Abstract
Pathology Image Informatics Platform (PIIP) is an NCI/NIH sponsored project intended for managing, annotating, sharing, and quantitatively analyzing digital pathology imaging data. It expands on an existing, freely available pathology image viewer, Sedeen. The goal of this project is to develop and embed some commonly used image analysis applications into the Sedeen viewer to create a freely available resource for the digital pathology and cancer research communities. Thus far, new plugins have been developed and incorporated into the platform for out of focus detection, region of interest transformation, and IHC slide analysis. Our biomarker quantification and nuclear segmentation algorithms, written in MATLAB, have also been integrated into the viewer. This paper describes the viewing software and the mechanism to extend functionality by plugins, brief descriptions of which are provided as examples, to guide users who want to utilize this platform.
Keywords: Digital pathology, Whole Slide Images, Microscopy, Image Analysis, Software
I. Introduction
With the advent of whole slide digital scanners, histopathology slides can be digitized into very high-resolution digital images, realizing a new “big data” stream that can potentially rival “-omics data” in size and complexity (1–3). The first FDA approval for marketing a whole slide imaging system for primary diagnosis was granted in April 2017, and this will generate increased interest in using digital scanners by pathologists in hospital settings. Features extracted from whole slide images (WSIs) using sophisticated image analysis methods have already been shown to improve diagnosis (4) and have prognostic power in a number of applications (5). There is also growing interest in combining morphological information extracted from WSIs with genomic data and in vivo medical images to discover new biomarkers (6, 7).
There is a need for user-friendly software tools which can support the display, annotation, and analysis of different sources of image data. For researchers, the ability to use analysis algorithms that have been contributed by other researchers is also a key requirement.
Whilst other image analysis platforms exist, most have not been designed to handle digital pathology WSIs.
For example, Fiji (fiji.sc) is an image processing package aimed at providing scientific image analysis tools for life sciences researchers and it is built on ImageJ (imagej.nih.gov), which is an extensible, open source, Java-based program developed by the NIH. Fiji is popular for cell image analysis but it was not designed to view and process large tiled whole slide images.
It is necessary to crop images to a sub-region on import and/or to limit their resolution which means that the user has to process images tile by tile, losing the spatial context of each patch. This is a major limitation which makes Fiji incompatible with a clinical pathology workflow. OpenSlide (openslide.org) is a C library that allows application developers to parse various whole slide image formats. Although it provides a simple slide viewer, OpenSlide lacks support for annotations, analysis, and multi-modality images. QuPath (qupath.github.io) is a new and interesting software application for the digital pathology community which appears to provide good support for whole slide image viewing, annotations, and basic analysis. Extensions can be added via JavaScript and Groovy (a scripting language for Java); however, while these are widely used languages, they are less suited for image analysis in pathology because of the very high demands on memory and performance.
Sedeen is a pathology image viewer (http://pathcore.com/downloads/) which was initially developed with funding from the Ontario Cancer Research Institute. It was designed specifically to address the unmet need for a platform that could be used by the academic community to share both established and novel digital pathology visualization and analysis tools with pathologists and other imaging researchers (8). It is built in C++, which ensures good computational performance while also making it simple to integrate with common image analysis libraries such as ITK, VTK, and OpenCV.
The pathology informatics platform (PIIP) is a multi-institutional project supported by the NCI/ITCR, which is leveraging the Sedeen viewer framework (see Figure 1). It is a collaboration between five different participating academic institutions (University of Pennsylvania, University of Michigan, Ohio State University, Case Western Reserve University, University at Buffalo in the US and Sunnybrook Research Institute in Canada) and Pathcore, a for-profit company which has been providing free access to Sedeen to the academic community since 2010. See video 1 for a brief overview of the PIIP project and a demonstration of the Sedeen viewer.
Figure 1.
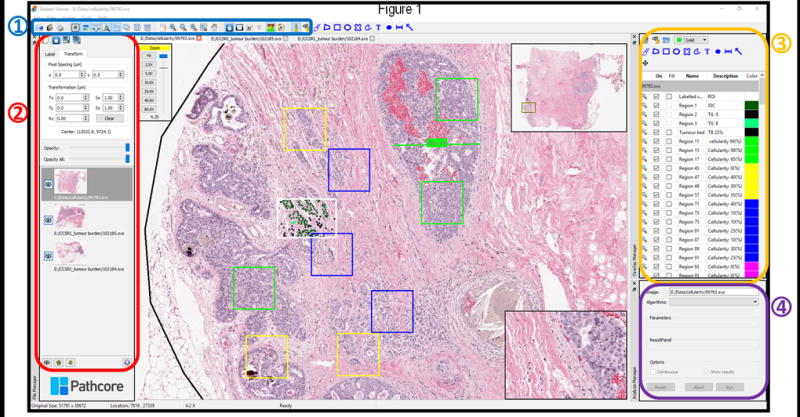
shows the user inter-face of Sedeen Viewer. Certain key aspects of Sedeen Viewer’s functionality will be highlighted here while further details of its use are described in the user manual. The Toolbar (1) provides access to controls for image navigation and allows the display layout (stacked, vertical, horizontal) to be customized. Screenshot, format conversion and cropping tools are also available. The File Manager panel (2) allows the order of images and their visibility to be controlled. Opacity of images and transformation parameters can also be modified which supports manual alignment. The Overlay Manager (3) provides tools for annotation and markup of images. Drawing tools include polygons, rulers, arrow, rectangles, ovals and freehand shapes. The properties of overlays (color, name and description) can be modified and reports on their relevant characteristics (e.g. area, length) can be displayed. Annotations can be exported in a human readable XML format, and can be imported into other programs and applications. The Analysis Manager (4) allows plugins generated with Pathcore SDK to be loaded into Sedeen. SDK generated plugins allow developers to collect parameters and display results of analysis in real-time.
II. Sedeen Viewer
Sedeen allows users to view images from multiple modalities (radiology and pathology in particular) and to perform visual overlays and registrations for biomarker validation (e.g. IHC and H&E) and multi-modality comparisons (H&E and MRI). Among other capabilities, Sedeen provides rich annotation capabilities, supports a wide range of whole slide formats, and is extensible though the Pathcore Software Development Kit (SDK), allowing researchers to share and validate novel cancer informatics tools. At present, each slide scanner vendor has a proprietary whole slide image format. Typically, an image is saved as a series of tiles at multiple resolution levels; this allows sub-regions of the image to be retrieved without having to load the whole image into memory at once. An essential feature of the Sedeen viewer is the ability to import multiple file formats, including Aperio SVS, Leica SCN, Hamamatsu NDPI, Zeiss MRXS, VisionTek SVSlide, other TIFF-based formats, and JPEG-2000.
Although the viewer is currently distributed as a Windows application, it has been designed to be portable to other platforms, e.g. MacOS and Linux, if required. Dr. Emily Patterson (Ohio State University), a human factors expert, has conducted a heuristic review of all the toolbars and menus on the Sedeen interface and will also provide input on the design of plugin interfaces (9). This will ensure that the software, including its plugins, is intuitive to users, which is important in collaborative research.
III. Software Development Kit
The Pathcore SDK is a tool for integrating quantitative image analysis algorithms with the Sedeen viewer. The SDK is written in C++ and uses an open-source tool, CMake (cmake.org), to manage the build process in way that is independent of both the operating system and the compiler used. The SDK API is fully documented and generated using Doxygenwhich is a standard tool for extracting documentation from source file comments. It is distributed with a set of tutorials and exemplar algorithms, which can be selected from a pull down menu at run time. Sedeen utilizes an on-the-fly image decoding strategy (directly from the disk), which is transparent to the clients and is very efficient. This allows client code to access arbitrary regions in an image without first loading the entire image into memory. The SDK simplifies access to WSI data by providing a set of format-independent data extraction routines, the ability to create analysis pipelines, and the ability to easily visualize analysis results. Algorithm designers can therefore develop algorithms much more easily by leveraging the capabilities of Sedeen and the image analysis architecture provided by the Pathcore SDK.
The Pathcore SDK also provides numerous widgets which allow user parameters to be collected from the analysis manager GUI and transferred to the analysis algorithms. The output from the algorithm can then be rendered within the Sedeen viewer in the form of an overlay, new annotations, or as text.
IV. PIIP Plug-ins
With funding from the ITCR program, groups at Case Western University, The Ohio State University and Sunnybrook Research Institute are implementing a variety of quantitative image analysis tools as plugins. Examples are described below and all of these plugins will be distributed with the next release of Sedeen. A github repository (https://github.com/sedeen-piip-plugins) contains the source code and user documentation for the plugins developed by the PIIP. Pathcore are providing support and are also working to expand the capabilities of the SDK to facilitate academic research, for example by developing methods for accessing MATLAB and Python code directly from the Sedeen Viewer.
IV.a. Biomarker Quantification
The co-PI (Madabhushi) and his team have developed a powerful tool for high-throughput biomarker quantification named Hierarchical Normalized Cuts (HNCuts) (10). The method combines a frequency-weighted version of the mean shift (MS) algorithm with the Normalized Cuts scheme to segment all the image pixels into representative classes. Unlike supervised segmentation strategies, our scheme only requires specification of a small representative sample of the target class to rapidly identify similar objects in the image.
The PIIP provides algorithms needed to compute user provided inputs (images, parameters, annotations) and reports results through the Sedeen Viewer’s GUI. In this case, users access the viewer’s annotation tools to define the sample color swatch, which can be accessed by the HNCuts plugin. Once processing is carried out, the segmentation and quantification results are rendered within the Sedeen Viewer as a color/contour overlay. This plugin also demonstrates how algorithms implemented in Matlab can be integrated into a plugin.
IV.b. Out of Focus Detection
As whole slide imaging becomes more prevalent and many centers, particularly in Europe and Canada, adopt digital pathology, the demand for scanning whole slides has increased. Although many modern scanners are equipped with software to automatically detect areas that are out of focus, and adjust the scanning parameters accordingly, large numbers of digital slides contain out of focus areas. According to one study conducted at a Dutch academic pathology lab, 5% of the cases had problems with scanning artifacts such as out of focus areas (11). In a large-scale digitization effort, this percentage is expected to increase considerably, as the large volume of cases make it impractical to visually identify problematic cases and then rescan them if necessary.
The goal of this plugin is to detect out of focus areas and report them to the user. In its implementation, a plugin was created using Pathcore SDK and integrated with the Sedeen Viewer. This plugin also demonstrates the use of the OpenCV API, a popular computer vision library, with the SDK. The plugin identifies regions that are out of focus and produces an overlay showing where these regions are located.
IV.c. Transforming and exporting annotations
One of the key differences between the Sedeen viewer and other platforms is that it provides tools for users to manually co-register images. This allows users to make direct comparisons between images acquired using different modalities or stained using different antigens. The ExportTransformedROI plugin extends the capabilities of Sedeen by allowing an annotation defined on a source image to be exported to a target image. If the source image and the target image have been aligned, then the transformations needed to achieve the alignment are also applied to the annotations.
V. Use case example: Analysis of multiple immunohistochemistry (IHC) slides
Serial sections cut from a tissue block and stained using different immunohistochemistry markers can be used to compare the relative expression of different antigen receptors in tumors. In the supplementary file, the workflow needed to spatially align two IHC images and then carry out a quantitative comparison of tissue staining over corresponding regions of interest is described. This demonstrates how Sedeen’s built in functionality can be combined with user contributed plugins to achieve a specific analysis task.
V. Discussion
The Sedeen Viewer has all of the advantages of a viewer built specifically for digital pathology, with a user interface that is intuitive for pathologists and biologists, whilst also providing a mechanism to expand its functionality through Pathcore SDK. The viewer is easy to download and install on Windows platforms and, in time, it will be made available for other platforms. We have thus far successfully demonstrated that image analytic algorithms from three different institutions (OSU, CWRU, SB) can be integrated within the PIIP. Our future plans involve enabling the seamless integration of analytic tools developed by other users into the platform. We envision a PIIP that will be dynamically populated with validated image analytic “apps” developed by the community and hence provide immense benefit to end users, including clinical and research centric pathologists and computational image analysis scientists. Pathcore has been providing free access to Sedeen since 2010, and is extending the capabilities of both the viewer and the Pathcore SDK in response to feedback from the PIIP project team. They are currently working on an improved mechanism to support the compilation and distribution of contributed plugins from the wider user community.
Other novel aspects of this project are also underway. Working with an expert on disease ontologies, Dr. Barry Smith, we have proposed a quantitative histopathology image ontology (QHIO) which will facilitate interoperability between histopathology datasets and methods used in pathological imaging and analysis. This ontology fosters data compatibility and provides a mechanism for linking algorithms with compatible datatypes. In (12), we demonstrate how the QHIO can be applied to the problem of hot spot detection in breast IHC WSIs. Human-computer interaction and user-experience is an integral part of the PIIP. We are working with a human-computer interaction expert, Dr. Emily Patterson, to define user interface design requirements. Based on her suggestions, several design decisions have already been modified to make the interface friendlier to users with different experience levels.
Supplementary Material
Acknowledgments
The project described was supported in part by Award Number U24CA199374 (PIs: Gurcan, Madabhushi, Martel) from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
This project is also supported by Pathcore, a digital pathology software company Pathcore has been supporting the research community since 2011 via the Sedeen Viewer and PIIP.
We are grateful to Craig Madho and Deyu Wang, at Pathcore for their help in the coordination of the project and facilitating the writing process.
Financial support:
This project is also supported in part by Pathcore Inc, Toronto, Canada
Footnotes
Conflicts-of-Interest:
Anant Madabhushi and Metin Gurcan are on the scientific advisory board of Inspirata, Inc.
Madabhushi has an equity stake in Elucid Bioimaging and Inspirata Inc. He is also a scientific advisory board member of Astrazeneca and has a sponsored research project with Philips.
Dan Hosseinzadeh is co-founder and CEO of Pathcore Inc. Anne Martel is co-founder and CSO of Pathcore.
Availability: PIIP project materials including a video describing its usage and applications, and links for the Sedeen Viewer, plug-ins, and user manuals are freely available through the project web page: http://pathiip.org.
References
- 1.Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B. Histopathological image analysis: a review. IEEE Rev. Biomed. Eng. 2009:147–71. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madabhushi A, Lee G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med. Image Anal. 2016:170–5. doi: 10.1016/j.media.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhargava R, Madabhushi A. Emerging Themes in Image Informatics and Molecular Analysis for Digital Pathology. Annu Rev Biomed Eng. Annual Reviews. 2016;18:387–412. doi: 10.1146/annurev-bioeng-112415-114722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fauzi MFA, Pennell M, Sahiner B, Chen W, Shana’ah A, Hemminger J, et al. Classification of follicular lymphoma: the effect of computer aid on pathologists grading. BMC Med Inform Decis Mak. 2015;15:115. doi: 10.1186/s12911-015-0235-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis JS, Ali S, Luo J, Thorstad WL, Madabhushi A, Madabhushi A. A quantitative histomorphometric classifier (QuHbIC) identifies aggressive versus indolent p16-positive oropharyngeal squamous cell carcinoma. Am J Surg Pathol. NIH Public Access. 2014;38:128–37. doi: 10.1097/PAS.0000000000000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singanamalli A, Sparks R, Rusu M, Shih N, Ziober A, Tomaszewski J, Rosen M, Feldman M, Madabhushi A. Identifying in vivo DCE MRI markers associated with Microvessel Architecture and Gleason Grades of Prostate Cancer—Preliminary Findings. Journal of Magnetic Resonance Imaging. doi: 10.1002/jmri.24975. [Epub ahead of print] (PMID:26110513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee G, Singanamalli A, Wang H, Feldman MD, Master SR, Shih NNC, et al. Supervised Multi-View Canonical Correlation Analysis (sMVCCA): Integrating Histologic and Proteomic Features for Predicting Recurrent Prostate Cancer. IEEE Trans Med Imaging. 2015;34:284–97. doi: 10.1109/TMI.2014.2355175. [DOI] [PubMed] [Google Scholar]
- 8.Clarke GM, Peressotti C, Constantinou P, Hosseinzadeh D, Martel A, Yaffe MJ. Comput Med Imaging Graph. Vol. 35. Elsevier Ltd; 2011. Increasing specimen coverage using digital whole-mount breast pathology: implementation, clinical feasibility and application in research; pp. 531–41. [DOI] [PubMed] [Google Scholar]
- 9.Mount-Campbell A, Hosseinzadeh D, Gurcan M, Patterson E. Hum Factors Ergon Heal Care. New Orleans, LA: Applying Human Factors Engineering to Improve Usability and Workflow in Pathology Informatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janowczyk A, Chandran S, Singh R, Sasaroli D, Coukos G, Feldman MD, et al. High-Throughput Biomarker Segmentation on Ovarian Cancer Tissue Microarrays via Hierarchical Normalized Cuts. IEEE Trans Biomed Eng. 2012;59:1240–52. doi: 10.1109/TBME.2011.2179546. [DOI] [PubMed] [Google Scholar]
- 11.Stathonikos N, Veta M, Huisman A, van Diest PJ. J Pathol Inform. Vol. 4. Medknow Publications and Media Pvt. Ltd.; 2013. Going fully digital: Perspective of a Dutch academic pathology lab; p. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gurcan MN, Tomaszewski J, Overton JA, Doyle S, Ruttenberg A, Smith B. Developing the Quantitative Histopathology Image Ontology (QHIO): A case study using the hot spot detection problem. Journal of Biomedical Informatics. 2017;66:129–135. doi: 10.1016/j.jbi.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


