Abstract
Over the last decade, our appreciation for the contribution of resident gut microorganisms—the gut microbiota—to human health has surged. However, progress is limited by the sheer diversity and complexity of these microbial communities. Compounding the challenge, the majority of our commensal microorganisms are not close relatives of Escherichia coli or other model organisms and have eluded culturing and manipulation in the laboratory. In this Review, we discuss how over a century of study of the readily cultured, genetically tractable human gut Bacteroides has revealed important insights into the biochemistry, genomics and ecology that make a gut bacterium a gut bacterium. While genome and metagenome sequences are being produced at breakneck speed, the Bacteroides provide a significant ‘jump-start’ on uncovering the guiding principles that govern microbiota–host and inter-bacterial associations in the gut that will probably extend to many other members of this ecosystem.
Microbial life thrives in the anaerobic environment of the human gastrointestinal tract, which represents one of the densest microbial communities known in nature. With a collective microbial gene count (the ‘microbiome’) exceeding that of the host by more than 100-fold, the microorganisms living within our gut (the ‘microbiota’) in many ways represent an additional metabolic organ that contributes substantially to our physiology—including nutrient absorption, metabolism, disease resistance and susceptibility, and response to xenobiotics1–5. More than a decade’s worth of 16S rRNA sequencing and metagenomic analyses of hundreds of human gut microbiomes collected over numerous geographical locations have shown that each individual harbours a unique assemblage of microbial life that first establishes itself when we are born, changes markedly over the first two to three years of life, and then remains fairly stable throughout our adult lives6–9. These communities include representatives from all three domains of life, with hundreds of species and strains occupying and competing for limited space and resources.
Most healthy adult microbiota are dominated by just two bacterial phyla—the Gram-positive Firmicutes (many genera) and the Gram-negative Bacteroidetes (primarily Bacteroides, Alistipes, Parabacteroides, and Prevotella)—that together comprise the majority of the bacterial taxa in the gut of many individuals10 (Fig. 1). Other taxonomic groups include the Proteobacteria, Actinobacteria, Fusobacteria, Verrucomicrobia, methanogenic archaea, Eukarya (protists and fungi), and other more transient colonizers8,10,11. The taxa found in healthy individuals tend to be most similar among family members (for example, parents and their children) and most dissimilar across cultures and geographical space (for example, Western versus non-Western societies)9,12 (Fig. 1). While there are many external factors that help to explain this pattern, the precise underlying mechanisms that account for interpersonal microbiota variation are still largely mysterious— and while the majority of our gut microbial inhabitants have been cultured, most are not yet amenable to genetic manipulation, making efforts to answer questions about how they operate much more challenging13–17.
Figure 1. Global distributions and abundances of human gut microorganisms at the phylum and genus levels.
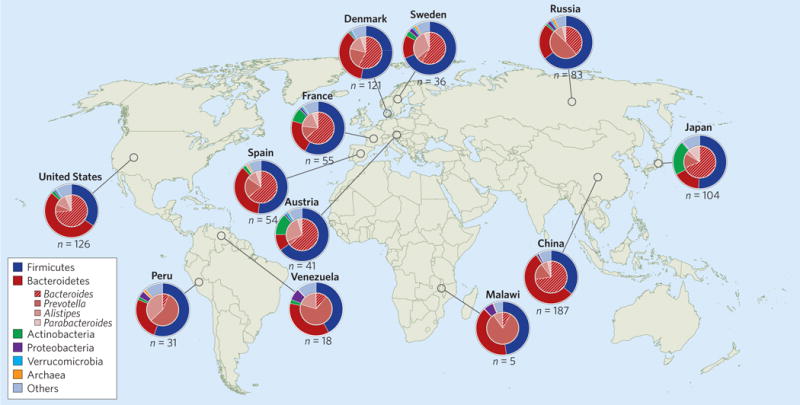
Of the gut microbial taxa represented among healthy adult human populations, the Gram-positive Firmicutes and the Gram-negative Bacteroidetes appear to be universal. While different microbiota can appear similar at the phylum level (for example, Sweden versus Russia), they often differ significantly at lower taxonomic levels. In this figure, each pie chart represents an average per cohort, with outer pies depicting phylum-level taxa and Archaea, and inner pies depicting genus-level taxa within Bacteroidetes. Inter-personal variation within each country is also significant. Data adapted from ref. 147.
Fortunately, some of the most abundant species in our gut are both readily cultured and genetically tractable. As a prominent genus within the Bacteroidetes phylum (Fig. 1), the obligately anaerobic Bacteroides have become a major focus of human gut microbiology over the course of a century since their discovery18–20. They live and grow exclusively in the gastrointestinal tracts of mammals, suggesting strong adaptation to the gut environment21. As commensals and mutualists, they can establish stable, long-term associations with their human hosts and confer numerous health benefits8. Because they are major constituents of the microbiota, are widespread across human populations, are highly adapted to life in the gut, and form a foundational part of the microbial food webs in the gut, the Bacteroides are an ideal model group from which to discover the fundamental principles that determine how our core gut bacteria colonize and persist not just over the life of their host, but over vast evolutionary timescales.
This Review will use the Bacteroides as a window into the obscure world of the gut microbiota. We will examine the qualities that enable them to tolerate and persist through various environmental changes or insults, assess how they interact to establish cooperative or antagonistic relationships with their neighbours, and discuss the complicated relationships our commensals can have with their host that may ultimately result in either immunological tolerance or disease. The Bacteroides field enjoys a multiple-decade ‘jump-start’ over our understanding of other gut microorganisms: this Review highlights the general principles that have emerged and probably apply to much of the microbiome.
Bacteroides reveal microbiota biochemistry
Gut-associated microbial communities are compositionally distinct from other natural microbial communities and are a product of the unique environment in which they evolved22. The human intestinal tract reaches an average of 7.5–8.5 m in length in most adult humans, presenting more than 32 m2 of surface area (when accounting for intestinal villi) for host–microbial interaction23,24. The gut is divided into two major sections: the small intestine (between the pyloric and ileocecal sphincters), where most host nutrient absorption occurs, and the large intestine, where the vast majority of gut microorganisms reside. As invading microorganisms pass through the intestinal tract, they face a gauntlet of environmental challenges to sustained colonization25. Firstly, a strong pH gradient spans from the proximal small intestine (pH ~2–5) to the distal colon (pH ~6 –7)26; secondly, a three-dimensional oxygen gradient occurs from the proximal small intestine and epithelium (highest O2 concentration) to the large intestine and lumen (lowest O2 concentration)27; thirdly, the host immune system presents several added challenges to colonizers, with host secretory immunoglobulin A (sIgA) and antimicrobial peptides continuously secreted from the epithelium into the small intestine, and even more so during inflammation28,29; and lastly, constant peristalsis of the intestinal smooth muscles ensures that any ingested microorganism must experience and survive this maelstrom of challenges to successfully colonize the distal gut. It is perhaps a result of the comparatively harsher conditions in the small intestine that the bulk of bacterial growth occurs in the large intestine, where host nutrient absorption is minimal, luminal pH is fairly neutral and oxygen levels are in the sub-micromolar range. Studies of Bacteroides have revealed multiple biochemical mechanisms to adapt and persist within this dynamic environment3,30.
To help cope with these challenges, Bacteroides can actively refine the gut environment to make it more hospitable to themselves and other microorganisms. For instance, many encode a cytochrome bd oxidase that is hypothesized to reduce intracellular oxygen levels, and by extension, gut oxygen levels, thereby permitting the growth of strict anaerobes that are otherwise killed by the presence of O2 (ref. 31–33). This ability to tolerate and reduce oxygen levels would likely aid Bacteroides in spreading to new hosts and is perhaps a major reason why they are so widespread among mammals21,34,35. They can also alter the nutritional landscape of the gut by promoting physiological changes in their host to induce the production of certain food sources, such as fucosylated glycoproteins, or by liberating fucose and sialic acid residues from glycoproteins that can be consumed by other microorganisms, including pathogens36,37. Many of these Bacteroides-dependent changes to the gut environment have been identified by comparing germfree animals that lack microorganisms of their own, with gnotobiotic counterparts (born germfree but later colonized with microorganisms) mono-associated with individual Bacteroides strains and mutants.
Although one might imagine life in the gut as replete with an excess of food choices for microorganisms, in the colon (where bacterial densities are highest), the simple, readily accessible sugars have largely already been consumed or absorbed38. Among the remains are complex, long-chain polysaccharides that are not freely absorbed and cannot be digested by the human enzymatic repertoire. For many bacteria, these complex polysaccharides are similarly intractable and cannot be transported across membranes. However, early studies established Bacteroides as adept glycan degraders, with an unusual ability to recognize and metabolize more than a dozen plant- and host-derived polysaccharides39,40. Bacteroides species accomplish this through gene clusters, which appear unique to Bacteroidetes, termed polysaccharide utilization loci (PULs). PULs can determine which metabolic niches Bacteroides can occupy, and can even determine their biogeographical locations within the intestinal tract25,41. PULs are classified as such because they contain homologues of susC and susD from the classic starch utilization system42,43 (Fig. 2a), as well as other components (glycoside hydrolases, polysaccharide lyases, glycosyltransferases, carbohydrate esterases) that are important for the breakdown of a wide variety of either plant- or host-derived glycans44. Extracellular polysaccharides are bound by outer membrane lipoproteins (SusD-, SusE- and SusF-like) and cleaved by SusG-like glycoside hydrolases into smaller oligosaccharides, which are transported through SusC-like channels into the periplasm in a TonB-dependent manner, where they are further degraded by SusA- and SusB-like enzymes into simple sugars that are taken into the cytoplasm through inner membrane permeases45 (Fig. 2a). Other lineages of gut bacteria, including the Gram-positive Actinobacteria, have far less sophisticated pathways to degrade and import dietary polysaccharides, due in large part to the absence of an outer membrane barrier. Bifidobacterium breve, for instance, uses multi-domain, cell-surface-anchored enzymes (rather than a cascade of separate enzymes) that can bind to and degrade polymers such as starch, pullulan and glycogen46.
Figure 2. Consumption and production of polysaccharides in Bacteroides.
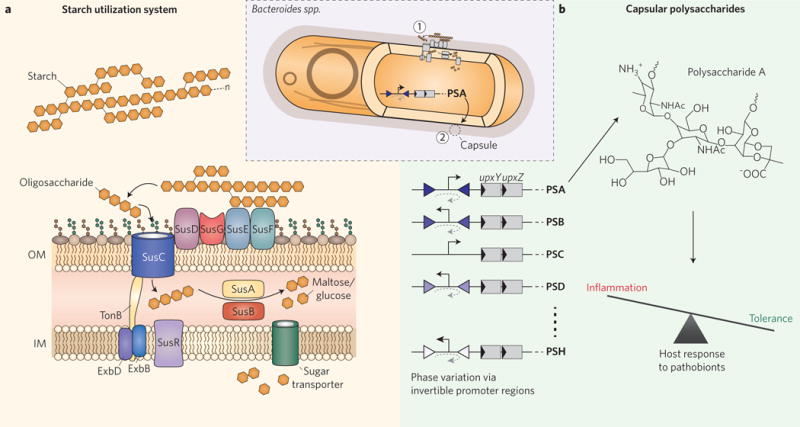
a, The starch utilization system. Starch-binding lipoproteins in the outer membrane (SusDEF) bind to and immobilize large extracellular starch polymers, which are then broken into smaller oligosaccharides by the α-amylase SusG. The TonB-dependent transporter SusC carries these oligosaccharides into the periplasm, where the α-amylase SusA and the α-glucosidase SusB break them down further into disaccharides (maltose) or monosaccharides (glucose) for import into the cytoplasm through a sugar-transporting permease. The presence of maltose in the periplasm triggers the regulator SusR to promote the expression of additional Sus components. Bacteroides typically encode 80 or more PULs within their genomes. OM, outer membrane; IM, inner membrane; n, variable polysaccharide size. The starch utilization system is indicated by (1) in the inset. b, Capsule biosynthesis and host protection. Capsular polysaccharide biosynthesis in Bacteroides, such as B. fragilis, is regulated by invertible promoter regions (inverted triangles), trans-locus inhibitors (UpxZ, where x is a, b, c, and so on, depending on the locus), and cis-acting transcriptional anti-terminators (UpxY). Polysaccharide A can promote immunological tolerance to pathobionts such as Helicobacter hepaticus, thus protecting the host from pathogen-induced colitis. The cell capsule is indicated by (2) in the inset.
Comparative genomic analyses have identified dozens of distinct PULs in each species of Bacteroides. For example, Bacteroides thetaiotaomicron, the first member of the Bacteroides to have its genome sequenced, dedicates nearly a fifth of its 6.26 Mbp genome to 88 distinct PULs44,47,48. Rather than expressing all PULs concomitantly to capture whatever polysaccharides might be available in their immediate surroundings, Bacteroides tightly regulate their expression via a multitude of SusR-like regulators, hybrid two-component systems and trans-envelope signalling pathways involving extra-cytoplasmic σ/anti-σ factors47,49–53. Perhaps as a consequence of the need to partially degrade long polysaccharide chains prior to transport across the outer membrane, B. thetaiotaomicron and other gut Bacteroidetes have managed to couple transcription of appropriate PULs with the abundance of their corresponding polysaccharide breakdown intermediates, rather than the abundance of the extracellular polysaccharides themselves54. This allows gut bacteria to rapidly adapt to changes in nutrient availability and ease the process of switching between carbon sources54. This form of regulation represents a departure from classical feedback control in which the first committed step of a pathway is regulated by the product of the final step of the pathway. Whether this strategy—coupling transcriptional regulation to the rate of intermediary metabolite breakdown via a non-allosteric mechanism—is a common feature of gut bacteria remains unknown; however, other variations on this paradigm exist within Bacteroides themselves55. Other gut anaerobes, such as Gram-positive Firmicutes, degrade polysaccharides via multi-enzyme complexes called cellulosomes and amylosomes, but their regulation appears tied to intact polysaccharides56,57.
Because PUL activation is a product of the gut environment, a diet rich in plant-derived polysaccharides and glycoproteins will lead to a temporary upregulation of PULs to degrade those dietary nutrients. However, when those substrates are scarce, such as during periods of fasting or on certain low-fibre diets (as in a high-sugar, high-fat Western diet), glycophilic Bacteroides switch their transcriptional profile to consume host-derived glycans instead58. These temporary switches are probably a major factor that allows certain bacteria to initially colonize the gut during infancy and persist there over the course of our lifetime. While Bacteroides can subsist primarily on host glycans during the relatively short period of host infancy, they can ultimately go extinct when forced to subsist on a low-fibre diet over the timespan of multiple host generations, according to recent work using animal models59.
Bacteroides are not merely adept at degrading polysaccharides; they assemble them, as well. Their cell surface is coated with a multitude of glycoproteins and capsular polysaccharides that can extend hundreds of nanometres from the cell surface and contribute to the overall fitness of these bacteria in their natural environment50,60. These surface structures mark the interface between individual members of the microbiota (and their host), and thus are likely to play an important role during microorganism–microorganism and microorganism–host interactions in the gut. This is certainly the case for numerous pathogens whose capsules aid in immune system evasion via mechanisms that include disruption of neutrophil chemotaxis (for example, Salmonella typhi61) and evasion of host complement system (for example, Klebsiella pneumoniae62). By contrast, Bacteroides use a process called phase variation that controls the expression of their capsular polysaccharide biosynthetic loci in an on/off manner63 (Fig. 2b). Bacteroides fragilis, for example, produces distinct capsular polysaccharides from eight biosynthetic loci, whose expression is tightly regulated by invertible promoter regions and trans-locus inhibitors that limit simultaneous expression of multiple loci, thereby allowing isogenic populations of B. fragilis to collectively display a wide array of surface architectures63–67 (Fig. 2b). This contrasts with oral Bacteroidetes, which do not phase vary their capsules, suggesting that this process is important to life in the gut66. The precise ecological role of phase variation in this environment remains unclear; however, Bacteroides seem to require the ability to produce a minimum of one capsular polysaccharide to persist within the mammalian gut, as an acapsular B. fragilis mutant is rapidly outcompeted by the wildtype in gnotobiotic mice68. Phase variation and expression of multiple capsular polysaccharides can provide a degree of protection to cells by minimizing binding of sIgA, as well as offering a more general physical protection to the cell69. However, the capsule can also be a hindrance to the cell by acting as a diffusion barrier to large nutrients, such as branched dietary polysaccharides50. To counteract this diffusion barrier, Bacteroides encode surface-exposed lipoproteins (SusD-, SusE- and SusF-like homologues) that bind to and immobilize large polysaccharide chains for degradation and import into the cell49. The capsular composition of gut bacteria also has a major impact on the host. One zwitterionic capsular polysaccharide in particular from B. fragilis, polysaccharide A (PSA), has been shown to promote immunological tolerance in gnotobiotic mice and even suppress the development of experimental colitis induced by the pathobiont Helicobacter hepaticus70–72 (Fig. 2b).
Bacteroides and the ecology of the microbiome
The microbial food webs in our gut are largely built from the nutrients we consume11,45,73–75. Thus, host diet plays a fundamental role in determining the composition of our gut microbiota, including during infancy6,76–78. Most infant diets generally consist of human breast milk and/or bovine milk-based formula, which are composed of a complex mixture of simple sugars (for example, lactose, galactose, glucose, fucose, sialic acid, and N-acetylglucosamine) and complex oligosaccharides, often with appended fucose or sialic acid moieties79,80. Given the link between our diet and microbiota, there is a growing interest in identifying dietary factors that can function as prebiotics to help promote a healthy gut microbiota for patients suffering from diseases such as inflammatory bowel disease (IBD), or infants suffering from malnutrition. A recent study that analysed the breast milk of post-partum mothers in Malawi found a link between low levels of sialylated milk oligosaccharides and higher degrees of growth stunting in infants81. By transplanting the micro-biota from stunted Malawian infants into germfree mice (or a subset of cultured microorganisms into germfree piglets) fed a prototypical Malawian diet supplemented with or without sialylated bovine milk oligosaccharides (S-BMO), Charbonneau and colleagues observed microbiota-mediated augmentations in lean body mass, bone density and host nutrient absorption compared to controls. Out of all species present, B. fragilis and Escherichia coli showed the strongest transcriptional responses to S-BMO, with B. fragilis being its primary metabolizer (breaking sialyllactose into sialic acid, galactose and glucose) and E. coli being a secondary consumer of these mono-saccharides. Surprisingly, none of the bacteria that showed a different transcriptional response to the presence of S-BMO belonged to the genus Bifidobacterium, members of which have long been classified as probiotics due to their positive associations with human health81–83. Notably, colonizing germfree mice with B. fragilis and E. coli alone on the S-BMO-supplemented Malawian diet did not recapitulate the growth augmentations observed in the presence of the complete community, suggesting that more intricate microbial interactions and food webs are required and underscoring the need to mechanistically understand these interactions to better design effective prebiotic and probiotic regimes81. As model gut commensals, the Bacteroides could provide important tools for developing a new generation of probiotics (see Box 1).
Box 1. Emerging tools to study the microbiome.
Studying multispecies microbial communities can be exceedingly challenging given their characteristic degree of biological complexity. For this reason, there is an ever-growing demand for the development of improved tools with which to dissect our gut microbiota. Here, we highlight some of the tools developed or improved upon in recent years, which have facilitated the discovery of much of what we now understand about our microbiomes.
Gnotobiotics
The gut microbiome field benefits greatly from the use of gnotobiotic (known life) animal models—particularly gnotobiotic mice. These animals enable researchers to conduct a wide array of experiments that cannot be performed on human subjects. Gnotobiotic mice are born germfree (without any microorganisms of their own); from this empty stage, researchers can precisely control the composition of their microbiota by building simplified, defined communities consisting of known species and mutants within these species, and can perform time series assays comparing multiple conditions and controls in parallel. Defined communities can readily be quantified by quantitative PCR or techniques such as COmmunity PROfiling by Sequencing (COPRO-Seq), which uses highly parallel sequencing of genomic DNA extracted from faecal samples, followed by mapping to reference genomes134,135. Important caveats of gnotobiotic animal studies have been reviewed elsewhere, and include their altered anatomy and underdeveloped immune system, and the possibility that adult germfree mice receiving a transplanted microbiota may lack microbial exposure during important developmental periods. Mice that receive a microbiota from human donors generally do not exactly mirror their donor due to diet, anatomy or other differences136,137.
High-throughput culturing and sequencing
While human gut microbial communities can be transplanted into germfree mice, their biological complexity makes it difficult to parse the contributions of key species to an ecological function or host phenotype137. For these and other reasons, it is often desirable to simplify and define the members of a community in order to combine them in customizable ways depending on the specific goals of a study. High-throughput anaerobic culturing is a method that creates arrayed libraries of individual species from a given donor13,14,16,138. Combining this technique with gnotobiotics opens the door to identifying specific commensals that cause or exacerbate disease phenotypes or are responsible for specific host or microbiota traits. For instance, by combining gnotobiotics with arrayed anaerobic culturing of whole human gut communities from patients with IBD, researchers were able to identify individual strains with high levels of IgA coating (assessed by flow cytometry) using 16S sequencing-based IgA-Seq139. Gnotobiotic mice colonized with defined communities of these high-IgA-associated strains were found to be more susceptible to colitis139.
Commensal genetics and fitness
Though most of our microbiota is currently culturable, only a fraction is amenable to genetic manipulation, making it fundamentally more difficult to dissect how our individual gut microorganisms function in their environment. Of those that are culturable and genetically tractable, the Bacteroides have given us some of our most important glimpses into microbial life in our gut. Building on the pioneering work of Abigail Salyers140, new tools combine high-throughput genetic screens, as in the transposon-based INSeq method, with gnotobiotics in order to explore functional requirements of microorganisms in the gut89,141. Targeted gene deletion studies can then be applied to verify and validate the phenotypes of important mutants, thereby providing invaluable steps towards uncovering not only the location of key genomic regions within gut commensals, but also the function of the gene products encoded therein. Systems biological approaches have also been developed recently for use in Bacteroides and other gut bacteria, including inducible expression systems that can be turned on or off both in vitro and in gnotobiotic animals upon sensing specific signals142,143. Enhancements and modifications to these systems will be critical for efforts to monitor the gut environment using biosensors, or even to deliver drug therapies in a more targeted manner144.
Microscopy and microbiota visualization
The mammalian gut is a fascinatingly complex microbial environment, but for practical reasons we rarely get to peek inside. This severely limits our understanding of the spatial distributions and physical interactions between microorganisms within our gut. However, advances in immunofluorescence labelling, cellular substrate-modifying ‘click’ chemistry, cross-section and whole-body imaging and image analysis software have begun to shed new light on these problems145,146. These methods will be critical for studying spatial co-localization between species within the gut (that is, niche sharing/exclusion), identifying mucosa-associated species that may interact physically with host cells and effects of diet, community composition, antibiotics, inflammation, or infection on the microbial landscape in the gut.
Ecological interactions are common throughout the microbiota in which members of these communities respond transcriptionally and metabolically to the presence and activity of one another. Reductionist approaches involving gnotobiotic mice have helped to uncover these responses (see Box 1). For instance, colonizing germfree mice with a single representative of the Bacteroidetes (B. thetaiotaomicron) and a single representative of the Firmicutes (Eubacterium rectale) showed that in response to the presence of one another, B. thetaiotaomicron upregulates certain PULs for catabolism of host glycans (genes which E. rectale lacks), while E. rectale upregulates the expression of genes for fermenting amino acids and importing simple sugars84. Similar responses occur in co-colonization experiments involving B. thetaiotaomicron and the probiotic Firmicutes, Lactobacillus casei, or the probiotic Actinobacterium, Bifidobacterium longum, suggesting that transcriptional responses provide a valuable window into niche segregation in the gut for both genetically tractable and intractable members of this community85.
Some microorganisms directly depend on others for resources that they themselves cannot produce. A class of metabolic cofactors called corrinoids (the most famous of which is vitamin B12) is a central component of the fitness landscape in the gut, as they are required in many cases for methionine synthesis and other metabolic pathways86,87. Several gut-resident Firmicutes, Actinobacteria and Proteobacteria provide these cofactors to the rest of the community by encoding the genes for their de novo synthesis. However, many members of the microbiota, including the Bacteroidetes, lack some or all of the genes for the synthesis of these large and complex metabolites, and instead rely exclusively on corrinoid transporters to extract them from the extracellular milieu88,89. A key corrinoid transport system was identified in B. thetaiotaomicron using a transposon-based screening method called INsertion Sequencing (INSeq). Transposon mutants harbouring insertions within this corrinoid transporter locus of B. thetaiotaomicron are unable to compete with wildtype cells in gnotobiotic mice89. Moreover, the requirement for this system in more complex gut microbial communities strongly correlates with the abundance of a corrinoid-producing commensal, Ruminococcus obeum (that is, as the abundance of R. obeum decreases, the fitness of corrinoid transporter mutants also decreases). This suggests that the presence of corrinoid-producing microorganisms in the gut can affect community structure overall, and Bacteroides abundance in particular, and highlights how the genetic tractability of Bacteroides enables identification of interactions between gut bacteria, the molecular ‘currency’ of these interactions, and the specific proteins involved89 (see Box 1).
While members of different phyla clearly compete for shared resources in the gut, the stiffest competition appears to occur between close evolutionary relatives that share many of the same genes and functional capacities within the gut ecosystem. In the most extreme case, human gut Bacteroides are unable to super-colonize a mono-associated gnotobiotic mouse that already carries members of that species, suggesting that all potential niches for the invading bacteria are occupied by its established sister cells41 (Fig. 3a). However, a species of Bacteroides can easily colonize gnotobiotic mice that already carry a different species of Bacteroides, suggesting limited niche overlap (Fig. 3b). Using an elegant genetic screen, Lee and colleagues identified a unique form of PUL, termed commensal colonization factor (CCF), which is required for resident bacteria to prevent invasion by other members of the same species41 (Fig. 3c). In B. fragilis, a ccf mutant displayed reduced colonization of colonic crypts compared to wildtype and reduced horizontal transmission to other gnotobiotic mice, suggesting that the crypts can serve as a microbial reservoir that seeds the gut lumen, allowing long-term colonization of the gut41 (Fig. 3c). However, general mechanisms by which niche occupation in the gut takes place remain unclear. One possibility is niche specialization, in which each community member actively fills a distinct niche by expressing some unique set of genes, allowing for a specialized function within the community. Another possibility is incomplete niche overlap: by harbouring a partially overlapping assemblage of genes shared by many members of the community, the metabolic capacity of each species is slightly different, thus reducing direct competition between any two neighbouring species and instead making each neighbour a partial competitor for shared resources. These two explanations are not mutually exclusive, but rather fall along a continuum of influence that depends on environmental context. In general, the simpler a community is in terms of biological diversity, the more likely that niche specialization is to be important. However, in more complex communities consisting of dozens to hundreds of species, incomplete niche overlap is likely to become more important.
Figure 3. Commensal colonization of colonic crypts.
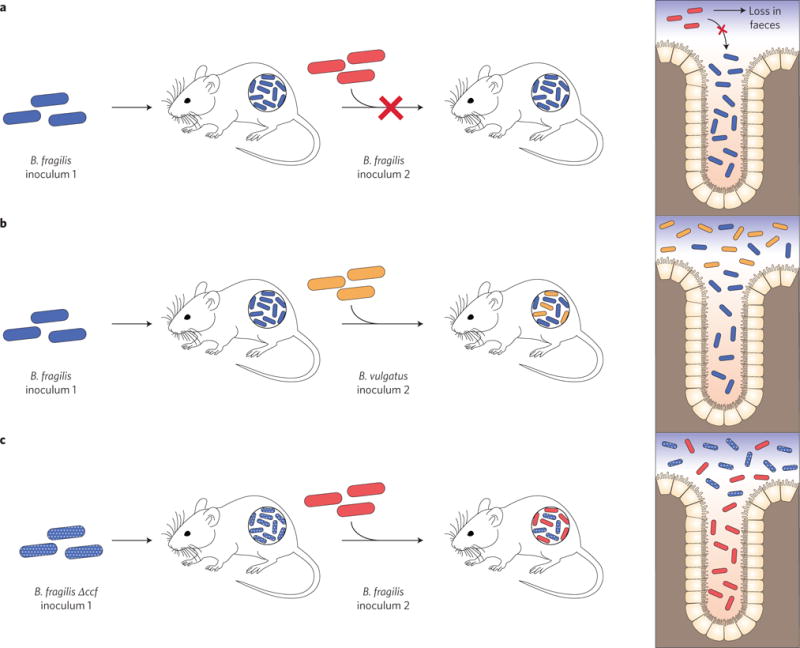
a, Colonization exclusion. Stable colonization of germfree mice with B. fragilis prevents the subsequent colonization by isogenic B. fragilis sister cells, suggesting that the initial population of B. fragilis has occupied all niches available to it and are not subject to forces of random displacement. b, Co-colonization. Stable colonization of germfree mice with B. fragilis does not prevent invasion by another species of Bacteroides, suggesting limited niche overlap between the two species. c, Niche displacement. Stable colonization of germfree mice with a B. fragilis mutant lacking genes for a specialized PUL named commensal colonization factor (CCF) can be disrupted by subsequent colonization by wildtype B. fragilis cells. B. fragilis ccf mutants are defective in their ability to colonize deep within the colonic crypts, which are thought to serve as a microbial reservoir that seeds the gut lumen and promotes long-term colonization of the gut. Figure based on data from ref. 41.
The line between cooperator and competitor in the gut is far from clear and many species appear to play both roles simultaneously. For instance, we know that some Bacteroides species compete with one another for dietary polysaccharides, such as inulin and xylan in co-culture experiments, yet share some of their polysaccharide harvesting machinery (for example, glycoside hydrolases) with neighbouring species either through the secretion of outer membrane vesicles (OMV)90 or via the extracellular activity of the cell-tethered enzymes themselves91 (Fig. 4a). OMV secretion by members of the Bacteroidetes is a well-documented phenomenon, but its ecological impact is still uncertain as the genes responsible for OMV formation have yet to be identified92, 93. Surprisingly, these OMVs can serve as shuttles for public goods between genetically distinct members of the microbiota, thereby allowing other species to expand their palate for carbon sources without directly encoding the genes to do so (Fig. 4a). OMVs can also serve as delivery vessels between microorganisms and their host, as is the case for the anti-inflammatory microbial compound PSA, produced by B. fragilis70,71,94. Recently, an OMV-independent mechanism for nutrient sharing has been observed between related species of Bacteroides. Bacteroides ovatus encodes two extracellular inulin glycoside hydrolases that are dispensable for its own growth on inulin, but required to promote the growth of Bacteroides vulgatus, which lacks these enzymes91. B. ovatus appears to benefit reciprocally from B. vulgatus through an unknown mechanism91. These polysaccharide-sharing mechanisms suggest that cooperation among members of the gut microbiota (within and probably beyond Bacteroides) is not merely limited to production and consumption of metabolic waste products and help to shed light on the processes governing ecological function and stability in the gut.
Figure 4. Mechanisms of inter-bacterial cooperation and antagonism among Bacteroides.
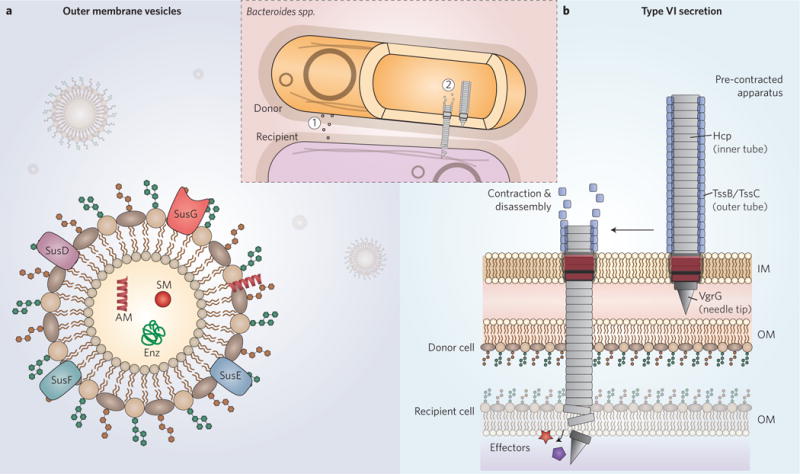
a, Outer membrane vesicles. OMVs are generated and released by Bacteroides both in vitro and in vivo. They can act as carriers of public goods, including Sus-like components, small molecule (SM) cofactors, anti-inflammatory capsule components, such as PSA, antimicrobial (AM) proteins, such as BSAP-1, and other enzymes (Enz). Outer membrane vesicles are indicated by (1) in the inset. b, Bacteroidetes type VI secretion systems. The bacterial T6SS is a contractile molecular lancet that rapidly delivers toxic effector proteins into neighbouring cells in a manner dependent on cell-to-cell contact. The outer tube, which consists of TssB/TssC multimers in a high-energy state, contracts to a low-energy state that forces the rigid inner tube, composed of Hcp multimers, out of the donor cell and into a nearby recipient cell. The needle tip can carry bound effector proteins that mediate cell damage and death in susceptible recipient cells lacking cognate immunity proteins. The Bacteroidetes T6SS lacks a subset of components found in Proteobacterial T6SSs, and carries novel components of unknown function or structure (not depicted). IM, inner membrane; OM, outer membrane. The T6SS is indicated by (2) in the inset.
Some interactions between gut microorganisms, however, appear to be purely antagonistic, or even territorial via the secretion of toxic proteins. Bacteriocins are freely diffusible secreted toxins with a variety of catalytic functions including pore formation, nucleic acid degradation and inhibition of cell wall synthesis, and are typically produced by and target closely related strains and species95. For example, strains of the Firmicutes pathobiont Enterococcus faecalis carrying a conjugative, bacteriocin-encoding plasmid can displace susceptible strains of E. faecalis in mice, implying a role for bacteriocins in niche displacement in the gut96. Strains of Bacteroides also produce and secrete antimicrobial proteins, such as Bacteroidales secreted antimicrobial protein 1, or BSAP-1 (B. fragilis; loaded into OMVs; Fig. 4a) and BSAP-2 (Bacteroides uniformis), which contain membrane attack complex/perforin (MACPF) domains and target closely related strains in vitro and in vivo97,98. To resist sister-cell killing, bacteriocin producers can directly encode immunity factors that protect against cytotoxicity, or in the case of Bacteroides BSAP producers, encode a non-targeted orthologue of the toxin receptor adjacent to the toxin genes themselves98,99. In this way, toxin/immunity mechanisms can readily spread by horizontal gene transfer.
The high bacterial cell densities in the gut are likely to substantially enhance resource competition among neighbouring cells. Many gut Bacteroidetes seem to have found a way to reduce local competition for shared resources between non-genetically identical cells, in part through the use of type VI secretion systems (T6SSs)100,101 (Fig. 4b). These multi-protein machines rapidly inject antibacterial toxins called effectors into adjacent cells upon direct cell-to-cell contact102. The T6SSs of Bacteroidetes are evolutionarily distinct from those of the well-studied Proteobacteria (Bacteroidetes are missing at least five core components and encode several novel components of unknown function), and collectively segregate into three distinct architectures, two of which are present on integrative conjugative elements that probably facilitate their lateral spread to other species101,103. The effector repertoires among gut Bacteroidetes vary widely from species to species and even among strains of the same species. For instance, B. fragilis strains each encode 2 of at least 12 putative effectors in conserved positions within the T6SS locus103,104. To date, four of these effectors have been confirmed to have antibacterial properties, though their mechanisms of toxicity and cellular targets are unknown104,105. T6SS effectors have been best studied in Proteobacteria, such as Pseudomonas aeruginosa and Vibrio cholerae, and can have wide-ranging targets, including Gram-negative peptidoglycan (amidases, glycosidases, muramidases), cell membranes (phospholipases, pore formation), nucleic acids (DNases), and dinucleotides (glycohydrolases)102,106. While some putative effectors within members of the Bacteroidetes are also predicted to have these toxic activities based on shared homology, others have no known or predicted function, or recognizable domains103,104. T6S-mediated antagonism appears to be a constant feature of life in the gut. Using mathematical modelling based on experimental measurements, B. fragilis was recently shown collectively to inject its effectors more than 109 times per minute per gram of gut content in gnotobiotic mice104. In contrast to diffusible bacteriocins or host-secreted antimicrobial peptides and sIgA, the requirement of direct contact limits the target range of this system to cells in the immediate vicinity. Thus, it is possible that T6SSs serve as a means for gut bacteria to limit local competition for shared resources while preserving the overall biodiversity in the gut, thereby allowing T6SS ‘donor’ bacteria to continue to benefit from diffusible metabolites produced by more distant cells that are out of range of the T6SS needle. Importantly, T6SS activity not only directly benefits the donor bacterium, but can also protect the host from enteric pathogens. Hecht and colleagues recently demonstrated that colonization of specific pathogen-free mice by enterotoxigenic B. fragilis could be inhibited in a T6SS-dependent manner by a commensal strain of B. fragilis encoding the T6SS effector Bte2 (Bacteroides type VI effector 2), thereby reducing disease severity107.
Bacteroides modulation of the host
The human gastrointestinal tract is in perpetual contact with microorganisms. Collectively, these microorganisms have profound impacts on the host immune system, whether in the context of pathogenesis by infectious agents, inflammatory bowel diseases, cancer or autoimmunity. In large part, what distinguishes friend from foe within the microbiota is determined by these interactions with host immunity, and often depends on a multitude of extrinsic factors. The Bacteroides illustrate how one bacterial group can wear three different hats—commensal, mutualist or even pathobiont— depending on details of biogeographical location within the host, microbiota composition and the availability of certain nutrients.
The impact of the gut microbiota on immune system development is perhaps most apparent from studies contrasting immunity in conventional animals and germfree counterparts. Germfree animals not only have an underdeveloped immune system—marked by smaller immune-associated tissues such as Peyer’s patches, lamina propria and mesenteric lymph nodes, and an imbalance between T helper 1 (Th1) and Th2 cells in favour of the latter, as well as reduced levels of sIgA in the gut, among other phenotypes—but are also more susceptible to infection by enteric pathogens, such as Shigella flexneri and Salmonella typhimurium108–111. Colonization of germfree mice by Bacteroides corrects many of these immune defects, suggesting important interactions between Bacteroides and the host112. One such interaction is mediated by the capsular polysaccharide component PSA from B. fragilis, as discussed above70,94,113 (Fig. 2b). A recent study has identified additional gut commensals that produce zwitterionic capsular polysaccharides, including a strain of Bacteroides cellulosilyticus, with similar anti-inflammatory properties to PSA114.
Other Bacteroides surface structures have also been shown to have immunomodulatory effects. Vatanen, Kostic and d’Hennezel et al. linked the prevalence of type 1 diabetes (T1D) in Estonian and Finnish children to their relatively high abundance of Bacteroides, as compared with Russian children who have a low prevalence of T1D and a higher abundance of Bifidobacteria. Using in vitro assays and animal models, they demonstrated that the lipopolysaccharide (LPS) from the most prominent Bacteroides species among their Finnish subjects, Bacteroides dorei, promotes immunological tolerance and fails to protect non-obese diabetic mice from developing autoimmune diabetes, as compared with LPS from E. coli115. Taken together, their data suggest the onset of certain immunological diseases can be influenced by microbiota composition during infancy115. Gram-positive organisms (for example, human gut Firmicutes) lack LPS but instead present a thick peptidoglycan layer to the host; the role of Gram-positive microbiota variation in host immunomodulation is not well understood, though Clostridium strains have been found to promote regulatory T cell accumulation and colitis resistance in mice116.
Although Bacteroides reveal important lessons about the role of microorganisms in promoting health, these species are also implicated in important human diseases. The intestinal pathogen enterohemorrhagic E. coli (EHEC) is known to form attaching and effacing lesions along the gut epithelium, leading to severe diarrhoea117,118. Surprisingly, its pathogenicity is enhanced by otherwise beneficial microorganisms, such as the gut commensals B. thetaiotaomicron and B. vulgatus, which cleave fucose, sialic acid moieties and other sugars from mucosal glycoproteins that are then consumed by EHEC, leading to enhanced expression of its virulence genes119–121. Similar effects have been observed using gnotobiotic models involving the pathogens S. typhimurium and Clostridium difficile following treatment with antibiotics37. As antibiotics can modulate the microbiota in a broad and imprecise manner, their consumption disturbs the stability and balance of the gut ecosystem, thereby making it easier for these pathogens to invade122. The diminished microbial competition within antibiotic-treated animals enables these pathogens to gain easier access to fucose and sialic acid moieties liberated by Bacteroides.
The mucosal barrier that keeps the microbiota from contacting host tissue is critical and can become compromised during certain situations, including intestinal surgery, tissue damage from Crohn’s disease or ulcerative colitis, development of diverticulitis, or rupturing of the appendix20,123–125. When this occurs, patients are at higher risk of developing intra-abdominal abscesses that must be drained and treated with antibiotics. Studies dating back more than a century noted the overabundance of anaerobic bacteria isolated from abscesses compared with aerobes20,126,127. While the nomenclature of those early days differed from that of today (the type species for Bacteroides, B. fragilis, was initially named ‘Bacillus fragilis’ in 1898 (ref. 20), and the genus name ‘Bacteroides’ was only coined two decades later128), it stands to reason from more recent studies that the majority of those anaerobes were indeed members of the Bacteroides, with B. fragilis being the most prominent of the anaerobes and E. coli being the most prominent of the aerobes33,123,129. It is for this reason that B. fragilis has long been considered a human pathogen130. However, when kept in their normal gut luminal environment, gut microorganisms such as B. fragilis are more helpful to their host than harmful70.
In order to persist, our gut microorganisms must be able to withstand periods of inflammation. Major factors within the inflamed gut include secreted host antimicrobial factors such as the peptidoglycan-targeting lectin RegIIIγ (regenerating islet-derived protein 3-γ) and the LPS-targeting peptide hLL-37 (or hCAP-18, human cathelicidin antimicrobial peptide 18) (refs 124 and 131). As many antimicrobial peptides carry positive charges, they are drawn to the negatively charged outer membranes of Gram-negative bacteria, whereupon they can mediate cell damage132. Over the course of evolutionary history, the Bacteroides have become highly resistant to host antimicrobial peptides through the direct modification of their LPS133. They accomplish this using a specialized phosphatase, LpxF, which can reduce the negative charge on the outer membrane, thereby making the cell less attractive to cationic antimicrobial peptides133. This mechanism serves to illustrate a key strategy to maintaining stable, long-term colonization of the human gut and resilience in the face of inflammation.
Future perspectives
The sheer biological complexity of the human gut microbiota presents a daunting challenge to the scientific community and creates numerous barriers to our ability to study, understand and manipulate these communities in a precise manner. One of the major impediments to progress is a lack of appropriate genetic tools to dissect our microorganisms to determine how they work at the cellular and molecular level. Because members of the Bacteroides are culturable and amenable to genetic manipulation, they have emerged as an ideal model group of human gut bacteria. The lessons that have been learned from decades of studying Bacteroides can inform our understanding of other members of our gut microbiota and their impacts on the health of their host. Understanding how Bacteroides avoid triggering host immune pathways or avoid assault by antimicrobial molecules, how they break down nutrients from food particles or host mucus, how they respond to changes in host diet and other perturbations, or how they interact with each other and other members of their community may ultimately point us to the fundamental, underlying mechanisms of microbial physiology in the gut. A firm grasp of these fundamentals may enable great strides in the development of targeted therapeutics that alter the microbiota of patients to combat the multitude of diseases, infections and disorders linked to the gut microbiota.
Acknowledgments
We thank W. Schofield and B. Lim for careful reading of the manuscript. Support for this work was provided by the National Institutes of Health (GM105456 and GM118159), the Pew Scholars Program, and the Burroughs Wellcome Fund to A.L.G. A.G.W. is supported by a fellowship from the Gruber Foundation.
Footnotes
Author contributions
A.G.W. drafted the manuscript and prepared figures. A.G.W. and A.L.G. edited the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing financial interests.
References
- 1.Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164:337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 2.Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 3.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 6.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faith JJ, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Browne HP, et al. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533:543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rettedal EA, Gumpert H, Sommer MO. Cultivation-based multiplex phenotyping of human gut microbiota allows targeted recovery of previously uncultured bacteria. Nat Commun. 2014;5:4714. doi: 10.1038/ncomms5714. [DOI] [PubMed] [Google Scholar]
- 15.Sommer MO. Advancing gut microbiome research using cultivation. Curr Opin Microbiol. 2015;27:127–132. doi: 10.1016/j.mib.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Goodman AL, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci USA. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagier JCC, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Distaso A. Contribution à l’étude sur l’intoxication intestinale. Centralb f Bakteriol. 1912;62:433. [Google Scholar]
- 19.Eggerth AH, Gagnon BH. The Bacteroides of human feces. J Bacteriol. 1933;25:389–413. doi: 10.1128/jb.25.4.389-413.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veillon MH, Zuber H. Recherches sur quelques microbes strictement anaérobies et leur rôle en pathologie. Arch Med Exp Anat Pathol. 1898;10:517–545. [Google Scholar]
- 21.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kraus D. Concepts of Modern Biology. Globe Book Company; 1993. [Google Scholar]
- 24.Helander HF, Fändriks L. Surface area of the digestive tract – revisited. Scand J Gastroenterol. 2014;49:681–689. doi: 10.3109/00365521.2014.898326. [DOI] [PubMed] [Google Scholar]
- 25.Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2015;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans DF, Pye G, Bramley R, Clark AG, Dyson TJ. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut. 1988;29:1035–1041. doi: 10.1136/gut.29.8.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albenberg L, Esipova TV, Judge CP, Bittinger K. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055–1063. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stecher B, Maier L, Hardt WDD. ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol. 2013;11:277–284. doi: 10.1038/nrmicro2989. [DOI] [PubMed] [Google Scholar]
- 29.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurokawa K, Itoh T, Kuwahara T, Oshima K. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baughn AD, Malamy MH. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature. 2004;427:441–444. doi: 10.1038/nature02285. [DOI] [PubMed] [Google Scholar]
- 32.Meehan BM, Baughn AD, Gallegos R, Malamy MH. Inactivation of a single gene enables microaerobic growth of the obligate anaerobe Bacteroides fragilis. Proc Natl Acad Sci USA. 2012;109:12153–12158. doi: 10.1073/pnas.1203796109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rocha ER, Smith CJ. Ferritin-like family proteins in the anaerobe Bacteroides fragilis: when an oxygen storm is coming, take your iron to the shelter. Biometals. 2013;26:577–591. doi: 10.1007/s10534-013-9650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smalley D, Rocha ER, Smith CJ. Aerobic-type ribonucleotide reductase in the anaerobe Bacteroides fragilis. J Bacteriol. 2002;184:895–903. doi: 10.1128/jb.184.4.895-903.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rocha ER, Selby T, Coleman JP, Smith JC. Oxidative stress response in an anaerobe, Bacteroides fragilis: a role for catalase in protection against hydrogen peroxide. J Bacteriol. 1996;178:6895–6903. doi: 10.1128/jb.178.23.6895-6903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host–microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 37.Ng KM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright EM, Martín MG, Turk E. Intestinal absorption in health and disease—sugars. Best Pract Res Clin Gastroenterol. 2003;17:943–956. doi: 10.1016/s1521-6918(03)00107-0. [DOI] [PubMed] [Google Scholar]
- 39.Salyers AA, Vercellotti JR, West SE. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl Environ Microbiol. 1977;33:319–322. doi: 10.1128/aem.33.2.319-322.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McNeil NI. The contribution of the large intestine to energy supplies in man. Am J Clin Nutr. 1984;39:338–342. doi: 10.1093/ajcn/39.2.338. [DOI] [PubMed] [Google Scholar]
- 41.Lee SM, et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson KL, Salyers AA. Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer membrane starch-binding sites and periplasmic starch-degrading enzymes. J Bacteriol. 1989;171:3192–3198. doi: 10.1128/jb.171.6.3192-3198.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson KL, Salyers AA. Genetic evidence that outer membrane binding of starch is required for starch utilization by Bacteroides thetaiotaomicron. J Bacteriol. 1989;171:3199–3204. doi: 10.1128/jb.171.6.3199-3204.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Motherway M, et al. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2008;74:6271–6279. doi: 10.1128/AEM.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu J, et al. A genomic view of the human–Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 48.Xu J, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cameron EA, et al. Multifunctional nutrient-binding proteins adapt human symbiotic bacteria for glycan competition in the gut by separately promoting enhanced sensing and catalysis. mBio. 2014;5:e01441–14. doi: 10.1128/mBio.01441-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martens EC, Roth R, Heuser JE, Gordon JI. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J Biol Chem. 2009;284:18445–18457. doi: 10.1074/jbc.M109.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lynch JB, Sonnenburg JL. Prioritization of a plant polysaccharide over a mucus carbohydrate is enforced by a Bacteroides hybrid two-component system. Mol Microbiol. 2012;85:478–491. doi: 10.1111/j.1365-2958.2012.08123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sonnenburg ED, et al. A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proc Natl Acad Sci USA. 2006;103:8834–8839. doi: 10.1073/pnas.0603249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Townsend GE, Raghavan V, Zwir I, Groisman EA. Intramolecular arrangement of sensor and regulator overcomes relaxed specificity in hybrid two-component systems. Proc Natl Acad Sci USA. 2013;110:161–169. doi: 10.1073/pnas.1212102110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raghavan V, Lowe EC, Townsend GE, Bolam DN, Groisman EA. Tuning transcription of nutrient utilization genes to catabolic rate promotes growth in a gut bacterium. Mol Microbiol. 2014;93:1010–1025. doi: 10.1111/mmi.12714. [DOI] [PubMed] [Google Scholar]
- 55.Schwalm ND, Townsend GE, Groisman EA. Multiple signals govern utilization of a polysaccharide in the gut bacterium Bacteroides thetaiotaomicron. mBio. 2016;7:e01342–16. doi: 10.1128/mBio.01342-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cann I, Bernardi RC, Mackie RI. Cellulose degradation in the human gut: Ruminococcus champanellensis expands the cellulosome paradigm. Environ Microbiol. 2016;18:307–310. doi: 10.1111/1462-2920.13152. [DOI] [PubMed] [Google Scholar]
- 57.Artzi L, Bayer EA, Moraïs S. Cellulosomes: bacterial nanomachines for dismantling plant polysaccharides. Nat Rev Microbiol. 2016;15:83–95. doi: 10.1038/nrmicro.2016.164. [DOI] [PubMed] [Google Scholar]
- 58.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 59.Sonnenburg ED, et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pumbwe L, Skilbeck CA, Wexler HM. The Bacteroides fragilis cell envelope: quarterback, linebacker, coach-or all three? Anaerobe. 2006;12:211–220. doi: 10.1016/j.anaerobe.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 61.Wangdi T, Lee CY, Spees AM, Yu C, Kingsbury DD. The Vi capsular polysaccharide enables Salmonella enterica serovar typhi to evade microbe-guided neutrophil chemotaxis. PLoS Pathog. 2014;10:e1004306. doi: 10.1371/journal.ppat.1004306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Doorduijn DJ, Rooijakkers SH, van Schaik W, Bardoel BW. Complement resistance mechanisms of Klebsiella pneumoniae. Immunobiology. 2016;221:1102–1109. doi: 10.1016/j.imbio.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 63.Krinos CM, et al. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature. 2001;414:555–558. doi: 10.1038/35107092. [DOI] [PubMed] [Google Scholar]
- 64.Chatzidaki-Livanis M, Weinacht KG, Comstock LE. Trans locus inhibitors limit concomitant polysaccharide synthesis in the human gut symbiont Bacteroides fragilis. Proc Natl Acad Sci USA. 2010;107:11976–11980. doi: 10.1073/pnas.1005039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Comstock LE, Kasper DL. Bacterial glycans: key mediators of diverse host immune responses. Cell. 2006;126:847–850. doi: 10.1016/j.cell.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 66.Coyne MJ, Comstock LE. Niche-specific features of the intestinal Bacteroidales. J Bacteriol. 2008;190:736–742. doi: 10.1128/JB.01559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fletcher CM, Coyne MJ, Bentley DL, Villa OF, Comstock LE. Phase-variable expression of a family of glycoproteins imparts a dynamic surface to a symbiont in its human intestinal ecosystem. Proc Natl Acad Sci USA. 2007;104:2413–2418. doi: 10.1073/pnas.0608797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Coyne MJ, Chatzidaki-Livanis M, Paoletti LC, Comstock LE. Role of glycan synthesis in colonization of the mammalian gut by the bacterial symbiont Bacteroides fragilis. Proc Natl Acad Sci USA. 2008;105:13099–13104. doi: 10.1073/pnas.0804220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 70.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 71.Shen Y, et al. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2012;12:509–520. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chu H, et al. Gene–microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116–1120. doi: 10.1126/science.aad9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 74.Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sonnenburg ED, Sonnenburg JL. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. 2014;20:779–786. doi: 10.1016/j.cmet.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harmsen HJ, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr. 2000;30:61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 77.Marcobal A, et al. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58:5334–5340. doi: 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marcobal A, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 80.Ninonuevo MR, Park Y, Yin H, Zhang J. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54:7471–7480. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- 81.Charbonneau MR, et al. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell. 2016;164:859–871. doi: 10.1016/j.cell.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 83.Saez-Lara MJ, Gomez-Llorente C. The role of probiotic lactic acid bacteria and bifidobacteria in the prevention and treatment of inflammatory bowel disease and other related diseases: a systematic review of randomized human clinical trials. Biomed Res Int. 2015;2015:505878. doi: 10.1155/2015/505878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mahowald MA, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA. 2009;106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sonnenburg JL, Chen CTL, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4:e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Allen RH, Stabler SP. Identification and quantitation of cobalamin and cobalamin analogues in human feces. Am J Clin Nutr. 2008;87:1324–1335. doi: 10.1093/ajcn/87.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Degnan PH, Taga ME, Goodman AL. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 2014;20:769–778. doi: 10.1016/j.cmet.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Degnan PH, Barry NA, Mok KC, Taga ME, Goodman AL. Human gut microbes use multiple transporters to distinguish vitamin B12 analogs and compete in the gut. Cell Host Microbe. 2014;15:47–57. doi: 10.1016/j.chom.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goodman AL, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rakoff-Nahoum S, Coyne MJ, Comstock LE. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol. 2014;24:40–49. doi: 10.1016/j.cub.2013.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rakoff-Nahoum S, Foster KR, Comstock LE. The evolution of cooperation within the gut microbiota. Nature. 2016;533:255–259. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Elhenawy W, Debelyy MO, Feldman MF. Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. mBio. 2014;5:e00909–14. doi: 10.1128/mBio.00909-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 95.Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005;3:777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 96.Kommineni S, et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526:719–722. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chatzidaki-Livanis M, Coyne MJ, Comstock LE. An antimicrobial protein of the gut symbiont Bacteroides fragilis with a MACPF domain of host immune proteins. Mol Microbiol. 2014;94:1361–1374. doi: 10.1111/mmi.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roelofs KG, Coyne MJ, Gentyala RR, Chatzidaki-Livanis M, Comstock LE. Bacteroidales secreted antimicrobial proteins target surface molecules necessary for gut colonization and mediate competition in vivo. mBio. 2016;7:e01055–16. doi: 10.1128/mBio.01055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tomita H, Fujimoto S, Tanimoto K, Ike Y. Cloning and genetic and sequence analyses of the bacteriocin 21 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pPD1. J Bacteriol. 1997;179:7843–7855. doi: 10.1128/jb.179.24.7843-7855.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Russell AB, et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe. 2014;16:227–236. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Coyne MJ, Zitomersky NL, McGuire AM, Earl AM, Comstock LE. Evidence of extensive DNA transfer between Bacteroidales species within the human gut. mBio. 2014;5:14. doi: 10.1128/mBio.01305-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol. 2014;12:137–148. doi: 10.1038/nrmicro3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Coyne MJ, Roelofs KG, Comstock LE. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics. 2016;17:58. doi: 10.1186/s12864-016-2377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wexler AG, et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci USA. 2016;113:3639–3644. doi: 10.1073/pnas.1525637113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci USA. 2016;113:3627–3632. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Whitney JC, et al. An interbacterial NAD(P)+ glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell. 2015;163:607–619. doi: 10.1016/j.cell.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hecht AL, et al. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep. 2016;17:1281–1291. doi: 10.15252/embr.201642282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Moreau MC, Ducluzeau R, Guy-Grand D. Increase in the population of duodenal immunoglobulin A plasmocytes in axenic mice associated with different living or dead bacterial strains of intestinal origin. Infect Immun. 1978;21:532–539. doi: 10.1128/iai.21.2.532-539.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Macpherson AJ, Harris NL. Opinion: interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 110.Sprinz H, et al. The response of the germfree guinea pig to oral bacterial challenge with Escherichia coli and Shigella flexneri. Am J Pathol. 1961;39:681–695. [PMC free article] [PubMed] [Google Scholar]
- 111.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Neff CP, et al. Diverse intestinal bacteria contain putative zwitterionic capsular polysaccharides with anti-inflammatory properties. Cell Host Microbe. 2016;20:535–547. doi: 10.1016/j.chom.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vatanen T, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Atarashi K, et al. Induction of colonic regulatory T Cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rosenshine I, Ruschkowski S, Stein M. A pathogenic bacterium triggers epithelial signals to form a functional bacterial receptor that mediates actin pseudopod formation. EMBO J. 1996;15:2613–2624. [PMC free article] [PubMed] [Google Scholar]
- 118.Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 119.Curtis MM, et al. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe. 2014;16:759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pacheco AR, et al. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huang YLL, Chassard C, Hausmann M, von Itzstein M, Hennet T. Sialic acid catabolism drives intestinal inflammation and microbial dysbiosis in mice. Nat Commun. 2015;6:8141. doi: 10.1038/ncomms9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rivera-Chávez F, et al. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe. 2016;19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.McClean KL, Sheehan GJ, Harding GK. Intraabdominal infection: a review. Clin Infect Dis. 1994;19:100–116. doi: 10.1093/clinids/19.1.100. [DOI] [PubMed] [Google Scholar]
- 124.Brook I, Frazier EH. Aerobic and anaerobic microbiology in intra-abdominal infections associated with diverticulitis. J Med Microbiol. 2000;49:827–830. doi: 10.1099/0022-1317-49-9-827. [DOI] [PubMed] [Google Scholar]
- 125.Vaishnava S, et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Runeberg B. Studien über die bei peritonealen infektionem appendikularen ursprungs vorkommenden sauerstofftoleranten, mit besonderer berücksichtigung ihrer bedeutung für die pathogenese derartiger peritonitiden. Arb a d Path Inst d Univ Helsinfors. 1908;1:271. [Google Scholar]
- 127.Heyde M. Bakteriologische und experimentalle untersuchungen zur aetiologie der wurmfortsatzentzündung (mit besondere berücksichtigung der anaeroben bakterien) Beitr z klin Chir. 1911;76:1. [Google Scholar]
- 128.Castellani A, Chalmers AJ. Manual of Tropical Medicine. Bailliere, Tindall & Cox; 1919. [Google Scholar]
- 129.Shah HN. The Genus Bacteroides and Related Taxa. Springer; 1992. [Google Scholar]
- 130.Polk BF, Kasper DL. Bacteroides fragilis subspecies in clinical isolates. Annu Int Med. 1977;86:569–571. doi: 10.7326/0003-4819-86-5-569. [DOI] [PubMed] [Google Scholar]
- 131.Dürr UHN, Sudheendra US, Ramamoorthy A. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim Biophys Acta. 2006;1758:1408–1425. doi: 10.1016/j.bbamem.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 132.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 133.Cullen TW, et al. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347:170–175. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.McNulty NP, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Arrieta MCC, Walter J, Finlay BB. Human microbiota-associated mice: a model with challenges. Cell Host Microbe. 2016;19:575–578. doi: 10.1016/j.chom.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 137.Baxter NT, Zackular JP, Chen GY, Schloss PD. Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome. 2014;2:20. doi: 10.1186/2049-2618-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lagier JCC, et al. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Palm NW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Salyers AA, Bonheyo G, Shoemaker NB. Starting a new genetic system: lessons from Bacteroides. Methods. 2000;20:35–46. doi: 10.1006/meth.1999.0903. [DOI] [PubMed] [Google Scholar]
- 141.Wu M, et al. Genetic determinants of in vivo fitness and diet responsiveness in multiple human gut Bacteroides. Science. 2015;350:aac5992. doi: 10.1126/science.aac5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mimee M, Tucker AC, Voigt CA, Lu TK. Programming a human commensal bacterium, Bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Syst. 2015;1:62–71. doi: 10.1016/j.cels.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kotula JW, et al. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc Natl Acad Sci USA. 2014;111:4838–4843. doi: 10.1073/pnas.1321321111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Biteen JS, et al. Tools for the Microbiome: Nano and Beyond. ACS Nano; 2015. [DOI] [PubMed] [Google Scholar]
- 145.Earle KA, et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe. 2015;18:478–488. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Geva-Zatorsky N, et al. In vivo imaging and tracking of host–microbiota interactions via metabolic labeling of gut anaerobic bacteria. Nat Med. 2015;21:1091–1100. doi: 10.1038/nm.3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nishijima S, et al. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 2016;23:125–133. doi: 10.1093/dnares/dsw002. [DOI] [PMC free article] [PubMed] [Google Scholar]


