Abstract
Advanced age is associated with reduced within-network functional connectivity, particularly within the default mode network. Most studies to date have examined age differences in functional connectivity via static indices that are computed over the entire blood-oxygen level dependent time-series. Little is known about the effects of age on short-term temporal dynamics of functional connectivity. Here we examined age differences in dynamic connectivity as well as associations between connectivity, metabolic risk, and cognitive performance in healthy adults (N=168; age 18 to 83). A sliding-window k-means clustering approach was used to assess dynamic connectivity from resting-state fMRI data. Three out of eight dynamic connectivity profiles were associated with age. Furthermore, metabolic risk was associated with the relative amount of time allocated to two of these profiles. Finally, the relative amount of time allocated to a dynamic connectivity profile marked by heightened connectivity between default mode and medial temporal regions was positively associated with executive functions. Thus, dynamic connectivity analyses can enrich understanding of age-related differences beyond what is revealed by static analyses.
Keywords: resting state fMRI, cognitive aging, executive function, functional connectivity, default network
1. Introduction
Aging affects many aspects of brain structure and function and is associated with cognitive decline (see Kennedy & Raz, 2015; Fjell et al., 2014 for recent reviews). Understanding age differences in the brain functional organization is an important step in elucidating neural mechanisms of cognitive aging, and since its introduction, resting state functional magnetic resonance imaging (rs-fMRI; (Biswal et al., 1995; Raichle et al., 2001), has been applied to assess age-related differences in brain network functioning. In rs-fMRI analyses, configuration and strength of functional organization is commonly inferred from spatial patterns of temporal correlations between low-frequency fluctuations in blood-oxygen-level dependent (BOLD) signals of different brain regions, termed “functional connectivity” (Biswal et al., 1995; Cordes et al., 2001; Lowe et al., 1998). Many resting state networks (RSNs) have been identified (Beckmann et al., 2005; Damoiseaux et al., 2006; De Luca et al., 2006; Fox et al., 2005), and the spatial configurations of these RSNs have been comparable to spatial configurations of networks observed during task performance (Calhoun et al., 2008; Smith et al., 2009).
Of the multiple known RSNs, the Default Mode Network (DMN), which is more active during wakeful, task-free rest and less active during overt task engagement, has received extensive attention (Buckner et al., 2008; Greicius et al., 2003; Raichle et al., 2001; Shulman et al., 1997). DMN activity is associated with episodic memory and future planning (Buckner et al., 2008), and DMN connectivity at rest predicts subsequent memory performance (Sala-Llonch et al., 2012; Wang et al., 2010). Furthermore, resting DMN connectivity is negatively related to age (Andrews-Hanna et al., 2007; Damoiseaux et al., 2008; Wu et al., 2011), and, in comparison to younger participants, older adults exhibit lower task-related deactivation of DMN regions (Grady et al., 2006; Lustig et al., 2003; Sambataro et al., 2010). Thus, advanced age may be linked to reduced flexibility in response to task demands. Whole-brain functional connectivity analysis has also revealed age-associated breakdown of communication within RSNs and elevated communication between RSNs (Chan et al., 2014), suggesting age-related de-differentiation of brain organization.
An important common feature, and possible limitation, of most rs-fMRI studies in healthy adults is reliance on functional connectivity indices calculated from an entire scan session. Potentially important information about within-scan temporal changes in functional connectivity may be lost in this aggregation (Allen et al., 2014; Chang and Glover, 2010; Sakoglu et al., 2010). The assessment of dynamic functional connectivity in rs-fMRI (see Hutchinson et al., 2013, for methodological review) has revealed that individuals may transition between different dynamic whole-brain connectivity profiles (often called ‘states’) characterized by distinct connectivity patterns (Allen et al., 2014). The dynamic connectivity profiles reveal variability in functional brain organization over time. This variability may reflect changes in neural activity related to cognitive and sensorimotor operations, as well as non-neuronal factors such as systemic physiological changes or spontaneous head motion. Previous work suggests that variability in hub region multi-network participation is lower (Schaefer et al., 2014), and variability within DMN dynamic functional connectivity is higher (Madhyastha and Grabowski, 2014) in older compared to younger adults. Furthermore, stability of functional connectivity increases with age in some regions (e.g. middle frontal gyrus), while decreasing in others (e.g. supramarginal gyrus) (Yin et al., 2016). These patterns of age differences suggest that dynamic properties of brain networks may reflect neural phenomena relevant to age-related functional declines.
Despite growing interest in connectivity dynamics, investigations of lifespan age differences therein remain scarce. Therefore, the present study aimed to determine age-related differences in dynamic functional connectivity and their relation to cognitive performance in healthy adults. We hypothesized age differences in patterns of DMN dynamic connectivity as static connectivity differences within this network have been previously observed (Andrews-Hanna et al., 2007; Damoiseaux et al., 2008; Wu et al., 2011) and increased DMN variability has been noted in older adults (Madhyastha and Grabowski, 2014). Specifically, we hypothesized that time allocation among specific dynamic connectivity profiles would be age dependent and that older adults would devote less time to profiles dominated by strong connectivity within DMN, and between DMN and other networks, compared to younger counterparts. We also hypothesized that time share of specific profiles would be related to cognitive performance, with more “youthful” patterns of dynamic connectivity linked to higher cognitive scores, analogous to associations observed in extant studies of static DMN connectivity and cognition (Sala-Llonch et al., 2012; Wang et al., 2010). Considering reported age-related decreases in dynamic hub-region network variability (Garrett et al., 2010; Schaefer et al., 2014) and increased variability in DMN component intercorrelation (Madhyastha and Grabowski, 2014), we also expected our dynamic analysis to capture age differences in the rate of switching between connectivity profiles. Specifically, we hypothesized older adults would switch profiles at a lower rate, which might reflect less-than-optimal cognitive processing.
2. Methods
2.1 Participants
Structural and functional MRI data were available for 168 adults (61 men, 107 women; 18–83 years, M = 48.8, SD = 18.0), recruited from the Metro Detroit, Michigan area through advertisements in newspapers and flyers. Participants were enrolled in an ongoing longitudinal study, in which the resting state sequence was introduced after some had undergone more than one wave of testing. Therefore, complete baseline cognitive testing data corresponding to the time of baseline resting state functional MRI acquisition were available only for a subsample of 91 participants (33 men, 58 women) with age range 18–78 years (M = 42.2, SD = 17.6). The Wayne State University Institutional Review Board approved the study and signed informed consent was obtained from all participants. Debriefing followed the experiment. Participants spoke English as their first language and were deemed right-hand dominant after scoring over 75% on the Edinburgh Inventory (Oldfield, 1971). They were screened for neurological, psychiatric, cardiovascular, and endocrine diseases, diabetes, cancer, and a history of loss of consciousness greater than 5 minutes. Participants were also screened for dementia (Mini-Mental State Exam, MMSE ≥ 26; (Folstein et al., 1975) and depression (Center for Epidemiological Study Depression questionnaire, CES-D ≤16; (Radloff, 1977). A metabolic risk score was computed as the sum of standardized indicators of metabolic syndrome (Grundy et al., 2004): waist-hip-ratio, blood triglycerides, systolic blood pressure, fasting blood glucose and high-density lipoprotein (reverse-coded). See previous report for details (Damoiseaux et al., 2016). Metabolic health indicators were available for 151 of the 168 participants, and 84 of the 91 participants with cognitive data.
2.2 Assessment of Cognitive Performance
Cognitive tests, described in previous publications (e.g., Raz et al., 2009), were administered across four sessions within a three-month window around the MRI session. We performed confirmatory factor analysis (CFA) to determine the main cognitive constructs. The CFA model consisted of three latent factors: processing speed, with letter comparison and pattern comparison scores as indicators; memory, with Woodcock-Johnson-R Memory for Names (WJR memory, immediate and delayed) scores as indicators; and executive functioning, with the following indicators: Stroop, Wisconsin Card Sorting Test, size judgment span, listening span, spatial recall, and Cattell Culture Fair Test (form 3B, tests 1, 2, 3, 4). Analyses were conducted with Mplus 6.0 (Muthén and Muthén), and composite factor scores were calculated for each latent factor (see Damoiseaux et.al., 2016 for a detailed description of the CFA).
2.3 MRI Data Acquisition
Imaging was performed at the Wayne State University MRI research facility on a 3-Tesla Siemens Verio (Siemens Medical AG, Erlangen, Germany) full-body magnet with a 12-channel head coil. For the resting-state functional scan, 200 volumes of 43 axial slices were acquired using a T2*-weighted echo planar sequence: repetition time (TR) = 2500 ms, echo time (TE) = 30 ms, flip angle = 90°, pixel bandwidth = 2298 Hz/pixel, GRAPPA accele ration factor PE = 2, field-of-view = 210 mm, matrix size = 64 × 64, voxel size = 3.3 × 3.3 × 3.3 mm. Participants were instructed to remain still with eyes open. For the anatomical scan, a 3D T1-weighted magnetization-prepared rapid gradient-echo (MPRAGE) sequence was acquired: TR = 1680 ms; TE = 3.51 ms; inversion time (TI) = 900 ms; flip angle = 9.0°, pixel bandwidth = 180 Hz/pixel, GRAPP A = 2; field-of-view = 256 mm, matrix size = 384 × 384, voxel size 0.67 × 0.67 × 1.34 mm.
2.4 Preprocessing
Image preprocessing was carried out with the FMRIB Software Library (FSL, version 5.0; (Smith et al., 2004). Resting state processing included removal of the first four image volumes, motion correction (Jenkinson et al., 2002), removal of non-brain structures (Smith, 2002), spatial smoothing (6 mm FWHM), 4D grand-mean scaling, and high-pass temporal filtering (Gaussian-weighted least-squares straight line fitting, sigma=150.0 s). The scan was then aligned with the corresponding high resolution T1 and subsequently registered to 3 mm isotropic MNI152 space with affine linear registration (Jenkinson et al., 2002). Translation and rotation time courses were regressed from the images to attenuate the influence of head motion on results. Participants with more than 3 mm absolute head displacement during the scan were not included in the analysis. Global signal contribution was removed.
2.5 ROI Generation and Organization
175 ROIs were defined from a group-level parcellation generated with spatially-constrained normalized-cut spectral clustering (Craddock et al., 2012) restricted to a gray matter mask segmented from an MNI image using FSL’s FAST (Zhang et al., 2001). Of the available ROIs, 172 were used for subsequent analysis after removal of brain stem ROIs. Mean ROI time-series were calculated and assigned a unit variance before covariance estimations. Therefore, subsequent covariance calculations were tantamount to correlation.
Static whole-brain covariance matrices were computed for all participants, Fisher z-transformed, and averaged. A weighted, undirected graph was constructed with ROIs as nodes and the top 10% of positive connections, in terms of strength, as edges. ROIs were organized into non-overlapping communities with Infomap to define network structure (Lancichinetti and Fortunato, 2009; Rosvall and Bergstrom, 2008). To ensure that the network structure was derived from reliable, strong connectivity, only the top 10% of positive connections were included in the Infomap community detection. Global signal regression, which was applied as a preprocessing step, may induce anti-correlation between networks (Murphy et al., 2009), rendering the interpretation of negative correlations difficult. The Infomap analysis converged on eight communities (further referred to as RSNs), which after visual inspection were labeled as DMN, Sensorimotor (SM), Visual (Vis.), Dorsal/Lateral Temporal (dLTL), Ventral/Medial Temporal (vMTL), Cerebellar (CB), Subcortical (SC), and Orbitofrontal (OF) (Figure 1). DMN included medial prefrontal, posterior & anterior cingulate, precuneus, inferior parietal, dorsolateral prefrontal, and rostro-lateral prefrontal cortex. SM included premotor, primary motor, and primary somatosensory cortex. dLTL included Wernicke’s area, Broca’s area, superior & middle temporal, auditory, parietal, and secondary somatosensory cortex. vMTL included inferior temporal, fusiform, and parahippocampal cortex, hippocampus, and amygdala. SC included thalamus, basal ganglia, and mammillary bodies. CB corresponded to the cerebellum, OF to the orbitofrontal cortex, and Vis. to the occipital cortex.
Figure 1.
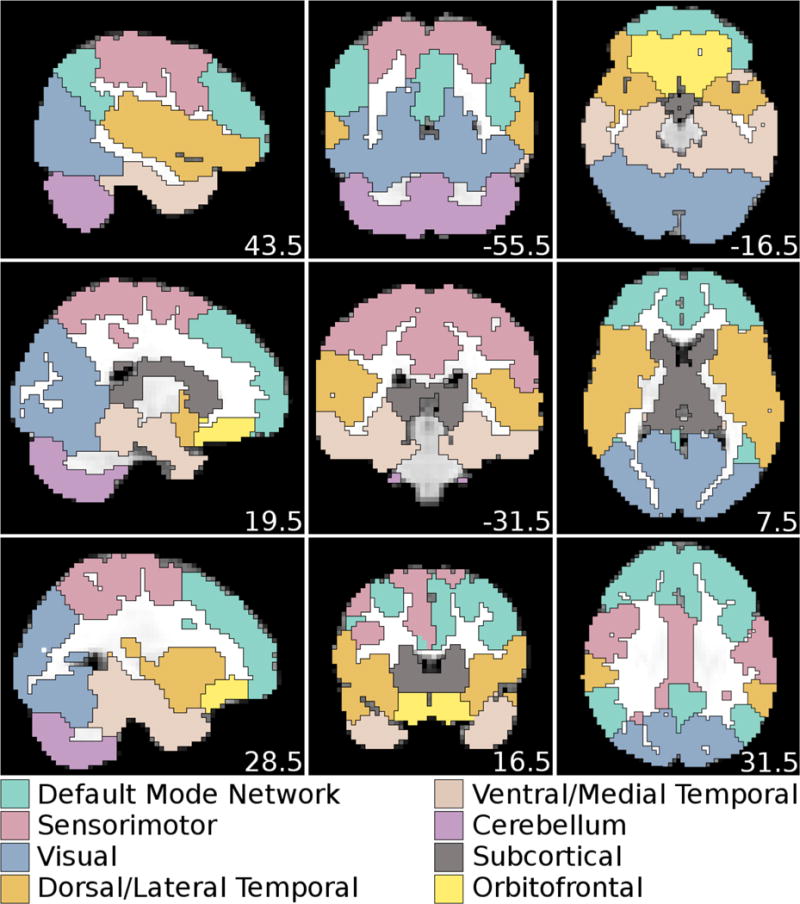
ROI network structure after running Infomap on the static FC graph of the 90th percentile of positive connections. Eight ROI subgraphs were detected by Infomap. Coordinates are in MNI space.
2.6 Static Connectivity Analysis
Average static within- and between-network connectivity, calculated as average covariance between ROIs in the same RSN or between ROIs in different RSNs, was computed for each participant. In addition, connectivity within the DMN and between the DMN and other networks was assessed. Multiple linear regression was used to evaluate possible relationships between static network connectivity and age, gender, and metabolic risk. Based on previous findings (La Joie et al., 2014; Wang et al., 2010), we also evaluated the associations of within-DMN connectivity and DMN-vMTL connectivity with cognitive performance, while controlling for age, gender, and metabolic risk.
2.7 Dynamic Covariance Calculation
Dynamic connectivity was assessed via sliding-window k-means clustering, after Allen et al. (2014). Covariance matrices were estimated from regularized precision matrices (Smith et al., 2011; Varoquaux et al., 2010) computed from windowed segments of the mean ROI time-series (Tukey window; width = 28 TR or 70 seconds, alpha = 0.2, step-size = 7 TR). Window width and step-size were chosen based on published research (Hutchison et al., 2013b; Shirer et al., 2012), with the aim of reducing autocorrelation between successive windows, and retaining adequate power to detect dynamic connectivity patterns. Regularization was carried out via group sparse covariance estimation (Varoquaux et al., 2010) with NiLearn (based on the scikit-learn package; (Abraham et al., 2014; Pedregosa et al., 2011). Regularization was optimized for each participant independently; the estimator was fit on a participant’s set of covariance matrices by assessing the likelihood of unseen matrices through leave-one-out cross-validation. Matrices were Fisher z-transformed after estimation.
We applied k-means clustering (Lloyd, 1982), using L1 distance (Aggarwal et al., 2001), to the set of all dynamic covariance matrices to identify consistent and differing dynamic connectivity patterns. K-means analysis was repeated with the number of clusters (k) ranging from 2 to 9, and run 500 times at each level of k to escape local minima. The final number of clusters, k = 8, was determined by the gap statistic (Tibshirani et al., 2001) with 50 generated reference samples. After determining k, the data were bootstrap-resampled 50 times and clustered the same way as the original data to assess stability of clusters via Jaccard similarity (Hennig, 2007). For each cluster in the original set, the similarity value between that cluster and the most similar cluster in the bootstrapped set was recorded. Maximum similarities over all 50 resampling rounds were averaged for each cluster (Table S1).
2.8 Dynamic Connectivity Profile Quantification
Modularity was calculated twice for each dynamic connectivity profile (Rubinov and Sporns, 2011): once using the static network definition, and once with a network structure determined by applying Infomap to the dynamic centroid. Modularity of a profile given the static network definition was interpreted as representing proximity of the modular structure of the dynamic profile to static structure. Modularity given dynamic network structure was interpreted as indication that a modular structure was present even if dynamic and static structures did not correspond.
Average whole-brain within- and between-network connectivity, as well as average connectivity within the DMN and between the DMN and the rest of the brain, were compared between each dynamic connectivity profile and the static profile with independent samples t-tests (Table 1). Independent samples t-tests were also used to compare each connection in the dynamic connectivity profiles to the corresponding connection in the static profile to assess individual ROI pairing connectivity differences (Figure S1). Dynamic connectivity profiles were also assessed qualitatively by plotting radial tree graphs of the top 10% of positive connections of the centroids (Figure 2).
Table 1.
Within- and between-network average connectivity differences between the static connectivity profile and dynamic connectivity profiles. Two-tailed independent samples t-tests. Negative t values indicate that the average connectivity was greater in the dynamic profile.
| Overall Within | Overall Between | DMN Within | DMN Between | |
|---|---|---|---|---|
| Static Average Connectivity | 0.2601 | −0.0265 | 0.2074 | −0.0293 |
|
| ||||
| Profile 1 Average Connectivity | 0.4205 | −0.0332 | 0.3199 | −0.0575 |
| t(410) | −15.12 | 4.9274 | −8.3144 | 7.7877 |
| p | **<.0001 | **<.0001 | **<.0001 | **<.0001 |
|
| ||||
| Profile 2 Average Connectivity | 0.2371 | −0.0234 | 0.1786 | −0.0261 |
| t(1485) | 4.5141 | −5.1539 | 4.7751 | −1.9272 |
| p | **<.0001 | **<.0001 | **<.0001 | 0.0549 |
|
| ||||
| Profile 3 Average Connectivity | 0.3048 | −0.0278 | 0.2417 | −0.0395 |
| t(775) | −6.8210 | 1.6469 | −4.2732 | 4.2302 |
| p | **<.0001 | 0.1008 | **<.0001 | **<.0001 |
|
| ||||
| Profile 6 Average Connectivity | 0.3582 | −0.0298 | 0.3123 | −0.0262 |
| t(417) | −10.4580 | 3.0395 | −9.4912 | −1.0227 |
| p | **<.0001 | **0.0026 | **<.0001 | 0.3076 |
|
| ||||
| Profile 7 Average Connectivity | 0.2668 | −0.0252 | 0.2492 | −0.0224 |
| t(984) | −1.1698 | −2.0326 | −6.0450 | −3.7135 |
| p | 0.2431 | *0.0430 | **<.0001 | **0.0002 |
|
| ||||
| Profile 8 Average Connectivity | 0.3011 | −0.0277 | 0.2624 | −0.0220 |
| t(627) | −5.7559 | 1.3968 | −5.9295 | −3.1728 |
| p | **<.0001 | 0.1638 | **<.0001 | **0.0017 |
Significant at α = 0.05
Significant after experiment-wise FDR correction
Figure 2.
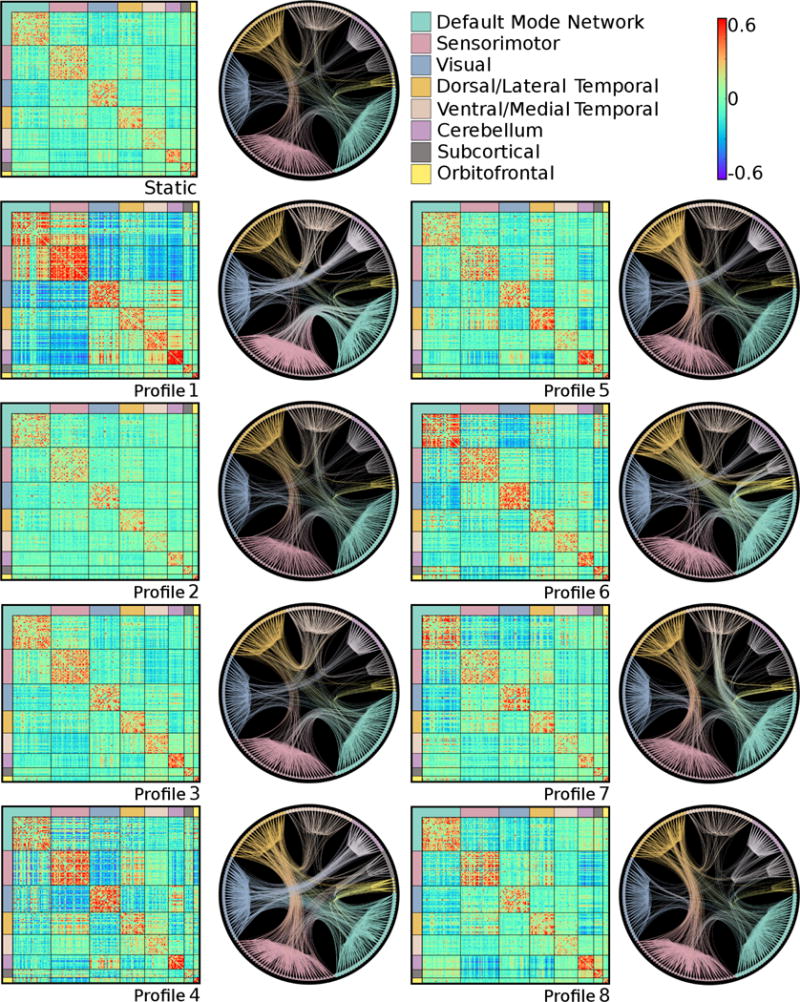
Functional connectivity for the static connectivity profile and each dynamic connectivity profile. Matrices represent average positive and negative connectivity between ROIs for all participants (static only) or average positive and negative connectivity between ROIs for all windows that correspond to a dynamic profile. Radial tree graphs represent the 90th percentile of positive connections.
2.9 Associating Dynamic Connectivity with Age, Gender, and Metabolic Risk
Having a portion of a scan allocated to a dynamic connectivity profile was treated for each participant as a binary outcome – 1 for having and 0 for not having. We used logistic regression to investigate the impact of age, gender, and metabolic risk on that outcome for each profile. The number of unique connectivity profiles and transitions between profiles per participant was also noted. Linear regression assessed the associations between age and metabolic risk with the number of unique dynamic profiles and the relationship between age and number of transitions.
Vectors indicating to which dynamic connectivity profile each windowed covariance matrix belonged were created for each participant. The amount of time allocated to each dynamic connectivity profile was then calculated and multiple linear regression was performed to examine the influences of age, gender, and metabolic risk on the relative amount of time allocated to a profile. Participants who did not have a portion of their scan correspond to a profile were not included in that regression; therefore, each analysis drew a different subsample of the participant pool.
2.10 Associating Dynamic Connectivity with Cognitive Performance
The event of allocating any time to a particular profile was related to cognitive performance in the subset of 91 participants while controlling for age, gender, and metabolic risk via ANCOVA. For participants who had some portion of their scan correspond to a profile, the relative amount of time allocated to that profile was related to cognitive performance constructs via general linear models with age, gender, and metabolic risk included. Only connectivity profiles 2, 3, and 7 were evaluated because too few participants with metabolic and cognitive data had connectivity patterns related to the other profiles (n ≪ 40).
2.11 Packages, Visualizations, and Multiple Comparison Correction
In-house Python (2.7.6) and R (3.3.1) scripts, and SPSS (23.0), were used for statistical analyses, FSLVIEW was used for ROI RSN structure visualization, graph-tool (Peixoto, 2014) was used for radial tree graph visualization, and matplotlib (Hunter, 2007) was used for all other visualizations. Experiment-wise False Discovery Rate correction (q=5%) was applied. Matrices comparing each static connection to the corresponding dynamic connections were not included in this FDR correction, and were instead Bonferroni corrected (α = 3.39E-6) on a per-matrix basis.
3. Results
3.1 Static Functional Connectivity
Advanced age was associated with lower functional connectivity between DMN and vMTL (β = −0.282, p = 0.001), and greater connectivity between DMN and SM (β = 0.222, p = 0.01), while controlling for metabolic risk and gender. Women, regardless of age or network type, had lower average within (β = −0.239, p = 0.008) and between (β = −0.233, p = 0.01) network connectivity compared to men.
3.2 Dynamic Functional Connectivity and its Correlates
3.2.1 Connectivity profiles
Eight dynamic connectivity profiles identified in these data are depicted in Figure 2. Not every participant entered each profile, and the mean number of profiles per person was 3.12 (SD = 1.00). Dynamic connectivity profiles 2, 3, and 7 had high membership, whereas the rest had moderate to low membership, particularly profile 4 (Table S1). All connectivity profiles, except profile 5, exhibited moderate (greater than 0.5) to high (greater than 0.7) average Jaccard values, indicating cluster stability (Table S1). Less stable profiles could reflect important between-subject variability, but also intermediate transitional states between more stable profiles. No profile represented a dissolved network configuration as all had moderate modularity given a network structure optimized for the profile centroid (Table S1). Because of low membership and low stability, profiles 4 and 5 were removed from further analyses. Differences between the remaining dynamic connectivity profiles and the static profile in whole brain and DMN-specific within- and between-network connectivity are detailed in Table 1 and depicted in Figure S1.
3.2.2 Predictors of connectivity profile presence
Differences in presence or absence of a connectivity profile may indicate differences in brain network organization and efficiency. We therefore applied logistic regression to test age, gender, and metabolic risk as predictors of individual dynamic connectivity profile membership (Table S2). Older age was associated with lower odds of having profiles 2 (OR = .957, p = .005) and 7 (OR = .953, p < .0001), and higher odds of having profile 3 (OR = 1.028, p =.008). Neither metabolic risk nor gender were associated with any of the dynamic profiles. The age difference in odds of having profiles 3 and 7 is in accord with our finding of lower DMN-vMTL connectivity in older adults in the static functional connectivity analysis, because profiles 3 has low whereas profile 7 has high DMN-vMTL between-network connectivity (Figure 2). Since the connectivity pattern of profile 2 is most similar to the static connectivity pattern (Figure S1), lower odds of the observing profile 2 could be interpreted as an age-related deviation from typical brain integration.
3.2.3 Number of unique connectivity profiles and profile transitions
The number of unique dynamic connectivity profiles per individual was unrelated to age or metabolic risk (R2 = .004, p = .72; age β = −.07, p = .43; metabolic risk β = .006, p = .94). Moreover, we observed no association between the number of profile transitions and age (R2 = .005, age β = −.07, p = .35). This was contrary to our expectations that older adults would have lower transition rates.
3.2.4 Age, Gender, Metabolic Risk, and Relative Time Allocated to Dynamic Connectivity Profiles
Older age was associated with more time allocated to profile 8 (β = .357, p = .009), that evidenced lesser connectivity between DMN and SM, Vis., and dLTL, and greater DMN-CB and DMN-SC connectivity compared to the static profile (Figure S1). Furthermore, greater metabolic risk score was associated with more time allocated to profile 2 (β = .278, p = .007) and less time to profile 7 (β = −.350, p = .003). None of the other tested associations reached significance (see Table S3).
3.2.5 Cognitive Performance and Functional Connectivity
Whether an individual exhibited a specific dynamic profile or not was unrelated to cognitive performance after controlling for age, gender, and metabolic risk. However, the relative amount of time allocated to profile 7 was positively associated with executive functions score (β = .297, p = .013) for participants who had that pattern of connectivity. Thus, the more time participants allocated to a dynamic connectivity pattern with high DMN connectivity, the better their performance was on tasks of executive functioning outside of the scanner. None of the other tested associations between time allocated to a profile and cognitive function reached significance (Table S4). Furthermore, static DMN connectivity was unrelated to cognitive performance beyond age, gender, and metabolic risk (Tables S5 and S6).
3.2.6 Effect of Motion on Dynamic Functional Connectivity
Motion artifacts can remain in the signal time series after motion regression (Power et al., 2012), and dynamic connectivity could be influenced by head motion (Laumann et al., 2016). Assessment of the association between head motion and dynamic functional connectivity in our data revealed that mean framewise displacement (FD) was associated with odds of demonstrating any profile except profile 8, and with time allocated to profile 2 (Tables S7 & S8). Mean FD was unrelated to rate of switching between profiles (r =.14, p = .06). The observed age, gender, and metabolic risk effects remained largely the same after including mean FD into the models. Only the age-related odds of demonstrating profile 2 were reduced to a trend level and age-related odds of demonstrating profile 3 were no longer significant (Table S7).
4. Discussion
We examined age-related differences in dynamic functional connectivity, and contrasted the results to findings obtained from a static connectivity analysis. In line with previous research, we found lower static functional connectivity between default mode regions and ventral and medial regions of the temporal lobe in older participants (Andrews-Hanna et al., 2007; Damoiseaux et al., 2016). This lower static DMN-vMTL connectivity may represent an age-related reduction in communication within putative memory systems (Ranganath and Ritchey, 2012). Projections from MTL regions to posterior DMN regions are part of a presumed posterior memory system, which is thought to be involved in episodic memory function (Kahn et al., 2008; Ranganath and Ritchey, 2012). Our finding of gender differences in static whole brain within- and between-network connectivity is in accord with previous reports of gender differences across functional connectivity measures (Gong et al., 2011; Tomasi and Volkow, 2012). Because of the association between gender and functional connectivity, we included gender in the models but the results remained significant after this control. No gender differences were observed in the dynamic connectivity measures.
4.1 Dynamic Resting Functional Connectivity
After partitioning the total BOLD time series into discrete segments, we found that older participants were less likely to allocate windowed time segments to a dynamic connectivity profile similar to the static state (profile 2) and a profile with high DMN-vMTL connectivity (profile 7) but more likely to allocate them to a profile with low DMN-vMTL connectivity (profile 3). These findings suggest that the lower connectivity observed in the static state can be explained by both lower and higher odds of having certain transient connectivity profiles.
Connectivity profile 2 has the highest membership of all profiles (i.e. present in 132 participants and accounted for 31.4% of all windows; Table S1) and is most similar to the static state (Figure S1). Therefore, this connectivity pattern may reflect a kind of “ground state” with other profiles possibly reflecting deviations from it that arise due to cognition, movement in the scanner (Laumann et al., 2016; Power et al., 2012; Van Dijk et al., 2012), sleep (Allen et al., 2014), or respiration and variations in arterial blood flow (Chang and Glover, 2010; Wise et al., 2004). The lower odds of having certain profiles in older adults could reflect known age-related differences observed using static connectivity approaches (Andrews-Hanna et al., 2007; Campbell et al., 2013; Damoiseaux et al., 2008; Damoiseaux et al., 2016), with the age differences observed in profile 3 and 7 providing additional information. The lower DMN-vMTL connectivity of profile 3 could indicate disconnection of posterior memory regions, which appears more prevalent in older adults. Conversely, connectivity in profile 7 possibly represents a strengthening or integration of anterior and posterior memory regions as it exhibits greater connectivity between not just DMN and vMTL, but also between DMN and OF, and vMTL and OF regions (Figure S1). Projections from MTL to OF regions are considered part of an anterior memory system (Ranganath and Ritchey, 2012).
Profile 7 is of interest in the context of cognitive aging. Association of the profile with lesser metabolic risk and better performance on executive functions suggests that modulation of the DMN by vMTL activity may be important in mitigating decline in typical age-sensitive cognitive domains (Yuan and Raz, 2014) and resisting negative influence of metabolic risk on cognition (Dahle et al., 2009). Both are aspects of aging that are increasingly viewed as mutually related determinants of late-life development (Allan et al., 2016). This is in accord with the extant reports that age-related DMN alterations at rest may be related to age-sensitive cognitive skills (Andrews-Hanna et al., 2007; Damoiseaux et al., 2008). Low membership of profile 7, therefore, may represent a marker for cognitive impairment. An unknown mechanism related to aging or metabolic risk might be preventing maintenance of this connectivity pattern at rest. Conversely, time allocation to profile 7 could be an indicator of cognitive reserve (Stern, 2002). Either way, longitudinal analysis is necessary to determine the relationship between change in cognitive performance and maintenance of dynamic connectivity patterns like profile 7.
Our results indicate that although individuals, regardless of age, can traverse multiple dynamic connectivity profiles, the odds of allocating time to a specific connectivity pattern vary with age. However, contrary to our expectations, we did not observe any association between older age and rate of switching between dynamic connectivity profiles. Because dynamic hub-region network variability decreases with age (Garrett et al., 2010; Schaefer et al., 2014), we hypothesized that functional connectivity patterns would be more similar across shorter periods for older adults. Contrary to this expectation, we found no association between age and the rate of profile switching. Possible explanations for this discrepancy may include differences in analysis approach and sample characteristics, such as exact age range and participant health, between the studies. It is also important to note that, regardless of the observed age-related differences, most of the associations tested did not reveal significant differences. Therefore, the stability of age differences in dynamic functional connectivity remains unclear and their magnitude may be relatively modest.
4.2 Limitations
Some of the identified connectivity profiles (e.g., profile 4) had low membership, a fact that may confound interpretation. It is possible that clusters with low membership are generalizable and that scanning conditions could be keeping most participants from entering those profiles. Those profiles, however, may also reflect artifacts that confound individual variability in connectivity.
One of the limitations of k-means clustering is the “curse of dimensionality.” As clustering algorithms are forced to classify a vast number of connections associated with whole-brain connectivity, dimensionality of a space grows, and empty space between data points increases exponentially. Such disproportionate increase in sparsity can jeopardize the effectiveness of k-means analysis, thus casting doubt on whether obtained clusters optimally represent a true configuration. Most clusters in the present analysis were stable and can therefore be interpreted as meaningful even if not representative of the “true” dynamic connectivity. Still, future analyses could assess dynamic interactions between select regions rather than attempting to quantify the whole brain to overcome dimensionality woes.
Furthermore, the use of group LASSO regression (Varoquaux et al., 2010) to enforce covariance sparsity on participants individually comes with caveats. Group LASSO forces the same sparsity structure on the set of input covariance matrices. Each participant therefore is assigned their own sparsity structure. Because we expect dynamic connectivity profiles to reveal different connectivity patterns, the “true” underlying sparsity structure between windows may differ. Therefore, applying LASSO is associated with a trade-off: attenuation of covariance estimation error comes with reduction of differences between connectivity profiles – i.e., LASSO might artificially make dynamic profiles more similar.
Simulation studies have raised concerns regarding the utility of sliding window approaches to assessing dynamic functional connectivity (Hindriks et al., 2016; Shakil et al., 2016). Future analysis is needed to establish optimal parameters and use of this approach. Dynamic patterns could also be contaminated by head motion (Laumann et al., 2016), and motion artifacts remain in the signal time course even after adjustment via motion regression (Power et al., 2012), therefore it is possible that static age-related connectivity differences and the age-related odds of having a specific dynamic connectivity profile could be related to the same motion artifacts. To address this possibility, we assessed the association between mean FD and odds of demonstrating a profile, time allocated to a profile, and rate of switching between profiles. We found that motion was associated with the odds of demonstrating, and the time allocated to, certain profiles. However, observed age, gender, and metabolic risk effects remained largely the same. Thus, although motion is associated with dynamic FC, it does not fully explain the relationship between dynamic FC, age, and metabolic risk.
It is also suggested that dynamic connectivity reflects systematic and reoccurring patterns of cerebral blood flow that might represent optimized global metabolic processing (Zalesky et al., 2014). However, given that machine learning classifiers can reliably discern between connectivity profiles that arise due to different task demands (Shirer et al., 2012), it is likely that unsupervised algorithms produce meaningful connectivity clusters that provide interesting information about the brain at rest that relates to cognitive functioning.
In this study, we used resting state functional MRI data to assess the effect of age on dynamic functional connectivity. It is possible that dynamic functional connectivity assessed during task-evoked functional MRI could yield slightly different results. As observed in the extant literature, static functional connectivity differences exist across different conditions (Shirer et al., 2012) although network connectivity structure remains similar (Smith et al., 2009). Therefore, our prediction regarding the effect of age on DMN connectivity would hold for task-evoked MRI data, even though specific profiles may look different. Nevertheless, more research comparing functional MRI data acquired under different conditions is needed to examine the effect of behavioral context.
5. Conclusion
We observed age-related differences in configuration of dynamic whole-brain functional connectivity patterns at rest. Advanced age, elevated metabolic risk, and low executive function scores were associated with lesser likelihood of a dynamic profile that is characterized by heightened functional connectivity between DMN and vMTL. Lower membership in specific dynamic profiles may denote a potential marker for age-related cognitive decline. Our results indicate that dynamic analysis can capture nuanced differential age-related FC patterns that are obscured by aggregation of BOLD data over the whole time-series.
Supplementary Material
Highlights.
Age differences in dynamic connectivity profiles were found in healthy adults.
Dynamic profile with high DMN connectivity was linked to executive functions.
Dynamic connectivity was associated with metabolic risk factors.
Acknowledgments
This work was supported by grant R37 AG011230 from the National Institutes of Health to Naftali Raz. We thank Cheryl Dahle, Andrew Bender, Ana Daugherty, and Yiqin Yang for assistance in data collection and management.
Abbreviations
- DMN
Default Mode Network
- SM
Sensorimotor Network
- Vis.
Visual Network
- dLTL
Dorsal Lateral Temporal Network
- vMTL
Ventral Medial Temporal Network
- CB
Cerebellum
- SC
Subcortical Network
- OF
Orbitofrontal Cortex
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham A, Pedregosa F, Eickenberg M, Gervais P, Mueller A, Kossaifi J, Gramfort A, Thirion B, Varoquaux G. Machine learning for neuroimaging with scikit-learn. Front Neuroinform. 2014;8:14. [Google Scholar]
- Aggarwal CC, Hinneburg A, Keim DA. On the surprising behavior of distance metrics in high dimensional space. Lect Notes Comput Sc. 2001;1973:420–434. [Google Scholar]
- Allan JL, McMinn D, Daly M. A Bidirectional Relationship between Executive Function and Health Behavior: Evidence, Implications, and Future Directions. Front Neurosci-Switz. 2016;10 doi: 10.3389/fnins.2016.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD. Tracking Whole-Brain Connectivity Dynamics in the Resting State. Cereb Cortex. 2014;24(3):663–676. doi: 10.1093/cercor/bhs352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews-Hanna JR, Snyder AZ, Vincent JL, Lustig C, Head D, Raichle ME, Buckner RL. Disruption of large-scale brain systems in advanced aging. Neuron. 2007;56(5):924–935. doi: 10.1016/j.neuron.2007.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos T Roy Soc B. 2005;360(1457):1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional Connectivity in the Motor Cortex of Resting Human Brain Using Echo-Planar Mri. Magnet Reson Med. 1995;34(4):537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Andrews-Hanna JR, Schacter DL. The brain’s default network – Anatomy, function, and relevance to disease. Ann Ny Acad Sci. 2008;1124:1–38. doi: 10.1196/annals.1440.011. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Kiehl KA, Pearlson GD. Modulation of temporally coherent brain networks estimated using ICA at rest and during cognitive tasks. Hum Brain Mapp. 2008;29(7):828–838. doi: 10.1002/hbm.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KL, Grigg O, Saverino C, Churchill N, Grady CL. Age differences in the intrinsic functional connectivity of default network subsystems. Front Aging Neurosci. 2013;5 doi: 10.3389/fnagi.2013.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MY, Park DC, Savalia NK, Petersen SE, Wig GS. Decreased segregation of brain systems across the healthy adult lifespan. P Natl Acad Sci USA. 2014;111(46):E4997–E5006. doi: 10.1073/pnas.1415122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH. Time-frequency dynamics of resting-state brain connectivity measured with fMRI. Neuroimage. 2010;50(1):81–98. doi: 10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes D, Haughton VM, Arfanakis K, Carew JD, Turski PA, Moritz CH, Quigley MA, Meyerand ME. Frequencies contributing to functional connectivity in the cerebral cortex in “resting-state” data. Am J Neuroradiol. 2001;22(7):1326–1333. [PMC free article] [PubMed] [Google Scholar]
- Craddock RC, James GA, Holtzheimer PE, Hu XPP, Mayberg HS. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum Brain Mapp. 2012;33(8):1914–1928. doi: 10.1002/hbm.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahle CL, Jacobs BS, Raz N. Aging, Vascular Risk, and Cognition: Blood Glucose, Pulse Pressure, and Cognitive Performance in Healthy Adults. Psychol Aging. 2009;24(1):154–162. doi: 10.1037/a0014283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JS, Beckmann CF, Arigita EJS, Barkhof F, Scheltens P, Stam CJ, Smith SM, Rombouts SARB. Reduced resting-state brain activity in the “default network” in normal aging. Cereb Cortex. 2008;18(8):1856–1864. doi: 10.1093/cercor/bhm207. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SARB, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. Consistent resting-state networks across healthy subjects. P Natl Acad Sci USA. 2006;103(37):13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JS, Viviano RP, Yuan P, Raz N. Differential effect of age on posterior and anterior hippocampal functional connectivity. Neuroimage. 2016;133:468–476. doi: 10.1016/j.neuroimage.2016.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M, Beckmann CF, De Stefano N, Matthews PM, Smith SM. fMRI resting state networks define distinct modes of long-distance interactions in the human brain. Neuroimage. 2006;29(4):1359–1367. doi: 10.1016/j.neuroimage.2005.08.035. [DOI] [PubMed] [Google Scholar]
- Fjell AM, McEvoy L, Holland D, Dale AM, Walhovd KB, Initi, A.s.D.N. What is normal in normal aging? Effects of aging, amyloid and Alzheimer’s disease on the cerebral cortex and the hippocampus. Prog Neurobiol. 2014;117:20–40. doi: 10.1016/j.pneurobio.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, Mchugh PR. Mini-Mental State – Practical Method for Grading Cognitive State of Patients for Clinician. J Psychiat Res. 1975;12(3):189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. P Natl Acad Sci USA. 2005;102(27):9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett DD, Kovacevic N, McIntosh AR, Grady CL. Blood Oxygen Level-Dependent Signal Variability Is More than Just Noise. J Neurosci. 2010;30(14):4914–4921. doi: 10.1523/JNEUROSCI.5166-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady CL, Springer MV, Hongwanishkul D, McIntosh AR, Winocur G. Age-related changes in brain activity across the adult lifespan. J Cognitive Neurosci. 2006;18(2):227–241. doi: 10.1162/089892906775783705. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: A network analysis of the default mode hypothesis. P Natl Acad Sci USA. 2003;100(1):253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy SM, Brewer HB, Cleeman JI, Smith SC, Lenfant C, Participants C. Definition of metabolic syndrome – Report of the National Heart, Lung, and Blood Institute/American Heart Association Conference on Scientific Issues Related to Definition. Circulation. 2004;109(3):433–438. doi: 10.1161/01.CIR.0000111245.75752.C6. [DOI] [PubMed] [Google Scholar]
- Hennig C. Cluster-wise assessment of cluster stability. Comput Stat Data An. 2007;52(1):258–271. [Google Scholar]
- Hindriks R, Adhikari MH, Murayama Y, Ganzetti M, Mantini D, Logothetis NK, Deco G. Can sliding-window correlations reveal dynamic functional connectivity in resting-state fMRI? Neuroimage. 2016;127:242–256. doi: 10.1016/j.neuroimage.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JD. Matplotlib: A 2D graphics environment. Comput Sci Eng. 2007;9(3):90–95. [Google Scholar]
- Hutchison RM, Womelsdorf T, Allen EA, Bandettini PA, Calhoun VD, Corbetta M, Penna S, Duyn JH, Glover GH, Gonzalez-Castillo J, Handwerker DA, Keilholz S, Kiviniemi V, Leopold DA, Pasquale F, Sporns O, Walter M, Chang C. Dynamic functional connectivity: Promise, issues, and interpretations. Neuroimage. 2013a;80:360–378. doi: 10.1016/j.neuroimage.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Gati JS, Everling S, Menon RS. Resting-state networks show dynamic functional connectivity in awake humans and anesthetized macaques. Hum Brain Mapp. 2013b;34(9):2154–2177. doi: 10.1002/hbm.22058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17(2):825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Kahn I, Andrews-Hanna JR, Vincent JL, Snyder AZ, Buckner RL. Distinct cortical anatomy linked to subregions of the medial temporal lobe revealed by intrinsic functional connectivity. J Neurophysiol. 2008;100(1):129–139. doi: 10.1152/jn.00077.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy K, Raz N. Normal aging of the brain, Brain Mapping: An Encyclopedic Reference. Academic Press; Elsevier Cambridge: 2015. pp. 603–617. [Google Scholar]
- La Joie R, Landeau B, Perrotin A, Bejanin A, Egret S, Pelerin A, Mezenge F, Belliard S, de la Sayette V, Eustache F, Desgranges B, Chetelat G. Intrinsic Connectivity Identifies the Hippocampus as a Main Crossroad between Alzheimer’s and Semantic Dementia-Targeted Networks. Neuron. 2014;81(6):1417–1428. doi: 10.1016/j.neuron.2014.01.026. [DOI] [PubMed] [Google Scholar]
- Lancichinetti A, Fortunato S. Community detection algorithms: A comparative analysis. Phys Rev E. 2009;80(5) doi: 10.1103/PhysRevE.80.056117. [DOI] [PubMed] [Google Scholar]
- Laumann TO, Snyder AZ, Mitra A, Gordon EM, Gratton C, Adeyemo B, Gilmore AW, Nelson SM, Berg JJ, Greene DJ, McCarthy JE, Tagliazucchi E, Laufs H, Schlaggar BL, Dosenbach NU, Petersen SE. On the Stability of BOLD fMRI Correlations. Cereb Cortex. 2016 doi: 10.1093/cercor/bhw265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd SP. Least-Squares Quantization in Pcm. Ieee T Inform Theory. 1982;28(2):129–137. [Google Scholar]
- Lowe MJ, Mock BJ, Sorenson JA. Functional connectivity in single and multislice echoplanar imaging using resting-state fluctuations. Neuroimage. 1998;7(2):119–132. doi: 10.1006/nimg.1997.0315. [DOI] [PubMed] [Google Scholar]
- Lustig C, Snyder AZ, Bhakta M, O’Brien KC, McAvoy M, Raichle ME, Morris JC, Buckner RL. Functional deactivations: Change with age and dementia of the Alzheimer type. P Natl Acad Sci USA. 2003;100(24):14504–14509. doi: 10.1073/pnas.2235925100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhyastha TM, Grabowski TJ. Age-related differences in the dynamic architecture of intrinsic networks. Brain Connect. 2014;4(4):231–241. doi: 10.1089/brain.2013.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Birn RM, Handwerker DA, Jones TB, Bandettini PA. The impact of global signal regression on resting state correlations: are anti-correlated networks introduced? Neuroimage. 2009;44(3):893–905. doi: 10.1016/j.neuroimage.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthén LK, Muthén BO. Mplus user’s guide 1998–2010 [Google Scholar]
- Oldfield RC. The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia. 1971;9(1):97–113. doi: 10.1016/0028-3932(71)90067-4. [DOI] [PubMed] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- Peixoto TP. The Graph-Tool Python Library. Figshare; 2014. [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59(3):2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff LS. The CES-D scale: A self-report depression scale for research in the general population. Applied psychological measurement. 1977;1(3):385–401. [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. P Natl Acad Sci USA. 2001;98(2):676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganath C, Ritchey M. Two cortical systems for memory-guided behaviour. Nature Reviews Neuroscience. 2012;13(10):713–726. doi: 10.1038/nrn3338. [DOI] [PubMed] [Google Scholar]
- Raz N, Rodrigue KM, Kennedy KM, Land S. Genetic and Vascular Modifiers of Age-Sensitive Cognitive Skills: Effects of COMT, BDNF, ApoE, and Hypertension. Neuropsychology. 2009;23(1):105–116. doi: 10.1037/a0013487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosvall M, Bergstrom CT. Maps of random walks on complex networks reveal community structure. P Natl Acad Sci USA. 2008;105(4):1118–1123. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Weight-conserving characterization of complex functional brain networks. Neuroimage. 2011;56(4):2068–2079. doi: 10.1016/j.neuroimage.2011.03.069. [DOI] [PubMed] [Google Scholar]
- Sakoglu U, Pearlson GD, Kiehl KA, Wang YM, Michael AM, Calhoun VD. A method for evaluating dynamic functional network connectivity and task-modulation: application to schizophrenia. Magn Reson Mater Phy. 2010;23(5–6):351–366. doi: 10.1007/s10334-010-0197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Llonch R, Pena-Gomez C, Arenaza-Urquijo EM, Vidal-Pineiro D, Bargallo N, Junque C, Bartres-Faz D. Brain connectivity during resting state and subsequent working memory task predicts behavioural performance. Cortex. 2012;48(9):1187–1196. doi: 10.1016/j.cortex.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Sambataro F, Murty VP, Callicott JH, Tan HY, Das S, Weinberger DR, Mattay VS. Age-related alterations in default mode network: Impact on working memory performance. Neurobiol Aging. 2010;31(5):839–852. doi: 10.1016/j.neurobiolaging.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Margulies DS, Lohmann G, Gorgolewski KJ, Smallwood J, Kiebel SJ, Villringer A. Dynamic network participation of functional connectivity hubs assessed by resting-state fMRI. Front Hum Neurosci. 2014;8 doi: 10.3389/fnhum.2014.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakil S, Lee CH, Keilholz SD. Evaluation of sliding window correlation performance for characterizing dynamic functional connectivity and brain states. Neuroimage. 2016;133:111–128. doi: 10.1016/j.neuroimage.2016.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirer WR, Ryali S, Rykhlevskaia E, Menon V, Greicius MD. Decoding Subject-Driven Cognitive States with Whole-Brain Connectivity Patterns. Cereb Cortex. 2012;22(1):158–165. doi: 10.1093/cercor/bhr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman GL, Fiez JA, Corbetta M, Buckner RL, Miezin FM, Raichle ME, Petersen SE. Common Blood Flow Changes across Visual Tasks: II. Decreases in Cerebral Cortex. J Cogn Neurosci. 1997;9(5):648–663. doi: 10.1162/jocn.1997.9.5.648. [DOI] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF. Correspondence of the brain’s functional architecture during activation and rest. P Natl Acad Sci USA. 2009;106(31):13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang YY, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, Ramsey JD, Woolrich MW. Network modelling methods for FMRI. Neuroimage. 2011;54(2):875–891. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- Stern Y. What is cognitive reserve? Theory and research application of the reserve concept. J Int Neuropsych Soc. 2002;8(3):448–460. [PubMed] [Google Scholar]
- Tibshirani R, Walther G, Hastie T. Estimating the number of clusters in a data set via the gap statistic. J Roy Stat Soc B. 2001;63:411–423. [Google Scholar]
- Van Dijk KRA, Sabuncu MR, Buckner RL. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage. 2012;59(1):431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoquaux G, Gramfort A, Poline JB, Thirion B. Brain covariance selection: better individual functional connectivity models using population prior, Advances in Neural Information Processing Systems. 2010. pp. 2334–2342. [Google Scholar]
- Wang L, LaViolette P, O’Keefe K, Putcha D, Bakkour A, Van Dijk KRA, Pihlajamaki M, Dickerson BC, Sperling RA. Intrinsic connectivity between the hippocampus and posteromedial cortex predicts memory performance in cognitively intact older individuals. Neuroimage. 2010;51(2):910–917. doi: 10.1016/j.neuroimage.2010.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RG, Ide K, Poulin MJ, Tracey I. Resting fluctuations in arterial carbon dioxide induce significant low frequency variations in BOLD signal. Neuroimage. 2004;21(4):1652–1664. doi: 10.1016/j.neuroimage.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Wu JT, Wu HZ, Yan CG, Chen WX, Zhang HY, He Y, Yang HS. Aging-related changes in the default mode network and its anti-correlated networks: A resting-state fMRI study. Neurosci Lett. 2011;504(1):62–67. doi: 10.1016/j.neulet.2011.08.059. [DOI] [PubMed] [Google Scholar]
- Yin D, Liu WJ, Zeljic K, Wang ZW, Lv Q, Fan MX, Cheng WH, Wang Z. Dissociable Changes of Frontal and Parietal Cortices in Inherent Functional Flexibility across the Human Life Span. J Neurosci. 2016;36(39):10060–10074. doi: 10.1523/JNEUROSCI.1476-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Raz N. Prefrontal cortex and executive functions in healthy adults: A meta-analysis of structural neuroimaging studies. Neurosci Biobehav R. 2014;42:180–192. doi: 10.1016/j.neubiorev.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Cocchi L, Gollo LL, Breakspear M. Time-resolved resting-state brain networks. P Natl Acad Sci USA. 2014;111(28):10341–10346. doi: 10.1073/pnas.1400181111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YY, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. Ieee T Med Imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


