Abstract
We present NDEx 2.0, the latest release of the Network Data Exchange (NDEx) (1,2) online data commons (www.ndexbio.org) and the ways in which it can be used to (1) improve the quality and abundance of biological networks relevant to the cancer research community, (2) provide a medium for collaboration involving networks, and (3) facilitate the review and dissemination of networks. We describe innovations addressing the challenges of an online data commons: scalability, data integration, data standardization, control of content and format by authors, and decentralized mechanisms for review. The practical use of NDEx is presented in the context of a novel strategy to foster network-oriented communities of interest in cancer research by adapting methods from academic publishing and social media.
Keywords: Network, Collaboration, Publishing, Database, Cytoscape
Introduction
NDEx 2.0 is the latest release of the Network Data Exchange (NDEx) (1,2) online resource (www.ndexbio.org), a framework in which users can store, share, access, and disseminate networks. In this article, we discuss workflows and methods used by NDEx in the context of its role as an online data commons and its mission to foster the emergence of network-centered communities of scientists involved in all aspects of disease biology, from basic research to personalized medicine.
Highlights of innovations in NDEx 2.0 include:
A novel, modular network data exchange standard, CX, developed in collaboration with the Cytoscape (3) project.
A total redesign of the NDEx server for scalability and input/output speed, supporting large communities, large networks, and high access rates.
A scoring system for network annotation combined with search engine prioritization of high-scoring networks, rewarding authors for compliance with best annotation practices.
"Network Sets", a facility for managing and publishing collections of networks.
Finding Networks
In NDEx 2.0, every page in the website includes a search interface featuring a menu of search examples (See Video 1). The interface conforms to standard query practices, enabling networks to be found by specific document attributes such as labels, title, author or description and by biological attributes such as tissue or organism. Networks can also be found based on the identifiers and names associated with network nodes (e.g. "TP53", "P04637", "ENSG00000141510", or "GO:0006915").
Because NDEx is an open scientific commons, finding networks presents a novel challenge: search would be more effective if networks were required to be copiously annotated using standard vocabularies, but NDEx must also promote the contribution of content by minimizing the burden on authors and respecting their decisions when designing networks. In response to the diversity of identifier systems and common aliases used in both networks and queries, we have improved search recall using domain-specific "term expansion" where NDEx pre-processes queries to add aliases for genes, proteins, and other entities. To encourage but not impose the addition of annotations and the use of standard vocabularies, we have implemented an NDEx server algorithm that evaluates each network for compliance with annotation best practices and provides a score as immediate feedback to the author. A higher annotation score rewards the author's network with a better ranking in search results, thus promoting "search engine optimization" for networks.
Accessing Networks
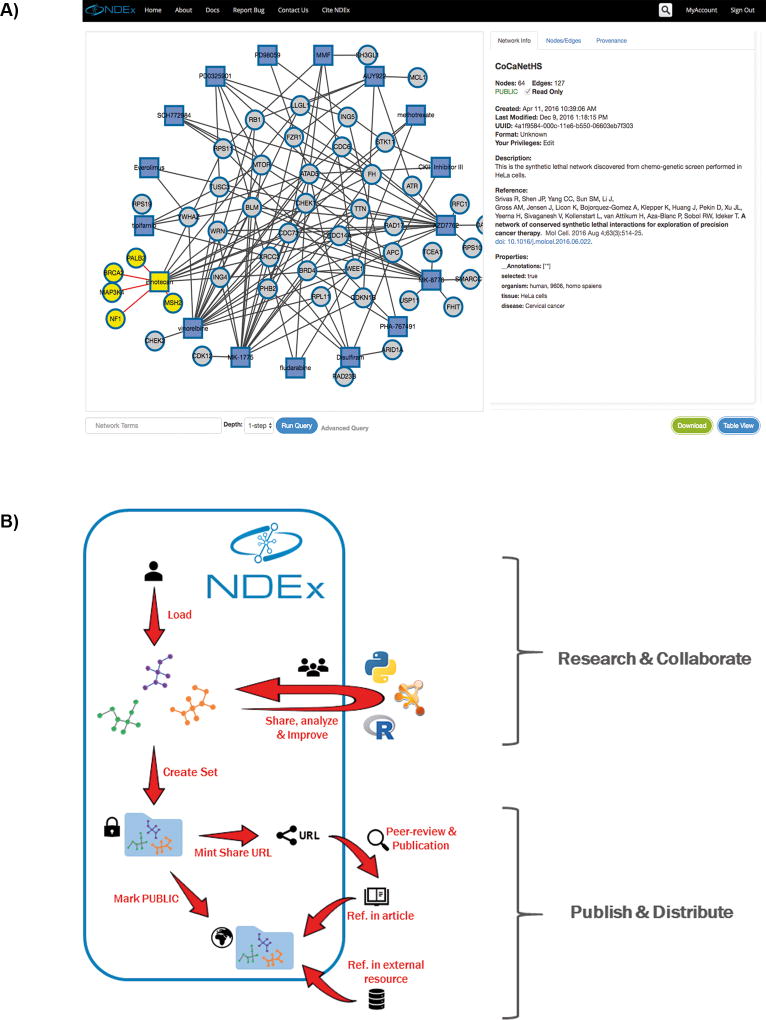
Every network in NDEx is assigned a stable, universally unique identifier (UUID) that can be used to access the network data or to link to a corresponding "network page" at the NDEx website (Fig. 1A). Authors are encouraged to create a separate network for each version, annotated with its version identifier, supporting reproducible access to specific data. Alternatively, a UUID may identify a dynamically updated public network, where the network essentially serves as an online database resource maintained by its authors, as is the case for the “RAS Machine”(4), described later in this article.
Figure 1. Collaboration and dissemination via NDEx.
A) Any network in NDEx can be explored using the interactive graphic panel on the left hand side and info section on the right. Additional tabs in the info section allow users to explore in detail nodes and edges and the network provenance history. B) Schematic representation of an NDEx workflow for collaboration and publication: see text for a detailed description.
Scripts and other applications can find, download, create or update NDEx networks using their UUIDs and a programmatic web access interface, a REST API(5). For example, when the public NDEx web application presents networks in either a graphic visualization or as tables of nodes and edges, it uses this API retrieve the network data. The API can also be used to retrieve a small sample of a network, enabling the interface to handle NDEx networks too large for practical display.
All networks are initially private, accessible only to the individual scientist or organization that owns them. For pre-publication collaboration, network owners can grant access to other NDEx users or groups of users via a permission management interface that can be reached from each network page. In NDEx 2.0, users can also generate a special "shareable URL" to grant access to a private network, whether or not the recipient is a registered NDEx user. A primary use of shareable URLs in NDEx is to streamline the submission and peer-review of networks supporting publications by providing editors and reviewers with instant, interactive, and anonymous access to the networks. Finally, users and organizations can make their networks publically accessible for broad dissemination, can designate them as stable "read-only" resources, and can select them for display on their NDEx account homepage using the "showcase" facility.
Reviewing Networks
When searching NDEx, users want to find networks that they deem credible, backed by evidence and informed analysis. This need is addressed by features that facilitate the review and recommendation of networks by researchers, following the successful models of academic peer review and of user reviews in Internet media. This strategy promotes the growth of NDEx by providing both motivation for authors and benefit to users. The introduction of "Network Sets" in NDEx 2.0 enables users to create and manage named collections of networks with stable UUIDs, making these collections destinations in their own right. Individuals and organizations can publish documented collections of networks, collections that can even include public networks owned by other users. An individual might choose to create a specialized collection such as "Cell adhesion pathways in hepatocytes", sharing their expert selections with the community. In the same way, organizations such as academic publishers or leading laboratories can publish sets of networks that they select in formal review processes, conferring recognition to the authors and helping researchers locate useful, well-founded networks with confidence.
An NDEx Workflow for Collaboration and Publication
The following example (Fig. 1B) describes a workflow that starts when a team of scientists uploads a network to NDEx directly from an analysis script. The network is initially private, invisible and inaccessible to other users. The authors share it with a group of collaborators who use NDEx as a data hub as they analyze and improve the network. They import it into Cytoscape via the CyNDEx App(6) (See Video 1) that uses the same REST API used by the initial analysis script. They then add a layout, choose a graphic style, and use a heat-diffusion algorithm to highlight a subnetwork of interest. Communication between NDEx and Cytoscape is mediated by the CX exchange format in which each "aspect" of the network is expressed in a distinct module. In this case, CX facilitates subsequent reuse of the network because the formatting of the network is separated from the scientific content. The improved network is re-uploaded to NDEx, and included in a Network Set supporting an upcoming publication. When the manuscript is submitted for publication, editors and reviewers anonymously access the Network Set via a shareable URL. On publication, the authors make their networks publically accessible, allowing online journal readers to immediately view the networks, copy them to their private accounts, or use them in applications. Databases and websites can also reference the Network Set by its URL, providing alternative dissemination channels. The Cancer Cell Map Initiative (CCMI) (7,8), a multi-institution NCI center for systems biology, has adopted NDEx for these purposes, facilitating collaboration by researchers, data sharing between applications, and access to CCMI networks by other scientists.
The team of scientists also maintains a large, dynamically updated network of inferred molecular relationships, providing an up-to-date data resource without the need for a specialized web portal. The RAS Machine, a software agent developed under the DARPA Big Mechanism program (9), reads literature daily, adds new knowledge to its model of RAS signaling, and publishes the changes as an update to its public NDEx network.
Another collaborating institution then uses the dynamically updated network as input to their web-based genomic analysis tool, creating a channel from the team's output to the users of the application. The Cancer-Related Analysis of VAriants Toolkit (CRAVAT) (10) is a web application using NDEx, accessing pathway networks for enrichment analysis of lists of mutated genes.
Conclusion and Future Work
NDEx 2.0 is a significant step in the evolution of the NDEx data commons, supporting its mission to foster network-centered communities of scientists. Recent NDEx innovations have addressed challenges of scalability, data integration, and support for annotation and review of networks. In future work, we will explore community building via Internet media techniques, such as user reviews of networks, recommendation algorithms, and in-system messaging. We will facilitate the association of Digital Object Identifiers (DOIs) (11) with published networks, integrate network-authoring tools, and recruit disease and mechanism experts as authors and reviewers. We conclude by inviting readers to be pioneering users of NDEx as authors, reviewers, publishers, creators of communities of interest, and as developers of NDEx-enabled workflows, applications, and analysis pipelines.
Acknowledgments
Financial Support: NIH ITCR U24 CA1884427; DARPA W911NF-14-1-0397
We thank Dan Carlin, other members of the Ideker laboratory, and the Sorger Lab at Harvard Medical School for their early adoption of the NDEx platform and their valuable feedback.
Footnotes
Conflict of Interest Disclosure Statement:
Trey Ideker is co-founder of Data4Cure, Inc. and has an equity interest. Trey Ideker has an equity interest in Ideaya BioSciences, Inc. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. All of the other authors declare no potential conflict of interest.
References
- 1.Pillich RT, Chen J, Rynkov V, Welker D, Pratt D. NDEx: A Community Resource for Sharing and Publishing of Biological Networks. Methods Mol Biol. 2017;1558:271–301. doi: 10.1007/978-1-4939-6783-4_13. [DOI] [PubMed] [Google Scholar]
- 2.Pratt D, Chen J, Welker D, Rivas R, Pillich R, Rynkov V, et al. NDEx, the Network Data Exchange. Cell Syst. 2015;1:302–5. doi: 10.1016/j.cels.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The RAS Machine [Internet] NDEx: The RAS Machine. [cited 2017 Feb 27]. Available from: http://www.ndexbio.org/#/newNetwork/50e3dff7-133e-11e6-a039-06603eb7f303.
- 5.Fielding RT, Taylor RN. ACM Trans Internet Technol. Vol. 2. New York, NY, USA: ACM; 2002. Principled Design of the Modern Web Architecture; pp. 115–50. [Google Scholar]
- 6.NDEx Cytoscape App Tutorial [Internet] The NDEx Project. 2014 [cited 2017 Feb 24]. Available from: http://home.ndexbio.org/ndex-cyapp-tutorial-alt/
- 7.Krogan NJ, Lippman S, Agard DA, Ashworth A, Ideker T. The cancer cell map initiative: defining the hallmark networks of cancer. Mol Cell. 2015;58:690–8. doi: 10.1016/j.molcel.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cell Maps FAQ - Cancer Cell Map Initiative [Internet] Cancer Cell Map Initiative. [cited 2017 Feb 21]. Available from: http://www.ccmi.org/cell-maps/cell-maps-faq/
- 9.Cohen PR. DARPA’s Big Mechanism program. Phys Biol. 2015;12:045008. doi: 10.1088/1478-3975/12/4/045008. [DOI] [PubMed] [Google Scholar]
- 10.Douville C, Carter H, Kim R, Niknafs N, Diekhans M, Stenson PD, et al. CRAVAT: cancer-related analysis of variants toolkit. Bioinformatics. 2013;29:647–8. doi: 10.1093/bioinformatics/btt017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paskin N. Encyclopedia of library and information sciences. Vol. 3. Taylor & Francis; England: 2010. Digital object identifier (DOI) system; pp. 1586–92. [Google Scholar]