Abstract
FoxP3 conditions the transcriptional signature and functional facets of regulatory T (Treg) cells. Its mechanism of action, whether as an activator or a repressor, remains unclear. Chromatin analysis shows that FoxP3 binds active enhancer elements, not repressed chromatin, around loci over- and under-expressed in Treg cells. We evaluated the impact of a FoxP3 mutation panel on transcriptional activity and interactions with DNA, transcriptional cofactors and chromatin. Computational integration, confirmed by biochemical interaction and size analyses, showed that FoxP3 exists in distinct multimolecular complexes. It is active and primarily an activator when complexed with the transcriptional co-factors RELA, IKZF2, and KAT5. In contrast, FoxP3 is inactive when complexed to EZH2, YY1 and IKZF3, where super-resolution microscopy shows this complex to partition to a peripheral region of the nucleus. Thus, FoxP3 acts in multimodal fashion to directly activate or repress transcription, in a context- and partner-dependent manner, to govern Treg cell phenotypes.
INTRODUCTION
Regulatory T (Treg) cells are of central importance in immunological tolerance and in the control of inflammatory processes 1–3. FoxP3, a transcription factor (TF) of the Forkhead family, is specifically expressed in Treg cells, is essential for their differentiation and function, and is the defining factor of the lineage 2,4. FoxP3 loss-of-function leads to Treg deficiency and to devastating multi-organ inflammation in scurfy mice and human IPEX patients.
Treg cells share a core transcriptional signature, genes that are over- or under-expressed in Treg cells relative to their naive CD4+ T cell counterparts (Tconv) 5–9. This Treg signature encodes a number of molecules that mediate Treg suppressor activity (IL-10, CTLA-4), but also includes transcripts typically induced (or repressed) upon T cell activation, in keeping with the self-reactive nature of the T cell receptor in many Treg cells. Much of this signature is controlled by FoxP3. An important aspect of FoxP3 function in maintaining Treg identity is the suppression of effector cytokines produced by activated Tconv cells such as IL-2, IL-4 or IL-17 10,11. Beyond this shared signature, Treg transcriptomes are further modified and adapted to their location and function. For instance, transcripts controlled by the nuclear receptor PPARγ promote metabolic adaptation to adipose tissue and are uniquely found in Treg cells that reside there 12.
FoxP3 contains several structural modules, a short zinc finger (ZF) of unknown function, a leucine zipper (LZ) domain for homo- or heterodimerization 13,14. The C-terminal Forkhead domain (FKH) is the primary DNA-binding domain, but also interfaces with transcriptional co-regulators 15. The structure of the FKH domain is known 16,17, but the N-terminal region appears like an Intrinsically Disordered Protein 18 and has resisted structural determination 19. FoxP3 interacts physically with many other TFs 20 such as RUNX1, NFAT, EOS (IKZF4), IRF4, RORγ, RORα, HIF1α, STAT3, TCF1, and EZH2, and mass spectrometry analysis further identified a large set of FoxP3 interactors within multiprotein complexes 21. Some of the interactions with these co-factors have been mapped to various regions of the FoxP3 protein, in the FKH or N-terminal domains 15,22, and several tune Treg activity, modulating their ability to suppress particular T cell phenotypes or autoimmune diseases 22–25.
How these structural aspects are integrated, and how FoxP3 regulates its target genes, is incompletely understood. Its transcriptional effects are thought to reflect sequence-specific binding to enhancers that affect its targets. Accordingly, FoxP3 is detected on a number of sites in the genome, some in the close vicinity of Treg signature genes, although the majority of FoxP3-binding sites are nowhere near any relevant genes, perhaps reflecting long-range interactions 17 or structural roles. FoxP3 binding sites tend to be active enhancer elements 9 but are not exclusive to FoxP3 and, in its absence, are occupied by other factors 26. Whether FoxP3 is a repressor or an activator, or both, has been diversely interpreted over time. FoxP3 was initially considered to be a repressor 15,27–29, in part because attention focused mainly on Il2 as a target, but broader analysis of the Treg signature and to its effects on the chromatin of target genes led to a dual activator and repressor perspective 7,30. More recently a unifying model of FoxP3 action was proposed in which FoxP3 binds to enhancer elements that are generally open in CD4+ T cells, recruiting EZH2 and the PRC2 repressor complex 31. In this model, FoxP3 contributes to the up-regulation of Treg signature genes only indirectly (by repressing repressors). However, one can argue that this model is not easily compatible with transcriptional correlates of natural or engineered FoxP3 variants 8,25.
FoxP3’s mechanism of action is clearly a key question to be resolved. Comprehensive analysis of ChIPseq data sets including histone modifications, show FoxP3 and its co-factors as a direct activator but passive repressor. A resolution of the conundrum may stem from the different multimolecular complexes in which FoxP3 can partake. We thus asked how an array of mutations, spaced through the FoxP3 protein, affected its binding to a set of cofactors, and how this impacted on FoxP3’s transcriptional properties. The results clearly identify two modes of operation for FoxP3, which correspond to distinct multimolecular complexes that segregate differentially within the nucleus.
RESULTS
FoxP3 binds active enhancers at both Treg -up and -down loci
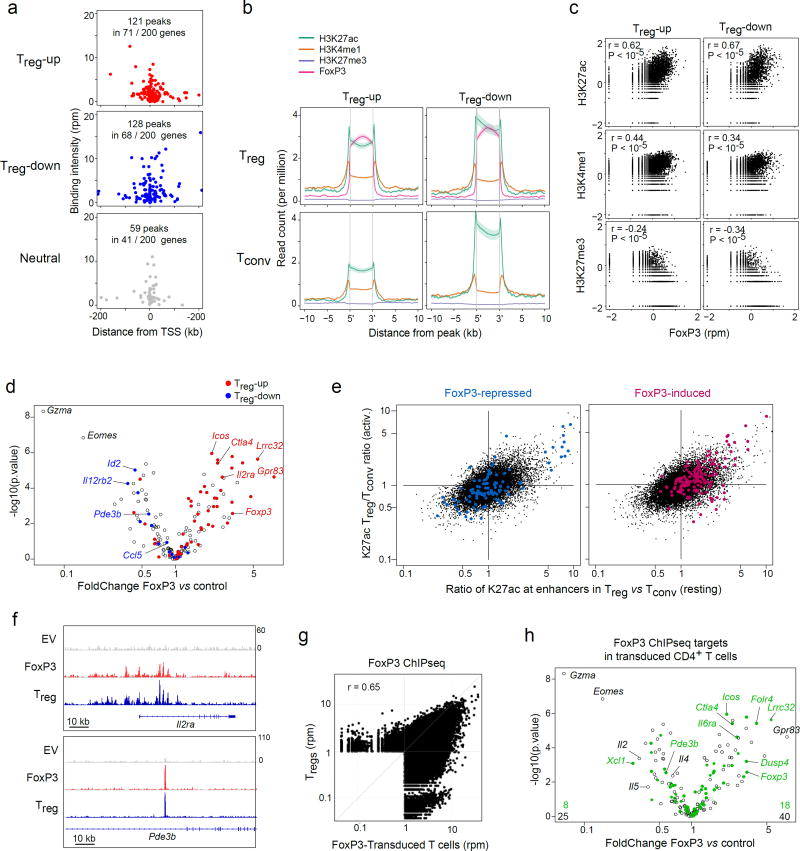
To begin to elucidate the mode of action of FoxP3 in specifying different components of the Treg signature, we re-analyzed several published chromatin immunoprecipitation (ChIPseq) datasets that define the position of FoxP3 and chromatin marks of enhancer activity or repression onto the mouse genome 9,32. We defined high-confidence FoxP3 binding sites in the genome as those replicated in two independent FoxP3 ChIPseq datasets 26,32, selecting 5,000 with the highest replicated signals, and parsed their distribution around loci encoding transcripts over- or under-expressed in Treg relative to Tconv (Treg -up and Treg -down, respectively, 200 with most extreme differences, FC>1.8). In keeping with previous conclusions 26, FoxP3 was equally represented in the vicinity of both Treg -up and -down signature genes, predominantly within 50 kb of their transcriptional startsite, evoking a direct action of FoxP3 (Fig. 1a; FoxP3 also bound some Treg -neutral loci, albeit at significantly lower frequency). To assess which type of regulatory element bound FoxP3 in the vicinity of these signature genes, we evaluated the histone marks at these FoxP3-ChIPseq peak regions, in both Treg and Tconv cells (Fig. 1b. As expected, FoxP3 binding sites in the vicinity of Treg -up signature genes carried marks of active enhancers (H3K27ac, H3K4me1), but not H3K27me3, associated with repressed chromatin. These H3K27ac signals were also present in Tconv cells but increased in Treg relative to Tconv, consistent with the notion that FoxP3 activates pre-existing enhancers 26. Genomewide, there was a positive correlation between FoxP3 binding and H3K27ac signals, but a negative correlation with H3K27me3 (Fig. 1c). All these observations are compatible with the notion that FoxP3 increases enhancer activity (or the frequency of cells in which the enhancers are active) around Treg -up genes. Less expected, however, was that FoxP3 binding sites around Treg -down signature loci also showed characteristics of active enhancers, in both Treg and Tconv cells, with no detectable increase in H3K27me3 signal that might be expected from repression. Thus, FoxP3 generally appears to bind to active enhancer elements.
Fig. 1. FoxP3 binds to active enhancers around both Treg -up and -down loci.
(a) FoxP3 binding peaks (replicated in ChIPseq dataset 26,32) in the vicinity of genes over- or under-expressed in Treg vs Tconv cells, or random neutral genes. (b) Density of chromatin marks in a 10 kb window around FoxP3 binding sites chromatin for Treg -up- or –down genes (per a). (c) Correlation of ChIPseq signal intensity for FoxP3 vs different chromatin marks in the vicinity of Treg -up- or –down genes (Pearson r and p-value). (d) 48 hrs after transduction of activated CD4+ T cells with a retroviral vector encoding FoxP3, or an empty vector control, transcripts were quantitated by Nanostring profiling. WT/EV expression FoldChange is plotted vs t.test p.value. Transcripts belonging to the classic Treg -up or - down signatures 7 are highlighted. (e) Ratio of H3K27ac intensity in Treg vs Tconv cells (genomewide data 31); x-axis: ratio in resting cells; y-axis: ratio in activated cells; red and blue highlights represent the positions of enhancers within 50 kb of genes that belong to the FoxP3-induced and -repressed genes defined by FoxP3 transduction. (f) Binding of FoxP3 around IL2ra and Pde3b in CD4+ T cells transduced with EV or FoxP3, with traces from ex vivo Treg cells 31 for reference (bottom row). (g) Genomewide comparison of FoxP3-binding in our FoxP3-transduced CD4+ T cells vs ex vivo Treg cells 31. Representative of biological duplicates. (h) Changes induced by FoxP3 transduction (same plot as d), where green highlights denote genes that bind FoxP3 (ChIPseq analysis in transduced cells); the average of biological quadruplicates.
We then tested the mechanism of FoxP3 action, using retroviral transduction of FoxP3 into CD4+ Tconv, a good setting to assess the direct effects of a TF, without the adaptations or redundancies that can occur in established cells in vivo. This approach was previously used for FoxP3 in several studies, and found to confer some transcriptional and functional aspects of Treg cells, but not all 6,7,33. We transduced FoxP3 into anti-CD3/28-activated FoxP3-negative cells from a Foxp3gfp mouse. The cells were sorted 72 hrs later, in a constant window of the THY1.1 reporter encoded in the retroviral vector, pre-calibrated to ensure a level of FoxP3 equivalent to that of ex vivo Treg cells, and thus to avoid over-expression artefacts by excluding cells grossly over-expressing FoxP3. Gene expression was profiled in these sorted cells using Nanostring, with a custom codeset (transcripts typical of the Treg signature, of tissue Treg cells, and major TFs and effectors of activated Tconv cells; Supplementary Table 1). Relative to control-transduced cells (empty vector, EV), we observed robust induction or repression of two genesets (Fig. 1d). Induced transcripts strongly overlapped the set of genes over-expressed in the classic Treg signature (red highlights). This overlap was not complete, as expected, because a sizeable segment of the Treg signature is independent of FoxP3 6,7. Most of the induced transcripts corresponded to genes with enhancers normally more active in ex vivo Treg than in Tconv cells, as reflected by H3K27ac marks 9 (Fig. 1e). Reproducing the paradox above, a proportion of the transcripts repressed by FoxP3 also corresponded to Treg -specific enhancers. We also verified the relevance of FoxP3 transduction by ChIPseq, pulling down the chromatin of cells transduced with HIS-tagged FoxP3. Clear peaks were observed, absent in the EV control, which corresponded well to previously mapped FoxP3 binding in Treg cells 26 (example loci in Fig. 1f, genome-wide Fig. 1g). A large proportion of the genes induced and repressed by FoxP3 transduction bound FoxP3 in transduced cells (88 and 89%, respectively; Fig. 1h). Thus, whether in ex vivo Treg cells or in transduced CD4+ T cells in which its action is more likely to be direct, FoxP3 seems to activate a sizeable fraction of the Treg -up signature, by binding to, and increasing the activity of, specific enhancers.
Determinants of FoxP3 transcriptional functions
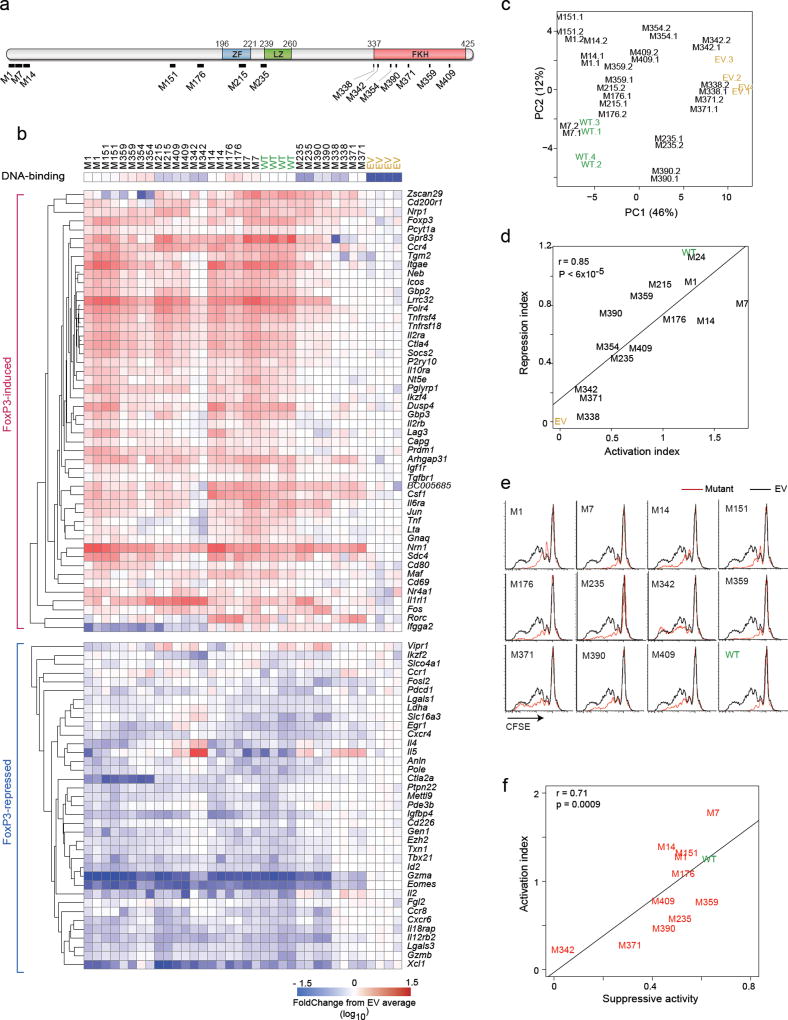
To gain further mechanistic insight on FoxP3 interaction with transcriptional cofactors to orchestrate transcriptional activation or repression, we engineered a set of 14 Alanine-replacement mutants that spanned different domains (Fig. 2a, Supplementary Table 2). Flow cytometric analysis of transduced cells, standardizing FoxP3 staining intensity against the co-transcribed THY1.1 reporter, showed for all mutants expression similar to that of wild-type Foxp3 (Supplementary Fig. 1a, Supplementary Table 3). Immuno-blotting of extracts from transduced cells showed that all mutant proteins were full-length (Supplementary Fig. 1b). Proper nuclear localization of the mutants was confirmed by immunofluorescence microscopy (Supplementary Fig. 1c). We then tested how the mutations affected DNA binding, in a solution capture assay using biotinylated oligo with a dimer of the canonical 5’-AAACA motif (Supplementary Fig. 1d). All mutants in the set were able to bind DNA, albeit with a partial reduction for a few (Supplementary Fig. 1e).
Fig. 2. Transcriptional activity of the FoxP3 mutants.
(a) Schematic of the 14 Alanine-replacement mutants of FoxP3 (details in Supplementary Table S2). (b) Transcriptional activity of the mutants. Activated CD4+ T cells transduced with retroviruses encoding mutant or wild-type FoxP3, or with empty vector and sorted at 72 hrs (matching expression of FoxP3 based on co-linear THY1.1) and profiled in biological duplicate (Nanostring); results are shown as FoldChange relative to the mean of EV controls. DNA-binding efficiency (from Supplementary Fig. 1e) is shown for reference. (c) Principal Component Analysis of data from b. Each point represents an individual experiment. (d) General activation or repression indices were computed for each mutant (average of FoldChanges for all FoxP3-induced or all FoxP3-repressed transcripts, respectively) and the two plotted. (e) Suppressive activity of CD4+ T cells transduced with wild-type or mutant FoxP3, as inhibition of division of CFSE-labeled Tconv (black: Tconv alone, red: after supplementation with transduced cells; data representative of 3 independent experiments). (f) Suppression index (from e, average of 3 experiments) plotted vs Activation Index (from d); Pearson correlation r and p-value.
How these mutations affect FoxP3’s ability to affect its transcriptional targets was tested in CD4+ T cells with the retroviral transduction and signature profiling system. The results of two independent transductions for each mutant are compiled and compared to wild-type and EV controls (Fig. 2b, Supplementary Table 4). A range of results were observed, some mutants yielding profiles similar to wild-type FoxP3, others with severely affected potential. Duplicates from independent experiments showed highly similar outcomes, as evidenced by Principal Components Analysis (Fig. 2c). There was no segregation according to the position of the mutations on the protein, although the most extreme effects were seen with FKH mutations also found in some IPEX mutants (M371, M338). Although a general gradient of FoxP3 activity was observed, there was also a great diversity of response patterns for individual targets (e.g. Nrn1 compared with Gpr83). Not all targets are equally affected by the panel of mutants, including some unique effects (the ability of M354 to repress Zscan29, which was instead induced by M7). There were some paradoxical effects, such as the induction by M342 and M338 of Il4 and Il5, which are normally repressed by wild-type FoxP3. Even the most extreme mutants maintained some activity, for instance the ability to repress Gzma or Eomes.
To relate more generally activating vs. repressive activity of each mutant, as denoted by mutant effects, we computed global activation and a repression indices, averaging overall effects on induced or repressed targets, respectively (Fig. 2d). These proved to be highly correlated, indicating that trans-activation and -repression activities of FoxP3 are generally governed by the same mechanisms, reminiscent of its binding to enhancer elements in the vicinity of both Treg -up and Treg -down signature genes.
To test for functional correlates of these transcriptional effects, we assessed the ability of cells transduced by FoxP3 or the set of mutants to inhibit the proliferation of CFSE-labeled Tconv cells stimulated by anti-CD3/CD28 beads 34. Clear suppressive activity was observed after transduction of wild-type FoxP3, and of several mutants, which was lost for M342 and M371 (Fig. 2e). The averaged suppression activity correlated well with the transcriptional activation index of the mutants (Fig. 2f; when assessed against transactivation of specific FoxP3 targets, suppressive activity correlated strongly with several effector molecules (Lrrc32, Il2ra) but not with non-canonical FoxP3 targets like Il1rl1, Rorc or Il4 (Supplementary Fig. 1f). These results indicated a great degree of variegation in the mechanism through which FoxP3 activates or represses its targets.
Mechanistic underpinning of transcriptional differences
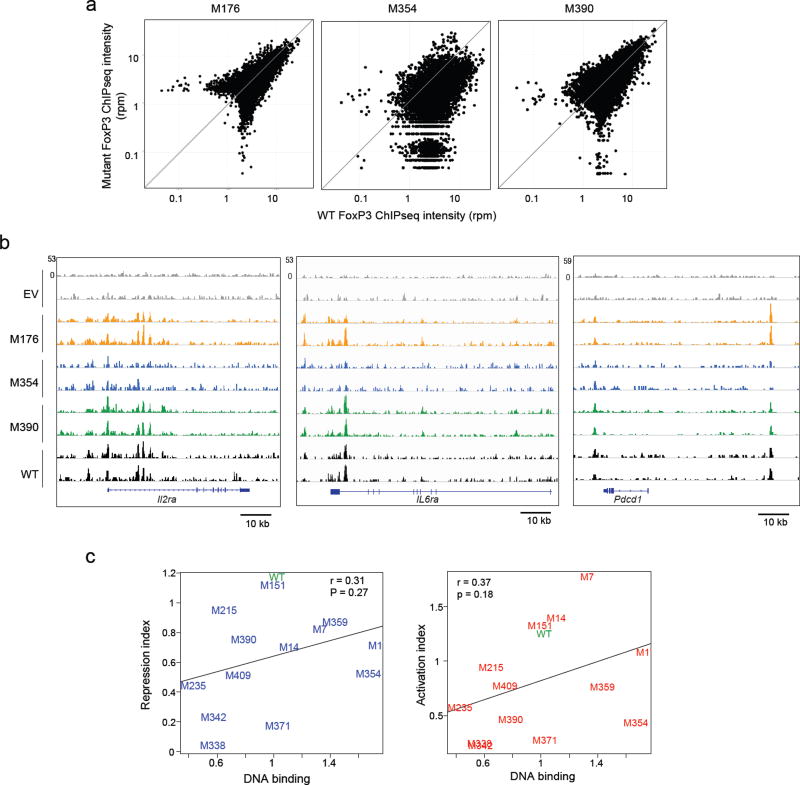
One mechanistic interpretation of the variable effect of the FoxP3 mutants on its targets was that each mutation altered the range of enhancer elements that FoxP3 binds across the genome. To test this hypothesis, we selected three mutations (one mild: M176; two harsher: M354 and M390) and performed FoxP3 ChIPseq in transduced CD4+ T cells. No clear relationship to transcriptional effects was found: the mild mutant M176 bound much like wild-type FoxP3 as did the more severe mutant M390, while binding of M354 was clearly perturbed (average intensity relative to wild-type of 0.54, 0.05–0.95 range 0.19 to 1.18 for individual peaks; Fig. 3a, b). We then correlated DNA-binding activity of the mutants on the canonical FKRE dimer (per Supplementary Fig. 1e) vs. their transcriptional output (activation and repression indices from Fig. 2d). Only limited correlation for both activation (r = 0.37, p= 0.16) and repression (r = 0.31, p= 0.27) was observed, several of the mutants with high DNA-binding potential having low activation or repression potential, and vice-versa (Fig. 3c). Together, these data suggest that DNA or chromatin binding by FoxP3 is important but not a primary determining factor in modulating its ability to activate or repress its target genes.
Fig. 3. Mutation effects on Foxp3 chromatin-binding.
ChIP-seq libraries were prepared from CD4+ T cells transduced with EV, wild-type, M176, M354 and M390 (6xHIS-tagged); (a) Genomewide comparison of wild-type and mutant profile (average of 2 replicates); (b) Representative traces in three FoxP3-binding regions (data from two independent experiments). (c) Correlation between DNA-binding activity of mutant FoxP3 (from Supplementary Fig. 1e) vs their transcriptional output (as the activation and repression indices from Fig. 2d); Pearson r and p-value.
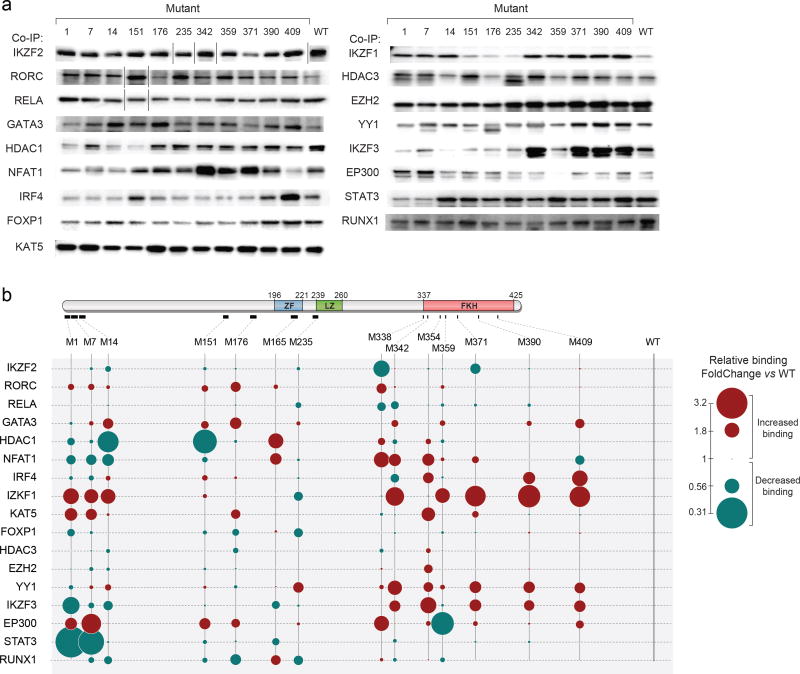
Given the lack of correlation between FoxP3 binding and transcriptional output we hypothesized that altered transcriptional activity of the mutant FoxP3 proteins might result from changed interactions with transcriptional cofactors. We thus tested the FoxP3 mutant panel in co-immunoprecipitation (co-IP) assays together with 17 known FoxP3 cofactors (listed and referenced in Supplementary Table 5). HEK293 cells were transfected with FLAG-tagged FoxP3 together with each cofactor (also tagged); nuclear extracts immunoprecipitated with anti-FLAG (FoxP3). In agreement with the literature, the cofactors were specifically and efficiently co-precipitated with FoxP3 (Supplementary Fig. 2a), in a manner that did not depend on DNA co-binding (not inhibited by DNAse treatment or Ethidium Bromide intercalation; Supplementary Fig. 2b, c), nor resulted from artefactual agglomeration after cell lysis (Supplementary Fig. 2d). When tested against the FoxP3 mutant panel (representative gels in Fig. 4a, quantitated in Fig. 4b and Supplementary Table 6), no simple pattern emerged and interactions with every cofactor proved unique. Even interactions with the related IKZF1, IKZF2 and IKZF3 proteins, three members of the IKAROS family, showed some similarities but were differentially affected by replacements in the N-terminal and FKH domains. In most cases, interactions with any one cofactor were influenced by mutations in several domains of FoxP3, with perhaps the exception of STAT3 and the dominant effect of N-terminal mutations. Many instances of enhanced binding were observed, most readily explained by relative displacement of a competing cofactor. The strongest of such contrasts were observed in the N-terminal region, where M1 and M7 dampened binding to IKZF3, NFAT1 and STAT3, while enhancing binding to KAT5 (aka TIP60), EP300, or IKZF1. Thus, there is great complexity in the determinism of FoxP3’s interactions with cofactors, in keeping with the flexibility conferred by the intrinsically disordered nature of much of the protein
Fig. 4. Cofactor-binding activity of the FoxP3 mutants.
After co-transduction of wild-type or mutant FoxP3 (FLAG-tagged) with each co-factor (GFP, HA or 6xHIS-tagged) into HEK293 cells, cell lysates were precipitated with anti-FLAG, and the co-precipitated TFs detected by immunoblot. (a) Representative co-immunoprecipitations. (each row is from a single representative experiment, cut for clarity to remove additional lanes on some gels). (b) Color-coded quantification (average of 2 to 5 such experiments), normalized to co-IP of each TF by wild-type FoxP3. Standard deviation and p-value of co-IP experiments shown in Supplementary Table 6.
Connecting transcription and interaction with DNA or cofactors
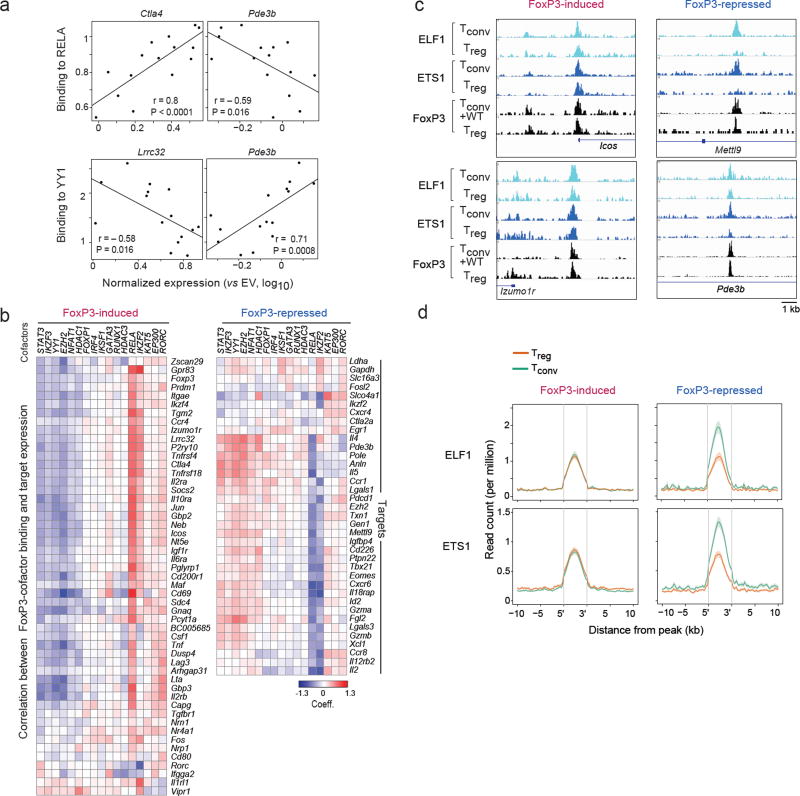
To understand the relationship between FoxP3’s cofactor interactions and transcriptional impact we correlated, across the mutant panel, their ability to bind individual co-factors and to induce or repress individual target genes (representative examples in Fig. 5a, overall heatmap in Fig. 5b). For induced transcripts, and with the exception of a few targets with more specific correlations like Rorc or Vipr1, a dominant pattern emerged: target activation correlated strongly with the mutants’ ability to interact with RELA or IKZF2, and to a lesser extent to KAT5, EP300 and GATA3. It correlated negatively with binding to IKZF3, YY1 and EZH2. This dichotomy suggests that FoxP3 may engage in two major types of interactions, which have activating or repressive properties. This interpretation is compatible with known biochemical activities: histone acetyltransferases KAT5 and EP300 generally activate transcription by acetylating histones and other TFs, and specifically FoxP3 20; the NF-κB pathway plays a strong positive role in Treg differentiation and function 35,36. Conversely, IKZF3, EZH2 and YY1 are known repressors, in general and in the context of FoxP3 31,37–40, and NFAT1 is a central player in the repression of Il2 15.
Fig. 5. Connecting cofactor interaction and transcriptional activity.
(a) Pearson correlation between FoxP3 mutants’ binding to RELA or YY1 (Y-axis) and their ability to induce expression of FoxP3-induced (Ctla4, Lrrc32) or –repressed (Pde3b) targets (X-axis, per Fig. 2b). (b) As in a, correlation between the mutants’ ability to activate each FoxP3-induced target and the ability to bind each cofactor; the heatmap shows the coefficient from linear model including DNA-binding as a covariate (hierarchically clustered, rows and columns). (c) Representative traces of ELF1 and ETS1 and FoxP3 binding at representative FoxP3-induced and –repressed signature loci. (d) Density of chromatin marks for ELF1 and ETS1 ChIPseq in a 10 kb window around FoxP3 binding sites for Treg -up or-down signature loci (data from 26).
The FoxP3-repressed transcripts again showed a paradoxical pattern. While one might have expected that FoxP3’s repressive association to negative cofactors would drive the downregulation, the exact opposite was observed: for most FoxP3-repressed transcripts, expression was positively correlated with binding to IKZF3, YY1 and EZH2, and was negatively correlated with binding to RELA (e.g. Pde3b in Fig. 5). In other words, the positively-acting FoxP3-RELA-IKZF2 complexes are the most effective at repression.
To account for this paradox, we considered how FoxP3 might be exerting its repressive effects. In general, transcriptional repressors can be active or passive 41,42: active by recruiting dominant inhibitors (Polycomb or NuRD), passive by competing against more effective transactivators. We hypothesized that the FoxP3-RELA complex might be displacing more effective transactivators at enhancers surrounding FoxP3-repressed loci, and analyzed published ChIPseq datasets 26 for differential binding in Treg vs. Tconv cells in the immediate vicinity of FoxP3 binding sites (per Fig. 1b). We found that binding of both ELF1 and ETS1 decreased in Treg cells at FoxP3-binding sites associated with FoxP3-repressed loci, but not at FoxP3-induced loci (Fig. 5c, d). Such patterns were not observed with other factors, in particular FOXO1 (increased in both types of loci).
These observations suggest that both induction and repression by FoxP3 are determined by the same molecular complexes, but that the outcome varies with the target genes and the other complexes that can bind to the corresponding enhancers in its absence.
FoxP3 in differentially active multimolecular complexes
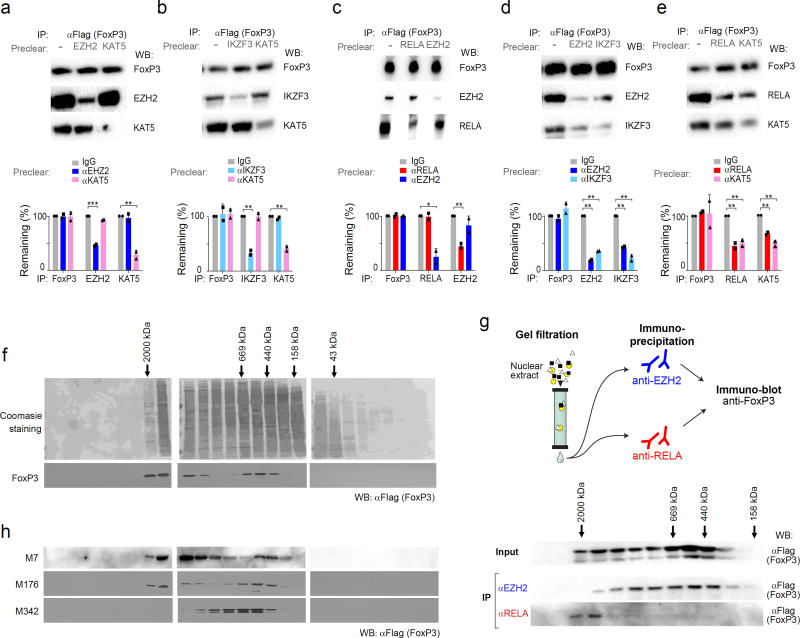
These results suggested that FoxP3 forms complexes that have diametrically opposite transcriptional activities. To tested this hypothesis in several ways. First, to test which co-factors belong, or not, to the same FoxP3-containing complexes, we performed FoxP3 co-IP experiments after preclearing the cell lysates with antibodies to one cofactor, before probing for a second. Preclearing with anti-EZH2 or -KAT5 did not significantly reduce the total amount of FoxP3, depleted as expected complexes containing EZH2, but did not affect KAT5-FoxP3 complexes (Fig. 6a). The converse was true after pre-clearing with anti-KAT5 (Fig. 6a) or with anti-RELA (Fig. 6b). Pre-clearing with anti-IKZF3 also failed to remove KAT5-FoxP3 complexes (Fig. 6c). These observations suggested that EZH2 and IKZF3 may belong to one complex, while RELA and KAT5 belong to another. This interpretation was confirmed as anti-RELA or KAT5 reciprocally depleted both FoxP3-containing complexes, as did EZH2 and IKZF3 (Fig. 6d,e). Thus, the associations suggested by transcriptional correlations (Fig. 5) were reproduced here as biochemical entities.
Fig. 6. FoxP3’s active and inactive cofactors form different complexes.
After co-transfection of wild-type FoxP3 with (a) EZH2 and KAT5 (aka TIP60); (b) RELA and EZH2; (c) IKZF3 and KAT5; (d) EZH2 and IKZF3; (e) RELA and KAT5, cell lysates were pre-cleared by capture with antibodies against indicated factors, before co-IP as in Fig. 4 (anti-FoxP3 IP, immunoblot with anti-cofactor as shown); blots representative of three independent experiments, combined in bar graphs below (Student’s t test p-value vs depletion by non-specific IgG; *: p< 0.05, ** < 0.005, *** < 0.001. (f) Gel filtration: Total cell lysate from FoxP3-transduced CD4+ T cells was fractionated by Superose 6 FPLC, and the fractions tested for FoxP3 content by PAGE immunoblotting; data representative of two independent experiments. (g) Gel filtration and co-IP: FPLC fractions from FoxP3-transduced CD4+ T cells were IPed with anti-RELA or -EZH2, and the co-precipitated FoxP3 detected by immunoblot; data representative of two independent experiments. (h) Total cell lysates from CD4+ T cells transduced with mutants (M7, M176 and M342) fractionated and probed as in f; data representative of biological replicates.
We then applied FPLC gel filtration to resolve FoxP3 molecular complexes in Foxp3-transduced CD4+ T cells. In accordance with a previous study 21, FoxP3 was detected in several locations between 200 and 2,000 kDa (Fig. 6f). Gel filtration fractions were each precipitated with anti-EZH2 or -RELA before immunoblotting for FoxP3 (Fig. 6g). The FoxP3/RELA complexes were found in the larger size range (1000–2,000 kDa), while FoxP3-EZH2 complexes were intermediate sized (~400–800 kDa). We tested the effect of some of the mutations on these complexes. The M7 and M176 mutants of FoxP3, which have only mild effects on transactivation, distributed much as did wild-type FoxP3 (Fig. 6h). The severe M342 mutation induced an abnormal distribution of FoxP3, lacking the larger, RELA-interacting, complexes. This inability to partake in RELA-containing complexes was consistent with M342’s inability to transactivate most RELA-dependent targets.
Different FoxP3 molecular complexes in different nuclear regions
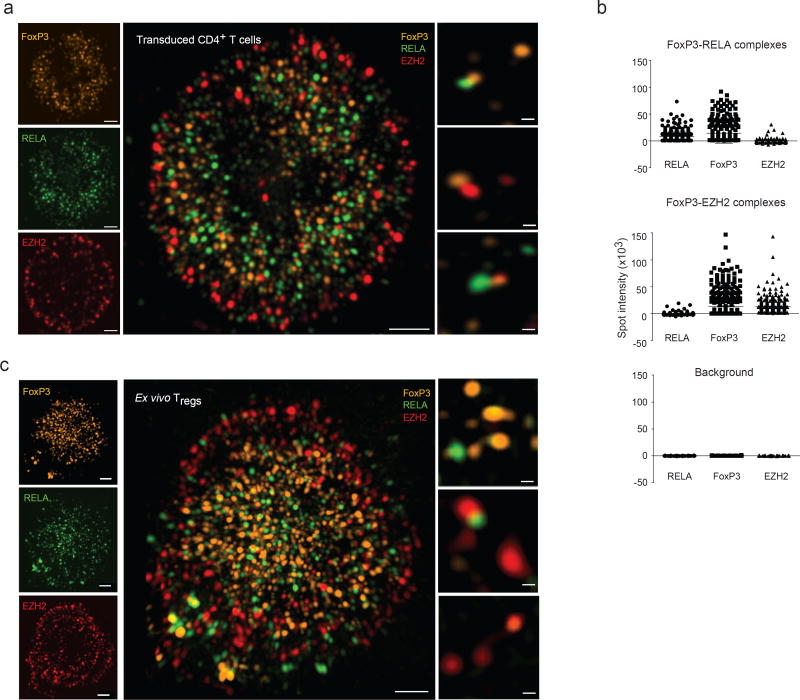
To further dissect this duality of FoxP3-containing multi-molecular complexes, we sought to visualize them within the nucleus. We used three-dimensional structured illumination microscopy (3D-SIM), which has the power to resolve colocalized proteins that would elude confocal microscopy, to visualize FoxP3 molecular complexes in transduced CD4+ T cells. FoxP3, RELA, and EZH2 could be detected in discrete microclusters of 1–200 nm apparent size (Fig. 7a, left and Supplementary video 1). FoxP3 and RELA tended to predominate at the center and EZH2 at the periphery of the nucleus. IKZF3, the other cofactor associated with repression, also localized to the nuclear periphery (Supplementary Fig. 3a). We then searched the 3D images for co-localization (Fig. 7a, center and right panels). In 48 nuclei (from 7 independent experiments) analyzed from these transduced CD4+ T cells, we identified 24,380 FoxP3 microclusters, of which 6% and 4.8% colocalized with RELA or EZH2, respectively (as a reference for detection of colocalization, 75% of FoxP3 stained with two secondary antibodies labeled with different fluorochromes appeared colocalized under our detection criteria). FoxP3-RELA clusters dominated in the nuclear center, and FoxP3-EZH2 in nuclear periphery. Indeed, colocalization of FoxP3 with RELA or EZH2 tended to be mutually exclusive, as only 0.06% of FoxP3 microclusters colocalized with both RELA and EZH2, vs 0.28% expected by chance (Fig. 7b). Essentially identical images were observed in ex vivo Treg cells (Fig. 7c). Thus, the differential engagement of FoxP3 with activating (RELA) or repressive cofactors (EZH2, IKZF3) detected biochemically corresponds to partitioning into different nuclear zones.
Fig. 7. Different FoxP3 molecular complexes distribute to different regions of the nucleus.
(a) FoxP3 was transduced into activated CD4+ T cells, and imaged with endogenous RELA and EZH2 by 3D-SIM. Left: overview of the nuclear distribution of the factors; center: higher magnification merge; right: representative co-localized microclusters, single z plane. Data representative of 50 cells in biological triplicates. (b) Co-localized spots containing FoxP3 and RELA (top), or FoxP3 and EZH2 (middle) were selected, and florescence intensity for three proteins measured in these voxels (bottom: background values outside the microclusters). Data combined from 5 independent experiments. (c) FoxP3, RELA and EZH2 were visualized in ex vivo Treg by 3D-SIM. Data representative of 30 cells in biological triplicates.
DISCUSSION
This study has tackled the enigmatic question of how FoxP3 operates as a transcription factor, associating dense mutagenesis with a systematic exploration of the determinism and functional impact of FoxP3’s interactions with its known cofactors. The results show that FoxP3’s operation as keyed by binding to active enhancers, with either a repressive or activating outcome depending on the target locus. This duality corresponds to FoxP3’s integration into different multimolecular complexes, which also control localization to different regions of the nucleus. FoxP3’s transcriptional activities were highly variegated, portraying an interaction hub with the flexibility to adjust to the broad span of physiological roles of Treg cells.
Many transcription factors are thought to have dual roles as transcriptional activators and repressors, in a gene- or context-dependent manner, but how they switch and balance the two functions has never been well established. In fact, the activator vs repressor debate is one of the oldest in molecular biology, going back to Englesberg’s “positive control” vs Jacob and Monod’s repressor models of transcriptional regulation in bacteria 43. The question remains open for eukaryotic TFs, even some as extensively studied as p53 44. Transcriptional repressors can be passive or active 41,42: passive by competing out activators for binding to DNA sequence motifs or to co-activators, by forming inert heterodimers with activators; active by recruiting inhibitory elements like histone deacetylases, histone or DNA methylases, displacing target loci into inactive chromatin configurations or nuclear localizations. Transcriptional activation can result from scaffold formation between enhancers and promoters into active transcriptional hubs, chromatin decompaction, or recruiting elongation factors to release stalled polymerases. How does FoxP3’s function relate in these schemes of transcriptional regulation? Clearly, many of the present observations are not compatible with an interpretation in which FoxP3 would primarily be an active repressor via recruitment of EZH2, and transactivation being largely indirect 31: both induction and repression by FoxP3 appears quickly after transduction, both induced and repressed loci are enriched in FoxP3-binding sites, and the finely variegated effects of FoxP3 mutations is not readily compatible with indirect effects. The association of FoxP3 with active enhancers around both activated and repressed loci, suggests that FoxP3 locates enhancers and modulates their activity, consistent with earlier conclusions 26.
Once FoxP3 is bound to an enhancer, the potential for interaction with different cofactors seemed to be the dominant driver of its functional outcome, judging from the correlation between co-factor binding and target transactivation. Transactivation by FoxP3 was, for the most part, positively correlated with the ability of mutant FoxP3 to form complexes with RELA, IKZF2, EP300 or KAT5, and negatively correlated to EZH2, YY1, IKZF3, NFAT1 or STAT3. As discussed above, these results make biochemical and functional sense 15,20,31,37–40. This genetic evidence for distinct FoxP3 complexes with differential transactivation potential was directly confirmed by biochemical co-immmunoprecipitation and fractionation experiments. We thus propose a model in which FoxP3 can be alternatively assembled in different complexes. It usually potentiates an enhancer when together with RELA/KAT5/EP300, possibly through acetylation-mediated BRD4/p-TEFβ activation. It is inactive when complexed with IKZF3/YY1/EZH2, leading to repression by recruitment of the NuRD and Polycomb assemblies and displacement away from active regions of the nucleus 45. The outcome of FoxP3’s activity would thus result from the balance between these two complexes, varying with genomic location (different enhancers being more or less favored by each complex), and also influenced more generally by changes in the cell state, organismal location or environmental cues, possibly via post-translational modifications 29.
One might have expected that FoxP3 repressor complexes operate dominantly on repressed loci: better binding to EZH2/YY1/IKZF3 would lead to better repression. Yet the exact opposite was observed, binding of FoxP3 mutants to EZH2, YY1 or IKZF3 correlated positively to the expression of FoxP3-repressed targets (and negatively correlation to RELA or IKZF2 binding). In other words, FoxP3/RELA/IKZF2 complexes were now the better inhibitors of FoxP3-repressed targets. One possible explanation of this paradox is that repression by FoxP3 is mostly indirect, via induction of repressive feedback factors. This interpretation is consistent with the close correlation between induction and repression indices, but doesn’t fit with the rapid effect upon transduction, or the binding of FoxP3 in the vicinity of repressed loci. Rather, we propose that the FoxP3/RELA/KAT5 complex, as a passive repressor, interferes with stronger activating complexes. These loci appear repressed, but simply because they are less efficiently activated. There is precedent for the CREL-FoxP3 complex being reported as repressive 46. Supporting this notion, higher level of ELF1 and ETS1 signals in Tconv dropped down only at FoxP3 repressed loci in Treg (no difference at FoxP3 induced loci). This behavior may not be exclusive to ELK1 and ETS1 (displacement of AP1 is also possible 15,47), but these provide plausible candidates for the displacement that would accompany passive repression
Beyond this “dominant theme” of the RELA and EZH2-containing complexes, however, several FoxP3 targets likely obeyed different modes of transactivation, some even appearing independent of DNA binding (we cannot rule out that these results reflect squelching, or de-repression by dominant-negative variants 48). Some of the “atypical” responses to mutations make sense, like the coordinated de-repression of Il4 and Il5, which are co-regulated in Th2 cells. Interestingly, atypically-regulated targets include Il1rl1 and Rorc, whose expression in Treg cells is largely restricted to tissue- Treg cells 3. This observation suggests that FoxP3 has an inherent ability to activate these transcripts, which is revealed, or de-inhibited, by cofactors induced in Treg cells by tissue localization cues.
In conclusion, this study uncovered a multimodal operation of FoxP3 and the complexes it belongs to: it acts most frequently as an activator, on other loci as a passive repressor by tuning down enhancer activity, and as an active repressor by association with the major repressor complexes. Importantly, we do not know how rapidly tunable these complexes are in individual Treg cells, and it will be important to determine whether the “population averages” observed here reflect frequencies of binary states, or whether they fluctuate rapidly in every cell. This view of FoxP3 as a multimodal interaction hub is consistent with the fine-tuning Treg transcriptional programs needed to adjust to the broad span of Treg physiology.
ONLINE METHODS
Mice and cells
C57/BL/6J and Foxp3gfp mice and mutants were bred in an SPF facility at Harvard Medical School, under Institutional Animal Care and Use Committee protocol 02954. Plat-E cells were obtained from Cell Biolab INC. and tested negative for mycoplasma contamination.
Cloning and mutagenesis
To construct the FoxP3 alanine scan library, FoxP3 cDNA was amplified from Treg cDNA and inserted into the MSCV-IRES-THY1.1 retroviral vector 7 with N-terminal FLAG tag motif (FLAG-FoxP3Met1). From this template, alanine replacement mutations were generated by substituting groups of 6 amino acids for M1 (M1PNPRP6), M7 (A7KPMAPS13), M14 (L14ALGPSP20), M151 (G151INVAS156), M176 (P176RKDSN181), M215 (K215HCQADH221) and M235 (R235EVVQS240) or each individual amino acid within the FKH domain including M328 (P338), M342 (Y342), M354 (P354), M359 (T359), M371 (F371), M390 (S390) and M409 (D409). Directed mutagenesis used the QuikChange XLII SiteDirected Mutagenesis Kit (Agilent, Santa Clara, CA). All coding sequences in the mutant plasmids were verified by Sanger sequencing. To generate 6xHIS tagged FoxP3, mFoxP3 cDNA was cloned into pENTR-Topo (ThermoFisher) and transferred into pDEST40 (Invitrogen) by Gateway cloning. To express FoxP3 co-factors, cDNAs encoding human IKZF1, IKZF3, IKZF2, YY1, HDAC1, HDAC3, STAT3, IRF4, RUNX1, EP300, EZH2, KAT5, RELA, RORC and GATA3 were obtained from DF/HCC DNA Resource Core and cloned into pDest vectors; GFP tag: pDest-N1 (Addgene), 6xHIS tag: pDest40 (Invitrogen), V5-6xHIS: MSCV-Attr1-ccdb-Attr2-V5-6xHIS-IRES-GFP by Gateway cloning. Mouse cDNA encoding Foxp1 and Nfat1 were obtained from Addgene, and expressed similarly. For FoxP3 ChIP-seq experiment with anti-V5/anti-6XHIS, cDNAs from wild-type, M176, M354 and M390 plasmids were re-cloned into MSCV-Attr1- ccdb-Attr2-V5-6xHIS-IRES-GFP vector, which generated by insertion of Attr1-ccdb-Attr2-V5-6xHIS.into MSCV-IRES-GFP vector.
Retroviral production and titration
For the production of retroviral particles, Plat-E packaging cells 49, were transfected with TransIT-293 (Mirus Bio LLC.) per the manufacturer’s protocol. Briefly, complete culture medium was changed with DMEM with 10% FCS before 6 hr transfection, and cells were transfected with 6ug of each retroviral plasmid and 4ug of Gag, Pol and Env packaging plasmid (pCL-ECO, Addgene) together with 40ul of TransIT-293 in 1ml of Opti-mem (Gibco). After 12 hr of transfection, culture medium was replaced with complete DMEM supplemented with 10% FCS, 3 mM L-glutamine and 100 U/ml penicillin-streptomycin and cells were cultured for a further 48 hr at 32°C. Viral particles were harvested, filtered on a 45um filter (Millipore) and stored at −80°C before use. To titer retroviral stocks, NIH3T3 cells were infected with diluted stock and analyzed for THY1.1 expression by flow cytometry. Viral titer was calculated as = {(F × Cn) /V} × DF, where F: frequency of GFP-positive cells determined by flow cytometry; Cn: The total number of target cells infected; V: volume of the inoculum.; DF: virus dilution factor. Throughout experiments, 1 X 107/ml of viral particles were used.
Preparation and activation of mouse CD4+ T cells
Splenocytes were prepared from spleen of 6~8 weeks old C57BL/6 mice by mechanical dissociation. CD4+ T cells were negatively purified by magnetic selection: cells were stained with PE conjugated antibody cocktail [anti-CD11b (M1/70), anti-CD11c (N418), anti-CD19 (6D5), anti-CD8α (53–6.7), anti-CD25 (PC61), anti-NK1.1 (PK136) anti-GR1 (8C5) and anti-TER119 (TER-119); all from BioLegend] for 20 min. After washing with PBS, stained cells were captured with anti-PE microbead (130-048-801; Miltenyi Biotec) for 20 min on ice and purified by MACS LD columns (Miltenyi) followed by manufacturer’s protocol. After isolation, purity was determined by flow cytometry ( >97%). Then, purified cells (0.5~1 X 105 cells / well) were plated in round bottom 96 well plate (Sigma, #CLS3799) and activated with anti- CD3/CD28 beads (Invitrogen, #11452) at 1 cell per 1/2 bead ratio for 36 hr in 200ul of complete RPMI medium supplemented with 10% FCS, 3 mM L-glutamine, 10mM Sodium pyruvate, 10mM Non-essential amino acids, 100 U/ml penicillin-streptomycin, 50 μM β-ME and 50U/ml human IL-2 (Peprotech). For viral infection, culture medium was removed and activated cells were spin-infected with 200 ul/well of each viral supernatant containing 50 U/ml of IL-2 and 8ug/ml of Polybrene (Sigma) for 2 hr at 32°C and 2000rpm, then plates were incubated at 32°C for 6 more hours. Medium was exchanged with fresh culture medium containing 50U/ml of hIL-2 at 37°C until harvest.
ChIP-seq
For FoxP3 ChIP-seq, we used a previously published procedure, with minor modification 50. Activated CD4+ T cells transduced with FoxP3 or mutants were sorted according to THY1.1 reporter expression to select cells with normal levels of FoxP3 expression (matching ex vivo Treg cells) as above. These cells (7×106 cells/ sample) were cross-linked with 1% of formaldehyde for 10 min and lysed on ice in RIPA buffer [10mM Tris- HCl (pH 8.0), 1mM EDTA (pH 8.0), 140mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulphate (SDS) and 0.1% sodium deoxycholate] supplemented with complete protease inhibitor cocktail (Roche). Chromatin was sheared using a AFA™ Focused-ultrasonicator (Covaris) for 9 min (duty cycle 2%, intensity 3, cycle/burst 200) and the sheared material was cleared by a 10 min centrifugation at 13,000 rpm at 4°C. The cleared material was IPed with the Abs combination to maximize IP efficiency by targeting different parts of FoxP3 protein (5ug of anti-V5; Invitrogen, and 5ug of anti-6XHIS) conjugated with magnetic Protein- G beads overnight at 4°. Beads were sequentially washed with 1ml of washing buffers (5 times for each buffer, 10 min for each washing) including ice-cold RIPA, high-salt RIPA [10mM Tris-HCl (pH 8.0), 1mM EDTA (pH 8.0), 500mM NaCl, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate], LiCl [10mM Tris-HCl (pH 8.0), 1mM EDTA (pH 8.0), 250mM LiCl, 0.5% NP-40 and 0.5% sodium deoxycholate] and TE [10mM Tris-HCl (pH 8.0) and 1mM EDTA (pH 8.0)]. Chromatin was eluted from the beads, treated with 1µg DNase-free RNase (Roche) for 30 min at 37°C and with Proteinase K (Roche) for 2 hr at 37°C followed by reversing crosslinks by incubating at 65°C for 16 hr. Reverse cross-linked DNA was purified with SPRI beads (Agencourt AMPure XP beads, Beckman Coulter) and sequential steps of end-repair, A-base addition, adaptor-ligation and PCR amplification (15 cycles) were performed to prepare the ChIP-seq library for each sample. ChIP-seq libraries were size-selected for 200–500bp fragments with SPRI beads. Equivalent amounts of barcoded libraries were pooled and sequenced on a NextSeq 500 (Illumina) instrument. To control for background noise, we prepared anti-V5/6XHIS IPed samples using chromatin from EV transduced T cells.
Short reads (50bp, single end) were aligned to the mouse reference genome (mm10) using the bowtie aligner 51. Reads with multiple alignments were removed with samtools (v1.1) and de-duplicated with picard (v1.130). To identify peaks from ChIP-seq reads, we used the HOMER package makeTagDirectory followed by the findPeaks command with the ‘histone’ parameter. To visualize individual ChIP-seq data on Integrative Genomics Viewer (IGV) 52, we converted bam output files from picard into normalized bigwig format using the bamCoverage function in deepTools (v1.6) with options – fragmentLength 200 – normalizeUsingRPKM 53. HOMER-generated peak files for H3K27ac, H3K4me1, H3K27me3 and FoxP3 were used to analyze with the ROSE algorithm described previously 54, wherein FoxP3 peaks are stitched together if they are located within 10 kb of each other. Line plots as in Fig. 1b were generated using ngs.plot 55.
To derive a robust list of FoxP3 chromatin binding sites from previous ChIPseq data, raw fastq files from 26,32 were mapped to the mm10 reference genome using bowtie 51. Peaks from both studies were called using HOMER findPeaks function with an FDR of 1% using the parameter -style factor and respective input controls, recovering 9,685 26 and 7,751 32 FoxP3 peaks. Intersection of peaks were derived from both datasets using the BEDtools intersect function with a 50% reciprocal overlap requirement, yielding 5,047 robust FoxP3 peaks.
Evaluation of FoxP3 expression
For immunohistochemistry, HEK cells were seeded on glass coverslips and transfected with EV, wild-type or each mutant plasmids by TransIT®-293 (Mirus). After 72 hr, cells were fixed with cold 100% Me-OH for 20 min at −20 °C, then washed three times with PBS. Cells were blocked with 5% BSA in PBS for 1 hr, incubated with anti-FLAG (1 to 200 dilution) overnight at 4°C, followed by staining with Donkey-anti-mouse-IgG-Cy5 (1 to 1000 dilution) for 1 hr at room temperature (Jackson ImmunoResearch Laboratory; 715-095-150 or 715-175-150). DAPI (100nM, Invitrogen) was used for nuclear counterstaining. Images were acquired on a Zeiss Axio M1 fluorescence microscope. For flow cytometric analysis of FoxP3 and THY1.1 expression in transduced primary T cells after infection with EV, wild-type or each mutant, cells were stained with anti-THY1.1 (OX-7), fixed/permeabilized with 1X Fixation/Permeabilization buffer (eBioscience) overnight at 4°C, stained with anti-FoxP3 (eBioscience, #14-5773) in 1X Permeabilization buffer for 1 hr at 4°C and analyzed on an LSRII (Becton Dickinson). For FoxP3 immunoblotting, cells were sorted by THY1.1 expression and nuclear proteins were extracted with nuclear extraction kit (Active motif: 40010). These lysates were resolved by SDS-PAGE and subjected to immunoblotting with anti-FLAG and HRP-conjugated anti-mouse-IgG (Jackson ImmunoResearch Laboratory).
DNA binding assay
Activated CD4+ T cells (2 X 106) were transduced to express FLAG-tagged FoxP3 or mutants, harvested, stained and sorted for a window of colinear THY1.1 reporter expression determined to ensure a match with FoxP3 levels in normal Treg cells. After sorting, nuclear extracts were prepared (Active motif: 40010), and total protein in each sample quantified by BCA protein assay (ThermoFisher, 23225). A 25 bp double-stranded biotinylated oligonucleotide encompassing two copies of the canonical FoxP3 motif (5’-CAAGGTAAACAAGAGTAAACAAGTC-3’) 56 or a control oligo with a scrambled motif (5’-CAAGACGCGAGCGATGCCTAGGGTC-3’) was used as probes. Protein-DNA binding was analyzed with Episeeker DNA-protein binding assay kit (Abcam, ab117139) followed by manufacture’s protocol. Briefly, 40ng of probe and 50ug of each extract were incubated on streptavidin-coated plate, which was washed with 1X washing buffer and FLAG-tagged FoxP3 detected with by anti-FLAG (Sigma; F1804) and HRP-conjugated anti-mouse-IgG (Jackson ImmunoResearch Laboratory) with TMB as substrate. Binding was assessed by reading absorbance on a micro-plate reader at 450nm.
Transcriptional analysis
For Nanostring profiling, the Nanostring nCounter (Nanostring Technologies, Seattle, WA) system was used to digitally count transcripts in multiplex reaction. Briefly, 5ul of lysate (1000 cells/ul) in RLT buffer was hybridized for 18h with a custom codeset (Supplementary Table 1), washed and quantitated using the nCounter Analysis System at maximum counting (555 images per sample). For processing, each data was normalized relative to External RNA Control Consortium (ERCC) positive and negative spike-in probes, background from negative-control probes was subtracted, and the values then normalized relative to average expression of control genes (Actb, Gapdh, Hprt, Ms4a1, Rp119 and Tbp).
In vitro suppression assay
CD4+Foxp3− Tconv from Foxp3gfp mice were isolated and labeled with 10umol/L CFSE (Invitrogen) in RPMI 1640 at 106/mL at 37° for 20 min. CD4+ T cells infected with retroviral vector encoding wild-type or mutant FoxP3, or empty control vector, were sorted after 72 hr for equivalent levels of the collinear THY1.1 retroviral reporter, and co-cultured with CFSE labeled Tconv at the ratio 1:1 in the presence of anti-CD3/CD28 beads. Proliferation was measured as CFSE dilution after 72hrs of co-culture. Suppression Index was calculated by (PIEV-PIx)/PIEV, where PI: Proliferation Index determined simply as % of proliferated Tconv (all proliferated peaks included).
Computational analyses
To identify robust FoxP3 binding in chromatin (Fig. 1), we combined by rank intensity two available FoxP3 ChIPseq datasets 26,32, retaining peak regions that were called in the top 5000 of peak intensity (integrated read count per million) in both datasets. Treg -up, Treg - down genes in Fig. 1a–c are the 200-most overexpressed in Treg and Tconv cells, respectively, “neutral” genes were 200 randomly picked genes (150 iterations averaged in Fig. 1a) among transcripts with Treg / Tconv FoldChange >0.9 and <1.1.
To analyze the significance of transactivation or transrepression by FoxP3 in transduced cells, a simple FoldChange was computed between the mean for all wild-type FoxP3 transduced cells vs all empty-vector controls (n=4 or 5), and a point-wise p.value derived from a t.test between the log of these values. Treg signature genes were those identified by microarray analyses in 7 and 57. dd To focus the analysis of mutation effects on a robust set of transactivated or repressed transcripts, genes up-regulated by FoxP3 (Fig. 1d) were selected on the basis of the volcano plot of as those with a FoldChange >1.8 (or <0.55) and −log10 (p.value) >1.3 after transduction with wild-type FoxP3. We also included in this analysis a few genes transcripts changed by 3-fold or more in at least one mutant (average of two determinations). PCA analysis (Fig. 2c) was performed on expression values for these selected genes in cells transduced by all mutants, the princomp function in R, and the barplot for the first 2 components plotted (Fig. 2c)
From this selection, a general “Repression index” (Fig. 2d) was computed for each mutant by averaging the FoldChanges for all FoxP3-repressed transcripts (equivalently weighting each transcript by first normalizing to the mean FoldChange for the transcript), and similarly deriving a “Activation index” from the FoxP3 up-regulated geneset.
To relate suppressive activity of cells transduced with WT or mutants FoxP3 with their transcriptional programs, a Pearson correlation coefficient was calculated between in vitro suppressive index and the Activation and Repression indices (Fig. 2f), or versus against expression of each FoxP3-induced or -repressed transcript (as FoldChange vs EV control; Supplementary Fig. 1f).
To relate DNA-binding activity of the mutants FoxP3 with their ability to affect transcription (Fig. 3c), a Pearson correlation coefficient was calculated between DNA- binding ability (normalized to wild-type activity) and the Activation or Repression indices derived from Nanostring profiling of transduced cells.
To relate the transactivation/transrepression activity of the FoxP3 mutants with their ability to bind cofactors a linear model was fit between target genes expression in the transduced cells (as FoldChange relative to EV control) and the factor-binding results (normalized to wild-type FoxP3 values), both log-transformed, adding the DNA-binding ability as an additional covariate to test whether the patterns might indirectly reflect DNA-binding activity (in practice, omitting the DNA binding term yielded very similar coefficients.)
Gel filtration chromatography
Activated CD4+ T cells transduced with FoxP3 or mutants (1×109 cells/sample) were lysed in NP-40 lysis butter (20mM Tris-HCl, 2mM EDTA, 1% NP-40 10% glycerol, 150mM NaCl, 1mM PMSF and 1X Proteinase inhibitor cocktail; Roche, #04693116001) for 2 hr with gentle vortexing at 4°C. The lysates were sedimented at 15,000 rpm for 30 min at 4°C to remove aggregates, and the supernatant injected into an FPLC Superose 6 column (10 X 300mm, GE Healthcare Life Science) at a flow rate of 400ul/min in PBS. Twenty seven fractions of 400 ul were collected, immediately concentrated with filter centrifugation (Centricon 10,000 KDa cutoff, Millipore) down to 50ul, resolved by SDS-PAGE and probed by sequential immunoblotting for FoxP3 (eBioscience, #14-5773). FoxP3 containing fractions were incubated with anti-RELA (Abcam, #ab7970) or anti-EZH2 (Active motif, #AC22), revolved by SDS-PAGE and immunoblotted with anti-FoxP3.
Co-immunoprecipitation and immunoblotting
HEK293 cells (1 X 107) were co-transfected as above with plasmids including FLAG- tagged wild-type or mutant FoxP3 and another TF (tagged with HA, 6xHIS or GFP). About 48 hr after the transfection, cells were harvested and lysed in NP-40 lysis butter (20mM Tris-HCl, 2mM EDTA, 1% NP-40 10% glycerol, 150mM NaCl, 1mM PMSF and 1X Proteinase inhibitor cocktail) for 20 min. Cell lysates were centrifuged at 13,000 rpm, 4°C for 30 min, and protein concentration was determined by Bradford protein assay (Bio-rad). Approximately 1mg of protein lysate were incubated with slow rotation for 4 hr at 4°C with anti-FLAG pre-bound to Protein G-coupled magnetic beads (Life Technologies) complex (per sample, 10ug of anti- FLAG and 50ul of Protein G suspension complex prepared by overnight incubation at 4°C, washed with lysis buffer twice just before use).
After washing with lysis buffer, proteins were eluted from the beads in 30 ul of 1X Laemmli Sample Buffer for 10 min at 100°C, resolved on a 7~10% SDS polyacrylamide gel and electroblotted onto PVDF membrane (Bio-rad). Membranes were blocked with 5% skim milk in 1X TBS buffer (50 mM Tris-Cl, pH 7.5. and 150 mM NaCl) for 1 hr at room temperature and probed with each primary antibody overnight at 4°C (αFLAG: Sigma, #F1804, αFoxP3: eBioscience, #14-5773, αHA: Sigma, #11583816001, αGFP: Abcam, #ab290, αFoxp1: Abcam, #ab16645 and α6XHIS: Abcam, #ab9108). After 5 washes with 1X TBS supplemented with 0.05% Tween-20, membranes were incubated with HRP-conjugated secondary antibody and developed with ECL prime Western Blotting Detection Reagent (GE Healthcare). Chemiluminescent images were acquired by ChemiDoc™ XRS+ System (Biorad) and analyzed using AlphaView Software (version 3.2.2; ProteinSimple)
To exclude the possibility of DNA contamination mediated non-specific interaction, indicated amounts of protein lysate were incubated with anti-FLAG/Bead complex in the presence of 10ug/ml Ethidium bromide, or the immunoprecipitates bound to the beads were treated with 1ug/ml of RNA free DNaseI (Roche) for 30 min at 37°C.
For the preclearing experiments, HEK293 cells (1 X 107) were triply transfected to express FLAG-tagged FoxP3 together with 2 other cofactors (either of RELA-GFP, KAT5-6XHIS, IZKF3-GFP or 6XHIS and EZH2-HA) together with FoxP3. 48 hr after transfection protein lysate was first precleared with specific antibody (control IgG, anti-RELA, anti-EZH2, anti-6XHIS for KAT5 or anti-GFP for IKZF3; 30ug/sample) for 12 hr at 4°C and precleared proteins were removed by inducing with Protein-G bead conjugated with magnetic bead (200ul/sample) for 2 hr at 4°C. Subsequently, precleared lysate was used for immuno- precipitation with anti-FLAG to pull FoxP3 complexes down. IPed protein was immuno- blotted with specific antibodies (anti-FoxP3, anti-6xHIS, anti-GFP or anti-HA) as above.
Super-resolution microscopy (3D-SIM)
Activated CD4+ T cells transduced with FoxP3 or mutants were immobilized on anti- CD3 (1ug/ml) coated coverslips (1.5um), fixed with 4% formaldehyde for 10 min at room temperature and washed 5 times with PBS. Fixed cells were permeabilized with ice-cold 100% Me-OH at 4°C for 20 min and washed ten times with PBS. Cells were stained with purified rat-anti-FoxP3 (1;200 dilution), rabbit-anti-EZH2 (1:200), mouse-anti-RELA (1:100) and rabbit- anti-Ikzf3 (1:200) for 1 hr at room temperature, washed and followed by staining with Donkey-anti-mouse-IgG-alexa488, Donkey-anti-rat-IgG-alexa594 and Donkey-anti-mouse- IgG-alexa647 (all secondary antibodies; 1:2000 dilution, Invitrogen) for 1 hr at room temperature. 3D-SIM data was collected on a DeltaVision OMX V4 Blaze system (GE Healthcare) equipped with a 60× / 1.42 N.A. Plan Apo oil immersion objective lens (Olympus), and three Edge 5.5 sCMOS cameras (PCO). Alexa488 was exited with a 488nm laser and a 528/48 emission filter, Alexa594 with a 571nm laser and 609/37 emission filter and Alexa647 with 645nm laser with 683/40 emission filter. Z-stacks of ~2 microns were acquired with a z-step of 125 nm and with 15 raw images per plane (five phases, three angles). Spherical aberration was minimized using immersion oil matching 58. Super-resolution images were computationally reconstructed from the raw datasets with a channel-specific measured optical transfer function (OTF) and a Wiener filter constant of 0.001 using CUDA-accelerated 3D-SIM reconstruction code based on Gustafsson et al 59. Subsequently, 3D-SIM images were analyzed with the Imaris Bitplane program. The spot function in Imaris automatically located FoxP3, EZH2 and RELA protein based on size and intensity thresholds. Each spots were represented by a sphere of arbitrary size (X, Y and Z) determined by Imaris (default setting, automatic mode). To determine the colocalization from SIM data, we first verified channel alignment and the absence of chromatic aberration with 0.2um diameter TetaSpeck microsphere beads (Invitrogen) stained throughout with multiple fluorescent dyes. Clusters were called as colocalized when their centers were less than 100 nm apart (there is uncertainty in the actual position of an object in SIM, owing to the length of the molecules and of the antibody pair used for detection). As positive control, we imaged Treg cells with FoxP3 primary antibody, then simultaneously counterstained with Alexa 488- and Alexa 568- conjugated secondary antibodies (~75 % of FoxP3 spots were called in both wavelengths). To calculate spot intensity of co-localized spots, RELA or EZH2 spots that overlapped with FoxP3, voxels were further determined with the centered fluorescence intensity of each FoxP3 spot based on Imaris default settings. As controls, the same numbers of FoxP3 or RELA spots were randomly selected to calculate their EZH2/RELA or FoxP3/EZH2 intensity.
Supplementary Material
Acknowledgments
We thank Drs A. Arvey, L. Chen, T. Chatila, K. Struhl, J. Waters and A. Rudensky for insightful discussions: K. Hattori, C. Araneo and A. Rhoads for help with mice, cell sorting, profiling and software, the HMS Cell Biology Microscopy Facility and L. Shao for 3D-SIM. This work was supported by NIH grant AI116834 and a Sponsored Research Agreement from GSK, and HKK by NRF fellowship 357-2011-1C00084.
Footnotes
AUTHOR CONTRIBUTIONS
HK.K. and HM.C. performed the experiments; HK.K., HM.C., C.B. and D.M. designed the study, analyzed/interpreted the data, HK.K., C.B. and D.M. wrote the manuscript.
Accession codes
FoxP3 ChIPseq datasets are deposited as GSE102281.
References
- 1.Sakaguchi S. Naturally arising CD4+ regulatory T cells for immunologic self-tolerance and negative control of immune responses. Annu Rev. Immunol. 2004;22:531–562. doi: 10.1146/annurev.immunol.21.120601.141122. [DOI] [PubMed] [Google Scholar]
- 2.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panduro M, Benoist C, Mathis D. Tissue Tregs. Annu. Rev Immunol. 2016;34:609–633. doi: 10.1146/annurev-immunol-032712-095948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ziegler SF. FOXP3: of mice and men. Annu Rev. Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
- 5.Fontenot JD, et al. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 6.Sugimoto N, et al. Foxp3-dependent and -independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNA microarray analysis. Int. Immunol. 2006;18:1197–1209. doi: 10.1093/intimm/dxl060. [DOI] [PubMed] [Google Scholar]
- 7.Hill JA, et al. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Ferraro A, et al. Interindividual variation in human T regulatory cells. Proc Natl Acad Sci U S A. 2014;111:E1111–E1120. doi: 10.1073/pnas.1401343111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arvey A, et al. Genetic and epigenetic variation in the lineage specification of regulatory T cells. Elife. 2015;4:e07571. doi: 10.7554/eLife.07571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–770. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- 11.Gavin MA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 12.Cipolletta D, et al. PPAR-gamma is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopes JE, et al. Analysis of FOXP3 reveals multiple domains required for its function as a transcriptional repressor. J Immunol. 2006;177:3133–3142. doi: 10.4049/jimmunol.177.5.3133. [DOI] [PubMed] [Google Scholar]
- 14.Li B, et al. FOXP3 is a homo-oligomer and a component of a supramolecular regulatory complex disabled in the human XLAAD/IPEX autoimmune disease. Int. Immunol. 2007;19:825–835. doi: 10.1093/intimm/dxm043. [DOI] [PubMed] [Google Scholar]
- 15.Wu Y, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 16.Bandukwala HS, et al. Structure of a domain-swapped FOXP3 dimer on DNA and its function in regulatory T cells. Immunity. 2011;34:479–491. doi: 10.1016/j.immuni.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, et al. DNA binding by FOXP3 domain-swapped dimer suggests mechanisms of long-range chromosomal interactions. Nucleic Acids Res. 2015;43:1268–1282. doi: 10.1093/nar/gku1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersen KG, Nissen JK, Betz AG. Comparative Genomics Reveals Key Gain-of-Function Events in Foxp3 during Regulatory T Cell Evolution. Front Immunol. 2012;3:113. doi: 10.3389/fimmu.2012.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao Y, et al. Histone acetyltransferase mediated regulation of FOXP3 acetylation and Treg function. Curr. Opin. Immunol. 2010;22:583–591. doi: 10.1016/j.coi.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudra D, et al. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat Immunol. 2012;13:1010–1019. doi: 10.1038/ni.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bettini ML, et al. Loss of epigenetic modification driven by the Foxp3 transcription factor leads to regulatory T cell insufficiency. Immunity. 2012;36:717–730. doi: 10.1016/j.immuni.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng Y, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaudhry A, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darce J, et al. An N-terminal mutation of the Foxp3 transcription factor alleviates arthritis but exacerbates diabetes. Immunity. 2012;36:731–741. doi: 10.1016/j.immuni.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samstein RM, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schubert LA, et al. Scurfin (FOXP3) acts as a repressor of transcription and regulates T cell activation. J. Biol. Chem. 2001;276:37672–37679. doi: 10.1074/jbc.M104521200. [DOI] [PubMed] [Google Scholar]
- 28.Bettelli E, Dastrange M, Oukka M. Foxp3 interacts with nuclear factor of activated T cells and NF-kappa B to repress cytokine gene expression and effector functions of T helper cells. Proc Natl Acad Sci U S A. 2005;102:5138–5143. doi: 10.1073/pnas.0501675102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, et al. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc Natl Acad Sci U S A. 2007;104:4571–4576. doi: 10.1073/pnas.0700298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C, et al. Transcriptional regulation by Foxp3 is associated with direct promoter occupancy and modulation of histone acetylation. J Biol. Chem. 2006;281:36828–36834. doi: 10.1074/jbc.M608848200. [DOI] [PubMed] [Google Scholar]
- 31.Arvey A, et al. Inflammation-induced repression of chromatin bound by the transcription factor Foxp3 in regulatory T cells. Nat Immunol. 2014;15:580–587. doi: 10.1038/ni.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitagawa Y, et al. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat Immunol. 2017;18:173–183. doi: 10.1038/ni.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 34.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 35.Luo CT, Li MO. Transcriptional control of regulatory T cell development and function. Trends Immunol. 2013;34:531–539. doi: 10.1016/j.it.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benoist C, Mathis D. Treg cells, life history, and diversity. In: Mathis D, Rudensky A, editors. Immune Tolerance. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2013. pp. 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quintana FJ, et al. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat. Immunol. 2012;13:770–777. doi: 10.1038/ni.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong Y, et al. Polycomb antagonizes p300/CREB-binding protein-associated factor to silence FOXP3 in a Kruppel-like factor-dependent manner. J. Biol. Chem. 2012;287:34372–34385. doi: 10.1074/jbc.M111.325332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 40.Hwang SS, et al. YY1 inhibits differentiation and function of regulatory T cells by blocking Foxp3 expression and activity. Nat Commun. 2016;7:10789. doi: 10.1038/ncomms10789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cowell IG. Repression versus activation in the control of gene transcription. Trends Biochem. Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 42.Thiel G, Lietz M, Hohl M. How mammalian transcriptional repressors work. Eur J Biochem. 2004;271:2855–2862. doi: 10.1111/j.1432-1033.2004.04174.x. [DOI] [PubMed] [Google Scholar]
- 43.Hahn S. Ellis Englesberg and the discovery of positive control in gene regulation. Genetics. 2014;198:455–460. doi: 10.1534/genetics.114.167361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fischer M, Steiner L, Engeland K. The transcription factor p53: not a repressor, solely an activator. Cell Cycle. 2014;13:3037–3058. doi: 10.4161/15384101.2014.949083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mekhail K, Moazed D. The nuclear envelope in genome organization, expression and stability. Nat Rev Mol Cell Biol. 2010;11:317–328. doi: 10.1038/nrm2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loizou L, Andersen KG, Betz AG. Foxp3 interacts with c-Rel to mediate NF-kappaB repression. PLoS. ONE. 2011;6:e18670. doi: 10.1371/journal.pone.0018670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee SM, Gao B, Fang D. FoxP3 maintains Treg unresponsiveness by selectively inhibiting the promoter DNA-binding activity of AP-1. Blood. 2008;111:3599–3606. doi: 10.1182/blood-2007-09-115014. [DOI] [PubMed] [Google Scholar]
- 48.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 49.Fu W, et al. A multiply redundant genetic switch 'locks in' the transcriptional signature of regulatory T cells. Nat Immunol. 2012;13:972–980. doi: 10.1038/ni.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blecher-Gonen R, et al. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat Protoc. 2013;8:539–554. doi: 10.1038/nprot.2013.023. [DOI] [PubMed] [Google Scholar]
- 51.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson JT, et al. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramirez F, et al. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC. Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koh KP, Sundrud MS, Rao A. Domain requirements and sequence specificity of DNA binding for the forkhead transcription factor FOXP3. PLoS. ONE. 2009;4:e8109. doi: 10.1371/journal.pone.0008109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wakamatsu E, Mathis D, Benoist C. Convergent and divergent effects of costimulatory molecules in conventional and regulatory CD4+ T cells. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1220688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hiraoka Y, Agard DA, Sedat JW. Temporal and spatial coordination of chromosome movement, spindle formation, and nuclear envelope breakdown during prometaphase in Drosophila melanogaster embryos. J Cell Biol. 1990;111:2815–2828. doi: 10.1083/jcb.111.6.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gustafsson MG, et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 2008;94:4957–4970. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.