Abstract
Generation of the lipid messenger phosphatidylinositol-3,4,5-trisphosphate (PtdIns(3,4,5)P3) is crucial for development, cell growth and survival, and motility, and it becomes dysfunctional in many diseases including cancers. Here we reveal a mechanism for PtdIns(3,4,5)P3 generation by scaffolded phosphoinositide kinases. In this pathway, class I phosphatidylinositol-3-OH kinase (PI(3)K) is assembled by IQGAP1 with PI(4)KIIIα and PIPKIα, which sequentially generate PtdIns(3,4,5)P3 from phosphatidylinositol. By scaffolding these kinases into functional proximity, the PtdIns(4,5)P2 generated is selectively used by PI(3)K for PtdIns(3,4,5)P3 generation, which then signals to PDK1 and Akt that are also in the complex. Moreover, multiple receptor types stimulate the assembly of this IQGAP1–PI(3)K signalling complex. Blockade of IQGAP1 interaction with PIPKIα or PI(3)K inhibited PtdIns(3,4,5)P3 generation and signalling, and selectively diminished cancer cell survival, revealing a target for cancer chemotherapy.
Phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2) and PtdIns (3,4,5)P3 are lipid messengers that regulate most cellular functions1,2. PtdIns(4,5)P2 serves as a substrate for phospholipases and class I PI(3)Ks to generate additional messengers3,4. Besides, PtdIns(4,5)P2 directly binds to its effector proteins and regulates their targeting and activities5. The majority of PtdIns(4,5)P2 is made by phosphorylation at the 5 hydroxyl of the myo-inositol ring of PtdIns(4)P by type I phosphatidylinositol phosphate kinases (PIPKIs). In humans, there are three PIPKI isoforms (α, β and γ) with multiple splice variants2,6. Stimulation of receptor tyrosine kinases (RTKs), G-protein-coupled receptors (GPCRs) and integrins leads to activation of PI(3)Ks, which utilize PtdIns(4,5)P2 as a substrate to generate PtdIns(3,4,5)P3 (ref. 7). Although PI(3)Ks catalyse phosphorylation of PtdIns(4,5)P2 at the 3 hydroxyl in vitro8 and in vivo9, the cellular source of PtdIns(4,5)P2 used as the substrate has not been studied. It is hypothesized that a pre-existing plasma membrane pool of PtdIns(4,5)P2 is used by PI(3)Ks to synthesize PtdIns(3,4,5)P3 (ref. 10), as the cellular PtdIns(4,5)P2 content is orders of magnitude higher than that of PtdIns(3,4,5)P3 (ref. 10). Yet, the majority of membrane PtdIns(4,5)P2 is sequestered11 and little is present on endosomal compartments where PtdIns(3,4,5)P3 is generated after receptor internalization12. This suggests that de novo synthesis of PtdIns(4,5)P2 by PIPKIs is required for efficient PtdIns(3,4,5)P3 synthesis. In support, depletion of a single PIPKI isoform has no significant impact on cellular PtdIns(4,5)P2 levels13,14, while Akt phosphorylation, a readout of PI(3)K activity, is reduced following inhibition of a PIPKI15. These suggest that a locally organized specific pool of PtdIns(4,5)P2 is responsible for PtdIns(3,4,5)P3 synthesis.
IQ-motif-containing GTPase-activating protein 1 (IQGAP1) is a multidomain protein that scaffolds multiple signalling pathways to regulate a plethora of cell functions16,17. The scaffold role of IQGAP1 in the mitogen-activated protein kinase (MAPK) pathway is best characterized. MAPK pathway components, such as Raf, MEK and Erk, directly interact with IQGAP1, which brings these kinases in close proximity to facilitate their sequential phosphorylation18,19. Additional scaffold roles for IQGAP1 in GPCR, Wnt and integrin signalling were also identified16. These versatile roles of IQGAP1 are in part supported by its abundance. Notably, an unbiased quantitative study reveals that the messenger RNA and protein copy number of IQGAP1 is at least two orders of magnitude higher than that of its interacting proteins20. This is consistent with IQGAP1 integrating multiple signalling complexes and pathways. Although IQGAP1 is reported to modulate Akt under stress conditions21,22, the molecular mechanism by which IQGAP1 controls Akt is not known.
Here, we show an IQGAP1 complex that scaffolds the entire PI(3)K–Akt pathway and controls agonist-stimulated PtdIns(3,4,5)P3 synthesis and signalling. IQGAP1 interacts with PIPKIα and PI(3)K, bringing the phosphoinositide kinases in close proximity, which allows for the PtdIns(4,5)P2 produced by PIPKIα to be channelled to PI(3)K for PtdIns(3,4,5)P3 synthesis. Further, IQGAP1 assembles phosphatidylinositol 4-kinase III alpha (PI(4)KIIIα), which generates PtdIns(4)P from PtdIns, and the PtdIns(3,4,5)P3 effectors phosphoinositide-dependent kinase 1 (PDK1) and Akt. This IQGAP1-mediated pathway is responsible for PtdIns(3,4,5)P3 synthesis stimulated by integrin, RTK or GPCR activation. Further, blockade of the IQGAP1 interaction with either PIPKIα or PI(3)K inhibited PtdIns(3,4,5)P3 synthesis and Akt activation, which blocks cancer cell proliferation and survival. Collectively, these results establish that agonist-stimulated PtdIns(3,4,5)P3 synthesis requires spatiotemporal organization of PIPKIα and PI(3)K by IQGAP1.
RESULTS
PI(3)K–Akt pathway requires PI(4)KIIIα, PIPKIα and IQGAP1
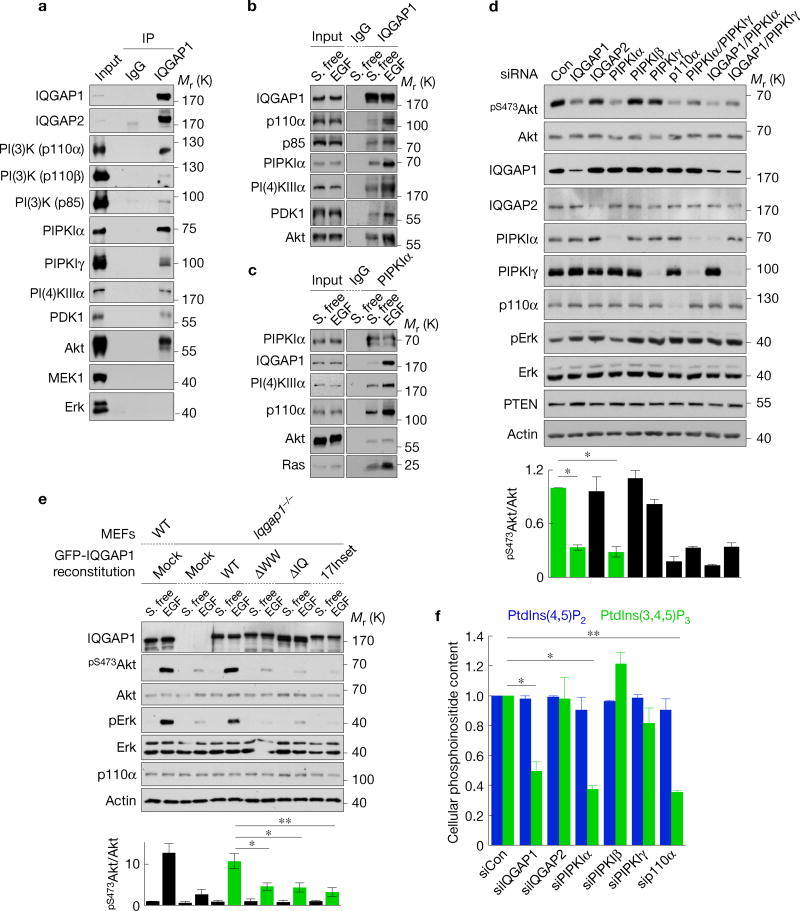
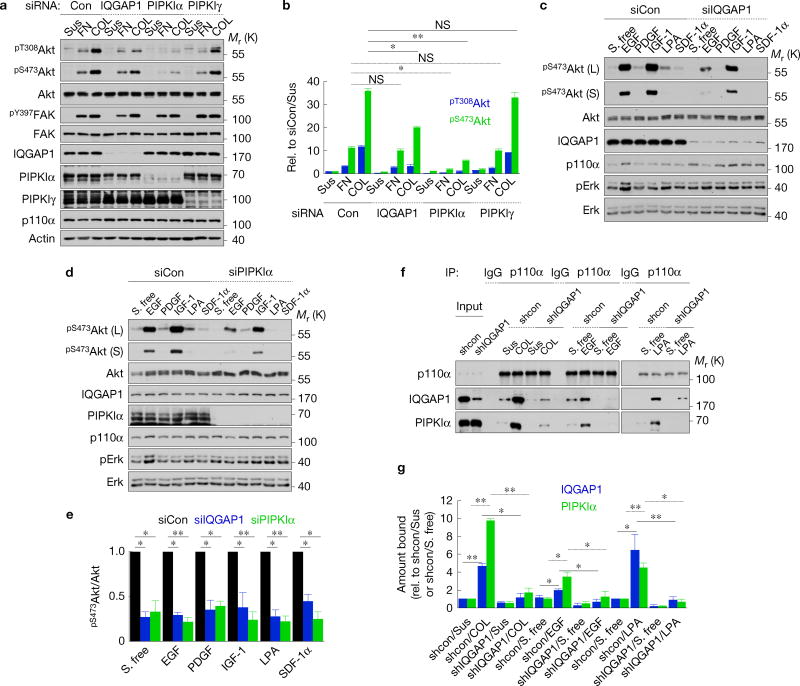
To define roles with phosphoinositide kinases, IQGAP1-associated proteins were analysed by immunoprecipitation. This demonstrated that PI(4)KIIIα, PIPKIα and PI(3)K associate with IQGAP1 (Fig. 1a). IQGAP1 also associated with the PI(3)K downstream effectors of PtdIns(3,4,5)P3 PDK1 and Akt. In addition, IQGAP2 and PIPKIγ associated with IQGAP1, confirming known interactors23,24. MEK1 and Erk associate with IQGAP1 in mouse embryonic fibroblasts (MEFs)25 and IQGAP1 is required for Erk phosphorylation (Fig. 1e) but these interactions were not detected in the cancer cells examined here, suggesting that these interactions are cell type specific. IQGAP1 associations with PI(4)KIIIα, PIPKIα, PI(3)K, PDK1 and Akt were increased by EGF stimulation (Fig. 1b,c), indicating that these interactions are enhanced by agonist stimulation. Increased Ras association was also detected, consistent with its interaction with and activation of PI(3)K (Fig. 1c).
Figure 1.
PIPKIα and IQGAP1 are required for PtdIns(3,4,5)P3 generation. (a) IQGAP1-associated proteins were analysed by immunoprecipitation in Hs578T cells. IgG, nonspecific immunoglobulin. (b,c) Changes in IQGAP1- (b) and PIPKIα- (c) associated proteins following EGF stimulation were analysed by immunoprecipitation. S. free, serum free. (d) Immunoblotting after RNAi of the indicated proteins in Hs578T cells (top), mean ± standard deviation (s.d.) of n = 3 independent experiments (bottom). P = 0.021, 0.016. Con, non-targeting siRNA. *P < 0.05; **P < 0.01; NS, not significant (Student’s t-test). (e) Iqgap1−/− mouse embryonic fibroblasts (MEFs) were reexpressed with the indicated IQGAP1 proteins (Supplementary Fig. 2a) and immunoblotted after EGF treatment (top), mean ± s.d. of n=3 independent experiments (bottom). P = 0.020, 0.015, 0.0083. (f) PtdIns(4,5)P2 and PtdIns(3,4,5)P3 contents after knockdown of the indicated proteins in Hs578T, mean ± s.d. of n=3 independent experiments. P=0.01, 0.01, 0.001. Source data for d,e,f can be found in Supplementary Table 1. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
The associations of all components of the PI(3)K pathway required for PtdIns(3,4,5)P3 generation and Akt activation suggest that PI(3)K signalling may be regulated by an IQGAP1 scaffold. To explore whether IQGAP1 modulates PI(3)K signalling, protein expression was depletedby RNA-mediated interference (RNAi) and PI(3)K activation was measured by Akt phosphorylation, which mirrors PtdIns(3,4,5)P3 generation3 (Fig. 1d and Supplementary Fig. 1a,g). Loss of IQGAP1, but not IQGAP2, significantly reduced Akt phosphorylation. PIPKIα was the only PIPKI isoform whose knockdown decreased Akt phosphorylation (Fig. 1d). These results are not due to off-target effects of short interfering RNAs (siRNAs) as Iqgap1-null MEFs also have reduced EGF-stimulated Akt phosphorylation that is rescued by reexpression of wild-type (WT) IQGAP1 (Fig. 1e). Significantly, PIPKIα kinase activity was required, as ectopic expression of WT PIPKIα, but not the kinase-dead mutant, increased Akt phosphorylation (Supplementary Fig. 1b,c,e). Expression of PIPKIγ did not impact Akt phosphorylation, indicating specificity for PIPKI isoforms. Knockdown of PI(4)KIIIα also significantly reduced Akt phosphorylation (Supplementary Fig. 1g). These results were replicated in multiple cell lines including MDA-MB-231, Hs578T, MCF-7 and HaCaT. These combined data support a model such that PI(4)KIIIα and PIPKIα via association with IQGAP1 generate a PtdIns(4,5)P2 pool that flows into the PI(3)K pathway for PtdIns(3,4,5)P3 synthesis and Akt activation (Supplementary Fig. 1e).
IQGAP1 and PIPKIα loss diminished Akt activation. This was not due to increased expression of the phosphatase and tensin homologue (PTEN), which dephosphorylates PtdIns(3,4,5)P3 to PtdIns(4,5)P2 (ref. 26; Fig. 1d). As PI(4)KIIIα and PIPKIα loss could reduce cellular PtdIns(4,5)P2 levels, diminishing its availability for PI(3)K generation of the PtdIns(3,4,5)P3 required for Akt activation9, PtdIns(4)P, PtdIns(4,5)P2 and PtdIns(3,4,5)P3 content was measured. Loss of PI(4)KIIIα, PIPKIα or IQGAP1 significantly reduced PtdIns(3,4,5)P3 levels (Fig. 1f and Supplementary Fig. 1h). Yet, PtdIns(3)P, PtdIns(4)P and PtdIns(4,5)P2 did not change (Fig. 1f and Supplementary Fig. 1f), indicating that diminished PtdIns(3,4,5)P3 generation or Akt phos-phorylation is not due to lack of PtdIns(4)P or PtdIns(4,5)P2. Ectopic expression of IQGAP1 or PIPKIα did not increase PtdIns(4,5)P2 but dramatically increased PtdIns(3,4,5)P3 (Supplementary Fig. 1d). Collectively these data suggest that PtdIns(4,5)P2 generation by PIPKIα leads to PtdIns(3,4,5)P3 synthesis and Akt activation that is regulated by IQGAP1.
IQGAP1 mediates a PIPKIα and PI(3)K complex
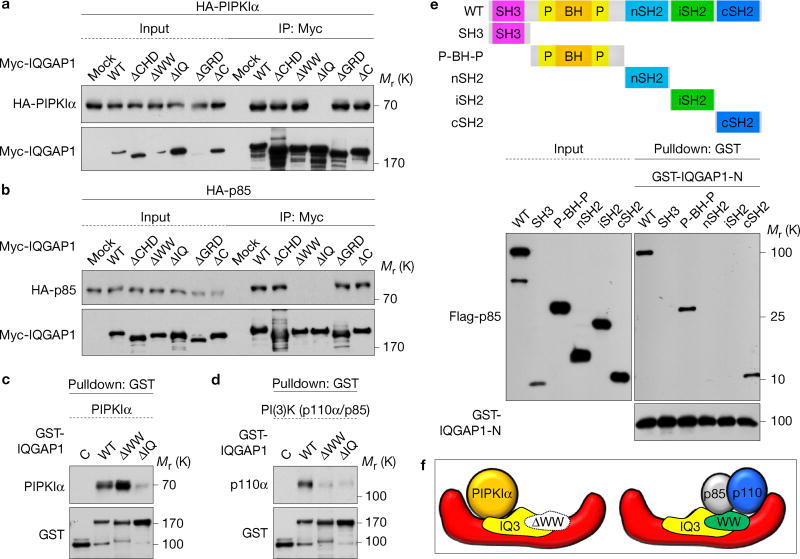
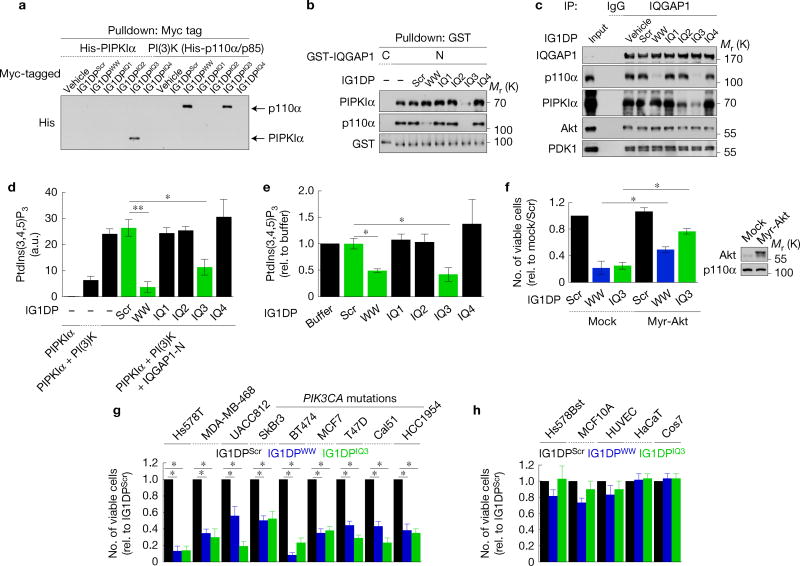
The PIPKIα- and PI(3)K-binding sites on IQGAP1 were mapped (Supplementary Fig. 2a)24,27. Deletion of the IQ domain abrogated PIPKIα co-immunoprecipitation and binding, indicating that the IQ domain is required, while interaction with PI(3)K required both the WW and IQ domains (Fig. 2a–d). To define IQGAP1-binding sites on PI(3)K, p85α domain constructswereused28.IQGAP1-Nboundtothe P-BH-P and cSH2 domains of p85α (Fig. 2e).
Figure 2.
PIPKIα and PI(3)K interact on the WW and IQ domains of IQGAP1. (a,b) PIPKIα and the p85 subunit of PI(3)K-binding sites on IQGAP1 were mapped with the indicated truncation mutants (Supplementary Fig. 2a) by immunoprecipitation. (c,d) In vitro binding experiments with the indicated recombinant proteins, representative of at least three experiments. (e) IQGAP1 binding sites were mapped in vitro with the indicated bovine p85α fragments. (f) A model summary of in vitro binding experiments. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
The IQ domain of IQGAP1 contains four structurally conserved IQ motifs (IQ1—IQ4)27. Polypeptides for WW and each IQ motif were fused to GST (Supplementary Fig. 2d) for binding assay. PIPKIα specifically bound to GST-IQ3, which blocked PIPKIα interaction with IQGAP1-N (Supplementary Fig. 2e–g). PI(3)K consisting of p110α and p85α bound to GST-WW and -IQ3 and either peptide blocked IQGAP1 interaction with PI(3)K (Supplementary Fig. 2h–j). In summary, PIPKIα and PI(3)K could bind distinct regions on IQGAP1 (Fig. 2f). Consistently, ectopic expression of the IQ domain that binds PIPKIα and PI(3)K reduced Akt phosphorylation (Supplementary Fig. 6b). Also, theWWandIQdomains were required for EGF-stimulated Akt activation (Fig. 1e).
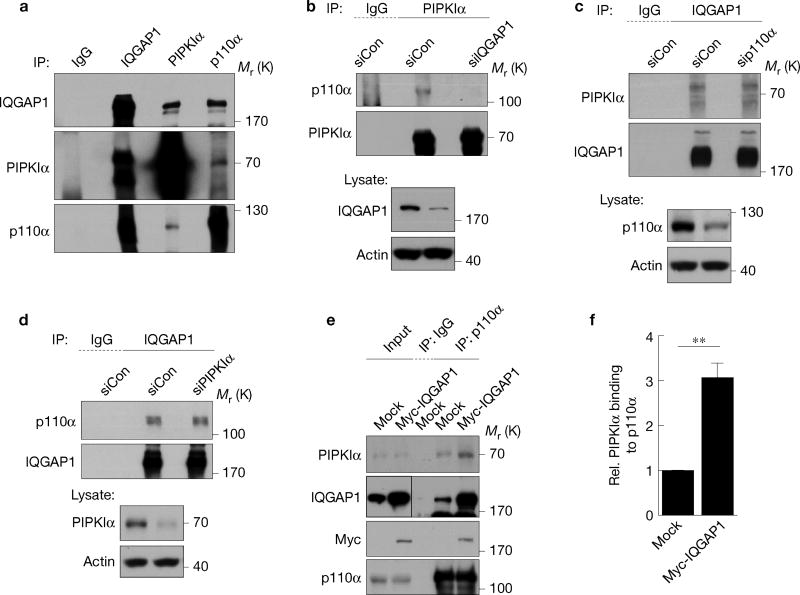
Endogenous IQGAP1, PIPKIα and PI(3)K form a complex in vivo (Fig. 3a) and loss of IQGAP1 eliminated the PI(3)K interaction with PIPKIα (Fig. 3b) indicating that IQGAP1 is necessary for the phosphoinositide kinases’ interaction. Loss of PIPKIα or PI(3)K (p110α) had no impact on the interaction of the other kinase with IQGAP1 (Fig. 3c,d). Overexpression of IQGAP1 significantly increased the PIPKIα co-immunoprecipitation with p110α and the generation of PtdIns(3,4,5)P3 (Fig. 3e,f and Supplementary Fig. 1e). As PIPKIα and PI(3)K do not bind to one another (Supplementary Fig. 2b,c), these data indicate that IQGAP1 scaffolds PIPKIα and PI(3)K, bringing them in proximity and enhancing generation of PtdIns(3,4,5)P3 in vivo.
Figure 3.
IQGAP1 physically links PIPKIα to PI(3)K. (a) The indicated endogenous proteins were immunoprecipitated from Hs578T cell lysates. Associated proteins were analysed by immunoblotting. (b–d) Hs578T cells were transfected with non-targeting (siCon), IQGAP1, p110α or PIPKIα siRNA for 48 h. The indicated proteins were immunoprecipitated and associated molecules were analysed by immunoblotting. (e) Hs578T cells were transfected with empty vector (Mock) or Myc-IQGAP1 for 48 h. p110α was immunoprecipitated and associated PIPKIα was analysed by immunoblotting. (f) Immunoblots for e were quantified and the graph is shown as mean ± s.d. of n=3 independent experiments. Rel., relative. **P = 0.028. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
IQGAP1 regulates PtdIns(3,4,5)P3 synthesis
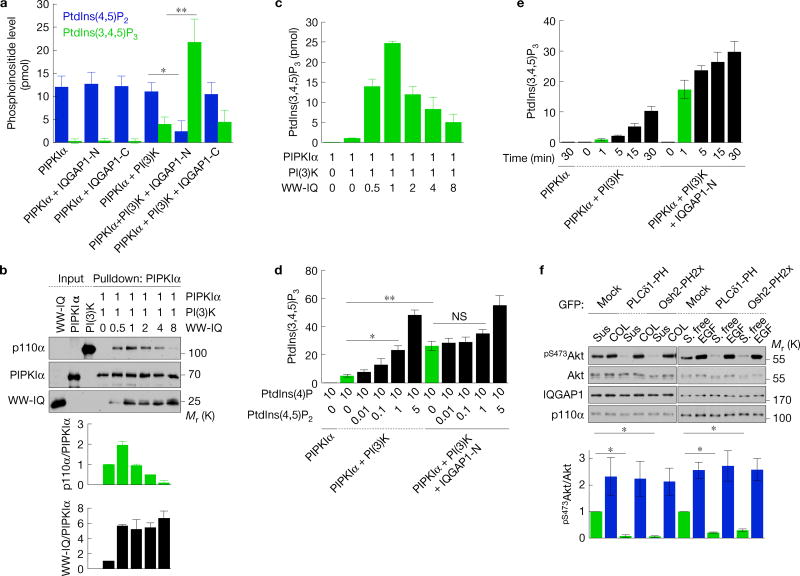
The scaffolding of PIPKIα and PI(3)K by IQGAP1 (Fig. 3) positions these kinases to coordinate enzymatic processivity. To test this, in vitro kinase assays were performed using PtdIns(4)P-containing liposomes. PIPKIα phosphorylates PtdIns(4)P to generate PtdIns(4,5)P2, which is then phosphorylated by PI(3)K to PtdIns(3,4,5)P3. PIPKIα alone or with IQGAP1 did not generate PtdIns(3,4,5)P3. PIPKIα in combination with PI(3)K generated PtdIns(3,4,5)P3 and this was significantly enhanced (~4.5-fold) by adding IQGAP1-N (Fig. 4a). These data indicate that IQGAP1-N, which scaffolds PIPKIα and PI(3)K, enhances PtdIns(3,4,5)P3 synthesis. Importantly, PtdIns(4,5)P2 accumulation was reduced by added IQGAP1-N, suggesting that PtdIns(4,5)P2 generated by PIPKIα is utilized by PI(3)K for PtdIns(3,4,5)P3 synthesis. As IQGAP1-N does not enhance enzyme activity of PIPKIα or PI(3)K (Fig. 4a and Supplementary Fig. 3a), the data indicate that PtdIns(4,5)P2 produced by PIPKIα is selectively passed to PI(3)K for PtdIns(3,4,5)P3 synthesis in the scaffold.
Figure 4.
PtdIns(4,5)P2 produced by PIPKIα is channelled to PI(3)K for PtdIns(3,4,5)P3 synthesis. (a,d,e) PtdIns(4,5)P2 and/or PtdIns(3,4,5)P3 measurement of in vitro kinase assays with PIPKIα, PI(3)K, IQGAP1-N or -C in the indicated combinations. Liposomes containing 10 mol% PtdIns(4)P (for a,e) with the indicated amount of PtdIns(4,5)P2 (for d) were used as substrates, mean ± s.d. of n=4 independent experiments. (b) In vitro binding experiments with PIPKIα, PI(3)K (p110α/p85α) and the WW-IQ fragment (Supplementary Fig. 2a), mean ± s.d. of n=3 independent experiments. The molar ratio of recombinant proteins used is shown. (c) PtdIns(3,4,5)P3 measurement of in vitro kinase assays with PIPKIα, PI(3)K and WW-IQ in the indicated combinations, mean ± s.d. of three experiments. (f) Immunoblotting after replating on type I collagen (COL) for 45 min or 20 ng ml−1 EGF treatment for 15 min in Hs578T cells expressing the indicated PH domains (top), mean ± s.d. of n=3 independent experiments (bottom). P=0.0068, 0.001. Two-tailed unpaired Student’s t-tests were used for statistical analysis (*,P < 0.05; **,P < 0.01; NS, not significant). Sus, suspension. Source data for a–f can be found in Supplementary Table 1. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
To further examine whether IQGAP1 mediates concerted PtdIns(3,4,5)P3 synthesis by scaffolding these kinases in vitro, the WW-IQ fragment was used as a minimal scaffold (Supplementary Fig. 2a). PIPKIα, PI(3)K and WW-IQ formed a ternary complex that peaks at a 1:1:1 ratio, but the complex was diminished with increasing WW-IQ (Fig. 4b). Consistently, addition of WW-IQ enhanced PtdIns(3,4,5)P3 synthesis from PtdIns(4)P and this peaked when the ratio was 1:1:1, but diminished at a higher ratio of WW-IQ (Fig. 4c). Collectively, these data signify that each IQGAP1 binds a single PIPKIα and PI(3)K, and PIPKIα and PI(3)K require scaffolding by IQGAP1 for efficient PtdIns(3,4,5)P3 synthesis.
In a concerted mechanism, the IQGAP1-tethered PI(3)K would selectively utilize the PtdIns(4,5)P2 generated from PIPKIα. To test this, in vitro kinase assays were performed with PIPKIα and PI(3)K using liposomes containing PtdIns(4)P or with increasing amounts of PtdIns(4,5)P2 (Fig. 4d). Without IQGAP1, PtdIns(3,4,5)P3 synthesis increased proportionally with the PtdIns(4,5)P2 molar percentage, suggesting that PtdIns(3,4,5)P3 is largely generated from the PtdIns(4,5)P2 in the liposomes. AdditionofIQGAP1 with PIPKIα and PI(3)K increased synthesis of PtdIns(3,4,5)P3 ~8-fold (compare green bars),with onlyamodest increase when PtdIns(4,5)P2 was added. This indicates that the PtdIns(3,4,5)P3 is preferentially generated from the PtdIns(4,5)P2 synthesized by PIPKIα. Most significant, the PIPKIα– PI(3)K–IQGAP1 complex synthesized PtdIns(3,4,5)P3 ~20-fold more rapidly than PIPKIα or PI(3)K alone (Fig. 4e, green bars), confirming that IQGAP1-bound PI(3)K preferentially utilizes the PtdIns(4,5)P2 generated by PIPKIα.
To explore a concerted mechanism in vivo, PtdIns(3)P-, PtdIns(4)P-and PtdIns(4,5)P2-binding proteins were used to sequester free lipid. PtdIns(3)P-, PtdIns(4)P- and PtdIns(4,5)P2-specific binding do-mains29–31 were stably expressed andtheimpactoncollagen- and EGF-stimulated PtdIns(3,4,5)P3 synthesis and Akt phosphorylation was quantified. The PtdIns(4)P (Osh2-PH2x)- and PtdIns(4,5)P2 (PLCδ1-PH)-binding proteins, but not the PtdIns(3)P-binding protein (Hrs-FYVE2x), blocked PtdIns(3,4,5)P3 synthesis and Akt activation in suspended and serum-starved cells but not following stimulation by collagen or EGF (Fig. 4f and Supplementary Fig. 3b–d). These results are consistent with a previous report using neomycin, which is a PtdIns(4,5)P2 sequestration reagent. Neomycin reduced Akt and Akt downstream effectors’ activity in unstimulated conditions, whereas neomycin had no impact on insulin-stimulated conditions32. This indicates that at least two distinct pathways of PtdIns(3,4,5)P3 synthesis exist in cells. One is a canonical pathway, where PtdIns(3,4,5)P3 is generated from a pool of membrane PtdIns(4,5)P2 that is accessible to the binding domains and is inhibited by sequestration of PtdIns(4)P or PtdIns(4,5)P2 (Supplementary Fig. 3b). In the IQGAP1-mediated pathway, PtdIns(4)P and PtdIns(4,5)P2 sequestration does not impact PtdIns(3,4,5)P3 synthesis as the binding domains would not access the channelling substrates in the IQGAP1-scaffolded complex, consistent with the data (Fig. 4f and Supplementary Fig. 3d).
If PIPKIα and PI(3)K are functionally linked by IQGAP1 then separating the PIPKIα and PI(3)K binding sites on IQGAP1 could uncouple PtdIns(4,5)P2 and PtdIns(3,4,5)P3 synthesis. This concept was assessed by inserting 17 amino acids between the WW domain and the IQ3 motif (Supplementary Fig. 4a). Consistent with this hypothesis, a 17-residue insert mutant reduced EGF-stimulated PtdIns(3,4,5)P3 synthesis and Akt phosphorylation (Fig. 1e and Supplementary Fig. 5b). Yet, the quantity of PIPKIα, PI(3)K, PDK1 and Akt1 that co-immunoprecipitated withtheinsertIQGAP1 mutant did not change (Supplementary Fig. 5b–e). This supports a model where IQGAP1 scaffolds PIPKIα and PI(3)K and the close proximity is required for the sequential phosphorylation of PtdIns(4)P to PtdIns(3,4,5)P3.
The above data indicate that the IQGAP1-PI(3)K scaffold functional assembly on a membrane compartment would be linked to a biological role. During cell migration, PI(3)K activation and PtdIns(3,4,5)P3 synthesis are localized to the leading edge of migrating cells33–35 and IQGAP1 is required for migration24. To examine a physiological role of the IQGAP1-mediated PtdIns(3,4,5)P3 synthesis, cells migrating into scratch wounds were immunostained (Supplementary Fig. 3e,f). PIPKIα co-localized with PtdIns(3,4,5)P3 at the leading edges in control cells, whereas PIPKIα and PtdIns(3,4,5)P3 at the leading edges were lost by IQGAP1 knockdown. These data demonstrate that the IQGAP1–PI(3)K scaffold synthesizes PtdIns(3,4,5)P3 at the leading edge of migrating cells.
Stimulated PtdIns(3,4,5)P3 synthesis needs IQGAP1
EGF stimulated IQGAP1 interactions with PI(3)K pathway components (Fig. 1b,c). To further investigate receptor regulation of the IQGAP1-mediated PtdIns(3,4,5)P3 synthesis pathway, control, IQGAP1 and PIPKIα knockdown cells were treated with agonists for a number of membrane receptor classes. Fibronectin or collagen receptor-mediated Akt activation and PtdIns(3,4,5)P3 generation were significantly reduced by IQGAP1 or PIPKIα loss (Fig. 5a,b and Supplementary Fig. 5a–c). Focal adhesion kinase (FAK) phosphory-lation is reported to regulate integrin-mediated PI(3)K activation36, but FAK phosphorylation remained unchanged following knockdown of IQGAP1 and PIPKIα, consistent with the complex functioning downstream of FAK (Fig. 5a and Supplementary Fig. 5d,e). IQGAP1 and PIPKIα loss reduced Akt phosphorylation and PtdIns(3,4,5)P3 generation by ligands for multiple RTKs and GPCRs (Fig. 5c–e and Supplementary Fig. 5c). This is not due to a loss of EGF receptor signalling following IQGAP1 knockdown (Supplementary Fig. 5f). To investigate how IQGAP1 and PIPKIα regulate the PI(3)K–Akt pathway downstream of receptors, IQGAP1 complex formation was quantified following agonist stimulation. The interaction of the PI(3)K p110α subunit with IQGAP1 and PIPKIα was enhanced by activation of all three receptor types (Fig. 1b,c,f,g). These data support a model where receptor activation stimulates IQGAP1–PIPKIα–PI(3)K assembly, which ensures efficient PtdIns(3,4,5)P3 synthesis and Akt activation.
Figure 5.
Membrane receptor signalling activates the IQGAP1-mediated pathway. (a,b) Immunoblotting after replating on fibronectin (FN) or COL for 30 min in MDA-MB-231 cells transfected with siRNAs against the indicated proteins (a), mean ± s.d. of n = 3 independent experiments (b). P = 0.82, 0.011, 0.95 (FN), 0.0036, 0.0001, 0.88 (COL). (c–e) Immunoblotting after treating with the indicated agonists (10 ng ml−1 EGF, 30 ng ml−1 PDGF, 20 ng ml−1 IGF-1, 15 µM LPA, or 30 ng ml−1SDF-1α) for 15 min in MDA-MB-231 cells transfected with siRNAs against control, IQGAP1 or PIPKIα (c,d), mean ± s.d. of n = 3 independent experiments (e). P = 0.011, 0.001 (S. free), 0.012, 0.001 (EGF), 0.0069, 0.001 (PDGF), 0.021, 0.001 (IGF-1), 0.0042, 0.009 (LPA), 0.001, 0.001 (SDF-1α). (f,g) Interactions of the p110α subunit of PI(3)K with IQGAP1 and PIPKIα were analysed by immunoprecipitation in Hs578T cells expressing shRNAs against the indicated proteins (f), mean ± s.d. of n = 3 independent experiments (g). P = 0.001, 0.001, 0.011, 0.001, 0.042, 0.020, 0.026, 0.020, 0.016, 0.001, 0.001, 0.012. Paired Student’s t-tests were used for statistical analysis (*, P < 0.05; **, P < 0.01; NS, not significant). Source data for b,e,g can be found in Supplementary Table 1. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
IQGAP1-derived peptides block PtdIns(3,4,5)P3 synthesis
In vitro the WW domain and IQ3 motif mediate the interaction of IQGAP1 with PIPKIα and PI(3)K (Supplementary Fig. 2). To expand on this, peptides corresponding to the WW domain and IQ motifs were synthesized and their impact on the PI(3)K–Akt pathway examined. These IQGAP1-derived peptides (IG1DPs) were made membrane-permeable by adding eight arginine residues to the amino terminus (Supplementary Fig. 6a)19. Consistent with the GST-fusion peptide data (Supplementary Fig. 2), IG1DPIQ3 bound to PIPKIα, and both IG1DPWW and IG1DPIQ3 bound to PI(3)K in vitro and blocked interactions with IQGAP1 in vitro and in vivo (Fig. 6a–c). The IQGAP1-mediated PtdIns(3,4,5)P3 synthesis was blocked by IG1DPWW and IG1DPIQ3 in vitro and in cells grown in normal culture (Fig. 6d,e). Activation of Akt was also diminished (Supplementary Fig. 6c–e). Overexpression of PI(3)K and PIPKIα increased PtdIns(3,4,5)P3 synthesis and this also required IQGAP1 (Supplementary Figs 1d,e and 6c). Treatment with both IG1DPWW and IG1DPIQ3 did not further inhibit Akt activation, consistent with blockade of the same pathway.
Figure 6.
IG1DPs inhibit cancer cell survival by blocking PtdIns(3,4,5)P3 synthesis. (a) In vitro binding assays of IG1DPs with PIPKIα or PI(3)K. (b,c) Effects of IG1DPs on IQGAP1 interactions with PIPKIα and PI(3)K were analysed by in vitro binding assay (b) or immunoprecipitation in Hs578T cells (c). (d,e) Effects of IG1DPs on PtdIns(3,4,5)P3 synthesis were analysed in vitro (d) or in Hs578T cells (e) of n = 3 independent experiments. P = 0.005, 0.042 (c), 0.011, 0.036 (e). (f) Viability of MDA-MB-231 cells transfected with a constitutively active Akt1 after 30µM IG1DP treatment, mean ± s.d. of n=3 independent experiments. P=0.013, 0.02. (g,h) Cell viability was measured after treating with 20µM IG1DPs in the indicated cells for 48 h, except 30 µM for T47D and Cal51 cells, mean ± s.d. of n = 3 independent experiments. P = 0.012, 0.015 (Hs578T), 0.024, 0.022 (MDA-MB-468), 0.034, 0.020 (UACC812), 0.027, 0.026 (SkBr3), 0.010, 0.015 (BT474), 0.020, 0.020 (MCF-7), 0.034, 0.028 (T47D), 0.033, 0.020 (Cal51), 0.022, 0.020 (HCC1954). Paired Student’s t-tests were used for statistical analysis (*, P < 0.05; **, P < 0.01; NS, not significant). Source data for c,e–h can be found in Supplementary Table 1. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
IQGAP1 peptides selectively kill cancer cells
The PI(3)K–Akt pathway regulates survival of many cell types3,7. To examine the roles of IG1DPs in cell survival, a variety of breast cancer cells and normal cells were treated with IG1DPs and cell viability was quantified. IG1DPWW and IG1DPIQ3 reduced the number of viable cells in breast cancer cell lines (Fig. 6f,g) with little or no effect on survival of normal cells (Fig. 6h). Expression of a constitutively active Akt137 partially rescued cell death by IG1DPIQ3 and IG1DPWW (Fig. 6f), suggesting that cell death induced by IG1DPs is not solely attributable to blockade of Akt. Akt-independent signalling downstream of PI(3)K, such as the PDK1-mediated pathway, is also reported to contribute to survival38. IQGAP1 modulates Rac1 and Cdc42 (refs 27,39) and Rac1 and Cdc42 activate p110β of PI(3)K40, but constitutively active Rac1 or Cdc42 did not rescue cell death by IG1DPs (Supplementary Fig. 7).
IQGAP1 modulates Ras-dependent Erk pathways via the WW and IQ domains18,19,22,25,41. Consistently, depletion of IQGAP1 reduced Erk phosphorylation and the WW and IQ domains are required for Erk phosphorylation in MEFs (Fig. 1e). However, knockdown of IQGAP1 in MDA-MB-231 and Hs578T cells had no impact on Erk phosphorylation (Figs 1d and 5c,d and Supplementary Fig. 5a,f). Also, overexpressionoftheIQ domainortreating with IG1DPs (WWorIQ3 peptides) had no impact on Erk phosphorylation in MDA-MB-231 and Hs578T cells (Fig. 7d and Supplementary Fig. 6b). Further, we could not co-immunoprecipitate Erk and MEK1 with IQGAP1 from those cells (Fig. 1a). These data indicate that IQGAP1 scaffolds the MAP kinase pathway only in certain cells (MEFs, MCF-7 and Hek293 cells25,42,43) but not in the cancer cells that we have tested in this study. This suggests that these cancer cells become addicted to the IQGAP1–PI(3)K pathway for survival and the MAPK pathway is not modulated by IQGAP1 in these cells. In support, a recent study indicates that WW peptide in a skin cancer model had no impact on Erk activity44.
Figure 7.
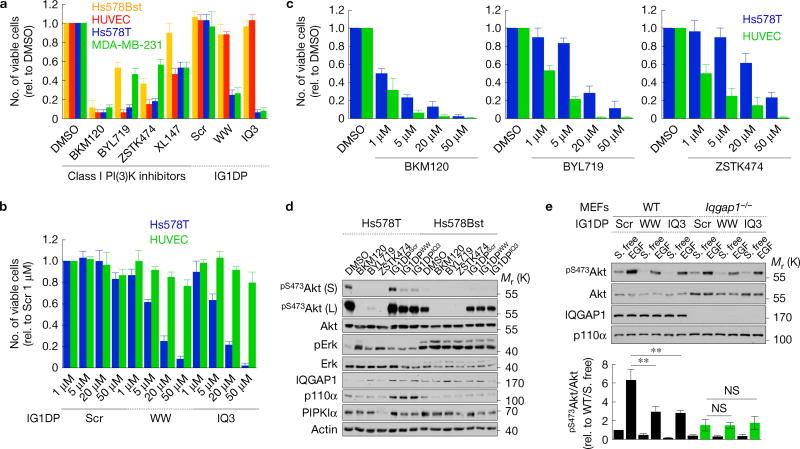
Inhibition of IQGAP1-mediated PtdIns(3,4,5)P3 synthesis is a mechanism for targeted cancer therapy. (a) Viability after IG1DP or PI(3)K inhibitor treatment in Hs578Bst, HUVEC, Hs578T (20 µM) and MDA-MB-231 cells (30 µM), mean ± s.d. of n = 3 independent experiments. (b,c) Viability of Hs578T and HUVEC cells after 1–50 µM IG1DP or PI(3)K inhibitor treatment, mean ± s.d. of n=3 independent experiments. (d) Immunoblots after treating with dimethylsulfoxide (DMSO; 5 µM), BKM120 (1 µM), BYL719 (5 µM), ZSTK474 (5 µM) or IG1DPs (20 µM) in Hs578T and Hs578Bst cells, representative of n = 3 independent experiments. (e) WT or Iqgap1−/− MEFs treated with 40 µM of the indicated peptides were stimulated with 20 ng ml−1 EGF for 15 min. Cell lysates were analysed by immunoblotting with the indicated antibodies (top). pS473Akt immunoblots were quantified and the graph is shown as mean ± s.d. of n=3 independent experiments (bottom). P = 0.030, 0.021. Paired Student’s t-tests were used for statistical analysis (*, P < 0.05; **, P < 0.01; NS, not significant). Source data for a–c,e can be found in Supplementary Table 1. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
To investigate the combined roles of PI(3)K in PtdIns(3,4,5)P3 generation compared with IQGAP1-selective PtdIns(3,4,5)P3 generation, Hs578Bst, HUVEC (normal), Hs578T and MDA-MB-231 (breast cancer) cells were treated with small-molecule PI(3)K inhibitors that are currently in clinical trials45 or the IG1DPs and cell viability was measured (Fig. 7). The PI(3)K inhibitors used in this study have submicromolar IC50, but in many cancer cell lines 1–10µM concentration is needed to effectively block the PI(3)K–Akt signalling pathway46,47. We used 1–5µM of PI(3)K inhibitors and at these concentrations observed cell killing. However, in some cell lines (including MDA-MB-231, MDA-MB-468 and Cal51), PI(3)K inhibitors showed an antiproliferative effect without killing. Under these conditions, PI(3)K inhibitors fully blocked the PI(3)K–Akt pathway (Fig. 7d) resulting in cell death or suppressing proliferation in both normal and cancer cells (Fig. 7a–c). In contrast, IG1DPs had a selective effect on cancer cells (Fig. 7a–d).
IG1DPs inhibit only a fraction of PtdIns(3,4,5)P3 generation in cells (Fig. 7d), consistent with the data indicating that PtdIns(3,4,5)P3 generation involves multiple pathways. IQGAP1 is required for IG1DPs inhibition of Akt activation as Iqgap1-null MEFs were not impacted by IG1DPs (Fig. 7e), validating the specificity of IG1DPs toward blockade of the IQGAP1–PI(3)K scaffold. The data indicate that the canonical PtdIns(3,4,5)P3 synthesis pathway is responsible for PtdIns(3,4,5)P3 synthesis from an accessible pool of PtdIns(4)P and PtdIns(4,5)P2, whereas the IQGAP1 pathway is for receptor-stimulated synthesis. IG1DPs inhibit only the IQGAP1 pathway, while PI(3)K inhibitors inhibit both. This is supported by data showing that IG1DPs partially reduced Akt phosphorylation in Hs578T breast cancer cells, but not in Hs578Bst normal mammary epithelial cells, leading to a selective antiproliferative effect on these cancer cells. In contrast, small-molecule PI(3)K inhibitors blocked both the IQGAP1-dependent and -independent pathways, leading to nonspecific loss of cells (Fig. 7a–d).
IQGAP1 is required for insulin signalling in vivo
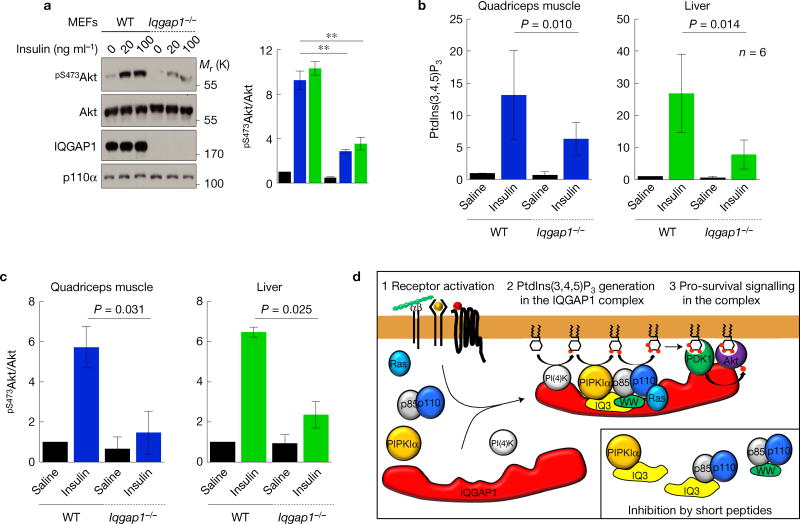
The PI(3)K–Akt pathway plays a key role in insulin signalling48. To assess the role of IQGAP1 in the insulin signalling, Akt phosphorylation was quantified in WT and Iqgap1-null MEFs stimulated by insulin. Akt activation was significantly reduced in Iqgap1-null MEFs (Fig. 8a). To establish the in vivo role, insulin or saline was injected into WT or Iqgap1-null mice. This demonstrated that insulin-stimulated PtdIns(3,4,5)P3 synthesis and Akt activation were significantly reduced in classic insulin-stimulated tissues, namely skeletal muscle and liver in the Iqgap1-null compared with WT mice (Fig. 8b,c). These data demonstrate that IQGAP1 is required for maximal activation of the PI(3)K–Akt pathway downstream of the insulin receptor in cells and mice.
Figure 8.
Insulin-stimulated PtdIns(3,4,5)P3 synthesis requires IQGAP1. (a) Insulin-stimulated pS473Akt was measured in MEFs, mean ± s.d. of n = 3 independent experiments. (b) WT or Iqgap1−/− mice were injected with insulin and the indicated tissues were analysed for PtdIns(3,4,5)P3. P value of paired t-test result is shown (n = 6 animals). P = 0.010, 0.014. (c) pS473Akt immunoblots with lysates from tissues were quantified (n = 3 samples). (d) A model for the IQGAP1-mediated PtdIns(3,4,5)P3 generation and downstream signalling. Source data for a–c can be found in Supplementary Table 1. Unprocessed original scans of blots are shown in Supplementary Fig. 7.
DISCUSSION
Here we define a concerted mechanism for agonist-stimulated PtdIns(3,4,5)P3 generation and signalling that is key for survival of some cancer cells. Agonist-activated PI(3)Ks use PtdIns(4,5)P2 as a substrate tosynthesize PtdIns(3,4,5)P3 andweshow thatIQGAP1 spatially organizes PtdIns(4,5)P2 and PtdIns(3,4,5)P3 producing kinases into a scaffolded complex. In this complex, PIPKIα and PI(3)K bind to the WW and IQ domains of IQGAP1. PIPKIα generates PtdIns(4,5)P2 that is channelled to PI(3)K, facilitating efficient PtdIns(3,4,5)P3 synthesis. The PtdIns(3,4,5)P3 in turn activates downstream effectors such as PDK1 and Akt that are also in the complex (Fig. 8d). The type IIIα PI(4)K, which generates PtdIns(4)P, is also required for Akt activation (Supplementary Fig. 1g,h), and is associated with IQGAP1 (Fig. 1a–c). IQGAP1 assembles all phosphoinositide kinases required for sequential phosphorylation of PtdIns to PtdIns(3,4,5)P3 and the effectors regulated by PtdIns(3,4,5)P3. Growth factors, extracellular matrix and other agonists stimulate the assembly and signalling of the IQGAP1–PI(3)K scaffold. This complex, summarized in Fig. 8d, has the intrinsic activities to spatially generate PtdIns(3,4,5)P3 from PtdIns leading to activation of PDK1 and Akt. Many receptors following agonist activation are rapidly internalized and continue signalling from endosomal compartments12,49, although these endosomal compartments contain little PtdIns(4)P or PtdIns(4,5)P2 (refs 1,50,51). As PtdIns is present in all internal membranes50,51, the IQGAP1–PI(3)K scaffold reveals a clear mechanism for efficient PtdIns(3,4,5)P3 generation and signalling from these compartments. Our results also support an IQGAP1-independent pathway(s) for PI(3)K generation of PtdIns(3,4,5)P3. The last pathways are sensitive to PtdIns(4)P- and PtdIns(4,5)P2-binding proteins, suggesting that they work on the accessible substrates in membranes. The PI(3)K pathway(s) that are independent of IQGAP1 may explain the survival of Iqgap1-null mice52. Significantly, mice lacking IQGAP1 are resistant to the development of some tumours19,27,39, consistent with a role in the PI(3)K pathway.
Alteration of the PI(3)K–Akt pathway is linked to many human diseases including cancers45,53,54. IQGAP1 is overexpressed in many cancers55 and correlated with tumorigenesis56,57. Recently, inhibition of PIPKIα was shown to significantly reduce Akt activation in prostate cancers15 and selectively kills cancer cells. This study supports our conclusion that PIPKIα directly integrates with the IQGAP1–PI(3)K pathway to regulate cell survival. Although the IQGAP1 pathway is present in normal and neoplastic cells, some cancer cells appear to depend onthis pathway for survival. These findings provide a potential target for cancer therapies. Unlike conventional PI(3)K inhibitors by ATP competition45, IG1DPs inhibit the IQGAP1-scaffolded kinases’ assembly and block Akt activation leading to selective cancer cell death. This suggests that some cancer cells become addicted to the IQGAP1-PI(3)K pathway for their survival58.
Insulin-stimulated PI(3)K signalling is implicated in many pathophysiological conditions including diabetes, cardiac function, ageing and others48,59–61. IQGAP1 is required for full insulin stimulation of PI(3)K and Akt activation that is important for many aspects of insulin function61. This establishes an in vivo role for the IQGAP1-scaffolded phosphoinositide kinases and effectors beyond cancers. IQGAP1 binding to the p85 subunit via the cSH2 domain (Fig. 2e) suggests a mechanism for activation of class I PI(3)Ks within the scaffold. As binding of the cSH2 domain of the p85 subunit to the catalytic p110 subunit suppresses kinase activity of PI(3)K, the interaction of the cSH2 domain with IQGAP1 could relieve the intramolecular inhibition activating PI(3)K in the IQGAP1 complex62. The interactions with both the P-BH-P and cSH2 may link multiple isoforms of class I PI(3)Ks explaining the diversity of agonist and receptors that require IQGAP1 for the PI(3)K signalling62.
In summary, here we have revealed a remarkable mechanism for PtdIns(3,4,5)P3 generation and signalling. This is achieved by the scaffold IQGAP1, where PI(4)KIIIa, PIPKIa and PI(3)K are linked for sequential phosphorylation of PtdIns for robust and efficient PtdIns(3,4,5)P3 synthesis that leads to the activation of associated effectors.
METHODS
Cell culture and constructs
MDA-MB-231, MDA-MB-468, HEK 293, Hs578T, Cal51, UACC812, SkBr3, HaCaT, Cos7 and MEF cells were purchased from ATCC and maintained in DMEM supplemented with 10% fetal bovine serum (Gibco). MCF-763, HUVEC64 (a gift from A. Rapraeger, University of Wisconsin-Madison, USA), MCF10A65 and Hs578Bst66 cells were maintained as described. All the cell lines used in this study are routinely tested for mycoplasma contamination and mycoplasma-negative cells were used. No cell lines used in this study were found in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. The cell lines were not authenticated. For cell viability assays, cells were switched to DMEM supplemented with 10% fetal bovine serum at least 24 h before assay. The PIPKIα and IQGAP1 constructs used for this work have been described previously24,67. The WW-IQ region was amplified using 5′-GATGGAT CCGGTGAAACTTACCACAGTGATCTTGCT-3′ (Forward) and 5′-GATAAG CTTCTAATCCTCAGCATTGATGAGAGTCTTGTA-3′ (Reverse) primers and cloned in pET28a vector (Novagen) within BamHI and HindIII enzyme sites. Bovine p85oc constructs (Fig. 2e) (gifts from D. Anderson, University of Saskatchewan, Canada) were described previously28. p85 (Fig. 2b),p100α, constitutively active Akt1 (Fig. 6f), Rac1 and Cdc42 (Supplementary Fig. 6f) constructs used in the study were purchased from Addgene and subcloned in pcDNA3.1 vector (Invitrogen). The GFP-PH domain constructs (gifts from T Balla, National Institutes of Health, USA) (Fig. 4f and Supplementary Fig. 3c,d) were described previously29,68–70. The 17Insert IQGAP1 construct (Fig. 1e and Supplementary Fig. 4) was generated by a series of site-directed mutagenesis polymerase chain reactions to insert 5′-C ATCTCTGGGGTGACTGCCCATATAACCGAGAACAGCTGTGGCTGGTC GAC-3′. Constructs were transfected in cells by a lipid-based delivery system from Mirus (MDA-MB-231 cells) or Invitrogen (other cell lines) according to the manufacturer’s instructions. Typically, 3–9 µg of DNA and 6–12 µl of lipid were used for transfecting in 6-well plates. In all overexpression experiments, GFP construct was transfected in parallel to monitor the transfection efficiency. Cells having at least 70% transfection efficiency were used for further analysis.
Stable cell line generation
To generate stable MDA-MB-231 and Hs578T cell lines expressing shRNAs against human IQGAP1, PIPKIoc or vector control, a pLL3.7 vector-based lentiviral delivery system was used. pLL3.7 lentiviral vector was cloned to express shRNAs against IQGAP171 and PIPKα72. After infection, cells were passaged at least 5 times before sorting GFP-positive cells in a cell sorter to select cells expressing shRNAs as pLL3.7 vector also expresses GFP bicistronically73. For generation of stable MEFs, cells were infected with retrovirus for 24 h. Then, cells expressing GFP-IQGAP1 were first selected for GFP expression, and then further sorted by expression level. For generation of stable Hs578T cells expressing GFP-tagged PH domains, cells transfected with lipid were selected by 1.2mgml−1 neomycin. Then, GFP-positive cells were further selected in a cell sorter.
Antibodies and siRNAs
All antibody information can be found in Supplementary Table 2. Polyclonal and monoclonal antibodies against total and PIPKIoc and PIPKIγ were produced as described previously74,75. Pooled siRNAs used in the study were obtained from Dharmacon (Fig. 1d). Single siRNAs targeting IQGAP1 (either 5′-G GAAAGCUCUGGCAAUUUAUU-3′ or 5′-GAACGUGGCUUAUGAGUACUU-3′) and PIPKIα (either 5′-AAGTTGGAGCACTCTTGG-3′ or 5′-GCACATTATCC CTACCTTA-3′) were used elsewhere71,72. siRNA targeting the 3′UTR of PIPKIα (5′-UGACUCCUGGAAGAAUACUCCUGUA-3′) was purchased from Thermo Fisher Scientific. Non-targeting siRNA (Dharmacon) was used as a control. siRNAs were delivered to cells by RNAiMAX reagent (Thermo Fisher Scientific) and knockdown efficiency was determined by immunoblotting. Knockdown efficiency greater than 80% was required to observe phenotypic changes in the study. This is consistent with a recent report that at least 80% knockdown of IQGAP1 is required to observe reduced Erk activity44.
Replating assay on extracellular matrix
To measure Akt and FAK phosphorylation in response to integrin activation on type I collagen and fibronectin (Figs 4f and 5a,b,f,g and Supplementary Figs 3d and 5a–e) replating assays were performed. Briefly, cells were serum starved for at least 4h and then lifted by treating with trypsin-EDTA for a short time (less than 1 min). Cells were treated with serum-free DMEM containing 0.2% bovine serum albumin and pelleted to remove trypsin– EDTA. For the remaining steps, cells were maintained in serum-free DMEM containing 0.2% bovine serum albumin. Cells were suspended at least for 1 h at 37°C by rotation before replating on tissue culture plates coated with 10 µg ml−1 type I collagen (Sigma Aldrich) or fibronectin (Thermo Fisher Scientific). After incubation for 15 to 60 min, cells were lysed and further analysed by immunoblotting or immunoprecipitation.
Cell viability assay
Cells were plated on 6-well plates at 30–50% confluency at the time of treatment. Cells were changed with fresh media containing class I PI(3)K inhibitors or IQGAP1-derived peptides at the indicated concentrations for the indicated times (24 to 72 h) (Figs 6f–h and 7a–e and Supplementary Fig. 6c–f). Dead cells floating in the culture medium were removed by washing twice with PBS. Residual PBS was completely removed and cells were lifted by incubating with trypsin-EDTA for 10 m. Cells were stained with 0.4% trypan blue solution (Thermo Fisher Scientific) and counted with a haemocytometer. The number of trypan blue-negative cells was counted and used for the statistical analysis.
Immunoprecipitation and immunoblotting
Cells were lysed in a buffer containing 1% Brij58, 150 mM NaCl, 20 mM HEPES, pH 7.4,2 mM MgCl2, 2 mM CaCl2,1 mM Na3VO4, 1 mM Na2MoO4 and protease inhibitors. The protein concentration of lysates was measured by the BCA method (Pierce) and equal amounts of protein were used for further analysis. All antibodies were diluted in a 1:1,000 ratio for immunoblotting. For immunoprecipitation, 0.5 to 1 mg of proteins were incubated with 1 µgof antibodies at 4°C for 8 hand then incubated with a 50% slurry of Protein G Sepharose (GE Life Sciences) for another 2h. After washing 5 times with lysis buffer, the protein complex was eluted with SDS sample buffer. For immunoblotting, 5 to 20 µg of proteins were loaded. After developing immunoblots, the film was scanned using a transmitted light scanner (resolution: 600 dpi). Protein bands were quantified using ImageJ, and statistical analysis of the data was performed with Microsoft Excel. The statistical analysis was performed using data from at least three independent experiments.
In vitro binding assay
Recombinant proteins were expressed in the BL21 E. coli strain. GST-tagged proteins were then purified with GST Sepharose 4B (GE Life Sciences) and His-tagged proteins were purified with His-Bind Resin (Novagen). Recombinant PI(3)K (p110a and p85α) proteins were purchased from Echelon Biosciences. GST-tagged proteins were incubated with glutathione beads before binding assays. The binding assay was performed in the lysis buffer used for immunoprecipitation by adding 10 nM to 5 µM of His-tagged proteins and 20 µl of GST-tagged protein-bound glutathione beads. After incubation for 1 h at 25 °C, unbound proteins were washed out and the protein complex was analysed by immunoblotting. For the triple binding assay with recombinant GST-IQGAP1, His-PIPKIα and PI(3)K (His-p110oc and untagged p85oc), PIPKIα was pulled down with an anti-PIPKIα antibody pre-bound on Protein G Sepharose beads. For binding assays with p85α, wild-type or deletion mutants of bovine p85α28 were expressed in HEK293 cells and cell lysates were used for binding experiments with recombinant IQGAP1 proteins.
Immunofluorescence microscopy
Glass coverslips were coated with 10µg ml−1 collagen, fibronectin or 10% serum before seeding cells. Cells were grown on coverslips placed inside 6-well plates until experimental manipulation. Immunostaining of PtdIns(3,4,5)P3 was performed as previously described28,76,77 with modifications. Briefly, cells grown in medium were rapidly fixed by adding an equal volume of 8% paraformaldehyde and 0.5% glutaraldehyde to medium for 15 min at room temperature. After a 30 min wash with PBS containing 50 mM NH4Cl, cells were permeabilized and blocked with a solution of buffer A (20 mM PIPES, pH 6.8, 135 mM NaCl, 5 mM KCl) containing 0.5% saponin and 5 vol% FBS for 45 min at room temperature. Primary antibodies were incubated in a solution of buffer A containing 0.1% saponin and 5 vol% of FBS for 12hat4°C. Concentrations of 2–4 µg ml−1 primary antibodies were used. After a 30 min wash with buffer A, fluorophore-conjugated secondary antibodies were incubated in a solution of buffer A containing 0.1% saponin and 5 vol% of FBS for 1 h at room temperature. Then, cells were washed with buffer A for 45 m at room temperature before post-fixation with 2% paraformaldehyde and 0.125% glutaraldehyde for 10 m at room temperature. Coverslips were washed 5 times with PBS containing 50 mM NH4Cl and once with distilled water. Fluorescence microscopy was performed using a 40 or 60 × plan-fluor objective on a Nikon Eclipse TE2000U equipped with a Photometrics CoolSNAP ES CCD (charge-coupled device) camera. Images were captured using MetaMorph v6.3 (Molecular Devices). Images were exported to Photoshop CS2 (Adobe) for final processing and assembly.
Cellular phosphoinositide measurement
Lipid extraction and cellular phospho-inositide measurement were performed using a kit from Echelon Biosciences according to the manufacturer’s instructions. Briefly, cells were lysed in ice-cold trichloroacetic acid (TCA, 0.5 M). Pellets were washed in 5% TCA/1mM EDTA and neutral lipids were extracted with CH3OH/CHCl3 (2:1) and discarded. Acidic lipids were extracted with CH3OH/CHCl3/HCl (80:40:1), recovered by phase-split. Recovered lipids were dried and resuspended for competitive ELISA analysis. PtdIns(3)P, PtdIns(4)P, PtdIns(4,5)P2 and PtdIns(3,4,5)P3 levels were quantified by competitive ELISA (Echelon Biosciences). For measuring PtdIns(4)P and PtdIns(4,5)P2, cells were harvested from 10 cm plates. For PtdIns(3)P and PtdIns(3,4,5)P3, cells were harvested from 15 cm plates.
In vitro phosphoinositide kinase assay
In vitro phosphoinositide kinase assays were performed as described previously74,78 with modification. Briefly, 0.02–0.1 µM of recombinant IQGAP1, PIPKIα and PI(3)K were incubated with 250 µM phosphoinositide in liposomes in a kinase buffer (50mM Tris HCl, pH 8.0, 10mM MgCl2, 0.5mM EDTA). The liposomes containing 10 mol% of phosphoinositides were generated as described previously24. Kinase reaction was initiated by adding 20 µM ATP and incubated for 30m at 25°C. A 50 µl reaction was terminated by adding 100 µl 1M HCl and 200µl CHCl3/CH3OH (1:1). Extracted lipids were dried and PtdIns(4,5)P2 and PtdIns(3,4,5)P3 levels were quantified by competitive ELISA (Echelon Biosciences) as above.
PtdIns(4)P liposomes that were used as a substrate contain 30% PC, 30% PE, 30% PS and 10% PtdIns(4)P in a molar ratio. Liposomes containing PtdIns(4,5)P2 were made by decreasing the PC content. For example, liposomes containing 5% PtdIns(4,5)P2 were generated with 25% PC, 30% PE, 30% PS, 10% PtdIns(4)P and 5% PtdIns(4,5)P2. All phospholipids (natural, porcine brain) were purchased from Avanti Polar Lipids. Mixed phospholipids were dried and rehydrated with 50mM Tris, pH 8.0 for kinase assays. Hydrated lipids were subjected to at least 5 cycles of freeze thawing in liquid nitrogen followed by 1min bath sonication and extrusion through a 0.1 µm filter.
PtdIns(4)P and PtdIns(4,5)P2 lipids used in kinase assays (Fig. 4 and Supplementary Fig. 3a) were from porcine brain (Avanti Polar Lipids) and their purity was extensively tested for the presence of contamination with PtdIns(3)P or PtdIns(3,4,5)P3. The identity and purity of brain PtdIns(4)P was tested using mass spectrometry, proton NMR, phosphorus NMR, TLC, and oxidation by UV/VIS. In addition to confirming the identity of brain PtdIns(4)P, the mass spectrometry also confirms the absence of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 species as their m/z are not present in the release spectra. As for the presence or absence of PtdIns(3)P, phosphorus NMR was used. There are distinctly different shifts for the phosphorus peaks in PtdIns(3)P and PtdIns(4)P. PtdIns(3)P has two peaks, one at approximately 0.1 and another at approximately −0.75. PtdIns(4)P contains two peaks, one around −0.45 and the second at around 2.5. These shifts can fluctuate slightly, but the pattern is consistent across synthetic and natural PIPs. Brain PtdIns(4,5)P2 was analysed by mass spectrometry, TLC, proton NMR, phosphorus NMR, and oxidation by UV/VIS. The phosphorus NMR peaks for PtdIns(4,5)P2 are at approximately 2.4, 1.7 and −0.4. The shifts for PtdIns(3,4,5)P3 are typically 0.7, 0.0 and 0.8. None of these overlaps with the peaks present in the PtdIns(4,5)P2 species.
PtdIns(3,4,5)P3 measurement in mouse tissues
Male 16–19-week-old wild-type or Iqgap1−/− mice (129 background) were fasted for4 h and received an intraperitoneal injection of saline or insulin (5Ukg−1). Mice were euthanized by CO2, and 15 min post-injection, tissues (liver and quadriceps muscle) were harvested and immediately frozen in a dry-ice ethanol bath and stored on dry ice until being placed in a −80°C degree freezer. Lipids were extracted from mouse tissues with the TCA extraction method as above. Lipids extracted from equal weights (approximately 100 mg) of mouse tissues were analysed for PtdIns(3,4,5)P3 using a competitive ELISA (Echelon Biosciences) and normalized by weight of tissue. Statistical analysis was performed with 6 paired cases.
Mice were bred in the animal facility at the National Institutes of Health (NIH) and maintained according to NIH guidelines. The studies were performed with approval of the NIH Animal Care and Use Committee.
Peptide production and treatment in cells
Octo-arginine-conjugated WW domain and IQ motif peptides were synthesized by Genscript. Peptides were dissolved in water to a 2.5–5mM working solution. Small-molecule class I PI(3)K inhibitors were purchased from Selleckchem and dissolved in dimethylsulfoxide to a 5mM working solution. Peptide and PI(3)K inhibitors were added directly into the tissue culture medium at the concentrations indicated. Dimethyl sulfoxide was used as a vehicle control.
Statistics and reproducibility
ImageJ software was used to quantify the intensity of immunoblotting images. Non-saturating exposure of immunoblot films was used for quantification with the appropriate loading controls as standards. Two-tailed unpaired t-tests were used for pairwise significance unless otherwise indicated. For consistency in these comparisons, the following denotes significance in figures: *,P <0.05; **,P <0.01. We note that no power calculations were used. Sample sizes were determined on the basis of previously published experiments where differences were observed. Each experiment was repeated independently at least three times and sample sizes and number of repeats are defined in each figure legend. We used 3–7 cases for statistical analysis. No samples were excluded. Investigators were blinded to allocation during experiments and outcome assessment.
Data availability
Source data for Figs 1d–f, 3f, 4a–f, 5b,e–g, 6c–h, 7a–c,e, 8a–c and Supplementary Figs 1a,d–h, 3a,d,f, 4b, 5b–f and 6c,e,f have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author on request.
Supplementary Material
Acknowledgments
We would like to thank T. Balla for the PH domain constructs, and all members of the R.A.A. and D.B.S. laboratories, A. Rapreager, P. Lambert, M. Sussman and R. Kimple (University of Wisconsin-Madison) for helpful discussions. This work was supported by National Institutes of Health (NIH) grants to R.A.A., the Intramural Research Program of the NIH to D.B.S. and American Heart Association fellowships to S.C.
Footnotes
Note: Supplementary Information is available in the online version of the paper
AUTHOR CONTRIBUTIONS
S.C. and R.A.A. designed experiments. S.C., A.C.H., S.S. and N.T. performed experiments and analysed data. S.C., A.C.H., D.B.S. and R.A.A. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Di Paolo G, DeCamilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 2.Sun Y, Thapa N, Hedman AC, Anderson RA. Phosphatidylinositol 4,5-bisphosphate: targeted production and signaling. Bioessays. 2013;35:513–522. doi: 10.1002/bies.201200171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 4.Kadamur G, Ross EM. Mammalian phospholipase C. Annu. Rev. Physiol. 2013;75:127–154. doi: 10.1146/annurev-physiol-030212-183750. [DOI] [PubMed] [Google Scholar]
- 5.Choi S, Thapa N, Tan X, Hedman AC, Anderson RA. PIP kinases define PI4, 5P signaling specificity by association with effectors. Biochim. Biophys. Acta. 2015;1851:711–723. doi: 10.1016/j.bbalip.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Bout I, Divecha N. PIP5K-driven PtdIns(4,5)P2 synthesis: regulation, cellular functions. J. Cell Sci. 2009;122:3837–3850. doi: 10.1242/jcs.056127. [DOI] [PubMed] [Google Scholar]
- 7.Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 2010;11:329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- 8.Auger KR, Serunian LA, Soltoff SP, Libby P, Cantley LC. PDGF-dependent tyrosine phosphorylation stimulates production of novel polyphosphoinositides in intact cells. Cell. 1989;57:167–175. doi: 10.1016/0092-8674(89)90182-7. [DOI] [PubMed] [Google Scholar]
- 9.Stephens LR, Hughes KT, Irvine RF. Pathway of phosphatidylinositol(3,4,5)-trisphosphate synthesis in activated neutrophils. Nature. 1991;351:33–39. doi: 10.1038/351033a0. [DOI] [PubMed] [Google Scholar]
- 10.Stephens LR, Jackson TR, Hawkins PT. Agonist-stimulated synthesis of phosphatidylinositol(3,4,5)-trisphosphate: a new intracellular signalling system? Biochim. Biophys. Acta. 1993;1179:27–75. doi: 10.1016/0167-4889(93)90072-w. [DOI] [PubMed] [Google Scholar]
- 11.McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 12.Sun Y, Hedman AC, Tan X, Schill NJ, Anderson RA. Endosomal type Igamma PIP 5-kinase controls EGF receptor lysosomal sorting. Dev. Cell. 2013;25:144–155. doi: 10.1016/j.devcel.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao YS, et al. Essential and unique roles of PIP5K-γ and -α in Fcgamma receptor-mediated phagocytosis. J. Cell Biol. 2009;184:281–296. doi: 10.1083/jcb.200806121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling K, et al. Type I gamma phosphatidylinositol phosphate kinase modulates adherens junction and E-cadherin trafficking via a direct interaction with mu 1B adaptin. J. Cell Biol. 2007;176:343–353. doi: 10.1083/jcb.200606023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Semenas J, et al. The role of PI3K/AKT-related PIP5K1α and the discovery of its selective inhibitor for treatment of advanced prostate cancer. Proc. Natl Acad. Sci. USA. 2014;111:E3689–E3698. doi: 10.1073/pnas.1405801111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith JM, Hedman AC, Sacks DB. IQGAPs choreograph cellular signaling from the membrane to the nucleus. Trends Cell Biol. 2015;25:171–184. doi: 10.1016/j.tcb.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hedman AC, Smith JM, Sacks DB. The biology of IQGAP proteins: beyond the cytoskeleton. EMBO Rep. 2015;16:427–446. doi: 10.15252/embr.201439834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren JG, Li Z, Sacks DB. IQGAP1 modulates activation of B-Raf. Proc. Natl Acad. Sci. USA. 2007;104:10465–10469. doi: 10.1073/pnas.0611308104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jameson KL, et al. IQGAP1 scaffold-kinase interaction blockade selectively targets RAS-MAP kinase-driven tumors. Nat. Med. 2013;19:626–630. doi: 10.1038/nm.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 21.Sbroggio M, et al. ERK1/2 activation in heart is controlled by melusin, focal adhesion kinase and the scaffold protein IQGAP1. J. Cell Sci. 2011;124:3515–3524. doi: 10.1242/jcs.091140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sbroggio M, et al. IQGAP1 regulates ERK1/2 and AKT signalling in the heart and sustains functional remodelling upon pressure overload. Cardiovasc. Res. 2011;91:456–464. doi: 10.1093/cvr/cvr103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt VA, Chiariello CS, Capilla E, Miller F, Bahou WF. Development of hepatocellular carcinoma in Iqgap2-deficient mice is IQGAP1 dependent. Mol. Cell. Biol. 2008;28:1489–1502. doi: 10.1128/MCB.01090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi S, et al. IQGAP1 is a novel phosphatidylinositol 4,5 bisphosphate effector in regulation of directional cell migration. EMBO J. 2013;32:2617–2630. doi: 10.1038/emboj.2013.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy M, Li Z, Sacks DB. IQGAP1 is a scaffold for mitogen-activated protein kinase signaling. Mol. Cell. Biol. 2005;25:7940–7952. doi: 10.1128/MCB.25.18.7940-7952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat. Rev. Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- 27.Li Z, Sacks DB. Elucidation of the interaction of calmodulin with the IQ motifs of IQGAP1. J. Biol. Chem. 2013;278:4347–4352. doi: 10.1074/jbc.M208579200. [DOI] [PubMed] [Google Scholar]
- 28.Chagpar RB, et al. Direct positive regulation of PTEN by the p85 subunit of phosphatidylinositol 3-kinase. Proc. Natl Acad. Sci. USA. 2009;107:5471–5476. doi: 10.1073/pnas.0908899107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammond GR, et al. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 2012;337:727–730. doi: 10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balla A, et al. Maintenance of hormone-sensitive phosphoinositide pools in the plasma membrane requires phosphatidylinositol 4-kinase IIIα. Mol. Biol. Cell. 2008;19:711–721. doi: 10.1091/mbc.E07-07-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stenmark H, Aasland R, Driscoll PC. The phosphatidylinositol 3-phosphate-binding FYVE finger. FEBS Lett. 2002;513:77–84. doi: 10.1016/s0014-5793(01)03308-7. [DOI] [PubMed] [Google Scholar]
- 32.Shimaya A, Kovacina KS, Roth RA. On the mechanism for neomycin reversal of wortmannin inhibition of insulin stimulation of glucose uptake. J. Biol. Chem. 2004;279:55277–55282. doi: 10.1074/jbc.M411540200. [DOI] [PubMed] [Google Scholar]
- 33.Devreotes P, Janetopoulos C. Eukaryotic chemotaxis: distinctions between directional sensing and polarization. J. Biol. Chem. 2003;278:20445–20448. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- 34.Huang YE, et al. Receptor-mediated regulation of PI3Ks confines PI(3,4,5)P3 to the leading edge of chemotaxing cells. Mol. Biol. Cell. 2003;14:1913–1922. doi: 10.1091/mbc.E02-10-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iijima M, Huang YE, Devreotes P. Temporal and spatial regulation of chemotaxis. Dev. Cell. 2002;3:469–478. doi: 10.1016/s1534-5807(02)00292-7. [DOI] [PubMed] [Google Scholar]
- 36.Chen HC, Appeddu PA, Isoda H, Guan JL. Phosphorylation of tyrosine 397 in focal adhesion kinase is required for binding phosphatidylinositol 3-kinase. J Biol. Chem. 1996;271:26329–26334. doi: 10.1074/jbc.271.42.26329. [DOI] [PubMed] [Google Scholar]
- 37.Kohn AD, Takeuchi F, Roth RA. Akt, a pleckstrin homology domain containing kinase, is activated primarily by phosphorylation. J. Biol. Chem. 1996;271:21920–21926. doi: 10.1074/jbc.271.36.21920. [DOI] [PubMed] [Google Scholar]
- 38.Tan J, et al. PDK1 signaling toward PLK1-MYC activation confers oncogenic transformation, tumor-initiating cell activation, and resistance to mTOR-targeted therapy. Cancer Discov. 2013;3:1156–1171. doi: 10.1158/2159-8290.CD-12-0595. [DOI] [PubMed] [Google Scholar]
- 39.Mataraza JM, et al. IQGAP1 promotes cell motility and invasion. J. Biol. Chem. 2003;278:41237–41245. doi: 10.1074/jbc.M304838200. [DOI] [PubMed] [Google Scholar]
- 40.Fritsch R, et al. RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell. 2013;153:1050–1063. doi: 10.1016/j.cell.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsunaga H, Kubota K, Inoue T, Isono F, Ando O. IQGAP1 selectively interacts with K-Ras but not with H-Ras and modulates K-Ras function. Biochem. Biophys. Res. Commun. 2014;444:360–364. doi: 10.1016/j.bbrc.2014.01.041. [DOI] [PubMed] [Google Scholar]
- 42.McNulty DE, Li Z, White CD, Sacks DB, Annan RS. MAPK scaffold IQGAP1 binds the EGF receptor and modulates its activation. J. Biol. Chem. 2011;286:15010–15021. doi: 10.1074/jbc.M111.227694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy M, Li Z, Sacks DB. IQGAP1 binds ERK2 and modulates its activity. J. Biol. Chem. 2004;279:17329–17337. doi: 10.1074/jbc.M308405200. [DOI] [PubMed] [Google Scholar]
- 44.Monteleon CL, et al. IQGAP1 and IQGAP3 serve individually essential roles in normal epidermal homeostasis and tumor progression. J. Invest. Dermatol. 2015;135:2258–2265. doi: 10.1038/jid.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat. Rev. Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 46.Vora SR, et al. CDK 4/6 inhibitors sensitize PIK3CA mutant breast cancer to PI3K inhibitors. Cancer Cell. 2014;26:136–149. doi: 10.1016/j.ccr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pei Y, et al. An animal model of MYC-driven medulloblastoma. Cancer Cell. 2011;21:155–167. doi: 10.1016/j.ccr.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat. Rev. Mol. Cell Biol. 2006;7:85–96. doi: 10.1038/nrm1837. [DOI] [PubMed] [Google Scholar]
- 49.Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 2009;10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li S, Wang Q, Chakladar A, Bronson RT, Bernards A. Gastric hyperplasia in mice lacking the putative Cdc42 effector IQGAP1. Mol. Cell. Biol. 2000;20:697–701. doi: 10.1128/mcb.20.2.697-701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo J, Manning BD, Cantley LC. Targeting the PI3K–Akt pathway in human cancer: rationale and promise. Cancer Cell. 2003;4:257–262. doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- 54.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 2009;8:627–644. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White CD, Brown MD, Sacks DB. IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett. 2009;583:1817–1824. doi: 10.1016/j.febslet.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feigin ME, Xue B, Hammell MC, Muthuswamy SK. G-protein-coupled receptor GPR161 is overexpressed in breast cancer and is a promoter of cell proliferation and invasion. Proc. Natl Acad. Sci. USA. 2014;111:4191–4196. doi: 10.1073/pnas.1320239111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jadeski L, Mataraza JM, Jeong HW, Li Z, Sacks DB. IQGAP1 stimulates proliferation and enhances tumorigenesis of human breast epithelial cells. J. Biol. Chem. 2008;283:1008–1017. doi: 10.1074/jbc.M708466200. [DOI] [PubMed] [Google Scholar]
- 58.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68:3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 59.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alessi DR, Downes CP. The role of PI 3-kinase in insulin action. Biochim. Biophys. Acta. 1998;1436:151–164. doi: 10.1016/s0005-2760(98)00133-7. [DOI] [PubMed] [Google Scholar]
- 61.Yao H, Han X, Han X. The cardioprotection of the insulin-mediated PI3K/Akt/mTOR signaling pathway. Am. J. Cardiovasc. Drugs. 2014;14:433–442. doi: 10.1007/s40256-014-0089-9. [DOI] [PubMed] [Google Scholar]
- 62.Burke JE, Williams RL. Synergy in activating class I PI3Ks. Trends Biochem. Sci. 2015;40:88–100. doi: 10.1016/j.tibs.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 63.Soule HD, Vazguez J, Long A, Albert S, Brennan M. A human cell line from a pleural effusion derived from a breast carcinoma. J. Natl Cancer Inst. 1973;51:1409–1416. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- 64.Beauvais DM, Ell BJ, McWhorter AR, Rapraeger AC. Syndecan-1 regulates αvβ3 and αvβ5 integrin activation during angiogenesis and is blocked by synstatin, a novel peptide inhibitor. J. Exp. Med. 2009;206:691–705. doi: 10.1084/jem.20081278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soule HD, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 66.Hackett AJ, et al. Two syngeneic cell lines from human breast tissue: the aneuploid mammary epithelial (Hs578T) and the diploid myoepithelial (Hs578Bst) cell lines. J. Natl Cancer Inst. 1977;58:1795–1806. doi: 10.1093/jnci/58.6.1795. [DOI] [PubMed] [Google Scholar]
- 67.Loijens JC, Anderson RA. Type I phosphatidylinositol-4-phosphate 5-kinases are distinct members of this novel lipid kinase family. J. Biol. Chem. 1996;271:32937–32943. doi: 10.1074/jbc.271.51.32937. [DOI] [PubMed] [Google Scholar]
- 68.Kim YJ, Guzman-Hernandez ML, Balla T. A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev. Cell. 2011;21:813–824. doi: 10.1016/j.devcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Balla A, Tuymetova G, Tsiomenko A, Varnai P, Balla T. A plasma membrane pool of phosphatidylinositol 4-phosphate is generated by phosphatidylinositol 4-kinase type-III α: studies with the PH domains of the oxysterol binding protein and FAPP1. Mol. Biol. Cell. 2005;16:1282–1295. doi: 10.1091/mbc.E04-07-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Varnai P, Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sakurai-Yageta M, et al. The interaction of IQGAP1 with the exocyst complex is required for tumor cell invasion downstream of Cdc42 and RhoA. J. Cell Biol. 2008;181:985–998. doi: 10.1083/jcb.200709076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mellman D, et al. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- 73.Thapa N, et al. Phosphoinositide signaling regulates the exocyst complex and polarized integrin trafficking in directionally migrating cells. Dev. Cell. 2012;22:116–130. doi: 10.1016/j.devcel.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schill NJ, Anderson RA. Two novel phosphatidylinositol-4-phosphate 5-kinase type Iγ splice variants expressed in human cells display distinctive cellular targeting. Biochem. J. 2009;422:473–482. doi: 10.1042/BJ20090638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Doughman RL, Firestone AJ, Wojtasiak ML, Bunce MW, Anderson RA. Membrane ruffling requires coordination between type Iα phosphatidylinositol phosphate kinase and Rac signaling. J. Biol. Chem. 2003;278:23036–23045. doi: 10.1074/jbc.M211397200. [DOI] [PubMed] [Google Scholar]
- 76.Hammond GR, Schiavo G, Irvine RF. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2) Biochem. J. 2009;422:23–35. doi: 10.1042/BJ20090428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sharma VP, DesMarais V, Sumners C, Shaw G, Narang A. Immunostaining evidence for PI(4,5)P2 localization at the leading edge of chemoattractant-stimulated HL-60 cells. J. Leukoc. Biol. 2008;84:440–447. doi: 10.1189/jlb.0907636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bazenet CE, Anderson RA. Phosphatidylinositol-4-phosphate 5-kinases from human erythrocytes. Methods Enzymol. 1992;209:189–202. doi: 10.1016/0076-6879(92)09023-v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for Figs 1d–f, 3f, 4a–f, 5b,e–g, 6c–h, 7a–c,e, 8a–c and Supplementary Figs 1a,d–h, 3a,d,f, 4b, 5b–f and 6c,e,f have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author on request.