Abstract
The origin of metazoans, one of the major transitions in evolution, remains mysterious. While many key aspects of metazoan origins can be reconstructed by comparing living organisms within a robust phylogenetic framework, uncertainty regarding the evolutionary relationships among sponges, ctenophores and the remainder of metazoan diversity has proven to be a major barrier. Comparative morphology and some phylogenomic analyses support the view that sponges represent the sister lineage to the rest of the metazoans, while other competing phylogenomic analyses strongly support ctenophores, a phylum of carnivorous, gelatinous marine organisms, as the sister lineage. Here, we explore why different phylogenomic studies yield different answers and discuss the implications of the two alternative hypotheses for understanding the origin of metazoan multicellularity. Reconstruction of ancient evolutionary radiations, like the one that gave rise to metazoans, is devilishly difficult and will likely require broader sampling of sponge and ctenophore genomes, improved analytical strategies, and critical analyses of the phylogenetic distribution and molecular mechanisms underlying apparently conserved traits. Rather than staking out positions in favor of the ctenophore-sister or the sponge-sister hypothesis, we submit that research programs aimed at understanding the biology of the first metazoans should embrace the uncertainty surrounding early metazoan evolution in their experimental designs.
Keywords: Ctenophora, Porifera, animal origins, phylogenomics, model, ambiguity, transition, unicellular/multicellular, trait evolution, Urmetazoan, myocyte, neuron
“Science works on the frontier between knowledge and ignorance. We’re not afraid to admit what we don’t know. There’s no shame in that. The only shame is to pretend that we have all the answers.”
—Neil deGrasse Tyson, Cosmos: A Spacetime Odyssey, 2014(http://channel.nationalgeographic.com/cosmos-a-spacetime-odyssey/videos/when-knowledge-conquered-fear/)
Introduction
Knowledge of phylogenetic relationships is critical for understanding the evolution of genes, gene regulatory pathways, and traits, as well as for generating and testing evolutionary hypotheses about life’s major transitions. To generate phylogenies, gene or protein sequence alignments can be interrogated to separate phylogenetic signal (sequence changes that reflect the evolutionary relationships among species) from noise. In the post-genomics era, the analysis of hundreds or thousands of genes using powerful models and computational algorithms has dramatically improved the accuracy with which the evolutionary relationships among living organisms can be deduced, but certain key branches have remained recalcitrant and sparked controversy [1–3].
Arguably the biggest controversy in metazoan phylogenetics concerns the evolutionary relationships among three lineages that originated near the base of the metazoan tree (Figure 1). The first lineage comprises the vast majority of metazoans and includes placozoans (an enigmatic group of organisms with a very simple organization), cnidarians (such as jellyfish and sea anemones), and all bilaterally symmetrical animals (including humans, fruit flies and worms). The remaining two lineages – the major actors in the controversy – are sponges and ctenophores, a marine phylum of gelatinous metazoans that bear distinctive “combs” of cilia, earning them the moniker “comb jellies.” The debate centers on which of these two, sponges or ctenophores, is the sister lineage (see Glossary) to the rest of the metazoans (Figure 1). The uncertainty stemming from this controversy has major implications for understanding the origin of metazoan multicellularity and development, deciphering the biology of early metazoans [4, 5], and unraveling the evolution of many marquee metazoan traits, including muscles and the nervous system [6–8].
Figure 1. Representatives of sponges and ctenophores, the two metazoan phyla at the center of the controversy.
Left: Stove-pipe sponge Aplysina archeri (source: https://en.wikipedia.org/wiki/Sponge#/media/File:Aplysina_archeri_(Stove-pipe_Sponge-pink_variation).jpg). Right: the ctenophore Mnemiopsis leidyi (from: Stefan Siebert, Brown University; https://phys.org/news/2013-12-aquatic-jelly-evolutionary-position.html).
In this minireview, we discuss the historical background of the sponge - ctenophore controversy, where it stands now, and its implications for understanding and studying the evolution of key metazoan traits. As short branches at the base of ancient evolutionary radiations are challenging to resolve, we argue that research programs aimed at deciphering the cellular and molecular foundations of metazoan multicellularity and development should embrace the uncertainty surrounding early metazoan evolution.
A brief historical perspective
Pre-molecular era efforts to reconstruct evolutionary relationships among metazoan phyla were largely based on their cellular and morphological characteristics [9, 10]. In those phylogenies, sponges were invariably placed as the sister branch to the rest of the metazoans, and ctenophores were thought to represent either the sister lineage to cnidarians [9] or to bilaterians (see Glossary) [10]. In fact, even the notion that sponges are metazoans was debated early on [11, 12], with later leading opinions arguing that sponges should be confined to Parazoa (see Glossary), metazoans of the “cellular grade of construction,” leaving the rest of metazoans in the “tissue grade of construction” Eumetazoa (see Glossary) [13]. Our modern classification of sponges as metazoans is based on phylogenomics, comparative genomics, and findings of conserved processes during embryogenesis [14, 15]. Sponges, like the rest of metazoans, produce differentiated sperm and egg, exhibit conserved developmental gene expression patterns, have epithelia, contain a suite of metazoan-specific genes (for example, Wnt), and develop through a clonal process of serial cell division [14–17]. Sponge collar cells, which bear a single flagellum surrounded by a collar of microvilli, resemble the cell morphology of the choanoflagellates ([18, 19] but see [20]), the closest living relatives of metazoans [21], which was interpreted as further support for the notion that sponges are the sister lineage to the rest of metazoans.
Despite eliciting early interest from zoologists and embryologists [13], the study of ctenophores escaped the attention of most cell and molecular biologists until relatively recently. The unusual challenges of working with ctenophores, including the requirement of marine laboratory facilities and the small size of their research community, meant that they were excluded from most early molecular phylogenies. Perhaps most importantly, early examinations of their morphological characteristics revealed them to be well differentiated [22], inspiring some to infer a close relationship to the bilaterians [13], while others argued for a sister group relationship with cnidarians based on the similarities of the two phyla in adult morphology [9].
Early molecular phylogenies based on a single or a few genes tended to place sponges as the sister lineage to the rest of metazoans [23, 24] and reject the close affinity of ctenophores with cnidarians, instead placing ctenophores as the sister to a clade comprised of cnidarians, placozoans and bilaterians [24–26]. In hindsight, these early molecular efforts were underpowered [24, 25, 27–29] and their results weakly supported and highly varied. For example, some early analyses favored now-obsolete scenarios such as metazoan polyphyly caused by the grouping of cnidarians with ciliates and fungi [30] or the inference of a clade comprised by sponges, ctenophores, cnidarians, and placozoans that was sister to that of bilaterians [31]. At the dawn of the 21ST century, it was unclear whether the observed volatility in metazoan relationships was due to the use of small amounts of molecular sequence data, the poor fit of phylogenetic models to the sequence data, or the genuine lack of phylogenetic signal that might be expected in the early phase of the metazoan evolutionary radiation, setting the stage (and expectations) that the then-nascent genomics revolution would come to the rescue.
Enter the phylogenomics era
Over the last decade, the remarkable advances in DNA sequencing technologies led to the sequencing of the first sponge [15] and ctenophore [7, 32] genomes as well as the availability of large amounts of transcriptome data [32–38]. This orders-of-magnitude increase in data, coupled with considerable developments at the interface of theoretical phylogenetics with computer science, culminated in faster software and enabled the use of new and more sophisticated strategies for phylogenomic inference. Consequently, molecular phylogenies of early-branching metazoan phyla were no longer based on analyses of just one or a handful of genes, but of hundreds of them.
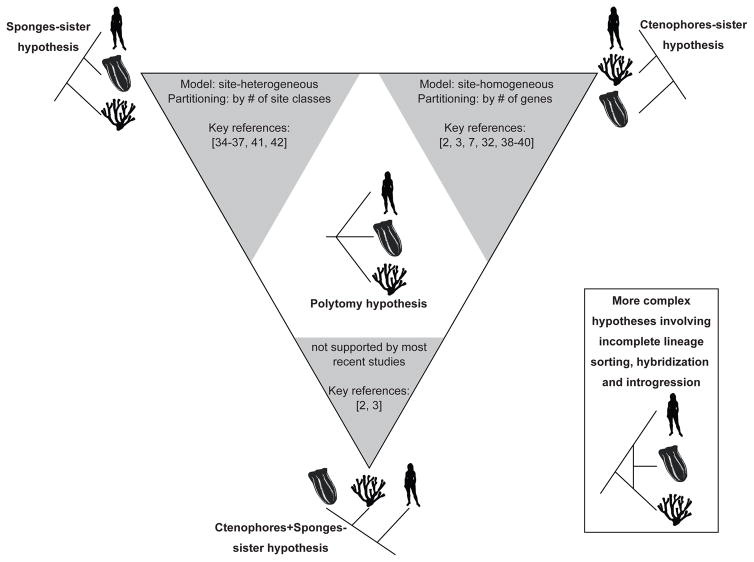
An early example of using phylogenomics to address long-standing questions about the metazoan tree was a 2008 study by Dunn and coworkers [33]; this was the first study suggesting that ctenophores – not sponges – were the sister lineage to the rest of the metazoans. Since then, more than a dozen conflicting phylogenomic analyses have offered support for the ctenophores-sister hypothesis [2, 3, 7, 32, 38–40], the sponges-sister hypothesis [34–37, 41, 42] or (much more rarely) neither [43] (Figure 2).
Figure 2. Different models of amino acid evolution favor different hypotheses for the evolutionary relationships between sponges, ctenophores, and other metazoans.
Three alternative hypotheses are shown on each of the corners of the triangle: sponges as the sister lineage to the rest of the metazoans (top left), ctenophores as the sister lineage (top right), or a clade of sponges + ctenophores as the sister lineage to the rest of the metazoans (represented by human). The grey triangles summarize some of the key characteristics of phylogenomic studies supporting or rejecting each hypothesis (e.g., application of site-homogeneous models tends to favor the ctenophores-sister hypothesis, whereas application of the CAT site-heterogeneous model tends to favor the sponges-sister hypothesis). The polytomy hypothesis, under which all three lineages originated at the same time, is displayed inside the triangle. The inset depicts more complex phylogenetic scenarios, such as hybridization, introgression, and incomplete lineage sorting of ancestral polymorphisms (see Glossary). Although evidence in support of either polytomy or a more complex scenario is lacking, had it occurred, it remains questionable whether it would be detectable with existing data and methodologies.
Why is it that, despite a decade of ever-increasing amounts of data and phylogenomic analyses, we have yet to reach consensus? In general, several different biological and analytical factors can conspire to make a particular branch recalcitrant to resolution (see Box 1). Although several such factors are likely at play, the controversy can be whittled down to two key issues: model selection and analytical strategy. The first concerns how one models sequence evolution in phylogenomic inference. Briefly, the standard “site-homogeneous” models (see Glossary) of protein evolution, assume that all sites in a given protein sequence alignment have evolved under the same substitution process, whereas site-heterogeneous models (see Glossary) evaluate and identify the best-fit substitutional process for each site of the protein sequence alignment. Site-heterogeneous models fit biological data better than site-homogeneous models [44, 45], but are computationally much more expensive [46]. In the context of phylogenomic analyses, where data matrices contain hundreds to thousands of protein sequence alignments, researchers typically resort to one of two strategies: they either use a site-homogeneous model for each gene in the data matrix (this partitioning of the data matrix is known to improve the fit of models to the data) or a site-heterogeneous model across the entire data matrix.
Box 1. Why are some evolutionary relationships so controversial?
Although many parts of the tree of life have been robustly and reproducibly resolved using many independent types of data and strategies, others have proven more challenging. The most heavily debated phylogenetic controversies center around short branches at the base of ancient evolutionary radiations [1]. This is largely because the resolution of such short, deep branches is highly vulnerable to analytical artifacts. As the taxa compared are only distantly related, their gene sequences are highly divergent (or altogether absent in some taxa), reducing the accuracy of orthology inference as well as of multiple sequence alignment of these putative orthologous sequences, while also increasing the amount of sequence data missing. Distantly related taxa also differ from each other in ways that influence evolutionary substitution rates (e.g., generation time, population size, mutation rate, and GC content), reducing the fit between models of sequence evolution and the data at hand. Poor fit between the model of evolution and the sequence data being analyzed can lead to long branch attraction (LBA), a phenomenon in which taxa whose sequence data have experienced the largest amounts of change are artifactually grouped together, irrespective of their true evolutionary relationships [74].
Variation in evolutionary rates across taxa also means that the same set of genes may be fast-evolving in some taxa but slowly-evolving in others, which can also lead to LBA. In the context of the metazoan phylogeny, the branches leading to sponges and ctenophores, which by virtue of being among the first to diverge are also some of the longest, are particularly prone to LBA. Sampling of additional sponge and ctenophore taxa could potentially help “break” these long branches and ameliorate LBA [75], a strategy employed in all recent phylogenomic studies on the controversy. However, while all major sponge lineages appear to be ancient (> 500 million years old), the last common ancestor of extant ctenophores is thought to be much younger [26], making LBA amelioration strategies based on taxon sampling potentially less effective.
Finally, short branches at the base of evolutionary radiations, including the one at the center of the controversy about metazoan origins, are hotspots for lineage sorting of ancestral polymorphisms [76] as well as for hybridization and introgression [77], each of which can produce gene histories that differ from those of their species. Such events are commonplace in the metazoan phylogeny [49, 76, 77]. Although evidence of lineage sorting, introgression or hybridization in the early history of metazoans is lacking, had it occurred, it remains questionable whether it would be detectable with existing data and methodologies (Figure 2).
In the case of the metazoan controversy, analyses employing site-homogeneous models of protein evolution with partitioning (a different model for each gene) tend to recover ctenophores as the sister branch ([2, 3, 7, 32, 33, 38–40], but see [35, 42]). In contrast, analyses employing site-heterogeneous models (and in particular the CAT model [47]) typically recover sponges as the sister branch ([34–37, 41, 42], but see [38, 40, 48]). At this time, there is no consensus as to which of these two strategies is more likely to yield an accurate phylogeny.
The second issue (or set of issues) concerns choices made during phylogenomic data matrix assembly, ranging from strategies for identifying orthologs, to the level of tolerance for missing data, and to the choice of outgroups and rules for excluding taxa [7, 35, 37, 38, 40–42]. Here, it is less clear how these choices influence the recovery of either sponges or ctenophores as the sister lineage. Although the strategies and models employed by the latest studies represent the field’s state-of-the-art, the absence of independent types of data for testing the validity of either of the two alternative phylogenetic hypotheses makes it hard to determine the optimal strategy. Thus, we argue that we can’t yet confidently infer which of the two hypotheses – sponges-sister or ctenophores-sister - is the best supported.
How can we gain greater confidence in our reconstructions of the earliest stages of metazoan diversification? Taking a cue from well-established branches of the tree of life, we should expect that the “correct” resolution should be robustly supported by independent sources of data, experimental designs, and methodologies. In these early days of the post-genomics era, much can be and remains to be done. To date, phylogenomicists have had only one sponge genome [15], two ctenophore genomes [7, 32], and a handful of transcriptomes [32–38] from each at their disposal; clearly more genomes from diverse sponges, ctenophores, and metazoan outgroup taxa are needed. Sequencing more ctenophore genomes may, for example, uncover one or more new lineages that have experienced slower evolutionary rates, which would be valuable for investigating whether the ctenophores-sister placement is an artifact stemming from the fast evolutionary rates of the two sequenced ctenophore genomes. Likewise, genomic sequencing of additional outgroup taxa may reveal the extent to which the current selection of taxa skews analyses toward either sponges-sister or ctenophores-sister in phylogenomic analyses (see Box 1).
Additional genomes may also catalyze phylogenomic method development, either by making the branch “easier” to resolve or by facilitating more clear demarcation of the strengths and weaknesses of the different strategies in data-rich, taxon-rich data matrices. More genomes could also facilitate the discovery of independent types of data, such as rare genomic changes, that shed light on early metazoan diversification. So far, the only type of rare genomic change analyzed has been gene content, with one study favoring ctenophores-sister [7] and a reanalysis favoring sponges-first [42]. While a step in the right direction, gene content can be even more sensitive than linear sequence data to variation in organismal lifestyle [48], raising concerns about its utility for resolving ancient radiations, such as the metazoan one.
Finally, the quest for a neatly bifurcating tree may mask the true history of early branching metazoan phyla. It is possible that more complex phylogenetic scenarios involving hybridization (see Glossary), introgression (see Glossary), or incomplete lineage sorting (see Glossary) were part of the early phyletic diversification of metazoans (Figure 2), despite the fact that these processes cannot be robustly detected using existing data and methodologies. This would imply that the evolutionary history of traits encoded by genes that experienced any of these processes may be genuinely different from that implied by the species phylogeny [49], regardless of whether it is sponges or ctenophores that diverged first.
Implications of the controversy for reconstructing the Urmetazoan
While reconstructing the relationships among metazoan phyla is a formidable challenge, it is essential for understanding the origins of modern metazoan diversity and the evolutionary processes that gave rise to it. Notwithstanding the current ambiguities regarding sponges and ctenophores, the robustness of other parts of the metazoan and eukaryotic phylogeny mean that much can yet be inferred about the biology of the last common ancestor of animals, the Urmetazoan (see Glossary).
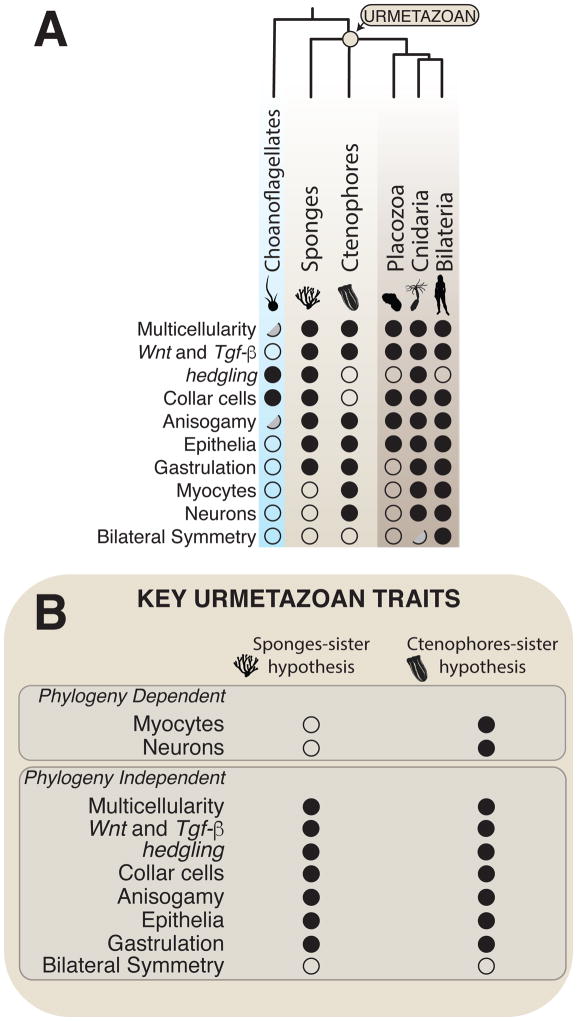
Many traits can be mapped to the Urmetazoan with near certainty, regardless of whether sponges or ctenophores are the sister lineage (Figure 3). Among these are the pan-metazoan homologies (see Glossary), homologous traits found in nearly all metazoans, and by inference, in their last common ancestor, the Urmetazoan. These Urmetazoan traits include some metazoan-specific traits predicted to have evolved along the metazoan stem lineage, including obligate, clonal multicellularity, spermatogenesis and oogenesis (i.e., anisogamy [4]), metazoan-specific developmental signaling pathways (e.g., Wnt and TGFbeta [50]), epithelia, and, potentially, Vasa- and Piwi-driven regulation of stem cell multipotency [4, 51–54]. Moreover, the Urmetazoan also contained several more ancient traits that are today conserved in metazoans and in some of their closest relatives, including phagotrophy, genes involved in metazoan cell signaling, cell adhesion, and transcriptional regulation (e.g., Brachyury, cadherins, integrins, and receptor tyrosine kinases [4, 55–58]), and cells bearing a single apical flagellum/cilium (e.g., found today on metazoan sperm and epithelial cells [4, 56, 58]).
Figure 3. Much can be inferred about the Urmetazoan, despite the ongoing phylogenetic controversy.
A. Distribution of illustrative traits in diverse metazoans (brown/tan shading) and their closest living relatives, the choanoflagellates (blue shading). The base of the metazoan tree is depicted as a polytomy to emphasize current uncertainty about the relative placement of sponges and ctenophores. Each trait is indicated as being either present in a clade (black circles), not detected in a clade (white circles), or as having been detected in an intermediate form in a subset of taxa within the lineage (half-gray circles). For example, some choanoflagellates produce an intermediate multicellular form, a “colony,” in which all cells have apparently equivalent morphology. How traits are coded (i.e., present vs. absent for intermediate or convergent traits) influences inferences about their presence or absence in the Urmetazoan. B. Controversies regarding the phylogenetic relationships among sponges, ctenophores, and other metazoans shape inferences about the biology of the Urmetazoan. Inferences about the presence (black circles) or apparent absence (white circles) of myocytes and neurons in the Urmetazoan are “Phylogeny Dependent” and are contingent on whether ctenophores or sponges are the sister group to all other metazoans. In contrast, the presence or absence from the Urmetazoan of “Phylogeny Independent” traits can be reasonably inferred, regardless of the branch order of sponges and ctenophores.
Several other traits, while absent from either sponges or ctenophores, can also be traced back to the Urmetazoan because they are conserved in diverse metazoans and in one or more of their protozoan relatives. For example, the conservation of the cadherin-repeat encoding gene hedgling in choanoflagellates, sponges, and cnidarians suggests that it was present in the Urmetazoan, despite its absence from the genomes of ctenophores, placozoans, and bilaterians [e.g., 59, 60]. Similarly, the collar cells found today in choanoflagellates, sponges, cnidarians and many bilaterians likely appeared in the Urmetazoan, despite their absence from ctenophores and most ecdysozoans [19].
Reconstructing the ancestry of other traits can be more challenging. If a trait is absent from non-metazoans and also not present in either sponges or ctenophores, inferences about its ancestry are contingent upon and await the resolution of the metazoan phylogeny (Figure 3). For some of these traits, ancestral reconstruction is further complicated by differing interpretations of their homology relationships. One trait that illustrates and suffers from both of these complications is the neuron, a cell type present in ctenophores and apparently lacking in sponges and placozoans (but see [61]). Historically, neurons in ctenophores, cnidarians, and bilaterians have been inferred to be homologous, with their homology inspiring some zoologists to place these lineages in the Neuralia clade [54]. Not only do neurons from ctenophores, cnidarians, and bilaterians produce a conserved set of diagnostic neuropeptides (as revealed by antibody staining), but the genomes of ctenophores and cnidarians encode homologs of diverse proteins that have been shown in bilaterians to be required for neuronal fate, neuronal patterning, and the formation of synapses, subcellular structures that allow the passage of chemical or electrical signals between neurons [7, 32, 62–66]. Nonetheless, ctenophore genomes appear to lack a number of classically-defined bilaterian neuronal genes (e.g., neuroligin) and don’t express other neuronal genes in a neuron-specific manner [32, 62]. Those who emphasize the similarities among ctenophore and other metazoan neurons infer that they have a shared ancestry and are homologous, while some who have focused on the differences argue for independent origins.
Given that the controversy surrounding the sister lineage to the rest of metazoans is far from settled, how do we make progress in reconstructing the ancestry of traits that are not conserved in all metazoans? We favor an explicit discussion about the implications of the two leading alternatives (ctenophores-sister vs. sponges-sister) to inferences about trait presence or absence in the Urmetazoan (Figure 3B). For the example of neurons, under the sponges-sister hypothesis, we might reasonably infer that the Urmetazoan lacked neurons and that neurons subsequently evolved in the stem lineage leading to ctenophores and all other metazoans, although it is not possible to rule out an Urmetazoan origin of neurons, only for them to be lost in sponges. (Note that under both of these scenarios, placozoans are presumed to have lost neurons (Figure 3A).
Under the ctenophore-first hypothesis, one must both explain the absence of neurons from sponges and placozoans and take account of the fact that some features of the ctenophore nervous system resemble those from cnidarians and bilaterians, while others are quite different. One recent hypothesis focuses on the differences and suggests that neurons in ctenophores, cnidarians, and bilaterians might have evolved convergently rather than being homologous cell types that evolved through descent with modification [32, 62]. A seemingly more likely scenario, regardless of whether ctenophores or sponges are the sister to all other metazoans, is that the last common ancestor of ctenophores and cnidarians/bilaterians may have had a rudimentary nervous system that provided the genetic and cellular building blocks, including neurons, of modern nervous systems. This ancestral nervous system may have then been independently elaborated upon in the ctenophore and cnidarian/bilaterian lineages [66–68], yielding divergent nervous systems in the extant organisms. According to this scenario, if ctenophores diverged first, the sponge lineage would presumably have lost the ancestral nervous system, akin to the more recent and incontrovertible losses of neurons in placozoans and the parasitic myxozoans [69].
This picture of a rudimentary ancestral nervous system being elaborated in some lineages and lost in others parallels our understanding of the evolution of other genetically complex traits, such as body appendages, the heart, and eyes [70]. For example, the complex eyes of molluscs, arthropods and vertebrates were thought to have originated independently until the discovery that they all develop under the control of a conserved master regulatory gene, Pax6, and the subsequent inference that they each derive from an ancestral photoreceptor system [71, 72]. To unravel the evolutionary histories of ensembles of complex traits, such as the nervous system, we must not only resolve the metazoan phylogeny, but also test our assumptions about trait homology at multiple levels (i.e., genetic, cellular ultrastructure, and system function [73]) by generating detailed insights into the molecular and cellular mechanisms that give rise to these traits in diverse early-branching metazoans.
Science is an iterative and, dare we say, evolutionary process. We now understand that sponges are metazoans, and that ciliates are not. Moreover, we are gaining remarkable insights into the cell and molecular biology of organisms that lived and died over 600 million years ago, leaving hardly a trace in the fossil record. We know that they produced eggs and sperm, became multicellular through serial cell division, initiated the production of many differentiated cell types and tissues during gastrulation and likely fed on bacteria using specialized collar cells. But our window through time to the Urmetazoan remains obscured by uncertainty regarding whether sponges or ctenophores are the sister lineage to all other metazoans. While we concur with the sentiment in the opening quote that “we’re not afraid to admit what we don’t know” and we revel in the insights that are currently possible regarding the dawn of metazoan life (Figure 3), we eagerly anticipate the future breakthroughs that will allow us to move from ignorance to knowledge in our quest to reconstruct animal origins.
Acknowledgments
We thank Thibaut Brunet, Pawel Burkhardt, Casey Dunn, Babcock Hall, Joe Ryan, Xing-Xing Shen, Monika Sigg, and Max Telford for constructive feedback on earlier drafts of this manuscript. Research in the King laboratory is supported by the Howard Hughes Medical Institute, the National Institutes of Health (R01GM099533), and the Gordon and Betty Moore Foundation. Research in the Rokas laboratory is supported by the National Science Foundation (DEB-1442113), the Burroughs Wellcome Fund, and the March of Dimes Prematurity Research Center Ohio Collaborative.
Glossary
- Bilateria
The monophyletic group (clade) of metazoan phyla with a bilateral axis of symmetry; this clade includes all extant metazoan phyla except sponges, ctenophores, cnidarians, and placozoans.
- Eumetazoa
A hypothetical subkingdom of metazoans that includes all phyla that exhibit a “tissue grade of construction”; it includes all extant metazoan phyla, except sponges and placozoans (which belong to the subkingdom Parazoa) [13].
- Hybridization and Introgression
Hybridization occurs when two organisms from different, typically closely related, species mate and produce offspring. When the hybrids mate back with members of one of the parent species, genes (or genetic regions) from the other parent species can cross the species barrier through a process called introgression. Hybridization and introgression can lead to genes whose histories differ from the history of their species, complicating inference of phylogenetic relationships from gene sequence data.
- Incomplete lineage sorting of ancestral polymorphisms
the retention of two or more alleles from an ancestral population in descendant species following successive speciation events. Incomplete lineage sorting is usually followed by random allele fixation in the descendant species, which can result in gene histories that differ from the history of their species.
- Pan-metazoan homologies
Homologous traits found in all or nearly all extant metazoan phyla. Pan-metazoan homologies include metazoan synapomorphies (shared, derived traits) as well as more ancient, pre-metazoan traits that have been conserved in all metazoans.
- Parazoa
A now-refuted, paraphyletic subkingdom of metazoans comprised of sponges and placozoans to the exclusion of the rest of the metazoan phyla [13].
- Site-heterogeneous models of sequence evolution
These models allow different amino acid positions in a protein alignment to have their own substitution models [47]; they are inspired by the observation that certain positions of protein sequence alignments tolerate substitutions between only a specific subset of the 20 amino acids (e.g., only substitutions between the two negatively charged amino acids may be tolerated for the protein to retain its function). This is important because sites that show such behavior tend to flip-flop between the permissible amino acids across the taxa included in the phylogeny; thus, they lack phylogenetic information but give the appearance of doing so, contributing to phylogenetic error. As site-heterogeneous models are specifically tailored to individual amino acid positions, they can in principle reduce the negative impact of such sites on phylogenetic inference much better than site-homogeneous sites ([41], but see [46]).
- Site-homogeneous models of sequence evolution
The standard models of protein sequence evolution; these models are constructed from empirically derived amino acid substitution matrices. As these models assume that all amino acid positions in a given protein sequence alignment have evolved under the same substitution process, all positions in the alignment use the same amino acid exchange rate matrix. Different site-homogeneous models may be used for different gene alignments, or data partitions, within a concatenated data matrix.
- Sister lineage/branch
a lineage that is the closest relative of another lineage and vice versa. In the context of the metazoan controversy, the ctenophore-sister hypothesis proposes that ctenophores are the closest relatives to the rest of the metazoans (and vice versa), whereas the sponges-sister hypothesis proposes that sponges are the closest relatives to the rest of the metazoans (and vice versa).
- Urmetazoan
The last common ancestor of all metazoans.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rokas A, Carroll SB. Bushes in the tree of life. PLoS Biol. 2006;4:e352. doi: 10.1371/journal.pbio.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen XX, Hittinger CT, Rokas A. Contentious relationships in phylogenomic studies can be driven by a handful of genes. Nature Ecol Evol. 2017;1:0126. doi: 10.1038/s41559-017-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arcila D, Ortí G, Vari R, Armbruster JW, Stiassny MLJ, Ko KD, Sabaj MH, Lundberg J, Revell LJ, Betancur-R R. Genome-wide interrogation advances resolution of recalcitrant groups in the tree of life. Nature Ecol Evol. 2017;1:0020. doi: 10.1038/s41559-016-0020. [DOI] [PubMed] [Google Scholar]
- 4.Richter DJ, King N. The genomic and cellular foundations of animal origins. Ann Rev Genet. 2013;47:509–537. doi: 10.1146/annurev-genet-111212-133456. [DOI] [PubMed] [Google Scholar]
- 5.Schierwater B, Holland PWH, Miller DJ, Stadler PF, Wiegmann BM, Wörheide G, Wray GA, DeSalle R. Never ending analysis of a century old evolutionary debate: “Unringing” the Urmetazoon bell. Front Ecol Evol. 2016;4:5. doi: 10.3389/fevo.2016.00005. [DOI] [Google Scholar]
- 6.Dohrmann M, Worheide G. Novel scenarios of early animal evolution--is it time to rewrite textbooks? Integr Comp Biol. 2013;53:503–511. doi: 10.1093/icb/ict008. [DOI] [PubMed] [Google Scholar]
- 7.Ryan JF, Pang K, Schnitzler CE, Nguyen AD, Moreland RT, Simmons DK, Koch BJ, Francis WR, Havlak P, Program NCS, et al. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 2013;342:1242592. doi: 10.1126/science.1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn CW, Leys SP, Haddock SH. The hidden biology of sponges and ctenophores. Trends Ecol Evol. 2015;30:282–291. doi: 10.1016/j.tree.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Brusca RC, Brusca GJ. Invertebrates. Sunderland, Mass: Sinauer; 1990. [Google Scholar]
- 10.Nielsen C. Animal Evolution. 2. Oxford: Oxford University Press; 2001. [Google Scholar]
- 11.Saville-Kent W. A Manual of the Infusoria. 1–3. London: David Bogue; 1880–1882. [Google Scholar]
- 12.Haeckel E. Memoirs: The gastraea-theory, the phylogenetic classification of the animal kingdom and the homology of the germ-lamellæ. J Cell Sci. 1874;s2–14:223–247. [Google Scholar]
- 13.Hyman LH. The Invertebrates: Protozoa through Ctenophora. 1. I. New York: McGraw-Hill; 1940. [Google Scholar]
- 14.Leys SP, Degnan BM. Embryogenesis and metamorphosis in a haplosclerid demosponge: gastrulation and transdifferentiation of larval ciliated cells to choanocytes. Invert Biol. 2002;121:171–189. [Google Scholar]
- 15.Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leininger S, Adamski M, Bergum B, Guder C, Liu J, Laplante M, Brate J, Hoffmann F, Fortunato S, Jordal S, et al. Developmental gene expression provides clues to relationships between sponge and eumetazoan body plans. Nat Commun. 2014;5:3905. doi: 10.1038/ncomms4905. [DOI] [PubMed] [Google Scholar]
- 17.Nakanishi N, Sogabe S, Degnan BM. Evolutionary origin of gastrulation: insights from sponge development. BMC Biol. 2014;12:26. doi: 10.1186/1741-7007-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James-Clark H. IV—Conclusive proofs of the animality of the ciliate sponges, and of their affinities with the Infusoria flagellata. J Nat Hist. 1867;19:13–18. [Google Scholar]
- 19.Brunet T, King N. The origin of animal multicellularity and cell differentiation. 2017 doi: 10.1101/161695. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mah JL, Christensen-Dalsgaard KK, Leys SP. Choanoflagellate and choanocyte collar-flagellar systems and the assumption of homology. Evol Dev. 2014;16:25–37. doi: 10.1111/ede.12060. [DOI] [PubMed] [Google Scholar]
- 21.King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harbison HR. On the classification and evolution of the Ctenophora. In: Conway Morris S, George JD, Gibson R, Platt HM, editors. The Origins and Relationships of Lower Invertebrates. Oxford: Oxford University Press; 1985. pp. 78–100. [Google Scholar]
- 23.Wainright PO, Hinkle G, Sogin ML, Stickel SK. Monophyletic origins of the metazoa: an evolutionary link with fungi. Science. 1993;260:340–342. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- 24.Collins AG. Evaluating multiple alternative hypotheses for the origin of Bilateria: an analysis of 18S rRNA molecular evidence. Proc Natl Acad Sci USA. 1998;95:15458–15463. doi: 10.1073/pnas.95.26.15458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medina M, Collins AG, Silberman JD, Sogin ML. Evaluating hypotheses of basal animal phylogeny using complete sequences of large and small subunit rRNA. Proc Natl Acad Sci USA. 2001;98:9707–9712. doi: 10.1073/pnas.171316998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Podar M, Haddock SH, Sogin ML, Harbison GR. A molecular phylogenetic framework for the phylum Ctenophora using 18S rRNA genes. Mol Phylogenet Evol. 2001;21:218–230. doi: 10.1006/mpev.2001.1036. [DOI] [PubMed] [Google Scholar]
- 27.Rodrigo AG, Bergquist PR, Bergquist PL, Reeves PR. Are sponges animals? An investigation into the vagaries of phylogenetic inference. In: van Soest RWM, van Kempen TMG, Braekman JC, editors. Sponges in Time and Space. Rotterdam: Balkema; 1994. pp. 47–54. [Google Scholar]
- 28.Abouheif E, Zardoya R, Meyer A. Limitations of metazoan 18S rRNA sequence data: implications for reconstructing a phylogeny of the animal kingdom and inferring the reality of the Cambrian explosion. J Mol Evol. 1998;47:394–405. doi: 10.1007/pl00006397. [DOI] [PubMed] [Google Scholar]
- 29.Rokas A, King N, Finnerty J, Carroll SB. Conflicting phylogenetic signals at the base of the metazoan tree. Evol Dev. 2003;5:346–359. doi: 10.1046/j.1525-142x.2003.03042.x. [DOI] [PubMed] [Google Scholar]
- 30.Field KG, Olsen GJ, Lane DJ, Giovannoni SJ, Ghiselin MT, Raff EC, Pace NR, Raff RA. Molecular phylogeny of the animal kingdom. Science. 1988;239:748–753. doi: 10.1126/science.3277277. [DOI] [PubMed] [Google Scholar]
- 31.Philippe H, Chenuil A, Adoutte A. Can the Cambrian explosion be inferred through molecular phylogeny? Development Suppl. 1994:15–25. [Google Scholar]
- 32.Moroz LL, Kocot KM, Citarella MR, Dosung S, Norekian TP, Povolotskaya IS, Grigorenko AP, Dailey C, Berezikov E, Buckley KM, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510:109–114. doi: 10.1038/nature13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452:745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 34.Philippe H, Derelle R, Lopez P, Pick K, Borchiellini C, Boury-Esnault N, Vacelet J, Renard E, Houliston E, Queinnec E, et al. Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19:706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- 35.Simion P, Philippe H, Baurain D, Jager M, Richter DJ, Di Franco A, Roure B, Satoh N, Queinnec E, Ereskovsky A, et al. A large and consistent phylogenomic dataset supports sponges as the sister group to all other animals. Curr Biol. 2017;27:958–967. doi: 10.1016/j.cub.2017.02.031. [DOI] [PubMed] [Google Scholar]
- 36.Pick KS, Philippe H, Schreiber F, Erpenbeck D, Jackson DJ, Wrede P, Wiens M, Alie A, Morgenstern B, Manuel M, et al. Improved phylogenomic taxon sampling noticeably affects nonbilaterian relationships. Mol Biol Evol. 2010;27:1983–1987. doi: 10.1093/molbev/msq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nosenko T, Schreiber F, Adamska M, Adamski M, Eitel M, Hammel J, Maldonado M, Muller WE, Nickel M, Schierwater B, et al. Deep metazoan phylogeny: when different genes tell different stories. Mol Phylogenet Evol. 2013;67:223–233. doi: 10.1016/j.ympev.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 38.Whelan NV, Kocot KM, Moroz LL, Halanych KM. Error, signal, and the placement of Ctenophora sister to all other animals. Proc Natl Acad Sci USA. 2015;112:5773–5778. doi: 10.1073/pnas.1503453112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hejnol A, Obst M, Stamatakis A, Ott M, Rouse GW, Edgecombe GD, Martinez P, Baguna J, Bailly X, Jondelius U, et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proc Roy Soc Ser B. 2009;276:4261–4270. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borowiec ML, Lee EK, Chiu JC, Plachetzki DC. Extracting phylogenetic signal and accounting for bias in whole-genome data sets supports the Ctenophora as sister to remaining Metazoa. BMC Genomics. 2015;16:987. doi: 10.1186/s12864-015-2146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Philippe H, Brinkmann H, Lavrov DV, Littlewood DT, Manuel M, Worheide G, Baurain D. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 2011;9:e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pisani D, Pett W, Dohrmann M, Feuda R, Rota-Stabelli O, Philippe H, Lartillot N, Worheide G. Genomic data do not support comb jellies as the sister group to all other animals. Proc Natl Acad Sci USA. 2015;112:15402–15407. doi: 10.1073/pnas.1518127112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schierwater B, Eitel M, Jakob W, Osigus HJ, Hadrys H, Dellaporta SL, Kolokotronis SO, Desalle R. Concatenated analysis sheds light on early metazoan evolution and fuels a modern “urmetazoon” hypothesis. PLoS Biol. 2009;7:e20. doi: 10.1371/journal.pbio.1000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halpern AL, Bruno WJ. Evolutionary distances for protein-coding sequences: modeling site-specific residue frequencies. Mol Biol Evol. 1998;15:910–917. doi: 10.1093/oxfordjournals.molbev.a025995. [DOI] [PubMed] [Google Scholar]
- 45.Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol. 2007;7 doi: 10.1186/1471-2148-7-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whelan NV, Halanych KM. Who let the CAT out of the bag? Accurately dealing with substitutional heterogeneity in phylogenomic analyses. Syst Biol. 2017;66:232–255. doi: 10.1093/sysbio/syw084. [DOI] [PubMed] [Google Scholar]
- 47.Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- 48.Chang ES, Neuhof M, Rubinstein ND, Diamant A, Philippe H, Huchon D, Cartwright P. Genomic insights into the evolutionary origin of Myxozoa within Cnidaria. Proc Natl Acad Sci USA. 2015;112:14912–14917. doi: 10.1073/pnas.1511468112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hahn MW, Nakhleh L. Irrational exuberance for resolved species trees. Evolution. 2016;70:7–17. doi: 10.1111/evo.12832. [DOI] [PubMed] [Google Scholar]
- 50.Babonis LS, Martindale MQ. Phylogenetic evidence for the modular evolution of metazoan signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2017;372 doi: 10.1098/rstb.2015.0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alie A, Hayashi T, Sugimura I, Manuel M, Sugano W, Mano A, Satoh N, Agata K, Funayama N. The ancestral gene repertoire of animal stem cells. Proc Natl Acad Sci USA. 2015;112:E7093–7100. doi: 10.1073/pnas.1514789112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alie A, Leclere L, Jager M, Dayraud C, Chang P, Le Guyader H, Queinnec E, Manuel M. Somatic stem cells express Piwi and Vasa genes in an adult ctenophore: ancient association of “germline genes” with stemness. Developmental Biology. 2011;350:183–197. doi: 10.1016/j.ydbio.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 53.Reitzel AM, Pang K, Martindale MQ. Developmental expression of “germline”- and “sex determination”-related genes in the ctenophore Mnemiopsis leidyi. Evodevo. 2016;7:17. doi: 10.1186/s13227-016-0051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nielsen C. Six major steps in animal evolution: are we derived sponge larvae? Evol Dev. 2008;10:241–257. doi: 10.1111/j.1525-142X.2008.00231.x. [DOI] [PubMed] [Google Scholar]
- 55.Abedin M, King N. The premetazoan ancestry of cadherins. Science. 2008;319:946–948. doi: 10.1126/science.1151084. [DOI] [PubMed] [Google Scholar]
- 56.Sebe-Pedros A, Ariza-Cosano A, Weirauch MT, Leininger S, Yang A, Torruella G, Adamski M, Adamska M, Hughes TR, Gomez-Skarmeta JL, et al. Early evolution of the T-box transcription factor family. Proc Natl Acad Sci USA. 2013;110:16050–16055. doi: 10.1073/pnas.1309748110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sebe-Pedros A, Roger AJ, Lang FB, King N, Ruiz-Trillo I. Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc Natl Acad Sci U S A. 2010;107:10142–10147. doi: 10.1073/pnas.1002257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suga H, Chen Z, de Mendoza A, Sebe-Pedros A, Brown MW, Kramer E, Carr M, Kerner P, Vervoort M, Sanchez-Pons N, et al. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat Commun. 2013;4:2325. doi: 10.1038/ncomms3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adamska M, Matus DQ, Adamski M, Green K, Rokhsar DS, Martindale MQ, Degnan BM. The evolutionary origin of hedgehog proteins. Curr Biol. 2007;17:R836–837. doi: 10.1016/j.cub.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 60.Nichols SA, Roberts BW, Richter DJ, Fairclough SR, King N. Origin of metazoan cadherin diversity and the antiquity of the classical cadherin/beta-catenin complex. Proc Natl Acad Sci USA. 2012;109:13046–13051. doi: 10.1073/pnas.1120685109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leys SP. Elements of a ‘nervous system’ in sponges. Journal of Experimental Biology. 2015;218:581–591. doi: 10.1242/jeb.110817. [DOI] [PubMed] [Google Scholar]
- 62.Moroz LL, Kohn AB. Unbiased view of synaptic and neuronal gene complement in ctenophores: Are there pan-neuronal and pan-synaptic genes across Metazoa? Integr Comp Biol. 2015;55:1028–1049. doi: 10.1093/icb/icv104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burkhardt P, Gronborg M, McDonald K, Sulur T, Wang Q, King N. Evolutionary insights into premetazoan functions of the neuronal protein homer. Mol Biol Evol. 2014;31:2342–2355. doi: 10.1093/molbev/msu178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jager M, Chiori R, Alie A, Dayraud C, Queinnec E, Manuel M. New insights on ctenophore neural anatomy: immunofluorescence study in Pleurobrachia pileus (Muller, 1776) J Exp Zool B Mol Dev Evol. 2011;316B:171–187. doi: 10.1002/jez.b.21386. [DOI] [PubMed] [Google Scholar]
- 65.Simmons DK, Pang K, Martindale MQ. Lim homeobox genes in the Ctenophore Mnemiopsis leidyi: the evolution of neural cell type specification. Evodevo. 2012;3:2. doi: 10.1186/2041-9139-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ryan JF. Did the ctenophore nervous system evolve independently? Zoology (Jena) 2014;117:225–226. doi: 10.1016/j.zool.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 67.Jekely G, Paps J, Nielsen C. The phylogenetic position of ctenophores and the origin(s) of nervous systems. Evodevo. 2015;6:1. doi: 10.1186/2041-9139-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liebeskind BJ, Hillis DM, Zakon HH. Convergence of ion channel genome content in early animal evolution. Proc Natl Acad Sci USA. 2015;112:E846–851. doi: 10.1073/pnas.1501195112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ryan JF, Chiodin M. Where is my mind? How sponges and placozoans may have lost neural cell types. Philos Trans R Soc Lond B Biol Sci. 2015;370 doi: 10.1098/rstb.2015.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carroll SB, Grenier JK, Weatherbee SD. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. 2. Malden, MA: Blackwell Pub; 2005. [Google Scholar]
- 71.Halder G, Callaerts P, Gehring WJ. New perspectives on eye evolution. Curr Opin Genet Dev. 1995;5:602–609. doi: 10.1016/0959-437x(95)80029-8. [DOI] [PubMed] [Google Scholar]
- 72.Randel N, Jekely G. Phototaxis and the origin of visual eyes. Philos Trans R Soc Lond B Biol Sci. 2016;371:20150042. doi: 10.1098/rstb.2015.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abouheif E, Akam M, Dickinson WJ, Holland PW, Meyer A, Patel NH, Raff RA, Roth VL, Wray GA. Homology and developmental genes. Trends Genet. 1997;13:432–433. doi: 10.1016/s0168-9525(97)01271-7. [DOI] [PubMed] [Google Scholar]
- 74.Anderson FE, Swofford DL. Should we be worried about long-branch attraction in real data sets? Investigations using metazoan 18S rDNA. Mol Phylogenet Evol. 2004;33:440–451. doi: 10.1016/j.ympev.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 75.Graybeal A. Is it better to add taxa or characters to a difficult phylogenetic problem? Syst Biol. 1998;47:9–17. doi: 10.1080/106351598260996. [DOI] [PubMed] [Google Scholar]
- 76.Degnan JH, Rosenberg NA. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol. 2009;24:332–340. doi: 10.1016/j.tree.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 77.Nakhleh L. Computational approaches to species phylogeny inference and gene tree reconciliation. Trends Ecol Evol. 2013;28:719–728. doi: 10.1016/j.tree.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]