Abstract
Triplication of chromosome 21 (trisomy 21) results in Down syndrome (DS), the most common live-born human aneuploidy. Individuals with DS have a unique facial appearance that can include form changes and altered variability. Using 3D photogrammatic images, 3D coordinate locations of 20 anatomical landmarks, and Euclidean Distance Matrix Analysis methods, we quantitatively test the hypothesis that children with DS (n = 55) exhibit facial form and variance differences relative to two different age-matched (4–12 yrs.) control samples of euploid individuals: biological siblings of individuals with DS (n = 55) and euploid individuals without a sibling with DS (n = 55). Approximately 36% of measurements differ significantly between DS and DS-sibling samples, whereas 46% differ significantly between DS and unrelated control samples. Nearly 14% of measurements differ significantly in variance between DS and DS sibling samples, while 18% of measurements differ significantly in variance between DS and unrelated euploid control samples. Of those measures that showed a significant difference in variance, all were relatively increased in the sample of DS individuals. These results indicate that faces of children with DS are quantitatively more similar to their siblings than to unrelated euploid individuals and exhibit consistent, but slightly increased variation with most individuals falling within the range of normal variation established by euploid samples. These observations provide indirect evidence of the strength of the genetic underpinnings of the resemblance between relatives and the resistance of craniofacial development to genetic perturbations caused by trisomy 21, while underscoring the complexity of the genotype-phenotype map.
Keywords: Down syndrome, human facial variation, gene-dosage imbalance, morphometric, Euclidean Distance Matrix Analysis (EDMA)
INTRODUCTION
Down syndrome (DS) is caused by inheritance of three copies of the genes on human chromosome 21 (HSA21) and occurs in all ethnic backgrounds and socioeconomic classes worldwide, at a frequency of about 1:700 live births [Azman et al. 2007; CDCP 2006; Kuppermann et al. 2006]. HSA21 genes are over-expressed as a consequence of triplication of chromosome 21 (i.e., trisomy 21), resulting in gene-dosage imbalance [Reeves et al. 2001]. Broad developmental, anatomical, and health consequences have been quantified in humans and several types of DS mouse models across multiple investigations [Hill et al. 2007; McElyea et al. 2016; Parsons et al. 2007; Richtsmeier et al. 2000; Richtsmeier et al. 2002b; Roper et al. 2009; Starbuck et al. 2014a]. However, the details of how dosage imbalance of HSA21 genes affects morphogenesis are not well understood.
Individuals with DS invariably exhibit a characteristic facial morphology and impaired cognition, although the severity of these manifestations varies from person to person [Megarbane et al. 2009; Roper and Reeves 2006]. Facial features associated with DS can include epicanthic folds, oblique palpebral fissures, a depressed nasal bridge, an upturned nose, reduced mandibular and maxillary size, midfacial retrusion (hypoplasia), impaired craniofacial growth, a relatively short face, an open-mouthed facial posture that may include a protruding tongue, and numerous other anatomical changes [Pueschel 2000]. While the face is affected in all people with DS, the degree of anatomical change in osseous and soft-tissue craniofacial characteristics induced by trisomy 21 during morphogenesis and subsequent growth varies on a person by person basis [Alio et al. 2008; Yahya-Graison et al. 2007].
Reports of increased phenotypic variation in individuals with DS are often based on limited qualitative assessments [Bersu 1980; Dunlap et al. 1986]. Although qualitative labels are useful for identifying and grouping facial features of individuals with DS, such labels may be poorly defined and subject to experience level of the person assessing features. These factors may influence data collection, analysis, and could potentially obscure true patterns of anatomical change associated with trisomy 21 and its impact on morphogenesis and growth. Many studies that have attempted to quantify variance associated with samples of individuals with DS have used only a few relatively simple measures of the palate, teeth, and dermatoglyphics [e.g. Barden 1980; Shapiro 1975; Shapiro et al. 1967]. Moreover, many early investigations of craniofacial morphology and variance in samples of individuals with DS were based on two-dimensional analyses from lateral or frontal cephalograms that cannot adequately capture variation and surface topography in three dimensions [e.g., Frostad 1971; Kisling 1966; O’Riordan 1979]. Interestingly, in one three-dimensional study of facial morphology Italian subjects with DS were found to have smaller facial size and mean z-scores falling outside the normal interval when compared to normal sex-, ethnic-, and age-matched controls [Sforza et al. 2005]. An additional investigation found differences in variance between Italian and Northern Sudanese subjects with DS, suggesting that patterns of variation may be influences by ethnic background [Sforza et al. 2015].
It has been argued that trisomy 21 alters facial morphology sufficiently to obscure family resemblance completely [Opitz and Gilbert-Barness 1990], although previous quantitative attempts to evaluate facial similarity between individuals with DS and their siblings were inconclusive [Shaner et al. 2001]. In samples of unaffected individuals, the resemblance between relatives is easily recognized by most observers, and different studies have shown that facial dimensions of both osseous and soft tissue are heritable [Baydas et al. 2007; Sherwood et al. 2008]. Therefore, we might expect family resemblance in facial appearance between individuals with DS and their euploid siblings since they share, on average, 50% of their genetic variation.
The purpose of this investigation is to quantitatively evaluate patterns of facial form and variance differences using three-dimensional images of individuals with DS, euploid siblings of individuals with DS, and unrelated euploid controls. We hypothesize that the face of a person with DS will resemble his or her unaffected family members for some quantitative facial traits and will resemble other, unrelated persons with DS for other traits. Our expectation is that faces of age-matched children with DS and their siblings exhibit fewer significant form and variance differences relative to the faces of age-matched, unrelated euploid children. Furthermore, we expect stronger, more distinct patterns of variance differences to be present between age-matched children with DS and unrelated euploid controls.
MATERIALS AND METHODS
Down Syndrome and Euploid Samples
We employ a three-sample study design to understand differences in facial morphology between samples of: 1) individuals with Down syndrome (hereafter referred to as the DS sample; n = 55); 2) euploid siblings of individuals with DS (hereafter referred to as the DSsib sample; n = 55); and 3) unrelated euploid individuals (hereafter referred to as the EU sample; n = 55). Each sample includes individuals ranging in age from 4 to 12 years that are age-matched across groups. Individuals reported by parents to have DS from mosaicism, translocation, or mosaic translocation were excluded from analysis. Sex ratios between age-matched samples are similar but not identical (sample 1: 45% male, 55% female; sample 2: 48% male, 52% female; sample 3: 38% male, 62% female). Precise matching by age and reported ethnicity was not possible, but the majority of individuals in each sample are Caucasian. Since ethnic differences have been found in previous publications on DS [Sforza et al. 2015], the inability to precisely match ethnicity may influence the results. All individuals were recruited under protocols reviewed and approved by a duly constituted ethics committee (the Pennsylvania State University IRB # 23283 and #36627).
Facial Image Acquisition and Data Collection
Three-dimensional facial images of each individual were acquired using the 3dMD photogrammatic system (3dMD, Atlanta, GA). This system acquires multiple images of an individual’s face simultaneously and uses algorithms developed by 3dMD to automatically merge two-dimensional images into a single three-dimensional surface. Images were acquired in <1.5 milliseconds while each individual sat in an upright position and displayed a neutral facial expression. Previous studies have found that 3dMD surface images are accurate three-dimensional representations of facial topography and that three-dimensional locations of anatomical landmarks recorded from 3dMD images are accurately located and highly repeatable [Aldridge et al. 2005; Weinberg et al. 2006; Wong et al. 2008].
To assess measurement error of anatomical landmark placement, coordinates for twenty anatomical soft-tissue landmarks were collected repeatedly from surface images of five individuals drawn randomly from the overall sample (Supplemental Fig. 1). Using 3dMDpatient (v4.0) software, each image was landmarked on five separate occasions, with at least 24 hours between landmarking sessions to avoid landmark placement memory bias. Standard deviations of landmark coordinate locations along the x, y, and z axes were averaged to calculate mean measurement error [Aldridge et al. 2005; Valeri et al. 1998]. Mean measurement error local to each landmark was estimated to be 0.29mm (0.26mm along the x-dimension, 0.30mm along the y-dimension, and 0.31mm along the z-dimension) and is considered sufficiently low for the purposes of this study.
Following assessment of measurement error, anatomical landmark coordinates were recorded from each image on two separate occasions with at least 24 hours between landmarking trials as done in previous investigations [Starbuck et al. 2011; Starbuck et al. 2013; Starbuck et al. 2014b]. After inspection of the data for gross landmarking errors (e.g., mislabeling right and left side landmarks), landmark coordinates were averaged from the two separate digitizing episodes to further minimize any potential effects of measurement error for each individual and these average measures were used in analyses.
Analysis of Facial Morphology
Two tests were used to determine whether statistically significant differences in facial form exist between the samples. The first is a test for global form similarity using Euclidean Distance Matrix Analysis (EDMA) [Lele and Richtsmeier 1991; Lele and Richtsmeier 2001]. The size and shape of each individual was quantified using a form matrix (FM), consisting of the Euclidean distances between all pairs of landmarks. With 20 landmarks, there are 190 unique inter-landmark linear distances, and a mean form matrix (FM) was computed for each sample [Lele and Richtsmeier 1995; Richtsmeier et al. 2005; Richtsmeier et al. 2002a]. Differences in mean form are compared using a form difference matrix (FDM), defined as a matrix of the ratios of all homologous linear distances between the sample mean forms. For example, to compare the DS and DSsib mean forms: FDM(DS, DSsib)ij = FM(DS)ij/FM(DSsib)ij, for every landmark pair i,j, where divisions are elementwise and 0/0=0. The null hypothesis is that the mean forms are the same, which would result in a FDM consisting of "1s" in all of the off-diagonal elements. The degree of the “overall” difference in form is measured by the statistic T = max/min(FDM). When two mean forms are identical (as expected under the null hypothesis), T=1; T will increase as two forms become more different. The null hypothesis is evaluated using nonparametric bootstrapping (10,000 resamples) [Lele and Richtsmeier 1991; Lele and Richtsmeier 2001]. We used a p-value of ≤ 0.05 to indicate rejection of the null hypothesis for the global EDMA test.
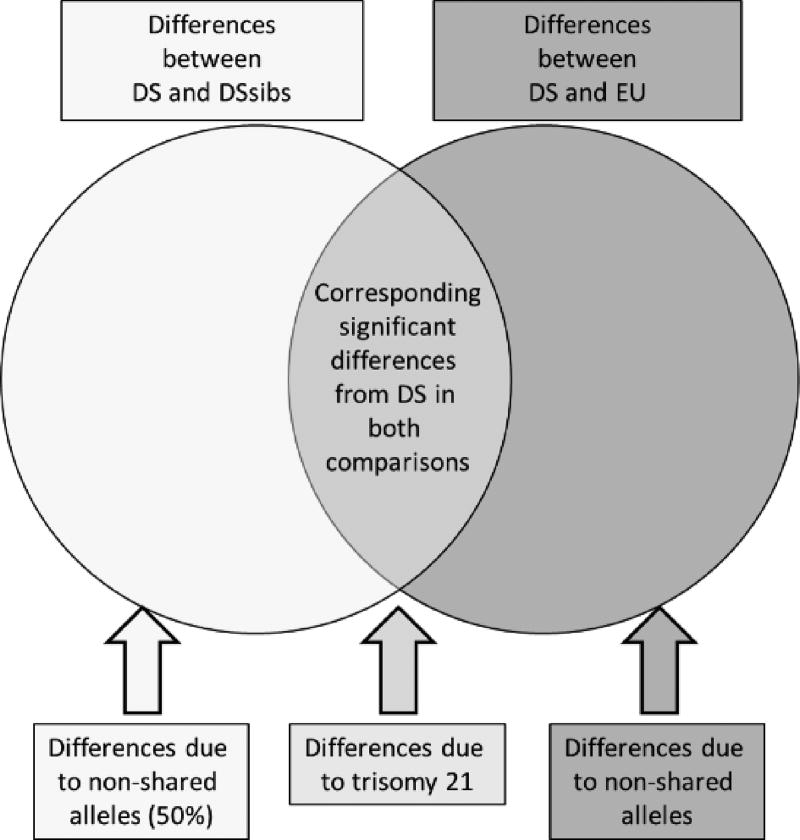
Localized differences in form (i.e., those involving specific linear distances) between samples were evaluated by computing confidence intervals for the ratios of homologous linear distances from each sample. Nonparametric bootstrapping was used to estimate 90% marginal confidence intervals (10,000 resamples) [Lele and Richtsmeier 1995; Lele and Richtsmeier 2001]. Global and local form analyses were carried out to estimate differences between the DS, DSsib, and EU samples. Homologous measurements that significantly differed between the DS sample and both euploid samples are attributed to the effects of trisomy 21 (Fig. 1).
Figure. 1.
Diagram of how morphological differences are attributed to the effects of trisomy 21 and genetic relatedness (or lack thereof) among the Down syndrome (DS), Down syndrome sibling (DSsib), and unrelated euploid (EU) samples. A diagram of differences from two sample comparisons of the DS sample compared to DSsibs (left) and DS compared to EU (right) are shown. The DS sample is common to each two-sample analysis. Significant measurements are divided into those that overlap among two-sample comparisons and those that do not. Homologous significant measurements from each two-sample comparison that included the DS sample are attributed to the effects of trisomy 21 on facial morphology. These corresponding differences were subtracted to remove the effects of DS on facial morphology and to explore morphological similarity between DSsibs, who share approximately 50% of alleles, and unrelated EU individuals. Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/journal/10.1002/(ISSN)1552-4833.
To assess intra- and inter-specific facial variance and differences in facial variance, we conducted an EDMA-based ordination procedure known as principal coordinates analysis (PCOORD) [Gower 1966; Reyment et al. 1984; Richtsmeier et al. 1998], which included individuals from all three samples (DS, DSsib, and EU). PCOORD summarizes the observed variation in form by projecting the samples into a low-dimensional space (i.e., two or three dimensions, compared to the number of interlandmark distances, which is 190). The purpose of this projection is to summarize complex patterns of variation in a way that makes them easier to interpret and visualize. Individuals that lie close together in the low-dimensional space are relatively similar in overall form, while more distant individuals are more dissimilar. The overall spread of samples of individuals in high-dimensional space provides a visual indication of relative differences in sample mean forms and variance.
To implement PCOORD, we first represent the form of an individual by computing all of the unique linear distances among the three-dimensional facial landmarks. These measurements are stored as a form matrix (FM) for each individual [Lele and Richtsmeier 2001]. Next, a form difference matrix (FDM) was estimated for each pair of individuals. A measure of overall dissimilarity between any two individuals was then computed from their FDM and, called FΩ. For example, for individuals A and B, over all landmark pairs i,j. If A and B have identical forms, FΩ(A,B) = 0; otherwise, FΩ becomes increasingly positive as A and B become more different. All of the pairwise FΩ statistics were placed into a square, symmetric dissimilarity matrix called FΩ. For example, if there are three individuals called A, B, and C, then
This matrix serves as the basis for the PCOORD analysis. After double-centering the matrix, so that its rows and columns all sum to zero [Gower 1966], it is subjected to eigenanalysis, which projects the individuals into a low-dimensional space. The space is defined with respect to a series of mutually-orthogonal principal coordinate axes. The square roots of the resulting eigenvalues describe the lengths of the principal axes, and the eigenvectors are the “scores” that describe where individual subjects fall along each axis.
The positioning of individuals along the principal axes can be interpreted in terms of original facial anatomy by estimating and examining correlations between eigenvector coefficients (scores on the axis) and the original interlandmark distances. Absolutely high correlations indicate that a given facial measurement is important for explaining the distribution of facial shapes along a particular axis. High positive correlations mean that an individual's position at the positive end of the axis is associated with large measures for those linear distances. Conversely, high negative correlations indicate that an individual's position at the positive end of the axis is associated with small measures for those linear distances.
Differences in the variances of interlandmark measures were assessed by a variant of the Hall-Martin test [Hall and Martin 1988] that compares sample statistics using bootstrap confidence intervals. All computations were performed using the MIBoot program [Cole III 2002]. For each measurement, a marginal confidence interval was calculated for the difference in sample variances. Each distance was first ln-transformed, and then the sample variances (s2) and their difference (s2diff) were computed; for example: s2diff(DS, DSsib) = s2(DS) − s2(DSsib). Parametric bootstrapping (with 10,000 random resamples) was used to compute a 90% confidence interval for each distance’s s2diff statistic. Under the null hypothesis of equal variances, the difference is expected to be 0; however, if the bootstrap confidence interval excludes 0, the null hypothesis is rejected for that interlandmark distance. Using this method, within-sample variances were estimated for all linear distances and compared across samples to determine inter-sample similarity of variance of facial measures. As with the previous EDMA form analysis, measurements with a variance that differed significantly between the DS sample and both euploid samples were attributed to the effects of trisomy 21 (Fig. 1).
RESULTS
DS Facial Morphology is More Similar to Siblings than Unrelated Euploid Controls
The global form comparison revealed that the DS and DSsib samples differ significantly (p-value = 0.01) (Table I). Form difference confidence intervals reveal that approximately 36% (69/190) of linear distances differ significantly between the DS and DSsib samples (Fig. 2A). These distances are, on average, 10% smaller (range 7–19% smaller) in the DS sample relative to the DSsib sample. The upper-facial landmarks, glabella, nasion, endocanthion, and exocanthion, were most frequently involved in linear distances that differ significantly between the DS and DSsib samples, although midfacial landmarks pronasale and subnasale were also repeatedly involved in significant differences in facial morphology between these samples.
Table I.
Down syndrome (DS), DS sibling (DSsib), and unrelated euploid (EU) facial form difference results.
| Samples Compared | Global Form Difference Results |
Confidence interval results for local linear distance (LD) form differences | ||
|---|---|---|---|---|
| Percentage of significantly different LDs |
Number of significantly smaller LDs | Number of significantly larger LDs |
||
| DS compared to DSsib | p-value = 0.01* | 69/190 (36%) | 69/190 (Avg. 10% smaller in DS; Range 7–19% smaller in DS) | 0/190 |
| DS compared to EU | p-value = 0.001* | 88/190 (46%) | 88/190 (Avg. 9% smaller in DS; Range 6–18% smaller in DS) | 0/190 |
| DSsib compared to EU | p-value = 0.124 | 1/190 (<1%) | 0/190 | 1/190 |
indicates that testing for global differences in form for each craniofacial region was statistically significant
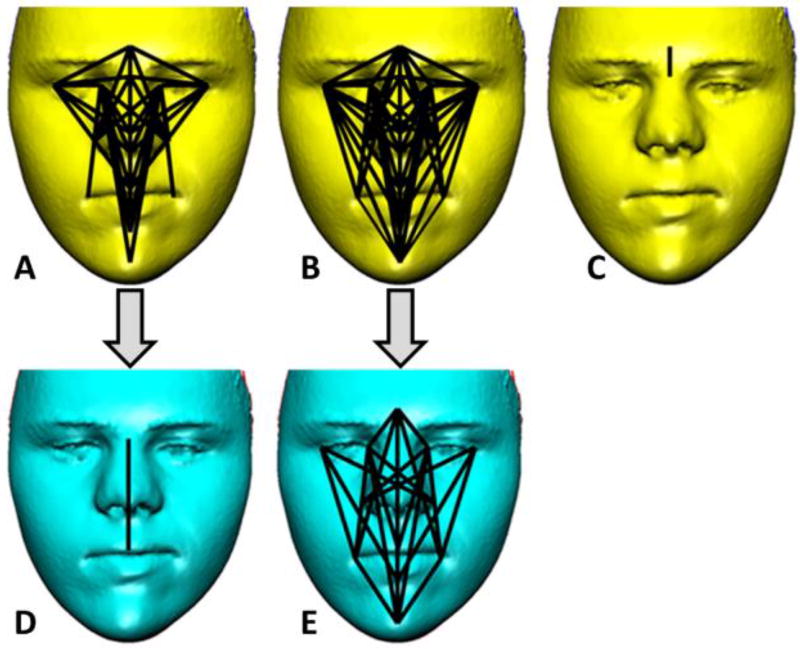
Figure. 2.
Facial form differences between Down syndrome (DS), DS sibling (DSsib), and unrelated euploid (EU) samples. A) Linear distances (LDs) that significantly differ between the DS and DSsib samples (69/190 or 36%). The LDs are all 7–19% smaller in the DS sample relative to the DSsib sample. B) The LDs that significantly differ between the DS sample and the EU sample (88/190 or 46%) are shown. All LDs are 6–18% smaller in the DS sample relative to the EU sample. C) The LD that differs significantly between the DSsib sample and the EU sample (1/190 or <1%) are shown. D) For the DS and DSsib comparison one significant LD remains after subtracting 68 LDs that were common to A and B and attributed to the effects of trisomy 21 (see Fig. 3). E) For the DS and EU comparison 20 significant LDs remain after subtracting 68 LDs that were common to A and B and attributed to the effects of trisomy 21. Facial images shown have been modified to remove identifiable features. Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/journal/10.1002/(ISSN)1552-4833.
A test of global difference in morphology revealed that the faces of the DS and EU samples differ significantly (p-value = 0.001) (Table I). Approximately 46% (88/190) of the confidence intervals indicated significant differences between EU individuals and those with DS (Fig. 2B). Similar to the comparison of DS and DSsib samples, all significant linear distances were an average of 9% smaller (range 6–18% smaller) in the DS sample, relative to homologous EU sample facial measures. The pattern of landmarks involved in significant linear distance differences between the DS and EU samples is similar to the differences defined between DS and DSsib samples above. The landmarks glabella, nasion, endocanthion, and exocanthion of the upper face contribute most frequently to the significant inter-sample differences, followed by midfacial landmarks pronasale and subnasale.
Globally, we failed to reject the null hypothesis of similarity in overall facial form between the DSsib and EU samples (p = 0.124) (Table I). Locally, only a single linear distance (1/190) representing an upper facial measure between glabella and nasion was significantly different between the two samples of euploid individuals (Fig. 2C).
The pattern of differences in facial morphology between the DS sample and each of the two euploid samples (DSsibs and EU) is similar (i.e., Fig. 2A and 2B). Sixty-eight of the 69 linear distances that were significantly different between the DS sample and the DSsib sample were also significantly different between the DS sample and the EU sample. Since we used two samples of euploid individuals, one that comprises full siblings of individuals with DS, the other comprised of unrelated euploid individuals, the source of the differences in facial morphology between individuals with DS and the two samples of euploid individuals is attributed to the genetic perturbation of trisomy 21 (Fig. 1). We consider those homologous linear distances showing significant differences between the samples of DS individuals and both of the euploid samples (68 linear distances) as trisomy 21-dependent. After removing the trisomy 21-dependent measures from the comparison of the DS and DSsib samples to explore patterns of differences not associated with having an extra copy of HSA21, only a single linear distance measuring midfacial height is significantly different between samples (Fig. 2D). This indicates overall similarity in facial form between individuals with DS and siblings of individuals with DS when the effects of trisomy 21 are removed. When the trisomy 21-dependent measures (i.e., the shared 68 significant linear distances) are removed from the comparison of DS and unrelated EU individuals, 20 linear distances are found to be significantly different between the DS and EU samples (Fig. 2E). This indicates substantial differences in facial form between these two samples in addition to those due to trisomy 21.
Down Syndrome Facial Variance is Elevated Relative to Euploid Controls
Collectively, principal coordinates (PCs) 1 and 2 account for nearly 60% of the variation in squared distances between subjects when all three samples are considered together. The distribution of individuals along PC1 and PC2 is shown in Figure 3, with each sample formspace outlined as a convex hull. The range of form-space occupied by the sample of DS individuals is larger relative to the space occupied by the two samples of euploid individuals (i.e., DSsibs and EU) along PC1 and PC2. The range of form-space occupied by the two euploid samples is similar, and both euploid samples almost completely overlap in high dimensional space.
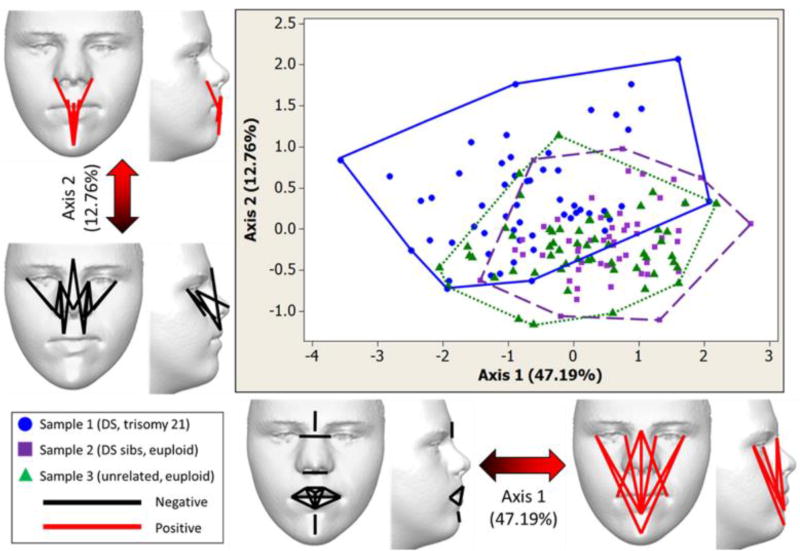
Figure 3.
Principal coordinate analysis plot of the first two principal axes (PCs) for the Down syndrome (DS), Down syndrome siblings (DSsibs), and unrelated euploid (EU) samples and morphological differences associated with each PC. PCs 1 and 2 explain the majority of facial variation among samples (59.95%). Although samples overlap, much of the DS sample (circles, solid outline) is found along the negative end of PC1 relative to the euploid samples (DSsibs depicted as squares and dotted outline; EU depicted as triangles and dashed outline). Along both PCs the sample outlines suggest that the range of formspace occupied by the DS sample is larger than the two euploid samples. The range of formspace occupied by the two euploid samples overlaps and is similar. Linear distances that are strongly correlated with the negative and positive ends of the PC axes are depicted on faces. Facial images shown have been modified to remove identifiable features. Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/journal/10.1002/(ISSN)1552-4833.
Although all three samples overlap, individuals in the DS sample span the length of PC1, while most of the euploid individuals are located at the positive end of PC1 (Fig. 3). PC1 accounts for 47.19% of facial variation across the three samples. The negative end of PC1 is associated with relatively small size for intercanthal width, nasal width, lower face height, and numerous measures of the mouth. In contrast, the positive end of PC1 is associated with relatively large size for vertical measures of the lower face and midface, associated with the outer and inner eye commissures, nasal wings, philtrum, mouth, and chin (Fig. 3).
PC2 accounts for 12.76% of the facial variation across the three samples (Fig. 3B). As with PC1, individuals with DS occupy a greater area than euploid individuals due to increased facial variation in the DS sample. Although all three samples occupy negative and positive ends of PC2, the DS sample stretches further into the positive end of this axis, while the euploid samples reach further into the negative end of this axis. The negative end of PC2 is associated with reduced relative height measures of the midface and nose, while the positive end of PC2 is associated with relative increases in vertical measures of the cutaneous upper lip and lower face (Fig. 3).
Comparisons of facial variances using bootstrap confidence intervals revealed that approximately 14% (27/190) of linear distances show significantly increased variance in the DS sample, relative to the DSsib sample (Table II). These significant differences can be divided into those that are increased (23/27 i.e., 89%) or decreased (4/27 i.e., 11%) in the DS sample relative to euploid siblings. Those measurements whose variances are significantly increased exhibit an 84% average increase in variance (range of 58–125% increase) in the DS sample relative to the DSsib sample (Fig. 4A). Those metrics whose variances are significantly decreased exhibit a 40% mean decrease in variance (range of 33–48%) in the DSsib sample relative to the DS sample. The mid-and lower-face landmarks sublabiale, subalare, nasion, pogonion, and subnasale were most frequently involved in linear distances with variances that significantly differ between the DS and DSsib samples.
Table II.
Down syndrome (DS), DS sibling (DSsib), and unrelated euploid (EU) facial variance results.
| Samples Compared | Confidence interval results for local linear distance (LD) variance differences | ||
|---|---|---|---|
| Percentage of significantly different LDs |
Number of LDS with significantly larger variances |
Number of LDS with significantly smaller variances |
|
| DS compared to DSsib | 27/190 (14%) | 23/190 (Avg. 84% variance increase in DS; Range 58–125% variance increase in DS) | 4/190 (Avg. 40% variance increase in EU; Range 33–48% variance increase in EU) |
| DS compared to EU | 34/190 (18%) | 34/190 (Avg. 74% variance increase in DS; Range 14–130% variance increase in DS) | 0/190 |
| DSsib compared to EU | 10/190 (5%) | 5/190 (Average 65% variance increase in DSsib; Range 60–70% variance increase in DSsib) | 5/190 (Avg. 41% variance increase in EU; Range 34–50% variance increase in EU) |
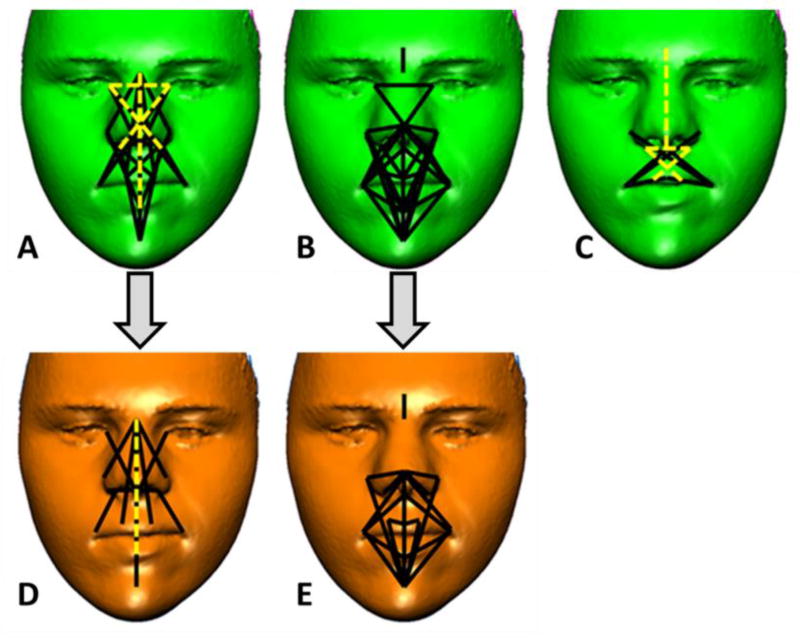
Figure 4.
Differences in facial variance between Down syndrome (DS), Down syndrome sibling (DSsib), and unrelated euploid (EU) samples. A) Linear distances (LDs) with variances that significantly differ (27/190 or 14%) between the DS and DSsib samples. LDs depicted with solid lines have significantly higher variances in the DS sample, and LDs depicted with dashed lines have significantly higher variances in the DSsib or EU sample. B) The LDs with variances that significantly differ between the DS and EU samples (34/190 or 18%) are shown. All LDs exhibit significantly higher variance in the DS sample relative to the EU sample (depicted as solid lines). C) The LDs with variances that significantly differ between the DSsib and the EU samples (10/190 or 5%) are shown. The LDs depicted with solid lines have significantly higher variances in the DSsib sample, and LDs depicted with dashed lines have significantly higher variances in the EU sample. D) For the DS and DSsib comparison 8 significant LDs remain after subtracting 19 LDs that were common to A and B and attributed to the effects of trisomy 21 (see Fig. 3). E) For the DS and EU comparison 15 significant LDs remain after subtracting 19 LDs that were common to A and B and attributed to the effects of trisomy 21. Facial images shown have been modified to remove identifiable features. Color figure can be viewed in the online issue, which is available at http://onlinelibrary.wiley.com/journal/10.1002/(ISSN)1552-4833.
Nearly 18% (34/190) of linear distances have significantly unequal variances between the DS and EU samples (Table II) and the variance is always relatively increased in the DS sample by an average of 74% (range of 14–130%) (Fig. 4B). The midface landmarks subalare, crista philtra, pronasale, alar curvature, and chelion were most frequently involved in linear distances with variances that are significantly increased in the DS sample relative to EU samples.
When the two euploid samples (DSsib and EU) are compared, relatively few differences in variance were found, and because of the small number of differences, biological interpretations of them should be made with caution. Approximately 5% (10/190) of the linear distances have variances that differ significantly between the DSsib and EU samples (Table II). These significant differences can be divided into those that are increased (5/10 i.e., 50%) or decreased (5/10 i.e., 50%) in the DSsib sample relative to EU sample. Those measurements whose variances are significantly increased exhibit a 65% average increase (range of 60–70%) in the DSsib sample relative to the EU sample (Fig. 4C). Those metrics whose variances are significantly decreased exhibit a 41% mean decrease in variance (range of 34–50%) in the EU sample relative to the DSsib sample. The landmarks chelion and subnasale were most frequently involved in linear distances with variances that significantly differ between the DSsib and EU samples
The set of significant differences in facial variance between the DS and DSsibs samples, and between the DS and EU samples, included 19 homologous linear measures with variances that were significantly different in each two-sample comparison involving the DS sample (i.e., Fig. 4A and 4B). Similar to the form analysis presented above, the 19 homologous facial measures that showed increased variance in DS individuals relative to both euploid samples were removed from a diagram of significant results (Fig. 4D and 4E). The use of related and unrelated euploid control samples allowed us to indirectly differentiate the effects of trisomy 21 and background genes (shared between siblings or not shared in unrelated euploid controls) on patterns of differences in facial variance. After removing homologous linear distances with significantly different variances that were attributed to the effects of trisomy 21, the remaining dimensions of the face with increased variance in individuals with DS relative to DSsibs include measures of facial height and midfacial measures of the nose and philtrum (Fig. 4D), while remaining significant variance differences between the DS and EU samples are localized to the lower face around the mouth, chin, philtrum, inferior border of the nose, and nose width (Fig. 4E). This lack of correspondence in patterns of significant variance differences between DS and DSsibs, and DS and EU, after removing effects attributed to trisomy 21 (i.e., homologous significant differences between two-sample comparisons), provides indirect evidence that genetic relatedness or lack thereof influences patterns and localization of variance differences between samples.
DISCUSSION
Craniofacial morphology is the result of a complex developmental program directed by manifold interactions of underlying genes and the environment. The roles of some of these genes in developmental processes are known, while others are yet to be discovered. Facial morphogenesis requires the correct spatiotemporal deployment of gene products, neural crest cells, and other cells to develop facial prominences, which in turn must form, grow, and merge according to a species-specific Bauplan [Brugmann et al. 2006; Feng et al. 2009]. Developmental deviations, such as those caused by trisomy 21 and gene-dosage imbalance, can lead to potentially detrimental birth defects and craniofacial anatomical changes by acting as a perturbation and modifying developmental morphogenetic pathways [Young et al. 2014]. Epigenetic effects may also influence facial form, but this topic is beyond the score of this paper which does not take into account any possible epigenetic effects of trisomy 21 on facial development.
The majority of studies assessing DS facial characteristics have done so by comparing and contrasting individuals with DS to unrelated “normal” or “typical” (i.e., euploid) individuals [Farkas et al. 2001b; Farkas et al. 2002a; Farkas et al. 2002b; Farkas et al. 1985; Ferrario 2004; Ferrario et al. 2004a; Ferrario et al. 2004b; Sforza et al. 2005; Sforza et al. 2011], thus confounding differences due to shared or unshared background genes with differences caused by trisomy 21. Although siblings share, on average, 50% of their genes, it has been argued that trisomy 21 alters facial morphology to such an extent that family resemblance is obscured [Opitz and Gilbert-Barness 1990]. Research designs that compare a genetically perturbed human population to unrelated control groups are unable to partition variation due to differences in genetic background from those due the genetic perturbation being investigated, thereby influencing the amount of confidence in the results and the ability of follow-up studies to replicate results from previous investigations. Our inclusion of control samples of age-matched siblings of individuals with DS and unrelated euploid individuals enabled the analytic separation of facial variation due to trisomy 21 from variation due to differences in shared ‘background’ genes among siblings (about 50%) and unshared background genes in unrelated euploid individuals.
Our results indicate that the faces of individuals with DS differ significantly from those of siblings of individuals with DS and unrelated euploid individuals in consistent ways, reflecting the effects of trisomy 21 upon typical facial development. The majority of facial measures that differed significantly between the DS and both euploid samples (DSsibs and EU) were smaller in the DS sample, thereby indicating the influence of triplicated HSA21 genes on facial features. It has been reported that faces of individuals with DS exhibit reduced facial dimensions including biocular distance, nasal height, width and protrusion, lip height and mouth width [Farkas et al. 2001a; Farkas et al. 2001b; Farkas et al. 1985; Farkas et al. 1991; Ferrario 2004; Ferrario et al. 2004a; Ferrario et al. 2004b; Sforza et al. 2005; Sforza et al. 2004; Sforza et al. 2011]. However, most facial measures of individuals with DS lie within the normal range established by previous investigations and those that do not tend to become more similar to measures from typical individuals as children mature into adults [Farkas et al. 2002a; Farkas et al. 2002b]. We found that many linear dimensions (i.e., 36–46%) of the face corresponding to those listed above are significantly reduced in children with DS, but overall a larger percentage of linear measures failed to reach statistical significance. This is true for variance differences as well (14–18% differed significantly). Only a single measurement was significantly different between the two samples of euploid individuals (i.e., DSsibs and EU). This result provides further evidence that the morphological differences found between the DS sample and each euploid sample are driven by the 1.5-fold increase in expression of triplicated HSA21 genes.
Removal of 68 corresponding facial measures that significantly differed in the form comparison of DS to both euploid samples leaves only a single facial measure identified as significantly different between the sample of individuals with DS and the sample of siblings of individuals with DS, whereas 20 facial measurements define morphological differences unique to the comparison of age-matched individuals with DS and the unrelated euploid sample (Fig. 2D and 2E). Overall, the patterns of these results suggest that, after accounting for the effects of trisomy 21 on facial features, the faces of individuals with DS and siblings of individuals with DS are quantitatively more similar to one another than the faces of individuals with DS and unrelated euploid faces. We attribute this to the approximate 50% shared allelic background between the DS and DS siblings and propose that, despite having an extra copy of HSA21, there is a quantifiable familial resemblance between individuals with DS and their euploid siblings.
All samples overlap in multivariate form space (Fig. 3), but the sample of DS individuals partially separates from the two euploid samples. The facial morphology of some individuals with DS establishes the extreme negative end of PC1 and the extreme positive end of PC2, thereby representing facial variation outside of the typical range established by the two euploid control samples (i.e., siblings of individuals with DS and unrelated euploid individuals). The sample of individuals with DS exhibits a wider range of facial variation as evidenced by the extensive spread of individuals with DS relative to euploid samples, which overlap almost completely along the PC axes (Fig. 3). Approximately 14% (DSsib) or 18% (EU) of facial variance measures significantly differ when the sample of individuals with DS is compared to siblings or unrelated individuals (Fig. 4A and 4B), respectively, resulting in unique patterns of midfacial and lower facial variance differences that remain even after significant effects attributed to trisomy 21 are removed (Fig. 1). These results suggest that when DS and euploid individuals are siblings, the main differences in facial variance are localized to the midface, whereas when DS and euploid individuals are unrelated, the main differences in variance are localized to the lower face and the inferior portion of the midface, including the philtrum. Although trisomy 21 influences all facial regions, comparisons of age-matched samples that either share or do not share approximately 50% of their alleles, on average, yield different patterns of differences.
It has been reported that anatomy influenced by trisomy 21 is more variable than that of typical populations [Farkas et al. 1985; Sforza et al. 2004]. We found an approximate 2.8- to 3.6-fold increase (i.e., 14/5 to 18/5) in percentages of significant facial variance differences associated with the DS sample. Generally, these results concur with assertions from previous investigations, but our results differ because we have localized variance differences to particular facial regions, thereby illustrating that variance differences are not distributed evenly throughout the entire face.
Our quantitative analysis of multivariate formspace shows that the majority of DS faces fall within the distribution of euploid faces (Fig. 3), despite the fact that the DS sample is consistently more variable than both euploid samples along PC axes. This result suggests that the means of many facial measures may be significantly reduced in populations of individuals with DS and variances of some facial measures may be increased beyond the range typically associated with euploid populations (see Fig. 5). This result may help investigators model variation associated with mild and more extreme degrees of dysmorphology exhibited by some individuals with trisomy 21, based on how closely they fall to the mean or extreme tails of the population distribution. Future studies should address whether these patterns of form and variance differences remain consistent in older individuals as faces mature after puberty and into adulthood.
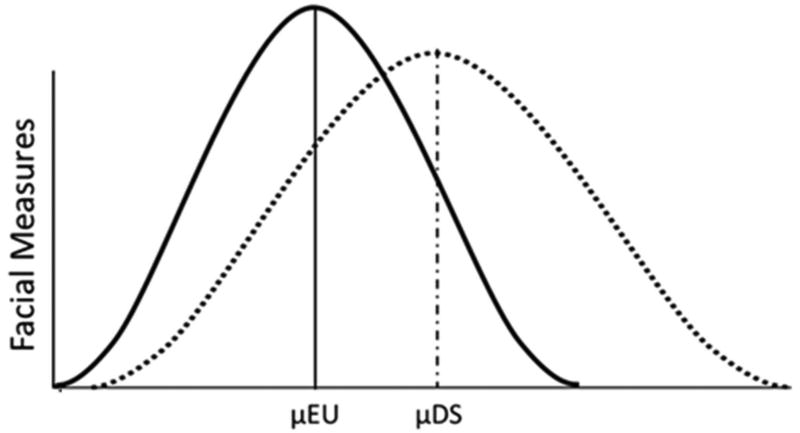
Figure 5.
Model of differences in distribution of facial measures for euploid (EU) and Down syndrome (DS) populations. The EU (solid) and DS (dotted) distributions are shown overlaid upon each other. Facial measures are represented along the y-axis. Along the x-axis each population has a mean trait value (µ), with most measures being reduced in the DS population due to impaired or reduced facial growth. The DS population is more variable as shown by the left tail of the DS distribution encapsulating most of the euploid distribution while simultaneously expanding outside of the typical EU range of variation and into a region of unique morphological variation represented by the right-side of the distribution tail – a region associated with the anatomical differences and “unique” constellation of facial phenotypic characteristics associated with DS. In the right tail of the DS distribution, those individuals just beyond the euploid range of variation may have a mild facial phenotype while those individuals further outside of the EU distribution may exhibit relatively more dysmorphology and perhaps more severe anatomical and health issues associated with the craniofacial complex.
Trisomy 21 is an extreme form of copy number variation whereby multiple triplicated genes potentially interact with each other and genes on other chromosomes using complex regulatory mechanisms to alter developmental trajectories and facial morphogenesis. Here, we have shown that faces of children with DS exhibit global and local form and variance differences relative to typical faces. However, patterns of differences are unique and vary based upon effects attributed to trisomy 21, genetic relatedness of control groups, and portion of the face affected. These observations provide indirect evidence of the strength of the genetic underpinnings of the resemblance between relatives and the resistance of craniofacial development to trisomy 21, while underscoring the complexity of the genotype-phenotype map. Though our study clearly shows that trisomy 21 influences facial form, the combination of the effects of those alleles shared by siblings is apparent in our quantitative assessment of the resemblance between relatives: DS and DS sibs differ from each other less than DS individuals differ from unrelated members of the euploid population.
Our results have implications for research design, particularly for the choice of control group (e.g., genetically related or unrelated) when comparing potentially heritable phenotypic measures. Patterns of facial form and variance differences are influenced by decisions about the type of control group to use for statistical comparison (i.e., siblings vs. unrelated individuals). Control group decisions and differences across studies may help explain heterogeneous results reported in literature on DS and some of the differences found between human and DS mouse model studies. This is because human-based studies typically compare genetically unrelated samples to each other, while many mouse-based studies compare cohorts of inbred strains that are genetically related to each other.
Supplementary Material
Supplemental Fig. 1. Anatomical soft-tissue facial landmarks collected. 3dMDpatient (v4.0) was used to record the three-dimensional coordinates of 20 standard landmarks (circles) from each image on two separate occasions. Eight medial and six bilateral landmarks were used in this study: 1) glabella (g), 2) nasion (n), 3) pronasale (prn), 4) subnasale (sn), 5) labiale superius (ls), 6) labiale inferius (li), 7) sublabiale (Sl), 8) pogonion (SPg), 9) endocanthion (en), 10) exocanthion (ex), 11) alar curvature (ac), 12) subalare (sbal), 13) crista philtra landmark (cpl), and 14) chelion (ch). Landmark definitions can be found at http://getahead.psu.edu/. Facial image shown has been modified to remove identifiable features.
Acknowledgments
The authors acknowledge support from the following grants: NIDCR/NIH: R01-HD038384 (RHR), R01-DE018500, R01-DE018500-S1, R01-DE022988, P01HD078233 (JTR); NSF: DGE-053135 (JMS), BCS-1061563 (JMS); BCS-0725227 (JTR).
References
- Aldridge K, Boyadjiev SA, Capone GT, DeLeon VB, Richtsmeier JT. Precision and error of three-dimensional phenotypic measures acquired from 3dMD photogrammetric images. Am J Med Genet A. 2005;138A(3):247–253. doi: 10.1002/ajmg.a.30959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alio JJ, Lorenzo J, Iglesias C. Cranial base growth in patients with Down syndrome: a longitudinal study. Am J Orthod Dentofacial Orthop. 2008;133(5):729–737. doi: 10.1016/j.ajodo.2006.03.036. [DOI] [PubMed] [Google Scholar]
- Azman BZ, Ankathil R, Siti Mariam I, Suhaida MA, Norhashimah M, Tarmizi AB, Nor Atifah MA, Kannan TP, Zilfalil BA. Cytogenetic and clinical profile of Down syndrome in Northeast Malaysia. Singapore medical journal. 2007;48(6):550–554. [PubMed] [Google Scholar]
- Barden HS. Fluctuating dental asymmetry: a measure of developmental instability in Down syndrome. Am J Phys Anthropol. 1980;52(2):169–173. doi: 10.1002/ajpa.1330520203. [DOI] [PubMed] [Google Scholar]
- Baydas B, Erdem A, Yavuz I, Ceylan I. Heritability of facial proportions and soft-tissue profile characteristics in Turkish Anatolian siblings. Am J Orthod Dentofacial Orthop. 2007;131(4):504–509. doi: 10.1016/j.ajodo.2005.05.055. [DOI] [PubMed] [Google Scholar]
- Bersu ET. Anatomical analysis of the developmental effects of aneuploidy in man: the Down syndrome. Am J Med Genet. 1980;5(4):399–420. doi: 10.1002/ajmg.1320050411. [DOI] [PubMed] [Google Scholar]
- Brugmann SA, Kim J, Helms JA. Looking different: understanding diversity in facial form. Am J Med Genet A. 2006;140(23):2521–2529. doi: 10.1002/ajmg.a.31361. [DOI] [PubMed] [Google Scholar]
- CDCP. Center for Disease Control and Prevention: Improved national prevalence estimates for 18 selected major birth defects – United States, 1999—2001. Morbidity and Mortality Weekly Report. 2006;54(51&52):1301–1305. [PubMed] [Google Scholar]
- Cole T., III MIBoot Windows-Based Software for Bootstrap-Based Comparison of Morphological Integration Patterns. Richtsmeier Lab. 2002 Available at: < getahead.psu.edu>.
- Dunlap SS, Aziz MA, Rosenbaum KN. Comparative anatomical analysis of human trisomies 13, 18, and 21: I. The forelimb. Teratology. 1986;33(2):159–186. doi: 10.1002/tera.1420330204. [DOI] [PubMed] [Google Scholar]
- Farkas LG, Katic MJ, Forrest CR. Surface Anatomy of the Face in Down’s Syndrome; Anthropometric Proportion Indices in the Craniofacial Regions. The Journal of Craniofacial Surgery. 2001a;12(6):519–526. doi: 10.1097/00001665-200111000-00003. [DOI] [PubMed] [Google Scholar]
- Farkas LG, Katic MJ, Forrest CR, Litsas L. Surface Anatomy of the Face in Down’s Syndrome; Linear and Angular Measurements in the Craniofacial Regions. The Journal of Craniofacial Surgery. 2001b;12(4):373–380. doi: 10.1097/00001665-200107000-00011. [DOI] [PubMed] [Google Scholar]
- Farkas LG, Katic MJ, Forrest CR. Age-related changes in anthropometric measurements in the craniofacial regions and in height in Down's syndrome. J Craniofac Surg. 2002a;13(5):614–622. doi: 10.1097/00001665-200209000-00004. [DOI] [PubMed] [Google Scholar]
- Farkas LG, Katic MJ, Forrest CR. Surface anatomy of the face in Down's syndrome: age-related changes of anthropometric proportion indices in the craniofacial regions. J Craniofac Surg. 2002b;13(3):368–374. doi: 10.1097/00001665-200205000-00002. [DOI] [PubMed] [Google Scholar]
- Farkas LG, Munro IR, Kolar JC. Abnormal measurements and disproportions in the face of Down's syndrome patients: preliminary report of an anthropometric study. Plast Reconstr Surg. 1985;75(2):159–169. doi: 10.1097/00006534-198502000-00002. [DOI] [PubMed] [Google Scholar]
- Farkas LG, Posnick JC, Hreczko T. Anthropometry of the head and face in 95 Down syndrome patients. Prog Clin Biol Res. 1991;373:53–97. [PubMed] [Google Scholar]
- Feng W, Leach SM, Tipney H, Phang T, Geraci M, Spritz RA, Hunter LE, Williams T. Spatial and temporal analysis of gene expression during growth and fusion of the mouse facial prominences. PLoS One. 2009;4(12):e8066. doi: 10.1371/journal.pone.0008066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrario VF, Dellavia C, Zanotti G, Sforza C. Soft Tissue Facial Anthropometry in Down Syndrome Subjects. Journal of Craniofacial Surgery. 2004;15(3):528–532. doi: 10.1097/00001665-200405000-00037. [DOI] [PubMed] [Google Scholar]
- Ferrario VF, Dellavia C, Colombo A, Sforza C. Three-dimensional assessment of nose and lip morphology in subjects with down syndrome. Ann Plast Surg. 2004a;53(6):577–583. doi: 10.1097/01.sap.0000130702.51499.6b. [DOI] [PubMed] [Google Scholar]
- Ferrario VF, Dellavia C, Zanotti G, Sforza C. Soft tissue facial anthropometry in Down syndrome subjects. J Craniofac Surg. 2004b;15(3):528–532. doi: 10.1097/00001665-200405000-00037. [DOI] [PubMed] [Google Scholar]
- Frostad WA, Cleall JF, Melosky LC. Craniofacial Complex in the Trisomy 21 Syndrome (Down’s Syndrome) Archs oral Biol. 1971;16:707–722. doi: 10.1016/0003-9969(71)90116-6. [DOI] [PubMed] [Google Scholar]
- Gower JC. Some Distance Properties of Latent Root and Vector Methods Used in Multivariate Analysis. Biometrika. 1966;53:325–338. [Google Scholar]
- Hall P, Martin M. On the bootstrap and two-sample problems. Austral Journal of Statistics. 1988;30A:179–192. [Google Scholar]
- Hill CA, Reeves RH, Richtsmeier JT. Effects of aneuploidy on skull growth in a mouse model of Down syndrome. J Anat. 2007;210(4):394–405. doi: 10.1111/j.1469-7580.2007.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisling E. Cranial Morphology in Down’s Syndrome: A Comparative Roentgencephalometric Study in Adult Males. Copenhagen: Royal Danish Dental College Copenhagen; 1966. [Google Scholar]
- Kuppermann M, Learman LA, Gates E, Gregorich SE, Nease RF, Lewis J, Washington AE. Beyond race or ethnicity and socioeconomic status: predictors of prenatal testing for Down syndrome. Obstet Gynecol. 2006;107:1087–1097. doi: 10.1097/01.AOG.0000214953.90248.db. [DOI] [PubMed] [Google Scholar]
- Lele S, Richtsmeier JT. Euclidean distance matrix analysis: a coordinate-free approach for comparing biological shapes using landmark data. Am J Phys Anthropol. 1991;86(3):415–427. doi: 10.1002/ajpa.1330860307. [DOI] [PubMed] [Google Scholar]
- Lele S, Richtsmeier JT. Euclidean distance matrix analysis: confidence intervals for form and growth differences. Am J Phys Anthropol. 1995;98(1):73–86. doi: 10.1002/ajpa.1330980107. [DOI] [PubMed] [Google Scholar]
- Lele SR, Richtsmeier JT. An Invariant Approach to Statistical Analyses of Shape. New York: Chapman and Hall/CRC; 2001. p. 308. [Google Scholar]
- McElyea SD, Starbuck JM, Tumbleson-Brink DM, Harrington E, Blazek JD, Ghoneima A, Kula K, Roper RJ. Influence of prenatal EGCG treatment and Dyrk1a dosage reduction on craniofacial features associated with Down syndrome. Human molecular genetics. 2016;25(22):4856–4869. doi: 10.1093/hmg/ddw309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megarbane A, Ravel A, Mircher C, Sturtz F, Grattau Y, Rethore MO, Delabar JM, Mobley WC. The 50th anniversary of the discovery of trisomy 21: the past, present, and future of research and treatment of Down syndrome. Genet Med. 2009;11(9):611–616. doi: 10.1097/GIM.0b013e3181b2e34c. [DOI] [PubMed] [Google Scholar]
- O’Riordan MWaGFW. Dimensional and Proportional Characteristics of the Face in Down’s Syndrome. Journal of Dentistry for the Handicapped. 1979;4(1):6–9. [PubMed] [Google Scholar]
- Opitz JM, Gilbert-Barness EF. Reflections on the pathogenesis of Down syndrome. Am J Med Genet Suppl. 1990;7:38–51. doi: 10.1002/ajmg.1320370707. [DOI] [PubMed] [Google Scholar]
- Parsons T, Ryan TM, Reeves RH, Richtsmeier JT. Microstructure of trabecular bone in a mouse model for Down syndrome. Anat Rec (Hoboken) 2007;290(4):414–421. doi: 10.1002/ar.20494. [DOI] [PubMed] [Google Scholar]
- Pueschel SM. A parent's guide to Down syndrome: towards a brighter future. New York: Paul H Brookes Pub Co.; 2000. [Google Scholar]
- Reeves RH, Baxter LL, Richtsmeier JT. Too much of a good thing: mechanisms of gene action in Down syndrome. Trends Genet. 2001;17(2):83–88. doi: 10.1016/s0168-9525(00)02172-7. [DOI] [PubMed] [Google Scholar]
- Reyment RA, Blackith RE, Campbell NA. Multivariate morphometrics. London: Academic Press; 1984. [Google Scholar]
- Richtsmeier J, Cole T, III, Lele S. Variation: A Central Concept in Biology. Boston: Elsevier Academic Press; 2005. Landmark morphometrics and the analysis of variation. [Google Scholar]
- Richtsmeier JT, Baxter LL, Reeves RH. Parallels of craniofacial maldevelopment in Down syndrome and Ts65Dn mice. Dev Dyn. 2000;217(2):137–145. doi: 10.1002/(SICI)1097-0177(200002)217:2<137::AID-DVDY1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Richtsmeier JT, Cole TM, 3rd, Krovitz G, Valeri CJ, Lele S. Preoperative morphology and development in sagittal synostosis. Journal of craniofacial genetics and developmental biology. 1998;18(2):64–78. [PubMed] [Google Scholar]
- Richtsmeier JT, DeLeon VB, Lele SR. The promise of geometric morphometrics. Am J Phys Anthropol Suppl. 2002a;35:63–91. doi: 10.1002/ajpa.10174. [DOI] [PubMed] [Google Scholar]
- Richtsmeier JT, Zumwalt A, Carlson EJ, Epstein CJ, Reeves RH. Craniofacial phenotypes in segmentally trisomic mouse models for Down syndrome. Am J Med Genet. 2002b;107(4):317–324. doi: 10.1002/ajmg.10175. [DOI] [PubMed] [Google Scholar]
- Roper RJ, Reeves RH. Understanding the basis for Down syndrome phenotypes. PLoS Genet. 2006;2(3):231–236. doi: 10.1371/journal.pgen.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper RJ, VanHorn JF, Cain CC, Reeves RH. A neural crest deficit in Down syndrome mice is associated with deficient mitotic response to Sonic hedgehog. Mech Dev. 2009;126(3–4):212–219. doi: 10.1016/j.mod.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sforza C, Dellavia C, Dolci C, Donetti E, Ferrario VF. A quantitative three-dimensional assessment of abnormal variations in the facial soft tissues of individuals with Down syndrome. Cleft Palate Craniofac J. 2005;42(4):410–416. doi: 10.1597/04-005.1. [DOI] [PubMed] [Google Scholar]
- Sforza C, Dellavia C, Zanotti G, Tartaglia GM, Ferrario VF. Soft tissue facial areas and volumes in subjects with Down syndrome. Am J Med Genet A. 2004;130A(3):234–239. doi: 10.1002/ajmg.a.30253. [DOI] [PubMed] [Google Scholar]
- Sforza C, Elamin F, Rosati R, Lucchini MA, Tommasi DG, Ferrario VF. Three-dimensional assessment of nose and lip morphology in North Sudanese subjects with Down syndrome. Angle Orthod. 2011;81(1):107–114. doi: 10.2319/042510-222.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sforza C, Dolci C, Dellavia C, Gibelli DM, Tartaglia GM, Elamin F. Abnormal Variations in the Facial Soft Tissues of Individuals With Down Syndrome: Sudan Versus Italy. Cleft Palate Craniofac J. 2015;52(5):588–596. doi: 10.1597/14-082. [DOI] [PubMed] [Google Scholar]
- Shaner DJ, Peterson AE, Beattie OB, Bamforth JS. Soft tissue facial resemblance in families and syndrome-affected individuals. Am J Med Genet. 2001;102(4):330–341. doi: 10.1002/ajmg.1491. [DOI] [PubMed] [Google Scholar]
- Shapiro B. Amplified Developmental Instability in Down’s Syndrome. Annals of Human Genetics, London. 1975;38:429–437. doi: 10.1111/j.1469-1809.1975.tb00632.x. [DOI] [PubMed] [Google Scholar]
- Shapiro BL, Gorlin RJ, Redman RS, Bruhl HH. The palate and Down's syndrome. N Engl J Med. 1967;276(26):1460–1463. doi: 10.1056/NEJM196706292762603. [DOI] [PubMed] [Google Scholar]
- Sherwood RJ, Duren DL, Demerath EW, Czerwinski SA, Siervogel RM, Towne B. Quantitative genetics of modern human cranial variation. J Hum Evol. 2008;54(6):909–914. doi: 10.1016/j.jhevol.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starbuck J, Reeves RH, Richtsmeier J. Morphological integration of soft-tissue facial morphology in Down Syndrome and siblings. Am J Phys Anthropol. 2011;146(4):560–568. doi: 10.1002/ajpa.21583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starbuck JM, Cole TM, 3rd, Reeves RH, Richtsmeier JT. Trisomy 21 and facial developmental instability. Am J Phys Anthropol. 2013;151(1):49–57. doi: 10.1002/ajpa.22255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starbuck JM, Dutka T, Ratliff TS, Reeves RH, Richtsmeier JT. Overlapping trisomies for human chromosome 21 orthologs produce similar effects on skull and brain morphology of Dp(16)1Yey and Ts65Dn mice. Am J Med Genet A. 2014a;164A(8):1981–1990. doi: 10.1002/ajmg.a.36594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starbuck JM, Ghoneima A, Kula K. Facial soft-tissue asymmetry in three-dimensional cone-beam computed tomography images of children with surgically corrected unilateral clefts. J Craniofac Surg. 2014b;25(2):476–480. doi: 10.1097/SCS.0000000000000619. [DOI] [PubMed] [Google Scholar]
- Valeri CJ, Cole TM, 3rd, Lele S, Richtsmeier JT. Capturing data from three-dimensional surfaces using fuzzy landmarks. Am J Phys Anthropol. 1998;107(1):113–124. doi: 10.1002/(SICI)1096-8644(199809)107:1<113::AID-AJPA9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Weinberg SM, Naidoo S, Govier DP, Martin RA, Kane AA, Marazita ML. Anthropometric precision and accuracy of digital three-dimensional photogrammetry: comparing the Genex and 3dMD imaging systems with one another and with direct anthropometry. J Craniofac Surg. 2006;17(3):477–483. doi: 10.1097/00001665-200605000-00015. [DOI] [PubMed] [Google Scholar]
- Wong JY, Oh AK, Ohta E, Hunt AT, Rogers GF, Mulliken JB, Deutsch CK. Validity and reliability of craniofacial anthropometric measurement of 3D digital photogrammetric images. Cleft Palate Craniofac J. 2008;45(3):232–239. doi: 10.1597/06-175. [DOI] [PubMed] [Google Scholar]
- Yahya-Graison EA, Aubert J, Dauphinot L, Rivals I, Prieur M, Golfier G, Rossier J, Personnaz L, Creau N, Blehaut H, Robin S, Delabar JM, Potier MC. Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes. The American Journal of Human Genetics. 2007;81:475–491. doi: 10.1086/520000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NM, Hu D, Lainoff AJ, Smith FJ, Diaz R, Tucker AS, Trainor PA, Schneider RA, Hallgrimsson B, Marcucio RS. Embryonic bauplans and the developmental origins of facial diversity and constraint. Development. 2014;141(5):1059–1063. doi: 10.1242/dev.099994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. Anatomical soft-tissue facial landmarks collected. 3dMDpatient (v4.0) was used to record the three-dimensional coordinates of 20 standard landmarks (circles) from each image on two separate occasions. Eight medial and six bilateral landmarks were used in this study: 1) glabella (g), 2) nasion (n), 3) pronasale (prn), 4) subnasale (sn), 5) labiale superius (ls), 6) labiale inferius (li), 7) sublabiale (Sl), 8) pogonion (SPg), 9) endocanthion (en), 10) exocanthion (ex), 11) alar curvature (ac), 12) subalare (sbal), 13) crista philtra landmark (cpl), and 14) chelion (ch). Landmark definitions can be found at http://getahead.psu.edu/. Facial image shown has been modified to remove identifiable features.