Summary
Antiretroviral therapy (ART) suppresses viral replication in HIV-infected individuals, but does not eliminate the reservoir of latently infected cells. Recent work identified PD-1+ follicular helper T cells (Tfh) as an important cellular compartment for viral persistence. Here, using ART-treated, SIV-infected rhesus macaques, we show that CTLA-4+PD-1− memory CD4+ T cells, which share phenotypic markers with regulatory T cells, were enriched in SIV-DNA in blood, lymph nodes (LN), spleen, and gut, and contained replication-competent and infectious virus. In contrast to PD-1+ Tfh, SIV-enriched CTLA-4+PD-1− CD4+ T cells were found outside the B-cell follicle of the LN, predicted the size of the persistent viral reservoir during ART, and significantly increased their contribution to the SIV reservoir with prolonged ART-mediated viral suppression. We have shown that CTLA-4+PD-1− memory CD4+ T cells are a previously unrecognized component of the SIV and HIV reservoir that should be therapeutically targeted for a functional HIV-1 cure.
eTOC Blurb
HIV persists in T follicular-helper cells within the lymph node during antiretroviral therapy, but decays with time. McGary et al. identify the persistence of replication-competent SIV and HIV outside the lymph node follicle in a unique subset of CTLA-4+PD-1- memory CD4+ T-cells that share features with regulatory T-cells.
Introduction
The ability of antiretroviral therapy (ART) to effectively suppress HIV-1 replication has dramatically reduced HIV morbidity and mortality (Bhaskaran et al., 2008; Cooper, 2008). Despite this success, HIV-infected individuals must remain on ART for their lifetime due to the persistence of latently infected cells containing transcriptionally silent, integrated provirus, which allows them to evade immune detection (Chun et al., 1997a; Chun et al., 1997b; Finzi et al., 1997; Finzi et al., 1999; Wong et al., 1997). A fraction of these latently infected cells contain proviruses that are replication competent, constituting the latent viral reservoir that is responsible for the rebound of viremia upon treatment interruption (Chun et al., 1999; Davey et al., 1999). Therefore, strategies that target and eliminate latently infected cells are critically needed to achieve a functional cure for HIV.
Identifying cellular subsets that preferentially harbor proviral DNA may facilitate the specific targeting of latent reservoirs. Resting memory CD4+ T cells are a well-characterized cellular reservoir, with numerous data suggesting the enrichment of proviral DNA within central, transitional, effector, and stem cell memory cells (Buzon, 2014; Chomont et al., 2009; Soriano-Sarabia et al., 2014); however, even among these memory subpopulations, there is a diversity of functional CD4+ T cell subsets, characterized by their distinct signature cytokines and immunological properties. Additionally, these subsets of memory CD4+ T cells are highly heterogeneous in their expression of surface markers, thus necessitating the identification of additional markers that more strictly define latently infected cells.
Recently, Banga et al. demonstrated that CD4+ T cells expressing programmed cell death protein-1 (PD-1) in lymph nodes (LN), which are largely composed of follicular helper T cells (Tfh), constitute an important source of persistent replication-competent virus in ART-treated, aviremic individuals (Banga et al., 2016). In that study, the contribution of PD-1+ CD4+ T cells to the persistent reservoir progressively decreased with increased length of ART; this finding suggests that other cell subsets, apart from PD-1+ Tfh cells, may contribute to the magnitude of the pool of latently infected cells. In addition to PD-1, other co-inhibitory receptors (Co-IRs) could maintain CD4+ T cells in a resting state (Kassu et al., 2010; Wherry, 2011). Virus-specific CD4+ T cells upregulate multiple Co-IRs, including PD-1, cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), and T cell Ig domain and mucin domain 3 (TIM-3), in the setting of HIV and SIV infection (D'souza et al., 2007; Jones et al., 2008; Kassu et al., 2010; Kaufmann et al., 2007). Consistent with this model, Fromentin et al. showed that CD4+ T cells co-expressing three Co-IRs (PD-1, TIGIT, and LAG-3) from the blood of ART-suppressed, HIV-infected individuals are enriched in proviral DNA when compared to subsets that included a single Co-IR (Fromentin et al., 2016).
Using ART-treated, SIV-infected rhesus macaques (RMs), we identified CTLA-4+PD-1− memory CD4+ T cells as a previously unrecognized component of the SIV reservoir. CTLA-4+PD-1− memory CD4+ T cells, a subset comprised predominantly of regulatory T cells (Tregs), are enriched in SIV DNA in multiple tissue compartments and contain robust amounts of replication-competent and infectious virus. In contrast to PD-1+ Tfh, SIV-enriched CTLA-4+PD-1− Treg cells localize outside the B-cell follicle of the LN; predict the size of the persistent viral reservoir during ART; and increase their contribution to the viral DNA pool with prolonged ART-mediated viral suppression. Finally, as in SIV-infected RMs, HIV-DNA is harbored by CTLA-4+PD-1− T cells outside the B-cell follicle of the LN in ART-treated, HIV infected patients. As such, CTLA-4 should be considered as an additional target when designing immunotherapies aimed at purging the viral reservoir.
Results
Expression of CTLA-4 defines a unique subset of virally enriched CD4+ T cells during ART in multiple tissues of SIV-infected RMs
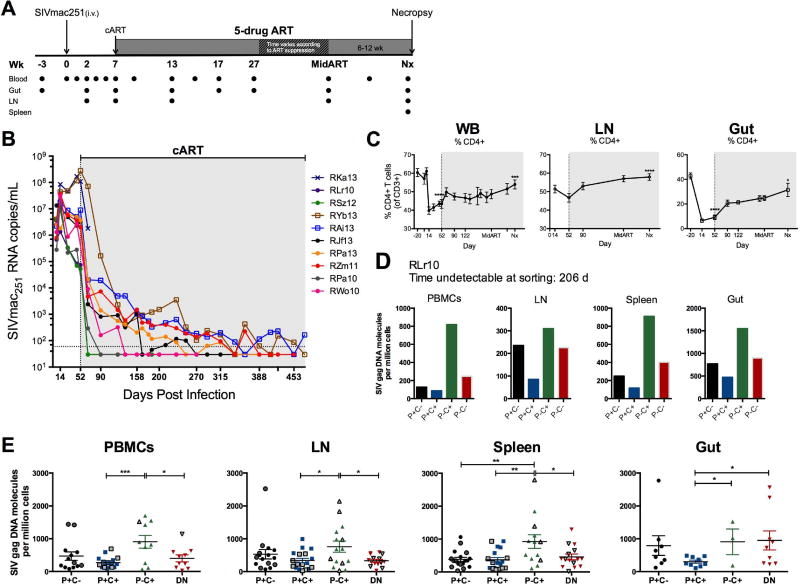
Ten RMs were infected intravenously with SIVmac251 (Figure 1A) and, at 52 days post infection, treated with ART (PMPA, FTC, raltegravir, and ritonavir-boosted darunavir; Table S1) for up to 14 months. RKa13 experienced rapid disease progression and was euthanized ten days into ART. Overall, the combined ART regimen was effective in suppressing plasma viremia (> 99.94% reduction from pre-ART, Figure S1A), with 7 of 9 RMs demonstrating durable virus suppression by 290 days of ART with a single viral blip in only one of these animals. The two RMs with the highest pre-ART viral loads similarly had undetectable viremia after 288 and 317 days of ART, though both showed two viral blips (< 500 copies/mL). At necropsy, 8 of 9 RMs had undetectable viral loads for an average of 125±76 days, thus ensuring that measurements of cell-associated SIV DNA were performed in the absence of measurable, ongoing viral replication in plasma (Figure 1B). Consistent with HIV-infected humans (Autran et al., 1997; Kaufmann et al., 2000), ART successfully elevated both CD4+ T cell frequencies (Figure 1C) and absolute counts (data not shown) in the blood and tissues of the SIV-infected RMs, although to frequencies that were significantly lower compared to pre-infection (Figure 1C). The efficacy of ART in suppressing viral replication was also evident in tissues, with a 61.6% reduction in the amount of SIV DNA found in LN as compared to pre-ART (Figure S1B, S1C).
Figure 1. CTLA-4+PD-1− memory CD4+ T cells harbor higher amounts of SIV DNA following ART-mediated, viral load suppression.
(A)Study design. Ten RMs were infected i.v. with 1000 TCID50 SIVmac251 (day 0), and at 7 weeks post-infection, initiated ART (PMPA, FTC, raltegravir, and ritonavir-boosted darunavir). All animals were maintained on ART regimen until plasma viremia was undetectable for at least 3 months. Peripheral blood (WB), rectal biopsy (Gut), and lymph node (LN) biopsies were collected at the indicated time points and multiple organs were harvested at elective necropsy. Sorting of memory CD4+ T cells by Co-IR expression was performed at two time points during ART: first, at Mid ART (approximately 1 month following undetectable viremia); and second, at necropsy. (B) Plasma viral loads are shown for the 10 individual RMs, quantified using the standard qRT-PCR assay (limit of detection, LOD, of 60 SIV RNA copies/mL of plasma represented by the horizontal dotted line). Undetectable measurements are plotted as one-half of the LOD (30 copies/mL). (C) Frequencies (of live CD3+ T cells) of CD4+ T cells were longitudinally measured in WB, LN, and gut biopsies. The gray shaded area represents time on ART; Nx represents the measured values from animal necropsy. Repeated-measures analyses were performed using a means model (SAS Mixed Procedure, version 9.4) to determine statistical significance, with indicated tests of significance representing comparison to pre-SIV infection (WB, Gut) or pre-ART initiation (LN). (D) Representative SIV DNA quantities in the PBMCs, LN, spleen, and gut tissues for an individual RM (RLr10) after 206 days of viral load suppression (n=9). (E) Cell-associated SIVGAG DNA was quantified from CTLA-4 and PD-1-sorted subsets of 9 ART-treated, SIV-infected RMs at least three months following the first undetectable viral load (163 ± 42 d). Data from subsets with less than 10,000 sorted cells were excluded when undetectable. Sample averages are indicated by the horizontal bar on each graph (± SEM), and t-tests were used to compare amounts of DNA between subsets (P, PD-1; C, CTLA-4). *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001. Please also see figure S2.
T cell homeostatic proliferation, which has been shown to be a major mechanism involved in HIV persistence, can induce the expression of several Co-IRs such as PD-1, CTLA-4, and others (Chomont et al., 2009; Hatano et al., 2013). To define the contribution of memory T cells expressing these Co-IRs to the pool of latently infected cells as well as to identify their anatomic localization, we measured cell-associated SIVGAG DNA in CD95+ memory CD4+ T cells sorted based on their expression of Co-IRs (Figure S2A) from the blood, LN, spleen, and gut after a minimum of 3 months (and up to 7 months) following their first undetectable viral load. Although CTLA-4 functions as an inhibitory receptor through its engagement on the outer cell surface with its ligands CD80 and CD86, it is located primarily in endosomal vesicles from where it cycles continuously to and from the cell surface; For this reason, intracellular CTLA-4 expression is routinely used to measure accumulation and expression of CTLA-4 molecules (Linsley et al., 1996; Read et al., 2000; Teft et al., 2006). Since cell permeabilization was incompatible with several analyses, we performed enhanced surface staining (ESS; see Experimental Procedures) to visualize surface CTLA-4 prior to sorting based on its expression. ESS captures rapidly upregulated surface CTLA-4 expression and reflects the amounts of intracellular CTLA-4 prior to stimulation, as previously shown (Kaufmann et al., 2007); we confirmed these observations by comparing ex vivo surface, ex vivo intracellular (ICS) and ESS staining (Figure S3). Of note, this short stimulation does not affect the kinetics and frequencies of cells expressing PD-1 (Figure S4). We found a consistent enrichment of SIV DNA in CTLA-4+PD-1− memory CD4+ T cells in the ART-treated, SIV-infected RMs at necropsy, with the average frequency of infection among the different tissues ranging from 760 to 923 copies of SIV per million CTLA-4+PD-1− cells (Figure 1D, E). In fact, CTLA-4+PD-1− memory CD4+ T cells harbored significantly more SIV DNA when compared to both PD-1+CTLA-4+ and PD-1−CTLA-4− cells in the blood, LN, and spleen (p<0.05 for all comparisons), and compared to PD-1+CTLA-4+ cells in the gut (p=0.0198) following an extended period (average of 125±76 days) of undetectable viremia (Figure 1E). The increased viral content was specific for SIV-DNA, since CTLA-4+PD-1− T cells harbored comparable (in the spleen) or lower (in LN and gut) amounts of SIV-RNA than CTLA-4− cells (Figure S5). Amounts of SIV DNA did not differ between TIM-3+ and TIM-3− populations within each CTLA-4 and PD-1-expressing subset (data not shown), confirming previous findings (Fromentin et al., 2016). These results demonstrate that CTLA-4+PD-1− cells constitute a subset of memory CD4+ T cells enriched in viral DNA that persist during ART-mediated viral suppression in several lymphoid tissues.
CTLA-4+PD-1− CD4+ T cells include a high frequency of regulatory T cells
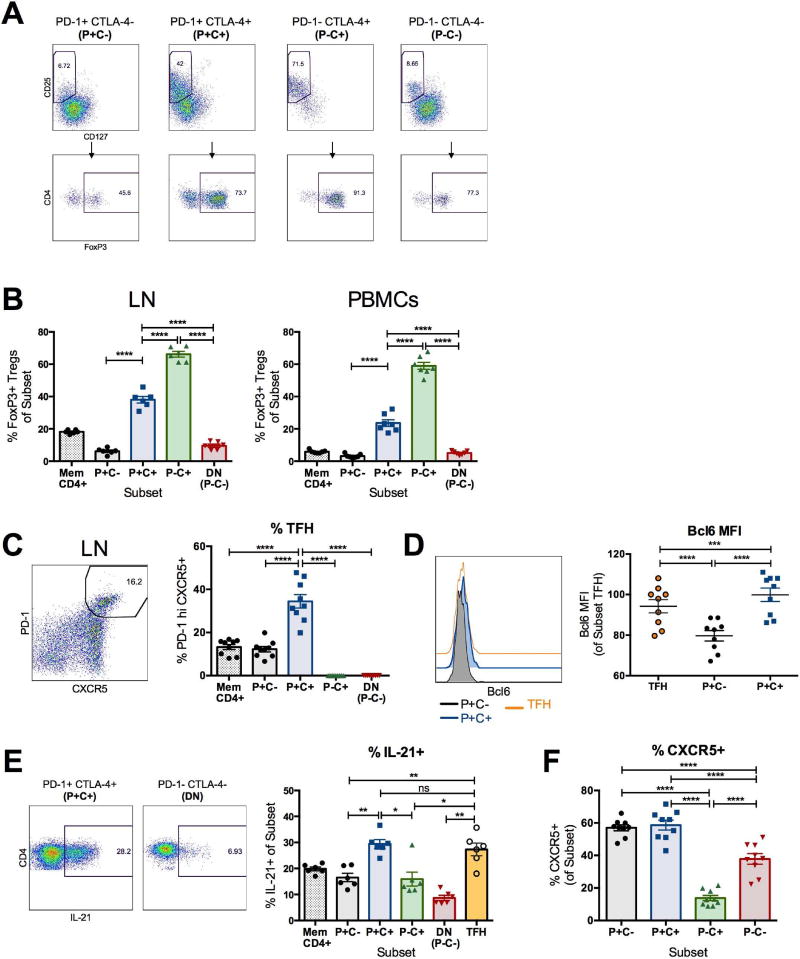
CTLA-4 is one of the markers that defines regulatory T cells (Treg) (Read et al., 2000; Wing et al., 2008); hence, we investigated in ex vivo conditions the overlap between the above defined CTLA-4+PD-1− T cells and CTLA-4+ Treg cells (Figure 2A). We found that, in ART-treated, SIV-infected RMs, CTLA-4+PD-1− memory CD4+ T cells were significantly enriched in CD25+CD127Io cells which also expressed Forkhead Box P3 (FoxP3), the master transcription factor of Treg, when compared to bulk memory CD4+ T cells as well as PD-1+CTLA-4−, PD-1+CTLA-4+, and PD-1−CTLA-4− memory CD4+ T cell subsets (Figure 2B) (Anderson et al., 2008; Seddiki et al., 2006). In fact, Treg cells were the dominant functional subset, as they comprised 66.2±1.8% and 59.0±2.2% of CTLA-4+PD-1− memory CD4+ T cells in the LN and PBMCs, respectively. We then monitored the expression of the chromatin modifier SATB1, an epigenetic modifier whose expression inhibits FoxP3 transcriptional activity (Beyer et al., 2011). We found that CTLA-4+ CD4+ T cells included significantly higher frequencies of FoxP3+SATB1− cells as compared to CTLA-4− CD4+ T cells (p<0.0001), consistent with the FoxP3 locus being transcriptionally active (Figure S6A) and confirmed by the expression of CTLA-4 and CD25 (two FoxP3 targets) in these cells. Even when gating on CD25+CD127− cells, we found a significant enrichment in CTLA-4+PD-1− CD4+ T cells as compared to the PD-1+CTLA-4− and PD-1−CTLA-4− subsets, for several markers previously shown to be expressed on subsets of Tregs and to promote Treg cell stability and suppressive activity. These markers include latency associated peptide (Chen et al., 2008); the transcription factor Helios (Baine et al., 2013; Ross et al., 2014); the plasma membrane protein CD39 (Borsellino et al., 2007; Gu et al., 2017); and the chemokine receptor CCR6 (Kitamura et al., 2010; Xu et al., 2010) (Figure S6A,B; p<0.001 for all markers). Thus, CTLA-4+PD-1− memory CD4+ T cells include functional Treg cells and represent a subset that contributes to SIV persistence during ART in multiple anatomic sites.
Figure 2. CTLA-4+PD-1− CD4+ T cells overlap with regulatory T cells.
(A) Representative flow plots of regulatory T cells (Treg), defined as CD25+CD127Io and FoxP3+, within CTLA-4 and PD-1-expressing memory CD4+ T cell subsets are shown from the LN of an ART-treated, SIV-infected RM at necropsy. The results shown are from unstimulated lymphocytes, where CTLA-4 expression was measured intracellularly; similar results were obtained following PMA and lonomycin stimulation (not shown). (B) The frequencies of FoxP3+ Treg cells within memory CD4+ T cell subsets are shown from the LN and PBMCs of 6 ART-treated, SIV-infected RMs. (C) Representative flow plot demonstrating the gating for follicular helper cells (Tfh) within the LN of an ART-treated, SIV-infected RM, defined as PD-1hi CXCR5+ cells. Frequencies of Tfh cells were quantified in the LN of 9 ART-treated, SIV-infected RMs in the absence of stimulation at day 90 post-SIV infection (38–41 days on ART). (D) The representative histograms and aggregate mean fluorescence intensity (MFI) of Bcl6 within LN PD-1-expressing memory CD4+ T cell subsets ex vivo are shown at day 90 post-SIV infection (n=9). (E) Representative flow plots of IL-21 production by PD-1+CTLA-4+ and PD-1−CTLA-4− (DN) memory CD4+ T cells (n=6). Production of IL-21 by each CTLA-4 and PD-1-expressing subset was measured from the LN of 6 ART-treated, SIV-infected RMs following a 3 hour PMA and lonomycin stimulation. (F) Frequencies of CXCR5+ cells were quantified between each CTLA-4 and PD-1-expressing subset in the LN of 9 ART-treated, SIV-infected RMs at day 90 post-SIV infection (38–41 days post-ART initiation). The results shown are from unstimulated lymphocytes. Averaged data are presented as the mean ± SEM, and ANOVAs using Tukey’s adjustment for multiple comparisons were used to compare differences between subsets. *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001. Please also see figure S6.
Recent reports have demonstrated that Tfh cells harbor heightened amounts of HIV-1 DNA in the LN of ART-suppressed individuals (Banga et al., 2016), and that productive persistent SIV infection in elite controller RMs, but not typical progressors, is markedly restricted to the Tfh subset in the B cell follicle (Fukazawa et al., 2015). In line with these reports, we identified LN Tfh cells phenotypically as CXCR5+PD-1hi memory CD4+ T cells and functionally by their production of IL-21 (Figure 2C, 2E) (Chtanova et al., 2004; Haynes et al., 2007; Petrovas et al., 2012). PD-1+CTLA-4+ memory CD4+ T cells included significantly higher frequencies of Tfh cells and contained higher amounts of Bcl6, the master transcriptional regulator of Tfh differentiation, than PD-1+CTLA-4− cells in the LN (Figure 2C, 2D; p<0.0001). Consistent with this result, PD-1+CTLA-4+ memory CD4+ T cells also produced higher amounts of IL-21 when compared to the other subsets, and to quantities comparable to those produced by CXCR5+PD-1hi Tfh in the LN (Figure 2E). Notably, CTLA-4+PD-1− memory CD4+ T cells included significantly fewer CXCR5+ cells when compared to the other subsets (including CTLA-4−PD-1−), suggesting that this Treg cell-enriched subset was less susceptible to trafficking into germinal centers (Figure 2F). Altogether, these results highlight the fact that two functional T helper subsets, i.e. Treg cells and Tfh cells, are targeted by SIV to establish viral persistence.
CTLA-4+PD-T− T cells enriched in virus are localized outside the lymph node follicle and contribute to viral persistence during ART-mediated viral suppression
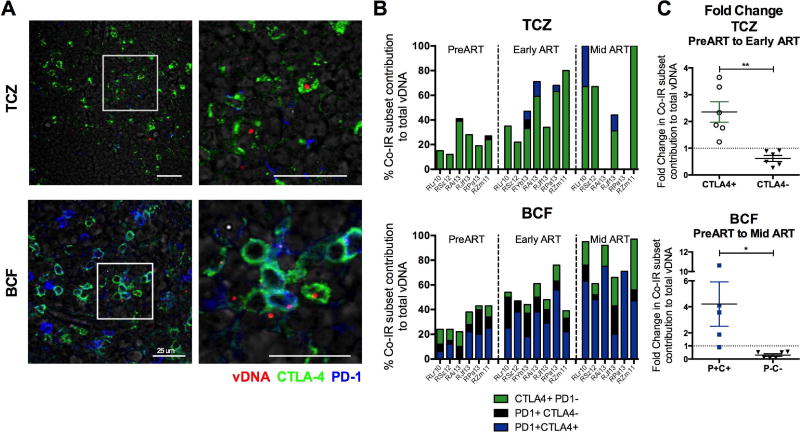
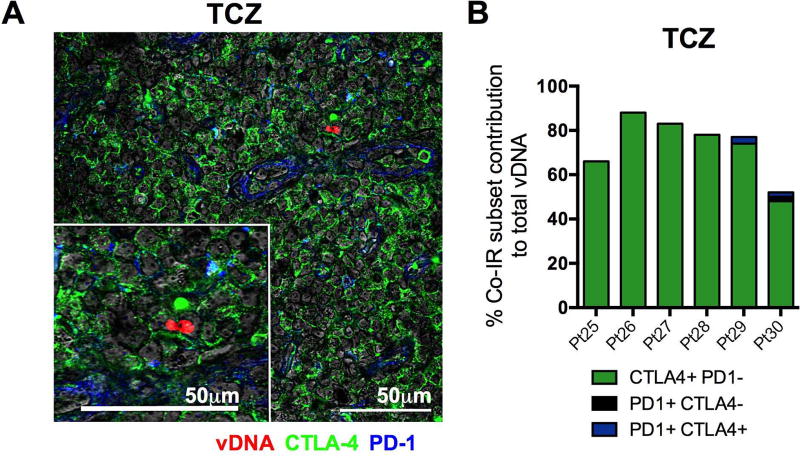
We used an in situ hybridization assay that combines the detection of viral DNA (vDNA) in fixed tissue sections with phenotypic analysis using immunofluorescence and confocal microscopy (Deleage et al., 2016). This approach allowed us to determine the cellular and anatomic localization of SIV DNA in the LN of RMs (Figure 3A). Prior to ART initiation, vDNA in the T cell zone (TCZ) was found predominantly in CTLA-4+PD-1− cells (Figure 3B), although the majority of vDNA+ cells in both the TCZ and the B cell follicle lacked expression of Co-IRs at this time point. Notably, following ART initiation and suppression of viral load, CTLA-4+ subsets exclusively harbored vDNA in the TCZ (Figure 3A, 3B). The localization of CTLA-4+PD-1− vDNA+ cells outside the B cell follicle was also confirmed in the spleen (data not shown). When compared to pre-ART, the proportion of vDNA+ cells expressing CTLA-4 in the LN TCZ had already increased 2.4-fold early after ART initiation (average of 1.2 months on ART; Figure 3C); this frequency further increased to an average of 3.0-fold with longer exposure to ART (average of 9.2 months on ART; Mid ART; data not shown). In contrast, the contribution of CTLA-4-vDNA+ cells was reduced with exposure to ART by 38.2% (early ART) and 32.4% (Mid ART) in the TCZ. Thus, among SIV-infected memory CD4+ T cells in the TCZ, those expressing CTLA-4 persist significantly longer during ART than CTLA-4− subsets (p=0.0014). Consistent with published reports, the majority of vDNA+ cells in the B cell follicle at early and Mid ART were PD-1+CTLA-4+, the subset which predominantly includes Tfh cells, corresponding to a 2.5-fold (p=0.0267; data not shown) and 4.2-fold (p=0.0163; Figure 3C) increase when compared to their contribution pre-ART to vDNA+ cells, respectively (Banga et al., 2016). Altogether, our results demonstrate the critical and increasing contribution of the CTLA-4+PD-1− T cell subset to viral persistence in several lymphoid tissues.
Figure 3. SIV DNA is found outside the B cell follicle in CTLA-4+PD-1− cells.
(A) Immunofluorescence staining for CTLA-4 (green) and PD-1 (blue) combined with DNAscope hybridization for SIV vDNA (red) in the LN T cell zone (TCZ) and B cell follicle of a representative RM (RJf13). Representative images are shown for each anatomic location within the LN following ART (RJf13: D315 p.i.) in the absence of detectable plasma viremia (n=7). Scale bars = 25 µm. The white box highlights the expanded view on the right. (B) Quantitative image analysis for the TCZ and B cell follicle of the LN demonstrating the fraction of SIV vDNA+ cells that express CTLA-4 and/or PD-1 in SIV-infected RMs (n=7) at 3 separate time points: PreART, Early ART (90 days post-SIV infection, 38–41 days post-ART initiation), and Mid ART (average time since last undetectable plasma viral load being approximately 80 ± 40 d). (C) Fold change in the fraction of vDNA+ cells in the TCZ that express CTLA-4, or lack its expression, was calculated using the DNAscope quantification of 6 RMs between the Early ART and PreART time points. Fold change is also shown for the fraction of vDNA+ cells in the B cell follicle that express PD-1 and CTLA-4 (P+C+) and those that lack their expression (P−C−) using the DNAscope quantification of 6 RMs between the Mid ART and PreART time points. Data are presented as mean ± SEM, with a dashed line corresponding to FC=1 (no change).
CTLA-4+PD-1− memory CD4+ T cells harbor replication competent and infectious SIV
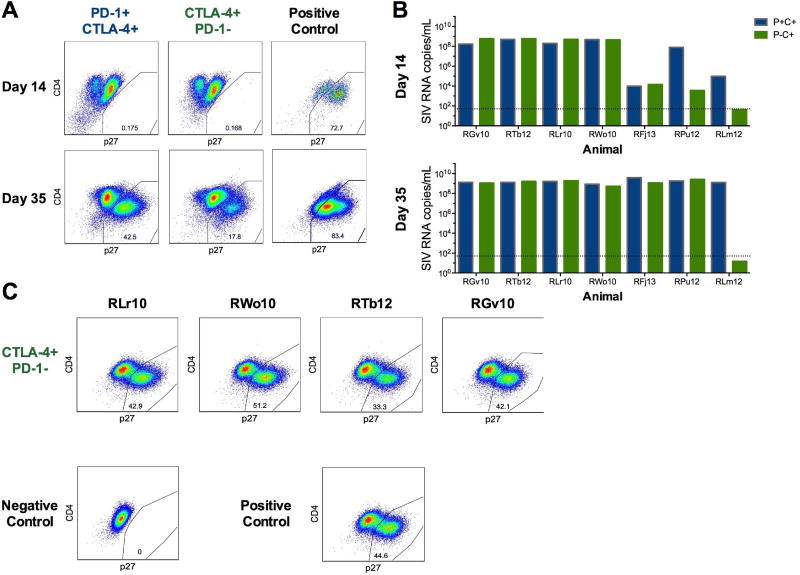
To confirm that CTLA-4+PD-1− memory CD4+ T cells contribute to the persistent SIV reservoir, we measured the production of replication competent virus in these cells using a modified viral outgrowth assay previously described for SIV infection (Micci et al., 2015). Memory CD4+ T cell subsets were sorted based on their single or dual expression of CTLA-4 and PD-1 from the LN of seven ART-treated, SIV-infected RMs after an average of 4.7 months of undetectable plasma viremia; cells were subsequently co-cultured with CEMx174 cells to measure productive viral infection. Frequencies of CEMx174 cells expressing p27 and the quantity of SIVGAG RNA released in the supernatant were measured at day 14 and 35 of co-culture. We compared the amounts of SIV replication competent virus from CTLA-4+PD-1− and PD-1+CTLA-4+ memory CD4+ T cells, as these two subsets contributed the most to viral persistence (Figure 3). Replication competent virus, as identified by the presence of p27+ cells, was detected in CTLA-4+PD-1− memory CD4+ T cells from 6 of 7 RMs, and from 7 RMs in PD-1+CTLA-4+ cells by day 35 of co-culture (representative p27 staining in Figure 4A). A 6-log exponential increase in SIV RNA was found in the co-culture supernatants of both subsets by day 35 (Figure 4B); with the amount of SIV RNA ranging between 108 and 109 in both CTLA-4+PD-1+ and CTLA-4+PD-1− cells, we could not identify a statistically significant difference between these two subsets. Finally, supernatants collected from the VOA assay from 4 RMs were spiked on CEMx174 cells to determine the presence of infectious virus. In all 4 RMs, supernatants collected from the CTLA-4+PD-1− cells (as well as from the PD-1+CTLA-4+, data not shown) infected a high frequency of CEMx174 cells already at day 7 of culture (Figure 4C).
Figure 4. CTLA-4+PD-1− memory CD4+ T cells harbor replication competent SIV.
(A) Memory CD4+ T cells from ART-treated, SIV-infected RM LNs were stimulated for three hours using PMA and lonomycin (see Methods), sorted based on CTLA-4 and PD-1 expression (see Figure S2A), and co-cultured with SIV-permissive CEMx174 cells for 35 days (n=7). Representative flow plots of intracellular SIV GAG p27 are shown for days 14 and 35 of co-culture within PD-1+CTLA-4+ and CTLA-4+PD-1− subsets from an individual ART-treated, SIV-infected RM (RJf13) (n=7). The right column shows SIV GAG p27 staining for a positive control (CEMx174 cells infected directly with SIVmac). (B) SIVGAG RNA was quantified at days 14 and 35 from the supernatant of the individual co-cultures. Each set of columns represents the SIV RNA data from the PD-1+CTLA-4+ (blue) and CTLA-4+PD-1− (green) memory CD4+ T cell subsets from an individual RM (n=7). Due to cell number limitations, only PD-1+CTLA-4+ memory CD4+ T cells had replicate wells; the SIV RNA data for this subset represents the average of the three replicate wells for animals RFj13, RPu12, and RLm12. (C) Uninfected CEMx174 cells were spiked using the clarified co-culture supernatant at day 35 (see Methods). Representative flow cytometry gates of intracellular SIV GAG p27 in CEMx174 cells at day 7 corresponding to the co-cultured CTLA-4+PD-1− subsets for four RMs (RLr10, RWo10, RTb12, and RGv10) as well as the positive and negative control.
Altogether, these results demonstrate that CTLA-4+PD-1− memory CD4+ T cells harbor replication competent and infectious SIV in ART-treated, SIV-infected RMs.
Seeding of virus in CTLA-4+PD-1− memory CD4+ T cells predicts viral persistence
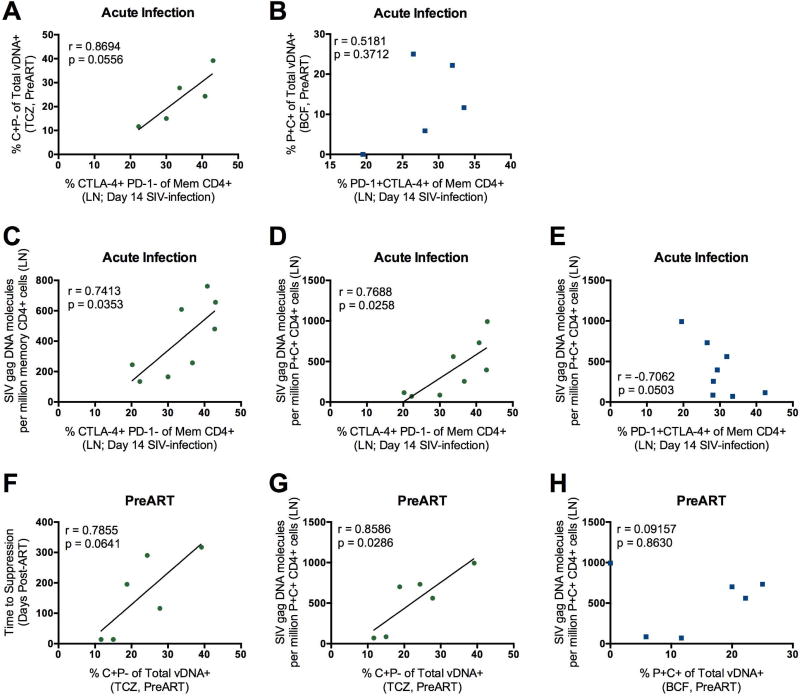
To investigate the relationship between the number of CTLA-4+PD-1− memory CD4+ T cells and viral persistence, we monitored the frequencies of these cells in the LN during SIV infection. We found a positive correlation (Figure 5A) between the frequencies (by flow cytometry) of CTLA-4+PD-1− memory CD4+ T cells in the LN at peak SIV infection (day 14 p.i) and the fraction (by DNA scope) of SIV-DNA+ CTLA-4+PD-1− cells in the LN TCZ pre-ART (day 52 p.i.); this association was specific for CTLA-4+PD-1− memory CD4+ T cells as frequencies of PD-1+CTLA-4+ Tfh-like cells did not show this correlation (Figure 5B). We next tested the hypothesis that the frequencies of CTLA-4+PD-1− memory CD4+ T cells in the LN at acute infection could predict SIV persistence at later time points on ART. We found that the frequencies of CTLA-4+PD-1− memory CD4+ T cells at peak SIV infection correlated with the amount of SIV DNA (by ddPCR) within memory CD4+ (Figure 5C) and memory PD-1+CTLA-4+ CD4+ T cells (Figure 5D) in the LN at necropsy, thus up to 14 months after ART initiation. These same correlations were not found with PD-1+CTLA-4+ memory CD4+ T cells (Figure 5E and data not shown).
Figure 5. Frequencies CTLA-4+PD-1− memory CD4+ T cells and their viral seeding predicts viral persistence during ART.
Correlations between the frequencies of CTLA-4+PD-1−(A) or PD-1+CTLA-4+ (B) memory CD4+ T cells in the LN at the peak of SIV infection (day 14) and the fraction of viral DNA (vDNA) found within C+P− in the TCZ (A) and P+C+ in the B cell follicle (B) prior to ART initiation by DNAscope. Only 5 SIV-infected RMs had measurements to relate in this analysis; all are shown. The frequencies of CTLA-4+PD-1− memory CD4+ T cells in the LN at the peak of SIV infection (day 14) are significantly associated with the frequencies of bulk memory (C; p=0.0353) and memory PD-1+CTLA-4+ (D; p=0.0258) CD4+ T cells harboring SIVG/AG DNA in the LN at necropsy (average time of ART-mediated viral suppression: 125 ± 76 days; n=8). (E) Correlation between the frequency of PD-1+CTLA-4+ memory CD4+ T cells in the LN at day 14 and the frequencies of memory PD-1+CTLA-4+ CD4+ T cells harboring SIV DNA in the LN at necropsy. The fraction of SIV DNA+ cells expressing CTLA-4+PD-1− in the TCZ preART initiation was positively correlated with time to viral load suppression (F; p=0.0641) and quantity of SIV DNA in PD-1+CTLA-4+ memory CD4+ T cells in the LN (G; p=0.0286) at necropsy. (H) The correlation is also shown between the fraction of SIV DNA+cells expressing PD-1+CTLA-4+ in the B cell follicle PreART initiation and the frequencies of memory PD-1+CTLA-4+ CD4+ T cells harboring SIV DNA in the LN at necropsy. All statistical analyses were performed using Pearson product-moment correlation tests.
We also found a positive association between the fraction of SIV-DNA+ CTLA-4+PD-1− cells in the TCZ pre-ART and the time to viral load suppression following ART (Figure 5F), further confirming the relationship between viral seeding in CTLA-4+PD-1− cells and viral persistence. Additionally, there was a significant positive correlation between the frequencies of CTLA-4+PD-1− SIV-DNA+ cells in the TCZ pre-ART and the amount of SIV-DNA in PD-1+CTLA-4+ memory CD4+ T cells in the LN at necropsy (Figure 5G). In contrast, the seeding of virus in PD-1+CTLA-4+ cells in the B cell follicle prior to ART initiation did not predict SIV DNA content in this LN subset at necropsy (Figure 5H). Altogether, these data are consistent with CTLA-4+PD-1− Treg cell-like memory CD4+ T cells uniquely contributing to the establishment and maintenance of the viral reservoir through their early seeding and long-term persistence.
CTLA-4+PD-1− CD4+ T cells express high quantities of cell survival molecules
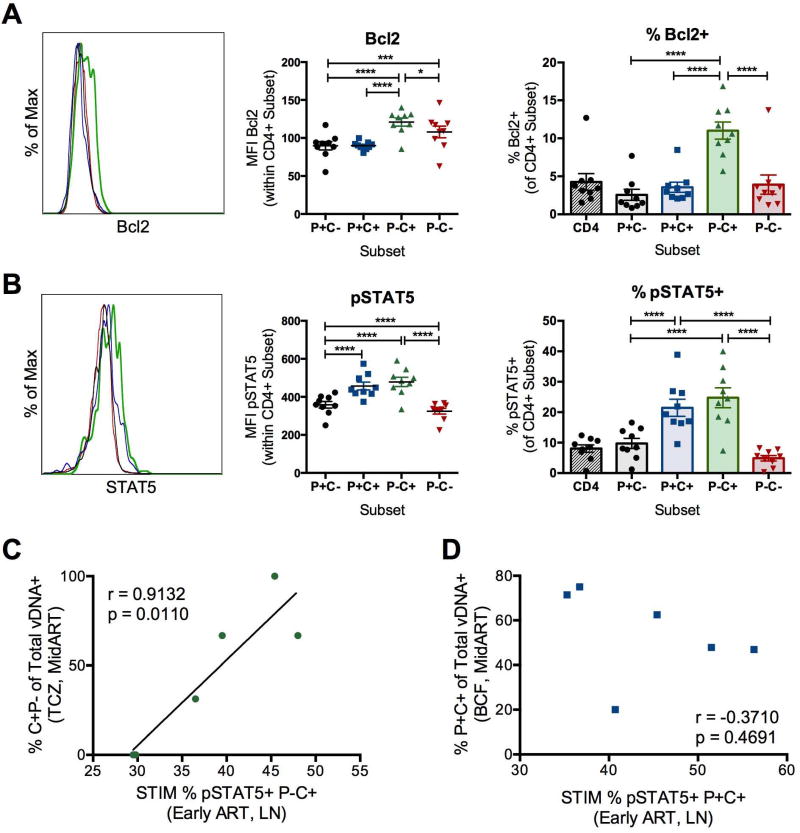
Memory CD4+ T cells harbor latent HIV infection due to their long half-lives and potential for homeostatic proliferation (Chomont et al., 2009). We found that CTLA-4+PD-1− memory CD4+ T cells contained significantly higher amounts of the anti-apoptotic molecule Bcl2 ex vivo, both by mean fluorescence intensity (MFI) and frequency of Bcl2+ cells, when compared to the other subsets (Figure 6A). Consistent with the increased concentration of Bcl2, CTLA-4+PD-1− CD4+ T cells also contained significantly higher concentrations of the phosphorylated form of STAT5 (pSTAT5) as compared to CTLA-4− cells (Figure 6B). Activation of pSTAT5 is triggered by the cytokines IL-2, IL-7, and IL-15, all known to contribute to HIV persistence, and results in increased survival of CD4+ T central memory cells (Riou et al., 2007). Altogether, these results indicate that the increased persistence of virally infected CTLA-4+PD-1− CD4+ T cells during ART is associated with their elevated expression of functional markers of cell survival and homeostatic proliferation. Consistent with a mechanistic link between increased capacity for survival and viral persistence, we found that the frequencies of CTLA-4+PD-1− CD4+ T cells expressing pSTAT5 early after ART were significantly correlated to the fraction of CTLA-4+PD-1− vDNA+ cells in the TCZ at a later ART time point (Mid ART; Figure 6C); however, this relationship was not seen in PD-1+CTLA-4+ CD4+ T cells in the LN, suggesting that distinct mechanisms control the persistence of these two subsets (Figure 6D). These data support a mechanism where CTLA-4+PD-1− memory CD4+ T cells are seeded early during SIV infection (Figure 5A), and, due to their increased capacity for cell survival (Figure 6A, 6B), persist during ART to increasingly contribute to the latent viral reservoir.
Figure 6. CTLA-4+PD-1− CD4+ T cells demonstrate increased potential for survival and homeostatic proliferation.
The representative mean fluorescence intensity (MFI) of Bcl2 (A) and pSTAT5 (B) expression is shown for CTLA-4 and PD-1-expressing memory CD4+ T cells from the LN of an individual ART-treated, SIV-infected RM. Aggregate MFI data are then shown for each molecule for 9 ART-treated, SIV-infected RMs. Frequencies of Bcl2+ (A) and pSTAT5+ (B) cells were quantified in the LN of 9 ART-treated, SIV-infected RMs at day 90 post-SIV infection (38–41 days post-ART initiation). Of note, Bcl2 and pSTAT5 expression were determined ex vivo, with CTLA-4 expression measured intracellularly. Averaged data are presented as the mean ± SEM, and ANOVAs using Tukey’s adjustment for multiple comparisons were used to compare differences between subsets. *, p<0.05; ***, p<0.001; ****, p<0.0001. Correlations are shown between the frequencies of CTLA-4+PD-1− (C) or PD-1+CTLA-4+ (D) memory CD4+ T cells in the LN during early ART expressing pSTAT5 and the fraction of viral DNA (vDNA) found within C+P− in the TCZ (C) and P+C+ in the B cell follicle (D) at MidART by DNAscope (MidART average time since last undetectable plasma viremia being approximately 80 ± 40 d). Statistical analyses were performed using Pearson product-moment correlation tests.
CTLA-4+PD-1− T cells enriched in virus localize outside the lymph node follicle and contribute to viral persistence in ART-treated, HIV-infected individuals
CTLA-4+PD-1− CD4+ T cells harbored the large majority of vDNA outside the B cell follicle in the LN of ART-suppressed, SIV-infected RMs (Figure 3). To increase the clinical relevance of our findings, we investigated the presence of cells harboring HIV-DNA outside the B cell follicle and in CTLA-4+PD-1− CD4+ T cells in ART-treated, HIV-infected subjects. To this aim, we combined DNAscope staining for the detection of HIV-DNA with phenotypic staining for CTLA-4 and/or PD-1 expression (Deleage et al., 2016) in LN tissues collected from six HIV-infected individuals on ART for an average of 37.8 months (range of 20.8–52.3 months), and with undetectable viremia for at least 15.6 months (Table S2). Consistent with the data generated in RMs, we identified HIV-DNA+ CD4+ T cells in the TCZ and found that the vast majority of these cells (72.9±5.8%) express CTLA-4+PD-1− (Figure 7A, 7B). CTLA-4−PD-1− CD4+ T cells contributed the remaining 26.2±5.4% of HIV-DNA+ cells. Notably, and despite being abundant in the B cell follicle, we could not identify PD-1+ CD4+ T cells harboring HIV-DNA in the TCZ (Figure 7B). Thus, CTLA-4+PD-1− memory CD4+ T cells harboring HIV DNA persist outside the B cell follicle of the LN and contribute to the long-term latent viral reservoir in HIV-infected individuals on ART.
Figure 7. HIV DNA is also found outside the B-cell follicle in CTLA-4+PD-1− cells in ART-suppressed HIV-infected individuals.
(A) Representative image (Pt. 27) of immunofluorescence staining for CTLA-4 (green) and PD-1 (blue) combined with DNAscope hybridization for HIV vDNA (red) in the LN T cell zone (TCZ); Magnification X600. (B) Quantitative image analysis for the TCZ of the LN demonstrating the fraction of HIV vDNA+ cells that express CTLA-4 and/or PD-1 in ART-treated HIV-infected individuals (n=6) with undetectable plasma viral loads (average duration of aviremia is 31.4 months; range of 15.650.5 months).
Discussion
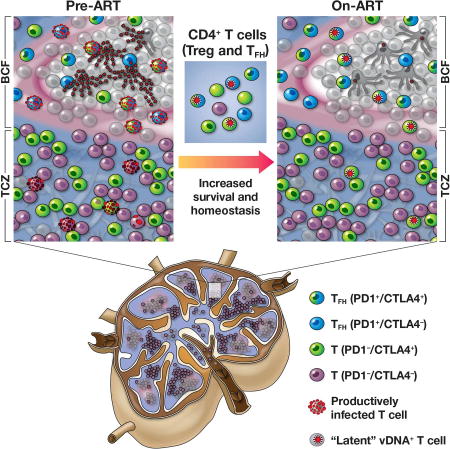
Reducing the persistent HIV and SIV reservoir remains an essential milestone for the development of a functional cure for HIV-1 infection; this goal has been markedly hindered by a poor identification of the CD4+ T cell subsets that harbor persistent replication competent virus, as well as the anatomic location of these cells. Our findings highlight the previously underappreciated contribution of CTLA-4+PD-1− memory CD4+ T cells to the persistent, replication competent, and infectious viral reservoir. Indeed, the frequency of CTLA-4+PD-1− memory CD4+ T cells harboring SIV DNA (but not SIV RNA) was higher than in other memory CD4+ T cell populations in PBMCs, LN, spleen, and gut, following prolonged (up to 14 months) ART treatment. Treg cells constitute a major proportion of these cells, and they persist outside of the B cell follicle, as in situ analysis of SIV-DNA+ cells in the LN demonstrated the persistence of virus exclusively in CTLA-4+ cells in the TCZ following ART-mediated viral suppression. Importantly, the seeding of virus in this cell subset uniquely predicted the size of the reservoir under ART, and the contribution of CTLA-4+PD-1− memory CD4+ T cells to the SIV reservoir significantly increased with prolonged ART-mediated viral suppression. Moreover, in addition to Tfh cells in the B cell follicle, we confirmed the persistence of HIV-DNA in CTLA-4+PD-1− CD4+ T cells in the TCZ of LN obtained from HIV-infected subjects on ART. These findings support the presence of latent HIV genomes in a previously unrecognized subset of cells characterized by their expression of CTLA-4, within an anatomic site distinct from Tfh cells. Thus, CTLA-4+PD-1− CD4+ Treg cells represent a bonafide SIV and HIV reservoir that should be targeted for a functional HIV-1 cure.
This unique enrichment of SIV-DNA in CTLA-4+PD-1− CD4+ cells may result from a number of different mechanisms. First, it may indicate their preferential infectivity, as demonstrated by recent work showing that the frequency of memory CD4+ T cells carrying integrated HIV-DNA in ART-naïve, HIV-infected subjects is up to 18-fold higher in CTLA-4+ as compared to CTLA-4− cells (El-Far et al., 2015). Previous studies have also demonstrated high quantities of CTLA-4 on HIV-specific CD4+ T cells (Kaufmann et al., 2007; Paris et al., 2015), which are preferential targets for HIV (Douek et al., 2002). The enrichment of CTLA-4+PD-1− memory CD4+ T cells may also arise from their increased expression of molecules that will confer long term survival (Riou et al., 2007). Indeed, we found higher concentrations of Bcl2 and pSTAT5 within CTLA-4+PD-1− memory CD4+ T cells over time, which support an increased persistence of this subset that preferentially harbors viral DNA. Others have also shown that Treg cells can resist apoptosis (Pandiyan et al., 2004). Notably, Bach2, which has been previously shown to promote the differentiation and survival of FoxP3+ Treg cells (Kim et al., 2014), was found to be a frequent integration site for HIV (Maldarelli et al., 2014), which is consistent with our model of increased infectivity and persistence of Treg cells.
Previous studies have demonstrated that Tfh cells are highly enriched in HIV-DNA in both viremic and aviremic individuals (Banga et al., 2016; Lindqvist et al., 2012; Perreau et al., 2013). Consistent with these studies, we found that PD-1+CTLA-4+ cells, the subset that contributes most to Tfh cells, were the dominant contributors to the viral DNA pool in the B cell follicle in the LN during ART; however, we also found persistent virus outside of the follicle in the TCZ in CTLA-4+PD-1− cells, which phenotypically overlap with Treg. Herein we reveal their high viral DNA content in multiple tissues, their localization outside the B cell follicle in lymphoid tissues, and their growing contribution to viral persistence during ART. These findings extend on prior studies that showed higher amounts of HIV-1 DNA in Treg cells from the blood of ART-treated, HIV-infected individuals (Jiao et al., 2015; Tran et al., 2008). The fact that these cells remain PD-1- (a molecule upregulated in activated T cells and in T cells undergoing homeostatic proliferation) strongly supports their nature as Treg cells. Furthermore, CTLA-4+PD-1− cells showed the lowest frequency of cells expressing the proliferation marker Ki-67 (not shown) when compared to other subsets of Co-IR+ CD4+ T cells. Treg cell migration and expansion in tissue sites of HIV replication, including LN, has been suggested to inhibit antiviral immune responses during HIV infection (Estes et al., 2006; Kinter et al., 2007; Moreno-Fernandez et al., 2012). Thus, it is possible that CTLA-4+PD-1− memory CD4+ T cells are seeded with virus during acute SIV infection while functioning as Treg cells to dampen T cell activation. In support of this mechanism, we found that, prior to ART, the amount of CD4+ T cell proliferation negatively correlated with the frequency of CTLA-4+PD-1− memory CD4+ T cells in the LN, suggesting that this CTLA-4 subset expands to reduce immune activation in the absence of ART. Moreover, HIV infection leads to changes in several cytokines that can influence the differentiation and survival of Treg cells, including IL-2, TGF-β, TNF-α, and IL-1 (Havlir et al., 2001; Orsilles et al., 2006). Thus, changes in the number and susceptibility to infection of Treg cells could also result from the increased inflammatory cytokine environment associated with HIV replication. Treg cells, in turn, also produce cytokines that will impede the development of T cell effector function and homeostasis (Fahlen et al., 2005; Ito et al., 2008), thus expanding the mechanisms by which Treg cells could contribute to viral persistence.
Low number of cells (analysis involved relatively small subsets of memory CD4+ T cells) hampered our capacity to perform a quantitative VOA; we therefore elected to perform a qualitative VOA. Although we cannot compare the frequency of cells harboring replication competent virus between CTLA-4+PD-1− and CTLA-4+PD-1+ T cells, our ability to rescue replication competent virus from CTLA-4+PD-1− CD4+ T cells confirm that these cells harbor measurable amounts of replication competent virus. Additionally, our VOA results were limited to ART-suppressed RMs, whom are limited in their time on ART as compared to long-term ART-suppressed, HIV-infected humans. Future studies will be critical for quantifying the amount of replication-competent virus in CTLA-4+PD-1− Treg cells from long-term virally-suppressed individuals in relation to other Co-IR-expressing subsets.
Co-inhibitory blockades of CTLA-4 have produced meaningful antitumor effects in clinical cancer trials (Sledzinska et al., 2015). Moreover, CTLA-4 blockade has also been explored as a monotherapy in viremic, SIV-infected RMs, where its use was associated with increased viral reactivation (Cecchinato et al., 2008; Hryniewicz et al., 2006). Importantly, a recent case report demonstrates the ability of ipilimumab (anti-CTLA-4 antibody) to increase cell-associated HIV-1 RNA in an ART-suppressed, HIV-infected melanoma patient (Wightman et al., 2015). This case study, by revealing the impact of targeting extracellular CTLA-4, supports the capacity of immunotherapeutic blockades to bind this transiently expressed molecule. These studies also support the capacity of CTLA-4 blockade to target latently infected cells and facilitate reactivation of virus, two actions necessary for the elimination of the latent reservoir. Further studies are needed in non-human primates to determine if CTLA-4 blockade, alone or combined with PD-1 blockade, is effective and well-tolerated in reducing the viral reservoir.
In summary, our study identifies CTLA-4+PD-1− memory CD4+ T cells as a unique subset harboring replication competent SIV DNA that persists outside the B-cell follicle despite ART-mediated viral suppression. By demonstrating the phenotypic overlap between CTLA-4+PD-1− memory CD4+ T cells and Treg cells, and the presence of these cells in ART-treated, HIV-infected subjects, our results highlight that HIV is able to establish and maintain viral persistence through the specific targeting of another CD4+ T cell subset critical for the regulation of the adaptive immune system, in addition to PD-1+ Tfh cells.
Experimental Procedures
Animals, SIV-infection, and antiretroviral therapy
Ten female Indian rhesus macaques (RMs; Macaca mulatta), all housed at the Yerkes National Primate Research Center, Atlanta, GA, were included in this study. All RMs were HLA*B07− and HLA*B17− with the following being HLA*A01+: RSz12, RLr10, and RZm11. Prior to study assignment, all RMs are screen for SIV, Cercopithecine herpesvirus 1 (B virus), Simian-T-lymphotropic virus (STLV), respiratory syncytial virus (RSV), tuberculosis (TB), and dewormed. After experimental infection, animals are isolation housed, to avoid SIV superinfection, in metal wire cages at an ambient temperature of 72±e°F. RMs are fed a diet consisting of jumbo biscuits supplemented with 15% protein; half an orange per day; and produce enrichment five times per week. All procedures are approved by the Emory University Institutional Animal Care and Use Committee (IACUC) and animal care facilities are accredited by the U.S. Department of Agriculture (USDA) and the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International.
All RMs were infected intravenously with 1000 TCID50 of SIVmac251 (Figure 1A). Approximately 7 weeks post-infection, all animals initiated an antiretroviral therapy (ART) regimen consisting of tenofovir (PMPA; 20–25 mg/kg/d, s.c.), emtricitabine (FTC; 30–50 mg/kg/d, s.c.), raltegravir (100–150 mg/bid, oral), darunavir (400–700 mg/bid, oral), and ritonavir (50 mg/bid, oral), which was maintained daily for the duration of the study (Table S1). To enhance control of viral load, this regimen was intensified with the addition of maraviroc (100–150 mg/bid, oral) 21–24 weeks after ART initiation in 7 RMs. Animals remained on ART until plasma viremia was undetectable (limit of detection: 60 copies viral RNA/mL) for at least 3 months (5.0±1.4 months from first undetectable plasma viral load). Following persistent viral suppression, animals underwent elective necropsy, with the exception of one RM who exhibited rapid disease progression and had to be sacrificed following ten days of ART (RKa13; Table S1). Longitudinal analysis accounts for all RMs, while cell-associated DNA and RNA quantitation includes only the 9 animals which reached study completion.
Study Approval
All animal experimentations were conducted following guidelines established by the Animal Welfare Act and the NIH for housing and care of laboratory animals and performed in accordance with Institutional regulations after review and approval by the Institutional Animal Care and Usage Committees (IACUC; Permit number: 2002089) at the Yerkes National Primate Research Center (YNPRC). Anesthesia was administered prior to performing any procedure, and proper steps were taken to minimize the suffering of the animals in this study.
Sample collection and processing
Blood, lymph node (LN), and rectal biopsies (Gut) were performed longitudinally and at necropsy. Blood samples were used for a complete blood count and routine chemical analysis, and plasma was separated by centrifugation within 1 hr of phlebotomy. Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood by density gradient centrifugation. To obtain rectal biopsy punches, an anoscope was placed a short distance into the rectum for the collection of up to 20 pinch biopsies with biopsy forceps. Gut-derived lymphocytes were digested with 1 mg/mL collagenase for 2 hours at 37°C, and then passed through a 70- µm cell strainer to remove residual tissue fragments. Intestinal samples, which included segments of the rectum, colon, ileum, and jejunum, were collected from necropsied animals into RPMI 1640 medium supplemented with 10%FBS, 100 IU/mL penicillin, and 100 µg/mL streptomycin. Intestinal samples were cut into small pieces, and digested with 1 mg/mL collagenase by shaking for 2 hr at 37°C. To enrich for lymphocytes, filtered samples were layered over a discontinuous Percoll density gradient (GE Life Sciences), centrifuged and washed in PBS prior to staining and/or sorting. For LN biopsies, the skin over the axillary or inguinal region was clipped and surgically prepped. An incision was then made in the skin over the LN, which was exposed by blunt dissection and excised over clamps. LNs were then homogenized and passed through a 70-um cell strainer to isolate lymphocytes. All samples were processed, fixed (1% paraformaldehyde), and analyzed within 24 hours of collection.
Determination of viral load RNA
Quantitative real-time RT-PCR was performed to determine SIV plasma viral load as previously described (Amara et al., 2001).
Flow cytometric analysis
Fourteen-parameter flow cytometric analysis was performed on peripheral blood, LN, and gut-derived cells according to standard procedures using a panel of monoclonal antibodies that we and others have shown to be cross-reactive with RMs (Micci et al., 2015). The following antibodies were used at predetermined optimal concentrations: anti-CD3-APC-Cy7 (clone SP34–2), anti-Ki-67-Alexa700 (clone B56), anti-CD95-PE-Cy5 (clone DX2), anti-CCR7-PE-Cy7 (clone 3D12), anti-CTLA-4-BV421 (clone BNI3), all from BD Biosciences; anti-CD4-BV605 (clone OKT4), anti-PD-1-PE-Cy7 (clone EH12.2H7), anti-CD95-BV605 (clone DX2), anti-CD25-BV711 (clone BC96), and anti-FoxP3-APC (clone 150D) from Biolegend; anti-CD8-Qdot705 (clone 3B5) and Aqua Live/Dead amine dye-AmCyan from Invitrogen; anti-CD28-ECD (clone CD28.2) from Beckman Coulter; anti-CD127-PECy5 (clone eBioRDR5) and anti-CXCR5-PerCp-Cy5.5 (clone MU5UBEE) from eBioscience; and anti-TIM-3-PE (clone 344823) from R&D. To determine the ex vivo phenotype of CTLA-4+ CD4+ T cells, CTLA-4 staining has been performed intracellularly in unstimulated T cells. To detect the expression of CTLA-4 on the cell surface with improved sensitivity, as required for sorting and viral reservoir measurements, mononuclear cells were stimulated for 3 hours at 37°C with phor bol myristate acetate (PMA; 80 ng/mL) and ionomycin (500 ng/mL), and CTLA-4 antibody was added to the stimulation media at the start of stimulation. To detect the expression of FoxP3 intracellularly, mononuclear cells were fixed and permeabilized with FoxP3 Fix/Perm solution (Tonbo), and subsequently stained intracellularly for FoxP3. Flow cytometric acquisition was performed on at least 100,000 CD3+ T cells on an LSRII cytometer driven by the FACS DiVa software, or at least 10,000 CD3+ T cells for gut-derived cells. The data acquired were analyzed using FlowJo software (version 9.8.5; TreeStar).
Flow cytometry cell sorting
Mononuclear cells isolated from blood, LN, spleen, and pooled gut mucosal tissues (including right and left colon, ileum, jejunum, and rectum) were stimulated for 3 hours with PMA and lonomycin, as described above, during which anti-CTLA-4 antibody was added with the stimulation media, and subsequently stained with anti-CD3, anti-CD4, anti-CD8, anti-CD28, anti-CD95, anti-PD-1, anti-CTLA-4 and anti-TIM-3. Memory CD4+ T cells were then sorted based on their expression of PD-1, CTLA-4, and TIM-3 using a FACS Ariall (BD Biosciences). With the exception of gut mucosal tissues, memory CD4+ T cells sorted at necropsy were first separated into PD-1 positive and negative populations. These subsets were then sorted into four subsets based on their expression of CTLA-4 and TIM-3 (Figure S2). Memory CD4+ T cells sorted during the study, and from gut mucosal tissues at necropsy, were sorted by their PD-1 and CTLA-4 expression alone, due to a lower quantity of starting material. Sorted CD4+ T cell subsets were on average >96% pure as determined by post-sorting flow cytometry analysis.
Quantitation of cell-associated SIV DNA and RNA
Cellular DNA and RNA were extracted from at least 10,000 sorted memory CD4+ T cells lysed in RLT Plus buffer (Qiagen) and isolated using the AllPrep DNA/RNA Mini Kit (Qiagen) per the manufacturer’s instructions. cDNA was synthesized from extracted RNA, and quantification of SIVmac GAG DNA and cDNA was performed on samples using the Q×100™ Droplet Digital™ PCR system (Bio-rad). Total SIV DNA and RNA were quantified for these samples using SIVmac GAG primers and probes, and normalized to the GAPDH gene (see Key Resource Table). Data was analyzed using the Quantasoft analysis software 1.3.2.0 (Bio-rad).
Intracellular cytokine staining
The production of IL-21 by Co-IR-expressing subsets was determined by examining the frequency of memory CD4+ T cells that produced IL-21 following a 3 hour in vitro stimulation with PMA and ionomycin (see above). PBMC- and LN-derived cells, isolated as described above, were resuspended to 3×106 cells/mL in complete RPMI 1640 media containing PMA, ionomycin, Brefeldin A, and BD Golgistop. Cells were then incubated at 37°C for 3 hours. Stimulation was stopped by washing cells with PBS. Following a 30 minute surface stain, cells were fixed and permeabilized prior to staining intracellularly with anti-IL-21-PE (clone 3A3-N2.1; BD Biosciences) for 45 minutes at room temperature. Following staining, cells were washed and fixed in PBS containing 1% paraformaldehyde (PFA), and acquired on a BD LSRII cytometer.
SIV DNA in situ hybridization and immunofluorescent detection for confocal phenotypic analysis
Viral DNA detection (DNAscope) was performed and validated as described previously (Deleage et al., 2016). Briefly, following HIER (Pretreat 2 step; ACD) and proteinase digestion (Pretreat 3 step, ACD), the slides were incubated overnight (between 18–21 hours) at 40°C with HIV clade B sense probe or SIVmac239 sense probe. Amplification steps were performed according to the ACD protocol with the exception that all wash steps used a 0.5X wash buffer. Amplification reagents from the RNAscope 2.5 HD Brown Detection Kits were used for Tyramide Signal Amplification (TSA™) Plus Cy3.5 immunofluorescence detection. We combined DNAscope detection (TSA™ Plus Cy3.5) with immunofluorescence incubating overnight at RT: rabbit polyclonal anti-PD-1 (1:200; Cat#HPA035981; SIGMA) and goat polyclonal anti-hCTLA-4 (1:200; Cat#AF-386-PB; R&D Systems). Slides were washed, incubated with secondary donkey anti-goat IgG-Alexa 488 and anti-rabbit IgG-Alexa 647 (all from Molecular Probes/ThermoFisher Scientific) for 1 hr at room temperature, and washed 2 times for 5 min in TBS + tween (0.05% v/v). To decrease auto-fluorescence, the tissues were incubated with Sudan Black solution [0.1% in 80% ethanol (ENG Scientific, Inc.) in 1× TBS]; for 20–30 min at room temperature and then washed, counterstained with DAPI (RTU; ACD) for 10 min, washed, and cover slipped with #1.5 GOLD SEAL® cover glass (EMS) using Prolong® Gold reagent (Invitrogen).
To quantify the number of vDNA+ cells and proportion that were PD-1+CTLA-4−, PD-1+CTLA-4+, PD-1−CTLA-4+, and PD-1−CTLA-4−, high magnification confocal images were collected from regions of interests and manually counted (depending of the tissue size, 5 to 10 B cell follicles and 5 to 10 within the T cell zone) using an Olympus FV10i confocal microscope using a 60x phase contrast oil-immersion objective (NA 1.35) imaging using sequential mode to separately capture the fluorescence from the different fluorochromes at an image resolution of 1024×1024 pixels. Absolute numbers of viral DNA+ (vDNA+) cells varied by time assessed: Pre-ART, 37–240 vDNA+ cells; Early ART, 23–236 vDNA+ cells; and MidART, 15–67 vDNA+ cells.
Determination of replication competent and infectious virus in memory CD4+ T cells during ART
Cryopreserved LN cells from ART-treated, SIV-infected RMs were sorted to high purity into memory CD4+ T cell subsets based on their expression of CTLA-4 and PD-1. Due to limited cryopreserved LN cell numbers, 7 RM LN were sorted: 3 SIVmac251-infected RM followed in this study (RJf13, 108 days with undetectable plasma viremia; RLr10, 206 days undetectable; RWo10, 153 days undetectable), and 4 SIVmac239-infected RMs from another study (RPu12, 106 days undetectable; RLm12, 165 days undetectable; RGv10, 136 days undetectable; RTb12, 106 days undetectable). Purified CTLA-4 and PD-1-expressing memory CD4+ T cell subsets were stimulated for 6 hours with anti-CD3 and anti-CD28, and then co-cultured at a 1:1 ratio with the CEMx174 cell line, a fusion product between the human CEM T cell line with human B cell line 721.174 (NIH AIDS Research and Reference Reagent Program), with starting cell numbers ranging from 1.5 to 2.5 × 105 memory CD4+ T cells per well. Co-cultures were maintained in complete RPMI 1640 media supplemented with 10%FBS, 100 1U/mL penicillin, 100 ug/mL streptomycin and IL-2 (100 U/mL) at 37°C under 5% CO2. Cells were split and fed with fresh media containing IL-2 weekly (days 14, 21, 28, and 35), and harvested at days 14 and 35. The presence of replication competent virus was detected by positive SIV GAG p27 expression within the CD4+ cell population, using flow cytometry (Fukazawa et al., 2015; Micci et al., 2015), and by quantifying SIVGAG RNA in the supernatant using qRT-PCR (see Experimental Procedures above). 200 µ L of clarified co-culture supernatant at day 35 from the wells containing the CTLA-4+PD-1− was used to spike 5 × 105 uninfected CEMx174 cells. Only uninfected (negative control) or in vitro SIV-infected (positive control) CEMx174 cells were also used. Cultures were maintained for 7 days under the same conditions as the co-culture (see above) and the presence of replication competent virus was detected by positive SIV GAG p27 expression within the CD4+ T cell population, using flow cytometry.
Assessment of STAT5 phosphorylation
The amount of phosphorylated STAT5 within CTLA-4 and PD-1-expressing subsets was measured by phospho-flow cytometry in ART-treated, SIV-infected RM LN samples, both ex vivo and following a three hour in vitro stimulation with PMA and ionomycin (see above). A minimum of 106 cells were stained with anti-CD3, anti-CD4, anti-CD8, anti-CD20 (clone 2H7; BD Biosciences), anti-PD-1, anti-CTLA-4 and a viability stain (Live/Dead Fixable Aqua, Invitrogen) for 20 minutes at 4°C. Samples were fixed for 10 mi nutes with 4% PFA at 37°C, permeabilized for 20 minutes with Perm Buffer III (BD Biosciences) on ice, and then stained intracellularly with anti-pSTAT5 (clone 47/Stat5(pY694); BD Biosciences) for 30 minutes at room temperature. Following staining, cells were washed and fixed in 1% PFA, and acquired on a BD LSR Fortessa.
Patient population
Six HIV-seropositive subjects on suppressive ART for an average of 37.8 months (Table S2) were consented for a study approved by the Emory University Institutional Review Board. Individuals were receiving various combination antiretroviral regimens containing nucleoside reverse transcriptase inhibitors, a protease inhibitor, and/or integrase inhibitor. All subjects had undetectable viremia for at least 15.6 months (limit of detection: 50 copies/mL) before undergoing elective lymph node (LN) biopsies. LN sections were formalin-fixed and paraffin-embedded prior to performing DNAscope analysis (see above).
Statistical Analysis
Repeated-measures analyses for each memory CD4+ T cell subset outcome were performed with a means model via the SAS MIXED Procedure (version 9.4; SAS Institute, Cary, NC), providing separate estimates of the means by time on study. The statistical model included one predictor (time on study) with up to 17 categorical levels, and was fit separately for each anatomic location. A compound-symmetric variance-covariance form in repeated measurements was assumed for each outcome and robust estimates of the standard errors of parameters were used to perform statistical tests and construct 95% confidence intervals. The model-based means are unbiased with unbalanced and missing data, so long as the missing data was non-informative (missing at random, MAR). Three specific statistical tests were done within the framework of the mixed effects linear model for each anatomic location. All statistical tests were two-sided and unadjusted for multiple comparisons. A p-value ≤0.05 was considered statistically significant for each of the specific statistical comparisons. Results from these studies also focused on the magnitude of the differences for each outcome as reflected by confidence intervals, consistency of findings, and biological significance. Similar repeated-measures analyses were performed for log10 SIV DNA over time by anatomic location. Comparisons of cell-associated DNA were calculated using t-tests and Mann-Whitney U tests, respectively, as determined by sample distribution (normal or nonnormal). DNA measurements were excluded for samples in which <10,000 cells were sorted and values fell outside the assay limit of detection. Longitudinal comparisons of cell-associated DNA were calculated using paired t-tests. Pearson product-moment correlation coefficients were used to estimate linear associations for SIV DNA data. Data showing continuous outcomes are represented as mean ± SEM. Analyses of SIV DNA were conducted using GraphPad Prism 6.0.
Supplementary Material
Highlights.
CTLA-4+PD-1- memory CD4+ T-cells are enriched in SIV DNA across multiple tissues
Persistently infected CTLA-4+PD-1- T-cells localize outside the follicle on ART
CTLA-4+PD-1- T-cells, which share Treg features, harbor replication-competent virus
Seeding of CTLA-4+PD-1- memory CD4+ T-cells predicts viral persistence during ART
Acknowledgments
We thank Stephanie Ehnert, Christopher Souder, and all the animal care and veterinary staff at the YNPRC, as well as Barbara Cervasi at the Emory Flow Cytometry Core. We also thank David Favre and Shari Gordon at GSK, and David Margolis at UNC Chapel Hill, for helpful discussion; Mathias Lichterfeld and Sean Harrington of the Ragon Institute for technical assistance; Romas Geleziunas (Gilead), Guenter Kraus (Johnson & Johnson, and Daria Hazuda (Merck) for supplying the antiretroviral drugs. The SIVmac251 used to infect the RMs was kindly provided by Chris Miller. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID: 174xCEM Cells from Dr. Peter Cresswell. Dr. Paiardini is supported by the NIAID, NIH under award numbers 104278, 116379, 116171, and 110334, and by amfAR under award numbers 109354-59-RGRL. This work was also supported by ORIP/OD P51OD011132 (to the YNPRC), P30AI50409 (to the Emory CFAR), and in part with federal funds from the National Cancer Institute (NIH Contract HHSN261200800001E). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have declared that no conflict of interest exists.
Author contributions: C.S.M, G.S., R.P.S. and M.P. conceived the study; C.S.M., J.H., S.P., E.S.R., and S.J. performed sample collection and processing; C.D. and J.D.E. performed DNAScope analyses; C.S.M., L.K.C., S.P.R., C.B., J.H., and L.M. performed cell survival phenotyping and viral outgrowth assays; K.E. performed statistical analysis; V.M. provided human specimens; C.S.M., J.D.E., R.P.S., and M.P. wrote the manuscript with inputs from all authors.
References
- Amara RR, Villinger F, Altman JD, Lydy SL, O’Neil SP, Staprans SI, Montefiori DC, Xu Y, Herndon JG, Wyatt LS, et al. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science. 2001;292:69–74. doi: 10.1126/science.1058915. [DOI] [PubMed] [Google Scholar]
- Anderson A, Martens CL, Hendrix R, Stempora LL, Miller WP, Hamby K, Russell M, Strobert E, Blazar BR, Pearson TC, et al. Expanded nonhuman primate tregs exhibit a unique gene expression signature and potently downregulate alloimmune responses. American Journal of Transplantation. 2008;8:2252–2264. doi: 10.1111/j.1600-6143.2008.02376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autran B, Carcelain G, Li TS, Blanc C, Mathez D, Tubiana R, Katlama C, Debre P, Leibowitch J. Positive effects of combined antiretroviral therapy on CD4+ T cell homeostasis and function in advanced HIV disease. Science. 1997;277:112–116. doi: 10.1126/science.277.5322.112. [DOI] [PubMed] [Google Scholar]
- Baine I, Basu S, Ames R, Sellers RS, Macian F. Helios induces epigenetic silencing of IL2 gene expression in regulatory T cells. J Immunol. 2013;190:1008–1016. doi: 10.4049/jimmunol.1200792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, Corpataux JM, de Leval L, Pantaleo G, Perreau M. PD-1 and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med. 2016 doi: 10.1038/nm.4113. [DOI] [PubMed] [Google Scholar]
- Beyer M, Thabet Y, Muller RU, Sadlon T, Classen S, Lahl K, Basu S, Zhou X, Bailey-Bucktrout SL, Krebs W, et al. Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat Immunol. 2011;12:898–907. doi: 10.1038/ni.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskaran K, Hamouda O, Sannes M, Boufassa F, Johnson AM, Lambert PC, Porter K, Collaboration C. Changes in the risk of death after HIV seroconversion compared with mortality in the general population. Jama-J Am Med Assoc. 2008;300:51–59. doi: 10.1001/jama.300.1.51. [DOI] [PubMed] [Google Scholar]
- Borsellino G, Kleinewietfeld M, Di Mitri D, Sternjak A, Diamantini A, Giometto R, Hopner S, Centonze D, Bernardi G, Dell’Acqua ML, et al. Expression of ectonucleotidase CD39 by Foxp3+ Treg cells: hydrolysis of extracellular ATP and immune suppression. Blood. 2007;110:1225–1232. doi: 10.1182/blood-2006-12-064527. [DOI] [PubMed] [Google Scholar]
- Buzon MJ, Sun H, Li C, Shaw A, Seiss K, Ouyang Z, Martin-Gayo E, Leng J, Henrich TJ, Li JZ, Pereyra F, Zurakowski R, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M. HIV-1 Persistence in CD4+ T cells with stem cell-like properties. Nat Med. 2014:1–4. doi: 10.1038/nm.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchinato V, Tryniszewska E, Ma ZM, Vaccari M, Boasso A, Tsai WP, Petrovas C, Fuchs D, Heraud JM, Venzon D, et al. Immune activation driven by CTLA-4 blockade augments viral replication at mucosal sites in simian immunodeficiency virus infection. J Immunol. 2008;180:5439–5447. doi: 10.4049/jimmunol.180.8.5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ML, Yan BS, Bando Y, Kuchroo VK, Weiner HL. Latency-associated peptide identifies a novel CD4+CD25+ regulatory T cell subset with TGFbeta-mediated function and enhanced suppression of experimental autoimmune encephalomyelitis. J Immunol. 2008;180:7327–7337. doi: 10.4049/jimmunol.180.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893–U892. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS, Mackay CR. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J Immunol. 2004;173:68–78. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- Chun TW, Carruth L, Finzi D, Shen XF, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 Infection. Nature. 1997a;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- Chun TW, Davey RT, Jr, Engel D, Lane HC, Fauci AS. Re-emergence of HIV after stopping therapy. Nature. 1999;401:874–875. doi: 10.1038/44755. [DOI] [PubMed] [Google Scholar]
- Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proceedings of the National Academy of Sciences of the United States of America. 1997b;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DA. Life and death in the cART era. Lancet. 2008;372:266–267. doi: 10.1016/S0140-6736(08)61086-7. [DOI] [PubMed] [Google Scholar]
- D’souza M, Fontenot A, Mack D, Lozupone C, Dillon S, Meditz A, Wilson C, Connick E, Palmer B. Programmed death 1 expression on HIV-specific CD4(+) T cells is driven by viral replication and associated with T cell dysfunction. J Immunol. 2007;179:1979–1987. doi: 10.4049/jimmunol.179.3.1979. [DOI] [PubMed] [Google Scholar]
- Davey RT, Bhat N, Yoder C, Chun TW, Metcalf JA, Dewar R, Natarajan V, Lempicki RA, Adelsberger JW, Millers KD, et al. HIV-1 and T cell dynamics after interruption of highly active antiretroviral therapy (HAART) in patients with a history of sustained viral suppression. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:15109–15114. doi: 10.1073/pnas.96.26.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleage C, Wietgrefe SW, Del Prete G, Morcock DR, Hao XP, Piatak M, Jr, Bess J, Anderson JL, Perkey KE, Reilly C, et al. Defining HIV and SIV Reservoirs in Lymphoid Tissues. Pathogens & immunity. 2016;1:68–106. doi: 10.20411/pai.v1i1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, et al. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- El-Far M, Ancuta P, Routy JP, Zhang Y, Bakeman W, Bordi R, DaFonseca S, Said EA, Gosselin A, Tep TS, et al. Nef promotes evasion of human immunodeficiency virus type 1-infected cells from the CTLA-4-mediated inhibition of T-cell activation. The Journal of general virology. 2015;96:1463–1477. doi: 10.1099/vir.0.000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes JD, Li Q, Reynolds MR, Wietgrefe S, Duan L, Schacker T, Picker LJ, Watkins DI, Lifson JD, Reilly C, et al. Premature induction of an immunosuppressive regulatory T cell response during acute simian immunodeficiency virus infection. The Journal of infectious diseases. 2006;193:703–712. doi: 10.1086/500368. [DOI] [PubMed] [Google Scholar]
- Fahlen L, Read S, Gorelik L, Hurst SD, Coffman RL, Flavell RA, Powrie F. T cells that cannot respond to TGF-beta escape control by CD4(+)CD25(+) regulatory T cells. J Exp Med. 2005;201:737–746. doi: 10.1084/jem.20040685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, et al. Latent infection of CD4(+) T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- Fromentin R, Bakeman W, Lawani MB, Khoury G, Hartogensis W, DaFonseca S, Killian M, Epling L, Hoh R, Sinclair E, et al. CD4+ T Cells Expressing PD-1, TIGIT and LAG-3 Contribute to HIV Persistence during ART. PLoS pathogens. 2016;12:e1005761. doi: 10.1371/journal.ppat.1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa Y, Lum R, Okoye AA, Park H, Matsuda K, Bae JY, Hagen SI, Shoemaker R, Deleage C, Lucero C, et al. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med. 2015;21:132–139. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Ni X, Pan X, Lu H, Lu Y, Zhao J, Guo Zheng S, Hippen KL, Wang X, Lu L. Human CD39hi regulatory T cells present stronger stability and function under inflammatory conditions. Cell Mol Immunol. 2017;14:521–528. doi: 10.1038/cmi.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano H, Jain V, Hunt PW, Lee TH, Sinclair E, Do TD, Hoh R, Martin JN, McCune JM, Hecht F, et al. Cell-based measures of viral persistence are associated with immune activation and programmed cell death protein 1 (PD-1)-expressing CD4+ T cells. The Journal of infectious diseases. 2013;208:50–56. doi: 10.1093/infdis/jis630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havlir DV, Torriani FJ, Schrier RD, Huang JY, Lederman MM, Chervenak KA, Boom WH. Serum interleukin-6 (IL-6), IL-10, tumor necrosis factor (TNF) alpha, soluble type II TNF receptor, and transforming growth factor beta levels in human immunodeficiency virus type 1-infected individuals with Mycobacterium avium complex disease. Journal of clinical microbiology. 2001;39:298–303. doi: 10.1128/JCM.39.1.298-303.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes NM, Allen CD, Lesley R, Ansel KM, Killeen N, Cyster JG. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J Immunol. 2007;179:5099–5108. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- Hryniewicz A, Boasso A, Edghill-Smith Y, Vaccari M, Fuchs D, Venzon D, Nacsa J, Betts MR, Tsai WP, Heraud JM, et al. CTLA-4 blockade decreases TGF-beta, IDO, and viral RNA expression in tissues of SIVmac251-infected macaques. Blood. 2006;108:3834–3842. doi: 10.1182/blood-2006-04-010637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Hanabuchi S, Wang YH, Park WR, Arima K, Bover L, Qin FX, Gilliet M, Liu YJ. Two functional subsets of FOXP3+ regulatory T cells in human thymus and periphery. Immunity. 2008;28:870–880. doi: 10.1016/j.immuni.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao YM, Liu CE, Luo LJ, Zhu WJ, Zhang T, Zhang LG, Su LS, Li HJ, Wu H. CD4+CD25+CD127 regulatory cells play multiple roles in maintaining HIV-1 p24 production in patients on long-term treatment: HIV-1 p24-producing cells and suppression of anti-HIV immunity. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases. 2015;37:42–49. doi: 10.1016/j.ijid.2015.06.008. [DOI] [PubMed] [Google Scholar]
- Jones RB, Ndhlovu LC, Barbour JD, Sheth PM, Jha AR, Long BR, Wong JC, Satkunarajah M, Schweneker M, Chapman JM, et al. Tim-3 expression defines a novel population of dysfunctional T cells with highly elevated frequencies in progressive HIV-1 infection. J Exp Med. 2008;205:2763–2779. doi: 10.1084/jem.20081398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassu A, Marcus RA, D’Souza MB, Kelly-McKnight EA, Golden-Mason L, Akkina R, Fontenot AP, Wilson CC, Palmer BE. Regulation of Virus-Specific CD4(+) T Cell Function by Multiple Costimulatory Receptors during Chronic HIV Infection. J Immunol. 2010;185:3007–3018. doi: 10.4049/jimmunol.1000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann DE, Kavanagh DG, Pereyra F, Zaunders JJ, Mackey EW, Miura T, Palmer S, Brockman M, Rathod A, Piechocka-Trocha A, et al. Upregulation of CTLA-4 by HIV-specific CD4(+) T cells correlates with disease progression and defines a reversible immune dysfunction. Nat Immunol. 2007;8:1246–1254. doi: 10.1038/ni1515. [DOI] [PubMed] [Google Scholar]
- Kaufmann GR, Zaunders JJ, Cunningham P, Kelleher AD, Grey P, Smith D, Carr A, Cooper DA. Rapid restoration of CD4 T cell subsets in subjects receiving antiretroviral therapy during primary HIV-1 infection. Aids. 2000;14:2643–2651. doi: 10.1097/00002030-200012010-00003. [DOI] [PubMed] [Google Scholar]
- Kim EH, Gasper DJ, Lee SH, Plisch EH, Svaren J, Suresh M. Bach2 regulates homeostasis of Foxp3+ regulatory T cells and protects against fatal lung disease in mice. J Immunol. 2014;192:985–995. doi: 10.4049/jimmunol.1302378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinter A, McNally J, Riggin L, Jackson R, Roby G, Fauci AS. Suppression of HIV-specific T cell activity by lymph node CD25+ regulatory T cells from HIV-infected individuals. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3390–3395. doi: 10.1073/pnas.0611423104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K, Farber JM, Kelsall BL. CCR6 marks regulatory T cells as a colon-tropic, IL-10-producing phenotype. J Immunol. 2010;185:3295–3304. doi: 10.4049/jimmunol.1001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist M, van Lunzen J, Soghoian DZ, Kuhl BD, Ranasinghe S, Kranias G, Flanders MD, Cutler S, Yudanin N, Muller MI, et al. Expansion of HIV-specific T follicular helper cells in chronic HIV infection. The Journal of clinical investigation. 2012;122:3271–3280. doi: 10.1172/JCI64314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsley PS, Bradshaw J, Greene J, Peach R, Bennett KL, Mittler RS. Intracellular trafficking of CTLA-4 and focal localization towards sites of TCR engagement. Immunity. 1996;4:535–543. doi: 10.1016/s1074-7613(00)80480-x. [DOI] [PubMed] [Google Scholar]
- Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micci L, Ryan ES, Fromentin R, Bosinger SE, Harper JL, He T, Paganini S, Easley KA, Chahroudi A, Benne C, et al. Interleukin-21 combined with ART reduces inflammation and viral reservoir in SIV-infected macaques. The Journal of clinical investigation. 2015;125:4497–4513. doi: 10.1172/JCI81400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Fernandez ME, Presicce P, Chougnet CA. Homeostasis and function of regulatory T cells in HIV/SIV infection. Journal of virology. 2012;86:10262–10269. doi: 10.1128/JVI.00993-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsilles MA, Pieri E, Cooke P, Caula C. IL-2 and IL-10 serum levels in HIV-1- infected patients with or without active antiretroviral therapy. APMIS. 2006;114:55–60. doi: 10.1111/j.1600-0463.2006.apm_108.x. [DOI] [PubMed] [Google Scholar]
- Pandiyan P, Gartner D, Soezeri O, Radbruch A, Schulze-Osthoff K, Brunner-Weinzierl MC. CD152 (CTLA-4) determines the unequal resistance of Th1 and Th2 cells against activation-induced cell death by a mechanism requiring PI3 kinase function. J Exp Med. 2004;199:831–842. doi: 10.1084/jem.20031058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris RM, Petrovas C, Ferrando-Martinez S, Moysi E, Boswell KL, Archer E, Yamamoto T, Ambrozak D, Casazza JP, Haubrich R, et al. Selective Loss of Early Differentiated, Highly Functional PD1high CD4 T Cells with HIV Progression. PloS one. 2015;10:e0144767. doi: 10.1371/journal.pone.0144767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreau M, Savoye AL, De Crignis E, Corpataux JM, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med. 2013;210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovas C, Yamamoto T, Gerner MY, Boswell KL, Wloka K, Smith EC, Ambrozak DR, Sandler NG, Timmer KJ, Sun X, et al. CD4 T follicular helper cell dynamics during SIV infection. The Journal of clinical investigation. 2012;122:3281–3294. doi: 10.1172/JCI63039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read S, Malmstrom V, Powrie F. Cytotoxic T lymphocyte-associated antigen 4 plays an essential role in the function of CD25(+)CD4(+) regulatory cells that control intestinal inflammation. J Exp Med. 2000;192:295–302. doi: 10.1084/jem.192.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou C, Yassine-Diab B, Van grevenynghe J, Somogyi R, Greller LD, Gagnon D, Gimmig S, Wilkinson P, Shi Y, Cameron MJ, et al. Convergence of TCR and cytokine signaling leads to FOXO3a phosphorylation and drives the survival of CD4+ central memory T cells. J Exp Med. 2007;204:79–91. doi: 10.1084/jem.20061681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross EM, Bourges D, Hogan TV, Gleeson PA, van Driel IR. Helios defines T cells being driven to tolerance in the periphery and thymus. European journal of immunology. 2014;44:2048–2058. doi: 10.1002/eji.201343999. [DOI] [PubMed] [Google Scholar]
- Seddiki N, Santner-Nanan B, Martinson J, Zaunders J, Sasson S, Landay A, Solomon M, Selby W, Alexander SI, Nanan R, et al. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J Exp Med. 2006;203:1693–1700. doi: 10.1084/jem.20060468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledzinska A, Menger L, Bergerhoff K, Peggs KS, Quezada SA. Negative immune checkpoints on T lymphocytes and their relevance to cancer immunotherapy. Molecular oncology. 2015;9:1936–1965. doi: 10.1016/j.molonc.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano-Sarabia N, Bateson RE, Dahl NP, Crooks AM, Kuruc JD, Margolis DM, Archin NM. Quantitation of replication-competent HIV-1 in populations of resting CD4+ T cells. Journal of virology. 2014;88:14070–14077. doi: 10.1128/JVI.01900-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teft WA, Kirchhof MG, Madrenas J. A molecular perspective of CTLA-4 function. Annual review of immunology. 2006;24:65–97. doi: 10.1146/annurev.immunol.24.021605.090535. [DOI] [PubMed] [Google Scholar]
- Tran TA, de Goer de Herve MG, Hendel-Chavez H, Dembele B, Le Nevot E, Abbed K, Pallier C, Goujard C, Gasnault J, Delfraissy JF, et al. Resting regulatory CD4 T cells: a site of HIV persistence in patients on long-term effective antiretroviral therapy. PloS one. 2008;3:e3305. doi: 10.1371/journal.pone.0003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- Wightman F, Solomon A, Kumar SS, Urriola N, Gallagher K, Hiener B, Palmer S, McNeil C, Garsia R, Lewin SR. Effect of ipilimumab on the HIV reservoir in an HIV-infected individual with metastatic melanoma. Aids. 2015;29:504–506. doi: 10.1097/QAD.0000000000000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, Nomura T, Sakaguchi S. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322:271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- Xu L, Xu W, Qiu S, Xiong S. Enrichment of CCR6+Foxp3+ regulatory T cells in the tumor mass correlates with impaired CD8+ T cell function and poor prognosis of breast cancer. Clinical immunology. 135:466–475. doi: 10.1016/j.clim.2010.01.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.