL PVA (PBS-PVA) three times and then fixed in 4% paraformaldehyde containing 1 ml/mL PVA for 15 min at room temperature. After washing three times with PBS-PBA, blastocysts were permeabilized with PBS-PVA containing 1% Triton X-100 for 30 min, washed in PBS-PVA containing 0.1% Triton X-100 (washing buffer; WB), and blocked in PBS-PVA supplemented with 10% normal donkey serum. Embryos were incubated with primary antibodies (rabbit anti-SOX2 antibody, BioGenex Cat# NU579-UC and mouse anti-human nuclei antibody, Millipore Cat# MAB1281) overnight at 4°C. After repeated washes in WB, the embryos were incubated with secondary antibodies (Alexa Fluor 568 anti-rabbit IgG and Alexa Fluor 488 anti-mouse IgG, Invitrogen) at room temperature for 1 hr. Then, blastocysts were counter-stained with 10 μg/ml Hoechst 33342 at room temperature for 20 min. Stained blastocysts were mounted on a glass slide containing ProLong Gold antifade solution (Invitrogen), covered by a coverslip, and imaged using an inverted fluorescence microscope.
Pig In Vivo Embryo Recovery and Transfer
This work was conducted in two commercial pig farms located in Southeastern Spain (Murcia, Spain), and in the pig experimental unit of the University of Murcia (Murcia, Spain).
Superovulation and Detection of Estrus
Weaning was used to synchronize the estrus in donors and recipients. Only sows with a weaning-to-estrus interval of 3–4 days were selected as donors and recipients. The superovulation of donors was induced by the intramuscular administration of 1000 IU equine chorionic gonadotropin (eCG; Foligon, Intervet, Boxmeer, the Netherlands) 24 hr after weaning. Estrus was checked twice per day by exposing sows to a mature boar (nose-to-nose contact) and applying manual back pressure. Females that exhibited a standing estrous reflex were considered to be in estrus. Only sows with clear signs of estrus at 48–72 hr post-eCG administration were further intramuscularly administered with 750 IU of human chorionic gonadotropin (Veterin Corion, Divasa, Farmavic S.A., Barcelona, Spain) at the onset of estrus.
Insemination of Donors
The donors were post-cervically inseminated at 6 and 24 hr after the onset of estrus. The insemination doses (1.5 × 109 spermatozoa in 45 mL) were prepared from sperm-rich fractions of the ejaculates extended in Beltsville thawing solution extender (Pursel and Johnson, 1975) and were stored for a maximum of 72 hr at 18°C.
Embryo Recovery and Evaluation
The collection of embryos was performed in a specifically designed surgical room located on the farm. The donors were subjected to a mid-ventral laparotomy on Day 2 of the estrous cycle (Day 0: onset of estrus). The donors were sedated with azaperone (2 mg/kg body weight, intramuscular; Stresnil, Lab. Dr. Esteve, Barcelona, Spain). General anesthesia was induced using sodium thiopental (7 mg/kg body weight, intravenous; Tiobarbital 1g, B. Braun VetCare SA, Barcelona, Spain) and was maintained with isoflurane (3.5%–5%; Isoflo; Lab. Abbot, Madrid, Spain). After exposure of the genital tract, the corpora lutea on the ovaries were counted. Zygotes were collected by flushing each oviduct with 20 mL of protein-free embryo recovery medium consisting of Tyrode’s lactate (TL)-HEPES-polyvinyl alcohol (PVA)-medium (TL-HEPES-PVA)(Funahashi et al., 2000) with some modifications (Martinez et al., 2014). Collected embryos were washed three times in TL-HEPES-PVA, placed in sterile Eppendorf tubes containing 1.5 mL of the same medium and transported in a thermostatically controlled incubator at 39°C to our laboratory at the University of Murcia within 1 hr after collection. Embryos were then evaluated for morphology under a stereomicroscope at a magnification of 60 x. In vivo collected embryos used in this study include zygotes with a single cell and two visible polar bodies, 2–4 cell embryos, morulae, and blastocysts.
In Vitro Embryo Culture and Assessment of In Vitro Embryo Development
Zygotes were transferred (40 zygotes per well) to a 4-well multidish (Nunc) containing 500 μL of glucose-free NCSU-23 medium (Petters and Wells, 1993) that was supplemented with 0.3 mM pyruvate and 4.5 mM lactate for 24 hr and then changed to fresh NCSU-23 medium containing 5.5 mM glucose for an additional 5 days. Cultures were performed at 39°C, 5% CO2 in air and 95%–97% relative humidity. At Day 5, embryo culture wells were supplemented with 10% (v/v) fetal calf serum (FCS, Sigma-Aldrich Quimica S.A., Madrid, Spain; cat. no. F7524; lot no. 120M3395). In vitro embryo development was evaluated under a stereomicro-scope at 24 hr and 6 days of culture to determine cleavage and blastocyst formation rates, respectively. An embryo that had cleaved to the 2- to 4-cell stage was defined as cleaved, and an embryo with a well-defined blastocoel and an inner cell mass and trophoblast totally discernible was defined as a blastocyst. Blastocysts were injected as described above.
Culture of hiPSC-Injected Blastocysts
Immediately after hiPSC injection, the blastocysts were incubated in 500 μL of medium used for the culture of hiPSCs for 3–4 hr and then changed to NCSU-23 medium and hiPSC medium (1:1) for an additional 20–22 hr.
Surgical Embryo Transfer
Injected blastocysts were loaded into a Gynétics embryo transfer catheter (Gynétics Medical Products N.V., Lomel, Belgium) connected to a 1mL syringe for transfer into the recipients. The embryo transfer medium was NCSU-23 supplemented with 10 mM HEPES, 0.4% (v/v) BSA and 10% (v/v) FCS. The embryo transfer catheter was loaded with air bubbles that separated the 30 μL drop of medium that contained the embryo from two drops of medium before and after the embryo. All transfers were conducted in asynchronous (−24 hr to embryo collection) recipients. One hour before the transfer, each recipient received a single intramuscular injection of a long-acting amoxicillin suspension (Clamoxyl LA; Pfizer, Madrid, Spain) at a dose of 15 mg/kg. The transfers were conducted using the same procedure described previously for embryo collection. The embryos were transferred to the tip of a uterine horn (15 to 20 cm from the uterotubal junction) with the embryo transfer catheter inserted through the uterine wall, which was previously punctured with a blunt Adson forcep. Post transfer, all recipients were evaluated daily for behavioral changes, including signs of estrus beginning at 12 days post transfer. Pregnancy was diagnosed by transabdominal ultrasonography (Logiq Book XP, General Electric, Solingen, Germany) on Days 20 to 22 post-transfer. All pregnant sows were deeply anesthetized on days 23 to 25 post-transfer and subsequently euthanized by using a captive bolt pistol. Immediately, a midline longitudinal incision was made between the posterior pair of nipples and the ovaries and uterus were located. The cervix and the ovarian stalks were occluded with transfixing ligatures and the reproductive tract removed. The genital tract was then placed in water tight plastic bags kept on ice and transported to the laboratory within 20 min. Once in the laboratory the uterus was opened and fetuses removed from the placenta tissues and numbered in sequential order. Fetuses were individually measured and weighed. Afterward, each fetus was checked for fluorescence emission using an epifluorescence stereomicroscope (Nikon SMZ 18, Japan).
Pig Parthenogenetic Embryo Transfer
This work was conducted at the University of California Davis animal facilities.
Estrus Induction and Synchronization
Estrus and ovulation was induced by intramuscular administration of 400 I.U equine chorionic gonadotropin and 800 I.U. of human chorionic gonadotropin. (PG600; Merck Animal Heath, Summit, NJ, USA) followed 72 hr later by an intramuscular administration of 750 I.U. of hCG (Choluron, Merck Animal Health, Summit, NJ, USA). Estrus was checked twice per day by exposing sows to a mature boar (nose-to-nose contact) and applying manual back pressure. Females that exhibited a standing estrous reflex were used as recipients of surgical embryo transfer 5 to 6 days after hCG administration. Prior to surgical procedure the animals were fasted (food and water) for 24 hr. Anesthesia was induced by intramuscular administration of 2 mg/kg of Telazol (Zoetis, Kalamazoo, MI, USA) prior to intubation and then maintained at a surgical plane by inhalation of Isoflurane (0.5%–5% as need to maintain anesthesia). The Ovary and uterus was exposed by a ventral medial laparotomy. The embryos were transferred to the tip of a uterine horn (15 to 20 cm from the uterotubal junction) with the embryo transfer catheter inserted through the uterine wall, which was previously punctured with a blunt needle. Pregnancy was diagnosed by transabdominal ultrasonography (WED-2000AV, Welld, Shenzhen, China) 17 to 20 days after embryo transfer. On days 23 to 28 of gestation all pregnant gilts were deeply anesthetized by intramuscular administration of 2 mg/kg of Telazol (Zoetis, Kalamazoo, MI, USA) and subsequently euthanized by intracardiac administration of 2.25 mL/kg euthanasia solution (Fatal Plus Solution, Vortex Pharmaceutical Ltd, Dearborn, MI, USA). Then, the reproductive tract was removed and transported to the laboratory within 20 min. Once in the laboratory the uterus was opened and fetuses removed from the placenta tissues and numbered in sequential order. Fetuses were individually measured and weighed. Afterward, each fetus was checked for fluorescence emission using an epifluorescence stereomicroscope (Zeiss, Germany).
Immunocytochemistry
Between day 21–28 of gestation, surrogates were euthanized and embryos were dissected and immerse in paraformaldehyde and incubated at 4°C during 4 hr for small-sized embryos and overnight for normal-sized embryos. After overnight cryoprotection in 30% sucrose solution (Sigma), the embryos were embedded in OCT compound (Sakura Finetek) and frozen in dry ice. Sections (10 μm thick for small-sized embryos and 20 μm for normal-sized embryos) of the different embryos were cut on a Leica cryostat. For immunohistochemistry, we used standard staining procedures and antigen retrieval solution (HistoVT one, Nacali Tesque, INC) according to manufacturer’s instructions. The primary antibodies used were rabbit anti-monomeric Kusabira-Orange 2 (MBL Code # PM051M, 1:500), rabbit anti-GFP (MBL Code# 598, 1:500), rat anti-cytokeratin 8 (TROMA-I, DSHB Antibody Registry ID: AB_531826, 1:20), mouse anti-epithelial antigen (Dako Cat# M0804, 1:200), Mouse monoclonal anti-Ep-CAM (Santa Cruz Cat# sc-25308, 1:50), mouse anti-actin α-smooth muscle (Sigma Cat# A5228, 1:200), Rabbit polyclonal anti-Histone H3 (tri methyl K9) (Abcam Cat# ab8898, 1:50), Rabbit polyclonal anti-Histone H4 (tri methyl K20) (Abcam Cat# ab9053, 1:50), Mouse monoclonal anti-Oct3/4 (C-10) (Santa Cruz Cat# sc-5279, 1:200), Rat anti-Mouse CD31(Clone MEC13.3) (BD PharMingen Cat# 553370, 1:200), Mouse anti-Tubulin β3 (Clone TUJ1) (Biolegend Cat# 801202, 1:500).
QUANTIFICATION AND STATISTICAL ANALYSIS
For Figure 1E, qPCR for quantifying rat contribution in rat-mouse chimera was performed using SYBR Green PCR Master Mix (Applied Biosystems) and total genomic DNAs isolated from different tissues of the chimera, mouse tail tip and DAC2 rat ESCs. The data were analyzed using the ΔΔCT method, which were first normalized to the values of the mouse specific primers. A rat specific primer was used for detecting rat cells. The levels of chimerism were determined based on the values generated from serial dilutions of rat:mouse genomic DNA. The primers used for genomic qPCR are listed in Table S2. For Figures 1E, S2C, and S3E data are presented as mean (SD). For Figure 1B, n represents number of embryo transfers.
Supplementary Material
Figure S1. Chimeric Contribution of Rat PSCs to Different Organs of Rat-Mouse Chimeras, Related to Figure 1
(A) Bright field (top) and fluorescence (bottom) images showing hKO-labeled rat ESCs (DAC8) contributed to different organs of a neonatal rat-mouse chimera. Control organs from a wild-type neonatal mouse were showing on the left.
(B) Representative fluorescence images showing hKO-labeled rat iPSCs (SDFF) contributed to different tissues in a 3-week-old rat-mouse chimera. Red, hKO-labeled rat cells; blue, DAPI. Scale bar is 100 μm.
(C) Bright field (left) and fluorescence (right) images showing an isolated neonatal mouse gallbladder contributed by hKO-labeled rat iPSCs (SDFE), as shown in Figure 1F.
Figure S2. CRISPR-Cas9-Mediated Rat-Mouse Blastocyst Complementation, Related to Figure 2
(A) Bright field images showing a wild type (P0, left) mouse and a Pdx1−/− (E18.5, right) mouse fetus generated by zygotic co-injection of Cas9 mRNA and Pdx1 sgRNA. Li, liver; St, stomach; Sp, spleen. Yellow-dotted line encircles the pancreas in the wild type, and indicates the lack of a pancreas in the Pdx1−/− fetus.
(B) Bright field (top) and fluorescence (bottom) images showing more chimeric contribution of rat cells in a Pdx1−/− mouse pancreas than a wild type mouse pancreas. Li, liver; St, stomach; Sp, spleen. Yellow-dotted line encircles the pancreases. Red, hKO-labeled rat cells.
(C) Glucose tolerance test results of adult (>7 months) Pdx1−/− and Pdx1+/− mice complemented with rat PSCs. Age-matched wild type mice and rats were included as controls. Error bars indicate s.d.
(D) Top, a representative bright field image showing a wild type (left) and Nkx2.5−/− (right) E10.5 mouse embryos Middle, rat ESC-derivatives rescued retarded growth of E10.5 Nkx2.5−/− mouse embryo and were enriched in the heart. H, heart. Bottom, an E10.5 Nkx2.5−/− embryo showing retarded growth with little to no rat cells contribution to the heart. Yellow-dotted line encircles the heart. Scale bar is 100μm.
(E) Top, a representative bright field image showing a wild type (left) and a Pax6−/− (right) E15.5 mouse fetuses. Bottom, fluorescence images showing more chimeric contribution of rat cells in the eye of Pax6−/− (right) than wild type (left) chimeras. WT+rPSCs, control rat-mouse chimera without Cas9/sgRNA injection. Red, hKO-labeled rat cells. Blue, DAPI. Scale bar is 100μm.
(F) Frequencies of wild type, homozygous, heterozygous mutants and mosaic mutant mice generated by zygotic co-injection of Cas9 mRNA and sgRNAs for Pdx1, Nkx2.5, or Pax6.
Figure S3. Teratoma Assay and Interspecies ICM Incorporation of Different Types of hiPSCs to Cattle and Pig Blastocysts, Related to Figure 4
(A) Representative images showing hematoxylin and eosin staining of histological sections derived from teratomas generated by 2iLD-hiPSCs, 4i-hiPSCs, NHSM-hiPSCs and FAC-hiPSCs. hiPSC-derived teratomas contained tissues from all three germlineages: endoderm (top), mesoderm (middle) and ectoderm (bottom). Scale bar is 100 μm.
(B) Laser-assisted microinjection of hiPSCs into a bovine blastocyst. Left, before injection laser beam was used to perforate the zona pellucida. Right, a blunt end pipette was used to transfer hiPSCs to the blastocoel. Red color indicates hKO-labeled hiPSCs (bottom).
(C) Representative immunofluorescence images showing ICM incorporation of naive and intermediate hiPSCs. Top, SOX2; Middle, HuNu, Bottom, merged images. Blue, DAPI. Scale bar is 100 μm.
(D) Number of hiPSCs remained in the cattle (left) and pig (right) blastocysts after injection of 10 hiPSCs followed by 2 days in vitro culture. Red line, average number of cells. Blue dot, number of ICM-incorporated hiPSCs in each blastocyst. BL, blastocyst.
(E) Comparison of several parameters of ICM incorporation for each type of hiPSCs between pig and cattle. Error bars indicate s.d.
Figure S4. Generation of Post-implantation Pig Embryos Derived from Blastocyst Injection of hiPSCs, Related to Figure 5
(A) Representative bright field images showing: a) freshly collected pig zygotes; b) in vitro derived pig blastocysts; c) hiPSCs are being injected into a pig blastocyst; d) Pig female reproductive tract at day-28 of pregnancy; e) a day 28 pig conceptus displaying the embryo proper and the amniotic and allantoic membranes; f) a magnified image of the day-28 pig embryo showing in e.
(B) Representative bright field images showing a normal size day-28 pig embryo (left) and growth retarded day-28 embryos (middle and right).
Figure S5. Chimeric Contribution of hiPSCs to Post-implantation Pig Embryos, Related to Figure 6
(A) Representative bright field (left), fluorescence (middle) and immunofluorescence (right) images showing the contribution of hKO-labeled NHSM-hiSPCs, GFP-labeled 2iLD-hiPSCs and FAC-hiPSCs to normal size day 28 pig embryos. Scale bar is 200 μm.
(B) Representative immunofluorescence images showing the contribution of FAC-hiPSCs to six additional normal size day 21–28 pig embryos (FAC #2–#7). Scale bar is 200 μm.
(C) Representative immunofluorescence images showing the chimeric contribution and differentiation of FAC-hiPSCs within a normal size, day 28 pig embryo (FAC #3). Embryo sections were stained with antibodies against GFP (green, left), EpiCAM, HNF3β, CK8 and SMA (red, middle). Right, merged images. Insets are higher magnification images of boxed regions. Scale bar is 100 μm.
Highlights.
Naive rat PSCs robustly contribute to live rat-mouse chimeras
A versatile CRISPR-Cas9 mediated interspecies blastocyst complementation system
Naive rodent PSCs show no chimeric contribution to post-implantation pig embryos
Chimerism is observed with some human iPSCs in post-implantation pig embryos
Acknowledgments
J.C.I.B. dedicates this paper to Dr. Rafael Matesanz, Director of the Spain’s National Organ Transplant Organization. Rafael’s work has helped save thousands of patients in need of an organ. He constitutes a relentless inspiration for those of us trying to understand and alleviate human disease. The authors are grateful to Xiomara Lucas, Maria Dolores Ortega, Moises Gonzalvez, Jose Antonio Godinez, and Jesus Gomis for their assistance throughout this work. We thank the staff of the Agropor S.A. and Porcisan S.A. piggeries (Murcia, Spain) for the help and excellent management of animals. We thank Joan Rowe, Bret McNabb, Aaron Prinz, and Kent Parker and their crews for excellent assistance with embryo transfers and pig care at UC Davis. We thank Mako Yamamoto for help with mouse embryo dissection. We would like to thank Uri Manor of the Salk Waitt Advanced Biophotonics Core for technical advice on imaging analysis. We would like to thank the Salk Stem Cell Core for providing cell culture reagents. We would like to thank May Schwarz and Peter Schwarz for administrative help. We thank David O’Keefe for critical reading and editing of the manuscript. This experimental study was supported by The Fundación Séneca (GERM 19892/GERM/15), Murcia, Spain. The MINECO is acknowledged for their grant-based support (BES-2013-064087 and BES-2013-064069) (to C.A.M and A.N.). P.J.R was supported by a UC Davis Academic Senate New Research grant. Work in the laboratory of J.C.I.B. was supported by the UCAM, Fundacion Dr. Pedro Guillen, G. Harold and Leila Y. Mathers Charitable Foundation, and The Moxie Foundation.
Footnotes
Supplemental Information includes five figures and six tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2016.12.036.
AUTHOR CONTRIBUTIONS
J.W. and J.C.I.B. conceived the study. J.W. generated and characterized all naive and intermediate hiPSC lines. K.S. generated and characterized primed hiPSCs. J.W. and T.H. generated rat iPSCs. J.W., A.P.-L., T.Y., M.M.V., D.O., A.O., P.R., C.R.E., J.W., and P.M.R. performed immunohistochemistry analyses of mouse and pig embryos. K.S., T.Y., E.S., A.P.-L., and M.M.V. performed genotyping, genomic PCR, and genomic qPCR analyses. A.S., M.S., and J.P.L. performed mouse Cas9/sgRNA injection, blastocyst injection, and embryo transfer. Y.S.B., M.S., and M.V. prepared hiPSCs, performed morulae and blastocyst injections, and analyzed hiPSC contribution to cattle and ppig ICMs. H.W. produced parthenogenetic pig embryos. D.A.S., Y.S.B., and M.V. produced cattle embryos. Work at UC Davis and University of Murcia was performed under the supervision of P.J.R. and E.A.M., respectively. E.A.M., M.A.G., C.C., I.P., C.A.M., S.S.B., A.N., and J.R. designed, coordinated, performed, and analyzed data related to pig embryo collection, embryo culture, blastocyst injection, embryo transfer, and embryo recover. E.N.D., J.L., I.G., P.G., T.B., M.L.M.-M., and J.M.C. coordinated work between Salk, and University of Murcia. J.W., P.J.R., and J.C.I.B. wrote the manuscript.
References
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, González F, Vassena R, Bilić J, Pekarik V, Tiscornia G, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- Abeydeera LR. In vitro production of embryos in swine. Theriogenology. 2002;57:256–273. doi: 10.1016/s0093-691x(01)00670-7. [DOI] [PubMed] [Google Scholar]
- Chen J, Lansford R, Stewart V, Young F, Alt FW. RAG-2-deficient blastocyst complementation: an assay of gene function in lymphocyte development. Proc Natl Acad Sci USA. 1993;90:4528–4532. doi: 10.1073/pnas.90.10.4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Los Angeles A, Ferrari F, Xi R, Fujiwara Y, Benvenisty N, Deng H, Hochedlinger K, Jaenisch R, Lee S, Leitch HG, et al. Hallmarks of pluripotency. Nature. 2015;525:469–478. doi: 10.1038/nature15515. [DOI] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funahashi H, Ekwall H, Rodriguez-Martinez H. Zona reaction in porcine oocytes fertilized in vivo and in vitro as seen with scanning electron microscopy. Biol Reprod. 2000;63:1437–1442. doi: 10.1095/biolreprod63.5.1437. [DOI] [PubMed] [Google Scholar]
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Gardner RL, Johnson MH. Investigation of early mammalian development using interspecific chimaeras between rat and mouse. Nat New Biol. 1973;246:86–89. doi: 10.1038/newbio246086a0. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Ikeo K. Pax 6: mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- Grupen CG. The evolution of porcine embryo in vitro production. Theriogenology. 2014;81:24–37. doi: 10.1016/j.theriogenology.2013.09.022. [DOI] [PubMed] [Google Scholar]
- Hansen PJ. Current and future assisted reproductive technologies for mammalian farm animals. In: Cliff Lamb G, DiLorenzo N, editors. Current and Future Reproductive Technologies and World Food Production. Springer; New York: 2014. pp. 1–22. [DOI] [PubMed] [Google Scholar]
- Hasler JF. Forty years of embryo transfer in cattle: a review focusing on the journal Theriogenology, the growth of the industry in North America, and personal reminisces. Theriogenology. 2014;81:152–169. doi: 10.1016/j.theriogenology.2013.09.010. [DOI] [PubMed] [Google Scholar]
- Hishida T, Nozaki Y, Nakachi Y, Mizuno Y, Okazaki Y, Ema M, Takahashi S, Nishimoto M, Okuda A. Indefinite self-renewal of ESCs through Myc/Max transcriptional complex-independent mechanisms. Cell Stem Cell. 2011;9:37–49. doi: 10.1016/j.stem.2011.04.020. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm P, Booth PJ, Schmidt MH, Greve T, Callesen H. High bovine blastocyst development in a static in vitro production system using SOFaa medium supplemented with sodium citrate and myoinositol with or without serum-proteins. Theriogenology. 1999;52:683–700. doi: 10.1016/S0093-691X(99)00162-4. [DOI] [PubMed] [Google Scholar]
- Huang Y, Osorno R, Tsakiridis A, Wilson V. In vivo differentiation potential of epiblast stem cells revealed by chimeric embryo formation. Cell Rep. 2012;2:1571–1578. doi: 10.1016/j.celrep.2012.10.022. [DOI] [PubMed] [Google Scholar]
- Irie N, Weinberger L, Tang WWC, Kobayashi T, Viukov S, Manor YS, Dietmann S, Hanna JH, Surani MA. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isotani A, Hatayama H, Kaseda K, Ikawa M, Okabe M. Formation of a thymus from rat ES cells in xenogeneic nude mouse↔rat ES chimeras. Genes Cells. 2011;16:397–405. doi: 10.1111/j.1365-2443.2011.01495.x. [DOI] [PubMed] [Google Scholar]
- Izpisúa-Belmonte JC, Brown JM, Crawley A, Duboule D, Tickle C. Hox-4 gene expression in mouse/chicken heterospecific grafts of signalling regions to limb buds reveals similarities in patterning mechanisms. Development. 1992;115:553–560. doi: 10.1242/dev.115.2.553. [DOI] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- King TJ, Dobrinsky JR, Zhu J, Finlayson HA, Bosma W, Harkness L, Ritchie WA, Travers A, McCorquodale C, Day BN, et al. Embryo development and establishment of pregnancy after embryo transfer in pigs: coping with limitations in the availability of viable embryos. Reproduction. 2002;123:507–515. [PubMed] [Google Scholar]
- Kobayashi T, Yamaguchi T, Hamanaka S, KatoItoh M, Yamazaki Y, Ibata M, Sato H, Lee YS, Usui J, Knisely AS, et al. Generation of rat pancreas in mouse by interspecific blastocyst injection of pluripotent stem cells. Cell. 2010;142:787–799. doi: 10.1016/j.cell.2010.07.039. [DOI] [PubMed] [Google Scholar]
- Kutner RH, Zhang X-Y, Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc. 2009;4:495–505. doi: 10.1038/nprot.2009.22. [DOI] [PubMed] [Google Scholar]
- Li P, Tong C, Mehrian-Shai R, Jia L, Wu N, Yan Y, Maxson RE, Schulze EN, Song H, Hsieh CL, et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- Martinez EA, Angel MA, Cuello C, Sanchez-Osorio J, Gomis J, Parrilla I, Vila J, Colina I, Diaz M, Reixach J, et al. Successful non-surgical deep uterine transfer of porcine morulae after 24 hour culture in a chemically defined medium. PLoS ONE. 2014;9:e104696. doi: 10.1371/journal.pone.0104696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, Hong H, Nakagawa M, Tanabe K, Tezuka K, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Parrish JJ, Susko-Parrish JL, Leibfried-Rutledge ML, Critser ES, Eye-stone WH, First NL. Bovine in vitro fertilization with frozen-thawed semen. Theriogenology. 1986;25:591–600. doi: 10.1016/0093-691x(86)90143-3. [DOI] [PubMed] [Google Scholar]
- Parrish JJ, Susko-Parrish J, Winer MA, First NL. Capacitation of bovine sperm by heparin. Biol Reprod. 1988;38:1171–1180. doi: 10.1095/biolreprod38.5.1171. [DOI] [PubMed] [Google Scholar]
- Petters RM, Wells KD. Culture of pig embryos. J Reprod Fertil Suppl. 1993;48:61–73. [PubMed] [Google Scholar]
- Pursel VG, Johnson LA. Freezing of boar spermatozoa: fertilizing capacity with concentrated semen and a new thawing procedure. J Anim Sci. 1975;40:99–102. doi: 10.2527/jas1975.40199x. [DOI] [PubMed] [Google Scholar]
- Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- Ross PJ, Ragina NP, Rodriguez RM, Iager AE, Siripattarapravat K, Lopez-Corrales N, Cibelli JB. Polycomb gene expression and histone H3 lysine 27 trimethylation changes during bovine preimplantation development. Reproduction. 2008;136:777–785. doi: 10.1530/REP-08-0045. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, et al. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Chen Z, Bartunkova S, Yamasaki N, Izumo S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development. 1999;126:1269–1280. doi: 10.1242/dev.126.6.1269. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RDG. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, Friedli M, He Y, Planet E, O’Neil RC, Markoulaki S, Pontis J, Wang H, Iouranova A, Imbeault M, et al. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell. 2016;19:502–515. doi: 10.1016/j.stem.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Ohinata Y. A modified EpiSC culture condition containing a GSK3 inhibitor can support germline-competent pluripotency in mice. PLoS ONE. 2014;9:e95329. doi: 10.1371/journal.pone.0095329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger L, Ayyash M, Novershtern N, Hanna JH. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol. 2016;17:155–169. doi: 10.1038/nrm.2015.28. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wu J, Izpisua Belmonte JC. Dynamic pluripotent stem cell states and their applications. Cell Stem Cell. 2015;17:509–525. doi: 10.1016/j.stem.2015.10.009. [DOI] [PubMed] [Google Scholar]
- Wu J, Izpisua Belmonte JC. Stem cells: a renaissance in human biology research. Cell. 2016;165:1572–1585. doi: 10.1016/j.cell.2016.05.043. [DOI] [PubMed] [Google Scholar]
- Wu J, Okamura D, Li M, Suzuki K, Luo C, Ma L, He Y, Li Z, Benner C, Tamura I, et al. An alternative pluripotent state confers interspecies chimaeric competency. Nature. 2015;521:316–321. doi: 10.1038/nature14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Greely HT, Jaenisch R, Nakauchi H, Rossant J, Belmonte JC. Stem cells and interspecies chimaeras. Nature. 2016;540:51–59. doi: 10.1038/nature20573. [DOI] [PubMed] [Google Scholar]
- Xiang AP, Mao FF, Li W-Q, Park D, Ma B-F, Wang T, Vallender TW, Vallender EJ, Zhang L, Lee J, et al. Extensive contribution of embryonic stem cells to the development of an evolutionarily divergent host. Hum Mol Genet. 2008;17:27–37. doi: 10.1093/hmg/ddm282. [DOI] [PubMed] [Google Scholar]
- Yang L, Güell M, Niu D, George H, Lesha E, Grishin D, Aach J, Shrock E, Xu W, Poci J, et al. Genome-wide inactivation of porcine endogenous retroviruses (PERVs) Science. 2015;350:1101–1104. doi: 10.1126/science.aad1191. [DOI] [PubMed] [Google Scholar]
- Yoshioka K, Noguchi M, Suzuki C. Production of piglets from in vitro-produced embryos following non-surgical transfer. Anim Reprod Sci. 2012;131:23–29. doi: 10.1016/j.anireprosci.2012.01.018. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Chimeric Contribution of Rat PSCs to Different Organs of Rat-Mouse Chimeras, Related to Figure 1
(A) Bright field (top) and fluorescence (bottom) images showing hKO-labeled rat ESCs (DAC8) contributed to different organs of a neonatal rat-mouse chimera. Control organs from a wild-type neonatal mouse were showing on the left.
(B) Representative fluorescence images showing hKO-labeled rat iPSCs (SDFF) contributed to different tissues in a 3-week-old rat-mouse chimera. Red, hKO-labeled rat cells; blue, DAPI. Scale bar is 100 μm.
(C) Bright field (left) and fluorescence (right) images showing an isolated neonatal mouse gallbladder contributed by hKO-labeled rat iPSCs (SDFE), as shown in Figure 1F.
Figure S2. CRISPR-Cas9-Mediated Rat-Mouse Blastocyst Complementation, Related to Figure 2
(A) Bright field images showing a wild type (P0, left) mouse and a Pdx1−/− (E18.5, right) mouse fetus generated by zygotic co-injection of Cas9 mRNA and Pdx1 sgRNA. Li, liver; St, stomach; Sp, spleen. Yellow-dotted line encircles the pancreas in the wild type, and indicates the lack of a pancreas in the Pdx1−/− fetus.
(B) Bright field (top) and fluorescence (bottom) images showing more chimeric contribution of rat cells in a Pdx1−/− mouse pancreas than a wild type mouse pancreas. Li, liver; St, stomach; Sp, spleen. Yellow-dotted line encircles the pancreases. Red, hKO-labeled rat cells.
(C) Glucose tolerance test results of adult (>7 months) Pdx1−/− and Pdx1+/− mice complemented with rat PSCs. Age-matched wild type mice and rats were included as controls. Error bars indicate s.d.
(D) Top, a representative bright field image showing a wild type (left) and Nkx2.5−/− (right) E10.5 mouse embryos Middle, rat ESC-derivatives rescued retarded growth of E10.5 Nkx2.5−/− mouse embryo and were enriched in the heart. H, heart. Bottom, an E10.5 Nkx2.5−/− embryo showing retarded growth with little to no rat cells contribution to the heart. Yellow-dotted line encircles the heart. Scale bar is 100μm.
(E) Top, a representative bright field image showing a wild type (left) and a Pax6−/− (right) E15.5 mouse fetuses. Bottom, fluorescence images showing more chimeric contribution of rat cells in the eye of Pax6−/− (right) than wild type (left) chimeras. WT+rPSCs, control rat-mouse chimera without Cas9/sgRNA injection. Red, hKO-labeled rat cells. Blue, DAPI. Scale bar is 100μm.
(F) Frequencies of wild type, homozygous, heterozygous mutants and mosaic mutant mice generated by zygotic co-injection of Cas9 mRNA and sgRNAs for Pdx1, Nkx2.5, or Pax6.
Figure S3. Teratoma Assay and Interspecies ICM Incorporation of Different Types of hiPSCs to Cattle and Pig Blastocysts, Related to Figure 4
(A) Representative images showing hematoxylin and eosin staining of histological sections derived from teratomas generated by 2iLD-hiPSCs, 4i-hiPSCs, NHSM-hiPSCs and FAC-hiPSCs. hiPSC-derived teratomas contained tissues from all three germlineages: endoderm (top), mesoderm (middle) and ectoderm (bottom). Scale bar is 100 μm.
(B) Laser-assisted microinjection of hiPSCs into a bovine blastocyst. Left, before injection laser beam was used to perforate the zona pellucida. Right, a blunt end pipette was used to transfer hiPSCs to the blastocoel. Red color indicates hKO-labeled hiPSCs (bottom).
(C) Representative immunofluorescence images showing ICM incorporation of naive and intermediate hiPSCs. Top, SOX2; Middle, HuNu, Bottom, merged images. Blue, DAPI. Scale bar is 100 μm.
(D) Number of hiPSCs remained in the cattle (left) and pig (right) blastocysts after injection of 10 hiPSCs followed by 2 days in vitro culture. Red line, average number of cells. Blue dot, number of ICM-incorporated hiPSCs in each blastocyst. BL, blastocyst.
(E) Comparison of several parameters of ICM incorporation for each type of hiPSCs between pig and cattle. Error bars indicate s.d.
Figure S4. Generation of Post-implantation Pig Embryos Derived from Blastocyst Injection of hiPSCs, Related to Figure 5
(A) Representative bright field images showing: a) freshly collected pig zygotes; b) in vitro derived pig blastocysts; c) hiPSCs are being injected into a pig blastocyst; d) Pig female reproductive tract at day-28 of pregnancy; e) a day 28 pig conceptus displaying the embryo proper and the amniotic and allantoic membranes; f) a magnified image of the day-28 pig embryo showing in e.
(B) Representative bright field images showing a normal size day-28 pig embryo (left) and growth retarded day-28 embryos (middle and right).
Figure S5. Chimeric Contribution of hiPSCs to Post-implantation Pig Embryos, Related to Figure 6
(A) Representative bright field (left), fluorescence (middle) and immunofluorescence (right) images showing the contribution of hKO-labeled NHSM-hiSPCs, GFP-labeled 2iLD-hiPSCs and FAC-hiPSCs to normal size day 28 pig embryos. Scale bar is 200 μm.
(B) Representative immunofluorescence images showing the contribution of FAC-hiPSCs to six additional normal size day 21–28 pig embryos (FAC #2–#7). Scale bar is 200 μm.
(C) Representative immunofluorescence images showing the chimeric contribution and differentiation of FAC-hiPSCs within a normal size, day 28 pig embryo (FAC #3). Embryo sections were stained with antibodies against GFP (green, left), EpiCAM, HNF3β, CK8 and SMA (red, middle). Right, merged images. Insets are higher magnification images of boxed regions. Scale bar is 100 μm.