SUMMARY
Previous studies suggested that expression of most yeast mRNAs is dominated by either transcription factor TFIID or SAGA. We reexamined the role of TFIID by rapid depletion of S. cerevisiae TFIID subunits and measurement of changes in nascent transcription. We find that transcription of nearly all mRNAs is strongly dependent on TFIID function. Degron-dependent depletion of Tafs 1,2,7,11, and 13 showed similar transcription decreases for genes in the Taf1-depleted, Taf1-enriched, TATA-containing and TATA-less gene classes. The magnitude of TFIID-dependence varies with growth conditions, although this variation is similar genome-wide. Many studies have suggested differences in gene regulatory mechanisms between TATA and TATA-less genes and these differences have been attributed in part to differential dependence on SAGA or TFIID. Our work indicates that TFIID participates in expression of nearly all yeast mRNAs and that differences in regulation between these two gene categories is due to other properties.
Graphical Abstract
INTRODUCTION
TFIID and SAGA are two widely used RNA polymerase II (Pol II) transcription factors that together regulate transcription of nearly all Pol II-transcribed genes (Hahn and Young, 2011). Yeast (y) TFIID is comprised of TATA binding protein (TBP) and 14 TBP associated factors (Tafs) (Berger et al., 2011; Sainsbury et al., 2015). Although transcription at TATA box-containing promoters can be reconstituted with only TBP and other basal transcription factors, the TFIID Tafs are thought to be critical for promoter recognition at the ~80% of eukaryotic promoters lacking a TATA box. TFIID is structurally organized into three lobes with lobe C (Tafs 1, 2, and 7) binding several promoter DNA elements found in higher eukaryotes (e.g., Inr, DPE, MTE) (Cianfrocco and Nogales, 2013; Louder et al., 2016; Theisen et al., 2010; Verrijzer et al., 1995). TFIID also binds transcription activators, acetylated nucleosomes, the universally required coactivator Mediator and probably other components of the basal transcription machinery (Müller and Tora, 2014). These functions allow TFIID to respond to regulatory inputs from both UAS elements and the chromatin state and to act as a platform for assembly of the transcription preinitiation complex (PIC).
SAGA contains 19 subunits and is structurally organized into 5 modules that function in histone acetylation and deubiquitylation, and in the binding of TBP and transcription activators (Hahn and Young, 2011). Five of the ySAGA subunits are shared with yTFIID and seven of the SAGA subunits, including four common Tafs, contain histone fold domains that likely form a structural module at the center of SAGA (Bieniossek et al., 2013; Grant et al., 1998; Han et al., 2014; Setiaputra et al., 2015). Although SAGA and TFIID share a set of subunits and both bind activators and TBP, SAGA has no known DNA binding activity or interactions with other components of the basal transcription machinery.
Earlier studies using the yeast S. cerevisiae, have classified the TFIID and SAGA-dependence of genes based on genome-wide mapping of TFIID binding and transcriptional changes observed in strains with mutations in TFIID and SAGA subunits. First, formaldehyde-based ChIP and ChIP-exo methods have mapped the relative ratio of the TFIID subunit Taf1 versus TBP at yeast promoter regions, identifying Taf-enriched versus Taf-depleted genes (Kuras et al., 2000; Li et al., 2000; Rhee and Pugh, 2012). Second, steady-state mRNA levels were measured before and after heat shock inactivation of temperature-sensitive Taf proteins (Huisinga and Pugh, 2004; Lee et al., 2000; Li et al., 2000; Shen et al., 2003). In parallel, steady state mRNA levels were measured in strains with mutations in SAGA subunits and in strains with both TFIID and SAGA mutations (Huisinga and Pugh, 2004; Lee et al., 2000; Shen et al., 2003). Combined, these studies suggested that TFIID and SAGA likely contribute to the expression of nearly all genes, but that expression of most genes is dominated by either TFIID or SAGA.
Many studies suggest that there are at least two broad classes of genes: housekeeping and highly-regulated genes (de Jonge et al., 2016; Eisenberg and Levanon, 2013; Huisinga and Pugh, 2004; Mencía et al., 2002; Ohtsuki et al., 1998; Zabidi and Stark, 2016). These two gene classes differ in their response to transcription activators, chromatin organization and modifications, and in promoter sequence elements. In yeast, promoters lacking a consensus TATA box are typically in the housekeeping class, have the +1 nucleosome adjacent to the site of PIC formation, are enriched in H4 acetylation, and nearly always classified as Taf1-enriched and TFIID-dominated (Basehoar et al., 2004; Huisinga and Pugh, 2004; Rhee and Pugh, 2012). On the other hand, TATA-containing promoters are often highly regulated by stress, are typically modulated by chromatin remodeling and/or covalent histone modifications and are often classified as Taf1-depleted and SAGA-dominated.
Recent results, however, have raised questions about the TFIID and SAGA specificity at these two classes of genes. First, a formaldehyde-independent method for mapping genome-wide binding termed ChEC-seq mapped apparent equal occupancy of TFIID at promoters of both gene classes (Grünberg et al., 2016). Second, mapping SAGA-dependent histone modifications showed that SAGA associates with all transcribed genes (Bonnet et al., 2014). Third, inactivation of SAGA was found to decrease Pol II recruitment and the rate of transcription from several genes previously characterized as TFIID-dominated (Bonnet et al., 2014). Finally, it was demonstrated that yeast mRNA levels are largely buffered to major transcriptional perturbations (Haimovich et al., 2013; Munchel et al., 2011; Sun et al., 2013; 2012). For example, inhibition of Pol II activity by several approaches led to a corresponding increase in mRNA stability for most genes. Therefore, using steady state mRNA levels to determine the role of widely used transcription factors is problematic. Based on these findings, we have reexamined the TFIID-dependence of yeast genes and find that, for cells grown in rich media, nearly all Pol II-transcribed genes are strongly dependent on TFIID. The magnitude of TFIID dependence can change in different growth conditions. However, even under different growth conditions, genes in the Taf1-depleted, Taf1-enriched, TATA-containing and TATA-less categories respond similarly to depletion of TFIID-specific subunits such as Taf1, 2, 7, 11 and 13. Our findings have important implications for the mechanism of gene regulation and the nature of the PIC at both housekeeping and highly regulated genes.
RESULTS
A dramatic decrease in genome-wide Pol II transcription upon TFIID inactivation
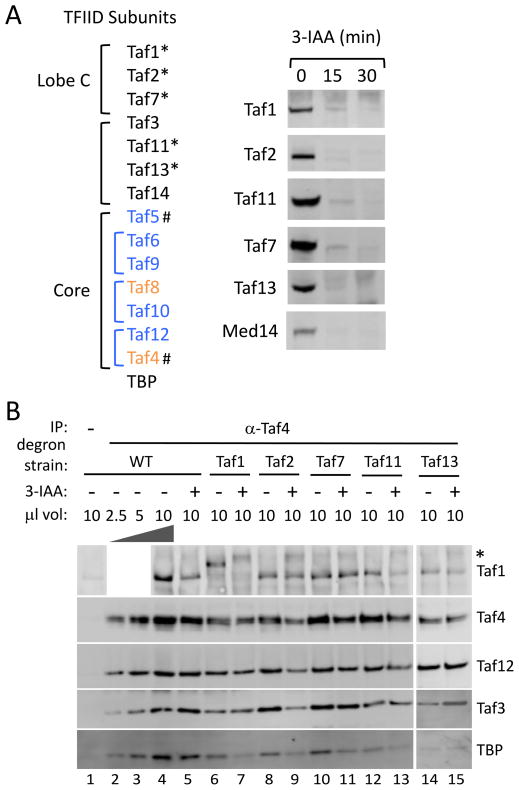
Since all TFIID subunits are essential for viability, previous studies in yeast utilized heat shock of temperature sensitive mutants to inactivate TFIID followed by steady state mRNA analysis (Huisinga and Pugh, 2004; Shen et al., 2003). As an alternative approach to TFIID inactivation, we used the auxin-inducible degron system (Nishimura et al., 2009) to rapidly deplete selected TFIID subunits. First, a yeast strain was constructed containing both a triple Flag epitope tag at the C-terminus of the Pol II subunit Rpb3 and constitutively expressing the plant-specific F-box protein OsTIR1. Next, the degron, auxin repressor protein IAA7, was integrated at the C-terminus of the TFIID-specific subunits Taf1, Taf2, Taf7, Taf11 and Taf13, (Cianfrocco et al., 2013; Hahn and Young, 2011; Leurent et al., 2004; 2002; Louder et al., 2016) (Fig 1A, left panel). Taf1, Taf11 and Taf13 have been implicated in TFIID-TBP binding (Anandapadamanaban et al., 2013; Lavigne et al., 1999; Shen et al., 2003). IAA7 was also fused to Med14, the Mediator subunit that connects the head, middle and tail modules (Plaschka et al., 2016; Tsai et al., 2014) with the reasoning that depletion of this subunit will inactivate Mediator. The control strain, designated as wild type, was identical except for lacking an IAA7 degron tag. Western blot analyses showed that all of these subunits fused to IAA7 were efficiently and rapidly degraded 30 min after addition of the auxin 3-IAA (Fig 1A, right panel). After auxin addition for 30 min, all strains were ≥88% viable, although prolonged exposure was lethal. Without 3-IAA, the Taf1,2,7, and 13 degron tagged strains grew normally at 25, 30 and 37 C°, while the Taf11-degron strain grew normally at 25 and 30 C° but slowly at 37 C°. Analysis of Pol II transcription in the absence of 3-IAA at 30 C° (see below) showed that wild type and all degron strains had highly similar transcription profiles across the 4,808 genes used for genome-wide analysis (Pearson correlation coefficient ≥0.88; Fig S1A).
Figure 1.
Auxin-dependent depletion of yeast TFIID subunits. (A) Listed at left are yeast TFIID subunits in lobe C, the shared or similar TFIID/SAGA core subunits and other auxiliary subunits. * indicates subunits tagged with the 3xV5-IAA7 degron tag and # indicates subunits tagged with FRB for anchor-away. Blue indicates subunits shared with SAGA and Orange indicates that SAGA contains a related but SAGA-specific subunit. Shown at right is Western analysis of cells treated with 3-IAA for the indicated time. Western blot was probed with anti V5 antibody. (B) Analysis of TFIID subunits after Taf depletion. After 30 min treatment with 3-IAA, whole cell extracts were made and immune precipitated with anti Taf4 antibody. Shown is quantitative Western analysis of the IP, probing for Tafs 1, 3, 4, 12 and TBP. * indicates a non-specific signal observed with the anti Taf1 antibody.
The integrity of TFIID after Taf depletion was monitored by immune precipitation of TFIID core subunit Taf4 followed by Western analysis of co precipitated proteins (Fig 1B). Our results show that, upon degradation of Taf1, 2, 7, 11, or 13, the tested TFIID subunits (Tafs 1, 12, 3 and TBP) still co precipitated with Taf4, showing no obvious disruption in subunit association of the remaining TFIID complex. We note that ~2-fold lower levels of TBP co precipitated upon depletion of Taf1 or Taf11, which is expected since both subunits have been implicated in TBP binding (Anandapadamanaban et al., 2013; Lavigne et al., 1999; Shen et al., 2003).
To monitor genome-wide nascent transcription we used native Pol II ChIP to analyze the position of Pol II-DNA complexes before and after 3-IAA addition. Exponentially growing cells in rich media were treated with either 500 μM 3-IAA or DMSO for 30 min. Cells were collected by filtration and quick frozen. For a spike-in control to allow comparison of different samples, frozen cells were mixed with 10% by weight of frozen S. pombe cells, also containing a Rpb3-triple flag tag. Cells were broken under conditions to preserve both chromatin and Pol II elongation complexes, treated with Micrococcal nuclease (MNase), and Pol II complexes isolated by IP. Short DNA fragments associated with Pol II were isolated and sequenced. Assays were performed in biological duplicate. The duplicates were highly similar, hence we performed all further analyses using data combined from the duplicates.
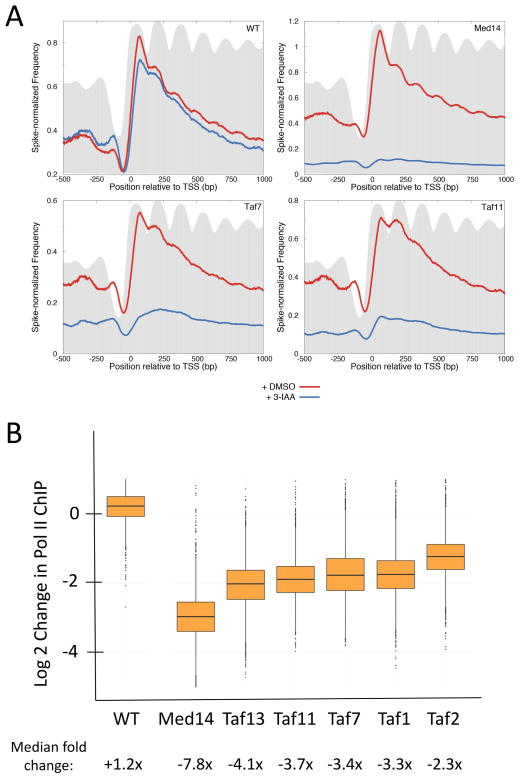
Next we analyzed metagene profiles of Pol II elongation complexes aligned relative to the transcription start site (TSS) and overlaid on histone H3 profiles generated from previously published histone H3 ChIP-seq data (Thurtle and Rine, 2014) (Fig 2A). As previously observed under these growth conditions (Churchman and Weissman, 2011), the average level of Pol II-DNA complexes is highest near the TSS and gradually declines toward the 3′ end of genes (Fig 2A, WT). Addition of either 3-IAA or DMSO to cells lacking a degron gave nearly identical profiles. In contrast, depletion of TFIID subunits TAF7 and TAF11 showed dramatic decreases in genome-wide transcription (Fig 2A, Taf7 and Taf11). Similarly, depletion of the Mediator subunit Med14 resulted in a dramatic decrease in genome-wide Pol II transcription, confirming the essential role of Mediator in transcription (Fig 2A, Med14).
Figure 2.
Genome-wide decreases in transcription upon depletion of TFIID and Mediator subunits. (A) Metagene profile of native Pol II ChIP in wild type and cells containing the indicated IAA7 degron fusion (4807 genes). Results are aligned at the major TSS (Park et al., 2014). Blue line = cells treated with 3-IAA for 30 min prior to harvest; red line = cells treated equivalently but with DMSO. Data are averages of biological duplicates with samples normalized by spike-in with S. pombe Rpb3-Flag cells. Grey shows metagene profile of histone H3 plotted using published H3 ChIP-seq data (Thurtle and Rine, 2014). The degron fused strains are labeled in the upper right corner of the different panels. (B) Box plots show the change in native Pol II ChIP on a log2 scale from 4807 genes comparing DMSO and 3-IAA addition. Median fold-changes in ChIP signals are indicated below. See also Fig S1 and Table S3.
Similar TFIID dependence of Taf-enriched and Taf-depleted genes
To quantitate transcriptional changes at individual genes, the levels of Pol II ChIP (25–140 bp DNA fragments) was averaged over the interval from 1–100 bp downstream from the TSS. This analysis showed that transcription from nearly all Pol II-transcribed genes is dependent on TFIID (Fig 2B; Table S3). The median fold changes in Pol II ChIP levels upon 3-IAA addition were: −7.8 (Med14), −4.1 (Taf13) −3.7 (Taf11) −3.4, Taf7, −3.3 (Taf1) and −2.3 (Taf2). We chose 1–100 bp from the TSS since it captured the highest peak of RNAPII downstream of TSS. We also performed the same analysis on tiling windows of 100 bp in the first 1000 bp downstream of the TSS (or transcription end site (TES), whichever was shorter) and found the fold changes in Pol II upon 3-IAA addition to be similar throughout the gene body (not shown).
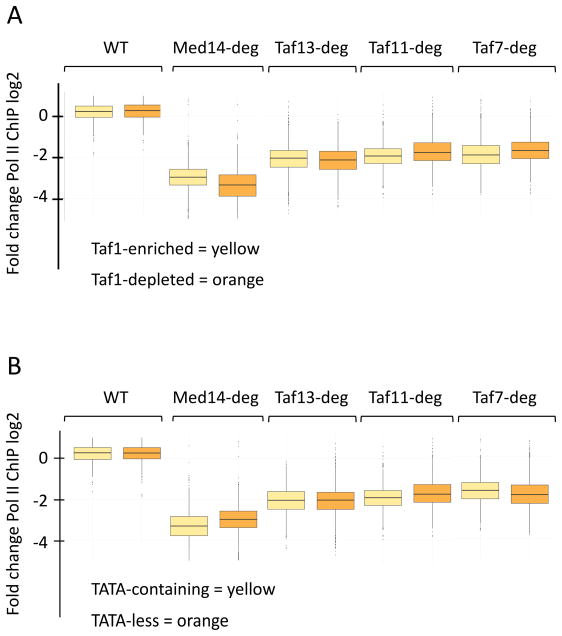
While nearly all genes show decreased transcription upon TFIID inactivation, Fig 2B shows that there are a small number of genes with little or no apparent expression changes upon depletion of individual Tafs. One possibility is that these non-responsive genes are enriched in genes previously characterized as Taf-depleted and/or SAGA-dominated (Huisinga and Pugh, 2004; Rhee and Pugh, 2012). To test this hypothesis, genes were grouped by the previously defined Taf1-enriched and Taf1-depleted categories and analyzed as above (Fig 3A). While the Taf1-depleted gene class shows slightly more sensitivity to Med14 depletion compared to the Taf1-enriched class, genes in both categories show very similar responses to depletion of Tafs 13, 11, and 7. This behavior was also observed for Taf1 and Taf2 (Fig S1B). Previous analyses showed that genes lacking a consensus TATA correlated with Taf1-enriched genes (Basehoar et al., 2004; Rhee and Pugh, 2012). However, in agreement with the above analysis, we found that both TATA-containing and TATA-less genes showed near-identical responses to Taf depletion (Fig 3B). We conclude that, for cells grown in rich media, transcription of nearly all Pol II–transcribed yeast genes is dependent on TFIID.
Figure 3.
TFIID functions at genes previously thought to be less dependent on TFIID. (A) Box plots of data from Fig 3 split into “Taf1-enriched” and “Taf1-depleted” categories (Rhee and Pugh, 2012). (B) Box plots of data from Fig 3 split into TATA-containing and TATA-less promoter categories (Rhee and Pugh, 2012).
Transcription of several Taf1-independent genes is dependent on other TFIID subunits
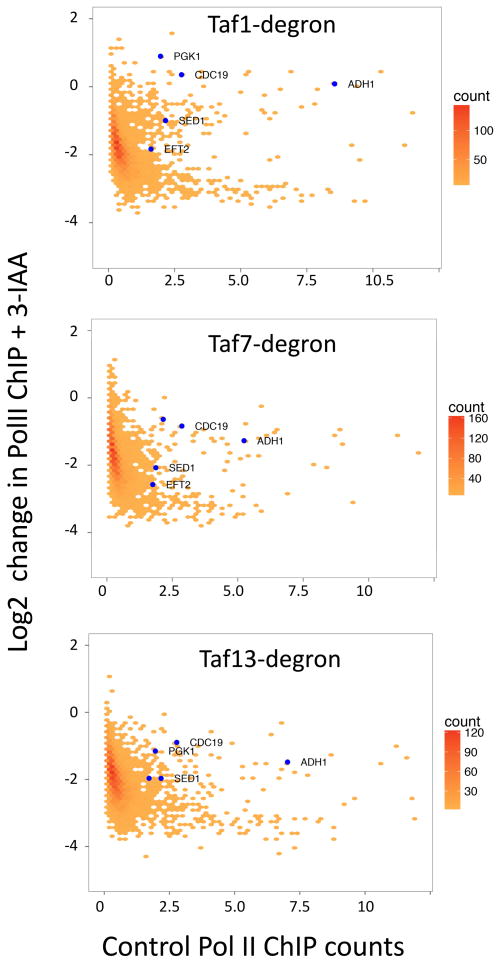
Early work on yeast TFIID function identified several individual genes with low ChIP ratios of Taf1/TBP and for which steady state mRNA levels were insensitive to Taf1 inactivation by heat shock (Kuras et al., 2000; Li et al., 2000). We analyzed our genome-wide data and found that several of these genes (PGK1, CDC19/PYK1, ADH1) were in fact insensitive to Taf1 depletion (Fig 4). However, all of these genes showed greater than a two-fold decrease in Pol II ChIP upon degradation of Taf13 showing that most transcription from these genes is TFIID-dependent.
Figure 4.
Several previously characterized Taf1-independent genes are dependent on other TFIID subunits. Plotted is a hexagonally binned 2d-histogram of Log2 fold change in Pol II ChIP upon 3-IAA addition vs. spike-normalized Pol II ChIP signal (a measure of transcription) averaged over the 1–100 bp from the TSS of each gene (4807 genes). The blue points are genes used in pre genome-wide studies to examine TFIID-dependence (Kuras et al., 2000; Li et al., 2000). See also Figs S1, S2 and Table S3.
To further verify the role of TFIID in transcription, we next examined the effect of Taf1, Taf7 and Med14 depletion on nascent transcription (Bonnet et al., 2014; Duffy et al., 2015; Sun et al., 2013). 30 min after 3-IAA or DMSO addition, RNAs were metabolically labeled for 6 min with 4-thio Uracil and nascent RNAs purified and quantitated by RT qPCR. Selected genes in each of three previously defined classes were analyzed: Taf1-enriched, TFIID-independent, and Taf1-depleted. In agreement with the Pol II ChIP analysis, expression of all genes tested except PGK1, ADH1, and CDC19 were highly sensitive to Taf1 and Taf7 depletion while transcription of all tested genes was dependent on Med14 (Fig S2). From these results we conclude that, while nearly all genes are sensitive to TFIID inactivation, there is a small set of genes that show little change upon inactivation of subunits in TFIID lobe C.
Comparison of genome-wide Taf dependence and TFIID binding
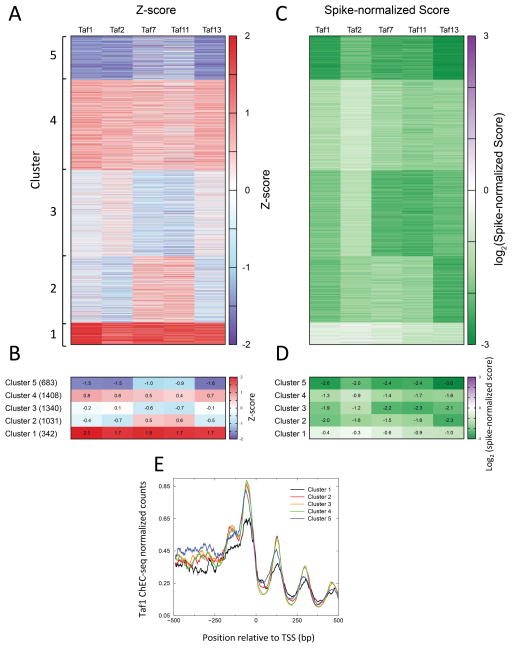
To compare global changes in transcription between the degron strains, we converted the change in Pol II ChIP signals to Z-scores. This ensured that the distribution of changes in Pol II ChIP upon addition of 3-IAA was similar across all five Taf degron strains. K-means clustering of the Z-scores was used to separate genes into 5 clusters (Fig 5A, B, Table S4). Cluster 1 (with 342 genes) has the lowest decrease in Pol II ChIP after depletion of Taf1, Taf2, Taf7, Taf11, and Taf13 while Cluster 5 (with 683 genes) has the largest decrease in Pol II ChIP after 3-IAA addition. Clusters 2, 3, and 4, which make up ~85% of the genes, have a mean Pol II ChIP change within 1 Z-score in all five experiments, implying that majority of genes have similar changes in all five degron strains.
Figure 5.
Analysis of genome-wide dependence on TFIID subunits. (A) Heatmap of genes ordered by clusters that were obtained after k-means clustering of the Z-scores from Taf-degron experiments. (B) The mean Z-scores of the 5 clusters for the 5 different Taf subunit degrons plotted as a heatmap. The mean values are indicated in each heatmap cell. (C) Same ordering of genes as (A), but the spike-normalized change in Pol II at the TSS is plotted. (D) The mean Spike-normalized change of the 5 clusters for the 5 different Taf subunit degrons plotted as a heatmap. (E) Average Taf1 ChEC-seq profiles (Grünberg et al., 2016) plotted for the 5 clusters shown in panel (A). See also Tables S3, S4, and S5.
To understand the magnitude of changes in transcription in the different clusters, we plotted a heat map of the changes in Pol II ChIP signals after Taf-degradation (Fig 5C) and the mean of these signals for each cluster (Fig 5D). We observed that even for Cluster 1, which features the lowest decrease in transcription upon Taf depletion, the mean Pol II ChIP score decreased for all five degron strains. In Cluster 2, which featured slightly different Z-scores for Taf7 and Taf11 compared with Tafs 1, 2 and 13, the log2 fold change in Pol II ChIP was between −1.5 and −2.3 (Fig 5D), indicating that the Z-scores were highlighting differences in experiments that translated to minor differences in the Pol II ChIP scores. We also checked for enrichment of Gene Ontology (GO) terms in the different clusters. We found many metabolic processes to be enriched in Cluster 1, and RNA processes including tRNA, rRNA and ncRNA metabolism enriched in Clusters 3 and 5. All GO terms with false discovery rate < 5% are listed in Table S5.
We next compared the levels of TFIID bound at Pol II promoters to the TFIID-dependence clusters. ChEC-seq was recently used to map genome-wide TFIID binding and we had found that TFIID was localized to nearly all yeast promoters (Grünberg et al., 2016). We next asked whether changes in Pol II ChIP signals observed in clusters 1–5 corresponded to sets of genes with different levels of TFIID. We analyzed the average distribution of Taf1 ChEC-seq signal across the clusters (Fig 5E) and found Cluster 1 to be the only outlier in Taf1 distribution, with Taf1 promoter binding being lower for Cluster 1 compared to other Clusters. It is important to note, however, that genes in Cluster 1 have significant levels of bound TFIID (Fig 5E). Cluster 1 features the weakest decrease in Pol II transcription upon degradation of the Tafs, implying that the genes in this cluster depend less on TFIID for transcription compared to the other clusters. We then asked how these clusters compare to previously identified Taf-enriched and Taf-depleted gene lists. We found Cluster 1 to have a 1.8-fold enrichment of Taf-depleted genes compared to all genes, indicating some correspondence with the published Taf depleted list. However, only 111 genes out of 870 total Taf-depleted genes were part of Cluster 1. Thus, we conclude that, upon Taf depletion, genes in Cluster 1 have smaller decreases in Pol II transcription compared to the global average and this group is enriched for genes that were defined as Taf-depleted in previous studies. Nevertheless, these genes are still TFIID-dependent.
Consistent with these findings, the genes with little or mild Taf1 and Taf7-dependence from Fig 4 and Fig S2 (PGK1, CDC19, ADH1) are found in Cluster 1. The heat map in Fig 5C reveals that genes in Cluster 1 seem less sensitive to depletion of Taf1, Taf2 and Taf7, suggesting that genes in Cluster 1 are less dependent on TFIID subunits in lobe C. Further analysis showed that genes with previously characterized strong TFIID-dependence (RPL25 and RPS5) are in Cluster 5, while most genes previously characterized as Taf1-depleted (e.g., SSB1, VTC1, SSH1, SED1, and ETF2) are clearly TFIID-dependent and found in clusters 2,3,4.
TFIID subunit Taf4 and TFIID/SAGA subunit Taf5 are required for genome-wide Pol II transcription
In an orthogonal approach to test TFIID-dependence of Pol II transcription, the TFIID-specific subunit Taf4 and shared TFIID/SAGA subunit Taf5 were depleted using the anchor-away method in which proteins are rapidly exported from the nucleus to the cytoplasm upon rapamycin addition. Taf4 and Taf5 were fused to FRB in a strain expressing RPL13A-FKBP12 to induce anchoring of Taf4-FRB or Taf5-FRB at ribosomes upon rapamycin treatment (Haruki et al., 2008). Both strains grew normally in the absence of rapamycin (Fig S3A) and both steady state and newly-synthesized mRNA levels are similar to wild-type at 16 assayed genes (Fig S3C,D). As observed for the Taf-degron strains, nuclear depletion of Taf4 or Taf5 for 30 min was not lethal (Fig S3B) but prolonged exposure was lethal. Fractionation of cells after rapamycin treatment showed efficient change in localization of the tagged subunit from the nucleus to the cytoplasm (Fig S3E). Under these conditions, the other TFIID subunits were stable but a portion mislocalized to the cytoplasm. For example, upon Taf5 anchor-away, Taf11 was mostly cytoplasmic while Taf4 stayed mostly nuclear and upon Taf4 anchor-away Taf5 and Taf11 were mostly, but not entirely, nuclear.
Preliminary time course experiments showed that strong transcriptional effects without any detectable growth delay were already observable following 15 min of rapamycin exposure. Thus, anchor-away strains growing in log phase were exposed to rapamycin for 30 min and pulse labeled with 4-thiouracil. Labeled cells were mixed in a fixed ratio with labeled S. pombe cells for spike-in normalization. Purified labeled (newly-synthesized) and total (steady-state) RNA were hybridized to Affymetrix microarrays containing probes for both S. cerevisiae and S. pombe transcriptome.
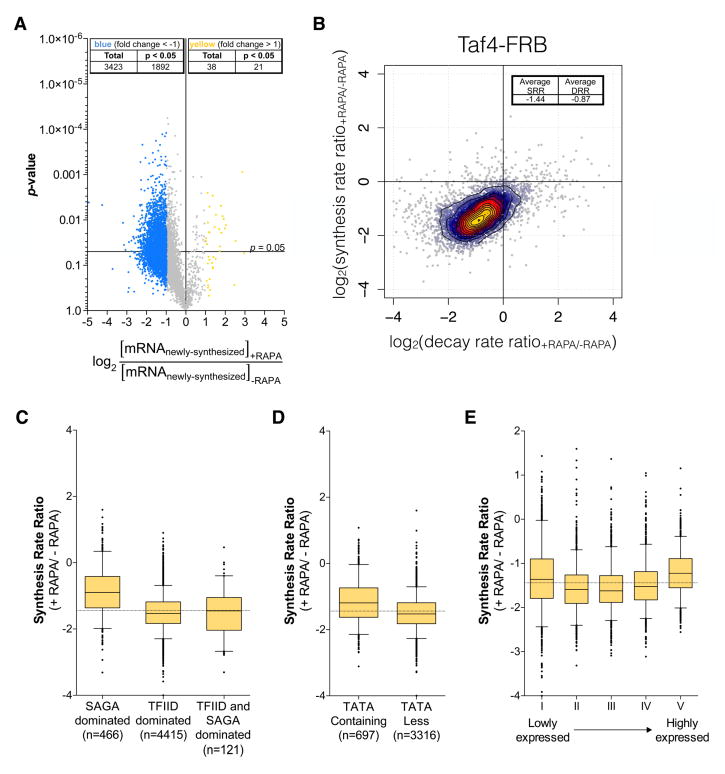
For all yeast genes with a positive signal on microarrays, we measured the ratio of mRNA levels between the TAF4-FRB strain treated and untreated with rapamycin. Newly-synthesized mRNA levels of a large number of genes (3423 out of 5657) were found to be decreased by at least two-fold whereas only 38 genes were upregulated by two-fold (Fig. 6A). We used comparative dynamic transcriptome analysis (cDTA) (Sun et al., 2013), to measure changes in both synthesis and decay rates upon nuclear depletion of Taf4 for this gene set. This analysis revealed that, for the vast majority of genes, a decrease in synthesis rate was compensated by a decrease in decay rates (Fig. 6B). However, this compensation was sub-optimal with a median decrease in synthesis rate by 2.7-fold and a median decrease of decay rate by 1.8-fold, leading to partial buffering of steady state mRNA levels.
Figure 6.
cDTA analysis of Taf4 anchor-away strain. (A) Volcano plot showing fold changes in newly-synthesized mRNA levels upon Taf4 nuclear depletion relative to their significance (p-value). 5657 genes were analyzed in two biological replicates and thresholds of 2-fold change and 0.05 p-values were considered (genes upregulated by 2-fold are indicated in yellow and genes downregulated by 2-fold are indicated in blue). (B) Changes in mRNA synthesis rates for 5434 genes upon rapamycin treatment in a TAF4-FRB strain were plotted against the changes of the decay rates. Log2 values of the average synthesis rate ratio and decay rate ratio are indicated. (C–E) Box plots showing the distribution of changes in synthesis rates upon Taf4 nuclear depletion according to the gene class (C, D) or to the gene expression levels (E) Dotted horizontal lines in panels C, D, E show the median of the synthesis rate ratio. See also Figs S3 and S4.
Additionally, we addressed whether changes in synthesis rates for different genes were correlated to their previously proposed dependence on either TFIID or SAGA or the existence of a TATA consensus sequence in their promoters. Although the transcription of all gene classes was affected by the nuclear depletion of Taf4, the overall synthesis rates of the genes previously designated as TFIID-dominated (or TATA-less genes) decreased more than that of previously designated SAGA-dominated (or TATA-containing) genes (Fig. 6C,D). “SAGA-dominated” gene class had a median fold change of −1.86 whereas the “TFIID-dominated” class had a median fold change of −2.88, suggesting that genes from this latter group are more sensitive to Taf4 depletion. Interestingly, the sensitivity of transcription to Taf4 depletion was not significantly correlated with the level of gene transcription (Fig. 6E and Fig. S4B).
Nuclear depletion of Taf5 had very similar effects on transcription as Taf4 depletion but with a broader effect which might be explained by a combined inactivation of both TFIID and SAGA (see Discussion) (Fig. S4C–I). Indeed, the newly-synthesized mRNA levels of 4238 genes (~75% of analyzed genes) were decreased by at least two-fold (Fig. S4D). However, as observed above with anchor-away of Taf4, our analysis showed that genes of the “TFIID-dominated” and TATA-less gene classes were more sensitive to Taf5 depletion (see Discussion). Nevertheless, our combined results using several different approaches demonstrate that TFIID is required for transcription of the vast majority of yeast genes regardless of previous gene classification.
Genome-wide TFIID dependence can vary with growth conditions
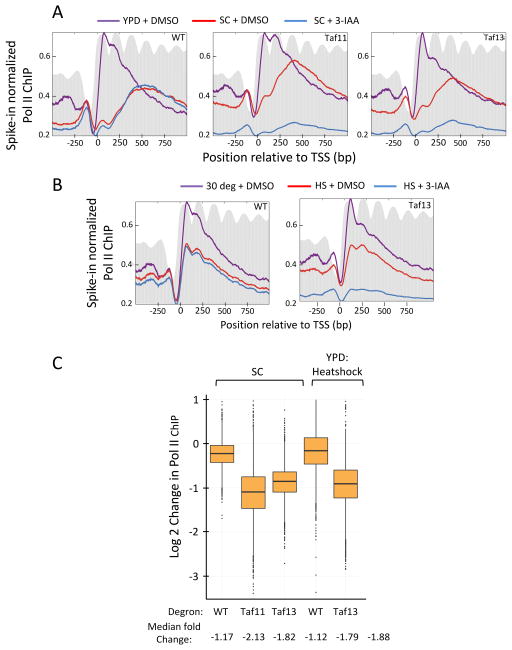
One possible reason for the differences in our findings compared to published studies is a change in cell growth conditions. For example, some previous studies cultured cells in synthetic media and all earlier studies of yeast Taf-dependence involved heat shock of temperature sensitive strains. To test whether these alternative growth conditions altered TFIID-dependence, we repeated the above TFIID depletion experiment with cells grown in synthetic complete glucose media (SC) and with YPD-grown cells that had been subjected to a 30 min heat shock followed by 3-IAA addition. The degron system worked efficiently under these two growth conditions with <10% Taf11 and 13 remaining after 30 min of 3-IAA treatment (Fig S5).
Surprisingly, the Pol II ChIP profile from wild type cells grown in SC (Fig 7A, left panel, red line) was different from YPD-grown cells (left panel, purple line), with the peak of Pol II density over the middle of the coding sequence in SC compared to the peak density of Pol II near the TSS in YPD. This suggests a difference in the rates of initiation and/or elongation in the two growth conditions (Ehrensberger et al., 2013). After Taf depletion in SC, genome-wide Pol II transcription decreased (Fig 7A, middle and right panels), but the magnitude of TFIID-dependence was somewhat lower compared to growth in rich media (Fig 7C). For example, the median Taf11 and Taf13-dependence was −3.7 and −4.1-fold in YPD compared with −2.1 and −1.8-fold in SC.
Figure 7.
TFIID-dependence of transcription in synthetic media and heat shock stress. (A) Metagene profile of native Pol II ChIP from cells grown in synthetic complete (SC) media and containing the indicated IAA7 degron fusion. Results are presented as in Fig 2. Purple line is Pol II ChIP data from YPD-grown cells (+DMSO) from Fig 2; Red and blue lines are cells grown in SC with or without 3-IAA for 30 min. (B) Same as in A but Pol II ChIP from cells under heat shock stress. Purple line is Pol II ChIP from YPD-grown cells (+DMSO) in Fig 2. Red and blue lines are from YPD-grown cells heat shocked for 30 min prior to 30 min addition of DMSO or 3-IAA as indicated. (C) Genome-wide changes in transcription upon TFIID subunit depletion from the data in panels A and B. Box plots show the change in native Pol II ChIP on a log2 scale from 4807 genes comparing DMSO and 3-IAA addition. For heat shock conditions, the plot indicates changes in Pol II ChIP in heat shocked WT and heat shocked degron-containing cells upon addition of 3-IAA. Median fold-changes in ChIP signals are indicated below. See also Figs S5, S6 and S7.
Heat shock of wild type cells results in a general ~1.3-fold decrease in genome-wide transcription, (Fig 7B, left panel, purple line vs red line). Depletion of Taf13 after the 30 min heat shock resulted in a further strong decrease in genome-wide Pol II transcription (Fig 7B, right panel). However, the magnitude of TFIID-dependence was again less than observed for growth at 30 deg (Fig 7C). Under heat shock stress, the median Taf13-dependence was −1.8-fold. However, TFIID dependence in both synthetic media and heat shock conditions were reflected nearly equally in both the SAGA and TFIID-dominated gene classes (Fig S6). For example, Taf13 depletion in synthetic media and heat shock conditions had near equal effects on Taf1-depleted and Taf1-enriched genes as well as genes with or without a consensus TATA box.
Genome-wide comparisons of transcription after Taf-degradation in the three growth conditions are shown in Fig S7. We found that relative Pol II ChIP signals after degradation of individual Tafs are highly correlated between cells grown in YPD and SC at 30 deg and in cells ± heat shock. Therefore, the overall magnitude of TFIID-dependence for genome-wide expression can vary with environmental conditions but the relative dependence on TFIID for expression of individual genes is similar in all tested growth conditions.
DISCUSSION
Previous studies suggested that transcription from most Pol II-transcribed genes can be classified as either dominated by the transcription factors TFIID or SAGA. Here we have used orthogonal approaches to deplete TFIID-specific subunits and measure effects on nascent Pol II transcription. In contrast to earlier conclusions, in rich media, we found a striking genome-wide dependence on TFIID with nearly all genes showing greater than 2-fold decreases in transcription upon TFIID inactivation. Importantly, defects in gene expression observed after degron-dependent Taf 1,2,7, 11 and 13 depletion were similar in the previously defined classes of “Taf1-depleted”, “Taf1-enriched”, TATA-containing and TATA-less genes. This genome-wide TFIID-dependence was observed under three different growth 15 conditions, although the magnitude of TFIID contribution to total Pol II transcription varied with environmental conditions.
Anchor-away depletion of both the TFIID-specific subunit Taf4 and the shared SAGA/TFIID subunit Taf5 also showed decreased transcription from all classes of genes. However, depletion of these two Tafs gave stronger effects at the “TFIID-dominated” and TATA-less gene classes. One possibility is that both of these Tafs are more important for transcription of the TATA-less gene class compared to TATA-containing genes. Alternatively, anchor-away depletion may not be as severe as the degron method or it may result in some partial TFIID and/or SAGA complexes that differentially function at the two gene classes. We attempted to generate an IAA7-tagged Taf4 strain to compare anchor-away and degron depletion of Taf4. However, we were unable to construct this strain, suggesting that the IAA7 tag is detrimental to Taf4 function. In any case, our combined results clearly show that transcription of nearly all genes of both classes is dependent on TFIID.
Several factors likely contribute to the differences between earlier work and the results reported here. First, formaldehyde crosslinking was used in earlier studies to map the location of TFIID subunits. Many studies have shown that, for most proteins in close contact with DNA, formaldehyde crosslinking is a reliable way to map protein-DNA locations. However, the factors Mediator, TFIID and SAGA all show striking differences in genome-wide location when comparing formaldehyde-based methods versus the nuclease-based ChEC-seq method and other functional assays. For example, ChEC-seq analysis of TFIID suggested that TFIID occupies both the “Taf1-depleted” and “Taf1-enriched” classes of genes to the same extent (Grünberg et al., 2016). Similarly, monitoring chromatin modifications and Pol II occupancy suggested that SAGA functions at all expressed genes (Bonnet et al., 2014). While the reasons for these differences in TFIID mapping are not understood, one possible explanation may be the proximity of nucleosomes to PICs at these two promoter types. Genome-wide mapping showed that PICs at TATA-less, “TFIID-dominated” yeast promoters are usually adjacent to the +1 nucleosome while PICs at TATA-containing promoters are usually located upstream from the +1 nucleosome (Rhee and Pugh, 2012). Perhaps protein-protein crosslinking of TFIID to the adjacent nucleosome contributes to stronger ChIP signals at TATA-less versus TATA-containing PICs. It is important to note that, at most so-called Taf1-depleted genes, low but detectable ChIP signals from Taf1 and other Tafs were detected (Kuras et al., 2000; Rhee and Pugh, 2011). A second difference with earlier work is the measurement of newly synthetized transcription instead of steady-state RNA levels. The cDTA analysis here clearly shows that, although some effects are already visible in total levels of RNA, inactivation of TFIID causes a genome-wide stabilization of steady state mRNAs, masking the effects of TFIID depletion. Finally, earlier studies used heat shock to inactivate temperature sensitive TFIID subunits followed by measurement of steady state mRNA levels. We show here that heat shock causes a general genome-wide decrease in Pol II transcription and that transcription under these repressed conditions is somewhat less dependent on TFIID.
Binding of TFIID to Pol II promoters is one of the first steps on the pathway to PIC formation. TFIID interacts with some transcription activators, binds promoter DNA and likely interacts with other basal transcription factors. In metazoans, TFIID (including its TBP subunit) interacts with promoter elements including TATA, Inr, MTE, and DPE. These latter interactions are thought to recruit and stabilize TBP to the ~80% of promoters lacking a consensus TATA. Since ChEC-seq mapping showed that TFIID is present at nearly all promoters, the promoter-binding function of TFIID likely explains the viability of yeast with TBP mutations that disrupt specific TBP-TATA binding (Kamenova et al., 2014). Paradoxically, no conserved promoter elements analogous to the Inr, MTE, and DPE have been identified in yeast so the basis for yeast TFIID promoter recognition is not clear. Lobe C in human TFIID interacts with these downstream promoter elements so it is expected that lobe C subunits, especially Taf1, should be critical for TFIID function (Louder et al., 2016). However, we found a small number of genes where transcription is less sensitive to Taf1, 2 and 7 depletions compared to dependence on Taf11 and Taf13 (Cluster 1). Thus, the downstream promoter DNA binding function of yeast TFIID may not be as critical as in metazoans. Alternatively, other partial TFIID assemblies, such as the core TFIID, the 7TAF or 8TAF complexes (Bieniossek et al., 2013; Trowitzsch et al., 2015; Wright et al., 2006), may carry out partial functions at these promoters, which are less sensitive to Taf1 and/or Taf7 depletions.
A popular view is that TFIID and SAGA can replace each other, each functioning independently of the other at different classes of genes. Results shown in the accompanying paper (Baptista et al, in press) also contradict this view as yeast SAGA was shown to localize to the UAS regions of nearly all yeast genes and to be important for nearly all Pol II transcription, regardless of TATA presence, or Taf1 enrichment measured by ChIP. Our combined results lead to the view that (i) most active yeast promoters have SAGA localized to UASs and TFIID localized at promoters and (ii) that SAGA and TFIID are not alternative factors but both make independent major contributions to expression of nearly all genes. It is important to note that we found newly synthetized transcription more sensitive to Mediator inactivation compared with TFIID inactivation. Therefore, in yeast there may be a low level of transcription at many genes that is TFIID-independent and derives from PICs assembled with TBP, but lacking Tafs. Since the magnitude of TFIID dependence varies with growth conditions, it may be that the level or function of TBP relative to Tafs also varies with environmental conditions. An alternative explanation is that auxin-dependent protein degradation is not complete and that more residual TFIID or partial TFIID complexes (see above) compared with Mediator remains after 3-IAA addition. These scenarios are also plausible when using anchor-away strains, leaving some residual subunits or partial complexes that could replace canonical complexes for transcriptional regulation by holo-TFIID as previously suggested in metazoans (Indra et al., 2005; Tatarakis et al., 2008; Wright et al., 2006).
Finally, many studies showed at least two broad classes of Pol II-transcribed genes: housekeeping and highly regulated genes. Highly regulated genes are usually TATA-containing, modulated by chromatin rearrangement and/or modifications, are sensitive to levels of transcription activators, and are repressed by the regulators NC2 and Mot1. In contrast, the housekeeping class is generally TATA-less, associated with high levels of H4 acetylation in promoter regions, are responsive to a different set of activators and generally activated (or derepressed) by an NC2/Mot1 interplay. These two gene classes are often characterized as TFIID or SAGA-dominated. Our new findings show that these two gene classes are not distinguished by TFIID and SAGA requirements as both these factors make important contributions to expression of nearly all genes. This suggests that some or all of the other properties described above are likely responsible for the differences in regulation observed between these different gene categories.
STAR METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-V5 | Invitrogen | Cat # 46-0705 |
| Anti-FLAG M2 Magnetic Beads | Sigma-Aldrich | Cat # M8823 |
| Rabbit polyclonal anti-Taf1 | Hahn lab | Rabbit # 3506 |
| Rabbit polyclonal anti-Taf4 | Hahn lab | Rabbit # 4022 |
| Rabbit polyclonal anti-Taf12 | Hahn lab | Rabbit # 3434 |
| Rabbit polyclonal anti-Taf3 | Hahn lab | Rabbit # 4045 |
| Rabbit polyclonal anti-TBP | Hahn lab | Rabbit # 7370 |
| Rabbit polyclonal anti-H3 | Abcam | Cat # ab1791 |
| Rabbit polyclonal anti-Taf4 | P. Anthony Weil | N/A |
| Rabbit polyclonal anti-Taf5 | P. Anthony Weil | N/A |
| Rabbit polyclonal anti-Taf11 | P. Anthony Weil | N/A |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| 3-Indoleacetic acid (3-IAA) | Sigma-Aldrich | I3750; CAS: 87-51-4 |
| Micrococcal Nuclease (MNase) | Worthington Biochemical Corporation | Cat # NFCB |
| cOmplete Mini, EDTA-free Protease inhibitor cocktail tablets | Roche | Cat # 11836170001 |
| 4-thiouracil | Sigma-Aldrich | Cat # 440736; CAS: 591-28-6 |
| MTSEA biotin-XX | Biotium | Cat # 90066 |
| Dynabeads MyOne Streptavidin C1 | Invitrogen, ThermoFisher | Cat # 65001 |
| AMPureXP beads | Beckman Coulter | Cat # A63881 |
| EZ-Link HPDP Biotin | ThermoFisher | Cat # 21341 |
| Rapamycin | Euromedex | Cat # SYN-1185 |
| Dyanabeads Protein G | Thermo Fisher Scientific | Cat # 10004D |
| 0.5 mm zirconia disruption beads | Research Products International | Cat # 9834 |
| Critical Commercial Assays | ||
| RiboPure yeast kit | Ambion, Life Technologies | Cat # AM1926 |
| RNeasy MinElute Cleanup Kit | Qiagen | Cat # 742040 |
| μMACS Streptavidin Kit | Miltenyi Biotec | Cat# 130-074-101 |
| Deposited Data | ||
| All sequencing datasets have been uploaded in GEO under accession GSE97081 | ||
| Western blot original images | Mendeley Data file | http://dx.doi.org/10.17632/gwtwc4ndpj.1 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| S. pombe: Strain background: 972h | Laboratory of Gerald Smith | ATCC: 24843 |
| S. cerevisiae: Strain background: BY4705 | Laboratory of Dan Gottschling | ATCC: 200869 |
| Yeast strains, see Table S2 | This paper | N/A |
| Recombinant DNA | ||
| Sequence-Based Reagents | ||
| Primers for RT-qPCR, see Table S1 | This paper | N/A |
| Software and Algorithms | ||
| Bowtie2 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml | N/A |
| Shell Scripts for Sequence Alignment | This paper | |
| Perl Scripts for analysis | This paper | |
| R Scripts for analysis | This paper | |
| Other | ||
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, Dr. Steven Hahn (shahn@fredhutch.org).
METHOD DETAILS
Strain Construction
S. cerevisiae and S. pombe strains were constructed using standard yeast methods. Proteins were chromosomally tagged by high efficiency yeast transformation and homologous recombination of PCR-amplified DNA. Plasmid pFA6a-3V5-IAA7-KanMX6 (from the Karstein Weiss laboratory, UC Berkeley - a gift from M. Miller, FredHutch) was used as the template for generating the IAA7 degron tags. The C-terminal tag contains three copies of the V5 epitope tag followed by the IAA7 degron (32 kDa total).
Yeast Cell Growth, Harvesting, and Breakage of Frozen Cells for Modified Pol II ChIP-seq For each yeast strain and condition, two liters of S. cerevisiae were grown at 30 deg in YPD (+ 100 μl Antifoam 204) to an absorbance of ~0.7–1.0. Cells were then induced for protein degradation by the addition of 10 ml 200X 3-IAA (500 μM final working concentration) dissolved in DMSO, or 10 ml DMSO for the no 3-IAA controls, for 30 minutes at 30 deg. Cells were rapidly harvested by vacuum filtration through a 90 mm diameter 0.45 micron nitrocellulose filter, followed by a 100 ml wash with the cell culture media. The cell paste was scraped off with a pre-chilled spatula and plunged into liquid nitrogen to flash freeze cells. For spike-in normalization, an 8 liter batch of S. pombe was grown in YE (+ Antifoam 204) to a similar absorbance and harvested as described above. 2.5 g S. cerevisiae were combined with 0.25 g S. pombe in a mortar pre-chilled on dry ice and liquid nitrogen. Frozen cells were lysed with a 4.5″ OD mortar and large pestle. Cells were ground for 1 min, liquid nitrogen was then added to the cells and ground for 1 min. The grinding and addition of liquid nitrogen was repeated for a total of 6 minutes. The frozen powdered ground cells were transferred to a cold conical tube and stored at −80 deg.
For the minimal media growth conditions, two liters of S. cerevisiae were grown at 30 deg as above, except in synthetic complete media containing 2% glucose and lacking isoleucine and valine (+ 100 μl Antifoam 204). For heat shock conditions, two liters of S. cerevisiae were grown at 30 deg in YPD as above, except when cells reached an absorbance of ~0.7–1.0 two liters of YPD prewarmed to 44 deg was added and cells were transferred to a 37 deg C shaker for 30 min before the addition of 3-IAA for 30 min. Cells were then harvested and processed as described above.
Chromatin Extract Preparation and MNase Digestion
0.5 g frozen cells were added to a pre-chilled 15 ml conical tube and kept on dry ice until all cell preps were weighed. Tubes containing frozen cells were placed on ice for 5 min to slowly warm. 2.5 ml cold lysis buffer + protease inhibitors (50 mM HEPES, 150 mM NaCl, 0.5 mM EDTA, 0.05% TritonX-100, pH 7.4, cOmplete Mini protease inhibitor tablet (Roche)) was added to the cells and the cells were allowed to resuspend with occasional mixing on ice for ~ 5–10 min. These resuspended, lysed cells were incubated in a 37 deg water bath for 5 min, then 2.5 mM CaCl2 and 5 μl 100 U/μl MNase were added and mixed by inversion. Chromatin was MNase digested for 5 min in a 37 deg C water bath, then the reaction was stopped by addition of EGTA to a final concentration of 12 mM. Reactions were incubated on ice for at least 10 minutes while other samples were processed. Extracts were spun at 3K x g for 2 min at 4 deg., then the supernatant transferred to a new tube and spun at 17K x g for 15 min at 4 deg. Supernatant was used immediately for IP.
IPs to Recover Elongation Complexes
1 ml of MNase-treated chromatin extract from above was incubated with 100 μl magnetic anti-Flag beads (prewashed 3 times in lysis buffer + 5 mg/ml BSA + 1.5 mM EDTA + cOmplete Mini protease inhibitor tablet Roche) overnight at 4 deg with rotation. Beads were washed 4 times with 1.5 ml lysis buffer + 1.5 mM EDTA and resuspended in 400 μl same buffer. DNA was eluted by the following: add 20 μg RNaseA and additional NaCl and EDTA so that the final concentrations increase by 100 mM (NaCl) and 10 mM (EDTA). Samples were incubated at 37 deg for 10 min with mixing. 0.5% SDS (final concentration) and 80 μg Proteinase K (Invitrogen) were added and samples incubated at 65 deg for 20 min with mixing. Supernatant was phenol/CHCl3/IAA extracted and DNA precipitated with 30 μg glycogen (Roche), 1/10 vol 3 M NaOAc, and 0.7 vol isopropanol. DNA was resuspended in 0.1X TE and quantitated by PicoGreen assay (Thermo Fisher).
Preparation of DNA Libraries for NGS
DNA isolated from IPs was prepared for sequencing as previously described (Skene and Henikoff, 2017), except for the following changes. End Repair and A-tailing was performed in a 20 μl volume containing up to 50 ng DNA and the following final concentrations: 1X T4 DNA ligase buffer (NEB #B0202S), 0.5 mM each dNTP, 0.25 mM ATP, 2.5% PEG 4000, 2.5 U T4 PNK (NEB #M0201S), 0.05 U T4 DNA polymerase (Invitrogen #18005025), and 0.05 U Taq DNA polymerase and reactions were incubated at 12 deg 15 min, 37 deg 15 min, 72 deg 20 min, then put on ice and immediately used in adapter ligation reactions. Adapter Ligation was performed in a 50 μl volume containing 100:1 adapter:insert molar ratios, 1X Rapid DNA ligase buffer (Enzymatics #B101L) and 3000 U DNA ligase (Enzymatics #L6030-HC-L) and reactions were incubated at 20 deg for 15 min. A single post-ligation cleanup was performed using 0.4X vol AMPure XP reagent (Beckman Coulter #A63881) and DNA eluted in 40 ul 0.1X TE. Library Enrichment was performed in a 25 μl volume containing 16.25 μl DNA from previous step and the following final concentrations: 1X KAPA buffer, 0.3 mM each dNTP, 0.5 μM each P5 and P7 PCR primer, and 0.5 U KAPA HS HIFI polymerase (#KK2502). DNA was amplified with the following program: 98 deg 45 sec, [98 deg 15 sec, ramp to 60 deg @ 1 deg/sec, 60 deg 20 sec, ramp to 98 deg @ 1 deg/sec] 8–10X, 72 deg 1 min. A post-PCR cleanup was performed using 1.4X vol AMPure XP reagent and DNA was eluted into 40 μl 0.1X TE. Libraries were sequenced on the Illumina HiSeq2500 platform using 25 bp paired-ends at the Fred Hutchinson Cancer Research Center Genomics Shared Resources facility.
Nascent RNA Labeling and Purification for RTqPCR
S. cerevisiae and S. pombe cells were grown and newly synthesized RNAs were labeled as previously described (Bonnet et al., 2014), except that S. cerevisiae strains at an A600 of 1.0 were treated with 500 μM 3-IAA or DMSO for 30 minutes immediately prior to RNA labeling. Labeled S. cerevisiae and S. pombe cells were mixed in a 3:1 ratio before total RNA was extracted using the RiboPure yeast kit (Ambion, Life Technologies). RNA was then biotinylated essentially as described (Duffy and Simon, 2016; Duffy et al., 2015) using 40 μg total RNA and 4 μg MTSEA biotin-XX (Biotium). Unreacted MTS-biotin was removed by phenol/CHCl3 extraction and RNA was precipitated by isopropanol precipitation. Biotinylated RNA was purified also as described (Duffy and Simon, 2016) using MyOne Streptavidin C1 Dynabeads (Invitrogen) and RNAs eluted into 50 μl streptavidin elution buffer (100 mM DTT, 20 mM HEPES 7.4, 1 mM EDTA, 100 mM NaCl, 0.05% Tween-20). At this point, 10% input RNA (4 μg) was diluted into 50 μl streptavidin elution buffer and processed the same as the labeled RNA samples. 50 μl each input and purified RNA was adjusted to 100 μl with nuclease-free water and purified on RNeasy columns (Qiagen) using the modified protocol as described (Duffy and Simon, 2016). RNAs were eluted into 14 μl nuclease-free water.
cDNA Synthesis and Quantitative PCR of Nascent RNA
Eleven microliters RNA was used to generate cDNA using Transcriptor (Roche), random hexamer primer, and the manufacturer’s instructions. cDNA was used either undiluted, 1/20, or 1/100 for quantitative PCR (qPCR) depending on the gene analyzed. Gene specific qPCR was performed as previously described (Herbig et al., 2010) except that primers were designed near the 5′ end of the gene and reactions were run on the QuantStudio5 Real-Time System (ABI). Relative amounts of transcript were normalized to S. pombe tubulin transcripts.
cDTA analysis
Comparative dynamic transcriptome analysis (cDTA) is an approach that allows analysis of newly-synthesized RNA in order to determine transcriptional defects at much higher sensitivity than those methods solemnly relying on evaluation of steady-state levels of RNA. This method relies on a short, non-perturbing metabolic labeling of newly-synthesized RNAs with 4-thiouracil for 6 min. After microarray hybridization or RNA sequencing, it is possible to determine synthesis and decays rates simultaneously for every transcript.
For each yeast strain and condition, 200mL of S. cerevisiae cells were grown in YPD medium at 30°C to an OD600 ≈ 0.8. At that point, the culture was divided into two equal volumes of 100mL each and to one of those cultures rapamycin was added (+RAPA) until a final concentration of 1μg/mL, for 30 min, to allow nuclear depletion of either Taf4 or Taf5. To the other half of the culture, the control samples (-RAPA/minus rapamycin), only the vehicle (DMSO) was added. Newly synthesized RNAs were labeled for 6 min by adding 4-thiouracil (Sigma-Aldrich) until a final concentration of 5mM. In parallel, wild-type S. pombe cells were similarly grown in YES medium, at 31°C, and labeled for 6 min to be used as a spike-in across all samples. Cells were immediately pelleted and flash-frozen in liquid N2 and stored at −80°C until further use. Immediately before freezing the cells, a small aliquot was collected for cell counting purposes. S. cerevisiae and S. pombe cells were mixed in a ratio of 3:1. Total RNAs were extracted using RiboPure yeast kit (Ambion, Life Technologies) according to the description provided by the manufacturer.
Prior to biotinylation, RNA samples were heated for 10 min at 60°C and cooled down on ice for another 5 min. Subsequently, RNA biotinylation was carried out on 200μg of total RNA using 200μL of 1mg/mL EZ-link HPDP- Biotin (Pierce) in 100μL of biotinylation buffer (100mM Tris- HCl at pH 7.5, 10mM EDTA) and adjusted to a final volume of 1000μL with DEPC-treated RNase-free water (Sigma-Aldrich) for 3 h at room temperature, protected from light and gentle agitation. After chloroform extraction and isopropanol precipitation (1/10 vol 5M NaCl and 2.5 vol isopropanol), purified RNAs were suspended in 100μL of DEPC-treated RNase-free water (Sigma-Aldrich).
Recovered RNA samples were incubated at 65°C for 10 min and allowed to cool down on ice for 5 min. Newly synthesized biotinylated RNAs were bound to 100μL of μMACS streptavidin microbeads (Miltenyi Biotec) for 90 min at room temperature with gentle shaking. Purification of labeled RNA was then carried out using μMACS streptavidin starting kit (Miltenyi Biotec). Columns were first equilibrated with 1mL of washing buffer (100mM Tris-HCl at pH 7.5, 10mM EDTA, 1M NaCl, 0.1% Tween20). Samples were passed through the columns twice and washed five times with increasing volumes of washing buffer (600, 700, 800, 900, and 1000μL). Ultimately, labeled RNAs were eluted twice with 200μL of 100mM DTT. Following ethanol precipitation (overnight precipitation in 1/10 vol of 3M NaOAc, 3 vol of 100% ethanol and 20μg of RNA-grade glycogen), RNAs were washed in ice-cold 70% ethanol and resuspended in 20μL of DEPC-treated RNase-free water (Sigma-Aldrich). The quality of the samples was ascertained and finally hybridized to GeneChip Yeast Genome 2.0 microarrays following the instructions from the supplier (Affymetrix). All experiments were done using two independent biological replicates.
Raw data were normalized to S. pombe signal and used to calculate fold-changes in total and newly-synthesized RNA levels, as represented in volcano plots. Further calculations of synthesis and decay rates, based on mathematical model as described in Sun et al., were performed using a pipeline and R/Bioconductor package publicly available (Schwalb et al., 2012).
Cell fractionation, growth curve and cell viability
150 ml cell culture (OD600 ≈ 0.5) was harvested and treated with 0.1M Tris, pH 9.4, 10mM DTT for 5 min at room temperature. Cells were pelleted and resuspended in YPD/S (YPD with 1M Sorbitol) and collected by centrifugation. Cells were resuspended in 1mL YPD/S and 750μL 2M sorbitol. Zymolyaze 100T was added until a final concentration of 0.3mg/mL. Cells were incubated at 30°C for 30min (progress of spheroplasting every was checked every 10min). After this treatment, spheroplasts were washed and resuspended in room temperature YPD/S for 30 min at 30°C. Cells were quickly cooled down in ice, centrifuged, washed and resuspended in 1.5 ml lysis buffer (20 mM K-phosphate pH 6.5, 0.5 mM MgCl2, 18% Ficoll, 1XPIC). Spheroplasts were lysed three times with a B dounce and shortly centrifuged at 4500g to remove unbroken cells. The precleared whole cell lysate (Total lysate fraction) was centrifuged at 21000g for 45 min resulting into a crude nuclear pellet (nuclear fraction) and post-nuclear supernatant cytosol fraction (cytoplasmic fraction). 200μl of each fraction was TCA precipitated, washed with ice-cold acetone and resuspended in SDS-sample buffer.
For growth curve analysis, the anchor-away (TAF4-FRB and TAF5-FRB) and parental strains were grown overnight and then diluted to OD600 ≈ 0.1 in pre-heated YPD (200mL). To the culture, rapamycin was added until a final concentration of 1μg/mL (+ RAPA). To the control samples (-RAPA), only the vehicle (DMSO) was added. Every 30min, OD600 was measured until saturation of the culture.
For cell viability, strains were grown until log-phase (OD600 ≈ 0.7). To the culture, rapamycin was added until a final concentration of 1μg/mL (+ RAPA). To the control samples (-RAPA), only the vehicle (DMSO) was added. After 30min an aliquot was collected, stained with methylene blue and counted using a neubauer chamber.
TFIID Immunoprecipitations and Western Blot Analysis
For each yeast strain, 400 ml of S. cerevisiae was grown at 30 deg in YPD to an absorbance of ~0.7–1.0. Cultures were then split into two 200 ml flasks and were treated by addition either 1 ml of 200X 3-indoleacetic acid (3-IAA) (500 μM final working concentration) dissolved in DMSO, or 1 ml DMSO for the no 3-IAA controls, for 30 minutes at 30 deg. Cells were collected by centrifugation, washed with 25 ml IP buffer (40 mM HEPES, 150 mM NaCl, 10% glycerol, 0.1% Tween-20) supplemented with 1 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonyl fluoride (PMSF), 0.31 mg/ml benzamidine, 0.3 μg leupeptin, 1.4 μg/ml pepstatin, and 2 μg/ml chymostatin, and then resuspended in ~750 μl of the same buffer. Cells were transferred to a 2 ml tube containing 1.5 ml 0.5 mm zirconia beads and small scale whole cell extracts were prepared in a mini bead beater-96 (BioSpec products) by shaking 5 times for 3 min each with 1 min chill in ice water between each lysis step. Lysates were clarified by a 3000 x g spin for 5 min followed by a high speed spin in a microfuge at 20K x g for 15 min. Concentrations of extracts were determined by Bradford assays. For IP experiments, 5 mg of extracts were incubated at 4°C overnight with 30 μl Protein G Dynabeads which had been conjugated and crosslinked with α-Taf4 antibody or no antibody as a control. For antibody conjugation, Protein G Dynabeads were washed twice with PBS, then resuspended in PBS keeping the beads at 30 mg/ml, and either 10 μl rabbit polyclonal α-Taf4 or 10 μl PBS then rotated at room temp for 45 min. Beads were washed 3X in PBS, 1X in 0.2M triethanolamine pH 8.2, then resuspended in 0.2M triethanolamine pH 8.2 containing 25 mM dimethyl pimelimidate (DMP), rotated at room temp for 30 min, then washed 1X in PBS. The DMP incubation and PBS wash were repeated for a total of 3 times. Beads were washed 1X in 0.1M ethanolamine and rotated for 5 min, then washed 1X in PBS. Beads were washed in 1M glycine pH 3.0 and rotated for 10 min at room temp and then repeated. Beads were finally washed 3X in IP buffer supplemented with 1 mM DTT and protease inhibitors listed above. Following the overnight IP step, beads were washed 3X with IP buffer, then proteins were eluted by incubating beads in 15 μl 1 M glycine pH 2.5 for 10 min at room temperature. The elution step was repeated and elutions combined. 10 μl 1M Tris pH 8.5 was added to the combined eluate. Eluted proteins were separated by SDS-PAGE and analyzed by Western blot using rabbit polyclonal antibodies. Protein signals were quantified using Odyssey scanner software (Li-Cor) by generating a standard curve using a titration from WT IP. Each protein analyzed was normalized to the amount of the Taf4 protein obtained by IP.
Yeast Viability Following 3-IAA Treatment
For each yeast strain, 15 ml of S. cerevisiae were grown at 30 deg in YPD to an absorbance of ~0.5–0.7. Cultures were split into two 5 ml cultures and were then induced for protein degradation by the addition of 25 μl 200X 3-IAA (500 μM final working concentration) dissolved in DMSO, or 25 μl DMSO for the no 3-IAA controls, for 30 minutes at 30 deg. Cultures were diluted in H2O and plated to YPD plates for recovery at 30 deg. Colonies were counted after 3 days, and the percent viability was calculated for cells treated with 3-IAA versus DMSO only treatment. Each strain was grown and assayed in duplicate.
QUANTIFICATION AND STATISTICAL ANALYSIS
Analysis of ChIP-Seq Data
Paired-end reads were independently aligned to S. cerevisiae reference genome (sacCer3) and S. pombe reference genome (ASM294v2.20). Coverage at each base-pair of the S. cerevisiae reference genome was calculated as number of reads of given length (25–140 bp for the analyses presented here) that mapped at that position normalized by the factor N:
Here 10,000 is an arbitrarily chosen number. The normalized coverage at each base-pair from each replicate was averaged when combining multiple replicates. TSS positions were obtained from (Park et al., 2014). Only those genes were considered which had no overlap in with other genes in the TSS - 50 bp to TSS + 50% of gene length interval, which resulted in a list of 4808 genes. For quantifying changes in Pol II levels upon 3-IAA addition to various strains, first, total normalized signal per bp in the interval TSS to TSS+100 bp was calculated for each gene in the +DMSO and +3-IAA datasets. Then, a log ratio of +3-IAA to +DMSO was calculated as the net effect of depletion of different factors in the degron strains. For nucleosome profiles shown in Fig. 2, 140–155 bp fragments from published H3 ChIP-seq paired-end data (SRA Accession: SRR1105662) was used and coverage at each position of a gene was normalized to the average reads over TSS ± 1000 bp interval of that gene.
For determining clusters of genes that change concordantly due to Taf degradation across the 5 Taf degron experiments (Taf1, Taf2, Taf7, Taf11, and Taf13), we first converted the log2 changes in spike-normalized Pol2 enrichment to Z-scores using the mean and standard deviation calculated by fitting a Gaussian function to the log2 distributions. We then performed k-means clustering on the Z-scores across five distributions using Cluster 3.0 (de Hoon et al., 2004) using Euclidean distance as the similarity metric and organizing the genes into 5 clusters. Published Taf1 ChEC-seq data (GEO: GSM2149798) (Grünberg et al., 2016) was used in Figure 4E.
DATA AND SOFTWARE AVAILABILITY
All sequencing datasets have been uploaded in GEO under accession GSE97081. Original Western blot images have been deposited at mendeley.com: http://dx.doi.org/10.17632/gwtwc4ndpj.1
Supplementary Material
Table S3 (Related to Figs 2, 7 and S1). Summary of Spike-normalized Pol II ChIP changes after 3-IAA addition.
Table S4 (related to Fig 5). TFIID-dependent Gene Clusters
Table S5 (related to Fig 5). TFIID-dependent GO terms analysis
Acknowledgments
We thank Steve Henikoff (FHCRC) for support, advice and encouragement throughout the course of this work, Matt Miller and Toshi Tsukiyama (FHCRC) for reagents, advice and assistance with the auxin degron system, Christine Codomo (FHCRC) for advice on construction of DNA sequencing libraries, Erin Duffy and Matt Simon (Yale) for advice on labeling and purification of nascent mRNA, Marc HT Timmers (UMC Utrecht, The Netherlands) for anchor-away strains and Sebastian Grünberg, Rafał Donczew and Steve Henikoff for comments on the manuscript. Supported by NIH grant GM053451 (S Hahn) and the Howard Hughes Medical Institute (to S. Henikoff). TB was funded by a Marie Curie Initial Training Networks (EU-FP7 PEOPLE-2013 program, PITN-GA-2013-606806, NR-NET) and Association pour la Recherche sur le Cancer fellowships. This study was also supported by the European Research Council (ERC) Advanced grant (ERC-2013-340551, Birtoaction, to LT), ANR grant ANR-15-CE11-0022 (SAGA2) and grant ANR-10-LABX-0030-INRT, a French State fund managed by the Agence Nationale de la Recherche under the frame program Investissements d’Avenir ANR-10-IDEX-0002-02 to IGBMC.
Footnotes
AUTHOR CONTRIBUTIONS
SH, LW, SR, TB, DD and LT conceived and designed the experiments, LW performed all auxin-degron experiments, 4 thioU labeling and RT qPCR analysis of degron-containing cells and TFIID IPs. SR performed the computational analysis of all auxin-degron experiments. TB performed experiments using anchor-away strains (4 thioU labeling, genome wide transcriptional analyses, computational analyses and cell fractionation experiments). All authors analyzed the results and wrote the manuscript.
Supplemental Information includes 7 figures and 5 tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anandapadamanaban M, Andresen C, Helander S, Ohyama Y, Siponen MI, Lundström P, Kokubo T, Ikura M, Moche M, Sunnerhagen M. High-resolution structure of TBP with TAF1 reveals anchoring patterns in transcriptional regulation. Nat Struct Mol Biol. 2013;20:1008–1014. doi: 10.1038/nsmb.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- Berger I, Blanco AG, Boelens R, Cavarelli J, Coll M, Folkers GE, Nie Y, Pogenberg V, Schultz P, Wilmanns M, et al. Structural insights into transcription complexes. Journal of Structural Biology. 2011;175:135–146. doi: 10.1016/j.jsb.2011.04.015. [DOI] [PubMed] [Google Scholar]
- Bieniossek C, Papai G, Schaffitzel C, Garzoni F, Chaillet M, Scheer E, Papadopoulos P, Tora L, Schultz P, Berger I. The architecture of human general transcription factor TFIID core complex. Nature. 2013;493:699–702. doi: 10.1038/nature11791. [DOI] [PubMed] [Google Scholar]
- Bonnet J, Wang CY, Baptista T, Vincent SD, Hsiao WC, Stierle M, Kao CF, Tora L, Devys D. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes Dev. 2014;28:1999–2012. doi: 10.1101/gad.250225.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfrocco MA, Nogales E. Regulatory interplay between TFIID’s conformational transitions and its modular interaction with core promoter DNA. Transcription. 2013;4 doi: 10.4161/trns.25291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfrocco MA, Kassavetis GA, Grob P, Fang J, Juven-Gershon T, Kadonaga JT, Nogales E. Human TFIID Binds to Core Promoter DNA in a Reorganized Structural State. Cell. 2013;152:120–131. doi: 10.1016/j.cell.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics (Oxford, England) 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- de Jonge WJ, O’Duibhir E, Lijnzaad P, van Leenen D, Groot Koerkamp MJ, Kemmeren P, Holstege FC. Molecular mechanisms that distinguish TFIID housekeeping from regulatable SAGA promoters. Embo J. 2016 doi: 10.15252/embj.201695621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy EE, Simon MD. Enriching s(4) U-RNA Using Methane Thiosulfonate (MTS) Chemistry. Curr Protoc Chem Biol. 2016;8:234–250. doi: 10.1002/cpch.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy EE, Rutenberg-Schoenberg M, Stark CD, Kitchen RR, Gerstein MB, Simon MD. Tracking Distinct RNA Populations Using Efficient and Reversible Covalent Chemistry. Mol Cell. 2015;59:858–866. doi: 10.1016/j.molcel.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrensberger AH, Kelly GP, Svejstrup JQ. Mechanistic interpretation of promoter-proximal peaks and RNAPII density maps. Cell. 2013;154:713–715. doi: 10.1016/j.cell.2013.07.032. [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. Human housekeeping genes, revisited. Trends Genet. 2013;29:569–574. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR, Workman JL. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell. 1998;94:45–53. doi: 10.1016/s0092-8674(00)81220-9. [DOI] [PubMed] [Google Scholar]
- Grünberg S, Henikoff S, Hahn S, Zentner GE. Mediator binding to UASs is broadly uncoupled from transcription and cooperative with TFIID recruitment to promoters. Embo J. 2016;35:2435–2446. doi: 10.15252/embj.201695020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S, Young ET. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics. 2011;189:705–736. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimovich G, Medina DA, Causse SZ, Garber M, Millán-Zambrano G, Barkai O, Chávez S, Pérez-Ortín JE, Darzacq X, Choder M. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013;153:1000–1011. doi: 10.1016/j.cell.2013.05.012. [DOI] [PubMed] [Google Scholar]
- Han Y, Luo J, Ranish J, Hahn S. Architecture of the Saccharomyces cerevisiae SAGA transcription coactivator complex. Embo J. 2014;33:2534–2546. doi: 10.15252/embj.201488638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Indra AK, Mohan WS, Frontini M, Scheer E, Messaddeq N, Metzger D, Tora L. TAF10 is required for the establishment of skin barrier function in foetal, but not in adult mouse epidermis. Developmental Biology. 2005;285:28–37. doi: 10.1016/j.ydbio.2005.05.043. [DOI] [PubMed] [Google Scholar]
- Kamenova I, Warfield L, Hahn S. Mutations on the DNA binding surface of TBP discriminate between yeast TATA and TATA-less gene transcription. Mol Cell Biol. 2014;34:2929–2943. doi: 10.1128/MCB.01685-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras L, Kosa P, Mencia M, Struhl K. TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000;288:1244–1248. doi: 10.1126/science.288.5469.1244. [DOI] [PubMed] [Google Scholar]
- Lavigne AC, Gangloff YG, Carré L, Mengus G, Birck C, Poch O, Romier C, Moras D, Davidson I. Synergistic Transcriptional Activation by TATA-Binding Protein and hTAFII28 Requires Specific Amino Acids of the hTAFII28 Histone Fold. Mol Cell Biol. 1999;19:5050–5060. doi: 10.1128/mcb.19.7.5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- Leurent C, Sanders SL, Demény MA, Garbett KA, Ruhlmann C, Weil PA, Tora L, Schultz P. Mapping key functional sites within yeast TFIID. Embo J. 2004;23:719–727. doi: 10.1038/sj.emboj.7600111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leurent C, Sanders S, Ruhlmann C, Mallouh V, Weil PA, Kirschner DB, Tora L, Schultz P. Mapping histone fold TAFs within yeast TFIID. Embo J. 2002;21:3424–3433. doi: 10.1093/emboj/cdf342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XY, Bhaumik SR, Green MR. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- Louder RK, He Y, López-Blanco JR, Fang J, Chacón P, Nogales E. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature. 2016;531:604–609. doi: 10.1038/nature17394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencía M, Moqtaderi Z, Geisberg JV, Kuras L, Struhl K. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol Cell. 2002;9:823–833. doi: 10.1016/s1097-2765(02)00490-2. [DOI] [PubMed] [Google Scholar]
- Munchel SE, Shultzaberger RK, Takizawa N, Weis K. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Molecular Biology of the Cell. 2011;22:2787–2795. doi: 10.1091/mbc.E11-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F, Tora L. Chromatin and DNA sequences in defining promoters for transcription initiation. Biochim Biophys Acta. 2014;1839:118–128. doi: 10.1016/j.bbagrm.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Meth. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Ohtsuki S, Levine M, Cai HN. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Morris AR, Battenhouse A, Iyer VR. Simultaneous mapping of transcript ends at single-nucleotide resolution and identification of widespread promoter-associated non-coding RNA governed by TATA elements. Nucleic Acids Res. 2014;42:3736–3749. doi: 10.1093/nar/gkt1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C, Nozawa K, Cramer P. Mediator Architecture and RNA Polymerase II Interaction. J Mol Biol. 2016;428:2569–2574. doi: 10.1016/j.jmb.2016.01.028. [DOI] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Comprehensive Genome-wide Protein-DNA Interactions Detected at Single-Nucleotide Resolution. Cell. 2011;147:1408–1419. doi: 10.1016/j.cell.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012 doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury S, Bernecky C, Cramer P. Structural basis of transcription initiation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:129–143. doi: 10.1038/nrm3952. [DOI] [PubMed] [Google Scholar]
- Schwalb B, Schulz D, Sun M, Zacher B, Dümcke S, Martin DE, Cramer P, Tresch A. Measurement of genome-wide RNA synthesis and decay rates with Dynamic Transcriptome Analysis (DTA) Bioinformatics (Oxford, England) 2012;28:884–885. doi: 10.1093/bioinformatics/bts052. [DOI] [PubMed] [Google Scholar]
- Setiaputra D, Ross JD, Lu S, Cheng DT, Dong MQ, Yip CK. Conformational flexibility and subunit arrangement of the modular yeast Spt-Ada-Gcn5 acetyltransferase complex. Journal of Biological Chemistry. 2015;290:10057–10070. doi: 10.1074/jbc.M114.624684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WC, Bhaumik SR, Causton HC, Simon I, Zhu X, Jennings EG, Wang TH, Young RA, Green MR. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. Embo J. 2003;22:3395–3402. doi: 10.1093/emboj/cdg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife. 2017;6:e21856. doi: 10.7554/eLife.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Schwalb B, Pirkl N, Maier KC, Schenk A, Failmezger H, Tresch A, Cramer P. Global analysis of eukaryotic mRNA degradation reveals xrn1-dependent buffering of transcript levels. Mol Cell. 2013;52:52–62. doi: 10.1016/j.molcel.2013.09.010. [DOI] [PubMed] [Google Scholar]
- Sun M, Schwalb B, Schulz D, Pirkl N, Etzold S, Larivière L, Maier KC, Seizl M, Tresch A, Cramer P. Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Res. 2012;22:1350–1359. doi: 10.1101/gr.130161.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarakis A, Margaritis T, Martinezjimenez C, Kouskouti A, Mohanii W, Haroniti A, Kafetzopoulos D, Tora L, Talianidis I. Dominant and Redundant Functions of TFIID Involved in the Regulation of Hepatic Genes. Mol Cell. 2008;31:531–543. doi: 10.1016/j.molcel.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Theisen JWM, Lim CY, Kadonaga JT. Three key subregions contribute to the function of the downstream RNA polymerase II core promoter. Mol Cell Biol. 2010;30:3471–3479. doi: 10.1128/MCB.00053-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurtle DM, Rine J. The molecular topography of silenced chromatin in Saccharomyces cerevisiae. Genes Dev. 2014;28:245–258. doi: 10.1101/gad.230532.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowitzsch S, Viola C, Scheer E, Conic S, Chavant V, Fournier M, Papai G, Ebong IO, Schaffitzel C, Zou J, et al. Cytoplasmic TAF2-TAF8-TAF10 complex provides evidence for nuclear holo-TFIID assembly from preformed submodules. Nat Commun. 2015;6:6011. doi: 10.1038/ncomms7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KL, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Asturias FJ. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell. 2014;157:1430–1444. doi: 10.1016/j.cell.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrijzer CP, Chen JL, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- Wright KJ, Marr MT, Tjian R. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc Natl Acad Sci USA. 2006;103:12347–12352. doi: 10.1073/pnas.0605499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabidi MA, Stark A. Regulatory Enhancer-Core-Promoter Communication via Transcription Factors and Cofactors. Trends Genet. 2016;32:801–814. doi: 10.1016/j.tig.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3 (Related to Figs 2, 7 and S1). Summary of Spike-normalized Pol II ChIP changes after 3-IAA addition.
Table S4 (related to Fig 5). TFIID-dependent Gene Clusters
Table S5 (related to Fig 5). TFIID-dependent GO terms analysis