SUMMARY
Protein partner of Sans-fille (PPS) and its human homolog DIDO mediate diverse chromatin activities including the regulation of stemness genes in embryonic stem cells and splicing in Drosophila. Here, we show that the PHD fingers of PPS and DIDO recognize the histone mark H3K4me3 in a pH-dependent manner: the binding is enhanced in high pH values but is decreased at low pH. Structural analysis reveals that the pH-dependency is due to the presence of a histidine residue in the K4me3-binding aromatic cage of PPS. The pH-dependent mechanism is conserved in DIDO but is lost in yeast Bye1. Acidification of cells leads to the accelerated efflux of endogenous DIDO, indicating the pH-dependent sensing of H3K4me3 in vivo. This novel mode for the recognition of H3K4me3 establishes the PHD fingers of PPS and DIDO as unique epigenetic readers and high pH sensors, and suggests a role for the histidine switch during mitosis.
Keywords: PTM, PHD finger, PPS, epigenetic, methylation, chromatin, histone binding
eTOC Blurb
Tencer et al. identified a novel pH-dependent mechanism by which epigenetic readers, the PHD fingers of PPS and DIDO, recognize the histone mark H3K4me3. The pH sensing ability might be necessary for normal biological processes and those characterized by altered cellular pH.
INTRODUCTION
Male-exon skipping splicing of the sex determination gene Sex-lethal (Sxl) in Drosophila relies on Protein partner of Sans-fille (PPS) (Johnson et al., 2010). PPS associates with spliceosomal RNAs including the U1 snRNP protein Sans-fille (SNF) to mediate Sxl splicing autoregulation (Johnson et al., 2010). Alternative splicing of the Sxl pre-mRNA determines gender during development in Drosophila, producing protein-encoding mRNAs in females but yielding inactive and truncated mRNAs in males. Once a functional Sxl protein is generated, it orchestrates female specific development and behavior through regulating downstream genetic programs. Bioinformatics and primary sequence analyses reveal a set of homologous to PPS proteins, including human DIDO (Death Inducer Obliterator 3) and PHF3 (PHD finger protein 3), and S. cerevisiae Bye1 (Bypass of Ess1). All three PPS homologs have been implicated in gene regulation, with DIDO regulating stemness genes in embryonic stem cells (Futterer et al., 2005; Gatchalian et al., 2013) and Bye1 directly associating with the RNA polymerase II in yeast (Kinkelin et al., 2013; Pinskaya et al., 2014). Aberrant functions of human DIDO and PHF3 are linked to cancer (Futterer et al., 2005; Pallasch et al., 2005).
Fly PPS and its homologs are characterized by similar domain architecture. PPS contains a plant homeodomain (PHD) finger, Brahma and Kismet (BRK), a transcription elongation factor S-II subunit M (TFSIIM) domain, and a spen paralogue and orthologue (SPOC) domain (Johnson et al., 2010) (Fig. 1a). Although the precise biological roles of the BRK, TFSIIM, and SPOC modules of PPS remain unclear, these structured regions are usually found in proteins involved in transcription and developmental signaling in higher eukaryotes (Kettenberger et al., 2003). A set of zinc-coordinating PHD fingers function as readers of post-translational modifications (PTMs) in histones, including methylated lysine residues in histone H3 (Musselman and Kutateladze, 2011; Musselman et al., 2012), and the PHD fingers of DIDO and Bye1 have been shown to interact with trimethylated lysine 4 of histone H3 (H3K4me3) (Gatchalian et al., 2013; Kinkelin et al., 2013). However the PHD finger of PHF3 does not recognize either modified or unmodified histone H3 tail (Gatchalian et al., 2013), and it is unclear whether PPS has the histone binding activity.
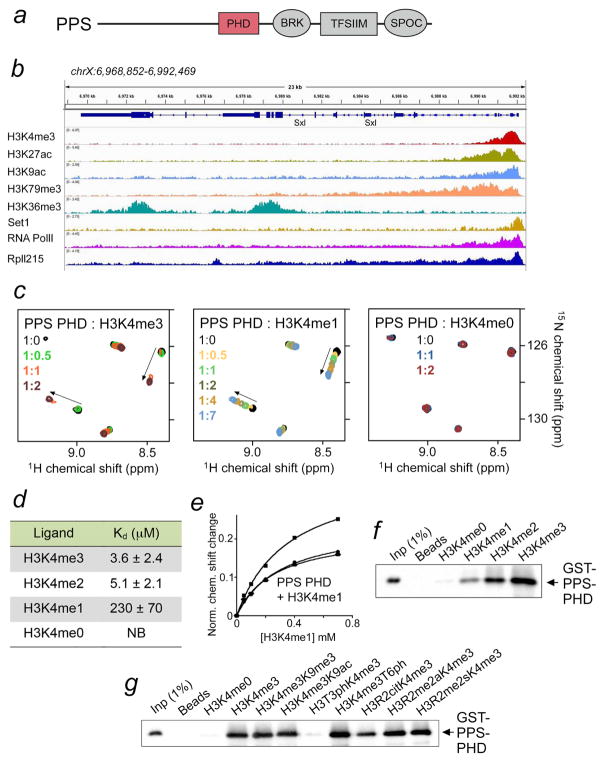
Figure 1. PPS-PHD selects for histone H3K4me3.
(a) PPS domain composition. (b) Snapshot of a 23 kb genomic region containing the Drosophila Sxl gene. Individual tracks for H3K4me3 (SRX331384), H3K27ac (SRX331371), H3K9ac (SRX287847), H3K79me3 (SRX331396), H3K36me3 (SRX287679), Set1 (SRX1794236), RNA PolII (SRX318801), and Rpll215 (SRX859009) ChIP-seq experiments were analyzed in IGV. (c) Superimposed 1H,15N HSQC NMR spectra of PPS-PHD recorded while indicated histone H3 peptides (1–12) were titrated in. Spectra are color coded according to the PPS-PHD:histone peptide molar ratio. Arrows indicate chemical shift changes. (d) Binding affinities of PPS-PHD to indicated histone peptides as measured by intrinsic tryptophan fluorescence (for H3K4me3 and H3K4me2) or NMR at pH 6.9. (e) Representative binding curves used to determine the Kd values by NMR. (f, g) Western blot analysis of pull-downs using GST-PPS-PHD and indicated biotinylated histone H3 peptides at pH 7.4.
Here, we detail the structural mechanism for the recognition of the modified histone H3 tail by the PHD finger of Drosophila PPS (PPS-PHD). We found that the interaction of PPS-PHD with H3K4me3 is pH-dependent and is increased in high pH values. Structural, biochemical and mutational analyses of PPS reveal that the pH-dependency is due to the presence of a histidine residue (H919) in the K4me3-binding aromatic cage of the protein that undergoes protonation in acidic environment. The pH-dependent behavior is conserved in the PHD finger of human DIDO but is lost in yeast Bye1. We discuss the potential implications of this previously uncharacterized pH sensing ability of epigenetic readers for normal biological processes and those characterized by altered cellular pH.
RESULTS AND DISCUSSION
Binding of PPS-PHD to H3K4me3/me2 is modulated by nearby PTMs
PPS has been shown to associate with unspliced Sxl pre-mRNA and initially occupy promoter and the first exon of the Sxl gene (Johnson et al., 2010); however, the molecular basis for this observation has not been established. Analysis of the String database suggests a network of functional partners for PPS, including the RNA polymerase II 215 kD (RpII215) subunit and a putative peptidyl-prolyl cis-trans isomerase Dodo, implying that the interaction of PPS with Sxl could be direct or indirect (data not shown). The site near the Sxl promoter where PPS binds overlaps with a number of epigenetic marks associated with active transcription, including histone H3K4me3, H3K27ac, H3K9ac and H3K79me3, and additionally the H3K4-specific methyltransferase Set1 and RpII215 are recruited to the same region (Fig. 1b). As PPS contains a PHD finger, we postulated that PPS occupies the Sxl promoter site at least in part due to the histone binding activity of this domain.
To determine whether the PHD finger of PPS binds to histone tails, we expressed it as 15N-labeled protein and tested in 1H,15N heteronuclear single quantum coherence (HSQC) NMR experiments (Fig. 1c). Titration of the H3K4me3 peptide (residues 1-12 of H3) to the PPS-PHD finger caused substantial chemical shift perturbations (CSPs) in the protein, indicating direct interaction. A number of resonances corresponding to the free state of PPS-PHD disappeared, while another set of crosspeaks, corresponding to the bound state, appeared and slightly moved (Fig. 1c, left panel). This pattern of CSPs is a characteristic of slow-to-intermediate exchange regime on the NMR timescale and denotes tight binding. In agreement, a 3.6 μM binding affinity of PPS-PHD for the H3K4me3 peptide was measured in fluorescence assays (Fig. 1d). This value is in the range of binding affinities exhibited by the majority of epigenetic readers (Andrews et al., 2016; Musselman et al., 2012; Taverna et al., 2007). A weaker interaction with the dissociation constant (Kd) of 5.1 μM was observed between PPS-PHD and H3K4me2 (Fig. 1d). Titration of the H3K4me1 peptide to the PPS-PHD NMR sample resulted in CSPs similar in direction to those induced by H3K4me3 (compare left and middle panels in Fig. 1c), however fast exchange regime indicated a significant decrease in binding affinity for the monomethylated species (Kd = 230 μM, as measured by NMR) (Fig. 1d, e). Interestingly, no CSPs in PPS-PHD were seen upon addition of the H3K4me0 peptide, inferring that PPS-PHD does not recognize unmethylated histone H3 tail at all (Fig. 1c, right panel). The progressive decrease in binding to the low methylation states of H3K4 was corroborated by pull-down experiments with a GST fusion construct of PPS-PHD and longer histone H3 peptides (residues 1-20 of H3) (Fig. 1f). Altogether, these results demonstrate that the PPS PHD finger tightly binds to H3K4me3 but is incapable of associating with unmodified H3. This behavior is somewhat unusual for a PHD finger and may have important functional consequences.
To assess the impact of other H3 PTMs on the interaction of PPS-PHD with H3K4me3, we examined GST-PPS-PHD and histone peptides harboring multiple PTMs in conjunction with H3K4me3 in pull-down experiments (Fig. 1g). The pull-down showed that phosphorylation of neighboring threonine 3 (T3) completely abrogates binding of PPS-PHD to H3K4me3. The effect of other modifications varied, with citrullination of R2 substantially reducing this interaction, phosphorylation of T6 slightly enhancing it, and acetylation or methylation of K9 and methylation of R2 yielding no significant changes (Fig. 1g).
Structural basis for the interaction of PPS-PHD with H3K4me3
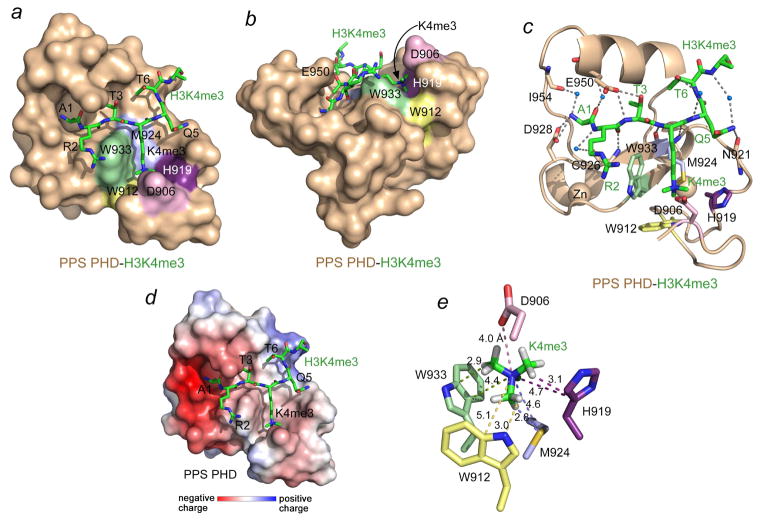
To gain mechanistic insight into the interaction, we obtained a 1.95 Å-resolution crystal structure of PPS-PHD in complex with H3K4me3 peptide (Fig. 2 and Table 1). The PPS-PHD finger adopts the canonical Cys4HisCys3 fold in which two zinc ions are coordinated in a cross-braced topology. The protein secondary structure elements include a double-stranded antiparallel β-sheet, a 310-helical turn, a central α-helix, and another short α-helix at the C-terminus (Fig. 2c). The H3K4me3 peptide occupies a large, primarily negatively charged binding site of the PHD finger and makes extensive contacts with the protein (Fig. 2a–d). It lies anti-parallel to and pairs with the β2 strand of the protein, forming distinctive β-sheet backbone hydrogen bonds among its R2 and K4me3 residues and C926 and M924 of PPS-PHD (Fig. 2c). In addition to the backbone-mediated intermolecular contacts, a set of hydrogen bonds involving the side chains of the peptide residues stabilize the complex: the hydroxyl group of T3 is hydrogen bonded to the carboxylate of E950, and the side chain amide of Q5 donates a hydrogen bond to the backbone carbonyl group of N921.
Figure 2. Structural basis for the recognition of H3K4me3 by PPS-PHD.
(a, b) PPS-PHD is depicted as a solid surface colored wheat with the H3K4me3 peptide shown as green sticks. (c) The ribbon diagram of the PPS-PHD-H3K4me3 complex. Dashed lines represent hydrogen bonds and blue spheres represent water molecules. (d) The electrostatic surface potential of PPS-PHD is colored blue and red for positive and negative charges, respectively. (e) A zoom-in view of the H3K4me3 binding pocket. The distances (in Å) are indicated by dashed lines and labeled.
Table 1.
Data collection and refinement statistics (molecular replacement)
| PPS Apo | PPS:H3K4me3 | |
|---|---|---|
| Data collection | ||
| Space group | P1211 | P1211 |
| Cell dimensions | ||
| a, b, c (Å) | 23.02, 41.06, 26.41 | 22.97, 44.22, 31.31 |
| α, β, γ (°) | 90.0, 98.43, 90.0 | 90.0, 94.77, 90.0 |
| Resolution (Å) | 26.12-1.40 (1.42-1.40)* | 31.2-1.95 (2.0-1.95)* |
| Rmerge (%) | 3.1 (8.7)* | 3.4 (5.9)* |
| I/σI | 55.2 (20.4)* | 37.9 (21.2)* |
| Completeness (%) | 99.3 (100.0)* | 99.3 (95.7)* |
| Redundancy | 10.0 (9.3)* | 3.8 (2.8)* |
| Refinement | ||
| Resolution (Å) | 26.1-1.40 | 31.2-1.95 |
| No. reflections | 9593 | 4556 |
| Rwork/Rfree | 13.6/16.0 | 14.5/18.6 |
| No. atoms | 566 | 608 |
| Protein/peptide | 494 | 527 |
| Water | 64 | 79 |
| Ligand/ion | 8 | 2 |
| B-factors | 9.5 | 11.2 |
| Protein | 8.1 | 10.2 |
| Ligand/ion | 10.0 | 7.6 |
| Water | 20.3 | 18.4 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | 0.006 |
| Bond angles (°) | 0.88 | 0.88 |
| Ramachandran quality | ||
| Favored (%) | 98.3 | 96.6 |
| Allowed (%) | 1.7 | 3.4 |
| Outliers (%) | 0 | 0 |
Values in parentheses are for highest-resolution shell. Data was collected from a single crystal.
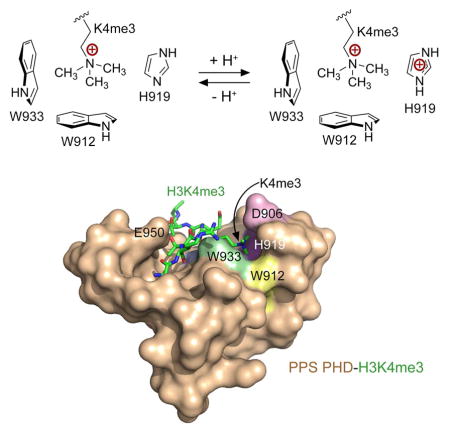
Two features of the H3K4me3-recognition mechanism distinguish PPS-PHD from other canonical PHD fingers, that is, the unique binding pocket for K4me3 and coordination of A1 (see below for further discussion). The K4me3-binding pocket in PPS-PHD contains three aromatic residues, W912, H919, and W933, one hydrophobic residue, M924, and one negatively charged residue, D906. The aromatic side chains form three walls of the binding pocket, whereas M924 lines the bottom, and D906 covers the bound K4me3 group as a lid (Fig. 2a, b). Such a unique arrangement of the binding site residues creates a fully closed cage around the positively charged trimethylammonium moiety of K4 (Fig. 2e). The short distances between the partially positively charged methyl groups of K4me3 and W933 indicate that W933 is the major driving force of the cation-π contact. Although the aromatic rings of W912 and H919 are tilted, they still appear to significantly contribute to the cation-π interactions. Both the negatively charged carboxyl group of D906 and the polarizable lone electron pair of the sulfur atom of M924 are likely involved in electrostatic contacts with the positively charged trimethylammonium group, locking this group in the center of the fully closed cage (Fig. 2e). In summary, the structure of the complex demonstrates that the K4me3 group is stabilized through cation-π interactions with the aromatic rings of W933, W912 and H919, and through hydrophobic, electrostatic, and van der Waals contacts.
A salt bridge and hydrogen bond with the carboxyl group of D928 and a water-mediated hydrogen bond with the backbone carbonyl oxygen atom of I954 restrain the positively charged N-terminal amino group of A1 (Fig. 2c). Further stabilization of A1 is achieved through the formation of a water-mediated hydrogen bond between its backbone carbonyl group and the backbone amide of E950, whose side chain carboxyl group is also engaged with the backbone amide of T3. Both methyl groups of A1 and T3 are buried in small cavities. The robust inhibitory effect of T3 phosphorylation on the interaction with H3K4me3 observed in pull-down assays could be explained in terms of steric hindrance and electrostatic repulsion between the bulky negatively charged phosphate group and the negatively charged carboxylic group of E950 (Figs. 1g and 2c). Interestingly, although the guanidino group of R2 appears to be unrestrained in the structure, citrullination of this residue substantially reduced binding of PPS-PHD in pull-down assays (Fig. 1g). Unlike the side chain of T3, the hydroxyl group of T6 could be phosphorylated without high steric or electrostatic penalty. In addition, T6 is bound in a positively charged shallow groove of PPS-PHD, which may account for a slight increase in the binding activity toward H3K4me3T6ph peptide in pull-down assays (Fig. 1g).
Recognition of both K4me3 and A1 of histone H3 is required
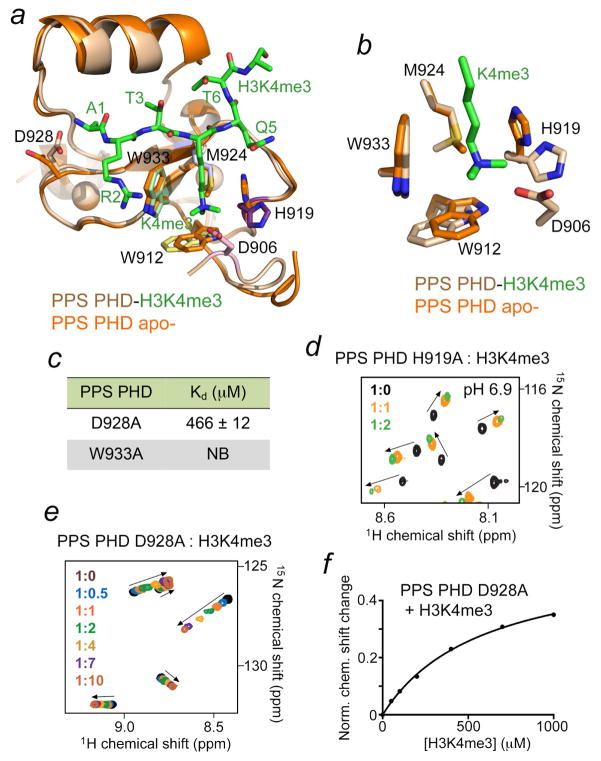
To establish the basis for the H3K4me3 recognition, we determined a 1.4 Å-resolution crystal structure of the PPS-PHD domain in the apo- state and compared it to the structure of PPS-PHD in the H3K4me3-bound state (Fig. 3a, b and Table 1). Overall, the structures superimpose with a root-mean-square-deviation of 0.4 Å, implying that binding of the peptide does not induce substantial conformational changes in the protein (Fig. 3a). A close examination of the K4me3-binding pocket in the apo- state reveals that it is essentially pre-arranged to accommodate the trimethylammonium group but some differences in position of the side chains of W912, H919 and M924 are evident (Fig. 3b). The aromatic moiety of H919 and, to a lesser extent, of W912 shifted outwards upon binding of the H3K4me3 peptide, and the methyl group of M924 flipped from occupying the binding cage in the apostate to pointing toward the hydrophobic core, thereby directing the lone electron pair of the sulfur atom toward the bound K4me3 group in the complex. Of note, because the PPS-PHD apo- construct that was crystallized lacks D906, we were unable to compare the position of this residue.
Figure 3. Unique binding mechanism of PPS-PHD.
(a, b) Structural overlay of the PPS-PHD finger in the apo-state and bound to H3K4me3 peptide. (c) Binding affinities of the mutant PPS-PHD to H3K4me3 peptide; NB-no binding. (d, e) Superimposed 1H,15N HSQC spectra of the PPS-PHD mutants, collected upon titration with the H3K4me3 peptide. Spectra are color coded according to the protein:peptide molar ratio. (f) Representative binding curve used to determine the Kd values by NMR.
A high significance of the binding site residues in PPS-PHD for the interaction with H3K4me3 was substantiated through mutagenesis. Replacement of W933 in PPS-PHD with an alanine abolished binding, and interaction of the H919A mutant was decreased (Fig. 3c, d). Interestingly, binding activity of the D928A mutant was reduced over 100-fold, suggesting that engagement of both A1 and K4me3 in the histone tail is necessary for the robust interaction of PPS-PHD with H3K4me3 (Fig. 3e, f).
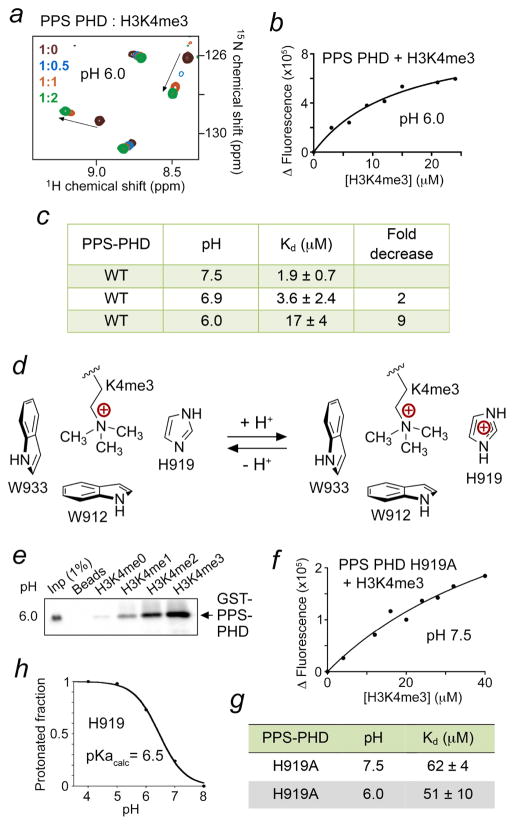
Binding of PPS-PHD to H3K4me3 is pH-dependent
Depending on the individual pKa value, the side chain of histidine residues in proteins can exist in protonated and deprotonated forms in a physiologically relevant pH. Because the aromatic cage in PPS-PHD contains one histidine residue, H919, we hypothesized that protonation of H919 would add a positive charge to the aromatic cage and therefore expel the positively charged K4me3 group of H3K4me3 peptide from the cage (Fig. 4). To explore this idea, we performed NMR titration experiments in a lower pH buffer of 6.0 and analyzed CSPs induced in PPS-PHD by the H3K4me3 peptide (Fig. 4a). Indeed, the pattern of CSPs indicated that PPS-PHD associates with the peptide significantly weaker in an acidic environment (pH 6.0), as compared to its tight binding at pH 6.9 (Fig. 4a and 1c). To quantitatively assess the effect of pH, we measured Kds for the PPS-PHD-H3K4me3 interaction at various pHs using intrinsic tryptophan fluorescence (Fig. 4b, c). In agreement with the NMR data, binding affinity of PPS-PHD was noticeably increased when pH of buffer was raised from 6.0 to 7.5. The PPS-PHD finger exhibited a 17 μM affinity for the H3K4me3 peptide at pH 6.0, however this interaction became ~9-fold tighter at pH 7.5 (Kd = 1.9 μM). Furthermore, pull-down assays performed at pH 7.4 and 6.0 (Figs. 1f and 4e) also indicated that GST-PPS-PHD associates with the low methylation states of H3K4 in a pH dependent manner. Collectively, our data demonstrate that pH mediates binding of PPS-PHD to the histone H3K4me3 tail (Fig. 4d).
Figure 4. Binding of PPS-PHD to H3K4me3 is pH-dependent.
(a) Superimposed 1H,15N HSQC spectra of PPS-PHD, recorded while the H3K4me3 peptide was titrated in at pH 6.0. (b, f) Representative binding curves used to determine the Kd values by fluorescence at indicated pH. (c, g) Summary of binding affinities of WT and mutated PPS-PHD to H3K4me3 peptide as measured by tryptophan fluorescence. (d) A diagram showing the effect of protonation of H919. (e) Western blot analysis of pull-downs using GST-PPS-PHD and indicated biotinylated histone H3 peptides at pH 6.0. (h) A titration curve used to calculate the pKa value of H919.
We next asked whether the pH sensitivity of PPS-PHD depends on H919. This histidine residue was mutated to an alanine, and binding affinity of the H919A mutant to the H3K4me3 peptide was measured at different pH values by fluorescence. Compared to the histone-binding activity of WT PPS-PHD, binding of H919A was substantially compromised in either pH, indicating that this residue is necessary for the interaction. However, the H919A mutant lost its pH-dependent behavior and associated with H3K4me3 to the same extent at pH 6.0 and pH 7.5, confirming that the pH-dependency is due to the presence of H919 in the binding pocket (Fig. 4f, g). Using Molecular Dynamics (MD) simulations and the structure of PPS-PHD, we computed pKa of His919 by sampling protein protonation states at all titratable sites in various pHs (Fig. 4h). The resulting titration curve and pKa of 6.5 at ionic strength of 0.1 M suggest that His919 exists in a nearly completely deprotonated state under physiological conditions (pH ~7.3), allowing H3K4me3 binding, whereas at pH 6.5, half of His919 is protonated and another half is deprotonated.
Unique features of the histone-binding mechanism
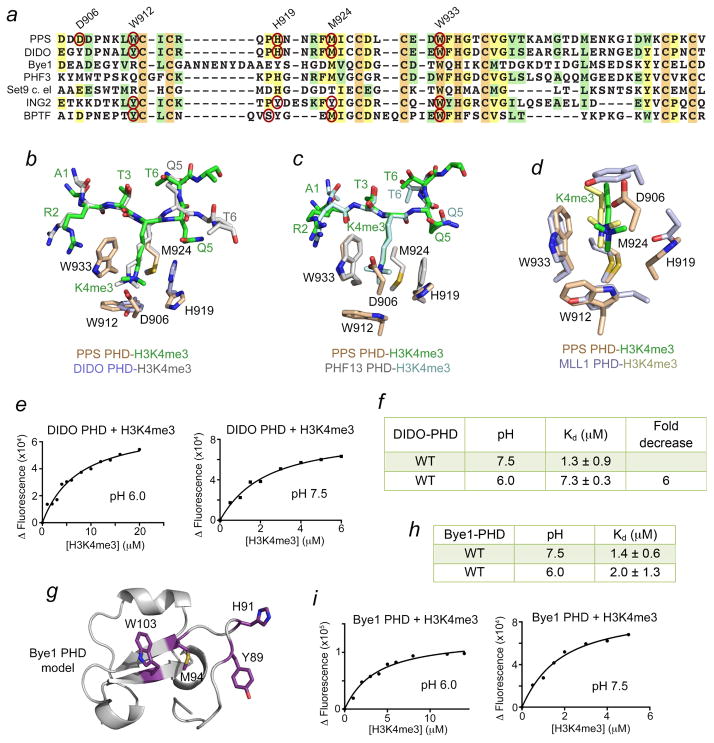
To assess whether the H3K4me3-recognition mechanism and pH dependency are conserved in other PHD fingers, we aligned amino acid sequences of the PHD fingers that have been reported to bind H3K4me3 (data not shown). Figure 5a shows the alignment of a small set of PHD fingers pertinent to this study, with the aromatic cage residues outlined with red ovals. Notably, all H3K4me3-binding PHD fingers contain an invariable tryptophan (W933 in PPS-PHD) as the wall-forming residue and a methionine (in rare instances, tyrosine or threonine) as the base of the cage. Another aromatic residue (W912 in PPS-PHD) located just N-terminal to the first zinc-coordinating cysteine often creates the second, orthogonal to tryptophan, wall of the cage. The residue forming the third, opposite to tryptophan, wall is the least conserved residue in the PHD fingers. In the classic PHD finger proteins, such as BPTF and ING2, this residue could be a tyrosine or serine(Li et al., 2006; Peña et al., 2006), but it is a histidine (H919) in PPS. The PHD fingers of three other proteins, including human DIDO, human PHF3, and worm Set9 also contain histidine in exactly the same position in the primary amino acid sequence as does PPS, whereas the PHD finger of yeast Bye1 has a tyrosine and a histidine nearby (Fig. 5a).
Figure 5. pH-sensitivity is conserved in DIDO-PHD but not in Bye1-PHD.
(a) Alignment of amino acid sequences of the PHD fingers: absolutely, moderately and weakly conserved residues are colored orange, pale yellow, and light green, respectively. The aromatic cage residues of the PHD fingers, for which the atomic-resolution structures of the complexes with H3K4me3 have been determined, are indicated by red ovals. (b–d) Structural overlays of the H3K4me3-bound PHD fingers of PPS (this work) and DIDO (PDB 4L7X), PHF13 (PDB 3O7A), and MLL1 (PDB 3LQJ). (e, i) Representative binding curves used to determine the Kds for DIDO and Bye1. (f, h) Binding affinities of DIDO-PHD and Bye1-PHD to H3K4me3 peptide at indicated pH. (g) A model of the Bye1-PHD finger fold generated with Phyre2.
The structural overlay of H3K4me3-bound PPS-PHD and DIDO-PHD reveals that the aromatic cage histidine adopts similar position in both complexes, suggesting that protonation of this residue in DIDO can also inhibit binding of H3K4me3 (Fig. 5b and see below for discussion). Unlike the DIDO complex, in which A1 of the peptide makes typical hydrogen bonds to several backbone carbonyl groups of the protein, in the PPS complex, A1 is stabilized via contacts with the side chain carboxyl group of D928 (Fig. 2c and ref. (Gatchalian et al., 2013)). The corresponding aspartate in DIDO instead restrains the guanidino moiety of R2 of the histone peptide (Gatchalian et al., 2013).
Further significant differences are observed in coordination of Q5 and T6 of the peptide. The H3K4me3 peptide in the PPS complex makes a sharp turn resulting in a less common conformation for the C-terminal part of the peptide, where T6 occupies the site, which is normally occupied by Q5. This conformation is stabilized by the water-mediated intra-peptide hydrogen bonds (Fig. 2c) and is also seen in the complex of the PHD finger of PHF13 (Chung et al., 2016) (Fig. 5c). In contrast to the PPS complex, in which T6 is more solvent exposed and its phosphorylation appears to slightly enhance binding to H3K4me3, in the DIDO complex, the hydroxyl group of T6 is restrained and T6ph eliminates binding of DIDO-PHD to H3K4me3. Lastly, the presence of the ‘lid’ residue, D906, which shields the bound K4me3 group, places PPS-PHD together with the PHD finger of MLL1 that uses an aromatic tyrosine residue as the ‘lid’ (Wang et al., 2010) to a category of readers distinguished by unique H3K4me3-caging mechanisms (Fig. 5d).
pH-sensitivity is conserved in DIDO-PHD but not in Bye1-PHD
To determine if binding of the DIDO PHD finger to H3K4me3 is pH-dependent, we purified WT DIDO-PHD and examined it by tryptophan fluorescence. We found that binding affinity of DIDO-PHD for the H3K4me3 peptide is 1.3 μM at pH 7.5, whereas this binding was reduced ~6 fold at pH 6.0 (Kd = 7.3 μM) (Fig. 5e, f). We concluded that much like histone-binding function of PPS-PHD, the histone-binding function of the DIDO PHD finger is modulated by changes in pH – it is enhanced in basic conditions but is decreased in acidic conditions.
Bye1, the yeast homolog of PPS and DIDO, contains a tyrosine residue in place of the histidine in PPS and DIDO (Fig. 5a). However a histidine is located in the +2 position with respect to the tyrosine in the Bye1 sequence. As the structure of the Bye1-PHD-H3K4me3 complex has not been determined, we postulated that if a histidine is present in the aromatic cage of Bye1-PHD, then similar to its human and fly counterparts it might exhibit pH dependence. If a tyrosine were in the binding pocket, then binding to H3K4me3 would be pH-insensitive. We note that computing the structure of Bye1-PHD in Phyre2 resulted in a model in which either Y89 or H91 could be part of the aromatic cage (Fig. 5g). We measured Kds for the interaction of Bye1-PHD with the H3K4me3 peptide in different conditions using tryptophan fluorescence. As shown in Figures 5h and 5i, Bye1-PHD was able to associate with the peptide similarly and independently of pH. These data suggest that the aromatic cage in Bye1-PHD likely contains the tyrosine residue rather than histidine. It will be of great interest to determine the structure of Bye1-PHD in complex with H3K4me3 to validate this biochemical finding.
Mitotic chromatin acidification promotes DIDO dissociation
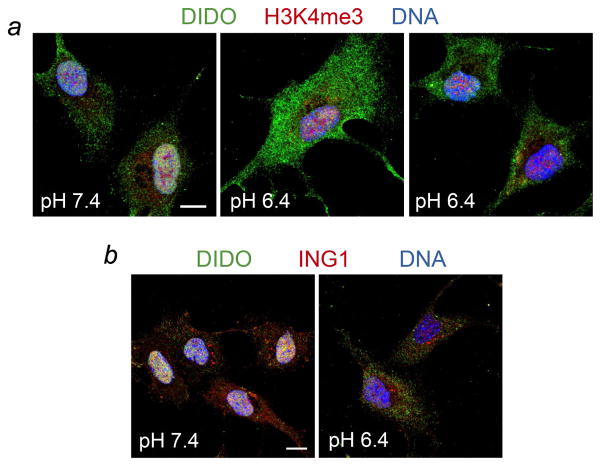
We have previously shown that DIDO associates with chromatin in interphase through binding of its PHD finger to H3K4me3 but translocates to the cytosol in mitosis (Gatchalian et al., 2013). To determine the effect of pH on the release of DIDO from mitotic chromatin we monitored subcellular localization of endogenous DIDO using confocal scanning laser microscopy. RPE-1 cells were first arrested in early mitosis by Haspin inhibition, and then washed and incubated in pre-warmed normal (pH 7.4) or acidic (pH 6.4) medium to equilibrate cytosolic pH and resume mitosis in the presence of nigericin, a membrane-perforating agent specific for H+/K+ (Fig. 6). The interphase nucleus is known to maintain pH approximately 0.3–0.5 units above that of the cytosol (Seksek and Bolard, 1996) but this pH gradient is dissipated by nigericin (Seksek and Bolard, 1996). We found that endogenous DIDO localizes to chromatin in cells that were treated at normal pH 7.4 (Fig. 6a, left panel). However, incubation of the cells in an acidic environment resulted in an efflux of DIDO from the nucleus even though H3K4me3 remained on chromatin (Fig. 6a, middle and right panels). The accelerated release of nuclear DIDO at pH 6.4 suggests the pH-dependent sensing of H3K4me3 in vivo and supports the pH dependent binding of DIDO in vitro. In contrast, ING1-PHD that has no histidine in its H3K4me3-binding aromatic cage remained attached to the H3K4me3-containing chromatin in the low pH conditions, corroborating the idea that the pH sensitivity is a specific feature of the DIDO-PHD.
Figure 6. Acidification promotes DIDO expulsion from the nucleus.
RPE-1 cells were arrested in early mitosis by inhibiting Haspin, washed, and released into control (pH 7.4) or acidic (pH 6.4) medium without inhibitor. The nuclear pH gradient was dissipated with nigericin. Mitosis was allowed to proceed for 10 minutes, cells were fixed and labeled for DIDO (all panels, green) and H3K4me3 (panel a, red) or ING1 (panel b, red). DNA was counterstained with DAPI (all panels, blue). Color separations are shown in Supplementary Figures S1 and S2. Scale bars, 10 μm.
Concluding remarks
Collectively, our results point to a unique mechanism by which the PPS PHD finger binds to H3K4me3. In the PPS-PHD complex, the K4me3 group is locked in the aromatic cage composed of three aromatic residues and a methionine and covered by the ‘lid’, a negatively charged aspartate, to complete the cage. The size of the aromatic cage is ideal to enclose tri- and dimethylated lysine, however is too large for the unmodified lysine, likely accounting for the inability of PPS-PHD to interact with H3K4me0. We further show that recognition of both K4me3 and A1 residues in the H3K4me3 peptide is required for the strong binding, as mutation of the PPS-PHD residues that are in contact with either A1 or K4me3 blocks this interaction.
Arguably, the most unusual aspect of the histone-binding mechanism of PPS-PHD is the pH-sensitivity. We found that interaction of PPS-PHD with H3K4me3 is pH-dependent, and is increased with increasing of pH. The pH-dependency is due to the presence of histidine in the aromatic cage of the PHD finger, which undergoes protonation in acidic environment. Generally speaking, histidine is fairly commonly found in active, catalytic, or binding sites of proteins, despite its relatively low natural abundance (~3%) and it is known to play a role of a pH sensor (He et al., 2008; He et al., 2009; Hom et al., 2007; Lee et al., 2005; Srivastava et al., 2007). The side chain of histidine, the imidazole group, has a pKa value of 6–7.5 in folded proteins. We calculated that H919 has a pKa of 6.5 (Fig. 4h). The fact that histidine exists in protonated and deprotonated forms in near physiologically relevant conditions means that even small fluctuations in pH lead to a substantial change in its ionization state and consequently perturb interactions involving not only histidine itself but also nearby residues.
The pH-dependent behavior is conserved in the PHD finger of human DIDO but is lost in the yeast homolog, Bye1-PHD. The 6-fold enhancement in the H3K4me3-binding activity of human DIDO-PHD at pH 7.5 compared to its binding at pH 6.0 can have significant biological implications. We found that the acidic environment accelerates the release of endogenous DIDO from the cell nucleus despite the fact that H3K4me3 persists. Such pH-dependence can mediate functions of DIDO in cells with abnormal nuclear pH and in normal cells during physiological processes that are accompanied by pH alterations. Cellular pH often fluctuates in response to cell growth, differentiation, and apoptosis and varies in abnormal cells during ischemia, inflammation and cancer (Casey et al., 2010). Growth factor-induced proliferation, cell migration, and cell cycle progression are associated with an increase in cellular pH, whereas acidification stimulates caspase-dependent and independent apoptosis (Srivastava et al., 2007). Acidic extracellular pH and elevated intracellular pH that promotes unrestricted proliferation and genomic instability are a hallmark of cancer (Webb et al., 2011).
Although much less is known regarding alterations in pH of the cell nucleus, growing evidence suggests that it may affect nuclear physiological processes. Measurement of nuclear pH in many types of cells shows that it is 0.3–0.5 pH units more alkaline compared to the cytosol (Seksek and Bolard, 1996). Recent reports indicate that changes in pH in conjunction with Ca2+ signaling in the nucleus likely influence the activity of transcription factors and ultimately gene expression (Hulikova and Swietach, 2016). Opposite changes in nuclear pH following blocking the Ca2+ release were observed in adult and neonatal myocytes, underscoring important contribution of diverse signaling pathways at different stages of development and differentiation (Hulikova and Swietach, 2016). A significant alkaline shift in internal pH has been detected during mitosis compared to pH in interphase in one-cell mouse embryos, whereas blocking cell division prevents alkalization (Amirand et al., 2000). Likewise, breaking down the nuclear envelope would lower the chromatin pH, promoting protonation of histidine and helping to exclude pH-sensors from mitotic chromosomes (Gatchalian et al., 2016; van Wely et al., 2017). It will be critical in future studies to establish the significance of the pH sensing by the epigenetic readers in normal processes and disease.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Email contact for reagent and resource sharing: tatiana.kutateladze@ucdenver.edu
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial Strains
The Drosophila PPS PHD finger constructs, aa 904–965, aa 908–965, and aa 910–964, were cloned from full-length PPS in pCaSpeR4 (a kind gift from Helen Salz) into a pDEST15 using Gateway cloning technology. Point mutants were generated using site-directed mutagenesis. Proteins were expressed in Rosetta2 (DE3) pLysS or BL21 (DE3) RIL in either Luria Broth or M19 minimal media supplemented with 50 μM ZnCl2 and 15NH4Cl (Sigma-Aldrich). Cells were induced with 0.2–0.5 mM IPTG (Gold biotechnology), grown for 16 hours at 16–18°C, and harvested by centrifugation.
METHOD DETAILS
Protein purification
Cells were lysed by sonication in lysis buffer containing 25 mM Tris pH 7.5, 150 mM NaCl, 3 mM dithiothreitol (DTT), 0.5% Triton X-100, and 5 mM MgCl2, and clarified by centrifugation. Proteins were purified on glutatahione Sepharose 4B beads (Thermo Fisher Sci) and the GST-tag was cleaved using either PreScission or Tobacco Etch Virus (TEV) protease for at least overnight at 4°C. Cleaved proteins were concentrated into 25 mM Tris pH 6.0–7.5, 150 mM NaCl, and 3 mM DTT. Unlabeled proteins were further purified by size exclusion chromatography using a HiPrep 16/60 Sephacryl S-100 column.
The DIDO-PHD (aa 266–325) and Bye1-PHD (aa 47–134) constructs were generated, and the proteins were purified as above.
NMR Spectroscopy
NMR experiments were performed at 298K on a Varian INOVA 600 MHz spectrometer equipped with a cryogenic probe. 1H,15N HSQC spectra of 0.1 mM uniformly 15N-labelled wild type or mutant PHD fingers were recorded in the presence of increasing concentrations of modified and unmodified histone H3 peptides (aa 1–12) (synthesized by Synpeptide). Kd value was calculated by a nonlinear least-squares analysis in Kaleidagraph using the equation:
where [L] is concentration of the peptide, [P] is concentration of the protein, Δδ is the observed chemical shift change, and Δδmax is the normalized chemical shift change at saturation. Normalized chemical shift changes were calculated using the equation , where Δδ is the change in chemical shift in parts per million (ppm).
Fluorescence spectroscopy
Spectra were recorded at 25 °C on a Fluoromax-3 spectrofluorometer (HORIBA). The samples containing 0.5–1 μM wild type or mutated PHD fingers in 20 mM phosphate (or 25 mM Tris) pH 6.0, 6.9 or 7.5, 150 mM NaCl, 3 mM DTT buffer and progressively increasing concentrations of the histone peptides were excited at 295 nm. Emission spectra were recorded between 320 and 360 nm with a 0.5 nm step size and a 0.5 s integration time and averaged over 3 scans. The Kd values were determined using a nonlinear least-squares analysis and the equation:
where [L] is the concentration of the histone peptide, [P] is the concentration of the protein, ΔI is the observed change of signal intensity, and ΔImax is the difference in signal intensity of the free and bound states of the protein. The Kd values were averaged over three separate experiments (two in Fig. 4f for pH 7.5), with error calculated as the standard deviation between the runs.
X-Ray crystallography
The PPS-PHD (aa 908–965) construct was used for structure determination of the apo- state. The protein (2.5 mg/mL) was crystallized at 18°C in 0.1 M HEPES pH 7.5, 0.2 M NaCl, 25% PEG 3,000 using the hanging drop method. Data collection was performed at the ALS 4.2.2 beamline, Berkeley. The structure was solved using molecular replacement and the structure of DIDO-PHD (PDB 4L7X) as a model. The PPS-PHD (aa 904–965) construct was used for structure determination of the complex. Purified PPS-PHD was concentrated to 8 mg/mL in 10 mM Tris pH 8.0 buffer, supplemented with 75 NaCl and 1 mM DTT, and incubated with the H3K4me3 (1–12 aa) peptide at a 1:2 molar ratio for 15 min on ice prior crystallization. Crystals of the PPS-PHD:H3K4me3 complex were grown using hanging-drop diffusion method at 4°C in 0.1 mM Tris pH 8.4 and 24% PEG 6,000. The diffraction data was collected on the UC Denver X-ray Crystallography core facility Rigaku Micromax 007 high frequency microfocus X-ray generator equipped with a Pilatus 200K 2D area detector. Indexing and scaling was completed using HKL3000. The phase solution was solved with Phenix.phaser using molecular replacement with the apo- PPS-PHD structure as the model. Model building was carried out with Coot and refinement was performed with Phenix.refine (Adams et al., 2010; Emsley et al., 2010). The final structure was validated with MolProbity (Chen et al., 2010). Crystallographic statistics for the PPS-PHD structures are shown in Table 1.
Pull-down assay
Peptide pull-downs were performed essentially as described (Rothbart et al., 2012) except that the assay buffer contained 50 mM Tris pH 6.0–7.4, 300 mM NaCl, and 0.1% NP-40, and 500 pmols of biotinylated histone peptides were loaded onto streptavidin coated magnetic beads before incubation with 40 pmols of GST fusion PPS-PHD. Bound protein was detected with rabbit GST antibody (homemade).
Molecular Dynamics simulations
Crystal structure of the apo- PPS-PHD was used in the CpHMD simulations with generalized Born (GB) implicit solvation as described (Mongan et al., 2004). To obtain the titration curves, MD simulations were performed at pH 0, 1, 2, 3, 4, 5, 6, 7 and 8 using AMBER16. The pKa values were determined by nonlinear least-squares analysis in Kaleidagraph using the following equation:
where fd is the deprotonated fraction.
Confocal scanning fluorescence microscopy
RPE-1 cells were seeded on glass coverslips and grown in DMEM/F12 containing 10% FCS. When reaching 50% confluence, cells were arrested in early mitosis with 2 μM CHR6494 (Tocris Bioscience, Bristol, UK) from a 500 μM stock in dimethylsulfoxide. Cells were then washed briefly and incubated for 10 minutes in prewarmed medium without CHR6494 to allow redistribution of nuclear proteins. To determine pH sensitivity, release medium was acidified to pH 6.4 by adding 25 mM 2-(N-morpholino) ethanesulfonic acid (MES) and the nuclear-cytosolic pH gradient dissipated with 5 μM nigericin (Seksek and Bolard, 1996). Non-acidified control cells were released into normal medium containing 5 μM nigericin. For immunofluorescence, cells were washed briefly in PBS, fixed in PBS containing 4% formaldehyde for 10 min and permeabilized for 10 min in PBS containing 0.5% Triton X-100. After blocking in PBS containing 1% bovine serum albumin (1 h, room temperature), cells were incubated with primary antibodies (1 h, room temperature), washed, incubated with secondary antibodies, washed again and mounted in Prolong Gold antifade (Life Technologies, Grand Island, NY). Primary antibodies against DIDO (AF6947) and ING1 (MAB57581) were from R&D Systems (Minneapolis, MN), and the antibodies against H3K4me3 (AB8580) were from Abcam (Cambridge, UK). DIDO was detected with Alexa488-labelled secondary antibodies, and all other proteins with Alexa594-labelled secondary antibodies. DNA was counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Confocal microscopy was performed using an IX81 microscope (Olympus, Tokio, Japan) with sequential excitation of fluorophores. Representative images corresponding to maximum projection of 0.5 μm confocal layers are shown in the figures.
DATA AND SOFTWARE AVAILABILITY
Software
Software used in this this study has been previously published as detailed in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| DIDO | R&D Systems | AF6947 |
| ING1 | R&D Systems | MAB57581 |
| H3K4me3 | Abcam | AB8580 |
| rabbit GST | homemade | |
| Bacterial and Virus Strains | ||
| Escherichia coli Rosetta2 (DE3) pLysS | Ali et al., 2012 | N/A |
| BL21 (DE3) RIL | Andrews et al., 2016 | N/A |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Dithiothreitol | Gold Biotechnology | 27565-41-9 |
| H3K4me3, H3K4me2, H3K4me1 and H3K4me0 peptides (1–12 aa) | Synpeptide | N/A |
| 15NH4Cl | Sigma-Aldrich | 299251 |
| IPTG | Gold biotechnology | I2481C100 |
| Glutatahione Sepharose 4B beads | Thermo Fisher Sci | 16101 |
| PreScission and TEV proteases | Home expressed | N/A |
| Critical Commercial Assays | ||
| Deposited Data | ||
| PPS-PHD in complex with H3K4me3 | This study | PDB: 5WLE |
| PPS-PHD, apo | This study | PDB: 5WLF |
| Experimental Models: Cell Lines | ||
| Human: RPE-1 | N/A | |
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| Recombinant DNA | ||
| pDEST15 | N/A | |
| Software and Algorithms | ||
| Phenix | Adams et al., 2010 | https://www.phenix-online.org/ |
| Coot | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| CpHMD | Mongan et al., 2004 | N/A |
| MOLProbity | Chen et al., 2010 | http://molprobity.biochem.duke.edu/ |
| Other | ||
Data Resources
Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 5WLE and 5WLF.
Supplementary Material
HIGLIGHTS.
The histone H3K4me3-recognizing PHD fingers of PPS and DIDO are high pH sensors
pH-Dependency is due to the presence of a histidine residue in the aromatic cage
The pH-dependent mechanism is conserved in human DIDO but is lost in yeast Bye1
Acidification of cells leads to the accelerated efflux of endogenous DIDO in vivo
Acknowledgments
We thank Jay Nix at beam line 4.2.2 of the ALS in Berkeley for help with X-ray crystallographic data collection, Helen Salz for kindly providing original DNA of PPS, and Forest Andrews and Ruben Rosas Ospina for help with experiments. This work was supported by NIH grants R01 GM106416, GM101664 and GM100907 to T.G.K., and GM110058 to B.D.S. K.H.W. is supported by MINECO grant SAF2016-75456-R from the Spanish government and A.H.T. is supported by the NIH grant T32AA007464.
Footnotes
AUTHOR CONTRIBUTIONS
A.H.T., J.G., B.J.K., A.K., Y.Z. and K.H.W. performed experiments and together with B.D.S. and T.G.K. analyzed the data. A.H.T., K.H.W. and T.G.K. wrote the manuscript with input from all authors. Lead contact: T.G.K.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amirand C, Mentre P, van de Geijn S, Waksmundzka M, Debey P. Intracellular pH in one-cell mouse embryo differs between subcellular compartments and between interphase and mitosis. Biology of the cell. 2000;92:409–419. doi: 10.1016/s0248-4900(00)01080-7. [DOI] [PubMed] [Google Scholar]
- Andrews FH, Strahl BD, Kutateladze TG. Insights into newly discovered marks and readers of epigenetic information. Nature chemical biology. 2016;12:662–668. doi: 10.1038/nchembio.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey JR, Grinstein S, Orlowski J. Sensors and regulators of intracellular pH. Nature reviews Molecular cell biology. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica Section D, Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HR, Xu C, Fuchs A, Mund A, Lange M, Staege H, Schubert T, Bian C, Dunkel I, Eberharter A, et al. PHF13 is a molecular reader and transcriptional co-regulator of H3K4me2/3. eLife. 2016;5 doi: 10.7554/eLife.10607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futterer A, Campanero MR, Leonardo E, Criado LM, Flores JM, Hernandez JM, San Miguel JF, Martinez AC. Dido gene expression alterations are implicated in the induction of hematological myeloid neoplasms. J Clin Invest. 2005;115:2351–2362. doi: 10.1172/JCI24177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatchalian J, Futterer A, Rothbart SB, Tong Q, Rincon-Arano H, Sanchez de Diego A, Groudine M, Strahl BD, Martinez AC, van Wely KH, Kutateladze TG. Dido3 PHD modulates cell differentiation and division. Cell reports. 2013;4:148–158. doi: 10.1016/j.celrep.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatchalian J, Gallardo CM, Shinsky SA, Ospina RR, Liendo AM, Krajewski K, Klein BJ, Andrews FH, Strahl BD, KHMvW, Kutateladze TG. Chromatin condensation and recruitment of PHD finger proteins to histone H3K4me3 are mutually exclusive. Nucleic acids research. 2016 doi: 10.1093/nar/gkw193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Haney RM, Vora M, Verkhusha VV, Stahelin RV, Kutateladze TG. Molecular mechanism of membrane targeting by the GRP1 PH domain. J Lipid Res. 2008;49:1807–1815. doi: 10.1194/jlr.M800150-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Vora M, Haney RM, Filonov GS, Musselman CA, Burd CG, Kutateladze AG, Verkhusha VV, Stahelin RV, Kutateladze TG. Membrane insertion of the FYVE domain is modulated by pH. Proteins. 2009;76:852–860. doi: 10.1002/prot.22392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom RA, Vora M, Regner M, Subach OM, Cho W, Verkhusha VV, Stahelin RV, Kutateladze TG. pH-dependent binding of the Epsin ENTH domain and the AP180 ANTH domain to PI(4,5)P2-containing bilayers. Journal of molecular biology. 2007;373:412–423. doi: 10.1016/j.jmb.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulikova A, Swietach P. Nuclear proton dynamics and interactions with calcium signaling. Journal of molecular and cellular cardiology. 2016;96:26–37. doi: 10.1016/j.yjmcc.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ML, Nagengast AA, Salz HK. PPS, a large multidomain protein, functions with sex-lethal to regulate alternative splicing in Drosophila. PLoS genetics. 2010;6:e1000872. doi: 10.1371/journal.pgen.1000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenberger H, Armache KJ, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- Kinkelin K, Wozniak GG, Rothbart SB, Lidschreiber M, Strahl BD, Cramer P. Structures of RNA polymerase II complexes with Bye1, a chromatin-binding PHF3/DIDO homologue. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:15277–15282. doi: 10.1073/pnas.1311010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SA, Eyeson R, Cheever ML, Geng J, Verkhusha VV, Burd C, Overduin M, Kutateladze TG. Targeting of the FYVE domain to endosomal membranes is regulated by a histidine switch. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13052–13057. doi: 10.1073/pnas.0503900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongan J, Case DA, McCammon JA. Constant pH molecular dynamics in generalized Born implicit solvent. Journal of computational chemistry. 2004;25:2038–2048. doi: 10.1002/jcc.20139. [DOI] [PubMed] [Google Scholar]
- Musselman CA, Kutateladze TG. Handpicking epigenetic marks with PHD fingers. Nucleic acids research. 2011;39:9061–9071. doi: 10.1093/nar/gkr613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musselman CA, Lalonde ME, Cote J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nature structural & molecular biology. 2012;19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallasch CP, Struss AK, Munnia A, Konig J, Steudel WI, Fischer U, Meese E. Autoantibodies against GLEA2 and PHF3 in glioblastoma: tumor-associated autoantibodies correlated with prolonged survival. International journal of cancer. 2005;117:456–459. doi: 10.1002/ijc.20929. [DOI] [PubMed] [Google Scholar]
- Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinskaya M, Ghavi-Helm Y, Mariotte-Labarre S, Morillon A, Soutourina J, Werner M. PHD and TFIIS-Like domains of the Bye1 transcription factor determine its multivalent genomic distribution. PloS one. 2014;9:e102464. doi: 10.1371/journal.pone.0102464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nature structural & molecular biology. 2012;19:1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seksek O, Bolard J. Nuclear pH gradient in mammalian cells revealed by laser microspectrofluorimetry. Journal of cell science. 1996;109(Pt 1):257–262. doi: 10.1242/jcs.109.1.257. [DOI] [PubMed] [Google Scholar]
- Srivastava J, Barber DL, Jacobson MP. Intracellular pH sensors: design principles and functional significance. Physiology. 2007;22:30–39. doi: 10.1152/physiol.00035.2006. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nature structural & molecular biology. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wely KH, Mora Gallardo C, Vann KR, Kutateladze TG. Epigenetic countermarks in mitotic chromosome condensation. Nucleus. 2017:1–6. doi: 10.1080/19491034.2016.1276144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Song J, Milne TA, Wang GG, Li H, Allis CD, Patel DJ. Pro Isomerization in MLL1 PHD3-Bromo Cassette Connects H3K4me Readout to CyP33 and HDAC-Mediated Repression. Cell. 2010;141:1183–1194. doi: 10.1016/j.cell.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb BA, Chimenti M, Jacobson MP, Barber DL. Dysregulated pH: a perfect storm for cancer progression. Nature reviews Cancer. 2011;11:671–677. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.