Abstract
The plant hormone ethylene plays various functions in plant growth, development and response to environmental stress. Ethylene is perceived by membrane-bound ethylene receptors, and among the homologous receptors in Arabidopsis, the ETR1 ethylene receptor plays a major role. The present study provides evidence demonstrating that Arabidopsis CPR5 functions as a novel ETR1 receptor-interacting protein in regulating ethylene response and signaling. Yeast split ubiquitin assays and bi-fluorescence complementation studies in plant cells indicated that CPR5 directly interacts with the ETR1 receptor. Genetic analyses indicated that mutant alleles of cpr5 can suppress ethylene insensitivity in both etr1-1 and etr1-2, but not in other dominant ethylene receptor mutants. Overexpression of Arabidopsis CPR5 either in transgenic Arabidopsis plants, or ectopically in tobacco, significantly enhanced ethylene sensitivity. These findings indicate that CPR5 plays a critical role in regulating ethylene signaling. CPR5 is localized to endomembrane structures and the nucleus, and is involved in various regulatory pathways, including pathogenesis, leaf senescence, and spontaneous cell death. This study provides evidence for a novel regulatory function played by CPR5 in the ethylene receptor signaling pathway in Arabidopsis.
INTRODUCTION
The gaseous phytohormone ethylene plays an important role in various plant processes, including seed germination, promotion of fruit ripening, organ senescence, abscission, apical hook formation, flowering and gravitropism (Abeles et al. 1992). Ethylene treatment can result in inhibition of dark-grown Arabidopsis seedling root and hypocotyl elongation, hypocotyl radial swelling, and an exaggerated apical hook (Bleecker et al. 1988; Guzman and Ecker 1990), termed the “triple response”. By screening for mutants exhibiting an altered “triple response” phenotype, many core components in the ethylene signal pathway have been isolated, including ethylene receptors, a MAPKKK protein CTR1, the endoplasmic reticulum (ER) membrane-localized protein, EIN2, and the transcription factor EIN3/EIL (Chang et al. 1993; Hua et al. 1995, 1998; Chao et al. 1997; Sakai et al. 1998; Alonso et al. 1999). Ethylene signaling starts with ethylene binding to the ER membrane-associated receptors, and finally leads to changes in gene expression within the nucleus (Guo and Ecker 2004).
In Arabidopsis thaliana, there are five homologous ethylene receptors (ETR1, ERS1, EIN4, ETR2, ERS2), which resemble the two-component histidine protein kinase family primarily found in prokaryotes (Chang et al. 1993; Hua et al. 1995, 1998; Sakai et al. 1998). These receptors are negative regulators of ethylene responses (Hua and Meyerowitz 1998; Hall and Bleecker 2003; Qu et al. 2007) that, upon binding ethylene with the help of a copper cofactor are inactivated, leading to triggering of the ethylene response; however, in the absence of ethylene, the active receptors block ethylene signaling (Rodriguez et al. 1999).
According to their structure similarities, these receptors can be divided into two subfamilies: subfamily I contains ETR1 and ERS1, and subfamily II is comprised of ETR2, ERS2 and EIN4. The subfamily I receptors have three N-terminal transmembrane domains, and a highly conserved histidine kinase domain, whereas the subfamily II receptors have four N-terminal transmembrane domains and a degenerated histidine kinase domain, and they may have Ser/Thr kinase activity (Gamble et al. 1998; Moussatche and Klee 2004; Chen et al. 2009). All five ethylene receptors contribute to ethylene signaling and are redundant in regulating ethylene responses, while subfamily I receptors, and especially ETR1, play a predominant role (Hua et al. 1995; Hua and Meyerowitz 1998; Hall and Bleecker 2003; Qu et al. 2007; Liu et al. 2010).
RTE1 (REVERSION TO ETHYLENE SENSITIVITY 1) was identified as a positive and upstream regulator of the ETR1 receptor, and this regulation is specific genetically (Resnick et al. 2006; Resnick et al. 2008; Rivarola et al. 2009). RTE1 is an activator of ETR1, which was also supported by the direct interaction of the two proteins and their co-localization (Dong et al. 2008, 2010). Conserved RTE1 homologs are widely distributed in higher eukaryotes (Resnick et al. 2006). Arabidopsis RTE1 encodes an integral membrane protein with three homologs in tomato, two of which were shown to regulate ethylene responses in this crop plant (Barry and Giovannoni 2006; Klee 2006; Ma et al. 2012). In Rosa hybrida, the expression of Rh-RTH1 was responsive to ethylene, and was also partially correlated with expression of Rh-ETR1 and Rh-ETR3 (Yu et al. 2010). In addition, it was reported that the rice RTE1 homolog (OsRTH1, Zhang et al. 2012) and the RTE-like genes (DCRTE1 and DCRTH1) of carnation (Yu et al. 2011) participate in the regulation of ethylene responses in seedling growth and flower senescence.
Arabidopsis RTE1 is localized to ER and Golgi membranes (Resnick et al. 2006; Zhou et al. 2007; Dong et al. 2008). Furthermore, based on a yeast two-hybrid screen, it was shown that the ER-localized cytochrome B5 (cb5) and lipid transfer protein LTP1 could directly interact with RTE1 and function in modulation of ethylene responses and signaling (Chang et al. 2014; Wang et al. 2016). The Arabidopsis Cb5 proteins are likely involved in electron transfer reactions, as in all eukaryotes. Both Cb5-D and LTP1 might act upstream of RTE1 in regulating the ETR1-mediated repression of ethylene responses. Both atcb5 and atltp1 mutants show increased ethylene sensitivity, whereas overexpression of Cb5-D or LTP1 conferred decreased ethylene sensitivity. It appears that both AtCb5 and AtLTP1 play positive roles in ethylene signaling and responses, probably by participating in the protein complex involved in regulation of the ethylene signal, via RTE1. These findings suggest that ETR1 receptor signaling might be tightly controlled by a protein complex containing RTE1 and the RTE1-binding proteins. It is conceivable that other unknown regulators might exist in the ETR1 receptor signaling pathway.
The Arabidopsis CPR5 (CONSTITUTIVE EXPRESSOR OF PATHOGENESIS-RELATED GENES 5) gene was isolated based on a screen for constitutive expressions of systemic acquired resistance (SAR) (Bowling et al. 1997; Boch et al. 1998). Recently, it was reported that the Arabidopsis CPR5 (hereafter CPR5) acts as a nucleoporin in controlling effector-triggered immunity (ETI) and programmed cell death (PCD) in plants (Gu et al. 2016). During plant growth and development, CPR5 regulates endoreduplication, cell division, cell expansion and spontaneous cell death (Kirik et al. 2001; Brininstool et al. 2008; Perazza et al. 2011). It was proposed that CPR5 participates in a complex interacting with cell cycle regulators, and plays a critical role in this process (Bao and Hua 2014). CPR5 also participates in plant stress responses, including thermotolerance (Wang et al. 2012), ABA signaling (Gao et al. 2011), cellular ROS status and/or signaling (Jing and Dijkwel 2008; Jing et al. 2008), and K+ homeostasis (Borghi et al. 2011). As a positive modulator, CPR5 regulates plant growth under physiological conditions and during stress by antagonizing SA-dependent growth inhibition through the unfolded protein response (UPR) pathway (Meng et al. 2017).
A role for CPR5 in ethylene signaling was suggested by earlier studies in which it was reported that exogenous application of the ethylene precursor, 1-aminocyclopropane-1-carboxylic acid (ACC), to Arabidopsis cpr5/hys1/old1 mutant seedlings gave an enhanced ethylene response (Jing et al. 2005, 2007; Aki et al. 2007). However, little is currently known concerning the underlying molecular mechanism by which CPR5 might act in the ethylene signaling pathway. In the present study, we provide data showing that AtCPR5 plays a critical role in the regulation of ethylene signaling, via direct association with the ETR1 receptor.
RESULTS
Screening for suppressors of etr1-2 ethylene insensitivity
Previously, a genetic screen for suppressors of ethylene insensitivity performed on the etr1-2 mutant identified a positive regulator of the ETR1 receptor (Resnick et al. 2006). In order to identify additional potential ETR1 regulators, a genetic screen was conducted using a new pool of ethyl methyl sulfonate (EMS)-mutagenized etr1-2 plants. To this end, dark-grown seedlings were treated with ethylene and screened for etr1-2 suppressor mutants that exhibited the “triple-response” phenotype. A mutant displaying this restored ethylene “triple-response” was chosen for in-depth analysis. To assess whether the mutation was intragenic or extragenic to the etr1-2 allele, this mutant was crossed to wild type (Col-0) and the resultant F1 progeny was analyzed for the etiolated seedling “triple response” phenotype on MS agar plates containing ACC (100 μM).
Due to the dominant nature of etr1-2, the resulting progeny of the F1 seedlings displayed an ethylene-insensitive phenotype, suggesting that the mutation was extragenic to etr1-2. We next twice backcrossed this mutant line to etr1-2 to remove extraneous mutations. A mapping population was made by crossing the mutant with a dominant ethylene insensitive etr1 allele, in the Arabidopsis ecotype Landsberg erect (Resnick et al. 2006). Using simple sequence length polymorphism (SSLP) markers, we mapped the extragenic mutation locus to a 68-kb interval on the bottom of chromosome 5. After sequencing the corresponding DNA fragments of 9 genes within this region, we identified a single nucleotide C-to-T missense mutation which codes for Pro535 in place of Ser535 in the open reading frame (ORF) of gene AT5G64930. This is the same mutation as in hys1-2 (Yoshida et al. 2002), an allele of CPR5 (Bowling et al. 1997). The etiolated mutant seedling is shown in Figure 1A (see etr1-2 hys1-2). The other genes within this region were sequenced but no further sequence differences were detected.
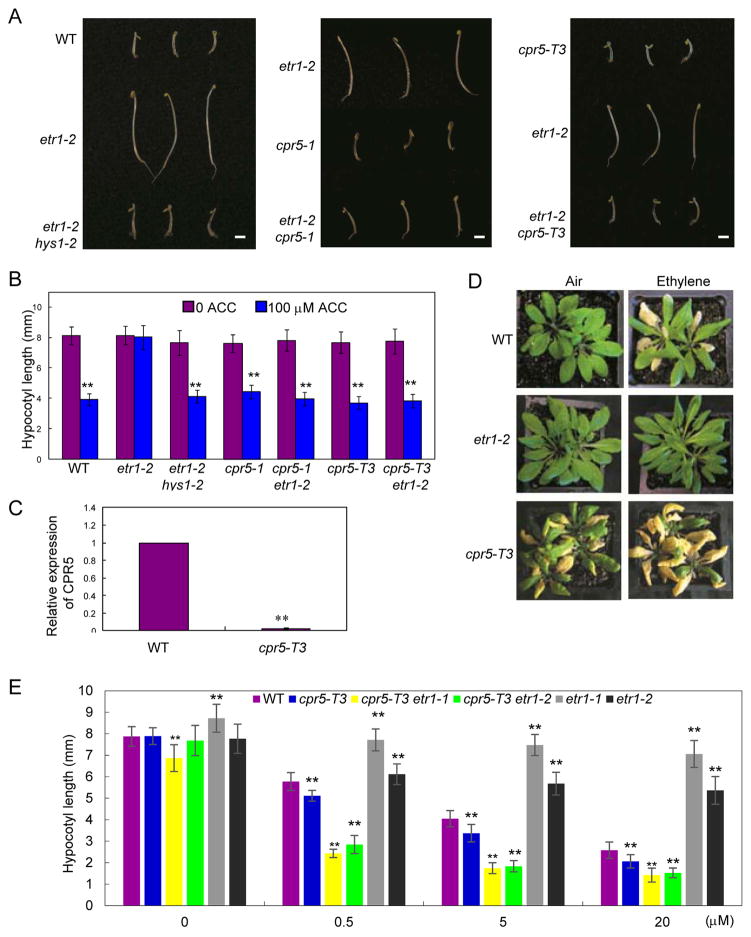
Figure 1. Ethylene sensitivity analysis of the cpr5 mutants.
(A) Representative 4-day-old etiolated of the wild type (WT), etr1-2 and etr1-2 hys1-2 germinated in the presence of the ethylene precursor ACC (100 μM). Bars= 3 mm. (B) Quantitative analysis of hypocotyl lengths of 4-day-old dark-grown seedlings of wild type (WT), etr1-2, etr1-2 hys1-2, cpr5-1, etr1-2 cpr5-1, cpr5-T3, and cpr5-T3 etr1-2 germinated in the absence or presence of the ethylene precursor ACC (100 μM). Values are mean ±SD, n = 25, **p < 0.01. (C) Relative expression of CPR5 in the wild type (WT) and cpr5-T3 by qRT-PCR. Values are means ±SD; **, P < 0.01. (D) Ethylene-induced leaf senescence in 4-week-old plants treated with or without 100 ppm of ethylene for 4 days. Leaf senescence is indicated by yellowing. (E) Quantitative analysis of hypocotyl lengths for the wild type (WT), cpr5-T3, cpr5-T3 etr1-1, cpr5-T3 etr1-2, etr1-1, and etr1-2 on 1/2×MS medium containing ACC (0, 0.5, 5, 20 μM). Values are mean ±SD, n = 30, **P<0.01.
We next tested a second allele by obtaining the cpr5-1 mutant described in Bowling et al. (1997), crossing it with etr1-2, and identifying the double mutant etr1-2 cpr5-1 in the F2 generation. Ethylene “triple response” analysis established the same suppression of ethylene insensitivity in these etr1-2 cpr5-1 plants (Figure 1A, B), providing further support that the cpr5 mutations could affect ETR1-mediated ethylene signaling.
Disruption of CPR5 significantly affects ethylene signaling
We next analyzed a cpr5 knockout allele (cpr5-T3) using a T-DNA insertion line from the Arabidopsis Biological Resource Center (ABRC, Ohio State University, Columbus). Quantitative RT-PCR revealed that CPR5 transcripts were barely detected in this T-DNA insertion mutant (Figure 1C). To examine the ethylene sensitivity of the cpr5-T3 mutant, gaseous ethylene was applied to these plants. Our assays indicated that cpr5-T3 had more yellow leaves compared to wild type following ethylene treatment for 3 days (Figure 1D). Here, etr1-2 plants were used as a control and they were always ethylene insensitive. Interestingly, senescent leaves were observed in the sealed box test system after 3 days, even without injection of exogenous ethylene, raising the possibility that cpr5-T3 plants could have enhanced ethylene sensitivity. Assays using dark-grown etiolated seedlings treated with ACC, at different concentrations, confirmed that cpr5-T3 seedlings had an enhanced ethylene response (Figure 1E). In the absence of ethylene treatment, cpr5-T3 seedlings failed to display a hypersensitivity response (Figure 1E).
To test whether cpr5-T3 could suppress the etr1-2 ethylene insensitivity, a genetic cross of cpr5-T3 with etr1-2 was performed and the double mutant, etr1-2 cpr5-T3, was identified from the F2 generation. As expected, the cpr5-T3 mutation could suppress the ethylene insensitivity of etr1-2 (Figure 1A, B). Taken together, these results support the notion that CPR5 plays a regulatory function in the ethylene response.
CPR5 overexpression enhances the ethylene response and signaling
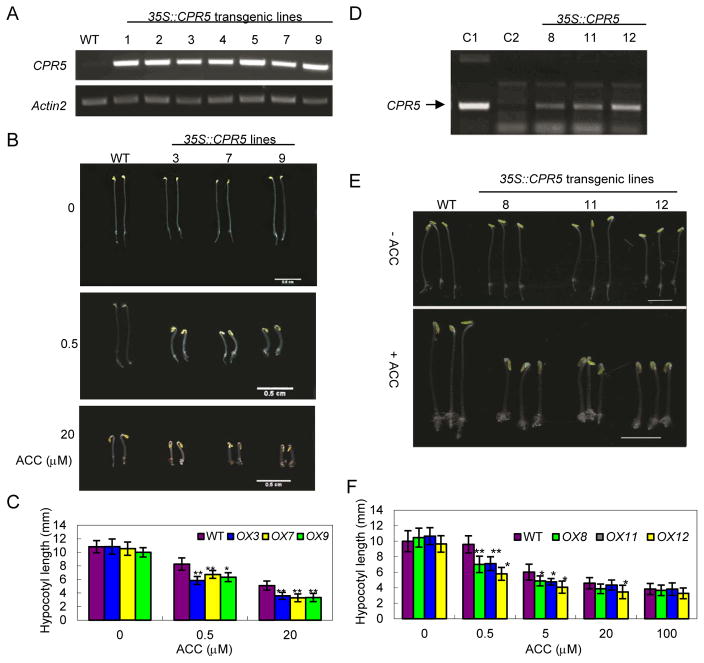
To advance our understanding of the role played by CPR5 in regulating ethylene responses, an approach of over-expressing CPR5 in transgenic Arabidopsis was employed. An Arabidopsis CPR5 full length cDNA was amplified by PCR and the resultant product was cloned under the strong CaMV35S promoter. A binary vector harboring p35S::CPR5 was transferred into Agrobacterium for flower dip transformation of Arabidopsis thaliana plants (Clough and Bent 1998). Seven transgenic lines were examined for CPR5 transcript levels by RT-PCR; all transgenic lines showed high levels of CPR5 transcripts (Figure 2A). Under treatment with different concentrations of ACC (0, 0.5, 20 μM), the etiolated seedlings of CPR5-overexpressing (OX) lines showed a significant difference compared to wild type plants, and all OX lines exhibited shorter hypocotyls (Figure 2B, C), indicating an increase in CPR5 transcripts conferred enhanced ethylene sensitivity.
Figure 2. Analysis of transgenic plants overexpressing CPR5.
(A) Semi-quantitative RT-PCR analysis for CPR5 transcripts in wild-type (WT) and CPR5-overexpression lines. Actin 2 was used as an internal control. (B) Ethylene sensitivity analysis showing representative 4-day old etiolated seedlings of the wild type (WT) and CPR5-overexpression lines treated with or without ACC (0, 0.5, 20 μM). Bars = 5 mm. (C) Quantitative analysis of hypocotyl lengths for the wild type (WT) and CPR5-overexpression lines on 1/2×MS medium containing ACC (0, 0.5, 20 μM). Values are mean ±SD, n = 20, **p<0.01. (D) Detection of Arabidopsis CPR5 transcripts in the plasmid DNA control (C1), wild-type tobacco leaves (C2), and the transgenic tobacco lines (#8, 11, 12) by semi-quantitative RT-PCR. The arrow indicates the position of PCR-amplified CPR5 fragments. (E) Ethylene sensitivity analysis showing representative 6-day old dark-grown seedlings the wild type (WT) and CPR5-overexpression transgenic tobacco lines treated with or without ACC (0.5 μM). Bars = 2 mm. (F) Quantitative analysis of hypocotyl lengths for the wild type (WT) and CPR5-overexpression transgenic tobacco lines (OX8, OX11, OX12) on 1/2MS medium containing ACC (0, 0.5, 5, 20, 100 μM). Values are mean ±SD (n = 30); *P<005; **P<0.01.
The p35S::CPR5 construct was also introduced into tobacco (Nicotiana tabacum) mediated by Agrobacterium-mediated transformation (Supplement S2). The CPR5 transcript levels were assayed by RT-PCR in three transgenic lines; all showed high levels (Figure 2D). The ethylene response of dark-grown wild type and CPR5-overexpression (OX) transgenic tobacco 6-day-old seedlings were analyzed in response to a range of ACC concentrations (0, 0.5, 5, 20, 100 μM) (Figure 2F) Interestingly, the hypocotyl length of CPR5-OX seedlings was significantly shorter than those of wild type seedlings, especially at low ACC concentrations (Figure 2E, F). This finding provides support for the notion that ectopic expression of Arabidopsis CPR5 can enhance ethylene sensitivity in tobacco plants.
Altered expression of downstream ERFs in cpr5-T3 and CPR5-OX plants supports a regulatory function of CPR5 in ethylene signaling
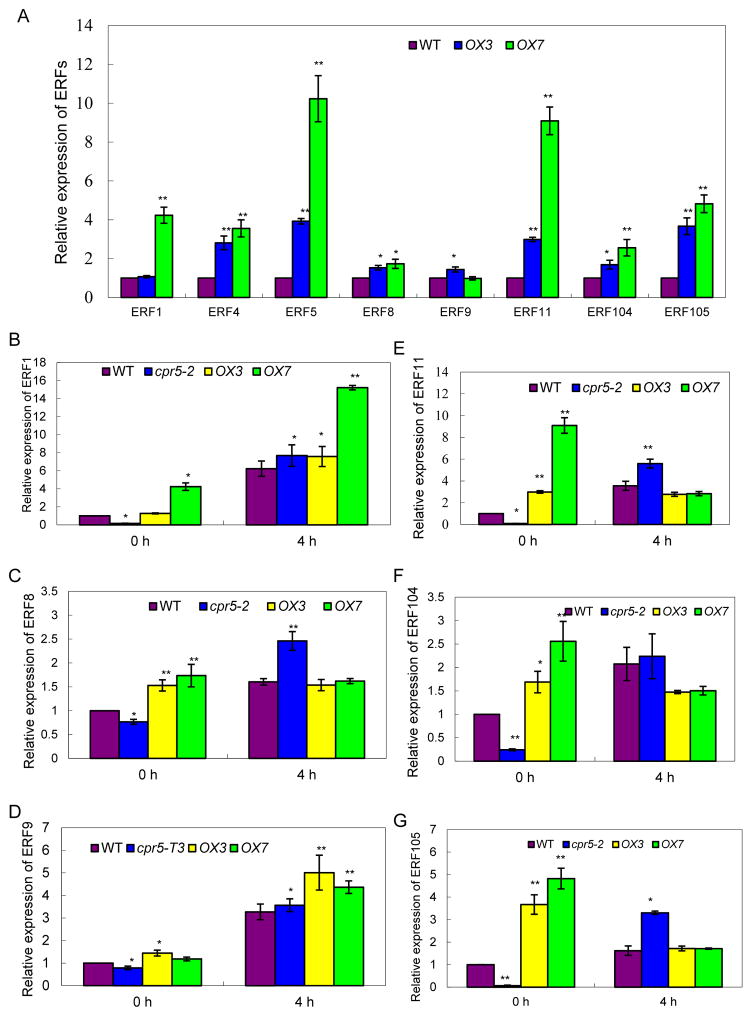
To examine the effect of CPR5-OX on the endogenous transcript levels of ethylene responsive factors (ERFs) in the Arabidopsis CPR5-OX transgenic plants, nine genes (ERF1, 2, 4, 5, 8, 9, 11, 104, and 105) were chosen for quantitative RT-PCR analysis. These ethylene-induced ERFs were chosen based on information from The Arabidopsis Information Resource (TAIR) (http://www.arabidopsis.org/) and Wang et al. (2016). Our assays indicated that the transcript levels for almost all the examined ERFs in the CPR5-OX (#OX3 and #OX7) were higher than those in the wild type, indicating that increased CPR5 transcript levels promote expression of downstream ERFs in these plants (Figure 3A).
Figure 3. Relative expression of Arabidopsis ERFs in cpr5-T3 and CPR5-overexpression lines using qRT-PCR.
(A) Relative expression of Arabidopsis ERF1, 4, 5, 8, 9, 11,104 and 105 in 10-day-old light-grown seedlings of the wild type (WT) and CPR5-overexpression lines (OX3, OX7), respectively. (B–G) Relative expression of Arabidopsis ERF1, 8, 9, 11, 104 and 105 in 10-day-old light-grown seedlings of the wild type (WT), cpr5-T3, and CPR5-overexpression lines (OX3, OX7), respectively. Plants were grown on 1/2×MS medium for 10 days, then treated with or without ACC (100 mM) for 4 h. Values are mean ±SD; *P<0.05, ** P <0.01.
In contrast to the elevated ERF transcript levels in the CPR5-OX transgenic plants, ERF expression in cpr5 plants was lower than in wild type plants, among all the examined ERFs when no exogenous ACC was applied (Figure 3B–G). Surprisingly, we found that in cpr5-T3 plants transcript levels for these ERFs were higher than those in the wild type plants, when exogenous ACC was applied (Figure 3B–G). These findings offer a clue as to why cpr5 plants exhibited enhanced ethylene sensitivity (Figure 1D, E) following application of exogenous ethylene or its precursor (ACC). In addition, we noted that exogenous application of ACC could only slightly promote ERF expression (ERF1 and ERF9) in CPR5-OX plants, implying that high levels of endogenous ERFs in these transgenic lines might restrain further induction of ERFs by exogenous ethylene.
Genetic analyses indicate that CPR5 mainly affects the ETR1 receptor
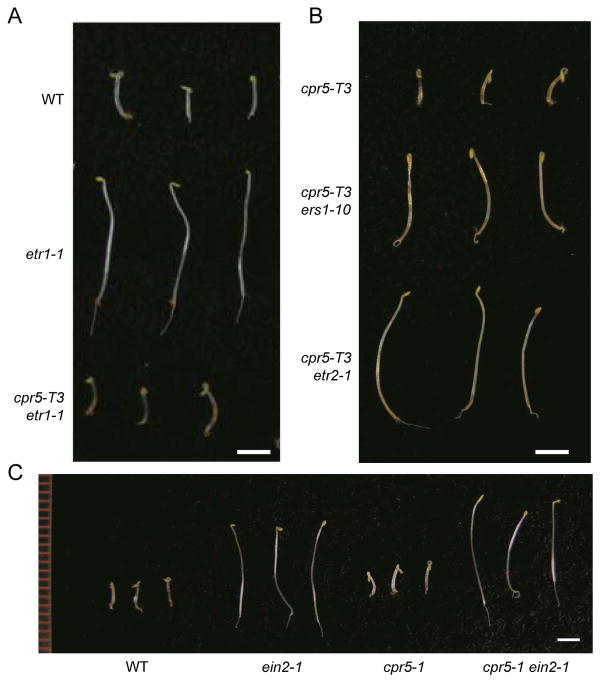
In order to examine whether the dominant ethylene-insensitive mutations in other ethylene receptor and downstream genes are suppressed by cpr5, genetic crosses of the ethylene insensitive mutants with cpr5-T3 were performed and the following double mutants were obtained; cpr5-T3 etr2-1, cpr5-T3 ers1-10 and cpr5-T3 ein2-1. Seedling “triple response” phenotypic analyses showed that cpr5-T3 could not suppress the ethylene insensitivity of etr2-1, ers1-10 or ein2-1 (Figure 4B, C). These findings suggested that CPR5 might regulate mainly ETR1 receptor signaling.
Figure 4. Ethylene “triple response” assays in Arabidopsis double mutants.
(A) Comparison of 4-day-old etiolated seedlings germinated in the presence of the ethylene precursor ACC (100 μM). Three representative seedlings of the wild type (WT) and the etr1-1 and cpr5-T3 etr1-1 mutants are shown. (B) Comparison of 4-day-old etiolated seedlings of the cpr5-T3, cpr5-T3 ers1-10 and cpr5-T3 etr2-1 mutants germinated in the presence of the ethylene precursor ACC (100 μM). Three representative seedlings of the mutants are shown. (C) Comparison of 4-day-old etiolated seedlings of the wild type (WT) and ein2-1, cpr5-1, and cpr5-1 ein2-1 germinated in the presence of the ethylene precursor ACC (100 μM). Scale bars = 2 mm.
We also tested whether CPR5 disruption could suppress another ETR1 mutant allele, etr1-1, a gain-of-function allele conferring ethylene insensitivity. A genetic cross of cpr5-T3 with etr1-1 was performed and the double mutant, etr1-1 cpr5-T3 was identified from an F2 population using molecular markers. Seedling “triple response” phenotypic analysis clearly confirmed that cpr5 could suppress the ethylene insensitivity of etr1-1 (Figure 4A). This result is different from that of rte1, which suppresses the ethylene insensitivity of etr1-2 but not that of etr1-1 (Resnick et al. 2006), indicating that CPR5 functions in ethylene signaling in a manner different from that of RTE1.
Molecular association of CPR5 with the ETR1 ethylene receptor
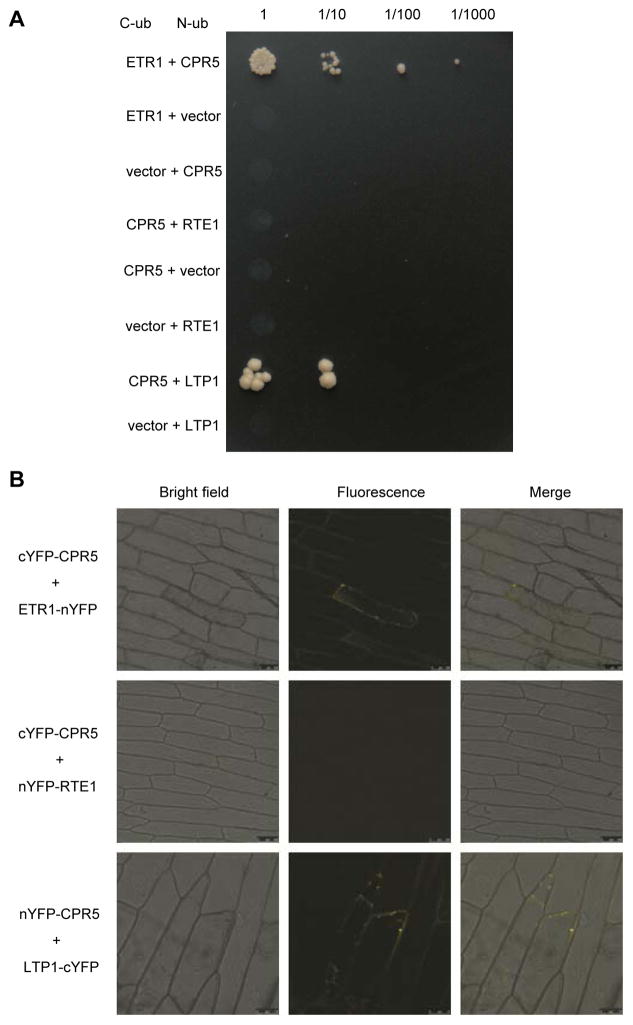
The fact that CPR5 functions in regulating ethylene signaling prompted us to examine whether CPR5 could directly interact with the ETR1 receptor. At first, we performed a yeast split-ubiquitin assays (Chang et al. 2014; Wang et al. 2016). In frame fusions of Cub-ETR1 and Nub-CPR5 constructs were verified by DNA sequencing, and protein interactions were examined on selective medium lacking adenine, histidine, leucine and tryptophan. Strong protein interactions were detected in yeast cells co-expressing Cub-ETR1 and Nub-CPR5, whereas no colonies survived on the selection medium if either construct was absent (Figure 5A). It is interesting to note that the protein interaction between CPR5 and RTE1 was not detected, while a positive interaction between CPR5 and LTP1 was observed.
Figure 5. Molecular association of Arabidopsis CPR5 and ethylene receptor ETR1.
(A) Molecular interaction of CPR5 and ETR1 in the yeast split-ubiquitin assay. CPR5 was paired with ETR1, RTE1, LTP1 or an empty vector. Yeast viability is shown on medium lacking histidine, alanine, leucine and tryptophan, while interaction is indicated by growth on the same selection medium. Undiluted and 1:10, 1:100, 1:1000 diluted liquid cultures were spotted on the indicated plates and incubated for 5 days at 30°C. (B) Molecular interaction of CPR5 and ETR1 in onion epidermal cells shown by BiFC assay. Constructs expressing the N- and C-terminal halves of YFP fused to the C-terminus of ETR1 or RTE1, and the N-terminus of CPR5 or LTP1, respectively, were co-infiltrated into onion epidermal cells. YFP fluorescence was detected by laser scanning confocal microscopy at 505–530 nm. Scale bars: 75 μm or 100 μm.
We next examined protein interaction between CPR5 and ETR1, in planta, using the bimolecular fluorescence complementation (BiFC) assay. The coding sequences of the YFP halves, cYFP and nYFP, were fused to the full-length coding sequences of CPR5 at the N-terminus and ETR1 at the C-terminus, respectively. In- frame fusion of the resultant constructs was verified by DNA sequencing. When cYFP-CPR5 and ETR1-nYFP fusions were transiently co-expressed in onion epidermal peel cells, a fluorescent signal was readily detected (Figure 5B). In our controls, fluorescence was not detected in cells if the cYFP-CPR5 and RTE1-nYFP fusions were co-expressed. In contrast, strong fluorescence was observed when the nYFP-CPR5 and LTP1-cYFP fusions were co-expressed. These BiFC assays indicated that CPR5 can physically interact with the ETR1 receptor, in vivo.
DISCUSSION
CPR5 is a novel ETR1 interacting protein
CPR5 is a membrane protein with five transmembrane helices at the C terminus and is associated with the endomembrane system, including the nuclear envelope and ER-associated large granules. It is known that in Arabidopsis, CPR5 is involved in a number of regulatory pathways, including the SAR pathway (Bowling et al. 1997; Gu et al. 2016), RPS2 signal transduction pathway (Boch et al. 1998), cellular ROS status and/or signaling (Jing and Dijkwel 2008; Jing et al. 2008), the unfolded protein response (UPR) pathway (Meng et al. 2017), and regulation of endoreduplication, cell division, cell expansion and spontaneous cell death (Kirik et al. 2001; Perazza et al. 2011). An earlier study reported that an ectopically expressed GFP-CPR5 fusion protein, driven by the strong 35S promoter, was localized to the nuclei of tobacco cells (Perazza et al. 2011), suggesting that CPR5 could be proteolytically cleaved and transported into the nucleus by an unknown mechanism. In this study, we show that CPR5 can physically interact with the ethylene receptor ETR1 (Figure 5A, B), which is localized predominantly to the ER membrane (Chen et al. 2002; Dong 2008; Grefen et al. 2008), and we provide evidence demonstrating that CPR5 plays a regulatory function in ETR1 receptor signaling. These findings implicate a new role for this ER-associated CPR5 protein.
CPR5 plays a key role in regulating ethylene signaling
Previously, enhanced ethylene responses of the Arabidopsis cpr5/hys1/old1 mutant seedlings were observed when exogenous ethylene precursor ACC was applied to the culture medium (Jing et al. 2005, 2007; Aki et al. 2007). In our study, various approaches, including cpr5 point mutations or gene knockout and CPR5-OX transgenic plants were used to probe the regulatory functions of CPR5 in ethylene response and signaling. These studies indicated that CPR5 plays a crucial role in ethylene response and signaling, based on the following: (1) the cpr5/hys1-2 point mutations could restore the ethylene sensitivity of etr1-2 in the double mutant (Figure 1A, B, E); (2) a T-DNA insertion in CPR5 gave rise to the cpr5-T3 mutant exhibiting hypersensitive to exogenous ACC (Figure 1D, E); and (3) transgenic plants over-expressing CPR5 either in Arabidopsis or ectopically in tobacco displayed enhanced ethylene sensitivity in the presence of exogenous ACC (Figure 2B, C, E, F). To gain insight into the underlying regulation, we examined expression of the downstream ERFs in the ethylene signaling pathway using quantitative RT-PCR. These assays revealed that, in the cpr5-T3 mutant, endogenous transcript levels of these downstream ERFs were lower than in wild type plants (Figure 3B–G), whereas the levels in the CPR5-OX transgenic lines were higher than in wild type (Figure 3A), suggesting that CPR5 functions as a key regulator in controlling expression of downstream ERFs that function in the ethylene signaling pathway.
It is also interesting to note that the levels of most of ERFs examined in the cpr5-T3 mutant were higher than in wild type plants when exogenous ACC was applied, although their endogenous levels were lower than in the wild type controls (Figure 3B–G). These findings provide insight into why the cpr5 mutant exhibits ethylene hypersensitivity in response to exogenous ethylene or its precursor (ACC) (Figure 1D, E). Currently, it is not known how high expressions of these ERFs might be modulated in the cpr5 mutant.
This study provides evidence showing the molecular interaction between CPR5 and the ETR1 receptor (Figure 5), indicating that CPR5 might directly regulate the ETR1 receptor signaling. As CPR5 could function as a nucleoporin (Gu et al. 2016), knockout of CPR5 might affect the nucleocytoplasmic transport of mRNAs and the ETR1 protein localization to ER. Assays by CPR5 over-expressing either in transgenic Arabidopsis or tobacco plants confirm further the regulatory function of CPR5 in ethylene signaling. It was observed that increase of the CPR5 transcripts enhances the ethylene sensitivity (Figure 2B, C, E, F). It is suggested that the over-produced CPR5 might sequester the ETR1 receptor protein, a negative factor in ethylene signaling, leading to higher expression of the downstream ERFs (Figure 3A) and enhanced ethylene sensitivity. However, the other regulation might also exist, for example, increase of the CPR5 levels might interfere with the unknown ethylene signaling component(s).
CPR5 functions differently from RTE1 in regulating ETR1 receptor signaling
In the past, limited information has been gained as to the molecular regulation of the ethylene receptors. Defects in RAN1 severely disrupt the formation of wild type ethylene receptors because of improper copper loading (Hirayama et al. 1999; Woeste and Kieber 2000). In ran1, the etr1-1 receptor is stable in the absence of copper and is locked into the repressor signaling state. Compared to RAN1, RTE1 is an activator of the ethylene signaling receptor, ETR1 (Resnick et al. 2006). RTE1 co-localizes with ETR1 at the ER and Golgi apparatus and they physically interact (Dong et al. 2008, 2010). It was proposed that RTE1 affects the conformation of the ETR1 ethylene-binding domain and/or the equilibrium state of ETR1, resulting in the promotion or stabilization of the signaling state of ETR1 (Resnick et al. 2008). However, it is still unclear why the rte1 mutant is capable of suppressing the etr1-2 but not the etr1-1 allele (Resnick et al. 2006). The etr1-2 encodes an Ala102-to-Thr substitution in the putative ethylene binding domain, whereas the etr1-1 mutation fully blocks ethylene binding (Schaller and Bleecker 1995; Hall et al. 1999), because of a Cys65-to-Tyr substitution in this mutant prevents association with the copper cofactor (Rodriguez et al. 1999). The etr1-1 mutant confers strong ethylene insensitivity, indicating that the signaling domain is essentially locked into an active conformation. In an earlier study, genetic analyses suggested that CPR5 is independent of ETR1 in controlling of leaf senescence (Jing et al. 2002). However, effect of the cpr5/old1 mutation on the ethylene sensitivity of the double mutant was not examined. In the present study, we show that cpr5 mutations can restore the ethylene sensitivity in both etr1-2 and etr1-1, indicating that CPR5 may be upstream of the ETR1 receptor in regulation of ethylene signaling. Based on a recent finding that CPR5 could act as a nucleoporin in controlling of the nucleocytoplasmic transport of mRNAs (Gu et al. 2016), it is likely that CPR5 might be required for ETR1 mRNA transport out of the nucleus for protein translation, or transport of ETR1 protein to the ER.
Genetic analyses using double mutants of cpr5-T3 with each of three other ethylene insensitive mutants (ers1-10, etr2-1, and ein2-1), showed that the ethylene insensitivity displayed by these mutants was not suppressed by the cpr5-T3, indicating that the ETR1 ethylene receptor might well be the main target of CPR5 in the ethylene signaling pathway.
MATERIALS AND METHODS
Mutant screening and map-based cloning
For mutagenesis, 200 mg of the etr1-2 seeds were washed once in a solution of 0.1% Tween 20 (Sigma Aldrich) and then incubated in a solution of 0.3% ethylmethanesulfonate (EMS) (Sigma Aldrich) for 12 hours with gentle agitation. After washing twice with sterile water the seeds were sown onto soil in a growth chamber. Seeds were collected from these M1 plants and plated onto 100 μM ACC plates for suppressor screening based on the seedlings “triple response” phenotype in response to ethylene. Potential suppressor mutants were identified and verified in the next generation. Confirmed mutants were crossed to wild type (Col-0) and to etr1-2 in order to determine whether the mutations were intragenic or extragenic and dominant or recessive, respectively. The selected suppressor mutants were backcrossed to etr1-2 twice to remove extraneous mutations.
For genetic mapping, the selected mutants were crossed with an etr1 mutant in the Landsberg erect (Ler) background and the F2 population was used for PCR-based mapping, as previously described (Resnick et al. 2006). Mutants were scored as homozygous for etr1-2 on the basis of the etiolated seedling “triple response” phenotype. A total of 526 individuals were selected for use in the simple sequence polymorphism (SSLP) marker-based mapping. Initial mapping of the selected suppressor mutant linked the mutation to the marker MVP7, MXK3 and MNA5 on the bottom of chromosome 5. Genomic DNA corresponding to candidate genes was PCR-amplified from mutant and wild type plants and sequenced to identify the mutation.
Genetic crosses and mutant genotyping
The Arabidopsis mutant cpr5-1 and the double mutant ein2-1 cpr5-1 were kindly provided by Dr. Xinnian Dong from Duke University (Bowling et al. 1997). The Arabidopsis T-DNA insertion line of CPR5 (salk_074631, assigned as cpr5-T3) was obtained from the Arabidopsis Biological Resource Center (ABRC, Ohio State University, Columbus). The cpr5-T3 mutant has a T-DNA insertion at exon 3 according to the ABRC. The double mutants cpr5-T3 etr2-1, cpr5-T3 ers1-10 and cpr5-T3 etr1-1 were generated by genetic crosses, and the F2 progeny from these crosses were screened by specific PCR markers, as previously described (Resnick et al. 2006; Wang et al. 2016).
Plant growth, transformation and ethylene response assays
Wild-type plants of Arabidopsis thaliana (ecotype Columbia (Col-0), or Landsberg erecta (Ler)) and tobacco (Nicotiana tabacum NC89) were grown in 1/2 MS (Murashige and Skoog) medium or soil in a controlled environment growth chamber at 21°C under 16 h light/8 h dark. Transgenic Arabidopsis plants were generated by the floral dip infiltration method (Clough and Bent 1998) mediated by Agrobacterium tumefaciens (strain GV3101). Transformed plants were selected by BASTA, and the homozygous transgenic lines from the T3 generation were used for gene expression analysis and phenotypic characterization.
For tobacco transformation, pieces of tobacco leaves were inoculated with Agrobacterium in liquid culture (MS + 1.0 mg/L 6-BA + 0.1 mg/L NAA) for 10 min with gentle shaking, and then cultured in darkness for 3 d. After washing, leaf discs were transferred to selection medium (MS +1.0 mg/L 6-BA + 0.1 mg/L NAA + 250 mg/L cefotaxime + 30 mg/L hygromycin). After propagation for ~30 d, transformed shoots were obtained. Hygromycin-resistant shoot buds were allowed to grow to ~2 cm in height and then plantlets were transferred into rooting medium (1/2 MS + 350 mg/L cefotaxime) for ~3 weeks before being transferred to soil. Transgenic seeds were collected 4–5 months later.
Infiltration of Agrobacterium tumefaciens (strain GV3101) was carried out, as previously described (Dong et al. 2010; Wang et al. 2016). In brief, 50 mL of Agrobacterium culture, in LB broth supplemented with antibiotics (50 mg/L kanamycin, 50 mg/L rifampicin), was precipitated, washed, and re-suspended in a solution containing 10 mM MgCl, 10 mM MES and 100 μM acetosyringone. Fresh onion peels were used for agroinfiltration. Generally, 20 independent agroinfiltrations were performed for each transformation construct.
The ethylene response assay of Arabidopsis seedlings was as previously described (Wang et al. 2016). In brief, seeds were surface sterilized and then sowed on 1/2 MS medium containing ACC at different concentrations (0, 0.5, 5, 20, or 100μM). After treatment at 4°C for 3 d, plates were moved to a growth chamber for 8 h under white light, then wrapped with aluminum foil and placed in a growth chamber for the indicated periods. Image J 1.48u software (http://rsb.info.nih.gov/ij/) was used to measure hypocotyl lengths. Statistical analyses were performed by one-way ANOVA with a 95% or 99 % confidence for significant difference.
For leaf senescence assays, ethylene treatment of adult plants was carried out in airtight clear acrylic chambers into which either ethylene gas or air was injected (Resnick et al. 2006). Plants were photographed after the chambers were placed in a growth room at 21°C for 3 d under 24-h lighting.
Yeast split-ubiquitin assays
For yeast split-ubiquitin assays, the open reading frame cDNA fragments of CPR5 and ETR1 were each PCR-amplified from existing templates with primers (CPR5-For (SfiI): ATTGGCCATTACGGCCATGGAAGCCCTCCTCCTCCCT; CPR5-Rev (SfiI): ATTGGCCATTACGGCCTCAAGCATAGTCAGACCCAC; ETR1-For (SfiI)-2: ATTGGCCATTACGGCCGAAGTCTGCAATTGTATTGA; ETR1-Rev (SfiI)-2: ATTGGCCGCCTCGGCCGGCATGCCCTCGTACAGTACCCG), cloned into a bait vector pPR3-N and a prey vector pBT3-STE through the restriction sites SfiI, and the resultant constructs, CPR5-pPR3-N and ETR1-pBT3-STE, were obtained, respectively. The vector construction of LTP1 or RTE1 for the yeast split-ubiquitin assay was as previously described (Wang et al. 2016). The insert sequences were verified by DNA sequencing, and protein interactions were examined, as previously described (Chang et al. 2014).
Constructs for BiFC assays
To generate constructs encoding the gene fusions of cYFP-CPR5 and nYFP-CPR5, the PCR fragment of cYFP or nYFP was first amplified, based on the template pSPYCE or pSPYNE vector (Walter et al. 2004) with primers (cYFP-For: GCTCTAGAGCGACAAGCAGAAGAACGGCATCA; cYFP-Rev: CGGGGTACCCCGCTTGTACAGCTCGTCCATGCCGAG; nYFP-For: GCTCTAGAGCATGGTGAGCAAGGGCGAG; and nGFP-Rev: CGGGGTACCCCGGGCCATGATATAGACGTTGTG) and cloned into the binary vector, pCambia1300-221-HA through restriction sites XbaI and KpnI. The coding sequence for full length CPR5 was PCR-amplified from an existing CPR5 cDNA template, using primers (Kpn I-CPR5 For: CGGGGTACCCCGATGGAAGCCCTCCTCCTCCCTCC, and BamHI-CPR5 Rev: CGGGATCCCGTCAAGCATAGTCAGACCCACCAT), and then cloned into the same vector through the restriction sites KpnI and BamHI. The insert sequences and the in frame fusions of cYFP-CPR5 and nYFP-CPR5 were verified by DNA sequencing. The same vectors containing the gene fusions of ETR1-nYFP, LTP1-cYFP, and nYFP-RTE1 were used, as previously described (Dong et al. 2010; Wang et al. 2016).
Fluorescence microscopy
Imaging of fluorescent proteins in onion epidermal cells was conducted using a laser scanning confocal microscope (Leica TCS SP5). The excitation wavelength for YFP was 488 nm, and the emission filter wavelengths were 505–530 nm for YFP. Pieces of fresh onion peels were directly mounted into water on a glass slide for visualization. For each experiment, at least 15 different samples were examined and experiments were repeated three times.
RNA isolation, semi-quantitative and quantitative PCR analyses
Total RNA was isolated from light-grown seedlings with TRIzol (Sigma), and the reverse transcription of RNA was performed using PrimeScript TM RT Enzyme Mix, according to the manufacturer’s recommendations (Takara Bio Inc, Otsu, Japan). Primers employed for Rt-PCR and quantitative RT-PCR are listed in Table S1. Quantitative RT-PCR analysis was performed on an Agilent Real-Time qPCR apparatus (Mx3000P system) using SYBR Premix ExTaqTM II (Takara Bio Inc, Otsu, Japan). Biological replicates for each set of experiments were carried out three times, and the mean value was normalized using Actin2 as the internal control.
Supplementary Material
Figure S1. Generation of transgenic tobacco plants over-expressing CPR5
Table S1. Primers used for qRT-PCR analysis
Acknowledgments
We thank the Experimental Center of Qingdao Agricultural University for confocal microscopy support (Leica TCS SP5). We thank Dr. Xinnian Dong of Duke University for cpr5-1 and cpr5-1 ein2-1 seeds. This work was supported by the National Natural Science Foundation of China (31370322) and the Shandong Development Program for Science and Technology (2015GNC110012) to CHD, the National Natural Science Foundation of China (31400247) to HP, and a grant from the National Institutes of Health 1R01GM071855 to CC.
Footnotes
AUTHOR CONTRIBUTIONS
F.W., L.W. and L.Q. performed most of the research and F.W. wrote the first draft of the manuscript. J.C. performed some protein interaction experiment and M.B.P. carried out the mutant screenings. H.P. and T.Z. revised the manuscript. C.C. and C.H.D. designed the experiments and revised the manuscript.
References
- Abeles FB, Morgan PW, Saltveit ME. Ethylene in Plant Biology, Second Edition. Academic Press, Inc; San Diego: 1992. [Google Scholar]
- Aki T, Konishi M, Kikuchi T, Fujimori T, Yoneyama T, Yanagisawa S. Distinct modulations of the hexokinase1-mediated glucose response and hexokinase1-independent processes by HYS1/CPR5 in Arabidopsis. J Exp Bot. 2007;58:3239–3248. doi: 10.1093/jxb/erm169. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- Bao Z, Hua J. Interaction of CPR5 with cell cycle regulators UVI4 and OSD1 in Arabidopsis. PLoS ONE. 2014;9:100347–101371. doi: 10.1371/journal.pone.0100347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry CS, Giovannoni JJ. Ripening in the tomato Green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethlene signaling. Proc Natl Acad Sci USA. 2006;103:7923–7928. doi: 10.1073/pnas.0602319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science. 1988;241:1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- Boch J, Verbsky ML, Robertson TL, Larkin JC, Kunkel BN. Analysis of resistance gene-mediated defense responses in Arabidopsis thaliana plants carrying a mutation in CPR5. Mol Plant Microbe Interact. 1998;11:1196–1206. [Google Scholar]
- Borghi M1, Rus A, Salt DE. Loss-of-function of Constitutive Expresser of Pathogenesis Related Genes5 affects potassium homeostasis in Arabidopsis thaliana. PLoS ONE. 2011;6:26360–101371. doi: 10.1371/journal.pone.0026360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell. 1997;9:1573–1584. doi: 10.1105/tpc.9.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brininstool G, Kasili R, Simmons LA, Kirik V, Hülskamp M, Larkin JC. Constitutive Expressor Of Pathogenesis-related Genes5 affects cell wall biogenesis and trichome development. BMC Plant Biol. 2008;8:58. doi: 10.1186/1471-2229-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: Similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Chang JH, Clay JM, Chang C. Association of cytochrome b5 with ETR1 ethylene receptor signaling through RTE1 in Arabidopsis. Plant J. 2014;77:558–567. doi: 10.1111/tpj.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENEINSENSITIVE3 and related proteins. Cell. 1997;89:1133–1144. doi: 10.1016/s0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- Chen YF, Randlett MD, Findell JL, Schaller GE. Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. J Biol Chem. 2002;277:19861–19866. doi: 10.1074/jbc.M201286200. [DOI] [PubMed] [Google Scholar]
- Chen T, Liu J, Lei G, Liu YF, Li ZG, Tao JJ, Hao YJ, Cao YR, Lin Q, Zhang WK, Ma B, Chen SY, Zhang JS. Effects of tobacco ethylene receptor mutations on receptor kinase activity, plant growth and stress responses. Plant Cell Physiol. 2009;50:1636–1650. doi: 10.1093/pcp/pcp107. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dong CH, Jang M, Scharein B, Malach A, Rivarola M, Liesch J, Groth G, Hwang I, Chang C. Molecular association of the Arabidopsis ETR1 ethylene receptor and a regulator of ethylene signaling, RTE1. J Biol Chem. 2010;285:40706–40713. doi: 10.1074/jbc.M110.146605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CH, Rivarola M, Resnick JS, Maggin BD, Chang C. Subcellular co-localization of Arabidopsis RTE1 and ETR1 supports a regulatory role for RTE1 in ETR1 ethylene signalling. Plant J. 2008;53:275–286. doi: 10.1111/j.1365-313X.2007.03339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble RL, Coonfield ML, Schaller GE. Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci USA. 1998;95:7825–7829. doi: 10.1073/pnas.95.13.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Zhang S, Wang C, Yang X, Wang Y, Su X, Du J, Yang C. Arabidopsis CPR5 independently regulates seed germination and postgermination arrest of development through LOX pathway and ABA signaling. PLoS ONE. 2011;6:19406–101371. doi: 10.1371/journal.pone.0019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Städele K, Růžička K, Obrdlik P, Harter K, Horák J. Subcellular localization and in vivo interaction of the Arabidopsis thaliana ethylene receptor family members. Mol Plant. 2008;1:308–320. doi: 10.1093/mp/ssm015. [DOI] [PubMed] [Google Scholar]
- Gu Y, Zebell SG, Liang Z, Wang S, Kang BH, Dong X. Nuclear pore permeabilization is a convergent signaling event in effector-triggered immunity. Cell. 2016;166:1526–1538. doi: 10.1016/j.cell.2016.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR. The ethylene signaling pathway: new insights. Curr Opin Plant Biol. 2004;7:40–49. doi: 10.1016/j.pbi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Guzman P, Ecker JR. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AE, Bleecker AB. Analysis of combinatorial loss-offunction mutants in the Arabidopsis ethylene receptors reveals that the ers1 etr1 double mutant has severe developmental defects that are EIN2 dependent. Plant Cell. 2003;15:2032–2041. doi: 10.1105/tpc.013060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AE, Chen QHG, Findell JL, Schaller GE, Bleecker AB. The relationship between ethylene binding and dominant insensitivity conferred by mutant forms of the ETR1 ethylene receptor. Plant Physiol. 1999;121:291–299. doi: 10.1104/pp.121.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Kieber JJ, Hirayama N, Kogan M, Guzman P, Nourizadeh S, Alonso JM, Dailey WP, Dancis A, Ecker JR. RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease-related copper transporter, is required for ethylene signaling in Arabidopsis. Cell. 1999;97:383–393. doi: 10.1016/s0092-8674(00)80747-3. [DOI] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz E. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Jing HC, Dijkwel PP. CPR5: A Jack of all trades in plants. Plant Signal Behav. 2008;3:562–563. doi: 10.4161/psb.3.8.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing HC, Schippers JHM, Hille J, Dijkwel PP. Ethyleneinduced leaf senescence depends on age-related changes and OLD genes in Arabidopsis. J Exp Bot. 2005;56:2915–2923. doi: 10.1093/jxb/eri287. [DOI] [PubMed] [Google Scholar]
- Jing HC, Anderson L, Sturre MJ, Hille J, Dijkwel PP. Arabidopsis CPR5 is a senescence-regulatory gene with pleiotropic functions as predicted by the evolutionary theory of senescence. J Exp Bot. 2007;58:3885–3894. doi: 10.1093/jxb/erm237. [DOI] [PubMed] [Google Scholar]
- Jing HC, Hebeler R, Oeljeklaus S, Sitek B, Stühler K, Meyer HE, Sturre MJ, Hille J, Warscheid B, Dijkwel PP. Early leaf senescence is associated with an altered cellular redox balance in Arabidopsis cpr5/old1 mutants. Plant Biol. 2008;10:85–98. doi: 10.1111/j.1438-8677.2008.00087.x. [DOI] [PubMed] [Google Scholar]
- Kirik V, Bouyer D, Schobinger U, Bechtold N, Herzog M, Bonneville JM, Hulskamp M. CPR5 is involved in cell proliferation and cell death control and encodes a novel transmembrane protein. Curr Biol. 2001;11:1891–1895. doi: 10.1016/s0960-9822(01)00590-5. [DOI] [PubMed] [Google Scholar]
- Perazza D, Laporte F, Balagué C, Chevalier F, Remo S, Bourge M, Larkin J, Herzog M, Vachon G. GeBP/GPL transcription factors regulate a subset of CPR5-dependent processes. Plant Physiol. 2011;157:1232–1242. doi: 10.1104/pp.111.179804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Xu C, Wen CK. Genetic and transformation studies reveal negative regulation of ERS1 ethylene receptor signaling in Arabidopsis. BMC Plant Biol. 2010;10:60. doi: 10.1186/1471-2229-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee H. Highly conserved proteins that modify plant ethylene responses. Proc Natl Acad Sci USA. 2006;103:7537–7538. doi: 10.1073/pnas.0602599103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Du W, Brandizzi F, Giovannoni JJ, Barry CS. Differential control of ethylene responses by GREEN-RIPE and GREEN-RIPE LIKE1 provides evidence for distinct ethylene signaling modules in tomato. Plant Physiol. 2012;160:1968–1984. doi: 10.1104/pp.112.205476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z, Ruberti C, Gong Z, Brandizzi F. CPR5 modulates salicylic acid and unfolded protein response to manage tradeoffs between plant growth and stress responses. Plant J. 2017;89:486–501. doi: 10.1111/tpj.13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussatche P, Klee HJ. Autophosphorylation activity of the Arabidopsis ethylene receptor multigene family. J Biol Chem. 2004;279:48734–48741. doi: 10.1074/jbc.M403100200. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhou X, Wen CK. Modulation of ethylene responses by OsRTH1 overexpression reveals the biological significance of ethylene in rice seedling growth and development. J Exp Bot. 2012;63:4151–4164. doi: 10.1093/jxb/ers098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Hall BP, Gao Z, Schaller GE. A strong constitutive ethylene-response phenotype conferred on Arabidopsis plants containing null mutations in the ethylene receptors ETR1 and ERS1. BMC Plant Biol. 2007;7:3. doi: 10.1186/1471-2229-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick JS, Wen CK, Shockey JA, Chang C. REVERSIONTO-ETHYLENE SENSITIVITY1, a conserved gene that regulates ethylene receptor function in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:7917–7922. doi: 10.1073/pnas.0602239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick JS, Rivarola M, Chang C. Involvement of RTE1 in conformational changes promoting ETR1 ethylene receptor signaling in Arabidopsis. Plant J. 2008;56:423–431. doi: 10.1111/j.1365-313X.2008.03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivarola M, McClellan C, Resnick J, Chang C. ETR1-specific mutations distinguish ETR1 from other Arabidopsis ethylene receptors as revealed genetic interaction with RTE1. Plant Physiol. 2009;150:547–551. doi: 10.1104/pp.109.138461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez FI, Esch JJ, Hall AE, Binder BM, Schaller GE, Bleecker AB. A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science. 1999;283:996–998. doi: 10.1126/science.283.5404.996. [DOI] [PubMed] [Google Scholar]
- Sakai H, Hua J, Chen QG, Chang C, Medrano LJ, Bleecker AB, Meyerowitz EM. ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:5812–5817. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller GE, Bleecker AB. Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science. 1995;270:1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, Harter K, Kudla JJ. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ye Q, Zhang M, Yang C. Involvement of Arabidopsis CPR5 in thermotolerance. Acta Physiol Plant. 2012;34:2093–2103. [Google Scholar]
- Wang H, Sun Y, Chang J, Zhang F, Pei H, Yi Y, Chang C, Dong CH. Regulatory function of Arabidopsis lipid transfer protein 1 (LTP1) in ethylene response and signaling. Plant Mol Biol. 2016;91:471–484. doi: 10.1007/s11103-016-0482-7. [DOI] [PubMed] [Google Scholar]
- Woeste KE, Kieber JJ. A strong loss-of-function mutation in RAN1 results in constitutive activation of the ethylene response pathway as well as a rosette-lethal phenotype. Plant Cell. 2000;12:443–455. doi: 10.1105/tpc.12.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Ito M, Nishida I, Watanabe A. Identification of a novel gene HYS1/CPR5 that has a repressive role in the induction of leaf senescence and pathogen-defence responses in Arabidopsis thaliana. Plant J. 2002;29:427–437. doi: 10.1046/j.0960-7412.2001.01228.x. [DOI] [PubMed] [Google Scholar]
- Yu Y, Wang J, Wang H, Zhang Z, Liu J. Relationship between Rh-RTH1 and ethylene receptor gene expression in response to ethylene in cut rose. Plant Cell Rep. 2010;29:895–904. doi: 10.1007/s00299-010-0875-z. [DOI] [PubMed] [Google Scholar]
- Yu Y, Wang H, Liu J, Fu Z, Wang J, Liu J. Transcriptional regulation of two RTE-like genes of carnation during flower senescence and upon ethylene exposure, wounding treatment and sucrose supply. Plant Biol. 2011;13:719–724. doi: 10.1111/j.1438-8677.2010.00441.x. [DOI] [PubMed] [Google Scholar]
- Zhou X, Liu Q, Xie F, Wen CK. RTE1 is a Golgi-associated and ETR1-dependent negative regulator of ethylene responses. Plant Physiol. 2007;145:75–86. doi: 10.1104/pp.107.104299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Generation of transgenic tobacco plants over-expressing CPR5
Table S1. Primers used for qRT-PCR analysis