Abstract
For deep-sea hydrothermal vent crabs, recent investigations have revealed some epibiotic bacteria, but no study has described the bacterial community associated with the gill and intestine. In this study, the microbiota attached to the gill and intestine of the hydrothermal vent crab Austinograea sp. and two shallow-water crab species (Eriocheir sinensis and Portunus trituberculatus) were compared by high-throughput sequencing of 16S rDNA genes. The highest and lowest diversity in bacterial communities were observed in the gill and intestine of Austinograea sp., respectively. Non-metric multidimensional scaling (NMDS) analysis indicated that Austinograea sp. harbored a distinct microbial community. Operational taxonomic units (OTUs) for phylum Fusobacteria, class Epsilonproteobacteria, and genera Leucothrix, Polaribacter, Fusibacter, etc. were dominant in Austinograea sp. Of these, Leucothrix, Sulfurospirillum, and Arcobacter may be involved in oxidizing reduced sulfur compounds and sulfur metabolism; Marinomonas, Polaribacter adapted to the low temperature, and Fusibacter and Psychrilyobacter may survive well under hypoxic conditions. Bacteria commonly present in seawater were dominant in the gill, whereas anaerobic bacteria showed strikingly high abundance in the intestine. Interestingly, Firmicutes and Epsilonproteobacteria may complement each other in Austinograea sp., forming an internal environment. The diversified microbial community of Austinograea sp. reveals adaptation to the hydrothermal vent environment.
Introduction
The deep-sea hydrothermal vent environment is one of the most variable ecosystems on Earth, and the fauna inhabiting this environment need special adaptations to tolerate the harsh conditions [1]. In such ecosystems, life is based on chemosynthetic primary production and widespread microbial-invertebrate associations [2]. The associations between hydrothermal vent invertebrates and chemosynthetic symbionts are diverse, including loose epibiosis and obligate endosymbiosis [2]. Decapod crustaceans usually exhibit a diverse range of bacterial associates such as chemoautotrophs and methanotrophs [3,4]. A number of studies suggest that some chemoautotrophic epibionts play a nutritional role in the hydrothermal shrimp, Rimicaris exoculata [5–8], and different microbial communities are hypothesized to participate in different inorganic metabolic pathways [9]. Epsilonproteobacteria also supply nutrition to the host by metabolizing sulfur compounds in some deep-sea hydrothermal vent invertebrates such as shrimps or crabs [3,4,9–16], and tubeworms [17].
In addition, invertebrate-associated bacteria in hydrothermal vents not only have host specificity, but also organ specificity within the host’s gut or gill chamber [10,11,18–23], consistent with their nutrient sources [4,24] and challenging hydrothermal vent environments [7,10]. Many studies have been conducted to explore the gill-associated microbiota in hydrothermal vent invertebrates. Hydrothermal vent mytilid mussels with vesicomyid clams and some provannid gastropods harbor symbiotic bacteria in their gills [18–20], and the hydrothermal shrimp, R. exoculata, also harbors epibiotic bacteria on the inner side of its enlarged gill chambers [21–23]. Besides the epibionts on the gill chamber, some microflora is gut specific and may be implicated in nutrition of hydrothermal vent shrimps [8,10,11,24]. Earlier observations have also elucidated the epibionts associated with the setae of deep-sea hydrothermal vent decapod [3,13–16,25,26]. In the crab Xenograpsus testudinatus inhabiting in shallow-water hydrothermal vents, the dominant bacteria also have both host and potential organ specificities, consistent with a potential trophic transfer between host and bacteria [27]. However, little is known about the microbial communities harbored in the gill and intestine of deep-sea hydrothermal vent crabs.
Austinograea sp., which belongs to the order Brachyura, family Bythograeidae, is the most common and abundant species and the top predator in hydrothermal vent [28]. All species have whitish, smooth and rounded carapaces, vestigial eye stalks and resemble each other in gross morphology [29]. Apart from the morphological description [30–33] and mitochondrial genomes [28,34], reports of these species are very limited and it is not clear how they adapt to the deep-sea hydrothermal vent environment. Transcriptomes of four tissues (eyestalk, gill, hepatopancreas and muscle) of Austinograea alayseae are performed and reveal the molecular basis of adaptive evolution [35], while no data are collected for the bacterial associations and their roles in crab adaptive evolution.
High-throughput sequencing (HTS) of 16S rDNA allows exploration of microbial diversity at an unprecedented scale [36]. Here, we described and characterized, for the first time, the composition of the microbial communities associated with the gill and intestine of the deep-sea crab Austinograea sp. by HTS of 16S rDNA genes. The Chinese mitten crab Eriocheir sinensis H. Milne Edwards, 1853 (Crustacea: Decapoda: Brachyura) is a fresh water crab species, however spawns and mates in saline water. The swimming crab Portunus trituberculatus Miers, 1876- (Crustacea: Decapoda: Brachyura), is a sea-dwelling crab inhabiting in the seafloor with sand or pebbles. These two shallow-water crab species, representing contrasting geochemical settings and depths to a certain extent, were used as comparisons to illustrate the diversity in the microbial communities harbored in the gill and intestine of Austinograea sp. The current study may present a good example for the interactions between microorganisms and invertebrate hosts in deep-sea hydrothermal vents.
Materials and methods
Ethics statement
The experiments were conducted in strict accordance with the guidelines of the Institutional Animal Care and Use Committee (IACUC) of the Chinese Academy of Sciences (No. 2011–2). This study was specifically approved by the Committee on the Ethics of Animal Experiments of the Institute of Oceanology at the Chinese Academy of Sciences. All efforts were made to minimize the suffering of the crabs.
Sample collection
Healthy Chinese mitten crabs (E. sinensis) and swimming crabs (P. trituberculatus) were collected from Liaohe River of Liaoning province and Qingdao coast of Shandong province, respectively. Deep-sea hydrothermal vent crabs were collected between two dive sites (#30/Desmos and #32/Pacmas, Table 1) in the South China Sea by the remotely operated vehicle (ROV) Faxian, which was deployed using the RV KEXUE. Desmos and Pacmas are the two fields in the Manus Basin, where low pH and high concentrations of H2S and SO42- were detected [37]. Other environment data and microbial communities in the sediments of these two sites have been published by Wang et al. [38]. Fresh intestines and gills of E. sinensis (72 ± 0.2 g) and P. trituberculatus (135 ± 0.2 g) were collected from four live individuals for genomic DNA extraction. Samples from Austinograea sp. were kept at -80°C before used for genetic analysis. Specifically, half of the gills (the other half was used for the transcriptome sequencing) and the separated entire intestine in each crab were dissected for further analyses and also four individuals (5.5 ± 0.1 g) were treated (S1 Fig).
Table 1. Details of the dive sites where deep-sea crabs were collected.
| Location | Sampling date | Coordinates | Depth | Salinity | Temperature |
|---|---|---|---|---|---|
| Desmos | 2015.6.9 | 151°52′50.084″E, 3°42′47.259″S | 1995 m | 35.67‰ | 1.01°C |
| Pacmas | 2015.6.11 | 151°40′09.137″E, 3°43′41.246″S | 1680 m | 35.67‰ | 1.15°C |
Genomic DNA extraction and PCR amplification
Total DNA from the gills and intestines were extracted by the phenol-chloroform method as described by Sambrook and Russell [39]. The concentration of DNA samples were diluted to 1 ng/μL using sterile water after measured by a spectrophotometer and their purity were determined by 1% agarose gel electrophoresis.
The 16S rDNA genes of hypervariable regions 16SV3-V4 were amplified using the following specific primers with the barcode: 341F (CCTAYGGGRBGCASCAG) and 806R (GGACTACNNGGGTATCTAAT). Barcodes added to the specific primer for distinguishing different samples were shown in S1 Table. PCR was performed in 30 μL volumes containing 15 μL of Phusion® High-Fidelity PCR Master Mix (New England Biolabs), 0.2 μmol of forward and reverse primers, and about 10 ng template DNA. Thermal cycling consisted of initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and elongation at 72°C for 30 s, and a final elongation of 5 min at 72°C. PCR products mixed with same volume of 6 × loading buffer (contained SYB green) were examined on 2% agarose gel. Samples with a bright main band between 400–450 bp were chosen for further experiments.
HTS of bacterial 16S rDNA
PCR products mixed in equidensity ratios were purified using the Qiagen Gel Extraction Kit (Qiagen, Germany). Sequencing libraries were generated using TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA) following the manufacturer's recommendations and index codes were added. The samples were then sequenced on an Illumina HiSeq2500 platform.
Statistical and bioinformatics analysis
Paired-end reads, merged using FLASH (V1.2.7, http://ccb.jhu.edu/software/FLASH/) [40], were assigned to samples based on their unique barcode. Quality filtering on the raw tags was performed under specific filtering conditions to obtain high-quality clean tags [41] according to the QIIME (V1.7.0, http://qiime.org/index.html) [42] quality controlled process. The effective tags were obtained by comparison with the reference database (Gold database, http://drive5.com/uchime/uchime_download.html) and removing the chimera sequences.
Sequences analyzed by Uparse software (Uparse v7.0.1001, http://drive5.com/uparse/) [43] with similarity not less than 97%, were assigned to the same Operational Taxonomic Units (OTUs). The representative sequence for each OTU was annotated by the RDP classifier (Version 2.2, http://sourceforge.net/projects/rdp-classifier/) algorithm to obtain taxonomic information [44]. Alpha diversity was applied in analyzing the complexity of species diversity for a sample through 6 indices, including Observed-species, Chao1, Shannon, Simpson, ACE, and Good’s-coverage. All these indices in our analyses were calculated with QIIME (Version 1.7.0) and displayed with R software (Version 2.15.3). The indices of Chao1 and ACE estimator were selected to identify community richness. Shannon and Simpson indices were used to identify community diversity whereas Good’s coverage was calculated to characterize the sequencing depth. The significant levels between all samples were detected by T-test, Wilcox-test and Tukey-test, and P < 0.05 meant significant different between two samples. Non-metric multidimensional scaling (NMDS) based on weighted UniFrac distance was shown to give a visual comparison of the pairwise UniFrac distance among samples. Phylogenetic analysis was conducted to exhibit the distribution of major microbial communities in different groups by using the software MUSCLE (Version 3.8.31, http://www.drive5.com/muscle/) [45]. The linear discriminant analysis (LDA) effect size (LEfSe) method (http://huttenhower.sph.harvard.edu/lefse/) [46] was performed to characterize the specialization of microorganisms from different groups. All group data were calculated by the average of four specimens from each species.
Results
Microbial relative abundance and diversity
In total, 227,785 (111,974 for gills and 115,811 for intestines) effective tags were obtained from Austinograea sp., E. sinensis, and P. trituberculatus by HTS. By the species associated bacterial composition analysis, 1,118 and 1,050 OTUs were collected from the gill and intestine of Austinograea sp., respectively, 1,309 and 967 OTUs were collected from E. sinensis, whereas 1,114 and 1,657 OTUs were obtained from P. trituberculatus, respectively (Table 2). The diversity indices, including the Shannon, Simpson, and community richness- Chao1 and ACE all revealed microbiota variations between the Austinograea sp. and two shallow-water crab species, and even between the gill and intestine from Austinograea sp. In all three crabs, the highest microbial diversity was observed in the gill of the hydrothermal vent crab (5.18 in Shannon index), and the lowest bacterial diversity was found in intestine of Austinograea sp. (3.87 in Shannon index).
Table 2. Similarity-based OTUs and species richness of the bacterial phylotypes in gills and intestines of three species.
| Sample name | Effective tags | OTU | Shannon | Simpson | Chao1 | ACE |
|---|---|---|---|---|---|---|
| AG | 37,199 | 1,118 | 5.18 | 0.90 | 763.53 | 787.64 |
| EG | 38,922 | 1,309 | 4.75 | 0.88 | 933.93 | 983.70 |
| PG | 35,854 | 1,114 | 4.73 | 0.82 | 741.11 | 790.25 |
| AI | 37,231 | 1,050 | 3.87 | 0.80 | 691.11 | 760.20 |
| EI | 36,589 | 967 | 4.48 | 0.87 | 619.89 | 660.45 |
| PI | 41,991 | 1,657 | 4.44 | 0.85 | 1210.51 | 1242.40 |
Abbreviations: for sample names, the first letter represents species- A for Austinograea sp., E for E. sinensis, P for P. trituberculatus; the second letter represents tissues- G for gill, I for intestine.
Composition of microbiota in the gills and intestines from three species of crabs
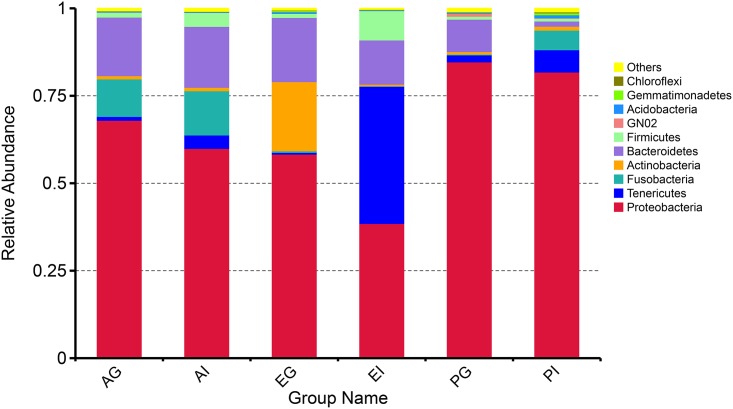
Based on the annotated information of the microbial 16S rDNA data obtained by HTS, 39 different phyla of bacteria were observed in the gills and intestines of three crab species. The top 10 phyla accounting for more than 99% of the microbial communities in each sample were selected to represent the relative abundance of various microbiota in different species (Fig 1). In the three crabs, Proteobacteria was the most predominant phylum in the gills and intestines of all specimens (68.0% in Austinograea sp., 58.4% in E. sinensis, and 84.7% in P. trituberculatus, respectively). In addition, the phyla Bacteroidetes, Tenericutes and Firmicutes were also found to be predominant microbes in the gills and intestines of the three crab species. Interestingly, a higher abundance of phylum Fusobacteria was observed in the gill and intestine of Austinograea sp. than in the other two crabs.
Fig 1. Relative abundance of predominant bacterial communities in the gills and intestines of Austinograea sp. and the other two shallow-water crab species at the phylum level.
A color-coded bar plot shows the average bacterial genus distribution in different samples.
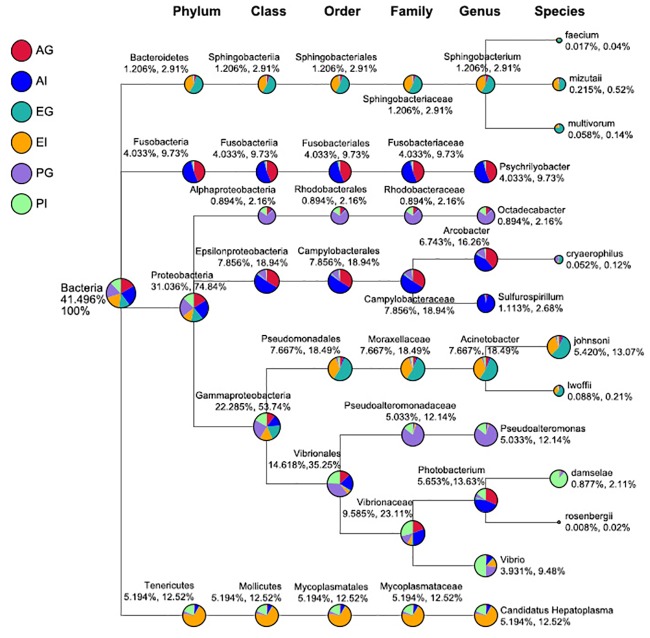
At the genus level, four predominant phyla could be assigned into ten different genera (Fig 2). In Austinograea sp., Psychrilyobacter (Fusobacteria), Arcobacter, and Photobacterium (Proteobacteria) were the dominant genera both in the gill and intestine, and Sulfurospirillum was predominant in the intestine. In E. sinensis, Sphingobacterium (Bacteroidetes) and Acinetobacter (Proteobacteria) were the dominant genera both in the gill and intestine, and Candidatus Hepatoplasma (Tenericutes) was predominant in the intestine. The most abundant sequences in P. trituberculatus were affiliated to Octadecabacter, Pseudoalteromonas, and Vibrio (Proteobacteria).
Fig 2. The taxonomic tree for bacteria in the gills and intestines of the three crab species.
The sector with different colors represents different samples. The size of the sector indicates the relative abundance of a sample in this taxon. The two digits under the taxon indicate the relative abundance percentage in all taxa and the selected taxon, respectively.
Differentiation of the microbiota in the gills and intestines between Austinograea sp. and two shallow-water crabs
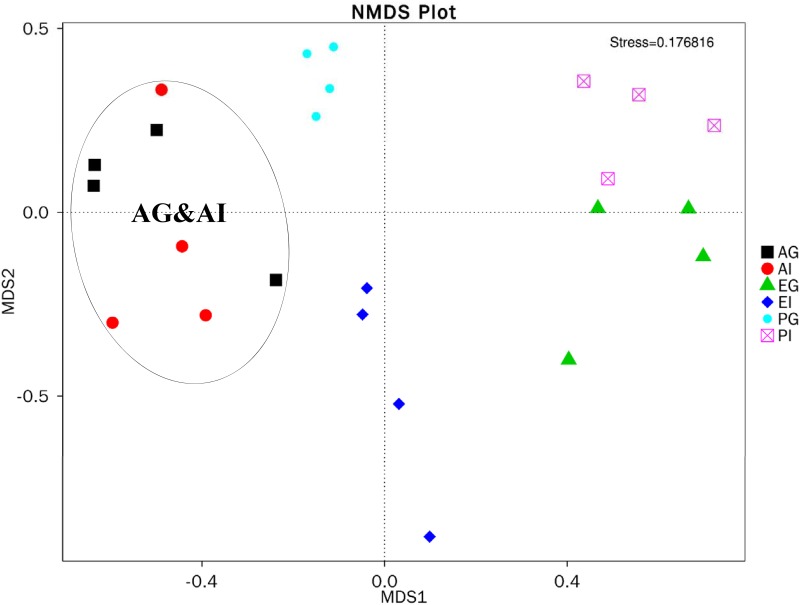
An NMDS plot was generated to compare the relationship of the bacterial communities in the gills and intestines between Austinograea sp. and two shallow-water crabs at the OTU level. Microbial communities in the gill and intestine from Austinograea sp. obviously separated from those in the other two crabs as shown in Fig 3 (P < 0.05). This means that samples from the hydrothermal vent possess bacterial communities different from those in the samples from the shallow water.
Fig 3. NMDS plot showing microbial community differences between the hydrothermal vent crab Austinograea sp. and the other two shallow-water crabs.
Each point represents a sample. The distance between points indicates the degree of difference. The significant levels between different samples are evaluated by Tukey-test. Sample of the same group are shown in the same color. Stress < 0.2 indicates the reliability of the NMDS analysis.
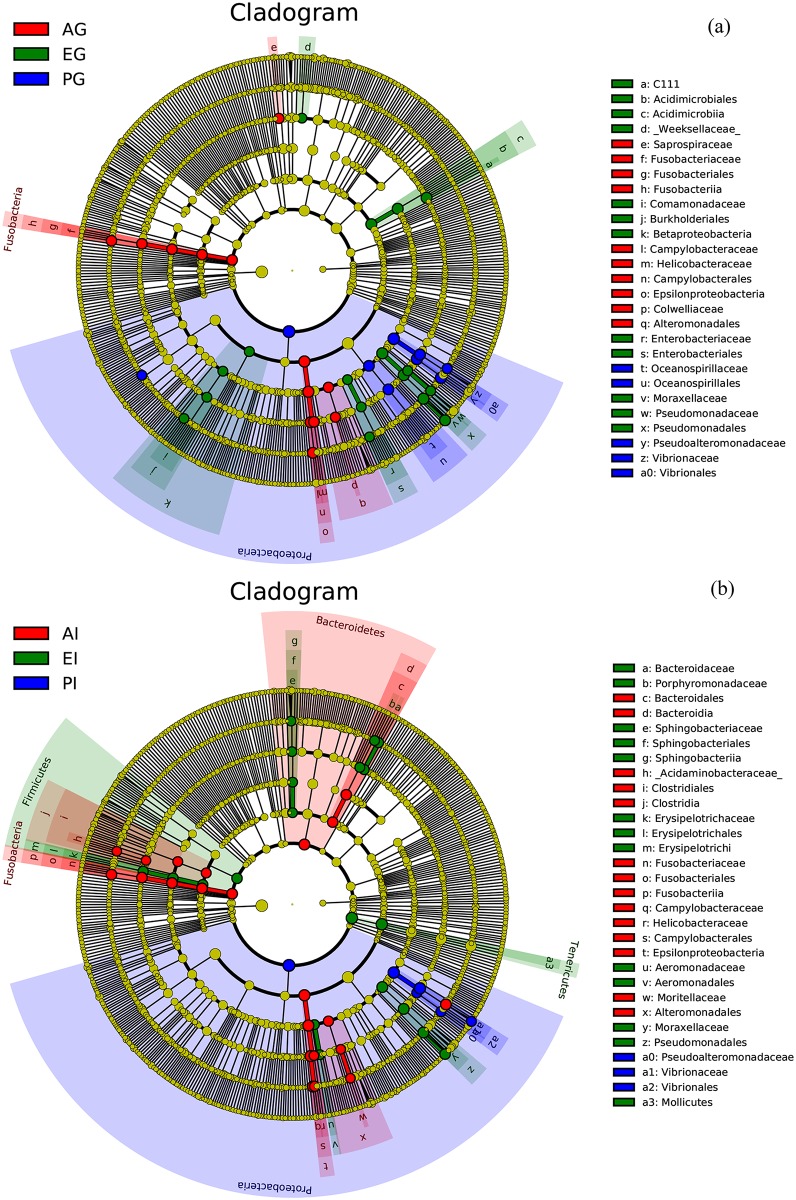
To understand the differences among the bacterial groups in the gill and intestine from the three species of crabs, LEfSe was used to perform the comparisons among three crab species. The cladograms exhibited the structure of the microbiota in the gills and intestines of different species and their predominant bacteria (Fig 4). The greatest differences in taxa between the gills (Fig 4a) and intestines (Fig 4b) from different species were displayed. Thirteen specific microbial taxa at different levels were discovered in the gill of Austinograea sp. (Fig 4a). The first dominant taxon was the phylum Proteobacteria, especially the class Epsilonproteobacteria. The second specific taxon was the phylum Fusobacteria. Twenty-two unique bacterial categories were found in the intestine of the hydrothermal vent crab (Fig 4b). Similar to the gill, Epsilonproteobacteria was also a specific class in the intestine of Austinograea sp. Meanwhile, some other bacterial taxa jointly constituted the unique microbial communities in the intestine of Austinograea sp., including the phyla Bacteroidetes and Fusobacteria. The phylum Firmicutes were also predominant in the intestine of the hydrothermal vent crab.
Fig 4. LEfSe identified the most differentially abundant taxa in the gills (a) and intestines (b) of the three crab species.
Red, green, and blue colors represent the bacterial communities in the gills or intestines of P. trituberculatus, E. sinensis, and Austinograea sp., respectively. The dots on the cladogram represent the specific bacterial taxa whose colors are the same as those of the corresponding groups. The size of dots is proportional to the relative abundance.
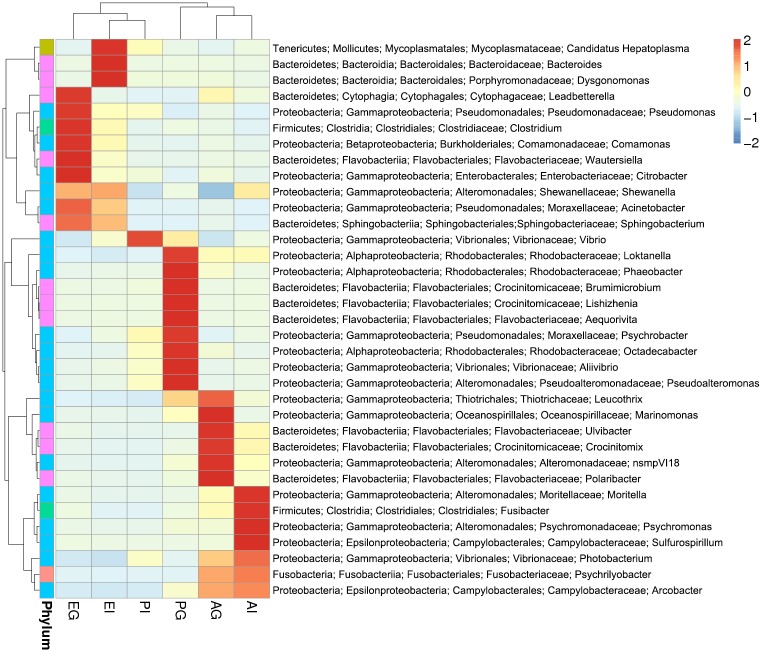
To further identify the specific bacterial taxa associated with the gill and intestine of hydrothermal vent crab clearly, a hierarchically clustered heatmap was selected to get an overview of the identified connections among the studied samples. Based on the bacterial 16S rDNA data and the microbial community abundance, the top 35 genera of bacteria from the gill and intestine of each species were used to construct the phylogenetic heatmap (Fig 5). Compared to the two shallow-water crabs, Austinograea sp. showed a higher abundance of the genera Leucothrix, Marinomonas, Ulvibacter, Crocinitomix, nsmpVl18, and Polaribacter in the gill; Moritella, Fusibacter, Psychromonas, and Sulfurospirillum in the intestine; and Photobacterium, Psychrilyobacter, and Arcobacter in both tissues.
Fig 5. Heatmap of the microbial composition in the gills and intestines of the three crab species at the genus level.
The heatmap indicates the relative abundance of each genus in different samples. The Y-axis clustering exhibits the variables and phylogenetic relationships of each bacterial species in different samples and the X-axis clustering indicates the phylogenetic relationships of these samples. The color intensity in the square grid represents the bacterial relative abundance, which is named as the z-value and generated by the relative abundance of bacterial species in each line after normalization treatment.
Discussion
The deep-sea hydrothermal vents are characterized by low oxygen, oligotrophic, high thermal gradient (2–350°C), and presence of various toxic chemical compounds, such as hydrogen sulfide (H2S), methane (CH4), and heavy metals [47,48]. The symbiotic microbes associated with the hydrothermal vent animals may be different from those in some shallow-water animals to support their hosts adaption to the extreme environment. In this study, we observed the diversity in bacterial communities associated with Austinograea sp. and compared it to that of shallow-water crabs, P. trituberculatus and E. sinensis. This is the first study to investigate the difference in the microbial composition and characteristics between deep-sea and shallow-water crabs.
Phylogenetic diversity and constitutive characteristics of the microbiota in the gill and intestine of Austinograea sp.
The variation in microbial communities in the gills and intestines between Austinograea sp. and two shallow-water crabs might be affected by their different habitat environments. The surroundings of P. trituberculatus and E. sinensis are moderate, while the habitat of Austinograea sp. is one of the most extreme environments on Earth, where bacterial diversity (7.20–8.27 for Shannon) may be much greater than other marine microbial diversity [38,49]. The highest diversity of microbial communities observed in the gill of Austinograea sp. may also be relevant to these challenging conditions [37]. Inconsistent with the gills, the intestine of Austinograea sp. had the lowest diversity among these three crab species. This might be due to the fact that the intestine is relatively independent from the external environment. On the other hand, hydrothermal vents are oligotrophic ecosystems [50]. For food sources apart from the particulate matter falling from above and drifting of food materials from the surrounding regions, Austinograea sp. might be mainly dependent on harbour endosymbiotic chemosynthetic bacteria for nutrition [2]. In addition, the microbial diversity in gill and intestine of Austinograea sp. were higher than that of shallow-water hydrothermal vent crabs [27]. These results indicate that environment and food sources influence the composition of the gill and intestinal microbiota.
Symbionts associated with sulfur metabolism in Austinograea sp.
Almost all vent-endemic animals are considered to have a close relationship with the endo- and/or epi- symbiotic chemoautotrophic microorganisms as the primary producers [51]. Chemoautotrophic Epsilonproteobacteria and Gammaproteobacteria are the predominant primary producers in both free-living and symbiotic microbial communities in global deep-sea hydrothermal fields [52,53]. Epsilonproteobacteria use sulfur compounds as both electron-donors and acceptors, and extend the energetically feasible habitats with versatile energy metabolisms. In this research, Epsilonproteobacteria, possibly involved in the oxidation-reduction of sulfur compounds and carbon fixation [9], were considered as a significant microbiota in the gill and intestine of Austinograea sp. unlike that in the shallow-water crabs. This finding agrees with the result of the comparison of genes participating in sulfur metabolism among the three crabs [35]. In the hydrothermal vent of the Manus Basin, Epsilonproteobacteria are the predominant bacterial class in the sediments [37] and gastropods [54]. Moreover, Epsilonproteobacteria are also found in the shallow-water hydrothermal vent crabs and sea water [27]. These results confirm that Epsilonproteobacteria is a common bacterial group in the hydrothermal systems [55].
The class Epsilonproteobacteria encompasses a single order Campylobacterales, which includes two families [56]. The genera Campylobacter, Arcobacter, Sulfurospirillum, and Thiovulum belong to the family Campylobacteraceae, whereas the genera Helicobacter and Wolinella form the family Helicobacteraceae. All the above-mentioned genera, except for Thiovulum and Wolinella, were found in the gill and/or intestine of Austinograea sp., suggesting a close relationship between these microbes and the habitat of Austinograea sp. Particularly, the genus Sulfurospirillum found in the intestine of Austinograea sp. with a very high abundance is the only organohalide-respiring Epsilonproteobacteria described so far. They can thrive in polluted habitats that include many toxic compounds [57]. Another bacterium involved in sulfur metabolism and inhabiting the gill and intestine of deep-sea crab was the genus Arcobacter, which is a novel sulfur-oxidizing Epsilonproteobacteria and an important member of the microbial communities at deep-sea vents [58]. The presence of these bacteria reflects the adaptation of the hydrothermal vent crab to extreme environments by associated microorganisms.
As a sulfur-oxidizer [12,59], the genus Leucothrix belonging to the class Gammaproteobacteria was observed to show higher abundance in the gill of Austinograea sp. in our study. This result indicates that the Leucothrix associated with Austinograea sp. may use reduced sulfur compounds as an energy source for their chemoheterotrophic growth and provide the host with nutrition [12,59]. Epsilonproteobacteria, as well as some Gammaproteobacteria form a significant microbiota in the gill and intestine of Austinograea sp., which is consistent with the discovery of gastropods in the hydrothermal vent of the Manus Basin [54]. These microorganisms are mainly involved in metabolism of sulfur or sulfur compounds. Otherwise, transcriptome analyses of these three crab species demonstrated a significantly higher amount of genes participating in sulfur metabolism in the Austinograea sp. than in the two shallow-water crab species [35]. All these discoveries reveal that sulfur and sulfur compounds should be the primary inorganic chemicals in the habitat of Austinograea sp.
Symbionts associated with cold-adaptation in Austinograea sp.
Besides the characteristics of chemoautotrophs, some deep-sea Gammaproteobacteria are psychrophilic [60]. Some psychrophilic bacteria dominant in the deep-sea crab Austinograea sp., were identified as Gammaproteobacteria, including the genera Marinomonas, Moritella, and Psychromonas. The genus Marinomonas comprises the Gram-negative bacterial strains distributed in different marine environments and even in cold environments, such as deep-sea sediments [58,61]. Some of these strains have proven to be an excellent source of metabolic enzymes, which are involved in carbohydrate metabolism, oxidation, desulfation, serine protease-producing and so on [62–66]. In this study, a higher abundance of the genus Marinomonas was observed in the gill of Austinograea sp., which suggests that gill surface-associated Marinomonas might be of interest for the production of enzymes for host metabolism [23].
The genus Moritella has been known to consist solely of psychrophilic species and has been studied as model microorganisms for low-temperature-adapted enzymes and piezophilic adaptation of marine bacteria to the deep sea [67]. Since Moritella species are also facultative anaerobes, Moritella strains were identified from the intestines of hydrothermal vent crab as expected [10]. The genus Psychromonas includes piezophilic, halophilic, and psychrophilic species that are widely distributed in aquatic environments and are an important component of polar and deep-sea microbiota [68–71]. They are chemoorganotrophs, and so, Austinograea sp. might obtain a significant portion of their energy from bacterial symbionts, making the symbiosis a true mutualism.
Members of the diverse bacterial phylum Bacteroidetes have colonized virtually all types of habitats on earth. According to Bergey's Manual of Systematic Bacteriology [72], Bacteroidetes comprises four classes: Bacteroidia, Flavobacteria, Sphingobacteria, and Cytophagia. Among these, the Bacteroidia is a normal component of the microbiota in animals especially in the gastrointestinal tract (GIT) [73], and the other three classes belong to the environmental Bacteroidetes. Interestingly, three genera Ulvibacter, Polaribacter, and Crocinitomix (Flavobacteria) were mainly discovered in the gill of the deep-sea hydrothermal vents crab Austinograea sp., and then in the intestine. As they are well known degraders of polymeric organic matter [72], these gill and intestinal bacteria could also help the host to gain energy from otherwise refractory carbohydrate sources, as symbionts.
Symbionts associated with hypoxia adaptation in Austinograea sp.
The deep sea is low of oxygen (O2) and the hydrothermal-vent crab itself has a specific adaptive mechanism for this extreme environment [1]. In addition, they are also dependent on symbiotic microorganisms to survive in hypoxic conditions. The phylum Fusobacteria contains facultative aerobic to obligate anaerobic organisms that ferment carbohydrates or amino acids and peptides to produce various organic acids [74]. These species occur in anoxic environments including sediments as well as in the oral or intestinal habitats of animals. The genus Psychrilyobacter belonging to the family Fusobacteriaceae, was a specific microbiota with a higher abundance in the gill and intestine of the deep-sea hydrothermal vent crab Austinograea sp. They can utilize sugars, amino acids and peptone as carbon sources and produce H2 and acetate as the major fermentation products [75].
The phylum Firmicutes is phenotypically, physiologically, and ecologically diverse and consists of at least 26 families and 223 genera [76]. The genus Fusibacter belonging to the family Clostridiales, is also a strictly anaerobic, thiosulfate-reducing bacterium and utilizes a limited number of carbohydrates to produce acetate, butyrate, CO2, and H2 as the end products from glucose fermentation [77]. In this study, a higher abundance of these bacteria in the deep-sea hydrothermal vent crab gill and intestine indicates that Austinograea sp. may be depend on these associated microorganisms for their metabolism to adapt to the anaerobic conditions in hydrothermal vents. Meanwhile, they can also use thiosulfate and sulfur as electron acceptors during glucose fermentation, with production of H2S [76]. Therefore, they complement each other with the Epsilonproteobacteria in Austinograea sp., forming an inner environment, allowing the crabs to adapt better to the extreme environment.
Conclusions
In conclusion, aquatic animals have a persistent, close relationship with the surroundings that they inhabit. In our study, the microbiota of the deep-sea hydrothermal vents crab, Austinograea sp., show differences from those of two shallow-water crabs suggesting the impact of the environment and food source on the deep-sea hydrothermal vents crab. Moreover, different bacterial abundances between the gill and intestine of Austinograea sp. indicate the existence of tissue-specific bacteria, and the specific association of bacteria with the host.
Supporting information
(JPG)
(DOCX)
Acknowledgments
The samples were collected by RV KEXUE. We greatly appreciated members working in the RV KEXUE for the sampling.
Data Availability
All data were deposited in Sequence Read Archive (SRA) database (http://www.ncbi.nlm.nih.gov/Traces/sra/) with accession numbers (SRX2986360 for Austinograea sp., SRX2986361 for E. sinensis and SRX2986363 for P. trituberculatus).
Funding Statement
This work was supported by the Scientific and Technological Innovation Project of Qingdao National Laboratory for Marine Science and Technology (grant No. 2015ASKJ02 to Prof. Zhaoxia Cui).
References
- 1.Hourdez S, Lallier FH. (2007) Adaptations to hypoxia in hydrothermal-vent and cold-seep invertebrates. Rev Environ Sci Bio 6: 143–159. [Google Scholar]
- 2.Dubilier N, Bergin C, Lott C. (2008) Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol 6:725–740. doi: 10.1038/nrmicro1992 [DOI] [PubMed] [Google Scholar]
- 3.Goffredi SK, Jones WJ, Erhlich H, Springer A, Vrijenhoek RC. (2008) Epibiotic bacteria associated with the recently discovered Yeti crab, Kiwa hirsuta. Environ Microbio 10:2623–2634. [DOI] [PubMed] [Google Scholar]
- 4.Ponsard J, Cambon-Bonavita MA, Zbinden M, Lepoint G, Joassin A, Corbari L, et al. (2013) Inorganic carbon fixation by chemosynthetic ectosymbionts and nutritional transfers to the hydrothermal vent host-shrimp Rimicaris exoculata. Isme Journal 7:96–109. doi: 10.1038/ismej.2012.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gebruk AV, Pimenov NV, Savvichev AS. (1993) Feeding specialization of bresiliid shrimps in the TAG site hydrothermal community. Mar Ecol Prog Ser 98:247–253. [Google Scholar]
- 6.Gebruk AV, Southward EC, Kennedy H, Southward AJ. (2000) Food sources, behaviour, and distribution of hydrothermal vent shrimps at the Mid-Atlantic Ridge. J Mar Biol Assoc UK 80:485–499. [Google Scholar]
- 7.Polz MF, Robinson JJ, Cavanaugh CM, Van Dover CL. (1998) Trophic ecology of massive shrimp aggregations at a Mid-Atlantic Ridge hydrothermal vent site. Limnol Oceanogr 43: 1631–1638. [Google Scholar]
- 8.Zbinden M, Cambon-Bonavita MA. (2003) Occurrence of Deferribacterales and Entomoplasmatales in the deep-sea Alvinocarid shrimp Rimicaris exoculata gut. Fems Microbiol Ecol 46:23–30. doi: 10.1016/S0168-6496(03)00176-4 [DOI] [PubMed] [Google Scholar]
- 9.Hügler M, Petersen JM, Dubilier N, Imhoff JF, Sievert SM. (2011) Pathways of carbon and energy metabolism of the epibiotic community associated with the deep-sea hydrothermal vent shrimp Rimicaris exoculata. PLoS One 6:e16018 doi: 10.1371/journal.pone.0016018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durand L, Zbinden M, Cueff-Gauchard V, Duperron S, Roussel EG, Shillito B, et al. (2009) Microbial diversity associated with the hydrothermal shrimp Rimicaris exoculata gut and occurrence of a resident microbial community. Fems Microbiol Ecol 71:291–303. doi: 10.1111/j.1574-6941.2009.00806.x [DOI] [PubMed] [Google Scholar]
- 11.Durand L, Roumagnac M, Cueff-Gauchard V, Jan C, Guri M, Tessier C, et al. (2015) Biogeographical distribution of Rimicaris exoculata resident gut epibiont communities along the Mid-Atlantic Ridge hydrothermal vent sites. Fems Microbiol Ecol 91:fiv101. [DOI] [PubMed] [Google Scholar]
- 12.Petersen JM, Ramette A, Lott C, Cambon-Bonavita MA, Zbinden M, Dubilier N. (2010) Dual symbiosis of the vent shrimp Rimicaris exoculata with filamentous gamma- and epsilonproteobacteria at four Mid-Atlantic Ridge hydrothermal vent fields. Environ Microbiol 12:2204–2218. doi: 10.1111/j.1462-2920.2009.02129.x [DOI] [PubMed] [Google Scholar]
- 13.Goffredi SK, Gregory A, Jones WJ, Morella NM, Sakamoto RI. (2014) Ontogenetic variation in epibiont community structure in the deep-sea yeti crab, Kiwa puravida: convergence among crustaceans. Mol Ecol 23:1457–1472. doi: 10.1111/mec.12439 [DOI] [PubMed] [Google Scholar]
- 14.Konishi M, Watsuji T, Nakagawa S, Hatada Y, Takai K, Toyofuku T. (2013) Effects of hydrogen sulfide on bacterial communities on the surface of galatheid crab, Shinkaia crosnieri, and in a bacterial mat cultured in rearing tanks. Microbes Environ 28:25–32. doi: 10.1264/jsme2.ME12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watsuji T, Nishizawa M, Morono Y, Hirayama H, Kawagucci S, Takahata N. (2012) Cell-specific thioautotrophic productivity of epsilon-proteobacterial epibionts associated with Shinkaia crosnieri. PLoS One 7:e46282 doi: 10.1371/journal.pone.0046282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zwirglmaier K, Reid W D K, Heywood J, Sweeting CJ, Wigham BD, Polunin NV, et al. (2015) Linking regional variation of epibiotic bacterial diversity and trophic ecology in a new species of Kiwaidae (Decapoda, Anomura) from East Scotia Ridge (Antarctica) hydrothermal vents. Microbiologyopen 4:136–150. doi: 10.1002/mbo3.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forget NL, Kim Juniper S. (2013) Free-living bacterial communities associated with tubeworm (Ridgeia piscesae) aggregations in contrasting diffuse flow hydrothermal vent habitats at the Main Endeavour Field, Juan de Fuca Ridge. Microbiologyopen 2:259–275. doi: 10.1002/mbo3.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bougerol M, Boutet I, LeGuen D, Jollivet D, Tanguy A. (2015) Transcriptomic response of the hydrothermal mussel Bathymodiolus azoricus in experimental exposure to heavy metals is modulated by the Pgm genotype and symbiont content. Mar Genom 21:63–73. [DOI] [PubMed] [Google Scholar]
- 19.Tame A, Yoshida T, Ohishi K, Maruyama T. (2015) Phagocytic activities of hemocytes from the deep-sea symbiotic mussels Bathymodiolus japonicus, B. platifrons, and B. septemdierum. Fish Shellfish Immun 45:146–156. [DOI] [PubMed] [Google Scholar]
- 20.Wentrup C, Wendeberg A, Huang JY, Borowski C, Dubilier N. (2013) Shift from widespread symbiont infection of host tissues to specific colonization of gills in juvenile deep-sea mussels. Isme Journal 7:1244–1247. doi: 10.1038/ismej.2013.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casanova B, Brunet M, Segonzac M. (1993) Impact of bacterial epibiosis on functional-morphology of shrimp associated with the Mid-Atlantic hydrothermal conditions. Cah Biol Mar 34:573–588. [Google Scholar]
- 22.Segonzac M, Desaintlaurent M, Casanova B. (1993) Enigma of the trophic adaptation of the shrimp Alvinocarididae in hydrothermal areas along the Mid-Atlantic Ridge. Cah Biol Mar 34:535–571. [Google Scholar]
- 23.Zbinden M, Le Bris N, Gaill F, Compère P. (2004) Distribution of bacteria and associated minerals in the gill chamber of the vent shrimp Rimicaris exoculata and related biogeochemical processes. Mar Ecol Prog Ser 284:237–251. [Google Scholar]
- 24.Van Dover CL, Fry B, Grassle JF, Humphris S, Rona PA. (1988) Feeding biology of the shrimp Rimicaris exoculata at hydrothermal vents on the Mid-Atlantic Ridge. Mar Biol 98:209–216. [Google Scholar]
- 25.Watsuji T, Nakagawa S, Tsuchida S, Toki T, Hirota A, Tsunogai U. (2010) Diversity and function of epibiotic microbial communities on the galatheid crab, Shinkaia crosnieri. Microbes Environ 25:288–294. [DOI] [PubMed] [Google Scholar]
- 26.Watsuji T, Yamamoto A, Takaki Y, Ueda K, Kawagucci S, Takai K. (2014) Diversity and methane oxidation of active epibiotic methanotrophs on live Shinkaia crosnieri. Isme Journal 8:1020–1031. doi: 10.1038/ismej.2013.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang SH, Chiang PW, Hsu TC, Kao SJ, Tang SL. (2016).Bacterial community associated with organs of shallow hydrothermal vent crab Xenograpsus testudinatus near Kuishan Island, Taiwan. PLoS One, 11: e0150597 doi: 10.1371/journal.pone.0150597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SJ, Lee KY, Ju SJ. (2013) Nuclear mitochondrial pseudogenes in Austinograea alayseae hydrothermal vent crabs (Crustacea: Bythograeidae): effects on DNA barcoding. Mol Ecol Resour 13:781–787. doi: 10.1111/1755-0998.12119 [DOI] [PubMed] [Google Scholar]
- 29.Tsuchida S, Hashimoto J. (2002) A new species of bythograeid crab, Austinograea rodriguezensis (Decapoda, brachyura), associated with active hydrothermal vents from the Indian Ocean. J Crustacean Biol 22:642–650. [Google Scholar]
- 30.Guinot D. (1989) Austinograea alayseae sp. nov., crabe hydrothermal decouvert dans le bassin de Lau, Pacifique sud-occidental (Crustacea, Decapoda, Brachyura). Bulletin du Museum National d'Histoire Naturelle Section A Zoologie Biologie et Ecologie Animales, 11:879–903. [Google Scholar]
- 31.Hessler RR, Martin JW. (1989) Austinograea-williamsi, new genus, new species, a hydrothermal vent crab (Decapoda, Bythograeidae) from the Mariana Back-Arc Basin, Western Pacific. J Crustacean Biol 9:645–661. [Google Scholar]
- 32.Tsuchida S, Fujikura K, Hashimoto J, Fujiwara Y, Hunt JC. (1997) Morphological characters in the bythograeid crab, Austinograea williamsi—heterochely, relative growth, gonopods morphology. JAMSTEC J Deep Sea Res 13:85–93. [Google Scholar]
- 33.Tsuchida S, Fujikura K. (2000) Heterochely, relative growth, and gonopod morphology in the bythograeid crab, Austinograea williamsi (Decapoda, Brachyura). J Crustacean Biol 20:407–414. [Google Scholar]
- 34.Kim SJ, Kim HS, Ju SJ. (2014) Mitochondrial genome of the hydrothermal vent crab Austinograea alayseae (Crustacea: Bythograeidae): genetic differences between individuals from Tofua Arc and Manus Basin. Mitochondrial DNA, 25:251–252. doi: 10.3109/19401736.2013.800489 [DOI] [PubMed] [Google Scholar]
- 35.Hui M, Song CW, Yiu Y, Li CL, Cui ZX. (2017) Exploring the molecular basis of adaptive evolution in hydrothermal vent crab Austinograea alayseae by transcriptome analysis. PLoS One 12:e0178417 doi: 10.1371/journal.pone.0178417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Logares R, Sunagawa S, Salazar G, Cornejo-Castillo FM, Ferrera I, Sarmento H, et al. (2014) Metagenomic 16S rDNA Illumina tags are a powerful alternative to amplicon sequencing to explore diversity and structure of microbial communities. Environ Microbiol 16:2659–2671. doi: 10.1111/1462-2920.12250 [DOI] [PubMed] [Google Scholar]
- 37.Gamo T, Okamura K, Charlou JL, Urabe T, Auzende JM, Ishibashi J, et al. (1997) Acidic and sulfate-rich hydrothermal fluid from the Manus back-arc basin, Papua New Guinea. Geology, 25:139–142. [Google Scholar]
- 38.Wang HL, Zhang J, Sun QL., Lian C, Sun L. (2017) A comparative study revealed first insights into the diversity and metabolisms of the microbial communities in the sediments of Pacmanus and Desmos hydrothermal fields. PLoS one, 12: e0181048 doi: 10.1371/journal.pone.0181048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Russell DW. (2001) Molecular cloning: a laboratory manual. New York: Cold Spring Harbor. [Google Scholar]
- 40.Magoč T, Salzberg SL. (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, et al. (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59. doi: 10.1038/nmeth.2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC. (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- 44.Wang Q, Garrity GM, Tiedje JM, Cole JR. (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ling Z, Liu X, Jia X, Cheng Y, Luo Y, Yuan L, et al. (2014) Impacts of infection with different toxigenic clostridium difficile strains on faecal microbiota in children. Sci Rep 4:7485 doi: 10.1038/srep07485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Little CTS, Vrijenhoek RC. (2003) Are hydrothermal vent animals living fossils? Trends Ecol Evol 18:582–588. [Google Scholar]
- 48.Van Dover CL. (2000) The ecology of deep-sea hydrothermal vents. Princeton: Princeton Univ. [Google Scholar]
- 49.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. (2006) Microbial diversity in the deep sea and the underexplored “rare biosphere”. P Natl A Sci 103(32): 12115–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jannasch HW. (1985) Review lecture: The chemosynthetic support of life and the microbial diversity at deep-sea hydrothermal vents. P Roy Soc B: Biol Sci 225: 277–297. [Google Scholar]
- 51.Jeanthon C. (2000) Molecular ecology of hydrothermal vent microbial communities. Anton Leeuw 77:117–133. [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto M, Takai K. (2011) Sulfur metabolisms in epsilon- and gamma- proteobacteria in deep-sea hydrothermal fields. Front Microbiol 2:192 doi: 10.3389/fmicb.2011.00192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Campbell BJ, Engel AS, Porter ML, Takai K. (2006) The versatile ε-proteobacteria: key players in sulphidic habitats. Nat Rev Microbiol 4:458–468. doi: 10.1038/nrmicro1414 [DOI] [PubMed] [Google Scholar]
- 54.Urakawa H., Dubilier N., Fujiwara Y., Cunningham D.E., Kojima S., Stahl D.A., 2005. Hydrothermal vent gastropods from the same family (Provannidae) harbour epsilon- and gamma-proteobacterial endosymbionts. Environmental Microbiology 7, 750–754. doi: 10.1111/j.1462-2920.2005.00753.x [DOI] [PubMed] [Google Scholar]
- 55.Garrity GM, Holt JG. (2015) Bergey's Manual of Systematics of Archaea and Bacteria In: Whitman WB (ed.). The road map to the Manual. New York: Springer; 1–70. [Google Scholar]
- 56.Goris T, Diekert G. (2016) Organohalide-respiring bacteria In: Adrian L and Löffler FE (eds.). The genus Sulfurospirillum. New York: Springer; 209–234. [Google Scholar]
- 57.Sievert SM, Wieringa E, Wirsen CO, Taylor CD. (2007) Growth and mechanism of filamentous-sulfur formation by Candidatus Arcobacter sulfidicus in opposing oxygen-sulfide gradients. Environ microbio 9:271–276. [DOI] [PubMed] [Google Scholar]
- 58.Grabovich MY, Muntyan MS, Lebedeva VY, Ustiyan VS, Dubinina GA. (1999) Lithoheterotrophic growth and electron transfer chain components of the filamentous gliding bacterium Leucothrix mucor DSM 2157 during oxidation of sulfur compounds. Fems Microbiol Lett 178:155–161. [Google Scholar]
- 59.Lauro FM, Allen MA, Wilkins D, Williams TJ, Cavicchioli R. (2011) Extremophiles handbook In: Horikoshi K (ed.). Psychrophiles: genetics, genomics, evolution. Tokyo: Springer, 865–890. [Google Scholar]
- 60.Bai X, Lai Q, Dong C, Li F, Shao Z. (2014) Marinomonas profundimaris sp. nov., isolated from deep-sea sediment sample of the Arctic Ocean. Anton Leeuw 106: 449–455. [DOI] [PubMed] [Google Scholar]
- 61.Dong C, Bai X, Lai Q, Xie Y, Chen X, Shao Z. (2014) Draft genome sequence of Marinomonas sp. strain D104, a polycyclic aromatic hydrocarbon-degrading bacterium from the deep-sea sediment of the Arctic ocean. Genome Announc 2:e01211–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ding H, Zeng Q, Zhou L, Yu Y, Chen B. (2017) Biochemical and structural insights into a novel thermostable β-1, 3-galactosidase from Marinomonas sp. BSi20414. Mar Drugs 15:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanchez-Amat A, Solano F, Lucas-Elío P. (2010) Finding new enzymes from bacterial physiology: a successful approach illustrated by the detection of novel oxidases in Marinomonas mediterranea. Marine drugs 8:519–541. doi: 10.3390/md8030519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X, Duan D, Xu J, Gao X, Fu X. (2015) Characterization of a novel alkaline arylsulfatase from Marinomonas sp. FW-1 and its application in the desulfation of red seaweed agar. J Ind Microbiol Biot 42:1353–1362. [DOI] [PubMed] [Google Scholar]
- 65.Yoo AY, Park JK. (2016) Isolation and characterization of a serine protease-producing marine bacterium Marinomonas arctica PT-1. Bioproc Biosyst Eng 39:307–314. [DOI] [PubMed] [Google Scholar]
- 66.Zhao W, Peng R, Xiong A, Fu X, Tian Y, Yao Q. (2012) Expression and characterization of a cold-active and xylose-stimulated β-glucosidase from Marinomonas MWYL1 in Escherichia coli. Mol biol rep 39:2937–2943. doi: 10.1007/s11033-011-1055-0 [DOI] [PubMed] [Google Scholar]
- 67.Urakawa H. (2014) The Prokaryotes-Gammaproteobacteria In: Rosenberg E et al. (eds.). The family Moritellaceae. Berlin: Springer; 477–489. [Google Scholar]
- 68.Auman AJ, Breezee JL, Gosink JJ, Kämpfer P, Staley JT. (2006) Psychromonas ingrahamii sp. nov., a novel gas vacuolate, psychrophilic bacterium isolated from Arctic polar sea ice. Int J Syst Evol Microbiol 56:1001–1007. doi: 10.1099/ijs.0.64068-0 [DOI] [PubMed] [Google Scholar]
- 69.Auman AJ, Breezee JL, Gosink JJ, Schumann P, Barnes CR, Kämpfer P, et al. (2010) Psychromonas boydii sp. nov., a gas-vacuolate, psychrophilic bacterium isolated from an Arctic sea-ice core. Int J Syst Evol Microbiol 60:84–92. doi: 10.1099/ijs.0.007773-0 [DOI] [PubMed] [Google Scholar]
- 70.Miyazaki M, Nogi Y, Fujiwara Y, Horikoshi K. (2008) Psychromonas japonica sp. nov., Psychromonas aquimarina sp. nov., Psychromonas macrocephali sp. nov. and Psychromonas ossibalaenae sp. nov., psychrotrophic bacteria isolated from sediment adjacent to sperm whale carcasses off Kagoshima, Japan. Int J Syst Evol Microbiol 58:1709–1714. doi: 10.1099/ijs.0.65744-0 [DOI] [PubMed] [Google Scholar]
- 71.Nogi Y, Kato C, Horikoshi K. (2002) Psychromonas kaikoae sp. nov., a novel piezophilic bacterium from the deepest cold-seep sediments in the Japan Trench. Int J Syst Evol Microbiol 52:1527–1532. doi: 10.1099/00207713-52-5-1527 [DOI] [PubMed] [Google Scholar]
- 72.Krieg NR. (2010) Bergey’s manual of systematic bacteriology In: Whitman WB (ed.). Bacteroidetes, New York: Springer; 25–469. [Google Scholar]
- 73.Thomas F, Hehemann JH, Rebuffet E, Czjzek M, Michel G. (2011) Environmental and gut bacteroidetes: the food connection. Front Microbiol 2:93 doi: 10.3389/fmicb.2011.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Staley JT, Whitman WB. (2010) Bergey’s Manual of Systematic Bacteriology In: Whitman WB (ed.). Fusobacteria. New York: Springer; 747–774. [Google Scholar]
- 75.Zhao JS, Manno D, Hawari J. (2009) Psychrilyobacter atlanticus gen. nov., sp. nov., a marine member of the phylum Fusobacteria that produces H2 and degrades nitramine explosives under low temperature conditions. Int J Syst Evol Micr 59:491–497. [DOI] [PubMed] [Google Scholar]
- 76.Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, et al. (2011) Bergey’s Manual of Systematic Bacteriology In: Whitman WB (ed.). The Firmicutes. New York: Springer; 1151–1155. [Google Scholar]
- 77.Ravot G, Garcia JL, Magot M, Ollivier B. (2015) Bergey's Manual of Systematics of Archaea and Bacteria In: Whitman WB (ed.). Fusibacter. New York: Springer; 1–5. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(JPG)
(DOCX)
Data Availability Statement
All data were deposited in Sequence Read Archive (SRA) database (http://www.ncbi.nlm.nih.gov/Traces/sra/) with accession numbers (SRX2986360 for Austinograea sp., SRX2986361 for E. sinensis and SRX2986363 for P. trituberculatus).