Abstract
Select CMV epitopes drive life-long CD8+ T cell memory inflation, but the extent of CD4 memory inflation is poorly studied. CD4+ T cells specific for human CMV (HCMV) are elevated in HIV+ HCMV+ subjects. To determine whether HCMV epitope-specific CD4+ T cell memory inflation occurs during HIV infection, we used HLA-DR7 tetramers loaded with the glycoprotein-B DYSNTHSTRYV (DYS) epitope to characterize circulating CD4+ T cells in co-infected, HLA-DR7+ long-term non-progressor HIV subjects with undetectable HCMV plasma viremia. DYS-specific CD4+ T cells were inflated among these HIV+ subjects compared to those from a HIV− HCMV+ HLA-DR7+ cohort, or to HLA-DR7-restricted CD4+ T cells from the HIV co-infected cohort that were specific for epitopes of HCMV phosphoprotein-65, tetanus toxoid precursor, Epstein-Barr virus nuclear antigen 2 or HIV gag protein. Inflated DYS-specific CD4+ T cells comprised effector memory or effector memory-RA+ subsets with restricted TCR-beta usage and nearly monoclonal CDR3 containing novel conserved amino acids. Expression of this near monoclonal TCR in a Jurkat cell transfection system validated fine DYS specificity. Inflated cells were polyfunctional, not senescent, and displayed high ex vivo levels of granzyme-B, CX3CR1, CD38 or HLA-DR, but were less often CD38+HLA-DR+ co-expressing. The inflation mechanism did not involve apoptosis suppression, increased proliferation or HIV gag cross-reactivity. Instead, the findings suggest that intermittent or chronic expression of epitopes such as DYS drive inflation of activated CD4+ T cells that home to endothelial cells and have the potential to mediate cytotoxicity and vascular disease.
Introduction
Classical CD4+ and CD8+ memory T cell responses against viruses expand during primary infection and contract to low magnitudes after infection resolution (1). However, CD8+ T cell responses to select epitopes of human (HCMV) (2, 3), rhesus (4), and murine cytomegalovirus (MCMV) (5–9) persist for decades at very high magnitudes after primary infection or during latency. This phenomenon is termed “memory inflation” and has been best characterized among CMV-specific CD8+ T cells that consist of mainly CD45RO+ CCR7− CD27− T cells (effector memory/TEM) and their CD45RA+ revertants, CD45RO− CCR7− CD27− T cells (effector memory-RA+/TEMRA) (8–12). CMV-specific CD8+ T cells express high levels of CX3CR1 that bind CX3CL1 (fractalkine), which is expressed on vascular endothelial cells (VECs), a major target of CMV latent infection (1).
Classical CMV-specific CD8+ T cells display an IL-7-receptor-alpha/CD127+ programmed cell death protein-1−, PD-1− phenotype (capable of homeostatic proliferation controlled by IL-7 and other cytokines), while inflated CMV-specific CD8+ T cells are CD127− PD-1− T cell immunoglobulin and ITIM domain/TIGIT− Granzyme B+ CX3CR1+ with evidence suggesting they are maintained by low-level exposure to persistent antigen from stochastic CMV reactivation (1, 13–16). These data suggest inflated responses are maintained through recurrent stimulation by peptide-MHC (17–19) produced by persistent, stochastic expression of specific CMV transcripts (20–22). These epitopes are presented to CMV-specific T cells by latent HCMV-infected, non-hematopoietic reservoirs, including VECs, lymph node (LN) stroma cells, and cells in the bone marrow and lungs (1, 23–25). Maintenance of inflated CMV-specific T cell responses might also depend on their longer telomeres that positively correlate with persistence (26), or on epitope cleavage by constitutive proteasomes (6, 27).
CMV-specific CD4+ T cells suppress HCMV lytic replication (28) and maintain CD8+ T cell inflation (29). HCMV lysate-specific CD4+ T cells persist at high magnitudes in HIV+ HCMV+ co-infection (30), which might be due to higher HCMV disease burden (31, 32). Yet it is not known whether CD4+ T cells specific to individual HCMV epitopes undergo memory inflation in co-infected subjects. Glycoprotein B/gB has the highest population prevalence of CD4 responses of any HCMV protein (33). gB polyprotein colocalizes to endosomes that process and present its class II epitopes directly from infected endothelial cells upon IFN-γ-induced HLA class II expression (28, 34, 35) without needing professional APCs. gB-loaded endosomes are also secreted as immunogenic exosomes that stimulate CD4+ memory T cells (36, 37). In HLA-DRB1*07:01 (DR7+) persons, the most immunogenic gB epitope is the extremely conserved DYSNTHSTRYV (DYS) epitope that is recognized by cytotoxic, CX3CR1+ CD4+ T cells (11, 38).
HIV+ HCMV+ co-infection is implicated in the emerging higher incidence of HCMV-related, non-AIDS comorbidities of cardiovascular diseases including hypertension, coronary artery disease, and stroke despite suppressive antiretroviral therapy (ART) (31, 39–43). These disease risks are further increased in co-infected subjects with elevated CD4+ T cell activation (CD38+HLA-DR+) (44), which are mostly CMV-reactive (45) and are reduced by anti-CMV therapy (46). Indeed, CMV-reactive CD4+ CX3CR1+ T cells have been proposed as potential mediators of these comorbidities (36, 47, 48). Increased magnitudes of CD4+ CX3CR1+ T cells positively correlate with arterial stiffness (49), and these populations significantly decrease in magnitude after anti-CMV therapy (50). However, the specific epitopes and activation phenotype of these CMV-reactive CD4+ CX3CR1+ T cells remain unknown.
We propose a model where HIV+ HCMV+ co-infection increases stochastic, nonproductive HCMV reactivation that drives CD4 memory inflation. We hypothesized that HLA-DR7-restricted DYS-specific (DYS+) CD4+ T cells from HIV+ HCMV+ DR7+ subjects undergo increased memory inflation compared to similar cells from HIV− HCMV+ DR7+ subjects, and these cells upregulate CX3CR1, CD38 and HLA-DR. To test this hypothesis, we studied the ex vivo frequencies among subjects, response magnitudes and properties of DYS+ CD4+ T cells both in HIV+ HCMV+ long-term non-progressors (to avoid confounding effects of HIV-induced subclinical HCMV expression) and in HIV− HCMV+ individuals using DR7-restricted DYS (DR7:DYS) tetramer because cytokine-based assays can underestimate actual T cell response magnitudes and the expression of phenotypic markers can change after re-stimulation (51). The threshold for inflation was arbitrarily set at 1% of circulating CD4+ T cells as there is no standard minimum in the literature.
Materials and Methods
Subjects
HIV+ and HIV− subjects were randomly recruited through the Vanderbilt Comprehensive Care Clinic (IRB 030005), Vanderbilt Stem-Cell Clinic (IRB 061215), and the Australian Red Cross (IRB 2011/02) after they signed consent forms authorized by respective IRBs. HCMV+ or HCMV− status was determined by CMV IgG serology. Class II HLA-typing was performed at the Institute for Immunology and Infectious Diseases (Perth, Western Australia), or at DCI Tissue Typing Laboratory (Nashville, TN). PBMCs were isolated, frozen and thawed as previously described (52).
Tetramers and peptides
The NIH Tetramer Core Facility (contract HHSN272201300006C) synthesized DRB1*07:01-restricted PE-, APC-, and BV421-conjugated HCMV gB217–227 | DYSNTHSTRYV; HCMV pp65177–191 | EPDVYYTSAFVFPTK (EPD) (53); Human CLIP87–101 | PVSKMRMATPLLMQA; tetanus toxoid (TT) precursor586–605 | LINSTKIYSYFPSVISKVNQ (LIN) (54) and HIV gag293–312 | FRDYVDRFYKTLRAEQASQE (FRD) (55) tetramers. DRB1*07:01:PRSPTVFYNIPPMPLPPSQL (DR7:PRS, EBV EBNA2276–295) tetramer was synthesized by Beranoya Research Institute (Seattle, WA) (56). A2:NLVPMVATV (A2:NLV, CMV pp65495–503) tetramer was synthesized as described (57). The NIH AIDS Reagent Program, Division of AIDS, NIAID, provided HCMV pp65 Peptide Pool (overlapping 15-mers; #11549), HIV-1 PTE Gag Peptide Pool (overlapping 15-mers; #12437), and CMV AD169 strain (#1910). Lyophilized DYS, EPD, FRD, PRS and LIN peptides, and 19 overlapping, high DR7-affinity HIV gag peptides from the HIV-1 PTE Gag Peptide Pool (predicted by NetMHCII 2.2 Server) were synthesized at ≥98% purity (GenScript).
Flow cytometry
Cryopreserved PBMCs were thawed, washed in PBS (Corning), and disentangled with Nuclease S7 (Roche). Depending on assay, PBMCs were left untouched or negatively-enriched for CD4+ T cells (Miltenyi Biotec). Using our modified version of a class II tetramer stain protocol (56), we first stained for dead cells (Life Technologies), and washed with human Ab serum (Corning). Next, we stained with pre-titrated tetramer volumes at 37°C (1 h), anti-CCR7 Ab at 37°C (20 min), and room temperature surface protein stain (20 min) and, if necessary, intracellular Ab stain (20 min) at room temperature after fixation and permeabilization (BD). mAbs included CCR7-BV421 (150503), CD3-BV711 (UCHT1), CD4-PerCP-Cy5.5 (RPA-T4), CD45RO-PE-CF594 (UCHL1), CD27-PE-Cy7 (M-T271), CD14-V500 (M5E2), CD19-V500 (HIB19), IFN-γ-FITC (B27), TNF-α-PE-Cy7 (MAb11), CD4-FITC (SK3), Bcl-2-PE (Bcl-2/100), Granzyme-B-FITC (GB11), CX3CR1-PE (2A9-1), Ki-67-PE-Cy7 (B56), CD57-FITC (NK-1), CD45RA-PE-Cy7 (L48), CD38-PE-Cy7 (HIT2), and HLA-DR-FITC (G46-6) that were ordered from BD; CD8-APC-AF750 (3B5) from Invitrogen; PD-1-PE (EH12.2H7) from BioLegend; TIGIT-PE-Cy7 (MBSA43) and CD28-PE-Cy7 (CD28.2) from eBiosciences; and custom CD127-PE-Cy5.5 (R34.34) from Beckman Coulter. Cells were sorted with FACSAria-IIIu (BD) or collected on LSR Fortessa (BD). FlowJo (v10.1r5; Tree Star) was used for analysis. Only background-subtracted, tetramer+ response magnitudes 3× > the respective CLIP tetramer response magnitudes were considered positive. Median fluorescence intensity (MFI) analyses were done only on samples stained and collected the same day.
ELISpot
IFN-γ ELISpot was conducted on a BioMek FXP high-throughput platform (Beckman Coulter) using the Human IFN-γ ELISpotBASIC (HRP) kit (Mabtech). Cryopreserved PBMCs were thawed and rested overnight in R10 media (52) before triplicate stimulations of 150,000–250,000 cells/well in MultiScreen-IP Filter Plates (Millipore) with no peptide, or with 0.001 μg/μl final concentrations of DYS, EPD, FRD, PRS or LIN epitope, or HCMV pp65 peptide pool, HIV-1 PTE Gag peptide pool, or anti-human CD3 (Mabtech). IFN-γ spot-forming units were developed using tetramethylbenzidine (Mabtech) and counted with ELISpot reader (AutoImmun Diagnostika) after drying. Positive spots = data > background mean + 3 times background SEM (38).
Bulk-cell TCR sequencing
TCRβ gene sequencing of DNA extracted (#DC6701; Promega) from bulk-sorted TEM or TEMRA DYS+ and DYS− CD4+ T cells was completed and bioinformatically analyzed by Adaptive Biotechnologies. CDR3 proportions, productive template fractions and clonalities, and V(D)J segments were analyzed on ImmunoSEQ Analyzer v3.0. Circos plots were generated using VDJtools and circlize (58, 59). The TCR CDR3 data have been deposited in the NCBI Sequence Read Archive repository (https://trace.ncbi.nlm.nih.gov/Traces/study/?go=home) under study accession number SRP113337 and in VDJDB (https://vdjdb.cdr3.net).
TCR artificial expression and stimulation by LCL-pulsed epitopes
TCRα and TCRβ CDR3 sequences of Subject 10027’s DYS+ sorted single-cells were determined using a previously published technique (60). Briefly, single cells were sorted into separate wells in a 96-well plate containing RT-PCR buffer using FACSAria-IIIu cell sorter (BD) for three rounds of PCR amplification using nested, barcoded, TCR-specific and Illumina Paired-End primers (60). Purified PCR products were sequenced on Illumina MiSeq platform. After sequence analyses, TCRα and TCRβ products with identical barcodes were selected for full TCR gene completion using the international ImMunoGeneTics database. Following published methods (61), the DYS-specific TCRα and TCRβ genes were cloned into pSELECT-GFPzeo plasmids (InvivoGen) and expressed in Jurkat cells (clone E6-1, TIB-152; American Type Culture Collection) along with pNFAT-Luciferase (Affymetrix). These cells and lymphoblastoid cell lines (LCLs) were maintained in R10 media (52). DYS or control epitopes were pulsed with the LCLs to stimulate the DYS-specific TCRs expressed on the Jurkat cells. Luciferase absolute light units were measured using FilterMax F5 Multi-Mode Microplate Reader (Molecular Devices). The TCRα and TCRβ CDR3 data have been deposited in the NCBI Sequence Read Archive repository (https://trace.ncbi.nlm.nih.gov/Traces/study/?go=home) under study accession number SRP113337 and in VDJDB (https://vdjdb.cdr3.net).
Droplet digital PCR (ddPCR)
ddPCRs were performed entirely using a QX200 AutoDG Droplet Digital PCR System (Bio-Rad). For both assays, plates of droplets of PCR mixture were automatically generated with an Auto-droplet generator/AutoDG and TaqMan oil for probes, heat-sealed and amplified with C1000 Touch thermal cycler. Droplets were read using a Droplet Reader. The magnitude of false-positive responses in no-template controls was 15% (data not shown). DNA concentrations±95% CI were determined from only wells with >12,000 droplets using QuantaSoft v1.7.4.0917 after manually setting the positive droplet threshold above the negative droplet signal of the no template controls in the same plate. Primers and probes concentrations: 900nM and 250nM, respectively. All PCRs were multiplexed with RPP30 housekeeping gene. HCMV DNA quantitation: 20μl PCR mixture was prepared using DNA, ddPCR SuperMix for Probes, water and these CMV primers and probes: IE1-specific forward primer 5′-TGAAGCGCCGCATTGA, IE2-specific reverse primer 5′-TGGCCCGTAGGTCATCCA, and IE1-specific probe 5′-6FAM-TCTGCATGAAGGTCTTTGCCCAGTACATCC-TAMRA. Thermocycling conditions: 50°C (2 min), 95°C (10 min), 40 cycles of 95°C (15 s) with 60°C (1 min) (2°C/s ramp rate). Modified HIV DNA quantitation (62): 20μl PCR mixture was prepared similarly, but using these HIV LTR primers and probes instead: forward primer 5′-AGCACTCAAGGCAAGCTTTA, reverse primer 5′-TGTACTGGGTCTCTCTGGTTAG, and probe 5′-FAM-GCAGTGGGTTCCCTAGTTAGCCAGAGAG-3IABkFQ. Thermocycling conditions: 95°C (10 min), 40 cycles of 94°C (30 s) with 60°C (1 min), and 98°C (10 min) (2°C/s ramp rate).
Statistics
GraphPad Prism v7.0a was used for non-parametric, two-tailed analyses of Wilcoxon matched-pairs signed rank test (paired), Mann-Whitney U test (non-paired), and Spearman’s rank correlation (ρ) (linear regression). *P≤0.05, **P≤0.01, ***P≤0.001.
Results
HLA-DR7-restricted DYS+ CD4+ T cells are inflated in HIV+ HCMV+ DR7+ compared to HIV− HCMV+ DR7+ subjects
Following the gating hierarchy in Supplemental Fig. 1, we verified tetramer specificity using DR7:CLIP tetramer stain of CD4-enriched PBMCs from time-point 1 (tp1; no ART) of 8 HIV+ HCMV+ DR7+ subjects (Table I, and Fig. 1A). We also confirmed the HLA-DR7 restriction of the response by staining CD4-enriched PBMCs from 7 co-infected subjects lacking HLA-DR7 allele i.e. DR7− (Supplemental Fig. 2A). To determine the HLA-DR7+ DYS+ CD4+ T cell response magnitude in the HIV+ HCMV+ DR7+ subjects, the tp1 CD4-enriched or untouched PBMCs were stained with DR7:DYS tetramer. We detected high DYS+ CD4+ T cell magnitudes of response in 7 subjects (0.43–17.91%), with 6 of them displaying inflated responses (Fig. 1A). To determine if DYS+ CD4+ T cell inflation was abrogated after ART-induced HIV suppression, we stained aviremic PBMCs from later time-points (9.5 years median time lapse) of 4 co-infected individuals (time-point 2, tp2, in Table I) with the tetramer and detected values ranging from 0.11–26.34% (Fig. 1B). Inflation magnitude did not correlate with age (possibly due to small sample size, which could be increased in future studies), HIV infection duration, nadir CD4 count or HIV load (data not shown).
Table I.
Characteristics of HIV+ HCMV+ DR7+ subjects.
| Subject ID | HLA-DRB1 alleles | Time-point | Age (years) | Years of HIV infection | CD4 T cell count/μl blood | HIV load (RNA copies/ml plasma) | Time on ART (years) |
|---|---|---|---|---|---|---|---|
| 10004 | 07:01, 03:01 | 1 | 58 | 22 | 203 | <50 | – |
| 2 | 62 | 26 | 181 | <50 | 2.17 | ||
| 10013 | 07:01, 04:08 | 1 | 47 | 22 | 420 | 5,354 | – |
| 2 | 60 | 35 | 380 | <50 | 5.0 | ||
| 10027 | 07:01, 03:01 | 1 | 66 | 14 | 378 | 7,340 | – |
| 2 | 75 | 23 | 977 | <50 | 7.17 | ||
| 10032 | 07:01, 03:01 | 1 | 46 | 5 | 543 | 2,837 | – |
| 2 | 56 | 15 | 886 | <50 | 4.42 | ||
| 10040 | 07:01, 13:01 | 1 | 47 | 14 | 1,161 | <50 | – |
| 10030 | 07:01, 09:01 | 1 | 60 | 13 | 856 | <50 | – |
| 10066 | 07:01, 07:01 | 1 | 52 | 14 | 1,063 | <50 | – |
| 10069 | 07:01, 08:04 | 1 | 46 | 6 | 903 | 2,470 | – |
FIGURE 1.
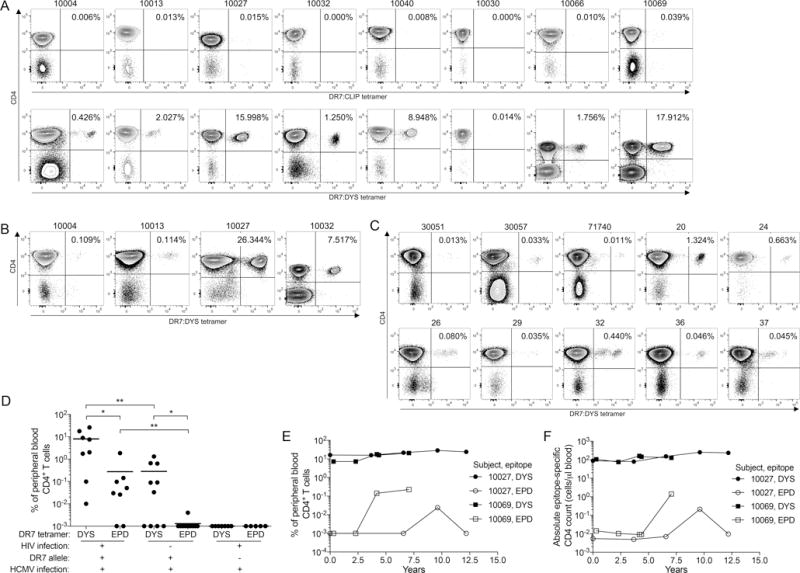
HLA-DR7-restricted HCMV glycoprotein B DYS-epitope specific CD4+ T cells undergo memory inflation in HIV+ HLA-DR7+ subjects. A–B: CD4-enriched or untouched peripheral blood mononuclear cells (PBMCs) from HIV+ DR7+ subjects were stained with DR7:CLIP or DR7:DYS tetramer for their (A) tp1 (no ART) (n=8), and (B) tp2 (on ART) samples (n=4). (C) DR7:DYS tetramer staining of HIV− DR7+ subjects’ PBMCs (n=10). (D) Response magnitude comparisons of DYS+ and EPD+ CD4+ T cells from HIV+ HCMV+ DR7+ (n=8; tp1 of Subject 10013, and tp2 of Subjects 10004, 10027 and 10032), HIV− HCMV+ DR7+ (n=10) and HIV+ HCMV+ DR7− (n=5–7) subjects determined simultaneously from the same samples per subject. E–F: Longitudinal (E) response magnitudes and (F) absolute counts of DYS+ and EPD+ CD4+ T cells from Subjects 10027 and 10069. Values in (D) represent ≥3 biological replicates with means, except for the HIV+ HCMV+ DR7− cohort with no replicates.
Importantly, we observed significantly lower magnitudes of DYS+ CD4+ T cells (0.01–1.32%) in 10 HIV− HCMV+ DR7+ subjects (Fig. 1C) compared to those of the HIV+ HCMV+ DR7+ individuals (median 0.06% vs. 4.76%, P=0.004; Fig. 1D). Samples from tp1 of Subject 10013, and tp2 of Subjects 10004, 10027 and 10032 were used in this and all future experiments unless otherwise indicated. CD4 counts of the HIV− cohort were unavailable for absolute DYS+ CD4 count comparison. To confirm that these cells are undergoing memory inflation, we analyzed DYS+ CD4+ T cells from five time-points of two HIV+ DR7+ subjects spanning a period of up to twelve years and detected stable magnitudes and absolute counts (Figs. 1E and 1F, respectively). Together, these findings identify inflated CD4+ T cells against HLA-DR7-restricted DYS epitope of HCMV gB in HIV+ HCMV+ co-infected subjects.
HLA-DR7-restricted CD4+ T cells responses to other persistent and non-persistent epitopes are low in HIV+ HCMV+ DR7+ subjects
Tetramer stains of CD4+ T cells specific for a DR7-restricted, highly conserved HCMV pp65 EPD epitope, which were absent in HIV+ HCMV+ DR7− subjects (Supplemental Fig. 2B), revealed significantly lower magnitude ranges in the HIV+ HCMV+ DR7+ cohort (0.01–1.87%, Supplemental Fig. 2C) compared to inflated DYS+ CD4+ T cells in this cohort (median 0.04% vs. 4.76%, P=0.02; Fig. 1D). We did not detect any memory inflation of these EPD+ CD4+ T cells in four longitudinal samples obtained from the two subjects with the highest DYS-specific inflation over a twelve-year or less period (Figs. 1E and 1F). A similar trend was observed in the HIV− HCMV+ DR7+ cohort between EPD+ (0.003–0.04%, Supplemental Fig. 2D) and DYS+ CD4+ T cells (median 0.001% vs. 0.06%, P=0.03; Fig. 1D). As observed for DYS+ CD4+ T cells, EPD+ CD4 response magnitudes were also significantly higher in HIV+ HCMV+ DR7+ than in HIV− HCMV+ DR7+ cohort (P=0.002; Fig. 1D), confirming a recent report using pp65 peptide pools instead (63). To determine whether the inflation could be due to generalized HIV-induced inflammation, we compared the magnitudes of CD4+ T cells specific for DR7-restricted TT, EBV or HIV epitopes from the HIV+ DR7+ subjects to their DYS+ CD4+ T cell counterparts. We observed that the magnitudes of these other epitopes were undetectable or very low compared to the inflated DYS-specific response (P=0.0078 for each comparison; Supplemental Fig. 2E). To evaluate potential CD8+ T cell inflation, we stained PBMCs from Subject 10027, who has the highest DYS+ CD4 inflation (26.34%) and carries a HLA-A2:01 allele with A2:NLV tetramer, but detected a magnitude of only 0.75% to this epitope (Supplemental Fig. 2F). Collectively, these results indicate that other DR7+ epitope-specific CD4+ T cells in most co-infected subjects are present at lower magnitudes than DYS+ CD4+ T cells.
DYS+ CD4+ T cells consist of TEM and/or TEMRA subsets
We determined the memory phenotype of DYS+ CD4+ T cells by measuring surface expression of memory markers CD45RO, CCR7 and CD27 to define TEM, TEMRA, central (TCM; CD45RO+ CCR7+ CD27+), transitional (TTM; CD45RO+ CCR7− CD27+), naïve (TNai; CD45RO− CCR7+ CD27+), and intermediate (TInt; CD45RO− CCR7− CD27+) subsets (Supplemental Fig. 1) (10). Compared to the non-DYS+ (DYS−) CD4+ T cells, DYS+ CD4+ T cells from HIV+ HCMV+ DR7+ subjects were biased toward TEM (46.6–97.97% vs. 6.1–51.9%, P=0.0156) and TEMRA (0.03–48.1% vs. 0.4–16.7%, P=0.0781) (Fig. 2A), and similar observations were made in the HIV− HCMV+ DR7+ cohort (Fig. 2B). Most CD45RO− DYS+ and DYS− CD4+ T cells were CD45RA+ as shown in Subject 10027 (Fig. 2C).
FIGURE 2.
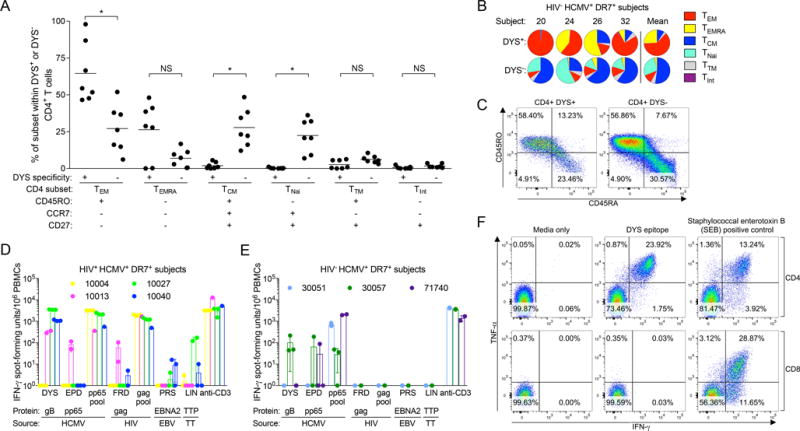
CD4+ T cells specific for DYS epitope consist of TEM and TEMRA subsets, and secrete cytokines upon DYS stimulation. (A) Response magnitude comparisons of DYS+ and DYS− CD4+ T cell subsets from HIV+ HCMV+ DR7+ subjects (n=7). PBMCs were stained with the tetramer and memory markers to identify the subsets. Plots show grand means and represent at least two biological replicates. (B) Normalized magnitudes of CD4 subsets within DYS+ or DYS− CD4+ T cells from HIV− HCMV+ DR7+ subjects with sufficient tetramer+ response for analyses. (C) CD45RO and CD45RA staining of Subject 10027’s PBMCs. D–E: Background-corrected IFN-γ ELISpot responses of PBMCs from (D) HIV+ HCMV+ DR7+ (n=4; tp1) and (E) HIV− HCMV+ DR7+ (n=3) subjects upon stimulation with 0.001 μg/ml of DYS, EPD, FRD, PRS, LIN epitopes, and of the following controls: HCMV pp65 overlapping 15-mer peptide pool, HIV’s gag overlapping 15-mer peptide pool and anti-human CD3. Data represent technical triplicates except in conditions without mean±SD. (F) Intracellular cytokine staining of Subject 10027’s PBMCs after DYS or SEB stimulation.
DYS-stimulated CD4+ T cells secrete IFN-γ and TNF-α
Most HIV+ HCMV+ DR7+ PBMC samples stimulated with DYS produced IFN-γ in high-throughput ELISpot, and only Subject 10013 responded to EPD (Fig. 2D), indicating that tetramer staining was more sensitive or that the cells were dysfunctional. Subject 10004 did not respond to either epitope possibly due to dysfunction or anergy. 1 of 3 screened HIV− HCMV+ DR7+ subjects responded to DYS stimulation (Fig. 2E). Responses to HIV FRD epitope, EBV PRS epitope or TT LIN epitope were relatively diminished in HIV+ DR7+ subjects compared to DYS-induced responses and not detected in HIV− DR7+ subjects (Figs. 2D and 2E, respectively). Dual IFN-γ and TNF-α intracellular cytokine staining of Subject 10027’s PBMCs, which produced the largest IFN-γ ELISpot response to DYS, confirmed that the ELISpot responses originated from CD4s and not CD8s, and suggested that these inflated cells were likely polyfunctional (Fig. 2F), as previously reported (11, 35).
Inflated DR7+ DYS+ CD4+ T cells have highly restricted TCRβ repertoires
TCR analyses were conducted only on subjects with adequate DYS+ CD4 magnitudes of response for bulk cell sorting and sequencing: HIV+ Subjects 10027, 10040, 10069 and 10032 (26.34%, 8.95%, 17.91% and 7.52%, respectively) and HIV− Subject 20 (1.32%) as a control. We observed highly restricted TCR-beta-variable (TCRβV) and -joining (TCRβJ) gene pairing in bulk-sorted TEM and TEMRA subsets of DYS+ CD4+ T cells compared to the more diverse DYS− counterparts in all subjects (Fig. 3, A – E). The dominant TCRβV and TCRβJ gene families of each individual’s DYS+ CD4+ T cells comprised 69.5% to 99.7% of the DYS+ CD4+ T cell repertoire, and were identical between their TEM and TEMRA subsets (Supplemental Fig. 3, A – E). However, the dominant TCRβV and TCRβJ gene families of DYS− CD4+ T cells were lower (10.42–56.46%) and different between TEM and TEMRA subsets (Supplemental Fig. 3, A – E). These findings indicate a strong TCRβ conservation among inflated DYS+ CD4+ T cells.
FIGURE 3.
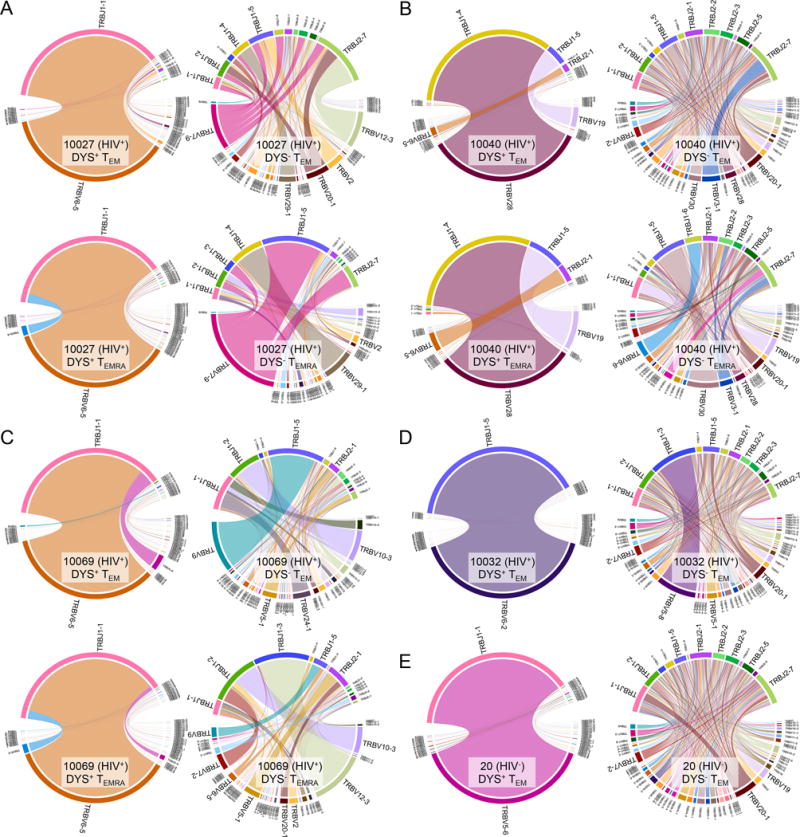
Productive TCRβ V and J gene pairs of bulk-sorted, inflated DYS+ CD4+ T cells are highly restricted. TCRβV and TCRβJ gene family pairings of TEM and/or TEMRA subsets of productive DYS+ and DYS− CD4 TCRs from Subjects (A) 10027, (B) 10040, (C) 10069, (D) 10032, and (E) 20. Data shown represent single experiments. V and J gene pairs are connected by stems between their arcs. Arc lengths reflect gene family proportions within the sample’s repertoire. High magnitude V and J gene families are emphasized.
Inflated, DR7+ DYS+ CD4+ T cells utilize nearly monoclonal CDR3s
We analyzed the CDR3 repertoires of productive V(D)J rearrangements (in-frame and without stop codons) of the bulk-sorted DYS+ CD4+ T cells and observed that they were dominated by specific clones with unique V(D)J rearrangements (69.41–99.64%, median=91.37%) (Fig. 4, A – E). Interestingly, we discovered that 97.29% and 7.6% of the productive DYS+ CD4+ TEM CDR3 repertoires of Subjects 20 (HIV−) and 10069 (HIV+), respectively, were identical. TEM CDR3 analysis of a HIV− HCMV− subject showed no clonal expansion (data not shown), suggesting that clonal expansion among DYS− TEM CDR3s might be tied to HCMV+ status. The DYS+ and DYS− CDR3 clonal dominance reflected their respective TCRβ gene-family distributions. DYS+ TEM and TEMRA dominant clones within each subject were identical, and this is likely a reflection of the reversible T cell differentiation from TEM (CD45RO+ CD45RA−) to TEMRA (CD45RO− CD45RA+) (11).
FIGURE 4.
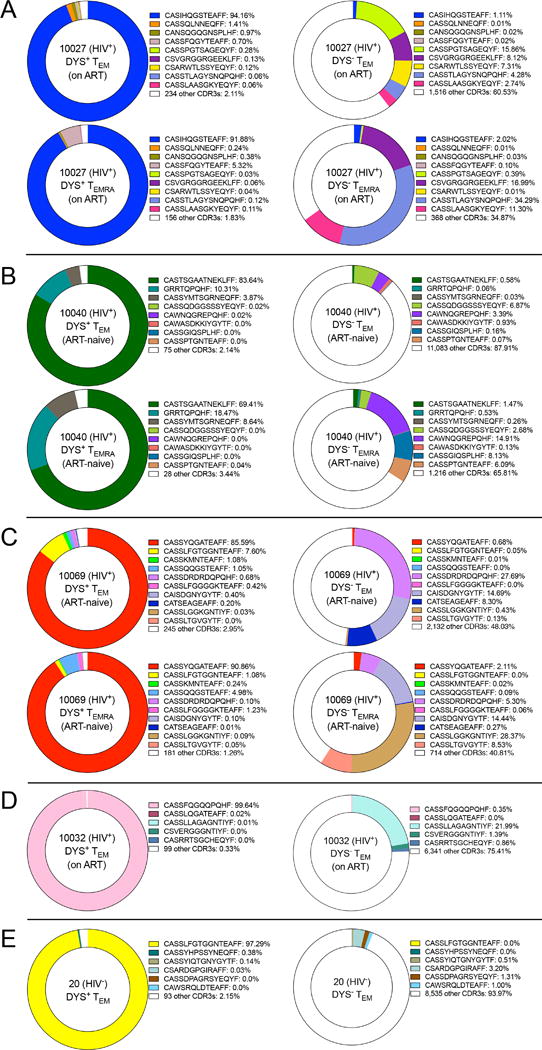
Inflated DYS+ CD4+ T cells have nearly monoclonal productive CDR3s. Productive and unique TCRβ CDR3 clones of DYS+ and DYS− TEM and/or TEMRA subsets from Subjects (A) 10027, (B) 10040, (C) 10069, (D) 10032 and (E) 20. Data represent single experiments.
In vivo stimulation of inflated cells involves NFAT-mediated cellular activation and proliferation upon TCR ligation by peptide-MHC. To confirm this activity and the accuracy of the clonal CDR3 sequence, we simulated the antigen presentation conditions for Subject 10027 using autologous B cell-derived LCLs and the DYS epitope to stimulate autologous DYS+ α:β TCR expressed on Jurkat cells with an NFAT-mediated luciferase reporter. Using single-cell sorting and TCR sequencing, we first determined the paired α:β TCR CDR3 sequences of autologous DYS+ CD4+ T cells to be TCRα CAGRSSNTGKLIF CDR3 (TCRαV25 and TCRαJ37), and TCRβ CASIHQGSTEAFF CDR3 (TCRβV6-5 and TCRβJ1-1) that matched Subject 10027’s nearly monoclonal CDR3 sequence (Fig. 5A). After TCR expression and stimulation with autologous, DYS-pulsed DR7+ LCLs, we detected a dose-dependent luciferase luminescence that was not present with no epitope, a different epitope (EPD) or a DR7− LCL (Fig. 5A). Subject 10069’s LCL (DR7, DR8) confirmed that DYS was presented by DR7 and not DR3, which was the other DRB1 allele of Subject 10027 (Fig. 5A). Clonality comparison revealed that DYS+ CD4+ TEM and TEMRA cells were significantly more clonal compared to DYS− counterparts, and almost monoclonal in Subject 10032 (P=0.0078; Fig. 5B). We verified the expectation of more productive V(D)J rearrangements within DYS+ compared to DYS− CD4+ T cells since the former were sorted based on specific HLA-restricted epitope recognition (P=0.0078; Fig. 5C). These results suggest that DYS+ CD4+ T cells are inflated via a highly clonal mechanism that likely involves DYS stimulation.
FIGURE 5.
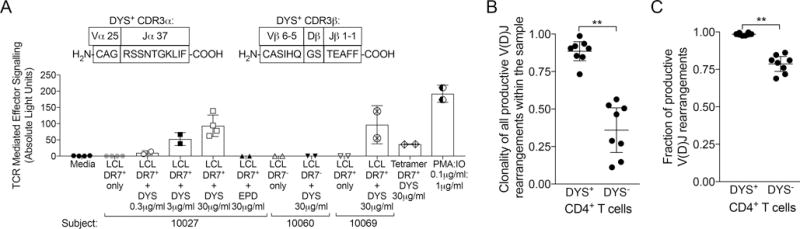
Expressed clonal TCR of inflated DYS+ CD4+ T cells recognize DR7-restricted DYS epitope. (A) Subject 10027’s DYS+ single cell α:β TCR CDR3 sequences, and NFAT-mediated luciferase luminescence response of DR7+ LCL-presented, serially diluted DYS epitope stimulation of DYS+ TCR. Subject 10027’s DYS+ α:β TCR gene sequences were determined from single cell sorting and expressed using plasmids in an NFAT-luciferase reporter Jurkat cell line for stimulation. Graph shows mean±SD: conditions with two data-points represent technical replicates, while those with four represent two biological replicates of two technical replicates. (B) Clonality comparison of productive CDR3s of DYS+ to DYS− CD4+ TEM and TEMRA subsets from Subjects 10027, 10040, 10069, 10032 and 20. Clonality fractions were bioinformatically determined after productive entropy normalization. Values near 1: more clonal. (C) Fractional comparison of productive V(D)J rearrangements of DYS+ to DYS− CD4+ TEM and TEMRA subsets from all five subjects. Values near 1: fewer out-of-frame sequences or stop codons. B–C: Data represent single experiments for each subject with mean±95% CI.
Different DYS+ CDR3 clones share conserved amino acid
We assessed whether potential amino acid conservation among the different, dominant DYS+ CDR3s of all subjects could explain their common recognition of DYS. Remarkably, our V(D)J alignments revealed two new conserved amino acids (serine (S), and threonine (T)) within the D-segments in addition to the published glutamine (Q) (64), all of which have polar, neutral side chains (Table II). We did not see a similar conservation among DYS− CDR3 clones (Table III). These findings indicate that amino acids with polar and neutral side chains might be critical in DYS recognition.
Table II.
Dominant CDR3βs of inflated DYS+ CD4+ T cells share conserved polar, neutral amino acids.
| Subject ID | CDR3 freq. | TCRβV family | V segment (start of CDR3) | N1 | D segment | N2 | J segment | TCRβJ family | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10027 | 94% | 6-5 | C | A | S | I | H | Q | G | S | T | E | A | F | F | 1-1 | ||||||||
| tgt | gcc | agc | atc | cat | caa | ggg | agc | act | gaa | gct | ttc | ttt | ||||||||||||
|
| ||||||||||||||||||||||||
| 10040 | 84% | 28 | C | A | S | T | S | G | A | A | T | N | E | K | L | F | F | 1-4 | ||||||
| tgt | gcc | agc | act | tca | ggg | gcg | gca | act | aat | gaa | aaa | ctg | ttt | ttt | ||||||||||
|
| ||||||||||||||||||||||||
| 10069 | 91% | 6-5 | C | A | S | S | Y | Q | G | A | T | E | A | F | F | 1-1 | ||||||||
| tgt | gcc | agc | agt | tat | cag | ggc | gcc | act | gaa | gct | ttc | ttt | ||||||||||||
|
| ||||||||||||||||||||||||
| 10032 | 99% | 6-2 | C | A | S | S | F | Q | G | Q | Q | P | Q | H | F | 1-5 | ||||||||
| tgt | gcc | agc | agt | ttc | cag | ggg | caa | cag | ccc | cag | cat | ttt | ||||||||||||
|
| ||||||||||||||||||||||||
| 20 | 96% | 5-6 | C | A | S | S | L | F | G | T | G | G | N | T | E | A | F | F | 1-1 | |||||
| tgt | gcc | agc | agc | ttg | ttc | ggg | aca | ggg | ggg | aac | act | gaa | gct | ttc | ttt | |||||||||
Conserved glutamine (Q), serine (S) and threonine (T) within the D-segment are underlined. The dominant CDR3s were identical for DYS+ CD4+ TEM and TEMRA in all subjects.
Table III.
Dominant CDR3βs of DYS− CD4+ T cells do not share conserved polar, neutral amino acids.
| Subject | CD4 subset | CDR3 Freq. | TCRβV family | V gene start of CDR3 | N1 | D gene | N2 | J gene | TCRβJ family | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10027 | TEM | 16% | 12-3 | C | A | S | S | P | G | T | S | A | G | E | Q | Y | F | 2-7 | |||||||
| tgt | gcc | agc | agt | ccg | ggg | act | agc | gcc | ggc | gag | cag | tac | ttc | ||||||||||||
| TEMRA | 34% | 7-9 | C | A | S | S | T | L | A | G | Y | S | N | Q | P | Q | H | F | 1-5 | ||||||
| tgt | gcc | agc | agc | acg | ttg | gcg | gga | tat | agc | aat | cag | ccc | cag | cat | ttt | ||||||||||
|
| |||||||||||||||||||||||||
| 10040 | TEM | 7% | 3-1 | C | A | S | S | Q | D | G | G | S | S | S | Y | E | Q | Y | F | 2-7 | |||||
| tgt | gcc | agc | agc | caa | gac | ggc | ggg | agt | agc | tcc | tac | gag | cag | tac | ttc | ||||||||||
| TEMRA | 15% | 30 | C | A | W | N | Q | G | R | E | P | Q | H | F | 1-5 | ||||||||||
| tgt | gcc | tgg | aat | cag | ggg | cgc | gag | ccc | cag | cat | ttt | ||||||||||||||
|
| |||||||||||||||||||||||||
| 10069 | TEM | 28% | 9 | C | A | S | S | D | R | D | R | D | Q | P | Q | H | F | 1-5 | |||||||
| tgt | gcc | agc | agc | gac | cgg | gac | agg | gat | cag | ccc | cag | cat | ttt | ||||||||||||
| TEMRA | 28% | 12-3 | C | A | S | S | L | G | G | K | G | N | T | I | Y | F | 1-3 | ||||||||
| tgt | gcc | agc | agc | ctg | ggg | ggg | aaa | gga | aac | acc | ata | tat | ttt | ||||||||||||
|
| |||||||||||||||||||||||||
| 10032 | TEM | 22% | 5-8 | C | A | S | S | L | L | A | G | A | G | N | T | I | Y | F | 1-3 | ||||||
| tgt | gcc | agc | agc | tta | cta | gca | ggg | gct | gga | aac | acc | ata | tat | ttt | |||||||||||
|
| |||||||||||||||||||||||||
| 20 | TEM | 3% | 20-1 | C | S | A | R | D | G | P | G | I | R | A | F | F | 1-1 | ||||||||
| tgc | agt | gct | aga | gat | ggt | cca | ggg | att | cga | gct | ttc | ttt | |||||||||||||
Glutamine (Q) within the D-segment is underlined.
Inflated DR7+ DYS+ CD4+ T cells are CD127− TIGIT− and Granzyme B+
We measured plasma HCMV DNA load of all subjects but detected no viral DNA despite being HCMV+, suggesting that inflation of these circulating cells was not due to ongoing HCMV replication in the blood. Other samples such as saliva and semen in which active HCMV replication has been reported were unavailable for testing. We next determined whether these cells displayed similar CD127− PD-1− TIGIT− granzyme B+ phenotype of inflated CMV-specific CD8+ T cells (1, 14, 16). For all onward comparison experiments of inflated DYS+ CD4+ T cells (n=7), we used HIV− HCMV− controls (n=10) to provide contrast with classical TEM cells and avoid other potential inflationary HCMV epitope-specific responses, and also because HIV+ HCMV− subjects are extremely rare. We focused on only TEM because DYS+ TEMRA was present in only 5 HIV+ HCMV+ DR7+ subjects. We compared CD127, PD-1, and TIGIT expressions on DYS+ CD4+ TEM to CD4+ TEM from HIV− HCMV− controls, and detected significantly lower CD127 (P=0.025), no difference in PD-1 (P=0.364), and significantly lower TIGIT (P=0.0001) among the inflated cells (Fig. 6, A – C). The dual IFN-γ and TNF-α secretions in Fig. 2F also suggest these cells are not exhausted. To further determine polyfunctionality, we compared ex vivo intracellular granzyme B levels of DYS+ CD4+ TEM from the HIV+ HCMV+ DR7+ subjects to controls, and detected significantly higher levels with DYS specificity (P=0.0001; Fig. 6D), confirming previous cytotoxicity (11, 28, 35, 38, 64) and polyfunctionality (11, 35) reports for DYS+ CD4+ T cells. B-cell lymphoma-2 (Bcl-2) protein MFI of inflated DYS+ CD4+ T cells were not different compared to controls (P=0.536; Fig. 6E). None of these protein levels correlated with the magnitude of DYS+ CD4 inflation (data not shown). These findings reveal that inflated DYS+ CD4+ T cells are CD127− PD-1+/− TIGIT− and Granzyme B+.
FIGURE 6.
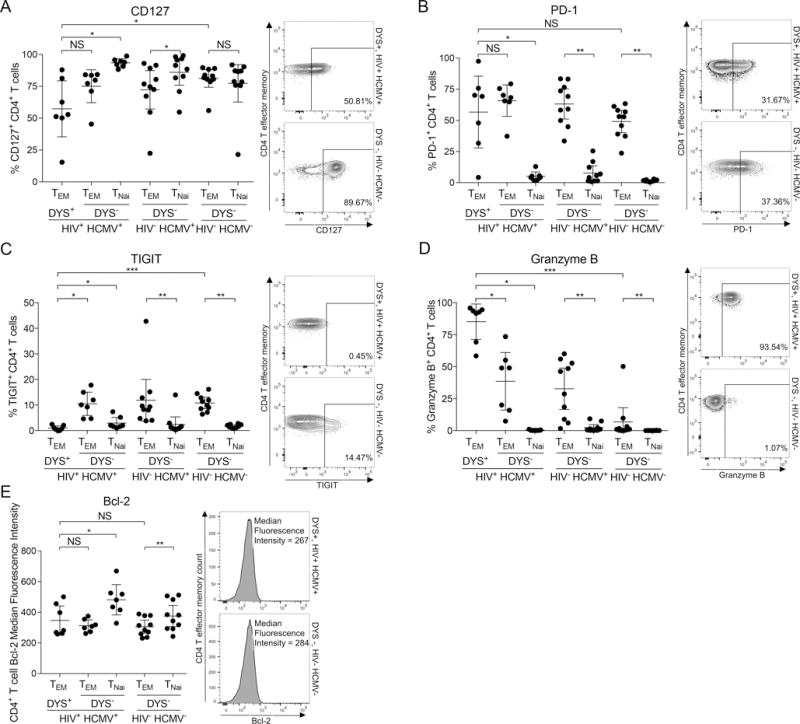
Inflated DYS+ CD4+ T cells have a CD127− TIGIT− Granzyme B+ phenotype. Ex vivo comparisons of (A) CD127, (B) PD-1, (C) TIGIT, (D) Granzyme B expressions or (E) Bcl-2 median fluorescence intensity (MFI) of DYS+ CD4+ TEM of HIV+ HCMV+ subjects to DYS− CD4+ TEM of HIV− HCMV− controls. PBMCs were stained with tetramer and mAbs for either surface PD-1, CD127 and TIGIT or intracellular granzyme B and Bcl-2 proteins. HIV+ HCMV+: n=7, HIV− HCMV+: n=10 and HIV− HCMV−: n=10. Graphs represent single experiments for each subject, with mean±95% CI for all subjects.
Inflated DR7+ DYS+ CD4+ T cells are CX3CR1high and are not undergoing higher proliferation
Latent HCMV reservoirs present endogenous DYS epitopes to DYS+ CD4+ T cells (28, 35, 65). Therefore we hypothesized that such reservoirs expand with HIV co-infection to cause memory inflation. While we could not directly measure HCMV reservoir size, we further hypothesized that expanded HCMV latent reservoirs, including VECs, would express more CX3CL1 and consequently correlate with higher expression of CX3CR1 on inflated DYS+ CD4+ T cells. Indeed, these cells had significantly higher CX3CR1 MFI compared to controls (P=0.0007; Fig. 7A), confirming previous reports (11). To determine whether effectual TCR stimulation by DYS-presenting latent reservoirs caused ongoing proliferation in vivo and by extension memory inflation, we measured ex vivo Ki-67+ levels within inflated DYS+ CD4 TEM and detected a slightly wider range of, but not significantly higher, magnitudes compared to controls (P=0.474; Fig. 7B), confirming studies on MCMV epitope-specific CD8+ T cell inflation (5, 9). Using CD57 and CD28 dual staining of Subject 10027’s tp2 PBMCs, we observed that <2% of CD4+ DYS+ T cells displayed the CD57+ CD28+/− phenotype for replicative senescence (Fig. 7C) (66). CX3CR1 and Ki-67 levels did not correlate with DYS+ CD4 inflation magnitudes (data not shown). These findings suggest that inflated DYS+ CD4 T cells might interact with HCMV reservoirs that express CX3CL1 and their inflation is not linked to increased ongoing proliferation.
FIGURE 7.
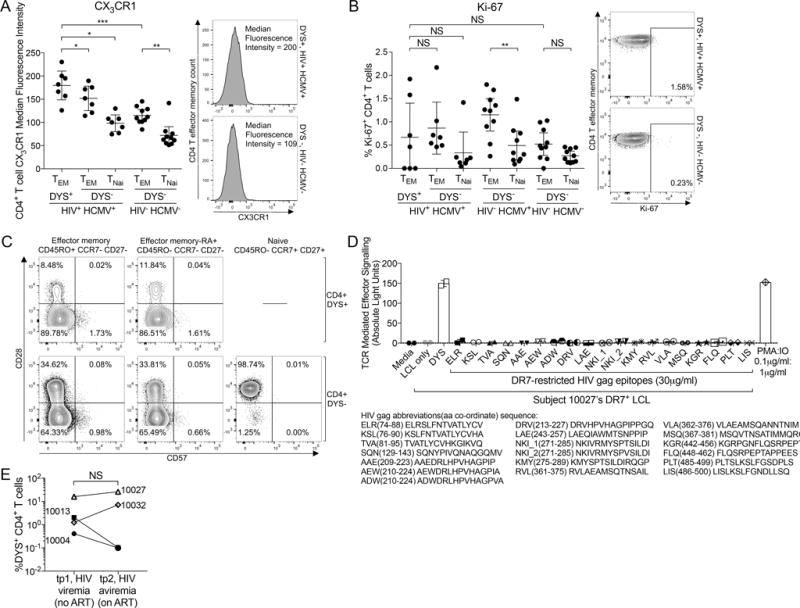
Inflated DYS+ CD4 T cells are CX3CR1high, not replicatively senescent and do not cross-react with HIV gag proteins. A–B: Ex vivo comparisons of (A) CX3CR1 MFI and (B) Ki-67 response magnitude of DYS+ CD4+ TEM of HIV+ HCMV+ subjects to DYS− CD4+ TEM of HIV− HCMV− controls. HIV+ HCMV+: n=7, HIV− HCMV+: n=10 and HIV− HCMV−: n=10. (C) CD57 and CD28 expressions of Subject 10027 DYS+ and DYS− CD4+ T cells. (D) Luciferase luminescence response of 19 DR7-presented HIV gag epitopes stimulation of Subject 10027’s DYS+ TCR expressed in the Jurkat cell line. (E) DYS+ CD4 magnitude change from tp1 (HIV viremia) to tp2 (HIV aviremia). A–B, E: graphs show mean±95% CI for all subjects and represent single experiments with no replicates except (E) showing technical replicates. D: data shows mean±SD of technical duplicates.
Inflation is not caused by DR7-restricted HIV gag epitope cross-reactivity
TCR cross-reactivity is ubiquitous and can occur between unrelated pathogens including HIV (gag) and influenza A virus (67). To assess cross-reactive TCR role in inflation, we repeated the HLA-epitope-TCR simulation experiment using 19 high-affinity DR7-restricted HIV gag epitopes instead, but detected no response (Fig. 7D). Also, HIV viremia was not associated with a significant increase in inflation compared to aviremia (P=0.625; Fig. 7E). These findings suggest that the inflation is not likely caused by cross-reactive HIV gag epitopes.
We also investigated whether HCMV might latently infect the inflated CMV-specific memory CD4+ T cells it induces. We optimized ddPCR quantitation of HCMV DNA using HCMV AD169 strain, but did not detect any HCMV DNA in DYS+ CD4+ T cells (Supplemental Figs. 4A and 4B).
Inflated DYS+ CD4+ T cells display elevated levels of CD38 or HLA-DR, but less often co-express CD38 and HLA-DR
We measured CD38 and HLA-DR dual and individual expression on inflated DYS+ CD4+ T cells. Although we observed no significant difference in their CD38+HLA-DR+ co-expression magnitude compared to controls (P=0.091; Fig. 8A), we observed significantly higher levels of CD38+HLA-DR+ co-expression on their DYS− TEM counterparts within the HIV+ HCMV+ cohort compared to controls (P=0.0001; Fig. 8A). Additionally, individual protein analyses revealed significantly higher levels (P=0.033 and P=0.033; Figs. 8B and 8C, respectively). Remarkably, there was a stepwise increase in the mean expressions from DYS− TEM of HIV− HCMV− to DYS− TEM of HIV− HCMV+ to DYS− and DYS+ TEM of HIV+ HCMV+ subjects. These protein levels on DYS+ CD4+ T cells did not correlate with the magnitudes of DYS+ CD4 inflation (data not shown). CD38 was elevated on naïve T cells as expected (68). Finally, we quantified the latent HIV DNA in Subject 10027’s DYS+ CD4+ T cells but did not detect any enrichment both without and with ART compared to the DYS− counterparts, despite the inflation (P=0.25 and P=0.031, respectively; Supplemental Fig. 4C). HIV was not detected in enriched EPD+ CD4+ T cells, but low cell numbers might have limited the sensitivity. Overall, these findings indicate that inflated DYS+ CD4+ T cells do contribute to the increased T cell activation associated with higher risk of HCMV-related non-AIDS comorbidities in HIV+ HCMV+ subjects, but further studies are required to define the specific subsets of activated cells that correlate most closely with these adverse outcomes.
FIGURE 8.
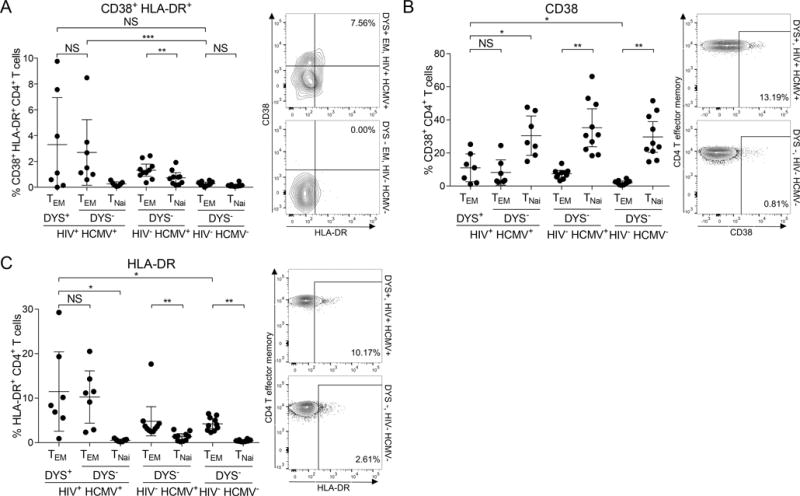
Inflated DYS+ CD4+ T cells have a wider but not significantly higher response magnitude of CD38+HLA-DR+ co-expression. Ex vivo comparisons of (A) CD38+HLA-DR+ (B) CD38 and (C) HLA-DR magnitudes on DYS+ CD4+ TEM of HIV+ HCMV+ subjects to DYS− CD4+ TEM of HIV− HCMV− controls. HIV+ HCMV+: n=7, HIV− HCMV+: n=10 and HIV− HCMV−: n=10. Graphs represent single experiments for each subject and mean±95% CI for all subjects.
Discussion
Here, we show memory inflation of HLA-DR7 restricted, HCMV epitope-specific CD4+ T cells in HCMV+ long-term non-progressor HIV subjects that could potentially contribute to the higher T cell activation associated with elevated risks of HCMV-related non-AIDS cardiovascular comorbidities in such co-infected patients. Ex vivo DR7:DYS tetramer stains revealed persistent, inflated percentages of HCMV’s DYS+ CD4+ T cells in our HIV+ HCMV+ DR7+ subjects that consisted of mostly TEM and TEMRA subsets, and secreted IFN-γ and TNF-α upon in vitro DYS stimulation of their nearly monoclonal TCR repertoires. The 28.75% DYS+ CD4 response magnitude of Subject 10027 measured in the fourth time-point of the longitudinal analyses is the largest reported CD4 response magnitude against DYS epitope to our knowledge, and is similar to the 24% magnitude of a DQ6-restricted pp6541–55 LLQTGIHVRVSQPSL-specific CD4 response (11), although the HIV status of the subject was not specified. It is not clear why Subject 10030 had an extremely low DYS+ CD4+ T cell magnitude. We doubt this was due to CMV sequence variation because the DYS epitope and adjacent residues involved in proteosomal cleavage are known to be completely conserved.
Our findings represent the first ex vivo and tetramer-based evidence for CD4 memory inflation in HIV+ subjects and it is striking in frequency and magnitude for the DYS epitope. It is quite remarkable that DR7+ CD4 responses to other HCMV, TT, EBV or HIV epitopes analyzed in the same cohort were significantly lower. The reduced magnitude of IFN-γ responses to stimulations by these additional epitopes was reflected in the lower absolute counts among the stimulated cells in the ICS assay. Therefore, we believe that the inflation of DYS-specific CD4+ T cells is more likely due to specific HLA-epitope-TCR interactions, and unlikely to be due to HIV-induced inflammation. This is also supported by the finding of a highly enriched CDR3 clonotype in HIV− Subject 20. Also, it does not appear that this is an intrinsic feature of persistent viruses, as EBV epitope-specific response were of low magnitude or absent within the same individuals. Although we were unable to measure these responses among the HIV− DR7+ cohort due to IRB restrictions on re-inviting the subjects, we believe their magnitudes will similarly be low or undetectable. The EPD+ CD4+ T cell response was of unusually high magnitude in the HIV+ Subject 10013 at both tp1 (1.87%) and tp2 (1.4%, data not shown). This individual was also the only subject in whom secreted IFN-γ was detected upon EPD stimulation. These findings suggest that the EPD epitope may drive a memory-inflated response in HIV, and we cannot exclude the possibility that EPD might not also drive an inflated rarely response in HIV− individuals. Interestingly, low-level DYS+ CD4+ T cell responses were detected in all HIV− HCMV+ DR7+ subjects with the exception of one subject with 1.32% magnitude of response. The observation that one HIV− subject had DYS+ CD4+ T cell inflation, which was predominantly TEM phenotype, illustrates that memory inflation with this epitope can occur in HIV− subjects. However, the prevalence and magnitude of memory inflation was substantially higher with HIV co-infection. Taken together the findings suggest that CD4+ memory inflation can occur in HIV negative individuals but HIV acts to increase the prevalence and magnitude.
It is not fully understood how CMV, and why only CMV, induces chronic memory inflation and why this property has been conserved in mice (5–9), rhesus macaques (4) and man (2, 3). The inflated DYS and comparatively lower EPD responses in five HIV co-infected subjects parallel recent reports of different epitopes from the same protein inducing both high- and low-magnitude responses (5, 8). Potential explanations for DYS-specific inflation include the translation of gB mRNA without HCMV replication (69), gB colocalization to endosomes and endogenous presentation (28, 35), and the secretion of such gB epitope-loaded endosomes as immunogenic exosomes (36, 37). Endogenous epitope processing and presentation has been demonstrated recently to drive CD8 memory inflation (6, 27). However, the low pp65 EPD-specific responses might be due to pp65 polyprotein absence in immunogenic exosomes (37). Yet, this mechanism does not explain the published DQ6-restricted pp6541–55 LLQTGIHVRVSQPSL epitope-induced inflation (11), suggesting that multiple factors underlie inflation. Differential gene expression patterns (70), and the presence of higher avidity TCRs specific for DYS might also play some role. It is important to note that cytotoxic CD4+ T cells in general are elevated in HIV infection (71). This may be due to low CCR5 expression, especially by CMV-specific CD4+ T cells, which protects such cells from HIV infection and might explain the lack of HIV DNA enrichment in our results (72).
Few studies have described the TCR repertoire of HLA class II-restricted epitope-specific CD4 responses based on tetramer sorted cells, and most were conducted in vitro or without TCR sequencing (55, 73–83). Notably, some of the repertoires of these single epitope-specific T cells are diverse with over six unique, dominant TCR gene families (55, 73, 80, 83). Therefore, to our knowledge, our work represents the first combination of ex vivo, class II tetramer-derived and deep sequencing-based identification of a nearly monoclonal TCR repertoire of inflated HLA-restricted epitope-specific CD4+ T cells at the resolution of the CDR3. We discovered three new DYS-specific TCRβV gene families: TCRβV6-2, TCRβV5-6 and TCRβV28 in addition to the published TCRβV6-5 (64). CDR3 sequencing confirmed that the inflations were driven by nearly monoclonal expansions, especially in Subject 10032 where 99.4% of all DYS+ TEM were a single clonotype. HIV− Subject 20’s DYS+ CD4+ T cell clonality indicates that clonal expansion to DYS was not unique to HIV+ subjects, a finding that again suggests that HIV co-infection is not necessary for, but rather increases the likelihood of, and amplifies DYS+ CD4+ T cell inflation. To determine if the inflation was unique to DYS, a comparison between bulk DYS+ CDR3s and those of EPD, LIN, PRS, or FRD epitope would have been sufficient; however, magnitudes of cells specific to these additional epitopes were too low for that analysis. CDR3 sequences within DYS− CD4 samples were generally polyclonal with a few exceptions. This noteworthy observation might be driven by clonal expansions induced by epitopes that overlap with DYS as observed in HCMV IE1 epitopes (84), or to other inflation-inducing epitopes of HCMV or other pathogens. The presence of the dominant DYS+ CDR3s within the DYS− repertoire at relatively lower magnitudes reflected potential loss of tetramer binding, while the reverse could reflect non-specific binding to the DYS tetramer. The in vitro HLA-DR7-presented DYS stimulation of the inflated DYS+ CD4 TCR in our Jurkat cell transfection system confirmed the specificity of the tetramer stain and accuracy of our bulk-cell and single-cell TCRα and TCRβ sequencing. We analyzed the dominant DYS+ CDR3 sequences from different subjects for amino acid conservation as: (i) there are no reports of such conservation within inflationary CD4+ T cell CDR3s, and (ii) even dominant clones of well-characterized, non-inflated HLA-A2:NLV CD8 responses from different individuals do not always contain conserved motifs (85). In addition to the published glutamine (64), we also discovered novel conservations of serine and threonine that preceded the germline glycine within the D-segment of the different, dominant DYS+ CD4 CDR3 clones. These amino acids are polar with neutral side chains that might serve as TCR binding residue sites for hydrogen bond formation with DYS and HLA-DR7. Further verification by crystallographic reconstruction of the DR7-DYS-TCR complex is required.
Inflation may be due to intermittent, subclinical CMV reactivation or expression of specific transcripts. We detected no HCMV DNA in plasma samples from this cohort. Future studies could investigate this reactivation through monitoring of other specimens such as saliva, semen, etc. Although we were unable to determine the exact mechanism by which HCMV stimulates inflated responses in our human subjects, we analyzed ex vivo protein expressions of DYS+ CD4+ T cells and observed similarities (CD127− PD-1+/− TIGIT− Granzyme B+) to those on inflated CMV-specific CD8+ T cells reported to be maintained by low-level exposure to antigens from stochastic HCMV reactivation (1, 11, 16, 35). PD-1 might not be an appropriate co-inhibitory protein to evaluate on DYS+ CD4+ T cells due to their low levels of CD28 (64), which has been recently shown to mediate PD-1 suppression of T cells (86, 87). The normal expression levels of anti-apoptotic Bcl-2 suggest that DYS+ CD4+ T cell inflation is not due to apoptosis suppression, but might be due to other maintenance mechanisms such as longer telomeres (26) that could offset the normal rate of apoptosis. The persistence of inflated DYS+ TEM and TEMRA subsets, despite the lack of CCR7+ DYS+ T cell thymic emigrants, is explained by reports that thymectomy does not affect memory T cell inflation or homeostasis (88).
Surprisingly, Ki-67 data suggests that the DYS+ CD4+ T cell inflations were not driven to significantly higher proliferation. While this observation might be due to cross-sectional sampling limitations, it does confirm findings in chronic MCMV models of inflation (5, 9). CD28 and CD57 analyses confirm that only a very limited number of the inflated cells are too senescent to replicate (66). The cause of inflation does not appear to involve DYS+ CD4+ TCR cross-reactivity with the DR7+ HIV gag epitopes either. But, cross-reactivity with other DR7+ HIV epitopes or to other inflated TCRs cannot be excluded. We observed increased inflation in two subjects when HIV replication was suppressed to undetectable levels with ART, suggesting that HIV replication is not required for maintenance of inflation.
The sites of HCMV latency are important (1) and HIV could alter the environment to help HCMV persist in long-lived non-hematopoietic cells in HCMV reservoir sites such as LNs and vascular endothelial cells (VECs). VECs can serve as latent HCMV reservoirs and also express the CX3CR1 ligand—fractalkine. Vascular homing might bring CX3CR1high DYS+ CD4+ T cells in close contact with these potential HCMV reservoirs, resulting in re-stimulation and inflation. Although we did not detect HCMV DNA in the inflated cells, it is possible that they passively disseminate HCMV from LNs to vascular endothelium without getting infected, as LN DCs do for HIV. Herpes viruses such as CMV are species-specific and cause life-long infection. Therefore, it is also possible that the inflated responses provide a degree of protective immunity against other infections, making them mutually beneficial to CMV and its host.
Although CD38+HLA-DR+ co-expression on the inflated cells was not significantly elevated compared to controls, a wider distribution was observed with inflation. Remarkably, a similar comparison of CD38+HLA-DR+ co-expression on the DYS− TEM counterpart of the inflated cells to controls produced a significant difference. These DYS− TEMs consist of clonal CDR3 expansions (Fig. 4) that are potentially induced by other inflationary epitopes. Consequently, it is plausible that analyses of CD4 responses against a collection of inflationary epitopes or in a larger number of subjects might yield a difference. Both statistical trends are not due to generalized, HIV-induced activation because other pathogen/antigen specific CD4+ T cells, including TT, are not necessarily more activated with HIV infection (45). This observation implies that inflated CD4+ T cells in these subjects could potentially contribute to the increased T cell activation associated with greater risks of HCMV-related non-AIDS cardiovascular comorbidities that continue to plague HIV+ subjects despite effective ART. The capacity of DYS+ CD4+ T cells to secrete granzyme B, IFN-γ and TNF-α might facilitate the development of these comorbidities (36, 47, 48). A larger cohort of HIV+ HCMV+ DR7+ subjects with varying magnitudes of DYS+ CD4+ T cells is needed to directly evaluate the correlation of their activation with disease outcomes. The stepwise increments in CD38+HLA-DR+ levels indicate that HCMV infection without HIV co-infection increases CD4+ TEM activation in general, and HIV co-infection further synergizes such activation. This elevated activation might be tied to an HIV-induced latent HCMV reservoir expansion, presenting potential unintended negative consequences of HCMV vaccine candidates that contain inflation-inducing epitopes for all individuals, especially HIV+ DR7+ subjects. Although we studied HIV long-term non-progressors, a previous study found that CMV lysate induces high levels of CMV-specific CD4+ T cells in HIV+ subjects with ART-induced HIV aviremia (30), suggesting that our findings may generalize to a broader range of HIV-infected patients.
In conclusion, we have shown that HIV+ HCMV+ co-infection boosts CD4 responses to HCMV gB’s DYS and pp65’s EPD epitopes, resulting in mostly memory-inflated DYS+ CD4+ T cells. To our knowledge, this is the first ex vivo evidence of both CD4+ T cell memory inflation against the DYSNTHSTRYV epitope in HIV+ subjects and nearly monoclonal CDR3 repertoire of inflated CD4+ T cells that contain novel conserved motifs. Although the underlying mechanism may be multifactorial, we hypothesize that increased low-level exposure and subsequent clonal expansion targeting the DYS epitope from stochastic HCMV reactivation or expression largely contributes to our observation. Our findings suggest that “memory inflation”-inducing epitopes might contribute to the immunopathogenesis of non-AIDS comorbidities and raise safety implications for CMV vaccines that contain inflation-inducing epitopes that should be considered in trials being planned in both HIV and non-HIV infected subjects. This work also suggests that the relative contributions of conventional and inflated CMV-specific T cell responses to protection of the host from infection or malignancy, vaccine responsiveness or co-morbidities of aging such as vascular disease should be considered separately.
Supplementary Material
Acknowledgments
We are very grateful to all donors, the Vanderbilt-Ingram Cancer Center for HIV− samples, and the Vanderbilt Flow Cytometry Core for data acquisition. In no particular order, we are grateful to Mark Watson, Ian James, Rita Smith, David Haas, Heather Long, John Koethe, Alec Redwood, Kaija Strautins, Kristina Williams, Katie White, Wannakuwatte Fernando, Katherine Konvinse, and Jessica Thomas for donor recruitment, blood draws and helpful discussions.
Funding: This work was supported by the NIH grants P30 AI110527 (to SAM), P01 AI030731 and R01 AI094019 (to DMK) and by the National Institute of General Medical Sciences grant T32 GM007347 (to COA).
Abbreviations
- DYS
DYSNTHSTRYV
- HCMV
human cytomegalovirus
- MCMV
murine cytomegalovirus
- LN
lymph node
- gB
glycoprotein B
- DR7+
HLA-DRB1*07:01
- EPD
EPDVYYTSAFVFPTK
- TT
tetanus toxoid
- LIN
LINSTKIYSYFPSVISKVNQ
- FRD
FRDYVDRFYKTLRAEQASQE
- PRS
PRSPTVFYNIPPMPLPPSQL
- MFI
median fluorescence intensity
- ddPCR
droplet digital PCR
- LCLs
lymphoblastoid cell lines
- TEM
effector memory
- TEMRA
effector memory-RA+
- VECs
vascular endothelial cells
- pp65
phosphoprotein-65
- ART
antiretroviral therapy
- tp1
time-point 1
- tp2
time-point 2
- TCRβV
TCR-beta-variable
- TCRβJ
TCR-beta-joining
Footnotes
Contributions: COA, MAP, SG, CW, PK, EJP, DMK, SAK and SAM designed research studies. COA, CH, DKJ, BGE, MHJ, SAK and SAM acquired samples. COA, SG, AC, LB, RG, CH, and DKJ conducted experiments. COA, SG, AC, and LB acquired data. COA, SG, WJM, EJP, SK and SAM analyzed data. COA, MAP, PK, DMK, SAK and SAM wrote the manuscript, and all authors edited it.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Klenerman P, Oxenius A. T cell responses to cytomegalovirus. Nat Rev Immunol. 2016;16:367–377. doi: 10.1038/nri.2016.38. [DOI] [PubMed] [Google Scholar]
- 2.Waller EC, Day E, Sissons JG, Wills MR. Dynamics of T cell memory in human cytomegalovirus infection. Med Microbiol Immunol. 2008;197:83–96. doi: 10.1007/s00430-008-0082-5. [DOI] [PubMed] [Google Scholar]
- 3.Klarenbeek PL, Remmerswaal EB, Berge IJ ten, Doorenspleet ME, van Schaik BD, Esveldt RE, Koch SD, ten Brinke A, van Kampen AH, Bemelman FJ, Tak PP, Baas F, de Vries N, van Lier RA. Deep sequencing of antiviral T-cell responses to HCMV and EBV in humans reveals a stable repertoire that is maintained for many years. PLoS Pathog. 2012;8:e1002889. doi: 10.1371/journal.ppat.1002889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cicin-Sain L, Sylwester AW, Hagen SI, Siess DC, Currier N, Legasse AW, Fischer MB, Koudelka CW, Axthelm MK, Nikolich-Zugich J, Picker LJ. Cytomegalovirus-specific T cell immunity is maintained in immunosenescent rhesus macaques. J Immunol. 2011;187:1722–1732. doi: 10.4049/jimmunol.1100560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolinger B, Sims S, Swadling L, O’Hara G, de Lara C, Baban D, Saghal N, Lee LN, Marchi E, Davis M, Newell E, Capone S, Folgori A, Barnes E, Klenerman P. Adenoviral Vector Vaccination Induces a Conserved Program of CD8(+) T Cell Memory Differentiation in Mouse and Man. Cell Rep. 2015;13:1578–1588. doi: 10.1016/j.celrep.2015.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dekhtiarenko I, Ratts RB, Blatnik R, Lee LN, Fischer S, Borkner L, Oduro JD, Marandu TF, Hoppe S, Ruzsics Z, Sonnemann JK, Mansouri M, Meyer C, Lemmermann NA, Holtappels R, Arens R, Klenerman P, Fruh K, Reddehase MJ, Riemer AB, Cicin-Sain L. Peptide Processing Is Critical for T-Cell Memory Inflation and May Be Optimized to Improve Immune Protection by CMV-Based Vaccine Vectors. PLoS Pathog. 2016;12:e1006072. doi: 10.1371/journal.ppat.1006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karrer U, Sierro S, Wagner M, Oxenius A, Hengel H, Koszinowski UH, Phillips RE, Klenerman P. Memory inflation: continuous accumulation of antiviral CD8+ T cells over time. J Immunol. 2003;170:2022–2029. doi: 10.4049/jimmunol.170.4.2022. [DOI] [PubMed] [Google Scholar]
- 8.Munks MW, Cho KS, Pinto AK, Sierro S, Klenerman P, Hill AB. Four distinct patterns of memory CD8 T cell responses to chronic murine cytomegalovirus infection. J Immunol. 2006;177:450–458. doi: 10.4049/jimmunol.177.1.450. [DOI] [PubMed] [Google Scholar]
- 9.Sierro S, Rothkopf R, Klenerman P. Evolution of diverse antiviral CD8+ T cell populations after murine cytomegalovirus infection. Eur J Immunol. 2005;35:1113–1123. doi: 10.1002/eji.200425534. [DOI] [PubMed] [Google Scholar]
- 10.Burgers WA, Riou C, Mlotshwa M, Maenetje P, de Assis Rosa D, Brenchley J, Mlisana K, Douek DC, Koup R, Roederer M, de Bruyn G, Karim SA, Williamson C, Gray CM, CAIS Team Association of HIV-specific and total CD8+ T memory phenotypes in subtype C HIV-1 infection with viral set point. J Immunol. 2009;182:4751–4761. doi: 10.4049/jimmunol.0803801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pachnio A, Ciaurriz M, Begum J, Lal N, Zuo J, Beggs A, Moss P. Cytomegalovirus Infection Leads to Development of High Frequencies of Cytotoxic Virus-Specific CD4+ T Cells Targeted to Vascular Endothelium. PLoS Pathog. 2016;12:e1005832. doi: 10.1371/journal.ppat.1005832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snyder CM, Cho KS, Bonnett EL, van Dommelen S, Shellam GR, Hill AB. Memory inflation during chronic viral infection is maintained by continuous production of short-lived, functional T cells. Immunity. 2008;29:650–659. doi: 10.1016/j.immuni.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conrad JA, Ramalingam RK, Smith RM, Barnett L, Lorey SL, Wei J, Simons BC, Sadagopal S, Meyer-Olson D, Kalams SA. Dominant clonotypes within HIV-specific T cell responses are programmed death-1high and CD127low and display reduced variant cross-reactivity. J Immunol. 2011;186:6871–6885. doi: 10.4049/jimmunol.1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Hara GA, Welten SP, Klenerman P, Arens R. Memory T cell inflation: understanding cause and effect. Trends Immunol. 2012;33:84–90. doi: 10.1016/j.it.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Okoye AA, Rohankhedkar M, Konfe AL, Abana CO, Reyes MD, Clock JA, Duell DM, Sylwester AW, Sammader P, Legasse AW, Park BS, Axthelm MK, Nikolich-Zugich J, Picker LJ. Effect of IL-7 Therapy on Naive and Memory T Cell Homeostasis in Aged Rhesus Macaques. J Immunol. 2015;195:4292–4305. doi: 10.4049/jimmunol.1500609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnston RJ, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, Park S, Javinal V, Chiu H, Irving B, Eaton DL, Grogan JL. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell. 2014;26:923–937. doi: 10.1016/j.ccell.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Gamadia LE, van Leeuwen EM, Remmerswaal EB, Yong SL, Surachno S, Wertheim-van Dillen PM, Ten Berge IJ, Van Lier RA. The size and phenotype of virus-specific T cell populations is determined by repetitive antigenic stimulation and environmental cytokines. J Immunol. 2004;172:6107–6114. doi: 10.4049/jimmunol.172.10.6107. [DOI] [PubMed] [Google Scholar]
- 18.Klenerman P, Hill A. T cells and viral persistence: lessons from diverse infections. Nat Immunol. 2005;6:873–879. doi: 10.1038/ni1241. [DOI] [PubMed] [Google Scholar]
- 19.Lang A, Brien JD, Nikolich-Zugich J. Inflation and long-term maintenance of CD8 T cells responding to a latent herpesvirus depend upon establishment of latency and presence of viral antigens. J Immunol. 2009;183:8077–8087. doi: 10.4049/jimmunol.0801117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holtappels R, Pahl-Seibert MF, Thomas D, Reddehase MJ. Enrichment of immediate-early 1 (m123/pp89) peptide-specific CD8 T cells in a pulmonary CD62L(lo) memory-effector cell pool during latent murine cytomegalovirus infection of the lungs. J Virol. 2000;74:11495–11503. doi: 10.1128/jvi.74.24.11495-11503.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seckert CK, Griessl M, Buttner JK, Scheller S, Simon CO, Kropp KA, Renzaho A, Kuhnapfel B, Grzimek NK, Reddehase MJ. Viral latency drives ‘memory inflation’: a unifying hypothesis linking two hallmarks of cytomegalovirus infection. Med Microbiol Immunol. 2012;201:551–566. doi: 10.1007/s00430-012-0273-y. [DOI] [PubMed] [Google Scholar]
- 22.Simon CO, Holtappels R, Tervo HM, Bohm V, Daubner T, Oehrlein-Karpi SA, Kuhnapfel B, Renzaho A, Strand D, Podlech J, Reddehase MJ, Grzimek NK. CD8 T cells control cytomegalovirus latency by epitope-specific sensing of transcriptional reactivation. J Virol. 2006;80:10436–10456. doi: 10.1128/JVI.01248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seckert CK, Schader SI, Ebert S, Thomas D, Freitag K, Renzaho A, Podlech J, Reddehase MJ, Holtappels R. Antigen-presenting cells of haematopoietic origin prime cytomegalovirus-specific CD8 T-cells but are not sufficient for driving memory inflation during viral latency. J Gen Virol. 2011;92:1994–2005. doi: 10.1099/vir.0.031815-0. [DOI] [PubMed] [Google Scholar]
- 24.Torti N, Walton SM, Brocker T, Rulicke T, Oxenius A. Non-hematopoietic cells in lymph nodes drive memory CD8 T cell inflation during murine cytomegalovirus infection. PLoS Pathog. 2011;7:e1002313. doi: 10.1371/journal.ppat.1002313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gordon CL, Miron M, Thome JJ, Matsuoka N, Weiner J, Rak MA, Igarashi S, Granot T, Lerner H, Goodrum F, Farber DL. Tissue reservoirs of antiviral T cell immunity in persistent human CMV infection. J Exp Med. 2017;214:651–667. doi: 10.1084/jem.20160758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Bryan JM, Woda M, Co M, Mathew A, Rothman AL. Telomere length dynamics in human memory T cells specific for viruses causing acute or latent infections. Immun Ageing. 2013;10:37. doi: 10.1186/1742-4933-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hutchinson S, Sims S, O’Hara G, Silk J, Gileadi U, Cerundolo V, Klenerman P. A dominant role for the immunoproteasome in CD8+ T cell responses to murine cytomegalovirus. PLoS One. 2011;6:e14646. doi: 10.1371/journal.pone.0014646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hegde NR, Dunn C, Lewinsohn DM, Jarvis MA, Nelson JA, Johnson DC. Endogenous human cytomegalovirus gB is presented efficiently by MHC class II molecules to CD4+ CTL. J Exp Med. 2005;202:1109–1119. doi: 10.1084/jem.20050162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walton SM, Torti N, Mandaric S, Oxenius A. T-cell help permits memory CD8(+) T-cell inflation during cytomegalovirus latency. Eur J Immunol. 2011;41:2248–2259. doi: 10.1002/eji.201141575. [DOI] [PubMed] [Google Scholar]
- 30.Komanduri KV, Donahoe SM, Moretto WJ, Schmidt DK, Gillespie G, Ogg GS, Roederer M, Nixon DF, McCune JM. Direct measurement of CD4+ and CD8+ T-cell responses to CMV in HIV-1-infected subjects. Virology. 2001;279:459–470. doi: 10.1006/viro.2000.0697. [DOI] [PubMed] [Google Scholar]
- 31.Lichtner M, Cicconi P, Vita S, Cozzi-Lepri A, Galli M, Lo Caputo S, Saracino A, De Luca A, Moioli M, Maggiolo F, Marchetti G, Vullo V, d’Arminio Monforte A, I.F. Study Cytomegalovirus coinfection is associated with an increased risk of severe non-AIDS-defining events in a large cohort of HIV-infected patients. J Infect Dis. 2015;211:178–186. doi: 10.1093/infdis/jiu417. [DOI] [PubMed] [Google Scholar]
- 32.Maidji E, Somsouk M, Rivera JM, Hunt PW, Stoddart CA. Replication of CMV in the gut of HIV-infected individuals and epithelial barrier dysfunction. PLoS Pathog. 2017;13:e1006202. doi: 10.1371/journal.ppat.1006202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, Sleath PR, Grabstein KH, Hosken NA, Kern F, Nelson JA, Picker LJ. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med. 2005;202:673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kambayashi T, Laufer TM. Atypical MHC class II-expressing antigen-presenting cells: can anything replace a dendritic cell? Nat Rev Immunol. 2014;14:719–730. doi: 10.1038/nri3754. [DOI] [PubMed] [Google Scholar]
- 35.Pachnio A, Zuo J, Ryan GB, Begum J, Moss PA. The Cellular Localization of Human Cytomegalovirus Glycoprotein Expression Greatly Influences the Frequency and Functional Phenotype of Specific CD4+ T Cell Responses. J Immunol. 2015;195:3803–3815. doi: 10.4049/jimmunol.1500696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broadley I, Pera A, Morrow G, Davies KA, Kern F. Expansions of Cytotoxic CD4+CD28- T Cells Drive Excess Cardiovascular Mortality in Rheumatoid Arthritis and Other Chronic Inflammatory Conditions and Are Triggered by CMV Infection. Front Immunol. 2017;8:195. doi: 10.3389/fimmu.2017.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker JD, Maier CL, Pober JS. Cytomegalovirus-infected human endothelial cells can stimulate allogeneic CD4+ memory T cells by releasing antigenic exosomes. J Immunol. 2009;182:1548–1559. doi: 10.4049/jimmunol.182.3.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elkington R, Shoukry NH, Walker S, Crough T, Fazou C, Kaur A, Walker CM, Khanna R. Cross-reactive recognition of human and primate cytomegalovirus sequences by human CD4 cytotoxic T lymphocytes specific for glycoprotein B and H. Eur J Immunol. 2004;34:3216–3226. doi: 10.1002/eji.200425203. [DOI] [PubMed] [Google Scholar]
- 39.Barbaro G, Fisher SD, Lipshultz SE. Pathogenesis of HIV-associated cardiovascular complications. Lancet Infect Dis. 2001;1:115–124. doi: 10.1016/S1473-3099(01)00067-6. [DOI] [PubMed] [Google Scholar]
- 40.Hsue PY, Lo JC, Franklin A, Bolger AF, Martin JN, Deeks SG, Waters DD. Progression of atherosclerosis as assessed by carotid intima-media thickness in patients with HIV infection. Circulation. 2004;109:1603–1608. doi: 10.1161/01.CIR.0000124480.32233.8A. [DOI] [PubMed] [Google Scholar]
- 41.Cheng J, Ke Q, Jin Z, Wang H, Kocher O, Morgan JP, Zhang J, Crumpacker CS. Cytomegalovirus infection causes an increase of arterial blood pressure. PLoS Pathog. 2009;5:e1000427. doi: 10.1371/journal.ppat.1000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wall NA, Chue CD, Edwards NC, Pankhurst T, Harper L, Steeds RP, Lauder S, Townend JN, Moss P, Ferro CJ. Cytomegalovirus seropositivity is associated with increased arterial stiffness in patients with chronic kidney disease. PLoS One. 2013;8:e55686. doi: 10.1371/journal.pone.0055686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slot MC, Kroon AA, Damoiseaux JG, Theunissen R, Houben AJ, de Leeuw PW, Tervaert JW. CD4+CD28null T Cells are related to previous cytomegalovirus infection but not to accelerated atherosclerosis in ANCA-associated vasculitis. Rheumatol Int. 2017 doi: 10.1007/s00296-016-3643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaplan RC, Sinclair E, Landay AL, Lurain N, Sharrett AR, Gange SJ, Xue X, Hunt P, Karim R, Kern DM, Hodis HN, Deeks SG. T cell activation and senescence predict subclinical carotid artery disease in HIV-infected women. J Infect Dis. 2011;203:452–463. doi: 10.1093/infdis/jiq071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith MZ, Bastidas S, Karrer U, Oxenius A. Impact of antigen specificity on CD4+ T cell activation in chronic HIV-1 infection. BMC Infect Dis. 2013;13:100. doi: 10.1186/1471-2334-13-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunt PW, Martin JN, Sinclair E, Epling L, Teague J, Jacobson MA, Tracy RP, Corey L, Deeks SG. Valganciclovir reduces T cell activation in HIV-infected individuals with incomplete CD4+ T cell recovery on antiretroviral therapy. J Infect Dis. 2011;203:1474–1483. doi: 10.1093/infdis/jir060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sacre K, Hunt PW, Hsue PY, Maidji E, Martin JN, Deeks SG, Autran B, McCune JM. A role for cytomegalovirus-specific CD4+CX3CR1+ T cells and cytomegalovirus-induced T-cell immunopathology in HIV-associated atherosclerosis. AIDS. 2012;26:805–814. doi: 10.1097/QAD.0b013e328351f780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van de Berg PJ, Yong SL, Remmerswaal EB, van Lier RA, ten Berge IJ. Cytomegalovirus-induced effector T cells cause endothelial cell damage. Clin Vaccine Immunol. 2012;19:772–779. doi: 10.1128/CVI.00011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chanouzas D, Dyall L, Dale J, Moss P, Morgan M, Harper L. CD4+CD28- T-cell expansions in ANCA-associated vasculitis and association with arterial stiffness: baseline data from a randomised controlled trial. Lancet. 2015;385(Suppl 1):S30. doi: 10.1016/S0140-6736(15)60345-2. [DOI] [PubMed] [Google Scholar]
- 50.Chanouzas D, Sagmeister M, Dyall L, Nightingale P, Ferro C, Moss P, Morgan M, Harper L. Role of cytomegalovirus in the expansion of CD4+CD28− T cells in patients with ANCA-associated vasculitis: a proof-of-concept, randomised controlled trial. The Lancet. 2017;389:S17. [Google Scholar]
- 51.Klenerman P, Cerundolo V, Dunbar PR. Tracking T cells with tetramers: new tales from new tools. Nat Rev Immunol. 2002;2:263–272. doi: 10.1038/nri777. [DOI] [PubMed] [Google Scholar]
- 52.Nicholas KJ, Flaherty DK, Smith RM, Sather DN, Kalams SA. Chronic HIV-1 Infection Impairs Superantigen-Induced Activation of Peripheral CD4+CXCR5+PD-1+ Cells, With Relative Preservation of Recall Antigen-Specific Responses. J Acquir Immune Defic Syndr. 2017;74:72–80. doi: 10.1097/QAI.0000000000001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Pira G, Bottone L, Ivaldi F, Pelizzoli R, Del Galdo F, Lozzi L, Bracci L, Loregian A, Palu G, De Palma R, Einsele H, Manca F. Identification of new Th peptides from the cytomegalovirus protein pp65 to design a peptide library for generation of CD4 T cell lines for cellular immunoreconstitution. Int Immunol. 2004;16:635–642. doi: 10.1093/intimm/dxh065. [DOI] [PubMed] [Google Scholar]
- 54.James EA, Bui J, Berger D, Huston L, Roti M, Kwok WW. Tetramer-guided epitope mapping reveals broad, individualized repertoires of tetanus toxin-specific CD4+ T cells and suggests HLA-based differences in epitope recognition. Int Immunol. 2007;19:1291–1301. doi: 10.1093/intimm/dxm099. [DOI] [PubMed] [Google Scholar]
- 55.Vingert B, Perez-Patrigeon S, Jeannin P, Lambotte O, Boufassa F, Lemaitre F, Kwok WW, Theodorou I, Delfraissy JF, Theze J, Chakrabarti LA, AEHCS Group HIV controller CD4+ T cells respond to minimal amounts of Gag antigen due to high TCR avidity. PLoS Pathog. 2010;6:e1000780. doi: 10.1371/journal.ppat.1000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Long HM, Chagoury OL, Leese AM, Ryan GB, James E, Morton LT, Abbott RJ, Sabbah S, Kwok W, Rickinson AB. MHC II tetramers visualize human CD4+ T cell responses to Epstein-Barr virus infection and demonstrate atypical kinetics of the nuclear antigen EBNA1 response. J Exp Med. 2013;210:933–949. doi: 10.1084/jem.20121437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leisner C, Loeth N, Lamberth K, Justesen S, Sylvester-Hvid C, Schmidt EG, Claesson M, Buus S, Stryhn A. One-pot, mix-and-read peptide-MHC tetramers. PLoS One. 2008;3:e1678. doi: 10.1371/journal.pone.0001678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 59.Shugay M, Bagaev DV, Turchaninova MA, Bolotin DA, Britanova OV, Putintseva EV, Pogorelyy MV, Nazarov VI, Zvyagin IV, Kirgizova VI, Kirgizov KI, Skorobogatova EV, Chudakov DM. VDJtools: Unifying Post-analysis of T Cell Receptor Repertoires. PLoS Comput Biol. 2015;11:e1004503. doi: 10.1371/journal.pcbi.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol. 2014;32:684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anmole G, Kuang XT, Toyoda M, Martin E, Shahid A, Le AQ, Markle T, Baraki B, Jones RB, Ostrowski MA, Ueno T, Brumme ZL, Brockman MA. A robust and scalable TCR-based reporter cell assay to measure HIV-1 Nef-mediated T cell immune evasion. J Immunol Methods. 2015;426:104–113. doi: 10.1016/j.jim.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 62.Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH, Spina CA, Woelk CH, Richman DD. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One. 2013;8:e55943. doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garg A, Trout R, Spector SA. Human Immunodeficiency Virus Type-1 Myeloid Derived Suppressor Cells Inhibit Cytomegalovirus Inflammation through Interleukin-27 and B7-H4. Sci Rep. 2017;7:44485. doi: 10.1038/srep44485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crompton L, Khan N, Khanna R, Nayak L, Moss PA. CD4+ T cells specific for glycoprotein B from cytomegalovirus exhibit extreme conservation of T-cell receptor usage between different individuals. Blood. 2008;111:2053–2061. doi: 10.1182/blood-2007-04-079863. [DOI] [PubMed] [Google Scholar]
- 65.Ventura C, Bisceglia H, Girerd-Chambaz Y, Burdin N, Chaux P. HLA-DR and HLA-DP restricted epitopes from human cytomegalovirus glycoprotein B recognized by CD4+ T-cell clones from chronically infected individuals. J Clin Immunol. 2012;32:1305–1316. doi: 10.1007/s10875-012-9732-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brenchley JM, Karandikar NJ, Betts MR, Ambrozak DR, Hill BJ, Crotty LE, Casazza JP, Kuruppu J, Migueles SA, Connors M, Roederer M, Douek DC, Koup RA. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood. 2003;101:2711–2720. doi: 10.1182/blood-2002-07-2103. [DOI] [PubMed] [Google Scholar]
- 67.Acierno PM, Newton DA, Brown EA, Maes LA, Baatz JE, Gattoni-Celli S. Cross-reactivity between HLA-A2-restricted FLU-M1:58–66 and HIV p17 GAG:77–85 epitopes in HIV-infected and uninfected individuals. J Transl Med. 2003;1:3. doi: 10.1186/1479-5876-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chattopadhyay PK, Roederer M. Good cell, bad cell: flow cytometry reveals T-cell subsets important in HIV disease. Cytometry A. 2010;77:614–622. doi: 10.1002/cyto.a.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smuda C, Bogner E, Radsak K. The human cytomegalovirus glycoprotein B gene (ORF UL55) is expressed early in the infectious cycle. J Gen Virol. 1997;78(Pt 8):1981–1992. doi: 10.1099/0022-1317-78-8-1981. [DOI] [PubMed] [Google Scholar]
- 70.Dekhtiarenko I, Jarvis MA, Ruzsics Z, Cicin-Sain L. The context of gene expression defines the immunodominance hierarchy of cytomegalovirus antigens. J Immunol. 2013;190:3399–3409. doi: 10.4049/jimmunol.1203173. [DOI] [PubMed] [Google Scholar]
- 71.Appay V, Zaunders JJ, Papagno L, Sutton J, Jaramillo A, Waters A, Easterbrook P, Grey P, Smith D, McMichael AJ, Cooper DA, Rowland-Jones SL, Kelleher AD. Characterization of CD4(+) CTLs ex vivo. J Immunol. 2002;168:5954–5958. doi: 10.4049/jimmunol.168.11.5954. [DOI] [PubMed] [Google Scholar]
- 72.Oswald-Richter K, Grill SM, Leelawong M, Tseng M, Kalams SA, Hulgan T, Haas DW, Unutmaz D. Identification of a CCR5-expressing T cell subset that is resistant to R5-tropic HIV infection. PLoS Pathog. 2007;3:e58. doi: 10.1371/journal.ppat.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ertelt JM, Johanns TM, Mysz MA, Nanton MR, Rowe JH, Aguilera MN, Way SS. Selective culling of high avidity antigen-specific CD4+ T cells after virulent Salmonella infection. Immunology. 2011;134:487–497. doi: 10.1111/j.1365-2567.2011.03510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Falta MT, Fontenot AP, Rosloniec EF, Crawford F, Roark CL, Bill J, Marrack P, Kappler J, Kotzin BL. Class II major histocompatibility complex-peptide tetramer staining in relation to functional avidity and T cell receptor diversity in the mouse CD4(+) T cell response to a rheumatoid arthritis-associated antigen. Arthritis Rheum. 2005;52:1885–1896. doi: 10.1002/art.21098. [DOI] [PubMed] [Google Scholar]
- 75.Kim C, Wilson T, Fischer KF, Williams MA. Sustained interactions between T cell receptors and antigens promote the differentiation of CD4(+) memory T cells. Immunity. 2013;39:508–520. doi: 10.1016/j.immuni.2013.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kimata M, Cullins DL, Brown ML, Brand DD, Rosloniec EF, Myers LK, Stuart JM, Kang AH. Characterization of inhibitory T cells induced by an analog of type II collagen in an HLA-DR1 humanized mouse model of autoimmune arthritis. Arthritis Res Ther. 2012;14:R107. doi: 10.1186/ar3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koehne G, Hasan A, Doubrovina E, Prockop S, Tyler E, Wasilewski G, O’Reilly RJ. Immunotherapy with Donor T Cells Sensitized with Overlapping Pentadecapeptides for Treatment of Persistent Cytomegalovirus Infection or Viremia. Biol Blood Marrow Transplant. 2015;21:1663–1678. doi: 10.1016/j.bbmt.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lanzer KG, Johnson LL, Woodland DL, Blackman MA. Impact of ageing on the response and repertoire of influenza virus-specific CD4 T cells. Immun Ageing. 2014;11:9. doi: 10.1186/1742-4933-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Legoux F, Gautreau L, Hesnard L, Leger A, Moyon M, Devilder MC, Bonneville M, Saulquin X. Characterization of the human CD4(+) T-cell repertoire specific for major histocompatibility class I-restricted antigens. Eur J Immunol. 2013;43:3244–3253. doi: 10.1002/eji.201343726. [DOI] [PubMed] [Google Scholar]
- 80.Nose H, Kubota R, Seth NP, Goon PK, Tanaka Y, Izumo S, Usuku K, Ohara Y, Wucherpfennig KW, Bangham CR, Osame M, Saito M. Ex vivo analysis of human T lymphotropic virus type 1-specific CD4+ cells by use of a major histocompatibility complex class II tetramer composed of a neurological disease-susceptibility allele and its immunodominant peptide. J Infect Dis. 2007;196:1761–1772. doi: 10.1086/522966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Petersen J, van Bergen J, Loh KL, Kooy-Winkelaar Y, Beringer DX, Thompson A, Bakker SF, Mulder CJ, Ladell K, McLaren JE, Price DA, Rossjohn J, Reid HH, Koning F. Determinants of gliadin-specific T cell selection in celiac disease. J Immunol. 2015;194:6112–6122. doi: 10.4049/jimmunol.1500161. [DOI] [PubMed] [Google Scholar]
- 82.Poli C, Raffin C, Dojcinovic D, Luescher I, Ayyoub M, Valmori D. MHC class II/ESO tetramer-based generation of in vitro primed anti-tumor T-helper lines for adoptive cell therapy of cancer. Haematologica. 2013;98:316–322. doi: 10.3324/haematol.2012.071712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sabatino JJ, Jr, Huang J, Zhu C, Evavold BD. High prevalence of low affinity peptide-MHC II tetramer-negative effectors during polyclonal CD4+ T cell responses. J Exp Med. 2011;208:81–90. doi: 10.1084/jem.20101574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Braendstrup P, Mortensen BK, Justesen S, Osterby T, Rasmussen M, Hansen AM, Christiansen CB, Hansen MB, Nielsen M, Vindelov L, Buus S, Stryhn A. Identification and HLA-tetramer-validation of human CD4+ and CD8+ T cell responses against HCMV proteins IE1 and IE2. PLoS One. 2014;9:e94892. doi: 10.1371/journal.pone.0094892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F, Ji X, Han A, Krams SM, Pettus C, Haas N, Arlehamn CSL, Sette A, Boyd SD, Scriba TJ, Martinez OM, Davis MM. Identifying specificity groups in the T cell receptor repertoire. Nature. 2017;547:94–98. doi: 10.1038/nature22976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hui E, Cheung J, Zhu J, Su X, Taylor MJ, Wallweber HA, Sasmal DK, Huang J, Kim JM, Mellman I, Vale RD. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science. 2017;355:1428–1433. doi: 10.1126/science.aaf1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kamphorst AO, Wieland A, Nasti T, Yang S, Zhang R, Barber DL, Konieczny BT, Daugherty CZ, Koenig L, Yu K, Sica GL, Sharpe AH, Freeman GJ, Blazar BR, Turka LA, Owonikoko TK, Pillai RN, Ramalingam SS, Araki K, Ahmed R. Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science. 2017;355:1423–1427. doi: 10.1126/science.aaf0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Okoye AA, Rohankhedkar M, Abana C, Pattenn A, Reyes M, Pexton C, Lum R, Sylwester A, Planer SL, Legasse A, Park BS, Piatak M, Jr, Lifson JD, Axthelm MK, Picker LJ. Naive T cells are dispensable for memory CD4+ T cell homeostasis in progressive simian immunodeficiency virus infection. J Exp Med. 2012;209:641–651. doi: 10.1084/jem.20112071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


