Abstract
Recently, a new type of limb-girdle muscular dystrophy (LGMD type 2Z) has been identified due to a missense mutation in POGLUT1 (protein O-glucosyltransferase- Rumi), an enzyme capable of adding glucose to a distinct serine residue of epidermal growth factor-like repeats containing a C-X-S-X-(P/A)-C consensus sequence such as Notch receptors. Affected patients demonstrate reduced Notch signaling, decreased muscle stem cell pool and hypoglycosylation of α–dystroglycan, leading to LGMD phenotype. Here we report the generation and characterization of an iPSC line (CSCRMi001-A) from a LGMD-2Z patient with missense mutation in POGLUT1 which can be used for in vitro disease modeling.
Resource Table
| Unique stem cell line identifier | CSCRMi001-A |
| Alternative name(s) of stem cell line | POGLUT1 c.699T > G (D233E)- II.5-3 |
| Institution | Center for Stem Cell and Regenerative Medicine (CSCRM), The University of Texas Health Science Center at Houston (UTHealth), Houston, TX 77030, USA |
| Contact information of distributor | Radbod.Darabi@uth.tmc.edu |
| Type of cell line | iPSC |
| Origin | Human skin fibroblasts |
| Additional origin info |
Age: 37 Sex: Male Ethnicity: From a consanguineous family member in Southern Spain |
| Cell Source | Fibroblasts |
| Method of reprogramming | Sendai Virus |
| Genetic Modification | No |
| Type of Modification | N/A |
| Associated disease | Limb-Girdle Muscular Dystrophy (LGMD) type 2Z (LGMD-2Z, OMIM#617232) |
| Gene/locus | Homozygous c.699T > G transversion mutation (D233E) in POGLUT1 (protein O-glucosyltransferase or Rumi) gene on Chromosome 3 |
| Method of modification | N/A |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | September, 2016 |
| Cell line repository/bank | N/A |
| Ethical approval | Reviewed and approved by The Human Embryonic Stem Cell Research Oversight Committee (SCRO) of UTHealth on November11, 2015. |
Resource utility
This iPSC line is from a new type of LGMD (LGMD2Z, OMIM#617232) which is caused by a mutation in POGLUT1 [1–3] and it serves as a unique tool for in vitro disease modeling, identification of involved pathways and therapeutic targets as well as gene correction approaches.
Resource Details
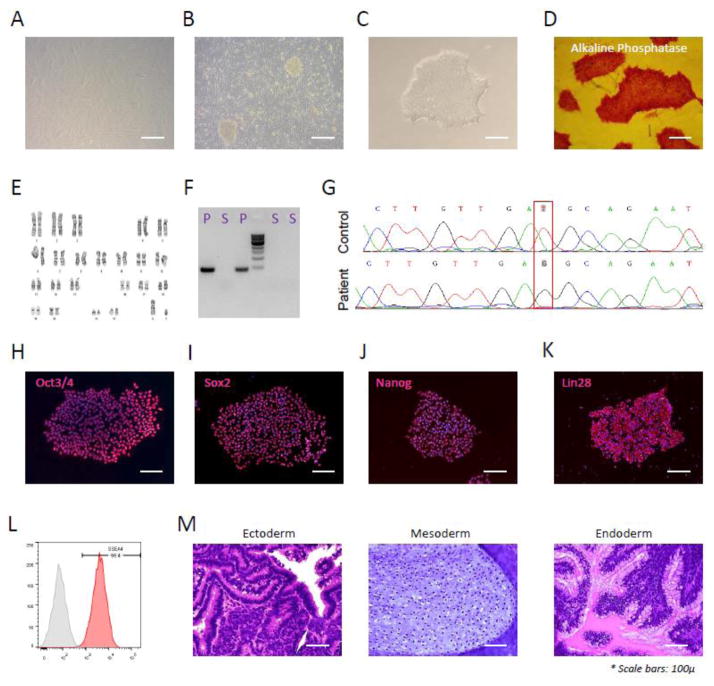
Skin fibroblasts were taken by a skin biopsy from a 37 years old male patient diagnosed with LGMD due to homozygous missense mutation (c.699T>G) in the protein O-glucosyltransferase 1 gene, POGLUT1 [1]. Samples were taken by our collaborator group in Spain after informed consent and approval by local ethic committee. After culture and expansion of fibroblasts, frozen cells were shipped to our facility in Houston, Texas for iPSC reprogramming. Fibroblasts were then thawed and after initial expansion (Fig. 1A), infected with Sendai virus containing reprogramming factors OCT4, SOX2, KLF4 and c-MYC [4]. Seven days after infection, fibroblasts were harvested and plated on irradiated mouse embryonic fibroblasts (MEFs) in human stem cell medium and were inspected daily for the emergence of iPSC colonies.
Figure 1.
As demonstrated, iPSC colonies (Fig. 1B) were detected 1 to 2 weeks later and individual colonies were harvested and transferred into Matrigel-coated plates containing mTeSR medium for subsequent feeder-free culture. The colony described here (CSCRMi001-A) was later selected for further characterization.
CSCRMi001-A showed the typical colony morphology of iPSCs (Fig. 1C) in undifferentiated feeder-free culture, positive for Alkaline Phosphatase (Fig. 1D), carried normal karyotype (Fig. 1E) and was negative for mycoplasma (Fig. 1F, S: iPSC DNA Sample, P: Positive control). STR analysis (Supplementary info) also further confirmed the identical genetic background of donor fibroblasts and described iPSC clone. To confirm the presence of missense mutation, DNA sample from described clone was sequenced for the affected region in POGLUT1 gene which confirmed a homozygous c.699T > G transversion as compared to control (Fig 1.G).
This clone was further characterized for pluripotency markers using immunofluorescent staining for OCT4, SOX2, NANOG and LIN28 (Fig. 1H–K, merged images counterstained with DAPI), which confirmed uniform expression of these markers. FACS analysis also confirmed high expression of the SSEA-4 (Fig. 1L).
Finally, to test multi-lineage differentiation potential, the teratoma assay was done, which confirmed the presence of ectodermal (neural rosette), mesodermal (cartilage) and endodermal (mucin- producing glands) tissues (as shown in Fig. 1M). These results demonstrated that the above-mentioned patient-derived iPSC clone (CSCRMi001-A) has the right donor genetic background along with the disease-causing missense mutation in POGLUT1 gene and demonstrates common pluripotency and differentiation characteristics of human iPSC cells.
Materials and Methods
iPSC reprogramming
Fibroblasts were reprogrammed using CytoTune®-iPS 2.0 Sendai Reprogramming Kit (ThermoFisher, Cat# A16517) according to manufacturer’s protocol. Briefly, cells were expanded for three days in DMEM medium supplemented with 10% fetal bovine serum (FBS) and were infected with reprogramming viruses according to the protocol. Seven days later, infected cells were harvested and plated on irradiated mouse embryonic fibroblasts (MEFs) in Matrigel coated plates containing human embryonic stem cell (ESC) medium. Medium was changed daily and the plates were monitored for emergence of reprogrammed iPSC colonies in the following two weeks. Finally, typical iPSC colonies were picked up and subcultured in feeder-free mTeSR medium in Matrigel coated plates. CSCRMi001-A was chosen for further characterization.
Mycoplasma detection
A pair of universal mycoplasma primers was used for detecting the mycoplasma’s 16s rRNA (listed in Table 2). PCR conditions were 94 °C 2 min, 63 °C 2 min, 72 °C 2 min; 40 cycles of 94 °C 1 min, 63 °C 1 min and 72 °C 2 min; 72 °C 10 min. The assay was performed in accordance to the method described before [5]. Samples were run along with positive control DNA as demonstrated in Figure 1E. A 500 bp band appears for mycoplasma-positive cells, and no band appears for mycoplasma-negative cells.
Table 2.
Reagents details
| Antibodies used for immunocytochemistry/flow-cytometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency Marker | Rabbit mAb anti-OCT4A | 1:400 | StemLight kit, Cell Signaling Technology, Cat# 9092 |
| Pluripotency Marker | Rabbit mAb anti-SOX2 | 1:400 | StemLight kit, Cell Signaling Technology, Cat# 9092 |
| Pluripotency Marker | Rabbit mAb anti-NANOG | 1:400 | StemLight kit, Cell Signaling Technology, Cat# 9092 |
| Pluripotency Marker | Rabbit mAb anti-LIN28A | 1:400 | StemLight kit, Cell Signaling Technology, Cat# 9092 |
| Pluripotency Marker | PE-conjugated mouse mAb anti-SSEA4 | 1:20 | ThermoFisher, Cat# 12-8843-41 |
| Secondary antibody | Goat anti-Rabbit IgG Alexa Fluor Plus 488 | 1:500 | ThermoFisher, Cat# A32731 |
| Isotype control antibody | Mouse IgG3 Isotype Control, PE | 1:20 | ThermoFisher, Cat# 12-4742-42 |
| Primers | |||
| Target | Forward/Reverse primer (5′-3′) | ||
| Mycoplasma detection | Mycoplasma 16S rRNA |
GGCGAATGGGTGAGTAACACG CGGATAACGCTTGCGACCTATG |
|
| POGLUT1 mutation fragment amplification | A 506-bp amplicon containing mutation region |
TGACCTGAACAACATACCCTTCA GCTAATGCTGGTTCATGGAACTT |
|
| POGLUT1 amplicon sequencing | A 20-bp region located 125-bp upstream of the mutation within the amplicon | GTCCTAGTCCTGCTCACCTT | |
POGLUT1 mutation detection
Genomic DNA was extracted and a pair of primers was designed to amplify the POGLUT1 mutation fragment from DNA. Another primer was used for DNA sequencing of the mutation region within the amplicon (all listed in Table 2). A normal control human iPSC DNA was used to compare sequencing results as demonstrated in Figure 1G.
Karyotyping
Karyotyping was done using classical Chromosome GTG-banding analysis based on international system for human cytogenetic nomenclature (ISCN).
Immunofluorescence staining
Staining was done using StemLight™ iPS Cell Reprogramming Antibody kit (Cell Signaling Technology, Cat# 9092). Briefly, iPSCs were fixed using 4% PFA and after treatment with Triton-X and blocking, were incubated with primary antibodies according to manufacturer’s protocol. Finally, after appropriate staining with secondary antibodies, they were counterstained for DAPI and used for microscopy.
Alkaline phosphatase (AP) staining
AP staining was done using AP staining kit (STEMGENT, Cat# 00-0009). Briefly, iPSCs were expanded and then fixed and incubated with AP staining solutions according to the manufacturer’s protocol. Cells were then washed using PBS solution, covered with mounting solution and used for microscopy.
Flow cytometry analysis
iPSCs were harvested using StemPro Accutase (ThermoFisher, Cat# A1110501) and after washing with PBS, were stained for a PE-conjugated SSEA-4 antibody (ThermoFisher, Cat# 12-8843-41) and were analyzed along with the appropriate PE isotype control (ThermoFisher, Cat# 12-4742-42) using a BD FACSAria cytometer.
Teratoma formation assay
For teratoma assay, iPSCs were harvested and resuspended in 50μl of Matrigel and injected into quadriceps femoris muscles of NSG immunodeficient mice (NOD-scid IL2Rgnull, The Jackson laboratory, Cat# 005557). Six weeks later and after teratoma formation, tumors were harvested and embedded into paraffin and used for H&E staining and histopathology.
Table 1.
Characterization and validation
| Classification | Test | Result | Data |
|---|---|---|---|
| Morphology | Photography | Normal | Figure 1 panel C |
| Phenotype | Immunocytochemistry | Expression of pluripotency markers: OCT4, NANOG, SOX2, LIN28 | Figure 1 panels H–K |
| Flow cytometry | SSEA-4: 99.4% | Figure 1 panel L | |
| Genotype | Karyotype (G-banding) and resolution | 46XY, Resolution 400 | Figure 1 panel E |
| Identity | Microsatellite PCR (mPCR) | Not performed | |
| STR analysis | 14 loci, Matched | submitted in archive with journal | |
| Mutation analysis (IF APPLICABLE) | Sequencing | c.699T > G transversion mutation in POGLUT1 | Figure 1 panel G |
| Southern Blot OR WGS | N/A | ||
| Microbiology and virology | Mycoplasma | Mycoplasma testing by PCR- Negative | Figure 1 panel F |
| Differentiation potential | Teratoma formation | Teratoma formation for proof of three germlayers formation | Figure 1 panel M |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | Not performed | |
| Genotype additional info (OPTIONAL) | Blood group genotyping | Not performed | |
| HLA tissue typing | Not performed |
Acknowledgments
This work was partially supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) of the National Institutes of Health (NIH) to R.D. under award number 1R01AR068293 and by the Health Institute Carlos III and FEDER to C. Paradas (FIS PI13-01739). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. STR DNA fingerprinting was done by the CCSG-funded Characterized Cell Line Core, NCI # CA016672 at the Characterized Cell Line Core Facility of the University of Texas MD Anderson Center. Authors also would like to thank the Human Stem Cell Core and the Baylor Genetics Laboratories of the Baylor College of Medicine at Houston for performing the karyotype analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Servian-Morilla E, et al. A POGLUT1 mutation causes a muscular dystrophy with reduced Notch signaling and satellite cell loss. EMBO Mol Med. 2016;8(11):1289–1309. doi: 10.15252/emmm.201505815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rana NA, et al. O-glucose trisaccharide is present at high but variable stoichiometry at multiple sites on mouse Notch1. J Biol Chem. 2011;286(36):31623–37. doi: 10.1074/jbc.M111.268243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez-Valdivia R, et al. Regulation of mammalian Notch signaling and embryonic development by the protein O-glucosyltransferase Rumi. Development. 2011;138(10):1925–34. doi: 10.1242/dev.060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seki T, Yuasa S, Fukuda K. Generation of induced pluripotent stem cells from a small amount of human peripheral blood using a combination of activated T cells and Sendai virus. Nat Protoc. 2012;7(4):718–28. doi: 10.1038/nprot.2012.015. [DOI] [PubMed] [Google Scholar]
- 5.Deutschmann SM, Kavermann H, Knack Y. Validation of a NAT-based Mycoplasma assay according European Pharmacopoiea. Biologicals. 2010;38(2):238–48. doi: 10.1016/j.biologicals.2009.11.004. [DOI] [PubMed] [Google Scholar]