Abstract
Early thymic progenitors (ETPs) are endowed with diverse potencies and can give rise to myeloid and lymphoid lineage progenitors. How the thymic environment guides ETP commitment and maturation towards a specific lineage remains obscure. We have previously shown that ETPs expressing the heteroreceptor (HR) comprising IL-4 receptor alpha (IL-4Rα) and IL-13 receptor alpha 1 (IL-13Rα1) give rise to myeloid but not T cells. Here we show that signaling through the HR inhibits ETP maturation to the T cell lineage but enacts commitment towards the myeloid cells. Indeed, HR-positive ETPs (HR+ETPs), but not HR-negative ETPs (HR−ETPs), exhibit activated STAT6 transcription factor which parallels with downregulation of Notch1, a critical factor for T cell development. Meanwhile, myeloid-specific transcription factor, C/EBPα, usually under the control of Notch1 is up-regulated. Furthermore, in vivo inhibition of STAT6 phosphorylation restores Notch1 expression in HR+ETPs which regain T-lineage potential. In addition, upon stimulation with IL-4 or IL-13 HR−ETPs expressing virally transduced HR, also exhibit STAT 6 phosphorylation and down-regulation of Notch1 leading to inhibition of lymphoid but not myeloid lineage potential. These observations indicate that environmental cytokines play a role in conditioning ETP lineage choice which would impact T cell development.
Introduction
Bone marrow (BM)-derived thymic settling progenitors (TSPs) (1) undergo a maturation process to give rise to a massive number of young thymocytes. Early on, TSPs were considered to be early T-cell lineage progenitors destined to give rise mostly to T cells (2). Later on, however, these progenitors were found to give rise to both lymphoid and myeloid cells (3, 4) and were referred to as early thymic progenitors (ETPs) to accommodate their multipotent attribute (3). Although the maturation process of ETPs is relatively well defined (5–7), the environmental trigger for ETP commitment remains largely unknown. Recent studies identified ETP subsets that could only differentiate to one specific lineage (8–10). A common feature associated with these “unipotent” subsets is expression of a cytokine receptor. For instance, we have previously reported that the unipotent attribute of an ETP subset identified in the thymus is tied to expression of the IL-13Rα1 chain (9), which is known to associate with IL-4Rα to form a functional heteroreceptor (HR) through which both IL-4 and IL-13 can signal (11–13). This HR-positive ETP subset (HR+ETP) is restricted to the myeloid lineage and gives rise to CD11b+ cells both in vitro when cultured on stromal cells and in vivo when intra-thymically injected into HR-deficient (HR−/−) mice (9). However, HR+ETPs do not to give rise to T cells either in vitro or in vivo upon intrathymic transfer (9). These observations point to a link between the HR and restriction of commitment to the myeloid lineage as the HR offers a responsive element to the thymic environment that could be triggered by both IL-4 and IL-13 cytokines. Given that cytokine signaling through the HR has been shown to play a role in the death of neonatal Th1 cells (12), the function of dendritic cells (14, 15) and the differentiation of macrophages (13), we postulate that the HR on ETPs plays an active role in their commitment to a specific lineage. Specifically, environmental IL-4 and IL-13 could trigger HR signaling and guide commitment to the myeloid lineage. This indeed proved to be correct as HR+ETPs display an active form of STAT6 transcription factor which plays a critical role in antagonizing Notch1 expression and commitment to the T-cell lineage. Interference with Notch1 enacted the myeloid pathway, hence commitment of the ETPs to CD11b myeloid cells. These observations point to a new role environmental IL-4/IL-13 and their HR plays in ETP maturation which would impact central tolerance and T cell development.
Materials and Methods
Mice
All animal experiments were done according to protocols approved by the University of Missouri Animal Care and Use Committee. C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). IL-13Rα1+/+-GFP and IL-13Rα1−/− C57BL/6 mice were previously described (9). Only female mice were used throughout the study. Animals were typically 6–8 weeks old at the time experiments were performed. All animals were maintained under specific pathogen–free conditions in individually ventilated cages and kept on a 12 h light-dark cycle with access to food and water ad libitum.
Flow Cytometry
Antibodies
Anti-CD3 (145-2C11), anti-CD4 (RM4-5), anti-CD8 (53-6.7), anti-CD25 (7D4), anti-CD44 (IM7), anti-CD45 (30-F11), anti-CD11b (M1/70), anti-CD117 (2B8), anti-CD127 (SB/199), anti-Id3 (S30-778), anti-pSTAT6Y641 (J71-773.58.11) and anti-Tcf1(S33-966) antibodies were purchased from BD Biosciences (San Jose, CA). Anti-Notch1 antibody (22E5) and anti-pERK1/2T202/Y204 (MILAN8R) were purchased from e-biosciences (San Diego, CA). Anti-Hes1 (7H11) and anti-C/EBPα (EP709Y) antibodies were from Abcam (Cambridge, MA). Anti-IL-13Rα1 antibody (1G3-A7) produced in our laboratory was previously described (13).
Antibody lineage (Lin) depletion cocktail
This kit which was purchased from Miltenyi Biotech includes antibodies against CD4 (L3T4), CD8α (Ly-2), CD11b (Mac-1), CD11c, CD19, B220 (CD45R), CD49b (DX5), CD105, MHCII+, Ter-119+, and TCRγ /δ.
Fluorochromes
Antibodies were directly conjugated to fluorescein isothiocyanate (FITC), phycoerythrin (PE), PE-Cy5, PE-Cy5.5, peridinin-chlorophyll-protein complex (PerCP)-Cy5.5, PE-Cy7, allophycocyanin (APC), APC-Cy7 (or APCeFluor780), or biotin. Biotinylated antibodies were revealed with Streptavidin PE.
Sample reading
Sample analysis utilized a Beckman Coulter CyAn (Brea, CA) and data were analysed using FlowJo version 10 (Tree Star). Dead cells were excluded using 7-aminoactinomycin D (7-AAD; EMD Biosciences) or Fixable Viability Dye (FVD) eFluor® 780 (ebioscience).
Cell sorting
ETPs
ETPs were isolated as previously described (9). In brief, thymi were harvested from either IL-13Rα1+/+-GFP or IL-13Rα1−/− C57BL/6 mice after perfusion with PBS, the CD4+ cells were eliminated by MACS using anti-CD4 microbeads. The ETPs were then isolated after depletion of Lin+ (CD8α+, CD11b+, CD11c+, CD19+, B220+, CD49b+, CD105+, MHCII+, Ter-119+, TCRγ/δ+) thymic cells. HR+ETPs (cKit+CD44+CD25−) were sorted from Lin− thymic cells of IL-13Rα1+/+-GFP reporter mice on the basis of GFP (IL-13Rα1) expression. HR−/PETPs represent the GFP− cells of the lin−cKit+CD44+CD25− thymic cells. These cells have the genetic potential for receptor expression because they are isolated from IL-13Rα1+/+-GFP reporter mice and as such referred to as HR−/PETPs. HR−/−ETPs were sorted from Lin− thymic cells of IL-13Rα1−/− mice on the basis of CD44, cKit and CD25 (cKit+CD44+CD25−).
DN1c thymocytes
Thymic cells from IL-13Rα1-GFP reporter mice were depleted of Lin+ cells and the HR+DN1c population was sorted as GFP+CD44+CD25−CD24+c-Kitint cells. HR−DN1c population was sorted as CD44+CD25−CD24+c-Kitint cells from Lin− thymic cells of IL-13Rα1−/− mice.
Sorting was performed on a Beckman Coulter MoFlo XDP (Brea, CA) cell sorter. Cell purity was routinely checked, and only sorts with a purity of >95% were used in this study.
OP9 and OP9-DL1 Cell Culture
OP9 and OP9-DL1 cultures were used as previously described (16) with slight modifications. Briefly, OP9 and OP9-DL1 stromal cells were plated 2 days before initiation of cultures at a concentration of 20,000 cells/ml in 24-well plates. Progenitors were then added at the indicated cell number per well. IL-7 was used at a final concentration of 1 ng/ml, Flt3 ligand at 5ng/ml, GM-CSF at 10ng/ml, IL-4 at 10ng/ml and IL-13 at 20ng/ml. Lymphoid progeny were evident at 3–10 days of OP9-DL1 cell culture and myeloid progeny were evident at day 3 of OP9 cell culture under these conditions.
Cloning of IL-13Rα1 and Tcf-1 into Retroviral Vectors
Total RNA was isolated from gut epithelial cells for IL-13Rα1 and thymocytes for Tcf-1, cDNA was made using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) according to manufacturer’s protocol. cDNA was used as a template to amplify IL-13Rα1 or Tcf-1 using gene specific primers. The amplified products were then cloned into Not I/Cla I restriction sites of the MSCV-IRES-Thy1.1(17) vector to generate MSCV-IL-13Rα1-IRES-Thy1.1 (HR-RV) and MSCV-Tcf-1-IRES-Thy1.1 (Tcf-1-RV), respectively. The primers used to clone IL-13Rα1 and Tcf-1 are:
IL-13Rα1
sense 5′-TAGTAGGCGGCCGCACCATGGCGCGGCCAGCGCTGCTGGGCGAG-3′
antisense 5′-TAGTAGATCGAT CCATCAAGGAGCTGCTTTCTTCAG-3′
Tcf-1
sense 5′-TAGTAGGCGGCCGCACCATGTACAAAGAGACTGTCTACT-3′
antisense 5′-AATAGTAGATCGATCCACTAGAGCACTGTCATCGGAAGG-3′
The constructs containing IL-13Rα1 or Tcf-1 genes were verified by automated sequencing.
Retroviral Transduction
Retroviral packaging was performed as described with slight modifications (17). Briefly, 293FT cells (Invitrogen) were transfected with either HR-RV or Tcf-1-RV or control vector MSCV-IRES-Thy1.1 (Empty-RV) along with the retroviral packaging vector (p-cECO) by Lipofectamine 2000. After 48 hr, viral supernatants were then mixed with polybrene (1μg/ml) and used to transduce ETPs. Retrovirus infected ETPs were then suspended for 48 hours in stimulation DMEM cocktail containing 1% penicillin/streptomycin, 20% fetal calf serum (FCS), L-glutamate (2 mM), IL-3 (10ng/ml), IL-6 (10ng/ml), SCF (20ng/ml) and Flt3-ligand (20ng/ml) before sorting.
Single cell analysis
ETPs (DN1a,b population) sorted from HR−/− mice were transduced with either HR-RV or Empty-RV and used in single cell culture on OP9-DL1 stromal cells. Single cells were dispensed in 96-well plates containing OP9-DL1 stromal cells using a Beckman Coulter MoFlo XDP (Brea, CA) cell sorter. The culture was then supplemented with 1 ng/ml IL-7 and 5 ng/ml Flt3 ligand without (NIL) or with 10 ng/ml IL-4 and 20 ng/ml IL-13. After 10 days, the resulting cultures were analysed for the expression of CD25 T-cell lineage marker by CD45+ live cells.
RT-PCR
Sorted HR+ETPs (DN1c), HR−/PETPs or HR−/−DN1c thymocytes were used to isolate RNA by Trizol extraction and isopropanol precipitation. RT and DNA amplification were performed on a StepOnePlus Instrument cycler (Applied Biosystems) using Power SYBR Green RNA-to-CT1-Step Kit (Applied Biosystems) according to the manufacturer’s instructions. RT-PCR was done with primers specific for:
Tcf1
sense 5′-CCAGTGTGCACCCTTCCTAT-3′
antisense 5′-AGCCCCACAGAGAAACTGAA-3′
Hes1
sense 5′-CGGCATTCCAAGCTAGAGAAGG-3′
antisense 5′-GGTAGGTCATGGCGTTGATCTG-3′
C/EBPα
sense 5′–AGCAACGAGTACCGGGTACG-3′
antisense 5′-GTTTGGCTTTATCTCGGCTC-3′
Notch1
sense 5′-GGACATGCAGAACAACAAGG-3′
antisense 5′-CAGTCTCATAGCTGCCCTCA-3′
Id3
sense 5′-AGCTTAGCCAGGTGGAAATCCT-3′
antisense 5′-TCAGCTGTCTGGATCGGGAG-3′
Deltex1
sense 5′-GAGGATGTGGTTCGGAGGTA-3′
antisense 5′-CCCTCATAGCCAGATGCTGT-3′
IL-7Rα
sense 5′-AGTCCGATCCATTCCCCATAA-3′
antisense 5′-ATTCTTGGGTTCTGGAGTTTCG-3′
Rag2
sense 5′–CACATCCACAAGCAGGAAGTACAC-3′
antisense 5′-GGTTCAGGGACATCTCCTACTAAG-3′
Ptcrα
sense 5′-CTGGCTCCACCCATCACACT-3′
antisense 5′-TGCCATTGCCAGCTGAGA-3′
CD25
sense 5′-AACCATAGTACCCAGTTGTCGG-3′
antisense 5′-TCCTAAGCAACGCATATAGACCA-3′
Gata3
sense 5′-GAGGTGGTGTCTGCATTCCAA-3′
antisense 5′-TTTCACAGCACTAGAGACCCTGTTA-3′
Lat
sense 5′-CTGTTGTCTCCTCTGCTCCTGT-3′
antisense 5′-CTCACTCTCAGGAACATTCACG-3′
Lck
sense 5′-CTAGTCCGGCTTTATGCAGTG-3′
antisense 5′-CCGAGGGAGTCTTGAGAAAAT-3′
Egr1
sense 5′-GAGGAGATGATGCTGCTGAG-3′
antisense 5′-TGCTGCTGCTGCTATTACC-3′
GAPDH
sense 5′-AACTTTGGCATTGTGGAAGG-3′
antisense 5′-GGATGCAGGGATGATGTTCT-3′
Relative transcript abundance was determined by using the comparative threshold cycle method using the StepOne software (Applied Biosystems) normalization with GAPDH. The forward and reverse primers are listed in supplementary documents. All samples were run in triplicate.
In vivo and in vitro inhibition of STAT6 and ERK1/2
SCH772984 ERK1/2 inhibitor (Selleckchem) and AS1517499 STAT6 inhibitor (Axon Medchem) were dissolved in 50μl DMSO/PBS (1vol/1vol) and administered to mice intraperitoneally. SCH772984 (12.5 mg/kg) was injected twice per day for 10 days, and AS1517499 (10 mg/kg) was given once a day for 5 days. DMSO/PBS with no inhibitor was used as control. For in vitro inhibition STAT6 and ERK1/2 inhibitors were used at 200nM and 10μM, respectively.
In vitro HR signaling assay
HR−ETPs (Lin− CD4− CD8− CD25− CD44+ c-Kit+) transduced with HR-RV were cultured for 48 hours in stimulation DMEM cocktail and sorted on the basis of IL-13Rα1 and Thy1.1 expression. Transduced (HR+) cells were then cultured in the presence or absence of different cytokines (10ng/ml IL-4, 20ng/ml IL-13, 10ng/ml IFN-γ or 10ng/ml IL-12) for either 1 hr (for phosphorylation of intracellular signaling molecules) or 24 hr (for expression of transcription factors). STAT6 or ERK1/2 inhibitors were used as above.
Statistical Analysis
Data were analyzed using either an unpaired, two-tailed Students t-test, or one-way ANOVA as indicated. All statistical analyses were performed using Prism software version 4.0c (GraphPad).
Results
HR expression restricts ETP commitment to the myeloid lineage
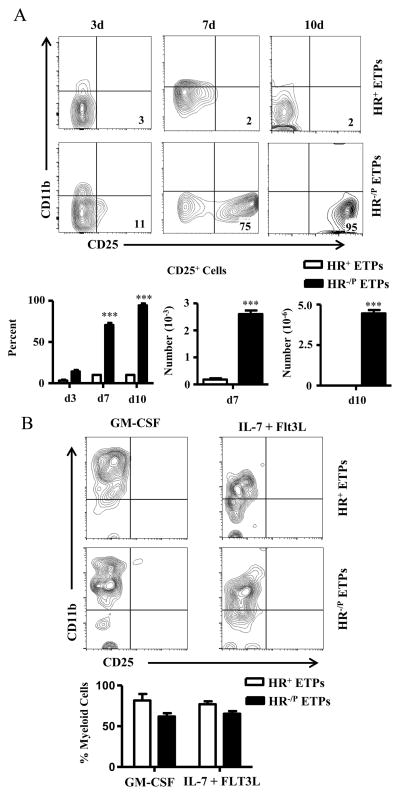
In our previous study, a short time culture on OP9-DL1 stromal cells indicated that HR-negative ETPs from HR+/+ mice, referred to as HR−/P due to their genetic potential for receptor expression, differentiate into the T-lineage cells while HR+ETPs do not (9). As the kinetics of ETP maturation are flexible (3, 18) the question remains open as to whether the HR influences ETP maturation in a time dependent manner. To test this premise, ETPs were sorted into HR−/P and HR+ ETPs (Fig. S1), co-cultured on OP9-DL1 cells for extended time periods, and assessed for commitment to the T cell lineage. The results show that while HR−/PETP maturation on OP9-DL1 stromal cells, which support lymphoid differentiation (16), yielded T-lineage cells that reached optimal numbers by day 10, the HR+ETPs were unable to mature through the T-lineage pathway throughout the 10 day culture time period (Fig. 1A). Percentage and cell number data compiled from several experiments confirm that HR−/PETPs, but not HR+ETPs give rise to T cells (Fig. 1A). However, culture on OP9 stromal cells, which support myeloid differentiation (16) revealed that both HR+ETPs and HR−/PETPs commit to the myeloid lineage in a short 3-day time period (Fig. 1B). Also, the findings are reproducible as similar results were obtained with several repeat experiments (Fig. 1B). Overall, these kinetic studies reveal that the differentiation program of HR+ETPs is fixed to the myeloid lineage while the maturation of HR−/PETP remain flexible and the progenitors are able to mature along the lymphoid or myeloid lineage pathways.
Figure 1. HR+ETPs are fixed to myeloid lineage maturation.
Thymic cells from HR+/+ mice expressing GFP under the IL-13Rα1 promoter were depleted of lineage-positive cells and the CD25−CD44+c-Kit+ ETPs (Lin− CD4− CD8− CD25− CD44+ c-Kit+) were sorted and further separated into (GFP+) HR+ and (GFP−) HR−/P (which have the genetic potential to up-regulate the HR) by flow cytometry. (A) HR+ and HR−/PETPs (3 x 103 cells per well) were cultured on OP9-DL1 stromal cells in the presence of IL-7+Flt3L for the indicated days and maturation into the T cell lineage was assessed by expression of CD25. All cells were gated on CD45+ and 7-AAD− to exclude stromal and dead cells. Contour plots are representative experiments and the numbers indicate cell percentages. The bar graphs show the percentage (left panel) and number (right panel) of CD25+ cells for the indicated time points compiled from 4 independent experiments. Each bar represents the mean ± SD for each population. ***p<0.001 as analysed by two-tailed, unpaired Student’s t-test. (B) HR+ and HR−/PETPs were cultured (3 x 103 cells per well) on OP9 stromal cells for 3 days in the presence of the indicated growth factors and maturation into myeloid lineages was assessed by CD11b expression by cells gated on CD45+ and 7-AAD−. Contour plots are representative experiments and the numbers indicate cell percentages. The bar graphs show compiled data from 4 independent experiments. The percent cells represent the mean ± SD for each population.
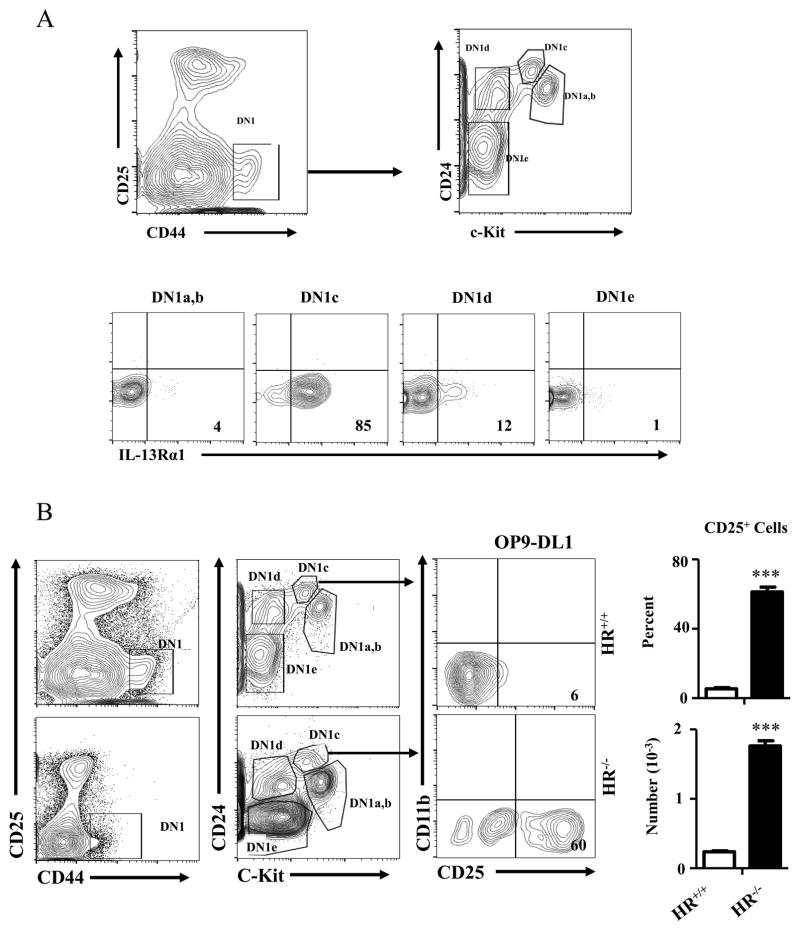
It has previously been shown that thymic myeloid DCs originate from a double negative 1c (DN1c) population which unusually expresses intermediate levels of c-Kit (c-Kitint) and lineage-specific markers (19). It was then suggested that the DN1c population represents thymic seeding precursors for DCs (TSPDC) that are devoid of T cell potential (20). Given that DN1c cells have been shown to mature along the T-cell lineage pathway (21), it is likely that the DN1c population encompasses precursors with T cell potential (21) as well as TSPDC (19, 20). Studies were then performed to determine the DN1 subset with which HR+ETPs are associated. The findings demonstrate that HR+ETPs belong only to the DN1c population (Fig. 2A). Given that HR+ETPs are Lin− cells which do not express CD11c or CD8 surface markers, we conclude they do not represent TSPDC (20), especially that the latter are restricted to CD11c CD8αDCs while HR+ETPs give rise to CD11b myeloid cells.
Figure 2. HR expression influences T-lineage commitment of early thymic progenitors.
(A) Thymic cells from IL-13Rα1-GFP reporter mice were depleted of lineage-positive (Lin+) cells and the Lin−CD4−CD8− cells were analysed for CD25 and CD44 expression. The CD25−CD44+ cells (referred to as DN1 cells) were further assessed for c-Kit and CD24 expression to distinguish the different subsets within the DN1 population. As anticipated the following subsets emerged: DN1a,b (CD24+cKithi), DN1c (CD24+cKitint), DN1d (CD24+cKit−) and DN1e (CD24−c-Kit−) (upper panels). GFP-based analysis demonstrates that only the DN1c subset displays HR expression (lower panels). (B) Lin−CD4−CD8− thymic cells were isolated from HR+/+ as well as HR−/− mice and stained with anti-CD44, CD25, CD24 and c-Kit antibodies. CD44+CD25−CD24+c-KitintDN1c progenitors from both HR+/+ as well as HR−/− were then sorted and cultured (3 x 103 cells per well) on OP9-DL1 stromal cells in the presence of IL-7+Flt3L for 7 days and maturation into the T cell lineage was assessed by expression of CD25. Contour plots show data from a representative experiment and the bar graphs show the percentage (upper panel) and number (lower panel) of CD25+ cells compiled from 3 independent experiments. Each bar represents the mean ± SD for each population. The numbers in the contour plots represent cell percentages. ***p<0.001 as analysed by two-tailed, unpaired Student’s t-test.
Furthermore, DN1c progenitors from HR−/− mice express genes essential for T-cell lineage-commitment (Supplementary Fig. S2) and upon culture on stromal cells differentiate to the T-cell lineage significantly relative to HR+/+DN1c (Fig. 2B), again reconciling the T-cell potential previously reported for DN1c cells (21). On the other hand, since HR+DN1c progenitors (HR+ETPs) commit to the myeloid lineage (Fig. 1B) but not to the T-cell lineage (Fig. 2B), while HR−/−DN1c and HR−/PETPs remain flexible and can differentiate to the myeloid (Fig. 1B) as well as the T cell lineage (Fig. 2B), it is logical to credit the HR with an active role in the commitment of HR+ETPs to the myeloid but not the T-cell lineage. Perhaps signaling through the HR turns on the myeloid pathway or turn off the T-cell lineage commitment.
Commitment to the T cell lineage is shut-down in HR+ETPs
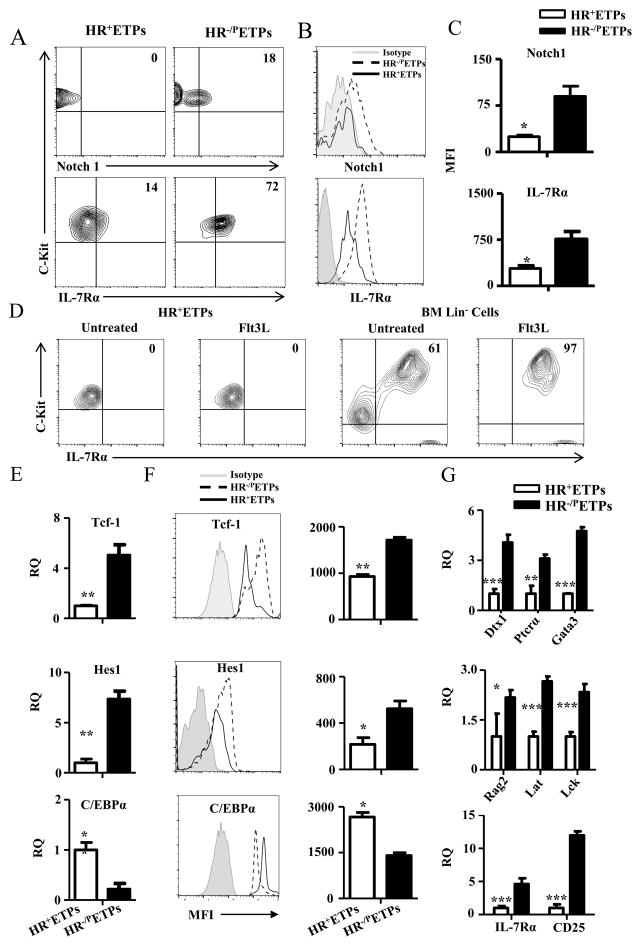
Given that HR+ETPs are fixed toward the myeloid lineage one would envision that the receptor functions to turn on the myeloid but not the lymphoid signaling pathway. Alternatively, signaling through the HR would reinforce blockade of ETP differentiation program towards the T-cell lineage. Notch1 and IL-7 receptor signaling represent major pathways for commitment of ETPs to the T-cell lineage (22–27). We then set up ex vivo experiments to test for expression of Notch1 and IL-7Rα in HR+ETPs in comparison to HR−/P ETPs. The results show that CD25−CD44+c-Kit+GFP+ (HR+) ETPs display minimal Notch1 expression in comparison to CD25−CD44+c-Kit+GFP− (HR−/P) ETPs (Fig. 3A and B). Similarly, IL-7Rα expression was significantly lower in HR+ETPs. The reduction in both Notch1 and IL-7Rα expression is statistically significant as indicated by compiled results from several independent experiments (Fig. 3C). In addition, Notch1 downregulation seems to be critical for inhibition of T cell lineage development as Flt3L (28) was able to induce IL-7Rα expression in Lin− BM cells but not in HR+ETPs (Fig. 3D). These findings led us to question why HR+ ETPs do not express Notch1 and whether it is the cause of inhibition of commitment to the T-lineage.
Figure 3. HR+ETPs display impaired expression of factors required for T cell lineage maturation.
(A, B) Thymic cells from IL-13Rα1-GFP reporter mice were depleted of Lin+ cells and the Lin−CD4−CD8− were stained with antibodies to CD25, CD44, c-Kit, and Notch1 or IL7Rα. The CD25−CD44+c-Kit+GFP+ (HR+ETPs) and CD25−CD44+c-Kit+GFP− (HR−/PETPs) were analysed for Notch1 and IL-7Rα expression. (A) Shows contour plots and (B) shows histograms for Notch1 and IL-7Rα expression. The numbers in the contour plots represent cell percentages. (C) Shows MFI data for Notch1 and IL-7Rα expression compiled from 3 independent experiments. *p<0.05 as analysed by a paired t-test. (D) GFP sorted HR+ETPs and MACS purified Lin− BM control cells were cultured in vitro with or without Flt3L for 72hrs, and the cells gated on CD45 were analysed for c-Kit and IL-7Rα expression. The numbers represent cell percentages. (E) HR+ETPs isolated from IL-13Rα1-GFP reporter mice were analysed for expression Tcf-1, Hes1 and C/EBPα by real-time PCR in comparison to HR−/PETPs counterparts. The data shows mRNA expression (RQ) relative to GAPDH housekeeping gene. (F) Shows MFI for Tcf-1, Hes1 and C/EBPα protein expression by thymic cells gated on CD25−CD44+c-Kit+GFP+(HR+ETPs) and CD25−CD44+c-Kit+GFP− (HR−/PETPs). The histograms illustrate data from a representative experiment and the bar graphs show compiled results from 3 independent experiments. *p<0.05 and **p<0.01 as determined by a paired t-test. (G) Shows mRNA expression relative to GAPDH for the indicated genes in sorted HR+ and HR−/PETPs. The bars represent the mean ± SD data compiled from 3 independent experiments. *p<0.05, **p<0.01 and ***p<0.001 as determined by two-tailed, unpaired Student’s t-test.
It has previously been shows that during commitment to the T-cell lineage, Notch1 signaling leads to up-regulation of Tcf-1 (29) and Hes1 (7) transcription factors. Moreover, Hes1 constrains the expression of C/EBP-α, a transcription factor that promotes myeloid development (7, 30). We then sought to determine whether HR+ETPs would display a reverse expression pattern for these molecules. The findings indicate that Tcf-1 and Hes1 were significantly down-regulated at both the mRNA and protein levels in HR+ relative to HR−/P ETPs (Fig. 3E and F). In contrast, C/EBP-α is increased in HR+ETPs which bodes well with the downregulation of its antagonist, Hes1. Also, the Notch1 target molecules, Ptcrα and Deltex1, as well as the Tcf-1 controlled genes, Gata3, Lat and Lck, were down-regulated in HR+ ETPs (Fig. 3G). Finally, IL-7Rα and CD25, while up-regulated in HR−/P ETPs, remain at minimal levels (Fig. 3G). In all, the findings indicate that the signaling pathways and transcription machinery required for commitment to the T-cell lineage are shut down in HR+ ETPs.
Restoration of HR expression nullifies the potential of ETPs to commit to the T cell lineage
HR−/− unlike HR+/+ ETPs are able to commit to the T-cell lineage perhaps because the cells lack HR expression and Notch1 inhibition is not operative. If this is the case, then restoration of HR expression should nullify maturation towards the T-cell lineage. To test this postulate, we set up a series of experiments to assert the involvement of the HR in T cell development. To this end, IL-13Rα1 was cloned into a retroviral vector carrying Thy1.1 marker and the resulting vector, referred to as HR-RV, was transduced into HR−/−ETPs and the ETPs were assessed for maturation into the T cells as well as the myeloid, lineage. Initially, we ensured that HR-RV is efficient in transducing ETPs and in driving HR expression. Indeed, HR-RV is able to transduce 46% of HR−/−ETPs which is similar to the 58% transduction rate observed with the vector carrying Thy1.1 without IL-13Rα1 (Empty-RV) (Fig. S3). More importantly, 96% of the ETPs transduced with HR-RV express the HR on the cells surface as detected by anti-IL-13Rα1 antibody (13). Subsequently, the HR-RV transduced ETPs were cultured on OP9-DL1or OP9 stromal cells and tested for maturation to lymphoid and myeloid lineages in the presence of IL-4 and IL-13 cytokines. The results show that in the absence of cytokines HR-transduced ETPs remain able to commit to the T-cell lineage to the same extent as the Empty-RV transduced ETPs (Fig. 4A). This is understandable as the HR-RV transduced ETPs, unlike freshly isolated HR+ETPs (from HR+/+ mice), have never been exposed to IL-4 and IL-13 cytokines. This assumption has been proven correct as exposure of HR-RV transduced ETPs to IL-4 or IL-13 reduces maturation to the T cell lineage in a significant manner relative to Empty-RV transduced ETPs as determined by cell percentage and number (Fig. 4A). This likely reflects interference with their T-cell lineage potential rather than proliferation or survival as the total number of live CD45+ cells is similar in all OP9-DL1 cultures settings (Fig. 4A). Furthermore, the same HR-RV cells cultured on OP9 stromal cells proliferate and mature towards the myeloid lineage upon addition of IL-4 or IL-13 cytokines to the same extent as the GM-CSF positive control (Fig. 4B). Note that the cytokines can drive myeloid potential without GM-CSF perhaps serving as growth factors in the OP9 culture system. Given that the HR−/− ETPs include DN1a, b and c, all of which can commit to the T-cell lineage ( Fig. 2B), HR expression and cytokine signaling through it restores inhibition of the potential of ETPs to commit to the T-cell lineage. The interference of IL-4 and IL-13 cytokine signaling through the HR with maturation to the T cell lineage occurs even when HR transduction used HR−/− DN1a, b ETPs. Indeed, single cell culture of these ETPs on OP9-DL1 cells shows that T-cell lineage potential is inhibited by IL-4 and IL-13 cytokines (Table 1). In fact, while 100% of single cells display T-cell lineage potential with IL-7+Flt3L, only 13–18% had such a potential when IL-4 and IL-13 were added to the culture. The inhibition of T-cell lineage potential by the cytokines is dependent on the HR, as 100% of empty-RV transduced HR−/− DN1a, b ETPs display T-cell lineage potential in the presence of either cytokine (Table 1). It is possible that the 13–18% single cells (Table 1), as well as the 22–24% of bulk ETPs (Fig. 4A), that displayed T-cell lineage in the presence of cytokines is related to lower expression of the HR. In all, IL-4 and IL-13 signaling through the HR inhibits T-cell lineage potential in multipotent ETPs.
Figure 4. Restoration of HR expression in ETPs nullifies T cell lineage potential.
HR−ETPs (Lin−CD4−CD8−CD25−CD44+c-Kit+) which include DN1a,b and DN1c were isolated from HR−/− mice and transduced with HR-RV or empty-RV. Anti-IL-13Rα1 antibody was used to sort HR-transduced ETP (HR-RV) and anti-Thy1.1 antibody was utilized to isolate ETP harboring the empty vector not carrying the HR (Empty-RV). Only ETPs with 96% purity or higher were used for subsequent experiments. (A) The sorted ETPs (0.7 x 103 cells per well) were cultured on OP9-DL1 cells for 7 days in the presence of IL-7 and Flt3L. IL-4 (10ng/ml) and IL-13 (20ng/ml) cytokines were added at the beginning of the culture to induce HR signaling. Culture without cytokine addition (NIL) was included for control purposes. (B) HR-RV transduced ETPs from the preparation above were cultured (0.7 x 103 cells per well) on OP9 cells for 7 days in the presence of no cytokine or growth factors (NIL), GM-CSF growth factor (10ng/ml) alone (positive control), and IL-4 (10ng/ml) or IL-13 (20ng/ml) alone without GM-CSF. Maturation into myeloid (CD11b) and lymphoid (CD25) cells was measured by CD11b and CD25 expression on CD45+ 7-AAD− live cells. Contour plots show data from a representative experiment with the numbers within the quadrants representing the percentages of CD25+ (A) and CD11b+ (B) cells. The number on top of each contour plot represents the total number ± SD of CD45+ live cells at the end of the culture compiled from 3 independent experiments. The bar graphs show the percent and number of the indicated cells compiled from 3 independent experiments. Each bar represents the mean ± SD for each population. ***p<0.001 as analysed by two-tailed, unpaired Student’s t-test.
Table 1.
IL-4 and IL-13 inhibit ETP T-cell lineage potential
| T-cell potential (%) | |||
|---|---|---|---|
| Single ETP | NIL | IL-4 | IL-13 |
| HR-RV | 100 (32) | 18 (38) | 13 (30) |
| Empty-RV | 100 (36) | 100 (31) | 100 (35) |
DN1a, b ETP cells from HR−/− mice were transduced with either HR-RV or Empty-RV and single cells were sorted and cultured on OP9-DL1 stromal cells in the presence of IL-7+Flt3L alone (Nil) or with addition of IL-4 or IL-13 as described in Materials and Methods. After 10 days the cultures were analyzed for the expression of CD25 T-cell lineage marker on CD45+ live cells. The numbers represent the percentage of single progenitor cells that gave rise to T-lineage cells. The percentages are normalized based on the number of productive colonies (150 or more CD45+cells in a well) raised from single cells, excluding wells that did not support growth. The values in the parentheses represent the numbers of productive colonies. Plating efficiencies were 70 %.
HR+ETPs display activated STAT6 which parallels with up-regulation of Notch1 pathway inhibitors
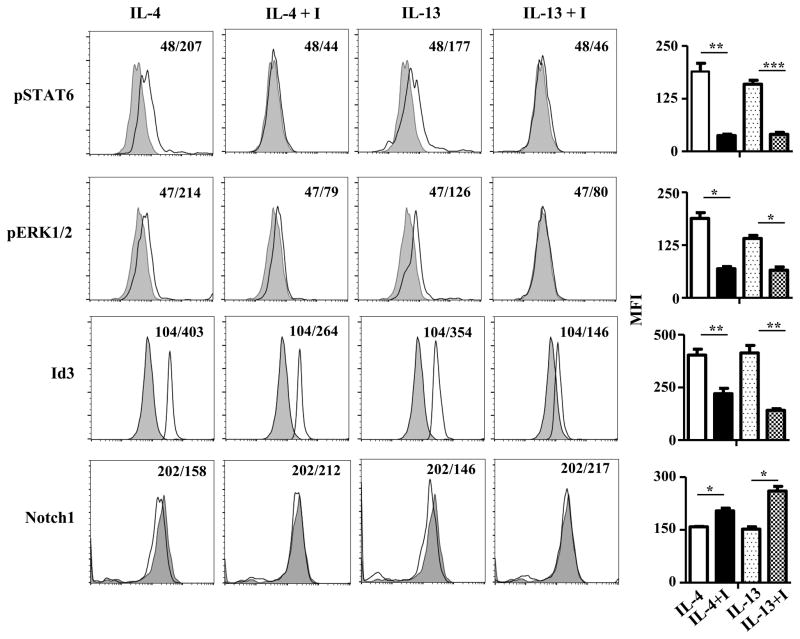
Despite the fact that the HR is involved in allergic inflammation (15, 31), differentiation of macrophages (13), death of neonatal Th1 cells (12, 14), regulation of IL-12 production by dendritic cells (14, 32), our understanding of HR signaling is in its infancy (33) and much less is known on how the receptor operates ETP maturation. Per analogy to IL-4R signaling in Th2 cells we sought that STAT6 activation may be operative in HR+ETPs. This indeed proved to be correct as ex vivo phosphorylation of STAT6Y641 was significantly increased in HR+ versus HR−/P ETPs (Fig. 5A). As STAT6 activation has been shown to trigger the ERK1/2-Egr1 pathway in other cell types (34), this function would be relevant for HR+ETPs as Egr1 can up-regulate expression of Id3, a factor involved in the repression of Notch1 transcription (35, 36). This indeed was the case as phosphorylation of ERK1/2T202/Y204 as well as expression of Egr1 and Id3 transcription factor were significantly increased in HR+ relative to HR−/PETPs (Fig. 5A). Also, the CT value for Notch1 transcription was much higher in HR+ versus HR−/PETPs and the relative mRNA quantity for Notch1 was significantly lower in HR+ relative to HR−/P ETPs (Fig. 5B). This data suggests that HR-driven STAT6 activation likely triggers downregulation of Notch1 at the transcriptional level.
Figure 5. HR+ETPs display elevated phosphorylation of STAT6 transcription factor.
(A) Thymic cells from IL-13Rα1-GFP reporter mice were depleted of Lin+ cells and the CD25−CD44+c-Kit+GFP+ (HR+ETPs) cells were analysed for STAT6 (pSTAT6) and ERK1/2 (pERK1/2) phosphorylation as well as expression of Egr1 and Id3 transcription factors ex vivo in comparison to CD25−CD44+c-Kit+GFP− (HR−/PETPs) counterparts. The histograms show a representative experiment and the bar graphs show compiled MFI results from 3 independent experiments. *p<0.05 and **p<0.01 as determined by a paired t-test. (B) HR+ETPs and HR−/PETPs sorted from IL-13Rα1-GFP reporter mice were analysed for Notch1 transcription ex vivo. The plot illustrates a representative experiment for Notch1 amplification while the bar graph show relative mRNA quantification (RQ) data compiled from 4 independent experiments. ***p<0.001 as determined by two-tailed, unpaired Student’s t-test.
HR signaling causes STAT6 activation leading to Notch1 downregulation and interference with ETP commitment to the T-cell lineage
To ensure that signaling through the HR is responsible for STAT6 activation, down-regulation of Notch1 expression and interference with ETP maturation towards the T cell lineage, HR−/− ETPs were transduced with HR-RV, treated with cytokine (IL-4 or IL-13) alone or in a combination with STAT6 inhibitor and assessed for phosphorylation of STAT6/ERK1/2 and expression of Id3. The findings indicate that IL-4 or IL-13 alone induces STAT6 and ERK1/2 phosphorylation as well as Id3 expression and Notch1 downregulation (Fig. 6). However, while the addition of STAT6 inhibitor reduced STAT6 and ERK1/2 phosphorylation as well as Id3 expression, Notch1 remained optimal (Fig. 6). Compiled results from several experiments indicate that cytokine induced STAT6 signaling and its consequence on ERK1/2 phosphorylation and Id3 expression are statistically significant (Fig. 6), further confirming that signaling through the HR utilizes STAT6 activation to down regulate Notch1 expression. In fact, in vitro blockade of STAT6 or ERK1/2 activation in Lin− thymocytes rescues Notch1 surface expression on HR+ETPs in a significant manner (Fig. S4). More importantly, inhibition of STAT6 phosphorylation in vivo nullifies activation of its substrate ERK1/2 leading to downregulation of Id3, the master inhibitor of Notch1 transcription (Fig. 7A). Similarly, inhibition of ERK1/2 led to Id3 downregulation without affecting STAT6 phosphorylation indicating that STAT6-driven Notch1 downregulation operates through the function of ERK1/2 and Id3 transcription factors. In fact, inhibition of either STAT6 or ERK1/2 restores Notch1 expression further confirming the involvement of this pathway in Notch1 downregulation (Fig. 7B). The data are statistically significant as indicated by the p values obtained from results compiled from several experiments.
Figure 6. IL-4 and IL-13 signal STAT6 activation and Notch1 downregulation in HR-RV transduced ETPs.
HR−ETPs (Lin−CD4−CD8−CD25−CD44+c-Kit+) isolated from HR−/− mice were transduced with HR-RV, sorted on the basis of IL-13Rα1 and Thy1.1 expression and cultured in the presence (open histograms) or absence (shaded histograms) of 10ng/ml IL-4, or 20 ng/ml IL-13 alone or in a combination with STAT6 inhibitors. Phosphorylation of STAT6 and ERK1/2 were analyzed after 1 hour while Id3 and Notch1 expression was measured after 24 and 48 hour incubation, respectively. The histograms show a representative experiment with the numbers indicating representative MFI. The bar graphs show mean ± SD MFI results compiled from three independent experiments. *p<0.05, **p<0.01, and ***p<0.001 as determined by a paired t-test.
Figure 7. In vivo inhibition of STAT6 or ERK1/2 signaling restores Notch1 expression in HR+ETPs.
(A) IL-13Rα1-GFP reporter mice were given i.p. STAT6 or ERK1/2 inhibitor and the Lin−CD4−CD8− thymic cells were isolated and stained with antibodies to CD25, CD44, c-Kit, pSTAT6, pERK1/2 and Id3. The histograms show a representative experiment for STAT6 and ERK1/2 phosphorylation as well as Id3 expression by CD25−CD44+ c-Kit+ HR+ (GFP+) ETPs. The bar graphs show the mean ± SD MFI results compiled from 3 independent experiments. *p<0.05 and **p<0.01 as determined by a paired t-test. (B) The HR+ETPs from (A) were also analysed for Notch1 transcription by quantitative PCR. The plot shows data from a representative Notch1 amplification experiment and the bars show relative mRNA quantification (RQ) data compiled from compiled from 2 independent experiments. **p<0.01 as determined by one way ANOVA.
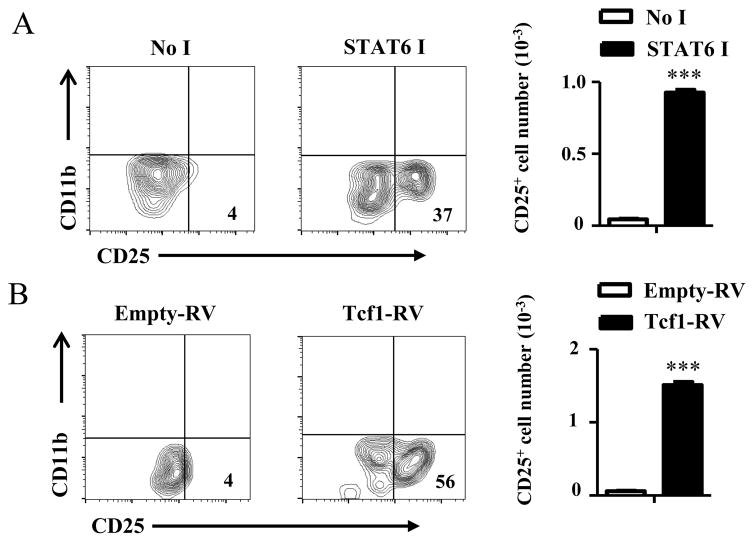
Interestingly, in vivo STAT6 inhibition rescues HR+ETP maturation to the T cell lineage as the percentage of ETP-derived CD25+ cells rose from a background level of 4% in the absence of inhibitor to 37% in the mice recipient of STAT6 inhibitor (Fig. 8A). The results are statistically significant as determined by cell number compiled from several experiments (Fig. 8A). Since thymocyte progression from ETP to the DN2 (CD25+) stage relies on Notch1 mediated induction of its downstream transcriptional target Tcf-1 transcription factor (29), it is logical to envision that forced expression of Tcf-1 would rescue commitment to the T cell lineage by HR+ETPs. To test this premise, Tcf-1 was cloned into a retroviral vector (Tcf-1-RV), transduced into HR+ETPs and the cells were tested for maturation into the T cell lineage upon culture on OP9-DL1 stromal cells. The results show that the percentage of ETP-derived CD25+ cells rose from a background level of 4% in the empty-RV transduced HR+ETPs to 56% in HR+ETPs recipients of forced Tcf-1 expression (Fig. 8B). This is statistically significant as determined by cell number compiled from several experiments (Fig. 8B). In all, the HR in ETPs signals Notch1 inhibition and negates commitment to the T cell lineage through activation of STAT6 transcription factor.
Figure 8. HR signaling negates ETP commitment to the T cell lineage.
(A) IL-13Rα1-GFP reporter mice were given STAT6 inhibitor (STAT6 I) or diluent (No I) and 5 days later HR+ETPs were sorted and cultured (3 x 103 cells per well) on OP9-DL1 cells in the presence of IL-7+ Flt3L. Ten days later the cultures were assessed for commitment to the T-cell lineage by measuring CD25 expression on 7-AAD−CD45+CD11b− cells. (B) HR+ETPs were isolated from IL-13Rα1-GFP reporter mice, transduced with Tcf-1-RV or Empty RV and cultured (0.7 x 103 cells per well) on OP9-DL1 cells in the presence of IL-7+Flt3L for 10 days. Commitment to the T-cell lineage was assessed by measuring CD25 expression on 7-AAD−CD45+CD11b− cells.
In all contour plots the numbers indicate the percent of CD25+ cells and bar graphs show the number of CD25+ cells compiled from 3 independent experiments. Each bar represents the mean ± SD for each population. ***p<0.001 as determined by two-tailed, unpaired Student’s t-test.
Discussion
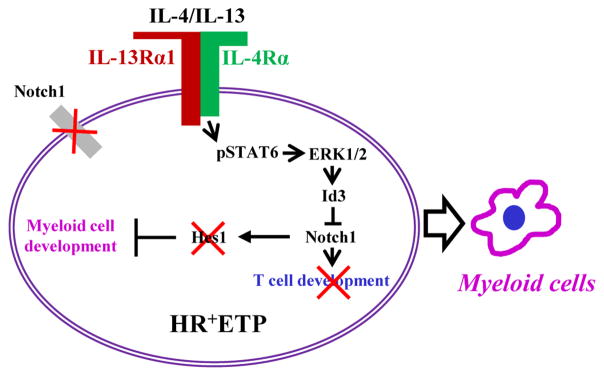
This study demonstrates that environmental IL-4 and IL-13 guide ETP maturation by signaling through their IL-4Rα/IL-13Rα HR. Indeed, ETPs from HR−/− mice which arise in an environment where both IL-4 and IL-13 are unable to signal through the HR commit to both the myeloid and lymphoid lineages. In contrast, ETPs from HR+/+ mice which arise in an environment where both IL-4 and IL-13 can signal through their HR commit only to the myeloid lineage. Given that ETPs with a history of IL-7 receptor (IL-7R) expression are also restricted to a single lineage choice (8), it is logical to envision that the HR and IL-7R serve as responsive element to environmental cytokines and influence ETP lineage choice. The study presented in this report, while demonstrate that signaling through the HR restricts commitment to the T-cell lineage, also sheds light on the mechanism by which such a restriction comes about. Indeed, HR+ETPs which would be physiologically exposed to environmental IL-4/IL-13 in vivo exhibit an active form of STAT6 while their HR− counterparts do not. Interestingly, Notch1, a critical factor for ETPs maturation to the T cell lineage (22, 37) was significantly downregulated in HR+ relative to HR−ETPs. These observations pointed to an active blockade of ETP commitment potential to the T cell lineage which is set off by IL-4/IL-13 interaction with the HR (Fig. 9). As a consequence of STAT6 activation the ERK1/2-Egr1 pathway is put in motion leading to induction of Id3 which represses Notch1 transcription, hence inhibition of commitment potential to the T cell lineage (Fig. 9). Also, downregulation of Notch1 frees expression of C/EBPα, a critical factor in ETP commitment to the myeloid lineage (7).
Figure 9.
Schematic diagram illustrating the mechanism underlying inhibition of T-cell lineage potential in HR+ETPs.
Prior reports indicated that IL-7Rα increases survival and sustains progenitor commitment to lymphoid lineages (8, 38). This function must avoid STAT6 activation to preserve Notch1 expression and elude Id3 transcription. It is therefore conceivable that different cytokine receptors utilize distinct signaling pathways to reinforce lineage fate. This bodes well with an earlier observation indicating that IL-12Rβ sustains commitment of T cell progenitors towards the myeloid lineage (39).
The HR is perhaps tasked to instruct lineage commitment by its ligands IL-4 and IL-13 cytokines. The puzzle here is where do the cytokines come from and how are they tied to ETP maturation? It has previously been shown that type 2 innate lymphoid cells (ILC2) (40) which can be generated in the thymus from DN1/DN2 thymocytes (41) produce type II cytokines including IL-13 (15). Similarly, NKT cells which also arise in the thymus can produce IL-4 (42). Thus, it is possible that the type 2 cytokines from these cells signal through the HR to guide ETP commitment. Alternatively, HR signaling could have been initiated during travel of thymus settling progenitors (TSP) from the bone marrow to the thymus in conduits where the cytokines would be readily available. Whatever the source of the ligand or the timing of receptor triggering, the findings suggest that environmental rather than genetic programs control ETP commitment. Specifically, whereas clonal distribution of lymphocyte specificity is genetically programmed by random Ag receptor rearrangement, ETP commitment seems to rely on ligand receptor expression and ligand availability. From another perspective, it would be logical to envision that multipotent ETPs (3, 4) would not have been subjected to ligand/receptor interactions while presumed unipotent ETPs (8, 9) have been driven to acquire such an attribute by those environmental factors.
The essence of the findings is that signaling by environmental IL-4/IL-13 through the HR diverts ETP commitment from the T cell lineage to myeloid cells. Since myeloid cells could serve as APCs in T cell selection the diversion may impact central tolerance and shape the autoimmune repertoire.
Supplementary Material
Acknowledgments
Funding. This work was supported by grant RO1 NS057194 (to H.Z.) from the National Institutes of Health. M.M.M. was supported by T32 Training Grant GM008396 from the National Institute of General Medical Sciences.
Abbreviations
- 7-AAD
7-aminoactinomycin D
- BM
bone marrow
- DC
dendritic cells
- DN
double negative
- ETPs
early thymic progenitors
- HR
IL-4Rα/IL-13Rα1 heteroreceptor
- Lin
lineage
- TSP
thymic settling progenitors
References
- 1.Wu L, Antica M, Johnson GR, Scollay R, Shortman K. Developmental potential of the earliest precursor cells from the adult mouse thymus. J Exp Med. 1991;174:1617–1627. doi: 10.1084/jem.174.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, Meraz A, Bhandoola A. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 3.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 4.Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 5.Petrie HT, Zuniga-Pflucker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- 6.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Obaldia ME, Bell JJ, Wang X, Harly C, Yashiro-Ohtani Y, DeLong JH, Zlotoff DA, Sultana DA, Pear WS, Bhandoola A. T cell development requires constraint of the myeloid regulator C/EBP-alpha by the Notch target and transcriptional repressor Hes1. Nat Immunol. 2013;14:1277–1284. doi: 10.1038/ni.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlenner SM, Madan V, Busch K, Tietz A, Laufle C, Costa C, Blum C, Fehling HJ, Rodewald HR. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–436. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Haymaker CL, Guloglu FB, Cascio JA, Hardaway JC, Dhakal M, Wan X, Hoeman CM, Zaghouani S, Rowland LM, Tartar DM, VanMorlan AM, Zaghouani H. Bone marrow-derived IL-13Ralpha1-positive thymic progenitors are restricted to the myeloid lineage. J Immunol. 2012;188:3208–3216. doi: 10.4049/jimmunol.1103316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krueger A, von Boehmer H. Identification of a T lineage-committed progenitor in adult blood. Immunity. 2007;26:105–116. doi: 10.1016/j.immuni.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Obiri NI, Leland P, Murata T, Debinski W, Puri RK. The IL-13 receptor structure differs on various cell types and may share more than one component with IL-4 receptor. J Immunol. 1997;158:756–764. [PubMed] [Google Scholar]
- 12.Li L, Lee HH, Bell JJ, Gregg RK, Ellis JS, Gessner A, Zaghouani H. IL-4 utilizes an alternative receptor to drive apoptosis of Th1 cells and skews neonatal immunity toward Th2. Immunity. 2004;20:429–440. doi: 10.1016/s1074-7613(04)00072-x. [DOI] [PubMed] [Google Scholar]
- 13.Dhakal M, Hardaway JC, Guloglu FB, Miller MM, Hoeman CM, Zaghouani AA, Wan X, Rowland LM, Cascio JA, Sherman MP, Zaghouani H. IL-13Ralpha1 is a surface marker for M2 macrophages influencing their differentiation and function. Eur J Immunol. 2014;44:842–855. doi: 10.1002/eji.201343755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhakal M, Miller MM, Zaghouani AA, Sherman MP, Zaghouani H. Neonatal Basophils Stifle the Function of Early-Life Dendritic Cells To Curtail Th1 Immunity in Newborn Mice. J Immunol. 2015;195:507–518. doi: 10.4049/jimmunol.1500027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halim TY, Hwang YY, Scanlon ST, Zaghouani H, Garbi N, Fallon PG, McKenzie AN. Group 2 innate lymphoid cells license dendritic cells to potentiate memory TH2 cell responses. Nat Immunol. 2016;17:57–64. doi: 10.1038/ni.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 17.Wan X, Guloglu FB, VanMorlan AM, Rowland LM, Jain R, Haymaker CL, Cascio JA, Dhakal M, Hoeman CM, Tartar DM, Zaghouani H. Mechanisms underlying antigen-specific tolerance of stable and convertible Th17 cells during suppression of autoimmune diabetes. Diabetes. 2012;61:2054–2065. doi: 10.2337/db11-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyszkiewicz M, Zietara N, Fohse L, Puchalka J, Diestelhorst J, Witzlau K, Prinz I, Schambach A, Krueger A. Limited niche availability suppresses murine intrathymic dendritic-cell development from noncommitted progenitors. Blood. 2015;125:457–464. doi: 10.1182/blood-2014-07-592667. [DOI] [PubMed] [Google Scholar]
- 19.Luche H, Ardouin L, Teo P, See P, Henri S, Merad M, Ginhoux F, Malissen B. The earliest intrathymic precursors of CD8alpha(+) thymic dendritic cells correspond to myeloid-type double-negative 1c cells. Eur J Immunol. 2011;41:2165–2175. doi: 10.1002/eji.201141728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krueger A. A missing link in thymic dendritic cell development. Eur J Immunol. 2011;41:2145–2147. doi: 10.1002/eji.201141850. [DOI] [PubMed] [Google Scholar]
- 21.Porritt HE, Rumfelt LL, Tabrizifard S, Schmitt TM, Zuniga-Pflucker JC, Petrie HT. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 2004;20:735–745. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Tan JB, Visan I, Yuan JS, Guidos CJ. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nat Immunol. 2005;6:671–679. doi: 10.1038/ni1217. [DOI] [PubMed] [Google Scholar]
- 23.Maillard I, Schwarz BA, Sambandam A, Fang T, Shestova O, Xu L, Bhandoola A, Pear WS. Notch-dependent T-lineage commitment occurs at extrathymic sites following bone marrow transplantation. Blood. 2006;107:3511–3519. doi: 10.1182/blood-2005-08-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson A, MacDonald HR, Radtke F. Notch 1-deficient common lymphoid precursors adopt a B cell fate in the thymus. J Exp Med. 2001;194:1003–1012. doi: 10.1084/jem.194.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radtke F, Ferrero I, Wilson A, Lees R, Aguet M, MacDonald HR. Notch1 deficiency dissociates the intrathymic development of dendritic cells and T cells. J Exp Med. 2000;191:1085–1094. doi: 10.1084/jem.191.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan JT, Dudl E, LeRoy E, Murray R, Sprent J, Weinberg KI, Surh CD. IL-7 is critical for homeostatic proliferation and survival of naive T cells. Proc Natl Acad Sci U S A. 2001;98:8732–8737. doi: 10.1073/pnas.161126098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacDonald HR, Wilson A, Radtke F. Notch1 and T-cell development: insights from conditional knockout mice. Trends Immunol. 2001;22:155–160. doi: 10.1016/s1471-4906(00)01828-7. [DOI] [PubMed] [Google Scholar]
- 28.Mazzucchelli R, Durum SK. Interleukin-7 receptor expression: intelligent design. Nat Rev Immunol. 2007;7:144–154. doi: 10.1038/nri2023. [DOI] [PubMed] [Google Scholar]
- 29.Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, Bhandoola A. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 31.Munitz A, Brandt EB, Mingler M, Finkelman FD, Rothenberg ME. Distinct roles for IL-13 and IL-4 via IL-13 receptor alpha1 and the type II IL-4 receptor in asthma pathogenesis. Proc Natl Acad Sci U S A. 2008;105:7240–7245. doi: 10.1073/pnas.0802465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee HH, Hoeman CM, Hardaway JC, Guloglu FB, Ellis JS, Jain R, Divekar R, Tartar DM, Haymaker CL, Zaghouani H. Delayed maturation of an IL-12-producing dendritic cell subset explains the early Th2 bias in neonatal immunity. J Exp Med. 2008;205:2269–2280. doi: 10.1084/jem.20071371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wills-Karp M, Finkelman FD. Untangling the complex web of IL-4- and IL-13-mediated signaling pathways. Sci Signal. 2008;1:pe55. doi: 10.1126/scisignal.1.51.pe55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee PJ, Zhang X, Shan P, Ma B, Lee CG, Homer RJ, Zhu Z, Rincon M, Mossman BT, Elias JA. ERK1/2 mitogen-activated protein kinase selectively mediates IL-13-induced lung inflammation and remodeling in vivo. J Clin Invest. 2006;116:163–173. doi: 10.1172/JCI25711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nedjic J, Aifantis I. RNA-binding proteins come out of the shadows. Nat Immunol. 2010;11:697–698. doi: 10.1038/ni0810-697. [DOI] [PubMed] [Google Scholar]
- 36.Yashiro-Ohtani Y, He Y, Ohtani T, Jones ME, Shestova O, Xu L, Fang TC, Chiang MY, Intlekofer AM, Blacklow SC, Zhuang Y, Pear WS. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009;23:1665–1676. doi: 10.1101/gad.1793709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, Pear WS, Bhandoola A. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol. 2005;6:663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- 38.Akashi K, Kondo M, von Freeden-Jeffry U, Murray R, Weissman IL. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell. 1997;89:1033–1041. doi: 10.1016/s0092-8674(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 39.King AG, Kondo M, Scherer DC, Weissman IL. Lineage infidelity in myeloid cells with TCR gene rearrangement: a latent developmental potential of proT cells revealed by ectopic cytokine receptor signaling. Proc Natl Acad Sci U S A. 2002;99:4508–4513. doi: 10.1073/pnas.072087899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walker JA, Barlow JL, McKenzie AN. Innate lymphoid cells--how did we miss them? Nat Rev Immunol. 2013;13:75–87. doi: 10.1038/nri3349. [DOI] [PubMed] [Google Scholar]
- 41.Wong SH, Walker JA, Jolin HE, Drynan LF, Hams E, Camelo A, Barlow JL, Neill DR, Panova V, Koch U, Radtke F, Hardman CS, Hwang YY, Fallon PG, McKenzie AN. Transcription factor RORalpha is critical for nuocyte development. Nat Immunol. 2012;13:229–236. doi: 10.1038/ni.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14:1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.