Abstract
The putative cannabinoid receptor GPR55 has been shown to play a tumor-promoting role in various cancers, and is involved in many physiological and pathological processes of the gastrointestinal (GI) tract. While the cannabinoid receptor 1 (CB1) has been reported to suppress intestinal tumor growth, the role of GPR55 in the development of GI cancers is unclear. We, therefore, aimed at elucidating the role of GPR55 in colorectal cancer (CRC), the third most common cancer worldwide.
Using azoxymethane (AOM)- and dextran sulfate sodium (DSS)-driven CRC mouse models, we found that GPR55 plays a tumor-promoting role that involves alterations of leukocyte populations, i.e. myeloid-derived suppressor cells and T lymphocytes, within the tumor tissues. Concomitantly, expression levels of COX-2 and STAT3 were reduced in tumor tissue of GPR55 knockout mice, indicating reduced presence of tumor-promoting factors. By employing the experimental CRC models to CB1 knockout and CB1/GPR55 double knockout mice, we can further show that GPR55 plays an opposing role to CB1. We report that GPR55 and CB1 mRNA expression are differentially regulated in the experimental models and in a cohort of 86 CRC patients. Epigenetic methylation of CNR1 and GPR55 was also differentially regulated in human CRC tissue compared to control samples.
Collectively, our data suggest that GPR55 and CB1 play differential roles in colon carcinogenesis where the former seems to act as oncogene and the latter as tumor suppressor.
Keywords: GPR55, CB1, CNR1, colorectal carcinogenesis
Introduction
Despite benefits from early screening, colorectal cancer (CRC) remains a leading cause among cancer-related deaths emphasizing the need for new treatment options.1 Recent studies indicate that in addition to traditional chemotherapy, immunotherapy may become a valuable option of treatment against CRC.2 Knowledge on leukocytes, inflammatory mediators and their receptors within the tumor microenvironment is therefore crucial in order to explore and design new immunotherapies. Although CRC can be traced back to a familial basis such as adenomatous polyposis in some cases, the majority of incidences are sporadic cases and likely caused by environmental components and by inflammation of the colon (reviewed in 3). For instance, cyclooxygenase (COX)-2-derived mediators, transcription factors like STAT3 and NF-κB, and cytokines involved in intestinal inflammation, are now regarded as driving factors in colon carcinogenesis3, suggesting that molecules involved in gastrointestinal (GI) homeostasis are also critical in the development of GI cancer.
In this context, the endocannabinoid system is of great interest as it plays a prominent role in physiological and pathophysiological processes of the GI tract4. Cannabinoid (CB) receptors, in particular CB1, but also other cannabinoid-responsive G protein-coupled receptors (GPCRs), such as the G protein-coupled receptor 55 (GPR55), are expressed in the GI tract and they are fundamentally involved e.g. in the regulation of motility and inflammation4,5. In experimental models of colitis, CB1 and CB2 receptors exert anti-inflammatory properties6,7 while GPR55 has been linked to pro-inflammatory processes8. Genetic deletion of the CB1 receptor provokes a strong increase in intestinal tumors in Apcmin/+ mice9. The role of GPR55 in GI carcinogenesis, however, has not been fully elucidated so far.
Activity of GPR55 has been shown to be modulated by the endocannabinoids anandamide and virodhamine10. In turn, CB1 may regulate the signaling properties of GPR5511. Thus, despite sharing little homology with the classical CB receptors12, GPR55 can be considered part of an expanded endocannabinoid system. Contrary to CB receptors, GPR55 initiates excitatory rather than inhibitory effects by signaling through Gα12/13 and Gq proteins13, which are known to promote carcinogenesis14. Its structure is closely related to other cancer-relevant GPCRs, such as GPR35, GPR92 and GPR2315. GPR55 expression has been demonstrated in several types of tumor cell lines, such as breast, prostate, ovarian and colon cancer cells16–18. In vivo, the receptor promotes carcinogenesis in a mouse model of skin cancer19. With the exception of cholangiocarcinoma20, a majority of studies propose a pro-oncogenic role for GPR5516–19. Beside cancer cells, GPR55 is expressed in various types of leukocytes, such as macrophages8, neutrophils21, and lymphocytes22. Recently, we showed that the receptor drives intestinal inflammation via mechanisms involving leukocyte infiltration8. We have further elucidated an involvement of GPR55 in migration and adhesion of colon cancer cells and in liver metastasis18. These effects were sensitive to the GPR55 antagonist CID1602004611,18. We have also found that levels of the endogenous ligand, L-α-lysophosphatidylinositol (LPI)23 were higher in blood samples of colon cancer patients than in control individuals18. Other groups demonstrated an important role of the LPI-GPR55 axis in breast cancer metastasis24,25. Because of its potentially pro-carcinogenic behavior and its actions within the endocannabinoid system contrary to those of CB18, we systemically explored the role of GPR55 in GPR55 knockout (GPR55-/-) and wild-type mice using a mouse model of azoxymethane (AOM)- and dextran sulfate sodium (DSS)-induced CRC that displays robust infiltration of inflammatory cells, a feature not only seen in colorectal tumors connected to inflammatory bowel diseases (IBD) but also in CRC not associated with IBD3. We also created CB1/GPR55 double knockout (CB1-/-GPR55-/-) mice and performed experiments in these mice.
Our findings reveal that tumor burden in the colon of GPR55-/- mice was lower than in wild-types and this was in contrast to what we observed in CB1 knockouts (CB1-/-) which displayed higher tumor burden in the colon. In the CB1-/-GPR55-/- mice, these effects were compensated as their tumor burden did not differ from that of their wild-type littermates. Unexpectedly, pharmacological manipulation of GPR55 in colon cancer cell lines failed to show effects on proliferation. However, in comparison with wild-type littermates, tumors of GPR55-/- mice revealed down-regulation of COX-2 and STAT3, increased influx of cytotoxic CD8+ T-cells and decreased infiltration of myeloid-derived suppressor cells (MDSC; a heterogeneous population of immature myeloid progenitor cells that suppresses immune responses), indicating that a change in the tumor microenvironment of GPR55-/- mice may be involved in the inhibitory effects on carcinogenesis.
Materials and Methods
Patient data
For analyzing the influence of GPR55 expression on CRC patient clinical outcome, we made use of a publicly available data set generated on an Affymetrix U133 Plus 2.0 chip platform (GSE39582). This data set contains whole transcriptome data of 566 CRC patients as previously described26. For analysis of these data we used a visualization software [R2: microarray analysis and visualization platform (http://r2.amc.nl)]26. Nine patients lacked survival data and were omitted automatically by the software, resulting in overall 557 available samples. Differences in 10-years relapse-free survival between groups of high and low GPR55 expression were tested by using a cut-off point determined automatically by the software. Survival was illustrated by a Kaplan-Meier curve, and differences in survival between dichotomized groups were assessed with the log-rank test.
DNA methylation and transcriptomics data were obtained from CRC patients as part of the OncoTrack project (www.oncotrack.eu). Written informed consent was obtained from all patients. Ethical approval was granted by the ethics committee of the Medical University of Graz (23-015 ex 10/11).
The methylation status of CNR1 and GPR55 in CRC samples and adjacent healthy mucosa was assessed using the Illumina 450K Infinium Human Methylation BeadChip27. mRNA expression of CB1 and GPR55 was assessed according to standardized procedures. Normalization was done using the RPKM method27,28.
Mice
GPR55-/- mice were acquired from the Mutant Mouse Resource & Research Center and bred in house19. CB1-/- breeding pairs were kindly provided by A. Zimmer. CB1/GPR55 double knockout (CB1-/-GPR55-/-) mice were created by crossing the two strains. Experimental procedures were approved by the Austrian Federal Ministry of Science, Research and Economy (protocol number: BMWF-66.010/0112-WF/II/3b/2014) and performed in strict accordance with international guidelines. All efforts were made to minimize suffering. Experiments were started when mice were 5-7 weeks old and weighed about 20 g.
Mouse model of colitis-associated CRC
Colitis-associated CRC was induced in CB1-/-, GPR55-/-, CB1-/-GPR55-/- mice, and respective wild-type littermates on a C57Bl/6 background as described29. AOM was obtained from Sigma (Vienna, Austria), and DSS from MP Biomedicals (Illkirch, France). Mice were sacrificed on day 80 for tissue collection and on day 108 for analysis of leukocytes recruited into the tumors. Colon tissue was also collected from age-matched healthy mice. For the pharmacological approach, CD1 mice - a strain that is more susceptible to this model - were obtained from Charles River (Germany). Either 5 mg/kg CID16020046 (GPR55 antagonist; ChemDiv, San Diego, CA, USA) or dimethyl sulfoxide (vehicle control; VWR, Vienna, Austria) diluted in PBS (Pan-Biotech, Aidenbach, Germany) were applied s.c. on days 22-28 and 38-46. Tumors were evaluated on day 100. In graphs that show data from the mouse model experiments, each data point corresponds to tumors obtained from one individual mouse.
Mouse model of spontaneous tumor progression
Spontaneous tumor progression was induced in CB1-/- and GPR55-/- mice, and C57Bl/6 wild-type littermates as described29. Tumors were evaluated 6 months (CB1) or 8 months (GPR55) after the last AOM injection.
Cell culture
Colon cancer cell lines HCT116, SW480, SW620, HT29 and DLD-1 were obtained from Interlab Cell Line Collection (Genova, Italy), CaCo-2 from ATCC (Manassas, VA, USA). HCT116 and HT29 were maintained in McCoy’s 5A, SW480, SW620, DLD-1, and CaCo-2 in DMEM, both supplemented with 10% FBS (all Life Technologies, Vienna, Austria), 2 mM L-glutamine and 1% penicillin/streptomycin (both PAA Laboratories, Pasching, Austria), at 37°C, 5% CO2 in a humidified atmosphere. Cells stably overexpressing GPR55 were generated by transfecting SW480 with pcDNA3.1 coding for GPR55 with an N-terminal hemagglutinin tag (3xHA-GPR55), as described30. Transfected cells were selected with 4 mg/ml G418 (Life Technologies) and grown to confluence. Cells were then sorted on a FACSAria (BD Biosciences, Franklin Lakes, USA) after staining with anti-HA (Covance #MMS-101P) and goat anti-mouse Alexa 488 (Thermo Scientific #A-21121) antibodies. Sorted cells were maintained in DMEM supplemented with 0.5 mg/ml G418.
Cell viability was assessed after treatment with LPI (GPR55 agonist; Sigma) or CID16020046 as previously described18.
Cell lines were authenticated by DNA short-tandem repeat analysis by the Cell Culture Facility of the Center for Medical Research at the Medical University of Graz (Austria).
RNA extraction, reverse transcription to cDNA, and qRT-PCR
RNA was extracted from tissue using TRIzol (Life Technologies) and from cultured cells with RNeasy Kit (Qiagen, Hilden, Germany). Samples were treated with DNA-free DNA Removal Kit (Life Technologies) and 2µg of total RNA were then reverse transcribed to cDNA using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, USA). Quantification of gene expression was carried out by real-time PCR using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, Vienna, Austria). Primers were acquired from Bio-Rad (listed in Supplementary Table 1) and relative gene expression was assessed according to the ΔΔCq-method.
Protein extraction, Western blotting, and cytokine analysis
For protein extraction and Western blotting, protocols from Cell Signaling Technology (CST) were followed. Antibodies used were: COX-2 (ab15191, 1:1000), NF-κB (CST 4764, 1:1000), STAT3 (CST 4904, 1:1000), p(Tyr705)-STAT3 (CST 9145, 1:1000), PCNA (Dako M0879, 1:1000), Bax (CST 2772, 1:1000), Bcl-xL (CST 2764, 1:1000), β-actin (Sigma A5316, 1:7500), and GAPDH (CST 5174, 1:1000); goat-anti-mouse (#115-036-062) or goat-anti-rabbit antibody (#111-036-045, both Jackson ImmunoResearch, 1:5000). Detection was performed on a ChemiDoc Touch Imaging System using Clarity Western ECL Blotting Substrate (both Bio-Rad). Immunoblot images were analyzed with Image Lab 5.2 software (Bio-Rad). Band intensities of proteins of interest were normalized against band intensities of loading controls and ratios are presented in arbitrary units. GAPDH was found to be stably expressed over different tissues, but β-actin gave better stability for comparison between tumors of GPR55-/- and wild-type mice. Cytokine expression was measured using the ProcartaPlex Multiplex Immunoassay (affymetrix eBioscience, Vienna, Austria).
Immunohistochemistry and in situ hybridization
Colon sections were stained as described previously8. The following antibodies were used: COX-2 (ab15191, 1:2000), STAT3 (CST 4904, 1:500), and CD3 (ab5690, 1:1000). Antibody binding was visualized with ImmPACT NovaRed (Vector Laboratories) and sections were counterstained with hematoxylin. Images were taken with a high resolution digital camera (Olympus DP 50) and analyzed by Cell^A imaging software (Olympus, Vienna, Austria). Only contrast and brightness of images were adjusted.
For cellular detection of GPR55 mRNA in colonic sections, RNAscope® 2.5 Chromogenic Assay with RNAscope® mouse GPR55 probe (#318231; ACD Advanced Cell Diagnostics, Hayward, CA, USA) was used according to the manufacturer’s instructions.
Mass spectrometry
Prostanoids from mouse tissue were analyzed by liquid chromatography-mass spectrometry (LC-MS/MS) as previously described31.
Isolation of recruited leukocytes and flow cytometry
Tumors were excised from the colon, cut into small pieces with a scalpel and washed with HBSS containing 20 mM HEPES and penicillin/streptomycin (P/S) 4 x 10 min at room temperature. Intraepithelial immunocytes were extracted by incubation with HBSS containing 10 mM EDTA, 2.5% FBS, and P/S for 4 x 20 min at 37°C on a rotating device. The tissue was washed with RPMI and digested with RPMI supplemented with 5% FBS, 10mM HEPES, P/S, and 200 U/ml collagenase type II for 1 hour at 37°C (all Life Technologies). Isolated cells were passed through a 40µm cell strainer, washed with PBS once, and leukocytes were stained for flow cytometry. Cells were stained with the following antibodies (all BD, Vienna, Austria) for 1 hour at 4°C: CD45-APC (1:200, clone: 30-F11), CD3e-BV510 (1:100, clone 145-2C11), CD4-PE-Cy7 (1:500, clone RM4-5), CD8a-APC-H7 (1:100, clone 53-6.7), CD11b-FITC (1:200, clone M1/70), Ly6G-BV421 (1:500, clone 1A8), Ly6C-PE-Cy7 (1:200, clone AL-21), CD16/32 (1:100, clone 2.4G2). Samples were analyzed on a BD FACS Canto II flow cytometer. Analysis was done with FlowJo 4.0 software.
Demethylation and bisulfite sequencing
Demethylation of colon cancer cell lines was induced as previously described32. Briefly, cells were treated with 5-aza-2’-deoxycytidine (Sigma) for 3 days, DNA was extracted using Gentra Puregene Cell Kit and bisulfite conversion was performed with the EpiTect Bisulfite Kit (both Qiagen). Primers (listed in Supplementary Table 2) spanning three regions of interest determined from methylation data obtained from CRC patients, i.e. chr2:231774770 in the gene body (“body”), chr2:231789465 in the 5’-untranslated region (“5’-UTR”), and chr2:231790813, upstream of the transcription start site (“TSS”) were designed using Methyl Primer Express (Life Technologies) with hg19 as a reference. The regions of interest were amplified using the HotStar Taq® Master Mix Kit (Qiagen) and sequenced using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Life Technologies) according to the manufacturer’s protocol. The sequencing reactions were purified using Sephadex G50 Superfine (Sigma) and sequenced on an ABI 3130xl Genetic Analyzer. Sequences were analyzed using the SeqScape software (Life Technologies) and CpG methylation was calculated as a function of the area under the curve of the C and the T traces.
Statistical analysis
Statistical analysis of in vivo and in vitro experiments was performed with GraphPad Prism 4.0 (GraphPad Software, La Jolla, CA, USA). Treatment groups were compared using unpaired two-tailed student’s t-test unless stated otherwise. Welch’s correction was applied if variances were unequal.
Results
Loss of GPR55 reduces tumor burden
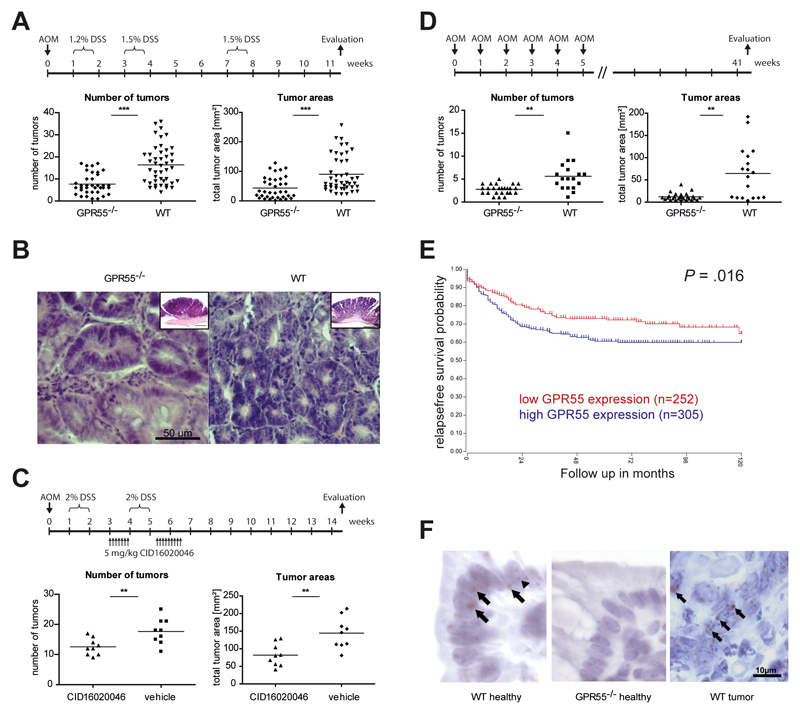
In a mouse model of AOM+DSS-induced colitis-associated CRC, GPR55-/- mice showed ~50% less tumors than their wild-type littermates. Accordingly, the average total tumor areas were also reduced by ~50% in GPR55-/- mice (Fig. 1A). Histological evaluation of the tumors revealed that in both GPR55-/- and wild-type mice, criteria of tubular-villous adenomas with low and high grade intraepithelial neoplasia occurred (Fig. 1B). In GPR55-/- mice, the nuclear-cytoplasmic ratio was changed towards a higher nucleus size, however, nuclei were located predominantly at the cell base. Wild-type mice showed enlarged nuclear sizes accompanied by hyperchromasia and irregularity of tumor cell shape and nuclear shape. Nuclei were largely apically located. The cytoplasm appeared strongly basophilic. Nuclear clearing was observed particularly in wild-type mice and, albeit to a much lesser degree, also in GPR55-/- mice. No sign of invasion was detected in either group. In general, GPR55-/- mice exhibited a more low grade form of neoplasia compared to the low to high grade form of tumors in the wild-type mice.
Figure 1. GPR55 promotes colorectal cancer.
(A) In a model of colitis-associated CRC, GPR55-/- mice (n=36) had less tumors and smaller total tumor areas than their wild-type littermates (WT, n=44). (B) H&E staining from sections of tumors in the colon induced by AOM+DSS in GPR55-/- (left panel) and wild-type mice (right panel). Images are representative of 4 mice of each cohort. Inserts in the right upper corner of the images show adenomas in GPR55-/- and wild-type mice in lower magnification (size bars: 500 µm). (C) Pharmacological inhibition of GPR55 with CID16020046 (5 mg/kg) reduced tumor numbers and areas compared to vehicle treatment (n=9 for both groups). (D) Tumor numbers and areas were reduced in GPR55-/- mice (n=27) compared to wild-type mice (n=18) in a model of spontaneous tumor progression. (E) In CRC patients, high GPR55 mRNA expression in tumor tissue was significantly associated with reduced relapse-free survival (P=.016, log-rank test). (F) Cellular detection of GPR55 mRNA with RNAscope ® 2.5 Chromogenic Assay using 3,3'-diaminobenzidine (DAB) as a chromogene (brown color). Sections were counterstained with hematoxylin (blue). Sections of colonic mucosa of healthy wild-type (WT) mice showed GPR55 expression (left panel), while the staining was absent in knockout (GPR55-/-) controls (middle panel). Tumors of wild-type mice expressed GPR55 mRNA in various cells (right panel). Arrows point at DAB precipitates at the sites of GPR55 mRNA expression in cells. Arrowheads denote heterochromatin (blue) in cell nuclei. Symbols depict data from individual mice, and bars show the mean. ** P < .01, *** P < .001
Pharmacological inhibition of GPR55 with the antagonist CID16020046 also led to a decreased tumor burden in colitis-associated CRC in wild-type mice. Treatment with CID16020046 (5 mg/kg) caused a reduction in tumor numbers and areas compared to vehicle control treatment (Fig 1C). Since chronic inflammation is the driving force of the colitis-associated CRC model and as GPR55 has recently been shown to play a pro-inflammatory role in colitis8, we subjected mice also to an inflammation-independent model of repeated AOM administration. In this model of spontaneous tumor progression, GPR55-/- mice again had less tumors and smaller total tumor areas than wild-type littermates (Fig. 1D).
For human cross validation, we analyzed a publicly available clinical CRC expression data set (n=557) and found a significant (P = .016) association between high GPR55 expression and shortened relapse-free survival (Fig. 1E). These findings suggest that GPR55 plays a tumor-promoting role in colon carcinogenesis.
GPR55 modulates the tumor microenvironment
To investigate the mechanism through which GPR55 exerts its pro-tumorigenic function we first performed proliferation assays with CRC cell lines. Although all cell lines examined expressed GPR55 (Supp Fig. 1A), we did not find any effects of the agonist LPI or the antagonist CID16020046 on cell proliferation (Supp Fig. 1 B,C), cell cycle distribution or apoptosis (data not shown). Since our in situ hybridization data indicated that GPR55 was not only present on epithelial cells but also on cells of the lamina propria and of lymph follicles from healthy murine colonic mucosa as well as in various cells within the tumor tissue of AOM+DSS treated mice (Fig. 1F and Supp Fig. 2), we focused on the analysis of collected tissues.
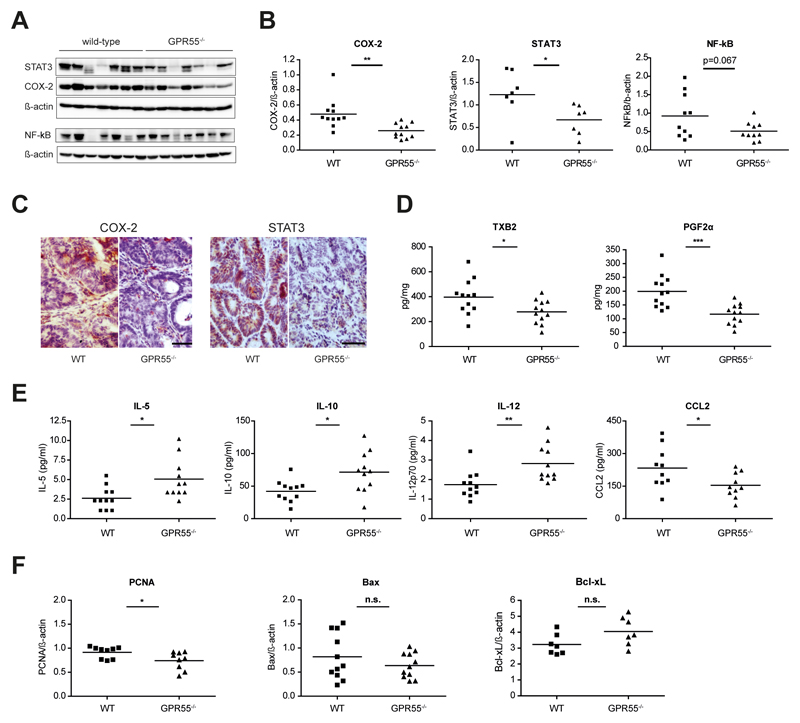
Prostaglandin H2 synthesizing enzyme cyclooxygenase 2 (COX-2) as well as transcription factors NF-κB and STAT3 have been described to constitute some of the most important pathways in inflammation-induced cancer3,33,34. Indeed, we observed that COX-2 and STAT3 expression were increased in non-tumor and even more in tumor tissue as compared to healthy control colon (Supp Fig. 3). Comparison of tumor tissue collected from GPR55-/- mice and wild-type littermates revealed that COX-2 and STAT3 expression were reduced in tumors of GPR55-/- mice (Fig. 2 A-C). Notably, reduction of COX-2 and STAT3 expression was not limited to immunocytes but rather also occurred in tumor cells of GPR55-/- mice (Fig. 2C). Reduced levels of NF-κB and phosphorylated STAT3 were also observed although statistical significance was slightly missed (p=0.067 and p=0.08, respectively, Fig. 2 A,B and Supp Fig. 4). Down-stream effector molecules of COX-2, i.e. thromboxane A2 (as determined by measurement of the primary metabolite thromboxane B2 (TXB2)) and prostaglandin F2α (PGF2α), were also found to be expressed at a lower level in tumor tissue of GPR55-/- mice (Fig. 2D). Cytokine analysis further showed that interleukins 5, -10, and -12 (IL-5, IL-10, IL-12) were elevated in tumors of GPR55-/- mice, whereas the expression of myeloid cells-recruiting chemokine monocyte chemoattractant protein-1 (MCP-1/CCL2) was diminished (Fig. 2E). Analysis of the cell proliferation marker proliferating-cell-nuclear-antigen (PCNA) showed a slight decrease in tumors of GPR55-/- mice, whereas apoptosis markers Bax and Bcl-xL were not significantly altered (Fig. 2F).
Figure 2. Tumors of GPR55-/- mice display alterations in the microenvironment.
(A) Western blotting revealed reduced expression levels of STAT3, COX-2, and NF-κB in tumors of GPR55-/- mice compared to wild-type (WT) mice. Images were obtained by stripping and re-probing of the membrane and are representative of 3 independent blots. (B) Statistical analysis of data obtained by Western blotting. (C) Sections of colonic tumors of GPR55-/- and wild-type mice were stained (brown color) for COX-2 and STAT3 expression showing clear reductions in the GPR55-/- mice. Representative images are shown (scale bar, 50 µm). (D) Thromboxane B2 (TXB2) and prostaglandin F2α (PGF2α) were found at lower levels in GPR55-/- tumors as assessed by mass spectrometry. (E) Interleukin -5, -10, and -12 (IL-5, IL-10, IL-12) expression was increased in GPR55-/- tumors compared to wild-type tumors, whereas monocyte chemoattractant protein-1 (MCP-1/CCL2) expression was decreased. (F) Expression levels of proliferation marker PCNA were slightly reduced in tumors of GPR55-/- mice whereas apoptotic markers Bax and Bcl-xL were not significantly (n.s.) altered. * P < .05, ** P < .01, *** P < .001, n=7-12
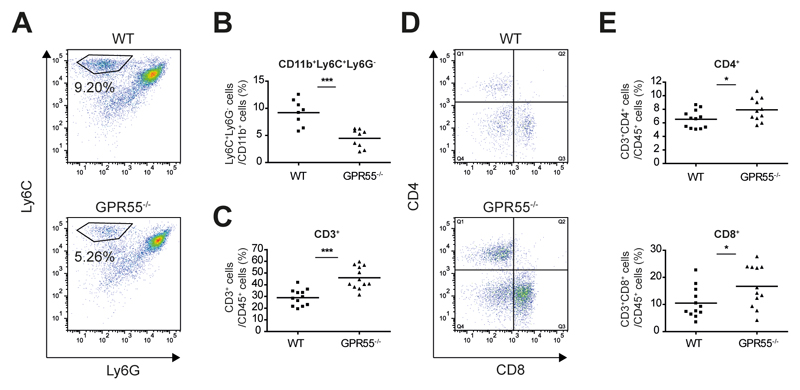
Together, these observations indicate that GPR55 might be involved in the immunomodulation of the tumor microenvironment. We thus analyzed the leukocyte populations recruited into tumors of wild-type and GPR55-/- mice. Since it has been recently reported that deletion of CCL2 reduced carcinogenesis in colitis-associated CRC via reduced infiltration of MDSCs35, we examined this cell population in our model. Indeed, we found that tumors of GPR55-/- mice showed reduced numbers of CD11b+Ly6C+Ly6G- cells (Fig. 3 A,B), which have been described as monocytic MDSCs35,36. MDSCs are reported to contribute to colonic tumor growth in colitis-associated CRC via T cell inhibition36. Accordingly, tumors of GPR55-/- mice displayed strongly enriched numbers of CD3+ leukocytes (Fig. 3C). Further analysis of the CD3+ cell population revealed an increase in both CD4+ and CD8+ T cells (Fig. 3 D,E). To double check for the presence of T cells, colonic sections were also stained immunohistochemically for CD3 (Supp Fig. 5). GPR55 has been reported to be expressed by various leukocytes21,22,37 and GPR55 antagonism has been shown to modulate the migratory behavior of cells8,18,21. Therefore, we examined whether the above mentioned alterations in the tumor microenvironment could be caused by “per se” effects of the genetic deletion of GPR55. As shown in Supp Fig. 6A, however, the amount of leukocytes recruited into the tumors did not differ between GPR55-/- and wild-type mice. Expression levels of COX-2 and STAT3 in healthy control colon were not statistically different, either (Supp Fig. 6 B,C).
Figure 3. GPR55 modulates the tumor microenvironment through alteration of recruited leukocytes.
Leukocytes were extracted from tumors of GPR55-/- and wild-type (WT) mice and analyzed by flow cytometry. (A) Viable cells were gated from an FSC/SSC plot and CD11b+ cells were further analyzed for their Ly6C/Ly6G expression. (B) A subpopulation of CD11b+ cells, i.e. Ly6C+Ly6G- cells was significantly reduced in tumors of GPR55-/- mice. (C-E) Following a leukocyte gate (CD45+), cells were further gated for CD3. (C) Tumors of GPR55-/- mice showed a strong increase of CD3+ leukocytes. (D, E) CD3+ cells were further analyzed for their CD4 and CD8 expression. CD4+ cells (Q1) and CD8+ cells (Q3) were more abundant in tumors of GPR55-/- mice than in tumors of wild-type mice. Each dot represents the leukocytes extracted from tumors of one mouse, and bars are the means. * P < .05, *** P < .001, n=9-12
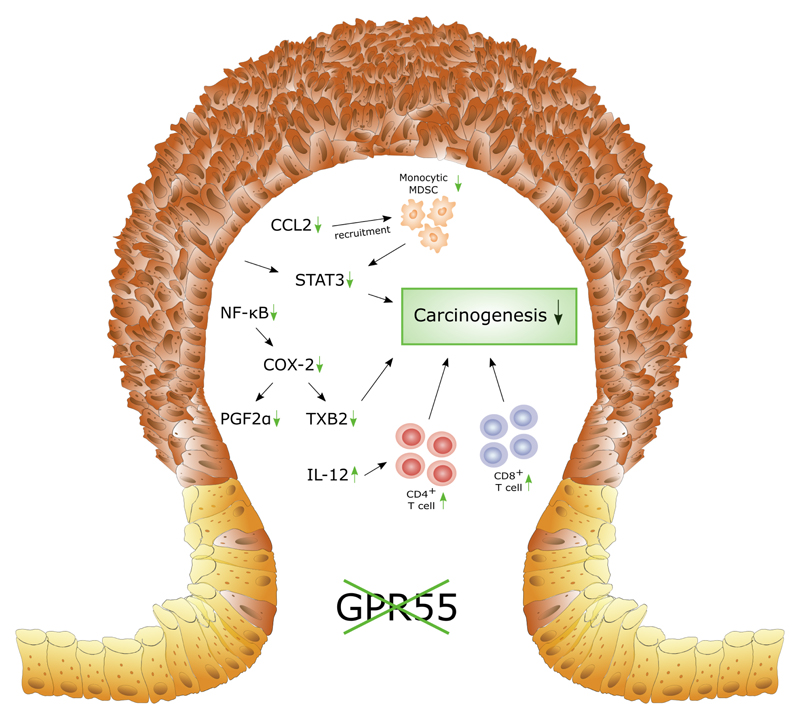
Taken together, our data thus indicate that GPR55 probably alters the composition of the leukocyte population that is recruited during colorectal carcinogenesis resulting in the formation of a tumor-promoting microenvironment that leads to increased expression of tumor-promoting molecules (Fig. 4).
Figure 4. Deletion of GPR55 alters the tumor microenvironment in murine colitis-associated CRC.
This scheme summarizes our findings of modulations that could lead to reduced colorectal carcinogenesis in GPR55-/- mice. Alterations in tumors of GPR55-/- mice compared to wild-type littermates are indicated by green arrows. Firstly, reduced numbers of monocytic myeloid-derived suppressor cells (MDSC) were observed which is likely a consequence of reduced CCL2 expression 35. Therefore, STAT3 expression in turn could be diminished partly by reduced CCL2-MDSC signaling 35, although other cells, e.g. neoplastic cells, could also play a role 43. Secondly, we observed reduced expression levels of transcription factor NF-κB, and of cyclooxygenase-2 (COX-2) as well as of its effector molecules prostaglandin F2α (PGF2α) and thromboxane B2 (TXB2). Both prostanoids have been shown to promote carcinogenesis 41,42. Thirdly, infiltration of both TH cells (CD4+) and cytotoxic T cells (CD8+) was increased in tumors of GPR55-/- mice. Increased recruitment of T cells has been shown to be associated with a favorable clinical outcome in CRC 40 and might thus also contribute to reduced carcinogenesis in our model.
CB1 plays a contrary role to GPR55 in CRC
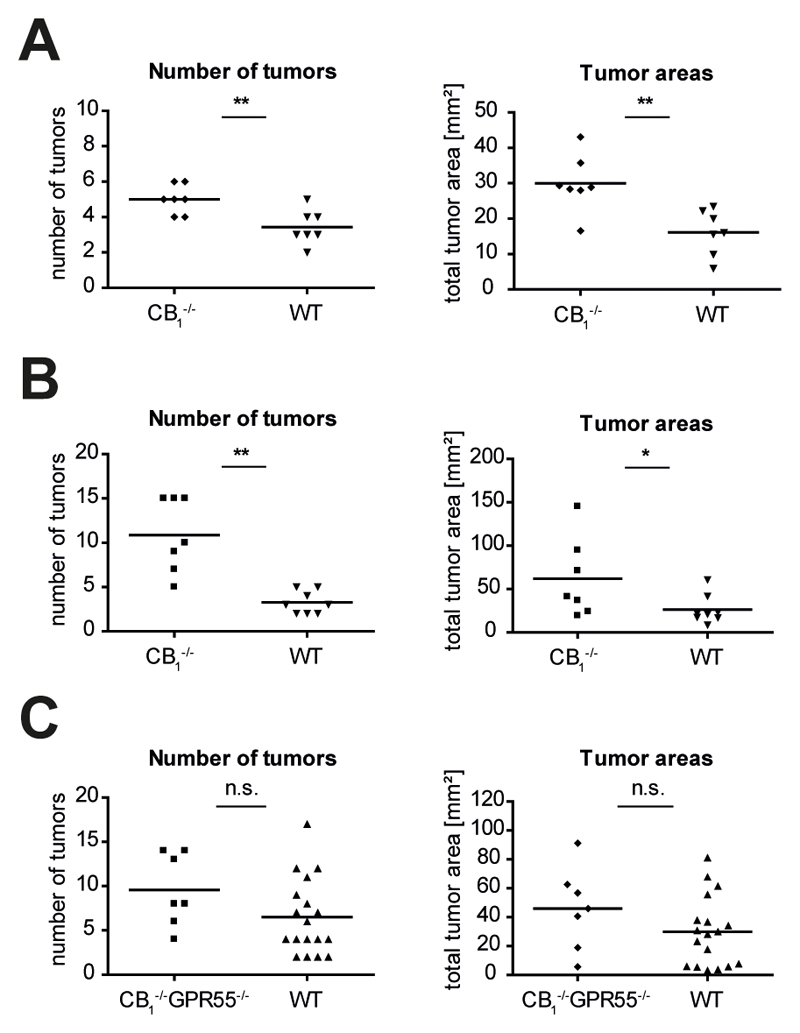
Since CB1 has been shown to play a tumor-suppressing role in a genetic mouse model of intestinal cancer9 in which polyps developed mainly in the small intestine (Apcmin/+ mice), we were interested to see whether CB1 would also be protective in our models of CRC. Indeed, we found that CB1-/- mice developed significantly more tumors and, accordingly, had larger tumor areas than wild-type littermates in both models of colon cancer, i.e. in the model of spontaneous tumor progression and in the colitis-associated CRC model (Fig. 5 A,B). These data suggest that CB1 plays a contrary role to GPR55 in CRC. To further support this notion, we applied the colitis-associated CRC model to CB1/GPR55 double knockout mice and wild-type littermates. As shown in Fig. 5C, there was no difference in tumor burden, i.e. neither in tumor numbers nor in tumor areas, between CB1-/-GPR55-/- mice and wild-type littermates, suggesting that the tumor-promoting role of GPR55 is indeed opposed by the tumor-suppressive role of CB1.
Figure 5. CB1 appears to be tumor-suppressive and opposing the tumor-promoting role of GPR55 in CRC.
(A) In the murine colon cancer model of spontaneous tumor progression, CB1-/- mice (n=7) developed more tumors and had larger tumor areas than wild-type littermates (WT, n=7). (B) Correspondingly, CB1-/- mice (n=7) also had increased tumor numbers and total tumor areas in the model of colitis-associated CRC compared to wild-type littermates (n=8). Here, because of high mortality in male CB1-/- mice after DSS application, only females were used. (C) When CB1/GPR55 double knockout (CB1-/-GPR55-/-) mice (n=7) were subjected to the colitis-associated CRC model they showed a tumor burden equal to their wild-type littermates (n=18). Data show values obtained from individual mice, and bars are the means. * P < .05, ** P < .01, n.s. not significant
CB1 and GPR55 are differentially regulated in murine colon carcinogenesis as well as in CRC patients
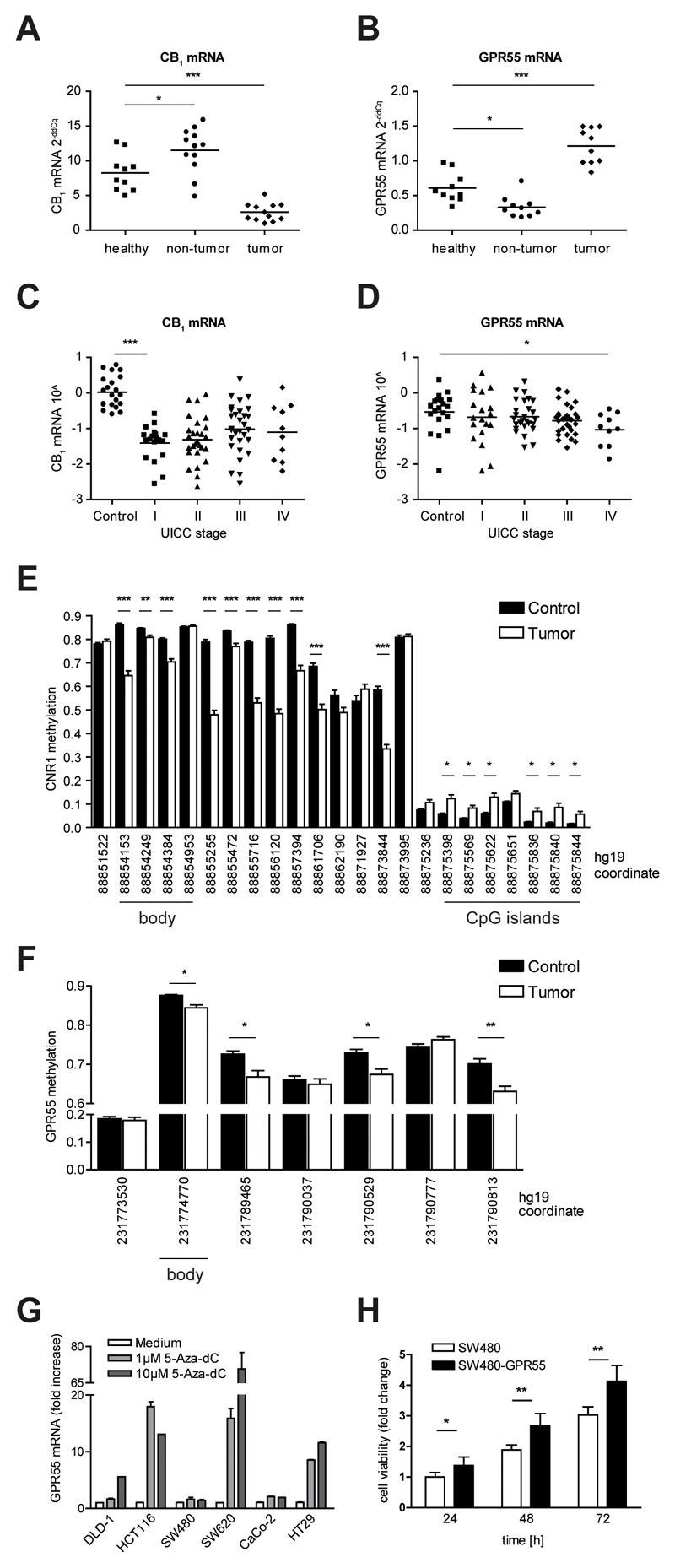
Wang et al recently reported that the tumor-suppressive role of CB1 could be diminished in CRC because of epigenetic hypermethylation of the CNR1 promoter that results in reduced transcription9. Following up on this finding, we examined expression levels of CB1 and GPR55 mRNA in tissue collected from the colitis-associated CRC mouse model and from a cohort of 86 CRC patients. In the mouse model of colitis-associated CRC we observed that in tumor tissue, CB1 expression was indeed strongly reduced as compared to healthy control colon. The colonic non-tumor tissue, i.e. tissue affected by multiple exposures to DSS but devoid of neoplastic lesions, showed an increase in CB1 mRNA levels as compared to healthy colon (Fig. 6A). On the contrary, GPR55 mRNA levels were decreased in non-tumor tissue and increased in tumor tissue compared to colon tissue of healthy control mice (Fig. 6B). In CRC patients, CB1 mRNA levels were drastically reduced in tumors of TNM stage I as compared to control tissue. With increased severity of disease, however, CB1 mRNA levels increased again (Fig. 6C). GPR55 mRNA levels, on the other hand, decreased with increasing stage (Fig. 6D). Taken together, our data indicate a differential regulation for the transcription of CB1 and GPR55 in murine and human CRC.
Figure 6. CB1 and GPR55 mRNA expression and DNA methylation are regulated in a differential manner.
(A, B) mRNA expression levels were determined by qRT-PCR in the colon of healthy control mice (healthy) and in colon tissue collected from mice subjected to the colitis-associated CRC model (non-tumor and tumor), reference gene: Hprt. (A) CB1 mRNA was found to be up-regulated in non-tumor tissue and down-regulated in tumor tissue compared to healthy colon tissue. (B) GPR55 mRNA, in contrast, was down-regulated in non-tumor tissue compared to healthy control colon whereas it was up-regulated in tumors. Statistical analysis for graphs A and B was performed using one-way ANOVA and Tukey’s posthoc test. * P < .05, *** P < .001 (C-F) Data of CRC patients were obtained from OncoTrack. (C, D) mRNA expression levels were obtained by RNAseq. Tumors (n=86) were TNM staged according to the guidelines of the Union internationale contre le cancer (UICC). Control samples (n=21) were obtained from tumor-adjacent non-neoplastic colon of CRC patients. Statistical analysis was done by one-way ANOVA followed by Tukey’s posthoc test. * P < .05, *** P < .001 (C) CB1 mRNA was down-regulated in tumors but increased with disease severity. (D) In contrast, GPR55 mRNA decreased with stage. (E, F) DNA methylation status was assessed in tumor samples (n=81, white bars) and in tumor-free control tissue (n=67, black bars). Each set of bars shows the mean + SEM degree of methylation in one CpG. The hg19 coordinate of each CpG is indicated by the x-axis label. Statistical analysis was performed with a regularized t-test that was conducted using the limma package in R as described in detail elsewhere27; P values were adjusted for multiple-testing using the false-discovery method. * P < .01, ** P < .001, *** P < .0001 (E) In tumor samples, CNR1 methylation was increased at CpG islands surrounding the promoter region whereas it was decreased in the body of the gene. (F) GPR55 lacks CpG islands and rather exhibits a global decrease in methylation in tumor samples. (G) GPR55 mRNA levels were increased in DLD-1, HCT116, SW620, and HT29 cells after treatment with 5-Aza-dC. Data shown are means + SEM of three independent experiments, reference gene: HPRT1. (H) Stable over-expression of GPR55 led to a growth advantage of SW480 cells. Data shown are means + SD of six independent experiments. Statistical analysis was done by one-way ANOVA and Tukey’s posthoc test. * P < .05, ** P < .01
Epigenetic changes in CNR1 and GPR55 in CRC patients
To further elucidate the aforementioned findings, we analyzed the methylation status of CNR1 and GPR55 in our cohort of CRC patients. In line with the observations of Wang et al9, we found that CNR1 has a CpG island-associated promoter (Fig. 6E) that is characterized by low DNA methylation in healthy colonic mucosa controls (fraction of methylated cells [β]<0.11) with six probes significantly hypermethylated in CRC samples (Δβ>0.04, P < .01). These CpGs are located at -755 to +268 with regard to the transcription start site (hg19 coordinates 88875844 to 88875398). Additionally, we observed differences within 5’-UTR and gene body probes in open sea regions: in controls these regions of CNR1 are characterized by much higher methylation (β>0.54); in CRC, however, ten probes in this region are substantially hypomethylated compared with control (Δβ=0.04-0.32, P < .001). This phenotype is consistent with the idea that CNR1 is actively transcribed in both CRC and healthy controls, but that transcription is being nuanced toward down-regulation in CRC samples. In contrast, GPR55 does not have CpG islands and exhibits a rather global decrease in methylation (Fig. 6F) with four probes being significantly less methylated in CRC samples (Δβ=0.03-0.07, P < .01).
Epigenetic changes in GPR55 in colon cancer cell lines
DNA demethylating agents, e.g. 5-aza-2’-deoxycytidine (5-aza-dC, “decitabine”) have been approved by the US Food and Drug Administration for the treatment of myelodysplastic syndrome and are currently being investigated for the treatment of solid tumors, including gastrointestinal cancer38. To examine the effect of epigenetic demethylation on the expression of GPR55, we treated six colon cancer cell lines with 5-aza-dC and analyzed the methylation status of GPR55 by bisulfite sequencing before and after treatment. GPR55 was heavily methylated in all CRC cell lines (Supp Fig. 7). SW480 cells, however, showed a slightly lower degree of methylation at CpGs in the gene body and 5’-UTR. Intriguingly, this cell line also expressed ~55-fold more GPR55 mRNA (see Supp Fig. 1A). Treatment with 5-aza-dC caused a reduction of methylation and a concomitant increase of mRNA expression in 4 of 6 cell lines, i.e. DLD-1, HCT116, SW620, and HT29 cells. The methylation status of SW480 and Caco-2 cells was not altered upon treatment and, accordingly, GPR55 mRNA expression levels were unchanged (Fig. 6G).
These findings prompted us to investigate whether overexpression of GPR55 in colon cancer cells could lead to a proliferative effect. Thus, we stably overexpressed the receptor in SW480 cells. Cell viability of native and GPR55-overexpressing SW480 (SW480-GPR55) cells was assessed after 24, 48, and 72 hours and revealed a significant growth advantage of SW480-GPR55 cells (Fig. 6H). These findings corroborate our hypothesis that GPR55 has a pro-tumorigenic role in CRC.
Discussion
A tumor-promoting function has been established for GPR55 and its endogenous ligand LPI in a variety of cancers, including breast25, prostate and ovarian cancer17, squamous cell carcinoma19, and glioblastoma16. Recently, we have reported GPR55-mediated effects in adhesion and migration of colon cancer cells, as well as in experimental liver metastasis18. The role of GPR55 in colorectal carcinogenesis, however, has not been elucidated yet.
Using the AOM+DSS-driven colitis-associated CRC model, we show that genetic deletion of GPR55 leads to reduced tumor burden in mice. Pharmacological inhibition of GPR55 with CID16020046 gave results that were similar to the genetic approach.
In the current study, we focused on investigating the effects of genetic ablation of GPR55 in colorectal carcinogenesis. As summarized in Figure 4, we found that tumors of GPR55-/- mice exhibited modifications in the tumor microenvironment that were characterized by altered leukocyte populations in the tumors and a concomitantly reduced expression of tumor-promoting factors. For instance, reduced expression of the myeloid cell-recruiting chemokine CCL2 was observed. Deletion of CCL2 has been reported to reduce the recruitment of myeloid-derived suppressor cells (MDSCs) in colitis-associated CRC35. In line with this finding, we observed reduced numbers of monocytic MDSCs in GPR55-/- tumors. MDSCs suppress immune responses mediated by CD4+ and CD8+ T cells by inhibiting T cell proliferation, migration and function39. We observed increased numbers of both CD4+ and CD8+ T cells in tumors of GPR55-/- mice which could be a consequence of reduced amounts of MDSCs in the microenvironment. Importantly, CD3+ lymphocytes have been shown to be beneficial in CRC patients since increased numbers of CD3+ cells were associated with an increased disease-free survival40. Furthermore, we found that COX-2, a promising target in the prevention of CRC34, and COX-2-derived mediators PGF2α and TXB2, which have both been attributed tumor-promoting functions41,42, were expressed at lower levels in tumors of GPR55-/- mice. STAT3 signaling, which has been established to drive colon carcinogenesis 33,43, was also reduced in tumors of GPR55-/- mice, which could partly be due to diminished CCL2-MDSC signaling35. Cytokine analysis further revealed increased levels of IL-5, IL-10, and IL-12 in tumors of GPR55-/- mice, which might be caused by increased numbers of T cells present in these tumors. While the role of IL-5 in CRC has not been fully clarified yet, clinical trials using recombinant human IL-10 (ClinicalTrials.gov identifier: NCT02009449) and IL-12 (NCT01417546) in solid tumors including CRC are currently ongoing. Taken together, our data suggest that abrogation of GPR55 indirectly reduces the expression of COX-2, STAT3, and PCNA in tumor cells resulting in reduced tumor growth, i.e. smaller tumor burden and lower grade neoplasia in GPR55-/- mice. This could explain why GPR55-/- mice had less tumors in our model. Other factors, however, might also play a role and remain to be investigated.
Since it has been postulated that the LPI-GPR55 axis enhances cancer cell proliferation44, we performed cell viability assays on two CRC cell lines, one with high GPR55 mRNA expression (SW480 cells) and one with low GPR55 mRNA expression (HCT116 cells). Interestingly, however, we did not detect any effect of the agonist LPI or the antagonist CID16020046 on the viability of either cell line. A possible explanation hereof could lie in the purported cross-regulation of CB1 and GPR55 meaning that GPR55 signaling was inhibited when CB1 and GPR55 were co-expressed in HEK293 cells11. HCT116 and SW480 cells express both CB19 and GPR55 and, indeed, we observed that SW480 cells that stably overexpressed GPR55 had a growth advantage compared to native SW480 cells. Whether CB1 actually inhibits GPR55 signaling in CRC cells remains to be established.
Although GPR55 is not considered a classical cannabinoid receptor but owing to its responsiveness to certain (endo-)cannabinoids, it can be regarded as part of an extended endocannabinoid system13. Since the endocannabinoid system is crucially involved in GI homeostasis and disorders4, we aimed at further investigating the role and regulation of CB1 in CRC. Here, we report that genetic deletion of CB1 led to an increased tumor burden in both our experimental models, i.e. in colitis-associated and spontaneous colon cancer, corroborating the findings of Wang et al9 who observed a detrimental effect of CB1 deletion in a genetic model of colon cancer (ApcMin/+ mice). When CB1/GPR55 double knockout mice were subjected to the colitis-associated CRC model, no differences in tumor burden were found as compared to wild-type littermates. These are the first in vivo data that show that GPR55 and CB1 regulate colorectal carcinogenesis in an opposing manner.
Receptor expression analysis of murine tissue further revealed a differential regulation of CB1 and GPR55. While CB1 appeared up-regulated in inflamed non-tumor tissue and down-regulated in tumor lesions, GPR55 was found to be down-regulated in non-tumor tissue and up-regulated in tumors when compared to healthy control colon. These findings corroborate the hypothesis that colonic tumor growth is facilitated by the absence of tumor-suppressing CB1 and the presence of tumor-promoting GPR55.
Since down-regulation of CB1 expression in tumors has been shown to be a result of epigenetic hypermethylation of CpGs in the promoter region of CNR1 in CRC patients9, we assessed the methylation status of GPR55 and CNR1 in a cohort of 86 CRC patients. In accordance with previous findings9, DNA methylation analysis revealed a strong increase of promoter cytosine methylation in the CpG islands of CNR1 in CRC samples compared to control tissue. Contrarily, a decrease in methylation of GPR55 was found in tumor samples. Alterations in DNA methylation are ubiquitous in human cancers, with promoter CpG island hypermethylation occurring in the context of a global decrease in methylation45. Increased promoter cytosine methylation has been established to silence gene expression. CNR1 promoter hypermethylation, therefore, is very likely to contribute to colon carcinogenesis through silencing of tumor-suppressing CB1. GPR55 on the other hand does not have CpG islands in the promoter region and is, perhaps, subject to the genome-wide hypomethylation frequently observed in cancer46.
In accordance with the above mentioned promoter hypermethylation, mRNA expression analysis revealed a strong down-regulation of CB1 in CRC samples compared to control tissue. Interestingly, CB1 expression levels were found increased with disease severity. GPR55 expression, in contrast, decreased with CRC stage in this cohort. These findings differ from previous reports stating that GPR55 mRNA expression is increased in several human tumors, e.g. pancreatic neoplasias and glioblastomas16. GPR55 expression levels, however, have also been shown to correlate with aggressiveness16 and, indeed as shown in Fig. 1E, high GPR55 expression was associated with a significantly reduced disease-free survival in CRC, confirming the tumor-promoting role of GPR55 in CRC.
In conclusion, our data suggest that GPR55 plays an opposing role to CB1 in colon carcinogenesis with GPR55 acting as tumor-promoter and CB1 as tumor-suppressor. Pharmacological activation or blockade of GPR55 in cancer cell lines had no effect on proliferation. Rather, GPR55 may be responsible for alterations in the leukocyte composition of the tumor microenvironment to promote tumor growth. However, as GPR55-overexpressing SW480 cells showed a growth advantage compared to native SW480 cells, direct effects of GPR55 on tumor cells in situ are conceivable. Our observations are of particular importance when targeting of the endocannabinoid system for future therapy of colorectal cancer is considered.
Supplementary Material
Novelty and Impact.
Preclinical evidence suggests that activation of cannabinoid receptor 1 (CB1) has antitumor effects in colorectal cancer (CRC). GPR55 shares endogenous and synthetic ligands with CB1 and was found to promote CRC in this study by mechanisms that involve modulation of tumor-promoting factors. Additionally, differential regulation of DNA methylation and expression of CB1 and GPR55 was observed in human CRC patients, suggesting opposing roles of two cannabinoid-responsive receptors.
Acknowledgments
PhD candidate Carina Hasenoehrl received funding from the Austrian Science Fund (grants P25633 and KLI 521-B31) and was trained within the frame of the PhD Program Molecular Medicine of the Medical University of Graz.
Abbreviations
- AOM
azoxymethane
- CB
Cannabinoid receptor
- CD
cluster of differentiation
- COX-2
cyclooxygenase-2
- CRC
colorectal cancer
- DSS
dextran sulfate sodium
- FBS
fetal bovine serum
- GI
gastrointestinal
- GPR55
G protein-coupled receptor 55
- HBSS
Hank’s balanced salt solution
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- IL
interleukin
- LPI
L-α-lysophosphatidylinositol
- MDSC
myeloid-derived suppressor cell
- NF-κB
nuclear factor ‘kappa-light-chain-enhancer’ of activated B-cells
- PBS
phosphate-buffered saline
- PGF2α
prostaglandin F2alpha
- RPKM
reads per kilobase per million mapped reads
- STAT3
signal transducer and activator of transcription 3
- TXB2
thromboxane B2
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Author contribution
Rudolf Schicho and Rufina Schuligoi designed and supervised the study, CS, JH, and SB provided and analyzed the OncoTrack data, LB performed the bioinformatic analysis of the patient DNA methylation data. DT and NF performed the mass spectrometry, RG and EH designed and performed the bisulfite sequencing. MP performed the survival analysis, EMS designed the leukocyte analysis. CH, DF, TB, and MG performed the in vivo and in vitro experiments. CH drafted the manuscript, performed statistical analysis and interpreted the data. All authors participated in the writing of the manuscript.
References
- 1.Rabeneck L, Horton S, Zauber AG, Earle C. Colorectal Cancer. In: Gelband H, Jha P, Sankaranarayanan R, Horton S, editors. Cancer: Disease Control Priorities. Third Edition. Vol. 3. Washington (DC): International Bank for Reconstruction and Development / The World Bank; 2015. [Google Scholar]
- 2.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terzić J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–2114.e5. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 4.Hasenoehrl C, Taschler U, Storr M, Schicho R. The gastrointestinal tract - a central organ of cannabinoid signaling in health and disease. Neurogastroenterol Motil. 2016;28:1765–80. doi: 10.1111/nmo.12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Izzo AA, Sharkey KA. Cannabinoids and the gut: new developments and emerging concepts. Pharmacol Ther. 2010;126:21–38. doi: 10.1016/j.pharmthera.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Massa F, Marsicano G, Hermana H, Cannich A, Monory K, Cravatt BF, Ferri GL, Sibaev A, Storr M, Lutz B, Hermann H, et al. The endogenous cannabinoid system protects against colonic inflammation. J Clin Invest. 2004;113:1202–9. doi: 10.1172/JCI19465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Storr MA, Keenan CM, Emmerdinger D, Zhang H, Yüce B, Sibaev A, Massa F, Buckley NE, Lutz B, Göke B, Brand S, et al. Targeting endocannabinoid degradation protects against experimental colitis in mice: Involvement of CB1 and CB2 receptors. J Mol Med. 2008;86:925–36. doi: 10.1007/s00109-008-0359-6. [DOI] [PubMed] [Google Scholar]
- 8.Stančić A, Jandl K, Hasenöhrl C, Reichmann F, Marsche G, Schuligoi R, Heinemann A, Storr M, Schicho R. The GPR55 antagonist CID16020046 protects against intestinal inflammation. Neurogastroenterol Motil. 2015;27:1432–45. doi: 10.1111/nmo.12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D, Wang H, Ning W, Backlund MG, Dey SK, DuBois RN. Loss of cannabinoid receptor 1 accelerates intestinal tumor growth. Cancer Res. 2008;68:6468–76. doi: 10.1158/0008-5472.CAN-08-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharir H, Console-Bram L, Mundy C, Popoff SN, Kapur A, Abood ME. The endocannabinoids anandamide and virodhamine modulate the activity of the candidate cannabinoid receptor GPR55. J Neuroimmune Pharmacol. 2012;7:856–65. doi: 10.1007/s11481-012-9351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kargl J, Balenga N, Parzmair GP, Brown AJ, Heinemann A, Waldhoer M. The cannabinoid receptor CB1 modulates the signaling properties of the lysophosphatidylinositol receptor GPR55. J Biol Chem. 2012;287:44234–48. doi: 10.1074/jbc.M112.364109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker D, Pryce G, Davies WL, Hiley CR. In silico patent searching reveals a new cannabinoid receptor. Trends Pharmacol Sci. 2006;27:1–4. doi: 10.1016/j.tips.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Lauckner JE, Jensen JB, Chen H-YY, Lu H-CC, Hille B, Mackie K. GPR55 is a cannabinoid receptor that increases intracellular calcium and inhibits M current. Proc Natl Acad Sci U S A. 2008;105:2699–704. doi: 10.1073/pnas.0711278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juneja J, Casey PJ. Role of G12 proteins in oncogenesis and metastasis. Br J Pharmacol. 2009;158:32–40. doi: 10.1111/j.1476-5381.2009.00180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–72. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 16.Andradas C, Caffarel MM, Pérez-Gómez E, Salazar M, Lorente M, Velasco G, Guzmán M, Sánchez C, Perez-Gomez E, Salazar M, Lorente M, et al. The orphan G protein-coupled receptor GPR55 promotes cancer cell proliferation via ERK. Oncogene. 2010;30:245–52. doi: 10.1038/onc.2010.402. [DOI] [PubMed] [Google Scholar]
- 17.Piñeiro R, Maffucci T, Falasca M. The putative cannabinoid receptor GPR55 defines a novel autocrine loop in cancer cell proliferation. Oncogene. 2011;30:142–52. doi: 10.1038/onc.2010.417. [DOI] [PubMed] [Google Scholar]
- 18.Kargl J, Andersen L, Hasenohrl C, Feuersinger D, Stancic A, Fauland A, Magnes C, El-Heliebi A, Lax S, Uranitsch S, Haybaeck J, et al. GPR55 promotes migration and adhesion of colon cancer cells indicating a role in metastasis. Br J Pharmacol. 2016;173:142–54. doi: 10.1111/bph.13345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pérez-Gómez E, Andradas C, Flores JM, Quintanilla M, Paramio JM, Guzmán M, Sánchez C, Perez-Gomez E, Andradas C, Flores JM, Quintanilla M, et al. The orphan receptor GPR55 drives skin carcinogenesis and is upregulated in human squamous cell carcinomas. Oncogene. 2012;32:2534–42. doi: 10.1038/onc.2012.278. [DOI] [PubMed] [Google Scholar]
- 20.Huang L, Ramirez JC, Frampton Ga, Golden LE, Quinn MA, Pae HY, Horvat D, Liang LJ, DeMorrow S. Anandamide exerts its antiproliferative actions on cholangiocarcinoma by activation of the GPR55 receptor. Lab Invest. 2011;91:1007–17. doi: 10.1038/labinvest.2011.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balenga NA, Aflaki E, Kargl J, Platzer W, Schröder R, Blättermann S, Kostenis E, Brown AJ, Heinemann A, Waldhoer M. GPR55 regulates cannabinoid 2 receptor-mediated responses in human neutrophils. Cell Res. 2011;21:1452–69. doi: 10.1038/cr.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henstridge CM, Balenga NA, Kargl J, Andradas C, Brown AJ, Irving AJ, Sanchez C, Waldhoer M. Minireview: recent developments in the physiology and pathology of the lysophosphatidylinositol-sensitive receptor GPR55. Mol Endocrinol. 2011;25:1835–48. doi: 10.1210/me.2011-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oka S, Nakajima K, Yamashita A, Kishimoto S, Sugiura T. Identification of GPR55 as a lysophosphatidylinositol receptor. Biochem Biophys Res Commun. 2007;362:928–34. doi: 10.1016/j.bbrc.2007.08.078. [DOI] [PubMed] [Google Scholar]
- 24.Andradas C, Blasco-Benito S, Castillo-Lluva S, Dillenburg-Pilla P, Diez-Alarcia R, Juanes-Garcia A, Garcia-Taboada E, Hernando-Llorente R, Soriano J, Hamann S, Wenners A, et al. Activation of the orphan receptor GPR55 by lysophosphatidylinositol promotes metastasis in triple-negative breast cancer. Oncotarget. 2016;7:47565–75. doi: 10.18632/oncotarget.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ford LA, Roelofs AJ, Anavi-Goffer S, Mowat L, Simpson DG, Irving AJ, Rogers MJ, Rajnicek AM, Ross RA. A role for L-α-lysophosphatidylinositol and GPR55 in the modulation of migration, orientation and polarization of human breast cancer cells. Br J Pharmacol. 2010;160:762–71. doi: 10.1111/j.1476-5381.2010.00743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marisa L, de Reyniès A, Duval A, Selves J, Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D, Ayadi M, Kirzin S, et al. Gene Expression Classification of Colon Cancer into Molecular Subtypes: Characterization, Validation, and Prognostic Value. PLoS Med. 2013;10:e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schütte M, Risch T, Abdavi-Azar N, Boehnke K, Schumacher D, Keil M, Yildiriman R, Jandrasits C, Borodina T, Amstislavskiy V, Worth CL, et al. Molecular dissection of colorectal cancer in pre-clinical models identifies biomarkers predicting sensitivity to EGFR inhibitors. Nat Commun. 2017;8:14262. doi: 10.1038/ncomms14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 29.Neufert C, Becker C, Neurath MF. An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat Protoc. 2007;2:1998–2004. doi: 10.1038/nprot.2007.279. [DOI] [PubMed] [Google Scholar]
- 30.Henstridge CM, Balenga NA, Ford LA, Ross RA, Waldhoer M, Irving AJ. The GPR55 ligand L-alpha-lysophosphatidylinositol promotes RhoA-dependent Ca2+ signaling and NFAT activation. FASEB J. 2009;23:183–93. doi: 10.1096/fj.08-108670. [DOI] [PubMed] [Google Scholar]
- 31.Radnai B, Sturm EM, Stančić A, Jandl K, Labocha S, Ferreirós N, Grill M, Hasenoehrl C, Gorkiewicz G, Marsche G, Heinemann A, et al. Eosinophils contribute to intestinal inflammation via chemoattractant receptor-homologous molecule expressed on Th2 cells, CRTH2, in experimental Crohn’s disease. J Crohn’s Colitis. 2016;10:1087–95. doi: 10.1093/ecco-jcc/jjw061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heitzer E, Artl M, Filipits M, Resel M, Graf R, Weißenbacher B, Lax S, Gnant M, Wrba F, Greil R, Dietze O, et al. Differential survival trends of stage II colorectal cancer patients relate to promoter methylation status of PCDH10, SPARC, and UCHL1. Mod Pathol. 2014;27:906–15. doi: 10.1038/modpathol.2013.204. [DOI] [PubMed] [Google Scholar]
- 33.Grivennikov S, Karin E, Terzic J, Mucida D, Yu G-Y, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, Karin M. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–13. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peddareddigari VG, Wang D, Dubois RN. The tumor microenvironment in colorectal carcinogenesis. Cancer Microenviron. 2010;3:149–66. doi: 10.1007/s12307-010-0038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chun E, Lavoie S, Michaud M, Gallini CA, Kim J, Soucy G, Odze R, Glickman JN, Garrett WS. CCL2 Promotes Colorectal Carcinogenesis by Enhancing Polymorphonuclear Myeloid-Derived Suppressor Cell Population and Function. Cell Rep. 2015;12:244–57. doi: 10.1016/j.celrep.2015.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katoh H, Wang D, Daikoku T, Sun H, Dey SK, DuBois RN. CXCR2-Expressing Myeloid-Derived Suppressor Cells Are Essential to Promote Colitis-Associated Tumorigenesis. Cancer Cell. 2013;24:631–44. doi: 10.1016/j.ccr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiurchiù V, Lanuti M, De Bardi M, Battistini L, Maccarrone M, De Bardi M, Battistini L, Maccarrone M. The differential characterization of GPR55 receptor in human peripheral blood reveals a distinctive expression in monocytes and NK cells and a proinflammatory role in these innate cells. Int Immunol. 2015;27:153–60. doi: 10.1093/intimm/dxu097. [DOI] [PubMed] [Google Scholar]
- 38.Nervi C, De Marinis E, Codacci-Pisanelli G. Epigenetic treatment of solid tumours: a review of clinical trials. Clin Epigenetics. 2015;7:127. doi: 10.1186/s13148-015-0157-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindau D, Gielen P, Kroesen M, Wesseling P, Adema GJ. The immunosuppressive tumour network: Myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. Immunology. 2013;138:105–15. doi: 10.1111/imm.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosoline M, Camus M, Berger A, Wind P, Zinzindohoué F, et al. Type, Density, and Location of Immune Cells Within Human Colorectal Tumors Predict Clinical Outcome. Science. 2006;313:1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 41.Li H, Liu K, Boardman LA, Zhao Y, Wang L, Sheng Y, Oi N, Limburg PJ, Bode AM, Dong Z. Circulating Prostaglandin Biosynthesis in Colorectal Cancer and Potential Clinical Significance. EBioMedicine. 2015;2:165–71. doi: 10.1016/j.ebiom.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keightley MC, Sales KJ, Jabbour HN. PGF2alpha-F-prostanoid receptor signalling via ADAMTS1 modulates epithelial cell invasion and endothelial cell function in endometrial cancer. BMC Cancer. 2010;10:488. doi: 10.1186/1471-2407-10-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez-Lopez E, Flashner-Abramson E, Shalapour S, Zhong Z, Taniguchi K, Levitzki A, Karin M. Targeting colorectal cancer via its microenvironment by inhibiting IGF-1 receptor-insulin receptor substrate and STAT3 signaling. Oncogene. 2015;35:1–11. doi: 10.1038/onc.2015.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Falasca M, Ferro R. Role of the lysophosphatidylinositol/GPR55 axis in cancer. Adv Biol Regul. 2016;60:88–93. doi: 10.1016/j.jbior.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 45.Jones PA, Baylin SB. The Epigenomics of Cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–59. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.