Abstract
Background
Minimally invasive specific biomarkers of neurodegenerative diseases (NDs) would facilitate patient selection and disease progression monitoring. We describe the assessment of circulating brain-enriched microRNAs as potential biomarkers for Alzheimer’s disease (AD), frontotemporal dementia (FTD), Parkinson’s disease (PD), and amyotrophic lateral sclerosis (ALS).
Methods
In this case-control study, the plasma samples were collected from 250 research participants with a clinical diagnosis of AD, FTD, PD, and ALS, as well as from age- and sex-matched control subjects (n = 50 for each group), recruited from 2003 to 2015 at the University of Pennsylvania Health System, including the Alzheimer’s Disease Center, the Parkinson’s Disease and Movement Disorders Center, the Frontotemporal Degeneration Center, and the Amyotrophic Lateral Sclerosis Clinic. Each group was randomly divided into training and confirmation sets of equal size. To evaluate the potential of circulating microRNAs enriched in specific brain regions affected by NDs and present in synapses as biomarkers of NDs, the levels of 37 brain-enriched and inflammation-associated microRNAs in the plasma of all participants were measured using individual qRT-PCR. A “microRNA pair” approach was used for data normalization.
Results
MicroRNA pairs and their combinations (classifiers) capable of differentiating NDs from control and from each other were defined using independently and jointly analyzed training and confirmation datasets. AD, PD, FTD, and ALS are differentiated from control with accuracy of 0.89, 0.90, 0.88, and 0.83 (AUCs, 0.96, 0.96, 0.94, and 0.93), respectively; NDs are differentiated from each other with accuracy ranging from 0.77 (AUC, 0.87) for AD vs. FTD to 0.93 (AUC, 0.98) for AD vs. ALS. The data further indicate sex dependence of some microRNA markers. The average increase in accuracy in distinguishing ND from control for all and male/female groups is 0.06; the largest increase is for ALS, from 0.83 for all participants to 0.92/0.98 for male/female participants.
Conclusions
The work presented here suggests the possibility of developing microRNA-based diagnostics for detection and differentiation of NDs. Larger multicenter clinical studies are needed to further evaluate circulating brain-enriched microRNAs as biomarkers for NDs and to investigate their association with other ND biomarkers in clinical trial settings.
Electronic supplementary material
The online version of this article (doi:10.1186/s13195-017-0316-0) contains supplementary material, which is available to authorized users.
Keywords: Alzheimer’s disease, Frontotemporal dementia, Parkinson’s disease, Amyotrophic lateral sclerosis, microRNA, Blood-based biomarkers
Background
Neurodegenerative diseases (NDs) have become a serious medical and social problem in developed countries, owing to significantly increased lifespans. For example, Alzheimer’s disease (AD) is now the fifth leading cause of death for those aged 65 years and older in the United States [1]. Despite intense research and numerous clinical trials, no effective treatments are currently available for NDs. One key element in the development of new therapies is the availability of diagnostic tools to help identify disease early, stratify patients for clinical trials, and monitor disease progression during treatment. In this respect, the most advanced diagnostic approaches for NDs, such as analysis of proteins in cerebrospinal fluid (CSF) and various imaging techniques [2, 3], are highly promising but not suitable for primary screening and monitoring purposes, owing to their invasiveness and high cost. Minimally invasive, cost-effective biomarkers of NDs would be very helpful in the advancement of ND diagnosis and treatment [4–8]. Several factors complicate the search for such biomarkers [7, 9]. First, mechanisms of ND initiation and development are not well understood; second, NDs may develop without any signs or symptoms for 10–20 years prior to clinical manifestation; and third, commonly occurring comorbidities and symptomatic overlap, such as cognitive impairment, complicate the differential diagnosis of NDs (see [10, 11] for detailed discussion of the need and the roadmap for the development of blood-based biomarkers for AD and other dementias).
Recently, several groups have proposed the use of microRNAs (miRNAs) circulating in plasma or serum for ND detection [12–23]. miRNAs are small molecules (~22 nucleotides) that play important roles in gene regulation by binding to complementary regions of messenger transcripts and repressing their translation or regulating their degradation [24, 25]. On the basis of sequence complementarity analysis, an individual miRNA can bind to and regulate > 100 messenger RNAs (mRNAs), and an mRNA can be regulated by multiple miRNAs; thus, as potential biomarkers, miRNAs are reflective of multiple cellular processes. Over 2000 miRNAs have been discovered in human cells to date, and many of these miRNAs are specific to or overexpressed in certain organs, tissues, and cells [25–30]. Some miRNAs, including those that are cell-specific, can be enriched in particular cellular compartments, such as neurites and synapses [31–37]. miRNAs can be secreted or excreted into the extracellular space [38–41] and are detectable in plasma and serum [14, 42–44].
Intracellular concentrations and rates of secretion of miRNAs can be dramatically affected by physiological and pathological cellular processes [39, 45–47]. In the case of mild cognitive impairment (MCI), a heterogeneous syndrome characteristic of early stages of various NDs, we have previously shown that the condition can be detected by analysis of miRNAs enriched in synapses of brain regions affected in early AD, such as the hippocampus [14, 48]. The concentration of a brain-enriched miRNA in bodily fluids depends on intrinsic factors such as its expression; its intracellular localization; and disease-associated changes in expression, metabolism, and secretion. It also depends on extrinsic factors such as blood supply to a particular brain area, changes in blood-brain barrier permeability, and miRNA stability in circulation.
To compensate for the impact of factors unrelated to a specific ND and to account for smaller changes in miRNA concentrations accompanying slowly developing pathologies (as compared with those in acute diseases such as stroke), other brain-enriched miRNAs can be used for normalization of the miRNA biomarkers. This miRNA “pair” approach (see below) has led to the discovery of miR-132 and miR-134 families of miRNA biomarker pairs capable of differentiating MCI from age-matched controls with approximately 0.90 accuracy [48, 49]. Interestingly, these miRNA pairs proved less effective as biomarkers for detecting AD dementia stages [49], possibly owing to the loss of synapses and death of neurons in the hippocampus causing the amount of hippocampal synaptic miRNAs to decrease in plasma as the disease progresses.
The present study was designed to evaluate circulating brain-enriched miRNAs as potential biomarkers for detection and differentiation of AD, Parkinson’s disease (PD), frontotemporal lobar degeneration (frontotemporal dementia [FTD]), and amyotrophic lateral sclerosis (ALS).
Methods
Clinical diagnosis and plasma samples
Thirty-seven miRNAs were analyzed in two 0.5-ml aliquots (1 ml total) of K2-ethylenediaminetetraacetic acid-treated frozen plasma samples from 250 research participants (n = 50 for each ND) recruited and clinically evaluated at the University of Pennsylvania Health System, including the Alzheimer’s Disease Center (in 2003–2014), the Parkinson’s Disease and Movement Disorders Center (in 2010–2015), the Frontotemporal Degeneration Center (in 2010–2013), and the Amyotrophic Lateral Sclerosis Clinic (in 2010–2014). Fifty samples from age- and sex-matched control subjects were collected in 2009–2015. The ND conditions were clinically diagnosed according to published procedures [50–58]; demographic characteristics of the study groups, including control subjects, are summarized in Table 1 and Additional file 1. The study participants in the AD group were selected on the basis of AD phenotype and tau and amyloid biomarkers as determined by CSF analysis. The biofluid samples were collected and processed on the day they were obtained following the same standard operating procedures in all specialty clinical practices at the Center for Neurodegenerative Disease Research, University of Pennsylvania [59]. Blood samples were centrifuged at 3000 × g for 15 minutes at 4 °C and aliquoted into 2.0-ml polypropylene cryovials (Corning Life Sciences, Corning, NY, USA). The aliquots were stored at −80 °C. De-identified samples were sent to the Asuragen laboratory (Austin, TX, USA) for RNA isolation and qRT-PCR (see below). Clinical information, including demographics, diagnosis and cognitive scores, and identifiers to match the samples, were sent to the DiamiR principal investigator, who kept the information blinded from the laboratory personnel.
Table 1.
Cohort demographics and clinical data
| Diagnosis | Subgroups | No. of subjects | Age (years) | Sex (M/F) | MMSE [NR] | CSF Aβ42 | CSF t-tau | CSF p-tau | Years from diagnosis | Years of education |
|---|---|---|---|---|---|---|---|---|---|---|
| Control | N/A | 50 | 64.06 ± 9.8 (25–83) | 24/26 | 29.3 ± 1.0 | N/A | N/A | N/A | N/A | 16.44 ± 2.7 |
| AD | N/A | 50 | 67.8 ± 10.4 (53–87) | 24/26 | 21.0 ± 5.6 | 145.7 ± 34.8 | 116.2 ± 68 | 45 ± 23.7 | 3.6 ± 2.2 | 16.5 ± 2.3 |
| PD | Total | 50 | 66.76 ± 7.7 (49–82) | 35/15 | 28.9 ± 1.1 [11] | 261.8 ± 49.8 | 43 ± 13.7 | 20.9 ± 7.4 | 10 ± 5.1 | 16.54 ± 2.2 |
| Dementia | 15 | 72.33 ± 6.1 (61–82) | 11/4 | 27 [1] | N/A | N/A | N/A | 11 ± 3.9 | 16.33 ± 2.8 | |
| MCI | 8 | 68.37 ± 7.0 (54–77) | 5/3 | 29 ± 0.8 [4] | N/A | N/A | N/A | 10.4 ± 7.4 | 16.25 ± 1.7 | |
| FTD | Total | 50 | 63.14 ± 6.9 (46–76) | 28/22 | 24.1 ± 5.3 [46] | 271.4 ± 52.3 | 62.2 ± 27.9 | 16.5 ± 7.8 | 3.2 ± 2.3 | 15.96 ± 3.1 |
| bv | 23 | 61.69 ± 6.3 (46–74) | 15/8 | 25.3 ± 4.4 | 278.6 ± 52.3 | 57.2 ± 22.5 | 14.5 ± 4.6 | 2.7 ± 2.2 | 16.43 ± 3.0 | |
| PPA/Lgpen | 1 | 70 | 1/0 | 22 | 2 | 18 | ||||
| PPA/PNFA | 8 | 67.75 ± 4.6 (61–75) | 3/5 | 19.6 ± 9.18 [5] | 269.9 ± 63.5 | 75.2 ± 35.2 | 14.6 ± 4.3 | 4.5 ± 2.1 | 15.2 ± 3.84 | |
| PPA/SD | 8 | 61.25 ± 8.4 (49–76) | 4/4 | 22.1 ± 5.19 | 286 ± 48.4 | 79.9 ± 36.6 | 15.9 ± 4.5 | 4 ± 3.0 | 17 ± 2.82 | |
| PSP | 10 | 63.6 ± 7.51 (54–75) | 5/5 | 25.6 ± 4.4 | 244.7 ± 46.0 | 50.6 ± 19.5 | 14.5 ± 4.6 | 2.7 ± 1.6 | 14.4 ± 2.99 | |
| ALS | Total | 50 | 59.64 ± 10.8 (29–83) | 36/14 | 24.8 ± 7.8 [16] | 240.4 ± 76.3 | 71.1 ± 50.7 | 13.8 ± 6.8 | 1.8 ± 1.8 | 14.71 ± 2.9 |
| With FTD | 5 | 65.4 ± 9.18 (49–70) | 4/1 | 14. 3 ± 12.0 [3] | 222.4 ± 88.8 | 107 ± 62.1 | 19 ± 10.1 | 4 ± 4.4 | 15.8 ± 2.3 | |
| With MCI | 3 | 57 ± 5.2 (54–63) | 2/1 | 30 [1] | 340 ± 3 | 66 ± 32 | 11.3 ± 0.6 | 1 ± 1 | 13.5 ± 7.8 |
Abbreviations: Aβ 42 Amyloid-β 42, AD Alzheimer’s disease, ALS Amyotrophic lateral sclerosis, FTD Frontotemporal dementia, PD Parkinson’s disease, M/F Male/female, MMSE Mini Mental State Examination, NR Number of research participants with Mini Mental State Examination reported (MMSE is reported for all participants in a subgroup [e.g., control] if no NR value is given), N/A Not available, bv Behavioral variant, PPA/Lgpen Primary progressive aphasia, logopenic variant, PPA/PNFA Primary progressive aphasia/progressive nonfluent aphasia, PPA/SD Primary progressive aphasia/semantic dementia, PSP Progressive supranuclear palsy, CSF Cerebrospinal fluid, MCI Mild cognitive impairment, p-tau Phosphorylated tau, t-tau Total tau
All groups were randomly divided into two sets of equal size (25 samples each, balanced by age, sex, and comorbidities) and used in training and confirmation experiments. The PD and ALS groups had fewer female participants, and the ALS group was younger than other ND and control groups. Age may affect miRNA expression, and thus differences in the age composition of different groups may contribute to the differentiation of the groups with specific miRNAs. In the present study, however, the average ages were not significantly different between various groups (see Additional file 1); we found that the exclusion of several outliers did not change the results. Correlations between miRNA ratios and age in all pathology cohorts are low (r < 0.2) and have low statistical significance (p > 0.04). To determine the source of miRNA ratio differences in disease groups and the control group, we performed linear regression analysis of miRNA ratios using disease group identity and age as covariates. p Values for the significance of the disease group identity for all miRNA pairs selected were < 0.0001, indicating that the differentiation of two groups depended on diagnosis. Only in the ALS group did some miRNA pairs correlate with age with p < 0.05. Age adjustment for these pairs slightly increased the accuracy (1–2%); age adjustment for all other pairs either did not change or slightly improved the accuracy (by < 2%). Similarly, to determine the effect of sex on differentiation of PD and ALS (the two NDs with smaller numbers of female participants; see Table 1 and Additional file 1) from control subjects, we performed a linear regression analysis of miRNA ratios using sex as a covariate. For most pairs, the effect of sex adjustment was insignificant. Additional stratified analysis of all NDs (described in the “miRNA biomarker sex-dependent effects” subsection of the Results section below) revealed miRNA pairs differentiating male and female participants separately better than all (male and female) participants.
RNA isolation and qRT-PCR
RNA was extracted from 1 ml of plasma using a TRIzol treatment (Life Technologies, Carlsbad, CA, USA) and silica (Ambion Glass Fiber Microcolumn; Fisher Scientific, Pittsburgh, PA, USA) binding protocol (http://asuragen.com/wp-content/uploads/2016/05/biomarkers.pdf). Single-target qRT-PCR was performed using the TaqMan® Reverse Transcription Kit and miRNA-specific stem-loop primers (Applied Biosystems, Foster City, CA, USA). The RT step for miRNA measurements was performed in triplicate and each RT reaction was then used separately for PCR. 2µl plasma equivalents were present in the final PCR. (Additional file 2 presents the qRT-PCR variability for each tested miRNA.) Placental RNA was used as a positive control and no template as a negative control. Calibration curves for each miRNA were generated to facilitate comparing and combining of training and confirmation datasets. Quality control of miRNA preparations was performed by testing two ubiquitous miRNAs, miR-16 and miR-27a, in each plasma preparation; all samples with values within 2 SD of the average value were qualified as acceptable for analysis. The average SDs of miR-16 and miR-27a were 1.40 and 1.01 in the training and confirmation sets, respectively. miRNAs with cycle threshold (C t) > 37 were excluded from the analysis of a respective sample. Although small RNA yield was not directly measured after RNA extraction, the control data reported in Additional file 2 provide a good indicator of miRNA yield.
Statistical analyses
In our approach, an effective biomarker is a ratio (pair) of two brain-enriched miRNAs. Software developed at DiamiR [48, 49] was used for all calculations, including determination of miRNA pairs capable of differentiating NDs from controls and from each other on the basis of their concentration ratios (∆C t) in plasma. The application was designed using .NET technology with a set of .NET statistical packages. The selection of miRNA pairs as well as their combinations was based on the AUC and p values as follows. For every pair, ROC curves were constructed to calculate the area under the ROC curve (AUC). In the training set analysis, pairs with AUC ≥ 0.75 for AD, PD, and ALS, as well as AUC ≥ 0.70 for FTD, were initially accepted. Training and confirmation sets were of the same size. A lower cutoff for FTD was used because of high heterogeneity of the group (presence of several FTD syndromes). In the combined set, there were 23 behavioral variant frontotemporal dementia (bvFTD), 17 primary progressive aphasia (PPA), and 10 progressive supranuclear palsy (PSP) cases. The acceptance p value threshold was set on the basis of significance of differences between any two experimental groups, calculated by the Mann-Whitney U test with Bonferroni correction. Validation of miRNA pair selection and testing for overfitting were performed using random forest classification with bootstrapping. For 37 miRNAs used, the total number of pairs was 666 (36 × 37/2). The ratios A/B and B/A are equivalent as potential biomarkers because they have the same variance, difference of means, correlation coefficients, p values, and ROC parameters. In cases where disease was compared with control, the ratio whose numerical value was larger in disease samples was selected. According to the Bonferroni correction, statistically significant results should then have a p value of 0.05/666 = 7.5 × 10−5. Because we were using highly correlated miRNAs for creating pairs (C t Spearman’s rank correlation coefficient, ≥ 0.8), the number of pairs analyzed was reduced to about 160, and thus the threshold for statistical significance rose to ~ 3.0 × 10−4. Pair sensitivity and specificity are reported for the cutoff points on the ROC curves that provided the best overall accuracy. For selecting effective pair combinations, miRNA classifiers, a stepwise algorithm for logistic regression using a linear model with no interaction [60] was applied. The miRNA classifiers having AUC ≥ 0.9 for AD, PD, and ALS, as well as AUC ≥ 0.8 for FTD, in the combined (training + confirmation) set were accepted.
Results
miRNA selection
In this work, we performed targeted preselection of miRNAs present in synapses, enriched in different brain regions, and also detectable in plasma. The levels of all preselected miRNAs in plasma samples were determined by qRT-PCR. The miRNAs analyzed in the study are listed in Additional file 3 and included (1) miRNAs present in synapses [33, 35, 36, 61, 62] and enriched in different brain regions affected by the target pathologies [26–30, 63–67]; (2) miRNAs associated with inflammatory processes [68–71]; (3) miR-206 highly enriched in muscle tissue and in cerebellum [68, 72, 73]; (4) ubiquitous apoptosis-associated miR-16 [74]; and (5) miR-451, which is more effectively excreted from pathologic than normal cells [39]. As shown in Additional file 3, certain brain-enriched miRNAs are expressed in several brain regions; typically, the levels of these miRNAs in specific brain regions (shown in bold in Additional file 3) are significantly higher than in others. Potential roles of many of the miRNAs listed in Additional file 3 in NDs are being investigated by several groups [62, 75–88].
ND detection
As described above, each group (AD, FTD, PD, ALS, and age- and sex-matched control group) was divided in two equal sets and analyzed independently in the training and confirmation studies. miRNA pairs and their combinations (miRNA classifiers) capable of differentiating each ND from controls with the highest accuracy were assessed in the training set and verified in the confirmation set analysis, followed by the analysis of the combined dataset (Figs. 1 and 2 and Additional file 4). Combining two or more qRT-PCR experiments is often challenging because of possible technical variations in RNA extraction, RT and/or PCR kits, and instrumental fluctuations. Furthermore, samples for the confirmation study were kept frozen for an additional 6 months. Nonetheless, certain miRNA pairs and classifiers effectively differentiated AD, FTD, PD, and ALS from controls in both training and confirmation studies (Additional file 4). The accuracy of ND detection in the combined dataset by miRNA classifiers for AD, PD, FTD, and ALS was 0.89, 0.90, 0.88, and 0.83, respectively. The comparison of the differentiation obtained with individual miRNAs and with miRNA pairs is reported in Additional file 5. Only muscle-enriched miR-206, whose average concentration is about eight times higher in patients with ALS than in control subjects, differentiates these two groups with accuracy similar to that of miRNA pairs. In all other cases, the use of miRNA pairs produces much better differentiation between NDs and controls than individual miRNAs.
Fig. 1.
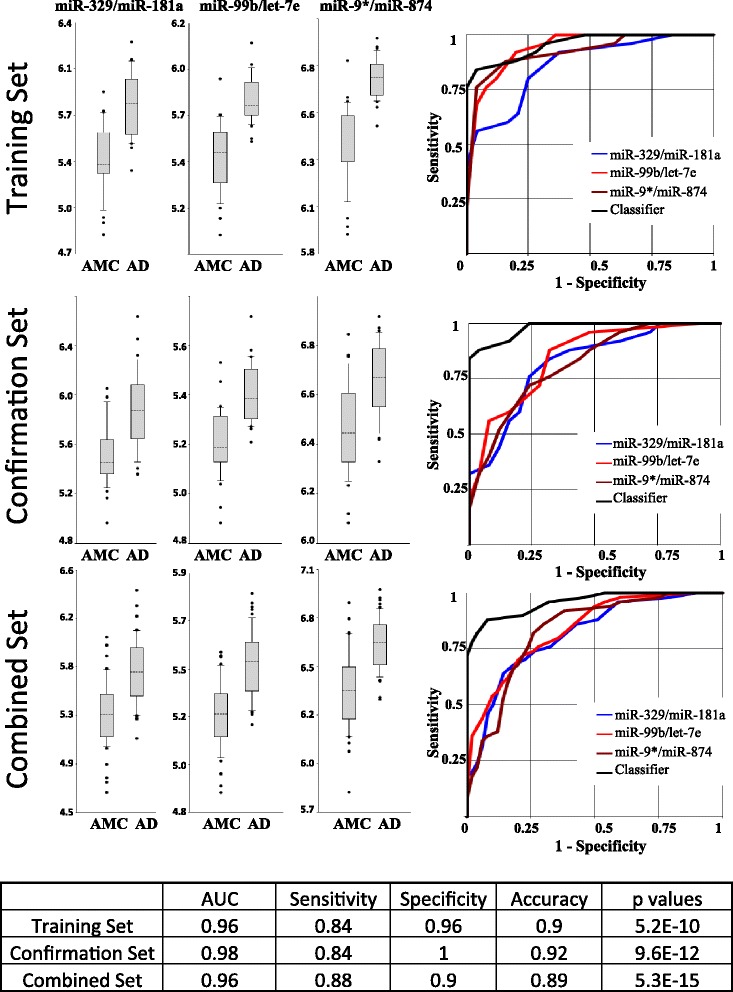
Differentiation of Alzheimer’s disease (AD) from control by select microRNA (miRNA, miR) pairs and their combination. The performance of three miRNA pairs and resulting signature in the training, confirmation, and combined sets is shown. The table at the bottom of the figure indicates performance of the miRNA classifier in the three sets. The area under the ROC curve (AUC) is presented; sensitivity, specificity, and accuracy of each biomarker/normalizer pair were calculated at the cutoff point with the highest accuracy. In the box-and-whisker plots, the ratios are calculated as 2−ΔCt × 100, and the results are presented in log10 scale. The upper and lower limits of the boxes and the lines inside the boxes indicate the 75th and 25th percentiles and the median, respectively. The upper and lower horizontal bars denote the 90th and 10th percentiles, respectively. The points indicate assay values located outside 80% of data. AMC Age- and sex-matched controls
Fig. 2.

Differentiation of neurodegenerative diseases (NDs) from control in combined set by select microRNA (miR) pairs and their combinations. a Frontotemporal dementia (FTD) vs. control. b Parkinson’s disease (PD) vs. control. c Amyotrophic lateral sclerosis (ALS) vs. control. AUC is area under ROC curve
ND differentiation
The same approach was used for establishing miRNA biomarkers capable of differentiating the four NDs (AD, PD, FTD, and ALS) from each other. Figure 3 and Additional file 6 present miRNA classifiers effectively distinguishing two NDs in the training and confirmation studies as well as in the combined dataset. The overall accuracy in the range of 0.75–0.93 was obtained. The lowest accuracy (0.77 with most effective miRNA biomarkers) was observed for differentiation of AD and FTD, for which partial anatomic overlap between the affected brain regions could be larger than between other NDs. The miRNA pairs most effectively differentiating disease groups from each other and from control are listed in Additional file 7.
Fig. 3.

Differentiation of neurodegenerative diseases (NDs) in combined set by select microRNA (miR) pairs and their combinations. a Alzheimer’s disease (AD) vs. frontotemporal dementia (FTD). b Alzheimer’s disease (AD) vs. Parkinson’s disease (PD). c AD vs. amyotrophic lateral sclerosis (ALS). d FTD vs. PD. e FTD vs. ALS. f PD vs. ALS
ND subgroup differentiation
Table 1 summarizes comorbidities and subgroups of NDs included in the study. Although the sample size of individual subgroups was insufficient for obtaining statistically significant results, promising miRNA biomarker candidates capable of distinguishing FTD syndromes were identified. Data presented in Additional file 8A for differentiation between bvFTD, PPA, and PSP with accuracy in the range of 0.70–0.87 warrant further studies of larger patient cohorts. Additional file 8B and C present data on differentiation of PD with dementia from other patients with PD and on differentiation of ALS-FTD from other patients with ALS, respectively.
miRNA biomarker sex-dependent effects
In our other studies focused on distinguishing very mild/mild cognitive impairment from control, we observed sex-specific differences in plasma concentrations of some brain-enriched miRNAs in patients and controls (unpublished data). Thus, in this study, we performed the additional stratified analysis for the four pathologies (AD, FTD, PD, and ALS) and control in the combined dataset as follows:
All (male and female) participants in each disease group vs. all control participants
Male participants in each disease group vs. male control participants
Female participants vs. female control participants
Table 2, section A, shows the performance of effective miRNA classifiers for differentiating each ND from control in all (male and female), male, and female groups. There are no significant differences in AUC and accuracy between these groups, which indicates that these miRNAs behave similarly in controls and male and female patients. However, other tested miRNAs demonstrated sex-associated effects, and miRNA pairs that included these miRNAs more effectively differentiated male or female participants from sex-matched controls. Table 2, section B, presents the most effective miRNA classifiers with additional miRNA biomarker candidates for differentiating AD, FTD, PD, and ALS vs. control separately in males and females. In this case, both AUC and accuracy were significantly higher than effective miRNA classifiers for all (male plus female) participants. In addition, we compared male and female subgroups in each disease group. The data presented in Additional file 9 demonstrate that certain miRNA pairs are capable of differentiating sex-specific subgroups from each other in every disease group with 66–75% accuracy. These data demonstrate sex-specific patterns in levels of circulating brain-enriched miRNAs in both patient and control samples and suggest that sex-specific assays can potentially yield higher accuracy in diagnosing NDs.
Table 2.
Sex-dependent effect in differentiation of neurodegenerative disease s from control by microRNA classifiers
| AD-CNTR | FTD-CNTR | PD-CNTR | ALS-CNTR | ||
|---|---|---|---|---|---|
| A. miRNA classifiers most effectively differentiating NDs from control in all (male + female) participants | |||||
| miR-99b/miR-181a and miR-9*/miR-874 and miR-7/miR-16 | miR-335/let-7e and miR-99b/let-7e and miR-9*/miR-181a | miR-9*/miR-129-3p and miR-99b/miR-874 and miR-9*/miR-411 | miR-206/miR-31 and miR-206/miR-125b and miR-99/miR-338-3p | ||
| Male + female | AUC | 0.97 | 0.94 | 0.96 | 0.95 |
| Accuracy | 0.91 | 0.91 | 0.86 | 0.83 | |
| Male | AUC | 0.98 | 0.98 | 0.96 | 0.94 |
| Accuracy | 0.92 | 0.94 | 0.88 | 0.9 | |
| Female | AUC | 0.97 | 0.91 | 0.96 | 0.95 |
| Accuracy | 0.9 | 0.88 | 0.85 | 0.83 | |
| B. miRNA classifiers most effectively differentiating NDs from control in male and female groups | |||||
| Male | miR-99b/miR-181a and miR-125/miR-874 and miR-9*/miR-29a | miR-335/let-7e and miR-99b/let-7e and miR-9*/miR-181a | miR-9*/miR-129-3p and miR-99b/miR-146a and miR-9*/miR-204 | miR-206/miR-155 and miR-9*/miR-129-3p and miR-335/miR-338-3p | |
| AUC | 0.99 | 0.98 | 0.98 | 0.99 | |
| Accuracy | 0.94 | 0.94 | 0.92 | 0.92 | |
| Female | miR-99b/miR-181a and miR-9*/miR-874 and miR-7/miR-451 | miR-491/let-7e and miR-107/miR-9 and miR-28/miR-181a | miR-9*/miR-29a and miR-99b/miR-874 and miR-491/let-7e | miR-206/miR-7 ph2008and miR-9*/miR-125b and miR-491/miR-204 | |
| AUC | 0.99 | 0.98 | 0.99 | 0.99 | |
| Accuracy | 0.96 | 0.92 | 0.95 | 0.98 | |
Abbreviations: AD Alzheimer’s disease, FTD Frontotemporal dementia, miR MicroRNA, ND Neurodegenerative disease, PD Parkinson’s disease, CNTR Control
Discussion
Previously, we proposed a novel approach to the development of biomarkers in the area of neurodegenerative, neurodevelopmental, and neurological diseases based on analysis of brain-enriched miRNAs circulating in plasma, and we demonstrated the viability of the concept for early AD detection with the discovery of two miRNA families capable of detecting MCI and pre-MCI with high accuracy [48, 49]. In the present study, we used this approach for the detection and differentiation of four NDs: AD, PD, FTD, and ALS.
The most common approaches of searching for circulating miRNA biomarkers to detect a specific pathology are based on the analysis of as many plasma/serum miRNAs as technically feasible using miRNA arrays or next-generation sequencing, followed by qRT-PCR of the identified candidates, and normalization of miRNA(s) whose concentration significantly changes in a pathology sample per minimally variable miRNA or average of all miRNAs tested. There are several disadvantages to these approaches: (1) their sensitivity is significantly lower than that of qRT-PCR, and thus many brain-enriched miRNAs are not reliably detected in plasma, which precludes their further analysis; (2) on one hand, the variability of these methods is high, and many potential candidate miRNA biomarkers are not confirmed by qRT-PCR, and on the other hand, some promising biomarkers are not selected as candidates; and (3) many potential normalizer miRNAs are not uniformly expressed in various pathologies. The latter consideration is especially important in NDs in elderly people because there is a high chance of comorbidities as well as use of various medications. For example, plasma concentration of miR-16, which is widely used as a normalizer in other indications, is changed in patients with AD ([89, 90] and our unpublished data). Other considerations are potential changes of brain blood supply or blood-brain barrier permeability, which would affect concentrations of brain-enriched miRNAs in plasma. Thus, in our earlier studies, we developed a different approach based on miRNA pairs, consisting of one miRNA enriched in synapses of a brain region affected by the disease and another miRNA enriched in a different brain region or cell type, such as glial cells. In effective miRNA pairs, miRNAs are frequently highly correlated [49], decreasing intersubject variability. By combining two or three effective miRNA pairs into a single miRNA classifier, we achieve greater accuracy. Other groups have also identified miRNA pairs as effective biomarkers in the context of cancer diagnosis and prognosis [91–93].
The following factors, which may complicate the search for miRNA biomarkers, should be considered: (1) data on miRNA enrichment in the brain and the different brain regions is still limited, and some brain-enriched miRNAs can be expressed, although at different levels, in several brain regions; (2) ND progression can substantially change both underlying processes, such as synapse dysfunction and destruction in early disease stages and neuronal death in late disease stages, and brain regions involved in a disease due to the expansion of pathologic processes to new brain regions; (3) NDs exhibit heterogeneous clinical symptoms and different brain pathologies (in this respect, analysis of larger cohorts of pathologically homogeneous ND groups would be valuable); and (4) development of neurodegeneration can result in changes in other tissues and organ systems, such as the muscle fibers in ALS.
The data obtained in the present study support our minimally invasive approach to the detection of NDs based on the analysis of circulating brain-enriched miRNA in plasma. Individual miRNA pairs tested in this study classified diseases with accuracies > 0.80, and combinations of several miRNA pairs demonstrated accuracies of up to 0.90. Some miRNA pairs proved to be effective in detecting more than one ND, such as AD and FTD, which partially affect overlapping brain regions. Effective classifiers for ALS detection in addition to brain-enriched miRNAs included muscle-enriched miR-206, which is also highly expressed in cerebellum [72, 94].
As expected, miRNA classifiers for AD dementia were different from the optimal miRNA classifiers for MCI and pre-MCI [48, 49]. MCI is a heterogeneous syndrome characteristic of many NDs. During the progression from MCI to the dementia stage of AD, the ratio of miRNA levels in bodily fluids can change as a result of several factors. Hippocampal synapse and neurite degeneration during early stages of the disease leads to reduced numbers of synapses and neurites in later stages of AD, resulting in diminished amounts of miRNAs being excreted from synapses and neurites of the hippocampus; in addition, because of the increased neuronal cell death that occurs during the later stages of AD, the concentration of neuronal cell body miRNAs in bodily fluids increases. As the disease progresses, new brain regions become involved in the pathology, compromising the reliability of the normalizer miRNAs. This results in numerator miRNAs of effective biomarker pairs for early AD (e.g., miR-128a and miR-874) becoming denominators in effective pairs for later disease stages. Concurrently, because the pathology expands to new brain regions (e.g., cortex and midbrain), synaptic miRNAs from those regions, such as miR-107 and miR-9*, become good biomarker numerators (Additional file 4A). These events can be of informative value for disease monitoring. For example, changes in relative concentrations of miRNAs enriched in different brain regions or different cell types (e.g., neurons and glial cells) may be used as an indicator of disease progression.
Our analysis further revealed miRNA classifiers differentiating AD from FTD with accuracy > 0.75 and all other NDs from each other with accuracy in the range of 0.80–0.90, a highly promising outcome for such heterogeneous diseases, which are in many cases accompanied by other related comorbidities.
Recent data indicate that brain aging and AD development are sex-dependent phenomena [95–98]. Analysis of the data presented here reveals sex dependence of certain brain-enriched miRNAs, which distinguished NDs from control in sex-specific subsets with significantly better accuracy than in the total (female plus male) sets (Table 2, section B). The effect is particularly notable for female participants, such as that accuracy is 0.98 for female and 0.83 for all patients with ALS. More detailed, larger studies are needed to further substantiate this observation and to investigate potential physiological mechanisms underlying this phenomenon. In particular, it has to be determined whether these sex-dependent differences are due to differential expression, secretion, or excretion of some brain-enriched miRNAs, intrinsic sex-dependent differences associated with NDs, or both.
It is worth noting that the approach of the present study was based on analysis of brain-enriched miRNAs in the total pool of miRNAs extracted from plasma. Recently, exosomes, and in particular neuronally derived exosomes, have been studied as potential biomarkers of NDs [99–101]. The promise of exosomes as biomarkers of pathology is due largely to their protein content and membrane composition because there is a high degree of variability in miRNA distribution across exosome populations, and contradictory data exist on the fraction of miRNAs circulating in exosomes as compared with other forms of cell-free miRNAs in the bloodstream [47, 102, 103].
Limitations
A limitation of this study is that all participants were recruited at a single clinical site. Larger, multicenter studies are needed to further evaluate the utility of the approach described herein. A further limitation of the study is that the blood samples were collected from symptomatic patients only. Longitudinal studies are needed to assess the prognostic value of the biomarker candidates in presymptomatic patients.
Conclusions
We report promising data on differentiation of four NDs from control and from each other by brain-enriched miRNA classifiers detectable in plasma. This work builds on our earlier studies on brain-enriched miRNAs as biomarkers of MCI [48, 49]. miRNAs hold strong potential as effective and patient-friendly biomarkers, and several miRNA-based assays are currently used in oncology clinical practice as part of “rule-in/rule-out” diagnostic panels. Future work will address the utility and scope of application of circulating brain-enriched miRNAs as biomarkers of NDs.
Additional files
Age and sex distribution in AD, FTD, PD, and ALS groups and control. (PDF 171 kb)
qRT-PCR controls and median data. (XLSX 17 kb)
miRNAs analyzed in the study. (PDF 57 kb)
miRNA pairs and classifiers differentiating NDs from control in training, confirmation, and combined sets. (PDF 225 kb)
Comparison of the best accuracy and AUC obtained for each disease group vs. control using individual miRNAs and miRNA pairs. (XLSX 11 kb)
miRNA pairs and classifiers differentiating NDs from each other in training, confirmation, and combined sets. (PDF 291 kb)
miRNA pairs most effectively differentiating different groups. (XLSX 18 kb)
Differentiation of ND subgroups from other research participants with the same ND. (PDF 89 kb)
Differentiation of male (M) and female (F) participants within each disease group and control by miRNA pairs. (XLSX 11 kb)
Acknowledgements
We thank members of the Center for Neurodegenerative Disease Research at the University of Pennsylvania who contributed to this work, as well as the many patients studied and their families, for making the reported research possible. We also thank Drs. Michael Kiefer and Gaspar Taroncher-Oldenburg for their help with editing the manuscript.
Funding
This work was funded by the National Institute on Aging, Small Business Innovation Research Program (grant number 2R44AG044860-02 awarded to DiamiR). Support for the work at the University of Pennsylvania was provided by grants from the National Institute on Aging and the National Institute of Neurological Disorders and Stroke (grant numbers P30-AG10124, P01-AG17586, and P50-NS053488). The funding sources had no role in the design and conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
Availability of data and materials
All raw data (C t values in all samples for all miRNAs tested) are available from the authors upon request.
Abbreviations
- Aβ42
Amyloid-β 42
- AD
Alzheimer’s disease
- ALS
Amyotrophic lateral sclerosis
- AMC
Age- and sex-matched controls
- bvFTD
Behavioral variant frontotemporal dementia
- CNTR
Control
- CSF
Cerebrospinal fluid
- Ct
Cycle threshold
- FTD
Frontotemporal dementia
- MCI
Mild cognitive impairment
- miRNA/miR
MicroRNA
- MMSE
Mini Mental State Examination
- mRNA
Messenger RNA
- N/A
Not available
- ND
Neurodegenerative disease
- PD
Parkinson’s disease
- PPA
Primary progressive aphasia
- PPA/Lgpen
Primary progressive aphasia, logopenic variant
- PPA/PNFA
Primary progressive aphasia/progressive nonfluent aphasia
- PPA/SD
Primary progressive aphasia/semantic dementia
- PSP
Progressive supranuclear palsy, p-tau, Phosphorylated tau
- t-tau
Total tau
Authors’ contributions
KSS and SRU had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. JBT, DI, MG, DW, HIH, ACP, DAW, LFM, LBE, and JQT acquired data. KSS, VGT, JBT, JQT, and SRU analyzed and interpreted data. KSS, JBT, JQT, and SRU drafted the manuscript. All authors critically revised the manuscript for important intellectual content. VGT and SRU performed the statistical analysis. SRU and JQT obtained funding. KSS, SRU, JBT, and JQT supervised the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Patient recruitment and clinical data and blood collection were performed at the Alzheimer’s Disease Center, the Parkinson’s Disease and Movement Disorders Center, the Frontotemporal Degeneration Center, and the Amyotrophic Lateral Sclerosis Clinic according to protocols approved by the institutional review board at the University of Pennsylvania. Written informed consent was obtained from all study participants and/or their authorized representatives.
Consent for publication
Not applicable.
Competing interests
KSS, VGT, and SRU are employees and shareholders of DiamiR. JBT, DI, MG, DW, HAH, ACP, DAW, LFM, LBE, and JQT declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s13195-017-0316-0) contains supplementary material, which is available to authorized users.
References
- 1.Heron M. Deaths: Leading Causes for 2014. Natl Vital Stat Rep. 2016;65(5):1–96. [PubMed] [Google Scholar]
- 2.Ahmed RM, Paterson RW, Warren JD, Zetterberg H, O’Brien JT, Fox NC, et al. Biomarkers in dementia: clinical utility and new directions. J Neurol Neurosurg Psychiatry. 2014;85(12):1426–34. doi: 10.1136/jnnp-2014-307662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Cedarbaum J, et al. 2014 Update of the Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2015;11(6):e1–120. doi: 10.1016/j.jalz.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnett JH, Lewis L, Blackwell AD, Taylor M. Early intervention in Alzheimer’s disease: a health economic study of the effects of diagnostic timing. BMC Neurol. 2014;14:101. doi: 10.1186/1471-2377-14-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen-Plotkin AS. Unbiased approaches to biomarker discovery in neurodegenerative diseases. Neuron. 2014;84(3):594–607. doi: 10.1016/j.neuron.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agrawal M, Biswas A. Molecular diagnostics of neurodegenerative disorders. Front Mol Biosci. 2015;2:54. doi: 10.3389/fmolb.2015.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kovacs GG. Molecular pathological classification of neurodegenerative diseases: turning towards precision medicine. Int J Mol Sci. 2016;17(2):189–221. doi: 10.3390/ijms17020189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Roeck EE, Engelborghs S, Dierckx E. Next generation brain health depends on early Alzheimer disease diagnosis: from a timely diagnosis to future population screening. J Am Med Dir Assoc. 2016;17(5):452–3. doi: 10.1016/j.jamda.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Vos SJ, Verhey F, Frölich L, Kornhuber J, Wiltfang J, Maier W, et al. Prevalence and prognosis of Alzheimer’s disease at the mild cognitive impairment stage. Brain. 2015;138(Pt 5):1327–38. doi: 10.1093/brain/awv029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snyder HM, Carrillo MC, Grodstein F, Henriksen K, Jeromin A, Lovestone S, et al. Developing novel blood-based biomarkers for Alzheimer’s disease. Alzheimers Dement. 2014;10(1):109–14. doi: 10.1016/j.jalz.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Bryant SE, Mielke MM, Rissman RA, Lista S, Vanderstichele H, Zetterberg H, et al. Blood-based biomarkers in Alzheimer disease: current state of the science and a novel collaborative paradigm for advancing from discovery to clinic. Alzheimers Dement. 2017;13(1):45–58. doi: 10.1016/j.jalz.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salta E, De Strooper B. Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol. 2012;11(2):189–200. doi: 10.1016/S1474-4422(11)70286-1. [DOI] [PubMed] [Google Scholar]
- 13.Dorval V, Nelson PT, Hébert SS. Circulating microRNAs in Alzheimer’s disease: the search for novel biomarkers. Front Mol Neurosci. 2013;6:24. doi: 10.3389/fnmol.2013.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheinerman KS, Umansky SR. Circulating cell-free microRNA as biomarkers for screening, diagnosis and monitoring of neurodegenerative diseases and other neurologic pathologies. Front Cell Neurosci. 2013;7:150. doi: 10.3389/fncel.2013.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar P, Dezso Z, MacKenzie C, Oestreicher J, Agoulnik S, Byrne M, et al. Circulating miRNA biomarkers for Alzheimer’s disease. PLoS One. 2013;8(7):e69807. doi: 10.1371/journal.pone.0069807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhatnagar S, Chertkow H, Schipper HM, Yuan Z, Shetty V, Jenkins S, et al. Increased microRNA-34c abundance in Alzheimer’s disease circulating blood plasma. Front Mol Neurosci. 2014;7:2. doi: 10.3389/fnmol.2014.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi I, Hama Y, Matsushima M, Hirotani M, Kano T, Hohzen H, et al. Identification of plasma microRNAs as a biomarker of sporadic amyotrophic lateral sclerosis. Mol Brain. 2015;8(1):67. doi: 10.1186/s13041-015-0161-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mushtaq G, Greig NH, Anwar F, Zamzami MA, Choudhry H, Shaik MM, et al. miRNAs as circulating biomarkers for Alzheimer’s disease and Parkinson’s disease. Med Chem. 2016;12(3):217–25. doi: 10.2174/1573406411666151030112140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon H, Flores LF, Kim J. MicroRNAs in brain cholesterol metabolism and their implications for Alzheimer’s disease. Biochim Biophys Acta. 2016;1861(12 Pt B):2139–47. doi: 10.1016/j.bbalip.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu HZ, Ong KL, Seeher K, Armstrong NJ, Thalamuthu A, Brodaty H, et al. Circulating microRNAs as biomarkers of Alzheimer’s disease: a systematic review. J Alzheimers Dis. 2016;49:755–66. doi: 10.3233/JAD-150619. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Yang R, Hu BL, Lu P, Zhou LL, He ZY, et al. Reduced circulating levels of miR-433 and miR-133b are potential biomarkers for Parkinson’s disease. Front Cell Neurosci. 2017;11:170. doi: 10.3389/fncel.2017.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lusardi TA, Phillips JI, Wiedrick JT, Harrington CA, Lind B, Lapidus JA, et al. MicroRNAs in human cerebrospinal fluid as biomarkers for Alzheimer’s disease. J Alzheimers Dis. 2017;55:1223–33. doi: 10.3233/JAD-160835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagaraj S, Laskowska-Kaszub K, Dębski KJ, Wojsiat J, Dąbrowski M, Gabryelewicz T, et al. Profile of 6 microRNA in blood plasma distinguish early stage Alzheimer’s disease patients from non-demented subjects. Oncotarget. 2017;8:16122–43. doi: 10.18632/oncotarget.15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hua YJ, Tang ZY, Tu K, Zhu L, Li YX, Xie L, et al. Identification and target prediction of miRNAs specifically expressed in rat neural tissue. BMC Genomics. 2009;10:214. doi: 10.1186/1471-2164-10-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129(7):1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 2008;14(1):35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo Z, Maki M, Ding R, Yang Y, Zhang B, Xiong L. Genome-wide survey of tissue-specific microRNA and transcription factor regulatory networks in 12 tissues. Sci Rep. 2014;4:5150. doi: 10.1038/srep05150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C, et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44(8):3865–77. doi: 10.1093/nar/gkw116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439(7074):283–9. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 32.Kye MJ, Liu T, Levy SF, Xu NL, Groves BB, Bonneau R, et al. Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA. 2007;13(8):1224–34. doi: 10.1261/rna.480407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lugli G, Torvik VI, Larson J, Smalheiser NR. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J Neurochem. 2008;106(2):650–61. doi: 10.1111/j.1471-4159.2008.05413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cougot N, Bhattacharyya SN, Tapia-Arancibia L, Bordonné R, Filipowicz W, Bertrand E, et al. Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J Neurosci. 2008;28(51):13793–804. doi: 10.1523/JNEUROSCI.4155-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10(12):842–9. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- 36.Bicker S, Lackinger M, Weiß K, Schratt G. MicroRNA-132, -134, and -138: a microRNA troika rules in neuronal dendrites. Cell Mol Life Sci. 2014;71(20):3987–4005. doi: 10.1007/s00018-014-1671-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smalheiser NR. The RNA-centred view of the synapse: non-coding RNAs and synaptic plasticity. Philos Trans R Soc Lond B Biol Sci. 2014;369(1652):20130504. doi: 10.1098/rstb.2013.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34(Database issue):D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5(10):e13515. doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiland M, Gao XH, Zhou L, Mi QS. Small RNAs have a large impact: circulating microRNAs as biomarkers for human diseases. RNA Biol. 2012;9(6):850–9. doi: 10.4161/rna.20378. [DOI] [PubMed] [Google Scholar]
- 41.Hoy AM, Buck AH. Extracellular small RNAs: what, where, why? Biochem Soc Trans. 2012;40(4):886–90. doi: 10.1042/BST20120019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burgos K, Malenica I, Metpally R, Courtright A, Rakela B, Beach T, et al. Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS One. 2014;9(5):e94839. doi: 10.1371/journal.pone.0094839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Ieva A, Butz H, Niamah M, Rotondo F, De Rosa S, Sav A, et al. MicroRNAs as biomarkers in pituitary tumors. Neurosurgery. 2014;75(2):181–9. doi: 10.1227/NEU.0000000000000369. [DOI] [PubMed] [Google Scholar]
- 44.Sun XY, Zhang J, Niu W, Guo W, Song HT, Li HY, et al. A preliminary analysis of microRNA as potential clinical biomarker for schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2015;168B(3):170–8. doi: 10.1002/ajmg.b.32292. [DOI] [PubMed] [Google Scholar]
- 45.Palma J, Yaddanapudi SC, Pigati L, Havens MA, Jeong S, Weiner GA, et al. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 2012;40(18):9125–38. doi: 10.1093/nar/gks656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012;40(21):10937–49. doi: 10.1093/nar/gks832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014;111(41):14888–93. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheinerman KS, Tsivinsky VG, Abdullah L, Crawford F, Umansky SR. Plasma microRNA biomarkers for detection of mild cognitive impairment: biomarker validation study. Aging (Albany NY) 2013;5(12):925–38. doi: 10.18632/aging.100624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheinerman KS, Tsivinsky VG, Crawford F, Mullan MJ, Abdullah L, Umansky SR. Plasma microRNA biomarkers for detection of mild cognitive impairment. Aging (Albany NY) 2012;4(9):590–605. doi: 10.18632/aging.100486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKeith IG, Dickson DW, Lowe J, Emre M, O’Brien JT, Feldman H, et al. Diagnosis and management of dementia with Lewy bodies: third report of the DLB Consortium. Neurology. 2005;65(12):1863–72. doi: 10.1212/01.wnl.0000187889.17253.b1. [DOI] [PubMed] [Google Scholar]
- 51.Emre M, Aarsland D, Brown R, Burn DJ, Duyckaerts C, Mizuno Y, et al. Clinical diagnostic criteria for dementia associated with Parkinson’s disease. Mov Disord. 2007;22(12):1689–707. doi: 10.1002/mds.21507. [DOI] [PubMed] [Google Scholar]
- 52.Strong MJ, Grace GM, Freedman M, Lomen-Hoerth C, Woolley S, Goldstein LH, et al. Consensus criteria for the diagnosis of frontotemporal cognitive and behavioural syndromes in amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2009;10(3):131–46. doi: 10.1080/17482960802654364. [DOI] [PubMed] [Google Scholar]
- 53.Mackenzie IR, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, et al. Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: an update. Acta Neuropathol. 2010;119(1):1–4. doi: 10.1007/s00401-009-0612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rascovsky K, Hodges JR, Knopman D, Mendez MF, Kramer JH, Neuhaus J, et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134(Pt 9):2456–77. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gorno-Tempini ML, Hillis AE, Weintraub S, Kertesz A, Mendez M, Cappa SF, et al. Classification of primary progressive aphasia and its variants. Neurology. 2011;76(11):1006–14. doi: 10.1212/WNL.0b013e31821103e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toledo JB, Brettschneider J, Grossman M, Arnold SE, Hu WT, Xie SX, et al. CSF biomarkers cutoffs: the importance of coincident neuropathological diseases. Acta Neuropathol. 2012;124(1):23–35. doi: 10.1007/s00401-012-0983-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arnold SE, Toledo JB, Appleby DH, Xie SX, Wang LS, Baek Y, et al. Comparative survey of the topographical distribution of signature molecular lesions in major neurodegenerative diseases. J Comp Neurol. 2013;521(18):4339–55. doi: 10.1002/cne.23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Irwin DJ, Cairns NJ, Grossman M, McMillan CT, Lee EB, Van Deerlin VM, et al. Frontotemporal lobar degeneration: defining phenotypic diversity through personalized medicine. Acta Neuropathol. 2015;129(4):469–91. doi: 10.1007/s00401-014-1380-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toledo JB, Van Deerlin VM, Lee EB, Suh E, Baek Y, Robinson JL, et al. A platform for discovery: the University of Pennsylvania Integrated Neurodegenerative Disease Biobank. Alzheimers Dement. 2014;10(4):477–84.e1. doi: 10.1016/j.jalz.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kayano M, Higaki S, Satoh JI, Matsumoto K, Matsubara E, Takikawa O, et al. Plasma microRNA biomarker detection for mild cognitive impairment using differential correlation analysis. Biomark Res. 2016;4:22. doi: 10.1186/s40364-016-0076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fiore R, Schratt G. MicroRNAs in synapse development: tiny molecules to remember. Expert Opin Biol Ther. 2007;7(12):1823–31. doi: 10.1517/14712598.7.12.1823. [DOI] [PubMed] [Google Scholar]
- 62.Lau P, de Strooper B. Dysregulated microRNAs in neurodegenerative disorders. Semin Cell Dev Biol. 2010;21(7):768–73. doi: 10.1016/j.semcdb.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 63.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M, et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A. 2004;101(26):9740–4. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jovičić A, Roshan R, Moisoi N, Pradervand S, Moser R, Pillai B, et al. Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. J Neurosci. 2013;33(12):5127–37. doi: 10.1523/JNEUROSCI.0600-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martini P, Sales G, Brugiolo M, Gandaglia A, Naso F, De Pittà C, et al. Tissue-specific expression and regulatory networks of pig microRNAome. PLoS One. 2014;9(4):e89755. doi: 10.1371/journal.pone.0089755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith A, Calley J, Mathur S, Qian HR, Wu H, Farmen M, et al. The rat microRNA body atlas; evaluation of the microRNA content of rat organs through deep sequencing and characterization of pancreas enriched miRNAs as biomarkers of pancreatic toxicity in the rat and dog. BMC Genomics. 2016;17:694. doi: 10.1186/s12864-016-2956-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Penso-Dolfin L, Swofford R, Johnson J, Alföldi J, Lindblad-Toh K, Swarbreck D, et al. An improved microRNA annotation of the canine genome. PLoS One. 2016;11(4):e0153453. doi: 10.1371/journal.pone.0153453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hruska-Plochan M, Li B, Kyburz D, Krützfeld J, Landmesser U, Aguzzi A, et al. New and emerging roles of small RNAs in neurodegeneration, muscle, cardiovascular and inflammatory diseases. Swiss Med Wkly. 2015;145:w14192. doi: 10.4414/smw.2015.14192. [DOI] [PubMed] [Google Scholar]
- 69.Alexander M, O’Connell RM. Noncoding RNAs and chronic inflammation: micro-managing the fire within. Bioessays. 2015;37(9):1005–15. doi: 10.1002/bies.201500054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Su W, Aloi MS, Garden GA. MicroRNAs mediating CNS inflammation: small regulators with powerful potential. Brain Behav Immun. 2016;52:1–8. doi: 10.1016/j.bbi.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lukiw WJ, Alexandrov PN, Zhao Y, Hill JM, Bhattacharjee S. Spreading of Alzheimer’s disease inflammatory signaling through soluble micro-RNA. Neuroreport. 2012;23(10):621–6. doi: 10.1097/WNR.0b013e32835542b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Panwalkar P, Moiyadi A, Goel A, Shetty P, Goel N, Sridhar E, et al. miR-206, a cerebellum enriched miRNA is downregulated in all medulloblastoma subgroups and its overexpression is necessary for growth inhibition of medulloblastoma cells. J Mol Neurosci. 2015;56(3):673–80. doi: 10.1007/s12031-015-0548-z. [DOI] [PubMed] [Google Scholar]
- 73.Amirouche A, Jahnke VE, Lunde JA, Koulmann N, Freyssenet DG, Jasmin BJ. Muscle-specific microRNA-206 targets multiple components in dystrophic skeletal muscle representing beneficial adaptations. Am J Physiol Cell Physiol. 2017;312(3):C209–21. doi: 10.1152/ajpcell.00185.2016. [DOI] [PubMed] [Google Scholar]
- 74.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17(2):215–20. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 75.Qiu L, Tan EK, Zeng L. microRNAs and Neurodegenerative Diseases. Adv Exp Med Biol. 2015;888:85–105. doi: 10.1007/978-3-319-22671-2_6. [DOI] [PubMed] [Google Scholar]
- 76.Yang L, Xiong Y, Hu XF, Du YH. MicroRNA-323 regulates ischemia/reperfusion injury-induced neuronal cell death by targeting BRI3. Int J Clin Exp Pathol. 2015;8(9):10725–33. [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao Y, Pogue AI, Lukiw WJ. MicroRNA (miRNA) Signaling in the human CNS in sporadic Alzheimer’s disease (AD) - novel and unique pathological features. Int J Mol Sci. 2015;16(12):30105–16. doi: 10.3390/ijms161226223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Salta E, De Strooper B. microRNA-132: a key noncoding RNA operating in the cellular phase of Alzheimer’s disease. FASEB J. 2017;31(2):424–33. doi: 10.1096/fj.201601308. [DOI] [PubMed] [Google Scholar]
- 79.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 80.Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27–41. doi: 10.3233/JAD-2008-14103. [DOI] [PubMed] [Google Scholar]
- 81.Satoh J. MicroRNAs and their therapeutic potential for human diseases: aberrant microRNA expression in Alzheimer’s disease brains. J Pharmacol Sci. 2010;114:269–75. doi: 10.1254/jphs.10R11FM. [DOI] [PubMed] [Google Scholar]
- 82.Tang Y, Liu D, Zhang L, Ingvarsson S, Chen H. Quantitative analysis of miRNA expression in seven human foetal and adult organs. PLoS One. 2011;6:e28730. doi: 10.1371/journal.pone.0028730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lau P, Frigerio CS, De Strooper B. Variance in the identification of microRNAs deregulated in Alzheimer’s disease and possible role of lincRNAs in the pathology: the need of larger datasets. Ageing Res Rev. 2014;17:43–53. doi: 10.1016/j.arr.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 84.Briggs CE, Wang Y, Kong B, Woo TU, Iyer LK, Sonntag KC. Midbrain dopamine neurons in Parkinson’s disease exhibit a dysregulated miRNA and target-gene network. Brain Res. 1618;2015:111–21. doi: 10.1016/j.brainres.2015.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koenig EM, Fisher C, Bernard H, Wolenski FS, Gerrein J, Carsillo M, et al. The beagle dog microRNA tissue atlas: identifying translatable biomarkers of organ toxicity. BMC Genomics. 2016;17:649. doi: 10.1186/s12864-016-2958-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Puthiyedth N, Riveros C, Berretta R, Moscato P. Identification of differentially expressed genes through integrated study of Alzheimer’s disease affected brain regions. PLoS One. 2016;11:e0152342. doi: 10.1371/journal.pone.0152342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kumar S, Vijayan M, Bhatti JS, Reddy PH. MicroRNAs as peripheral biomarkers in aging and age-related diseases. Prog Mol Biol Transl Sci. 2017;146:47–94. doi: 10.1016/bs.pmbts.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 88.Reddy PH, Williams J, Smith F, Bhatti JS, Kumar S, Vijayan M, et al. MicroRNAs, aging, cellular senescence, and Alzheimer’s disease. Prog Mol Biol Transl Sci. 2017;146:127–71. doi: 10.1016/bs.pmbts.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 89.Müller M, Kuiperij HB, Claassen JA, Küsters B, Verbeek MM. MicroRNAs in Alzheimer’s disease: differential expression in hippocampus and cell-free cerebrospinal fluid. Neurobiol Aging. 2014;35:152–8. doi: 10.1016/j.neurobiolaging.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 90.Parsi S, Smith PY, Goupil C, Dorval V, Hébert SS. Preclinical evaluation of miR-15/107 family members as multifactorial drug targets for Alzheimer’s disease. Mol Ther Nucleic Acids. 2015;4:e256. doi: 10.1038/mtna.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, Calabrò E, Croce CM, Pastorino U, Sozzi G. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A. 2011;108:3713–8. doi: 10.1073/pnas.1100048108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lehmann TP, Korski K, Gryczka R, Ibbs M, Thieleman A, Grodecka-Gazdecka S, Jagodziński PP. Relative levels of let-7a, miR-17, miR-27b, miR-125a, miR-125b and miR-206 as potential molecular markers to evaluate grade, receptor status and molecular type in breast cancer. Mol Med Rep. 2015;12:4692–702. doi: 10.3892/mmr.2015.4002. [DOI] [PubMed] [Google Scholar]
- 93.Fortunato O, Verri C, Pastorino U, Sozzi G, Boeri M. MicroRNA profile of lung tumor tissues is associated with a high risk plasma miRNA signature. Microarrays (Basel) 2016;5:18. doi: 10.3390/microarrays5030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Olsen L, Klausen M, Helboe L, Nielsen FC, Werge T. MicroRNAs show mutually exclusive expression patterns in the brain of adult male rats. PLoS One. 2009;4(10):e7225. doi: 10.1371/journal.pone.0007225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ronquillo JG, Baer MR, Lester WT. Sex-specific patterns and differences in dementia and Alzheimer’s disease using informatics approaches. J Women Aging. 2016;28(5):403–11. doi: 10.1080/08952841.2015.1018038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martínez-Pinilla E, Ordóñez C, Del Valle E, Navarro A, Tolivia J. Regional and gender study of neuronal density in brain during aging and in Alzheimer’s disease. Front Aging Neurosci. 2016;8:213. doi: 10.3389/fnagi.2016.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guebel DV, Torres NV. Sexual dimorphism and aging in the human hippocampus: identification, validation, and impact of differentially expressed genes by factorial microarray and network analysis. Front Aging Neurosci. 2016;8:229. doi: 10.3389/fnagi.2016.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Snyder HM, Asthana S, Bain L, Brinton R, Craft S, Dubal DB, et al. Sex biology contributions to vulnerability to Alzheimer’s disease: a think tank convened by the Women’s Alzheimer’s Research Initiative. Alzheimers Dement. 2016;12(11):1186–96. doi: 10.1016/j.jalz.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Winston CN, Goetzl EJ, Akers JC, Carter BS, Rockenstein EM, Galasko D, et al. Prediction of conversion from mild cognitive impairment to dementia with neuronally derived blood exosome protein profile. Alzheimers Dement (Amst) 2016;3:63–72. doi: 10.1016/j.dadm.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chivet M, Hemming F, Pernet-Gallay K, Fraboulet S, Sadoul R. Emerging role of neuronal exosomes in the central nervous system. Front Physiol. 2012;3:145. doi: 10.3389/fphys.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fiandaca MS, Kapogiannis D, Mapstone M, Boxer A, Eitan E, Schwartz JB, et al. Identification of preclinical Alzheimer’s disease by a profile of pathogenic proteins in neurally derived blood exosomes: a case-control study. Alzheimers Dement. 2015;11(6):600–7.e1. doi: 10.1016/j.jalz.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39(16):7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhao L, Yu J, Wang J, Li H, Che J, Cao B. Isolation and Identification of miRNAs in exosomes derived from serum of colon cancer patients. J Cancer. 2017;8(7):1145–52. doi: 10.7150/jca.18026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Age and sex distribution in AD, FTD, PD, and ALS groups and control. (PDF 171 kb)
qRT-PCR controls and median data. (XLSX 17 kb)
miRNAs analyzed in the study. (PDF 57 kb)
miRNA pairs and classifiers differentiating NDs from control in training, confirmation, and combined sets. (PDF 225 kb)
Comparison of the best accuracy and AUC obtained for each disease group vs. control using individual miRNAs and miRNA pairs. (XLSX 11 kb)
miRNA pairs and classifiers differentiating NDs from each other in training, confirmation, and combined sets. (PDF 291 kb)
miRNA pairs most effectively differentiating different groups. (XLSX 18 kb)
Differentiation of ND subgroups from other research participants with the same ND. (PDF 89 kb)
Differentiation of male (M) and female (F) participants within each disease group and control by miRNA pairs. (XLSX 11 kb)
Data Availability Statement
All raw data (C t values in all samples for all miRNAs tested) are available from the authors upon request.


