Abstract
To maintain stable genomes and to avoid cancer and aging, cells need to repair a multitude of deleterious DNA lesions, which arise constantly in every cell. Processes that support genome integrity in normal cells, however, allow cancer cells to develop resistance to radiation and DNA damaging chemotherapeutics. Chemical inhibition of the key DNA repair proteins and pharmacologically induced synthetic lethality have become instrumental in both dissecting the complex DNA repair networks and as promising anticancer agents. The difficulty in capitalizing on synthetically lethal interactions in cancer cells is that many potential targets do not possess well-defined small molecule binding determinates. In this review we will discuss several successful campaigns to identify and leverage small-molecule inhibitors of the DNA repair proteins, from PARP1, a paradigm case for clinically successful small molecule inhibitors, to coveted new targets, such as RAD51 recombinase, RAD52 DNA repair protein, MRE11 nuclease and WRN DNA helicase.
Introduction
DNA replication, repair, and recombination proteins form complex and agile networks. These networks organize the participating proteins into molecular machines that act on different substrates and channel them to different outcomes. Some of these machines display the capacity to accurately repair DNA damage or reestablish damaged DNA replication forks without the loss of genetic information. Under other circumstances, action of the same molecular machines destabilizes the genome, which can lead to cancer, or cause accumulation of toxic repair intermediates, which can lead to cell death. Moreover, variations on the very same processes that support genome integrity in normal cells, allow cancer cells to acquire a more aggressive character and facilitate the emergence of resistance to radiation and DNA damaging chemotherapeutics (Jeggo and Lobrich, 2015). A comprehensive understanding of the molecular events that draw otherwise normal DNA repair intermediates of the accurate DNA repair mechanisms into “rogue” mechanisms that lead to genome destabilization and cell death is essential, but is complicated due to the multiple roles and intricate regulation of the DNA repair proteins.
Since the 1940s genetic interactions in which the combined effect of two gene mutations is not simply additive, have been used to dissect molecular pathways (Dobzhansky, 1946). Negative (synthetically lethal and synthetically sick) and positive (alleviating) genetic interactions have been successfully used to establish relationships between various DNA repair proteins. Synthetic lethality here is an extreme case of a genetic interaction, where two individual viable mutations, when combined, result in a lethal phenotype. In 1997 Hartwell and colleagues (Hartwell et al., 1997) first proposed to use synthetic lethality as an anticancer therapeutic strategy to be employed in cancers that have genetic defects in DNA repair proteins, and also in cancers that are “addicted” to a particular DNA repair mechanism for robust DNA repair and replication. In treatment of such cancers, a defect in a DNA repair gene is combined with a chemical inhibition of an enzymatic activity or interactions of a DNA repair protein that is critical for survival of cancerous cells, but is less important for the survival of normal cells. The goal is to avoid or to minimize the toxicity associated with radiation and DNA damaging chemotherapies that remain a standard of care. In addition to their potential as anticancer therapeutics, specific inhibitors of DNA repair proteins attenuate a selected enzymatic activity or interaction only during the analysis, which permits a direct comparison with the functional state simply by removing the inhibitor. Therefore, pharmacological inhibition offers valuable tools for the dissection of the complex DNA repair networks that employ multifunctional proteins. Moreover, in some cases (as will be exemplified below by a sub-class of PARP inhibitors and by inhibitors of the helicase activity of WRN helicase/nuclease) inhibiting one activity of a multifunctional DNA repair enzyme may trap it on the DNA repair intermediate, preventing access by compensatory alternative mechanisms, and thereby leading to specific toxicity exceeding that of the enzyme depletion.
In this review we will discuss the state of the art in DNA repair inhibitors and their progression from research tools for dissecting the DNA repair pathways to the advancement of personalized cancer treatments, as well as how the inhibitors developed as anticancer treatments, are improving our understanding of the complex and interconnecting DNA repair networks. Figure 1 summarizes the action of the inhibitors discussed in this review.
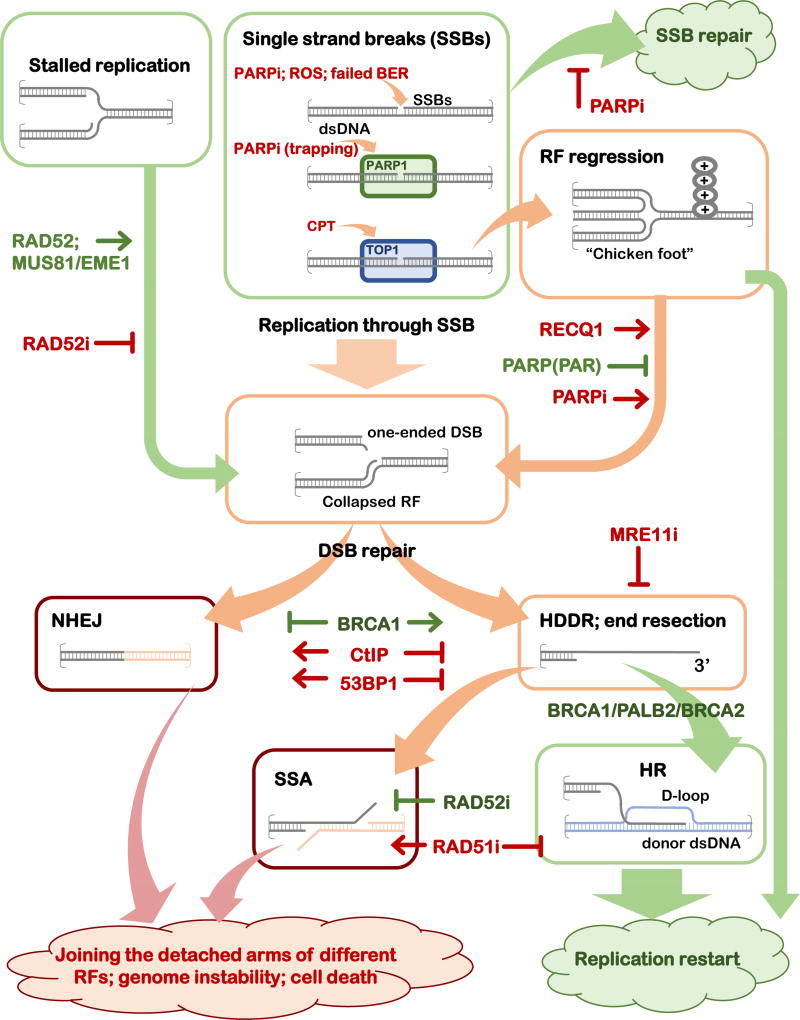
Figure 1. Roles of the DNA repair inhibitors in altering the outcomes of the DNA repair events in the presence of DNA replication.
Schematic representation of the progression of a DNA lesion (a single-strand break, SSB, or stalled replication fork, RF) through various DNA repair mechanisms leading to DNA repair and restoration of replication fork, or to genome destabilizing events. The intermediates and mechanisms that are likely to lead to accurate repair if processed through an appropriate mechanism are depicted in green boxes, the intermediates that can lead to either positive (replication fork restoration) or negative (genome instability and cell death) outcomes are in orange boxes, and the intermediates and mechanisms that funnel the DNA repair intermediates into the genome destabilizing outcomes are shown in red boxes. The key regulators (proteins and inhibitors) that affect the outcome of each event are shown.
Synthetic lethality and other synthetic interactions in targeting DNA repair
Several types of synthetic interactions can be considered when approaching DNA repair (Zinovyev et al., 2013), with the simplest being between-pathway synthetic lethality, which assumes that the two genes act in parallel pathways that compensate for one another in carrying out an essential cellular function. Another type of synthetic interaction is an in-pathway synthetic lethality (Zinovyev et al., 2013), whereby a genetic defect blocks a progression of a DNA repair intermediate through a repair pathway containing reversible steps, while the second mutation or chemical inhibition blocks the reverse step and thereby causes accumulation of a cytotoxic DNA structures of nucleoprotein complexes, or channels these structures to “rogue”, mutagenic DNA repair mechanisms.
A functional healthy cell, or even a cell containing a heterozygous mutation in an important tumor suppressor is an “antifragile system” (a system that readily tolerates and improves with both environmental and internal stresses) (Taleb, 2012). In contrast, DNA repair systems in cancerous cells are often robust at the expense of antifragility. This robustness allows cancerous cells to strive despite the defects in an important DNA repair pathway and to efficiently replicate their genomes, which is a prerequisite for rapid proliferation. This robustness also represents an Achilles’ hill of the cancerous cell that can be exploited for therapeutic purposes.
In 2014, the first drug targeting DNA repair defects, Lynparza (olaparib), a poly-ADP-ribose polymerase (PARP) inhibitor was approved by the FDA for treatment of advanced ovarian cancer associated with defective BRCA genes. Olaparib and the whole extended family of PARP inhibitors became a paradigm for targeting synthetic interactions in DNA repair deficient tumors. The action of these drugs has also highlighted pharmacologically induced synthetic lethality as a research and therapeutic tool.
BRCA1, BRCA2, and PARP1
BRCA1 and BRCA2 are tumor suppressors that are commonly mutated or downregulated in hereditary and sporadic breast and ovarian cancers. These tumor suppressors play diverse roles in cellular DNA metabolism. Both BRCA1 and BRCA2 are involved in homologous recombination (HR) (Prakash et al., 2015), which repairs the DNA double strand breaks (DSBs), DNA and protein-DNA cross-links, as well as collapsed replication forks without loss of information by using an intact homologous dsDNA, usually a sister chromatid, as a template (Couedel et al., 2004; Heyer, 2015; Jasin and Rothstein, 2013; Kowalczykowski, 2015; Moynahan and Jasin, 2010).
BRCA1
regulates the DNA repair pathway choice at DSBs by promoting homology directed DNA repair (HDDR) over non-homologous end joining (NHEJ), and further by promoting HR over mutagenic single-strand annealing (SSA) (Kass and Jasin, 2010; Prakash et al., 2015). Outside of DNA repair, BRCA1 functions at stalled or defective transcription start sites (Hill et al., 2014). In conjunction with BRCA1, BRCA2 also prevents RNA-DNA hybrid accumulation to prevent transcriptional stress (Bhatia et al., 2014; Hatchi et al., 2015).
BRCA2
is a recombination mediator. It facilitates assembly of the RAD51 nucleoprotein filament on ssDNA, thereby promoting the central step in HR, which culminates with the exchange of the DNA strands between damaged and template DNA molecules (Couedel et al., 2004; Jensen et al., 2010; Liu et al., 2010; Prakash et al., 2015; Thorslund et al., 2010). Along with the core HR machinery (RAD51 recombinase, the five human RAD51 paralogs, and RAD52), BRCA1 and BRCA2 contribute to stabilization of the distressed DNA replication forks, presumably by protecting the nascent ssDNA from excessive degradation by the MRE11 nuclease (see (Kolinjivadi et al., 2017) for review).
Mutations in BRCA1 and BRCA2 genes that render the respective protein dysfunctional or absent are associated with an increased risk in developing breast and ovarian cancers, and to lesser extent prostate, pancreas, and stomach cancers to name a few (Cavanagh and Rogers, 2015; Kobayashi et al., 2013). This is because impaired BRCA1 or BRCA2 function is associated with compromised HR repair and genomic instability (Konishi et al., 2011; Yata et al., 2014). Susceptible to the radiation, platinum-based chemotherapy and PARP inhibitors at first, BRCA-deficient tumors often develop drug resistance by restoring HR function or by developing compensatory mechanisms that allow the cancer cell to repair the damage and to proliferate.
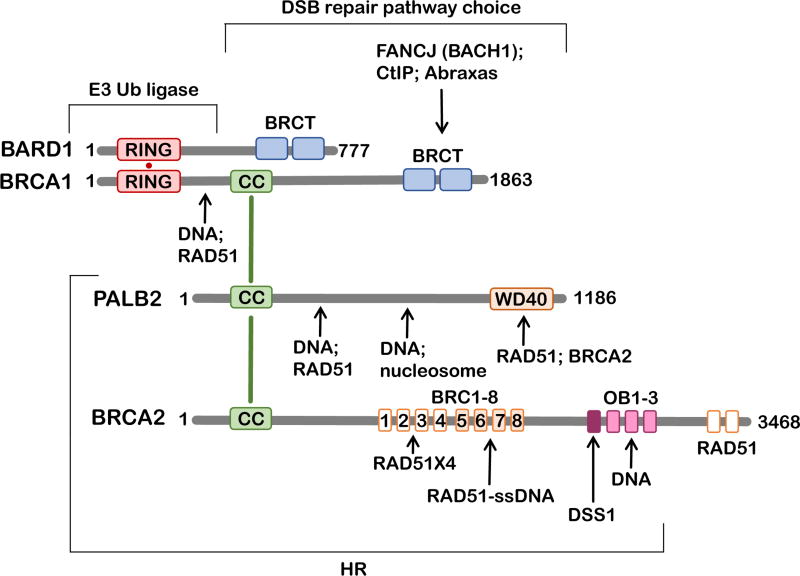
Because of the modular, multifunctional nature of both BRCA1 and BRCA2 proteins (Figure 2), it remains unclear which of the numerous activities and interactions endow the tumor suppressor function, and which promote the emergence of drug resistance. Understanding the roles of BRCA1 and BRCA2 is impossible without understanding their incorporation into the tumor suppressor network (current understanding of the BRCA1-BRCA2 interacting network is reviewed elsewhere (Jiang and Greenberg, 2015)). Thirteen different tumor suppressor proteins interact with BRCA1 and BRCA2. Most commonly found in a 1:1 complex with BRCA1 is a tumor suppressor BARD1 (Wu et al., 1996). BRCA1 and BARD1 form a heterodimer, which functions as an E3 ubiquitin ligase. The targets of BRCA1/BADR1 E3 activity include BRCA1 itself, CtIP and histone H2A (Thakar et al., 2010; Yu et al., 2006). The interaction between BRCA1 and BARD1 enhances the stability of both proteins, masks the nuclear export signal, increases the E3 ubiquitin ligase activity in vitro and enhances DNA damage site recognition. The main stabilizing partner of BRCA2 is DSS1, a small highly acidic protein, which is found in stoichiometric complexes with BRCA2 in the crystal structure (Yang et al., 2002) and in cells (Li et al., 2006). Both BRCA1 and BRCA2 interact with the tumor suppressor PALB2 (Park et al., 2014) and the BRCA1-PALB2-BRCA2 interaction mediates the DSB repair pathway choice promoting HR over SSA pathway that depends on the RAD52 protein function (Anantha et al., 2017).
Figure 2. BRCA-interacting network.
Schematic representation of the primary structures of BRCA1, BRCA2 and their interacting partners BARD1 and PALB2. Structural domains and regions of significance to this review are shown as rectangles: RING domains (red), CC – coiled coil domains (green), WD40 domain (orange), BRCT domains (blue), BRC motifs 1 – 8 (orange), OB folds 1 – 3 and DSS1-interacting region (purple), BRCA2 C-terminal RAD51-interacting domains (orange). Arrows point to the respective interaction sites. Brackets highlight regions important for the stated cellular function.
While the roles of BRCA1 and BRCA2 in transcription and chromatin remodeling are likely to play a role in tumorigenesis, it was the function of these tumor suppressors in HR that inspired the development of PARP inhibitors.
PARP1
(Poly (ADPribose) polymerase1) is a salient member of a large PARP family of enzymes that modify acceptor proteins with ADP-ribose modifications (Ame et al., 2004). PARP1 catalyzes the polymerization of ADP-ribose by employing NAD+ substrates, in which the ADP-D-ribosyl group of NAD+ is transferred to an acceptor carboxylate group on a target DNA repair protein (PARylation) or on the enzyme itself (auto-PARylation), and further ADP-ribosyl groups are transferred to the 2’-position of the terminal adenosine moiety, building up a negatively charged branched polymer with an average chain length of 20 – 30 units. PARP1 is an important player in the base excision repair (BER) and is a general sensor of DNA damage. It recognizes single-strand DNA breaks that arise due to ineffective BER, sugar damage by the reactive oxygen species, or from abortive topoisomerase 1 (TOP1) activity (Caldecott, 2008). PARP1 has catalytic, DNA-binding and protein-recruitment domains. Its enzymatic activity is activated by binding of PARP1 to different types of DNA lesions including double-strand breaks, single-strand breaks, DNA crosslinks, and stalled replication forks (Krishnakumar and Kraus, 2010). PARP1 activation facilities chromatin reorganization around the DNA lesion and subsequent DNA repair. Importantly for DNA repair and for understanding of the function of pharmacological inhibitors, PARP1 eventually auto-PARylates. The negative charge endowed by PAR chains then promotes PARP1 dissociation from the DNA.
PARP inhibitors
Because its activity converts DNA breaks into intracellular signals that activate DNA repair programs and/or regulates the cell death options, PARP1 was proposed as a potential anticancer drug target whose modulation may sensitize cancerous cells to DNA damaging agents (Decker and Muller, 2002). The initial thinking was that inhibition of PARP1 will leave single strand DNA breaks (SSBs), which may be converted into one-ended double strand breaks (DSBs) during replication, if unrepaired (Ashworth, 2008) (Figure 1). PARP1 was also shown to play a role in slowing the progression of replication forks in the presence of DNA damage and their subsequent repair by HR (Sugimura et al., 2008). Thus PARP1 inhibition was expected to be more toxic to rapidly proliferating cancer cells than to the normal cells, and to widen a therapeutic window for treatment with the DNA damaging chemotherapeutic drugs.
Because unrepaired single strand breaks lead, by the way of collapsed replication forks, to one-ended DSBs, which have to be repaired by HR (Figure 1), PARP1 inhibition was expected to be especially toxic to cancerous cells harboring HR defects, and in particular in cells lacking BRCA1 or BRCA2 tumor suppressor. In HR proficient cells, such DSBs are accurately and efficiently repaired. In the absence of HR (such as in BRCA-deficient cells), however, these one-ended DSBs are left behind. One would expect that broken arms of different damaged replication forks can then be mended together by error prone repair pathways such as SSA or NHEJ without regard for continuity of the original chromosome, leading to gross genomic instability and eventually lethality (Ashworth, 2008; De Lorenzo et al., 2013) (Figure 1). Notably, PARP inhibitor treatment was found to stimulate NHEJ in the HR-deficient cells (Patel et al., 2011).
As expected, PARP1 inhibition confers synthetic lethality with biallelic defects in BRCA1, BRCA2 or PALB2 commonly found in familial and sporadic breast and ovarian cancers (Bryant et al., 2005; Farmer et al., 2005). The possibility to take advantage of this pharmacologically induced synthetic lethality was tested in clinical trials since 2003 (Yap et al., 2011), and a decade later PARP inhibitors olaparib (2014) and later rucaparib (2016) were approved for the treatment of advanced, chemotherapy resistant ovarian cancer in patients with BRCA1 or BRCA2 germline mutations, and niraparib (2017) for treatment of platinum-sensitive recurrent ovarian, fallopian tube, and primary peritoneal cancers (see (Murata et al., 2016) for a comprehensive review on PARP inhibitors currently in various stages of clinical trials).
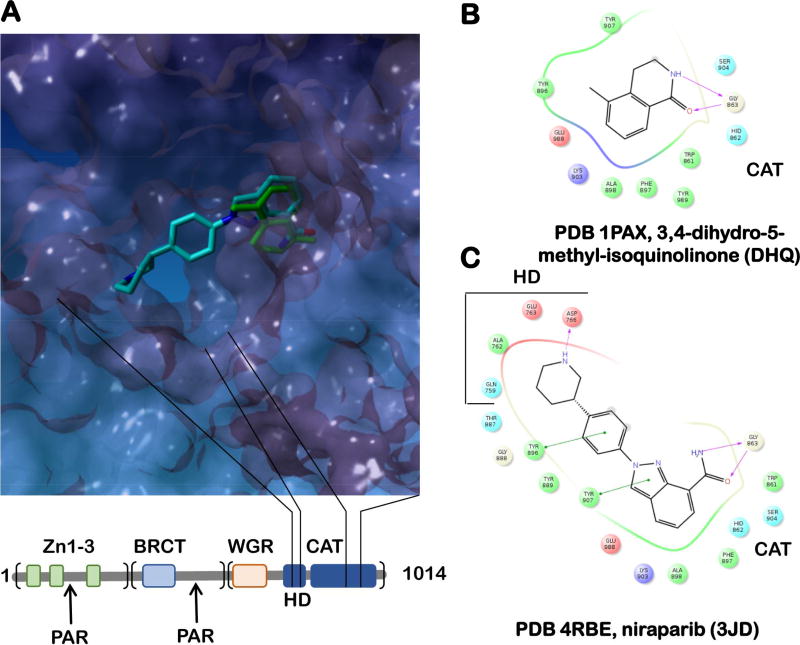
The first physiologically specific PARP inhibitors were derivatives of benzamide, a nicotinamide analog (Purnell and Whish, 1980). The best understood, first generation PARP inhibitors bind to the PARP catalytic domain with low nM affinity; the quinazolinone class of PARP inhibitors occupy both the nicotinamide-ribose binding site and the adenosine-ribose binding site of NAD+ thus exploiting the unique nature of the PARP bi-substrate pocket (Figure 3). Subsequently, the first generation of clinical PARP inhibitors was followed by the second generation of inhibitors (reviewed in (Lord and Ashworth, 2017)). Rather than acting simply by inhibiting the catalytic activity of PARP1, some of these compounds act by inhibiting dissociation of PARP1 itself from the site of DNA damage. “Trapping” of PARP1 on DNA results in a highly cytotoxic protein-DNA complex, which impedes DNA replication and may prevent access of the downstream DNA repair proteins to the site of the damage. This trapping activity was shown to vary widely between different PARP inhibitors; niraparib being the strongest and veliparib being the weakest poisons, respectively. The relationship between effectiveness of each PARP inhibitor in vitro, its ability to trap PARP1 on damaged DNA and the in vivo efficacy is not trivial. Thus, one must deconvolute at least two dimensions of structure activity relationship (SAR), binding affinity to the target and the less understood trapping of the PARP1-DNA complex. Another dimension in SAR is specificity of the available PARP inhibitors. While all successful compounds display similar inhibition of PARP1 and PARP2 enzymes in vitro, their effectiveness in cells, as well as their potential anti-proliferative activities outside of PARP inhibition vary (Chuang et al., 2012). This has been explained by the difference in PARP1 trapping, and also by differences in the off-target effects. Targeting promiscuity can be detrimental by causing unintended toxicity, or beneficial, if the non-canonical targets enhance drug efficacy. An in vitro study conducted on a series of 185 small-molecule PARP inhibitors and the catalytic domains of 13 out of 17 human PARP enzymes suggested a potential promiscuous binding to several PARP family members for even such clinically successful inhibitors as olaparib and rucaparib (Wahlberg et al., 2012). Niraparib and veliparib, on the other hand, have over 100-fold specificity for PARP1 and PARP2 over other PARP enzymes (Thorsell et al., 2017). Figure 3 shows one of the determinants for such selectivity – niraparib binds within the nicotinamide binding pocket in the ADP-ribosyl transferase catalytic site and also makes contacts with the regulatory subdomains of PARP1 and PARP2 (E763 & D766 in PARP1). Target maps for the four clinically successful inhibitors olaparib, veliparib, niraparib, and rucaparib, were recently developed using a mass spectroscopy based chemical proteomics assay (Knezevic et al., 2016). Despite the existence of many NAD+-binding proteins in the cell, PARP inhibitors as a class displayed high target selectivity towards PARP1, PARP2 and several of their interacting partners. However, several other unexpected targets were identified. Rucaparib, for example, inhibits hexose-6-phosphate dehydrogenase, whose loss of function may result in apoptosis of cancerous cells thus augmenting the rucaparib’s efficacy.
Figure 3. PARP inhibitors.
A. The overlap of a nicotineamide analog DHQ (green; PDB 1PAX) bound to the active site of PARP1 and the second generation PARP1 inhibitor niraparib (light blue; PDB 1PAX), which occupies both the nicotinamide-ribose binding pocket and the adenosine-ribose binding pocket of ADP-ribosyl transferase catalytic site, making additional contacts with E763 and D766 of the regulatory α-helical sub-domain (HD) (Thorsell et al., 2017). PARP1 ADP-ribosyl transferase catalytic domain (CAT; PDB 1PAX) is shown as a surface representation. The primary structure of PARP1 is shown below the active site image. The bead-on-a-string architecture of PARP1 (Langelier and Pascal, 2013) includes the N-terminal DNA binding region (the three Zn-finger domains are shown as green rectangles), the middle region containing a BRCT domain and the main auto-PARylation site (Altmeyer et al., 2009), and the C-terminal region that contains the WGR domain responsible for the interdomain contacts and the cross-talk between the DNA-binding/damage recognition domains, the regulatory HD domain as well as the CAT domain. B. and C. The ligand maps for DHQ and niraparib, respectively.
Lessons from PARP resistance
While approximately 15% of ovarian cancer patients with BRCA-deficient tumors remain disease free for more than 5 years from the beginning of the olaparib therapy (Ledermann et al., 2016), many patients develop PARP inhibitor resistance within the first year, which limits the effectiveness of the therapy (Incorvaia et al., 2017; Lupo and Trusolino, 2014; Murata et al., 2016). Several mechanisms were suggested for the emergence of resistance to PARP inhibitors. The wide spectrum of responses to PARP inhibitor treatment stems from the range of BRCA defects and the genetic backgrounds of BRCA-deficient cancers; the understanding of these mechanisms is essential for our understanding of the complex and agile network of the DNA repair mechanisms, their intended action and their failure, but remains limited (Incorvaia et al., 2017). Tumor cells that acquired resistance to PARP inhibitor were shown to accumulate frame shift mutations that correct the mutated BRCA1 or BRCA2 genes, restoring the protein expression and the HR function (Edwards et al., 2008; Sakai et al., 2008; Swisher et al., 2008). HR function can also be restored in BRCA1-mutated cells by the loss of 53BP1 (p53 binding protein 1) function, whose activity prevents DNA end resection in the absence of BRCA1, which is the first step in HR (Bouwman et al., 2010; Bunting et al., 2010). Overexpression of microRNA miR-622 in BRCA1-deficient high-grade serous ovarian carcinomas restores HR by downregulating the Ku complex and thereby shifts the DSB repair pathway choice from NHEJ to HR in a mechanism not dissimilar to the loss of 53BP1 function (Choi et al., 2016).
Another resistance mechanism is related to the function of BRCA hypomorphs – mutant proteins that retain residual activity and are able to compensate, at least partially, for the absence of the wild type protein. Of particular interest is BRCA1185delAG, which predisposes carriers to early-onset breast and ovarian cancer (Friedman et al., 1995; Struewing et al., 1996), promotes resistance to PARP inhibitors and cisplatin (Wang et al., 2016). While the 185delAG mutation was presumed to completely obliterate BRCA1 expression due to a mutation-induced stop codon, BRCA1185delAG alleles were translated from an alternative site downstream of the stop codon. The resulting protein lacking the RING domain was unable to dimerize with BARD1 and displayed no ubiquitin ligase activity. It, however, retained its ability to interact with DNA and HR proteins. It is important to note here that the full spectrum of ubiquitin ligase, transcription-related and HR-related activities of BRCA1 defines its function as a tumor suppressor, while the subset of these functions expected to be most critical for DNA repair and for supporting DNA replication seem to facilitate the emergence of drug resistance. Several investigations have attempted to clarify this point. A high throughput study of patient-derived BRCA1 missense variants in mice, for example, confined all deleterious variants that confer PARP inhibitor resistance to the RING and BRCT domain of BRCA1, which are involved in BARD1 interaction/E3 ubiquitin ligase activity and in the interaction with BACH1 helicase, CtIP and Abraxas, respectively (Bouwman et al., 2013). Another study showed that resistance to olaparib in the BRCA1-deficient triple negative breast cancer cells correlates well with the HR activity of each BRCA1 variant with the RING, coiled coil (CC) and BRCT domain mutants failing to confer resistance to PARP inhibitors (Anantha et al., 2017). It is important to note, however, that these mutants are different from the BRCA1185delAG discussed above, which produces a stable, HR proficient protein despite missing RING domain.
Finally, overexpression of RAD51 recombinase, which carries out the DNA strand exchange reaction at the central step of HR potentiates resistance of the triple negative breast cancer cells to PARP inhibitors (Liu et al., 2016).
PARP1 and RECQ1 DNA helicase
In addition to their clinical application, PARP inhibitors have proven valuable research tools to chart various DNA repair mechanisms regulated by PARylation. DNA repair helicase RECQ1 is an example of a DNA repair protein whose activity is regulated by PARP1, more specifically through PARP1 auto-PARylation (as described below). Human RECQ1 plays a key role in the recovery of DNA replication forks, which are slowed down and reversed into so-called “chicken foot” structures as a consequence of replication stress elicited by topoisomerase I (TOP1) inhibitors (Berti et al., 2013). Similar to PARP1 inhibition discussed above, TOP1 inhibition leaves behind DNA lesions that, if left unmended, are converted into replication-mediated DSBs (Pommier et al., 2003). Natural alkaloid camptothecin (CPT) and its clinical derivatives (such as irinotecan and topotecan, FDA approved drugs for treatment of colorectal carcinomas and ovarian cancers, respectively) act by trapping the TOP1 cleavage complex, a catalytic intermediate of the supercoiled DNA relaxation reaction in which TOP1 is covalently linked to the DNA (Fox et al., 2003). The inhibition of TOP1 relaxation activity with low doses of CPT slows replication forks by promoting accumulation of positive supercoils ahead of the replication fork, thereby delaying the appearance of the replication-mediated DSBs; PARP1 activity is necessary for this replication fork reversal (Koster et al., 2007; Ray Chaudhuri et al., 2012). An elegant combination of cellular studies in the presence of olaparib and CPT, single-fiber DNA analysis and in vitro biochemistry unraveled an unexpected mechanism of RECQ1 activity and regulation at the regressed DNA replication forks (Berti et al., 2013): RECQ1 has a unique ability among five human RecQ-family DNA helicases to unidirectionally drive chicken-foot conversion back to the fork structure. This activity, however, needs to be controlled to allow time for removal of TOP1 from the unreplicated DNA and mending the lesions. Unrestricted RECQ1 activity restores replication forks before the repair process is completed resulting in the replication-induced DSBs. RECQ1, again uniquely among human RecQ-family helicases, is prevented from engaging the chicken-foot structures by PARylated PARP1: the two proteins form a complex that keeps RECQ1 off the DNA. PARP-inhibited cells do not slow replication forks and do not accumulate reversed forks after the CPT treatment, instead accumulating the DSBs. Because low concentrations of olaparib and CPT were sufficient to induce replication-mediated DSBs, this study suggest a possibility of combining PARP and TOP1 inhibitors and offered RECQ1 as a potential new therapeutic target. If, for example, the regressed replication forks are restored by HR in the absence of RECQ1, the RECQ1 inhibition may significantly exacerbate the effectiveness of TOP1 inhibitors in the HR-deficient cells.
The caveat of targeting the DNA branch-migrating enzymes, like RECQ1, is that the cell has several DNA motors capable of both regression and restoration of replication forks. One such motor is RAD54 (Bugreev et al., 2011), a SNF2/SWI2 family ATP-dependent DNA translocase with several important roles in HR and genome maintenance (Ceballos and Heyer, 2011).
A pilot HTS screen of the Broad Institute validation library (3000 compounds) using a FRET-based assay for branch migration of Holliday junctions by RAD54 yielded several promising RAD54 inhibitors (Deakyne et al., 2013). The assay showed high robustness and consistency (Z’ of 0.825 – 0.875) and will be valuable for identification of inhibitors of other branch-migrating enzymes. One of the identified RAD54 inhibitors, streptonigrin was subjected to further biochemical analysis. Interestingly, streptonigrin inhibits branch migration by interfering with the RAD54 ATPase activity through a direct interaction and generation of reactive oxygen species, but has little or no effect on the RAD54-DNA interaction, or the RAD54 ability to stimulate the RAD51-mediated DNA strand exchange (Deakyne et al., 2013).
Inhibitors of RecQ-family DNA helicases
WRN (Werner syndrome protein), a RecQ-family human DNA helicase and nuclease, is an example of an enzyme whose chemical inhibition and protein depletion have dramatically different consequences (Aggarwal et al., 2013a; Aggarwal et al., 2013b; Aggarwal et al., 2011). Along with other RecQ family helicases, WRN is considered a caretaker of replication and genome integrity (Larsen and Hickson, 2013). Because of its role in response to replication stress, WRN was proposed as a potential target for small-molecule inhibition. NSC 19630 (1-(propoxymethyl)-maleimide) has emerged from a screen for small molecule inhibitors of WRN helicase activity (Aggarwal et al., 2011). NSC 19630 and its improved derivative NSC 617145 (Aggarwal et al., 2013b) are specific inhibitors of WRN DNA helicase (DNA duplex separation) activity, with IC50 values of 20 µM and 230 nM, respectively, but not of related DNA helicases. These small molecules also have minimal effect on other activities of WRN, such as DNA binding and nuclease activity, and may trap WRN on the DNA. WRN inhibition with NSC 19630 or NSC 617145 results in the accumulation of stalled replication forks, impaired growth and cell proliferation and apoptosis, sensitized cancer cells to the G-quadruplex-binding compound telomestatin and PARP inhibition. WRN inhibition also sensitizes cancer cells to the DNA cross-linking agent mitomycin C (MMC). The effect is especially pronounced in cells harboring Fanconi Anemia associated mutations.
The Fanconi Anemia (FA) pathway orchestrates repair of the interstrand DNA crosslinks (ICLs) and protein-DNA crosslinks in higher eukaryotes, thus enabling an unperturbed DNA replication and RNA transcription (Kottemann and Smogorzewska, 2013; Wang et al., 2015). This intricate multistep DNA repair mechanism deals with DNA damage arising due to exogenous insults, such as treatment with cross-linking agents (including cancer chemotherapeutic agents MMC, cisplatin and melphalan), as well as ICLs elicited by endogenous aldehydes arising from lipid peroxidation and ethanol metabolism or nitric oxide (Huang and Li, 2013). The danger of ICLs to DNA replication and RNA transcription, both of which involve transient separation of the DNA duplex, necessitates their repair at all stages of the cell cycle. The interconnecting networks of molecular events associated with HR and FA convolutes both prediction and interpretation of the synthetic interactions between their key players. An attempt to identify pharmacological inhibitors of FA pathway using a cell-based HTS assay yielded a series of compounds that lacked specificity with respect to the target, but nevertheless showed promise in sensitizing cisplatin-resistant tumor cells with functional FA pathway to cisplatin (Jacquemont et al., 2012).
Notably, depletion of WRN by RNAi did not produce the same effect on the MMC sensitivity of FA cells, suggesting that the sensitization is WRN-dependent and that in the physical absence of WRN, other helicases may compensate for the WRN function in DNA damage response in the absence of FA pathway (Aggarwal et al., 2013b). Such compensatory molecular processes may be prevented in the WRN-inhibited cells if WRN helicase remains associated with the DNA and blocking the access similar to the inhibitor-trapped trapped PARP1. It is also possible that blocking the helicase activity of WRN misappropriates its nuclease activity on a particular set of DNA substrates important for the ICL repair. A mechanistic understanding of how the pharmacological inhibition of the WRN helicase activity promotes the MMC-induced cytotoxicity in the FA cells will be instrumental in establishing the compensatory mechanisms that allow FA cells to repair ICLs. BRCA1, BRCA2 (FANCD1) along with other HR proteins, such as PALB2 (FANCN), RAD51 (FANCR), RAD51D (FANCO), etc. are the key players in the FA pathway. To date, the effects of WRN inhibition were only tested in the background of defective FANCA and FANCD2, whose protein products are key activators of the FA ICL repair pathway. It would be interesting to see if WRN inhibition has the same synthetic phenotype with defects in other FA genes, especially those shared between FA and HR pathways. The biochemical and structural understanding of NSC 19630 and NSC 617145 mediated WRN inhibition and its specificity will inform the design of other inhibitors targeting DNA helicases.
BLM (Bloom’s syndrome protein), is another RecQ-family human DNA helicase that has been successfully targeted with small molecules (Rosenthal et al., 2013; Rosenthal et al., 2010). BLM unwinds a broad range of DNA structures, and as an integral component of a multiprotein complex containing RMI1, RMI2, and topoisomerase IIIα promotes genome stability through dissolution of double Holliday junctions in HR (Larsen and Hickson, 2013). An HTS screen for the inhibitors of BLM yielded ML216 (1-(4-fluoro-3-(trifluoromethyl)phenyl)-3-(5-(pyridin-4-yl)-1,3,4-thiadiazol-2-yl)urea) (Table 2), the first small-molecule that specifically inhibits BLM helicase with micromolar efficiency (Rosenthal et al., 2010). ML216 and its improved derivatives act by interfering the BLM-ssDNA interaction (Rosenthal et al., 2013). The specificity of ML216 derivatives for BLM over related DNA helicases (compound 33, for example was 6.5-fold more effective against BLM than WRN, and more than 50-fold than against RECQ1), as well as their ability to induce sister chromatid exchanges (cellular characteristic of BLM-deficient cells), and its selective antiproliferative activity in BLM-positive cells makes ML216 a powerful molecular probe for dissecting multiple interconnected cellular functions of BLM.
Table 2.
Newly-developed small molecule modulators of DNA repair proteins
| Inhibitor |
Source Library and Screening Methodology |
In vitro Potency | Mechanism of Action | Cellular Outcome |
|---|---|---|---|---|
| Inhibitors of WRN DNA helicase | ||||
| NSC 19630 (CAS 72835-26-8) | National Cancer Institute Diversity Set library; HTS assay for the inhibition of the DNA unwinding activity. | Inhibits WRN helicase activity with an IC50 of 20 µM (Aggarwal et al., 2011). | Specifically inhibits WRN helicase activity, but not its nuclease activity | At 3 µM inhibits cell proliferation, induces apoptosis, accumulation of DSBs and formation of stalled replication forks; sensitizes cells to G-quadruplex-binding compound telomestatin, or PARP inhibitor (Aggarwal et al., 2011). |
| NSC 617145 (CAS 203115-63-3) | Structural analog of NSC19630. | Inhibited WRN helicase activity with an IC50 of 230 nM (Aggarwal et al., 2013b). | Specifically inhibits WRN helicase activity, but not its nuclease activity; likely traps WRN on the DNA substrate | Inhibits cell proliferation; sensitizes FA cells to cross-linking agent MMC (Aggarwal et al., 2013). |
| Inhibitor of BLM DNA helicase | ||||
| ML216 (CAS 1430213-30-1) | Molecular Libraries Small Molecule Repository library; fluorescent donor/quencher-based HTS helicase assay (Rosenthal et al., 2010) | Inhibition of the forked DNA unwinding by truncated (IC50 = 1.2 µM) and full length (IC50 = 3.39 µM) BLM helicase (Rosenthal et al., 2013; Rosenthal et al., 2010). | Inhibits helicase activity of BLM (Rosenthal et al., 2013; Rosenthal et al., 2010). | Inhibits proliferation of BLM- expressing fibroblast cells (PSNF5) over BLM-deficient cells (PSNG13), enhances sister chromatid exchange in PSNF5 cells (Rosenthal et al., 2013; Rosenthal et al., 2010). |
| Inhibitors of MRE11 endo- and exonuclease | ||||
| Mirin (CAS 299953-00-7) | ChemBridge DIVERSet™ collection; forward chemical genetics screen for compounds that inhibit the MRN/DSB-mediated ATM activation in cell-free extracts derived from Xenopus laevis eggs (Dupre et al., 2008) | Inhibits MRN-mediated ATM activation (inhibition of H2AX phosphorylation) with IC50 of 66 µM (Dupre et al., 2008). Specifically inhibits exonuclease activity of MRE11, but not its endonuclease activity (Dupre et al., 2008; Shibata et al., 2014). | Binding of mirin in the active site of MRE11 blocks DNA phosphate backbone rotation, which interferes with exonuclease activity (Shibata et al., 2014). Inhibits exonuclease activity of MRE11; inhibits MRN/DSB-mediated ATM activation without affecting ATM protein kinase activity; inhibits the MRN-dependent autophosphorylation of ATM at Ser1982 in response to DSBs (Dupre et al., 2008). | Inhibits MRN-dependent ATM activation similarly to MRN depletion, abolishes the G2/M checkpoint and homology-directed DNA repair in mammalian cells (Dupre et al., 2008). Inhibits dsDNA end resection in A549 cells with estimated IC50 of 200–300 µM (Shibata et al., 2014). |
| PFM01/SML1735 (CAS 1558598-41-6); PFM03 | Focused chemical library of mirin derivatives; the library is described in (Shibata et al., 2014) | Specifically inhibit endonuclease, but not exonuclease activity of MRE11 (Shibata et al., 2014). | Binding of PFM01 and PFM03 near the dimer interface blocks the ssDNA binding path towards the catalytic metal ions and disrupts endonuclease activity. | Inhibit dsDNA end resection in A549 cells with estimated IC50 of 50–75 µM (Shibata et al., 2014). |
| PFM39/SML1839 (CAS 1310744-67-2) | Specifically inhibits exonuclease activity of MRE11 (Shibata et al., 2014). | Binds in the active site similar to mirin. Inhibits exonuclease activity. | Inhibits dsDNA end resection in A549 cells with estimated IC50 of 50–75 µM (Shibata et al., 2014). | |
| Inhibitors of RAD52 | ||||
| 6-hydroxyDL- dopa (CAS 21373-30-8) | Sigma Lopac Library; fluorescence polarization anisotropy based HTS assay for disruptors of the RAD52-ssDNA interaction (Chandramouly et al., 2015). | Inhibits ssDNA binding by RAD52 with IC50 of 1.1 µM and by RAD521-209 with IC50 of 1.6 µM; disrupts RAD52 oligomers (Chandramouly et al., 2015). | Disrupts RAD52 oligomerization (Chandramouly et al., 2015). | Inhibits proliferation, reduces viability and increases apoptosis of BRCA1 deficient TNBC cell line MDA-MB-436. (Chandramouly et al., 2015). |
| D-103 | Broad’s diversity-oriented synthesis (DOS) library and Molecular Libraries Probe Center Network (MPLCN) library; fluorescence quenching based assay for inhibitors of the ssDNA annealing activity of RAD52 (Huang et al., 2016). National Cancer | Inhibits ssDNA annealing by RAD52 with IC50 of 5 µM, inhibits D-loop formation with IC50 of 8 µM; binds to RAD52 with Kd of 25.8 µM (measured by SPR) (Huang et al., 2016). | Inhibits RAD52-mediated ssDNA annealing (Huang et al., 2016). | Suppresses growth of BRCA1- and BRCA2-deficient cells (at ~ 2.5 µM concentration) and inhibits RAD52-mediated SSA in human U2OS cells. Selective towards SSA over (Huang et al., 2016). |
| D-G23 | Inhibits ssDNA annealing by RAD52 with IC50 of 5.6 µM, inhibits D-loop formation with IC50 of 7.2 µM; binds to RAD52 with Kd of 34 µM (Huang et al., 2016). | Suppresses growth of BRCA1- and BRCA2-deficient cells (at >10 µM concentration) (Huang et al., 2016). | ||
| AICAR (CAS 2627-69-2); AICAR 5’ phosphate (ZMP) | Institute drug-like compounds library and FDA approved drugs library; in silico screen (Sullivan et al., 2016) |
1 µM of ZMP reduced the RAD52-ssDNA interaction similar to 25 µM F79 aptamer (Sullivan et al., 2016). | Disrupt the RAD52-ssDNA interaction (Sullivan et al., 2016). | AICAR (20 µM) reduces growth of BRCA1-mutated HCC1937 cells and BRCA2-mutated Capan-1 cells; targets intracellular RAD52 (Sullivan et al., 2016) |
| (−)-Epigallocate chin, EGC (CAS 490-46-0) | MicroSource Spectrum Collection; FRET-based HTS assay for disruptors of the RAD52-ssDNA interaction (Hengel et al., 2016) | Inhibits ssDNA binding IC50 = 1.8 µM and inhibits RAD52-mediated annealing activity IC50 = 4.9 µM (Hengel et al., 2016). | Binds specifically to RAD52 (NMR); disrupts the RAD52-ssDNA interaction and the annealing activity of RAD52 (Hengel et al., 2016). | Kills BRCA2-depleted cells and recapitulates RAD52 depletion with respect to the RAD52-MUS81/EME1-mediated DSB formation at stalled replication forks in the absence of checkpoint (Hengel et al., 2016). |
| NP-004255 (CAS 23094-69-1) | AnalytiCon Natural Products library; in silico screen (Hengel et al., 2016) | Inhibits ssDNA binding IC50 = 1.5 µM; ssDNA binding inhibition | Binds specifically to RAD52 and disrupts the RAD52-ssDNA interaction (Hengel et al., 2016). | Not tested |
| Modulators of RAD51 recombinase | ||||
| DIDS (CAS 67483-13-0) | Program of Scientific Research on Priority Areas, Cancer Japan; RAD51-mediated DNA strand exchange reaction (Ishida et al., 2009). | Inhibits DNA strand exchange activity of RAD51 with IC50 of 5 µM (Ishida et al., 2009). | Binds directly to RAD51 with Kd of 2 µM. Inhibits ssDNA binding, dsDNA binding, and inhibits joint molecule formation in DNA strand exchange assays. Stimulates ATP hydrolysis. (Ishida et al., 2009) | Toxic in human cultured cells. Binds to outer cell membrane and inhibits Cl-channels (Ishida et al., 2009). |
| B02 (CAS 1290541-46-6) | NIH Small Molecule Repository; FRET-based DNA strand exchange assay (Huang et al., 2011). | Inhibits RAD51-mediated DNA strand exchange reaction with IC50 of 27.4 µM (Huang et al., 2011), | Inhibits RAD51-mediated strand invasion (Huang et al., 2011), | Enhances sensitivity of cancer cells to IR, MMC and cisplatin (Huang et al., 2012). Increases apoptosis and reduces viability of multiple myeloma cells (Alagpulinsa et al., 2014). Enhances the effect of cisplatin on triple negative breast cancer cells in a mouse xenograft model (Huang and Mazin, 2014) |
| RI-1 (CAS 415713-60-9) | ChemBridge DIVERSet™ collection; fluorescence polarization anisotropy based HTS assay for the RAD51 nucleoprotein filament formation (Jayathilaka et al., 2008) (Budke et al., 2012) | Inhibits biochemical activities of RAD51 (ssDNA binding and D-loop formation) with IC50 in the 5–30 µM range (Budke et al., 2012) | Contains a chloromaleimide group that acts as a Michael acceptor and reacts with cysteine 319 of RAD51 at the monomer-monomer interface near the ATP active site. | Inhibits HR, disrupts the DNA damage-induced RAD51 foci formation, and sensitizes human cancer cells HeLa, MCF-7 and U2OS to MMC (Budke et al., 2012). Inhibits HR-mediated DSB repair with IC50 of 13.1 µM (Lv et al., 2016a). |
| RI-2 (CAS 1417162-36-7) | RI-1 analog lacking the reactive a chloromaleimide group (Budke et al., 2013) | Inhibits the RAD51-mediated D-loop formation with IC50 of 11 µM (Budke et al., 2013; Lv et al., 2016a). | Binds to the same pocket at the monomer-monomer interface of the RAD51 nucleoprotein filament as RI-1; inhibits biochemical activities of RAD51. | Inhibits HR, sensitizes human cancer cells U2OS, PC-3, and MCF-7 to radiation (Budke et al., 2013). Inhibits HR-mediated DSB repair with IC50 of 3 µM (Lv et al., 2016a). |
| RS-1 (CAS 312756-74-4) | ChemBridge DIVERSet™ collection; fluorescence polarization anisotropy based HTS assay for the RAD51 nucleoprotein filament formation (Jayathilaka et al., 2008) | RS-1 is a stimulator of RAD51 activity with an active range of 48–107 nM (Jayathilaka et al., 2008) | Stabilizes the RAD51nucleoprotein filament and stimulates RAD51 biochemical activities (Jayathilaka et al., 2008). | Inhances resistance to cisplatin at ~7.5 µM (Jayathilaka et al., 2008). |
| Chicago Skye Blue (CSB) (CAS 2610-05-1) | Prestwick Chemical Library® of mostly approved drugs; electrophoresis-based DNA strand exchange assay (Normand et al., 2014) | Inhibits the RAD51-mediated DNA stand exchange reaction with IC50 of ~ 400 nM, Inhibits ssDNA binding by RAD52 with IC50 of 1.2 µM (Normand et al., 2014). | Prevents RAD51 nucleoprotein filament formation by interfering with the RAD51 binding to ssDNA (Normand et al., 2014). | Not tested |
| Inhibitor of the RAD54 DNA branch migration activity | ||||
| Streptonigrin (SN) (CAS 3930-19-6) | Broad Institute validation library; FRET-based assay for branch migration of Holliday junctions. | Inhibits branch migration of model Holliday junctions with IC50 of 16.5 µM, inhibits ATP hydrolysis with IC50 of 14.4 µM (Deakyne et al., 2013). | Binds directly to RAD54 with a Kd of 9.1 µM; inhibits the ATPase and DNA branch migration activity of RAD54 (Deakyne et al., 2013). | Has been previously reported to have a broad range antitumor activity; elicits DNA damage by generating reactive oxygen species (Harris et al., 1965). |
MRE11 separation of function inhibitors
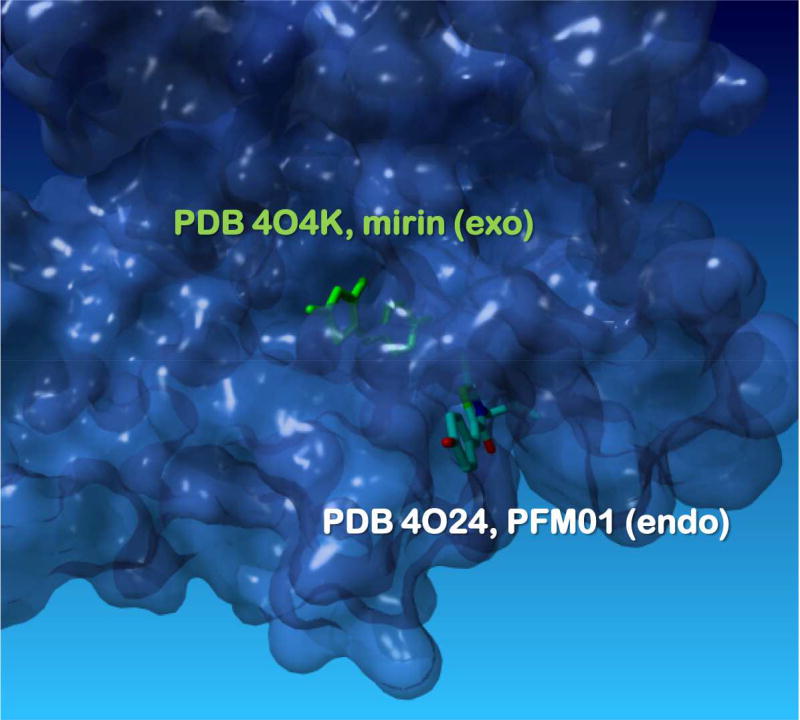
As a constituent of the MRE11-RAD50-NBS1 (MRN) complex, MRE11 nuclease participates in sensing, protecting and repairing the DSBs (reviewed in (Cejka, 2015; Symington, 2014, 2016)). MRE11 has 3’-5’ exonuclease activity and an endonuclease activity directed to ssDNA and hairpin dsDNA. Both activities play roles in the DSB resection, a process that irreversibly commits DSB to homology-directed DNA repair (HDDR). MRN complex also activates ATM kinase in response to DSBs. The first MRE11 inhibitor, mirin (6-(4-hydroxyphenyl)-2-thioxo-2,3-dihydro-4(1H)-pyrimidinone) emerged from a forward chemical genetics screen for compounds that inhibit the MRN/DSB-mediated ATM activation in cell-free extracts derived from Xenopus laevis eggs (Dupre et al., 2008). Mirin inhibits ATM activation by MRN complex, exonuclease activity of MRE11 and HDDR in human cells. It has been widely used as a research reagent. To dissect the exact function of MRE11 at DSBs, it was essential to separate its two nuclease activities. The inhibitors that are selective towards endo- or exonuclease activity were developed by leveraging a focused chemical library of mirin derivatives and a structural understanding of the MRE11 active site in the presence of mirin (Shibata et al., 2014). Two N-alkylated mirin derivatives with a rhodanine ring, PFM01, which has an isobutyl chain, and PFM03, which has a sec-butyl chain on the nitrogen, bound in a distinct position in the active site disrupting the ssDNA binding path towards the catalytic metal ions. Curiously, while these compounds were derivatives of mirin, their binding site observed in the crystal structures was very different from that of mirin (Figure 4). PFM39, a differently substituted mirin derivative was, like mirin a selective exonuclease inhibitor and bound in the same site as mirin. All four inhibitors prevented DSB end resection in human cells, but conferred distinct DNA repair phenotypes: normal DSB repair was observed in the presence of PFM01 or PFM03, while PFM39 and mirin interfered with slow, HR-mediated DSB repair. The DSB repair in the presence of PFM01 or PFM03, inhibitors of MRE11 endonuclease, proceeded through NHEJ (a situation similar to CtIP protein depletion) (Shibata et al., 2014). Studies enabled by the selective MRE11 inhibitors allowed the authors to better understand the mechanisms underlying the DSB repair pathway choice. This understanding culminated in a model whereby MRE11 first utilizes the endonuclease activity to create a nick on the 5’ strand when an attempt to repair DSB by NHEJ fails; the resection then proceeds bi-directionally, MRE11 digests in 3’-5’ direction moving towards the dsDNA end, while EXO1/BLM 5’-3’ exonuclease/helicase activity produces a long ssDNA overhang poised for HR.
Figure 4. MRE11 separation of function inhibitors.
Surface representation of the MRE11 nuclease active site with bound mirin (green, PDB: KO4K), an exonuclease inhibitor and PFM01 (elemental colors, from PDB: 4O24), an endonuclease inhibitor (Shibata et al., 2014). See text for the details of the inhibition mechanisms.
Inhibitors and synthetically lethal interactions involving the RAD52 DNA repair protein
The approval of PARP inhibitors mixed with recent evidence of bourgeoning drug resistance has inspired the search for other targets whose inhibition would be synthetically lethal with defective HR (Kelley and Fishel, 2008; Powell and Kachnic, 2008; Price and Monteiro, 2010; Shaheen et al., 2011; Wu and Brosh, 2010). Human DNA repair protein RAD52 has emerged as one of the most coveted targets. RAD52 depletion is synthetically lethal with bialellic mutations or deletions in HR-associated genome caretakers BRCA1, BRCA2, or PALB2 proteins (Cramer-Morales et al., 2013; Feng et al., 2011; Lok et al., 2012). RAD52 is a 46 kDa protein that forms oligomers, binds ssDNA, dsDNA, anneals complementary substrates and mediates several protein-protein interactions (Hanamshet et al., 2016). Why tumor suppressors BRCA1, BRCA2, or PALB2 and RAD52 are synthetically linked remains unclear largely due to the poorly understood cellular role of human RAD52 (reviewed in (Hanamshet et al., 2016)).
Similar to its yeast counterpart, human RAD52 promotes cell viability by supporting distressed replication in the absence of the functional checkpoint (Doe et al., 2004; Murfuni et al., 2013) and during break induced replication (BIR, an HR-related mechanism that functions outside of the S phase) (Bhowmick et al., 2016; Sotiriou et al., 2016). While this review was in preparation, a new biochemical activity of RAD52 has been reported, namely its ability to mediate an “inverse strand exchange” reaction where a dsDNA-bound RAD52 promotes swapping of one of the DNA strands with homologous ssDNA or ssRNA (Mazina et al., 2017). This activity may play a role in the RNA-templated DSB repair. The role of human RAD52 in HR, however, may only be peripheral – while RAD52 supports the survival of BRCA-deficient cells, it does not compensate for BRCA2 deficiency with respect to HR. Deletion of mammalian RAD52 has only a mild effect on HR (Rijkers et al., 1998; Yamaguchi-Iwai et al., 1998; Yanez and Porter, 2002). Mouse embryonic stem cells depleted of RAD52 show similar sensitivity to methyl methanesulfonate (MMS, an alkylating agent that stalls DNA replication forks ultimately leading to DSBs formation) and ionizing radiation (a direct source of DSBs) as wild-type ES cells (Rijkers et al., 1998). While adding to the mystery of the vertebrate RAD52 function, these observations have elevated RAD52 to a popular target of small-molecule inhibitors. Because synthetic lethality is expected when both copies of the tumor suppressor gene are defective or inactivated, but not in the heterozygous cells, and because RAD52 depletion has minimal effect on the BRCA-proficient cells, RAD52 inhibitors can be used to selectively kill cancerous cells and thereby minimize the toxicity associated with radiation and chemotherapies. This situation is similar to PARP inhibition, except it is unclear what type of synthetic lethality (in-pathway or between-pathways) is taken advantage of and which biochemical activity of RAD52 should be targeted. Earlier, we proposed that the cellular functions of RAD52 likely depend on its unique mode of ssDNA binding, where the ssDNA is wrapped around the heptameric protein ring (Grimme et al., 2010; Honda et al., 2011). Such a binding mode offers at least two targetable features, the integrity of the ring-shaped RAD52 oligomer and a narrow (though somewhat featureless) ssDNA binding groove. Several groups including ours have discovered small-molecule inhibitors of these two activities (Chandramouly et al., 2015; Hengel et al., 2016; Huang et al., 2016; Sullivan et al., 2016). These various studies screened different libraries, yielding hits with distinctly disparate chemical space from one another (Table 2). The first study by Chandramouly and colleagues used a fluorescence-based ssDNA binding assay to identify 6-hydroxyDL-dopa, which interferes with RAD52 oligomerization, inhibits RAD52 foci formation in murine hematopoietic cells deficient in BRCA1, and kills BRCA-deficient cancer cells (Chandramouly et al., 2015). Using a different fluorescence-based screen, Huang et al., identified two compounds, D-103 and D-G23 that inhibit the ssDNA annealing activity of RAD52, suppresses growth of BRCA-deficient cells, and at high concentrations inhibits SSA in human cells (Huang et al., 2016). A molecular docking of two compound libraries (total of ~141,000 compounds) into the site within the RAD52 DNA-binding domain that contains the ssDNA binding residues was used to identify several additional disrupters of the RAD52-ssDNA interaction (Sullivan et al., 2016). Among these, 5-Aminoimidazole-4-carboxamide ribonucleotide (AICAR) 5’ phosphate (ZMP) specifically inhibited the RAD52-ssDNA interactions. Cell permeating AICAR, which undergoes phosphorylation in the cytoplasm preferentially killed BRCA1- and BRCA2-mutated cells (Sullivan et al., 2016). Using a biophysical assay that reports on the ssDNA binding by RAD52, we identified several small-molecules including 1 (a natural green tea product (-)-Epigallocatechin, EGC), which was specific to RAD52, inhibited ssDNA binding and annealing by RAD52 in vitro, killed BRCA2-depleted cells and recapitulated RAD52 depletion with respect to RAD52-MUS81/EME1 activity that promotes replication recovery in the absence of checkpoint (Hengel et al., 2016). A robust SAR was developed based on the computationally determined RAD52-ligand complexes. We applied our virtual screening compound placement protocol, which was designed to enrich the true positive hits (the docked validated hits) and decrease the false negatives and false positives, to a larger in silico library. The natural product (a macrocycle) compound NP-00425 that emerged from the in silico screen yielded a promising hit that binds to RAD52 and competes with ssDNA binding. The validated SAR and the computational workflow will provide a strong foundation for the discovery of novel antineoplastic therapeutics for such an extraordinary difficult target as an extended ssDNA binding site of RAD52 protein ring (Hengel et al., 2016). Inadvertently, this study identified an unexpected novel synthetic interaction: depletion of MUS81 exacerbated cell death in the BRCA2-depleted cells after the replication stress.
Modulators of the RAD51 recombinase
The central player in HR is the RAD51 DNA strand exchange protein (aka recombinase); its active form is a nucleoprotein filament assembled on the ssDNA generated at the sites of DNA damage by the DSB resection machinery (reviewed in (Symington, 2014)). Formation of the RAD51 filament is controlled positively by BRCA2, which is the main recombination mediator (Prakash et al., 2015), and negatively by antirecombinogenic DNA helicases (Daley et al., 2014) and by the heteroduplex rejection machinery (Spies and Fishel, 2015). The RAD51 nucleoprotein filament promotes HR by exchanging strands between the damaged and the intact DNA molecules. It also has additional HR-independent function at damaged replication forks: it protects the fork from excessive degradation by the MRE11 nuclease activity (Kolinjivadi et al., 2017; Schlacher et al., 2011; Schlacher et al., 2012). The two types of the RAD51 nucleoprotein filament may be identical or very different with respect to their dynamics and function, and it has been suggested that it may be possible to selectively target the HR functions of the RAD51 nucleoprotein filament and not its activities supporting DNA replication. RAD51 has been an attractive target to developing small-molecule inhibitors for several reasons. First of all, HR and its central player, RAD51 recombinase, play an important role in repair of replication stalling lesions that are caused by the DNA damaging cancer treatments. Second, inhibiting the central, but not the initial step in HR, after the DSB is committed to be repaired through a homology directed mechanism prevents it from repair by NHEJ. Finally, RAD51 overexpression has been observed in a range of different cancers, and the associated resistance to radiation and chemotherapy was overcome by reduction in RAD51 expression (reviewed in (Klein, 2008) and (Budke et al., 2016)).
The three major groups of compounds that modulate RAD51 activity have been identified to date: compounds that interfere with the RAD51 nucleoprotein filament formation, a compound that stimulated RAD51 nucleoprotein filament and compounds that interfere with the D-loop formation. In 2009, DIDS (4,4’-diisothiocyanostilbene-2,2’-disulfonic acid, a known inhibitor of ionic channels) was found to inhibit the DNA binding activity of RAD51 and the DNA strand exchange activity by competing the DNA (Ishida et al., 2009). Although DIDS was efficiently imported into the nuclei of cultured cells, it appeared toxic (Ishida et al., 2009). In 2011 Huang and colleagues identified B02 compound ((E)-3-benzyl-2-(2-(pyridin-3-yl) vinyl) quinazolin-4(3H)-one) that specifically inhibited RAD51 binding to ssDNA and the RAD51-mediated DNA strand exchange activity (Huang et al., 2011). B02 conferred sensitivity to ionizing radiation, MMC and cisplatin (Huang et al., 2012), and significantly enhanced the therapeutic effect of cisplatin on triple negative breast cancers in mouse xenografts (Huang and Mazin, 2014), while a combination of B02 and doxyrubicin reduced HR, increased apoptosis, and reduced viability in multiple myeloma cells (Alagpulinsa et al., 2014). B02 has proven useful in the analysis of the DNA repair networks in triple negative breast cancers, an extremely aggressive breast cancer sub-type that lacks expression of the human EGF receptor-2, progesterone receptor, and estrogen receptor. Weigmans and colleagues found that kinase signaling is rewired in the triple negative breast cancers, and that inhibition of RAD51 with B02 can be combined with a PARP inhibitor and with inhibiting p38 kinase into an effective treatment (Wiegmans et al., 2016). A more recent screen of an existing drug library identified Chicago Sky Blue (CSB, 6,6'-[(3,3-dimethoxy[1,1'-biphenyl]-4,4'-diyl)bis(azo)]bis[4-amino-5-hydroxy-1,3-naphtalenedisulphonic acid]), which inhibited RAD51 ssDNA binding and strand exchange activity with sub-µM IC50 values (Normand et al., 2014).
An interesting set of RAD51 modulators has emerged from screening the ChemBridge DIVERSet™ (Budke et al., 2012). RI-1 contains a chloromaleimide group that functions as a Michael acceptor that reacts with the thiol group on cysteine 319 residue at the RAD51 monomer-monomer interface, forming a covalent bond providing stable irreversible inhibition of RAD51, and sensitization of multiple cancer cell lines to MMC (Budke et al., 2012). RI-2, a RI-1 analog that retained the inhibitory activity of RI-1 without its undesirable reactivity has subsequently emerged from an extensive SAR study (Budke et al., 2013). The same screen that yielded RI-1 also yielded RS-1, the only RAD51 activator identified thus far (Jayathilaka et al., 2008). A screen for compounds that inhibit the RAD51-mediated DNA strand exchange reaction, but not the nucleoprotein filament formation per se resulted in 9, (Lv et al., 2016b).
An alternative to inhibiting the RAD51-DNA interaction is to disrupt the RAD51-BRCA2 complex, and in particular the interaction between RAD51 and the BRC1–4 motifs of BRCA2. Two regions of BRCA2 are responsible for the interaction with RAD51. The first region consists of 8 BRC repeats, where BRC repeats 1 through 4 bind free RAD51 to facilitate the RAD51 loading onto the RPA (replication protein A) coated ssDNA, while BRC repeats 5 through 8 bind the RAD51 nucleoprotein filament (Carreira and Kowalczykowski, 2011). Collectively the interaction between BRCA2 BRC motifs and RAD51 promotes the DNA strand exchange reaction (Jensen et al., 2010; Liu et al., 2010; Shivji et al., 2006; Thorslund et al., 2010). An additional RAD51 binding site at the C-terminus is involved in both HR and replication fork stabilization (Kim et al., 2014; Saeki et al., 2006).
A small-molecule inhibitor that competes with the RAD51-BRC repeat interaction should effectively prevent the central step in HR thereby sensitizing cells to radiation and DNA damaging chemotherapies. Recently, a fragment based approach was successfully employed to identify a several promising fragments that bind to the hydrophobic pocket on the RAD51 surface that accommodates the aromatic residues of BRC4-derived peptide (Scott et al., 2015). The identified small molecule fragments can potentially be grown and/or linked to yield a more potent, specific inhibitor with drug-like properties (Scott et al., 2015). These endeavors will depend on the structural information on the RAD51-BRC peptide interaction (Pellegrini et al., 2002; Subramanyam et al., 2013) and the development of an experimental system compatible with the fragment analysis (Moschetti et al., 2016).
Challenges in developing inhibitors/modulators of DNA repair
The success of pharmacologically induced synthetic lethality between PARP inhibition and HR defects suggests that the concept of synthetic lethality in DNA repair can be further exploited to identify new targets for anticancer therapy and new adjuvants to treatments with ionizing radiation and DNA damaging chemotherapeutics.
One of the major obstacles to discovering and developing small molecule effectors that act on DNA repair systems is related to the nature of attempting to disrupt macromolecular interactions (Scott et al., 2015). Indeed, the development of specific and potent inhibitors targeting protein-protein interactions (PPI) and protein-DNA interactions (PNI) is one of the most challenging areas of modern medicinal chemistry. Although much more research has been focused on disruption of PPIs than PNIs, the fundamental challenges for development of low molecular weight modulators of these processes is highly similar. Essentially, one often is focused on targeting conserved macromolecular interactions which are shallow, yet extended grooves, which are partially solvated, and distinct from classical enzyme and receptor pockets. In recent years there has been significant traction using peptidomimetics to disrupt PPIs (Arkin et al., 2014). Although PPI lead compounds typically have lower than desirable ligand efficiency (LE; binding energy/heavy atom) and LLE (pIC50-pLogP) values, a number of clinical leads have been designed with acceptable drug-like parameters. Nevertheless, even if one finds promising leads, the question of specificity is ever looming, due to the similarities and promiscuity of many protein binding domains. In most cases, it is desired to identify a lead compound that competes with the native macromolecule-macromolecule interaction, yet retains high LE (>0.3 kcal/mol) and high LLE (>5). Being “fat and flat” is tied to poor pharmacological outcomes. Thus, the generally poor LE and LLE metrics from leads that result from PPI and PNI screening campaigns are simply the function of the nature of these protein-protein and protein-nucleic acid interfaces. Nevertheless, several campaigns described above yielded compounds with generally favorable ADME properties. For example, the BLM helicase inhibitor ML216 and its improved analog 33 display good microsomal stability, Log P, and plasma stability. These compounds, however, show poor aqueous solubility and cell permeability and therefore will require further optimization (Rosenthal et al., 2013) In addition to peptidomemetics, macrocyles have shown promise in providing a distinct solution to the problem effectively competing with macromolecular-macromolecular interactions. Johannes and colleagues (Johannes et al., 2017) recently employed a DNA-encoded library of non-natural peptides in a screening campaign against Mcl-1, a pro-apoptotic protein. Importantly, structures of the initial hits allowed the researchers to link the terminal ends of the ligand, to form a macrocylic analog, which led to a binding enhancement of more than an order of magnitude. More importantly, the final optimized macrocyle provided an impressive ~ 3 orders of magnitude selectivity in binding relative to the structurally similar Bcl-2. Additionally, we used structure based screening methods and molecular dynamics simulations to predict and validate that a macrocyle natural product (NP-004255) would effectively compete with ssDNA in binding to RAD52 protein (Hengel et al., 2016).
Conclusions
Inhibitors and modulators of DNA repair discussed in this review represent a broad range of targetable enzymatic activities and macromolecular interactions, all of which revealed something new, not only with regard to DNA repair pathways and synthetic interactions, but also with regard to expanding the repertoire of druggable targets. While the most successful compounds targeting DNA repair defects are the PARP inhibitors, which inhibit the ADP-ribose polymerization activity of PARP1 in its conventional enzyme active site, other types of targets are gaining traction. Of particular note are the advances made in specifically inhibiting the protein-ssDNA interaction by RAD51 and RAD52 DNA repair proteins, which represent formidable challenges; both proteins bind ssDNA in a sequence independent manner in an environment in which scores of other ssDNA binding proteins are present in the cell. Targeting PPIs in DNA repair is also gaining traction, exemplified in this review by the RAD52 oligomerization inhibitor and the fragment-based approach to targeting the RAD51-BRCA2 interaction. Both RAD51 and RAD52 are coveted anticancer drug targets, and the development of the small molecule modulators of their specific activities will be instrumental in parsing out the involvement of these activities and interaction in various DNA repair mechanisms. Finally, the ultimate scientific value of pharmacological inhibitors of DNA repair enzymes and proteins comes from their ability to parse the myriad of specific activities in their targets. In this light, selective inhibitors of MRE11 exo- and endonuclease, and small molecules that specifically prevent RAD51 strand invasion activity (D-loop formation inhibitors) offer a promise in similar discoveries for other multifunctional DNA repair enzymes.
Table 1.
Small molecule inhibitors of DNA repair proteins used in clinical applications
| Inhibitor | In vitroPotency | Mechanism of Action | Cellular Outcome | Clinical Applications |
|---|---|---|---|---|
| Inhibitors of Poly(ADP-ribose)-polymerase, PARP1 | ||||
| Benzamide (CAS 55-21-0) | IC50 = 3.3 µM (Rankin et al., 1989) | A substrate analog, competes with NAD+ | 3.5 µM selective inhibition of poly(ADP-ribose) metabolism (Rankin et al., 1989) | |
| Olaparib (Lynparza) (CAS 763113-22-0) | IC50(PARP1) = 5 nM IC50(PARP2) = 1 nM (Menear et al., 2008) IC50(PARP1) = 1.4 nM IC50(PARP2) = 12.3 nM (Thorsell et al., 2017) | Potent inhibitor of PARP1 and PARP2 (15–20x specificity over other PARP family enzymes); binds within nicotinamide binding pocket in the ADP-ribosyl transferase catalytic site | Synthetically lethal with HR defects; sensitizes cells to radiation and DNA damaging agents by inducing replication-mediated DSBs | 2014 – FDA approved for treatment of germline BRCA-mutated advanced ovarian cancer |
| Rucaparib (Clovis -AG014699, PF-01367338 Pfizer) (CAS 549868-92-9) | Ki(PARP1) = 1.4 nM (Thomas et al., 2007) IC50(PARP1) = 3.2 nM IC50(PARP2) = 28.2 nM (Thorsell et al., 2017) | Potent inhibitor of PARP1 and PARP2 (15–20x specificity over other PARP family enzymes); binds within nicotinamide binding pocket in the ADP-ribosyl transferase catalytic site | Synthetically lethal with HR defects; sensitizes cells to radiation and DNA damaging agents by inducing replication-mediated DSBs | 2016 – FDA approved for treatment of germline BRCA-mutated advanced ovarian cancer |
| Niraparib (MK- 827 Tesaro) CAS 1038915-60- 4) | IC50 PARP-1 = 3.8 nM; IC50 PARP-2= 2.1 nM (Jones et al., 2009) IC50(PARP1) = 16.7 nM IC50(PARP2) = 15.3 nM (Thorsell et al., 2017) | Selective inhibitor of PARP1 and PARP2 (>100x specificity over other PARP family enzymes); binds within nicotinamide binding pocket in the ADP-ribosyl transferase catalytic site and makes contacts with the regulatory subdomains (E763 & D766 in Figure 2); Efficiently traps PARP1 on the damage-containing DNA | Inhibits parylation with EC50 of 4 nM (Jones et al., 2009) Synthetically lethal with HR defects; sensitizes cells to radiation and DNA damaging agents by inducing replication-mediated DSBs Also inhibits hexo-6-phosphate dehydrogenase (Knezevic et al., 2016) | 2017 – FDA approved for treatment of platinum-sensitive recurrent ovarian, fallopian tube, and primary peritoneal cancers |
| Veliparib (ABT-888 Abbvie) (CAS 912444-00-9) | Ki PARP-1= 5.2 nM; Ki PARP-2 = 2.9 nM (Donawho et al., 2007) IC50(PARP1) = 3.3 nM IC50(PARP2) = 17.5 nM (Thorsell et al., 2017) | Selective inhibitor of PARP1 and PARP2 (>100x specificity over other PARP family enzymes); binds within nicotinamide binding pocket in the ADP-ribosyl transferase catalytic site and makes contacts with the regulatory subdomains; Efficiently traps PARP1 on the damage-containing DNA | Inhibits SSB and DSB repair, potentiates activity of temozolomide, cisplatin, carboplatin for variety of tumors including glioma and breast carcinoma (Wagner, 2015). | |
| TOP1 (Topoisomerase 1) inhibitors | ||||
| Camptothecin (CPT) (CAS 7689-03-4) | Bind at the enzyme-DNA interface; trap a catalytic intermediate of supercoiled DNA and TOP1 (Fox et al., 2003) | TOP1-DNA complexes are converted into replication mediated DSBs; low doses stall replication (Pommier et al., 2006) | ||
| Irinotecan (CAS 100286-90-6) | 1996 – FDA approved for treatment of metastatic carcinoma of the colon or rectum whose disease has recurred or progressed following initial flurouracil treatment 2015 – FDA approved for treatment in combination with fluorouracil and leucovorin, of advanced (metastatic) pancreatic cancer | |||
| Topotecan (Hycamtin® GlaxoSmithKline) (CAS 123948-87-8) | 2006 – FDA approved for treatment in combination with cisplatin for treatment of stage IVB recurrent or persistent cervical cancer | |||
Approval of drugs targeting poly-ADP-ribose polymerase is inspiring campaigns to identify new targets among DNA repair proteins and new inhibitors for personalized cancer treatments. This review discusses the state-of-the-art in DNA repair inhibitors and how these small molecules are improving our understanding of the complex and interconnecting DNA repair processes.
Acknowledgments
We gratefully acknowledge the support by the NIH R01 GM108617 and by the University of Iowa Holden Comprehensive Cancer Center Collaborative Pilot Grant to M.S., and by the NIH R01-GM097373 to M.A.S. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Drs. Todd Washington and Miles Pufall (University of Iowa), Dr. Robert Brosh (NIH) and Dr. Alexander Mazin (Drexel University) for critical reading of the manuscript and for valuable discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aggarwal M, Banerjee T, Sommers JA, Brosh RM., Jr Targeting an Achilles’ heel of cancer with a WRN helicase inhibitor. Cell cycle (Georgetown, Tex) 2013a;12:3329–3335. doi: 10.4161/cc.26320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal M, Banerjee T, Sommers JA, Iannascoli C, Pichierri P, Shoemaker RH, Brosh RM., Jr Werner syndrome helicase has a critical role in DNA damage responses in the absence of a functional fanconi anemia pathway. Cancer research. 2013b;73:5497–5507. doi: 10.1158/0008-5472.CAN-12-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal M, Sommers JA, Shoemaker RH, Brosh RM., Jr Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1525–1530. doi: 10.1073/pnas.1006423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagpulinsa DA, Ayyadevara S, Shmookler Reis RJ. A Small-Molecule Inhibitor of RAD51 Reduces Homologous Recombination and Sensitizes Multiple Myeloma Cells to Doxorubicin. Frontiers in oncology. 2014;4:289. doi: 10.3389/fonc.2014.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic acids research. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. BioEssays : news and reviews in molecular, cellular and developmental biology. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Anantha RW, Simhadri S, Foo TK, Miao S, Liu J, Shen Z, Ganesan S, Xia B. Functional and mutational landscapes of BRCA1 for homology-directed repair and therapy resistance. eLife. 2017;6 doi: 10.7554/eLife.21350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkin MR, Tang Y, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chemistry & biology. 2014;21:1102–1114. doi: 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashworth A. A synthetic lethal therapeutic approach: Poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J Clin Oncol. 2008;26:3785–3790. doi: 10.1200/JCO.2008.16.0812. [DOI] [PubMed] [Google Scholar]
- Berti M, Ray Chaudhuri A, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, Odreman F, Glatter T, Graziano S, Mendoza-Maldonado R, et al. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nature structural & molecular biology. 2013;20:347–354. doi: 10.1038/nsmb.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia V, Barroso SI, Garcia-Rubio ML, Tumini E, Herrera-Moyano E, Aguilera A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362. doi: 10.1038/nature13374. -+ [DOI] [PubMed] [Google Scholar]
- Bhowmick R, Minocherhomji S, Hickson ID. RAD52 Facilitates Mitotic DNA Synthesis Following Replication Stress. Mol Cell. 2016;64:1117–1126. doi: 10.1016/j.molcel.2016.10.037. [DOI] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nature structural & molecular biology. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, van der Gulden H, van der Heijden I, Drost R, Klijn CN, Prasetyanti P, Pieterse M, Wientjens E, Seibler J, Hogervorst FB, et al. A high-throughput functional complementation assay for classification of BRCA1 missense variants. Cancer discovery. 2013;3:1142–1155. doi: 10.1158/2159-8290.CD-13-0094. [DOI] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Budke B, Kalin JH, Pawlowski M, Zelivianskaia AS, Wu M, Kozikowski AP, Connell PP. An optimized RAD51 inhibitor that disrupts homologous recombination without requiring Michael acceptor reactivity. Journal of medicinal chemistry. 2013;56:254–263. doi: 10.1021/jm301565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budke B, Logan HL, Kalin JH, Zelivianskaia AS, Cameron McGuire W, Miller LL, Stark JM, Kozikowski AP, Bishop DK, Connell PP. RI-1: a chemical inhibitor of RAD51 that disrupts homologous recombination in human cells. Nucleic acids research. 2012;40:7347–7357. doi: 10.1093/nar/gks353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budke B, Lv W, Kozikowski AP, Connell PP. Recent Developments Using Small Molecules to Target RAD51: How to Best Modulate RAD51 for Anticancer Therapy? ChemMedChem. 2016;11:2468–2473. doi: 10.1002/cmdc.201600426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev DV, Rossi MJ, Mazin AV. Cooperation of RAD51 and RAD54 in regression of a model replication fork. Nucleic acids research. 2011;39:2153–2164. doi: 10.1093/nar/gkq1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW. Single-strand break repair and genetic disease. Nature reviews Genetics. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- Carreira A, Kowalczykowski SC. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc Natl Acad Sci U S A. 2011;108:10448–10453. doi: 10.1073/pnas.1106971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh H, Rogers KM. The role of BRCA1 and BRCA2 mutations in prostate, pancreatic and stomach cancers. Hereditary cancer in clinical practice. 2015;13:16. doi: 10.1186/s13053-015-0038-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos SJ, Heyer WD. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochimica et biophysica acta. 2011;1809:509–523. doi: 10.1016/j.bbagrm.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P. DNA End Resection: Nucleases Team Up with the Right Partners to Initiate Homologous Recombination. The Journal of biological chemistry. 2015;290:22931–22938. doi: 10.1074/jbc.R115.675942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouly G, McDevitt S, Sullivan K, Kent T, Luz A, Glickman JF, Andrake M, Skorski T, Pomerantz RT. Small-Molecule Disruption of RAD52 Rings as a Mechanism for Precision Medicine in BRCA-Deficient Cancers. Chemistry & biology. 2015;22:1491–1504. doi: 10.1016/j.chembiol.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YE, Meghani K, Brault ME, Leclerc L, He YZJ, Day TA, Elias KM, Drapkin R, Weinstock DM, Dao F, et al. Platinum and PARP Inhibitor Resistance Due to Overexpression of MicroRNA-622 in BRCA1-Mutant Ovarian Cancer. Cell reports. 2016;14:429–439. doi: 10.1016/j.celrep.2015.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HC, Kapuriya N, Kulp SK, Chen CS, Shapiro CL. Differential anti-proliferative activities of poly(ADP-ribose) polymerase (PARP) inhibitors in triple-negative breast cancer cells. Breast cancer research and treatment. 2012;134:649–659. doi: 10.1007/s10549-012-2106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couedel C, Mills KD, Barchi M, Shen L, Olshen A, Johnson RD, Nussenzweig A, Essers J, Kanaar R, Li GC, et al. Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes & development. 2004;18:1293–1304. doi: 10.1101/gad.1209204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer-Morales K, Nieborowska-Skorska M, Scheibner K, Padget M, Irvine DA, Sliwinski T, Haas K, Lee J, Geng H, Roy D, et al. Personalized synthetic lethality induced by targeting RAD52 in leukemias identified by gene mutation and expression profile. Blood. 2013;122:1293–1304. doi: 10.1182/blood-2013-05-501072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Gaines WA, Kwon Y, Sung P. Regulation of DNA Pairing in Homologous Recombination. Cold Spring Harbor perspectives in biology. 2014 doi: 10.1101/cshperspect.a017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lorenzo SB, Patel AG, Hurley RM, Kaufmann SH. The Elephant and the Blind Men: Making Sense of PARP Inhibitors in Homologous Recombination Deficient Tumor Cells. Frontiers in oncology. 2013;3:228. doi: 10.3389/fonc.2013.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakyne JS, Huang F, Negri J, Tolliday N, Cocklin S, Mazin AV. Analysis of the activities of RAD54, a SWI2/SNF2 protein, using a specific small-molecule inhibitor. The Journal of biological chemistry. 2013;288:31567–31580. doi: 10.1074/jbc.M113.502195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker P, Muller S. Modulating poly (ADP-ribose) polymerase activity: potential for the prevention and therapy of pathogenic situations involving DNA damage and oxidative stress. Current pharmaceutical biotechnology. 2002;3:275–283. doi: 10.2174/1389201023378265. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics of natural populations; recombination and variability in populations of Drosophila pseudoobscura. Genetics. 1946;31:269–290. doi: 10.1093/genetics/31.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CL, Osman F, Dixon J, Whitby MC. DNA repair by a Rad22-Mus81-dependent pathway that is independent of Rhp51. Nucleic acids research. 2004;32:5570–5581. doi: 10.1093/nar/gkh853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donawho CK, Luo Y, Luo Y, Penning TD, Bauch JL, Bouska JJ, Bontcheva-Diaz VD, Cox BF, DeWeese TL, Dillehay LE, et al. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee J-H, Nicolette ML, Kopelovich L, Jasin M, Baer R, Paull TT, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol. 2008;4:119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Feng Z, Scott SP, Bussen W, Sharma GG, Guo G, Pandita TK, Powell SN. Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:686–691. doi: 10.1073/pnas.1010959107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox BM, Xiao X, Antony S, Kohlhagen G, Pommier Y, Staker BL, Stewart L, Cushman M. Design, synthesis, and biological evaluation of cytotoxic 11-alkenylindenoisoquinoline topoisomerase I inhibitors and indenoisoquinoline-camptothecin hybrids. Journal of medicinal chemistry. 2003;46:3275–3282. doi: 10.1021/jm0300476. [DOI] [PubMed] [Google Scholar]
- Friedman LS, Szabo CI, Ostermeyer EA, Dowd P, Butler L, Park T, Lee MK, Goode EL, Rowell SE, King MC. Novel inherited mutations and variable expressivity of BRCA1 alleles, including the founder mutation 185delAG in Ashkenazi Jewish families. American journal of human genetics. 1995;57:1284–1297. [PMC free article] [PubMed] [Google Scholar]
- Grimme JM, Honda M, Wright R, Okuno Y, Rothenberg E, Mazin AV, Ha T, Spies M. Human Rad52 binds and wraps single-stranded DNA and mediates annealing via two hRad52-ssDNA complexes. Nucleic acids research. 2010 doi: 10.1093/nar/gkp1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanamshet K, Mazina OM, Mazin AV. Reappearance from Obscurity: Mammalian Rad52 in Homologous Recombination. Genes. 2016;7 doi: 10.3390/genes7090063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MN, Medrek TJ, Golomb FM, Gumport SL, Postel AH, Wright JC. CHEMOTHERAPY WITH STREPTONIGRIN IN ADVANCED CANCER. Cancer. 1965;18:49–57. doi: 10.1002/1097-0142(196501)18:1<49::aid-cncr2820180109>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science (New York, NY) 1997;278:1064–1068. doi: 10.1126/science.278.5340.1064. [DOI] [PubMed] [Google Scholar]
- Hatchi E, Skourti-Stathaki K, Ventz S, Pinello L, Yen A, Kamieniarz-Gdula K, Dimitrov S, Pathania S, McKinney KM, Eaton ML, et al. BRCA1 Recruitment to Transcriptional Pause Sites Is Required for R-Loop-Driven DNA Damage Repair. Mol Cell. 2015;57:636–647. doi: 10.1016/j.molcel.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengel SR, Malacaria E, Folly da Silva Constantino L, Bain FE, Diaz A, Koch BG, Yu L, Wu M, Pichierri P, Spies MA, et al. Small-molecule inhibitors identify the RAD52-ssDNA interaction as critical for recovery from replication stress and for survival of BRCA2 deficient cells. eLife. 2016;5:e14740. doi: 10.7554/eLife.14740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD. Regulation of recombination and genomic maintenance. Cold Spring Harbor perspectives in biology. 2015;7:a016501. doi: 10.1101/cshperspect.a016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SJ, Rolland T, Adelmant G, Xia XF, Owen MS, Dricot A, Zack TI, Sahni N, Jacob Y, Hao T, et al. Systematic screening reveals a role for BRCA1 in the response to transcription-associated DNA damage. Genes & development. 2014;28:1957–1975. doi: 10.1101/gad.241620.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M, Okuno Y, Yoo J, Ha T, Spies M. Tyrosine phosphorylation enhances RAD52-mediated annealing by modulating its DNA binding. The EMBO journal. 2011;30:3368–3382. doi: 10.1038/emboj.2011.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Goyal N, Sullivan K, Hanamshet K, Patel M, Mazina OM, Wang CX, An WF, Spoonamore J, Metkar S, et al. Targeting BRCA1- and BRCA2-deficient cells with RAD52 small molecule inhibitors. Nucleic acids research. 2016;44:4189–4199. doi: 10.1093/nar/gkw087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Mazin AV. A Small Molecule Inhibitor of Human RAD51 Potentiates Breast Cancer Cell Killing by Therapeutic Agents in Mouse Xenografts. PloS one. 2014;9 doi: 10.1371/journal.pone.0100993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Mazina OM, Zentner IJ, Cocklin S, Mazin AV. Inhibition of homologous recombination in human cells by targeting RAD51 recombinase. Journal of medicinal chemistry. 2012;55:3011–3020. doi: 10.1021/jm201173g. [DOI] [PubMed] [Google Scholar]
- Huang F, Motlekar NA, Burgwin CM, Napper AD, Diamond SL, Mazin AV. Identification of Specific Inhibitors of Human RAD51 Recombinase Using High-Throughput Screening. Acs Chem Biol. 2011;6:628–635. doi: 10.1021/cb100428c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li L. DNA crosslinking damage and cancer - a tale of friend and foe. Translational cancer research. 2013;2:144–154. doi: 10.3978/j.issn.2218-676X.2013.03.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incorvaia L, Passiglia F, Rizzo S, Galvano A, Listi A, Barraco N, Maragliano R, Calo V, Natoli C, Ciaccio M, et al. “Back to a false normality”: new intriguing mechanisms of resistance to PARP inhibitors. Oncotarget. 2017;8:23891–23904. doi: 10.18632/oncotarget.14409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Takizawa Y, Kainuma T, Inoue J, Mikawa T, Shibata T, Suzuki H, Tashiro S, Kurumizaka H. DIDS, a chemical compound that inhibits RAD51-mediated homologous pairing and strand exchange. Nucleic acids research. 2009;37:3367–3376. doi: 10.1093/nar/gkp200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont C, Simon JA, D’Andrea AD, Taniguchi T. Non-specific chemical inhibition of the Fanconi anemia pathway sensitizes cancer cells to cisplatin. Molecular cancer. 2012;11:26. doi: 10.1186/1476-4598-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harbor perspectives in biology. 2013;5:a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayathilaka K, Sheridan SD, Bold TD, Bochenska K, Logan HL, Weichselbaum RR, Bishop DK, Connell PP. A chemical compound that stimulates the human homologous recombination protein RAD51. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15848–15853. doi: 10.1073/pnas.0808046105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo PA, Lobrich M. How cancer cells hijack DNA double-strand break repair pathways to gain genomic instability. Biochemical Journal. 2015;471:1–11. doi: 10.1042/BJ20150582. [DOI] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Greenberg RA. Deciphering the BRCA1 Tumor Suppressor Network. The Journal of biological chemistry. 2015;290:17724–17732. doi: 10.1074/jbc.R115.667931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes JW, Bates S, Beigie C, Belmonte MA, Breen J, Cao S, Centrella PA, Clark MA, Cuozzo JW, Dumelin CE, et al. Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors. ACS medicinal chemistry letters. 2017;8:239–244. doi: 10.1021/acsmedchemlett.6b00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Altamura S, Boueres J, Ferrigno F, Fonsi M, Giomini C, Lamartina S, Monteagudo E, Ontoria JM, Orsale MV, et al. Discovery of 2-{4-[(3S)-piperidin-3-yl]phenyl}-2H-indazole-7-carboxamide (MK-4827): a novel oral poly(ADP-ribose)polymerase (PARP) inhibitor efficacious in BRCA-1 and −2 mutant tumors. Journal of medicinal chemistry. 2009;52:7170–7185. doi: 10.1021/jm901188v. [DOI] [PubMed] [Google Scholar]
- Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. Febs Lett. 2010;584:3703–3708. doi: 10.1016/j.febslet.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley MR, Fishel ML. DNA repair proteins as molecular targets for cancer therapeutics. Anticancer Agents Med Chem. 2008;8:417–425. doi: 10.2174/187152008784220294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TM, Son MY, Dodds S, Hu L, Hasty P. Deletion of BRCA2 exon 27 causes defects in response to both stalled and collapsed replication forks. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2014;766–767:66–72. doi: 10.1016/j.mrfmmm.2014.06.003. [DOI] [PubMed] [Google Scholar]
- Klein HL. The consequences of Rad51 overexpression for normal and tumor cells. DNA repair. 2008;7:686–693. doi: 10.1016/j.dnarep.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevic CE, Wright G, Remsing Rix LL, Kim W, Kuenzi BM, Luo Y, Watters JM, Koomen JM, Haura EB, Monteiro AN, et al. Proteome-wide Profiling of Clinical PARP Inhibitors Reveals Compound-Specific Secondary Targets. Cell chemical biology. 2016;23:1490–1503. doi: 10.1016/j.chembiol.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Ohno S, Sasaki Y, Matsuura M. Hereditary breast and ovarian cancer susceptibility genes (review) Oncology reports. 2013;30:1019–1029. doi: 10.3892/or.2013.2541. [DOI] [PubMed] [Google Scholar]
- Kolinjivadi AM, Sannino V, de Antoni A, Techer H, Baldi G, Costanzo V. Moonlighting at replication forks - a new life for homologous recombination proteins BRCA1, BRCA2 and RAD51. FEBS letters. 2017;591:1083–1100. doi: 10.1002/1873-3468.12556. [DOI] [PubMed] [Google Scholar]
- Konishi H, Mohseni M, Tamaki A, Garay JP, Croessmann S, Karnan S, Ota A, Wong HY, Konishi Y, Karakas B, et al. Mutation of a single allele of the cancer susceptibility gene BRCA1 leads to genomic instability in human breast epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17773–17778. doi: 10.1073/pnas.1110969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–363. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski SC. An Overview of the Molecular Mechanisms of Recombinational DNA Repair. Cold Spring Harbor perspectives in biology. 2015;7 doi: 10.1101/cshperspect.a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langelier M-F, Pascal JM. PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Current opinion in structural biology. 2013;23:134–143. doi: 10.1016/j.sbi.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen NB, Hickson ID. RecQ Helicases: Conserved Guardians of Genomic Integrity. Advances in experimental medicine and biology. 2013;767:161–184. doi: 10.1007/978-1-4614-5037-5_8. [DOI] [PubMed] [Google Scholar]
- Ledermann JA, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, Scott C, Meier W, Shapira-Frommer R, Safra T, et al. Overall survival in patients with platinum-sensitive recurrent serous ovarian cancer receiving olaparib maintenance monotherapy: an updated analysis from a randomised, placebo-controlled, double-blind, phase 2 trial. The Lancet Oncology. 2016;17:1579–1589. doi: 10.1016/S1470-2045(16)30376-X. [DOI] [PubMed] [Google Scholar]
- Li J, Zou C, Bai Y, Wazer DE, Band V, Gao Q. DSS1 is required for the stability of BRCA2. Oncogene. 2006;25:1186–1194. doi: 10.1038/sj.onc.1209153. [DOI] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nature structural & molecular biology. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Burness ML, Martin-Trevino R, Guy J, Bai S, Harouaka R, Brooks MD, Shang L, Fox A, Luther TK, et al. RAD51 Mediates Resistance of Cancer Stem Cells to PARP Inhibition in Triple-Negative Breast Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016 doi: 10.1158/1078-0432.CCR-15-1348. [DOI] [PubMed] [Google Scholar]
- Lok BH, Carley AC, Tchang B, Powell SN. RAD52 inactivation is synthetically lethal with deficiencies in BRCA1 and PALB2 in addition to BRCA2 through RAD51-mediated homologous recombination. Oncogene. 2012 doi: 10.1038/onc.2012.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science (New York, NY) 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo B, Trusolino L. Inhibition of poly(ADP-ribosyl)ation in cancer: old and new paradigms revisited. Biochimica et biophysica acta. 2014;1846:201–215. doi: 10.1016/j.bbcan.2014.07.004. [DOI] [PubMed] [Google Scholar]
- Lv W, Budke B, Kozikowski AP, Connell PP. Development of Small Molecules that Specifically Inhibit the D-loop Activity of RAD51. Int J Radiat Oncol. 2016a;96:S16–S17. doi: 10.1021/acs.jmedchem.5b01762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv W, Budke B, Pawlowski M, Connell PP, Kozikowski AP. Development of Small Molecules that Specifically Inhibit the D-loop Activity of RAD51. Journal of medicinal chemistry. 2016b;59:4511–4525. doi: 10.1021/acs.jmedchem.5b01762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazina OM, Keskin H, Hanamshet K, Storici F, Mazin AV. Rad52 Inverse Strand Exchange Drives RNA-Templated DNA Double-Strand Break Repair. Mol Cell. 2017;67:19–29. doi: 10.1016/j.molcel.2017.05.019. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menear KA, Adcock C, Boulter R, Cockcroft XL, Copsey L, Cranston A, Dillon KJ, Drzewiecki J, Garman S, Gomez S, et al. 4-[3-(4-cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phthalazin- 1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. Journal of medicinal chemistry. 2008;51:6581–6591. doi: 10.1021/jm8001263. [DOI] [PubMed] [Google Scholar]
- Moschetti T, Sharpe T, Fischer G, Marsh ME, Ng HK, Morgan M, Scott DE, Blundell TL, A RV, Skidmore J, et al. Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51. Journal of molecular biology. 2016;428:4589–4607. doi: 10.1016/j.jmb.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nature reviews Molecular cell biology. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S, Zhang C, Finch N, Zhang K, Campo L, Breuer EK. Predictors and Modulators of Synthetic Lethality: An Update on PARP Inhibitors and Personalized Medicine. Biomed Res Int. 2016 doi: 10.1155/2016/2346585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murfuni I, Basile G, Subramanyam S, Malacaria E, Bignami M, Spies M, Franchitto A, Pichierri P. Survival of the replication checkpoint deficient cells requires MUS81-RAD52 function. PLoS Genet. 2013;9:e1003910. doi: 10.1371/journal.pgen.1003910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand A, Riviere E, Renodon-Corniere A. Identification and characterization of human Rad51 inhibitors by screening of an existing drug library. Biochem Pharmacol. 2014;91:293–300. doi: 10.1016/j.bcp.2014.07.033. [DOI] [PubMed] [Google Scholar]
- Park JY, Zhang F, Andreassen PR. PALB2: the hub of a network of tumor suppressors involved in DNA damage responses. Biochimica et biophysica acta. 2014;1846:263–275. doi: 10.1016/j.bbcan.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3406–3411. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- Pommier Y, Barcelo JM, Rao VA, Sordet O, Jobson AG, Thibaut L, Miao ZH, Seiler JA, Zhang H, Marchand C, et al. Repair of topoisomerase I-mediated DNA damage. Progress in nucleic acid research and molecular biology. 2006;81:179–229. doi: 10.1016/S0079-6603(06)81005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Redon C, Rao VA, Seiler JA, Sordet O, Takemura H, Antony S, Meng L, Liao Z, Kohlhagen G, et al. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutation research. 2003;532:173–203. doi: 10.1016/j.mrfmmm.2003.08.016. [DOI] [PubMed] [Google Scholar]
- Powell SN, Kachnic LA. Therapeutic exploitation of tumor cell defects in homologous recombination. Anticancer Agents Med Chem. 2008;8:448–460. doi: 10.2174/187152008784220267. [DOI] [PubMed] [Google Scholar]
- Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harbor perspectives in biology. 2015;7:a016600. doi: 10.1101/cshperspect.a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M, Monteiro ANA. Fine tuning chemotherapy to match BRCA1 status. Biochemical Pharmacology. 2010;80:647–653. doi: 10.1016/j.bcp.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnell MR, Whish WJ. Novel inhibitors of poly(ADP-ribose) synthetase. The Biochemical journal. 1980;185:775–777. doi: 10.1042/bj1850775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin PW, Jacobson EL, Benjamin RC, Moss J, Jacobson MK. Quantitative studies of inhibitors of ADP-ribosylation in vitro and in vivo. The Journal of biological chemistry. 1989;264:4312–4317. [PubMed] [Google Scholar]
- Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, Cocito A, Costanzo V, Lopes M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nature structural & molecular biology. 2012;19:417–423. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- Rijkers T, Van Den Ouweland J, Morolli B, Rolink AG, Baarends WM, Van Sloun PP, Lohman PH, Pastink A. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal AS, Dexheimer TS, Gileadi O, Nguyen GH, Chu WK, Hickson ID, Jadhav A, Simeonov A, Maloney DJ. Synthesis and SAR studies of 5-(pyridin-4-yl)-1,3,4-thiadiazol-2-amine derivatives as potent inhibitors of Bloom Helicase. Bioorganic & medicinal chemistry letters. 2013;23 doi: 10.1016/j.bmcl.2013.1008.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal AS, Dexheimer TS, Nguyen G, Gileadi O, Vindigni A, Simeonov A, Jadhav A, Hickson I, Maloney DJ. Discovery of ML216, a Small Molecule Inhibitor of Bloom (BLM) Helicase. In Probe Reports from the NIH Molecular Libraries Program (Bethesda (MD): National Center for Biotechnology Information (US)) 2010 [PubMed] [Google Scholar]
- Saeki H, Siaud N, Christ N, Wiegant WW, van Buul PP, Han M, Zdzienicka MZ, Stark JM, Jasin M. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8768–8773. doi: 10.1073/pnas.0600298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, Villegas E, Jacquemont C, Farrugia DJ, Couch FJ, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–U1119. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DE, Coyne AG, Venkitaraman A, Blundell TL, Abell C, Hyvonen M. Small-molecule inhibitors that target protein-protein interactions in the RAD51 family of recombinases. ChemMedChem. 2015;10:296–303. doi: 10.1002/cmdc.201402428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen M, Allen C, Nickoloff JA, Hromas R. Synthetic lethality: exploiting the addiction of cancer to DNA repair. Blood. 2011;117:6074–6082. doi: 10.1182/blood-2011-01-313734. [DOI] [PubMed] [Google Scholar]
- Shibata A, Moiani D, Arvai Andrew S, Perry J, Harding Shane M, Genois M-M, Maity R, van Rossum-Fikkert S, Kertokalio A, Romoli F, et al. DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities. Mol Cell. 2014;53:7–18. doi: 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivji MK, Davies OR, Savill JM, Bates DL, Pellegrini L, Venkitaraman AR. A region of human BRCA2 containing multiple BRC repeats promotes RAD51-mediated strand exchange. Nucleic acids research. 2006;34:4000–4011. doi: 10.1093/nar/gkl505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou SK, Kamileri I, Lugli N, Evangelou K, Da-Re C, Huber F, Padayachy L, Tardy S, Nicati NL, Barriot S, et al. Mammalian RAD52 Functions in Break-Induced Replication Repair of Collapsed DNA Replication Forks. Mol Cell. 2016;64:1127–1134. doi: 10.1016/j.molcel.2016.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies M, Fishel R. Mismatch Repair during Homologous and Homeologous Recombination. Cold Spring Harbor perspectives in biology. 2015;7 doi: 10.1101/cshperspect.a022657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struewing JP, Abeliovich D, Peretz T, Avishai N, Kaback MM, Collins FS, Brody LC. The carrier frequency of the BRCA1 185delAG mutation is approximately 1 percent in Ashkenazi Jewish individuals (vol 11, pg 198, 1995) Nat Genet. 1996;12:110–110. doi: 10.1038/ng1095-198. [DOI] [PubMed] [Google Scholar]
- Subramanyam S, Jones WT, Spies M, Spies MA. Contributions of the RAD51 N-terminal domain to BRCA2-RAD51 interaction. Nucleic acids research. 2013;41:9020–9032. doi: 10.1093/nar/gkt691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura K, Takebayashi S, Taguchi H, Takeda S, Okumura K. PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. The Journal of cell biology. 2008;183:1203–1212. doi: 10.1083/jcb.200806068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K, Cramer-Morales K, McElroy DL, Ostrov DA, Haas K, Childers W, Hromas R, Skorski T. Identification of a Small Molecule Inhibitor of RAD52 by Structure-Based Selection. PloS one. 2016;11:e0147230. doi: 10.1371/journal.pone.0147230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher EM, Sakai W, Karlan BY, Wurz K, Urban N, Taniguchi T. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer research. 2008;68:2581–2586. doi: 10.1158/0008-5472.CAN-08-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS. End resection at double-strand breaks: mechanism and regulation. Cold Spring Harbor perspectives in biology. 2014;6 doi: 10.1101/cshperspect.a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS. Mechanism and regulation of DNA end resection in eukaryotes. Critical reviews in biochemistry and molecular biology. 2016;51:195–212. doi: 10.3109/10409238.2016.1172552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taleb NN. Antifragile : things that gain from disorder. 1. New York: Random House; 2012. [Google Scholar]
- Thakar A, Parvin J, Zlatanova J. BRCA1/BARD1 E3 ubiquitin ligase can modify histones H2A and H2B in the nucleosome particle. Journal of biomolecular structure & dynamics. 2010;27:399–406. doi: 10.1080/07391102.2010.10507326. [DOI] [PubMed] [Google Scholar]
- Thomas HD, Calabrese CR, Batey MA, Canan S, Hostomsky Z, Kyle S, Maegley KA, Newell DR, Skalitzky D, Wang LZ, et al. Preclinical selection of a novel poly(ADP-ribose) polymerase inhibitor for clinical trial. Molecular cancer therapeutics. 2007;6:945–956. doi: 10.1158/1535-7163.MCT-06-0552. [DOI] [PubMed] [Google Scholar]
- Thorsell AG, Ekblad T, Karlberg T, Low M, Pinto AF, Tresaugues L, Moche M, Cohen MS, Schuler H. Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors. Journal of medicinal chemistry. 2017;60:1262–1271. doi: 10.1021/acs.jmedchem.6b00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nature structural & molecular biology. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner LM. Profile of veliparib and its potential in the treatment of solid tumors. OncoTargets and therapy. 2015;8:1931–1939. doi: 10.2147/OTT.S69935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell A-G, Pol E, Frostell A, Ekblad T, Oncu D, et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotech. 2012;30:283–288. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- Wang AT, Kim T, Wagner JE, Conti BA, Lach FP, Huang AL, Molina H, Sanborn EM, Zierhut H, Cornes BK, et al. A Dominant Mutation in Human RAD51 Reveals Its Function in DNA Interstrand Crosslink Repair Independent of Homologous Recombination. Mol Cell. 2015;59:478–490. doi: 10.1016/j.molcel.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Krais JJ, Bernhardy AJ, Nicolas E, Cai KQ, Harrell MI, Kim HH, George E, Swisher EM, Simpkins F, et al. RING domain-deficient BRCA1 promotes PARP inhibitor and platinum resistance. The Journal of clinical investigation. 2016;126:3145–3157. doi: 10.1172/JCI87033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegmans AP, Miranda M, Wen SW, Al-Ejeh F, Moller A. RAD51 inhibition in triple negative breast cancer cells is challenged by compensatory survival signaling and requires rational combination therapy. Oncotarget. 2016;7:60087–60100. doi: 10.18632/oncotarget.11065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, Yang MC, Hwang LY, Bowcock AM, Baer R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- Wu Y, Brosh RM. Helicase-inactivating mutations as a basis for dominant negative phenotypes. Cell cycle (Georgetown, Tex) 2010;9:4080–4090. doi: 10.4161/cc.9.20.13667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Sonoda E, Buerstedde JM, Bezzubova O, Morrison C, Takata M, Shinohara A, Takeda S. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol Cell Biol. 1998;18:6430–6435. doi: 10.1128/mcb.18.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez RJ, Porter AC. Differential effects of Rad52p overexpression on gene targeting and extrachromosomal homologous recombination in a human cell line. Nucleic acids research. 2002;30:740–748. doi: 10.1093/nar/30.3.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, Pavletich NP. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science (New York, NY) 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- Yap TA, Sandhu SK, Carden CP, de Bono JS. Poly(ADP-ribose) polymerase (PARP) inhibitors: Exploiting a synthetic lethal strategy in the clinic. CA: a cancer journal for clinicians. 2011;61:31–49. doi: 10.3322/caac.20095. [DOI] [PubMed] [Google Scholar]
- Yata K, Bleuyard JY, Nakato R, Ralf C, Katou Y, Schwab RA, Niedzwiedz W, Shirahige K, Esashi F. BRCA2 coordinates the activities of cell-cycle kinases to promote genome stability. Cell reports. 2014;7:1547–1559. doi: 10.1016/j.celrep.2014.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Fu S, Lai M, Baer R, Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes & development. 2006;20:1721–1726. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinovyev A, Kuperstein I, Barillot E, Heyer WD. Synthetic lethality between gene defects affecting a single non-essential molecular pathway with reversible steps. PLoS computational biology. 2013;9:e1003016. doi: 10.1371/journal.pcbi.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]