Abstract
Background
Alterations in autophagy have been reported in hypertrophic cardiomyopathy (HCM) caused by Danon disease, Vici syndrome or LEOPARD syndrome, but not in HCM caused by mutations in genes encoding sarcomeric proteins, which account for most of HCM cases. MYBPC3, encoding cardiac myosin-binding protein C, is the most frequently mutated HCM gene.
Methods and Results
We evaluated autophagy in HCM patients carrying MYBPC3 mutations and in a Mybpc3-targeted knock-in (KI) HCM mouse model, as well as the effect of autophagy modulators on the development of cardiomyopathy in KI mice. Microtubule-associated protein 1 light chain 3 (LC3)-II protein levels were higher in HCM septal myectomies than in non-failing control hearts and in 60-week-old KI than wild-type (WT) mouse hearts. In contrast to WT, autophagic flux was blunted and associated with accumulation of residual bodies and glycogen in hearts of 60-week-old KI mice. We found that Akt-mTORC1 signaling was increased, and treatment with 2.24 mg/kgxd rapamycin or 40% caloric restriction for 9 weeks partially rescued cardiomyopathy or heart failure and restored autophagic flux in KI mice.
Conclusions
Altogether, we found that i) autophagy is altered in HCM patients with MYBPC3 mutations, ii) autophagy is impaired in Mybpc3-targeted KI mice and iii) activation of autophagy ameliorated the cardiac disease phenotype in this mouse model. We propose that activation of autophagy might be an attractive option alone or in combination with another therapy to rescue HCM caused by MYBPC3 mutations.
Keywords: Autophagy, caloric restriction, cardiomyopathy, cMyBP-C, hypertrophic cardiomyopathy, hypertrophy, MYBPC3, rapamycin
A well-controlled balance between protein synthesis and degradation is crucial for cellular homeostasis. The major pathways for degradation of cellular proteins are the ubiquitin-proteasome system (UPS) and the autophagy-lysosomal pathway (ALP).1 Autophagy is defined by the degradation of cellular material within the lysosome. It is a crucial process since it removes damaged proteins and organelles, supplies energy and maintains proper metabolism. Insufficient autophagy may lead to energy deficiency and proteotoxicity, while over-active autophagy can cause cell death. The genes and cellular processes that underlie autophagy are conserved from yeast to mammals and can be selective or nonselective. The most prevalent form of autophagy is called macroautophagy (hereafter autophagy), where a double-membrane vesicle, the phagophore, is formed and subsequently matures into an autophagosome, eventually fusing with a lysosome for degradation of its contents.2
Postmitotic cells such as neurons or cardiomyocytes are particularly dependent on energy and protein quality control. Whereas altered protein quality control mechanisms have been long correlated to neurological diseases,3 only a few cardiac diseases are known to be associated with defective autophagy. These include Danon disease,4, 5 LEOPARD syndrome,6 Vici syndrome,7, 8 desmin-related cardiomyopathy,9–11 diabetic cardiomyopathy,12 dilated cardiomyopathy (DCM) caused by lamin A/C (LMNA) mutations,13, 14 and left ventricular non-compaction (LVNC) caused by pleckstrin homology domain-containing family M, member 2 (PLEKHM2) mutations.15 In most of these cardiomyopathies there is a defect in a gene encoding a protein, which is involved in the ALP, either by acting directly on it or by inducing protein accumulation.
To the best of our knowledge, there is no evidence of altered autophagy in sarcomeropathy leading to hypertrophic cardiomyopathy (HCM) or DCM. HCM is an autosomal-dominant disorder, characterized by left ventricular hypertrophy (LVH) and diastolic dysfunction and has an estimated prevalence of 1:500 in the general population.16 The MYBPC3 gene, encoding cardiac myosin-binding protein-C (cMyBP-C), is frequently mutated in HCM, representing 40–50% of all HCM mutations.17, 18 cMyBP-C interacts with myosin, titin and actin and plays an important role in cardiac contraction and relaxation.17, 19–21 We previously reported impairment of the UPS and elevated protein levels of autophagic markers such as sequestosome-1 protein (p62), a marker for ubiquitinated protein aggregates, and microtubule-associated protein 1 light chain 3 (LC3)-II, an indicator of autophagosome number, in 60-week-old Mybpc3-targeted knock-in (KI) mice that develop LVH and cardiac dysfunction.22–26 These mice carry at the homozygous state the human c.772G>A MYBPC3 transition that results in a low level of mutant protein. In the present study, we investigated whether autophagy is altered in HCM patients and KI mice and whether activation of autophagy could ameliorate cardiomyopathy in KI mice.
Methods
Expanded Methods are available in the Data supplements.
Human samples
Human samples were obtained from septal myectomies of HCM patients carrying MYBPC3 mutations, from non-failing human heart tissue not suitable for transplantation or from donors that did not die from cardiac disease but of another cause (=non-failing, NF).27 All materials from patients and donors were taken with informed consent of the donors and with approval of the local ethical boards and according to the Declaration of Helsinki.
Animals
The investigation conformed to the guide for the care and use of laboratory animals published by the NIH (Publication No. 85-23, revised 2011, published by the National Research Council). The experimental procedures were in accordance with the German Law for the Protection of Animals and approved by the Authority for Health and Consumer Protection of the City State of Hamburg, Germany (no. 118/13 and 100/14).
Echocardiography
Transthoracic echocardiography was performed using the Vevo 2100 System (VisualSonics, Toronto, Canada) as described previously.25
Autophagic flux measurement
To measure the autophagic flux in vivo, mice were injected i.p. with 40 mg/kg leupeptin (Sigma Aldrich, L-8511) or sodium chloride (500 μL) as described previously.28
Experimental diet
Eleven-week-old KI and WT mice were kept on caloric restriction, rapamycin or control diet for 9 weeks. All diets were based on LabDiet 5LG6 (TestDiet) including Eudragit S100 (rapamycin coating material). Mice kept on caloric restriction were fed ~20% less in the first week and then ~40% less for the following 8 weeks than control mice. It was assumed that a 30 g mouse eats about 5 g per day. Mice were kept under tight observation and were regularly weighed. Mice on rapamycin diet received ~2.24 mg/kg rapamycin (Rapamycin Holdings™) encapsulated in Eudragit S100 daily. Echocardiography was performed at the beginning and at the end of the experiment. The autophagic flux was measured at the end of the experiment.
Electron Microscopy
Mouse hearts were initially fixed by perfusion with 1% paraformaldehyde/2% (vol/vol) glutaraldehyde in cardioplegic solution (50 mmol/L KCl, 5% dextrose in PBS) and next in 1% paraformaldehyde/2% (vol/vol) glutaraldehyde in 0.1 mol/L cacodylate buffer, pH 7.2. The heart was removed and immersed into the latter fixative (ice cold) and then left and right ventricular free walls and septa were isolated. Each region was divided into small fragments and fixed further in the same fixative at 4 °C, then postfixed/stained in 1% OSO4 (in water) before dehydration in acetone and embedding in epoxy resin. Ultrathin sections were counterstained with uranium and lead salts. Images were acquired on a Hitachi 7600 electron microscope equipped with an AMT digital camera.
Statistical analysis
Data were expressed as mean ± s.e.m. Statistical analyses were performed by one-way ANOVA plus Tukey’s or Dunnett’s post-test, or by unpaired Student’s t-test with GraphPad-Prism7 or by Welch’s ANOVA plus Tukey’s post-test with R v3.4.0, as indicated in the figure legends. A value of P < 0.05 was considered statistically significant.
Results
Autophagy is altered in HCM patients and KI mice
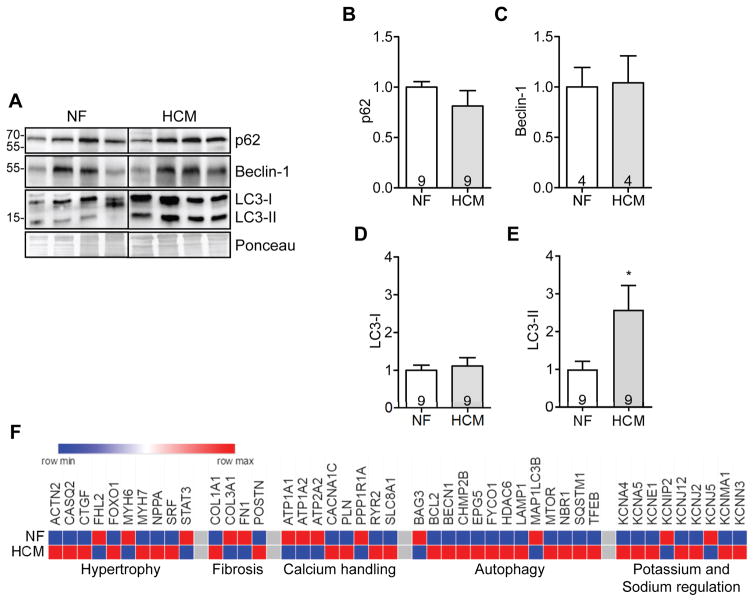
We evaluated p62, beclin-1 and LC3 protein levels in myectomy samples from patients carrying MYBPC3 mutations (Table S1). Whereas p62, beclin-1 and LC3-I levels did not differ between HCM and NF samples (Figure 1A to 1C), LC3-II protein levels were 2.6-fold higher in HCM (Figure 1A and 1E). We next quantified the expression of a customized panel of human genes regulated in heart failure, arrhythmias and autophagy in cardiac RNA pools of HCM and NF individuals. In addition to the commonly dysregulated genes in HCM, such as markers of hypertrophy or fibrosis and calcium/potassium handling proteins, the expression of several genes regulating autophagy was also altered in HCM, some being up-regulated (BCL2, BECN1, CHMP2B, EPG5, FYCO1, HDAC6, LAMP1, MTOR, NBR1, SQSTM1 and TFEB), others down-regulated (BAG3, MAP1LC3B,) when compared to NF (Figure 1F, Table S2).
Figure 1. Dysregulation of autophagy in HCM patients with MYBPC3 mutations.
Myectomy samples of HCM patients or heart samples from non-failing (NF) individuals were analyzed. A, Representative Western blots of p62, beclin-1 and LC3. Ponceau was used as loading control. Quantification of B, p62, C, beclin-1, D, LC3-I and E, LC3-II. Data are expressed as mean + s.e.m with *P<0.05 vs. NF, unpaired Student’s t-test. Number of individuals is indicated in the bars. F, Heatmap of selected genes comparing gene expression of proteins modulating hypertrophy, fibrosis, calcium handling, autophagy and potassium handling in NF and HCM (threshold <0.8- or >1.2-fold change to NF).
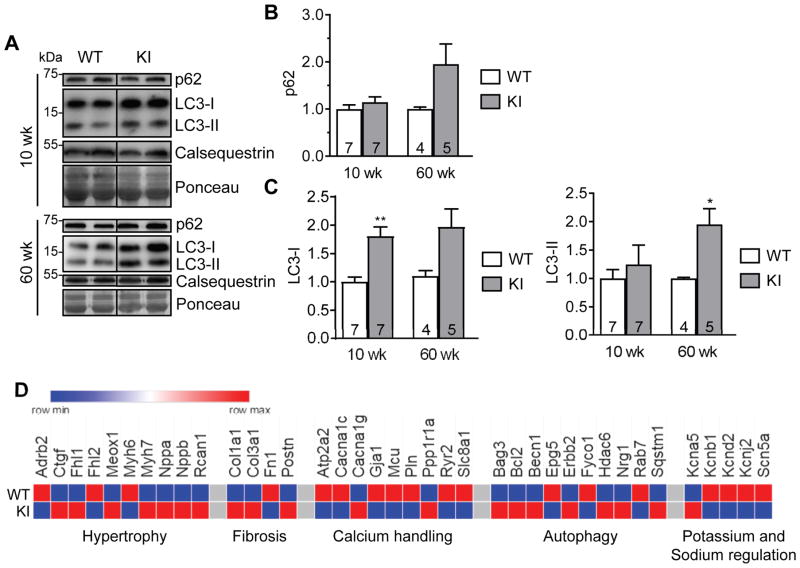
We then evaluated autophagic markers in 10-week-old and 60-week-old mice to explore changes during disease progression. We confirmed previously described higher p62 (non-significant) and LC3-II protein levels in 60-week-old,23 but not in 10-week-old KI mice (Figure 2A to 2C), suggesting that accumulation of p62 and LC3-II protein occurs late in the disease progression. Of note, however, LC3-I level was already higher in 10-week-old KI than WT mice (Figure 2A and 2C). We then quantified the expression of a customized panel of mouse genes associated with heart failure, arrhythmias and autophagy in ventricular RNA pools from 60-week-old KI and WT mice. KI exhibited dysregulated expression of proteins regulating hypertrophy, fibrosis, calcium handling, cardiac action potential, and autophagy (Bag3, Bcl2, Becn1, Epg5, Erbb2, Fyco1, Hdac6, Nrg1, Rab7 and Sqstm1; Figure 2D, Table S3).
Figure 2. Dysregulation of autophagy in KI mice.
Protein levels of p62 and LC3 in 10- and 60-week-old KI and WT mouse hearts. A, Representative Western blots of indicated proteins from mouse ventricular protein extracts (membrane-enriched fraction) of indicated ages. Calsequestrin and Ponceau were used as loading controls. Quantification of B, p62 and C, LC3-I and LC3-II protein levels normalized to Ponceau and related to WT. Data are expressed as mean + s.e.m. with *P<0.05 and **P<0.01 vs. WT, unpaired Student’s t-test (Welch’s test). Number of animals is indicated in the bars. D, Heatmap of selected genes (threshold <0.8 or >1.2 fold change to WT) comparing gene expression of hypertrophy, fibrosis, calcium handling, autophagy and potassium and sodium regulation between WT and KI mice.
Both human and mouse data suggest that autophagy is altered in HCM caused by MYBPC3 mutations.
Autophagic flux is impaired in KI mice
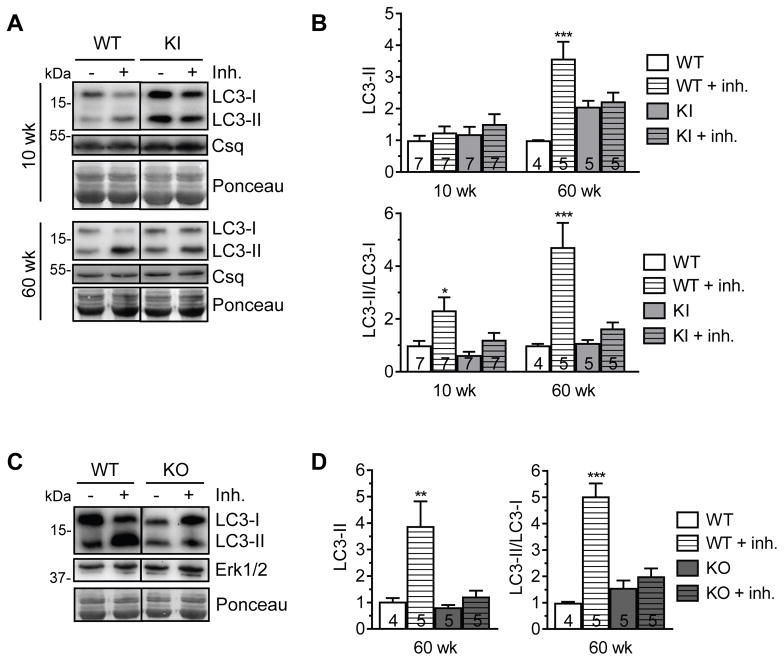
Measurement of basal levels of autophagic markers is not sufficient to conclude if autophagy is activated or impaired.29 Therefore, we next determined autophagic flux (macroautophagic activity) by evaluating LC3 turnover after injecting i.p. 40 mg/kg of the lysosomal protease inhibitor leupeptin in mice for 1 h. In hearts of 10-week-old mice, leupeptin treatment did not have any effect on the LC3-II level (Figure 3A and 3B), although the treatment worked in liver in both KI and WT mice (Figure S1). However, the LC3-II/LC3-I ratio was higher in WT than KI mice (Figure 3B). In hearts of 60-week-old mice, both LC3-II levels and the LC3-II/LC3-I ratio were markedly higher in leupeptin-treated than non-treated WT, whereas they did not differ between leupeptin-treated and non-treated KI mice (Figure 3A and 3B). This finding suggests an increased demand in autophagic activity in WT mice with aging, whereas the LC3 turnover was blunted in KI mice.
Figure 3. Impaired autophagic flux in KI and KO mice.
Evaluation of the autophagic flux in hearts of KI and WT mice. Either 40 mg/kg leupeptin (inh., inhibitor) or sodium chloride was injected i.p. into mice. After 1 h, hearts were extracted. A, Representative Western blots of indicated proteins from ventricular protein extracts of KI and WT mice of indicated ages. Calsequestrin and Ponceau were used as loading controls. B, Quantification of LC3-II (normalized to calsequestrin) and LC3-II/LC3-I ratio of KI and WT mice of indicated ages. C, Evaluation of the autophagic flux in hearts of KO and WT mice. Representative Western blots of indicated proteins from ventricular protein extracts of 60-week-old KO and WT mice. Ponceau and Erk1/2 were used as loading controls. D, Quantification of LC3-II (normalized to Erk1/2) of 60-week-old KO and WT mice. Quantifications are related to WT control. Data are expressed as mean + s.e.m. with *P<0.05, **P<0.01 and ***P<0.001 vs. corresponding control, one-way ANOVA (Welch’s test) plus Tukey’s post-test. Number of animals is indicated in the bars.
To examine whether the impaired autophagic flux in KI mice was due to the presence of mutant cMyBP-C or low level of cMyBP-C, we measured autophagic flux in 60-week-old Mybpc3-targeted knock-out (KO) mice that do not express any cMyBP-C but develop a similar cardiac disease phenotype as KI mice.30 LC3 turnover was blunted in KO mice to the same extent as in KI mice (Figure 3C and 3D). Although we cannot provide direct causality, these data suggest that low level of cMyBP-C rather than mutant cMyBP-C, in combination with pathological remodeling, induce impairment of autophagic flux.
Residual bodies and glycogen accumulate in KI mice
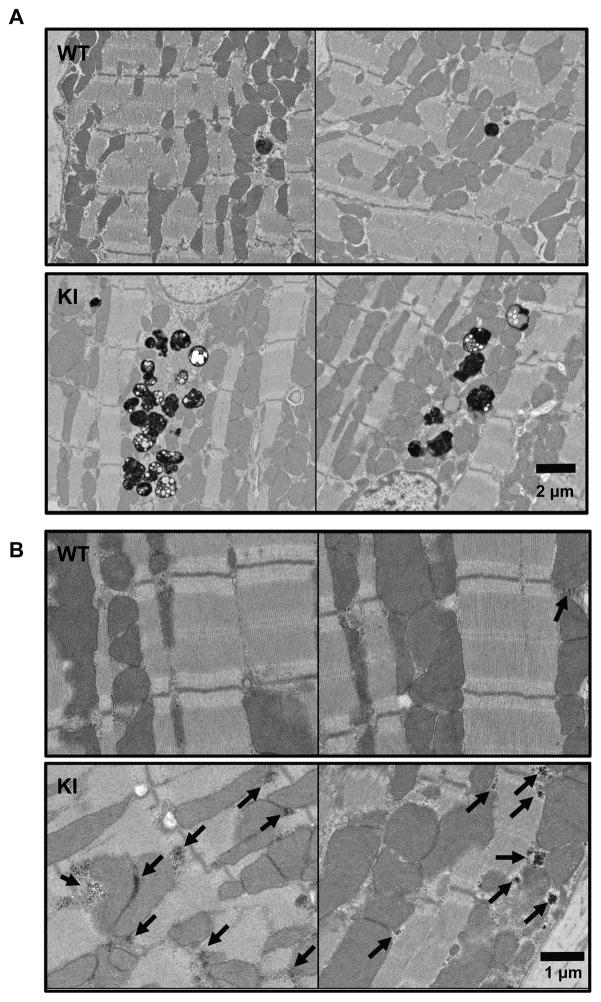
To assess autophagy-related ultrastructural differences between KI and WT mice, we analyzed osmium-stained cryosections from 60-week-old KI and WT mice using electron microscopy (Figure 4A). Terminal autolysosomes (residual bodies) containing cellular waste that was not broken down completely, probably resulting in lipofuscin or similar, markedly accumulated in KI compared to WT mice (Figure 4A, Figure S2). Lipofuscin vesicles usually accumulate with age, indicating an increase in cellular waste and/or deficiency in cellular waste degradation.31 Furthermore, we observed an accumulation of glycogen granula in KI mice (Figure 4B, Figure S3). Glycogen is degraded by the autophagic pathway, and an accumulation of glycogen granula is thought to be associated with impaired autophagy.32
Figure 4. Accumulation of residual bodies and glycogen granula in KI mice.
Electron microscope images of (osmium-stained) left ventricular tissues of 60-week-old WT and KI mice. Electron-dense structures like lipids stain dark. A, Residual bodies (black vesicular structures). B, Glycogen granula (indicated by arrows).
Lysosomes are functional in KI mice
To test if the autophagy impairment is induced by a decrease in number or compromised function of lysosomes, we assessed protein levels and activity of the lysosomal protease cathepsin D and protein levels of the lysosome-associated membrane protein 2 (LAMP-2). No differences in the levels of the different cathepsin D forms were detected between 60-week-old KI and WT mice (Figure S4A and S4B). Consistent with these data, the cathepsin D activity did also not differ between KI and WT (Figure S4C). Protein levels of LAMP-2 were unaltered in the KI mice as well (Figure S4D and S4E). These findings suggest that lysosomal degradation is not affected in KI mice.
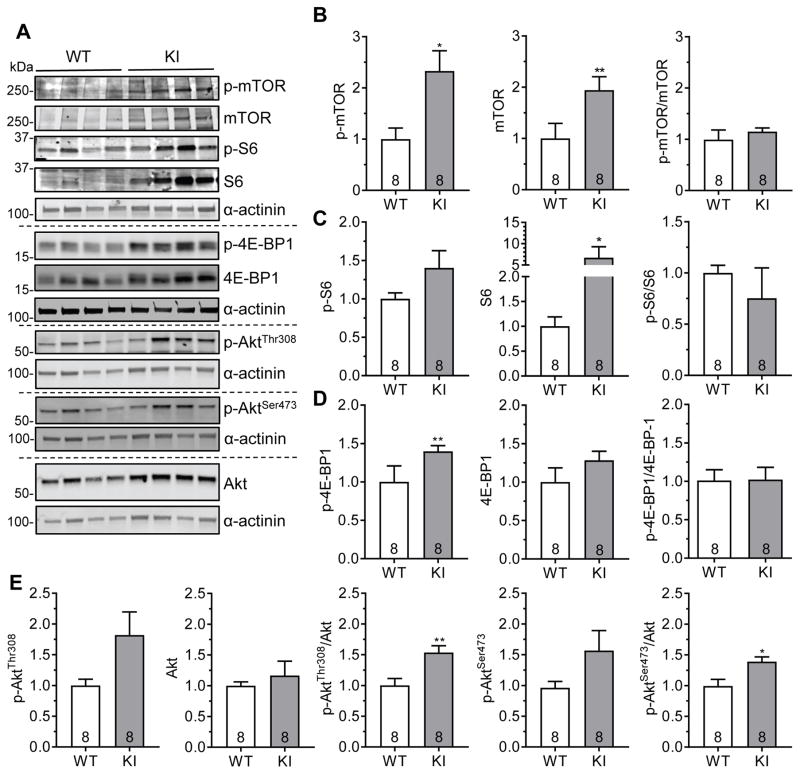
Akt-mTORC1 signaling is increased in KI mice
Mammalian target of rapamycin complex 1 (mTORC1) is a key negative regulator of autophagy and a recognized positive regulator of hypertrophy.33 Hence, we evaluated mTORC1 signaling in 60-week-old KI and WT mice (Figure 5). Levels of phosphorylated proteins (=activation) of mTOR (p-mTOR) and eukaryotic translation initiation factor 4E-binding protein 1 (p-4E-BP1), but not of ribosomal protein S6 (p-S6) were higher in KI than WT (Figure 5A to 5D). Levels of total mTOR and S6 (S6), but not of total 4E-BP1 were higher in KI than WT. Despite no difference in the ratio of phosphorylated-to-total proteins between the groups, increased p-mTOR and p-4E-BP1 levels suggest, at least in part, an increased mTORC1 signaling in KI mice. This increase in mTORC1 signaling was even more pronounced in KO (Figure S5) than in KI mice. The protein levels of Atg5 and Atg7, crucial autophagy enhancers, did not differ between KI, KO and WT mice (Figure S6).
Figure 5. Increased Akt-mTORC1 signaling in KI mice.
Protein levels of phosphorylated mTOR (p-mTOR), mTOR, phosphorylated S6 (p-S6), S6, phosphorylated 4E-BP1 (p-4E-BP1), 4E-BP1, phosphorylated Akt (p-AktThr308 and p-AktSer473) and Akt in 60-week-old KI and WT mouse hearts. A, Representative Western blots of indicated proteins from mouse ventricular protein extracts (cytosolic fraction). α-actinin was used as loading control. Quantification of B, p-mTOR, mTOR and p-mTOR/mTOR, C, p-S6, S6 and p-S6/S6, D, p-4E-BP1, 4E-BP1 and p-4E-BP1/4E-BP1 and E, p-AktThr308, Akt, p-AktThr308/Akt, p-AktSer473 and p-AktSer473/Akt. Protein levels were normalized to α-actinin and related to WT. Data are expressed as mean + s.e.m. with *P<0.05, **P<0.01 vs. WT, unpaired Student’s t-test. Number of animals is indicated in the bars.
We then evaluated which upstream pathways increased mTORC1 activity in KI mice (Figure 5E, Figure S7). Dual phosphorylation (=activation) of serine threonine kinase Akt/protein kinase B, evaluated by AktThr308/Akt and AktSer473/Akt ratios, was higher in KI than in WT mice (Figure 5E), whereas p-AMPK/AMPK, p-GSK3β, p-Erk1/2/Erk1/2 and p-p38/p38 ratios did not differ between KI and WT mice (Figure S7). These data suggest that activated mTORC1 results from activation of Akt signaling.
Rapamycin treatment or caloric restriction partially rescues cardiomyopathy in KI mice
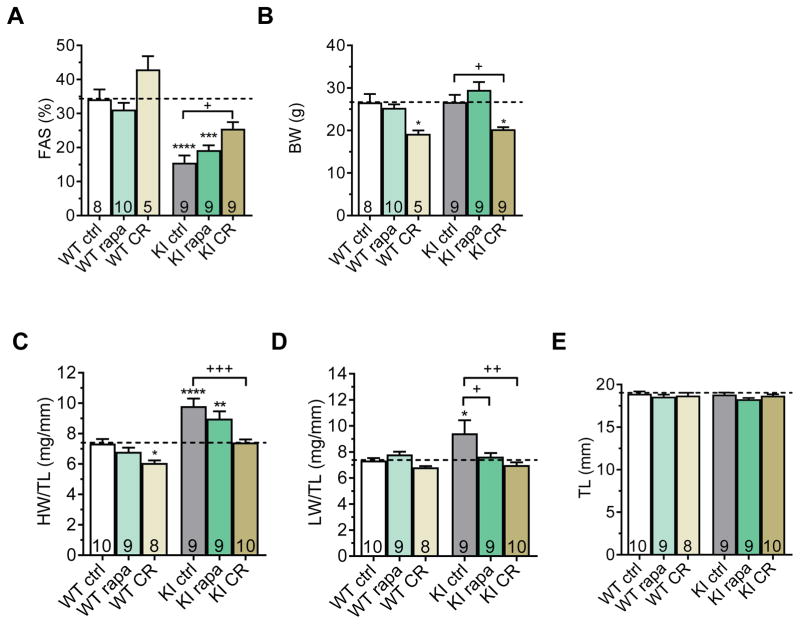
To activate autophagy in KI mice, we used rapamycin, an inhibitor of mTORC1, and caloric restriction (CR), which also decreases mTORC1 activity.34, 35 We evaluated whether these treatments could ameliorate cardiomyopathy in KI mice. Eleven-week-old KI and WT mice were subjected to a 9-week treatment with either 2.24 mg/kgxd rapamycin or 40% CR. At the beginning of the experiment, fractional area shortening (FAS) was lower and left ventricular mass-to-body weight ratio (LVM/BW) was higher in KI than WT mice, whereas BW did not differ between KI and WT, indicating systolic dysfunction and LVH (Figure 6A and 6B, Figure S8). At the end of the treatment, FAS did not significantly differ between rapamycin-treated and untreated WT and KI mice, whereas it was higher in CR-treated than untreated KI and WT mice (Figure 6A; Tables S4 and S5). The FAS difference of 10% between CR-treated and untreated KI mice was significant, suggesting partial amelioration of cardiac function in KI mice. As expected, BW was markedly lower in CR-treated than in untreated KI and WT mice, but was not affected by rapamycin treatment (Figure 6B). Heart weight-to-tibia length ratio (HW/TL) was ~30% higher in untreated KI than in untreated WT mice and ~24% lower in CR-treated KI than in untreated KI mice (Figure 6C), whereas TL did not differ between groups (Figure 6E). Lung weight-to-tibia length ratio (LW/TL) was higher in untreated KI than in untreated WT, indicating pulmonary edema induced by heart failure (Figure 6D). Both rapamycin and CR treatments lowered LW/BW in KI, which did not differ from WT in these conditions (Figure 6D), suggesting regression of heart failure in KI mice.
Figure 6. Partial rescue of cardiomyopathy by 9-week rapamycin treatment or caloric restriction in KI mice.
Determination of cardiac function by echocardiography and parameters of hypertrophy and heart failure in KI and WT mice after 9-week rapamycin treatment (rapa), 40% caloric restriction (CR) or control treatment (ctrl). Mice were fed with chow containing either 2.24 mg/kg rapamycin or coating material (control). Mice on caloric restriction were fed with 60% of control diet. A, Fractional area shortening (FAS). B, Body weight (BW). C, Heart weight-to-tibia length ratio (HW/TL). D, Lung weight-to-tibia length ratio (LW/TL) E, Tibia length (TL). Data are expressed as mean + s.e.m. with *P<0.05, **P<0.01, and ****P<0.0001 vs. WT ctrl, and +P<0.05, ++P<0.01 and +++P<0.001 vs. KI ctrl, one-way ANOVA plus Tukey’s post-test. Number of animals is indicated in the bars.
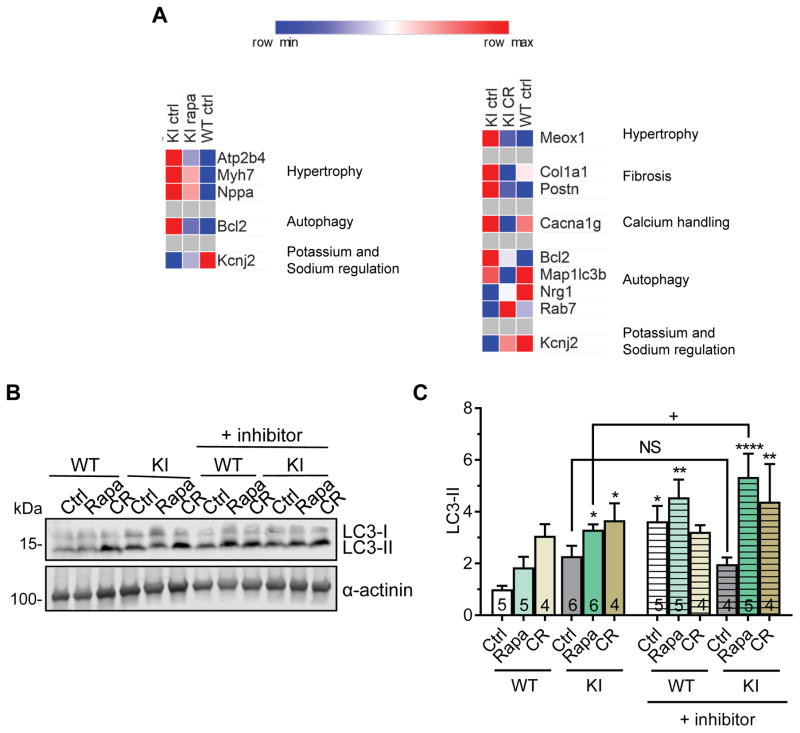
Both treatments normalized the higher Bcl2 mRNA levels and lower Kcnj2 mRNA levels in KI mice towards WT levels (Figure 7A, Table S6). In addition, rapamycin partially normalized the levels of markers of hypertrophy/heart failure (Atp2b4, Myh7, Nppa), while CR reversed the altered gene expression of hypertrophy and fibrosis markers (Meox1, Col1a1, Postn), the calcium handling protein Cacna1g and autophagy regulating genes (Map1lc3b, Nrg1 and Rab7; Figure 7A, Table S6). Furthermore, both treatments increased the LC3-II levels in both KI and WT mice, indicating activation of autophagy (Figure 7B and 7C). Finally, LC3-II levels increased after leupeptin in untreated WT, but not in KI mice, suggesting blunted LC3 turnover in KI mice (Figure 7B and 7C). The autophagic flux was restored in rapamycin-and CR-treated KI mice.
Figure 7. Gene expression analysis and autophagic flux in rapamycin-treated or calorie-restricted KI and WT mice.
KI and WT mice were treated for 9 weeks with either 2.24 mg/kg rapamycin (rapa), 40% caloric restriction (CR) or control treatment (ctrl). A, Heatmap of selected genes (threshold <0.8 or >1.2 fold change to KI ctrl) comparing gene expression of hypertrophy, fibrosis, calcium handling, autophagy and potassium and sodium regulation between KI ctrl, KI rapa or KI CR and WT ctrl mice. B, Representative Western blots of indicated proteins from mouse ventricular protein extracts (membrane-enriched fraction). α-actinin was used as loading control. C, LC3-II quantification (normalized to α-actinin) related to WT ctrl. Data are expressed as mean + s.e.m. with *P<0.05, **P<0.01 and ****P<0.0001 vs. WT ctrl, one-way ANOVA plus Dunnett’s post-test, and non-significant (NS) and +P<0.05 vs. indicated group (comparing with and without inhibitor), unpaired Student’s t-test. Number of animals is indicated in the bars.
Discussion
In this study, we investigated autophagy in cardiomyopathy associated with MYBPC3 mutations in human HCM septal myectomies and in a Mybpc3-targeted KI mouse model. Our major findings were: (1) autophagy is altered in MYBPC3 mutation-carrying HCM patients (2) autophagy is impaired in KI mice and (3) activation of autophagy by rapamycin or CR ameliorates cardiomyopathy and autophagic flux in KI mice.
LC3-II protein levels were higher in septal myectomies from HCM patients with MYBPC3 mutations, indicating an alteration of autophagy. Although we cannot conclude whether there is activation or inhibition of autophagy in patients, data obtained in KI mice argue for autophagy impairment. This was associated with dysregulated gene expression of several proteins regulating autophagy in both HCM patients and KI mice. LC3-II accumulation in the KI mouse hearts was progressive and accompanied by autophagic flux impairment with age. Furthermore, residual bodies and glycogen, which are both degraded by autophagy,31, 32 accumulated in KI mice. Glycogen accumulation was associated with LVH and was found in a number of diseases involving defective autophagy, e.g. Pompe, Danon and Fabry disease.36 In contrast to the UPS impairment, which was found only in aged KI mice with markedly low amounts of mutant cMyBP-C,23 autophagy impairment was common in both KI and KO mice, suggesting that cMyBP-C haploinsufficiency alone or in combination with cardiomyopathy is a trigger.
The involvement of autophagy in HCM patients and animal models with mutations in sarcomeric proteins has not been studied in depth. Only a few inherited cardiomyopathies are known to be associated with a defect in autophagy. Deficiency of the principal lysosomal membrane protein LAMP-2 causes Danon disease involving severe HCM.4, 5 LEOPARD syndrome, caused by mutations in PTPN11 (protein tyrosine phosphatase, non-receptor type 11) leads to increased phosphatidylinositol 3-kinase (PI3K) signaling associated with reduced autophagy and HCM.6 Lamp2-deficient mice (Danon disease) showed accumulation of autophagic vesicles,37 while PTPN11-targeted knock-in mice (LEOPARD syndrome)6 and PTEN-targeted knock-out38 showed increased mTORC1 signaling and decreased autophagic flux, all suggesting autophagy impairment. Vici syndrome, a rare autosomal-recessive inherited multisystem disorder involving cardiomyopathy7 is caused by mutations in EPG5, which encodes the ectopic P-granules autophagy protein 5, an essential protein for autophagic degradation.39 Defective autophagy has been also reported in desmin-related cardiomyopathy caused by αB-crystallin or desmin mutations and associated with the accumulation of cytotoxic misfolded proteins,9 in DCM caused by LMNA mutations13, 14 or mutations in BAG3,40, 41 encoding human BCL2 associated athanogene 3 gene involved in selective macroautophagy.42 LVNC caused by a PLEKHM2 mutation was associated with a defective ALP and impairment of autophagic flux in patients’ fibroblasts.15
In the present study, mTORC1 signaling was elevated in KI and KO mice. mTORC1 negatively regulates autophagy initiation, but can also inhibit autophagosome-lysosome fusion.43–45 In addition, mTORC1 negatively regulates transcription factor EB and thus inhibits transcription of autophagy and lysosomal genes.46 Out of the several signaling pathways that are known to activate mTOR, we found that Akt/PKB signaling was increased and likely contributed to mTORC1 activation in KI mice. This was associated with the up-regulation of Ctgf, encoding the connective tissue growth factor, which induced cardiomyocyte hypertrophy via Akt signaling,47 and Bcl2, encoding B-cell CLL/lymphoma 2 (BCL2), which can be up-regulated via Akt signaling (Figure 2D).48 Accumulation of BCL2 can sequester Beclin-1 or inhibit Bax/Bak-mediated apoptosis and thus inhibits autophagy.49, 50 Similarly, BCL2, CTGF and MTOR genes were up-regulated in HCM (Figure 1F), suggesting activated mTORC1 in HCM.
Treatment with rapamycin or caloric restriction to inhibit mTORC1 activity, and thereby activate autophagy partially rescued the cardiomyopathy phenotype or heart failure and restored the autophagic flux in KI mice. The mechanism of action is not fully certain and rapamycin and CR may affect other cellular functions besides autophagy. However, our customized transcriptome analysis indicates normalization of expression of markers of hypertrophy, fibrosis and also autophagy regulating genes. Specifically, the expression levels of both Bcl2 and Kcnj2, encoding the potassium inwardly-rectifying channel, subfamily J, member 2 (Kir2.1), were both markedly dysregulated in KI mice, and were partially normalized toward levels found in WT mice with either treatment. Our data are in agreement with previous findings showing that rapamycin administration to mice with PTPN11 mutation, which resulted in activation of PI3K pathway and increased mTORC1 activity, ameliorated cardiomyopathy.6 Similarly, rapamycin treatment reversed hypertrophy in a PTEN-deficient mouse model with increased mTORC1 activity.38 Moreover, rapamycin treatment or CR has been shown to have positive effects in different models of pressure overload-induced hypertrophy and age-related hypertrophy.6, 38
Up to now, there are only general treatments like calcium channel- and β-blockers, septal myectomy, ethanol ablation and heart transplantation available for human HCM. Here, we provide evidence that autophagy is defective in HCM patients and mice carrying MYBPC3 mutations and that activation of autophagy ameliorates cardiomyopathy in mice. We therefore propose that activation of autophagy might be an attractive option alone or in combination with another approach to rescue HCM induced by MYBPC3 mutations.
Supplementary Material
Clinical perspective.
What is new?
This is the first study to link altered autophagy to hypertrophic cardiomyopathy (HCM) in human and mice.
Myectomy samples from patients with MYBPC3 mutations had altered expression of both genes and proteins involved in autophagy, suggesting impaired autophagy in HCM.
When a MYBPC3 mutation was knocked into the mouse genome, the hearts were hypertrophied and autophagy markers accumulated over time, consistent with defective autophagic mechanisms.
Mammalian target of rapamycin complex-1 (mTORC1) is a key negative regulator of autophagy and increased mTORC1 activation was present in the knock in mice. This was associated with increased phosphorylation of mTORC1 by the serine threonine kinase Akt.
Both rapamycin and caloric restriction decrease mTORC1 activity. Both treatments led to a reduction in hypertrophy and the restoration of the defective autophagic flux in the knock in mice.
What are the clinical implications?
The MYBPC3 gene, encoding cardiac myosin-binding protein-C (cMyBP-C), is frequently mutated in HCM, representing 40–50% of all HCM mutations.
Up to now, there are only general treatments like calcium channel- and β-blockers, septal myectomy, ethanol ablation and heart transplantation available for HCM.
Our data suggest that activation of autophagy may hold promise as a treatment to explore to rescue HCM induced by MYBPC3 mutations.
Acknowledgments
We thank Julia Münch and Monica Patten (University Heart Center Hamburg, Hamburg, Germany) for patients’ recruitment, Giulia Mearini, Frederik Flenner and Felix Friedrich (UKE-Pharmacology, Hamburg, Germany) for help in preservation of human septal myectomies and database maintenance, Jutta Starbatty (UKE-Pharmacology, Hamburg, Germany) for protein preparations, and Konstantina Stathopoulou and Frederik Flenner (UKE-Pharmacology, Hamburg, Germany) for selection/validation of NanoString probe sequences.
Funding Sources: This work was supported by the Fondation Leducq (Research grant Nr. 11, CVD 04), the DZHK (German Centre for Cardiovascular Research; Grants B12-011 and B14-016), the German Ministry of Research Education (BMBF), the seventh Framework Program of the European Union (Health-F2-2009-241577-Big-Heart project), and the Netherlands Cardiovascular Research Initiative CVON2014-40 DOSIS, an initiative with support of the Dutch Heart Foundation.
Footnotes
Conflict of Interest Disclosures: None.
References
- 1.Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–6. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 2.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, Menzies FM, Moreau K, Narayanan U, Renna M, Siddiqi FH, Underwood BR, Winslow AR, Rubinsztein DC. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 3.Menzies FM, Fleming A, Rubinsztein DC. Compromised autophagy and neurodegenerative diseases. Nat Rev Neurosci. 2015;16:345–57. doi: 10.1038/nrn3961. [DOI] [PubMed] [Google Scholar]
- 4.Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, Mora M, Riggs JE, Oh SJ, Koga Y, Sue CM, Yamamoto A, Murakami N, Shanske S, Byrne E, Bonilla E, Nonaka I, DiMauro S, Hirano M. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–10. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 5.Danon MJ, Oh SJ, DiMauro S, Manaligod JR, Eastwood A, Naidu S, Schliselfeld LH. Lysosomal glycogen storage disease with normal acid maltase. Neurology. 1981;31:51–7. doi: 10.1212/wnl.31.1.51. [DOI] [PubMed] [Google Scholar]
- 6.Marin TM, Keith K, Davies B, Conner DA, Guha P, Kalaitzidis D, Wu X, Lauriol J, Wang B, Bauer M, Bronson R, Franchini KG, Neel BG, Kontaridis MI. Rapamycin reverses hypertrophic cardiomyopathy in a mouse model of LEOPARD syndrome-associated PTPN11 mutation. J Clin Invest. 2011;121:1026–43. doi: 10.1172/JCI44972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.del Campo M, Hall BD, Aeby A, Nassogne MC, Verloes A, Roche C, Gonzalez C, Sanchez H, Garcia-Alix A, Cabanas F, Escudero RM, Hernandez R, Quero J. Albinism and agenesis of the corpus callosum with profound developmental delay: Vici syndrome, evidence for autosomal recessive inheritance. Am J Med Genet. 1999;85:479–85. doi: 10.1002/(sici)1096-8628(19990827)85:5<479::aid-ajmg9>3.3.co;2-4. [DOI] [PubMed] [Google Scholar]
- 8.Cullup T, Dionisi-Vici C, Kho AL, Yau S, Mohammed S, Gautel M, Jungbluth H. Clinical utility gene card for: Vici Syndrome. Eur J Hum Genet. 2014;22:83–87. doi: 10.1038/ejhg.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLendon PM, Robbins J. Desmin-related cardiomyopathy: an unfolding story. Am J Physiol Heart Circ Physiol. 2011;301:H1220–8. doi: 10.1152/ajpheart.00601.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tannous P, Zhu H, Johnstone JL, Shelton JM, Rajasekaran NS, Benjamin IJ, Nguyen L, Gerard RD, Levine B, Rothermel BA, Hill JA. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc Natl Acad Sci U S A. 2008;105:9745–50. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanbe A, Osinska H, Saffitz JE, Glabe CG, Kayed R, Maloyan A, Robbins J. Desmin-related cardiomyopathy in transgenic mice: a cardiac amyloidosis. Proc Natl Acad Sci U S A. 2004;101:10132–6. doi: 10.1073/pnas.0401900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delbridge LM, Mellor KM, Taylor DJ, Gottlieb RA. Myocardial autophagic energy stress responses--macroautophagy, mitophagy, and glycophagy. Am J Physiol Heart Circ Physiol. 2015;308:H1194–204. doi: 10.1152/ajpheart.00002.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi JC, Muchir A, Wu W, Iwata S, Homma S, Morrow JP, Worman HJ. Temsirolimus activates autophagy and ameliorates cardiomyopathy caused by lamin A/C gene mutation. Sci Transl Med. 2012;4:144ra102. doi: 10.1126/scitranslmed.3003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramos FJ, Chen SC, Garelick MG, Dai DF, Liao CY, Schreiber KH, MacKay VL, An EH, Strong R, Ladiges WC, Rabinovitch PS, Kaeberlein M, Kennedy BK. Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci Transl Med. 2012;4:144ra103. doi: 10.1126/scitranslmed.3003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muhammad E, Levitas A, Singh SR, Braiman A, Ofir R, Etzion S, Sheffield VC, Etzion Y, Carrier L, Parvari R. PLEKHM2 mutation leads to abnormal localization of lysosomes, impaired autophagy flux and associates with recessive dilated cardiomyopathy and left ventricular noncompaction. Hum Mol Genet. 2015;24:7227–40. doi: 10.1093/hmg/ddv423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785–9. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 17.Carrier L, Mearini G, Stathopoulou K, Cuello F. Cardiac myosin-binding protein C (MYBPC3) in cardiac pathophysiology. Gene. 2015;573:188–97. doi: 10.1016/j.gene.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Behrens-Gawlik V, Mearini G, Gedicke-Hornung C, Richard P, Carrier L. MYBPC3 in hypertrophic cardiomyopathy: from mutation identification to RNA-based correction. Pflugers Arch. 2014;466:215–23. doi: 10.1007/s00424-013-1409-7. [DOI] [PubMed] [Google Scholar]
- 19.Cazorla O, Szilagyi S, Vignier N, Salazar G, Kramer E, Vassort G, Carrier L, Lacampagne A. Length and protein kinase A modulations of myocytes in cardiac myosin binding protein C-deficient mice. Cardiovasc Res. 2006;69:370–80. doi: 10.1016/j.cardiores.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Pohlmann L, Kroger I, Vignier N, Schlossarek S, Kramer E, Coirault C, Sultan KR, El-Armouche A, Winegrad S, Eschenhagen T, Carrier L. Cardiac myosin-binding protein C is required for complete relaxation in intact myocytes. Circ Res. 2007;101:928–38. doi: 10.1161/CIRCRESAHA.107.158774. [DOI] [PubMed] [Google Scholar]
- 21.Moss RL, Fitzsimons DP, Ralphe JC. Cardiac MyBP-C regulates the rate and force of contraction in mammalian myocardium. Circ Res. 2015;116:183–92. doi: 10.1161/CIRCRESAHA.116.300561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vignier N, Schlossarek S, Fraysse B, Mearini G, Kramer E, Pointu H, Mougenot N, Guiard J, Reimer R, Hohenberg H, Schwartz K, Vernet M, Eschenhagen T, Carrier L. Nonsense-mediated mRNA decay and ubiquitin-proteasome system regulate cardiac myosin-binding protein C mutant levels in cardiomyopathic mice. Circ Res. 2009;105:239–48. doi: 10.1161/CIRCRESAHA.109.201251. [DOI] [PubMed] [Google Scholar]
- 23.Schlossarek S, Englmann DR, Sultan KR, Sauer M, Eschenhagen T, Carrier L. Defective proteolytic systems in Mybpc3-targeted mice with cardiac hypertrophy. Basic Res Cardiol. 2012;107:235. doi: 10.1007/s00395-011-0235-3. [DOI] [PubMed] [Google Scholar]
- 24.Gedicke-Hornung C, Behrens-Gawlik V, Reischmann S, Geertz B, Stimpel D, Weinberger F, Schlossarek S, Precigout G, Braren I, Eschenhagen T, Mearini G, Lorain S, Voit T, Dreyfus PA, Garcia L, Carrier L. Rescue of cardiomyopathy through U7snRNA-mediated exon skipping in Mybpc3-targeted knock-in mice. EMBO Mol Med. 2013;5:1060–77. doi: 10.1002/emmm.201202168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mearini G, Stimpel D, Geertz B, Weinberger F, Kramer E, Schlossarek S, Mourot-Filiatre J, Stoehr A, Dutsch A, Wijnker PJ, Braren I, Katus HA, Muller OJ, Voit T, Eschenhagen T, Carrier L. Mybpc3 gene therapy for neonatal cardiomyopathy enables long-term disease prevention in mice. Nat Commun. 2014;5:5515. doi: 10.1038/ncomms6515. [DOI] [PubMed] [Google Scholar]
- 26.Mearini G, Stimpel D, Kramer E, Geertz B, Braren I, Gedicke-Hornung C, Precigout G, Muller OJ, Katus HA, Eschenhagen T, Voit T, Garcia L, Lorain S, Carrier L. Repair of Mybpc3 mRNA by 5′-trans-splicing in a Mouse Model of Hypertrophic Cardiomyopathy. Mol Ther Nucleic Acids. 2013;2:e102. doi: 10.1038/mtna.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thottakara T, Friedrich FW, Reischmann S, Braumann S, Schlossarek S, Kramer E, Juhr D, Schluter H, van der Velden J, Munch J, Patten M, Eschenhagen T, Moog-Lutz C, Carrier L. The E3 ubiquitin ligase Asb2beta is downregulated in a mouse model of hypertrophic cardiomyopathy and targets desmin for proteasomal degradation. J Mol Cell Cardiol. 2015;87:214–224. doi: 10.1016/j.yjmcc.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 28.Haspel J, Shaik RS, Ifedigbo E, Nakahira K, Dolinay T, Englert JA, Choi AM. Characterization of macroautophagic flux in vivo using a leupeptin-based assay. Autophagy. 2011;7:629–42. doi: 10.4161/auto.7.6.15100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carrier L, Knoll R, Vignier N, Keller DI, Bausero P, Prudhon B, Isnard R, Ambroisine ML, Fiszman M, Ross J, Jr, Schwartz K, Chien KR. Asymmetric septal hypertrophy in heterozygous cMyBP-C null mice. Cardiovasc Res. 2004;63:293–304. doi: 10.1016/j.cardiores.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 31.Brunk UT, Terman A. Lipofuscin: mechanisms of age-related accumulation and influence on cell function. Free Radic Biol Med. 2002;33:611–9. doi: 10.1016/s0891-5849(02)00959-0. [DOI] [PubMed] [Google Scholar]
- 32.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy. Microsc Res Tech. 2004;64:10–20. doi: 10.1002/jemt.20046. [DOI] [PubMed] [Google Scholar]
- 33.Sciarretta S, Volpe M, Sadoshima J. Mammalian target of rapamycin signaling in cardiac physiology and disease. Circ Res. 2014;114:549–64. doi: 10.1161/CIRCRESAHA.114.302022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–8. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 35.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, Guan JL, Oshiro N, Mizushima N. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–91. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arad M, Maron BJ, Gorham JM, Johnson WH, Jr, Saul JP, Perez-Atayde AR, Spirito P, Wright GB, Kanter RJ, Seidman CE, Seidman JG. Glycogen storage diseases presenting as hypertrophic cardiomyopathy. N Engl J Med. 2005;352:362–72. doi: 10.1056/NEJMoa033349. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lullmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–6. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 38.Xu X, Roe ND, Weiser-Evans MC, Ren J. Inhibition of mammalian target of rapamycin with rapamycin reverses hypertrophic cardiomyopathy in mice with cardiomyocyte-specific knockout of PTEN. Hypertension. 2014;63:729–39. doi: 10.1161/HYPERTENSIONAHA.113.02526. [DOI] [PubMed] [Google Scholar]
- 39.Cullup T, Kho AL, Dionisi-Vici C, Brandmeier B, Smith F, Urry Z, Simpson MA, Yau S, Bertini E, McClelland V, Al-Owain M, Koelker S, Koerner C, Hoffmann GF, Wijburg FA, ten Hoedt AE, Rogers RC, Manchester D, Miyata R, Hayashi M, et al. Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat Genet. 2013;45:83–7. doi: 10.1038/ng.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villard E, Perret C, Gary F, Proust C, Dilanian G, Hengstenberg C, Ruppert V, Arbustini E, Wichter T, Germain M, Dubourg O, Tavazzi L, Aumont MC, DeGroote P, Fauchier L, Trochu JN, Gibelin P, Aupetit JF, Stark K, Erdmann J, et al. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J. 2011;32:1065–76. doi: 10.1093/eurheartj/ehr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Norton N, Li D, Rieder MJ, Siegfried JD, Rampersaud E, Zuchner S, Mangos S, Gonzalez-Quintana J, Wang L, McGee S, Reiser J, Martin E, Nickerson DA, Hershberger RE. Genome-wide studies of copy number variation and exome sequencing identify rare variants in BAG3 as a cause of dilated cardiomyopathy. Am J Hum Genet. 2011;88:273–82. doi: 10.1016/j.ajhg.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim YC, Guan KL. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest. 2015;125:25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, Hailey DW, Oorschot V, Klumperman J, Baehrecke EH, Lenardo MJ. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–6. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puertollano R. mTOR and lysosome regulation. F1000Prime Rep. 2014;6:52. doi: 10.12703/P6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31:1095–108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayata N, Fujio Y, Yamamoto Y, Iwakura T, Obana M, Takai M, Mohri T, Nonen S, Maeda M, Azuma J. Connective tissue growth factor induces cardiac hypertrophy through Akt signaling. Biochem Biophys Res Commun. 2008;370:274–8. doi: 10.1016/j.bbrc.2008.03.100. [DOI] [PubMed] [Google Scholar]
- 48.Pugazhenthi S, Nesterova A, Sable C, Heidenreich KA, Boxer LM, Heasley LE, Reusch JE. Akt/protein kinase B up-regulates Bcl-2 expression through cAMP-response element-binding protein. J Biol Chem. 2000;275:10761–6. doi: 10.1074/jbc.275.15.10761. [DOI] [PubMed] [Google Scholar]
- 49.Lindqvist LM, Heinlein M, Huang DC, Vaux DL. Prosurvival Bcl-2 family members affect autophagy only indirectly, by inhibiting Bax and Bak. Proc Natl Acad Sci U S A. 2014;111:8512–7. doi: 10.1073/pnas.1406425111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zou H, Lai Y, Zhao X, Yan G, Ma D, Cardenes N, Shiva S, Liu Y, Bai X, Jiang Y, Jiang Y. Regulation of mammalian target of rapamycin complex 1 by Bcl-2 and Bcl-XL proteins. J Biol Chem. 2013;288:28824–30. doi: 10.1074/jbc.M113.505370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.